Abstract
Background
Podocalyxin-like 1 (PODXL) is an anti-adhesive transmembrane protein that has been demonstrated to be an independent factor of poor prognosis in colorectal cancer (CRC). The gene encoding PODXL is located to chromosome 7, which also harbours the gene for the epidermal growth factor receptor (EGFR). The aim of this study was to examine the associations between PODXL and EGFR expression in CRC in vitro and in vivo.
Methods
EGFR expression was analysed in tumours from three independent patient cohorts; cohort 1 (n = 533), cohort 2 (n = 259) and cohort 3 (n = 310), previously analysed for immunohistochemical PODXL expression and KRAS and BRAF mutations (cohort 1 and 3). Levels of EGFR and PODXL were determined by western blot in six different CRC cell lines.
Results
High expression of PODXL was significantly associated with high EGFR expression (p < 0.001) in all three cohorts, and with BRAF mutation (p < 0.001) in cohort 1 and 3. High EGFR expression correlated with BRAF mutation (p < 0.001) in cohort 1. High EGFR expression was associated with adverse clinicopathological factors and independently predicted a reduced 5-year overall survival (OS) in cohort 1 (HR 1.77; 95 % CI 1.27–2.46), cohort 2 (HR 1.58; 95 % CI 1.05–2.38) and cohort 3 (HR 1.83; 95 % CI 1.19–2.81). The highest risk of death within 5 years was observed in patients with tumours displaying high expression of both EGFR and PODXL in cohort 1 and 3 (HR 1.97; 95 % CI 1.18–3.28 and HR 3.56; 95 % CI 1.75–7.22, respectively). Western blot analysis showed a uniform expression of PODXL and EGFR in all six examined CRC cell lines.
Conclusions
The results from this study demonstrate that high expression of EGFR is an independent factor of poor prognosis in CRC. Moreover, strong links have been uncovered between expression of the recently proposed biomarker candidate PODXL with EGFR expression in CRC in vivo and in vitro, and with BRAF mutation in vivo. High expression of both PODXL and EGFR may also have a synergistic adverse effect on survival. These findings suggest a potential functional link in CRC between PODXL, EGFR and BRAF, all originating from chromosome 7, which may be highly relevant in the clinical setting and therefore merit future in-depth study.
Electronic supplementary material
The online version of this article (doi:10.1186/s12967-016-0882-0) contains supplementary material, which is available to authorized users.
Keywords: Podocalyxin-like, EGFR, BRAF, Colorectal cancer, Prognosis
Background
Podocalyxin-like protein (PODXL) is a transmembrane glycoprotein and member of the CD34 family [1]. PODXL regulates cell adhesion and is normally expressed by podocytes of the kidney, vascular endothelial cells and hematopoietic progenitor cells [2–4]. Moreover, PODXL has been shown to be overexpressed in a variety of malignancies, and associated with a more aggressive tumour phenotype and poor prognosis in breast, prostate, colorectal, ovarian, renal, pancreatic and bladder cancer, as well as in glioblastoma and astrocytoma [5–14]. The adverse prognostic role of PODXL expression in colorectal cancer (CRC) was first described only recently [7], and has since then been validated in several independent studies [15–17].
The exact mechanisms behind the role of PODXL in tumourigenesis are still unknown, but it has been demonstrated to be involved in epithelial-mesenchymal transition (EMT), a process in which epithelial cells obtain mesenchymal properties leading to increased migration, invasiveness and resistance to apoptosis [18]. In two recent studies, knockdown of PODXL in breast cancer cell lines resulted in impaired primary tumour growth and metastasis [19, 20].
The gene encoding PODXL is located to chromosome 7, which also harbours several other genes with important implications in CRC, e.g. the genes encoding the epidermal growth factor receptor (EGFR) and v-Raf murine sarcoma viral oncogene homolog B (BRAF). Of note, the PODXL and BRAF genes are located right next to each other at 7q32-33 and 7q34.
EGFR is a transmembrane receptor tyrosine kinase that plays an important role in CRC initiation and progression through the RAS–RAF–MEK- MAPK and the PI3K–PTEN–Akt signalling pathways. Overexpression of EGFR has been reported in 25–75 % of CRC [21].
The clinical significance of EGFR overexpression in CRC remains unclear. Whereas several studies have demonstrated a link between high EGFR expression and poor prognosis [22–26], other studies have not found EGFR expression to correlate with an adverse outcome [22, 27, 28]. However, due to its role in the progression of CRC, EGFR has become an interesting target for antitumoural therapy, and monoclonal anti-EGFR antibodies cetuximab and panitumumab are widely used in metastatic CRC [29].
The aim of this study was to investigate the relationship between PODXL and EGFR expression in CRC in vivo and in vitro. To this end, immunohistochemical expression of the proteins was compared in tumours from three different patient cohorts, and western blot analysis was performed on six different CRC cell lines.
Methods
Patients
Cohort 1 encompasses tumours from incident CRC cases in the population-based, prospective cohort Malmö Diet and Cancer Study (MDCS). Until end of follow-up 31 December 2008, 626 incident cases of CRC had been registered in the study population, and tumour tissue for tissue microarray (TMA) was available from 557 patients. The cohort has been described previously [7, 30, 31].
Cohort 2 is a consecutive, retrospective cohort comprising all patients who underwent surgery for CRC at Skåne University Hospital in Malmö, Sweden between 1 January 1990 and 31 December 1991, for whom archival tumour tissue was available (n = 270). The cohort has been described previously [15, 32, 33].
Cohort 3 consists of 337 patients who were surgically treated for CRC at the Central District Hospital in Västerås, Sweden between August 2000 and December 2003. TMAs were constructed from 320 patients. The cohort has been described previously [15, 32, 34].
Patient and tumour characteristics in the different cohorts are summarized in Additional file 1.
Approvals for the study were obtained from the Ethics Committees at Lund University (ref 51/90, 530/08 and 445/2007) and Uppsala University (ref 00/001).
PODXL and EGFR immunohistochemistry and evaluation
Formalin-fixed, paraffin-embedded CRC tissue blocks were used to construct TMAs as previously described [7], and immunohistochemical staining was performed on 4 μm TMA-sections.
PODXL expression has previously been analysed in all three cohorts [7, 15]. In brief, the affinity-purified polyclonal anti-PODXL antibody HPA 2110 (Atlas Antibodies, Stockholm, Sweden), diluted 1:250 was used, and staining was performed in an Autostainer Plus (Dako, Glostrup, Denmark) after automated pre-treatment with the PT-link system (Dako). PODXL expression was denoted as negative (0), weak cytoplasmic staining (1), moderate cytoplasmic staining (2), distinct membranous staining in ≤50 % of tumour cells (3) and distinct membranous staining in > 50 % of tumour cells (4), as previously described [7]. Based on the presence or absence of membranous staining, PODXL expression was dichotomised into low (0–2) or high (3–4).
