Summary
Fat and muscle lipolysis involves functional interactions of adipose triglyceride lipase (ATGL), α-β hydrolase domain-containing protein 5 (ABHD5), and tissue-specific perilipins 1 and 5 (PLIN1 and PLIN5). ABHD5 potently activates ATGL, but this lipase-promoting activity is suppressed when ABHD5 is bound to PLIN proteins on lipid droplets. In adipocytes, protein kinase A (PKA) phosphorylation of PLIN1 rapidly releases ABHD5 to activate ATGL, but mechanisms for rapid regulation of PLIN5-ABHD5 interaction in muscle are unknown. Here we identify synthetic ligands that release ABHD5 from PLIN1 or PLIN5 absent PKA activation and rapidly activate adipocyte and muscle lipolysis. Molecular imaging and affinity probe labeling demonstrated ABHD5 is directly targeted by these synthetic ligands and additionally revealed that ABHD5-PLIN interactions are regulated by endogenous ligands including long-chain acyl-CoA. Our results reveal a new locus of lipolysis control and suggest ABHD5 ligands might be developed into novel therapeutics that directly promote fat catabolism.
Graphical Abstract
eTOC summary
Lipolysis involves interactions between ATGL, ABHD5, and PLIN1 or PLIN5. Sanders et al. identify synthetic ligands that quickly release ABHD5 from PLIN proteins to directly activate fat and muscle lipolysis, bypassing canonical PKA-dependent signaling. Long-chain acyl-CoAs act as endogenous allosteric ligand of ABHD5 to suppress lipolysis.
Introduction
The storage and mobilization of lipid energy are fundamental processes of virtually all eukaryotic cells. Mobilization of fatty acids from stored triacylglycerol occurs on the surface of intracellular lipid droplets (LD), and is thought to be controlled by the functional interactions of three highly conserved proteins: adipose triglyceride lipase (ATGL, officially PNPLA2), α-β hydrolase domain-containing protein 5 (ABHD5, also known as CGI-58), and perilipin-1 (PLIN1) or perilipin-5 (PLIN5) (Young and Zechner, 2013). ATGL is responsible for the first and rate limiting step of triacylglycerol hydrolysis in adipocytes, skeletal muscle, and heart. ABHD5 is a critical upstream regulator of ATGL that potently increases the lipase activity of ATGL and possibly other lipases (Lass et al., 2006). Importantly, human mutations of ATGL and ABHD5 lead to an overlapping spectrum of diseases involving ectopic neutral lipid accumulation in several tissues (Caux et al., 2004; Fischer et al., 2007; Reilich et al., 2011; Schweiger et al., 2009).
Much of our understanding of acute lipolytic control derives from experiments in adipocytes, wherein lipolysis is manipulated by signals that converge on the cyclic AMP/protein kinase A (PKA) signaling pathway. In adipocytes, PLIN1 is a lipid droplet scaffold that is thought to be the major locus of lipolysis control. Under basal conditions, PLIN1 suppresses lipolysis by binding ABHD5, thereby preventing its activation of ATGL (Gandotra et al., 2011; Granneman et al., 2009a; Subramanian et al., 2004). Phosphorylation of PLIN1 by protein kinase A (PKA) simultaneously releases ABHD5, which activates ATGL and recruits phosphorylated hormone sensitive lipase (HSL) to the lipid droplet surface (Granneman et al., 2009a; Miyoshi et al., 2006). Presently, PKA-mediated signaling is the only known means of rapidly (seconds to minutes) activating adipocyte lipolysis. ABHD5 has also been implicated in intracellular lipolysis in tissues, such as skeletal muscle and heart, which use fatty acids internally as metabolic fuels. In these cells PLIN5 binds ABHD5 and suppresses basal and stimulated lipolysis (Pollak et al., 2015; Wolins et al., 2006); however, the regulation of ABHD5 by PLIN5 is poorly understood and might involve mechanisms that are independent of PKA (Granneman et al., 2009b).
To develop novel probes of intracellular lipolysis, we devised an assay that measured the interaction of ABHD5 with PLIN1 or PLIN5, and screened for compounds that would disrupt those interactions in the absence of PKA activation. We identified compounds that potently disrupted the interaction of ABHD5 with both PLIN1 and PLIN5, and stimulated lipolysis in fat cells and muscle. The synthetic compounds were found to be direct ligands of ABHD5 that rapidly regulated its interactions with PLIN1 and PLIN5. Use of these ligands in molecular imaging assays uncovered a pathway involving the dynamic regulation of ABHD5-PLIN interactions by endogenous long chain acyl-CoA. These results demonstrate that ABHD5 is subject to rapid direct allosteric regulation by endogenous and synthetic ligands, and suggest that this property might be exploited for therapeutic benefit.
Results
High throughput screening identifies compounds that disrupt the interaction of ABHD5 with PLIN1 and PLIN5
To develop novel probes of intracellular lipolysis, we devised an assay for compounds that would inhibit the interaction of ABHD5 with PLIN1 or PLIN5 in the absence of PKA activation. The assay was based upon luciferase complementation between ABHD5 and PLIN1 or PLIN5 (Granneman et al., 2009a; Granneman et al., 2009b; Remy and Michnick, 2006) (PubChem AID 493421 and AID 493057, Supplemental Information Tables S1 and S2 and Supplemental Methods) and was performed under conditions (no ATP or Mg++) in which PKA is inactive. Screening of >364,000 compounds from the NIH chemical library identified 5 compounds, representing 3 independent chemical scaffolds, that inhibited the interaction of ABHD5 with PLIN1 and PLIN5.
Shown in Figure 1A are representative members of two scaffolds, thiaza-tricyclo-ureas (TTU) represented by SR-4995 (CID 16016685), and sulfonyl piperazines (SPZ) represented by SR-4559 (CID 2674365). SR-4559 and SR-4995 prevented binding of ABHD5 to PLIN1 in luciferase complementation assays with IC50s of 510 +/− 60 nM and 200 +/− 30 nM (n=8), respectively (Figure 1B). As expected, compounds were fully active in preventing the interaction of ABHD5 with mutant PLIN1 lacking all 6 PKA sites (Figure S1A, S1B). To independently verify that compounds directly disrupt the interaction of ABHD5 with PLIN1 and confirm compound efficacy using human proteins, we identified a minimal fragment of human PLIN1 (amino acids 358–415) that binds ABHD5 in Cos7 cells. This fragment lacks PKA phosphorylation sites and localizes to lipid droplets by virtue of its interaction with ABHD5. In the resting state, the PLIN1 fragment and ABHD5 were colocalized on LD surfaces, and SR-4559 (1 µM final) triggered the release of the PLIN1 fragment within 1 minute of extracellular application (Figure 1C, Supplemental Information, video 1).
Figure 1.
Identification of compounds that disrupt the interaction of PLIN1 with ABHD5. A: Structure of the tricyclic thio urea (TTU) SR-4995 and the sulfonyl piperazine (SPZ) SR-4559. B: SR-4995 and SR-4559 blocked the interaction of PLIN1 with ABHD5 in the luciferase complementation assay (Binding). Values are means +/− s.e.m. of triplicate determinations and the experiment was replicated > 5 times. C: SR-4995 dissociates PLIN1(358–415)-EYFP from lipid droplet-targeted ECFP-ABHD5 in Cos7 cells within 2 minutes. Imaging results were replicated 9 times.
