Abstract
Previous studies indicate that prostate cancer antigen 3 (PCA3) is highly expressed in prostatic tumors. However, its clinical value has not been characterized. The aim of this study was to investigate the clinical value of the urine PCA3 test in the diagnosis of prostate cancer by pooling the published data. Clinical trials utilizing the urine PCA3 test for diagnosing prostate cancer were retrieved from PubMed and Embase. A total of 46 clinical trials including 12,295 subjects were included in this meta-analysis. The pooled sensitivity, specificity, positive likelihood ratio (+LR), negative likelihood ratio (−LR), diagnostic odds ratio (DOR) and area under the curve (AUC) were 0.65 (95% confidence interval [CI]: 0.63–0.66), 0.73 (95% CI: 0.72–0.74), 2.23 (95% CI: 1.91–2.62), 0.48 (95% CI: 0.44–0.52), 5.31 (95% CI: 4.19–6.73) and 0.75 (95% CI: 0.74–0.77), respectively. In conclusion, the urine PCA3 test has acceptable sensitivity and specificity for the diagnosis of prostate cancer and can be used as a non-invasive method for that purpose.
Prostate cancer is the most common cancer among men living in Western nations and now the most common malignancy (with a prevalence near that of bladder cancer) seen in urological clinics in China1. In the USA, prostate cancer is the most frequently diagnosed cancer and the second leading cause of cancer-related death in men, with an estimated 233,000 new cases and 29,480 deaths in 20142. In clinical practice, serum prostate-specific antigen (PSA), digital rectal examination (DRE), transrectal ultrasound, and biopsy are widely used for early detection. Although PSA improved prostate cancer detection in the early “PSA era,” it has many limitations, especially when PSA values are 4–10 ng/ml (the “gray zone”).
In 1999, Bussemakers et al. identified the DD3 gene (later also known as prostate cancer antigen 3, or PCA3), which is highly expressed in prostatic tumors3. A new diagnostic method uses polymerase chain reaction (PCR) to detect the over-expression of PCA3 mRNA in urine. This non-invasive urine biomarker has been evaluated in many clinical studies, in several of which it was combined with other markers to improve the diagnostic accuracy in order to further rule out aggressive cancer at biopsy. In 2012, the US Food and Drug Administration (FDA) approved the PROGENSA PCA3 assay, the first molecular test to help determine the need for repeat prostate biopsies in men with a previous negative biopsy (U.S. Food and Drug Administration Summary of Safety and Effectiveness Data: PROGENSA PCA3 Assay, 2012. Available at www.accessdata.fda.gov/cdrh_docs/pdf10/P100033b.pdf&U.S. Food and Drug Administration Medical Devices: PROGENSA PCA3 Assay, 2012. Available at www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm294907.htm).
We consider the current clinical evidence in investigating the diagnostic value of PCA3 in prostate cancer with both initial and repeat biopsy.
Methods
This meta-analysis was performed in accordance with PRISMA4 guidelines, which prefer reporting items from systematic reviews and meta-analyses.
Search strategy and study selection
A comprehensive, computerized literature search was performed in PubMed and Embase for work published through December 2014 using a combination of the following key words: [“prostatic neoplasm” or “prostate cancer”] AND [“PCA3” or “prostate cancer antigen3” or “dd3” or “upm3” or “aptima pca3”] AND [“diagnosis” or “sensitivity and specificity”]. Then, the reference sections of the identified publications were searched to identify additional potentially relevant articles. Studies included in our meta-analysis had to meet the following criteria: (1) case-control or cohort design; (2) diagnostic test using PCA3 itself or in combination with other biomarkers; and (3) prostate biopsy as the gold standard.
Data extraction and quality assessment
Data were extracted independently by 2 authors (Y.C. and C.L.) and then crosschecked. For each study, the following information was collected: last name of the first author, publication year, study design and ethnicity, age, PSA, sample size and the values of true positive (TP), false positive (FP), false negative (FN), true negative (TN), and area under the curve (AUC) (with 95% CI) if available. When more than one article was published using the same population, we selected the most recent or most informative report. Disagreements between the two authors were resolved by consensus. The quality of the selected studies was assessed using quality assessment of diagnostic accuracy studies (QUADAS)5. The QUADAS tool consists of a set of 14 questions, each of which is scored as yes, no, or unclear.
Statistical analysis
For each study, 2 × 2 tables for each test with TP, FP, FN, and TN results were extracted from the original scientific articles. Pooled estimates of sensitivity (Se) and specificity (Sp) and their 95% confidence intervals were calculated as the main outcome measures. Forest plots were used, and methodological heterogeneity was assessed during selection.
The threshold effect is a characteristic source of heterogeneity in the meta-analysis of diagnostic tests and arises when the included studies use different cut-off points to define a positive result of a diagnostic test. The analysis of the diagnostic threshold was assessed through the receiver operating characteristic (ROC) plane and Spearman’s correlation coefficient The ROC plane is the graphic representation of the pairs of Se and Sp, and it characteristically shows a curvilinear pattern if the threshold effect exists. Statistical heterogeneity was measured using the χ2 test and I2 scores. The I2 score was used as a measure of the inconsistency between studies in the meta-analysis and was interpreted as low (25–50%), moderate (51–75%), or high (>75%).
Data were analyzed using the statistical software package Metadisc, version 1.4. The results were synthesized and represented graphically in a forest plot. If heterogeneity was found, the meta-analysis was performed using a random effects model. If there was evidence of the threshold effect, the studies were combined to create a summarized ROC curve (SROC), to calculate an additional measurement of the accuracy of the technique (Q*) and to obtain the AUC.