For immunohistochemical analysis of EGFR, TMA-sections were automatically pre-treated using the PT-link system (DAKO, Glostrup, Denmark) and then stained in an Autostainer Plus (DAKO, Glostrup, Denmark) with the monoclonal antibody 31G7 (Zymed Laboratories Inc, San Francisco, CA, USA, diluted 1:25). For validatory purposes, TMAs from cohort 1 were also stained with the primary anti-EGFR antibody 3C6 (Ventana Medical Systems, Tucson, AZ, USA), stained in a BenchMark ULTRA (Ventana Medical Systems). Antigen retrieval was performed with protease1 (Ventana Medical Systems) for 8 min and antibody incubation time was 32 min in 36 °C. EGFR expression was determined by the intensity (0–3) of membranous staining in tumour cells in line with the scoring protocol proposed by Goldstein [21]. A score of 0–1 was considered low EGFR expression, and 2–3 indicated high EGFR expression.
Assessment of PODXL and EGFR expression was performed in the same way for all three cohorts by two independent observers (AL and KJ). Interobserver differences were discussed in order to reach consensus.
Analysis of KRAS and BRAF mutation status
The PyroMark Q24 system (Qiagen GmbH, Hilden, Germany) was used for pyrosequencing analysis of KRAS and BRAF mutations in DNA from 1 mm formalin-fixed, paraffin-embedded or fresh frozen tumour tissue cores taken from areas with >90 % tumour cells, as previously described [35]. In brief, DNA was isolated from tumour tissue using QIAamp MinElute spin columns (Qiagen) and DNA regions of interest were PCR-amplified (Veriti 96-Well Fast Thermal Cycler, Applied Biosystems Inc., Foster City, CA, USA).
Detection of mutations in KRAS codons 12 and 13 was performed using Therascreen KRAS Pyro Kit (Qiagen). Analysis of BRAF mutation hotspots in codons 600 and 601 was performed using previously published PCR primers (Richman, JCO 2009) and a novel BRAF sequencing primer (5′-TGATTTTGGTCTAGCTACA-3′) which was designed using the PyroMark Assay Design 2.0 software (Qiagen). All samples with a potential low-level mutation were re-analysed.
Cell culture
CRC cell lines Caco-2, RKO, SW480, SW620, HCT-116 and HT-29 were used and maintained in a humified atmosphere at 37 °C and 5 % carbon dioxide/95 % air. Caco-2 and RKO cell lines were grown in EMEM supplemented with 2 mM l-glutamine, 10 % FBS (fetal bovine serum) and 1XPEST (penicillin 90 IU/ml and streptomycin 90 µg/ml). SW480 and SW620 were maintained in DMEM with 4 mM l-glutamine, 4500 mg/L glucose, 1 mM sodium pyruvate, 10 % FBS and 1XPEST, while HCT-116 and HT-29 were grown in McCoy’s 5A medium supplemented with 1.5 mM l-glutamine, 2.2 g/L sodium bicarbonate, 10 % FBS and 1XPEST.
Western blot
The levels of EGFR and PODXL in cell lines were determined by western blot. To check for basal levels of the proteins, cells were harvested when still sub-confluent. For western blot analysis, cells were trypsinised and washed in PBS. Cell pellets were kept at −80 °C for at least 24 h after which they were lysed in RIPA buffer (Sigma-Aldrich Co, St Louis, MO, cat #R0278: 150 mM NaCl, 1.0 % IGEPAL® CA-630, 0.5 % sodium deoxycholate, 0.1 % SDS, 50 mM Tris, pH 8.0), supplemented with protease inhibitors and phos-STOP (Roche, Basel, Switzerland). Lysates were centrifuged at 4 °C for 10 min at 6200 g and supernatants collected. Protein concentrations were determined using the BCA Protein Assay (Thermo Scientific, Rockford, IL). Twenty µg of protein was separated on 4–15 % graded Mini-PROTEAN® TGX™ precast gels (Bio-Rad Laboratories Inc. Hercules, CA). For PODXL blots, an XCell SureLock® Mini-Cell (Thermo Scientific, Rockford, IL) wet transfer system was used (25 V for 2 h) with a transfer buffer containing 25 mM Tris base, 192 mM glycine, 20 % ethanol at a pH of 8.3. PVDF membranes were activated for 30 s in 99.5 % ethanol before transfer sandwich assembly. For EGFR blots, the Trans-Blot® Turbo™ Mini PVDF Transfer packs were used together with the Trans-Blot® Turbo™ Transfer System (Bio-Rad Laboratories Inc. Hercules, CA). Membranes were subsequently blocked with 5 % non-fat dry milk in TBS-tween 0.1 % and probed with the following antibodies overnight: EGFR (Cell Signaling Inc., Danvers, MA, cat#4267, dilution 1:1000), PODXL (Atlas Antibodies, cat#HPA002110, 1:500) and actin (Santa Cruz Biotechnology Inc., Dallas, TX, cat#sc-1616, 1:1000). Three sets of lysates were prepared and blotted for every cell line, and one representative experiment is shown.
Cell pellet array and immunocytochemistry
Cell lines were trypsinised and washed in PBS. Subsequently, cell pellets were fixed in formalin for at least 24 h followed by staining with Mayer’s haematoxylin for 5 min. Cells were washed once in PBS and dehydrated in graded ethanol series after which cell pellets were washed in molten paraffin several times. Cell pellets were arrayed in duplicate or triplicate 1.0 mm cores using a semi-automated arraying device (TMArrayer; Pathology Devices, Inc., Westminster, MD). Immunohistochemistry was performed on 5 µm sections using the same antibodies as for western blot with the following dilutions: EGFR, 1:100 and PODXL, 1:500. Images were captured at 20X using the cellSens entry software (version 1.8, Olympus).
Statistical analysis
The Chi square test was applied for comparison of PODXL expression with EGFR expression and molecular characteristics, and for comparison of EGFR expression with established prognostic clinicopathological factors. Kaplan–Meier analysis and log rank test were applied to illustrate differences in 5-year OS according to PODXL and EGFR expression. Cox’s proportional hazards regression was used for estimation of hazard ratios (HR) for death from CRC within 5 years, according to PODXL and EGFR expression in both univariable and multivariable analysis adjusted for age, sex, T-, N-, M-stage, differentiation grade and vascular invasion. Co-variables were entered into the multivariable analysis using backward selection where a p value of 0.05 decided entry and a p-value of 0.10 was used for removal. All tests were two-sided. A p value of 0.05 was considered significant. All statistical analyses were performed using SPSS version 20.0 (SPSS Inc, Chicago, IL).
Results
Overexpression of PODXL is associated with EGFR expression and BRAF mutation in colorectal cancer
EGFR could be analysed in 533/626 (85.1 %) cases in cohort 1, 259/270 (95.9 %) cases in cohort 2, and 310/337 (92.0 %) cases in cohort 3. Sample immunohistochemical images are shown in Fig. 1.