PLIN5 can be phosphorylated by PKA (Pollak et al., 2015); however, treatment of 3T3-L1 preadipocytes with forskolin/IBMX did not alter the ABHD5-PLIN5 interaction, as determined by Fluorescence Resonance Energy Transfer (FRET), in contrast to the disruption of the ABHD5-PLIN1 interaction (Figure S1C). Importantly, in brown adipocytes SR-4995 rapidly dissociated complexes of ABHD5 and PLIN5 (Figure S1D), as well as complexes of ABHD5 and a PLIN5 fragment (amino acids 360–417) that lacks PKA sites (Figure S1E).
Compounds rapidly stimulate lipolysis in adipocytes and muscle without activating phosphorylation of PLIN1 or HSL
We next tested the effects of compounds on lipolysis in cultured brown adipocytes. We found that SR-4995 and SR-4559 stimulated lipolysis with EC50s of about 4 µM, and produced maximal lipolytic rates that were > 75% of those produced by isoproterenol, a full beta adrenergic receptor agonist (Figure 2A). We also examined the temporal relation between compound application and intracellular fatty acid mobilization in live cells by fluorescence microscopy. In this assay, mobilized fatty acids trigger the translocation of EYFP-tagged SRC1 to the ligand binding domain of PPARα, which has been targeted to the lipid droplet surface by fusion to PLIN1 (Figure 2B) (Mottillo et al., 2012; Mottillo et al., 2014b). We found that compounds triggered lipolysis within 3 minutes, on a timeframe similar to the dissociation of PLIN1-ABHD5 complexes (Supplementary Information, video 2).
Figure 2.
TTU and SPZ ligands activate brown adipocyte lipolysis. A: SR-4995 and SR-4559 stimulate lipolysis with EC50 of 4–7 µM. In same assay isoproterenol (1 µM) released 28–32 nmol FFA/hr. Values are means +/− s.e.m. of triplicate determinations and the experiment was replicated > 5 times. B: SR-4559 rapidly stimulates adipocyte lipolysis, indicated by fluorescence increase on lipid droplets (arrows). Imaging results were replicated 4 times. C, D: Stimulation of lipolysis by SR-4995 and SR-4559 does not involve PKA-mediated phosphorylation. Brown adipocytes were treated with isoproterenol (10 nM), SR-4995 or SR-4559 (10 µM) for 30 minutes before determining fatty acid release (C), and phosphorylation of perilipin-1 (PLIN1) and hormone-sensitive lipase (HSL) with phospho-specific antibodies. Values are means +/− s.e.m. of triplicate wells. Experiment shown in C, D was replicated twice. E, F: SR-4995 stimulates lipolysis in mouse diaphragm ex vivo (E) without affecting phosphorylation of HSL (F). Values are means +/− s.e.m, n = 6.
As mentioned above, activation of cAMP-dependent PKA is the only known mechanism for rapid activation of adipocyte lipolysis. As expected, stimulation of lipolysis by the β-adrenergic agonist isoproterenol led to strong phosphorylation of PLIN1 and hormone sensitive lipase (HSL), as determined by phospho-specific antibodies. In contrast, equally effective concentrations of SR-4995 and SR-4559 triggered lipolysis without activating phosphorylation of either HSL or PLIN1 (Figure 2C and 2D). The similar efficacy of compounds in disrupting complexes of ABHD5 with PLIN1 or PLIN5 suggested that these agents might stimulate lipolysis in muscle, where PLIN5 is present, but PLIN1 is not. We found that SR-4995 (10 µM) and isoproterenol (10 µM) were equally effective in stimulating lipolysis in mouse diaphragm muscle ex vivo (Figure 2E). Importantly, SR-4995 stimulated muscle lipolysis in the absence of HSL phosphorylation, unlike isoproterenol (Figure 2F).
Compound stimulation of lipolysis requires ABHD5 and ATGL
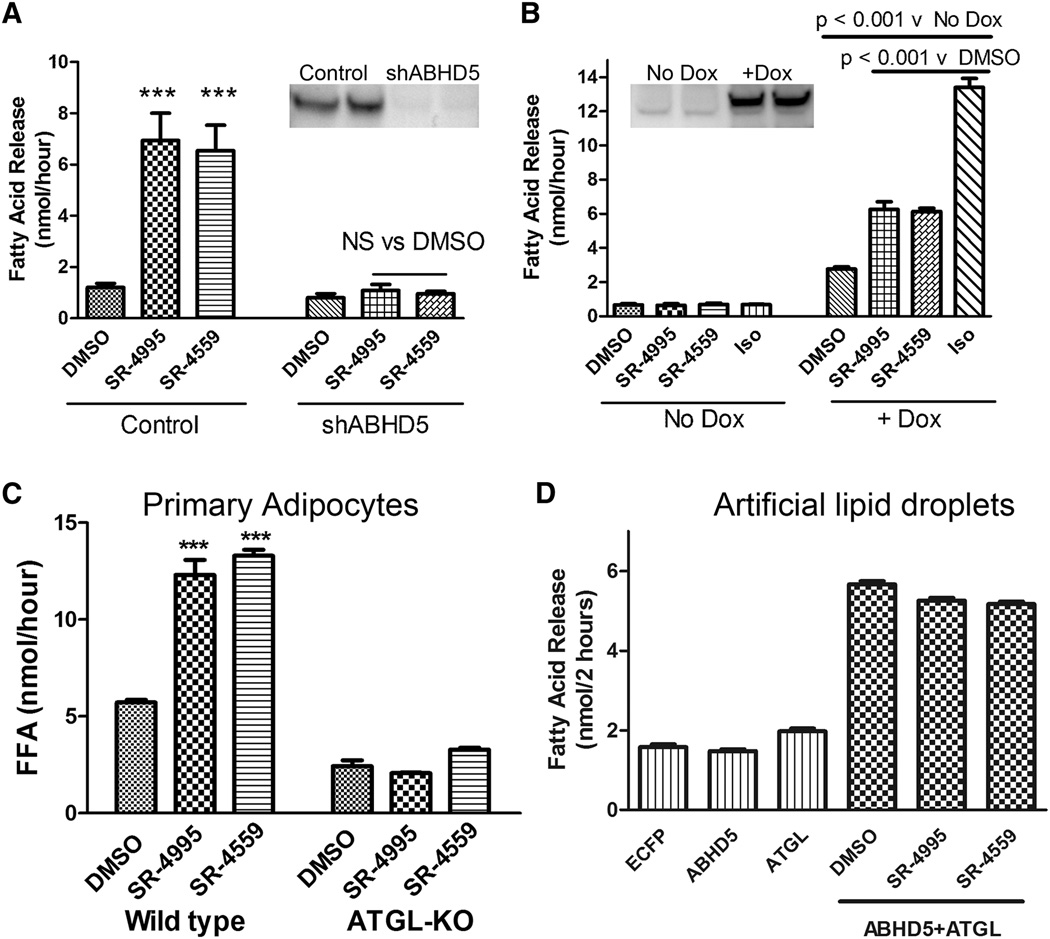
To verify the mechanism of compound action, we performed knockdown and rescue experiments with ABHD5. As shown in Figure 3A, SR-4995 and SR-4559 compounds (10 µM) increased lipolysis by ~7 fold in cultured brown adipocytes, and stable knockdown of ABHD5 by lentiviral shRNA (insert) abolished the ability of compounds to stimulate lipolysis. We transduced the knockdown cell line with a doxycycline-inducible ABHD5 construct that is resistant to the viral shRNA (Figure 3B). When tested in knockdown adipocytes, compounds were completely inactive in the absence of doxycycline, and the activity induced by isoproterenol was also reduced by ~85%. Reinstatement of ABHD5 expression with doxycycline restored the efficacy of the TTU and SPZ agonists as well as isoproterenol, establishing that stimulation of lipolysis by compounds requires ABHD5. Compounds SR-4995 and SR-4559 stimulated lipolysis in freshly-isolated mouse white adipocytes, and this effect was eliminated by adipocyte-specific knockout of ATGL (Figure 3C), the major lipase target of ABHD5 in adipocytes. We confirmed that ABHD5 potently increases the activity of ATGL against artificial lipid droplets in vitro (Figure 3D), and found that SR-4995 and SR-4559 did not modify this effect. These observations indicate that the compounds act by relieving inhibition by PLIN proteins, and that binding of compounds per se does not increase the ability of ABHD5 to activate ATGL.