Results
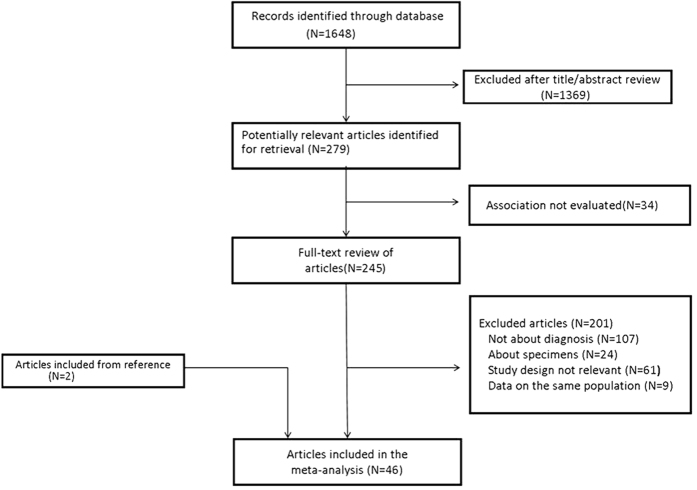
A total of 1,648 relevant references were obtained in our systematic search. The results and study selection process are shown in Fig. 1. There were 245 articles requiring full-text review, and 46 studies were included in the meta-analysis6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51. In addition, one study (by Hensen et al.52) that focused on combined initial prostate cancer biopsy in a North American and European multi-center cohort that overlapped in two studies13,34 was included in the stratified analysis of initial biopsy. Additionally, the study by Scattoni et al.48 included initial and repeat biopsy groups that were treated as two data sets. The quality of the selected studies on diagnostic testing was moderate to high according to the QUADAS scale (Table 1).
Figure 1. Literature search.
Table 1. Methodological quality of the 46 studies according to the QUADAS questionnaire.
| Study | Year | Country/region | Patients |
Test |
Result |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Author | Patients are representative of the question | Selection criteria described | Biopsy is performed in all patients | PCA3 assay described | Selection of controls | Number for cores per biopsy ≥10 | Blinded gold standard interpretation | Cut-off reported | ||
| Hessels | 2003 | Netherlands/Eur | Yes | Yes | No | Yes | Yes | No | No | Yes |
| Fradet | 2004 | Canada | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Tinzel | 2004 | Austria/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Groskopf | 2006 | US | Yes | Yes | Yes | Yes | No | Unclear | No | Yes |
| van Gils | 2007 | Netherlands/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| van Gils | 2007 | Netherlands/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Marks | 2007 | US, Canada | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Deras | 2008 | US, Canada | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Haese | 2008 | Europe | Yes | Yes | Yes | Yes | Yes | Unclear | Yes | Yes |
| Laxman | 2008 | US | Yes | Yes | Yes | Yes | No | Unclear | Yes | Yes |
| Ouyang | 2009 | US | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Shappell | 2009 | US | Yes | Yes | No | Yes | No | Unclear | No | Yes |
| Wang | 2009 | US | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Mearini | 2009 | Italy/Eur | Yes | Yes | Yes | Yes | No | Yes | No | Yes |
| Henderson | 2010 | UK/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Aubin | 2010 | US | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Auprich | 2010 | Europe | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Morotel | 2010 | Spain/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Nyberg | 2010 | Sweden/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Roobol | 2010 | Netherlands/Eur | Yes | Yes | Yes | Yes | Yes | Unclear | Yes | Yes |
| Rigau | 2010 | Spain/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Shen | 2010 | China/Asian | Yes | Yes | Yes | Yes | No | Unclear | Yes | Yes |
| Schilling | 2010 | Germany/Eur | Yes | Yes | Yes | Yes | Unclear | Yes | No | Yes |
| Rubio-Briones | 2011 | Spain/Eur | Yes | Yes | Yes | Yes | Yes | Unclear | Yes | Yes |
| Adam | 2011 | South Africa | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Cao | 2011 | China/Asian | Yes | Yes | Unclear | Yes | No | Yes | No | Yes |
| Ochiai | 2011 | Japan | Yes | Yes | Yes | Yes | Yes | Unclear | Yes | Yes |
| Perdona | 2011 | Italy/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Taille | 2011 | Europe | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Babera | 2012 | Italy/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No |
| CF Ng | 2012 | China/Asian | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No |
| Crawford | 2012 | US | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Pepe | 2012 | Italy/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Pepe | 2012 | Italy/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Sciarra | 2012 | Italy/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Wu | 2012 | US | Yes | Yes | Yes | Yes | No | Yes | No | Yes |
| Ferro | 2013 | Europe | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Goode | 2013 | US | Yes | Yes | Yes | Yes | No | Yes | No | Yes |
| Ochiai | 2013 | Japan | Yes | Yes | Yes | Yes | Yes | Unclear | Yes | Yes |
| Stephan | 2013 | Germany/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Perdona‘ | 2013 | Italy/EurCaucasian | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Salagierski | 2013 | Poland/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Scattoni | 2013 | Italy/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Chevli | 2013 | USA | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Giuseppe | 2014 | Italy/Eur | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Francesco | 2014 | Italy/Eur | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
Based on the studies described above, we retrieved data from 12,295 patients with PCA3 test results and prostate biopsy, of whom 4,225 were diagnosed with prostate cancer. All studies presented the sensitivity, specificity, and cut-off points (25 studies had a cut-off of PCA3 = 35), and most studies presented the ROC curve (Supplementary Table 1). Among the 46 trials, most were performed in the U.S. and Europe; 5 were performed in Asia (Table 1).
