Abstract
Chloroplast biogenesis is one of the most important subjects in plant biology. In this study, an Arabidopsis early chloroplast biogenesis mutant with a delayed pale-greening phenotype (dpg1) was isolated from a T-DNA insertion mutant collection. Both cotyledons and true leaves of dpg1 mutants were initially albino but gradually became pale green as the plant matured. Transmission electron microscopic observations revealed that the mutant displayed a delayed proplastid-to-chloroplast transition. Sequence and transcription analyses showed that AtDPG1 encodes a putatively chloroplast-localized protein containing three predicted transmembrane helices and that its expression depends on both light and developmental status. GUS staining for AtDPG1::GUS transgenic lines showed that this gene was widely expressed throughout the plant and that higher expression levels were predominantly found in green tissues during the early stages of Arabidopsis seedling development. Furthermore, quantitative real-time RT-PCR analyses revealed that a number of chloroplast- and nuclear-encoded genes involved in chlorophyll biosynthesis, photosynthesis and chloroplast development were substantially down-regulated in the dpg1 mutant. These data indicate that AtDPG1 plays an essential role in early chloroplast biogenesis, and its absence triggers chloroplast-to-nucleus retrograde signalling, which ultimately down-regulates the expression of nuclear genes encoding chloroplast-localized proteins.
The chloroplast is an essential organelle in plant cells and plays important roles in primary metabolism, such as CO2 fixation, manufacture of carbon skeletons and fatty acids, and synthesis of amino acids from inorganic nitrogen1. The products of such metabolism are very useful not only for the plant itself but also for most living organisms.
Chloroplasts are the product of serial endosymbiotic events and arose from a cyanobacterial ancestor that was engulfed by a eukaryote2,3. In higher plants, the biogenesis of chloroplasts is initiated from undifferentiated, non-photosynthetic proplastids in shoot meristems. As shoot meristematic cells begin to differentiate into mesophyll cells, proplastids coordinately differentiate into chloroplasts4,5. Although the chloroplasts in true leaves develop from meristematic proplastids as the leaf primordia emerge, chloroplasts in cotyledons develop from etioplasts that are already present in mesophyll tissue within the embryo6. During photomorphogenesis, light is an important environmental cue triggering chloroplast biogenesis, a complex process in which photosynthetic pigments biosynthesis, the import of nuclear-encoded proteins, and the building of thylakoid networks embedded with photosynthetic electron transport complexes are integrated to establish fully functional chloroplasts7,8.
The biogenesis of chloroplasts requires the coordinated expression and assembly of proteins encoded by both the nuclear and chloroplast genomes. Most of the chloroplast proteins are encoded by nuclear genes, translated in the cytosol as precursor proteins, and post-translationally imported into the chloroplast stroma9. From the stroma, some intermediate precursors are translocated into or across the thylakoid membrane10. Because the majority of the several thousands of chloroplast proteins are encoded by the nucleus, it is not surprising that numerous mutants that are disrupted in chloroplast biogenesis have mutations on nuclear-encoded genes with diverse biological functions, including biosynthesis of photosynthetic pigments, thylakoid biogenesis, lipid biosynthesis, protein import, photosystem assembly, protein maturation and degradation and plastid gene expression11,12,13,14,15,16,17,18. Therefore, chloroplast biogenesis is a complex process in higher plants, and identifying chloroplast biogenesis mutants and illuminating their molecular mechanisms will provide significant insight into the complex process of chloroplast biogenesis.
The coordinated expression of proteins encoded by the chloroplasts and of those encoded by the nucleus but destined for the chloroplasts is assumed to be regulated by anterograde and retrograde communication between the nucleus and the chloroplasts19. Anterograde mechanisms coordinate gene expression in chloroplasts in response to endogenous and environmental signals that are perceived by the nucleus. Retrograde mechanisms transmit signals that originate in the chloroplasts to regulate nuclear gene expression20. In recent decades, significant advances have occurred in understanding the complexity of chloroplast-to-nucleus retrograde signalling. For example, norflurazon (NF), an inhibitor of carotenoid biosynthesis, causes the photooxidative destruction of chloroplasts and leads to a markedly decreased expression of a large number of nuclear photosynthesis-related genes21,22. Changes in plastid gene expression (PGE) and the photosynthetic electron transport chain (PET) also trigger chloroplast-to-nucleus retrograde signalling19,21,23. Several recent studies have investigated the role of retrograde signalling in regulating the expression of nuclear genes during several stresses, including high light, drought, low temperature, heat, and excess ammonium24,25,26,27,28,29. Therefore, the disruption of the functional and developmental state of the chloroplast by endogenous factors or exogenous environmental cues generates a wide variety of chloroplast retrograde signals that regulate the expression of many nuclear genes encoding chloroplast-localized proteins.
To identify new genes required for the early steps of chloroplast biogenesis, we isolated several (22 lines) T-DNA insertion emb (embryo defective) mutants from the Arabidopsis Biological Resource Center and focused our subsequent analysis on those mutants exhibiting very low levels of chlorophyll. This report describes a mutant named delayed pale greening1 (dpg1), in which chloroplast biogenesis is clearly compromised and in which the transcription of chloroplast- and nuclear-encoded genes involved in chlorophyll biosynthesis, photosynthesis and chloroplast development is repressed. Our results indicate that AtDPG1 plays an essential role in early chloroplast biogenesis, its absence triggers chloroplast-to-nucleus retrograde signalling, ultimately down-regulating the expression of the nuclear genes encoding chloroplast-localized proteins.
Results
Identification of a new delayed pale-greening1 (dpg1) mutant in Arabidopsis thaliana
To identify the genes involved in early chloroplast biogenesis, a delayed pale-greening mutant, designated dpg1, was isolated from the Arabidopsis Biological Resource Center. The homozygous mutant was completely albino and unable to grow on solid medium lacking a supplemental carbon source (Supplementary Fig. S1). However, this mutant survived and continued to grow on 1/2 MS medium containing 1% sucrose. While germinating normally, the mutant plants had albino cotyledons that became pale green several days later (Fig. 1a–d). In later developmental stages, the true leaves of the dpg1 homozygous mutant were initially albino but gradually became pale green as the plant matured (Fig. 1c–g). When dpg1 plants were grown for 5 weeks on 1/2 MS medium containing 1% sucrose and then transferred to soil, some of them withered and died because they were unable to resist the dry stress. The surviving dpg1 plants looked weak and small, and their fertility was significantly reduced (Fig. 1h, Supplementary Table S1). At all growth stages, the mutant plants were greatly retarded in development, and they displayed a severe dwarf phenotype with small rosette leaves (Fig. 1a–h). Consistently with the phenotypes of dpg1 homozygous mutant plants, the contents of chlorophyll a, chlorophyll b, and total chlorophyll in dpg1 albino leaves were drastically lower than those in the wild type. After pale-greening, the contents of these pigments increased but were lower than those of the wild type (Fig. 1i). Additionally, within the immature siliques of the heterozygous dpg1 mutant, abnormal albino seeds that were randomly distributed along the length of the siliques were detected at a frequency of approximately 25% (Fig. 1j,k,m,n, Table 1). The segregation ratios suggested that the seed phenotype is associated with one recessive mutation and that the albino seeds represent dpg1 homozygous mutants. However, the seed colour was indistinguishable between the mutant and the wild type after seed desiccation (Fig. 1l).
Figure 1. Phenotypic analysis of the dpg1 mutant plants.
