Abstract
Sweet and umami tastes are perceived by T1r taste receptors in oral cavity. T1rs are class C G-protein coupled receptors (GPCRs), and the extracellular ligand binding domains (LBDs) of T1r1/T1r3 and T1r2/T1r3 heterodimers are responsible for binding of chemical substances eliciting umami or sweet taste. However, molecular analyses of T1r have been hampered due to the difficulties in recombinant expression and protein purification, and thus little is known about mechanisms for taste perception. Here we show the first molecular view of reception of a taste substance by a taste receptor, where the binding of the taste substance elicits a different conformational state of T1r2/T1r3 LBD heterodimer. Electron microscopy has showed a characteristic dimeric structure. Förster resonance energy transfer and X-ray solution scattering have revealed the transition of the dimerization manner of the ligand binding domains, from a widely spread to compactly organized state upon taste substance binding, which may correspond to distinct receptor functional states.
Taste sensation is evoked by specific interactions between taste substances and taste receptors residing in the plasma membranes of the taste cells in taste buds in the oral cavity1,2. One of these receptors is the taste receptor type 1, the T1r family, which is evolutionarily conserved in vertebrates, including fishes, birds, and mammals3. The heterodimer of T1r2 and T1r3 recognizes sweet taste substances such as sugars and artificial sweeteners, while the heterodimer of T1r1 and T1r3 recognizes umami taste substances such as l-glutamate4,5,6.
The T1r family proteins belong to the class C G-protein coupled receptor (GPCR) family7,8. The class C GPCR members function as constitutive homo- or heterodimers in the physiological state. The class C GPCR structure is characterized by the presence of a large extracellular domain upstream of the hepta-helical transmembrane region, which is commonly found among GPCRs. The extracellular domain consists of the ligand binding domain (LBD), responsible for primary agonist binding, followed by the cysteine rich domain (CRD), which mainly serves as a linker between the LBD and the transmembrane region (Fig. 1a). Ligand binding at the extracellular domain results in receptor activation and signal transmission to the heterotrimeric G-protein in the cytosol7,8. The receptor activation mechanism of the class A GPCR members, consisting solely of the transmembrane region, has been considered to occur via agonist binding, which changes the conformational dynamics of the protein by lowering the transition energy between the different states, and results in the transition towards the active-state conformation9. In contrast, the conformation of the transmembrane region of the class C GPCRs is considered to be allosterically regulated by agonist binding to the extracellular LBDs, probably through their conformational changes10,11,12,13. Accordingly, in the case of T1r, the major taste substances, including sugars and l-glutamate, are considered to target the LBD of T1r heterodimer14, and thus consequently induce the conformational change of the LBD.
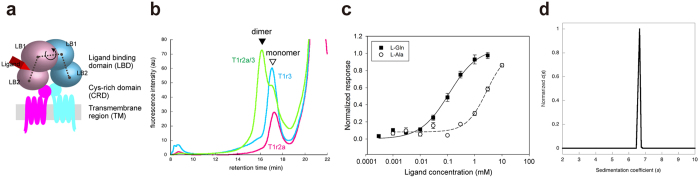
Figure 1. Taste Receptor T1r Proteins from Medaka Fish (mf).
(a) Schematic drawing of the overall architecture of class C GPCR, where the codebook vector of each domain in LBD (gray dot) and the protomer torsion angle (the arrow) were depicted. (b) FSEC analysis of mf T1R2aLBD, mf T1R3LBD, and co-expression of the T1R2a and T1r3 proteins. (c) Dose-response curves for l-alanine and l-glutamine by the full-length mf T1r2a/T1r3 receptor in HEK293 cells. The error bars are ± SEM of 4–34 independent determinations. (d) The c(s) distribution of the purified mf T1r2a/3LBD, obtained from the data analysis of SV-AUC experiments.
Due to the lack of structural information of T1r receptors, their functional mechanisms have so far been conjectured from the crystallographic observation on the other class C GPCR members. Crystal structures of the LBDs of metabotropic glutamate receptors (mGluRs) and GABAB receptor (GABABR) revealed the bilobal architecture; namely, the Venus-flytrap domain (VFTD), in which an agonist binds to the cleft between the two lobes, LB1 and LB215,16. Upon agonist binding, two types of conformational change were observed on the LBDs (Supplementary Table S1). One is the domain closure within the protomer, at the cleft between LB1 and LB2, which is referred to as the open- and closed-conformation. The other is the change between the two different forms of the dimer arrangement: the compact state with a smaller torsion angle (< ~ −30°) rotated around the axis between the LB1s of the two protomers (Fig. 1a), interpreted as the active (A)-state, and the widely spread states with a larger torsion angle (> ~ −50°), interpreted as the resting (R)-state (Supplementary Fig. S1)15. This view is basically compatible with the observations by Förster resonance energy transfer (FRET) of labeled mGluRs12,13. However, the actual conformations of the LBDs in the physiological state are still unknown, because available structural information is limited to those obtained in crystalline spaces. In fact, the dimer arrangements of homodimeric mGluR LBDs so far observed there were various regardless of the types of bound ligands, such as agonists, antagonists, or ligand-free15,17,18 (Supplementary Table S1), and a previous study also pointed out the possibility of biased trapping of certain conformations among dynamic conformational equilibrium by the crystal packing18. Moreover, a crystallographic analysis of the LBD of GABABR GBR1/GBR2 heterodimer revealed another mode of conformational change, where little significant dimer rearrangement is observed among apo-, antagonist bound-, and agonist bound-states16 (Supplementary Fig. S1 and Table S1). The current situation makes it obscure what kind of conformational change at the T1r LBD heterodimer is elicited by taste-substance binding.