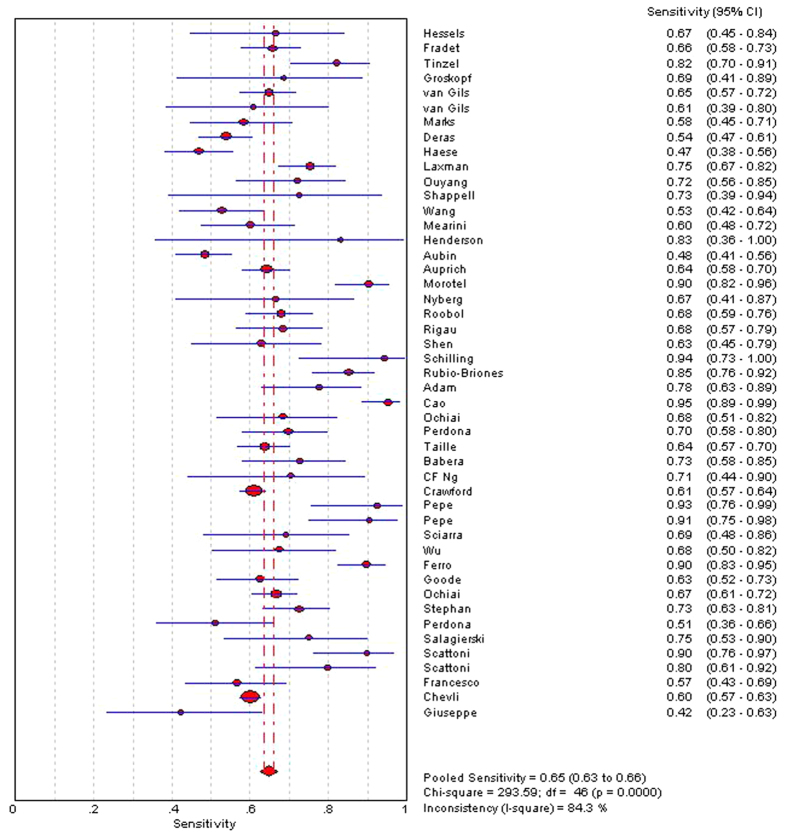
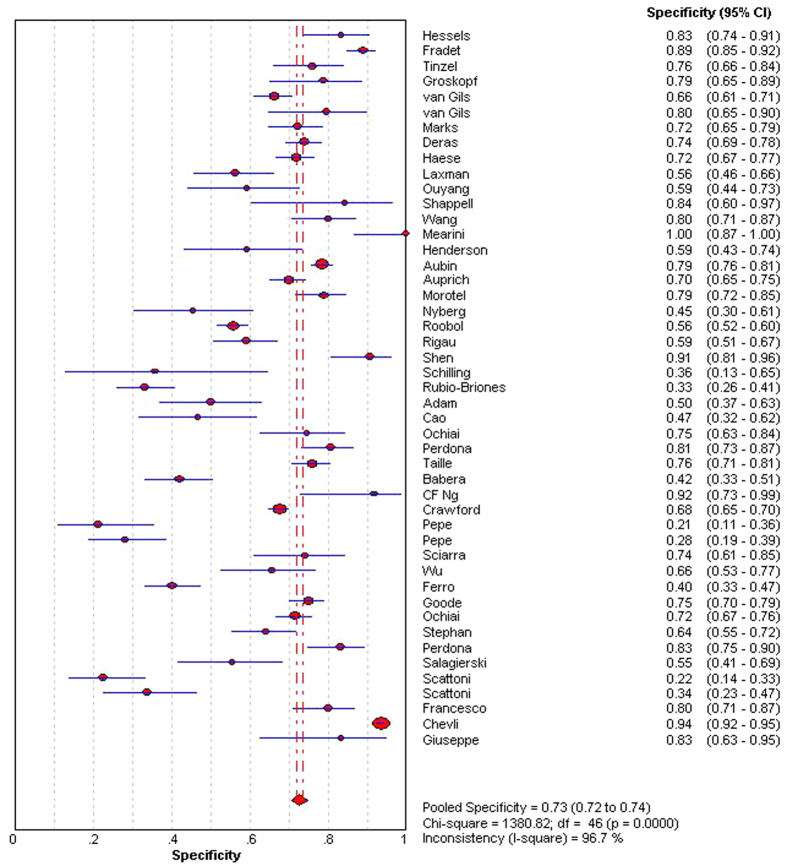
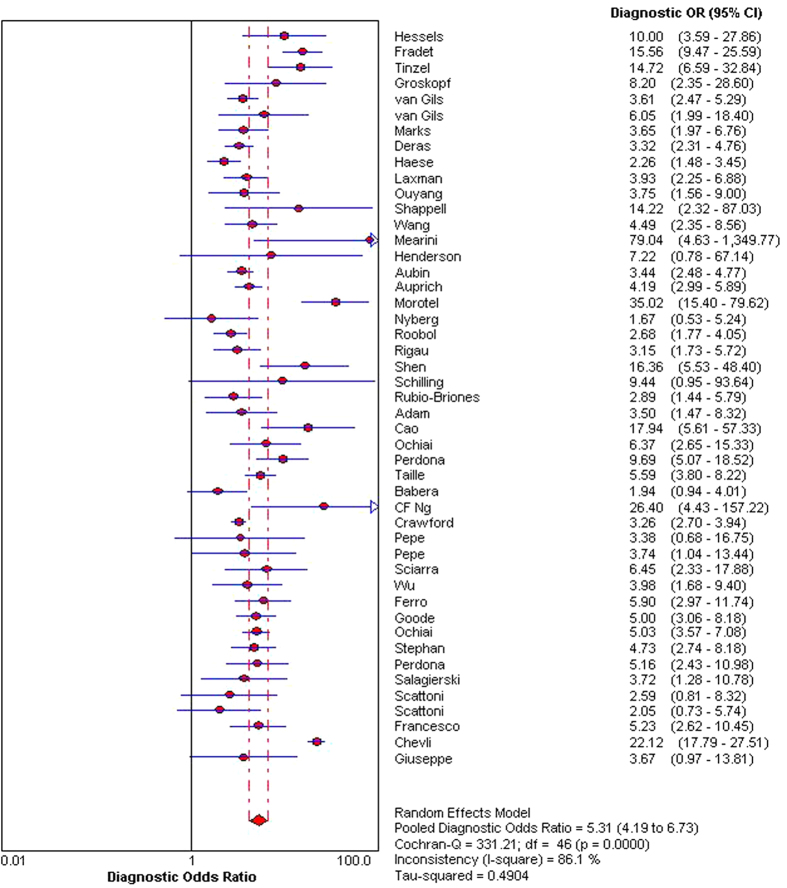
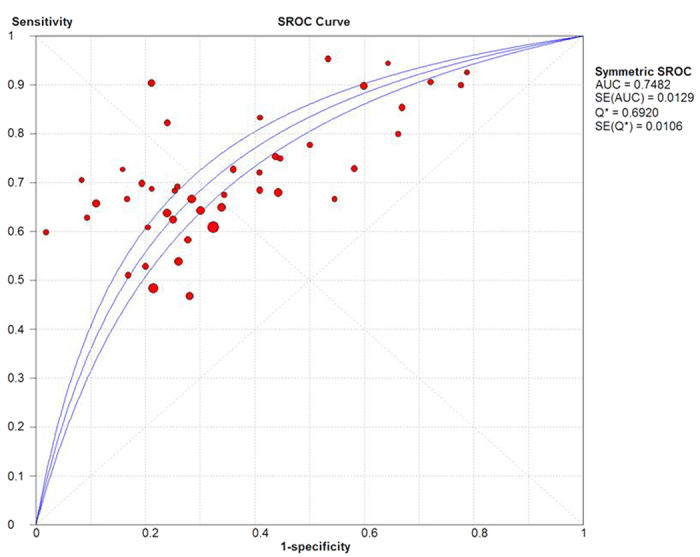
The indices of diagnostic validity obtained from the 2 × 2 tables showed that sensitivity ranged from 46.9% to 95%, and specificity ranged from 21.6% to 100%. In the 40 articles that presented the AUC, it ranged from 0.57 to 0.85. A meta-analysis was conducted using the 46 articles mentioned above. The ROC space showed a curvilinear trend, and Spearman’s correlation coefficient was 0.612 (P < 0.001), which suggests the existence of a threshold. There was a high degree of heterogeneity in sensitivity (χ2 = 271.39, P < 0.001), specificity (χ2 = 735.87, P < 0.001), and diagnostic OR (Cochran-Q = 137.22, P < 0.001); consequently, the diagnostic indices were calculated using a random effects model. Using a forest plot, the overall sensitivity, specificity, positive likelihood ratio , negative likelihood ratio and diagnostic OR were 0.65 (95% CI 0.63–0.66) (Fig. 2), 0.73 (95% CI 0.72–0.74) (Fig. 3), 2.23 (95% CI: 1.91–2.62), 0.48 (95% CI: 0.44–0.52) and 5.31 (95% CI: 4.19–6.73) (Fig. 4), respectively. We used a summary SROC to aggregate data and obtained a symmetrical curve with an AUC of 0.748 (Fig. 5) that represented the technique’s diagnostic performance.
Figure 2. Forest plot of pooled sensitivity.
Figure 3. Forest plot of pooled specificity.
Figure 4. Forest plot of pooled diagnostic OR.
Figure 5. SROC curve.
Discussion