(a) 5-day-old wild type. (b) 5-day-old dpg1. (c) 14-day-old wild type. (d) 14-day-old dpg1. (e) 21-day-old wild type. (f) 21-day-old dpg1. (g) 35-day-old dpg1. (h) 90-day-old dpg1. Scale bar = 5 mm. (i) The chlorophyll contents in the leaves of 21-day-old wild-type (Col-0), 21-day-old dpg1 (dpg1-I) and 35-day-old dpg1 (dpg1-II) plants. Chla, chlorophyll a; Chlb, chlorophyll b; Chl, total chlorophyll. Each value is the mean ± SE of three independent determinations, and different letters indicate significant differences at P < 0.05. (j) Green seeds in a 9-day-old wild-type silique. (k) Intermixed albino and green seeds in a 9-day-old dpg1 heterozygous silique. The asterisks indicate albino seeds. (l) Seeds in a 15-day-old dpg1 heterozygous silique. (m) A representative green embryo in a 9-day-old wild-type silique. (n) A representative albino embryo in a 9-day-old dpg1 heterozygous silique.
Table 1. Segregation of green and albino seeds in heterozygous dpg1 mutant.
| Total no. of seeds | Morphology |
Null hypothesis | χ2 | P | |
|---|---|---|---|---|---|
| Green | albino | ||||
| 902 | 683 | 219 | 3:1 | 0.25 | P > 0.05 |
Molecular characterization and complementation of dpg1
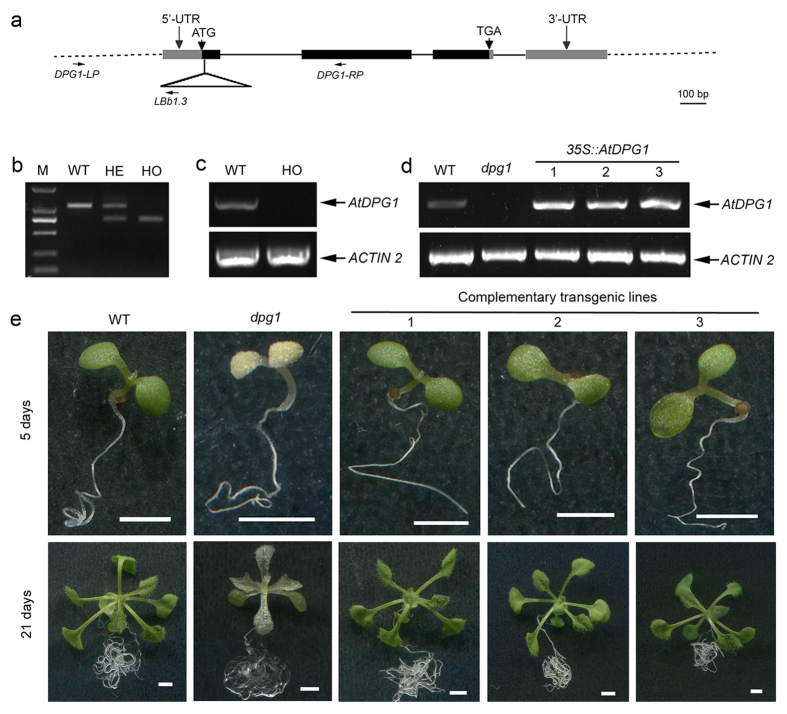
To explore the nature of the mutation, a heterozygous dpg1 mutant line (female parent) was backcrossed with the wild type (male parent). The F1 progeny displayed a normal green phenotype, suggesting that the mutation was recessive (data not shown). To determine whether the T-DNA insertion co-segregates with the delayed pale-greening phenotype, DNA from F2 progenies was used as a template for PCR analysis. Primers were designed to amplify a larger fragment from wild-type plants and a smaller fragment from homozygous mutants. In heterozygous mutants, both of the fragments were amplified (Fig. 2b), suggesting that the T-DNA insertion co-segregated with the mutant phenotype. Sequencing the T-DNA left border junction indicated that the T-DNA was inserted into the first exon of At1g49510 (Fig. 2a). Consistently with this result, no transcript of At1g49510 was detected in the homozygous lines (Fig. 2c). These results indicate that At1g49510 is a candidate gene for AtDPG1.
Figure 2. Molecular identification and phenotype rescue of the dpg1 mutant.
(a) Gene structure of At1g49510 and the position of the T-DNA insertion. Exons and introns are represented by boxes and lines, respectively. The insertion site of the T-DNA in dpg1 is indicated by a triangle. DPG1-LP, DPG1-RP and LBb1.3 are the primers that were used for genotyping PCR. (b) A representative genotyping PCR result of dpg1. Offspring of the heterozygous dpg1 were segregated into three genotypes: wild type (WT), heterozygous (HE) and homozygous (HO). (c) Semi-quantitative RT-PCR analysis of AtDPG1 in the wild type (WT) and homozygous dpg1 mutant (HO). The ACTIN2 gene was used as an internal control. (d) Semi-quantitative RT-PCR analysis of the AtDPG1 transcription levels in the wild type (WT), the homozygous dpg1 and three complementary transgenic lines (Lines 1, 2 and 3). The ACTIN2 gene was used as an internal control. (e) The phenotype of a dpg1 homozygous plant was rescued by transformation with a 35S::AtDPG1 transgene. Scale bar = 2 mm.
To further confirm that the AtDPG1 gene was At1g49510, we performed a complementation experiment. A 723-bp wild-type coding sequence (CDS) driven by the cauliflower mosaic virus 35S (CaMV 35S) promoter was introduced into dpg1 heterozygous plants. Then, 19 T1 transgenic plants were screened out. Among these plants, 8 were DPG1/dpg1 heterozygotes. The segregation rates of the offspring of these plants were far from 3:1 (green plants: albino plants; Supplementary Table S3). Furthermore, we determined the genotype of these offspring (T2 generation) and obtained dpg1/dpg1 homozygous plants from 3 T1 transgenic lines that carried fragments of the exogenous coding sequence. These plants displayed green cotyledons and true leaves after their germination (Fig. 2e). A control transformation of the wild-type with the 35S::AtDPG1 transgene further confirmed that the rescue was complete and that overexpression did not produce significant phenotypic changes (Supplementary Fig. S2). A semi-quantitative RT-PCR analysis also showed that the transcripts of At1g49510 were present in these plants (Fig. 2d). These results indicated that the 723-bp CDS fragment can successfully complement the mutated phenotype and that the AtDPG1 gene is At1g49510.