So far, not only the structural analyses but also the molecular functional analyses using the purified protein of T1rs have been completely hampered, due to the difficulties in the heterologous expression and purification even for the partial regions such as the LBD19. In this study, we found that the LBDs of T1r2 and T1r3 of medaka fish (Oryzias latipes, also known as Japanese rice fish), a representative model organism in vertebrates, can be expressed heterologously as a properly folded and functional heterodimeric protein, thus enabling various biophysical analyses. The solution-state structure analyses revealed the conformational change upon the taste substance binding to the T1r, under the condition devoid of any constraint derived from crystal packing.
Results
The ligand-binding domains of the T1r heterodimer from medaka fish exhibit proper recombinant expression
Chemosensory receptors, such as olfactory receptors and pheromone receptors, are known to have the specific chaperone systems in their native chemosensory cells, and the proper surface expression of the receptors in heterologous cells are only achieved in the presence of the systems20,21,22,23. However, in the case for taste receptor T1rs, such specific chaperone systems are unknown20, and indeed recombinant expression of mouse- or human-T1rLBDs displayed failure of folding and localization19. Thus we first performed extensive screening of T1r genes and expression conditions to find those exhibiting proper expression, using fluorescence-detection size-exclusion chromatography (FSEC)19,24. Most of the genes from different species, as well as the numerous variations of the expression conditions including host cells, resulted in unsuccessful protein production, as reported previously19. Nevertheless, the LBDs of T1r2a and T1r3 from medaka fish (mf)25 showed good secretion and a sharp FSEC elution peak corresponding to the dimer species, indicating proper folding and oligomerization, only when they were co-expressed in insect cells (Fig. 1b). mf T1r3LBD showed fair conservation with mammalian T1r3LBD (~37% similarity). On the other hand, LBD of mf T1r2a, one of the three T1r2 subtypes in medaka fish, showed moderate conservation with both mammalian T1r1LBD and T1r2LBD (37~39% similarity), reflecting the fact that the mfT1r2a/T1r3 heterodimer responds to a wide array of l-amino acids, but not to sugars or artificial sweeteners such as saccharin26.
Dose-response measurement of the full-length mf T1r2a and mf T1r3 heterodimer cloned in this study confirmed the similar EC50 value for l-alanine (2.70 ± 1.20 mM) to that reported previously26, and revealed an even higher affinity to l-glutamine, with the EC50 value of 100 ± 26.0 μM (Fig. 1c and Supplementary Table S2). Because medaka fish reportedly show preferences to the foods containing amino acids27, l-glutamine and alanine are considered to serve as taste substances to medaka fish. Therefore, the mf T1r2a/mf T1r3-LBD heterodimer and its amino-acid binding are expected to serve as the first molecular platform to assess the general structural and functional properties T1r LBD heterodimer, including those for taste substance binding.
T1r2aLBD and T1r3LBD form a stable heterodimer
mf T1r2aLBD and mfT1r3LBD were successfully co-purified after recombinant expression in insect cells (Supplementary Fig. S2). The purified protein exhibited a monodisperse distribution as confirmed by a sedimentation velocity analytical ultracentrifugation (SV-AUC) analysis. In the obtained c(s) distribution, a single peak with an estimated molecular weight of 108.5 kDa was observed (Fig. 1d). This result clearly indicated that the purified T1r2aLBD and T1r3LBD (53.5 and 55.2 kDa respectively, as estimated from the amino acid sequences) exclusively formed a stable heterodimer.
