Abstract
Bacteria play critical roles in peatland ecosystems. However, very little is known of how habitat heterogeneity affects the structure of the bacterial communities in these ecosystems. Here, we used amplicon sequencing of the 16S rRNA and nifH genes to investigate phylogenetic diversity and bacterial community composition in three different sub-Antarctic peat bog aquatic habitats: Sphagnum magellanicum interstitial water, and water from vegetated and non-vegetated pools. Total and putative nitrogen-fixing bacterial communities from Sphagnum interstitial water differed significantly from vegetated and non-vegetated pool communities (which were colonized by the same bacterial populations), probably as a result of differences in water chemistry and biotic interactions. Total bacterial communities from pools contained typically aquatic taxa, and were more dissimilar in composition and less species rich than those from Sphagnum interstitial waters (which were enriched in taxa typically from soils), probably reflecting the reduced connectivity between the former habitats. These results show that bacterial communities in peatland water habitats are highly diverse and structured by multiple concurrent factors.
Bacterial communities in peat bog ecosystems contribute significantly to nutrient cycling, to carbon sequestration and to greenhouse gas emissions1,2,3,4. However, we still have a limited knowledge of the diversity and spatial distribution of bacterial assemblages in peat bogs. As a result, we do not know, for example, how the composition of bacterial communities and their traits differ across peatland water bodies; although recent work has shown that bacterioplankton communities in peat bog pools are diverse and variable5. Understanding the diversity and spatial distribution of microbial communities across different peatland aquatic habitats is important, because these habitats are highly heterogeneous6; and habitat heterogeneity can lead to changes in biodiversity patterns, which provide information to many ecological and evolutionary questions, as well as to conservation planning7. The latter is especially crucial, as peat bog ecosystems may be highly vulnerable to climate change8.
Habitat differences between peatland water bodies may arise for a number of reasons. For example, peatlands can have contrasting nutrient conditions9, and it is well known that microbes often differ in their nutritional requirements10. Copiotrophic microbes have high nutritional requirements, while oligotrophic microbes are likely to outcompete copiotrophs in conditions of low nutrient availability10. Environmental parameters such as pH5 and DOC11 may also be important in determining microbial community composition in peatlands. The role of aquatic vegetation cannot be ignored either. The lack of surface vegetation allows the penetration of high levels of photosynthetically active radiation (PAR) and ultraviolet radiation, with consequences for nutrient cycling and primary production, which in turn, can promote compositional differences in microbial communities12. Furthermore, peatland vegetation is often dominated by Sphagnum mosses, and different Sphagnum species harbour different bacterial populations13.
Peatland water bodies also differ in connectivity, which influences microbial diversity patterns14,15,16. For instance, the total number of species in habitat patches connected by moderate dispersal should be higher than in a single large patch17. In addition to their effects on species richness, interconnected habitats may favour species with generalist traits. This should lead to a decline in community turnover because of increased homogenization of the metacommunity18. Consequently, understanding how habitat heterogeneity and differences in connectivity affect the distribution of bacterial communities and their traits is important to better understand the structure-function relationship in peat bog ecosystems and for conservation planning.
Here, we analysed the total and nitrogen-fixing bacterial communities found in three different aquatic habitats (vegetated and clear pools, and Sphagnum magellanicum interstitial water) within two peat bog complexes (Rancho Hambre and Valle de Andorra, Tierra del Fuego, Argentina), using high-throughput DNA sequencing of 16S rRNA and nifH genes. Biological nitrogen fixation of atmospheric nitrogen is an important process in peatlands, because the growth of Sphagnum mosses in peatlands is often N limited19. Vegetated and clear pools can be regarded as nutrient-poor island-like habitats embedded in a landscape of Sphagnum and fed essentially by rainfall, whereas Sphagnum interstitial waters are less nutrient-poor and exhibit a major degree of connectivity and are further linked to the surrounding landscape. Thus, we predict that the structure of both total bacterial and putative nitrogen-fixing communities will reflect the striking differences in nutrient status and connectivity between these habitats.