Chloroplast development in dpg1
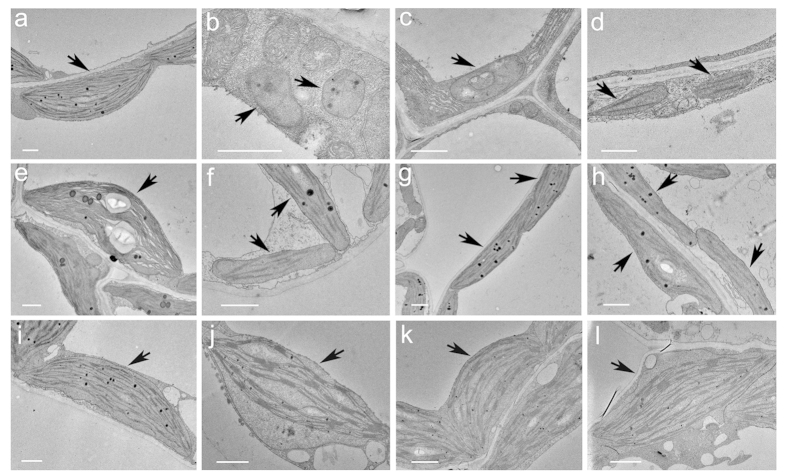
To examine the developmental status of the chloroplast in a dpg1 mutant, mesophyll cell plastids from 21- and 35-day-old mutant plants (exhibiting albino and pale-green leaves, respectively) were analysed by transmission electron microscopy. In wild-type plants, chloroplasts were crescent-shaped and contained well-developed thylakoid membranes with large grana stacks (Fig. 3a,e). However, the albino leaves of 21-day-old dpg1 seedlings contained many abnormal chloroplasts (Fig. 3b–d). According to their morphologies, these abnormal chloroplasts could be classified into three types. The first type was rounded, with almost no thylakoid membrane, and it appeared undifferentiated (Fig. 3b). The second type was oval-shaped, and no organized thylakoid membranes were observed except for a few circular internal membranes (Fig. 3c). The third type had very few but relatively continuous thylakoid membranes (Fig. 3d). The pale-green leaves of 35-day-old dpg1 seedlings contained differentiated chloroplasts that were elongated and had fewer thylakoid membranes than did chloroplasts of wild-type seedlings at the same growth stage (Fig. 3f–h). These chloroplasts resembled wild-type chloroplasts at an early stage of development. Some of the thylakoid membranes were arranged as grana stacks. Moreover, the chloroplast morphology, size, thylakoid abundance, and grana stacking were restored to those of the wild type in dpg1 plants expressing the 35S::AtDPG1 transgene (Fig. 3i–l). Altogether, these observations indicated that the delayed transition of proplastids to chloroplasts in dpg1 is consistent with the delayed pale-greening phenotype.
Figure 3. Ultrastructure of chloroplasts from wild-type, dpg1 and complementary plant leaves.
(a–d) Chloroplast structures from 21-day-old wild-type (a) and dpg1 (b–d) plants. (e–h) Chloroplast structures from 35-day-old wild-type (e) and dpg1 (f–h) plants. (i–l) Chloroplast structures from 21-day-old wild-type (i) and complementary (j–l) plants. The arrowheads indicate chloroplasts in the wild type, dpg1 and complementary plants. Scale bar = 1 μm.
AtDPG1 encodes a putatively chloroplast-localized protein that is highly conserved in various dicots
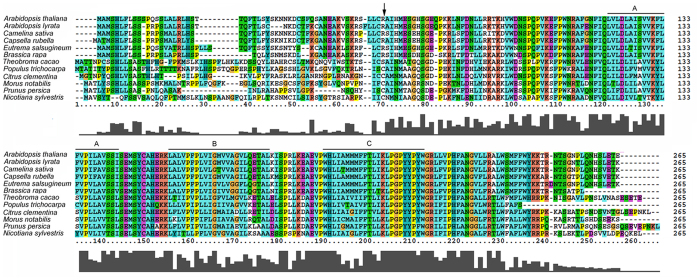
BLAST searches of the complete Arabidopsis thaliana sequence revealed that only one copy of the AtDPG1 gene is present in the nuclear genome, and it encodes a 240-amino acid protein with an apparent molecular mass of 27.38 kD and a theoretical pI of 10.27. The ChloroP 1.1, Predotar and iPSORT programs predicted that the AtDPG1 protein is targeted to the chloroplast and that its 55- amino-acid N-terminal region is likely to be a transit peptide (Fig. 4, Supplementary Table S4); this result was also reported in the SubCellular Database (http://suba.plantenergy.uwa.edu.au/flatfile.php?id). Consistently with this result, proteomic data have also indicated that AtDPG1 is most likely localized to the chloroplast envelope30. Moreover, trans-membrane helix prediction through TMHMM indicated the presence of 3 trans-membrane helices in AtDPG1 (Fig. 4, Supplementary Fig. S3). A search of the GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) revealed dozens of ESTs that are highly homologous to AtDPG1 in diversified species of dicots but not in monocots (Fig. 4). AtDPG1 is 93%, 89%, 87%, 83%, 79%, 66%, 63%, 59%, 55%, 53%, and 52% identical to the putative DPG1s of Arabidopsis lyrata (AlDPG1), Camelina sativa (CsDPG1), Capsella rubella (CrDPG1), Eutrema salsugineum (EsDPG1), Brassica rapa (BrDPG1), Populus trichocarpa (PtDPG1), Citrus clementina (CcDPG1), Prunus persica (PpDPG1), Theobroma cacao (TcDPG1), Morus notabilis (MnDPG1), and Nicotiana sylvestris (NsDPG1), respectively. To investigate the evolutionary relationship among DPG1 homologues, a phylogenetic analysis was performed. As shown in Supplementary Fig. S4, the proteins from Brassicaceae species and other dicots clustered in different subclades. Although there was a robust separation within these two big clades, some bootstrap percentages were low, thus making the analysis a less robust interpretation within Brassicaceae species and other dicots.
Figure 4. Multiple sequence alignment of AtDPG1 and its homologues.
The transmembrane domains (A–C) that were conserved among the DPG1 homologues are indicated. Protein sequences of DPG1 homologues from Arabidopsis lyrata (XP_002891514), Camelina sativa (XP_010479384), Capsella rubella (XP_006304157), Eutrema salsugineum (XP_006393250), Brassica rapa (XP_009124376), Theobroma cacao (XP_007042342), Populus trichocarpa (XP_006384460), Citrus clementina (XP_006423083), Morus notabilis (XP_010096165), Prunus persica (XP_007201809), and Nicotiana sylvestris (XP_009792237) were retrieved from GenBank. The downward arrow indicates the putative cleavage site of the transit peptide of AtDPG1 as predicted by ChloroP 1.1 program. Sequences were aligned with ClustalX 2.0 software.
Promoter activity analysis of the AtDPG1 gene
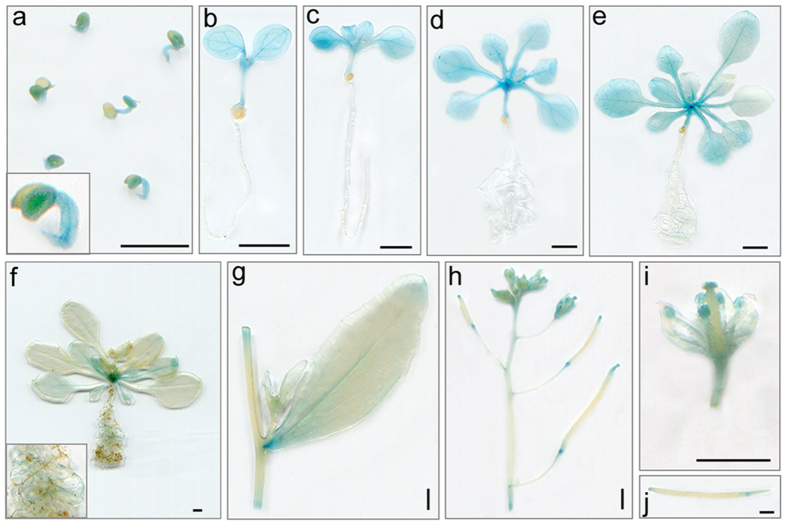
To investigate the possible physiological function of the AtDPG1 gene, the temporal and spatial expression patterns of the GUS gene driven by the AtDPG1 promoter were analysed in transgenic Arabidopsis plants. A 1,811-bp promoter fragment of the AtDPG1 gene was fused to the GUS reporter gene, and the expression cassette was introduced into wild-type Arabidopsis plants. According to the observed pattern, AtDPG1 was expressed very early in germinating seeds (Fig. 5a). In young seedlings, strong AtDPG1-GUS-driven expression appeared in the cotyledons, newly emerging true leaves and hypocotyls, but no detectable expression was observed in the roots (Fig. 5b,c). In vegetative plants, GUS staining was also observed in the green tissues, including cotyledons, true leaves and hypocotyls, but not in the roots (Fig. 5d,e). At a later stage (extended to the bolting stage), GUS expression was detected at a low level in the cotyledons, true leaves, cauline leaves and stems (Fig. 5f,g). Moreover, very weak GUS staining was also detected in partial roots (Fig. 5f). When the transgenic plants entered the reproductive stage, stronger GUS expression was detected in the inflorescence (Fig. 5h). Further examination of GUS expression showed that this gene was mainly expressed in the stamens and stigmas of open flowers (Fig. 5i). In addition, weak GUS activity was also found at the base and the tip of silique (Fig. 5h,j). GUS staining for transgenic lines showed that AtDPG1 was widely expressed throughout the plant and that higher expression levels were predominately found in green tissues during the early stages of Arabidopsis seedling development.
