Abstract
Squamous cell lung cancer (SCC) represents an area of unmet need in lung cancer research. For the last several years, therapeutic progress in SCC has lagged behind the now more common NSCLC histologic subtype of adenocarcinoma. However, recent efforts to define the complex biology underlying SCC have begun to bear fruit in a multitude of ways, including characterization of previously unknown genomic and signaling pathways, delineation of new potentially actionable molecular targets, and subsequent development of a large number of agents directed against unique SCC-associated molecular abnormalities. For the first time, SCC-specific prognostic gene signatures and predictive biomarkers of new therapeutic agents are emerging. In addition, recent and ongoing clinical trials, including the Lung-MAP master protocol, have been designed to facilitate approval of targeted therapy-biomarker combinations. In this comprehensive review we describe the current status of SCC therapeutics, recent advances in the understanding of SCC biology and prognostic gene signatures, and the development of innovative new clinical trials, all of which offer new hope for patients with advanced SCC.
Introduction
Squamous cell carcinoma of the lung (SCC), formerly the most common histologic subtype of non-small cell lung cancer (NSCLC), has steadily fallen in incidence over the last few decades, largely attributed to decreased smoking rates and changes to cigarette composition and filtering, which favor adenocarcinoma histology (1). Nevertheless, lung SCC remains a common malignancy overall, accounting for approximately 85,000 new cases in the USA each year and over 400,000 worldwide. The great majority of patients with SCC are current or former heavy smokers, in contrast to adenocarcinoma, where a growing proportion are never-smokers or former light smokers. (2,3) SCC remains highly associated with cigarette smoking; it is therefore not surprising that recent efforts to genomically characterize lung cancer, such as those of The Cancer Genome Atlas (TCGA) and others, have demonstrated that in general, SCC reflects the genomic complexity and high overall mutational load expected from tobacco carcinogenesis. As described below, genomically-defined subsets of SCC have now been identified, some of which have therapeutic implications for a growing number of developing targeted agents. In a similar fashion, despite multiple studies, there are currently no universally accepted prognostic gene signatures upon which to gauge risk of recurrence and subsequent death, or need for adjuvant chemotherapy in post-surgical patients with SCC.
While therapy of early stage SCC mimics that of other histologic subtypes of NSCLC, therapeutic options for advanced stage SCC in comparison with lung adenocarcinoma, in part due to discovery of “druggable” oncogene targets in never-smoker subsets of adenocarcinoma, such as those with activating mutations in the epidermal growth factor receptor (EGFR) or anaplastic lymphoma kinase (ALK) gene rearrangements (4). As of this writing, there is still no FDA-approved targeted therapy for advanced SCC, in which a biomarker is utilized to select patients most likely to benefit. Instead, the standard of care for frontline palliative systemic therapy remains platinum-based doublet chemotherapy, a clinical scenario that has not changed considerably for nearly two decades.
Here we describe recent advances in the molecular profiling of SCC, ongoing work to establish reliable prognostic gene signatures in early stage SCC, and new therapeutic approaches to advanced stage disease. Finally, unique perspectives are offered on how these developments will impact clinical care for the SCC patient and ultimately enhance patient outcomes.
Genomics of Lung SCC
Recent comprehensive genomic surveys have defined the genomic and epigenomic alterations driving lung SCC. Prior to these studies little was known about SCC genomics. However, several reports using single platform methods such as gene expression profiling, Single Nucleotide Polymorphism (SNP) arrays and focused DNA sequencing showed that the genetic alterations defining lung adenocarcinomas and SCC were distinct, likely explaining the lack of efficacy of targeted therapeutic agents in SCC which had been applied successfully in lung adenocarcinomas.
Lung SCC is defined by a strong genomic signature of tobacco use with most cohorts reporting a rate of tobacco exposure in excess of 90%(5). SCC displays a somatic mutation rate and spectrum comparable to that of patients with small cell lung cancer or other smoking-related cancers and is dissimilar to lung adenocarcinoma in which cancers from non-smokers harbor one-fifth to one-sixth the genomic alterations of a smoker’s cancer(6–9). This homogeneity is evident on a worldwide basis, as most genomic studies of lung SCC performed by investigators from North America, Europe and Asia have identified similar spectra of genomic alterations in their patient populations and similar subclasses of SCC. Further, the genomic alterations in lung SCC are strikingly similar to those found in Human Papilloma Virus (HPV) negative head and neck cancers(10, 11). The high mutation rate in SCC is likely to result in expression of a large complement of tumor antigens, and many of these are in the process of being defined in the context of immunotherapy trials.
