SUMMARY
Streptococcus pneumoniae inflicts a huge disease burden as the leading cause of community-acquired pneumonia and meningitis. Soon after mainstream antibiotic usage, multiresistant pneumococcal clones emerged and disseminated worldwide. Resistant clones are generated through adaptation to antibiotic pressures imposed while naturally residing within the human upper respiratory tract. Here, a huge array of related commensal streptococcal strains transfers core genomic and accessory resistance determinants to the highly transformable pneumococcus. β-Lactam resistance is the hallmark of pneumococcal adaptability, requiring multiple independent recombination events that are traceable to nonpneumococcal origins and stably perpetuated in multiresistant clonal complexes. Pneumococcal strains with elevated MICs of β-lactams are most often resistant to additional antibiotics. Basic underlying mechanisms of most pneumococcal resistances have been identified, although new insights that increase our understanding are continually provided. Although all pneumococcal infections can be successfully treated with antibiotics, the available choices are limited for some strains. Invasive pneumococcal disease data compiled during 1998 to 2013 through the population-based Active Bacterial Core surveillance program (U.S. population base of 30,600,000) demonstrate that targeting prevalent capsular serotypes with conjugate vaccines (7-valent and 13-valent vaccines implemented in 2000 and 2010, respectively) is extremely effective in reducing resistant infections. Nonetheless, resistant non-vaccine-serotype clones continue to emerge and expand.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) remains the leading cause of community-acquired pneumonia, meningitis, and bacteremia in children and adults (1) and the most common cause of otitis media in infants and young children. Globally, pneumonia remains the most common cause of death in children younger than 5 years of age, causing 1.6 million deaths annually (2). Pneumococcal disease continues to cause the most deaths among vaccine-preventable diseases according to the World Health Organization (WHO) (3). Persons at higher risk for invasive pneumococcal disease (IPD) (i.e., pneumococcus recovered from a normally sterile site) include children <2 years of age, adults ≥65 years of age, those with underlying chronic conditions (e.g., cardiovascular or pulmonary diseases, etc.), and those with immunosuppression (e.g., congenital immunodeficiency, human immunodeficiency virus [HIV] infection, leukemia, or systemic corticosteroid use, etc.) (4–6). Pneumococcal conjugate vaccines are effective against IPD and have had a significant direct effect on infants and young children as well as an indirect effect on those not targeted to receive the vaccine (7, 8).
Antimicrobial-resistant pneumococcal infections were documented as early as 1912, when optochin resistance in experimental mice was described (9). Acquired optochin resistance was seen in humans 5 years later (10). In 1939, treatment-acquired sulfonamide resistance was reported in a human case of pneumococcal meningitis (11). Penicillin-resistant pneumococci were also selected in laboratories (12, 13); however, it was not until 1965 that the first clinical isolate with reduced penicillin susceptibility was reported (14). During the 1970s and 1980s, pneumococci resistant to penicillin (MIC of ≥0.1 μg/ml), erythromycin, and trimethoprim-sulfamethoxazole (TMP-SMX) spread rapidly globally, including to Australia, Papua New Guinea, Israel, Spain, Poland, South Africa, and the United States (15–19). Tetracycline and chloramphenicol resistances were also identified, with rates varying by region and population (20). Finally, fluoroquinolone resistance has been documented at relatively low levels compared to those for the above-mentioned antibiotics (21).
Multidrug-resistant pneumococci, defined as strains resistant to three or more classes of antimicrobials, were first identified in children (22) via nosocomial transmission and are predominantly associated with pediatric serotypes, or serotypes associated with carriage and disease among the pediatric population (i.e., serotypes 6A, 6B, 9V, 14, 19A, 19F, and 23F) (20, 23–25). Among 21 European Union and European Economic Activity countries, multidrug resistance was observed among isolates of serotypes 19A, 14, 1, 19F, and 23F (26). In the United States, residual multidrug resistance is much less common after 14 years of conjugate vaccine use and more frequently seen among isolates of serotypes 15A, 15B, 15C, 6C, 23A, and 35B (data from Active Bacterial Core surveillance [ABCs], 2013 to 2015). Multiresistant serotype 19A isolates still show the highest MICs for β-lactams, macrolides, lincosamides, tetracycline, and co-trimoxazole (17 of 772 total ABCs isolates according to partial 2015 ABCs data), although the present frequency of multiresistant 19A is low compared to its frequency during 2003 to 2010.
IMPACT OF ANTIMICROBIAL RESISTANCE
In 2013, the U.S. Centers for Disease Control and Prevention (CDC) released the first national report on antibiotic resistance threats in the United States, underscoring their increasing importance (27). The CDC estimated that at least 2 million people acquired serious infections from pathogens that were antimicrobial resistant and that at least 23,000 people died as a result of antimicrobial-resistant infections annually in the United States (27). Antimicrobial resistance complicates treatment and can result in additional antibiotic courses and outpatient visits, excess hospitalizations, and work loss (27).
Specific to antibiotic-resistant pneumococcal pneumonia, a study by Reynolds et al. found that resistance led to 32,398 additional outpatient visits and 19,336 additional hospitalizations, accounting for $91 million (4%) in direct medical costs and $233 million (5%) in total costs, including work and productivity losses (28). A Canadian study found that increased costs associated with penicillin-resistant IPD in children ≤18 years of age admitted to two hospitals was due mainly to antibiotic choice (29). In adults, increased costs due to penicillin-nonsusceptible pneumonia and bacteremia were due to prolonged hospitalization and the use of more expensive antibiotics (30, 31).
Other studies have also examined the association between antibiotic-resistant IPD and clinical outcomes, with differing results, although no recent studies have been reported. Moroney et al. compared persons with bacteremic pneumonia with MICs of cefotaxime of ≥0.25 μg/ml with persons with less resistant bacteremic pneumonia; they found that the proportions of those who died did not differ significantly between the two groups (32). This finding was also documented by Plouffe et al. among pneumococcal bacteremia cases in 10 adult care hospitals in Franklin County, OH, but those authors found that the duration of hospitalization was significantly longer for patients with penicillin-nonsusceptible S. pneumoniae (PNSP) than for those with penicillin-susceptible disease (15.8 days versus 12.1 days; P = 0.05) (33). In contrast to previous studies that found no difference in mortality, both Metlay et al. and Feiken et al. demonstrated that there was an increased risk of mortality among persons infected with nonsusceptible and resistant S. pneumoniae (34, 35). Metlay et al. found that in-hospital mortality was significantly increased among cases with PNSP bacteremic pneumonia compared to those with penicillin-susceptible S. pneumoniae (relative risk, 2.1; 95% confidence interval, 1.0 to 4.3) (34), while in the study by Feiken et al., mortality was significantly increased in U.S. persons infected with pneumococci with penicillin MICs of ≥4.0 μg/ml after the fourth day of hospitalization (35).
DETECTION OF ANTIBIOTIC RESISTANCE
Determination of antimicrobial susceptibility is essential not only for treatment of an individual patient but also for tracking antimicrobial resistance patterns to inform antimicrobial guidance. Even though we can now identify pneumococci and many resistance patterns based upon genetic features, bacterial culture-based phenotypic susceptibility methods remain the gold-standard approach in clinical laboratories.
In the clinical setting, methods and interpretations used to assess antibiotic resistance in S. pneumoniae have been established by a number of professional bodies, such as the Clinical and Laboratory Standards Institute (CLSI), the British Society for Antimicrobial Chemotherapy (BSAC), and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (36). For some antibiotics, such as penicillin, defining resistance is a complex issue. Breakpoints are determined by a combination of the pharmacokinetic/pharmacodynamic properties of an antibiotic and patient outcome, and since antimicrobial resistance patterns continue to evolve over time, changes to breakpoints can also occur during the lifetime of an antibiotic. One example is the revised CLSI breakpoints for penicillin adopted in January 2008 (37, 38). This revision occurred when study data from patients with and without pneumococcal meningitis were reevaluated as a clinical response to penicillin was shown among nonmeningitis isolates despite reduced susceptibility in vitro. These changes in penicillin breakpoints for S. pneumoniae allow clinicians the choice of using penicillin instead of broad-spectrum antimicrobials to treat penicillin-susceptible nonmeningitis pneumococcal infections. These breakpoint differences should also be kept in mind when data from surveillance studies on pneumococcal penicillin resistance are compared. Clinically sensitive strains with 12- to >60-fold-higher MICs (0.12 to 2 μg/ml) than those for wild-type penicillin-sensitive strains (MIC = 0.01 μg/ml) could have profound advantages in the carriage reservoir.
Detection of S. pneumoniae typically relies on culture of clinical specimens and subsequent antibiotic susceptibility testing to guide treatment options. However, these methods are often slow, taking up to 48 h, and results are often negative due to prior antibiotic use before sampling or the tendency of S. pneumoniae to undergo autolysis. To improve detection from clinical specimens, PCR-based methods have been developed (39, 40). Knowledge of the molecular determinants of resistance to a number of antibiotics has also led to the development of a variety of molecular assays to detect the presence of resistance genes in pneumococcal isolates and also directly from clinical specimens (40–46). The majority of these assays are solely PCR based (40–43), although sequencing approaches and microarrays have also been used (45, 46). Several recent studies have also compared phenotypic drug susceptibility testing results with predictions based on whole-genome sequencing (WGS) data for a variety of bacterial pathogens (47), including S. pneumoniae (48, 49). Our CDC-based laboratory has developed a promising WGS-based “typing pipeline” for rapid and automated predictions of pneumococcal serotypes, MICs, genotypes, and additional features (50). Employing continually enhanced bioinformatic pipelines for querying WGS data will greatly expand the depth of laboratory-based strain surveillance efforts. For example, ARG-ANNOT (antibiotic resistance gene annotation) is a very useful bioinformatics tool that provides a periodically updated database (http://en.mediterranee-infection.com/article.php?laref=283&titer=arg-annot-) of known accessory resistance genes to screen bacterial whole-genome sequence data (51).
RISK FACTORS THAT CONTRIBUTE TO ACQUISITION OF RESISTANT INFECTIONS
Recent antibiotic use has been identified as the foremost risk factor for the development of resistance among IPD cases (52, 53), but other risk factors include age (particularly children under 5 years of age), female gender, hospitalization, living in an urban area, attending day care, pediatric serotypes (i.e., serotypes found commonly in children), HIV infection, and immunosuppression (53–55). Currently, >40% of isolates are penicillin resistant in several countries that lack significant conjugate vaccine coverage (56–62). Studies have found that previous use of β-lactam antibiotics (63), extremes of age (e.g., children <5 years of age and the elderly) (63–66), and child care attendance (in a carriage study using PNSP defined as an MIC of >0.06 μg/ml) (65) were associated with penicillin-nonsusceptible pneumococcal infections.
Fewer studies of the acquisition of multidrug resistance have been conducted; however, these studies have found that extremes of age (i.e., <5 years and >65 years of age), previous use of β-lactam antibiotics by patients with noninvasive disease, antibiotic use in the last month by patients with nasopharyngeal colonization, population density, geographic location, and pneumococcal seven-valent conjugate vaccine (PCV7) serotype are all independent risk factors (67–70). With the advent of HIV/AIDS in sub-Saharan Africa, risk factors for the acquisition of multidrug resistance in this immunocompromised group included extremes of age, PCV13 serotypes, pediatric serotypes, previous antibiotic use, previous hospital admission in the last 12 months, and tuberculosis treatment (63).
ANTIBIOTIC TREATMENT
Antibiotics have been a mainstay of IPD treatment and function by decreasing or eradicating the bacterial load (70, 71). Additionally, with severe pneumococcal disease, the inflammatory response needs to be controlled through antibiotics. For example, macrolides inhibit the production of pneumococcal virulence factors by macrolide-susceptible and macrolide-resistant pneumococci and have secondary anti-inflammatory properties to combat infection (72), including the control of neutrophil-mediated inflammation and inhibition of superoxide generation by neutrophils (73). Additionally, corticosteroids are also thought to be an adjunctive treatment for the early management of severe pneumococcal infections, including pneumococcal meningitis or sepsis (74). Unlike macrolides, corticosteroids are not as effective in controlling neutrophil-mediated inflammation (75) but might be best used in conjunction with β-lactams and macrolides to reduce morbidity and mortality associated with pneumococcal infection.
When an inappropriate antibiotic is selected and used for treatment, it can increase the risk of poor outcomes by leading to failed bacterial eradication, selection of resistant bacteria, and complications resulting from these resistant bacteria (72). Current guidelines recommend empirical, broad-spectrum antibiotic therapy for acute bacterial infections (76), with consideration of the common etiologic pathogens, probability of pneumococcus involvement, and antibiotic resistance trends in the local geographic area (71). For the treatment of bacteremic pneumococcal pneumonia in hospitalized children without underlying conditions, studies have shown that penicillin, ampicillin, or cefuroxime should be adequate treatment for those infections caused by isolates with penicillin MICs of ≤2 μg/ml (77). In children, oral monotherapy with amoxicillin, cefuroxime, or cefdinir should be effective after initial parenteral therapy (77). Macrolides can also be used for outpatient management of pneumococcal pneumonia, although breakthrough and/or sepsis meningitis has occurred due to resistant pneumococci (78–81). For adults with bacteremic pneumococcal pneumonia, studies have shown lower mortality rates for patients treated with a cephalosporin and a macrolide (82, 83), although no pediatric studies have replicated this finding. If the pneumococcal infection is resistant to penicillin (MIC of up to 2 μg/ml), then a third-generation cephalosporin or clindamycin can provide adequate treatment (77). Finally, if a pneumococcal isolate has a penicillin MIC of ≥4.0 μg/ml, both clindamycin and vancomycin are recommended (77), although this is a recommendation that might be questioned since most present-day invasive strains with this level of penicillin resistance are also clindamycin resistant (50; ABCs, unpublished data). A newer quinolone or linezolid might also be considered (84). While there was an increasing trend of penicillin resistance within pneumococci prior to conjugate vaccine implementation, high-dose parenteral penicillin and other parenteral antibiotics continue to be effective for pneumonia and bacteremia (85). In summary, at present, all antibiotic-resistant pneumococcal infections can be treated with antibiotics (85). Table 1 lists most known pneumococcal resistance features and causal genetic determinants.
TABLE 1.
Molecular mechanisms responsible for most observed cases of pneumococcal antibiotic resistance
| Antibiotic | Mechanism(s) |
|---|---|
| β-Lactams (penicillin and cephalosporins) | Mutations in penicillin-binding (transpeptidase) domains of pbp genes (primarily pbp2x, pbp2b, and pbp1a); mutations in aminoacyl-tRNA ligase gene (murM); mutations in other genes, including pdgA, ciaH-ciaR, and stkP |
| Macrolides | erm (23S rRNA methyltransferases) (ermB and rarely ermTR), mef-mediated efflux [mef(A) or mef(E)], mutations in 23S rRNA genes or L4 or L22 ribosomal protein genes (rplD and rplV, respectively) |
| Fluoroquinolones | Mutations in DNA gyrase (primarily gyrA) and/or topoisomerase IV genes (primarily parC), PmrA-mediated efflux |
| Tetracycline | Ribosomal protection proteins, primarily Tet(M) and rarely Tet(O) |
| Rifampin | Mutations in rpoB encoding the β-subunit of RNA polymerase |
| Chloramphenicol | Inactivation of chloramphenicol by cat-encoded chloramphenicol acetyltransferase |
| Trimethoprim-sulfamethoxazole | Mutations in the dihydrofolate reductase gene (folA) and dihydropteroate synthetase gene (folP) |
| Ketolides | Mutations in 23S rRNA or L4 or L22 ribosomal protein genes (rplD and rplV), ermB with deletion or mutation in leader sequence |
| Oxazolidinones | Mutations in 23S rRNA genes, deletions in L4 ribosomal protein gene rplD |
PNEUMOCOCCAL RESISTANCE TO β-LACTAM ANTIBIOTICS
Since the mass production of penicillin in the mid-1940s, treatment of pneumococcal infections has relied heavily upon penicillin and other β-lactam antibiotics, which are the most widely used and effective antibiotics against this species. Strains with reduced susceptibility to β-lactams were detected for the first time in 1967 (86), only about 20 years after penicillin was mass produced. Strains with higher penicillin MICs were observed during the late 1970s (22) and rapidly emerged and disseminated after this time (87, 88). In the United States, only 5% of 5,459 IPD isolates recovered during 1979 to 1987 were reported to be nonsusceptible (penicillin MIC of ≥0.1 μg/ml), with only 1 isolate being classified as resistant (MIC of ≥2 μg/ml) (89). This situation dramatically changed during the next few years in the United States. During 1993 to 1994, the percentage of nonsusceptible isolates was 14.1%, and 3.2% of these isolates were penicillin resistant with representation by a wider array of serotypes (90). Rates of IPD due to penicillin-nonsusceptible strains peaked in 1999, when such isolates accounted for 25.1% of all IPD isolates recovered in the United States. PCV7 serotypes accounted for ∼80% of penicillin-nonsusceptible IPD cases (55, 91).
Peptidoglycan Synthesis and Penicillin-Binding Proteins
Peptidoglycan is a major cell wall component found only in bacteria, constituting more than half of the Gram-positive bacterium dry weight. Peptidoglycan serves essential roles in cell expansion, maintenance of cell integrity, cell division, surface anchoring, and cellular diffusion. It follows that this structure is the target for nearly all commonly used and prospective antibiotics that target cell wall synthesis (307). Pneumococcal peptidoglycan is composed of strands of alternating glucosamine and N-acetylmuramic acid residues, directly cross-linked by transpeptidases between two N-acetylmuramic acid residues via short stem peptides of up to 5 amino acids (l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala) between the l-Lys of one stem and the penultimate d-Ala of an adjacent stem. Facilitated by the structural similarity of the β-lactam ring to the d-Ala-d-Ala terminus of the peptidoglycan stem peptide, β-lactams irreversibly bind transpeptidases at their active site. Binding of β-lactams to the transpeptidase active site of these penicillin-binding proteins (PBPs) thus blocks cross-linking of muropeptide chains to prevent cell wall synthesis (92). A second peptidoglycan cross-linking activity relevant to pneumococcal β-lactam resistance involves an addition to the stem through the substitution of the lysyl ε-amino group with l-Ala-l-Ala or l-Ser-l-Ala. These branches serve as a PBP cross-linking substrate to an adjacent stem via the fourth-position d-Ala. This branching activity is carried out by the aminoacyl ligases MurM and MurN, which add the first and second amino acids to the stem lysine, respectively (93).
β-Lactam resistance in pneumococcal disease isolates is due to combinations of altered PBPs that have decreased affinities for these antibiotics (94, 95). These strains invariably reveal profound changes in corresponding key PBP genes, and a very wide range of “resistant” PBP gene alleles have been documented (96–99). β-Lactamases, whether introduced via mobile genetic elements or expressed from the core genome, have never been observed within pneumococcal strains. It is remarkable that resistant PBP combinations expressed from the core genome somehow also serve their essential biosynthetic roles after structural alterations that prevent binding to analogs (β-lactams) of their normal substrates. Unlike the major beta-hemolytic streptococcal pathogens, the highly adaptable pneumococcus has rapidly reengineered these essential proteins required for cell growth and division in response to β-lactam selective pressure.
A seminal finding was that a combination of 3 cloned PBP gene alleles (pbp2b, pbp2x, and pbp1a) from a highly penicillin-resistant pneumococcal strain could be used in gene replacement experiments to transform a susceptible strain to the same high level of resistance (100). All three of these PBPs share a penicillin-sensitive N-terminal transpeptidase domain that contains three conserved motifs: SerXXLys, containing the active-site serine that is bound (acylated) by PBPs; SerXAsn; and LysSer(or Thr)Gly. In general, β-lactam-nonsusceptible pneumococci contain PBP gene substitutions that appear to affect the polarity, charge distribution, and flexibility of the region neighboring the active site to decrease PBP-binding affinities for penicillin and/or other β-lactam classes (101–103).
The involvement of these three PBPs in peptidoglycan expansion/cell shaping and cell division dictates that two (PBP2b and PBP2x) are also essential for cell viability (104), making it difficult to dissect specific steps in the acquisition of resistance. Although pbp1a can be inactivated under laboratory conditions, resulting in profound phenotypic defects (104), it undoubtedly serves irreplaceable roles in nature. The requirement of these PBPs for viability dictates that there are fitness costs associated with the development of pneumococcal β-lactam resistance. Nonetheless, the notion of reduced biological fitness within clinical isolates that are nonsusceptible to β-lactam antibiotics seems contradicted by the phenomenal disease burdens associated with individual resistant clonal complexes (CCs) that have emerged and thrived during the past 40 to 50 years (25, 105–112). It is likely that resistance-conferring mutations that profoundly affect cell fitness are usually rapidly lost in the population. In highly significant and rare instances, it also seems likely that these mutations are rapidly alleviated through compensatory chromosomal mutations. It is also clear that once stably established within a strain, successful resistance-conferring PBP alleles are readily disseminated among multiple pneumococcal clones.
A very large array of mutant transpeptidase combinations have been documented within the three PBP genes, resulting in various levels of reduced susceptibility to β-lactams. Since β-lactams each have differing affinities for individual PBPs (107, 113, 114), the variety of β-lactams consumed contributes to this great diversity. There are multiple chromosomal PBP gene alterations that are conserved within individual highly resistant, globally disseminated clones, and it is likely that β-lactam resistance has played a primary role in their global dissemination. The complex patterns of PBP gene changes observed, and the wide range of MICs of β-lactams represented, depict the amazing adaptability of this organism to various β-lactam selection intensities. Even though certain amino acid substitutions have been shown to be key to resistance, there is not a uniform and precise consensus of the specific contributing roles of the different substitutions within these three genes or of different allelic combinations. Resistance can have complex pathways involving additional PBP and non-PBP genes (115). In addition, bypass suppressor mutations can complicate assessments drawn from experiments with laboratory strains.
Nonetheless, certain key features of β-lactam resistance are consistent. A low level of β-lactam resistance results from alterations within the primary resistance PBPs PBP2x and PBP2b (114). PBP2x alterations effect low-level resistance to all or most β-lactams, while PBP2b mutations influence primarily penicillin resistance, consistent with its lack of binding to cephalosporins (113). An altered PBP1a is required for higher levels of β-lactam resistance, in combination with either or both of the primary PBPs. PBP2x mutations primarily affect expanded-spectrum cephalosporin resistance (107), while both PBP2x and PBP2b confer resistance to penicillins (116, 117). As one can surmise from large-scale surveillance data, nearly all highly β-lactam-resistant clinical isolates are resistant to both penicillin and cephalosporins (118). Clinical isolates of pneumococci with MICs of ≥0.5 μg/ml for penicillins or third-generation cephalosporins, such as ceftriaxone or cefotaxime, almost always contain profoundly altered pbp2b, pbp2x, and pbp1a genes. More subtly altered pbp2a genes have been reported for a smaller percentage of resistant clinical isolates, suggesting less typical involvement in the development of β-lactam resistance (119).