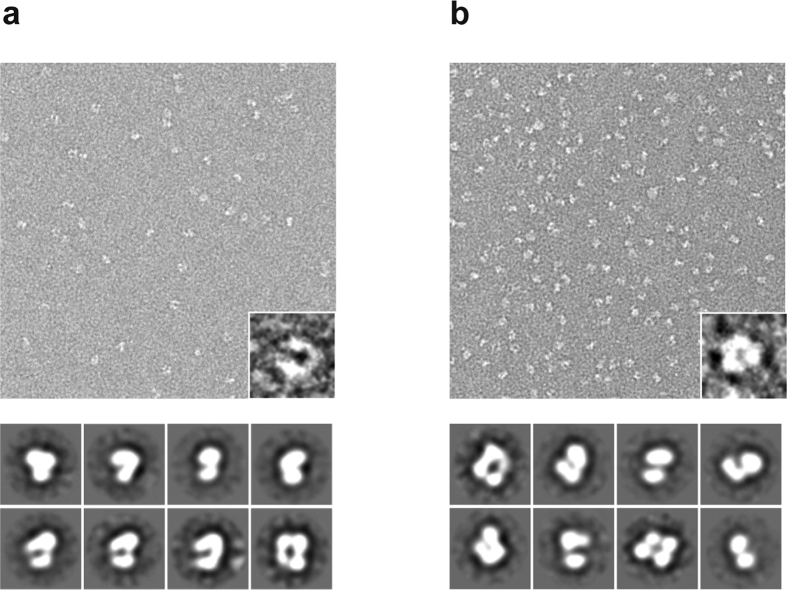
To visualize the structural organization of T1r2a/3LBD, we performed the electron microscopic observation of negatively stained T1r2a/3LBD (Fig. 2). The particles exhibit the presence of two segments, most likely corresponding to each protomer of T1r2a and T1r3 LBD proteins, both in the presence or absence of a taste substance l-glutamine. Two-dimensional averages of T1r2a/3LBD particle exhibited a bilobal feature, characteristic to VFTD. The approximate particle sizes were observed as ~95 × ~75 Å, which agrees with those for other class C GPCR LBD dimer structures. These observations indicated that the manner of dimerization of T1r2a/3LBD is similar to that observed on LBDs of other class C GPCRs, such as mGluRs.
Figure 2. Electron microscopic observation of T1r2a/3LBD.
The top panels are negative-staining raw particle images of the purified T1r2a/3LBD, with close-up views of representative particles in the insets. The bottom panels are the representative two-dimensional class averages of particles. (a) The l-glutamine-bound state. (b) The ligand-free state.
Conformational change of the ligand binding domains upon taste substance binding
So far, all the structural analyses of other class C GPCR LBDs were performed by crystallography, using deglycosylated samples in many cases15,16,17,18. In this study, we performed multiple structural analyses in both the presence and absence of a taste substance, using the glycosylated protein sample (Supplementary Fig. S2) in solution, which is closer to a physiological condition without any constraint derived from crystal packing.
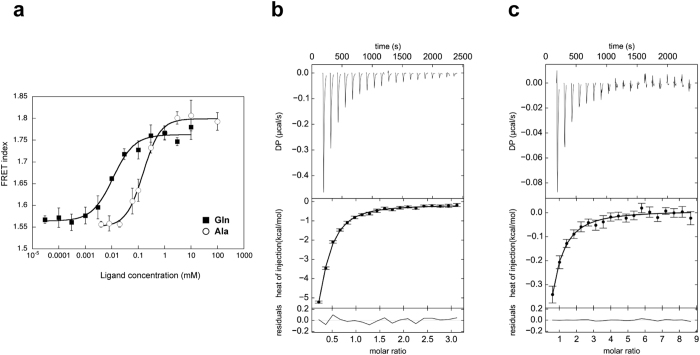
We first assessed whether the binding of a taste substance to the T1r LBD induces a conformational change of the protein. To answer this question, the T1r2a/3LBD heterodimer fused with a fluorescence protein, either Cerulean (a CFP variant) or Venus (a YFP variant)28, at their C-termini was subjected to Förster resonance energy transfer (FRET) analyses. In the presence of the taste substances l-glutamine and l-alanine, the labeled T1r2a/3LBD exhibited elevated FRET signals (Fig. 3a). The EC50 values for the FRET signal changes upon the l-glutamine and l-alanine titration were 12.7 ± 2.7 and 168 ± 19.0 μM, respectively (Supplementary Table S2). We confirmed that the FRET signal rise accompanies the taste substance binding, as the EC50 values for the former are close to the Kd values of l-glutamine and l-alanine to the non-labeled T1r2a/3LBD analyzed by isothermal titration calorimetry (Fig. 3b,c and Supplementary Table S2). These results indicated that taste substance binding to the T1r2a/3LBD induces a conformational change of the protein, in most likely which the C-termini of the LBDs of the two protomers come closer to each other.
Figure 3. Conformational change of T1r2a/3 LBD upon taste substance binding.