In lung adenocarcinoma much attention has been devoted to the concept of “driver oncogenes,” genomic alterations in kinase genes or other key mitogenic pathways which are required for ongoing tumor proliferation and on which the tumor is dependent. This concept has led to the clinical use of a number of kinase inhibitors in lung adenocarcinomas in genomically-selected patients and has improved outcomes for these individuals. In lung SCC recurrent alterations in kinase genes do not appear to be core genomic events with the most common genomic alterations being loss of TP53 and CDKN2A in the vast majority of cases(7–9). Other highly prevalent alterations that occur in a mutually exclusive manner are mutations of NFE2L2/KEAP1/CUL3, which activate a transcriptional program associated with response to oxidative stress, and truncating mutations of the NOTCH1 gene, a critical regulator of squamous cell differentiation(7, 8, 12, 13). SCCs of the lung and other organs are further defined by common amplification of 3q, a region containing SOX2, TP63 and PIK3CA and also by amplification of 7p11 and 8p12, regions harboring the EGFR and FGFR1 genes(3, 9, 14, 15). Highly recurrent tyrosine kinase mutations have not been reported in lung SCC, though mutations in FGFR2, FGFR3 and DDR2 have been described as potential therapeutic targets along with BAG4-FGFR1 and FGFR3-TACC3 fusions(16–19). Moreover, many SCC lung tumors display somatic alterations in one or more genes involved in PI3K/AKT signaling, though the functional consequences of many of these alterations remain unclear(7). Finally, genomic alterations in genes governing cellular immunity and immune evasion have been described including HLA-A, HLA-B, HLA-C, B2M, MICA, MICB, ULBP1 and ULBP2(7) (Table 1).
Table 1.
Commonly identified alterations in genomic studies of lung SCC
| Genetic Pathways and Alterations | Prevalence | Clinical Trials |
|---|---|---|
| RTK Amplification | >30% with EGFR and FGFR1 most common | EGFR mAbs, FGFR TKIs, FGFR mAbs, FGFR ligand traps |
| RTK mutations/fusions | Rare (<10% of cases), most common in FGFR2 and FGFR3 (FGFR3-TACC3), rare DDR2 mutations | FGFR TKIs, FGFR mAbs, FGFR ligand traps, dasatinib |
| RAS | 10–20%, most commonly loss of NF1 or RASA1, RAS mutations rare | MEK and ERK inhibitors, direct RAS inhibitors |
| PI3K | Common ~50% alterations in PIK3A, PTEN, PIK3R1 | PI3K and mTOR inhibitors |
| TP53 and CDKN2A/RB1 | Genomic loss in nearly all cases, amplification of CDK4/CDK6/CCND1 in CDKN2A intact tumors | CDK inhibitors? |
| Oxidative Stress Regulation | Common mutation of NFE2L2/KEAP1/CUL3 (25%) | PI3K inhibitors? |
| Differentiation | Common loss of NOTCH1; TP63 and SOX2 gain | ? |
| Immune evasion | Rare HLA and B2M mutations, <10% | Immune checkpoint inhibitors, vaccines |
FGFR kinases as genomically altered targets
With frequent focal amplification of FGFR1, recurrent activating mutations of FGFR2 and FGFR3 as well as FGFR1/3 fusion events the fibroblast growth factor receptor family represents the biggest and best studied class of “druggable” targets in lung SCC. Given the high recurrence of FGFR1 amplifications (10–15%) in lung SCC several groups have focused on the study of FGFR1 as a drug target in these tumors. A number of preclinical studies have shown that within the group of FGFR1-amplified SCC cell lines, a subgroup of cell lines is exquisitely sensitive to inactivation of FGFR1 signaling (3, 15). Consequently, in selected FGFR1-driven mouse xenograft models deactivation of FGFR1 leads to tumor shrinkage(20). Similar striking sensitivity to FGFR inhibition has been reported for a subset of tumors within the group of FGFR-mutant and FGFR-fusion positive samples in both pre-clinical and early clinical studies.
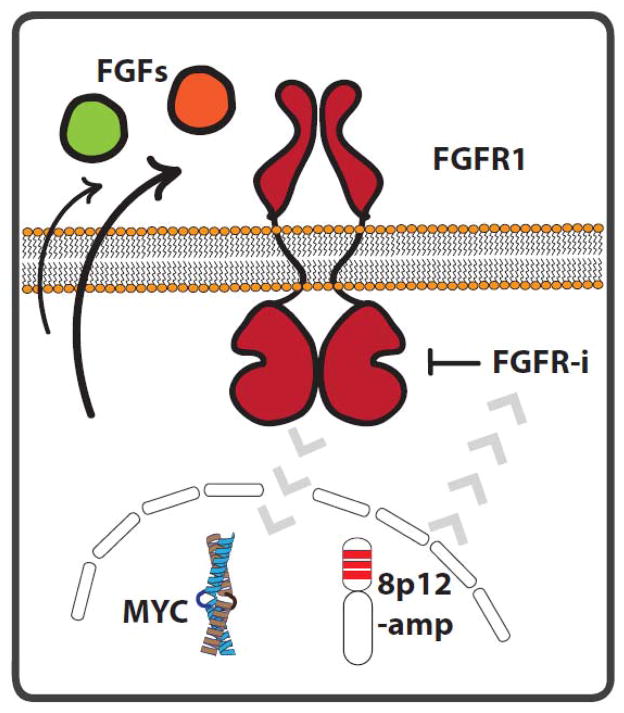
However, the modulators of FGFR1-dependency remain controversial and initial clinical data suggest that a minority of patients with FGFR1 amplification will derive clinical benefit from FGFR kinase inhibitors(21, 22). A number of studies have shown that the genomic pattern of the 8p12 locus amplifications is heterogeneous and that only a minority of tumors shows focal, high level amplification of FGFR1(3, 15, 23). The genomic complexity of the 8p12 locus together with the low resolution of routine FISH-based diagnostics for the detection of FGFR1 amplification might potentially lead to misclassification of tumors and subsequent underestimation of the activity of currently tested FGFR inhibitors. The difficulties with the precise determination of FGFR1 amplification status might also contribute to the fact that high mRNA and protein FGFR1 levels are found only in a subset of cells that are classified as FGFR1-amplified(3, 15, 24). This may be of importance as gene expression and protein levels of FGFR1 might correlate with the response rate to FGFR targeted drugs(24). Another source for modulators of FGFR1-dependency are FGFR ligands. It has been shown that FGFR1-amplified cells may be able to express and secrete a variety of FGFR ligands such as FGF-2 and FGF-9 that may be required to fully activate intracellular FGFR signaling(24, 25). An additional layer of complexity for the determination of FGFR1-dependency is the co-occurrence of FGFR1-amplifications with other genomic lesions such as MYC (Fig. 1). Recent evidence suggests that in FGFR1-amplified tumors high protein expression of the transcription factor MYC may be associated with pronounced response to FGFR inhibitors(23). However, a mechanistic link between the lineage-specific role of MYC in SCC tumors and FGFR1-dependency is currently missing.