Figure 5. Histochemical assays for the expression pattern of AtDPG1::GUS transgenic lines as visualized by GUS staining.
(a) Germinating seeds (36 h after sowing). (b) 5-day-old seedling. (c) 7-day-old seedling. (d) 14-day-old plant. (e) 21-day-old plant. (f) 35-day-old plant. (g) Stem and cauline leaf. (h) Inflorescence. (i) Fully opened flower. (j) Silique. Scale bar = 2 mm.
Expression pattern of the AtDPG1 gene
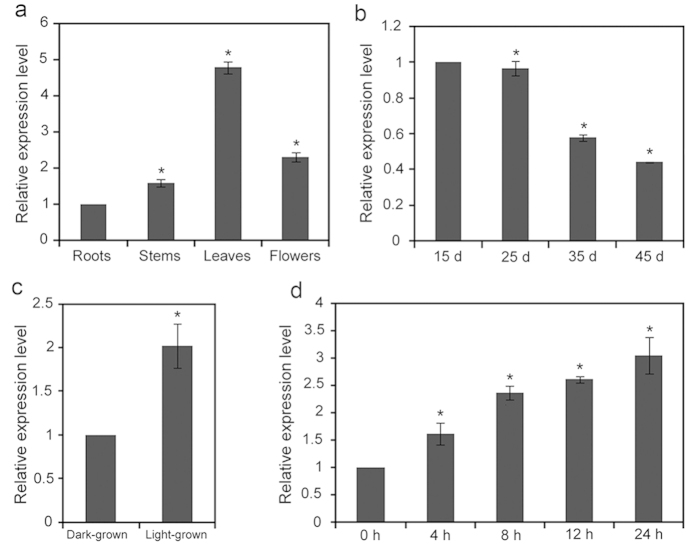
To elucidate the possible role of AtDPG1 in the course of plant growth and development, we examined its organ-specific expression. As shown in Fig. 6a, the leaves expressed the highest levels of AtDPG1 mRNA, whereas these levels in other organs of Arabidopsis plants were significantly lower. The roots accumulated the lowest level of all of the analysed tissues. To study the expression of AtDPG1 at different developmental stages, the levels of the AtDPG1 transcript in leaves ranging from 15 to 45 days old were evaluated. The expression level of AtDPG1 decreased as the age of the leaves increased. The AtDPG1 transcript content of leaves of 45-day-old plants was only approximately 40% of that detected in leaves of 15-day-old plants (Fig. 6b).
Figure 6. Expression pattern of the AtDPG1 gene.
(a) Quantitative real-time PCR analysis of AtDPG1 transcripts in different organs (roots, stems, leaves, and flowers) from 45-day-old wild-type Arabidopsis plants. The expression levels of AtDPG1 displayed a range of cycle Ct values ranging from 22.80 to 24.44 in different tissues. The roots expressed the lowest levels of AtDPG1 mRNA, with a Ct value of 24.44, and the AtDPG1 transcript level in roots was set to 1.0. (b) AtDPG1 transcript levels in the leaves of different developmental stages in wild-type Arabidopsis plants. The AtDPG1 transcript level in the leaves from 15-d-old seedlings was set to 1.0. (c) Transcript levels of AtDPG1 in 4-day-old wild-type light-grown and dark-grown seedlings. The AtDPG1 transcript level in the dark-grown seedlings was set to 1.0. (d) Transcript levels of AtDPG1 during the light-induced greening of etiolated wild-type seedlings. After growth in darkness for 4 days, the etiolated Arabidopsis seedlings were illuminated for 4, 8, 12, and 24 h. The AtDPG1 transcript level in the etiolated seedlings was set to 1.0. The bar graphs show the relative expression levels, as determined by the comparative CT method and normalized to the expression of the ACTIN2 gene. The error bars indicate the interval delimited by 2−(ΔΔCT ± SD). The asterisks indicate ΔCT values significantly different from those of the control in a Mann–Whitney U-test (P < 0.05).
Light induces the differentiation of non-photosynthetic proplastids into fully functional chloroplasts. To examine the effects of light on the expression of AtDPG1, we used real-time PCR to determine AtDPG1 expression in 4-d-old wild-type etiolated seedlings exposed to light for 4, 8, 12 and 24 h. As shown in Fig. 6c, the AtDPG1 transcript levels were high under light and much lower in the dark in 4-d-old etiolated seedlings. Furthermore, AtDPG1 expression was highly induced after illumination for 4 h and peaked after 24 h of illumination (Fig. 6d). These observations suggest that light plays a major role in regulating AtDPG1 expression.
Effects of the AtDPG1 mutation on the expression of chloroplast-encoded genes
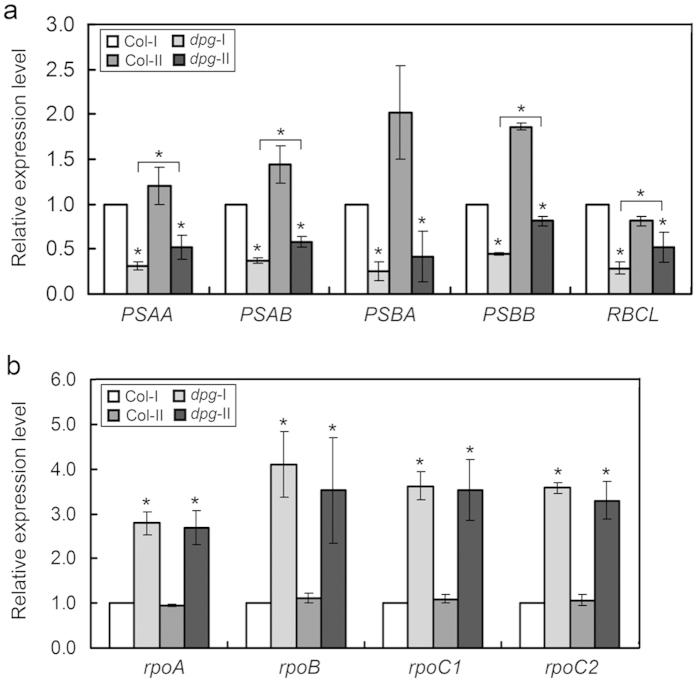
The expression of chloroplast-encoded genes is tightly associated with chloroplast developmental status. Because abnormal chloroplast development was observed in dpg1, we compared the transcript levels of various chloroplast-encoded genes between wild-type plants and dpg1 mutants. The five genes PSAA (encoding the PSAA protein, comprising the reaction centre for photosystem I), PSAB (encoding the D1 subunit of the photosystem I reaction centre), PSBA (encoding the D1 subunit of the photosystem II reaction centre core), PSBB (encoding the CP47 subunit of the photosystem II reaction centre), and RBCL (encoding the large subunit of Rubisco) were selected as plastid-encoded polymerase (PEP)-dependent genes. The expression levels of these genes were markedly lower in the mutant than in the wild type; however, the expression levels of most of these genes (PSAA, PSAB, PSBB, and RBCL) mildly increased as the albino leaves became pale green (Fig. 7a). Because the albino phenotype of dpg1 mutant was similar to that of mutants with a defect in PEP function31,32,33,34, dpg1 is probably a PEP-defective mutant. PEP is composed of plastid-encoded core subunits and a multiple nuclear-encoded sigma factor that confers promoter specificity to PEP. The core subunits of PEP are encoded by the plastid rpoA, rpoB, rpoC1, and rpoC2 genes, which are nucleus-encoded polymerase (NEP)-dependent genes. In the dpg1 mutant, the transcript levels of all four genes were several times greater than those of the wild type (Fig. 7b).