The clinical value for prostatic cancer diagnoses was not conclusive. According to the European Association of Urology and the National Comprehensive Cancer Network guidelines, the need for prostate biopsy should be determined on the basis of PSA and/or a suspicious DRE53,54. However, serum PSA levels can be elevated in benign conditions, and only 25% of men who are clinically suspected of having PCa will have a positive biopsy55,56. Thus, other biomarkers with high sensitivity and specificity are needed for screening.
The PCA3 gene was mapped to chromosome 9q21–22, in antisense orientation within intron 6 of the Prune homolog 2 gene (PRUNE2 or BMCC1), spanning a region of approximately 25 kb57,58. Ferreira et al. found that PCA3 may modulate PCa cell survival, and PCA3 expression is androgen-regulated via activation of AR-mediated signaling59. PCA3 is a non-coding, prostate-specific mRNA that is highly over-expressed in 95% of PCa cells, with a median 66-fold up-regulation compared with adjacent non-neoplastic cells60. Because PCA3 does not encode a protein, the only molecule that can be tested is the mRNA; PCA3 mRNA can be measured in urine sediment after DRE. A PCA3 score is the ratio of PCA3 mRNA to PSA mRNA multiplied by 1,000. Although we currently have a good understanding of the role of PCA3 in tumor genes and tissues, the picture is incomplete.
Several studies have indicated that the PCA3 test is useful in reducing the number of negative biopsies34,39,51, and more recently the FDA approved the PROGENSA PCA3 assay as a new test for prostate cancer.