Figure 1.
Schematic overview of potential modulators of cellular response to FGFR targeted drugs in FGFR1-amplified lung SCC. It has been shown that the chromosomal architecture of the 8p12 locus as well as the expression of c-MYC can modify (gray arrows) the cellular dependency on FGFR1 and therefore the efficacy of FGFR inhibitors in these tumors. Similarly, the secretion (black arrows) of FGF ligands (e.g. FGF2, FGF9) can perturb the activity of FGFR1 and modify the response to targeted inhibition of its kinase activity.
Analogous to other oncogenically driven lung tumors, feedback-loop mediated activation of resistance signaling may further complicate the ability to effectively treat patients with FGFR1-amplified tumors. Multiple studies have shown that EGFR and MET activation can facilitate adaptive resistance to FGFR inhibition in pre-clinical models (26–28). Overall, a major challenge for future initiatives will be the translation of the understanding of potential modulators of FGFR-dependency into routine clinical diagnostic for the enrichment of patients that might benefit from FGFR targeted drugs.
Prognostic Gene Signatures and the SPECS Project
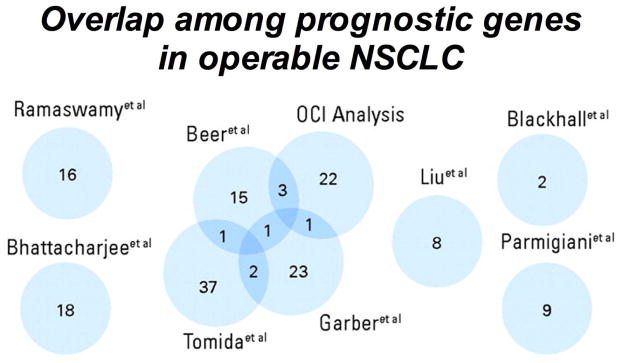
Prognostic factors for SCC have been mostly derived from surgically resected tumors in patients with early stage disease. In patients with advanced disease, treatments such as chemotherapy, radiotherapy, or targeted therapies may alter prognostic associations, and/or be predictive or combined prognostic/predictive. Of interest, among the numerous reports on genomic classifiers, there is surprisingly little overlap (29) (Fig. 2), and very few validation studies. Therefore, none of the prognostic classifiers are commonly used today in clinical practice. Additionally, studies reporting on prognostic factors are very heterogeneous regarding study populations and histology, which makes comparisons and validation even more difficult. Here we describe ongoing efforts to develop a validated prognostic classifier, being undertaken by a dedicated group of investigators who have established a “Squamous Lung Cancer Consortium” with the overall goal of validating existing (published and non-published) prognostic signatures within clinically well-defined SCC cohorts by using a standardized protocol for tissue processing, one centralized lab for RNA (and eventually DNA) extraction and with central histo-pathologic evaluation (Fig. 3). Once validated, the signatures can be used to develop clinically useful tests to differentiate patients with early stage SCC who have a poor prognosis versus a good prognosis.
Figure 2.
Venn diagram for degree of overlap of 158 candidate prognostic genes. Reference sources are shown. Reprinted from Lau et al. (29).