Involvement of Other Proteins in β-Lactam Resistance
Although the PBP genes pbp1a, pbp2b, and pbp2x have been clearly demonstrated to be required for high-level β-lactam resistance in naturally occurring clinical isolates, in some instances, low-level resistance is also dependent upon proteins that are not directly targeted by β-lactams. The scenario that different PBP allele combinations confer differing β-lactam resistance phenotypes is complicated by the finding that transformations using PBP genes from certain strains were not sufficient to transform wild-type strains to the same high level of resistance (120, 121). One study reported that strains exhibiting identical PBP transpeptidase domain sequences exhibited penicillin MICs ranging from 0.25 to 2.0 μg/ml (122).
Inactivation of the murM gene results in the lack of branching activity, resulting in the synthesis of peptidoglycan consisting of only linear muropeptides (93). The MurM aminoacyl ligase appears to be required for penicillin resistance since its inactivation within four different strains exhibiting penicillin MICs of 0.12 to 6 μg/ml resulted in a nearly complete loss of penicillin resistance (123). Besides this finding that suggested a direct role of aminoacyl ligase branching activity in penicillin resistance, highly modified murM genes have been revealed in some resistant pneumococcal strains, suggesting that murM-encoded branching activity evolves in a clone-specific manner to accommodate specific variant PBPs (124–127). Analysis of laboratory mutants with depleted levels of PBP2b revealed the increased incorporation of branched-stem peptides (128) theorized to mimic adaptation to PBP derivatives found in β-lactam-resistant strains that have low transpeptidase activity (126), and murM-null mutants required much higher PBP2b levels for continued growth than did wild-type murM strains (128). Despite these observations, most β-lactam-resistant strains appear to have unaltered murM genes (125). For example, the highly β-lactam-resistant emergent PMEN-14 clone and the closely related and highly successful serotype 19A switch variants of this clonal complex have a nonmosaic murM gene that is identical to that found in certain sensitive strains (49).
Inactivation of another gene shown to be nonessential in the laboratory setting, the peptidoglycan O-acetyltransferase encoded by the adr gene, also attenuates PBP variant-conferred penicillin resistance (129). A missense mutation within the pdgA-encoded peptidoglycan N-acetylglucosamine deacetylase (130) was involved in a late transformational step in achieving high-level penicillin resistance in PBP experiments employing donor chromosomal DNA from a highly penicillin-resistant pneumococcal strain.
StkP (Ser/Thr kinase) is involved in pneumococcal cell division (131) and has been shown to colocalize with PBP2x at the cell division site (132). Inactivation of stkP in a penicillin-resistant clinical isolate with altered PBPs abrogated penicillin resistance; however, a survey of penicillin-resistant isolates revealed no altered stkP alleles (133).
Mutations that can affect β-lactam resistance to a modest extent in the absence of PBP gene alterations are known. Alterations within the two-component sensor kinase CiaH have been found within β-lactam-resistant mutants selected in the laboratory and in nature (134). These mutations result in higher expression levels of genes upregulated by the transcriptional regulator CiaR, although the putative affected genes actually conferring β-lactam resistance are unknown. Mutations within cpoA, which encodes a glycosyltransferase required for the synthesis of diglycosyldiacylglycerols (135), conferred a modest level of piperacillin resistance (136) and were associated with reduced levels of PBP1a.
β-Lactams Influence the Nasopharyngeal Pneumococcal Strain Reservoir
Although current information indicates that high doses of parenteral β-lactams are currently effective against most penicillin-resistant pneumococci, carriage studies that reveal a high proportion of isolates with reduced β-lactam susceptibility suggest that β-lactam antibiotic use plays a significant role in the evolution of the species within its normal upper respiratory tract (URT) reservoir. This can be easily overlooked in the current clinical microbiology landscape, where typically a significant proportion of isolates have extensively remodeled PBP segments that allow for greatly increased penicillin MICs (≥0.12 to 2 μg/ml), which are now considered susceptible for nonmeningitis disease. This observation of stable genetic changes that have occurred in the postpenicillin era reflects extremely rapid and effective adaptation to the global selection pressure exerted by β-lactam antibiotics. Even in some relatively remote geographic areas, one can readily isolate from healthy individuals a high proportion of nasopharyngeal pneumococcal PBP gene mutants with elevated MICs of penicillin (137–139). Even in the absence of antibiotic usage data for a given region, these observations provide direct evidence of prior β-lactam selection pressure in the community or in individuals (140, 141) and suggest the fitness of these resultant emergent mutants in their natural reservoir. While pneumococcal isolates with penicillin MICs in the range of 0.12 to 2 μg/ml are considered resistant for meningitis, these values are not relevant for the treatment of other invasive infections (142). Nonetheless, treatment recommendations should take into account the need to prevent resistance from developing since there is little understanding of the evolution of these phenotypes in the carriage reservoir and how it shapes the evolution of the species as a whole. Patients with pneumococcal bacteremia who had been treated with either β-lactams or macrolides within the previous 6 months were highly likely to be infected with a penicillin-resistant strain, indicative of a prior causative selective effect (141).
The vast reservoir of related mitis group streptococci in the upper respiratory tract has repeatedly provided the genetic basis for the emergence of β-lactam-resistant pneumococcal strains. Although normally harmless, upper respiratory tract carriage of pneumococci, especially in the nasopharynx, is an obligatory step in the various types of infections caused by this major pathogen (143, 144). Pneumococcal carriage most frequently occurs in children, with very high carriage rates in certain undeveloped areas and lower rates in many other regions. The URT is highly colonized with multiple species of closely related members of the Streptococcus mitis group, with which pneumococci freely exchange core genetic loci that contribute to β-lactam resistance (145).
In this environment, microbial species are often subjected to various, and often nonlethal, concentrations of β-lactams. Pneumococci are naturally transformable by the uptake and chromosomal integration of DNA from other pneumococcal strains and other related nonpathogenic mitis group species. Compatible with the importance of recombination in the evolution of β-lactam-resistant pneumococci is the fact that even within neutral housekeeping genes, the majority of observed allelic changes have occurred through horizontal recombination events rather than through intrachromosomal mutation events (146). There is irrefutable observational evidence that β-lactam resistance in pneumococci has repeatedly occurred in nature through recombination events with highly related nonpathogenic fellow members of the Streptococcus mitis group. Since colonization with closely related nonpneumococcal species occurs in all humans, it is logical that β-lactam nonsusceptibility would arise first in such strains prior to its appearance in pneumococci. This prediction is supported by the remarkable observation that virtually all clinical isolates that display reduced penicillin susceptibility (MICs of >0.5 μg/ml) are characterized by one or more mosaic PBP1a, -2b, and -2x genes. Within these PBP genes, there are clearly discernible regions of sequence that clearly originated within nonpneumococcal mitis group species (96–100). In contrast, within basally penicillin-susceptible wild-type pneumococci, there is very little sequence variation between different alleles of the same PBP gene. Streptococcus mitis and Streptococcus oralis have been identified as PBP gene sequence donors for variant PBP2x, -2b, -1a, and -2a (108, 147, 148) found in certain PNSP strains. The observation of mosaic pneumococcal PBP genes is consistent with the finding that commensal mitis group nonpneumococcal species, which are also normally β-lactam susceptible, exhibit high β-lactam MICs in areas where rates of pneumococcal resistance are also high (149). It follows that optimal pneumococcal recipients for the development of β-lactam resistance would have optimal exposure to penicillin-nonsusceptible resistant DNA donors in the human URT. The pneumococcal recipient strain would be an efficient long-term colonizer and also highly transformable. These features, while obviously conducive for the development of the β-lactam nonsusceptible (NS) phenotype, are also conducive for the acquisition of other resistance determinants through horizontal transfer events.
There are relatively few highly resistant β-lactam-resistant pneumococcal lineages, although they constitute the vast majority of β-lactam-resistant (and multiple-drug-resistant [MDR]) pneumococci recovered from infections (25). Even incremental changes that increase β-lactam resistance are relatively rare, as evidenced by the long-observed low-level β-lactam resistance phenotypes of certain well-known common clonal complexes, such as serotype 19A/sequence type 199 (ST199), serotype 23A/ST338, and serotype 15A/ST63 (PMEN-25) (50). The relatively small number of β-lactam-resistant lineages may be due to the requirement for mutation events within multiple PBP genes. These PBP genes are not situated closely on the chromosome, presumably making cotransfer of mutated alleles from related resistant streptococcal strains relatively infrequent. These resultant mutated enzymes with poor binding affinity for an analog of their natural cell wall component substrate must somehow still carry out essential roles for normal growth and division. For these reasons, β-lactam resistance is an excellent measure of the adaptability of this species.
Our experiences attempting to analyze oropharyngeal specimens for pneumococcal species- and serotype-specific DNA sequences highlight the extremely high abundance and diversity of closely related homologs of pneumococcal genes (150), consistent with the nonpneumococcal mitis group streptococci providing a huge reservoir for selectable markers transferrable to pneumococci through homologous recombination. The initial acquisition of penicillin resistance in mitis group streptococci is presumably also a low-frequency event that requires multiple changes to occur within unlinked essential genes, and the mutational combinations that occur must be tolerated in order for a given strain to emerge to detectable numbers. Since there is much more genetic substrate for PBP gene changes to occur within the more common commensal nonpneumococcal mitis group members of the URT, the inevitable appearance of nonpneumococcal gene segments within β-lactam-nonsusceptible pneumococcal PBP genes is easier to reconcile. Although it is intuitive that many of the key point mutations that confer pneumococcal β-lactam resistance have in fact occurred in nonpneumococcal species, it is very difficult to quantitate the accumulation of point mutations within specific pneumococcal strains after the initial resistance-conferring recombination events from nonpneumococcal donors. Certain important point mutations have been localized to specific hot spots mapped on PBP crystal structures that have been frequently found in clinical isolates (101–103). For example, the Q552E and T550A PBP2x substitutions (117, 151, 152) are widespread in several different clones of resistant pneumococci. In our laboratory, through genomic analysis, we have localized 27 transpeptidase positions within the 3 key PBPs, where an amino acid change relative to a wild-type sensitive pneumococcal strain occurs within each of the ∼200 highly resistant (penicillin MIC of ≥4 μg/ml) pneumococcal clinical isolates that we have examined (Y. Li, B. J. Metcalf, S. Chochua, P. A. Hawkins, R. Gierke, T. Pilishvili, L. McGee, and B. W. Beall on behalf of the Active Bacterial Core surveillance team, unpublished data).
Dissection of a β-Lactam Resistance Pathway in the Laboratory
A recent study carefully employed genomic analysis of laboratory strains derived from stepwise transformations of a sensitive pneumococcal strain with genomic DNA from a naturally occurring highly penicillin-resistant S. mitis strain (124, 153). This study provided a glimpse of the genetic complexity of the transfer of β-lactam resistance from a highly resistant S. mitis strain to a pneumococcal donor within a controlled laboratory setting. In this study, a total of 78 different genes were affected by 36 different recombination events that occurred during the four consecutive transformation events that were required to confer high-level penicillin resistance (124). As expected, mosaic PBP genes were essential in the stepwise process. In particular, pbp2b and murM sequences from this S. mitis strain were essential for the final high level of penicillin resistance. Certain mosaic sequences were observed among a subset of pneumococcal clinical isolates, confirming the key role of S. mitis in the evolution of pneumococcal β-lactam resistance.
It is likely that next-generation genome sequencing (NGS) will be incorporated routinely for the analysis of pneumococcal disease isolates collected through national surveillance programs. This will lead to a more detailed understanding of the different pathways leading to pneumococcal β-lactam resistance. NGS will soon lead to bioinformatic approaches that will allow immediate deduction of the various MICs of different β-lactam antibiotics exhibited by this organism. From a practical clinical perspective, we have found that accurate predictions of MICs of 5 different β-lactam classes for the vast majority of invasive isolates can be simply predicted through an automated system that compiles 3 allele combinations of transpeptidase amino acid sequences of PBP1a, PBP2b, and PBP2x (50; our unpublished data).
Intrapneumococcal Exchange of β-Lactam Resistance Loci
Interspecies recombination events involving pneumococcal recipients result in the cotransformation of multiple unlinked genes, some of which are intrinsically required for resistance and some of which may or may not provide compensatory mutations that relieve deleterious effects on cell fitness (115). Although many different mosaic pneumococcal PBP genes have been associated with β-lactam resistance (48), existing multilocus sequence typing (MLST) data for genes that are under neutral selection indicate that most recombinant pneumococcal alleles reflect intraspecies recombination events. It is insightful that of the seven MLST targets used for typing of pneumococci, only one (ddl) is frequently associated with sequences of a nonpneumococcal origin and is due to its proximity to pbp2b causing it to be frequently cotransferred into pneumococci (154). Not coincidentally, nearly all such divergent ddl alleles are associated with a β-lactam NS phenotype.
Recombinational exchange events at the cps loci have been shown to have occurred often between pneumococcal strains (49, 98, 155, 156), providing a mechanism of immune escape from serotype-specific antibody. The first account of pneumococcal serotype switching was described in 1928 (157), leading to the discovery of the transforming substance in 1944 (158). Through transforming a sensitive strain with chromosomal DNA from a resistant strain and selecting for β-lactam resistance, another hitchhiking effect was observed: the cotransfer of the large cps locus and the closely flanking pbp1a and pbp2x genes (159). This experiment demonstrated the potential for the appearance of serotype-switching events through selection for β-lactam resistance. It logically follows that the reverse is also possible through immune selection of serotype-switching events. Soon after the implementation of PCV7, genotyping results from a set of invasive serotype 19A isolates suggested that a cotransformation event of this nature had occurred within a serotype 4 recipient strain, where an intermediately penicillin-resistant non-PCV7 strain served as a donor of the cps19A cps locus along with the flanking pbp1a and pbp2x alleles (160). This single cotransfer of both capsular serotype and mosaic PBP alleles was verified through genome sequence analysis of multiple progeny as well as identified donor and recipient strains (161). Multiple serotype 19A/CC695 strains of this nature were subsequently detected within many sites, all of which appeared to have occurred within the post-PCV7/pre-PCV13 period (112, 162). Subsequent genomic analysis demonstrated that the simultaneous transfer of multiple unlinked, and often large, recombinational fragments (up to at least 44 kb) had occurred simultaneously with the vaccine escape recombination event (163). At least five independent serotype switch events involving type 4 recipients and type 19A donors were identified by a combination of MLST and genomic analyses. It was found that one of these variants disseminated west across the United States (163). During the late 2000s, this serotype 19A/ST695 variant became the third most prevalent serotype 19A strain (112). Through NGS-based typing in the post-PCV13 period, we detected two of these independent variants among the few remaining serotype 19A clones (50). Our current NGS typing pipeline verified the identification of the originally described donors (161, 163) through recognition of specific MLST and PBP alleles within the two progeny strains (50). We detected linked cps19A and cotransferred pbp2x from the donor and in addition detected that a second unlinked recombination event had occurred, resulting in the cotransfer of pbp2b and ddl alleles that were also highly associated with the specific serotype 19A donor strain.
The ramifications of these and similar independent findings (164) are profound: one recombinational event (e.g., one that confers a different serotype and/or resistance feature) can unpredictably result in conferring additional unselected and chromosomally unlinked traits (165). It is likely that genomic traits that contributed to the success of the major serotype 4 CC695 in the pre-PCV7 era (111, 166) contributed to the success of the more resistant serotype 19A ST695 variant during the 2000s (112).
RESISTANCE TO MACROLIDES AND LINCOSAMIDES
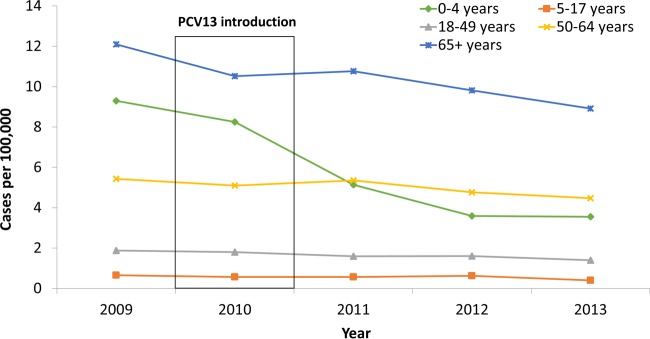
Macrolide antibiotics (e.g., erythromycin, clarithromycin, and azithromycin) have been widely used to treat community-acquired respiratory tract infections globally. In more recent years, resistance to macrolides in S. pneumoniae has increased substantially, and in many parts of the world today, macrolide-resistant pneumococci are now more common than penicillin-resistant S. pneumoniae (167). Erythromycin resistance has become more common in U.S. IPD isolates than penicillin resistance (MIC ≥2 μg/ml) since PCV13 implementation, due largely to the removal of penicillin-resistant strains (50). The most common IPD serotypes in children <5 years of age during 2013, in order of incidence, were serotypes 15B/15C (included together since they interconvert), 33F, 22F, and 35B and together comprised 48% of IPD cases (50). Nearly half (49%) of these isolates were erythromycin resistant (with serotype 35B isolates being predominantly penicillin nonsusceptible with an MIC of 2 μg/ml), predominantly due to the mef determinant (efflux mechanism described below). Both macrolide resistance proportions among isolates and resistance mechanisms vary considerably depending on the country and vaccine implementation. Reported proportions of erythromycin-resistant pneumococci are ∼15% in Latin American isolates, 30.2% in U.S. isolates (http://www.cdc.gov/abcs/reports-findings/survreports/spneu14.html), and as high as 80% among isolates in Southeast Asian countries (168). Besides conjugate vaccination status, these differences may reflect the variability in antibiotic prescribing rates among various countries. Although macrolide resistance accounted for far more IPD cases in 2009 than in 2013 (169), the proportion of erythromycin-resistant cases was actually lower (24.9% relative to 28.2% [see http://www.cdc.gov/abcs/reports-findings/surv-reports.html for ABCs data from 1997 to 2013]). It is also important to note that ∼28% of erythromycin-resistant cases during 2013 were also clindamycin resistant (almost all were constitutively resistant). Clindamycin resistance during 2013 was observed only among erythromycin-resistant strains. (Note that in the above-mentioned URLs, we refer to ABCs, the Centers for Disease Control and Prevention Active Bacterial Core surveillance program, a population-based laboratory and active surveillance system for bacterial pathogens at 10 U.S. sites.)
Macrolides are microbiostatic agents that inhibit bacterial protein synthesis through binding to the 23S rRNA component of the 50S ribosomal subunit. Macrolide resistance in S. pneumoniae is mediated by two major mechanisms: target modification and active efflux.
Target Modification
The most common form of target modification in macrolide-resistant pneumococci is ribosomal modification by 23S rRNA methylation, encoded primarily by the erm(B) gene. This methylation mostly confers constitutive high-level resistance to 14-, 15-, and 16-member macrolides as well as resistance to lincosamides and type B streptogramins (MLSB phenotype) (170). In a small percentage of isolates, erm(B) confers clindamycin resistance that is inducible by low concentrations of a macrolide.
Although rare, a methylase encoded by the erm(A) subclass gene erm(TR) has also been shown to confer MLSB resistance in pneumococci (171, 172). In pneumococci, Tn916 family transposons comprise most erm(B)-carrying mobile genetic elements. A number of Tn916 derivatives carrying erm(B) have been described (Tn1545, Tn3872, Tn6002, and Tn6003), and the tet(M) gene is typically also carried by these transposons (173). A large percentage of current macrolide-resistant S. pneumoniae isolates (31.5% from 2013 [unpublished ABCs data]) are therefore also resistant to tetracycline; however, some recent studies have shown Tn916-related elements where the tet(M) gene is present in a silent form (174). Other less commonly described target modifications are point mutations in domains II and V of 23S rRNA and in riboproteins L4 and L22 (175).
Efflux Mechanism
Lower-level erythromycin resistance affecting only 14- and 15-membered macrolides, but not lincosamides or streptogramins (M phenotype), is associated with efflux pumps. Active drug efflux is mediated by mef class genes, which include several variants: mef(A) and mef(E), which are the most common and share 90% sequence identity, and the rare variant mef(I), which has been described in only two clinical isolates from Italy (176). The genes for the three variants have been associated with different, but related, mobile genetic elements: the mef(A) gene on the defective transposon Tn1207.1 or the closely related transposon Tn1207.3 (177), mef(E) on the “macrolide efflux genetic assembly” (mega) element (178), and mef(I) on a nonmobile composite structure designated the 5216IQ complex (179).
Dual Mechanism for Macrolide Resistance
In the last decade, isolates with the dual resistance mechanism [both erm(B) and the mef gene] have been increasingly reported from the United States, Canada, South Africa, Mexico, and a number of countries in Asia and Europe (172, 180). The prevalence of isolates in the United States carrying both genes increased as a result of the diversification and expansion of lineages of Taiwan19F-14 (PMEN-14 clone) following conjugate vaccine introduction. This was especially true of the major serotype 19A ST320 variant prior to PCV13 implementation (162), as described in more detail below. This strain complex and the dual mechanism accounted for ∼47% of macrolide resistance among pediatric isolates during 2009 (50).
FLUOROQUINOLONE RESISTANCE
With increased rates of resistance to macrolide and β-lactam antibiotics among strains of S. pneumoniae, fluoroquinolones are now among the first choices for empirical treatment of respiratory tract infections and pneumonia in some countries. While the development of fluoroquinolone resistance has been linked with fluoroquinolone use (181), the rates of resistance in S. pneumoniae remain relatively low (<1% in the United States and <3% in Europe) (182), although higher rates have been reported in Asia (10.5%) (183, 184) and Canada (7.3%) (185). Resistance to fluoroquinolones can also develop during treatment, and there are several reports describing treatment failures in pneumococcal infections where fluoroquinolones were used (186, 187). These cases were primarily elderly patients with chronic lung disease, a patient population that is frequently exposed to fluoroquinolones and in which higher rates of resistance have been reported (188).
Fluoroquinolones target type II DNA topoisomerase enzymes (DNA gyrase and topoisomerase IV), which are vital for DNA supercoiling and chromosome segregation. Each of the enzymes consists of subunits that are structurally related to each other. The DNA gyrase subunits gyrA and gyrB are homologous to the parC and parE subunits of type IV topoisomerase. Fluoroquinolones inhibit DNA synthesis by binding to target sites within these proteins. Ciprofloxacin and levofloxacin target primarily topoisomerase IV (subunit ParC), while the principal target of moxifloxacin is DNA gyrase (subunit GyrA) (189, 190).
In pneumococci, two mechanisms that contribute to fluoroquinolone resistance have been identified, namely, target alteration and active efflux.
Target Alteration
Resistance mediated by target modification results from the alteration of the fluoroquinolone-binding site due to the stepwise accumulation of mutations in the quinolone resistance-determining region (QRDR) of the genes encoding the DNA gyrase (primarily gyrA) and DNA topoisomerase IV (primarily parC) subunits. Strains with mutations in only a single target enzyme often have susceptible phenotypes (first-step mutants) but present with an elevated risk of acquiring additional mutations during fluoroquinolone treatment, resulting in resistance (191). These so-called first-step mutants are considered precursors of resistant strains (191). Single mutations within the QRDRs of either parC or gyrA have also been frequently associated with clinically relevant fluoroquinolone resistance (192–194).
