Abstract
Congenital anomalies of the kidneys and urinary tract (CAKUT) are the most common cause of chronic kidney disease in the first three decades of life. Identification of single-gene mutations that cause CAKUT permits the first insights into related disease mechanisms. However, for most cases the underlying defect remains elusive. We identified a kindred with an autosomal-dominant form of CAKUT with predominant ureteropelvic junction obstruction. By whole exome sequencing, we identified a heterozygous truncating mutation (c.1010delG) of T-Box transcription factor 18 (TBX18) in seven affected members of the large kindred. A screen of additional families with CAKUT identified three families harboring two heterozygous TBX18 mutations (c.1570C>T and c.487A>G). TBX18 is essential for developmental specification of the ureteric mesenchyme and ureteric smooth muscle cells. We found that all three TBX18 altered proteins still dimerized with the wild-type protein but had prolonged protein half life and exhibited reduced transcriptional repression activity compared to wild-type TBX18. The p.Lys163Glu substitution altered an amino acid residue critical for TBX18-DNA interaction, resulting in impaired TBX18-DNA binding. These data indicate that dominant-negative TBX18 mutations cause human CAKUT by interference with TBX18 transcriptional repression, thus implicating ureter smooth muscle cell development in the pathogenesis of human CAKUT.
Introduction
Chronic kidney disease (CKD) takes a high toll on human health, requiring dialysis or kidney transplantation for survival. Congenital anomalies of the kidneys and urinary tract (CAKUT) are the most frequent cause of CKD in the first three decades of life, accounting for 40%–50% of all cases.1 CAKUT occurs in about 3 to 6 per 1,000 live births and constitutes 20%–30% of all birth defects identified in the neonatal period.2, 3, 4 No prophylaxis is available. Recent identification of monogenic mutations in genes that govern normal renal development permitted first insights into the pathomechanisms underlying some of these disorders in humans.5, 6, 7, 8, 9
It has been hypothesized that many forms of CAKUT in humans are probably caused by rare single-gene defects,10 based on multiple lines of evidence. (1) A large number of monogenic mouse models of CAKUT were reported,10 and many of the related genes participate in nephrogenesis.2, 11 (2) By devising a high-throughput exon sequencing technique,12 we previously showed that causative mutations in 12 different genes can be identified in about 10% of case subjects with CAKUT.6, 7, 8 However, extensive genetic heterogeneity has rendered gene discovery in CAKUT very difficult.7, 8, 10 Most known single genes that cause CAKUT if mutated are inherited in an autosomal-dominant manner and exhibit the clinical genetic features of incomplete penetrance and variable expressivity.7, 10
CAKUT in humans as well as in many mouse models might cover a wide spectrum of urinary tract malformations that ranges from mild to severe phenotypes including unilateral vesicoureteral reflux and bilateral renal agenesis. This spectrum can be found in different individuals of the same family with CAKUT, or between both kidneys of the same individual, even if the causative single-gene mutation is identical. Because this variability in anomalies of the kidneys and the lower urinary tract can be inseparable, the term “congenital anomalies of the kidneys and urinary tract” was coined.13 The phenotypic variability of CAKUT in the presence of an identical mutation might be due to stochastic gene dosage effects of the transcription factors involved. Alternatively, it might be caused by a so-called genetic “sequence,” in which the primary genetic defect causes urinary tract obstruction, which then mechanically leads to developmental defects like renal hypodysplasia.13
To gain further insights into the pathogenesis of CAKUT, we investigated a four-generation family by whole exome sequencing (WES) as well as 1,295 unrelated individuals with CAKUT. We here identify dominant-negative mutations in the transcription factor TBX18 gene as causing human CAKUT via lack of repression of TBX18 transcriptional activity, thereby implicating ureter smooth muscle cell development in the pathogenesis of human CAKUT.
Methods
Study Participants
After informed consent, we obtained clinical data, pedigree data, and blood samples from individuals with CAKUT from worldwide sources via a standardized questionnaire. Approval for human subjects’ research was obtained from the Institutional Review Boards of the University of Michigan, Boston Children’s Hospital, and Massachusetts General Hospital and from relevant local ethics review boards. Informed consent was obtained from the individuals and/or parents, as appropriate. The diagnosis of CAKUT was made by (pediatric) nephrologists and/or urologists based on relevant imaging.