(a) Dose-dependent FRET signal changes of the T1r2aLBD-Cerulean and T1r3LBD-Venus heterodimer for taste substance binding. The error bars are ± SEM of 3 independent determinations. (b,c) l-Glutamine (b) and l-alanine (c) binding to mfT1r2a/3LBD, measured by isothermal titration calorimetry. The upper and lower panels show the raw data and the integrated heat signals upon ligand injection, respectively, with binding isotherms fitted assuming 1 ligand: 1 heterodimer binding.
The order of the EC50 values of two amino acids was coincident with that for the receptor responses as observed above. Although the EC50 values for the FRET change by the labeled-LBD and the response by the full-length receptor have 8~16 fold differences, such deviations were often observed on the other receptors when the different assay conditions/methods were used, because the ligand efficacies are affected by the intrinsic receptor characteristics, the downstream signaling pathways, and so on29. For example, mGluR1 exhibited the Kd of 1.3 μM for the l-glutamate binding to the purified LBD measured by fluorescence change30, while EC50 of 4.6~22 μM for the l-glutamate response measured by Ca2+ -activated Cl− current in a Xenopus oocyte expression system (3.5~17 fold differences)31. Therefore, the results observed in this study suggested that the conformational transition is relevant to the receptor responses.
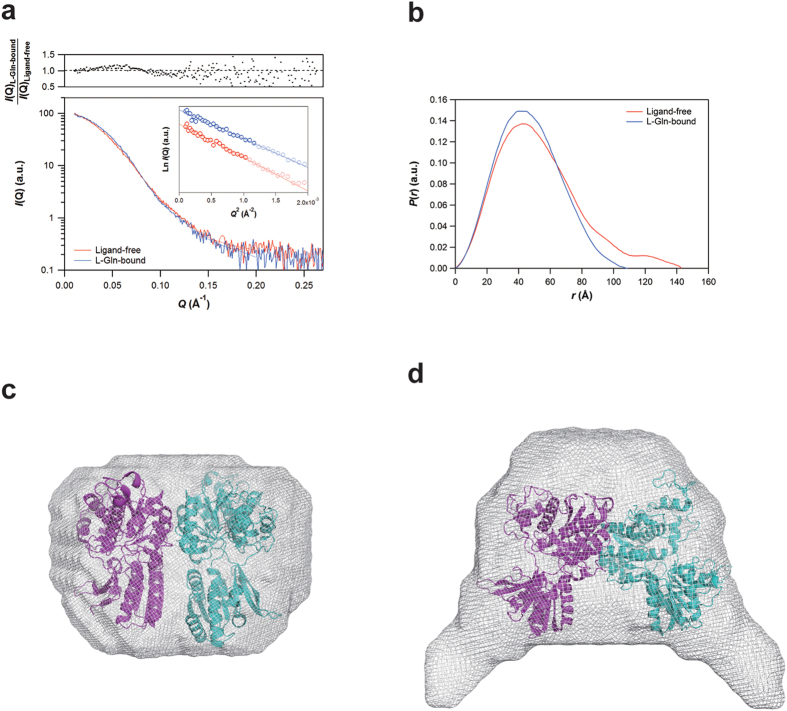
To analyze the conformational changes in further detail, the non-labeled T1r2a/3LBD, in the presence or absence of l-glutamine, was subjected to small-angle X-ray scattering (SAXS) analyses (Fig. 4). The molecular mass estimated on the bases of the forward scattering (121~123 kDa) as well as the Porod volume (144~150 kDa) was nearly constant irrespective of the presence of l-glutamine (Supplementary Table S3), exhibiting a fair agreement with the sum of those for T1r2aLBD and T1r3LBD determined by mass spectroscopy and SDS-PAGE (127 kDa; Supplementary Fig. S2).
Figure 4. Overall shapes of T1r2a/3 LBD in ligand-free and l-glutamine-bound states revealed by SAXS.
(a) SAXS curves of the ligand-free (red) and l-glutamine-bound (blue) forms of T1r2a/3 LBD. The inset indicates the Guinier plots of the ligand-free (red) and l-Gln-bound (blue) forms of T1r2a/3 LBD, for which the Guinier analyses were conducted by using the Q range (highlighted data points in the inset) from 0.01003 Å−1 to Qmax < 1.3/Rg. (b) Pair distribution functions, P(r), of the ligand-free (red) and l-gln-bound (blue) forms of T1r2a/3 LBD. Low-resolution models of the l-glutamine bound state (c) and the ligand-free state (d). The representative models were presented as smooth molecular envelopes, onto which the high-resolution models of the glutamate-bound A-state structure of mGluR1LBD (PDB 1EWK) and the ligand-free R-state of mGluR1LBD (PDB 1EWT) were superimposed. The theoretical SAXS curves of the restored models are shown by solid lines in panel (a) and are in good agreement with the experimental curves.