Studies focusing on the genetic basis of fluoroquinolone resistance in S. pneumoniae have reported that the most frequent mutations are in the parC S79 and gyrA S81 codons (192–194). Several other substitutions have been described in the QRDRs, but only a few have been reported to confer resistance through in vitro studies: gyrA-E85K, gyrA-Q118K, gyrB-E474K, parC-A63T, parC-D83N, parE-E474K, and parE-D435N or -H (193, 195, 196). Other frequently described substitutions are K137N in parC and I460V in parE, which are commonly found in susceptible strains and appear not to contribute to fluoroquinolone resistance (197). Our recent data for ABCs isolates from 2015 are in agreement with previous surveillance data that demonstrate the association of specific causative QRDR substitutions with a wide range of increased fluoroquinolone MICs (194), quite likely due to the added effects of separate mechanisms such as increased active efflux (198–202).
Efflux Pumps
A second mechanism contributing to nonsusceptibility to fluoroquinolones in some isolates is an increase in active efflux. Quinolones, like ciprofloxacin, that are small molecules seem to be more affected by active efflux than larger molecules such as moxifloxacin (199). In contrast to the mefA gene conferring macrolide resistance, the efflux mechanisms in fluoroquinolone resistance are not well characterized and have been reported mostly in isolates with low-level quinolone resistance. Overexpression of the ABC transporter genes patA and patB, which are also linked to stress responses, have been reported to confer efflux-mediated resistance to fluoroquinolones in pneumococci (199). Little is known about the expression regulation mechanism, but the efflux pump can be blocked by the plant alkaloid reserpine and, to a lesser degree, by verapamil (200). Efflux may not confer complete resistance but may be able to decrease the levels of intracellular fluoroquinolone to sublethal concentrations, fostering the occurrence of QRDR mutations (201).
Horizontal Gene Transfer and the Clonal Concept
In contrast to β-lactam resistance, the role of horizontal gene transfer and recombination in the evolution of fluoroquinolone resistance is uncertain. At least within the United States, where fluoroquinolone resistance has been rare for decades, it seems to have a modest role. Both intra- and interspecies transfers of fluoroquinolone resistance loci have been found to occur in vivo, but the impact of such events on fluoroquinolone resistance in the species might be small. In vitro models showed a higher frequency of recombination of QRDRs between viridans group streptococci and S. pneumoniae than of spontaneous mutations (202); however, this rate of recombination does not appear to be replicated in vivo (203). Reported studies addressing this question of recombination have estimated horizontal gene transfer in 0 to 11% of fluoroquinolone-resistant clinical isolates, and interestingly, this ratio appears to be higher for respiratory isolates than for invasive isolates (204–207).
Fluoroquinolone resistance has been documented in a number of international pneumococcal clones that have been associated with resistance to both penicillin and macrolides (208, 209). However, the role that clonal expansion plays in the increased frequency of fluoroquinolone resistance is controversial, with studies placing different significances on its importance. There has been little indication of clonal expansion of individual fluoroquinolone-resistant clones within the United States, where we screen for, but rarely recover, fluoroquinolone-resistant IPD isolates from an IPD surveillance population of >30 million individuals (http://www.cdc.gov/abcs/reports-findings/survreports/spneu14.html). The increased prevalence of levofloxacin resistance documented in Hong Kong between 1995 and 2001 was suggested to be associated with the spread of strains related to the Spain23F-1 clone. However, numerous studies have shown that clonal dissemination has not been a major contributor to the increase of fluoroquinolone resistance (209–211). Data on pneumococci resistant to levofloxacin from 25 countries, analyzed through the PROTEKT study (1999 to 2000), showed that while 34% were of the Spain23F-1 lineage, the majority of isolates were genetically unrelated (211). These reports suggest that during this period, both the emergence of newly resistant strains and the clonal dissemination of strains contributed to the spread of fluoroquinolone resistance.
RESISTANCE TO OTHER ANTIBIOTICS
Tetracycline Resistance
Tetracyclines are broad-spectrum bacteriostatic drugs previously used in clinical practice and shown to be active against S. pneumoniae. Nonsusceptibility to tetracyclines remains the most frequently observed resistance phenotype in some countries, perhaps reflecting patterns of previous antibiotic usage (212). In pneumococci, resistance to tetracycline occurs by the protection of the bacterial 30S ribosome subunit against antibiotic binding by the acquisition of tet genes (213, 214). The acquisition of tet(M) is the most common mechanism, with the tet(O) gene rarely being reported in pneumococci. In streptococci, tet(M) is most often located on mobile conjugative transposons of the Tn916-Tn1545 family and large composite structures like Tn5253 and Tn3872. A recent study discovered the oldest known examples of two different Tn916-like tet(M)-containing elements identified among S. pneumoniae isolates from 1967 and 1968 (212). These transposons often contain genes for resistance to other classes of antibiotics, such as erm(B), and the selection of these transposons by macrolide antibiotics could possibly explain the continued persistence of tetracycline resistance. Comparison of tet(M) sequences among 8 multidrug-resistant isolates representing 5 diverse species revealed a high degree of variation indicative of mosaicism traced to two distinct alleles (215). In pneumococci, there is evidence of both the clonal distribution of a small number of divergent selected alleles and the horizontal movement of mobile elements carrying the tet(M) gene (216, 217).
Rifampin Resistance
The use of rifampin in combination with either β-lactam antibiotics or vancomycin has been recommended for the treatment of meningitis caused by multidrug-resistant S. pneumoniae. Rifampin has been used in combination therapy for the treatment of tuberculosis and infections due to resistant staphylococci. It has also been used increasingly for prophylaxis against Neisseria meningitidis and Haemophilus influenzae type b exposure. The rates of resistance to rifampin reported among pneumococcal isolates are relatively low at present and vary between 0.1% and 1.5% (218, 219). We observed 5 rifampin-resistant (MIC of >2 μg/ml) isolates among a total of 2,932 (0.17%) IPD isolates recovered from the ABCs program in 2013 (our unpublished data). Rifampin resistance occurs due to alterations in the rpoB-encoded β-subunit of RNA polymerase and has been described in several bacterial species. In pneumococci, resistance has been linked primarily to mutations in regions I and II of rpoB (220). rpoB sequence data indicated past horizontal transfer from related mitis group strains in rifampin-resistant pneumococci, which is a common theme for certain pneumococcal resistances, such as resistance to β-lactams, co-trimoxazole, and rifampin, that rely upon alterations within essential core genome determinants.
Chloramphenicol Resistance
In S. pneumoniae, resistance to chloramphenicol is due to enzymatic inactivation of the antibiotic by the production of a chloramphenicol acetyltransferase (CAT), encoded by the cat gene. This cat gene is carried on the conjugative transposon Tn5253, a composite structure made up of the tetracycline resistance transposon, Tn5251, and Tn5252, which carries the cat gene (221). Chloramphenicol-resistant strains have been reported to contain sequences homologous to catpC194 and other flanking sequences from S. aureus plasmid pC194 (222).
Trimethoprim-Sulfamethoxazole (Co-Trimoxazole) Resistance
Trimethoprim and sulfamethoxazole antibiotics are broadly used in combination as the drug co-trimoxazole. Co-trimoxazole has been used as a treatment option for a range of pneumococcal diseases, particularly in children, because it is relatively inexpensive and generally effective. Resistance to co-trimoxazole has increased substantially worldwide, with recent studies showing rates ranging from 19% in Europe to 50% in Africa (in HIV-associated disease) to >60% in parts of Asia (22, 223, 224). Co-trimoxazole resistance is often accompanied by resistance to other antibiotics, especially penicillin. Trimethoprim resistance in pneumococci results from a single amino acid substitution (Ile100→Leu) in the dihydrofolate reductase (DHFR) protein (225) encoded by folA and is often associated with mosaic alleles. Additional mutations have been reported, which appear to modulate these alterations, affecting the affinity of DHFR for its natural substrates and thereby enhancing resistance (226). Resistance to sulfonamides is most often associated with localized 1- or 2-codon insertion mutations within the folP gene encoding dihydropteroate synthase (DHPS). Numerous studies have reported the occurrence of single- and/or multiple-amino-acid substitutions in DHPS of sulfonamide-resistant clinical pneumococcal isolates (227–229). In Africa, where sulfadoxine-pyrimethamine (Fansidar) therapy for malaria is common, studies have shown that its use contributes to increased co-trimoxazole resistance in pneumococci (230). Previous information revealed that both mutations (folA-I100L and folP insertion) are required for full co-trimoxazole (trimethoprim-sulfamethoxazole) resistance (MICs of >4/76 μg/ml) (231). We also find that most strains with intermediate co-trimoxazole resistance (MICs of 1/19 to 2/38 μg/ml) either contain a folP insertion or contain the folA-I100L mutation, with full resistance requiring both folA and folP mutant alleles (50).
Ketolide Resistance
Ketolides are a class of semisynthetic agents derived from erythromycin A and designed specifically to act against macrolide-resistant organisms. They bind to a secondary region on domain II of the 23S rRNA subunit and therefore have a stronger binding affinity for the ribosome, thereby maintaining activity against most erythromycin-resistant pneumococci. Telithromycin was the first ketolide drug approved for clinical use; however, safety issues have limited the clinical utility of this drug (232). Both cethromycin (ABT-773) and solithromycin (CEM-101), a novel fluoroketolide, have shown improved activity against macrolide-resistant as well as telithromycin-intermediate and telithromycin-resistant organisms (233–235). This enhanced potency shows promise for future clinical use for these compounds, subject to pharmacokinetic/pharmacodynamic, toxicity, and animal infection model study findings.
High-level telithromycin resistance in S. pneumoniae has been experimentally generated by mutations in domain II or V of 23S rRNA and ribosomal proteins L4 and L22 (236), and telithromycin-resistant mutants are easily generated in vitro from erm(B)-positive strains that exhibit mutations within the region upstream of erm(B) (237). In contrast, clinical telithromycin resistance in S. pneumoniae remains rare. Farrell et al. reported that among a global collection of 13,874 S. pneumoniae isolates (1999 to 2003), 10 were telithromycin resistant, with MICs of ≥4 μg/ml, and they all contained the erm(B) gene (238). Mutations in 23S rRNA, L4, and L22 have also been reported in clinical telithromycin-resistant isolates (239, 240), and a combination of mutated genes can result in higher telithromycin resistance than a mutation in only a single gene (241, 242). Wolter and colleagues demonstrated that erm(B) with a deletion in the leader sequence was responsible for high-level telithromycin resistance in a strain isolated in Canada in 2007 (243).
Oxazolidinone Resistance
Linezolid is the first antibiotic in the oxazolidinone drug class that was approved for clinical use in 2000 for the treatment of nosocomial and community-acquired pneumonia. Linezolid blocks protein synthesis by binding to the 50S subunit of the bacterial ribosome via interactions with the central loop segment of domain V of the 23S rRNA. To date, linezolid-nonsusceptible pneumococcal strains are rare (238, 244). Recent data from the U.S. LEADER and global ZAAPS surveillance systems show no linezolid-nonsusceptible isolates among 2,150 S. pneumoniae isolates tested in 2011 (245, 246), and a review of ABCs data revealed that of 45,165 isolates tested during 1997 to 2014, only 11 (0.02%) are recorded as being nonsusceptible to linezolid (our unpublished data). Nonsusceptibility to linezolid has also been rarely reported among clinical isolates of staphylococci and enterococci, and resistance in these organisms has been found to be conferred by mutations in domain V of 23S rRNA (247). For pneumococci, Wolter et al. (248) described two clinical isolates with decreased susceptibility to linezolid (MICs of 4 μg/ml) that contained 6-bp deletions in the rplD gene encoding the riboprotein L4. The rplD deletion alleles were also found to confer a novel mechanism of simultaneous resistance to macrolides, oxazolidinones, and chloramphenicol. A more recent study identified 2 additional linezolid-nonsusceptible pneumococci with mutations and deletions within the rplD gene from the United States (249) in the ABCs program. Whole-genome sequencing of linezolid-resistant laboratory-generated mutants also revealed a role in resistance for a 23S rRNA methyltransferase (spr0333) and for the ABC proteins PatA and PatB (250). A proteomic and transcriptomic screen suggested increased energy requirement needs associated with the burden of resistance in these laboratory-derived mutants (251).
Expanded-spectrum oxazolidinones like tedizolid, which is a protein synthesis inhibitor, are in clinical development for the treatment of Gram-positive infections. Tedizolid has demonstrated potent in vitro activity against penicillin-resistant S. pneumoniae, including linezolid-resistant strains (252).
Streptogramin Resistance
Quinupristin-dalfopristin is a 30:70 combination of a type B and a type A streptogramin. The two components target the late and early stages of bacterial protein synthesis, respectively, and thus have a synergistic inhibitory effect. Resistance to quinupristin-dalfopristin among Gram-positive cocci is rare. Two clinical isolates among 8,837 (0.02%) pneumococcal isolates were identified in 2001 to 2002 with MICs of 4 μg/ml. They both had a 5-amino-acid tandem duplication (RTAHI) in the L22 ribosomal protein gene (rplV) preventing synergistic ribosomal binding of the antibiotic (253).
CORESISTANCE
It has long been recognized that pneumococcal strains with elevated β-lactam MICs are most often resistant to additional antibiotics (22, 70, 90). The factors that lead to multidrug resistance are complex; however, key observations have been made over the years since this was initially observed (22). Pneumococci were uniformly sensitive to antibiotics prior to their introduction, and it follows that antimicrobial resistance has been directly linked to their usage. There are numerous studies describing that previous recent usage of β-lactams increases the risk for β-lactam-nonsusceptible systemic and nonsystemic pneumococcal infections (142, 254, 255). Suboptimal dosing for long periods of time increases the likelihood of carriage of β-lactam-nonsusceptible strains (256), and the presence of recent β-lactam selective pressure allows an advantageous environment for survival, spread, and subsequent infections caused by such strains (257).
The genesis of the β-lactam-resistant parental strains of the major MDR complexes likely required multiple unlinked and rare recombination events. Certain strains have been identified as being particularly efficient in recombination. In one study, it was found that serotype 3 and 18C strains were much less transformable with a selective marker than were serotype 6B, 14, 19F, 9V, and 23F strains (258). Not coincidentally, these 5 serotypes accounted for most penicillin-nonsusceptible IPD strains prior to PCV7 implementation, and penicillin-nonsusceptible isolates of serotypes 3 and 18C have rarely been identified within the ABCs system (our unpublished data). Interspecies and intraspecies genetic transformations probably play a very large role in most pneumococcal antimicrobial resistance mechanisms. Recombination events have been shown to encompass very large chromosomal fragments. In extreme instances, recombinational fragments of >70,000 bp in length have been implicated (259). Besides β-lactam resistance, resistance gene mosaicism has been demonstrated for several different core genome determinants, including those that encode resistance to trimethoprim, sulfonamides, and rifampin (109, 220, 225, 228).
A recent observational study identified a hyperrecombining set of pneumococcal strains based upon their higher degree of mosaicism within housekeeping genes and found a striking correlation of this strain set with resistance to β-lactams and other antibiotics (260). Unexpectedly, there was no strong association of these strains with resistant conjugate vaccine serotypes, suggesting that despite strong associations of only certain serotypes with β-lactam resistance, reemergence of resistance or other adaptations obtained through this increased propensity to incorporate foreign DNA are possible in the post-PCV13 era. Recently, an appendage on the surface of competent pneumococci was discovered and described as a type IV pilus required for genetic transformation (261). This structure, composed of the ComGC pilin, was shown to capture extracellular DNA with high efficiency in two divergent lineages and is likely to be expressed by most or all pneumococcal strains (261).
Efficient colonization allows for increased exposure to cocolonizing resistant bacterial strains, increasing the opportunity for interstrain homologous-recombination events that can lead to the incorporation of resistance determinants carried on the core genome (i.e., resistance to β-lactams, fluoroquinolones, and co-trimoxazole) and also selecting for integrative transposable elements (i.e., resistance to macrolides, lincosamides, tetracycline, and chloramphenicol). Subsequent continued colonization allows more exposure to sublethal concentrations of β-lactams, macrolides, and other antibiotics, selecting for the incremental accumulation of resistance-conferring genetic determinants. The ability to withstand a variety of selective agents allows still further opportunities as a recipient of additional accessory resistance genes from horizontal genetic transfer events. Besides selectively driving pneumococci to acquire specific resistance mechanisms, certain aminoglycosides, fluoroquinolones, and the DNA-damaging antibiotic mitomycin C actually trigger pneumococcal competence for genetic transformation through induction of the competence (com) regulon (262). The authors of that study proposed that pneumococcal competence is a stress response that seems analogous to the similarly induced SOS response that is lacking in pneumococci but very well characterized in Escherichia coli. Both responses result in increased levels of RecA, which is a key enzyme required for homologous recombination between host and recipient DNA. This finding gives further insight into the incompletely understood hazards of inappropriate antimicrobial treatments for all infectious diseases. For example, recent genomic evidence indicated that the use of tetracycline in the 1940s rapidly altered the normal population structure of the opportunistic pathogen Streptococcus agalactiae and “fixed” these strains within human hosts (263).
Serotypes such as serotypes 1, 5, and 7F are considered highly invasive since they are well represented in global IPD cases but are less commonly encountered in URT carriage in children (264). The possibility that isolates of these serotypes are not efficient colonizers is consistent with the observation that they are also represented primarily by strains that are uniformly sensitive to β-lactams and are also usually sensitive to other antibiotics. Not surprisingly, the majority of β-lactam-nonsusceptible strains in the pre-PCV7 era in the United States were of serotypes commonly carried by healthy children (serotypes 6B, 9V, 14, 19F, and 23F) and are still commonly carried in many unvaccinated or undervaccinated populations (264–267).
An increased selective advantage for dual macrolide and penicillin resistance was indicated by the much more rapid increase in the prevalences of invasive strains that were resistant to both antibiotics than of singly resistant strains revealed from U.S. surveillance data during the period from 1996 to 1999 (268). Coincidentally, the prevalence of strains resistant to erythromycin was increasing more rapidly among penicillin-resistant pneumococci, and resistance to penicillin was increasing only among erythromycin-resistant strains. During this period in the United States, much of this coresistance to erythromycin and penicillin may have originated from the singly erythromycin-resistant serotype 14 PMEN-9 strain (England14-9), since closely related penicillin- and erythromycin-resistant isolates within this clonal complex caused a large percentage of IPD cases during the pre-PCV7 implementation period (111, 166) (see the PMEN-9 complex in Fig. 1). Thus, it appears likely that emergent strains that are singly resistant to erythromycin or penicillin have a significant survival advantage in carriage and a better opportunity to survive and acquire additional resistance mechanisms. Notably, over a 20-year period (1994 to 2013), 5,807 (75.2%) of 7,724 ABCs isolates with penicillin MICs of ≥2 μg/ml were also resistant to erythromycin (unpublished ABCs data). Of 7,882 ABCs isolates with intermediate penicillin resistance (MICs of 0.12 to 1.0 μg/ml), 3,681 (46.7%) were erythromycin resistant. In contrast, only 3,760 (7.5%) of 49,940 penicillin-susceptible isolates were erythromycin resistant. While ∼24% (15,607/65,646) of the cumulative ABCs isolates were penicillin nonsusceptible, 22% (13,248/65,646) were erythromycin resistant. Of these 13,248 erythromycin-resistant isolates, 9,488 (71.6%) were penicillin nonsusceptible, which accounts for 60.8% of the total number of penicillin-nonsusceptible isolates.
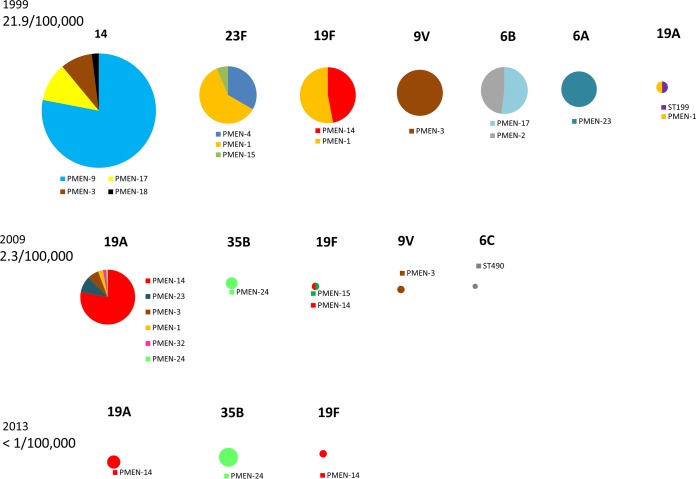
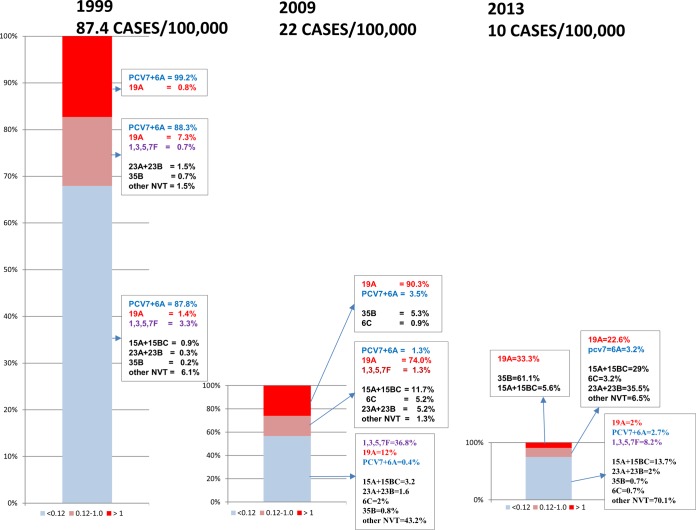
FIG 1.
Approximate numbers of cases per 100,000 individuals caused by penicillin resistance for each year (left). These data encompass all clonal complexes and serotypes associated with penicillin resistance (MIC of ≥2 μg/ml) in cases of pediatric IPD during 1999, 2009, and 2013. The circle diameters reflect relative IPD incidences.
β-Lactam-resistant pneumococci are also highly likely to be resistant to additional antibiotics such as lincosamides [most often coexpressed with macrolide resistance from erm(B)], tetracycline (tetM), and co-trimoxazole (core genome folA and folB mutations conferring resistance to trimethoprim and sulfonamides, respectively). This is also true of isolates with intermediate resistance levels. For example, nearly 80% of the ABCs penicillin-intermediate (MICs of 0.12 to 1.0 μg/ml) pediatric isolates from 2012 were resistant to at least one other antibiotic, with 52% being resistant to macrolides (unpublished ABCs data). Table 2 gives a general description of the genetic lineages that have comprised the majority of penicillin-resistant (MIC of ≥2 μg/ml) strains in the ABCs program since 1998 (Fig. 1). It is highly significant that almost all of these strains are coresistant to macrolides by virtue of containing the mefA-mefE and/or erm(B) determinant.
TABLE 2.