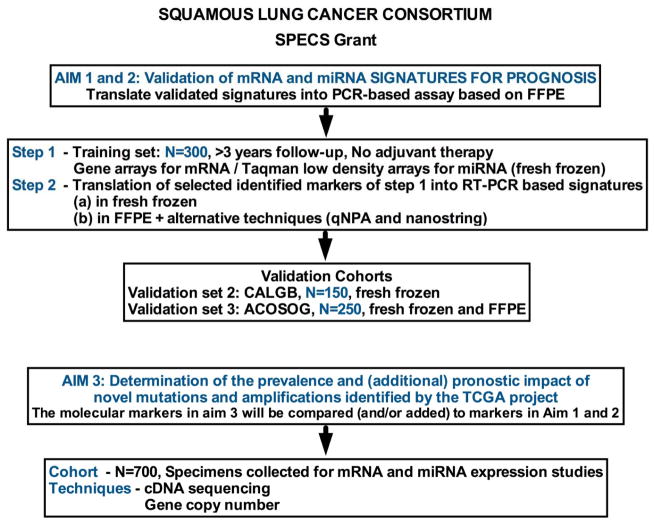
Figure 3.
Aims and research approach for the Squamous Lung Cancer SPECS Consortium.
The “Consortium”, which includes investigators from seven US/ Canadian institutions (University of Colorado, Mayo Clinic, University of Michigan, The Brigham and Women’s Hospital, University of California Davis, Washington University in St. Louis, Duke University and Princess Margaret Hospital in Toronto) was awarded the NCI SPECS (Strategic Partnering to Evaluate Cancer Signatures) grant. The SPECS project will determine if existing mRNA and miRNA prognostic signatures can distinguish between SCC patients with good prognosis versus poor prognosis first in a test set of 300 patients with early stage SCC (no adjuvant therapy and with a minimum of 3 years follow-up). Based on this evaluation and eventual development of “new signature (s)”, two validation sets have been identified and accepted for use: one a surgically treated SCC cohort (N=150) from the previous Cancer and Leukemia Group B (CALGB) and one from the American College of Surgeons Oncology Group (ACOSOG) (N=250), both cohorts today under the Alliance. The SPECS project includes also a validation of The Cancer Genome Atlas (TCGA) Project for SCC (7). Thus, it is the goal with the ongoing SPECS SCC Program to validate and eventually develop new prognostic classifier (s) based on standardized protocols and well-defined clinical cohorts and validate the prognostic association of the gene abnormalities found in the lung TCGA project and eventually identify new therapeutic targets.
Current Therapeutic Options for Lung SCC
Standard therapy
Patients diagnosed with metastatic or recurrent SCC of the lung are candidates for frontline systemic therapy given with a palliative (i.e., non-curative) intent. Unlike adenocarcinoma of the lung for which initial therapy is guided by the presence or absence of an increasing number of driver mutations, the standard of care for metastatic lung SCC is cytotoxic chemotherapy, most commonly a platinum-based doublet. Either cisplatin or carboplatin is used as the platinum backbone of these regimens, while agents like paclitaxel, nab-paclitaxel, docetaxel, or gemcitabine constitute the cytotoxic partner.
Phase III studies of cytotoxic therapy in NSCLC have shown differential outcomes for patients with SCC versus non-SCC cancers. In a phase III trial of cisplatin/pemetrexed versus cisplatin/gemcitabine in advanced NSCLC, patients with SCC histology were reported to have better survival with the gemcitabine-based doublet (median survival time 10.8 v 9.4 months, respectively) (30). Nab-paclitaxel, an albumin-bound nano-formulation of paclitaxel, was shown to have a higher rate of tumor response when combined with carboplatin versus standard paclitaxel/carboplatin (response rate ratio of 1.68, p<0.001) in the patient subset with SCC (31). Survival for the overall population was similar between the arms.
For patients with advanced NSCLC who complete four to six cycles of frontline platinum-doublet therapy and have documented stable or responding disease, maintenance therapy is an option, and is reported to improve progression-free survival in some patient subsets(32, 33). However, the role of maintenance therapy in those patients with SCC is less established. In the second line setting, agents such as docetaxel or erlotinib are considered reasonable therapeutic options, but these are not specifically approved for SCC. In the phase III BR-21 trial of erlotinib vs. placebo in the second/third line setting that included all histologic subtypes, the survival benefit for erlotinib was of equivalent magnitude in SCC and adenocarcinoma, and was even seen in a subset analysis of male, ever-smokers with SCC (34). Additionally, the US FDA recently approved ramicirumab, a VEGFR2-targeted monoclonal antibody, for use in combination with docetaxel in patients with advanced NSCLC progressing after primary platinum-based chemotherapy, regardless of tumor histology. This approval was based on the results of a phase III randomized trial (REVEL) that demonstrated a modest OS and Progression Free Survival (PFS) FS benefit for the addition of ramicirumab to docetaxel. (35)
It is notable that certain systemic therapies are specifically not recommended for use in patients with lung SCC. Specifically, the angiogenesis inhibitor bevacizumab and the multi-targeted antifolate pemetrexed are not approved for use in these patients due to either increased toxicity (in the case of bevacizumab) or decreased efficacy (in the case of pemetrexed) (36).