Results and Discussion
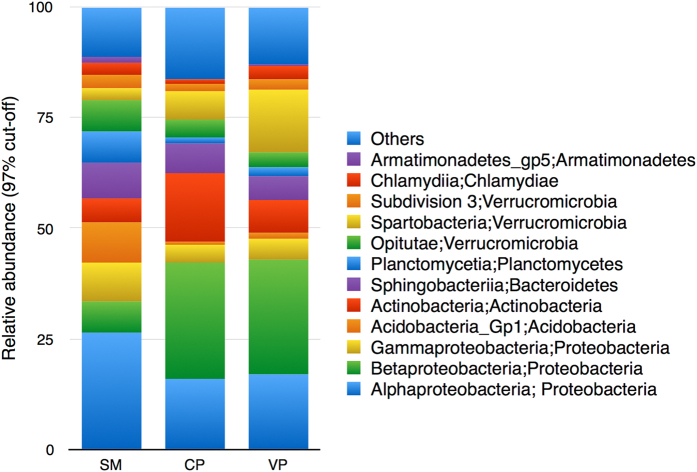
Rarefaction plots, Chao1 and Good’s coverage (95–100%) estimates indicated reasonable sequencing coverage, especially for nifH genes, (Supplementary Table S1, Supplementary Fig. S1); although more sequencing depth would be required to assess the true diversity of the samples. Overall, total bacteria and nitrogen-fixing taxa from Sphagnum interstitial waters were more diverse than those from pools (Supplementary Table S1, Supplementary Fig. S2). However, the differences were not statistically significant for putative nitrogen-fixing taxa. Most of the bacterial taxa found were members of the Proteobacteria, Verrucomicrobia, Actinobacteria, Bacteroidetes, Acidobacteria and Planctomycetes (Fig. 1), all of which are commonly found in peatland ecosystems1,5,20,21. Nevertheless, Betaproteobacteria and Actinobacteria dominated pool communities, while Sphagnum interstitial water contained a larger proportion of Alphaproteobacteria, Acidobacteria and Planctomycetes. This is in agreement with previous studies showing that members of these phyla are commonly found in association with Sphagnum mosses13,22.
Figure 1. Taxonomic information (class;phylum) based on 16S rRNA gene sequences (classified with confidence threshold of 80%) and expressed as fraction of total sequences.
The group Other encompasses unclassified sequences together with classes representing ≤1% of total sequences. CP, clear pools; VP, vegetated pools; SM, S. magellanicum interstitial waters.
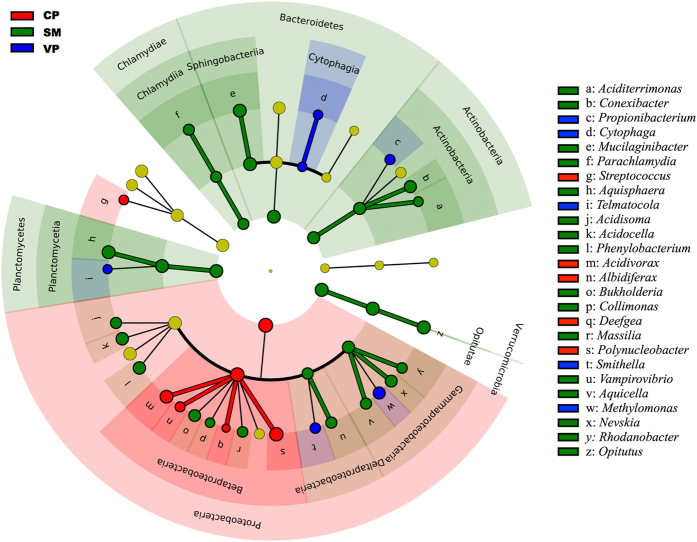
The differences in composition were also evident at the genus level. LEfSe analysis identified 26 bacterial genera as biomarkers (Fig. 2). Sphagnum interstitial water samples were enriched in genera such as Aciditerrimonas (Actinobacteria), Mucilaginibacter (Bacteroidetes) and Acidisoma (Alphaproteobacteria), whereas Propionibacterium (Actinobacteria), Polynucleobacter (Betaproteobacteria) and Methylomonas (Gammaproteobacteria) were overrepresented in pools. We do not have a complete understanding about the specific ecology of these microorganisms in peatland ecosystems. However, members of the genus Aciditerrimonas and Methylomonas have been reported as Sphagnum-associated methanotrophs4,23, which are an important sink for the methane produced in peatlands24. Mucilaginibacter spp. has the ability to use a broad range of heteropolysaccharides, such as xylan, laminarin, or pectin, in acidic and cold conditions1. Several bacterial strains affiliated to Acidisoma (e.g. A. sibiricum and A. tundrae) are aerobic, acidophilic, peat-inhabiting chemo-organotrophs that utilize a variety of sugars, polyalcohols and polysaccharides25. Fermenting bacteria such as Propionibacterium can degrade carbohydrates and polymeric compounds, therefore participating in carbon turnover26. Members of the genus Polynucleobacter can perform the assimilatory reduction of nitrate and assimilate sulphur and sulphate27. In regard to putative nitrogen-fixing microbes, most sequences were related to Proteobacteria, Firmicutes, Cyanobacteria and Verrucomicrobia (Supplementary Fig. S3). At the genus level, sequences closely related to Methylobacter (Gammaproteobacteria) and Desulfobulbus (Deltaproteobacteria) species dominated in pools, while sequences related to Bradyrhizobium (Alphaproteobacteria) and Burkholderia (Betaproteobacteria) species were overrepresented in Sphagnum interstitial waters. The nifH deduced amino acid sequences showed ≥96% similarity in BLAST analysis in all cases. Noteworthy, both Bradyrhizobium and Burkholderia species have been previously reported to associate with Sphagnum mosses28. In fact, Burkholderia strains have been found to be transmitted by Sphagnum mosses over their life cycle, highlighting the relevance of this association29. In all, our findings as well as others19,28,30, suggest that nitrogen fixation may be important in facilitating plant growth under ombrotrophic, nitrogen-limited conditions in bog ecosystems.