The radius of gyration (Rg) in the presence of glutamine (37.0 ± 0.5 Å) was smaller than that of the ligand-free protein (39.8 ± 0.6 Å; Supplementary Table S3). This difference in the Rg values amounted to ~7.6%, a comparable value to those analyzed by the previous SAXS measurements on the nucleotide-mediated conformational change of the myosin head domains (4.5~5.9%)32 or the open-close transition of phosphoglycerate kinase during catalysis (~8.0%)33. This result clearly indicates that a significant conformational change occurs between the two states.
The radical conformational switching was also supported by the pair distance distribution functions, P(r), which exhibited a ~30 Å reduction in the maximum dimension of particles (Dmax) upon glutamine binding (Fig. 4b and Supplementary Table S3). Judging from the crystal structures of other class C GPCR LBDs, the observed particle size difference seems not be solely attributed to the conformational change within a protomer, but to the rearrangement of the dimerization. For example, the open- (O) to close (C) transitions within the same dimerization state, such between PDB IDs 3KS9 to 1EWK (mGluR1) or 4MQE to 4MS3 (GABABR), results less than 3 Å differences, while the R- to A-state transitions of the dimerization state, such between 1EWT to 1EWK (mGluR1), makes more than 10 Å differences (Supplementary Fig. S3 and Table S1). The radical change in the Dmax of the ligand-free state is a consequence of the decrease and the increase in the probabilities at pair lengths of 30~50 Å and 80~140 Å, respectively. This rearrangement of the pair distribution produced a point of intersection between the ligand-free and the glutamine-bound P(r)s at a pair length of ~60 Å, which is closer to those between the R- and A-states P(r)s (~60 Å) than those between O-C transition P(r)s (~50 Å) (Supplementary Fig. S3). The observation further supported the dimerization rearrangement of T1r2a/3LBD heterodimer upon taste substance binding.
The different overall shapes of the ligand-free and the glutamine-bound T1r2a/3LBD were confirmed by restoring low-resolution models from the SAXS data. The l-glutamine bound T1rLBD took a flat round shape, resembling an A-state structure of class C GPCR LBDs (Fig. 4c). On the other hand, the ligand free T1rLBD displayed a change of the shape into a more extended state, which is reminiscent of that proposed for the A- to R-state transition (Fig. 4d). The results also implied that there at least is a dimer rearrangement between the two states.
All together, the results in this study indicated that the conformational equilibrium of T1r2a/3LBD moves from the widely spread state toward the compact state upon taste substance binding. The directions of conformational changes are generally consistent with those observed on the other class C GPCR LBD structures, and likely more resemble to that observed on mGluR-LBD with dimer rearrangement, rather than that observed on GABABR without significant dimer rearrangement.
Discussion
This study firstly presented the biophysical properties of the ligand binding domains of a taste receptor T1r heterodimer and their binding to a taste substance. All the analyses in this study were performed in solution using the glycosylated protein sample, which is closer to a physiological condition compared to those with the deglycosylated samples in the crystalline states, therefore the observed properties are expected to well reflect the native characteristics of T1rLBD.
The study revealed two different conformational equilibriums dependent on the existence of a taste substance, the widely spread state and the compact state. In particular, the results clearly indicated the correlation between the taste-substance binding (analyzed by ITC) and the conformational transition (analyzed by FRET and SAXS) of LBD, as both were occurred in the same concentration range of the ligands. Therefore the observations in this study provided solid evidence that the ligand binding induces the conformational change of the LBD, which has been long assumed but not yet actually demonstrated. Each LBD conformation may correspond to a distinct receptor functional state of T1r, and/or the transition between them might induce the transition between distinct functional states. The labeled points for detection of the conformational change in the FRET analysis were the C-termini of the LBDs, which are the direct connection points toward the CRD, followed by the transmembrane region. The crystallographic structure of the transmembrane region of mGluR1 revealed the tight interaction between the linker region connecting the CRD to the transmembrane region, and the extracellular loop 2 in the transmembrane region connecting to the transmembrane helix III, which is reportedly important for receptor activation in class A GPCRs34. Based on these observations, the results suggest a possible picture of the taste signal transduction by T1r: the taste substance binding induces the conformational change of LBDs so as to shift the relative positions of the C-termini of LBD heterodimers, that induce the positional shift of the CRD regions, and further induce the consequent conformational change of the downstream transmembrane regions for the cytosolic G-protein activation.
The tendency of the conformational change of T1r2a/3LBD observed in solution was similar to those generally observed on crystallographic analyses of other class C GPCR LBDs. Therefore the above presumed scheme for T1r signaling is also basically in line with those for the other class C GPCRs. On the other hand, the manner and extent of the observed conformational change of T1rLBD in the solution are unlikely to resemble to that of GABABR without dimer rearrangement, despite the fact that both T1r and GABABR are heterodimers, but more similar to those of homodimeric mGluR1 with dimer rearrangement.