Figure 3.
Stimulation of lipolysis by SR-4995 and SR-4559 depends on ABHD5 and ATGL. A: Stable knockdown of ABHD5 by shRNA (insert) eliminated stimulation of lipolysis by SR-4995 and SR-4559 (10 µM). B: Rescue of ABHD5 expression restores compound efficacy. In the absence of doxycycline (Dox), SR-4995 and SR-4559 were inactive and the efficacy of isoproterenol (Iso) was severely reduced. Induction of ABHD5 by Dox reinstated SR-4995 and SR-4559 activity, and increased responsiveness to isoproterenol by 5-fold. Experiments in A and B are means +/− s.e.m. of 3 independent experiments. C: SR-4995 and SR-4559 stimulated lipolysis in isolated white adipocytes, and this effect was abolished by adipocyte-specific knockout of ATGL. Values are means +/− s.e.m. of an experiment performed in triplicate and replicated twice. D: Compounds do not augment the ability of ABHD5 to stimulate lipase activity of ATGL using artificial substrates in vitro. Values are means +/− s.e.m. of a representative experiment that was repeated 3 times.
Compounds directly bind ABHD5
In theory, compounds that dissociate complexes could bind either ABHD5 or PLIN proteins; however, the similar potency of compounds in preventing formation of complexes of ABHD5 with PLIN1 or PLIN5 (Figure S1C and Figure S2) suggested that ABHD5 was the likely target. To determine whether ABHD5 was the direct binding target, we made use of an affinity probe, NBD-HE-HP, that was previously shown to covalently modify ABHD5 (Birner-Gruenberger and Hermetter, 2007). As shown in Figure 4A, the NBD-HE-HP affinity tag covalently modified ABHD5 in transfected Cos7 cells, and this labeling was blocked by co-incubation with TTU and SPZ ligands (50 µM). Importantly, SR-4995 blocked NBD-HE-HP binding to ABHD5 with a potency nearly identical to that observed for disrupting the interaction of ABHD5 with PLIN1 (Figures 1B and 4B). As expected (Birner-Gruenberger and Hermetter, 2007), the NBD-HE-HP affinity probe also labeled HSL expressed in transfected Cos7 cells; however, SR-4995 did not block NBD-HE-HP binding to HSL (Figure 4B) or ATGL (Figure S3), demonstrating the specificity of this ligand for binding to ABHD5.
Figure 4.
NBD-HE-HP affinity ligand identifies ABHD5 as the target of SR-4995 and SR-4559. A: Cos7 cells expressing ABHD5 were labeled with NBD-HE-HP (50 µM) in the presence of DMSO, SR-4995, or SR-4559 (50 µM) for the indicated times. Cellular proteins were resolved by SDS PAGE and labeled proteins imaged by laser-scanning fluorometry. B: NBD-HE-HP labeled both HSL and ABHD5 in transfected Cos7 cells, and SR-4995 potently blocked labeling of ABHD5, but not HSL. Experiments in A and B were each performed twice with the same result. C: Mass spectrometry of NBD-HE-HP-labeled ABHD5 identifies Y330 as the amino acid covalently modified by the affinity ligand. D: Mutation of Y330 to alanine (A) or phenylalanine (F) eliminates covalent modification of ABHD5 by NBD-HE-HP. Non, nontransfected cells. The experiment in D was repeated 3 times for the Y330F and twice for the Y330A mutation.
NBD-HE-HP was developed as an active site affinity label of serine hydrolases; however, because ABHD5 lacks the conserved catalytic serine seen in related α-β hydrolases (Caux et al., 2004; Schweiger et al., 2009; Yen and Farese, 2006), we reasoned that the site of modification and ligand interactions likely involves a novel binding domain. To determine the site of protein modification, we subjected purified NBD-HE-HP -labeled and unlabeled ABHD5 to LC/MS/MS analysis and observed 10 unique spectra demonstrating that Y330 is modified by the affinity ligand. Shown in Figure 4C are representative MS-2 spectra of peptide Thr321-Lys342 from unmodified (top) and NBD-HE-HP-labeled ABHD5 (bottom). The m/z difference of b9 and b10 ions in unlabeled and labeled ABHD5 unambiguously identified Y330 as the site of NBD-HE-HP modification, which was confirmed by the mass shift of the remaining b ions. As expected, mutation of Y330 to alanine or phenylalanine prevented tagging by NBD-HE-HP (Figure 4D). Mutation of Y330 to A or F, however, did not affect the ability of ABHD5 to promote lipolysis or to interact with PLIN1 (not shown). These data indicate that the SPZ and TTU ligands directly target ABHD5 and that the binding pocket overlaps with that occupied by NBD-HE-HP. The binding pocket likely includes the highly-conserved Y330, since it is directly modified by NBD-HE-HP, but this residue is not critical for binding of SR-4995 or SR-4559.