Fig. 1.

Sample immunohistochemical images. Immunohistochemical image of a colorectal tumour with high expression of both EGFR and PODXL. Note the subset of infiltrative cells with particularly strong membranous expression of both proteins
The intercorrelation between PODXL and other investigative factors are shown in Table 1. High expression of PODXL was significantly associated with high EGFR expression (p < 0.001) in all three cohorts, and with BRAF mutation (p < 0.001) and MSI (p < 0.001 and p = 0.021 respectively) in cohort 1 and 3. There was no significant correlation between PODXL expression and KRAS mutation (cohort 1 and 3). Information on BRAF status, MSI and KRAS mutation was not available in cohort 2.
Table 1.
Associations between PODXL expression and EGFR expression, BRAF, KRAS mutations and MSI status in colorectal cancer
| PODXL | Cohort 1 | p value | Cohort 2 | p value | Cohort 3 | p value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 0 | 1 | 2 | 3 | 4 | 0 | 1 | 2 | 3 | 4 | ||||
| N (%) | 268 (50.0) | 186 (34.7) | 10 (1.9) | 71 (13.2) | 1 (0.2) | 137 (50.7) | 67 (24.8) | 31 (11.5) | 25 (9.3) | 0 (0.0) | 197 (56.3) | 65 (18.6) | 29 (8.3) | 22 (6.3) | 3 (0.9) | |||
| EGFR | ||||||||||||||||||
| Low | 225 (87.5) | 141 (77.9) | 7 (70.0) | 34 (48.6) | 0 (0.0) | <0.001 | 106 (80.3) | 55 (84.6) | 22 (73.3) | 12 (50.0) | 0 (0.0) | 0.008 | 171 (89.5) | 55 (85.9) | 21 (72.4) | 10 (47.6) | 0 (0.0) | <0.001 |
| High | 32 (12.5) | 40 (22.1) | 3 (30.0) | 36 (51.4) | 1 (100.0) | 26 (19.7) | 10 (15.4) | 8 (26.7) | 12 (50.0) | 0 (0.0) | 20 (10.5) | 9 (14.1) | 8 (27.6) | 11 (52.4) | 3 (100.0) | |||
| Missing | 11 | 5 | 1 | 5 | 2 | 1 | 1 | 6 | 1 | 1 | ||||||||
| BRAF status | ||||||||||||||||||
| Wt | 227 (90.8) | 153 (85.5) | 4 (40.0) | 45 (70.3) | 0 (0.0) | <0.001 | N/A | N/A | N/A | N/A | N/A | 123 (89.8) | 33 (78.6) | 12 (60.0) | 13 (81.3) | 0 (0.0) | <0.001 | |
| Mutated | 23 (9.2) | 26 (14.5) | 6 (60.0) | 19 (29.7) | 1 (100.0) | N/A | N/A | N/A | N/A | N/A | 14 (10.2) | 9 (21.4) | 8 (40.0) | 3 (18.7) | 2 (100.0) | |||
| Missing | 18 | 7 | 7 | 60 | 23 | 9 | 6 | 1 | ||||||||||
| KRAS status | ||||||||||||||||||
| Wt | 153 (61.0) | 118 (65.9) | 9 (90.0) | 41 (64.1) | 1 (100.0) | 0.312 | N/A | N/A | N/A | N/A | N/A | 75 (58.1) | 29 (70.7) | 17 (85.0) | 8 (57.1) | 2 (100.0) | 0.095 | |
| Mutated | 98 (39.0) | 61 (34.1) | 1 (10.0) | 23 (35.9) | 0 (0.0) | N/A | N/A | N/A | N/A | N/A | 54 (41.9) | 12 (29.3) | 3 (15.0) | 6 (42.9) | 0 (0.0) | |||
| Missing | 17 | 7 | 7 | 68 | 24 | 9 | 8 | 1 | ||||||||||
| MSI status | ||||||||||||||||||
| MSS | 227 (88.7) | 146 (84.9) | 0 (0.0) | 52 (80.0) | 1 (100.0) | <0.001 | N/A | N/A | N/A | N/A | N/A | 170 (86.7) | 51 (81.0) | 20 (69.0) | 22 (100.0) | 2 (66.7) | 0.021 | |
| MSI | 29 (11.3) | 26 (15.1) | 9 (100.0) | 13 (20.0) | 0 (0.0) | N/A | N/A | N/A | N/A | N/A | 26 (13.3) | 12 (19.0) | 9 (31.0) | 0 (0.0) | 1 (33.3) | |||
| Missing | 12 | 14 | 1 | 6 | 1 | 2 | ||||||||||||
Associations of EGFR expression with established clinicopathological and investigative factors are shown in Table 2. In cohort 1 and 3, high EGFR expression was significantly associated with more advanced T-, N-, M-stage, low differentiation grade and vascular invasion. In cohort 2, there was a significant association between high EGFR expression and M-stage. Furthermore, high EGFR expression was significantly associated with BRAF mutation in cohort 1 (p < 0.001).
Table 2.