Figure 2. Least discriminant analysis (LDA) effect size taxonomic cladogram, based on 16S rRNA gene sequences, comparing all samples for the three habitats.
Significantly discriminant taxon nodes are coloured and branch areas are shaded according to the highest ranked group for that taxon. If the taxon is not significantly differentially represented among sample habitats, the corresponding node is coloured yellow.
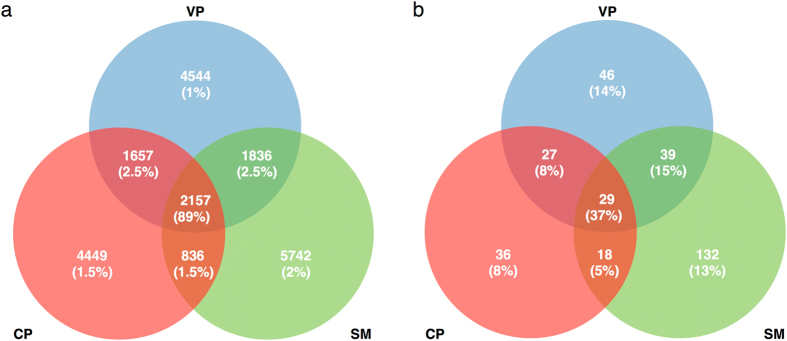
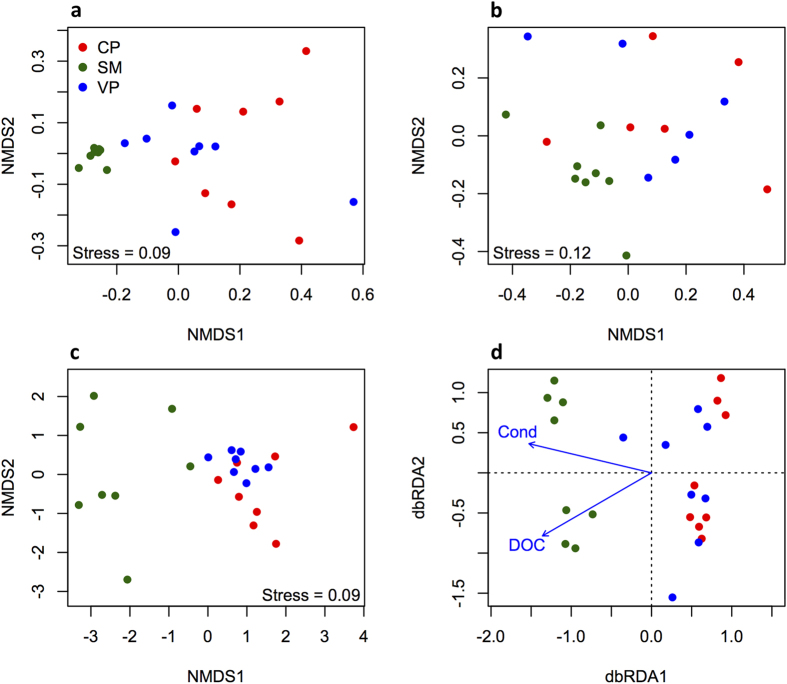
Relatively few bacterial OTUs were shared between all three communities (Fig. 3), but these shared OTUs accounted for 89% and 37% of the sequences for total and nitrogen-fixing bacterial communities, respectively. Interestingly, a large proportion of the taxa abundant in Sphagnum interstitial waters were rare in pools and vice versa (Fig. 4), suggesting a significant degree of habitat preference. Consequently, total bacterial communities, using Bray-Curtis dissimilarities, were found to differ in composition between Sphagnum interstitial water and pool samples (PERMANOVA: F2,23 = 3.43, R2 = 0.25, P = 0.001; Fig. 5a), but not between vegetated and clear pools (P > 0.5). Similar findings were obtained using weighted UniFrac dissimilarities (Supplementary Fig. S4a). Putative nitrogen-fixing bacterial communities gave comparable results to those of the total bacteria (Mantel r = 0.65, P = 0.001) (Fig. 5b, Supplementary Fig. S4b). Interestingly, total bacterial communities were considerably less variable in composition in Sphagnum interstitial water samples than in pool samples (Fig. 5a, Supplementary Figs S4a and S5a), which is interpreted as a sign of biotic homogenization. In contrast, nitrogen-fixing bacterial communities were equally variable between the three habitats (Fig. 5b, Supplementary Fig. S4b).