Whole Exome Sequencing
To identify a causative mutated gene for CAKUT, we investigated family members from a four-generation white Hispanic family in the USA with an autosomal-dominant form of CAKUT predominantly characterized by ureteropelvic junction obstruction (UPJO) and hydronephrosis (Table 1). DNA samples from three affected distant cousins (Figure 1A) were subjected to whole exome sequencing (WES) as established previously via Agilent SureSelect human exome capture arrays (Life Technologies) with next-generation sequencing (NGS) on an Illumina sequencing platform. Sequence reads were mapped against the human reference genome (NCBI build 37/hg19) using CLC Genomics Workbench (v.6.5.1) software (CLC bio). Mutation calling under an autosomal-dominant model was performed by geneticists and cell biologists, who had knowledge regarding clinical phenotypes, pedigree structure, genetic mapping, and WES evaluation (Table S1) and in line with proposed guidelines.14, 15 Sequence variants that remained after the WES evaluation process were examined for segregation in seven affected and seven unaffected family members.
Table 1.
Dominant TBX18 Mutations Detected in Individuals with Obstructive Uropathy and Other Forms of CAKUT
| Family-Individual | Ancestry | Nucleotide Alterationa | Exon (Zygosity) | Alteration in Coding Sequencea,b | Continuous AA Sequence Conservationc,d | Allelic Loss of Function (half-life/lucif/EMSA)a,e | Presenting Symptoms (at Age in Years) | Kidney Phenotype | Treatment | Serum Creatinine (mg/dl) (at Age in Years) |
|---|---|---|---|---|---|---|---|---|---|---|
| A3880-II1 | Hispanic (USA) | c.1010delG | 7 (het) | p.Gly337Valfs∗19 | protein truncating mutation | (+/+/−) | back pain (13) | bilateral upjo | left kidney surgery | 1.06 (52) |
| A3880-II4 | c.1010delG | p.Gly337Valfs∗19 | (+/+/−) | flank pain (43) | bilateral upjo | ureteral stent placement, unilateral nephrectomy | 0.8 (68) | |||
| A3880-III5 | c.1010delG | p.Gly337Valfs∗19 | (+/+/−) | flank pain (17) | left upjo | ureteral stent placement | 0.8 (42) | |||
| A3880-III9 | c.1010delG | p.Gly337Valfs∗19 | (+/+/−) | hematuria (49) | bilateral hydronephrosis | conservative | 0.68 (50) | |||
| A3880-IV1 | c.1010delG | p.Gly337Valfs∗19 | (+/+/−) | hydronephrosis (prenatal) | bilateral hydronephrosis | conservative | 0.36 (6) | |||
| A3880-IV3 | c.1010delG | p.Gly337Valfs∗19 | (+/+/−) | urinary tract infection (1) | bilateral uvjo | bilateral ureteral reimplantation, unilateral nephrectomy | 0.7 (37) | |||
| A3880-IV6 | c.1010delG | p.Gly337Valfs∗19 | (+/+/−) | flank pain (8) | unilateral upjo | unilateral nephrectomy | 0.98 (26) | |||
| A2900-I2 f | Albanian | c.1570C>T | 8 (het) | p.His524Tyr | Danio rerio | (+/+/−) | asymptomatic (28) | renal duplex and asymmetry | conservative | 0.8 (28) |
| A2900-II1 | hydronephrosis (prenatal) | megaureter and ureterocele | ureteral reimplantation | 0.4 (6) | ||||||
| A2900-II2 | asymptomatic (6) | renal asymmetry | conservative | 0.55 (6) | ||||||
| A2385-II1 | Albanian | c.1570C>T | 8 (het) | p.His524Tyr | Danio rerio | (+/+/−) | asymptomatic (13) | bilateral small kidneys | renal transplantation | ESKD (13) |
| ICH_006-D80240 | British | c.487A>G | 2 (het) | p.Lys163Glu | Danio rerio | (+/+/+) | bilateral small echogenic kidneys (prenatal) | left VUR III°, bilat. renal dysplasia | NA | ESKD |
Abbreviations are as follows: CKD, chronic kidney disease; ESKD, end-stage kidney disease; het, heterozygous; NA, not applicable; ND, no data; UPJO, ureteropelvic junction obstruction; UVJO, ureterovesical junction obstruction VUR, vesicoureteral reflux.
cDNA mutations are numbered according to human cDNA reference sequence GenBank: NM_001080508.1, isoform (TBX18), where +1 corresponds to the A of ATG start translation.