Synthetic ligands and long chain acyl-CoA converge to regulate the dynamic interaction of ABHD5 with PLIN proteins
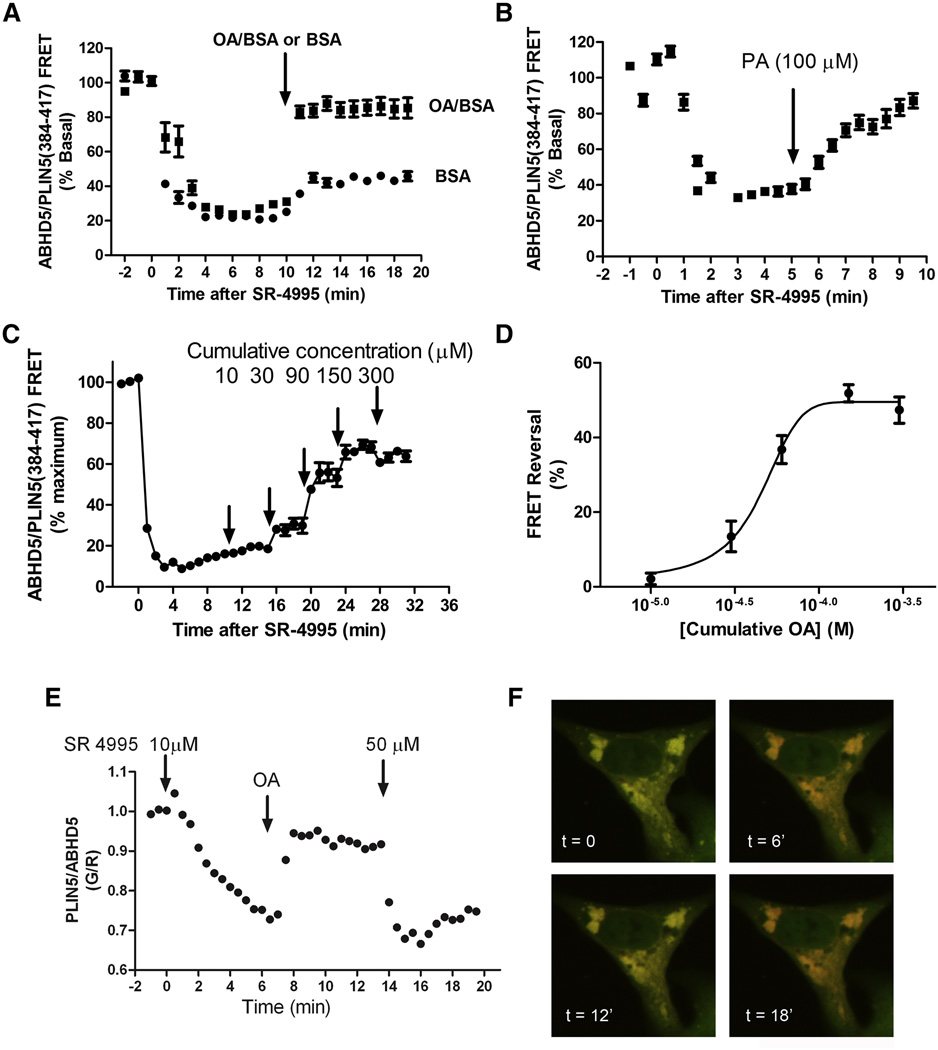
The demonstration that ABHD5 is sensitive to synthetic ligands exposes a new locus of lipolysis control that bypasses the PKA signaling pathway, and raises the question of whether synthetic ABHD5 ligands exploit binding sites used by endogenous ligands. We previously reported that external application of a mono-unsaturated fatty acid, oleic acid, increases the interaction of ABHD5 with PLIN5 (Granneman et al., 2009b) in transfected 293T cells, and this effect likely involved synthesis of long chain acyl-CoA. To assess the dynamic interaction of ABHD5 with PLIN proteins, we identified a minimal fragment of PLIN5, amino acids 384 to 417, that binds ABHD5, but is not otherwise targeted to lipid droplets. The release of PLIN5(384–417) from lipid droplets containing ABHD5 is a robust and dynamic assay that can be monitored by FRET in the whole cells or by the ratio of PLIN5/ABHD5 fluorescence at the lipid droplet surface. As shown in Figure 5A and Supplementary information video 3, SR-4995 rapidly dissociated complexes of ECFP-ABHD5 and PLIN5(384–417)-EYFP and application of oleic acid reassociated complexes within 2 minutes. Similar results were obtained with palmitic acid (Figure 5B), indicating that the effect was not specific to monounsaturated fatty acids. The effect of oleic acid was concentration-dependent and exhibited clear evidence of saturation (Figures 5C and 5D). Importantly, the effect of oleic acid on complex formation could be readily overcome by a higher concentration of SR-4995 (Figures 5E and 5F). Similar results were obtained using tagged human ABHD5 and human PLIN1(358–417) (Figure S4). Together, these observations indicate that synthetic ABHD5 ligands and long chain fatty acids, or a closely associated metabolite, converge on a common binding site in ABHD5 that dynamically regulates its association with PLIN proteins.
Figure 5.
Long chain fatty acids and synthetic compounds converge on a common mechanism to dynamically regulate interaction of ABHD5 and PLIN5. A: Cos7 cells expressing ECFP-ABHD5 and PLIN5(384–417)-EYFP were treated with SR-4995 (25 µM) at time = 0, then exposed to oleic acid (OA, 100 µM) or BSA carrier 10 minutes later. OA treatment reversed the effects of SR 4995 and rapidly promoted reappearance of ABHD5/PLIN5 complexes. Values are means +/− s.e.m, n = 15–16 cells. B: Palmitic acid reverses dissociation of ABHD5-PLIIN5 complexes induced by SR-4995 (25 µm). Values are means +/− s.e.m., n = 10–12 cells. C: Cells were exposed to increasing concentrations of OA, demonstrating that the effect of OA is concentration-dependent and saturable (D). Values are means +/− s.e.m., n = 6 cells. E: The effects of SR-4995 and OA are fully reversible. Association/dissociation kinetics were monitored by the ratio of PLIN5 fluorescence (pseudocolored green, G) to ABHD5 fluorescence (pseudocolored red, R) on lipid droplets. ABHD5-PLIN5(384–417) complexes were dissociated by 10 µM SR-4995, reassociated by 200 µM OA, then dissociated again by a higher concentration of SR-4995. Experiment was replicated three times with similar results. F) Merged confocal images of PLIN5 and ABHD5 channels demonstrating the reversible dissociation/association kinetics.