Figure 7. Effects of the AtDPG1 mutation on the expression of chloroplast-encoded genes.
(a) The expression levels of chloroplast-encoded PEP-dependent genes (PSAA, PSAB, PSBA, PSBB, and RBCL). (b) The expression levels of genes encoding PEP core subunits (rpoA, rpoB, rpoC1, and rpoC2). The terms dpg1-I and dpg1-II refer to the leaves that were albino or pale-green in 21-day-old and 35-day-old dpg1 mutants, respectively. The transcript level of each gene was set to 1.0 in the leaves from a 21-day-old wild type plant. The bar graphs show the relative expression levels, as determined by the comparative CT method and normalized to the expression of the ACTIN2 gene. Error bars indicate the interval delimited by 2−(ΔΔCT ± SD). The asterisks indicate ΔCT values significantly different from those of the control in a Mann–Whitney U-test (P < 0.05).
Effects of the AtDPG1 mutation on the expression of nuclear-encoded photosynthetic genes
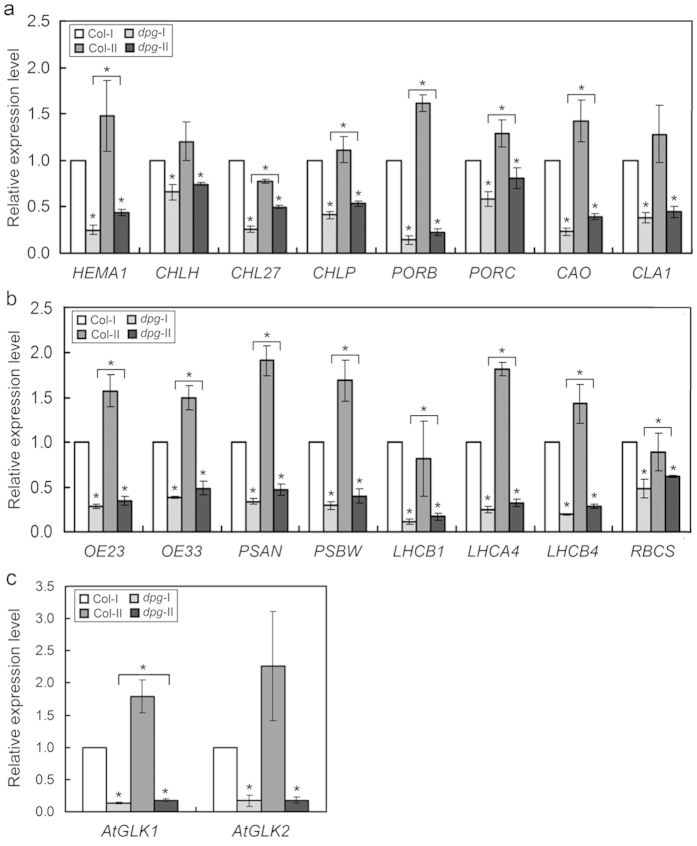
The developmental and functional status of chloroplasts controls the transcription of nuclear-encoded genes via retrograde signaling19,35. We next examined the transcription of nuclear-encoded genes associated with chlorophyll biosynthesis and photosynthesis in the dpg1 mutant. The transcription of nuclear-encoded genes that are critical for chlorophyll biosynthesis was first analysed, including genes encoding glutamyl-tRNA reductase (HEMA1), magnesium chelatase (CHLH), Mg-protoporphyrin IX monomethyl ester cyclase (CHL27), geranylgeranyl reductase (CHLP), protochlorophyllide oxidoreductase B (PORB), protochlorophyllide oxidoreductase C (PORC), chlorophyllide a oxygenase (CAO), and 1-deoxy-D-xylulose 5-phosphate synthase 1 (CLA1). These genes were down-regulated in the dpg1 mutant compared with the wild type; however, the expression levels of most of these genes (HEMA1, CHL27, CHLP, PORB, PORC, and CAO) modestly increased as the albino leaves became pale green (Fig. 8a). The transcription of nuclear-encoded photosynthetic genes was next examined, including genes encoding the proteins of the oxygen-evolving complex of photosystem II (OE23 and OE33), the N subunit of photosystem I (PSAN), the PSBW protein similar to the photosystem II reaction centre subunit W, the chlorophyll a/b binding proteins (LHCB1, LHCA4, and LHCB4), and the rubisco small subunit (RBCS). Similarly, the transcription levels of all eight genes in the dpg1 mutant were significantly lower than that in the wild-type plant; however, the expression levels of these genes mildly increased as the albino leaves became pale green (Fig. 8b). Previously, the GOLDEN2-LIKE (GLK) transcription factors AtGLK1 and AtGLK2 were identified as positive regulators of nuclear-encoded photosynthetic genes by binding to their promoter sequences8. Further analyses of the AtGLK1 and AtGLK2 transcript levels revealed that the two genes were significantly down-regulated in the dpg1 mutant compared with the wild type; however, the expression levels of the AtGLK1 gene modestly increased as the albino leaves became pale green (Fig. 8c).
Figure 8. Effects of the AtDPG1 mutation on the expression of nuclear-encoded photosynthetic genes.
(a) The expression levels of nuclear-encoded genes that are critical for chlorophyll biosynthesis (HEMA1, CHLH, CHL27, CHLP, PORB, PORC, CAO, and CLA1). (b) The expression levels of nuclear-encoded photosynthetic genes (OE23, OE33, PSAN, PSBW, LHCB1, LHCA4, LHCB4, and RBCS). (c) The expression levels of nuclear-encoded GLK transcription factor genes (AtGLK1 and AtGLK2). The terms dpg1-I and dpg1-II refer to the leaves that were albino or pale-green in 21-day-old and 35-day-old dpg1 mutants, respectively. The transcript level of each gene was set to 1.0 in the leaves from a 21-d-old wild type plant. The bar graphs show the relative expression levels, as determined by the comparative CT method and normalized with the expression of the ACTIN2 gene. The error bars indicate the interval delimited by 2–(ΔΔCT ± SD). The asterisks indicate ΔCT values significantly different from those of control in a Mann–Whitney U-test (P < 0.05).
Discussion
Chloroplasts contain several thousand different proteins, of which more than 95% are encoded by nuclear genes, synthesized in the cytosol as precursor proteins, and imported into the organelle36. Despite the discovery of a large fraction of genes involved in chloroplast development37, full understanding of the complex biogenesis of this organelle is lacking. In this study, the novel nuclear gene AtDPG1 was identified using a loss-of-function forward genetic approach. Although AtDPG1 has previously been isolated as EMBRYO DEFECTIVE 1273 (EMB1273) by Tzafrir et al.38, very limited information about the function of Arabidopsis AtDPG1 proteins is available. We first searched for members of this gene family in the GenBank database. The results revealed that the DPG1 protein is highly conserved in various dicots but not in monocots (Fig. 4), suggesting that the origination of DPG1 might have occurred after the separation of monocots and dicots. Consistently with the results of the sequence alignment, the iPSORT, ProteinProwler and ChloroP programs also yielded a high probability for chloroplast localization for DPG1 in higher plants (Supplementary Table S4). Proteomic data have also indicated that AtDPG1 is most likely localized to the chloroplast envelope30. Despite this evidence, the chloroplast localization of AtDPG1 needs to be experimentally verified in further investigations.