Investigational approaches
It is apparent that outcomes for patients with advanced lung SCC remain suboptimal, warranting diversification of targets and therapeutic options. Among these targets is the EGFR. It must be emphasized that in this SCC histologic subset, EGFR activating mutations are exceptionally uncommon, but that most cancers avidly express the wild type EGF receptor and a subset demonstrate EGFR amplification. A monoclonal antibody directed against EGFR - necitumumab – was evaluated specifically in lung SCC in a large phase III trial (SQUIRE) (37). In that study, 1093 patients with advanced SCC were randomized to gemcitabine-cisplatin with or without necitumumab. Treatment was given for up to six cycles. Subsequently, patients assigned to the necitumumab arm continued to receive maintenance necitumumab every three weeks until disease progression. Overall survival (OS) was significantly increased in necitumumab-treated patients (median survival time was 11.5 versus 9.9 months, HR 0.84, 95% CI 0.74–0.96). PFS was also significantly increased (HR 0.85, 95% CI 0.74–0.98). However, there were higher rates of grade 3 or greater toxicities seen in the necitumumab arm. Nevertheless, this was one of the first trials to show superior survival for a new agent when combined with chemotherapy versus chemotherapy alone in lung SCC.
In a Phase III trial focusing on patients with SCC, typically characterized by wild type EGFR, LUX-Lung 8, 795 patients with relapsed/refractory disease after first-line chemotherapy were randomized to either erlotinib or afatinib, an irreversible ErbB family blocker. The primary endpoint was PFS, while secondary endpoints included OS, objective response rate (ORR), and disease control rate (DCR). Median PFS was significantly higher for afatinib than erlotinib (2.4 vs 1.9 months; p=0.0427). The ORR was similar between the arms (4.8% vs. 3.0%, p=0.233), but DCR was significantly higher with afatinib than erlotinib (45.7% vs 36.8%; p=0.020) If OS results, currently pending, are positive for afatinib, this approach may prove to be another option for the SCC population (38).
Most recently, immunotherapy – particularly checkpoint inhibitor therapy with PD1 antibodies – has shown encouraging activity in patients with SCC. A more detailed description of immune checkpoint modulation is provided in a companion article from Soria and colleagues in this CCR Focus (37). Nivolumab, a humanized IgG4 antibody against PD1, was shown in a phase I dose-finding trial to have an overall response rate of 18% in a NSCLC subset which included patients with SCC. Interestingly, some of these responses appear to be durable (i.e., > 1 year in duration). Responses may be related to higher PDL1 expression in pretreatment tumor specimens, although data are mixed (39). In an updated analysis of this trial, one and two year survival rates of 42% and 14% were reported (40). Subsequently, nivolumab was tested in a phase III trial versus docetaxel in patients with advanced SCC progressing during or after platinum based therapy, with a primary endpoint of OS. According to a recent press release, a statistically significant OS benefit for patients receiving nivolumab was achieved. Specifically, nivolumab showed significantly superior OS as compared to docetaxel, with a 41% reduction in the risk of death (HR 0.59, p=0.00025]). The median OS was 9.2 months in the nivolumab arm (95% CI: 7.3, 13.3) and 6 months in the docetaxel arm (95% CI: 5.1, 7.3). These data led to the recent FDA approval of nivolumab for the treatment of metastatic lung SCC.
Pembrolizumab is another humanized IgG4 PD1 antibody (approved for use in melanoma) which was prospectively tested in patients with advanced solid tumors, including SCC (41, 42, 43). In a phase I trial (KEYNOTE1), pembrolizumab was given at 2 mg/kg every 3 weeks, 10 mg/kg every 3 weeks, or 10 mg/kg every two weeks until progression, death, or unacceptable toxicity. Tumor PDL1 was assessed by immunohistochemistry in archival specimens. A pooled analysis of 282 patients with treatment-naïve or previously treated advanced NSCLC was recently presented (41). The RECIST ORR was 21% in the overall study population; ORR was 18% in patients with SCC and 23% in patients with non-SCC. Response rates and PFS appeared to be higher for patients whose tumors more highly expressed PDL1; for instance, hazard ratio for PFS was 0.52 for patients with PD-L1 strong-positive versus PD-L1 weak-positive or negative tumors.