Figure 3.
Venn diagram showing the number of (a) shared total bacterial and (b) nitrogen-fixing OTUs. The percentage of sequences associated with OTUs is shown in parentheses. CP, clear pools; VP, vegetated pools; SM, S. magellanicum interstitial waters.
Figure 4. Taxonomic composition and abundance patterns of the two groups of 16S rRNA-derived OTUs that showed the largest changes in abundance between Sphagnum interstitial water and pools samples.
The group Other encompasses unclassified sequences together with orders representing ≤1% of total sequences. (a,b) OTUs that dominated pools (n = 169). (c,d) OTUs that dominated S. magellanicum interstitial waters (n = 200). Taxonomic data are presented as per cent contribution of each bacterial order to total sequences. OTUs were selected following the procedure described by Ruiz-González et al.15 using a mean distance >15. CP, clear pools; VP, vegetated pools; SM, S. magellanicum interstitial waters.
Figure 5.
Multidimensional scaling diagrams showing the degree of similarity (Bray-Curtis index) between (a) total bacterial communities, (b) putative nitrogen-fixing communities and (c) environmental conditions. (d) Redundancy analysis (RDA) biplot of total bacterial diversity and microenvironmental parameters. Only the environmental variables that significantly explained variability in microbial community structure are fitted to the ordination (arrows). The direction of the arrows indicates the direction of maximum change of that variable, whereas the length of the arrow is proportional to the rate of change. CP, clear pools; VP, vegetated pools; SM, S. magellanicum interstitial waters.
When differences in water chemistry were included in a non-metric multidimensional scaling ordination plot, Sphagnum interstitial water and pool samples grouped separately (PERMANOVA: F2,23 = 8.9, R2 = 0.45, P = 0.001; Fig. 5c). Conversely, no differences were found between vegetated and clear pools (P > 0.05). In addition, permutation dispersion showed that vegetated pools were more similar in chemical composition than Sphagnum interstitial water and clear pools (Supplementary Fig. S5b). A post-hoc ANOVA analysis found Sphagnum interstitial water samples having higher values of, phosphate, DOC and Chl a than pools (Supplementary Fig. 6). Using distance-based redundancy analysis we found that DOC and Conductivity explained 16% and 6% (P = 0.001) of the variation in total (Fig. 5d) and nitrogen-fixing (not shown) bacterial communities, respectively.
We propose three non-mutually exclusive explanations for the observed patterns in community composition, related to differences in (1) abiotic environmental conditions, (2) biotic interactions and (3) connectivity. Firstly, the fact that bacterial communities were different in the Sphagnum interstitial waters and the pools indicates a strong habitat effect, which is consistent with the concept of species sorting (e.g., ref. 31). Conductivity, a proxy for salinity, and DOC were the dominant drivers of the total bacterial communities (Fig. 5d). Previous studies have identified DOC to be a key driver for aquatic microorganisms32, and it is well established that different aquatic bacterial taxa have different preferences in terms of carbon utilization33. Similarly, salinity has been found to greatly influence bacterial diversity in aquatic ecosystems34, although to our knowledge, this is the first time it has been reported in peatland water bodies.
Secondly, only 6–16% of the total variation in bacterial community composition could be explained by conductivity and DOC, suggesting that other abiotic or biotic factors may be of greater importance35. For example, it is well known that Sphagnum mosses are colonized by species-specific microbial populations, which fulfil important functions (e.g. nutrient supply and pathogen defence) for moss growth and health13,30. For instance, Sphagnum fallax is colonized mainly by Verrucromicrobia and Planctomycetes, while Sphagnum magellanicum is dominated by Alphaproteobacteria and Gammaprotobacteria13. Both phyla were well represented in Sphagnum interstitial water samples (Fig. 1). Zooplankton, protists and viruses could also have a role in structuring these communities36. Different bacterial taxa are known to vary in their resistance to both grazing and viral lysis37 and mesocosm experiments have shown that predation by flagellates and ciliates can structure the composition of bacterial communities38. Interestingly, a number of studies have reported that Alphaproteobacteria are resistant to predation (e.g., ref. 39), whereas others have indicated that members of the Betaproteobacteria are vulnerable to grazing (ref. 33 and references therein). We note that Alphaproteobacteria dominated Sphagnum interstitial water samples, while Betaproteobacteria dominated pool communities (Fig. 1).