Associations of EGFR status with clinicopathological characteristics in three independent CRC patient cohorts
| EGFR | Cohort 1 | p value | Cohort 2 | p value | Cohort 3 | p value | |||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | ||||
| N (%) | 419 (78.6) | 114 (21.4) | 202 (78.0) | 57 (21.1) | 259 (83.5) | 51 (16.5) | |||
| Age | |||||||||
| Mean, median | 70.6, 71.4 | 69.9, 71.3 | 0.439 | 72.6, 73.5 | 70.7, 73.1 | 0.401 | 71.1, 73.0 | 71.5, 74.0 | 0.977 |
| Range | 51.3–85.6 | 49.8–83.5 | 37.6–92.1 | 37.8–93.3 | 36.0–94.0 | 50.0–87.0 | |||
| Sex | |||||||||
| Female | 219 (52.3) | 62 (54.4) | 0.688 | 98 (48.5) | 34 (59.6) | 0.138 | 129 (49.8) | 29 (56.9) | 0.358 |
| Male | 200 (47.7) | 52 (45.6) | 104 (51.5) | 23 (40.4) | 130 (50.2) | 22 (43.1) | |||
| Location | |||||||||
| Colon | 258 (61.9) | 76 (66.7) | 0.348 | 161 (79.7) | 48 (85.7) | 0.311 | 177 (68.3) | 30 (58.8) | 0.188 |
| Rectum | 159 (38.1) | 38 (33.3) | 41 (20.3) | 8 (14.3) | 82 (31.7) | 21 (41.2) | |||
| Missing | 2 | ||||||||
| T-stage | |||||||||
| T1 | 43 (10.9) | 1 (0.9) | <0.001 | 14 (7.1) | 4 (7.1) | 0.432 | 11 (4.2) | 0 (0.0) | 0.002 |
| T2 | 55 (13.9) | 7 (6.2) | 53 (26.8) | 11 (19.6) | 38 (14.7) | 1 (2.0) | |||
| T3 | 249 (62.9) | 75 (67.0) | 107 (54.0) | 33 (58.9) | 169 (65.3) | 37 (72.5) | |||
| T4 | 49 (12.4) | 29 (25.9) | 24 (14.3) | 8 (14.3) | 41 (15.8) | 13 (25.5) | |||
| Missing | 23 | 2 | 4 | 1 | |||||
| N-stage | |||||||||
| N0 | 232 (61.7) | 48 (43.2) | 0.002 | 129 (64.8) | 33 (61.1) | 0.331 | 164 (63.39 | 18 (35.3) | <0.001 |
| N1 | 87 (23.1) | 34 (30.6) | 50 (25.1) | 12 (22.2) | 53 (20.5) | 9 (17.6) | |||
| N2 | 57 (15.2) | 29 (26.1) | 20 (10.1) | 9 (16.7) | 42 (16.2) | 24 (47.1) | |||
| Missing | 43 | 3 | 3 | 3 | |||||
| M-stage | |||||||||
| M0 | 349 (84.5) | 82 (72.6) | 0.003 | 180 (89.6) | 40 (72.7) | 0.002 | 232 (89.6) | 39 (76.5) | 0.010 |
| M1 | 64 (15.5) | 31 (27.4) | 21 (10.4) | 15 (27.3) | 27 (10.4) | 12 (23.5) | |||
| Missing | 6 | 1 | 1 | 2 | |||||
| Diff. grade | |||||||||
| High | 28 (6.8) | 4 (3.6) | <0.001 | 16 (7.9) | 5 (8.8) | 0.402 | 8 (3.1) | 1 (2.0) | 0.006 |
| Intermediate | 310 (75.4) | 62 (55.4) | 138 (68.3) | 34 (59.6) | 202 (78.0) | 31 (60.8) | |||
| Low | 73 (17.8) | 46 (41.1) | 48 (23.8) | 18 (31.6) | 49 (18.9) | 19 (37.3) | |||
| Missing | 8 | 2 | |||||||
| Vasc. invasion | |||||||||
| No | 125 (53.2) | 27 (34.2) | 0.003 | 108 (54.8) | 24 (44.4) | 0.177 | 233 (90.0) | 38 (74.5) | 0.002 |
| Yes | 110 (46.8) | 52 (65.8) | 89 (45.2) | 30 (55.6) | 26 (10.0) | 13 (25.5) | |||
| Missing | 184 | 35 | 5 | 3 | |||||
| MSI status | |||||||||
| MSS | 333 (85.4) | 85 (81.0) | 0.266 | N/A | N/A | 214 (82.9) | 47 (92.2) | 0.098 | |
| MSI | 57 (14.6) | 20 (19.0) | N/A | N/A | 44 (17.1) | 4 (7.8) | |||
| Missing | 29 | 9 | 1 | ||||||
| KRAS status | |||||||||
| Wild-type | 253 (63.9) | 66 (61.1) | 0.596 | N/A | N/A | 108 (63.5) | 24 (70.6) | 0.433 | |
| Mutated | 119 (36.1) | 42 (38.9) | N/A | N/A | 62 (36.5) | 10 (29.4) | |||
| Missing | 23 | 6 | 89 | 17 | |||||
| BRAF status | |||||||||
| Wild-type | 348 (88.1) | 78 (72.2) | <0.001 | N/A | N/A | 152 | 27 | 0.147 | |
| Mutated | 47 (11.9) | 30 (27.8) | N/A | N/A | 27 | 9 | |||
| Missing | 24 | 6 | 80 | 15 | |||||
Kruskal–Wallis or Mann–Whitney U test applied for continuous variables
MSI microsatellite instability; MSS microsatellite stable
As further shown in Additional file 2, there was a very good correlation between EGFR protein expression assessed by both antibodies in cohort 1.
Overexpression of EGFR is associated with a poor prognosis, in particular in combination with PODXL overexpression
Kaplan–Meier analysis showed that high EGFR expression, in particular in combination with high PODXL expression, correlated with a reduced overall survival in all three cohorts (Fig. 2). As shown in Table 3, high EGFR expression was an independent predictor of a reduced 5-year OS in cohort 1 (HR 1.77; 95 % CI 1.27–2.46), cohort 2 (HR 1.58; 95 % CI 1.05–2.38) and cohort 3 (HR 1.83; 95 % CI 1.19–2.81). The highest risk of death within 5 years was observed in patients with tumours displaying high expression of both EGFR and PODXL in cohort 1 and 3 (unadjusted HR 1.97; 95 % CI 1.18–3.28 and HR 3.56; 95 % CI 1.75–7.22, respectively), remaining significant in adjusted analysis in cohort 3 (HR 3.71; 95 % CI 1.23–11.20). P values for term of interaction between EGFR and PODXL in cohort 1, 2 and 3 were 0.114, 0.690 and 0.147 respectively. In cohort 2, high PODXL expression was an independent prognostic factor in patients with tumours displaying low but not high EGFR expression (unadjusted HR 2.02; 95 % CI 1.02–4.02) and adjusted HR 2.59; 95 % CI 1.26–5.31).
Fig. 2.

Kaplan–Meier analysis. Kaplan-Meier estimates of 5-year OS according to combinations of PODXL and EGFR expression in cohort 1 (a), cohort 2 (b) and cohort 3 (c). Log rank p values correspond to pairwise comparisons of colorectal tumours with low expression of PODXL and EGFR with the other strata
Table 3.