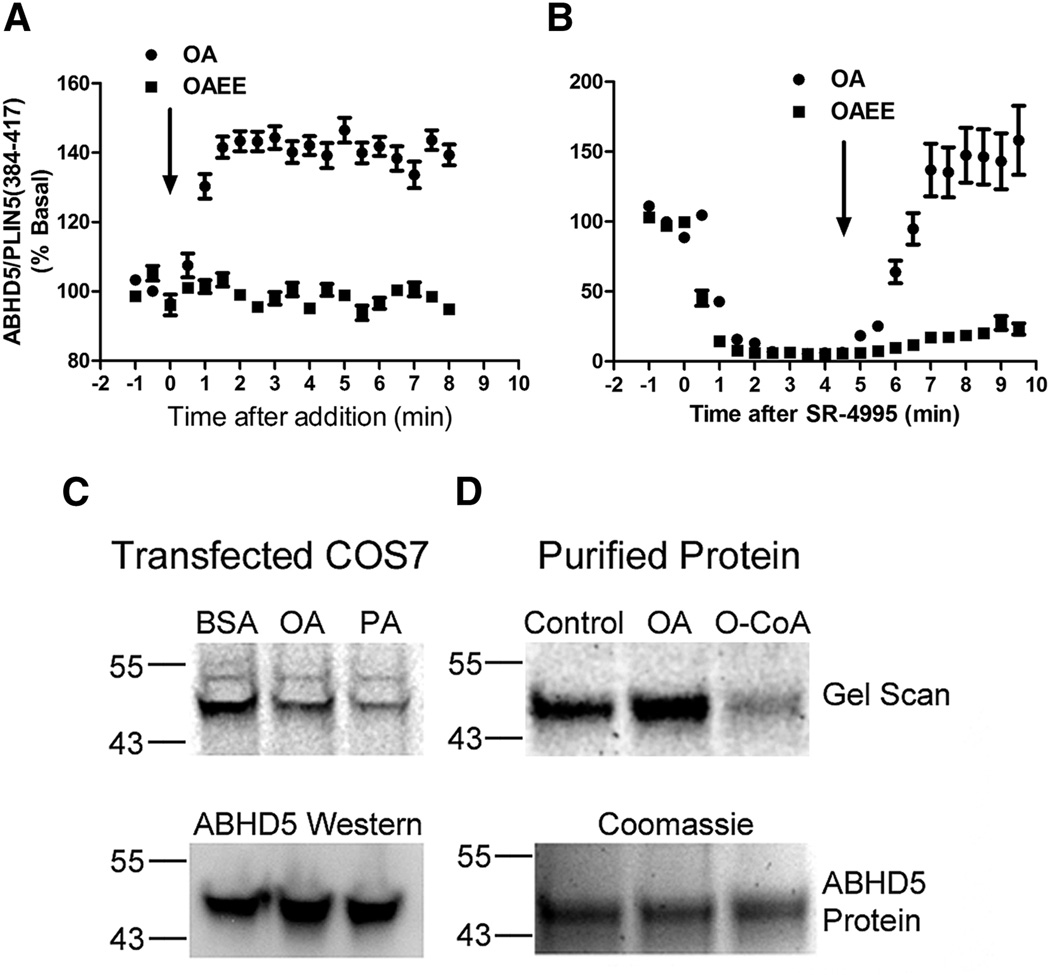
Based on previous work (Granneman et al., 2009b), we hypothesized that long chain acyl-CoA was a likely physiological ligand of ABHD5. Consistent with this hypothesis, we found that OA rapidly promoted the association of ABHD5 and PLIN5(384–417) in Cos7 cells cultured in the absence of OA supplementation (Figure 6A). As a control, we examined the effects of the ethyl ester of oleic acid (OAEE), which cannot directly form oleoyl-CoA without prior intracellular hydrolysis. We found that OAEE failed to trigger formation of ABHD5/PLIN5 complexes. Furthermore, OA, but not OAEE, rapidly reversed compound-induced dissociation of ABHD5/PLIN5 complexes (Figure 6B). OA rapidly reversed compound-induced dissociation in the presence of OAEE, indicating that the tested concentrations of OAEE are not toxic to COS7 cells (Figure S5). As expected, treatment of intact cells with oleic or palmitic acids (50 µM) suppressed NBD-HE-HP labeling of ABHD5 in transfected Cos7 cells (Figure 6C). Importantly, we found that oleoyl-CoA, but not oleic acid, effectively blocked NBD-HE-HP labeling of purified ABHD5 (Figure 6D), directly demonstrating that oleoyl-CoA, derived from oleic acid, is an endogenous ligand of ABHD5.
Figure 6.
Long chain acyl-CoA are ABHD5 ligands and mediate the effects of OA. A) Oleic acid (OA), but not oleic acid ethyl ester (OAEE), promotes the association of ABHD5 and PLIN5. Cos7 cells were transfected with fluorescently-tagged ABHD5 and PLIN5 constructs and cultured overnight without oleic acid supplementation. Cells were then exposed to oleic acid (OA, 300 µM) or oleic acid ethyl ester (OAEE, 300 µM). B) Oleic acid (OA), but not oleic acid ethyl ester (OAEE), reverses compound-induced complex dissociation. Cos7 cells expressing fluorescently-tagged ABHD5 and PLIN5 constructs were treated with SR-4995 (10 µM) at time = 0, then exposed to oleic acid (OA, 300 µM) or oleic acid ethyl ester (OAEE, 300 µM) 5 minutes later. For A and B values are means +/− s.e.m., n= 10–12 cells. C) Treatment of Cos7 cells with oleic acid (OA, 400 µM) or palmitic acid (PA, 400 µM) inhibited NBD-HE-HP labeling of ABHD5. D: NBD-HE-HP labeling of purified ABHD5 was inhibited by oleoyl-Coenzyme A (O-CoA, 50 µM), but not OA (50 µM). Experiments were performed at least two times with similar results.
Discussion
The canonical pathway for rapid regulation of lipolysis in adipocytes involves integration of extracellular signals and PKA-dependent phosphorylation of the key lipolytic effectors PLIN1 and HSL (Granneman et al., 2009a; Miyoshi et al., 2007; Miyoshi et al., 2006). Unlike white adipocytes, which export mobilized fatty acids in response to external signals, other tissues including muscle and brown adipose tissue mobilize fatty acids from intracellular lipid droplets for oxidation in situ. Thus, it might be expected that additional mechanisms exist for balancing fatty acid supply with demand that are independent of transmembrane signaling.
Using a novel protein interaction assay, we discovered compounds that potently disrupt the interaction of ABHD5 with PLIN1 or PLIN5. These synthetic ligands rapidly stimulated lipolysis in adipocytes (white and brown) and muscle in the absence of PKA-mediated phosphorylation of PLIN1 and HSL. Knockdown and rescue experiments demonstrated that compound activation of lipolysis is mediated by ABHD5 and requires ATGL. These results demonstrated that the synthetic compounds bypass receptor-mediated activation and directly dissociate ABHD5 from its inhibitory regulators, PLIN1 and PLIN5.
By taking advantage of the NBD-HE-HP affinity label we were able to show that our synthetic ligands directly bind ABHD5. Mass spectrometry of purified ABHD5 demonstrated that the docking site for NBD-HE-HP includes Y330 which lies within the highly conserved HYVYAD sequence of ABHD5. Although this sequence loosely resembles the H4XD (where X is any amino acid) motif observed in numerous hydrolases and acyl-transferases (Heath and Rock, 1998; Lewin et al., 1999), there are no data indicating involvement of this sequence in any enzymatic activity. However, the observation that SPZ and TTU ligands similarly disrupted ABHD5/PLIN complexes composed of human or mouse proteins indicates the binding pocket is well conserved, and raised the possibility that endogenous ligands might bind to ABHD5 in a manner that modulates its interactions with PLIN proteins.
Previous work showed that treating cells with oleic acid for 1–2 hours increases the interaction of ABHD5 and PLIN5, and this effect is inhibited by blocking long chain acyl-CoA synthesis with Triacsin C (Granneman et al., 2009b). The discovery of synthetic ABHD5 ligands and the development of fluorescent imaging tools allowed us to explore the physiological significance of ligand-regulated interactions between ABHD5 and PLIN proteins with high temporal and spatial resolution. We found that oleic acid treatment rapidly reversed complex dissociation that was induced by synthetic ligands. This effect was saturable, and was readily overcome by higher concentrations of synthetic ligand. Treatment of live cells with oleic acid or synthetic ligands effectively disrupted ABHD5 labeling by NBD-HE-HP, and competition binding experiments with purified ABHD5 demonstrated that oleoyl-CoA is a physiological metabolite that binds ABHD5. These results strongly indicate that ABHD5 contains a common binding site for synthetic and endogenous ligands. Furthermore, these ligands appear to be potent allosteric modulators that disrupt or promote the interaction of ABHD5 with PLIN proteins.