Thirdly, we suggest that the distinction between Sphagnum interstitial water and pools communities may be linked to differences in connectivity within these two habitats. Sphagnum interstitial water communities are more likely to be interconnected and further linked to terrestrial communities through hydrological networks, favouring microbial dispersal. Dispersal can lead to increased local species richness because it allows new species to enter communities18, leading to higher gamma diversity and reduced community dissimilarity17, patterns we observed in Sphagnum interstitial water samples. Similar findings have been reported in previous studies for invertebrates40, plants17, and bacteria and viruses41, suggesting that this may be a universal phenomenon. In addition, Sphagnum interstitial water samples showed a greater proportion of Acidobacteria and Gammaproteobacteria, phyla commonly found in terrestrial communities16. In contrast, clear and vegetated pools can be seen as discrete, unconnected patches isolated from the surrounding soils, which allows for the development of more dissimilar communities, even if environmental conditions are similar, as those of vegetated pools (Fig. 5c). This concept may be supported by the increase in abundance and diversity of common freshwater phyla such as Betaproteobacteria and Actinobacteria15,16,42 in pool communities relative to Sphagnum interstitial water communities. Indirect mechanisms such as dispersal-mediated trophic interactions can also generate apparent patterns of dispersal limitation in aquatic metacommunities43.
In summary, this study showed that species sorting, as a result of both abiotic and biotic interactions, plays a pivotal role in explaining differences in microbial (general and putative nitrogen-fixing bacteria) community diversity and composition across peatland water habitats. In contrast, within habitat differences were better explained by the degree of microbial dispersal, which is higher in Sphagnum interstitial water communities. This has implications for conservation planning, as the high turnover in species composition in pool communities suggests that maximizing protected area is the key to maintaining diversity in the long term. It remains to be elucidated how these changes in microbial structure and composition will affect ecosystem function.
Materials and Methods
Study site, sampling and chemical analysis
Two peatlands from Tierra del Fuego Province (Argentina) separated by a distance of 50 km were sampled in February 2014: Andorra peat bog (AN), located in the Andorra Valley (54°45′ S; 68°20′ W) and Rancho Hambre peat bog (RH), in Tierra Mayor Valley (54°44′ S; 67°49′ W) (Supplementary Fig. 7). Both peatlands are raised, nutrient-poor ombrotrophic peat bogs44. Samples were collected at twelve sampling points along a transect crossing each peat bog dome (n = 24). The sampling points included three different habitat types (four replicates each): interstitial water from Sphagnum magellanicum matrix (SM), clear pools (CP) and vegetated pools (VP), with the latter two representing patches within the Sphagnum matrix. Vegetated pools showed prolific growth of Sphagnum cf. fimbriatum and in some instances Drepanocladus uncinatus, while clear pools showed a mud bottom mainly formed by decaying S. magellanicum. Pools were sampled at the margins, while interstitial water from the Sphagnum matrix was collected by aseptically squeezing the mosses in situ. Sampled water (250 ml) was filtered through 0.22-μm sterile nitrocellulose membranes (Nalgene, Rochester, NY, USA), and membranes placed in RNAlater (Sigma-Aldrich, St. Louis, Mo, USA) and stored at 4 °C until further processing.
pH and conductivity were measured in situ using a Hach Sension 156 multiparametric probe (Hach, Loveland, CO, USA). Nutrient (ammonium, phosphate, total N and total P) concentrations were measured using a Hach DR2800 spectrophotometer and their corresponding reagent kits, following standard methods described in the Hach DR2800 spectrophotometer procedures manual (www.hach.com). Ammonium and phosphate concentrations were determined according to the salicylate (No. 8155) and ascorbic acid (No. 8048) Hach methods, respectively. Total N and total P were determined by performing an acid digestion with potassium persulphate and boric acid45 followed by nitrate and phosphate determinations by the cadmium reduction (No. 8192) and ascorbic acid (No. 8048) Hach methods, respecrively. Dissolved organic carbon (DOC) was determined using the high temperature Pt catalyst oxidation method (Shimadzu analyser TOC-5000A, SM5310B technique) following the recommendations of Sharp et al.46. Chlorophyll a (Chl a) concentration corrected for phaeopigments was determined spectrophotometrically (for details see ref. 14). Metadata and water chemistry values are shown in Supplementary Table S2.
DNA extraction and amplicon sequencing
DNA was extracted from half of each filter, cut into small pieces with a sterile blade, using a PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA). PCR was performed in a single-step PCR using HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA) with primer pairs 515F (5′-GTGYCAGCMGCCGCGGRA-3′) and 909R (5′-CCCCGYCAATTCMTTTRAG-3′) for the 16S rRNA genes47, and nifH1F (5′-TGYGAYCCNAARGCNGA-3′) and nifH2R (5′-ADNGCCATCATYTCNCC-3′) for the nifH genes48. Triplicate PCR products were pooled after amplification, mixed in equal concentrations with samples targeting a specific gene and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, MA, USA). Sequencing was carried out on an Illumina MiSeq2000 using a paired-end approach49 at the Molecular Research LP next generation sequencing service (http://www.mrdnalab.com).