Representative serotypes, common MLST types, and common resistance determinant patterns found within penicillin-resistant strains recovered through the ABCs program from individuals of all ages during 1998 to 2015
| Clonal complex reference strain (serotype, MLST type) | Associated serotype(s) | Common MLST type(s) | Predominant resistance-associated PBP transpeptidase profile(s)a | Common non-beta-lactam resistance determinant profile(s) |
|---|---|---|---|---|
| PMEN-1 (23F, ST81) | 23F, 19F, 19Aa | ST81, ST2346b | 15/12/18 | mef, cat, tetM, folA-I100L, folP insertion; ermB, cat, tetM, folA-I100L, folP insertion |
| PMEN-2 (6B, ST90) | 6B | ST90 | 34/57/56 | ermB, cat, tetM, folA-I100L, folP insertion; ermB, tetM, folA-I100L, folP insertion |
| PMEN-3 (9V, ST156) | 9V, 14, 19A,b 35B,b 11A,c 31c | ST156,ST166 | 15/12/18 | mef, folA-I100L, folP insertion; ermB, tetM, folA-I100L, folP insertion; folA-I100L, folP insertion |
| PMEN-4 (23F, ST37) | 23F | ST37 | 27/38/52 | mef, folA-I100L, folP insertion |
| PMEN-5 (14, ST18) | 14 | ST18 | Not available | ermB, cat, tetM, folA-I100L, folP insertion |
| PMEN-9 (14, ST9) | 14 | ST13 | 27/36/8 | mef, folA-I100L, folP insertion |
| PMEN-14 (19F, ST236) | 19F, 19A,b 3c | ST236, ST271, ST320,b ST1451b | 13/54/33, 13/11/16,b 13/11/33,b 13/14/26b | ermB, mef, tetM, folA-I100L, folP insertion |
| PMEN-15 (23F, ST242) | 23F, 6Ac | ST242 | 13/31/73, 13/31/146c | mef, tetM, folA-I100L, folP insertion; ermB, tetM, folP insertionb |
| PMEN-17 (6B, ST384) | 6B, 6A | ST384, ST147 | 41/36/8 | mef, tetM, folA-I100L, folP insertion |
| PMEN-18 (14, ST67) | 14 | 38/16/8, 38/16/36 | ermB, tetM, folA-I100L, folP insertion | |
| PMEN-23 (6A, ST376) | 6A, 19Ab | ST376, ST1339b | 42/42/8, 27/30/8b | mef, folA-I100L, folP insertion |
| PMEN-24 (35B, ST377) | 35B, 19Ab | ST558 | 4/7/7 | mef |
| PMEN-32 (14, ST230) | 19Aa | ST230, ST276 | 17/15/18, 17/7/8 | tetM, folA-I100L, folP insertion |
PBP profile data and matching transpeptidase sequence databases are available in reference 50.
First observed during the post-PCV7 period from 2002 to 2009.
First observed during the post-PCV13 period from 2012 to 2015.
FITNESS COSTS
The emergence and stability of antibiotic resistance are complex biological processes driven by various factors, including antibiotic use, the rate of resistant mutant formation, the fitness cost imposed, and the compensatory mechanisms to decrease this cost (269, 270). Compensatory mutations can occur in the same altered gene or in others contributing to the developed resistance mechanism, thereby retaining the same resistance level (271). This compensatory phenomenon can occur with or without antibiotic exposure. However, under antibiotic pressure, only those mutations that provide a competitive advantage will be selected, thereby allowing the survival and persistence of resistant strains (269).
In S. pneumoniae, fluoroquinolone-resistant strains have a low frequency due to the high fitness cost imposed by point mutations in the genes that encode type II topoisomerases (topoisomerase IV and DNA gyrase) (272, 273). However, the E85K change in gyrA identified as a high-cost mutation can be compensated for by the presence of a recombinant topoisomerase IV enzyme (273) in some laboratory mutants. These specific alterations acquired by interspecific recombination may be selected as they reduce the fitness cost associated with fluoroquinolone resistance mutations. Similarly, an increase in fitness cost and a loss of virulence have been described in laboratory strains where β-lactam resistance was conferred by mutant PBP gene alleles through transformation (274, 275). However, some clinical strains can also compensate for this fitness cost imposed by this resistance, producing a selective advantage for these strains with an increased potential to spread β-lactam resistance. In a transformation-based study employing resistant clinical strains as PBP gene sources, Albarracín Orio et al. (276) found that the fitness cost and cell division defects conferred by the given mutant pbp2b alleles were fully compensated for by the acquisition of the corresponding pbp2x and pbp1a alleles from the same strain. This suggests that pbp1a and pbp2x mutant alleles not only contribute to the development of higher β-lactam resistance levels but also are acquired for their compensatory effect on fitness, and this compensatory process may occur even without antibiotic pressure.
ANTIBIOTIC TOLERANCE
Antibiotic tolerance is described as the ability of bacteria to survive but not show any apparent growth in the presence of an antibacterial agent. Bacterial tolerance is not detected by routine in vitro susceptibility testing and has been proposed as a phenotype that could be a precursor to the phenotype of resistance (277). Tolerance was first described for β-lactam antibiotics, and the penicillin-tolerant phenotype was recognized in 1985 in eight clinical isolates of pneumococci (278). Tolerance to vancomycin has also been described in clinical isolates of S. pneumoniae (279–282) and has been linked with treatment failure (279). While the mechanism of tolerance has not been fully elucidated, it has been suggested that tolerance is due to a faulty system of regulation of autolysins or changes in the composition of the cell wall (283). Several studies have suggested that the molecular mechanism of tolerance to penicillin and vancomycin involves a defect in the activation of LytA, a murein hydrolase that mediates an endogenous process of death leading to cellular lysis (281, 284, 285). Sung et al. (285) also reported defective LytA autolysin and antimicrobial tolerance in clinical strains. Pep27, a 27-amino-acid secreted peptide that has an important role in controlling bacterial death, has also been suggested to play a role in the tolerance phenotype (281, 286).
VACCINES AND RESISTANCE
PCV-Driven Decrease of Antibiotic Resistance Has Targeted a Limited Array of Pneumococcal Clones
β-Lactam-resistant MDR clonal complexes originated within a surprisingly small number of strains, the most important of which are linked to historical, phenotypic, and genotypic data at the PMEN website (http://www.pneumogen.net/pmen/). These PMEN MDR CCs represented PCV7 serotypes that thrived and disseminated throughout the world during the 1980s and 1990s (25). Nonvaccinated or poorly vaccinated populations in countries with unrestricted antibiotic availability continue to have high proportions of highly resistant disease isolates within these clonal complexes (62, 287). As shown in Fig. 1 and Table 2, these clonal complexes have been responsible for the vast majority of penicillin-resistant pneumococcal strains recovered through the ABCs program since before PCV7 was introduced.
Only higher penicillin MICs (≥4 μg/ml) have been associated with treatment failures of nonmeningitis IPD cases (35). This level of resistance was associated with ∼8% of IPD cases in the United States immediately prior to PCV7 introduction (data not shown). Nearly half (49%) of these cases associated with clinically relevant penicillin resistance during 1999 were due to PCV7 serotype 14, while the remainder were accounted for primarily by PCV7 serotypes 23F (21%), 9V (10%), 19F (8%), 6B (6%), and 6A (3%) (data not shown). Previously recognized globally disseminated clones (25) were the apparent sources of major penicillin-resistant (MICs of ≥2 μg/liter) clonal complexes causing IPD within children in the United States immediately prior to PCV7 introduction (108, 109, 111, 166). Individual penicillin-resistant (MICs of ≥2 μg/ml) PCV7 serotypes were comprised primarily of 1 to 3 major genetic complexes (97, 105–111, 288, 289) represented by the PMEN reference strains shown in Table 2. The vaccine-related serotype 6A also exhibited significant penicillin resistance, due to strains highly related to PMEN-23 (110). It is likely that the dramatic emergence, proliferation, and spread of these multiresistant strains were due in part to antibiotic selective pressure. Their equally impressive decline in the United States was due to conjugate vaccines specifically targeting the predominant IPD-causing serotypes, which were largely comprised of these MDR clones.
Serotype 19A IPD
Figure 2 depicts the breakdown of pediatric IPD cases in the United States during 1999, 2009, and 2013 according to serotype class and penicillin susceptibility (2007 guidelines are used since they are consistent with the fact that the lower MIC values of 0.12 to 1 μg/ml are reflective of adaptation to penicillin and are relevant for meningitis cases). Lower MICs to indicate high resistance (≥2 μg/ml) are used since these values differ from the clinically significant MICs for nonmeningitis disease (≥4 μg/ml) by only a single dilution. An error margin of a single dilution factor is sometimes encountered in the clinical or surveillance laboratory, depending upon the precise MIC of individual strains. As discussed above, prior to PCV7 introduction, virtually all high-level penicillin resistance was due to serotypes 14, 23F, 19F, 9V, 6B, and 6A, with this level of resistance within serotype 19A isolates being rare (Fig. 1 and 2). In contrast, during the post-PCV7 years, high-level penicillin resistance was greatly reduced and was due primarily to serotype 19A. At the same time, the relative proportion of intermediately penicillin-resistant infections also increased, due primarily to the continued expansion of the serotype 19A/CC199 complex and the serotype 19A/CC695 switch variant (162). Although the emergence of highly resistant serotype 19A had a high impact, the rate of pediatric serotype 19A IPD plateaued during the mid- to late 2000s (112). In addition, despite a relatively small eroding effect of nonvaccine serotypes, the overall disease rate also stabilized, indicative of the durable protective effect of PCV7 (8).
FIG 2.
ABCs IPD rates in the population of individuals <5 years of age in 1999 (before PCV7 introduction), 2009 (9 years after PCV7 introduction), and 2013 (3 years after PCV13 introduction). The bright red portions indicate penicillin resistance (MICs of ≥2 μg/ml), lighter red indicates intermediate resistance (0.12 to 1.0 μg/ml), and gray indicates sensitivity (≤0.06 μg/ml). PCV13 types, besides those targeted by PCV7, are indicated in red and purple. Only nonvaccine types (NVT) that are found associated with intermediate or high penicillin resistance are specifically indicated in black boldface type.
It is noteworthy that serotype 19A isolates accounted for only a small proportion of penicillin resistance before PCV7 introduction (Fig. 1 and 2), although at the time, it was the most frequent non-PCV7-targeted serotype associated with high penicillin MICs. Besides its incidence in post-PCV7 IPD, serotype 19A was exceptional in its genetic diversity and resistance features (112, 162). Historical MLST data combined with data from genomic analysis indicated that multiple serotype 19A strains arose independently through serotype-switching events within various highly successful PCV7 serotype strains (49, 111, 155, 161) (Fig. 2). The most striking single change in post-PCV7 penicillin-resistant IPD in the United States was the total disappearance of the major resistant PMEN-9 clonal complex (Fig. 1). Of note, serotype 19A variants of this clonal complex did not appear post-PCV7, while several other serotype 19A variants of other PCV7 type clones emerged and proliferated (including the less penicillin-resistant ST695 serotype 19A variant of the nonresistant major PCV7 type 4 strain) (112, 162). All but one of these MDR serotype 19A variants disappeared as a cause of pediatric IPD in the post-PCV13 period, with the frequency of the remaining highly resistant serotype 19A variant of the PMEN-14 clonal complex being greatly reduced (50). These early results are reassuring in the sense that there are no indications of a structural serotype 19A serological variant that is not targeted by PCV13 in the same manner in which serotype 6C was not targeted by PCV7 and for many years was misidentified as serotype 6A (290, 291).
While individual PCV7 serotypes disappeared or drastically decreased soon after PCV7 introduction, much PCV7 clonal diversity was preserved within the emergent serotype 19A (Fig. 1). Throughout the post-PCV7 period, penicillin-resistant serotype 19A variants persisted, with multilocus sequence types being identical to or closely similar to (in order of abundance) the highly resistant clones PMEN-14, PMEN-23, PMEN-3, and PMEN-1 (Fig. 1). In addition, the intermediately resistant serotype 19A switch variant of a predominant antibiotic-sensitive serotype 4 clone (ST695) was first detected soon after PCV7 introduction and became one of the three most common serotype 19A invasive clonal complexes (described in more detail below). Additional minor penicillin-resistant “PCV7 genotypes” that were detected as serotype 19A variants during the post-PCV7 period included PMEN-26 (Colombia23F-26), PMEN-32 (Denmark14-32), PMEN-24, and PMEN-11 (CSR19A-11) (112, 162).
The most successful single clonal complex throughout the United States during the late 2000s was established by highly resistant serotype 19A ST320 strains within the PMEN-14 lineage (112); the emergence was first detected within the ABCs program among samples from pediatric IPD cases recovered in 2002 (160). Although tracking of this lineage throughout the 2000s documented its steady increase in frequency as a cause of IPD, this clonal complex was absent in pediatric IPD cases within the ABCs program during 1999 despite intensive strain surveillance (111, 166). The major fraction of the initial increase of the prevalence of serotype 19A IPD during the early 2000s was actually due to the moderately resistant CC199 (162), which was the main serotype 19A genotype in the pre-PCV7 period and rarely exhibited penicillin MICs of ≥0.5 μg/ml (111, 160, 166). CC199 rapidly emerged to become the single most abundant invasive clonal complex afflicting children during the early to mid-2000s (162). Although the ST199 genotype was the most common serotype 19A genotype prior to PCV7 implementation, the serotype 19A/CC199 incidence increased after PCV7 implementation, suggesting profound positive selection due to the removal of PCV7 serotypes from the carriage reservoir. From 2006 to 2010, there was also a pronounced shift in the serotype 19A genetic structure, where the prevalence of highly resistant PMEN-14-related serotype 19A strains (CC320) continued to increase with a concurrent decrease in the prevalence of CC199, suggesting that besides the expression of a nonvaccine serotype, high levels of antimicrobial resistance also strongly impacted serotype 19A emergence (112). The main CC320 genotype that became prevalent in serotype 19A strains appears to have originally originated within serotype 19F as a double-locus variant of PMEN-14 (ST236), although the most common serotype 19F genotypes (ST236 and ST271) were also documented within post-PCV7 resistant serotype 19A isolates by the ABCs program (49, 112, 162). These ST236 and ST271 serotype 19A variants originated through independent serotype switch events occurring within serotype 19F recipient strains (49). The serotype 19A/CC320 complex consisted of a high number of ST320 isolates and a plethora of less abundant single-locus variants, consistent with the notion of a single serotype 19A/ST320 strain becoming very abundant and gradually diversifying. Remarkably, throughout the post-PCV7 decade, it was still evident that PCV7 still offered sustained protection against most IPD (8, 292) (Fig. 1 and 2). Nonetheless, although the incidence of resistant infections decreased markedly, the proportion of IPD cases still associated with high penicillin MICs (≥4 μg/ml) actually increased from 8.2% to 9.1% during 1999 to 2009 due to the disproportionate relative incidence of serotype 19A/CC320 strains (50, 112, 162; unpublished ABCs data) (Fig. 1 and 2).
It is remarkable that virtually all serotype 19A IPD isolates surveyed during the past 15 years reflect genotypes that were likely to have originated within strains expressing other serotypes. Besides the variants that share genotypes that originated within PCV7 serotypes, this extends to the serotype 19A/CC199 complex and variants of other genotypes associated primarily with nonconjugate vaccine serotypes (112). The majority of nonconjugate vaccine serotype 15B and 15C isolates recovered in the pre-PCV7 period were of ST199, although these strains lack the intermediate penicillin resistance exhibited by the serotype 19A/CC199 complex. A serotype 19A variant complex of penicillin-intermediate and macrolide- and lincosamide-resistant PMEN-25 (Sweden 15A-25) was also observed during the 2000s (112). Since 2006, multiple serotype 19A switch variants of resistant serotype 35B/ST558 (PMEN-24 complex) have been recovered through the ABCs program (50, 112). The detection of such variants is consistent with naturally occurring immune pressure exerted against resistant serotype 35B and 15A strains that became more abundant in the PCV7 implementation period. Besides involving putative vaccine escape events, the serotype switch events in the post-PCV7 implementation period also involved non-PCV7-type parental strains. These events seem similar in nature to those that occurred before PCV7 introduction (156). During both the pre- and post-PCV7 periods, strains that were not targeted by pediatric vaccines and were also abundant in carriage served as donor and recipient partners in capsular locus switch events.
Current Moderately Successful Antibiotic-Resistant Nonvaccine Types
A pressing question that currently exists is whether any of the most predominant non-PCV-type resistant clonal complexes will emerge as major pathogens. Through the ABCs program, we observed statistically significant, but small, clonal shifts within strains of nonvaccine serotypes 35B, 15A, and 23A during the post-PCV7 era, where single intermediately penicillin-nonsusceptible clonal complexes within these serotypes appeared and modestly emerged (293, 294). Similar increases in prevalence within nonsusceptible serotype 15B/15C (ST3280) and 23B (ST1373) complexes have continued in post-PCV13 IPD (Fig. 2) (see reference 50 for detailed MLST-based data for all serotypes). In the post-PCV7 period, resistant serotype 35B made a moderate emergence, and resistant serotype 6C appeared for the first time to be associated with pediatric IPD (290, 291). Penicillin-nonsusceptible serotype 6C was more genetically complex, with 4 different CCs associated with penicillin nonsusceptibility (291). Interestingly, 3 of the 4 penicillin-nonsusceptible CCs have been commonly associated with the closely related serotype 6A (291), suggesting that much of the increase in the prevalence of serotype 6C IPD (mostly in adults, where it caused a significant disease burden) and the increased resistance originated from independent serotype switch events involving individual type 6A capsular locus recipients. Increasing proportions of clinical isolates within serotypes 35B and 6C in the United States with decreased susceptibility to β-lactams have been reported (295). Resistant serotype 35B (serotype 35B/ST558) IPD was virtually absent among children in the preconjugate vaccine era, but during the past decade, serotypes 35B, 6C, and additional serotypes have increased in both carriage (296, 297) and IPD cases representing all ages (50, 294, 295), potentially as a consequence of the removal of competing conjugate vaccine serotypes from the pediatric carriage reservoir. The PMEN-24 serotype 35B clonal complex has emerged to become the predominant pediatric β-lactam-resistant IPD serotype (50). Although the impact of serotype 35B on resistant IPD is currently small, the frequency of penicillin-resistant (MIC of ≥2 μg/ml) IPD in children due to this clonal complex is higher than what was attributed to serotype 19A in the pre-PCV7 era (Fig. 1). Despite the detected emergence of minor clonal complexes with various levels of β-lactam resistance, the continued strategy for targeting prevalent serotypes with conjugate vaccines is a proven success for sustained decreases in rates of IPD associated with all levels of antimicrobial resistance. These observations provide an obvious rationale for the continued monitoring of serotypes and resistance features of remaining IPD isolates in the post-PCV13 era. While PCV13 appears likely to provide sustained protection against resistant IPD (292), we are currently in a relatively early phase of monitoring, and there is a continued need to closely monitor serotype-specific IPD, especially cases associated with resistance.
Pneumococcal Conjugate Vaccines and Antibiotic Resistance
To summarize, pneumococcal conjugate vaccines serve as a powerful tool against antibiotic resistance. PCV7-type MDR clones peaked immediately before PCV7 implementation, causing the vast majority of IPD cases. By 2004, 4 years after PCV7 introduction in the United States, there were well-documented decreases in the rates of invasive disease caused by penicillin-nonsusceptible strains (6.3 to 2.7 cases per 100,000 individuals) and multidrug-nonsusceptible strains (4.1 to 1.7 cases per 100,000) (91). Vaccine-type resistant disease decreased by 87%, but a significant increase was seen in disease caused by serotype 19A, not included in PCV7 (91). After PCV7 introduction in 2000, the MDR PCV7-type clones virtually disappeared as causes of IPD cases associated with the vaccinated population (7, 8); however, certain non-PCV7 serotype variants of these strains emerged (50, 160, 166, 294). An updated analysis using ABCs data and the 2008 CLSI penicillin breakpoints also found similar declines (298); however, by 2007 to 2008, serotypes in PCV13, but not in PCV7, caused 78% to 97% of penicillin-nonsusceptible IPD, depending on age, with serotype 19A alone accounting for 82% of these cases (298). This increase was hypothesized to be multifactorial: (i) selective pressure from antibiotic use among children that led to subsequent transmission to adults (299–302), (ii) the possible emergence of clones that have advantageous traits for expansion, (iii) capsular switching (161), and (iv) the ability of serotype 19A to cause both colonization and invasive disease (162).
Similar to the peak of antimicrobial-nonsusceptible IPD seen prior to PCV7 introduction, the incidence of nonsusceptible IPD (particularly serotype 19A) increased prior to PCV13 introduction (8, 169) (Fig. 3). Over 100 countries have introduced PCV13 into their childhood immunization schedules. Data documenting the impact of PCV13 on antimicrobial-nonsusceptible pneumococci are of high interest. ABCs data were used to estimate trends of antimicrobial-nonsusceptible pneumococci 3 years after PCV13 introduction in the United States. During 2009 to 2013, the incidence of antimicrobial-nonsusceptible IPD (i.e., nonsusceptible to macrolides, cephalosporins, tetracyclines, penicillins, fluoroquinolones, and glycopeptides), antimicrobial-nonsusceptible IPD caused by all serotypes included in PCV13 but not in PCV7 (i.e., serotypes 1, 3, 5, 7F, and 19A), and multidrug-nonsusceptible IPD (i.e., nonsusceptibility to ≥3 antimicrobial classes) decreased in all age groups (Table 3) (169).
FIG 3.
Incidences (number of cases per 100,000 individuals) of antimicrobial-nonsusceptible (nonsusceptible to one or more classes of antimicrobials, including macrolides, cephalosporins, tetracyclines, penicillins, fluoroquinolones, and glycopeptides) IPD. (Data from Active Bacterial Core surveillance, 2009 to 2013.)
TABLE 3.
Incidences of antimicrobial-nonsusceptible IPD caused by all serotypes, serotypes included in PCV13 but not in PCV7, and multidrug-nonsusceptible IPD in the United Statesd
| Age group (yr) | Antimicrobial-nonsusceptible IPD caused by all serotypesa |
Antimicrobial-nonsusceptible IPD caused by serotypes in PCV13 but not in PCV7a,b |
Multidrug-nonsusceptible IPDa,c |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Rate (no. of cases per 100,000 individuals) |
% difference | Rate (no. of cases per 100,000 individuals) |
% difference | Rate (no. of cases per 100,000 individuals) |
% difference | ||||
| 2009 | 2013 | 2009 | 2013 | 2009 | 2013 | ||||
| <5 | 9.3 | 3.6 | −61.8 | 6.5 | 0.5 | −92.7 | 5.2 | 0.8 | −84.6 |
| 5–17 | 0.7 | 0.4 | −39.0 | 0.2 | 0.1 | −67.2 | 0.2 | 0.1 | −53.2 |
| 18–49 | 1.9 | 1.4 | −25.4 | 0.9 | 0.3 | −60.0 | 0.8 | 0.3 | −60.0 |
| 50–64 | 5.4 | 4.5 | −17.6 | 2.4 | 0.9 | −63.4 | 2.0 | 0.9 | −55.4 |
| ≥65 | 12.1 | 8.9 | −26.3 | 4.4 | 1.4 | −67.4 | 4.2 | 1.8 | −56.9 |
Nonsusceptible to any of the following antimicrobials: macrolides, cephalosporins, tetracyclines, penicillins, fluoroquinolones, and glycopeptides.
Serotypes in PCV13 but not in PCV7 include serotypes 1, 3, 5, 7F, and 19A. Serotype 6A was excluded due to cross-protection from serotype 6B, which was included in PCV7.
Nonsusceptible to ≥3 antimicrobial classes.
Data from Active Bacterial Core surveillance, 2009 to 2013.