Cox regression analysis of relative risks of death within 5 years according to EGFR and PODXL expression in colorectal cancer
| Cohort 1 | Cohort 2 | Cohort 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HR (95 % CI) | HR (95 % CI) | N (events) | HR (95 % CI) | HR (95 % CI) | N (events) | HR (95 % CI) | HR (95 % CI) | N (events) | |
| Unadjusted | Adjusted | Unadjusted | Adjusted | Unadjusted | Adjusted | ||||
| All | |||||||||
| EGFR low | 1.00 | 1.00 | 419 (146) | 1.00 | 1.00 | 202 (101) | 1.00 | 1.00 | 259 (88) |
| EGFR high | 1.82 (1.35–2.46) | 1.77 (1.27–2.46) | 114 (61) | 1.98 (1.39–2.84) | 1.58 (1.05–2.38) | 57 (43) | 2.60 (1.74–3.89) | 1.83 (1.19–2.81) | 51 (33) |
| All | |||||||||
| PODXL low | 1.00 | 1.00 | 464 (168) | 1.00 | 1.00 | 235 (123) | 1.00 | 1.00 | 291 (103) |
| PODXL high | 1.73 (1.21–2.46) | 1.16 (0.77–1.73) | 72 (38) | 2.27 (1.43–3.62) | 1.91 (1.11–3.27) | 25 (21) | 3.45 (2.13–5.58) | 1.23 (0.68–2.22) | 25 (20) |
| EGFR low | |||||||||
| PODXL low | 1.00 | 1.00 | 373 (128) | 1.00 | 1.00 | 183 (89) | 1.00 | 1.00 | 247 (82) |
| PODXL high | 1.06 (0.58–1.91) | 0.87 (0.45–1.66) | 34 (12) | 2.02 (1.02–4.02) | 2.59 (1.26–5.31) | 12 (9) | 1.62 (0.65–3.99) | 0.74 (0.28–1.95) | 10 (5) |
| EGFR high | |||||||||
| PODXL low | 1.00 | 1.00 | 75 (35) | 1.00 | 1.00 | 44 (31) | 1.00 | 1.00 | 37 (19) |
| PODXL high | 1.97 (1.18–3.28) | 1.38 (0.71–2.68) | 37 (26) | 1.60 (0.80–3.21) | 1.69 (0.74–3.84) | 12 (11) | 3.56 (1.75–7.22) | 3.71 (1.23–11.20) | 14 (14) |
Cohort 1 and 2 adjusted for age at surgery, sex, PODXL, EGFR, T-, N-, M-stage, differentiation grade and vascular invasion in multivariable analysis
Cohort 3 adjusted for age at surgery, sex, PODXL, EGFR, T-, N-, M-stage, differentiation grade, vascular invasion and neural invasion in multivariable analysis
PODXL and EGFR levels in colorectal cancer cell lines
As shown in Fig. 3, western blot analysis demonstrated that all cell lines with expression of PODXL also expressed EGFR, whereas the cell lines without PODXL expression did not. In cell lines derived from the same patient, EGFR and PODXL were expressed in the primary tumour cell line (SW480), but not in the metastatic derivative (SW620).
Fig. 3.
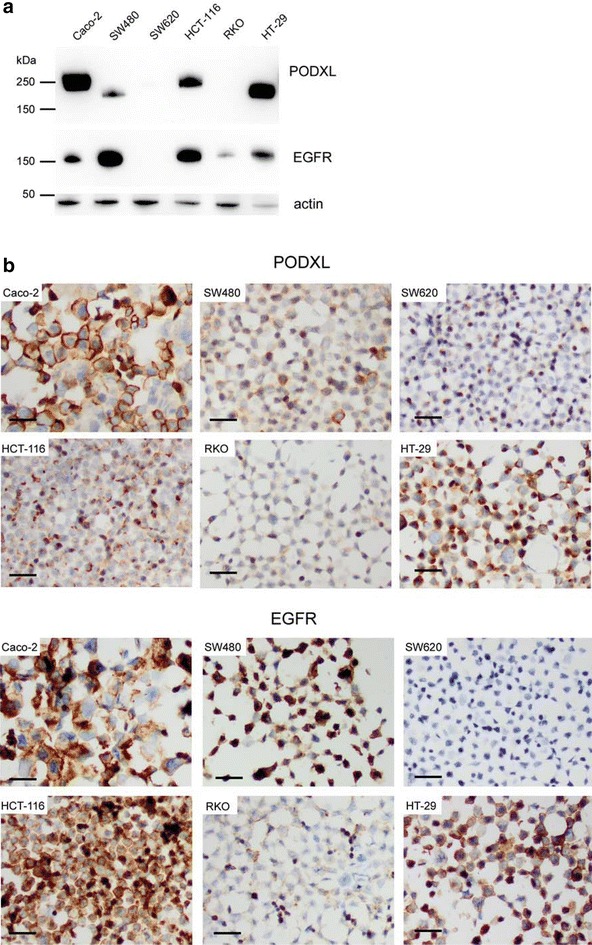
Western blot and immunocytochemical analysis of EGFR and PODXL in CRC cells. a Western blot and b immunocytochemical analysis of PODXL and EGFR protein levels in six different CRC cell lines; Caco-2, SW480, SW620, HCT-116, RKO and HT-29
Discussion
Chromosome 7 harbours several genes of importance in CRC, e.g. EGFR, PODXL and BRAF [36]. The results from the present study, based on analyses of tumours from more than 1100 patients, demonstrate, for the first time, strong significant associations between high protein expression of PODXL and EGFR in CRC. In the two largest examined cohorts, where data on BRAF mutation and MSI status was available for the majority of cases, PODXL expression was also found to correlate with BRAF mutation and MSI. The correlation between PODXL and EGFR was further demonstrated in vitro, with the proteins being uniformly expressed in six different CRC cell lines.
Moreover, the results from this study, based on immunohistochemical analysis of tumours from three independent patient cohorts, demonstrate that high EGFR protein expression is an independent negative prognostic factor in CRC patients. Despite diverging results in the literature regarding the prognostic significance of EGFR, these findings are in line with several previous studies, wherein EGFR protein expression has been associated with advanced disease stage [37, 38] and poor survival [39–42], hence adding further weight to the feasibility of EGFR as a negative prognostic biomarker in CRC.
Of note, the herein used anti-EGFR antibody was validated against another antibody, showing high concordance. These antibodies from Zymed and Ventana have also been demonstrated to perform better than others [43].
Expression of PODXL has previously been shown to be an independent adverse prognostic factor in all three herein investigated cohorts [7, 15]. In cohort 1 and 3, the worst prognosis was seen in patients with tumours displaying high expression of both EGFR and PODXL. This observation indicates that there may be a synergistic adverse prognostic effect of PODXL and EGFR, even if there was no significant interaction. Moreover, the results from cohort 2 differed somewhat in that high EGFR expression was an independent prognostic factor in PODXL low, but not high, tumours. Of note, in cohort 2, with exception for M-stage, there were no significant associations of EGFR expression with established unfavourable clinicopathological factors, whereas in cohort 1 and 3, high EGFR expression was significantly associated with more advanced T-, N- and M-stage, low differentiation grade and vascular invasion. In contrast to EGFR, high PODXL expression was found to correlate with more advanced N-stage, low differentiation grade and vascular invasion in cohort 2 [15].
Management of CRC has improved immensely over the past decades due to refined surgical techniques and optimal use of chemotherapy. The introduction of targeted therapies including bevacizumab and anti-EGFR antibodies has further improved outcome for patients with metastatic CRC. Disease stage is still the strongest prognostic factor, however, in this era of personalised medicine, new classification systems based on prognostic and predictive markers are needed to select the most efficient treatment for patients. In recent years, different research groups have proposed new classification systems based on gene expression in colorectal tumours [44–46]. Independently, all groups have indentified one subtype that is associated with EMT, poor differentiation and unfavorable prognosis. EMT is considered a critical step in the progression to metastasis, and PODXL plays an important role in this process [18].