The VFTD structures were also found in the amino-terminal domain (ATD) of the ionotropic glutamate receptors (iGluRs), one of the extracellular domains serving as a regulatory domain in some receptors. Crystallographic studies of iGluRs-ATD revealed the considerable diversity of the dimer arrangement between the different family members35,36,37. Therefore the VFTD architecture itself likely possesses a characteristic to provide diverse conformational possibilities.
In the case for class C GPCRs, the receptors discussed above have different structural organizations, such that mGluRs, GABABR, and T1r are homodimers with a CRD, a heterodimer lacking a CRD, and a heterodimer with a CRD, respectively. Therefore, the diversity on the conformational equilibriums for the VFTDs (LBDs) might reconcile these structural diversities within the class C GPCRs and be responsible for producing the same consequences at the transmembrane regions: cytosolic G-protein activation. It should be noted that the information for the other class C GPCR LBDs are mostly obtained in the crystalline states without the native glycosyl-chains. Further accumulation of information about the conformations of the member proteins under the conditions closer to the physiological states, including those in the full-length receptors, as well as the high-resolution crystallographic structure information, will be required for understanding the signal transduction mechanisms of the class C GPCR family, and their generalities or diversities.
Methods
Expression analysis
The T1r2aLBD and T1r3LBD proteins fused with GFPuv transiently expressed with sf9 cells were analyzed by fluorescence-detection size-exclusion chromatography (FSEC), basically as reported19. The samples were loaded either onto a Superose 6 10/300 GL column (GE Healthcare), and the elution profiles were detected using excitation (EX) and emission (EM) wavelengths of 395 nm and 507 nm, respectively. For more details see Supplementary information.
Measurement of the reactions of T1r2/T1r3 heterodimer to taste substances
The responses of T1r2a/T1r3 to taste substances were analyzed using the Flip-InTM 293 cell line (Life Technologies), which was established for stable expression of T1r2a, T1r3, and Gα16-gust4438. The Ca2+ flux assays were performed using a FLEX station 3 (Molecular Devices, LLC) for the cells loaded with 100 μl of Hank’s balanced salt solution (Sigma-Aldrich), containing 5 μM of the calcium indicator dye Fluo8 NW (AAT Bioquest).
The conformational change of T1r2a/T1r3LBD upon taste-substance binding was analyzed by Förster resonance energy transfer (FRET) measurement. The T1r2a LBD-Cerulean/T1r3LBD-Venus heterodimer protein was expressed by S2 cells, and purified from the culture medium using ANTI-FLAG M2 Affinity Gel (SIGMA). Fluorescence intensities were recorded at 277 K with a FluoroMax4 spectrofluorometer (Horiba). The sample was excited at 433 nm, and the emission at 526 nm and at 475 nm was recorded, for determination of the FRET index (Intensity at 526 nm/Intensity at 475 nm).
The binding of T1r2a/T1r3LBD to a taste substance was analyzed by isothermal titration calorimetry. T1r2a/3 LBD heterodimer protein was stably expressed by S2 cells, and purified as described above. The purified protein was loaded into the iTC200 cell (GE Healthcare), and the titration was performed at 298 K. For more details see Supplementary information.
Structure analysis
T1r2a/3 LBD heterodimer protein was stably expressed by S2 cells, and purified from the culture medium using ANTI-FLAG M2 Affinity Gel (SIGMA), followed by size-exclusion chromatography (HiLoad Superdex 200, 16/60 (GE Healthcare)).
The partial specific volume and the molecular weight of the protein were determined by sedimentation velocity analytical ultracentrifugation (SV-AUC) analysis. The samples with same protein concentration but different H2O/D2O ratios (100% H2O, 50% H2O/50% D2O, and 10% H2O/90% D2O) were subjected to the SV-AUC analysis performed at 293 K and 42,000 rpm, using a ProteomeLab XL-I analytical ultracentrifuge (Beckman Coulter) with an An-60 Ti rotor. The acquired data were analyzed as reported previously39. The specific volume was determined as 0.6787 cm3/g.
For electron microscopic observation, the protein sample was applied to a carbon-coated grid and negatively stained with 2% uranyl acetate. The sample grids were examined with a JEM-2100 electron microscope (JEOL) with a LaB6 gun operated at an accelerating voltage of 200 kV. Images were recorded on a slow-scan charge-coupled device (SSCCD) camera (MegaScan), at a final magnification of 65,000 and at defocus settings of 8,900 to 27,000 Å. The two dimensional class average of the particles was performed with the EMAN software suite40.