16S rRNA gene sequences were analysed in MOTHUR50, following a previously established pipeline51, using the Silva core set (http://www.arb-silva.de) for alignment. Reads were removed from further analysis if at least one of the following criteria was met: (i) reads shorter than 200 bp, (ii) number of ambiguous bases greater than 5, and (iii) presence of homopolymers with more than 8 bp. One mismatch to the sample-specific barcode and to the target-specific primer were allowed. After removal of chimeras and poor quality reads, a total of 3625057 sequences were obtained. Sequences were grouped into OTUs, defined using a 97% sequence similarity cut-off. Taxonomic affiliation was determined using the naive Bayesian rRNA classifier in MOTHUR; 80% confidence level. Singletons were removed and each sample was rarefied to 39908 sequences (the lowest number of sequences in any sample). A total of 21221 OTUs (957792 sequences) were retained for analysis.
Amplicons of the nifH genes were analysed using the FunGene pipeline (http://fungene.cme.msu.edu/FunGenePipeline/)52. Filtered reads were translated into amino acid sequences and clipped at 60 aa. Further analyses were carried out on amino acid sequences clustered at 95% similarity53. Representative sequences for each cluster were classified using the NCBI algorithm BLASTP. Singletons were removed and each sample was rarefied to 911 sequences. 327 OTUs (18220 sequences) from 20 samples, four samples did not give any amplification product, were kept for further analysis.
Statistical analysis
OTU richness and diversity indices (Shannon, Inverse Simpson and Pielou’s evenness), together with rarefaction curves, Chao1 and Good’s coverage estimates were calculated using MOTHUR. We applied mixed model ANOVA to determine significant differences in diversity and water chemistry between habitat types using the phia package54. In these analyses, we specified peat bog ID as a random factor. Abiotic data were standardized and pair-wise distances computed based on Euclidean distances. The community data matrices was Hellinger-transformed and the Bray-Curtis distance measure was used to generate a dissimilarity matrix. Weighted UniFrac dissimilarities were also obtained55. The structure of the bacterial community and the environmental variables were visualized using non-metric multidimensional scaling. The effect of abiotic data in explaining variation in bacterial community structure was assessed by distance-based redundancy analysis after forward selection of the best set of parameters that could explain the variation in community composition. A permutational analysis of variance was used to test for differences in composition between habitats, ‘adonis’ function (strata = peat bog ID) in vegan for R; whereas permutation dispersion was used to test for differences in their within-habitat dissimilarity, ‘betadisper’ function.
We used LEfSe analysis56 to explore the presence of taxonomic groups that can serve as biomarkers for different classes (habitats). Statistically significant groups are reported with high LDA (Linear Discriminant Analysis) scores, which characterize the degree of consistency in relative abundance between features (genera) together with their effect relevance in each class. Correlations between the two biotic distance matrices were tested using the ‘mantel’ function in the ecodist package for R.
Additional Information
How to cite this article: Oloo, F. et al. Habitat heterogeneity and connectivity shape microbial communities in South American petlands. Sci. Rep. 6, 25712; doi: 10.1038/srep25712 (2016).
Supplementary Material
Acknowledgments
Financial support was provided by the Genomics Research Institute (University of Pretoria, SA) and the ANPCyT (Argentina) through PICT 2012-0529. We thank the Dirección Provincial de Recursos Hídricos de Tierra del Fuego and the Centro Austral de Investigaciones Científicas (CADIC-CONICET) for logistic support.
Footnotes
Author Contributions A.V., M.V.Q., D.C. and G.M. designed research. F.O. and M.V.Q. performed research. F.O. and A.V. analysed the data. A.V. wrote the manuscript. All authors commented on the manuscript at all stages.