Lung Master Protocol in SCC (Lung-MAP, S1400)
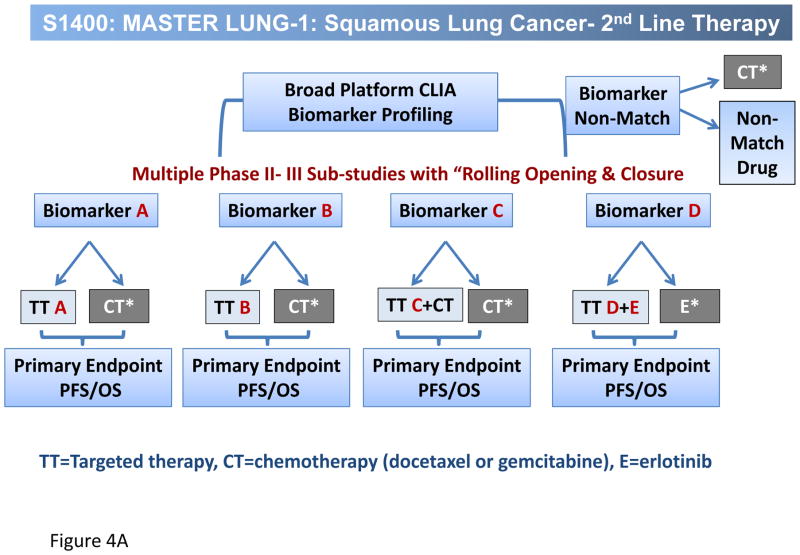
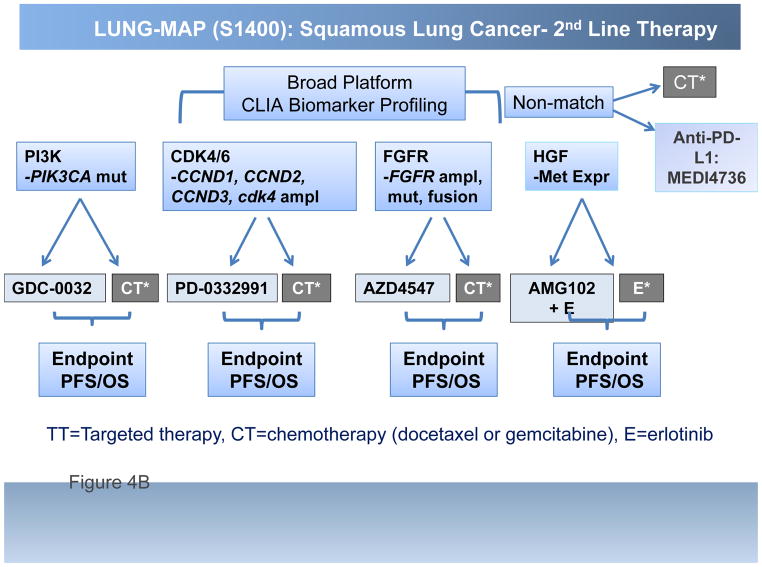
The Lung-MAP project, a multi-substudy master protocol designed to facilitate approval of targeted therapy-predictive biomarker combinations, represents a unique public-private partnership engaging the National Cancer Institute (NCI) and its Thoracic Malignancies Steering Committee (TMSC), the Foundation of the NIH (FNIH), the pharmaceutical industry, advocacy groups such as Friends of Cancer Research (FOCR), and most importantly, the Federal Drug Administration (FDA). The design is multiple simultaneously running Phase II/III trials, each capable of independently opening and/or closing without affecting the other substudies, in which patients eligible for 2nd line therapy for lung SCC have their cancers genomically screened through a next generation sequencing (NGS) platform (Foundation Medicine). Patients are then randomized into one of several sub-studies, each comparing an experimental targeted therapy with standard of care therapy, based on identification of candidate predictive biomarkers associated with each sub-study. Fig. 4 displays the overall schema for Lung-MAP (Fig. 4A) and the initial drug classes being tested, PI3K, FGFR, CDK 4/6, HGF and PD-L1 (Fig. 4B). Rapid turnaround time of NGS screening results, within 2 weeks, allows real time assignment into the appropriate sub-study. For those patients with cancers that do not “match” into a biomarker-driven sub-study, there is a ‘non-match” sub-study, in which a predictive biomarker is not yet of sufficient validation to utilize it in a drug-biomarker registration strategy. If successful, Lung-MAP will change the way new drugs are developed in lung cancer, and the approach will be extrapolated into other settings in lung cancer, and into other tumor types as well. Already, a master protocol design is being developed in ALK positive cancers based on the Lung-MAP design.
Figure 4.
A, Overall schema for Lung-MAP and B, the initial drug classes being tested, PI3K, FGFR, CDK 4/6, HGF and PD-L1. TT=Targeted therapy, CT=chemotherapy (docetaxel or gemcitabine), E=erlotinib
In summary, recent advances in understanding the underlying tumor biology of lung SCC, a subset of NSCLC for which progress has been modest at best over the last decade, have identified new “druggable” tumor targets and potential associated biomarkers. The ongoing SPECS project is seeking to validate prognostic gene signatures to better define subsets within lung SCC with differing natural histories and variable chances of relapse after surgical resection. Recent clinical trials dedicated to lung SCC are also showing promise. Finally, a SCC master protocol (Lung-MAP or S1400) is exploring a novel strategy designed to hasten approval of new targeted therapeutics and their companion diagnostics for this important subset of NSCLC.
Acknowledgments
D.R. Gandara and P.N. Lara Jr are supported by the NIH under award number P30CA93373. P.S. Hammerman is supported by the NIH under award number K08CA163677. M.L. Sos is supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grant #01ZX1406). F.R. Hirsch and D.R. Gandara are supported by the NCI/SPECS under award number UO1CA157715.