KRAS, NRAS and BRAF mutation status is used as predictive markers for response to treatment with monoclonal anti-EGFR antibodies. However, only approximately 40 % of CRC patients with tumours wild-type for KRAS, NRAS and BRAF benefit from such therapy [47–50], and patients who initially respond eventually become resistant to these drugs. Several potential mechanisms of acquired resistance to anti-EGFR drugs have been proposed, one of them being EMT. Evidence suggests that EGFR signalling can trigger EMT [51], but once EMT is established, signalling associated with EGFR activation is reduced [52]. Moreover, studies on non-small cell lung cancer (NSCLC) and CRC cell lines have shown that tumour cells that have undergone EMT are much less sensitive to anti-EGFR treatment [53, 54]. In a study by Buck et al., CRC cell lines derived from the same patient showed epithelial characteristics and sensitivity to EGFR tyrosine kinase inhibitor erlotinib in cells from the primary tumour, whereas tumour cells from the liver metastasis exhibited a mesenchymal phenotype and were not sensitive to erlotinib [54]. Interestingly, in our study, using the same cell lines, EGFR and PODXL were expressed in cells from the primary tumour, but not in the metastatic cell line. Thus, EMT, and possibly PODXL, may have a role in resistance to anti-EGFR drugs by activating alternative signalling pathways.
Moreover, previous in vitro studies have shown that expression of PODXL leads to recruitment of the Na +/H + Exchanger Regulatory Factor (NHERF) proteins to the apical domain of the epithelial cell [55]. NHERF-1 in turn has been shown to stabilise EGFR at the cell surface to restrict receptor downregulation, thus enhancing EGFR signalling [56]. Based on these results it would be of interest to investigate whether PODXL may affect the response to monoclonal anti-EGFR antibody therapy by limiting the downregulation of EGFR receptors through NHERF-1.
Conclusions
The results from this study demonstrate that high expression of EGFR is an independent factor of poor prognosis in CRC. Moreover, strong links have been uncovered between expression of the recently proposed biomarker candidate PODXL with EGFR expression in CRC in vivo and in vitro, and with BRAF mutation in vivo. High expression of both PODXL and EGFR may also have a synergistic adverse effect on survival. Taken together, these findings suggest a functional link in CRC between PODXL, EGFR and BRAF, all originating from chromosome 7. Future in-depth studies are warranted to further elucidate the mechanistic basis underlying these observations, which may be highly relevant in the clinical setting.
Authors’ contributions
AL performed the immunohistochemical scoring, statistical analysis and drafted the manuscript. BN constructed the TMAs, carried out the immunohistochemical stainings, and performed the mutation analyses. SL carried out the in vitro studies. MS designed the BRAF mutation assay and performed the mutation analyses together with EKa. EKu carried out the immunohistochemical stainings. HB is principal investigator of cohort 3. SW and JE collected clinical data in cohort 1. MU contributed with reagents and antibody validation. KJ conceived of the study and participated in its design and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Cancer Society, the Mrs Berta Kamprad Foundation, the Gunnar Nilsson Cancer Foundation, the Swedish Government Grant for Clinical Research, Lund University Faculty of Medicine and University Hospital Research Grants.
Competing interests
KJ is inventor on a patent related to the use of PODXL as a prognostic biomarker in CRC. The other authors declare no competing interests.
Additional files
10.1186/s12967-016-0882-0 Patient and tumour characteristics in the three cohorts.
10.1186/s12967-016-0882-0 Comparison of EGFR protein expression in CRC assessed by two different antibodies in cohort 1.
Contributor Information
Anna H. Larsson, Email: anna_h.larsson@med.lu.se
Sophie Lehn, Email: sophie.lehn@med.lu.se.
Sakarias Wangefjord, Email: sakarias.wangefjord@med.lu.se.
Emelie Karnevi, Email: emelie.karnevi@med.lu.se.
Eugenia Kuteeva, Email: eugenia.kuteeva@atlasantibodies.se.
Magnus Sundström, Email: magnus.sundstrom@igp.uu.se.
Björn Nodin, Email: bjorn.nodin@med.lu.se.
Mathias Uhlén, Email: mathias.uhlen@scilifelab.se.
Jakob Eberhard, Email: jakob.eberhard@med.lu.se.
Helgi Birgisson, Email: helgi.birgisson@surgsci.uu.se.
Karin Jirström, Email: karin.jirstrom@med.lu.se.
References
- 1.Sassetti C, Tangemann K, Singer MS, Kershaw DB, Rosen SD. Identification of podocalyxin-like protein as a high endothelial venule ligand for L-selectin: parallels to CD34. J Exp Med. 1998;187(12):1965–1975. doi: 10.1084/jem.187.12.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerjaschki D, Sharkey DJ, Farquhar MG. Identification and characterization of podocalyxin–the major sialoprotein of the renal glomerular epithelial cell. J Cell Biol. 1984;98(4):1591–1596. doi: 10.1083/jcb.98.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvat R, Hovorka A, Dekan G, Poczewski H, Kerjaschki D. Endothelial cell membranes contain podocalyxin–the major sialoprotein of visceral glomerular epithelial cells. J Cell Biol. 1986;102(2):484–491. doi: 10.1083/jcb.102.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyonnas R, Nielsen JS, Chelliah S, Drew E, Hara T, Miyajima A, McNagny KM. Podocalyxin is a CD34-related marker of murine hematopoietic stem cells and embryonic erythroid cells. Blood. 2005;105(11):4170–4178. doi: 10.1182/blood-2004-10-4077. [DOI] [PubMed] [Google Scholar]
- 5.Somasiri A, Nielsen JS, Makretsov N, McCoy ML, Prentice L, Gilks CB, Chia SK, Gelmon KA, Kershaw DB, Huntsman DG, et al. Overexpression of the anti-adhesin podocalyxin is an independent predictor of breast cancer progression. Cancer Res. 2004;64(15):5068–5073. doi: 10.1158/0008-5472.CAN-04-0240. [DOI] [PubMed] [Google Scholar]
- 6.Casey G, Neville PJ, Liu X, Plummer SJ, Cicek MS, Krumroy LM, Curran AP, McGreevy MR, Catalona WJ, Klein EA, et al. Podocalyxin variants and risk of prostate cancer and tumor aggressiveness. Hum Mol Genet. 2006;15(5):735–741. doi: 10.1093/hmg/ddi487. [DOI] [PubMed] [Google Scholar]