Physiologically, the regulation of ABHD5-PLIN interactions by long chain acyl-CoA provides an elegant negative feedback mechanism whereby fatty acid production is closely linked to its metabolism. This rapid regulation may be especially important in coupling fatty acid mobilization and oxidation in muscle and brown adipose tissue where lipid droplet and mitochondria are very closely associated and PLIN5 likely plays a role in forming a "metabolic synapse." PLIN5 is targeted to the lipid droplet surface that apposes mitochondria where it assembles the core lipolytic machinery by separately binding both ABHD5 and ATGL (Granneman et al., 2011; Kimmel and Sztalryd, 2014; Wang et al., 2011). Long chain acyl-CoA synthase 1 (ACSL1), which activates fatty acids released from lipid droplets, resides on the outer mitochondrial membrane and is well situated to both direct fatty acids toward oxidation (Ellis et al., 2010; Ellis et al., 2011) and modulate fatty acid supply via feedback inhibition that could involve ABHD5, ATGL, and HSL (Nagy et al., 2014; Severson and Hurley, 1984). SR-4995 did not block NBD-HE-HP labeling of HSL or ATGL, indicating that this activator does not affect lipolysis by direct interaction with these lipases (for example, by relieving acyl-CoA inhibition). Furthermore, synthetic compounds are inactive in the presence of HSL and ATGL if ABHD5 is absent.
The precise mechanism by which ABHD5 activates lipolysis is not understood. Our synthetic ligands did not promote the ATGL-activating properties of ABHD5 in biochemical assays that lacked PLIN proteins indicating that the activation of lipolysis in intact cells involves release of inhibition produced by PLIN binding. Conversely, our results indicate that long chain acyl-CoA can suppress lipolysis in part by promoting the sequestration of ABHD5 by PLIN proteins. Whether endogenous ligands exist that can dissociate ABHD5/PLIN complexes, as do our synthetic compounds, is currently under investigation. ABHD5 has no confirmed enzymatic activity, and the simplest model for activation entails direct allosteric activation of ATGL (Lass et al., 2006). It should be noted, however, that ABHD5 appears to activate lipases other than ATGL (Zierler et al., 2014), and it is conceivable that stimulation of lipolysis is mediated by other effects that ABHD5 exerts on LD surfaces. Regardless, the synthetic ABHD5 ligands we have discovered will likely provide new insights into ATGL-dependent and independent functions of ABHD5.
Finally, might ABHD5 ligands be developed into novel therapeutic entities? In general terms, the mass of triacylglycerol which accumulates in adipose tissues reflects the balance between energy storage and mobilization, and it is well established that persistent activation of adipocyte lipolysis by β3-adrenergic receptors triggers thermogenic responses that have potent anti-obesity and anti-diabetes properties in rodent models (Arch, 2002). Given that fatty acids are necessary and sufficient for brown adipocyte thermogenesis (Fedorenko et al., 2012; Guerra et al., 1998; Nicholls, 2006) and adipose tissue-specific overexpression of ATGL improves metabolic status of mice challenged with high fat diet (Ahmadian et al., 2009), ABHD5 ligands might provide a more direct means of promoting fat catabolism in brown and white adipose tissues that overcomes limitations associated with surface receptor expression (Arch, 2002). In addition, ABHD5 was recently shown to be a potent tumor suppressor in colorectal cancer (Ou et al., 2014). It has been proposed that loss of ABHD5 function promotes tumor invasiveness by reducing fatty acid oxidation and enhancing the production of growth-promoting metabolites from glycolysis. It would be predicted from these observations that activators of ABHD5, like those described here, might delay or prevent tumor progression by promoting oxidative metabolism.
In summary, high throughput compound screening identified ligands of ABHD5 that stimulate lipolysis in adipocytes and muscle, bypassing canonical PKA-dependent signaling. The synthetic ligands exposed an endogenous feedback pathway whereby long chain acyl-CoA rapidly modulated the interaction of ABHD5 with PLIN proteins. Our results demonstrate that ABHD5 is a ligand-regulated lipase activator, providing a foundation for the development of a new class of synthetic lipolysis modulators.
Methods
High throughput compound screening
Luciferase complementation constructs were generated as previously described (Granneman et al., 2009a; Granneman et al., 2009b; Granneman et al., 2011) and proteins were produced in 293T or Sf9 cells. High throughput compound screening was performed by the Molecular Screening Center at the Scripps Research Institute in Jupiter, Florida. The assays and primary screening methods and results are detailed in PubChem AID 493241 (ABHD5/PLIN5) and AID 493057 (ABHD5/PLIN1). The stepwise protocol used for the protein complementation HTS assays, screening strategy, and confirmatory counterscreening in the ABHD5/PLIN1–5 screening campaign are detailed in Supplemental Methods and in Supplemental Information Tables S1 and S2 .
Molecular and biochemical techniques for secondary analyses
Cellular lysates were prepared from transfected 293T cells (Granneman et al., 2009a; Granneman et al., 2009b) and complementation assays were performed in 96 well plates using 50 µl of each lysate and 1 µl of compounds dissolved in DMSO or DMSO vehicle. Luciferase activity was determined after incubation for 4 hours at room temperature. Immunoblotting was performed as previously described (Mottillo et al., 2012). In vitro lipolysis assays were performed using ABHD5 purified from Sf9 cells and ATGL overexpressed in Cos7 cells, as detailed (Schweiger et al., 2014). Cloning and mutagenesis were performed using PCR, and all constructs were confirmed by sequencing.
Analysis of lipolysis in brown adipocytes and muscle
Immortalized brown adipocytes were seeded in 96 well plates and differentiated as described (Mottillo et al., 2012). Three to four days after induction of differentiation, cells were washed and placed in HEPES-buffered Kreb’s Ringer buffer containing 1% bovine serum albumin (KRBB), and treated with DMSO (vehicle) or activators at concentrations specified in the figures. Results are reported as fatty acid efflux per well, each containing ~ 76,000 cells. Accumulated fatty acids were determined using WAKO kits adapted to fluorescence detection with Amplex Red, as described (Mottillo et al., 2012). Whole mouse diaphragms were removed, washed with PBS, divided into ~20 mg segments, and incubated in 500 µl of KRBB in the presence of vehicle, SR-4995 or isoproterenol at the indicated concentration. Thirty microliter aliquots were removed after 1, 2 and 3 hours and assayed for fatty acid release, as detailed above. Immunoblotting for PLIN1, HSL and ABHD5 was performed as previously described (Granneman et al., 2009a; Granneman et al., 2011). Phospho-HSL and phospho-PLIN1 antibodies were purchased from Cell Signaling (#4126) and Vala Sciences (#4856), respectively.
Stable knockdown and inducible rescue of ABHD5 in brown adipocytes
Lentiviral shRNA to Abhd5 was created from a plasmid purchased from Sigma (TRCN0000032737 NM_026179.1–1063s1c1), and used to stably transduce mouse brown adipocytes. Knockdown was confirmed by immunoblotting of differentiated adipocytes. To create an inducible rescue cell line we mutated the DNA sequence targeted by the shRNA while maintaining the wild type amino acid sequence of ABHD5, and tagged the construct with mCherry. Doxycycline-inducible expression was achieved by transferring the shRNA-resistant construct into pINDUCER20 (Meerbrey et al., 2011), and the resulting lentivirus was used to infect the brown adipocytes in which endogenous ABHD5 was stably knocked down.