Recent advances have included biomarkers such as HPG-1, AMACR, STAMP1, TMPRSS2, ERG, PHI, and P2PSA31,42,48,61,62,63. Some studies have assessed their efficacy by detecting these markers alone or in combination. Although several biomarkers may have specificity that is the same as or higher than that of PCA3, the non-invasive nature of the urine PCA3 test, which is performed after prostate massage, and its good diagnostic performance may make the PCA3 test a better choice for prostate cancer screening.
The current meta-analysis shows the clinical usefulness of this tumor marker in detecting prostate cancer with initial or repeat biopsy. We synthesized the current knowledge about early diagnosis of prostate cancer with PCA3 determination in urine samples. According to the data from 46 studies analyzed, specificity is 0.65 (range 47–95%), which is not adequate; sensitivity is 0.73, which is somewhat lower, and had a minimum value of 21%. The AUC of 0.75 obtained in the SROC curve suggests acceptable performance of the diagnostic test.
In the stratified analysis, a forest plot of initial biopsy showed overall sensitivity and specificity of 0.65 (95% CI 0.63–0.67) and 0.82 (95% CI 0.81–0.83), respectively, and a symmetrical curve with an AUC of 0.80 (95% CI 0.78–0.82) (Table 2). With repeat biopsy, these values dropped to 0.58 (95% CI 0.55–0.62), 0.69 (95% CI 0.67–0.71), and 0.68 (95% CI 0.67–0.70) (Table 2), respectively. Mixed biopsy showed values of 0.66 (95% CI 0.64–0.68), 0.68 (95% CI 0.67–0.69), and 0.75 (95% CI 0.74–0.76) (Table 2), respectively. These results suggest that PCA3 is potentially more suitable for initial prostate biopsy than repeat prostate biopsy. When we stratified the studies by PCA3 cut-off value, the overall sensitivity, specificity, and symmetrical curve AUC values were 0.63 (95% CI 0.62–0.65), 0.74 (95% CI 0.73–0.75), 0.74 (95% CI 0.73–0.76), respectively, for studies with a cut-off value ≠ 35 (Table 2) and 0.70 (95% CI 0.68–0.73), 0.67 (95% CI 0.65–0.69), and 0.77 (95% CI 0.75–0.79), respectively, for studies with a cut-off value = 35 (Table 2). Although the latter group showed better diagnostic performance, it has a greater range and more variable outcomes. This observation supports a cut-off of 35 for the standard value and clinical practice of many institutions. Comparing study designs, the overall sensitivity, specificity, and symmetrical curve AUC values were 0.63 (95% CI 0.61–0.66), 0.88 (95% CI 0.87–0.90), and 0.82 (95% CI 0.79–0.85) (Table 2), respectively, for case-control studies and 0.65 (95% CI 0.63–0.66), 0.73 (95% CI 0.72–0.74), and 0.75 (95% CI 0.74–0.76) (Table 2), respectively, for prospective studies. Because case-control studies typically enroll fewer patients and have greater heterogeneity, their quality is not as good as that of prospective studies.
Table 2. PCA3 stratified analysis.
| Data sets | Sensitivity (95% CI) | Specificity (95% CI) | Diagnostic OR (95% CI) | I2, %* | AUC (95% CI) | |
|---|---|---|---|---|---|---|
| Total | 43 | |||||
| Design | ||||||
| Case-control | 8 | 0.63 (0.61–0.66) | 0.88 (0.87–0.90) | 10.36 (5.51–21.25) | 36.9 | 0.82 (0.79–0.85) |
| Prospective | 39 | 0.65 (0.63–0.66) | 0.73 (0.72–0.74) | 5.31 (4.19–6.73) | 70.5 | 0.75 (0.74–0.76) |
| biopsy | ||||||
| Initial* | 14 | 0.65 (0.63–0.67) | 0.82 (0.81–0.83) | 8.14 (4.78–13.86) | 51.8 | 0.80 (0.78–0.82) |
| Repeated* | 11 | 0.58 (0.55–0.62) | 0.69 (0.67–0.71) | 3.19 (2.62–3.83) | 0 | 0.68 (0.67–0.70) |
| Mixed | 22 | 0.66 (0.64–0.68) | 0.68 (0.67–0.69) | 5.13 (3.99–6.60) | 76.9 | 0.75 (0.74–0.76) |
| cut-off value | ||||||
| Equal 35 | 26 | 0.63 (0.62–0.65) | 0.74 (0.73–0.75) | 4.75 (3.42–6.60) | 64.1 | 0.74 (0.73–0.76) |
| Not equal 35 | 21 | 0.70 (0.68–0.73) | 0.67 (0.65–0.69) | 6.22 (4.62–8.37) | 62.6 | 0.77 (0.76–0.79) |
Our meta-analysis has some limitations. First, the study numbers and the heterogeneity of their approaches influence the accuracy. Although the gold standard (biopsy) was used in all studies, the patient selection, lack of blinding, and different PCA3 cut-off values caused heterogeneity. Second, a potential publication bias may exist, although we tried to avoid this bias by expanding our searches in different databases and by conducting rigorous screening for studies. We evaluated the quality of the articles according to the QUADAS questionnaire. The quality of the studies in terms of diagnostic testing was moderate to high. The relatively small number of trials included in this meta-analysis and significant heterogeneity across the studies may make our conclusion conservative.
According to the current meta-analysis, the PCA3 test shows good diagnostic performance. However, it requires further exploration in well-designed and appropriately-powered trials to determine intermediate and long-term outcomes. Long-term observational studies of health outcomes are also subject to biases.