- 7.Larsson A, Johansson ME, Wangefjord S, Gaber A, Nodin B, Kucharzewska P, Welinder C, Belting M, Eberhard J, Johnsson A, et al. Overexpression of podocalyxin-like protein is an independent factor of poor prognosis in colorectal cancer. Br J Cancer. 2011;105(5):666–672. doi: 10.1038/bjc.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cipollone JA, Graves ML, Kobel M, Kalloger SE, Poon T, Gilks CB, McNagny KM, Roskelley CD. The anti-adhesive mucin podocalyxin may help initiate the transperitoneal metastasis of high grade serous ovarian carcinoma. Clin Exp Metastasis. 2012;29(3):239–252. doi: 10.1007/s10585-011-9446-0. [DOI] [PubMed] [Google Scholar]
- 9.Hsu YH, Lin WL, Hou YT, Pu YS, Shun CT, Chen CL, Wu YY, Chen JY, Chen TH, Jou TS. Podocalyxin EBP50 ezrin molecular complex enhances the metastatic potential of renal cell carcinoma through recruiting Rac1 guanine nucleotide exchange factor ARHGEF7. Am J Pathol. 2010;176(6):3050–3061. doi: 10.2353/ajpath.2010.090539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dallas MR, Chen SH, Streppel MM, Sharma S, Maitra A, Konstantopoulos K. Sialofucosylated podocalyxin is a functional E- and L-selectin ligand expressed by metastatic pancreatic cancer cells. Am J Physiol Cell Physiol. 2012;303(6):C616–C624. doi: 10.1152/ajpcell.00149.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binder ZA, Siu IM, Eberhart CG, Ap Rhys C, Bai RY, Staedtke V, Zhang H, Smoll NR, Piantadosi S, Piccirillo SG, et al. Podocalyxin-like protein is expressed in glioblastoma multiforme stem-like cells and is associated with poor outcome. PLoS One. 2013;8(10):e75945. doi: 10.1371/journal.pone.0075945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Yang L, Liao D, Chen Y, Wang W, Fang J. Podocalyxin regulates astrocytoma cell invasion and survival against temozolomide. Exp Ther Med. 2013;5(4):1025–1029. doi: 10.3892/etm.2013.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boman K, Larsson AH, Segersten U, Kuteeva E, Johannesson H, Nodin B, Eberhard J, Uhlen M, Malmstrom PU, Jirstrom K. Membranous expression of podocalyxin-like protein is an independent factor of poor prognosis in urothelial bladder cancer. Br J Cancer. 2013;108(11):2321–2328. doi: 10.1038/bjc.2013.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heby M, Elebro J, Nodin B, Jirstrom K, Eberhard J. Prognostic and predictive significance of podocalyxin-like protein expression in pancreatic and periampullary adenocarcinoma. BMC Clin Pathol. 2015;15:10. doi: 10.1186/s12907-015-0009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson A, Fridberg M, Gaber A, Nodin B, Leveen P, Jonsson G, Uhlen M, Birgisson H, Jirstrom K. Validation of podocalyxin-like protein as a biomarker of poor prognosis in colorectal cancer. BMC Cancer. 2012;12:282. doi: 10.1186/1471-2407-12-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsson AH, Nodin B, Syk I, Palmquist I, Uhlen M, Eberhard J, Jirstrom K. Podocalyxin-like protein expression in primary colorectal cancer and synchronous lymph node metastases. Diagn Pathol. 2013;8:109. doi: 10.1186/1746-1596-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaprio T, Fermer C, Hagstrom J, Mustonen H, Bockelman C, Nilsson O, Haglund C. Podocalyxin is a marker of poor prognosis in colorectal cancer. BMC Cancer. 2014;14:493. doi: 10.1186/1471-2407-14-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng X, Ezzati P, Wilkins JA. Requirement of podocalyxin in TGF-beta induced epithelial mesenchymal transition. PLoS One. 2011;6(4):e18715. doi: 10.1371/journal.pone.0018715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder KA, Hughes MR, Hedberg B, Brandon J, Hernaez DC, Bergqvist P, Cruz F, Po K, Graves ML, Turvey ME, et al. Podocalyxin enhances breast tumor growth and metastasis and is a target for monoclonal antibody therapy. Breast Cancer Res. 2015;17(1):46. doi: 10.1186/s13058-015-0562-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin CW, Sun MS, Liao MY, Chung CH, Chi YH, Chiou LT, Yu J, Lou KL, Wu HC. Podocalyxin-like 1 promotes invadopodia formation and metastasis through activation of Rac1/Cdc42/cortactin signaling in breast cancer cells. Carcinogenesis. 2014;35(11):2425–2435. doi: 10.1093/carcin/bgu139. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein NS, Armin M. Epidermal growth factor receptor immunohistochemical reactivity in patients with American Joint Committee on Cancer Stage IV colon adenocarcinoma: implications for a standardized scoring system. Cancer. 2001;92(5):1331–1346. doi: 10.1002/1097-0142(20010901)92:5<1331::AID-CNCR1455>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Spano JP, Fagard R, Soria JC, Rixe O, Khayat D, Milano G. Epidermal growth factor receptor signaling in colorectal cancer: preclinical data and therapeutic perspectives. Ann Oncol. 2005;16(2):189–194. doi: 10.1093/annonc/mdi057. [DOI] [PubMed] [Google Scholar]
- 23.Galizia G, Lieto E, Ferraraccio F, De Vita F, Castellano P, Orditura M, Imperatore V, La Mura A, La Manna G, Pinto M, et al. Prognostic significance of epidermal growth factor receptor expression in colon cancer patients undergoing curative surgery. Ann Surg Oncol. 2006;13(6):823–835. doi: 10.1245/ASO.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 24.Ljuslinder I, Melin B, Henriksson ML, Oberg A, Palmqvist R. Increased epidermal growth factor receptor expression at the invasive margin is a negative prognostic factor in colorectal cancer. Int J Cancer. 2011;128(9):2031–2037. doi: 10.1002/ijc.25559. [DOI] [PubMed] [Google Scholar]
- 25.Giralt J, de las Heras M, Cerezo L, Eraso A, Hermosilla E, Velez D, Lujan J, Espin E, Rosello J, Majo J, et al. The expression of epidermal growth factor receptor results in a worse prognosis for patients with rectal cancer treated with preoperative radiotherapy: a multicenter, retrospective analysis. Radiother Oncol. 2005;74(2):101–108. doi: 10.1016/j.radonc.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 26.Resnick MB, Routhier J, Konkin T, Sabo E, Pricolo VE. Epidermal growth factor receptor, c-MET, beta-catenin, and p53 expression as prognostic indicators in stage II colon cancer: a tissue microarray study. Clin Cancer Res. 2004;10(9):3069–3075. doi: 10.1158/1078-0432.CCR-03-0462. [DOI] [PubMed] [Google Scholar]
- 27.Yarom N, Jonker DJ. The role of the epidermal growth factor receptor in the mechanism and treatment of colorectal cancer. Discov Med. 2011;11(57):95–105. [PubMed] [Google Scholar]
- 28.Leung SP, Griffith OL, Masoudi H, Gown A, Jones S, Phang T, Wiseman SM. Clinical utility of type 1 growth factor receptor expression in colon cancer. Am J Surg. 2008;195(5):604–610. doi: 10.1016/j.amjsurg.2007.12.032. [DOI] [PubMed] [Google Scholar]
- 29.Van Cutsem E, Cervantes A, Nordlinger B, Arnold D. Group EGW: metastatic colorectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii1–iii9. doi: 10.1093/annonc/mdu260. [DOI] [PubMed] [Google Scholar]