SAXS experiments were performed at the SPring-8 beamline BL45XU41, or with the Nano-Viewer system (RIGAKU) equipped with a MicroMax-007HF X-ray generator (RIGAKU) with a Cu target (λ = 1.5418 Å) and a PILATUS 200 K detector (DECTRIS). The Rg values were estimated by the Guinier approximation, using the PRIMUS software42. P(r) functions were calculated by the GNOM software43 (Supplementary Table S3). Ab initio reconstructions of low-resolution models from the SAXS data were performed by using DAMMIF44 (Supplementary Table S4). Multiple reconstructions conducted independently to confirm the reproducibility were scored with the DAMAVER45 and DAMCLUST46 packages to obtain the most representative models as shown in Fig. 4. For more details see Supplementary information.
Additional Information
How to cite this article: Nango, E. et al. Taste substance binding elicits conformational change of taste receptor T1r heterodimer extracellular domains. Sci. Rep. 6, 25745; doi: 10.1038/srep25745 (2016).
Supplementary Material
Acknowledgments
We thank Naoshi Dohmae and the members of the Biomolecular Characterization Team, RIKEN, for N-terminal sequencing and mass spectroscopic analyses; Masanori Noda for the mass spectroscopic analysis; and Fumie Iwabuki, Noriko Matsuura, Maiko Tanaka, Naoko Ono, Takashi Yamada, and Junko Nakamura for technical assistance. Part of this work was conducted at the Institute for Molecular Science, with support from the Nanotechnology Platform Program (Molecule and Material Synthesis) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. The synchrotron radiation experiments were performed at BL45XU in SPring-8 with approval of RIKEN (Proposal No. 20120003). This work was supported by the Funding Program for Next Generation World-Leading Researchers (NEXT Program) from the Japan Society for the Promotion of Science (JSPS)/the Council for Science and Technology Policy (CSTP; Grant number LS130 to A.Y.), Strategic Programs for R&D from RIKEN, Japan (to A.Y.), the Targeted Proteins Research Program (TPRP; Grant number FEB2 to A.Y. and Y.Ku.) and a Grant-in-Aid (Grant Numbers 19687006 to A.Y., 25121722 and 26102530 to S.U.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Footnotes
A.Y., E.N., and Y.A. are inventors on a patent (Japan 2013-246300) related to a taste-substance evaluation method by use of the T1rLBD FRET assay conditions.
Author Contributions A.Y., E.N. and Y.A. conceived the study. A.Y. and Y.A. performed the gene cloning, screening, and protein expression. E.N., Y.A., N.N., A.N. and N.Y. performed protein purification. S.A., E.N., T.H., N.N., A.Y. and M.Y. performed the SAXS analysis. E.N., S.M.-Y., K.Y. and N.N. performed the electron microscopy. Y.Ku. and M.S. performed the receptor assays. S.U. and E.K. performed the sedimentation analysis and mass spectroscopy. E.N. performed the FRET analysis. T.M., S.U., N.A., N.Y., N.N. and Y.Ko. performed the isothermal titration calorimetry. All authors performed data analysis. A.Y., E.N., S.A., K.Y., N.N., Y.Ku and S.U. wrote the paper, together with input from all of the other authors.
References
- Chandrashekar J., Hoon M. A., Ryba N. J. & Zuker C. S. The receptors and cells for mammalian taste. Nature 444, 288–294 (2006). [DOI] [PubMed] [Google Scholar]
- Yarmolinsky D. A., Zuker C. S. & Ryba N. J. Common sense about taste: from mammals to insects. Cell 139, 234–244 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P. & Zhang J. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol. Biol. Evol. 23, 292–300 (2006). [DOI] [PubMed] [Google Scholar]
- Nelson G. et al. Mammalian sweet taste receptors. Cell 106, 381–390 (2001). [DOI] [PubMed] [Google Scholar]
- Nelson G. et al. An amino-acid taste receptor. Nature 416, 199–202 (2002). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Human receptors for sweet and umami taste. Proc. Natl. Acad. Sci. USA 99, 4692–4696 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin J. P., Galvez T. & Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol. Ther. 98, 325–354 (2003). [DOI] [PubMed] [Google Scholar]
- Brauner-Osborne H., Wellendorph P. & Jensen A. A. Structure, pharmacology and therapeutic prospects of family C G-protein coupled receptors. Curr. Drug Targets 8, 169–184 (2007). [DOI] [PubMed] [Google Scholar]
- Manglik A. & Kobilka B. The role of protein dynamics in GPCR function: insights from the β2AR and rhodopsin. Curr. Opin. Cell Biol. 27, 136–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateyama M., Abe H., Nakata H., Saito O. & Kubo Y. Ligand-induced rearrangement of the dimeric metabotropic glutamate receptor 1α. Nat. Struct. Mol. Biol. 11, 637–642 (2004). [DOI] [PubMed] [Google Scholar]
- Marcaggi P., Mutoh H., Dimitrov D., Beato M. & Knopfel T. Optical measurement of mGluR1 conformational changes reveals fast activation, slow deactivation, and sensitization. Proc. Natl. Acad. Sci. USA 106, 11388–11393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumazane E. et al. Illuminating the activation mechanisms and allosteric properties of metabotropic glutamate receptors. Proc. Natl. Acad. Sci. USA 110, E1416–1425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafabakhsh R., Levitz J. & Isacoff E. Y. Conformational dynamics of a class C G-protein-coupled receptor. Nature 524, 497–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc. Natl. Acad. Sci. USA 101, 14258–14263 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima N. et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407, 971–977 (2000). [DOI] [PubMed] [Google Scholar]