References
- Dedysh S. N. Cultivating uncultured bacteria from northern wetlands: Knowledge gained and remaining gaps. Front Microbiol 2, 184, doi: 10.3389/fmicb.2011.00184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larmola T. et al. The role of Sphagnum mosses in the methane cycling of a boreal mire. Ecology 91, 2356–2365 (2010). [DOI] [PubMed] [Google Scholar]
- Putkinen A. et al. Water dispersal of methanotrophic bacteria maintains functional methane oxidation in Sphagnum mosses. Front Microbiol 3, 15, doi: 10.3389/fmicb.2012.00015 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkinen A. et al. Peatland succession induces a shift in the community composition of Sphagnum-associated active methanotrophs. FEMS Microbiol Ecol 88, 596–611 (2014). [DOI] [PubMed] [Google Scholar]
- Quiroga M. V., Valverde A., Mataloni G. & Cowan D. Understanding diversity patterns in bacterioplankton communities from a sub-Antarctic peatland. Environ Microbiol Rep 7, 547–553 (2015). [DOI] [PubMed] [Google Scholar]
- Foster D. R., King G. A., Glaser P. H. & Wright H. E. Origin of string patterns in boreal peatlands. Nature 306, 256–258 (1983). [Google Scholar]
- McKnight M. W. et al. Putting beta-diversity on the map: Broad-scale congruence and coincidence in the extremes. Plos Biol 5, 2424–2432 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dise N. B. Peatland response to global change. Science 326, 810–811 (2009). [DOI] [PubMed] [Google Scholar]
- Daniels R. E. & Eddy A. In Handbook of European Sphagna. Institute of terrestrial Ecology (Cambrian News, 1985). [Google Scholar]
- Fierer N., Bradford M. A. & Jackson R. B. Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364 (2007). [DOI] [PubMed] [Google Scholar]
- Lin X. et al. Microbial community structure and activity linked to contrasting biogeochemical gradients in bog and fen environments of the glacial lake agassiz peatland. Appl Environ Microbiol 78, 7023–7031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder S., Berga M., Ostman O. & Szekely A. J. Temporal variation of beta-diversity and assembly mechanisms in a bacterial metacommunity. ISME J 6, 1107–1114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina A. et al. Sphagnum mosses harbour highly specific bacterial diversity during their whole lifecycle. ISME J 6, 802–813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mataloni G., González Garraza G. & Vinocur A. Landscape-driven environmental variability largely determines abiotic characteristics and phytoplankton patterns in peat bog pools (Tierra del Fuego, Argentina). Hydrobiologia 751, 105–125 (2015). [Google Scholar]
- Ruiz-González C., Niño-García J. P. & del Giorgio P. A. Terrestrial origin of bacterial communities in complex boreal freshwater networks. Ecol Lett 18, 1198–1206 (2015). [DOI] [PubMed] [Google Scholar]
- Crump B. C., Amaral-Zettler L. A. & Kling G. W. Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J 6, 1629–1639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka M. & Takamura N. Hydrologic connection between ponds positively affects macrophyte alpha and gamma diversity but negatively affects beta diversity. Ecology 93, 967–973 (2012). [DOI] [PubMed] [Google Scholar]
- Mouquet N. & Loreau M. Community patterns in source-sink metacommunities. Am Nat 162, 544–557 (2003). [DOI] [PubMed] [Google Scholar]
- Larmola T. et al. Methanotrophy induces nitrogen fixation during peatland development. Proc Nat Acad Sci USA 111, 734–739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tveit A., Schwacke R., Svenning M. M. & Urich T. Organic carbon transformations in high-Arctic peat soils: key functions and microorganisms. ISME J 7, 299–311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina A. et al. The Sphagnum microbiome supports bog ecosystem functioning under extreme conditions. Mol Ecol 23, 4498–4510 (2014). [DOI] [PubMed] [Google Scholar]
- Bragina A. et al. Similar diversity of Alphaproteobacteria and nitrogenase gene amplicons on two related Sphagnum mosses. Front Microbiol 2, 275, doi: 10.3389/fmicb.2011.00275 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kip N. et al. Ultra-deep pyrosequencing of pmoA amplicons confirms the prevalence of Methylomonas and Methylocystis in Sphagnum mosses from a Dutch peat bog. Environ Microbiol Rep 3, 667–673 (2011). [DOI] [PubMed] [Google Scholar]
- Kip N. et al. Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat Geosci 3, 617–621 (2010). [Google Scholar]
- Belova S. E., Pankratova T. A., Detkova E. N., Kaparullina E. N. & Dedysh S. N. Acidisoma tundrae gen. nov., sp. nov. and Acidisoma sibiricum sp. nov., two acidophilic, psychrotolerant members of the Alphaproteobacteria from acidic northern wetlands. Int J Syst Evol Microbiol 59, 2283–2290 (2009). [DOI] [PubMed] [Google Scholar]
- Wagner D., Kobabe S. & Liebner S. Bacterial community structure and carbon turnover in permafrost-affected soils of the Lena Delta, northeastern Siberia. Can J Microbiol 55, 73–83 (2009). [DOI] [PubMed] [Google Scholar]