Footnotes
Disclosure of Potential Conflicts of Interest
D.R. Gandara reports receiving speakers bureau honoraria from Boehringer Ingelheim, Bristol-Myers Squibb, Eli Lilly, Genentech, Merck, Novartis, and Synta Pharmaceuticals. P.S. Hammerman is a consultant/advisory board member for ARIAD Pharmaceuticals, AstraZeneca, Clovis Oncology, ImClone Systems, Janssen, and MolecularMD. M.L. Sos is a consultant/advisory board member for Blackfield. P.N. Lara Jr is a consultant/advisory board member for Lilly Oncology. F.R. Hirsch is a consultant/advisory board member for AstraZeneca, Bristol-Myers Squibb, Genentech, ImClone Systems/Eli Lilly, Novartis, and Pfizer. No other potential conflicts of interest were disclosed.
References
- 1.Thun MJ, Lally CA, Flannery JT, Calle EE, Flanders WD, Heath CW., Jr Cigarette smoking and changes in the histopathology of lung cancer. J Natl Cancer Inst. 1997;89:1580–6. doi: 10.1093/jnci/89.21.1580. [DOI] [PubMed] [Google Scholar]
- 2.Khuder SA, Mutgi AB. Effect of smoking cessation on major histologic types of lung cancer. Chest. 2001;120:1577–83. doi: 10.1378/chest.120.5.1577. [DOI] [PubMed] [Google Scholar]
- 3.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgensztern D, Waqar S, Subramanian J, Gao F, Govindan R. Improving survival for stage IV non-small cell lung cancer: a surveillance, epidemiology, and end results survey from 1990 to 2005. J Thorac Oncol. 2009;4:1524–9. doi: 10.1097/JTO.0b013e3181ba3634. [DOI] [PubMed] [Google Scholar]
- 5.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–5. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. Erratum in: Nature 2012, 491, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y, Hammerman PS, Kim J, Yoon JA, Lee Y, Sun JM, et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol. 2014;32:121–8. doi: 10.1200/JCO.2013.50.8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical Lung Cancer Genome Project (CLCGP); Network Genomic Medicine (NGM) A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seiwert TY, Burtness B, Weiss J, Gluck I, Eder JP, Pai SI, et al. A phase Ib study of MK-3475 in patients with human papillomavirus (HPV)-associated and non-HPV–associated head and neck (H/N) cancer. J Clin Oncol. 2014;32(suppl):5s. abstr 6011. [Google Scholar]
- 11.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, et al. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development. 2009;136:1751–9. doi: 10.1242/dev.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–73. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, Chande A, Tanaka KE, Stransky N, Greulich H, Gray NS, Meyerson M. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011:6e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerman PS, Sos ML, Ramos AH, Xu C, Dutt A, Zhou W, et al. Mutations in the DDR2 kinase gene identify anovel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73:5195–205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–47. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, Wang L, Li Y, Hu H, Shen L, Shen X, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res. 2014;20:4107–14. doi: 10.1158/1078-0432.CCR-14-0284. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Zhang L, Su X, Li M, Xie L, Malchers F, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18:6658–67. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 21.Bahleda R, Dienstmann R, Adamo B, Gazzah A, Infante JR, Zhong B, et al. Phase 1 study of JNJ-42756493, a pan-fibroblast growth factor receptor (FGFR) inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2014;32(suppl):5s. doi: 10.1200/JCO.2014.60.7341. abstr 2501. [DOI] [PubMed] [Google Scholar]
- 22.Nogova L, Sequist LV, Cassier PA, Hidalgo M, Delord J-P, Schuler MH, et al. Targeting FGFR1-amplified lung squamous cell carcinoma with the selective pan-FGFR inhibitor BGJ398. J Clin Oncol. 2014;32(suppl):5s. abstr 8034. [Google Scholar]
- 23.Malchers F, Dietlein F, Schöttle J, Lu X, Nogova L, Albus K, et al. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer Discov. 2014:4246–57. doi: 10.1158/2159-8290.CD-13-0323. [DOI] [PubMed] [Google Scholar]
- 24.Wynes MW, Hinz TK, Gao D, Martini M, Marek LA, Ware KE, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014:203299–309. doi: 10.1158/1078-0432.CCR-13-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harding TC, Long L, Palencia S, Zhang H, Sadra A, Hestir K, et al. Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Sci Transl Med. 2013;5:178ra39. doi: 10.1126/scitranslmed.3005414. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Mikse O, Liao RG, Li Y, Tan L, Janne PA, et al. Ligand-associated ERBB2/3 activation confers acquired resistance to FGFR inhibition in FGFR3-dependent cancer cells. Oncogene. 2014 Jun 9; doi: 10.1038/onc.2014.161. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terai H, Soejima K, Yasuda H, Nakayama S, Hamamoto J, Arai D, et al. Activation of the FGF2-FGFR1 autocrine pathway: a novel mechanism of acquired resistance to gefitinib in NSCLC. Mol Cancer Res. 2013;11:759–67. doi: 10.1158/1541-7786.MCR-12-0652. [DOI] [PubMed] [Google Scholar]