In dicotyledonous plants, the development of chloroplasts differs between cotyledons that initially act primarily as storage tissue and true leaves whose major function is photosynthesis. In cotyledons, plastids partially develop during embryogenesis, but this development arrests during seed maturation. When seeds germinate in the light, the plastids further develop into functional chloroplasts. In contrast to chloroplast development in cotyledons, in true leaves, chloroplasts develop directly from the proplastids in the shoot apex5,39. These differences have been detected in several mutants with chloroplast defects restricted either to cotyledons or to true leaves. For instance, the sco mutant group and the cyo1 and wco mutants have albino cotyledons but normal green true leaves40,41,42,43. In contrast, in the var2 and sg1 mutants, Arabidopsis cotyledons develop normally, but true leaves contain chlorophyll-deficient white sectors44,45. In our study, both the cotyledons and true leaves of dpg1 were initially albino and then gradually turned pale-green during development in dpg1 plants, although abnormal white embryos were observed during the early stages of embryogenesis (Fig. 1), suggesting that AtDPG1 plays an equally important role in early chloroplast development in the cotyledons and true leaves of Arabidopsis seedlings. A multiple-sequence alignment analysis showed that DPG1 is conserved in dicots but not in monocots. Given that the chloroplasts of monocots and dicots are highly similar except for some metabolic issues, DPG1 may play a specific role in chloroplast metabolism in dicots. The most striking feature of dpg1 was the delayed pale-greening of both its cotyledons and true leaves (Fig. 1). Furthermore, ultrastructural findings demonstrated that thylakoid membranes in the young leaves of dpg1 mutants were less abundant than in wild type, but their abundance significantly increased as the leaves matured (Fig. 3). GUS staining for AtDPG1::GUS transgenic lines showed that promoter of AtDPG1 is active in green tissues but not in roots during the early stages of Arabidopsis seedling development (Fig. 5b–e). Quantitative real-time RT-PCR analysis also revealed that AtDPG1 was expressed in leaves in a light-inducible way (Fig. 6c,d). Light is one of the most crucial environmental factors stimulating the differentiation of non-photosynthetic proplastids into photosynthetic chloroplasts. The increased accumulation of the AtDPG1 transcript after light illumination suggests that this gene may play an important role in the light-regulated chloroplast biogenesis. Our results also showed that the transcripts of both chloroplast- and nuclear-encoded photosynthetic genes were severely suppressed during the early stage in the dpg1 mutant; however, the expression levels of most of the genes mildly increased as the albino leaves became pale green (Figs 7a and 8). Altogether, these findings show that AtDPG1 plays an essential role in early stages of chloroplast biogenesis and that functional chloroplast biogenesis depends on leaves reaching a more mature developmental stage in the dpg1 mutant.
The young leaves of dpg1 mutants were initially albino but gradually turned to pale-green as the plant matured. There are two possible explanations for the seedling-stage-specific albino phenotype. The first explanation is that other proteins may partly compensate for the absence of AtDPG1 during later development stages. For example, the homozygous sig6-1 mutant plants had pale green cotyledons with drastically reduced chlorophyll content in 3-day-old Arabidopsis seedlings. However, the chlorophyll deficiency is limited to the young cotyledons and is restored to the wild-type level in 8-day-old seedlings46. Further studies have revealed that another late general sigma factor (possibly AtSIG1) is likely to have overlapping functions with AtSIG6 to compensate for its deficiency in sig6-1 mutants when 8 or more days old46,47. The second explanation is that AtDPG1 is not required for later developmental stages of chloroplast development in Arabidopsis. Quantitative real-time PCR analysis revealed that the expression level of AtDPG1 decreased as the age and developmental state of the leaves increased. The AtDPG1 transcript content of leaves in 45-day-old plants was only approximately 40% of that in 15-day-old plants (Fig. 6b). Although the leaves of 45-day-old plants displayed an approximately 5-fold increase in relative expression compared with that in root (Fig. 6a), there was a low absolute expression of AtDPG1, because GUS staining showed very weak expression of AtDPG1 in the roots (Fig. 5f). These results suggest that AtDPG1 plays specific roles in early stages of chloroplast development. The phenotype of dpg1 is similar but not identical to the formerly described mutant phenotype, which is characterized by young albino leaves with drastically reduced chlorophyll but nearly normal levels of chlorophyll in mature leaves. For instance, the Arabidopsis dg1 and ys1 mutants display a seedling-stage-specific albino or yellow seedling phenotype caused by delayed chloroplast development. Further studies have revealed that DG1 encodes a chloroplast-targeted PPR protein and that YS1 encodes a DYW protein that is required to edit rpoB transcripts32,48. Despite this evidence, only a few genes that are responsible for the seedling-stage-specific phenotype have been isolated, and their molecular mechanisms remain elusive. Therefore, elucidating the complex molecular mechanisms regulating chloroplast biogenesis in higher plants remains a major task for future research.
An analysis of chloroplast gene expression revealed that the transcript levels of PEP-dependent genes were significantly reduced in the dpg1 mutant (Fig. 7a), suggesting that the dpg1 mutant is severely impaired in PEP activity and that AtDPG1 may play an important role in the regulation of chloroplast gene expression. However, the results of expression analysis showed that the genes encoding PEP core subunits were dramatically induced in dpg1 (Fig. 7b). The increased transcript abundance of these genes may indicate a feedback mechanism to increase PEP levels to transcribe PEP-dependent genes. Even so, the accumulation of transcripts for PEP components did not result in the formation of functional PEP, because the expression of PEP-dependent genes decreased in the dpg1 mutant (Fig. 7a). A defect in polysome assembly has been shown to be accompanied by a decrease in the level of the RBCL mRNA49. In dpg1 mutant, the expression of RBCL drastically decreased (Fig. 7a), indicating that a loss-of-function mutation in AtDPG1 may affect the assembly of chloroplast ribosomes, eventually leading to the disruption of chloroplast translation during early chloroplast development.
The chloroplast developmental status controls a series of nuclear genes that encode chloroplast-localized proteins through retrograde signalling19,35,50,51. In the present study, we examined the transcription levels of nuclear-encoded genes that are associated with chlorophyll biosynthesis and photosynthesis in the dpg1 mutant. Quantitative real-time PCR analysis showed that the transcripts of these genes were severely suppressed during the early stage in the dpg1 mutant, but the expression levels of most of these genes mildly increased as the albino leaves became pale green (Fig. 8). The leaves of dpg1 were initially albino and then gradually turned pale-green during development in dpg1 plants (Fig. 1). Furthermore, ultrastructural observations showed that thylakoid membranes were less abundant in the albino leaves of mutant plants than in the wild type; however, their abundance gradually increased as the leaves became green (Fig. 3). It is likely that the retrograde signals derived from these impaired chloroplasts regulate the expression of nuclear genes encoding chloroplast-localized proteins. Several studies have shown that GOLDEN2-LIKE transcription factors (GLK) modulate chloroplast development52,53 and mediate retrograde signalling8,54 in response to the functional state of the chloroplast. The expression levels of AtGLK1 and AtGLK2 are tightly associated with the expression of nuclear photosynthesis genes in mutants that are defective in chloroplast structure and function18,54. Consistently with these findings, in the leaves of dpg1 plants, both AtGLKs (AtGLK1 and AtGLK2) had significantly reduced transcript levels (Fig. 8c). It is confirmed that chloroplast-to-nucleus retrograde signalling was evident in the dpg1 mutant. Furthermore, because AtGLKs regulate the expression of the LHCB gene family, as well as HEMA18,53, the decreased expression of these nuclear-encoded photosynthetic genes in the dpg1 mutant may occur through the regulation of AtGLKs expression.