- Geng Y., Bush M., Mosyak L., Wang F. & Fan Q. R. Structural mechanism of ligand activation in human GABAB receptor. Nature 504, 254–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya D., Kunishima N., Kamiya N., Jingami H. & Morikawa K. Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+. Proc. Natl. Acad. Sci. USA 99, 2660–2665 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto T., Tsuchiya D., Morikawa K. & Jingami H. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc. Natl. Acad. Sci. USA 104, 3759–3764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikawa Y. et al. GFP-based evaluation system of recombinant expression through the secretory pathway in insect cells and its application to the extracellular domains of class C GPCRs. Protein Sci. 20, 1720–1734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilegems E. et al. REEP2 enhances sweet receptor function by recruitment to lipid rafts. J. Neurosci. 30, 13774–13783 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S. & Matsunami H. Calreticulin chaperones regulate functional expression of vomeronasal type 2 pheromone receptors. Proc. Natl. Acad. Sci. USA 108, 16651–16656 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loconto J. et al. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell 112, 607–618 (2003). [DOI] [PubMed] [Google Scholar]
- Saito H., Kubota M., Roberts R. W., Chi Q. & Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691 (2004). [DOI] [PubMed] [Google Scholar]
- Kawate T. & Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- Ishimaru Y. et al. Two families of candidate taste receptors in fishes. Mech. Dev. 122, 1310–1321 (2005). [DOI] [PubMed] [Google Scholar]
- Oike H. et al. Characterization of ligands for fish taste receptors. J. Neurosci. 27, 5584–5592 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aihara Y. et al. Construction of a taste-blind medaka fish and quantitative assay of its preference-aversion behavior. Genes Brain. Behav. 7, 924–932 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushik S. V., Chen H., Thaler C., Puhl H. L. 3rd & Vogel S. S. Cerulean, Venus, and VenusY67C FRET reference standards. Biophys. J. 91, L99–L101 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange P. G. Agonist binding, agonist affinity and agonist efficacy at G protein-coupled receptors. Br. J. Pharmacol. 153, 1353–1363 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Moriyoshi E., Tsuchiya D. & Jingami H. Negative cooperativity of glutamate binding in the dimeric metabotropic glutamate receptor subtype 1. J. Biol. Chem. 279, 35526–35534 (2004). [DOI] [PubMed] [Google Scholar]
- Kubo Y., Miyashita T. & Murata Y. Structural basis for a Ca2+-sensing function of the metabotropic glutamate receptors. Science 279, 1722–1725 (1998). [DOI] [PubMed] [Google Scholar]
- Sugimoto Y. et al. Reverse conformational changes of the light chain-binding domain of myosin V and VI processive motor heads during and after hydrolysis of ATP by small-angle X-ray solution scattering. J. Mol. Biol. 392, 420–435 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerrad L. et al. A spring-loaded release mechanism regulates domain movement and catalysis in phosphoglycerate kinase. J. Biol. Chem. 286, 14040–14048 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. et al. Structure of a Class C GPCR Metabotropic Glutamate Receptor 1 Bound to an Allosteric Modulator. Science 344, 58–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R. et al. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 28, 1812–1823 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E., Simorowski N. & Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 475, 249–253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J., Schuck P., Jin R. & Mayer M. L. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat. Struct. Mol. Biol. 16, 631–638 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Ugawa S., Yamamura H., Imaizumi Y. & Shimada S. Functional interaction between T2R taste receptors and G-protein alpha subunits expressed in taste receptor cells. J. Neurosci. 23, 7376–7380 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. H., Balbo A., Zhao H., Ebel C. & Schuck P. Density contrast sedimentation velocity for the determination of protein partial-specific volumes. PLoS One 6, e26221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S. J., Baldwin P. R. & Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999). [DOI] [PubMed] [Google Scholar]
- Fujisawa T. et al. Small-angle X-ray scattering station at the SPring-8 RIKEN beamline. J. Appl. Cryst. 33, 797–800 (2000). [Google Scholar]
- Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J. & Svergun D. I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 (2003). [Google Scholar]
- Svergun D. I. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J. Appl. Cryst. 25, 495–503 (1992). [Google Scholar]
- Franke D. & Svergun D. I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V. V. & Svergun D. I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 36, 860–864 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov M. V. et al. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 45, 342–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.