- Boscaro V. et al. Polynucleobacter necessarius, a model for genome reduction in both free-living and symbiotic bacteria. Proc Natl Acad Sci USA 110, 18590–18595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina A., Berg C., Müller H., Moser D. & Berg G. Insights into functional bacterial diversity and its effects on Alpine bog ecosystem functioning. Sci Rep 3, 1955, doi: 10.1038/srep01955 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina A., Cardinale M., Berg C. & Berg G. Vertical transmission explains the specific Burkholderia pattern in Sphagnum mosses at multi-geographic scale. Front Microbiol 4, 394, doi: 10.3389/fmicb.2013.00394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opelt K., Berg C., Schönmann S., Eberl L. & Berg G. High specificity but contrasting biodiversity of Sphagnum-associated bacterial and plant communities in bog ecosystems independent of the geographical region. ISME J 1, 502–516 (2007). [DOI] [PubMed] [Google Scholar]
- Van der Gucht K. et al. The power of species sorting: local factors drive bacterial community composition over a wide range of spatial scales. Proc Natl Acad Sci USA 104, 20404–20409 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisner B. E., Peres-Neto P. R., Lindström E. S., Barnett A. & Longhi M. L. The role of environmental and spatial processes in structuring lake communities from bacteria to fish. Ecology 87, 2985–2991 (2006). [DOI] [PubMed] [Google Scholar]
- Newton R. J., Jones S. E., Eiler A., McMahon K. D. & Bertilsson S. A Guide to the Natural History of Freshwater Lake Bacteria. Microbiol Mol Biol Rev 75, 14–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. A. & Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104, 11436–11440 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaatinen K., Fritze H., Laine J. & Laiho R. Effects of short- and long-term water-level drawdown on the populations and activity of aerobic decomposers in a boreal peatland. Glob Change Biol 13, 491–510 (2007). [Google Scholar]
- Gasol J. M., Pedrós-Alió C. & Vaqué D. Regulation of bacterial assemblages in oligotrophic plankton systems: Results from experimental and emperical approaches. Anton Leeuw Int J G 81, 435–452 (2002). [DOI] [PubMed] [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3, 537–546 (2005). [DOI] [PubMed] [Google Scholar]
- Salcher M. M., Pernthaler J., Psenner R. & Posch T. Succession of bacterial grazing defense mechanisms against protistan predators in an experimental microbial community. Aquat Microb Ecol 38, 215–229 (2005). [Google Scholar]
- Langenheder S. & Jürgens K. Regulation of bacterial biomass and community structure by metazoan and protozoan predation. Limnol Oceanogr 46, 121–134 (2001). [Google Scholar]
- Stendera S. E. S. & Johnson R. K. Additive partitioning of aquatic invertebrate species diversity across multiple spatial scales. Freshwater Biol 50, 1360–1375 (2005). [Google Scholar]
- Declerck S. A. J., Winter C., Shurin J. B., Suttle C. A. & Matthews B. Effects of patch connectivity and heterogeneity on metacommunity structure of planktonic bacteria and viruses. ISME J 7, 533–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevecoeur S., Vincent W. F., Comte J. & Lovejoy C. Bacterial community structure across environmental gradients in permafrost thaw ponds: methanotroph-rich ecosystems. Front Microbiol 6, 192; 10.3389/fmicb.2015.00192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreydt D. et al. Dispersal-mediated trophic interactions can generate apparent patterns of dispersal limitation in aquatic metacommunities. Ecol Lett 15, 218–226 (2012). [DOI] [PubMed] [Google Scholar]
- Roig C. & Roig F. A., Consideraciones generales. In Los Turbales de la Patagonia, Bases para su Inventario y la Conservacion de su Biodiversidad. (Blanco D. E. & de la Balze V. M.) 5–21 (Fundacion Humedales, Wetlands International, Buenos Aires, 2004). [Google Scholar]
- APHA [American Public Health Association]. Standard methods for examination of water and wastewater, 21st edn. (WDF 2005).
- Sharp J. H. et al. Procedures subgroup report. Mar Chem 37, 37–50 (1993). [Google Scholar]
- Tamaki H. et al. Analysis of 16S rRNA amplicon sequencing options on the roche/454 next-generation titanium sequencing platform. PLoS ONE 6, e25263, doi: 10.1371/journal.pone.0025263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr J. P. & McReynolds L. A. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol 55, 2522–2526 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich J., Westcott S. L., Baxter N. T., Highlander S. K. & Schloss P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish J. A. et al. FunGene: the functional gene pipeline and repository. Front Microbiol 4, 291, doi: 10.3389/fmicb.2013.00291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton C. R. et al. Denitrifying and diazotrophic community responses to artificial warming in permafrost and tallgrass prairie soils. Front Microbiol 6, 746, doi: 10.3389/fmicb.2015.00746 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosario-Martinez H. phia: Post-Hoc Interactio Analysis. R package version 0.2–0. https://CRAN.R-project.org/package=phia (2015).
- Lozupone C. & Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71, 8228–8235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N. et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60, doi: 10.1186/gb-2011-12-6-r60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.