Induced knockout of ATGL in mouse adipocytes
Induced knockout of ATGL and analysis of adipocyte lipolysis was performed as described (Mottillo et al., 2014a). Briefly, B6N.129S–Pnpla2tm1Eek, B6N.129S–Tg(Adipoq-CreERT2)tm1Jgra double transgenic mice or single transgenic B6N.129S–Pnpla2tm1Eek (control) mice were treated with tamoxifen one week prior to adipocyte isolation and analysis of fatty acid release. Assay were performed in 200 µl of KRBB using 5% packed cell volume (~10,000 cells).
Imaging compound action in live cells
DNA constructs encoding human and mouse ABHD5 and PLIN proteins (and fragments) were amplified by PCR and cloned into the expression vectors ECFP-C1 and EYFP-N1, respectively. Cos7 cells were plated onto coverslips, and transfected and loaded with oleic acid, as previously described (Granneman et al., 2009a; Granneman et al., 2011). Prior to imaging, cells were washed once in KRBB and imaged at room temperature in KRBB. Compounds were added as 10X or 20X solutions dissolved in KRBB. Ratiometric imaging, including FRET, was performed using IPlabs software, as previously described (Granneman et al., 2009a; Granneman et al., 2011). FRET images of individual cells were imported into ImageJ to determine average net FRET values for each frame, which were then normalized to the mean value of three baseline images. For imaging of fatty acid production in live cells, we created a brown adipocyte cell line that expresses a fatty acid reporter system (Mottillo et al., 2014b) under the control of a doxycycline-sensitive promoter. In this system, PLIN1 is fused to the PPARα ligand binding domain (LBD), thus targeting the fatty acid-sensing domain of PPARα to lipid droplets. Mobilized fatty acids bound by the PPARα LBD are detected by the accumulation of EYFP-tagged SRC1 coactivator binding domain at the lipid droplet surface. All imaging experiments were performed on > 3 different coverslips and replicated in independent transfections.
Covalent labeling of ABHD5 with NBD-HE-HP and mass-spectrometry of labeled peptides
NBD-HE-HP affinity label was synthesized and purified as described (Birner-Gruenberger and Hermetter, 2007; Schmidinger et al., 2005). Cos7 cells transfected with PLIN1 and wild type or mutant ABHD5 were lipid loaded overnight with 200 µM oleic acid. One day after transfection, cells were labeled with 50 µM NBD in serum-free DMEM for one hour at 37°C. Cell s were rinsed with PBS and protein-matched aliquots of cell lysates were separated by SDS-PAGE. Two gels were run in parallel, one for fluorescent scanning and one for immunoblot detection of ABHD5. Gels for scanning were fixed (45% MeOH, 10% acetic acid) then visualized on a Typhoon 9410 Variable Mode Imager (GE Healthcare; excitation 488 nm, emission 520 nm).
LC/MS/MS characterization of NBD-HE-HP labeled ABHD5
293-T cells transfected with His-tagged ABHD5 were labeled with NBD in serum-free DMEM for 3 hours at 37°C. His-tagged ABHD5 was partially purified by His60 Ni2+ Superflow Resin column chromatography (Clontech), as described by the manufacturer. Labeled ABHD5 was further resolved by SDS-PAGE and gel bands containing labeled ABHD5 were subjected to LC/MS analysis (details in Supplemental Methods).
Effect of oleic acid and oleoyl-CoA on NBD-HE-HP labeling of ABHD5
ABHD5-transfected COS7 cells were incubated in serum-free medium for 2–3 hr to allow clearance of remaining oleic acid from overnight lipid loading. 400 µM oleic acid or 400 µM palmitic acid (complexed to BSA) was then added two minutes prior to labeling with 50 µM NBD-HE-HP. Cells were allowed to label for 2 hrs at 37°C. Parallel gels were run for scanning and for ABHD5 immunoblotting. Similar results were obtained when ABHD5 was co-transfected with Plin5 (not shown). For cell-free labeling of ABHD5, His-tagged ABHD5 was expressed in High-Five insect cells and purified by His60 Ni2+ Superflow Resin column chromatography (Clontech). Coomassie staining of SDS-PAGE gels indicated ABHD5-His protein was approximately 50% pure (not shown). Purified ABHD5 was resuspended in 100 mM potassium phosphate buffer (pH 7.0) containing artificial lipid droplets (PC:PI 3:1) and was pretreated for 15 min with 50 µM oleic acid or 50 µM oleoyl-CoA (Avanti) prior to the addition of 50 µM NBD-HE-HP. All ligands were dissolved in DMSO. Labeling then proceeded for 2 hr at 37°C, samples w ere separated by SDS-PAGE and gels were scanned as described above.
Supplementary Material
Highlights.
Synthetic ABHD5 ligands prevent ABHD5 interaction with PLIN1 or PLIN5
ABHD5 ligands stimulate lipolysis independently of PKA activation
Long chain acyl-CoAs bind ABHD5 and promote its interaction with PLIN proteins
Synthetic ABHD5 ligands might be exploited for therapeutic benefit
Acknowledgments
We thank Pierre Baillargeon and Lina DeLuca for compound management. We thank Anna Knapinska and Dmitriy Minond for transfering the ABHD5-Plin1/5 luciferase complementation assays to Scripps Florida and for the initial assay recapitulations. We thank Dr. Erin Kershaw (University of Pittsburgh) for providing floxed Pnpla2 mice.
Supported by National Institutes of Health Roadmap Initiative grant U54MH084512 (FM, PC, PH and WRR), and NIH grants DK62292, DK76629, DK091741, NS21061634 and Veterans Administration Grant 5IO-BX0001335 (JGG). The Wayne State University Proteomics Core is supported through the NIH Center Grant P30 ES06639, the NIH Cancer Center Support Grant P30 CA22453 and the NIH Shared Instrumentation Grant S10 OD 010700.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions to the research: HTS assays were designed by JGG. HTS screening was directed by PH and performed by FM and PC. Chemoinformatic analysis and medicinal chemistry were directed by WRR and performed by GH. Functional analyses and biological probe development were directed by JGG and performed by LM, MAS, XY, MA, HZ and JGG. MAS and JC performed affinity tagging and LC/MS/MS identification. JGG, WRR, PH, and MAS participated in writing the manuscript.
None of the authors of this manuscript has a financial interest related to this work.