- 28.Harbinski F, Craig VJ, Sanghavi S, Jeffery D, Liu L, Sheppard KA, et al. Rescue screens with secreted proteins reveal compensatory potential of receptor tyrosine kinases in driving cancer growth. Cancer Discov. 2012;2:948–59. doi: 10.1158/2159-8290.CD-12-0237. [DOI] [PubMed] [Google Scholar]
- 29.Lau SK, Boutros PC, Pintilie M, Blackhall FH, Zhu CQ, Strumpf D, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25:5562–9. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 30.Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, et al. PhaseIII study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–51. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 31.Socinski MA, Bondarenko I, Karaseva NA, Makhson AM, Vynnychenko I, Okamoto I, et al. Weekly nab-paclitaxel in combination with carboplatin versus solvent-based paclitaxel plus carboplatin as first-line therapy in patients with advanced non-small-cell lung cancer: final results of a phase III trial. J Clin Oncol. 2012;30:2055–62. doi: 10.1200/JCO.2011.39.5848. [DOI] [PubMed] [Google Scholar]
- 32.Fidias PM, Dakhil SR, Lyss AP, Loesch DM, Waterhouse DM, Bromund JL, et al. Phase III study of immediate compared with delayed docetaxel after front-line therapy with gemcitabine plus carboplatin in advanced non-small-cell lung cancer. J Clin Oncol. 2009;27:591–8. doi: 10.1200/JCO.2008.17.1405. [DOI] [PubMed] [Google Scholar]
- 33.Pérol M, Chouaid C, Pérol D, Barlési F, Gervais R, Westeel V, et al. Randomized, phase III study of gemcitabine or erlotinib maintenance therapy versus observation, with predefined second-line treatment, after cisplatin-gemcitabine induction chemotherapy in advanced non-small-cell lung cancer. J Clin Oncol. 2012;30:3516–24. doi: 10.1200/JCO.2011.39.9782. [DOI] [PubMed] [Google Scholar]
- 34.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. National Cancer Institute of Canada Clinical Trials Group. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–32. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 35.Garon EB, Ciuleanu TE, Arrieta O, Prabhash K, Syrigos KN, Goksel T, et al. Ramucirumab plus docetaxel versus placebo plus docetaxel for second-line treatment of stage IV non-small-cell lung cancer after disease progression on platinum-based therapy (REVEL): a multicentre, double-blind, randomised phase 3 trial. Lancet. 2014;384:665–73. doi: 10.1016/S0140-6736(14)60845-X. [DOI] [PubMed] [Google Scholar]
- 36.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–50. doi: 10.1056/NEJMoa061884. Erratum in: N Engl J Med 2007, 356, 318. [DOI] [PubMed] [Google Scholar]
- 37.Soria J-C, Marabelle A, Brahmer JR, Gettinger S. Immune checkpoint modulation for non–small cell lung cancer. Clin Cancer Res. 2015;21:xxx–xxx. doi: 10.1158/1078-0432.CCR-14-2959. [DOI] [PubMed] [Google Scholar]
- 38.Thatcher N, Hirsch FR, Szczesna A, Ciuleanu T-E, Szafranski W, Dediu M, et al. A randomized, multicenter, open-label, phase III study of gemcitabine-cisplatin (GC) chemotherapy plus necitumumab (IMC-11F8/LY3012211) versus GC alone in the first-line treatment of patients (pts) with stage IV squamous non-small cell lung cancer (sq-NSCLC) J Clin Oncol. 2014;32(suppl):5s. abstr 8008. [Google Scholar]
- 39.Goss G, Felip E, Cobo M, Lu S, Syrigos KN, Lee KH, et al. A randomized, open-label, phase III trial of afatinib (A) vs erlotinib (E) as second-line treatment of patients (pts) with advanced squamous cell carcinoma (SCC) of the lung following first-line platinum-based chemotherapy: LUX-Lung 8 (LL8) [abstract]. Proceedings of the 2014 ESMO Congress; 2014 Sep 26–30; Madrid, Spain. Lugano (Switzerland): European Society for Medical Oncology; 2014. Abstract nr 12220. [Google Scholar]
- 40.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Topalian SL, Sznol M, Brahmer JR, McDermott DF, Smith DC, Gettinger SN, et al. Nivolumab (anti-PD-1; BMS-936558; ONO-4538) in patients with advanced solid tumors: Survival and long-term safety in a phase I trial. J Clin Oncol. 2013;31(suppl) abstr 3002. [Google Scholar]
- 42.Patnaik A, Kang SP, Tolcher AW, Rasco DW, Papadopoulos KP, Beeram M, et al. Phase I study of MK-3475 (anti-PD-1 monoclonal antibody) in patients with advanced solid tumors. J Clin Oncol. 2012;30(suppl) doi: 10.1158/1078-0432.CCR-14-2607. abstr 2512. [DOI] [PubMed] [Google Scholar]
- 43.Garon EB, Gandhi L, Rizvi N, Hui R, Balmanoukian AS, Patnaik A, et al. Antitumor activity of pembrolizumab (Pembro; MK-3475) and correlation with programmed death ligand 1 (PD-L1) expression in a pooled analysis of patients (pts) with advanced non-small cell lung carcinoma (NSCLC) [abstract]. Proceedings of the 2014 ESMO Congress; 2014 Sep 26–30; Madrid, Spain. Lugano (Switzerland): European Society for Medical Oncology; 2014. Abstract nr LBA43. [Google Scholar]