All mutations had a PolyPhen-2 (PP2) humvar score of functional “deleteriousness” of 1.0.
Altered amino acid (aa) residues were conserved throughout vertebrates (see Figure S2).
All mutations were absent from 712 control individuals with renal ciliopathies, 448 individuals with steroid-resistant nephrotic syndrome, ∼13,000 healthy control alleles of the EVS, 2,577 control individuals of the 1000 Genomes project, and ∼112,000 control chromosomes of the ExAC server with the exception of p.His524Tyr, which is reported twice heterozygously in ∼120,000 controls.
Plus sign (+) indicates a defect of the allele in one of the three assays of TBX18 function: protein half-life (Figure 2B), luciferase assay (Figure 2C), or EMSA (Figure 3C).
Family A2900 included an unaffected sibling who does not carry the mutation.
Figure 1.
Identification of TBX18 Mutations in the Index Family A3880 and Three Additional Families with CAKUT
(A) Pedigree of index family A3880. Squares represent males, circles females, black symbols affected persons, and white symbols unaffected persons. All affected family members presented with obstructive uropathy. Pedigree is compatible with an autosomal-dominant mode of inheritance. Roman numerals denote generations. Individuals are numbered with Roman numerals if DNA was available for study. The arrow points to the proband IV3. WT denotes the wild-type allele. p.Gly337Valfs∗19 indicates the mutation c.1010 deletion of G in TBX18, leading to a frameshift mutation and introducing a premature stop codon 19 codons downstream. The mutation fully segregated heterozygously (WT/p.Gly337Valfs∗19) across all seven affected individuals examined and was absent from all seven unaffected family members available for study (WT/WT). Red circles indicate the persons selected for whole exome sequencing.
(B) Abdominal CT scan of the index individual A3880-IV3 revealing severe bilateral hydronephrosis (white asterisks) secondary to bilateral ureteral obstruction. The index individual presented during infancy and was diagnosed after evaluation of urinary tract infection. She underwent corrective surgery with multiple consecutive reconstructive operations to her lower urinary tract. She had no extrarenal manifestations.
(C) Prenatal renal US of individual A3880-IV1 with bilateral hydronephrosis at a gestational age of 28 weeks. Postnatally, he had unilateral pelviectasis (Figure S1A) with normal voiding cysturethrogram. At the age of 6.5 years there was left-sided renal hypodysplasia with no signs of obstruction (Figures S1B and S1C).
(D) Exon structure of human TBX18 cDNA and domain structure of the T-box domain 18 (TBX18) protein. TBX18 contains a nuclear localization signal (NLS, red); an engrailed homology-1 motif (EH1, blue), and a DNA-binding T-box domain (yellow). Start codon (ATG) and stop codon (TGA) are indicated.
(E) Chromatograms of heterozygous mutations detected in TBX18 (in relation to exons and protein domains) in the index family (red) and three additional families (black) with CAKUT. The index family’s heterozygous mutation c.1010delG leads to a frameshift and after 19 amino acids to a premature termination codon (p.Gly337Valfs∗19).
Whole Exome Sequencing Analysis
After WES, genetic variants were first filtered to retain only heterozygous, non-synonymous variants that were shared between the three affected relatives of family A3880 subjected to WES. Second, filtering was performed to retain only alleles with a minor allele frequency (MAF) < 0.1%, a widely accepted cutoff for autosomal-dominant disorders.16, 17 MAF was estimated using combined datasets incorporating all available data from the 1000 Genomes Project, the Exome Variant Server (EVS) project, dbSNP138, and the Exome Aggregation Consortium (ExAC). Third, observed sequence variants were analyzed with the UCSC Human Genome Bioinformatics Browser for the presence of paralogous genes, pseudogenes, or misalignments. Fourth, we scrutinized all variants with MAF < 0.1% within the sequence alignments of the CLC Genomic Workbench software program for poor sequence quality and for the presence of mismatches that indicate potential false alignments. Fifth, we employed web-based programs to assess variants for evolutionary conservation, to predict the impact of disease candidate variants on the encoded protein, and to predict whether these variants represented known disease-causing mutations. Finally, Sanger sequencing was performed to confirm the remaining variants in original DNA samples and to test for familial segregation of phenotype with genotype
Analysis of Copy-Number Variants
For two individuals from the index family (IV3 and IV6 from family A3880; Figure 1A), genome-wide genotyping for copy-number variant (CNV) analysis was performed on an Illumina platform as previously described.18 Genotype calls and quality-control analyses were performed with Genome Studio v.2010.3 (Illumina) and PLINK software. The CNV calls were determined with generalized genotyping methods implemented in the PennCNV program. CNVs were mapped to the human reference genome hg18 and annotated with UCSC RefGene and RefExon (CNVision program25).