Other countries have also reported similar reductions in antimicrobial-resistant IPD after pneumococcal conjugate vaccine introductions. In a South African clinical trial for a 9-valent conjugate vaccine, rates of penicillin-resistant IPD and trimethoprim-sulfamethoxazole-resistant IPD decreased by 67% and 56%, respectively (303). dos Santos et al. also found that penicillin and ceftriaxone nonsusceptibility decreased after PCV10 introduction in Brazil, although no trend was found for clindamycin, chloramphenicol, erythromycin, rifampin, tetracycline, or levofloxacin (304). In Colombia, penicillin resistance among invasive isolates decreased after PCV7 introduction (41.1% versus 14.2%; P = 0.02) (305). Data from Israel also showed a significant decrease in the proportion of isolates with penicillin MICs of ≥0.125 μg/ml (26.2% to 16.4%), although there was an increase in non-vaccine-type strains with penicillin MICs of ≥2 μg/ml, particularly among serotype 19A isolates, similarly to the United States (306). Overall, continued surveillance is needed to document the effects of PCV13 on serotype distribution, antimicrobial resistance, and replacement.
CONCLUSION
In conclusion, we have attempted a broad review of pneumococcal antimicrobial resistance, including the public health impact and risk factors for resistance; mechanisms of resistance, tolerance, and fitness; and the role of conjugate vaccines in decreasing antimicrobial resistance. With the advent of more advanced laboratory techniques, including whole-genome sequencing, and continued, high-quality surveillance of antimicrobial resistance, we can continue to further expand our understanding of this area. Conjugate vaccines have been demonstrated to be a powerful tool and should continue to be introduced in countries to decrease not only the burden of disease but also antimicrobial-resistant pneumococci.
ACKNOWLEDGMENTS
We are indebted to all of the hospitals and laboratories, the RDB Streptococcus Laboratory, and the RDB Epidemiology section that participate in the CDC Emerging Infections Program Network and the Active Bacterial Core surveillance program.
We declare no conflict of interest.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Biographies

Lindsay Kim is a medical epidemiologist in the CDC's Respiratory Diseases Branch, where she works on projects that are part of the Active Bacterial Core surveillance program, focusing on monitoring pneumococcal disease trends, evaluating the impact of pneumococcal vaccines, and informing vaccination policy. She also provides technical assistance to resource-poor countries on the epidemiology and prevention of Streptococcus pneumoniae. Prior to this, she completed her assignment in international tuberculosis through the CDC's Epidemic Intelligence Service during 2010 to 2012. Dr. Kim received her M.D. from Emory University and M.P.H. from the Johns Hopkins Bloomberg School of Public Health.

Lesley McGee received her B.S. honors in microbiology and biochemistry from University of Natal, Pietermaritzburg, and earned her doctorate in medical microbiology in 2002 from the University of Witwatersrand, Johannesburg, South Africa. After completing her Ph.D., she served on the faculty of Emory University's Rollins School of Public Health, Atlanta, GA, before joining the Respiratory Diseases Branch at the Centers for Disease Control and Prevention in 2008. She has worked extensively in the area of pneumococcal epidemiology and was instrumental in helping establish the Pneumococcal Molecular Epidemiology Network in 1997 for the global surveillance of S. pneumoniae. She currently works on projects that are part of the CDC's domestic ABC surveillance program and provides laboratory support to a number of global surveillance efforts and studies. Her research interests include streptococcal infections and disease surveillance, focusing on pathogen detection, antibiotic resistance, and molecular epidemiology and evolution.

Sara Tomczyk is an epidemiologist who recently completed her CDC Epidemic Intelligence Service assignment from 2013 to 2015 in the CDC's Respiratory Diseases Branch, where she worked on projects related to the Active Bacterial Core surveillance program, pneumococcal disease trends, the impact of the pneumococcal vaccines, and antibiotic use. She was also involved with various international capacity-building projects as they relate to respiratory disease and various disease outbreak investigations. She received her B.S.N./P.H.N. from the University of Minnesota and M.Sc. in Epidemiology at the London School of Hygiene and Tropical Medicine.

Bernard Beall has directed the Streptococcus Laboratory within the CDC's Respiratory Diseases Branch since 2004. He earned his B.S. in Biology (1982) and M.S. studying staphylococcal genetics (1985) at the University of Missouri—Kansas City. During his Ph.D. and early postdoctoral work (1985 to 1990) at the University of Kansas Medical Center, he studied aspects of bacterial cell division and sporulation. He continued postdoctoral study of sporulation at Emory University (1990 to 1993), where he was awarded an NIH Postdoctoral Individual National Research Service Award. For the past 22 years, he has worked within the CDC's Respiratory Diseases Branch, with his primary fascination involving invasive streptococcal strain surveillance as a participant in the CDC's Active Bacterial Core surveillance program (http://www.cdc.gov/abcs/index.html). Dr. Beall was elected a Fellow in the American Academy of Microbiology in 2014. He works within a collaborative research network to provide timely population-based invasive streptococcal strain characterization data useful for evaluating vaccines and antimicrobials.
REFERENCES
- 1.Lynch JP III, Zhanel GG. 2009. Streptococcus pneumoniae: epidemiology, risk factors, and strategies for prevention. Semin Respir Crit Care Med 30:189–209. doi: 10.1055/s-0029-1202938. [DOI] [PubMed] [Google Scholar]
- 2.Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, Jha P, Campbell H, Walker CF, Cibulskis R, Eisele T, Liu L, Mathers C, Child Health Epidemiology Reference Group of WHO and UNICEF. 2010. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. 2014. Global immunization data. World Health Organization, Geneva, Switzerland: http://www.who.int/immunization/monitoring_surveillance/global_immunization_data.pdf?ua=1 Accessed 17 September 2014. [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1993. Recommendations of the Advisory Committee on Immunization Practices (ACIP): use of vaccines and immunoglobulins in persons with altered immunocompetence. MMWR Morb Mortal Wkly Rep 42:1–18.8418395 [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1997. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Morb Mortal Wkly Rep 46:1–24.9011775 [Google Scholar]
- 6.Robinson KA, Baughman W, Rothrock G, Barrett NL, Pass M, Lexau C, Damaske B, Stefonek K, Barnes B, Patterson J, Zell ER, Schuchat A, Whitney CG, Active Bacterial Core Surveillance/Emerging Infections Program Network. 2001. Epidemiology of invasive Streptococcus pneumoniae infections in the United States, 1995-1998: opportunities for prevention in the conjugate vaccine era. JAMA 285:1729–1735. doi: 10.1001/jama.285.13.1729. [DOI] [PubMed] [Google Scholar]
- 7.Whitney CG, Pilishvili T, Farley MM, Schaffner W, Craig AS, Lynfield R, Nyquist AC, Gershman KA, Vazquez M, Bennett NM, Reingold A, Thomas A, Glode MP, Zell ER, Jorgensen JH, Beall B, Schuchat A. 2006. Effectiveness of seven-valent pneumococcal conjugate vaccine against invasive pneumococcal disease: a matched case-control study. Lancet 368:1495–1502. doi: 10.1016/S0140-6736(06)69637-2. [DOI] [PubMed] [Google Scholar]
- 8.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, Reingold A, Thomas A, Schaffner W, Craig AS, Smith PJ, Beall BW, Whitney CG, Moore MR, Active Bacterial Core Surveillance/Emerging Infections Program Network. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis 201:32–41. doi: 10.1086/648593. [DOI] [PubMed] [Google Scholar]
- 9.Morgenroth J, Kaufmann M. 1912. Arzneifestigkeit bei Bakterien (Pneumokokken). Z Immunitatsforsch Exp Ther 15:610. [Google Scholar]
- 10.Moore HF, Chesney AM. 1917. A study of ethylhydrocupreine (optochin) in the treatment of acute lobar pneumonia. Arch Intern Med 19:611. [Google Scholar]
- 11.Ross RW. 1939. Acquired tolerance of pneumococcus to M. & B. 693. Lancet i:1207–1208. [Google Scholar]
- 12.Eriksen KR. 1945. Studies on induced resistance to penicillin in a pneumococcus type 1. Acta Pathol Microbiol Scand 22:398–401. [DOI] [PubMed] [Google Scholar]
- 13.McKee CM, Houck CL. 1943. Induced resistance to penicillin of cultures of staphylococci, pneumococci, and streptococci. Proc Soc Exp Biol Med 53:33–34. doi: 10.3181/00379727-53-14172P. [DOI] [Google Scholar]
- 14.Kislak JW, Razavi LM, Daly AK, Finland M. 1965. Susceptibility of pneumococci to nine antibiotics. Am J Med Sci 250:261–268. doi: 10.1097/00000441-196509000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Anderson P, Betts R. 1989. Human adult immunogenicity of protein-coupled pneumococcal capsular antigens of serotypes prevalent in otitis media. Pediatr Infect Dis J 8(Suppl 1):S50–S53. [PubMed] [Google Scholar]
- 16.Henderson FW, Gilligan PH, Wait K, Goff DA. 1988. Nasopharyngeal carriage of antibiotic-resistant pneumococci by children in group day care. J Infect Dis 157:256–263. doi: 10.1093/infdis/157.2.256. [DOI] [PubMed] [Google Scholar]
- 17.Hansman D, Morris S. 1988. Pneumococcal carriage amongst children in Adelaide, South Australia. Epidemiol Infect 101:411–417. doi: 10.1017/S0950268800054364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klugman KP, Koornhof HJ, Kuhnle V. 1986. Clinical and nasopharyngeal isolates of unusual multiply resistant pneumococci. Am J Dis Child 140:1186–1190. [DOI] [PubMed] [Google Scholar]
- 19.Pérez JL, Linares J, Bosch J, López de Goicoechea MJ, Martín R. 1987. Antibiotic resistance of Streptococcus pneumoniae in childhood carriers. J Antimicrob Chemother 19:278–280. doi: 10.1093/jac/19.2.278. [DOI] [PubMed] [Google Scholar]
- 20.Klugman KP. 1990. Pneumococcal resistance to antibiotics. Clin Microbiol Rev 3:171–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones RN, Sader HS, Mendes RE, Flamm RK. 2013. Update on antimicrobial susceptibility trends among Streptococcus pneumoniae in the United States: report of ceftaroline activity from the SENTRY Antimicrobial Surveillance Program (1998-2011). Diagn Microbiol Infect Dis 75:107–109. doi: 10.1016/j.diagmicrobio.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs MR, Koornhof HJ, Robins-Browne RM, Stevenson CM, Vermaak ZA, Freiman I, Miller GB, Witcomb MA, Isaäcson M, Ward JI, Austrian R. 1978. Emergence of multiply resistant pneumococci. N Engl J Med 299:735–740. doi: 10.1056/NEJM197810052991402. [DOI] [PubMed] [Google Scholar]
- 23.Corso A, Severina EP, Petruk VF, Mauriz YR, Tomasz A. 1998. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb Drug Resist 4:325–337. doi: 10.1089/mdr.1998.4.325. [DOI] [PubMed] [Google Scholar]
- 24.Koornhof HJ, Wasas A, Klugman K. 1992. Antimicrobial resistance in Streptococcus pneumoniae: a South African perspective. Clin Infect Dis 15:84–94. doi: 10.1093/clinids/15.1.84. [DOI] [PubMed] [Google Scholar]
- 25.McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, Hakenbeck R, Hryniewicz W, Lefévre JC, Tomasz A, Klugman KP. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol 39:2565–2571. doi: 10.1128/JCM.39.7.2565-2571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.European Centre for Disease Prevention and Control. 2013. Annual epidemiological report 2012: reporting on 2010 surveillance data and 2011 epidemic intelligence data. European Centre for Disease Prevention and Control, Solna, Sweden. [Google Scholar]
- 27.Centers for Disease Control and Prevention. 2014. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 28.Reynolds CA, Finkelstein JA, Ray GT, Moore MR, Huang SS. 2014. Attributable healthcare utilization and cost of pneumonia due to drug-resistant Streptococcus pneumoniae: a cost analysis. Antimicrob Resist Infect Control 3:16. doi: 10.1186/2047-2994-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quach C, Weiss K, Moore D, Rubin E, McGeer A, Low DE. 2002. Clinical aspects and cost of invasive Streptococcus pneumoniae infections in children: resistant vs. susceptible strains. Int J Antimicrob Agents 20:113–118. doi: 10.1016/S0924-8579(02)00127-9. [DOI] [PubMed] [Google Scholar]
- 30.Einarsson S, Kristjansson M, Kristinsson KG, Kjartansson G, Jonsson S. 1998. Pneumonia caused by penicillin-non-susceptible and penicillin-susceptible pneumococci in adults: a case-control study. Scand J Infect Dis 30:253–256. doi: 10.1080/00365549850160882. [DOI] [PubMed] [Google Scholar]
- 31.Klepser ME, Klepser DG, Ernst EJ, Brooks J, Diekema DJ, Mozaffari E, Hendrickson J, Doern GV. 2003. Health care resource utilization associated with treatment of penicillin-susceptible and -nonsusceptible isolates of Streptococcus pneumoniae. Pharmacotherapy 23:349–359. doi: 10.1592/phco.23.3.349.32105. [DOI] [PubMed] [Google Scholar]
- 32.Moroney JF, Fiore AE, Harrison LH, Patterson JE, Farley MM, Jorgensen JH, Phelan M, Facklam RR, Cetron MS, Breiman RF, Kolczak M, Schuchat A. 2001. Clinical outcomes of bacteremic pneumococcal pneumonia in the era of antibiotic resistance. Clin Infect Dis 33:797–805. doi: 10.1086/322623. [DOI] [PubMed] [Google Scholar]
- 33.Plouffe JF, Breiman RF, Facklam RR. 1996. Bacteremia with Streptococcus pneumoniae. Implications for therapy and prevention. Franklin County Pneumonia Study Group. JAMA 275:194–198. [DOI] [PubMed] [Google Scholar]
- 34.Metlay JP, Hofmann J, Cetron MS, Fine MJ, Farley MM, Whitney C, Breiman RF. 2000. Impact of penicillin susceptibility on medical outcomes for adult patients with bacteremic pneumococcal pneumonia. Clin Infect Dis 30:520–528. doi: 10.1086/313716. [DOI] [PubMed] [Google Scholar]
- 35.Feikin DR, Schuchat A, Kolczak M, Barrett NL, Harrison LH, Lefkowitz L, McGeer A, Farley MM, Vugia DJ, Lexau C, Stefonek KR, Patterson JE, Jorgensen JH. 2000. Mortality from invasive pneumococcal pneumonia in the era of antibiotic resistance, 1995-1997. Am J Public Health 90:223–229. doi: 10.2105/AJPH.90.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edson DC, Glick T, Massey LD. 2006. Susceptibility testing practices for Streptococcus pneumoniae: results of a proficiency testing survey of clinical laboratories. Diagn Microbiol Infect Dis 55:225–230. doi: 10.1016/j.diagmicrobio.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; eighteenth informational supplement. CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 38.Centers for Disease Control and Prevention. 2008. Effects of new penicillin susceptibility breakpoints for Streptococcus pneumoniae—United States, 2006-2007. MMWR Morb Mortal Wkly Rep 57:1353–1355. [PubMed] [Google Scholar]
- 39.Carvalho MDG, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R, Sampson JS. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris KA, Turner P, Green EA, Hartley JC. 2008. Duplex real-time PCR assay for detection of Streptococcus pneumoniae in clinical samples and determination of penicillin susceptibility. J Clin Microbiol 46:2751–2758. doi: 10.1128/JCM.02462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasan V, du Plessis M, Beall BW, McGee L. 2011. Quadriplex real-time polymerase chain reaction (lytA, mef, erm, pbp2b(wt)) for pneumococcal detection and assessment of antibiotic susceptibility. Diagn Microbiol Infect Dis 71:453–456. doi: 10.1016/j.diagmicrobio.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 42.Fukushima KY, Yanagihara K, Hirakata Y, Sugahara K, Morinaga Y, Kohno S, Kamihira S. 2008. Rapid identification of penicillin and macrolide resistance genes and simultaneous quantification of Streptococcus pneumoniae in purulent sputum samples by use of a novel real-time multiplex PCR assay. J Clin Microbiol 46:2384–2388. doi: 10.1128/JCM.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kearns AM, Graham C, Burdess D, Heatherington J, Freeman R. 2002. Rapid real-time PCR for determination of penicillin susceptibility in pneumococcal meningitis, including culture-negative cases. J Clin Microbiol 40:682–684. doi: 10.1128/JCM.40.2.682-684.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zettler EW, Scheibe RM, Dias CA, Santafé P, Santos DS, Moreira JDS, Fritscher CC. 2006. Determination of penicillin resistance in Streptococcus pneumoniae isolates from southern Brazil by PCR. Int J Infect Dis 10:110–115. doi: 10.1016/j.ijid.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Cassone M, D'Andrea MM, Iannelli F, Oggioni MR, Rossolini GM, Pozzi G. 2006. DNA microarray for detection of macrolide resistance genes. Antimicrob Agents Chemother 50:2038–2041. doi: 10.1128/AAC.01574-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haanperä M, Huovinen P, Jalava J. 2005. Detection and quantification of macrolide resistance mutations at positions 2058 and 2059 of the 23S rRNA gene by pyrosequencing. Antimicrob Agents Chemother 49:457–460. doi: 10.1128/AAC.49.1.457-460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Köser CU, Ellington MJ, Peacock SJ. 2014. Whole-genome sequencing to control antimicrobial resistance. Trends Genet 30:401–407. doi: 10.1016/j.tig.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chewapreecha C, Marttinen P, Croucher NJ, Salter SJ, Harris SR, Mather AE, Hanage WP, Goldblatt D, Nosten FH, Turner C, Turner P, Bentley SD, Parkhill J. 2014. Comprehensive identification of single nucleotide polymorphisms associated with β-lactam resistance within pneumococcal mosaic genes. PLoS Genet 10:e1004547. doi: 10.1371/journal.pgen.1004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Croucher NJ, Chewapreecha C, Hanage WP, Harris SR, McGee L, van der Linden M, Song JH, Ko KS, de Lencastre H, Turner C, Yang F, Sá-Leão R, Beall B, Klugman KP, Parkhill J, Turner P, Bentley SD. 2014. Evidence for soft selective sweeps in the evolution of pneumococcal multidrug resistance and vaccine escape. Genome Biol Evol 6:1589–1602. doi: 10.1093/gbe/evu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metcalf BJ, Gertz RE, Gladstone RA, Walker H, Sherwood LK, Jackson D, Li Z, Law C, Hawkins PA, Chochua S, Sheth M, Rayamajhi N, Bentley SD, Kim L, Whitney CG, McGee L, Beall B. 2016. Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin Microbiol Infect 22:60.e9–60.e29. doi: 10.1016/j.cmi.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dowell SF, Schwartz B. 1997. Resistant pneumococci: protecting patients through judicious use of antibiotics. Am Fam Physician 55:1647–1654. [PubMed] [Google Scholar]
- 53.Levine OS, Farley M, Harrison LH, Lefkowitz L, McGeer A, Schwartz B. 1999. Risk factors for invasive pneumococcal disease in children: a population-based case-control study in North America. Pediatrics 103:E28. doi: 10.1542/peds.103.3.e28. [DOI] [PubMed] [Google Scholar]
- 54.Klugman KP. 2007. Risk factors for antibiotic resistance in Streptococcus pneumoniae. S Afr Med J 97:1129–1132. [PubMed] [Google Scholar]
- 55.Hofmann J, Cetron MS, Farley MM, Baughman WS, Facklam RR, Elliott JA, Deaver KA, Breiman RF. 1995. The prevalence of drug-resistant Streptococcus pneumoniae in Atlanta. N Engl J Med 333:481–486. doi: 10.1056/NEJM199508243330803. [DOI] [PubMed] [Google Scholar]
- 56.Navarro Torné A, Dias JG, Quinten C, Hruba F, Busana MC, Lopalco PL, Gauci AJ, Pastore-Celentano L, ECDC Country Experts for Pneumococcal Disease. 2014. European enhanced surveillance of invasive pneumococcal disease in 2010: data from 26 European countries in the post-heptavalent conjugate vaccine era. Vaccine 32:3644–3650. doi: 10.1016/j.vaccine.2014.04.066. [DOI] [PubMed] [Google Scholar]
- 57.Doern GV, Pfaller MA, Erwin ME, Brueggemann AB, Jones RN. 1998. The prevalence of fluoroquinolone resistance among clinically significant respiratory tract isolates of Streptococcus pneumoniae in the United States and Canada—1997 results from the SENTRY Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis 32:313–316. doi: 10.1016/S0732-8893(98)00081-9. [DOI] [PubMed] [Google Scholar]
- 58.Schmitz FJ, Verhoef J, Fluit AC. 1999. Comparative activity of 27 antimicrobial compounds against 698 Streptococcus pneumoniae isolates originating from 20 European university hospitals. SENTRY Participants Group. Eur J Clin Microbiol Infect Dis 18:450–453. doi: 10.1007/s100960050318. [DOI] [PubMed] [Google Scholar]
- 59.Doern GV, Pfaller MA, Kugler K, Freeman J, Jones RN. 1998. Prevalence of antimicrobial resistance among respiratory tract isolates of Streptococcus pneumoniae in North America: 1997 results from the SENTRY antimicrobial surveillance program. Clin Infect Dis 27:764–770. doi: 10.1086/514953. [DOI] [PubMed] [Google Scholar]
- 60.Song JH, Lee NY, Ichiyama S, Yoshida R, Hirakata Y, Fu W, Chongthaleong A, Aswapokee N, Chiu CH, Lalitha MK, Thomas K, Perera J, Yee TT, Jamal F, Warsa UC, Vinh BX, Jacobs MR, Appelbaum PC, Pai CH. 1999. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Clin Infect Dis 28:1206–1211. doi: 10.1086/514783. [DOI] [PubMed] [Google Scholar]
- 61.Fung CP, Hu BS, Lee SC, Liu PY, Jang TN, Leu HS, Kuo BI, Yen MY, Liu CY, Liu YC, Lau YJ, Yu KW. 2000. Antimicrobial resistance of Streptococcus pneumoniae isolated in Taiwan: an island-wide surveillance study between 1996 and 1997. J Antimicrob Chemother 45:49–55. doi: 10.1093/jac/45.1.49. [DOI] [PubMed] [Google Scholar]
- 62.Zhang B, Gertz RE Jr, Liu Z, Fu W, Beall B. 2012. Characterization of highly antimicrobial-resistant clinical pneumococcal isolates recovered in a Chinese hospital during 2009-2010. J Med Microbiol 61:42–48. doi: 10.1099/jmm.0.035675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crowther-Gibson P, Cohen C, Klugman KP, de Gouveia L, von Gottberg A, Group for Enteric Respiratory and Meningeal Disease Surveillance in South Africa. 2012. Risk factors for multidrug-resistant invasive pneumococcal disease in South Africa, a setting with high HIV prevalence, in the prevaccine era from 2003 to 2008. Antimicrob Agents Chemother 56:5088–5095. doi: 10.1128/AAC.06463-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nava JM, Bella F, Garau J, Lite J, Morera MA, Martí C, Fontanals D, Font B, Pineda V, Uriz S, Deulofeu F, Calderon A, Duran P, Grau M, Agudo A. 1994. Predictive factors for invasive disease due to penicillin-resistant Streptococcus pneumoniae: a population-based study. Clin Infect Dis 19:884–890. doi: 10.1093/clinids/19.5.884. [DOI] [PubMed] [Google Scholar]