- 30.Wangefjord S, Brandstedt J, Lindquist KE, Nodin B, Jirstrom K, Eberhard J. Associations of beta-catenin alterations and MSI screening status with expression of key cell cycle regulating proteins and survival from colorectal cancer. Diagn Pathol. 2013;8:10. doi: 10.1186/1746-1596-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandstedt J, Wangefjord S, Nodin B, Eberhard J, Sundstrom M, Manjer J, Jirstrom K. Associations of anthropometric factors with KRAS and BRAF mutation status of primary colorectal cancer in men and women: a cohort study. PLoS One. 2014;9(2):e98964. doi: 10.1371/journal.pone.0098964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birgisson H, Wallin U, Holmberg L, Glimelius B. Survival endpoints in colorectal cancer and the effect of second primary other cancer on disease free survival. BMC Cancer. 2011;11:438. doi: 10.1186/1471-2407-11-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hjelm B, Brennan DJ, Zendehrokh N, Eberhard J, Nodin B, Gaber A, Ponten F, Johannesson H, Smaragdi K, Frantz C, et al. High nuclear RBM3 expression is associated with an improved prognosis in colorectal cancer. Proteomics Clin Appl. 2011;5(11–12):624–635. doi: 10.1002/prca.201100020. [DOI] [PubMed] [Google Scholar]
- 34.Gaber A, Johansson M, Stenman UH, Hotakainen K, Ponten F, Glimelius B, Bjartell A, Jirstrom K, Birgisson H. High expression of tumour-associated trypsin inhibitor correlates with liver metastasis and poor prognosis in colorectal cancer. Br J Cancer. 2009;100(10):1540–1548. doi: 10.1038/sj.bjc.6605047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wangefjord S, Sundstrom M, Zendehrokh N, Lindquist KE, Nodin B, Jirstrom K, Eberhard J. Sex differences in the prognostic significance of KRAS codons 12 and 13, and BRAF mutations in colorectal cancer: a cohort study. Biol Sex Differ. 2013;4(1):17. doi: 10.1186/2042-6410-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Liang L, Fang JY, Xu J. Somatic gene copy number alterations in colorectal cancer: new quest for cancer drivers and biomarkers. Oncogene. 2015;35:2011–2019. doi: 10.1038/onc.2015.304. [DOI] [PubMed] [Google Scholar]
- 37.Abd El All HS, Mishriky AM, Mohamed FA. Epidermal growth factor receptor in colorectal carcinoma: correlation with clinico-pathological prognostic factors. Colorectal Dis. 2008;10(2):170–178. doi: 10.1111/j.1463-1318.2007.01306.x. [DOI] [PubMed] [Google Scholar]
- 38.Spano JP, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R, Attar A, Benichou J, Martin A, Morere JF, et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005;16(1):102–108. doi: 10.1093/annonc/mdi006. [DOI] [PubMed] [Google Scholar]
- 39.Galizia G, Lieto E, Ferraraccio F, De Vita F, Castellano P, Orditura M, Imperatore V, La Mura A, La Manna G, Pinto M, et al. Prognostic significance of epidermal growth factor receptor expression in colon cancer patients undergoing curative surgery. Ann Surg Oncol. 2006;13(6):823–835. doi: 10.1245/ASO.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 40.Ljuslinder I, Melin B, Henriksson ML, Oberg A, Palmqvist R. Increased epidermal growth factor receptor expression at the invasive margin is a negative prognostic factor in colorectal cancer. Int J Cancer. 2011;128(9):2031–2037. doi: 10.1002/ijc.25559. [DOI] [PubMed] [Google Scholar]
- 41.Resnick MB, Routhier J, Konkin T, Sabo E, Pricolo VE. Epidermal growth factor receptor, c-MET, beta-catenin, and p53 expression as prognostic indicators in stage II colon cancer: a tissue microarray study. Clin Cancer Res. 2004;10(9):3069–3075. doi: 10.1158/1078-0432.CCR-03-0462. [DOI] [PubMed] [Google Scholar]
- 42.Giralt J, de las Heras M, Cerezo L, Eraso A, Hermosilla E, Velez D, Lujan J, Espin E, Rossello J, Majó J, et al. The expression of epidermal growth factor receptor results in a worse prognosis for patients with rectal cancer treated with preoperative radiotherapy: a multicenter, retrospective analysis. Radiother Oncol. 2005;74(2):101–108. doi: 10.1016/j.radonc.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 43.Penault-Llorca F, Cayre A, Arnould L, Bibeau F, Bralet MP, Rochaix P, Savary J, Sabourin JC. Is there an immunohistochemical technique definitively valid in epidermal growth factor receptor assessment? Oncol Rep. 2006;16(6):1173–1179. [PubMed] [Google Scholar]
- 44.De Sousa EMF, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, de Jong JH, de Boer OJ, van Leersum R, Bijlsma MF, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 45.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA, Grotzinger C, Del Rio M, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12(6):594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 48.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 49.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 50.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 51.Barr S, Thomson S, Buck E, Russo S, Petti F, Sujka-Kwok I, Eyzaguirre A, Rosenfeld-Franklin M, Gibson NW, Miglarese M, et al. Bypassing cellular EGF receptor dependence through epithelial-to-mesenchymal-like transitions. Clin Exp Metastasis. 2008;25(6):685–693. doi: 10.1007/s10585-007-9121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clin Exp Metastasis. 2008;25(8):843–854. doi: 10.1007/s10585-008-9200-4. [DOI] [PubMed] [Google Scholar]
- 53.Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, Pham TQ, Soriano R, Stinson J, Seshagiri S, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11(24 Pt 1):8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 54.Buck E, Eyzaguirre A, Barr S, Thompson S, Sennello R, Young D, Iwata KK, Gibson NW, Cagnoni P, Haley JD. Loss of homotypic cell adhesion by epithelial-mesenchymal transition or mutation limits sensitivity to epidermal growth factor receptor inhibition. Mol Cancer Ther. 2007;6(2):532–541. doi: 10.1158/1535-7163.MCT-06-0462. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Li J, Straight SW, Kershaw DB. PDZ domain-mediated interaction of rabbit podocalyxin and Na(+)/H(+) exchange regulatory factor-2. Am J Physiol Renal Physiol. 2002;282(6):F1129–F1139. doi: 10.1152/ajprenal.00131.2001. [DOI] [PubMed] [Google Scholar]
- 56.Lazar CS, Cresson CM, Lauffenburger DA, Gill GN. The Na+/H+ exchanger regulatory factor stabilizes epidermal growth factor receptors at the cell surface. Mol Biol Cell. 2004;15(12):5470–5480. doi: 10.1091/mbc.E04-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]