Additional Information
How to cite this article: Cui, Y. et al. Evaluation of prostate cancer antigen 3 for detecting prostate cancer: a systematic review and meta-analysis. Sci. Rep. 6, 25776; doi: 10.1038/srep25776 (2016).
Supplementary Material
Acknowledgments
This study was supported by two Suzhou Science and Technology Foundation Grant–Leading Project for Medical Science (Nos SYSD2014114 and LCZX201511) and Suzhou Key Medical Center Foundation Grant (No. Szzx201506).
Footnotes
Author Contributions Conceived and designed the study strategy: Q.S.; acquisition of data, statistical analysis and interpretation of data: Y.C., W.Z.C. and Q.L.; drafting or revision of the manuscript: Q.S., H.S., C.L. and J.P.D.; reference collection and data management: Y.C. and J.F.X.; prepared the tables and figures: W.Z.C., Q.L. and J.P.D.; study supervision: Y.C. and Q.S. All authors reviewed the manuscript.
References
- Shao Q. et al. Prostate cancer in the senior men from rural areas in east district of China: contemporary management and 5-year outcomes at multi-institutional collaboration. Cancer Lett 315, 170–177, doi: 10.1016/j.canlet.2011.09.035 (2012). [DOI] [PubMed] [Google Scholar]
- Siegel R., Ma J., Zou Z. & Jemal A. Cancer statistics, 2014. CA Cancer J Clin 64, 9–29, doi: 10.3322/caac.21208 (2014). [DOI] [PubMed] [Google Scholar]
- Bussemakers M. J. et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 59, 5975–5979 (1999). [PubMed] [Google Scholar]
- Liberati A. et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 339, b2700, doi: 10.1136/bmj.b2700 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting P., Rutjes A. W., Reitsma J. B., Bossuyt P. M. & Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol 3, 25, doi: 10.1186/1471-2288-3-25 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessels D. et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol 44, 8–15; discussion 15–16 (2003). [DOI] [PubMed] [Google Scholar]
- Fradet Y. et al. uPM3, a new molecular urine test for the detection of prostate cancer. Urology 64, 311–315; discussion 315–316, doi: 10.1016/j.urology.2004.03.052 (2004). [DOI] [PubMed] [Google Scholar]
- Tinzl M., Marberger M. & Horvath S. & Chypre, C. DD3PCA3 RNA analysis in urine–a new perspective for detecting prostate cancer. Eur Urol 46, 182–186; discussion 187, doi: 10.1016/j.eururo.2004.06.004 (2004). [DOI] [PubMed] [Google Scholar]
- Groskopf J. et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem 52, 1089–1095, doi: 10.1373/clinchem.2005.063289 (2006). [DOI] [PubMed] [Google Scholar]
- van Gils M. P. et al. The time-resolved fluorescence-based PCA3 test on urinary sediments after digital rectal examination; a Dutch multicenter validation of the diagnostic performance. Clin Cancer Res 13, 939–943, doi: 10.1158/1078-0432.CCR-06-2679 (2007). [DOI] [PubMed] [Google Scholar]
- van Gils M. P. et al. Molecular PCA3 diagnostics on prostatic fluid. Prostate 67, 881–887, doi: 10.1002/pros.20564 (2007). [DOI] [PubMed] [Google Scholar]
- Marks L. S. et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology 69, 532–535, doi: 10.1016/j.urology.2006.12.014 (2007). [DOI] [PubMed] [Google Scholar]
- Deras I. L. et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J Urol 179, 1587–1592, doi: 10.1016/j.juro.2007.11.038 (2008). [DOI] [PubMed] [Google Scholar]
- Haese A. et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol 54, 1081–1088, doi: 10.1016/j.eururo.2008.06.071 (2008). [DOI] [PubMed] [Google Scholar]
- Laxman B. et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res 68, 645–649, doi: 10.1158/0008-5472.CAN-07-3224 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang B. et al. A duplex quantitative polymerase chain reaction assay based on quantification of alpha-methylacyl-CoA racemase transcripts and prostate cancer antigen 3 in urine sediments improved diagnostic accuracy for prostate cancer. J Urol 181, 2508–2513; discussion 2513–2504, doi: 10.1016/j.juro.2009.01.110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shappell S. B. et al. PCA3 urine mRNA testing for prostate carcinoma: patterns of use by community urologists and assay performance in reference laboratory setting. Urology 73, 363–368, doi: 10.1016/j.urology.2008.08.459 (2009). [DOI] [PubMed] [Google Scholar]
- Wang R., Chinnaiyan A. M., Dunn R. L., Wojno K. J. & Wei J. T. Rational approach to implementation of prostate cancer antigen 3 into clinical care. Cancer 115, 3879–3886, doi: 10.1002/cncr.24447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mearini E. et al. The combination of urine DD3(PCA3) mRNA and PSA mRNA as molecular markers of prostate cancer. Biomarkers 14, 235–243, doi: 10.1080/13547500902807306 (2009). [DOI] [PubMed] [Google Scholar]
- Henderson J. et al. The role of PCA3 testing in patients with a raised prostate-specific antigen level after Greenlight photoselective vaporization of the prostate. J Endourol 24, 1821–1824, doi: 10.1089/end.2010.0196 (2010). [DOI] [PubMed] [Google Scholar]