- 65.Wroe PC, Lee GM, Finkelstein JA, Pelton SI, Hanage WP, Lipsitch M, Stevenson AE, Rifas-Shiman SL, Kleinman K, Dutta-Linn MM, Hinrichsen VL, Lakoma M, Huang SS. 2012. Pneumococcal carriage and antibiotic resistance in young children before 13-valent conjugate vaccine. Pediatr Infect Dis J 31:249–254. doi: 10.1097/INF.0b013e31824214ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bédos JP, Chevret S, Chastang C, Geslin P, Régnier B. 1996. Epidemiological features of and risk factors for infection by Streptococcus pneumoniae strains with diminished susceptibility to penicillin: findings of a French survey. Clin Infect Dis 22:63–72. doi: 10.1093/clinids/22.1.63. [DOI] [PubMed] [Google Scholar]
- 67.Brandileone MC, Casagrande ST, Guerra ML, Zanella RC, Andrade AL, Di Fabio JL. 2006. Increase in numbers of β-lactam-resistant invasive Streptococcus pneumoniae in Brazil and the impact of conjugate vaccine coverage. J Med Microbiol 55:567–574. doi: 10.1099/jmm.0.46387-0. [DOI] [PubMed] [Google Scholar]
- 68.Clavo-Sánchez AJ, Girón-González JA, López-Prieto D, Canueto-Quintero J, Sánchez-Porto A, Vergara-Campos A, Marín-Casanova P, Córdoba-Doña JA. 1997. Multivariate analysis of risk factors for infection due to penicillin-resistant and multidrug-resistant Streptococcus pneumoniae: a multicenter study. Clin Infect Dis 24:1052–1059. doi: 10.1086/513628. [DOI] [PubMed] [Google Scholar]
- 69.Katsarolis I, Poulakou G, Analitis A, Matthaiopoulou I, Roilides E, Antachopoulos C, Kafetzis DA, Daikos GL, Vorou R, Koubaniou C, Pneumatikos I, Samonis G, Syriopoulou V, Giamarellou H, Kanellakopoulou K. 2009. Risk factors for nasopharyngeal carriage of drug-resistant Streptococcus pneumoniae: data from a nation-wide surveillance study in Greece. BMC Infect Dis 9:120. doi: 10.1186/1471-2334-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Eldere J, Mera RM, Miller LA, Poupard JA, Amrine-Madsen H. 2007. Risk factors for development of multiple-class resistance to Streptococcus pneumoniae strains in Belgium over a 10-year period: antimicrobial consumption, population density, and geographic location. Antimicrob Agents Chemother 51:3491–3497. doi: 10.1128/AAC.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jacobs MR. 2007. Clinical significance of antimicrobial resistance in Streptococcus pneumoniae. S Afr Med J 97:1133–1140. [PubMed] [Google Scholar]
- 72.File TM. 2009. The science of selecting antimicrobials for community-acquired pneumonia (CAP). J Manag Care Pharm 15:S5–S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feldman C, Anderson R. 2009. New insights into pneumococcal disease. Respirology 14:167–179. doi: 10.1111/j.1440-1843.2008.01422.x. [DOI] [PubMed] [Google Scholar]
- 74.Anderson R, Theron AJ, Feldman C. 1996. Membrane-stabilizing, anti-inflammatory interactions of macrolides with human neutrophils. Inflammation 20:693–705. doi: 10.1007/BF01488805. [DOI] [PubMed] [Google Scholar]
- 75.Barnes PJ. 2007. New molecular targets for the treatment of neutrophilic diseases. J Allergy Clin Immunol 119:1055–1062. doi: 10.1016/j.jaci.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 76.Mandell LA, Bartlett JG, Dowell SF, File TM Jr, Musher DM, Whitney C, Infectious Diseases Society of America. 2003. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults. Clin Infect Dis 37:1405–1433. doi: 10.1086/380488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaplan SL. 2004. Review of antibiotic resistance, antibiotic treatment and prevention of pneumococcal pneumonia. Paediatr Respir Rev 5:S153–S158. doi: 10.1016/S1526-0542(04)90030-9. [DOI] [PubMed] [Google Scholar]
- 78.Reid R Jr, Bradley JS, Hindler J. 1995. Pneumococcal meningitis during therapy of otitis media with clarithromycin. Pediatr Infect Dis J 14:1104–1105. doi: 10.1097/00006454-199512000-00016. [DOI] [PubMed] [Google Scholar]
- 79.Jackson MA, Burry VF, Olson LC, Duthie SE, Kearns GL. 1996. Breakthrough sepsis in macrolide-resistant pneumococcal infection. Pediatr Infect Dis J 15:1049–1051. doi: 10.1097/00006454-199611000-00026. [DOI] [PubMed] [Google Scholar]
- 80.Kelley MA, Weber DJ, Gilligan P, Cohen MS. 2000. Breakthrough pneumococcal bacteremia in patients being treated with azithromycin and clarithromycin. Clin Infect Dis 31:1008–1011. doi: 10.1086/318157. [DOI] [PubMed] [Google Scholar]
- 81.Bochud PY, Calandra T, Moreillon P, Baumgartner JD, Yersin B. 2001. Breakthrough Streptococcus pneumoniae meningitis during clarithromycin therapy for acute otitis media. Eur J Clin Microbiol Infect Dis 20:136–137. doi: 10.1007/PL00011244. [DOI] [PubMed] [Google Scholar]
- 82.Mufson MA, Stanek RJ. 1999. Bacteremic pneumococcal pneumonia in one American city: a 20-year longitudinal study, 1978-1997. Am J Med 107:34S–43S. [DOI] [PubMed] [Google Scholar]
- 83.Waterer GW, Somes GW, Wunderink RG. 2001. Monotherapy may be suboptimal for severe bacteremic pneumococcal pneumonia. Arch Intern Med 161:1837–1842. doi: 10.1001/archinte.161.15.1837. [DOI] [PubMed] [Google Scholar]
- 84.Kaplan SL, Patterson L, Edwards KM, Azimi PH, Bradley JS, Blumer JL, Tan TQ, Lobeck FG, Anderson DC, Linezolid Pediatric Pneumonia Study Group, Pharmacia and Upjohn. 2001. Linezolid for the treatment of community-acquired pneumonia in hospitalized children. Pediatr Infect Dis J 20:488–494. doi: 10.1097/00006454-200105000-00004. [DOI] [PubMed] [Google Scholar]
- 85.Jacobs MR. 2008. Antimicrobial-resistant Streptococcus pneumoniae: trends and management. Expert Rev Anti Infect Ther 6:619–635. doi: 10.1586/14787210.6.5.619. [DOI] [PubMed] [Google Scholar]
- 86.Hansman D, Bullen MM. 1967. A resistant pneumococcus. Lancet i:264–265. [DOI] [PubMed] [Google Scholar]
- 87.Liñares J, Pallares R, Alonso T, Perez JL, Ayats J, Gudiol F, Viladrich PF, Martin R. 1992. Trends in antimicrobial resistance of clinical isolates of Streptococcus pneumoniae in Bellvitge Hospital, Barcelona, Spain (1979-1990). Clin Infect Dis 15:99–105. doi: 10.1093/clinids/15.1.99. [DOI] [PubMed] [Google Scholar]
- 88.Michel J, Dickman D, Greenberg Z, Bergner-Rabinowitz S. 1983. Serotype distribution of penicillin-resistant pneumococci and their susceptibilities to seven antimicrobial agents. Antimicrob Agents Chemother 23:397–401. doi: 10.1128/AAC.23.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spika JS, Facklam RR, Plikaytis BD, Oxtoby MJ. 1991. Antimicrobial resistance of Streptococcus pneumoniae in the United States, 1979-1987. J Infect Dis 163:1273–1278. doi: 10.1093/infdis/163.6.1273. [DOI] [PubMed] [Google Scholar]
- 90.Butler JC, Hofmann J, Cetron MS, Elliott JA, Facklam RR, Breiman RF. 1996. The continued emergence of drug-resistant Streptococcus pneumoniae in the United States: an update from the Centers for Disease Control and Prevention's Pneumococcal Sentinel Surveillance System. J Infect Dis 174:986–993. doi: 10.1093/infdis/174.5.986. [DOI] [PubMed] [Google Scholar]
- 91.Kyaw MH, Lynfield R, Schaffner W, Craig AS, Hadler J, Reingold A, Thomas AR, Harrison LH, Bennett NM, Farley MM, Facklam RR, Jorgensen JH, Besser J, Zell ER, Schuchat A, Whitney CG, Active Bacterial Core Surveillance of the Emerging Infections Program Network. 2006. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N Engl J Med 354:1455–1463. doi: 10.1056/NEJMoa051642. [DOI] [PubMed] [Google Scholar]
- 92.Tipper DJ, Strominger JL. 1965. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A 54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Filipe SR, Pinho MG, Tomasz A. 2000. Characterization of the murMN operon involved in the synthesis of branched peptidoglycan peptides in Streptococcus pneumoniae. J Biol Chem 275:27768–27774. [DOI] [PubMed] [Google Scholar]
- 94.Percheson PB, Bryan LE. 1980. Penicillin-binding components of penicillin-susceptible and -resistant strains of Streptococcus pneumoniae. Antimicrob Agents Chemother 18:390–396. doi: 10.1128/AAC.18.3.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hakenbeck R, Tarpay M, Tomasz A. 1980. Multiple changes of penicillin-binding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 17:364–371. doi: 10.1128/AAC.17.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dowson CG, Hutchison A, Spratt BG. 1989. Extensive remodelling of the transpeptidase domain of penicillin binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol 3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 97.Dowson CG, Hutchison A, Brannigan JA, George RC, Hansman D, Liñares J, Tomasz A, Smith JM, Spratt BG. 1989. Horizontal transfer of penicillin binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci U S A 86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coffey TJ, Dowson CG, Daniels M, Zhou J, Martin C, Spratt BG, Musser JM. 1991. Horizontal gene transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes in natural populations of Streptococcus pneumoniae. Mol Microbiol 5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 99.Muñoz R, Dowson CG, Daniels M. 1992. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol Microbiol 6:2461–2465. [DOI] [PubMed] [Google Scholar]
- 100.Barcus VA, Ghanekar K, Yeo M, Coffey TJ, Dowson CG. 1995. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol Lett 126:299–303. doi: 10.1111/j.1574-6968.1995.tb07433.x. [DOI] [PubMed] [Google Scholar]
- 101.Gordon E, Mouz N, Duée E, Dideberg O. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J Mol Biol 299:477–485. doi: 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 102.Contreras-Martel C, Job V, Di Guilmi AM, Vernet T, Dideberg O, Dessen A. 2006. Crystal structure of penicillin-binding protein 1a (PBP1a) reveals a mutational hotspot implicated in β-lactam resistance in Streptococcus pneumoniae. J Mol Biol 355:684–696. doi: 10.1016/j.jmb.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 103.Contreras-Martel C, Dahout-Gonzalez C, Martins ADS, Kotnik M, Dessen A. 2009. PBP active site flexibility as the key mechanism for β-lactam resistance in pneumococci. J Mol Biol 387:899–909. doi: 10.1016/j.jmb.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 104.Kell CM, Sharma UK, Dowson CG, Town C, Balganesh TS, Spratt BG. 1993. Deletion analysis of the essentiality of penicillin-binding proteins 1A, 2B and 2X of Streptococcus pneumoniae. FEMS Microbiol Lett 106:171–175. doi: 10.1111/j.1574-6968.1993.tb05954.x. [DOI] [PubMed] [Google Scholar]
- 105.Pato MV, Carvalho CB, Tomasz A. 1995. Antibiotic susceptibility of Streptococcus pneumoniae isolates in Portugal. A multicenter study between 1989 and 1993. Microb Drug Resist 1:59–69. [DOI] [PubMed] [Google Scholar]
- 106.Muñoz R, Coffey TJ, Daniels M, Dowson CG, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser JM, Spratt BG, Tomasz A. 1991. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis 164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 107.Munoz R, Musser JM, Crain M, Briles DE, Marton A, Parkinson AJ, Sorensen U, Tomasz A. 1992. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterisation by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin Infect Dis 15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 108.McDougal LK, Rasheed JK, Biddle JW, Tenover FC. 1995. Identification of multiple clones of extended-spectrum cephalosporin-resistant Streptococcus pneumoniae isolates in the United States. Antimicrob Agents Chemother 39:2282–2288. doi: 10.1128/AAC.39.10.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gherardi G, Whitney CG, Facklam RR, Beall B. 2000. Major related sets of antibiotic-resistant pneumococci in the United States as determined by pulsed-field gel electrophoresis and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J Infect Dis 181:216–229. doi: 10.1086/315194. [DOI] [PubMed] [Google Scholar]
- 110.Richter SS, Heilmann KP, Coffman SL, Huynh HK, Brueggemann AB, Pfaller MA, Doern GV. 2002. The molecular epidemiology of penicillin-resistant Streptococcus pneumoniae in the United States, 1994-2000. Clin Infect Dis 34:330–339. doi: 10.1086/338065. [DOI] [PubMed] [Google Scholar]
- 111.Gertz RE Jr, McEllistrem MC, Boxrud DJ, Li Z, Sakota V, Thompson TA, Facklam RR, Besser JM, Harrison LH, Whitney CG, Beall B. 2003. Clonal distribution of invasive pneumococcal isolates from children and selected adults in the United States prior to 7-valent conjugate vaccine introduction. J Clin Microbiol 41:4194–4216. doi: 10.1128/JCM.41.9.4194-4216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beall BW, Gertz RE, Hulkower RL, Whitney CG, Moore MR, Brueggemann AB. 2011. Shifting genetic structure of invasive serotype 19A pneumococci in the United States. J Infect Dis 203:1360–1368. doi: 10.1093/infdis/jir052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hakenbeck R, Tornette S, Adkinson NF. 1987. Interaction of non-lytic b-lactams with penicillin-binding proteins in Streptococcus pneumoniae. J Gen Microbiol 133:755–760. [DOI] [PubMed] [Google Scholar]
- 114.Grebe T, Hakenbeck R. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob Agents Chemother 40:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hakenbeck R, Brückner R, Denapaite D, Maurer P. 2012. Molecular mechanisms of β-lactam resistance in Streptococcus pneumoniae. Future Microbiol 7:395–410. doi: 10.2217/fmb.12.2. [DOI] [PubMed] [Google Scholar]
- 116.Krauss J, van der Linden M, Grebe T, Hakenbeck R. 1996. Penicillin-binding proteins 2x and 2b as primary PBP-targets in Streptococcus pneumoniae. Microb Drug Resist 2:183–186. doi: 10.1089/mdr.1996.2.183. [DOI] [PubMed] [Google Scholar]
- 117.Coffey TJ, Daniels M, McDougal LK, Dowson CG, Tenover FC, Spratt BG. 1995. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob Agents Chemother 39:1306–1313. doi: 10.1128/AAC.39.6.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schrag SJ, McGee L, Whitney CG, Beall B, Craig AS, Choate ME, Jorgensen JH, Facklam RR, Klugman KP, Active Bacterial Core Surveillance Team. 2004. Emergence of Streptococcus pneumoniae with very-high-level resistance to penicillin. Antimicrob Agents Chemother 48:3016–3023. doi: 10.1128/AAC.48.8.3016-3023.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.du Plessis M, Smith AM, Klugman KP. 2000. Analysis of penicillin-binding protein lb and 2a genes from Streptococcus pneumoniae. Microb Drug Resist 6:127–131. doi: 10.1089/107662900419438. [DOI] [PubMed] [Google Scholar]
- 120.Hakenbeck R. 1999. Β-lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non-penicillin-binding proteins. Mol Microbiol 33:673–678. doi: 10.1046/j.1365-2958.1999.01521.x. [DOI] [PubMed] [Google Scholar]
- 121.Smith AM, Klugman KP. 2000. Non-penicillin-binding protein mediated high-level penicillin and cephalosporin resistance in a Hungarian clone of Streptococcus pneumoniae. Microb Drug Resist 6:105–110. doi: 10.1089/107662900419401. [DOI] [PubMed] [Google Scholar]
- 122.Chesnel L, Carapito R, Croizé J, Dideberg O, Vernet T, Zapun A. 2005. Identical penicillin-binding domains in penicillin-binding proteins of Streptococcus pneumoniae clinical isolates with different levels of β-lactam resistance. Antimicrob Agents Chemother 49:2895–2902. doi: 10.1128/AAC.49.7.2895-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Filipe SR, Tomasz A. 2000. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc Natl Acad Sci U S A 97:4891–4896. doi: 10.1073/pnas.080067697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sauerbier J, Maurer P, Rieger M, Hakenbeck R. 2012. Streptococcus pneumoniae R6 interspecies transformation: genetic analysis of penicillin resistance determinants and genome-wide recombination events. Mol Microbiol 86:692–706. doi: 10.1111/mmi.12009. [DOI] [PubMed] [Google Scholar]
- 125.Filipe SR, Severina E, Tomasz A. 2000. Distribution of the mosaic structured murM genes among natural populations of Streptococcus pneumoniae. J Bacteriol 182:6798–6805. doi: 10.1128/JB.182.23.6798-6805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garcia-Bustos J, Tomasz A. 1990. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc Natl Acad Sci U S A 87:5415–5419. doi: 10.1073/pnas.87.14.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smith AM, Klugman KP. 2001. Alterations in MurMN, a cell wall muropeptide branching enzyme, increase high-level penicillin and cephalosporin resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 45:2393–2396. doi: 10.1128/AAC.45.8.2393-2396.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Berg KH, Stamsås GA, Straume D, Håvarstein LS. 2013. Effects of low PBP2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. J Bacteriol 195:4342–4354. doi: 10.1128/JB.00184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Crisóstomo MI, Vollmer W, Kharat AS, Inhülsen S, Gehre F, Buckenmaier S, Tomasz A. 2006. Attenuation of penicillin resistance in a peptidoglycan O-acetyl transferase mutant of Streptococcus pneumoniae. Mol Microbiol 61:1497–1509. doi: 10.1111/j.1365-2958.2006.05340.x. [DOI] [PubMed] [Google Scholar]
- 130.Tait-Kamradt AG, Cronan M, Dougherty TJ. 2009. Comparative genome analysis of high-level penicillin resistance in Streptococcus pneumoniae. Microb Drug Resist 15:69–75. doi: 10.1089/mdr.2009.0891. [DOI] [PubMed] [Google Scholar]
- 131.Beilharz K, Nováková L, Fadda D, Branny P, Massidda O, Veening JW. 2012. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Natl Acad Sci U S A 109:E905–E913. doi: 10.1073/pnas.1119172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Morlot C, Bayle L, Jacq M, Fleurie A, Tourcier G, Galisson F, Vernet T, Grangeasse C, Di Guilmi AM. 2013. Interaction of penicillin-binding protein 2x and Ser/Thr protein kinase StkP, two key players in Streptococcus pneumoniae R6 morphogenesis. Mol Microbiol 90:88–102. doi: 10.1111/mmi.12348. [DOI] [PubMed] [Google Scholar]
- 133.Dias R, Félix D, Caniça M, Trombe MC. 2009. The highly conserved serine threonine kinase StkP of Streptococcus pneumoniae contributes to penicillin susceptibility independently from genes encoding penicillin-binding proteins. BMC Microbiol 9:121. doi: 10.1186/1471-2180-9-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Müller M, Marx P, Hakenbeck R, Brückner R. 2011. Effect of new alleles of the histidine kinase gene ciaH on the activity of the response regulator CiaR in Streptococcus pneumoniae R6. Microbiology 157:3104–3112. doi: 10.1099/mic.0.053157-0. [DOI] [PubMed] [Google Scholar]
- 135.Edman M, Berg S, Storm P, Wikström M, Vikström S, Ohman A, Wieslander A. 2003. Structural features of glycosyltransferases synthesizing major bilayer and nonbilayer-prone membrane lipids in Acholeplasma laidlawii and Streptococcus pneumoniae. J Biol Chem 278:8420–84288. doi: 10.1074/jbc.M211492200. [DOI] [PubMed] [Google Scholar]
- 136.Grebe T, Paik J, Hakenbeck R. 1997. A novel resistance mechanism against β-lactams in Streptococcus pneumoniae involves CpoA, a putative glycosyltransferase. J Bacteriol 179:3342–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Medina MJ, Greene CM, Gertz RE, Facklam RR, Jagero G, Hamel M, Shi YP, Slutsker L, Feikin DR, Beall B. 2005. Novel antibiotic-resistant pneumococcal strains recovered from the upper respiratory tracts of HIV-infected adults and their children in Kisumu, Kenya. Microb Drug Resist 11:9–17. doi: 10.1089/mdr.2005.11.9. [DOI] [PubMed] [Google Scholar]
- 138.Vallès X, Flannery B, Roca A, Mandomando I, Sigaúque B, Sanz S, Schuchat A, Levine M, Soriano-Gabarró M, Alonso P. 2006. Serotype distribution and antibiotic susceptibility of invasive and nasopharyngeal isolates of Streptococcus pneumoniae among children in rural Mozambique. Trop Med Int Health 11:358–366. doi: 10.1111/j.1365-3156.2006.01565.x. [DOI] [PubMed] [Google Scholar]
- 139.Schaumburg F, Alabi A, von Eiff C, Flamen A, Traore H, Grobusch MP, Peters G, Kremsner PG, van der Linden M. 2013. Streptococcus pneumoniae colonization in remote African Pygmies. Trans R Soc Trop Med Hyg 107:105–109. doi: 10.1093/trstmh/trs018. [DOI] [PubMed] [Google Scholar]
- 140.Arason VA, Kristinsson KG, Sigurdsson JA, Stefánsdóttir G, Mölstad S, Gudmundsson S. 1996. Do antimicrobials increase the carriage rate of penicillin resistant pneumococci in children? Cross sectional prevalence study. BMJ 313:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ruhe JJ, Hasbun R. 2003. Streptococcus pneumoniae bacteremia: duration of previous antibiotic use and association with penicillin resistance. Clin Infect Dis 36:1132–1138. doi: 10.1086/374556. [DOI] [PubMed] [Google Scholar]
- 142.Pallares R, Gudiol F, Liñares J, Ariza J, Rufi G, Murgui L, Dorca J, Viladrich PF. 1987. Risk factors and response to antibiotic therapy in adults with bacteremic pneumonia caused by penicillin-resistant pneumococci. N Engl J Med 317:18–22. doi: 10.1056/NEJM198707023170104. [DOI] [PubMed] [Google Scholar]
- 143.Gray BM, Converse GM III, Dillon HC Jr. 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis 142:923–933. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 144.Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D, Tung Y. 1997. Relationship between nasopharyngeal colonization and the development of otitis media in children. J Infect Dis 175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 145.Dowson CG. 2005. Genetic exchange in the respiratory tract, p 131–140. In Nataro JP, Cohen PS, Mobley HLT, Weiser JN (ed), Colonization of mucosal surfaces, 1st ed ASM Press, Washington, DC. [Google Scholar]