High-Throughput TBX18 Mutation Analysis
Massively parallel sequencing of TBX18 exons was performed in 1,589 individuals from 1,295 different families affected by CAKUT from different pediatric nephrology centers worldwide using microfluidic PCR (Fluidigm) and next-generation sequencing (MiSeq, Illumina) as described previously.7, 12, 19 Variants were filtered against public variant databases (EVS) and only novel heterozygous variants were considered, confirmed by Sanger sequencing, and tested for segregation with the CAKUT phenotype. Variants were also tested for absence from in-house control populations of 448 individuals with steroid-resistant nephrotic syndrome and 712 individuals with nephronophthisis.
Immunohistochemistry
Epitope retrieval on paraffin-embedded sections of mouse tissue was performed with OmniPrep Solution (pH 9.0) from ZYTOMED Systems. After quenching of endogenous peroxidases (10 min with 3% H2O2), the goat-derived polyclonal antibody (AB) TBX18 (C-20) (sc-17869 from Santa Cruz) was used for immunodetection at 1:100 dilutions. Blocking solution and peroxidase-conjugated anti-goat secondary AB were used from the VECTOR ImmPRESS reagent Kit. The VECTOR Peroxidase Substrate Kit ImmPACT DAB was used for the final peroxidase-mediated color reaction.
Cell Culture, Transient Transfections, and Reporter Assays
HEK293 cells were seeded at 20%–30% confluency in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal calf serum, grown overnight, and transfected by the calcium phosphate method. For reporter assays (Dual luciferase reporter assay system, Promega), HEK293 cells were seeded in 6-well dishes and transfected with constant amounts of reporter plasmids and 100 ng of pRL-TK Renilla Luciferase (kind gift from A. Gossler) for normalization. The total amount of expression plasmid was kept constant by adding empty pcDNA3. Per transfection, 500 ng of pGL3.Tbx18BS-luciferase reporter plasmid (which harbors two T-half sites as inverted repeats: 5′-AGGTGTGAAATCGCACCT-3′) and 500 ng of prK5.Myc.TBX18 expression plasmids were used. For competition experiments, the amount of prK5.Myc.TBX18_WT was held constant and prK5.Myc.TBX18_p.G337Vfs∗19 was added with increasing amounts. Firefly luciferase and Renilla luciferase activities were measured 24 hr after transfection. All transfections were performed in duplicate, and experiments were repeated at least three times. After normalization, the mean luciferase activities and standard deviations were plotted as “fold activation” when compared with the empty expression plasmid. p values were determined with the Student’s t test.
Immunofluorescence
Experiments in HEK293 cells were performed according to standard protocols. Primary antibodies used were rat anti-HA (3F10, Roche Applied Science) and mouse anti-myc (9E10, Sigma), both at 1:250 dilutions. Secondary antibodies were donkey anti-rat IgG (H+L) fluorescein isothiocyanate and donkey anti-mouse IgG(H+L) rhodamine (both Dianova) at 1:250 dilutions. Immunofluorescent detection of proteins was repeated at least three times, and representative examples were photographed on a Leica DM6000 microscope with DFC350FX camera (Leica).