- Aubin S. M. et al. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol 184, 1947–1952, doi: 10.1016/j.juro.2010.06.098 (2010). [DOI] [PubMed] [Google Scholar]
- Auprich M. et al. External validation of urinary PCA3-based nomograms to individually predict prostate biopsy outcome. Eur Urol 58, 727–732, doi: 10.1016/j.eururo.2010.06.038 (2010). [DOI] [PubMed] [Google Scholar]
- Morote J. et al. Behavior of the PCA3 gene in the urine of men with high grade prostatic intraepithelial neoplasia. World J Urol 28, 677–680, doi: 10.1007/s00345-010-0580-0 (2010). [DOI] [PubMed] [Google Scholar]
- Nyberg M. et al. PCA3 as a diagnostic marker for prostate cancer: a validation study on a Swedish patient population. Scand J Urol Nephrol 44, 378–383, doi: 10.3109/00365599.2010.521187 (2010). [DOI] [PubMed] [Google Scholar]
- Roobol M. J. et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. Eur Urol 58, 475–481, doi: 10.1016/j.eururo.2010.06.039 (2010). [DOI] [PubMed] [Google Scholar]
- Rigau M. et al. PSGR and PCA3 as biomarkers for the detection of prostate cancer in urine. Prostate 70, 1760–1767, doi: 10.1002/pros.21211 (2010). [DOI] [PubMed] [Google Scholar]
- Shen M. et al. The diagnostic value of PCA3 gene-based analysis of urine sediments after digital rectal examination for prostate cancer in a Chinese population. Exp Mol Pathol 90, 97–100, doi: 10.1016/j.yexmp.2010.10.009 (2011). [DOI] [PubMed] [Google Scholar]
- Schilling D. et al. Interpretation of the prostate cancer gene 3 in reference to the individual clinical background: implications for daily practice. Urol Int 85, 159–165, doi: 10.1159/000314078 (2010). [DOI] [PubMed] [Google Scholar]
- Rubio-Briones J. et al. [Outcomes of expanded use of PCA3 testing in a Spanish population with clinical suspicion of prostate cancer]. Actas Urol Esp 35, 589–596, doi: 10.1016/j.acuro.2011.04.001 (2011). [DOI] [PubMed] [Google Scholar]
- Adam A. et al. The role of the PCA3 assay in predicting prostate biopsy outcome in a South African setting. BJU Int 108, 1728–1733, doi: 10.1111/j.1464-410X.2011.10202.x (2011). [DOI] [PubMed] [Google Scholar]
- Cao D. L. et al. A multiplex model of combining gene-based, protein-based, and metabolite-based with positive and negative markers in urine for the early diagnosis of prostate cancer. Prostate 71, 700–710, doi: 10.1002/pros.21286 (2011). [DOI] [PubMed] [Google Scholar]
- Ochiai A. et al. Prostate cancer gene 3 urine assay for prostate cancer in Japanese men undergoing prostate biopsy. Int J Urol 18, 200–205, doi: 10.1111/j.1442-2042.2010.02711.x (2011). [DOI] [PubMed] [Google Scholar]
- Perdona S. et al. Prostate cancer detection in the “grey area” of prostate-specific antigen below 10 ng/ml: head-to-head comparison of the updated PCPT calculator and Chun’s nomogram, two risk estimators incorporating prostate cancer antigen 3. Eur Urol 59, 81–87, doi: 10.1016/j.eururo.2010.09.036 (2011). [DOI] [PubMed] [Google Scholar]
- de la Taille A. et al. Clinical evaluation of the PCA3 assay in guiding initial biopsy decisions. J Urol 185, 2119–2125, doi: 10.1016/j.juro.2011.01.075 (2011). [DOI] [PubMed] [Google Scholar]
- Barbera M., Pepe P., Paola Q. & Aragona F. PCA3 score accuracy in diagnosing prostate cancer at repeat biopsy: our experience in 177 patients. Arch Ital Urol Androl 84, 227–229 (2012). [PubMed] [Google Scholar]
- Ng C. F. et al. The role of urine prostate cancer antigen 3 mRNA levels in the diagnosis of prostate cancer among Hong Kong Chinese patients. Hong Kong Med J 18, 459–465 (2012). [PubMed] [Google Scholar]
- Crawford E. D. et al. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: a prospective study of 1,962 cases. J Urol 188, 1726–1731, doi: 10.1016/j.juro.2012.07.023 (2012). [DOI] [PubMed] [Google Scholar]
- Pepe P. & Aragona F. PCA3 score vs PSA free/total accuracy in prostate cancer diagnosis at repeat saturation biopsy. Anticancer Res 31, 4445–4449 (2011). [PubMed] [Google Scholar]
- Pepe P., Fraggetta F., Galia A., Skonieczny G. & Aragona F. PCA3 score and prostate cancer diagnosis at repeated saturation biopsy. Which cut-off: 20 or 35? Int Braz J Urol 38, 489–495 (2012). [DOI] [PubMed] [Google Scholar]
- Sciarra A. et al. Multiparametric magnetic resonance imaging of the prostate can improve the predictive value of the urinary prostate cancer antigen 3 test in patients with elevated prostate-specific antigen levels and a previous negative biopsy. BJU Int 110, 1661–1665, doi: 10.1111/j.1464-410X.2012.11146.x (2012). [DOI] [PubMed] [Google Scholar]
- Wu A. K., Reese A. C., Cooperberg M. R., Sadetsky N. & Shinohara K. Utility of PCA3 in patients undergoing repeat biopsy for prostate cancer. Prostate Cancer Prostatic Dis 15, 100–105, doi: 10.1038/pcan.2011.52 (2012). [DOI] [PubMed] [Google Scholar]