- 146.Feil EJ, Smith JM, Enright MC, Spratt BG. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dowson CG, Coffey TJ, Kell C, Whiley RA. 1993. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol Microbiol 9:635–643. doi: 10.1111/j.1365-2958.1993.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 148.Sibold C, Henrichsen J, König A, Martin C, Chalkley L, Hakenbeck R. 1994. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol Microbiol 12:1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 149.Doern GV, Ferraro MJ, Brueggemann AB, Ruoff KL. 1996. Emergence of high rates of antimicrobial resistance among viridans group streptococci in the United States. Antimicrob Agents Chemother 40:891–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Carvalho MDG, Pimenta FC, Moura I, Roundtree A, Gertz RE Jr, Li Z, Jagero G, Bigogo G, Junghae M, Conklin L, Feikin DR, Breiman RF, Whitney CG, Beall BW. 2013. Non-pneumococcal mitis-group streptococci confound detection of pneumococcal capsular serotype-specific loci in upper respiratory tract. PeerJ 1:e97. doi: 10.7717/peerj.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mouz N, Di Guilmi AM, Gordon E, Hakenbeck R, Dideberg O, Vernet T. 1999. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumoniae. Role in the specificity for β-lactam antibiotics. J Biol Chem 274:19175–19180. [DOI] [PubMed] [Google Scholar]
- 152.Pernot L, Chesnel L, Le Gouellec A, Croizé J, Vernet T, Dideberg O, Dessen A. 2004. A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to β-lactam antibiotics. J Biol Chem 279:16463–16470. doi: 10.1074/jbc.M313492200. [DOI] [PubMed] [Google Scholar]
- 153.Hakenbeck R, König A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high level beta-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol 180:1831–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Enright MC, Spratt BG. 1999. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol Biol Evol 16:1687–1695. doi: 10.1093/oxfordjournals.molbev.a026082. [DOI] [PubMed] [Google Scholar]
- 155.Wyres KL, Lambertsen LM, Croucher NJ, McGee L, von Gottberg A, Liñares J, Jacobs MR, Kristinsson KG, Beall BW, Klugman KP, Parkhill J, Hakenbeck R, Bentley SD, Brueggemann AB. 2013. Pneumococcal capsular switching: a historical perspective. J Infect Dis 207:439–449. doi: 10.1093/infdis/jis703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Coffey TJ, Enright MC, Daniels M, Morona JK, Morona R, Hryniewicz W, Paton JC, Spratt BG. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus leads to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol 27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 157.Griffith F. 1928. The significance of pneumococcal types. J Hyg (Lond) 27:113–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Trzciński K, Thompson CM, Lipsitch M. 2004. Single-step capsular transformation and acquisition of penicillin resistance in Streptococcus pneumoniae. J Bacteriol 186:3447–3452. doi: 10.1128/JB.186.11.3447-3452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pai R, Moore MR, Pilishvili T, Gertz RE, Whitney CG, Beall B, Active Bacterial Core Surveillance Team. 2005. Post vaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J Infect Dis 192:1988–1995. doi: 10.1086/498043. [DOI] [PubMed] [Google Scholar]
- 161.Brueggemann AB, Pai R, Crook DW, Beall B. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog 3:e168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Moore MR, Gertz RE Jr, Woodbury RL, Barkocy-Gallagher GA, Schaffner W, Lexau C, Gershman K, Reingold A, Farley M, Harrison LH, Hadler JL, Bennett NM, Thomas AR, McGee L, Pilishvili T, Brueggemann AB, Whitney CG, Jorgensen JH, Beall B. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J Infect Dis 197:1016–1027. doi: 10.1086/528996. [DOI] [PubMed] [Google Scholar]
- 163.Golubchik T, Brueggemann AB, Street T, Gertz RE Jr, Spencer CC, Ho T, Giannoulatou E, Link-Gelles R, Harding RM, Beall B, Peto TE, Moore MR, Donnelly P, Crook DW, Bowden R. 2012. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat Genet 44:352–355. doi: 10.1038/ng.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Porat N, Arguedas A, Spratt BG, Trefler R, Brilla E, Loaiza C, Godoy D, Bilek N, Dagan R. 2004. Emergence of penicillin-nonsusceptible Streptococcus pneumoniae clones expressing serotypes not present in the antipneumococcal conjugate vaccine. J Infect Dis 190:2154–2161. doi: 10.1086/425908. [DOI] [PubMed] [Google Scholar]
- 165.Hiller NL, Ahmed A, Powell E, Martin DP, Eutsey R, Earl J, Janto B, Boissy RJ, Hogg J, Barbadora K, Sampath R, Lonergan S, Post JC, Hu FZ, Ehrlich GD. 2010. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog 6:e1001108. doi: 10.1371/journal.ppat.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Beall B, McEllistrem MC, Gertz RE Jr, Wedel S, Boxrud DJ, Gonzalez AL, Medina MJ, Pai R, Thompson TA, Harrison LH, McGee L, Whitney CG, Active Bacterial Core Surveillance Team. 2006. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J Clin Microbiol 44:999–1017. doi: 10.1128/JCM.44.3.999-1017.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Felmingham D, Reinert RR, Hirakata Y, Rodloff A. 2002. Increasing prevalence of antimicrobial resistance among isolates of Streptococcus pneumoniae from the PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telithromycin. J Antimicrob Chemother 50(Suppl 1):25–37. doi: 10.1093/jac/dkf808. [DOI] [PubMed] [Google Scholar]
- 168.Felmingham D, Cantón R, Jenkins SG. 2007. Regional trends in β-lactam, macrolide, fluoroquinolone and telithromycin resistance among Streptococcus pneumoniae isolates 2001-2004. J Infect 55:111–118. doi: 10.1016/j.jinf.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 169.Tomczyk SM, Lynfield R, Schaffner W, Reingold A, Miller L, Petit S, Holtxman C, Zansky SM, Thomas A, Baumbach J, Harrison LH, Farley MM, Beall B, McGee L, Gierke R, Pondo T, Kim L. 7 February 2016. Prevention of antibiotic-nonsusceptible invasive pneumococcal disease with the 13-valent pneumococcal conjugate vaccine. Clin Infect Dis doi: 10.1093/cid/ciw067. [DOI] [PubMed] [Google Scholar]
- 170.Weisblum B. 1995. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Farrell DJ, Morrissey I, Bakker S, Felmingham D. 2002. Molecular characterization of macrolide resistance mechanisms among Streptococcus pneumoniae and Streptococcus pyogenes isolated from the PROTEKT 1999-2000 study. J Antimicrob Chemother 50:39–47. doi: 10.1093/jac/dkf806. [DOI] [PubMed] [Google Scholar]
- 172.Hawkins PA, Chochua S, Jackson D, Beall B, McGee L. 2015. Mobile elements and chromosomal changes associated with MLS resistance phenotypes of invasive pneumococci recovered in the United States. Microb Drug Resist 21:121–129. doi: 10.1089/mdr.2014.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Varaldo PE, Montanari MP, Giovanetti E. 2009. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob Agents Chemother 53:343–353. doi: 10.1128/AAC.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Montanari MP, Giovanetti E, Cochetti I. 2006. An unexpressed tet(M) gene is present in the vast majority of tetracycline-susceptible streptococci carrying erm(B), p 129 Abstr 46th Intersci Conf Antimicrob Agents Chemother. [Google Scholar]
- 175.Davies TA, Bush K, Sahm D, Evangelista A. 2005. Predominance of 23S rRNA mutants among non-erm, non-mef macrolide-resistant clinical isolates of Streptococcus pneumoniae collected in the United States in 1999-2000. Antimicrob Agents Chemother 49:3031–3033. doi: 10.1128/AAC.49.7.3031-3033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cochetti I, Vecchi M, Mingoia M, Tili E, Catania MR, Manzin A, Varaldo PE, Montanari MP. 2005. Molecular characterization of pneumococci with efflux-mediated erythromycin resistance and identification of a novel mef gene subclass, mef(I). Antimicrob Agents Chemother 49:4999–5006. doi: 10.1128/AAC.49.12.4999-5006.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Santagati M, Iannelli F, Cascone C, Campanile F, Oggioni MR, Stefani S, Pozzi G. 2003. The novel conjugative transposon Tn1207.3 carries the macrolide efflux gene mef(A) in Streptococcus pyogenes. Microb Drug Resist 9:243–247. doi: 10.1089/107662903322286445. [DOI] [PubMed] [Google Scholar]
- 178.Gay K, Stephens DS. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J Infect Dis 184:56–65. doi: 10.1086/321001. [DOI] [PubMed] [Google Scholar]
- 179.Mingoia M, Vecchi M, Cochetti I, Tili E, Vitali LA, Manzin A, Varaldo PE, Montanari MP. 2007. Composite structure of Streptococcus pneumoniae containing the erythromycin efflux resistance gene mef(I) and the chloramphenicol resistance gene catQ. Antimicrob Agents Chemother 51:3983–3987. doi: 10.1128/AAC.00790-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bley C, van der Linden M, Reinert RR. 2011. mef(A) is the predominant macrolide resistance determinant in Streptococcus pneumoniae and Streptococcus pyogenes in Germany. Int J Antimicrob Agents 37:425–431. doi: 10.1016/j.ijantimicag.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 181.Simoens S, Verhaegen J, van Bleyenbergh P, Peetermans WE, Decramer M. 2011. Consumption patterns and in vitro resistance of Streptococcus pneumoniae to fluoroquinolones. Antimicrob Agents Chemother 55:3051–3053. doi: 10.1128/AAC.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Domenech A, Tirado-Vélez JM, Fenoll A, Ardanuy C, Yuste J, Liñares J, de la Campa AG. 2014. Fluoroquinolone-resistant pneumococci: dynamics of serotypes and clones in Spain in 2012 compared with those from 2002 and 2006. Antimicrob Agents Chemother 58:2393–2399. doi: 10.1128/AAC.02669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ip M, Chau SS, Chi F, Cheuk ES, Ma H, Lai RW, Chan PK. 2007. Longitudinally tracking fluoroquinolone resistance and its determinants in penicillin-susceptible and -nonsusceptible Streptococcus pneumoniae isolates in Hong Kong, 2000 to 2005. Antimicrob Agents Chemother 51:2192–2194. doi: 10.1128/AAC.00139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang H, Chen M, Xu Y, Sun H, Yang Q, Hu Y, Cao B, Chu Y, Liu Y, Zhang R, Yu Y, Sun Z, Zhuo C, Ni Y, Hu B, Tan TY, Hsueh PR, Wang JH, Ko WC, Chen YH, Wahjono H. 2011. Antimicrobial susceptibility of bacterial pathogens associated with community-acquired respiratory tract infections in Asia: report from the Community-Acquired Respiratory Tract Infection Pathogen Surveillance (CARTIPS) study, 2009-2010. Int J Antimicrob Agents 38:376–383. doi: 10.1016/j.ijantimicag.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 185.Adam HJ, Hoban DJ, Gin AS, Zhanel GG. 2009. Association between fluoroquinolone usage and a dramatic rise in ciprofloxacin-resistant Streptococcus pneumoniae in Canada, 1997-2006. Int J Antimicrob Agents 34:82–85. doi: 10.1016/j.ijantimicag.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 186.Davidson R, Cavalcanti R, Brunton JL, Bast DJ, de Azavedo JC, Kibsey P, Fleming C, Low DE. 2002. Resistance to levofloxacin and failure of treatment of pneumococcal pneumonia. N Engl J Med 346:747–750. doi: 10.1056/NEJMoa012122. [DOI] [PubMed] [Google Scholar]
- 187.Anderson KB, Tan JS, File TM Jr, DiPersio JR, Willey BM, Low DE. 2003. Emergence of levofloxacin-resistant pneumococci in immunocompromised adults after therapy for community-acquired pneumonia. Clin Infect Dis 37:376–381. doi: 10.1086/376642. [DOI] [PubMed] [Google Scholar]
- 188.Ho PL, Yam WC, Cheung TK, Ng WW, Que TL, Tsang DN, Ng TK, Seto WH. 2001. Fluoroquinolone resistance among Streptococcus pneumoniae in Hong Kong linked to the Spanish 23F clone. Emerg Infect Dis 7:906–908. doi: 10.3201/eid0705.010526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pestova E, Millichap JJ, Siddiqui F, Noskin GA, Peterson LR. 2002. Non-PmrA-mediated multidrug resistance in Streptococcus pneumoniae. J Antimicrob Chemother 49:553–556. doi: 10.1093/jac/49.3.553. [DOI] [PubMed] [Google Scholar]
- 190.Smith HJ, Nichol KA, Hoban DJ, Zhanel GG. 2002. Dual activity of fluoroquinolones against Streptococcus pneumoniae: the facts behind the claims. J Antimicrob Chemother 49:893–895. doi: 10.1093/jac/dkf047. [DOI] [PubMed] [Google Scholar]
- 191.Lim S, Bast D, McGeer A, de Azavedo J, Low DE. 2003. Antimicrobial susceptibility breakpoints and first-step parC mutations in Streptococcus pneumoniae: redefining fluoroquinolone resistance. Emerg Infect Dis 9:833–837. doi: 10.3201/eid0907.020589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Janoir C, Zeller V, Kitzis MD, Moreau NJ, Gutmann L. 1996. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob Agents Chemother 40:2760–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pan XS, Fisher LM. 1996. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol 178:4060–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Brueggemann AB, Coffman SL, Rhomberg P, Huynh H, Almer L, Nilius A, Flamm R, Doern GV. 2002. Fluoroquinolone resistance in Streptococcus pneumoniae in United States since 1994-1995. Antimicrob Agents Chemother 46:680–688. doi: 10.1128/AAC.46.3.680-688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Weigel LM, Anderson GJ, Facklam RR, Tenover FC. 2001. Genetic analyses of mutations contributing to fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 45:3517–3523. doi: 10.1128/AAC.45.12.3517-3523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pletz MW, Fugit RV, McGee L, Glasheen JJ, Keller DL, Welte T, Klugman KP. 2006. Fluoroquinolone-resistant Streptococcus pneumoniae. Emerg Infect Dis 12:1462–1463. doi: 10.3201/eid1209.051400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Duesberg CB, Welte T, Pletz MW. 2007. The Lys137Asn mutation as surrogate marker for developing fluoroquinolone resistance in Streptococcus pneumoniae. J Chemother 19:750–751. doi: 10.1179/joc.2007.19.6.750. [DOI] [PubMed] [Google Scholar]
- 198.Zeller V, Janoir C, Kitzis MD, Gutmann L, Moreau NJ. 1997. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother 41:1973–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Baylay AJ, Ivens A, Piddock LJ. 2015. A novel gene amplification causes upregulation of the PatAB ABC transporter and fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 59:3098–3108. doi: 10.1128/AAC.04858-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Andersen CL, Holland IB, Jacq A. 2006. Verapamil, a Ca2+ channel inhibitor acts as a local anesthetic and induces the sigma E dependent extra-cytoplasmic stress response in E. coli. Biochim Biophys Acta 1758:1587–1595. doi: 10.1016/j.bbamem.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 201.Pletz MW, Michaylov N, Schumacher U, van der Linden M, Duesberg CB, Fuehner T, Klugman KP, Welte T, Makarewicz O. 2013. Antihypertensives suppress the emergence of fluoroquinolone-resistant mutants in pneumococci: an in vitro study. Int J Med Microbiol 303:176–181. doi: 10.1016/j.ijmm.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 202.Janoir C, Podglajen I, Kitzis MD, Poyart C, Gutmann L. 1999. In vitro exchange of fluoroquinolone resistance determinants between Streptococcus pneumoniae and viridans streptococci and genomic organization of the parE-parC region in S. mitis. J Infect Dis 180:555–558. doi: 10.1086/314888. [DOI] [PubMed] [Google Scholar]
- 203.Tankovic J, Perichon B, Duval J, Courvalin P. 1996. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother 40:2505–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Martín-Galiano AJ, Balsalobre L, Fenoll A, de la Campa AG. 2003. Genetic characterization of optochin-susceptible viridans group streptococci. Antimicrob Agents Chemother 47:3187–3194. doi: 10.1128/AAC.47.10.3187-3194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bast DJ, de Azavedo JC, Tam TY, Kilburn L, Duncan C, Mandell LA, Davidson RJ, Low DE. 2001. Interspecies recombination contributes minimally to fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 45:2631–2634. doi: 10.1128/AAC.45.9.2631-2634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pletz MW, McGee L, Beall B, Whitney CG, Klugman KP. 2005. Interspecies recombination in type II topoisomerase genes is not a major cause of fluoroquinolone resistance in invasive Streptococcus pneumoniae isolates in the United States. Antimicrob Agents Chemother 49:779–780. doi: 10.1128/AAC.49.2.779-780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Balsalobre L, Ferrándiz MJ, Liñares J, Tubau F, de la Campa AG. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob Agents Chemother 47:2072–2081. doi: 10.1128/AAC.47.7.2072-2081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pletz MW, McGee L, Jorgensen J, Beall B, Facklam RR, Whitney CG, Klugman KP. 2004. Levofloxacin-resistant invasive Streptococcus pneumoniae in the United States: evidence for clonal spread and the impact of conjugate pneumococcal vaccine. Antimicrob Agents Chemother 48:3491–3497. doi: 10.1128/AAC.48.9.3491-3497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Canton R, Morosini M, Enright MC, Morrissey I. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTEKT surveillance programme. J Antimicrob Chemother 52:944–952. doi: 10.1093/jac/dkg465. [DOI] [PubMed] [Google Scholar]
- 210.Hsueh PR, Teng LJ, Lee CM, Huang WK, Wu TL, Wan JH, Yang D, Shyr JM, Chuang YC, Yan JJ, Lu JJ, Wu JJ, Ko WC, Chang FY, Yang YC, Lau YJ, Liu YC, Leu HS, Liu CY, Luh KT, SMART Program 2001 Data. 2003. Telithromycin and quinupristine-dalfopristin resistance in clinical isolates of Streptococcus pyogenes: SMART Program 2001 data. Antimicrob Agents Chemother 47:2152–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Montanari MP, Tili E, Cochetti I, Mingoia M, Manzin A, Varaldo PE. 2004. Molecular characterization of clinical Streptococcus pneumoniae isolates with reduced susceptibility to fluoroquinolones emerging in Italy. Microb Drug Resist 10:209–217. doi: 10.1089/mdr.2004.10.209. [DOI] [PubMed] [Google Scholar]
- 212.Wyres KL, van Tonder A, Lambertsen LM, Hakenbeck R, Parkhill J, Bentley SD, Brueggemann AB. 2013. Evidence of antimicrobial resistance-conferring genetic elements among pneumococci isolated prior to 1974. BMC Genomics 14:500. doi: 10.1186/1471-2164-14-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Burdett V, Inamine J, Rajagopalan S. 1982. Heterogeneity of tetracycline resistance determinants in Streptococcus. J Bacteriol 149:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Widdowson CA, Klugman KP, Hanslo D. 1996. Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob Agents Chemother 40:2891–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Oggioni MR, Dowson CG, Smith JM, Provvedi R, Pozzi G. 1996. The tetracycline resistance gene tet(M) exhibits mosaic structure. Plasmid 35:156–163. doi: 10.1006/plas.1996.0018. [DOI] [PubMed] [Google Scholar]
- 216.Doherty N, Trzcinski K, Pickerill P, Zawadzki P, Dowson CG. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob Agents Chemother 44:2979–2984. doi: 10.1128/AAC.44.11.2979-2984.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dzierzanowska-Fangrat K, Semczuk K, Górska P, Giedrys-Kalemba S, Kochman M, Samet A, Tyski S, Dzierzanowska D, Trzciński K. 2006. Evidence for tetracycline resistance determinant tet(M) allele replacement in a Streptococcus pneumoniae population of limited geographical origin. Int J Antimicrob Agents 27:159–164. doi: 10.1016/j.ijantimicag.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 218.Doern GV, Brueggemann A, Holley HP Jr, Rauch AM. 1996. Antimicrobial resistance of Streptococcus pneumoniae recovered from outpatients in the United States during the winter months of 1994 to 1995: results of a 30-center national surveillance study. Antimicrob Agents Chemother 40:1208–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Marchese A, Mannelli S, Tonoli E, Gorlero F, Toni M, Schito GC. 2001. Prevalence of antimicrobial resistance in Streptococcus pneumoniae circulating in Italy: results of the Italian Epidemiological Observatory Survey (1997-1999). Microb Drug Resist 7:277–287. doi: 10.1089/10766290152652837. [DOI] [PubMed] [Google Scholar]
- 220.Ferrándiz MJ, Ardanuy C, Liñares J, Balsalobre L, García MT, de la Campa AG. 2011. New species genetic approach to identify strains of mitis group streptococci that are donors of rifampin resistance to Streptococcus pneumoniae. Antimicrob Agents Chemother 55:368–372. doi: 10.1128/AAC.00856-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ayoubi P, Kilic AO, Vijayakumar MN. 1991. Tn5253, the pneumococcal omega (cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J Bacteriol 173:1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Widdowson CA, Adrian PV, Klugman KP. 2000. Acquisition of chloramphenicol resistance by the linearization and integration of the entire staphylococcal plasmid pC194 into the chromosome of Streptococcus pneumoniae. Antimicrob Agents Chemother 44:393–395. doi: 10.1128/AAC.44.2.393-395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jones ME, Blosser-Middleton RS, Critchley IA, Karlowsky JA, Thornsberry C, Sahm DF. 2003. In vitro susceptibility of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis: a European multicenter study during 2000-2001. Clin Microbiol Infect 9:590–599. doi: 10.1046/j.1469-0691.2003.00573.x. [DOI] [PubMed] [Google Scholar]
- 224.Johnson DM, Stilwell MG, Fritsche TR, Jones RN. 2006. Emergence of multidrug-resistant Streptococcus pneumoniae: report from the SENTRY Antimicrobial Surveillance Program (1999-2003). Diagn Microbiol Infect Dis 56:69–74. doi: 10.1016/j.diagmicrobio.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 225.Adrian PV, Klugman KP. 1997. Mutations in the dihydrofolate reductase gene of trimethoprim-resistant isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 41:2406–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Maskell JP, Sefton AM, Hall LM. 2001. Multiple mutations modulate the function of dihydrofolate reductase in trimethoprim-resistant Streptococcus pneumoniae. Antimicrob Agents Chemother 45:1104–1108. doi: 10.1128/AAC.45.4.1104-1108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lopez P, Espinosa M, Greenberg B, Lacks SA. 1987. Sulfonamide resistance in Streptococcus pneumoniae: DNA sequence of the gene encoding dihydropteroate synthase and characterization of the enzyme. J Bacteriol 169:4320–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Maskell JP, Sefton AM, Hall LM. 1997. Mechanism of sulfonamide resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 41:2121–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Padayachee T, Klugman KP. 1999. Novel expansions of the gene encoding dihydropteroate synthase in trimethoprim-sulfamethoxazole-resistant Streptococcus pneumoniae. Antimicrob Agents Chemother 43:2225–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Feikin DR, Dowell SF, Nwanyanwu OC, Klugman KP, Kazembe PN, Barat LM, Graf C, Bloland PB, Ziba C, Huebner RE, Schwartz B. 2000. Increased carriage of trimethoprim/sulfamethoxazole-resistant Streptococcus pneumoniae in Malawian children after treatment for malaria with sulfadoxine/pyrimethamine. J Infect Dis 181:1501–1505. doi: 10.1086/315382. [DOI] [PubMed] [Google Scholar]