References
- Ahmadian M, Duncan RE, Varady KA, Frasson D, Hellerstein MK, Birkenfeld AL, Samuel VT, Shulman GI, Wang Y, Kang C, et al. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes. 2009;58:855–866. doi: 10.2337/db08-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arch JRS. β3-Adrenoceptor agonists: potential, pitfalls and progress. Eur J of Pharmacol. 2002;440:99–107. doi: 10.1016/s0014-2999(02)01421-8. [DOI] [PubMed] [Google Scholar]
- Birner-Gruenberger R, Hermetter A. Activity-based proteomics of lipolytic enzymes. Current drug discovery technologies. 2007;4:1–11. doi: 10.2174/157016307781115458. [DOI] [PubMed] [Google Scholar]
- Caux F, Selma ZB, Laroche L, Prud’homme JF, Fischer J. CGI-58/ABHD5 gene is mutated in Dorfman-Chanarin syndrome. American journal of medical genetics. Part A. 2004;129A:214. doi: 10.1002/ajmg.a.30228. [DOI] [PubMed] [Google Scholar]
- Ellis JM, Li LO, Wu PC, Koves TR, Ilkayeva O, Stevens RD, Watkins SM, Muoio DM, Coleman RA. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell metabolism. 2010;12:53–64. doi: 10.1016/j.cmet.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JM, Mentock SM, Depetrillo MA, Koves TR, Sen S, Watkins SM, Muoio DM, Cline GW, Taegtmeyer H, Shulman GI, et al. Mouse cardiac acyl coenzyme a synthetase 1 deficiency impairs Fatty Acid oxidation and induces cardiac hypertrophy. Molecular and cellular biology. 2011;31:1252–1262. doi: 10.1128/MCB.01085-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko A, Lishko PV, Kirichok Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell. 2012;151:400–413. doi: 10.1016/j.cell.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Lefevre C, Morava E, Mussini JM, Laforet P, Negre-Salvayre A, Lathrop M, Salvayre R. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nature genetics. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- Gandotra S, Lim K, Girousse A, Saudek V, O’Rahilly S, Savage DB. Human frame shift mutations affecting the carboxyl terminus of perilipin increase lipolysis by failing to sequester the adipose triglyceride lipase (ATGL) coactivator AB-hydrolase-containing 5 (ABHD5) J Biol Chem. 2011;286:34998–35006. doi: 10.1074/jbc.M111.278853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Krishnamoorthy R, Rathod M. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl) J Biol Chem. 2009a;284:34538–34544. doi: 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Mottillo EP, Zhu Z. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J Biol Chem. 2009b;284:3049–3057. doi: 10.1074/jbc.M808251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Mottillo EP, Zhu Z, Zhou L. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J Biol Chem. 2011;286:5126–5135. doi: 10.1074/jbc.M110.180711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Koza RA, Walsh K, Kurtz DM, Wood PA, Kozak LP. Abnormal nonshivering thermogenesis in mice with inherited defects of fatty acid oxidation. The Journal of clinical investigation. 1998;102:1724–1731. doi: 10.1172/JCI4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RJ, Rock CO. A conserved histidine is essential for glycerolipid acyltransferase catalysis. Journal of bacteriology. 1998;180:1425–1430. doi: 10.1128/jb.180.6.1425-1430.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel AR, Sztalryd C. Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Current opinion in lipidology. 2014;25:110–117. doi: 10.1097/MOL.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell metabolism. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Perfield JW, Souza SC, 2nd, Shen WJ, Zhang HH, Stancheva ZS, Kraemer FB, Obin MS, Greenberg AS. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J Biol Chem. 2007;282:996–1002. doi: 10.1074/jbc.M605770200. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Souza SC, Zhang HH, Strissel KJ, Christoffolete MA, Kovsan J, Rudich A, Kraemer FB, Bianco AC, Obin MS, et al. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J Biol Chem. 2006;281:15837–15844. doi: 10.1074/jbc.M601097200. [DOI] [PubMed] [Google Scholar]
- Mottillo EP, Balasubramanian P, Lee YH, Weng C, Kershaw EE, Granneman JG. Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic beta3-adrenergic receptor activation. Journal of lipid research. 2014a doi: 10.1194/jlr.M050005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottillo EP, Bloch AE, Leff T, Granneman JG. Lipolytic Products Activate Peroxisome Proliferator-activated Receptor (PPAR) α and δ in Brown Adipocytes to Match Fatty Acid Oxidation with Supply. Journal of Biological Chemistry. 2012;287:25038–25048. doi: 10.1074/jbc.M112.374041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottillo EP, Paul GM, Moore HP, Granneman JG. Use of fluorescence microscopy to probe intracellular lipolysis. Methods in enzymology. 2014b;538:263–278. doi: 10.1016/B978-0-12-800280-3.00015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy HM, Paar M, Heier C, Moustafa T, Hofer P, Haemmerle G, Lass A, Zechner R, Oberer M, Zimmermann R. Adipose triglyceride lipase activity is inhibited by long-chain acyl-coenzyme A. Biochim Biophys Acta. 2014;1841:588–594. doi: 10.1016/j.bbalip.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls DG. The physiological regulation of uncoupling proteins. Biochim Biophys Acta. 2006;1757:459–466. doi: 10.1016/j.bbabio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Ou J, Miao H, Ma Y, Guo F, Deng J, Wei X, Zhou J, Xie G, Shi H, Xue B, et al. Loss of abhd5 promotes colorectal tumor development and progression by inducing aerobic glycolysis and epithelial-mesenchymal transition. Cell reports. 2014;9:1798–1811. doi: 10.1016/j.celrep.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak NM, Jaeger D, Kolleritsch S, Zimmermann R, Zechner R, Lass A, Haemmerle G. The interplay of protein kinase A and perilipin 5 regulates cardiac lipolysis. J Biol Chem. 2015;290:1295–1306. doi: 10.1074/jbc.M114.604744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilich P, Horvath R, Krause S, Schramm N, Turnbull DM, Trenell M, Hollingsworth KG, Gorman GS, Hans VH, Reimann J, et al. The phenotypic spectrum of neutral lipid storage myopathy due to mutations in the PNPLA2 gene. Journal of neurology. 2011;258:1987–1997. doi: 10.1007/s00415-011-6055-4. [DOI] [PubMed] [Google Scholar]
- Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nature methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- Schmidinger H, Birner-Gruenberger R, Riesenhuber G, Saf R, Susani-Etzerodt H, Hermetter A. Novel fluorescent phosphonic acid esters for discrimination of lipases and esterases. Chembiochem : a European journal of chemical biology. 2005;6:1776–1781. doi: 10.1002/cbic.200500013. [DOI] [PubMed] [Google Scholar]
- Schweiger M, Eichmann TO, Taschler U, Zimmermann R, Zechner R, Lass A. Measurement of lipolysis. Methods in enzymology. 2014;538:171–193. doi: 10.1016/B978-0-12-800280-3.00010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. American journal of physiology. Endocrinology and metabolism. 2009;297:E289–E296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- Severson DL, Hurley B. Inhibition of the hormone-sensitive lipase in adipose tissue by long-chain fatty acyl coenzyme A. Lipids. 1984;19:134–138. doi: 10.1007/BF02534504. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, et al. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- Wang H, Sreenivasan U, Hu H, Saladino A, Polster BM, Lund LM, Gong DW, Stanley WC, Sztalryd C. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. Journal of lipid research. 2011;52:2159–2168. doi: 10.1194/jlr.M017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins NE, Quaynor BK, Skinner JR, Tzekov A, Croce MA, Gropler MC, Varma V, Yao-Borengasser A, Rasouli N, Kern PA, et al. OXPAT/PAT-1 Is a PPAR-Induced Lipid Droplet Protein That Promotes Fatty Acid Utilization. Diabetes. 2006;55:3418–3428. doi: 10.2337/db06-0399. [DOI] [PubMed] [Google Scholar]
- Yen CL, Farese RV., Jr Fat breakdown: a function for CGI-58 (ABHD5) provides a new piece of the puzzle. Cell metabolism. 2006;3:305–307. doi: 10.1016/j.cmet.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Young SG, Zechner R. Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes & development. 2013;27:459–484. doi: 10.1101/gad.209296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierler KA, Zechner R, Haemmerle G. Comparative gene identification-58/alpha/beta hydrolase domain 5: more than just an adipose triglyceride lipase activator. Current opinion in lipidology. 2014;25:102–109. doi: 10.1097/MOL.0000000000000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.