In summary, the novel nuclear gene AtDPG1 was identified, and its loss-of-function mutation resulted in defects in chloroplast structure and function. The expression of several nuclear-encoded genes involved in chlorophyll biosynthesis, photosynthesis and chloroplast development was substantially down-regulated in the dpg1 mutant, suggesting that chloroplast defects trigger a chloroplast-to-nucleus retrograde signal that may account for such transcriptional changes. Further studies of the biochemical properties of AtDPG1 will shed more light on the function of this protein in chloroplast biogenesis.
Methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used in all experiments. After 3 days of stratification in the dark at 4 °C, the surface-sterilized seeds were germinated on 1/2 MS medium supplemented with 1% (w/v) sucrose at 22 °C with a 16-h-light/8-h-dark cycle. The dpg1 (SALK_004621C) T-DNA insertion line was obtained from the Arabidopsis Biological Resource Center55. The T-DNA insertion site was identified by sequencing PCR products that were amplified from the mutant with a T-DNA primer (LBb1.3) and the gene-specific primers DPG1-LP and DPG1-RP. All of the primers used are listed in Supplementary Table S2.
Constructs and plant transformation
To construct the PDPG1::GUS fusion gene, a 1,811-bp DNA fragment upstream of the ATG start codon of the AtDPG1 gene (At1g49510) was amplified from Arabidopsis thaliana genomic DNA by PCR. The pair of primers used in the PCR was PDPG1-F and PDPG1-R (Hind III and Nco I sites were introduced). The specific PCR fragment was then inserted into binary vector pCAMBIA1301 between Hind III and Nco I sites, replacing the CaMV 35S promoter, to create the recombinant transcription unit PDPG1::GUS. For the construction of 35S::DPG1 unit, the full-length coding sequence (CDS) corresponding to the AtDPG1 gene (At1g49510) locus was cloned by using RT-PCR from Arabidopsis thaliana. The pair of primers used in the PCR was OEDPG1-F and OEDPG1-R (Nco I and BstE II sites were introduced). The specific PCR fragment was then inserted into binary vector pCAMBIA1301 between Nco I and BstE II sites, replacing the GUS gene, to create the recombinant transcription unit 35S::DPG1. The recombinant plasmids were then introduced into Agrobacterium tumefaciens strain GV3101 and transformed into wild-type Arabidopsis or heterozygous dpg1 mutant plants using the floral dip method56. The transformants were then screened on 1/2 MS medium containing 50 μg ml−1 hygromycin. All primers used are listed in Supplementary Table S2.
RNA extraction, cDNA synthesis, and gene expression analysis
Total RNA was extracted from plant tissues with TRI Reagents, this was followed by treatment with RNase-free DNase I (TaKaRa, Dalian, China) to degrade genomic DNA. First-strand cDNA was synthesized from 2.0 μg of total RNA using ImProm-IITM reverse transcriptase (Promega, Madison, WI, USA) in a 10.0-μl reaction following the manufacturer’s instructions with minor modifications. For semi-quantitative RT-PCR analysis, PCR mixes were subjected to 34 cycles for the AtDPG1 gene and 28 cycles for the ACTIN2 gene. Three independent biological repeats were used to generate the representative results shown here. For quantitative real-time PCR analysis, the reaction was performed using SYBR Green Perfect mix (TaKaRa, Dalian, China) on a CFX96 (Bio-Rad) following the manufacturer’s instructions. Gene expression was normalized to that of ACTIN2 by subtracting the CT value of ACTIN2 from the CT value of the gene of interest. Expression ratios were then obtained from the equation 2−ΔΔCT. For each sample, quantitative real-time PCR was performed with three technical replicates from three biological replicate samples. Efficiency-corrected ΔCT values were calculated, and ΔΔCT was used to quantify the relative differences in transcript accumulation. The primers for the genes of interest are listed in Supplementary Table S2.
Transmission electron microscopy (TEM) analysis
The leaves of Arabidopsis were fixed in 2.5% (w/v) glutaraldehyde in phosphate buffer (pH 7.4) overnight at 4 °C. Thereafter, the samples were rinsed thoroughly with the same buffer 3-6 times and post-fixed with 1% (w/v) osmium tetroxide for 2 hours at 4 °C. Then the samples were dehydrated in a graded ethanol series (v/v, 30%, 50%, 70%, 90%) and in 1:1 mixture of 90% ethanol and 90% acetone, at last in acetone 3 times, embedded in Epon812 and polymerized at 35 °C for 16 h, then 48 °C for 24 h, and 65 °C for 48 h. For observation, ultrathin sections of the samples were cut with a diamond knife and collected on 200-mesh copper grids. After contrasting with uranyl acetate and lead citrate, the grids were examined with a JEM-2100 transmission electron microscope.
Histochemical GUS staining
The histochemical GUS staining of homozygous T3 transgenic lines harbouring the PDPG1::GUS fusion gene was performed as previously described57. Images were recorded with a scanner (Epson perfection V30). At least 9 individual lines were analysed to produce the representative results shown here.
Analysis of the chlorophyll contents
The chlorophyll content was analysed as described previously with minor modifications58. Fresh plant tissues were homogenized in 80% acetone, and debris were removed by centrifugation. The absorbance of the supernatant at 663 and 645 nm was measured with a 723B spectrophotometer (Tianjin PURUISI Equipment Co. LTD, China). All of the measurements were repeated in three independent experiments.
Sequence alignment and phylogenetic analysis
The BLAST search program (http://blast.ncbi.nlm.nih.gov/) was used to search for the sequences of proteins homologous to AtDPG1. The multiple alignments were performed by using ClustalX 2.0 software59. To construct a phylogenetic tree, a clustal file was generated in the ClustalX 2.0 software using the protein sequences. The generated clustal file was downloaded and converted to MEGA file format using the MEGA5.1 software60. The generated MEGA file was then run in the MEGA5.1 software to construct the phylogenetic tree. The statistical parameters used to construct the phylogenetic tree were as follows: analysis: phylogenetic reconstruction, statistical method: Neighbor-joining, test of phylogeny: bootstrap method, no. of bootstrap replicates: 1000, substitution type: amino acids, model/methods: Jones-Taylor-Thornton (JTT) model, rates among sites: uniform rates, gaps/missing data treatment: partial deletion, site coverage cutoff: 95%.
Additional Information
How to cite this article: Liu, D. et al. The novel protein DELAYED PALE-GREENING1 is required for early chloroplast biogenesis in Arabidopsis thaliana. Sci. Rep. 6, 25742; doi: 10.1038/srep25742 (2016).
Supplementary Material
Acknowledgments
We would like to give our great thanks to Mrs. Lixia Ma for technical assistance, and the Arabidopsis Biological Resource Center at The Ohio State University for providing the T-DNA insertion line. This work was supported by the National Natural Science Foundation of China (Grant Nos 31100185 and 31560077) to D.L.
Footnotes
Author Contributions D.L. designed the experiments. D.L., W.L. and J.C. performed the experiments; D.L. prepared the manuscript.