- 231.Schmitz FJ, Perdikouli M, Beeck A, Verhoef J, Fluit AC, European SENTRY Participants . 2001. Resistance to trimethoprim-sulfamethoxazole and modifications in genes coding for dihydrofolate reductase and dihydropteroate synthase in European Streptococcus pneumoniae isolates. J Antimicrob Chemother 48:935–936. doi: 10.1093/jac/48.6.935. [DOI] [PubMed] [Google Scholar]
- 232.Lonks JR, Goldman DA. 2005. Telithromycin: a ketolide antibiotic for treatment of respiratory tract infections. Clin Infect Dis 40:1657–1664. doi: 10.1086/430067. [DOI] [PubMed] [Google Scholar]
- 233.McGhee P, Clark C, Kosowska-Shick KM, Nagai K, Dewasse B, Beachel L, Appelbaum PC. 2010. In vitro activity of CEM-101 against Streptococcus pneumoniae and Streptococcus pyogenes with defined macrolide resistance mechanisms. Antimicrob Agents Chemother 54:230–238. doi: 10.1128/AAC.01123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wilson DN. 2011. On the specificity of antibiotics targeting the large ribosomal subunit. Ann N Y Acad Sci 1241:1–16. doi: 10.1111/j.1749-6632.2011.06192.x. [DOI] [PubMed] [Google Scholar]
- 235.Patel SN, Pillai DR, Pong-Porter S, McGeer A, Green K, Low DE. 2009. In vitro activity of ceftaroline, ceftobiprole and cethromycin against clinical isolates of Streptococcus pneumoniae collected from across Canada between 2003 and 2008. J Antimicrob Chemother 64:659–660. doi: 10.1093/jac/dkp231. [DOI] [PubMed] [Google Scholar]
- 236.Leclercq R, Courvalin P. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob Agents Chemother 46:2727–2734. doi: 10.1128/AAC.46.9.2727-2734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Walsh F, Willcock J, Amyes S. 2003. High-level telithromycin resistance in laboratory-generated mutants of Streptococcus pneumoniae. J Antimicrob Chemother 52:345–353. doi: 10.1093/jac/dkg348. [DOI] [PubMed] [Google Scholar]
- 238.Farrell DJ, Morrissey I, Bakker S, Buckridge S, Felmingham D. 2004. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob Agents Chemother 48:3169–3171. doi: 10.1128/AAC.48.8.3169-3171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Reinert RR, van der Linden M, Al-Lahham A. 2005. Molecular characterization of the first telithromycin-resistant Streptococcus pneumoniae isolate in Germany. Antimicrob Agents Chemother 49:3520–3522. doi: 10.1128/AAC.49.8.3520-3522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Tait-Kamradt A, Davies T, Appelbaum PC, Depardieu F, Courvalin P, Petitpas J, Wondrack L, Walker A, Jacobs MR, Sutcliffe J. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob Agents Chemother 44:3395–3401. doi: 10.1128/AAC.44.12.3395-3401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Faccone D, Andres P, Galas M, Tokumoto M, Rosato A, Corso A. 2005. Emergence of a Streptococcus pneumoniae clinical isolate highly resistant to telithromycin and fluoroquinolones. J Clin Microbiol 43:5800–5803. doi: 10.1128/JCM.43.11.5800-5803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Pérez-Trallero E, Marimon JM, Iglesias L, Larruskain J. 2003. Fluoroquinolone and macrolide treatment failure in pneumococcal pneumonia and selection of multidrug-resistant isolates. Emerg Infect Dis 9:1159–1162. doi: 10.3201/eid0909.020810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wolter N, Smith AM, Low DE, Klugman KP. 2007. High-level telithromycin resistance in a clinical isolate of Streptococcus pneumoniae. Antimicrob Agents Chemother 51:1092–1095. doi: 10.1128/AAC.01153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Draghi DC, Sheehan DJ, Hogan P, Sahm DF. 2005. In vitro activity of linezolid against key Gram-positive organisms isolated in the United States: results of the LEADER 2004 surveillance program. Antimicrob Agents Chemother 49:5024–5032. doi: 10.1128/AAC.49.12.5024-5032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Flamm RK, Mendes RE, Ross JE, Sader HS, Jones RN. 2013. An international activity and spectrum analysis of linezolid: ZAAPS Program results for 2011. Diagn Microbiol Infect Dis 76:206–213. doi: 10.1016/j.diagmicrobio.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 246.Flamm RK, Mendes RE, Ross JE, Sader HS, Jones RN. 2013. Linezolid surveillance results for the United States: LEADER surveillance program 2011. Antimicrob Agents Chemother 57:1077–1081. doi: 10.1128/AAC.02112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Meka VG, Gold HS. 2004. Antimicrobial resistance to linezolid. Clin Infect Dis 39:1010–1015. doi: 10.1086/423841. [DOI] [PubMed] [Google Scholar]
- 248.Wolter N, Smith AM, Farrell DJ, Schaffner W, Moore M, Whitney CG, Jorgensen JH, Klugman KP. 2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob Agents Chemother 49:3554–3557. doi: 10.1128/AAC.49.8.3554-3557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Dong W, Chochua S, McGee L, Jackson D, Klugman KP, Vidal JE. 2014. Mutations within the rplD gene of linezolid-nonsusceptible Streptococcus pneumoniae strains isolated in the United States. Antimicrob Agents Chemother 58:2459–2462. doi: 10.1128/AAC.02630-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Feng J, Lupien A, Gingras H, Wasserscheid J, Dewar K, Légaré D, Ouellette M. 2009. Genome sequencing of linezolid-resistant Streptococcus pneumoniae mutants reveals novel mechanisms of resistance. Genome Res 19:1214–1223. doi: 10.1101/gr.089342.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Feng J, Billal DS, Lupien A, Racine G, Winstall E, Légaré D, Leprohon P, Ouellette M. 2011. Proteomic and transcriptomic analysis of linezolid resistance in Streptococcus pneumoniae. J Proteome Res 10:4439–4452. doi: 10.1021/pr200221s. [DOI] [PubMed] [Google Scholar]
- 252.Kisgen JJ, Mansour H, Unger NR, Childs LM. 2014. Tedizolid: a new oxazolidinone antimicrobial. Am J Health Syst Pharm 71:621–633. doi: 10.2146/ajhp130482. [DOI] [PubMed] [Google Scholar]
- 253.Jones RN, Farrell DJ, Morrissey I. 2003. Quinupristin-dalfopristin resistance in Streptococcus pneumoniae: novel L22 ribosomal protein mutation in two clinical isolates from the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother 47:2696–2698. doi: 10.1128/AAC.47.8.2696-2698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ford KL, Mason EO Jr, Kaplan SL, Lamberth LB, Tillman J. 1991. Factors associated with middle ear isolates of Streptococcus pneumoniae resistant to penicillin in a children's hospital. J Pediatr 119:941–944. doi: 10.1016/S0022-3476(05)83050-1. [DOI] [PubMed] [Google Scholar]
- 255.Tan QT, Mason EO Jr, Kaplan SL. 1993. Penicillin-resistant systemic pneumococcal infections in children: a retrospective case-control study. Pediatrics 92:761–767. [PubMed] [Google Scholar]
- 256.Guillemot D, Carbon C, Balkau B, Geslin P, Lecoeur H, Vauzelle-Kervroëdan F, Bouvenot G, Eschwége E. 1998. Low dosage and long treatment duration of β-lactam: risk factors for carriage of penicillin-resistant Streptococcus pneumoniae. JAMA 279:365–370. doi: 10.1001/jama.279.5.365. [DOI] [PubMed] [Google Scholar]
- 257.Reichler MR, Allphin AA, Breiman RF, Schreiber JR, Arnold JE, McDougal LK, Facklam RR, Boxerbaum B, May D, Walton RO. 1992. The spread of multiply resistant Streptococcus pneumoniae at a day care center in Ohio. J Infect Dis 166:1346–1353. doi: 10.1093/infdis/166.6.1346. [DOI] [PubMed] [Google Scholar]
- 258.Hsieh YC, Wang JT, Lee WS, Hsueh PR, Shao PL, Chang LY, Lu CY, Lee CY, Huang FY, Huang LM. 2006. Serotype competence and penicillin resistance in Streptococcus pneumoniae. Emerg Infect Dis 12:1709–1714. doi: 10.3201/eid1211.060414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hanage WP, Fraser C, Tang J, Connor TR, Corander J. 2009. Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 324:1454–1457. doi: 10.1126/science.1171908. [DOI] [PubMed] [Google Scholar]
- 261.Laurenceau R, Péhau-Arnaudet G, Baconnais S, Gault J, Malosse C, Dujeancourt A, Campo N, Chamot-Rooke J, Le Cam E, Claverys JP, Fronzes R. 2013. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog 9:e1003473. doi: 10.1371/journal.ppat.1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 263.Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, Spinali S, Perkins T, Lechat P, Dmytruk N, Sauvage E, Ma L, Romi B, Tichit M, Lopez-Sanchez MJ, Descorps-Declere S, Souche E, Buchrieser C, Trieu-Cuot P, Moszer I, Clermont D, Maione D, Bouchier C, McMillan DJ, Parkhill J, Telford JL, Dougan G, Walker MJ, DEVANI Consortium, Holden MT, Poyart C, Glaser P. 2014. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun 5:4544. doi: 10.1038/ncomms5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Mills RO, Twum-Danso K, Owusu-Agyei S, Donkor ES. 2015. Epidemiology of pneumococcal carriage in children under five years of age in Accra, Ghana. Infect Dis (Lond) 47:326–331. doi: 10.3109/00365548.2014.994185. [DOI] [PubMed] [Google Scholar]
- 265.Turner P, Turner C, Jankhot A, Helen N, Lee SJ, Day NP, White NJ, Nosten F, Goldblatt DA. 2012. Longitudinal study of Streptococcus pneumoniae carriage in a cohort of infants and their mothers on the Thailand-Myanmar border. PLoS One 7:e38271. doi: 10.1371/journal.pone.0038271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Dhoubhadel BG, Yasunami M, Nguyen HA, Suzuki M, Vu TH, Thi Thuy Nguyen A, Dang DA, Yoshida LM, Ariyoshi K. 2014. Bacterial load of pneumococcal serotypes correlates with their prevalence and multiple serotypes is associated with acute respiratory infections among children less than 5 years of age. PLoS One 9:e110777. doi: 10.1371/journal.pone.0110777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Otsuka T, Chang B, Shirai T, Iwaya A, Wada A, Yamanaka N, Okazaki M, SADO-study Working Group . 2013. Individual risk factors associated with nasopharyngeal colonization with Streptococcus pneumoniae and Haemophilus influenzae: a Japanese birth cohort study. Pediatr Infect Dis J 32:709–714. doi: 10.1097/INF.0b013e31828701ea. [DOI] [PubMed] [Google Scholar]
- 268.McCormick AW, Whitney CG, Farley MM, Lynfield R, Harrison LH, Bennett NM, Schaffner W, Reingold A, Hadler J, Cieslak P, Samore MH, Lipsitch M. 2003. Geographic diversity and temporal trends of antimicrobial resistance in Streptococcus pneumoniae in the United States. Nat Med 9:424–430. doi: 10.1038/nm839. [DOI] [PubMed] [Google Scholar]
- 269.Bjorkman J, Andersson DI. 2000. The cost of antibiotic resistance from a bacterial perspective. Drug Resist Updat 3:237–245. doi: 10.1054/drup.2000.0147. [DOI] [PubMed] [Google Scholar]
- 270.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 271.Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493. doi: 10.1016/S1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 272.Rozen DE, McGee L, Levin BR, Klugman KP. 2007. Fitness costs of fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 51:412–416. doi: 10.1128/AAC.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Balsalobre L, de la Campa AG. 2008. Fitness of Streptococcus pneumoniae fluoroquinolone-resistant strains with topoisomerase IV recombinant genes. Antimicrob Agents Chemother 52:822–830. doi: 10.1128/AAC.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Rieux V, Carbon C, Azoulay-Dupuis E. 2001. Complex relationship between acquisition of β-lactam resistance and loss of virulence in Streptococcus pneumoniae. J Infect Dis 184:66–72. doi: 10.1086/320992. [DOI] [PubMed] [Google Scholar]
- 275.Trzcinski K, Thompson CM, Gilbey AM, Dowson CG, Lipsitch M. 2006. Incremental increase in fitness cost with increased β-lactam resistance in pneumococci evaluated by competition in an infant rat nasal colonization model. J Infect Dis 193:1296–1303. doi: 10.1086/501367. [DOI] [PubMed] [Google Scholar]
- 276.Albarracín Orio AG, Piñas GE, Cortes PR, Cian MB, Echenique J. 2011. Compensatory evolution of pbp mutations restores the fitness cost imposed by β-lactam resistance in Streptococcus pneumoniae. PLoS Pathog 7:e1002000. doi: 10.1371/journal.ppat.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ortega M, Marco F, Soriano A, García E, Martínez JA, Mensa J. 2003. Lack of vancomycin tolerance in Streptococcus pneumoniae strains isolated in Barcelona, Spain, from 1999 to 2001. Antimicrob Agents Chemother 47:1976–1978. doi: 10.1128/AAC.47.6.1976-1978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Liu HH, Tomasz A. 1985. Penicillin tolerance in multiply drug-resistant natural isolates of Streptococcus pneumoniae. J Infect Dis 152:365–372. doi: 10.1093/infdis/152.2.365. [DOI] [PubMed] [Google Scholar]
- 279.McCullers JA, English BK, Novak R. 2000. Isolation and characterization of vancomycin-tolerant Streptococcus pneumoniae from the cerebrospinal fluid of a patient who developed recrudescent meningitis. J Infect Dis 181:369–373. doi: 10.1086/315216. [DOI] [PubMed] [Google Scholar]
- 280.Mitchell L, Tuomanen E. 2001. Vancomycin-tolerant Streptococcus pneumoniae and its clinical significance. Pediatr Infect Dis J 20:531–533. doi: 10.1097/00006454-200105000-00012. [DOI] [PubMed] [Google Scholar]
- 281.Olivares A, Trejo JO, Arellano-Galindo J, Zuñiga G, Escalona G, Vigueras JC, Marín P, Xicohtencatl J, Valencia P, Velázquez-Guadarrama N. 2011. pep27 and lytA in vancomycin-tolerant pneumococci. J Microbiol Biotechnol 21:1345–1351. doi: 10.4014/jmb.1105.05045. [DOI] [PubMed] [Google Scholar]
- 282.Mascher T, Zahner D, Merai M, Balmelle N, De Saizieu AB, Hakenbeck R. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J Bacteriol 185:60–70. doi: 10.1128/JB.185.1.60-70.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Bourgeois I, Pestel-Caron M, Lemeland JF, Pons JL, Caron F. 2007. Tolerance to the glycopeptides vancomycin and teicoplanin in coagulase-negative staphylococci. Antimicrob Agents Chemother 51:740–743. doi: 10.1128/AAC.00719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. 1999. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature 399:590–593. doi: 10.1038/21202. [DOI] [PubMed] [Google Scholar]
- 285.Sung H, Shin HB, Kim M-H, Lee K, Kim E-C, Song W, Jeong SH, Lee W-G, Park Y-J, Eliopoulos GM. 2006. Vancomycin-tolerant Streptococcus pneumoniae in Korea. J Clin Microbiol 44:3524–3528. doi: 10.1128/JCM.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Novak R, Charpentier E, Braun JS, Tuomanen E. 2000. Signal transduction by a death signal peptide: uncovering the mechanism of bacterial killing by penicillin. Mol Cell 5:49–57. doi: 10.1016/S1097-2765(00)80402-5. [DOI] [PubMed] [Google Scholar]
- 287.Chen Y, Deng W, Wang SM, Mo QM, Jia H, Wang Q, Li SG, Li X, Yao BD, Liu CJ, Zhan YQ, Ji C, Lopez AL, Wang XY. 2011. Burden of pneumonia and meningitis caused by Streptococcus pneumoniae in China among children under 5 years of age: a systematic literature review. PLoS One 6:e27333. doi: 10.1371/journal.pone.0027333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Shi ZY, Enright MC, Wilkinson P, Griffiths D, Spratt BG. 1998. Identification of the three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals by multilocus sequencing typing. J Clin Microbiol 36:3514–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Hall LM, Whiley RA, Duke B, George RC, Efstratiou A. 1996. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol 34:853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Park IH, Moore MR, Treanor JJ, Pelton SI, Pilishvili T, Beall BW, Shelly MA, Mahon BE, Nahm MH. 2008. Differential effects of pneumococcal vaccines against serotypes 6A and 6C. J Infect Dis 198:1818–1822. doi: 10.1086/593339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Carvalho MDG, Pimenta FC, Gertz RE Jr, Joshi HH, Trujillo AA, Keys LE, Findley J, Moura IS, Park IH, Hollingshead SK, Pilishvili T, Whitney CG, Nahm MH, Beall BW. 2009. PCR-based quantitation and clonal associations of the current prevalent invasive serogroup 6 pneumococcal serotype, 6C, in the United States: 1999, 2006-2007. J Clin Microbiol 47:554–559. doi: 10.1128/JCM.01919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Moore MR, Link-Gelles R, Schaffner W, Lynfield R, Lexau C, Bennett NM, Petit S, Zansky SM, Harrison LH, Reingold A, Miller L, Scherzinger K, Thomas A, Farley MM, Zell ER, Taylor TH Jr, Pondo T, Rodgers L, McGee L, Beall B, Jorgensen JH, Whitney CG. 2015. Effect of use of 13-valent pneumococcal conjugate vaccine in children on invasive pneumococcal disease in children and adults in the USA: analysis of multisite, population-based surveillance. Lancet Infect Dis 15:301–309. doi: 10.1016/S1473-3099(14)71081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Pai R, Gertz RE, Whitney CG, Beall B. 2005. Clonal association between Streptococcus pneumoniae serotype 23A within the United States and an internationally dispersed clone of 23F. J Clin Microbiol 43:5440–5444. doi: 10.1128/JCM.43.11.5440-5444.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Gertz RE Jr, Li Z, Pimenta FC, Jackson D, Juni BA, Lynfield R, Jorgensen JH, Carvalho MDG, Beall BW. 2010. Increased penicillin nonsusceptibility of nonvaccine-serotype invasive pneumococci other than serotypes 19A and 6A in post-7-valent conjugate vaccine era. J Infect Dis 201:770–775. doi: 10.1086/650496. [DOI] [PubMed] [Google Scholar]
- 295.Mendes RE, Biek D, Critchley IA, Farrell DJ, Sader HS, Jones RN. 2014. Decreased ceftriaxone susceptibility in emerging (35B and 6C) and persisting (19A) Streptococcus pneumoniae serotypes in the United States, 2011-2012: ceftaroline remains active in vitro among β-lactam agents. Antimicrob Agents Chemother 58:4923–4927. doi: 10.1128/AAC.02976-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Sharma D, Baughman W, Holst A, Thomas S, Jackson D, da Carvalho MDC, Beall B, Satola S, Jerris R, Jain S, Farley MM, Nuorti JP. 2013. Pneumococcal carriage and invasive disease in children before introduction of the 13-valent conjugate vaccine: comparison with the era before 7-valent conjugate vaccine. Pediatr Infect Dis J 32:e45–e53. doi: 10.1097/INF.0b013e3182788fdd. [DOI] [PubMed] [Google Scholar]
- 297.Desai AP, Sharma D, Crispell EK, Baughman W, Thomas S, Tunali A, Sherwood L, Zmitrovich A, Jerris R, Satola SW, Beall B, Moore MR, Jain S, Farley MM. 2015. Decline in pneumococcal nasopharyngeal carriage of vaccine serotypes after the introduction of the 13-valent conjugate vaccine in children in Atlanta, Georgia. Pediatr Infect Dis J 34:1168–1174. doi: 10.1097/INF.0000000000000849. [DOI] [PubMed] [Google Scholar]
- 298.Hampton LM, Farley MM, Schaffner W, Thomas A, Reingold A, Harrison LH, Lynfield R, Bennett NM, Petit S, Gershman K, Baumbach J, Beall B, Jorgensen J, Glennen A, Zell ER, Moore M. 2012. Prevention of antibiotic-nonsusceptible Streptococcus pneumoniae with conjugate vaccines. J Infect Dis 205:401–411. doi: 10.1093/infdis/jir755. [DOI] [PubMed] [Google Scholar]
- 299.Moore MR. 2009. Rethinking replacement and resistance. J Infect Dis 199:771–773. doi: 10.1086/597045. [DOI] [PubMed] [Google Scholar]
- 300.Hanage WP, Huang SS, Lipsitch M, Bishop CJ, Godoy D, Pelton SI, Goldstein R, Huot H, Finkelstein JA. 2007. Diversity and antibiotic resistance among nonvaccine serotypes of Streptococcus pneumoniae carriage isolates in the post-heptavalent conjugate vaccine era. J Infect Dis 195:347–352. doi: 10.1086/510249. [DOI] [PubMed] [Google Scholar]
- 301.Dagan R, Barkai G, Givon-Lavi N, Sharf AZ, Vardy D, Cohen T, Lipsitch M, Greenberg D. 2008. Seasonality of antibiotic-resistant Streptococcus pneumoniae that causes acute otitis media: a clue for an antibiotic-restriction policy? J Infect Dis 197:1094–1102. doi: 10.1086/528995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Van Effelterre T, Moore MR, Fierens F, Whitney CG, White L, Pelton SI, Hausdorff WP. 2010. A dynamic model of pneumococcal infection in the United States: implications for prevention through vaccination. Vaccine 28:3650–3660. doi: 10.1016/j.vaccine.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 303.Klugman KP, Madhi SA, Huebner RE, Kohberger R, Mbelle N, Pierce N, Vaccine Trialists Group . 2003. A trial of a 9-valent pneumococcal conjugate vaccine in children with and those without HIV infection. N Engl J Med 349:1341–1348. doi: 10.1056/NEJMoa035060. [DOI] [PubMed] [Google Scholar]
- 304.dos Santos SR, Passadore LF, Takagi EH, Fujii CM, Yoshioka CR, Gilio AE, Martinez MB. 2013. Serotype distribution of Streptococcus pneumoniae isolated from patients with invasive pneumococcal disease in Brazil before and after ten-pneumococcal conjugate vaccine implementation. Vaccine 31:6150–6154. doi: 10.1016/j.vaccine.2013.05.042. [DOI] [PubMed] [Google Scholar]
- 305.Parra EL, De La Hoz F, Díaz PL, Sanabria O, Realpe ME, Moreno J. 2013. Changes in Streptococcus pneumoniae serotype distribution in invasive disease and nasopharyngeal carriage after the heptavalent pneumococcal conjugate vaccine introduction in Bogota, Colombia. Vaccine 31:4033–4038. doi: 10.1016/j.vaccine.2013.04.074. [DOI] [PubMed] [Google Scholar]
- 306.Regev-Yochay G, Rahav G, Riesenberg K, Wiener-Well Y, Strahilevitz J, Stein M, Glikman D, Weber G, Potasman I, Dagan R, IAIPD Study Group . 2014. Initial effects of the National PCV7 Childhood Immunization Program on adult invasive pneumococcal disease in Israel. PLoS One 9:e88406. doi: 10.1371/journal.pone.0088406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Sham LT, Tsui HC, Land AD, Barendt SM, Winkler ME. 2012. Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Curr Opin Microbiol 15:194–203. doi: 10.1016/j.mib.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]