- Ferro M. et al. Prostate Health Index (Phi) and Prostate Cancer Antigen 3 (PCA3) significantly improve prostate cancer detection at initial biopsy in a total PSA range of 2-10 ng/ml. PLoS One 8, e67687, doi: 10.1371/journal.pone.0067687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode R. R., Marshall S. J., Duff M., Chevli E. & Chevli K. K. Use of PCA3 in detecting prostate cancer in initial and repeat prostate biopsy patients. Prostate 73, 48–53, doi: 10.1002/pros.22538 (2013). [DOI] [PubMed] [Google Scholar]
- Ochiai A. et al. Clinical utility of the prostate cancer gene 3 (PCA3) urine assay in Japanese men undergoing prostate biopsy. BJU Int 111, 928–933, doi: 10.1111/j.1464-410X.2012.11683.x (2013). [DOI] [PubMed] [Google Scholar]
- Stephan C. et al. Comparative assessment of urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion with the serum [-2]proprostate-specific antigen-based prostate health index for detection of prostate cancer. Clin Chem 59, 280–288, doi: 10.1373/clinchem.2012.195560 (2013). [DOI] [PubMed] [Google Scholar]
- Perdona S. et al. Prostate health index (phi) and prostate cancer antigen 3 (PCA3) significantly improve diagnostic accuracy in patients undergoing prostate biopsy. Prostate 73, 227–235, doi: 10.1002/pros.22561 (2013). [DOI] [PubMed] [Google Scholar]
- Salagierski M., Mulders P. & Schalken J. A. Predicting prostate biopsy outcome using a PCA3-based nomogram in a Polish cohort. Anticancer Res 33, 553–557 (2013). [PubMed] [Google Scholar]
- Scattoni V. et al. Head-to-head comparison of prostate health index and urinary PCA3 for predicting cancer at initial or repeat biopsy. J Urol 190, 496–501, doi: 10.1016/j.juro.2013.02.3184 (2013). [DOI] [PubMed] [Google Scholar]
- Chevli K. K. et al. Urinary PCA3 as a predictor of prostate cancer in a cohort of 3,073 men undergoing initial prostate biopsy. J Urol 191, 1743–1748, doi: 10.1016/j.juro.2013.12.005 (2014). [DOI] [PubMed] [Google Scholar]
- Porpiglia F. et al. The roles of multiparametric magnetic resonance imaging, PCA3 and prostate health index-which is the best predictor of prostate cancer after a negative biopsy? J Urol 192, 60–66, doi: 10.1016/j.juro.2014.01.030 (2014). [DOI] [PubMed] [Google Scholar]
- Albino G. et al. Evaluation of the diagnostic and predictive power of PCA3 in the prostate cancer. A different best cut-off in each different scenario. Preliminary results. Arch Ital Urol Androl 86, 306–310, doi: 10.4081/aiua.2014.4.306 (2014). [DOI] [PubMed] [Google Scholar]
- Hansen J. et al. Initial prostate biopsy: development and internal validation of a biopsy-specific nomogram based on the prostate cancer antigen 3 assay. Eur Urol 63, 201–209, doi: 10.1016/j.eururo.2012.07.030 (2013). [DOI] [PubMed] [Google Scholar]
- Heidenreich A. et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and treatment of clinically localised disease. Eur Urol 59, 61–71, doi: 10.1016/j.eururo.2010.10.039 (2011). [DOI] [PubMed] [Google Scholar]
- Mohler J. L. The 2010 NCCN clinical practice guidelines in oncology on prostate cancer. J Natl Compr Canc Netw 8, 145 (2010). [DOI] [PubMed] [Google Scholar]
- Greene K. L. et al. Prostate specific antigen best practice statement: 2009 update. J Urol 182, 2232–2241, doi: 10.1016/j.juro.2009.07.093 (2009). [DOI] [PubMed] [Google Scholar]
- Schalken J. A., Hessels D. & Verhaegh G. New targets for therapy in prostate cancer: differential display code 3 (DD3 (PCA3)), a highly prostate cancer-specific gene. Urology 62, 34–43 (2003). [DOI] [PubMed] [Google Scholar]
- Salagierski M. et al. Differential expression of PCA3 and its overlapping PRUNE2 transcript in prostate cancer. Prostate 70, 70–78, doi: 10.1002/pros.21040.(2010). [DOI] [PubMed] [Google Scholar]
- Clarke R. A. et al. New genomic structure for prostate cancer specific gene PCA3 within BMCC1: implications for prostate cancer detection and progression. PloS one 4, e4995, doi: 10.1371/journal.pone.0004995 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L. B. et al. PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling. BMC cancer 12, 507, doi: 10.1186/1471-2407-12-507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herness E. A. & Naz R. K. A novel human prostate-specific gene-1 (HPG-1): molecular cloning, sequencing, and its potential involvement in prostate carcinogenesis. Cancer research 63, 329–336 (2003). [PubMed] [Google Scholar]
- Luo J. et al. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res 62, 2220–2226 (2002). [PubMed] [Google Scholar]
- Korkmaz K. S. et al. Molecular cloning and characterization of STAMP1, a highly prostate-specific six transmembrane protein that is overexpressed in prostate cancer. J Biol Chem 277, 36689–36696, doi: 10.1074/jbc.M202414200 (2002). [DOI] [PubMed] [Google Scholar]
- Wang W. et al. Diagnostic ability of %p2PSA and prostate health index for aggressive prostate cancer: a meta-analysis. Sci Rep 4, 5012, doi: 10.1038/srep05012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.