References
- Staehelin L. A. & Newcomb E. H. Membrane structure and membranous organelles in Biochemistry and molecular biology of plants (eds Buchanan B. B., Gruissen W. & Jones R. L.) 1–50 (American Society of Plant Physiologists, 2000). [Google Scholar]
- Cavalier-Smith T. Only six kingdoms of life. Proc. Biol. Sci. 271, 1251–1262 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A., Weber A. P. & Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41, 147–168 (2007). [DOI] [PubMed] [Google Scholar]
- Valkov V. T. et al. Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control. Plant Physiol. 150, 2030–2044 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charuvi D. et al. Gain and loss of photosynthetic membranes during plastid differentiation in the shoot apex of Arabidopsis. Plant Cell 24, 1143–1157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T. & Langdale J. A. The making of a chloroplast. EMBO J. 28, 2861–2873 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vothknecht U. C. & Westhoff P. Biogenesis and origin of thylakoid membranes. Biochim. Biophys. Acta 1541, 91–101 (2001). [DOI] [PubMed] [Google Scholar]
- Waters M. T. et al. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. Chloroplast research in the genomic age. Trends Genet. 19, 47–56 (2003). [DOI] [PubMed] [Google Scholar]
- Di Cola A., Klostermann E. & Robinson C. The complexity of pathways for protein import into thylakoids: it’s not easy being green. Biochem. Soc. Trans. 33, 1024–1027 (2005). [DOI] [PubMed] [Google Scholar]
- Jarvis P. et al. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103 (1998). [DOI] [PubMed] [Google Scholar]
- Gutierrez-Nava M. D. L., Gillmor C. S., Jimenez L. F., Guevara-Garcia A. & Leon P. CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiol. 135, 471–482 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricova A., Quesada V. & Micol J. L. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol. 141, 942–956 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H. et al. The Arabidopsis spontaneous cell death1 gene, encoding a z-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signaling. Cell Res. 17, 458–470 (2007). [DOI] [PubMed] [Google Scholar]
- Qin G. et al. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17, 471–482 (2007). [DOI] [PubMed] [Google Scholar]
- Liu D. et al. cpSecA, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J. Exp. Bot. 61, 1655–1669 (2010). [DOI] [PubMed] [Google Scholar]
- Casanova-Saez R., Mateo-Bonmati E., Kangasjarvi S., Candela H. & Micol J. L. Arabidopsis ANGULATA10 is required for thylakoid biogenesis and mesophyll development. J. Exp. Bot. 65, 2391–2404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S. et al. Pale-green phenotype of atl31 atl6 double mutant leaves is caused by disruption of 5-aminolevulinic acid biosynthesis in Arabidopsis thaliana. Plos ONE 10, e0117662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Jung H. S., Koussevitzky S. & Chory J. Plastid-to-nucleus retrograde signalling. Annu. Rev. Plant Biol. 57, 739–759 (2006). [DOI] [PubMed] [Google Scholar]
- Woodson J. D. & Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. C., Sullivan J. A., Wang J. H., Jerome C. A. & MacLean D. Coordination of plastid and nuclear gene expression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 135–145 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru M. R., Zola J., Foudree A. & Rodermel S. R. Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiol. 150, 904–923 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey V., Wagner R., Brautigam K. & Pfannschmidt T. Photosynthetic redox control of nuclear gene expression. J. Exp. Bot. 56, 1491–1498 (2005). [DOI] [PubMed] [Google Scholar]
- Fernandez A. P. & Strand A. Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr. Opin. Plant Biol. 11, 509–513 (2008). [DOI] [PubMed] [Google Scholar]
- Estavillo G. M. et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. et al. Arabidopsis plastid AMOS1/EGY1 integrates abscisic acid signaling to regulate global gene expression response to ammonium stress. Plant Physiol. 160, 2040–2051 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. D. et al. Downregulation of chloroplast RPS1 negatively modulates nuclear heat-responsive expression of HsfA2 and its target genes in Arabidopsis. Plos Genet. 8, e1002669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Hou L., Li W. C., Cheng J. F. & Fu Y. Q. COR15B expression is affected by chloroplast functionality, and its function in response to salt stress in Arabidopsis thaliana. Biol. Plant 58, 667–675 (2014). [Google Scholar]
- Liu D., Li W. C., Cheng J. F. & Hou L. Expression analysis and functional characterization of a cold-responsive gene COR15A from Arabidopsis thaliana. Acta Physiol. Plant 36, 2421–2432 (2014). [Google Scholar]
- Ferro M. et al. AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9, 1063–1084 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O. A., Prikryl J. & Barkan A. Site-specific binding of a PPR protein defines and stabilizes 59 and 39 mRNA termini in chloroplasts. EMBO J. 28, 2042–2052 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. et al. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 58, 82–96 (2009). [DOI] [PubMed] [Google Scholar]
- Gao Z. P. et al. A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol. 157, 1733–1745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S., Schröter Y., Pfalz J. & Pfannschmidt T. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 157, 1043–1055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S. et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719 (2007). [PubMed] [Google Scholar]
- Cline K. & Dabney-Smith C. Plastid protein import and sorting: different paths to the same compartments. Curr. Opin. Plant Biol. 11, 585–592 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T. et al. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14, 354–362 (2004). [DOI] [PubMed] [Google Scholar]
- Tzafrir I. et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135, 1206–1220 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield S. G. & Briarty L. G. The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int. J. of Plant Sci. 157, 280–295 (1996). [Google Scholar]
- Albrecht V., Ingenfeld A. & Apel K. Characterization of the snowy cotyledon1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 60, 507–518 (2006). [DOI] [PubMed] [Google Scholar]
- Albrecht V., Ingenfeld A. & Apel K. Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol. Biol. 66, 599–608 (2008). [DOI] [PubMed] [Google Scholar]
- Albrecht V. et al. The cytoskeleton and the peroxisomal-targeted SNOWY COTYLEDON3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22, 3423–3438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H. et al. Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19, 3157–3169 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Jensen M. & Rodermel S. The yellow variegated mutant of Arabidopsis is plastid autonomous and delayed in chloroplast biogenesis. J. Hered. 90, 207–214 (1999). [DOI] [PubMed] [Google Scholar]
- Hu Z. et al. The tetratricopeptide repeat-containing protein slow green1 is required for chloroplast development in Arabidopsis. J. Exp. Bot. 65, 1111–1123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki Y. et al. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 42, 133–144 (2005). [DOI] [PubMed] [Google Scholar]
- Kanamaru K. et al. Plastidic RNA polymerase sigma factors in Arabidopsis. Plant Cell Physiol. 40, 832–842 (1999). [DOI] [PubMed] [Google Scholar]
- Chi W. et al. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147, 573–584 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5, 389–402 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M., Larkin R. M. & Chory J. Signal transduction between the chloroplasts and the nucleus. Plant Cell 14, S327–S338 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhang X. & Yang S. A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res. 19, 1205–1216 (2009). [DOI] [PubMed] [Google Scholar]
- Fitter D. W., Martin D. J., Copley M. J., Scotland R. W. & Langdale J. A. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 31, 713–727 (2002). [DOI] [PubMed] [Google Scholar]
- Waters M. T., Moylan E. C. & Langdale J. A. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 56, 432–444 (2008). [DOI] [PubMed] [Google Scholar]
- Kakizaki T. et al. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signalling. Plant Physiol. 151, 1339–1353 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003). [DOI] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Wang N. N., Shih M. C. & Li N. The GUS reporter-aided analysis of the promoter activities of Arabidopsis ACC synthase genes AtACS4, AtACS5, and AtACS7 induced by hormones and stresses. J. Exp. Bot. 56, 909–920 (2005). [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H. K. & Wellburn A. R. Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 11, 591–592 (1983). [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.