Half-Life Estimation
HEK293 cells were cultured and transfected as described above. At 48 hr post-transfection, 10 μg ml−1 cycloheximide (Sigma) was added and cells were incubated for the indicated time points. After incubation, cells were trypsinized and counted. Per 12,000 cells, 1 μl of lysis buffer (5 mM Tris [pH 7.5], 80 mM NaCl, 50 mM NaF, 1 mM MgCl2, 0.1% Nonidet P-40, 1 tablet/10 ml Complete Mini EDTA-free protease inhibitor Cocktail) was added. Protein levels were analyzed by immunoblotting with specific antibodies. The half-life was calculated with GraphPad Prism (v.4) with the following parameters: Nonlinear Regression (Curve Fit); Build-in model: Exponential; Equation: One phase decay.
Electrophoretic Mobility Shift Assay
Proteins used for electrophoretic mobility shift assay (EMSA) were generated from prK5.Myc.TBX18 expression constructs for TBX18 wild-type and mutant proteins using the SP6-coupled TNT Wheat Germ Extract System (Promega) for SP6-directed in vitro translation according to the supplier’s instructions. To generate a probe harboring two inverted T-half sites oligonucleotides BS.invF, 5′-GATCCACAGGTGTGAAATTTCACACCTACC-3′ and BS.invR, 5′-TCGAGGTAGGTGTGAAATTTCACACCTGTG-3′ were boiled for 5 min and cooled slowly down to room temperature to anneal. The annealed oligonucleotides were subcloned as BamHI/NcoI fragments into pKS vector (Stratagene) and released by BamHI/AseI as a 214 bp fragment. The fragment was dephosphorylated with Antarctic Phosphatase (New England Biolabs) for 1 hr at 37°C. The fragments were end-labeled with T4-PNK (New England Biolabs) in the presence of [γ-32P]ATP. Binding reactions for gel shift assays contained 2–5 μl of in vitro translated myc-tagged protein in a total volume of 20 μl of HEPES buffer (25 mM HEPES [pH 7.4], 10% glycerol, 75 mM NaCl, 0.25 mM EDTA, 1 mM MgCl2, 0.1% Nonidet P-40, 10 μg/ml BSA, 1 mM DTT) with 1 tablet/10 ml Complete Mini EDTA-free protease inhibitor Cocktail (Roche Applied Science) and 1.5 μg of double-stranded poly(dI-dC) (Roche). Reactions were preincubated for 20 min on ice before 15,000 counts of probe were added. For supershift experiments, 1.5 μl of anti-myc antibody (9E10, Sigma) was added to the lysate. Complexes were allowed to form at room temperature for 20 min, before the reactions were loaded on a native 4% polyacrylamide gel (0.5× Tris-borate-EDTA). Gels were run at 20 V/cm for 5 min and then decreased to 10 V/cm for 2–4 hr at 4°C before they were dried and exposed to imaging plate (Fujifilm).
Co-immunoprecipitation Assays
Myc- or HA-tagged prey proteins were produced by in vitro translation of full-length cDNAs cloned in pSP64 vectors (Promega TnT SP6). The binding reaction was performed in HEPES buffer (25 mM HEPES [pH 7.4], 10% glycerol, 75 mM NaCl, 1 mM MgCl2, 0.1% Nonidet P-40, 10 μg/ml BSA, 1 mM DTT) for 1 hr at 4°C. Protein G Sepharose beads (GE Healthcare) were preblocked with 1% BSA for 1 hr at room temperature. 2 μl of anti-Myc antibody and 25 μl of protein G Sepharose were added to the supernatant for 2 hr at 4°C, before the beads were washed three times with 500 μl of HEPES buffer. Proteins were eluted from the beads by boiling in loading buffer for 5 min and were analyzed by immunoblot with anti-myc-HRP (Abcam ab62928) and anti-HA-HRP (Abcam ab1265) antibodies, and 10% of the input was loaded as a control.
Results
TBX18 Mutations Cause CAKUT
To identify a causative mutated gene for CAKUT, we investigated a four-generation Hispanic family of ten individuals with congenital anomalies of the kidneys and urinary tract (CAKUT). Ureteropelvic junction obstruction (UPJO) was predominant together with hydronephrosis and/or vesicoureteral junction obstruction (Figures 1A–1C and S1; Table 1). Five of the seven affected individuals available for study required surgical intervention to relieve obstruction. Age at diagnosis ranged from the prenatal period to adulthood. None of the affected individuals had extra-renal malformations or syndromic features. There were seven unaffected family members available to the study that had normal urinary tract imaging. Pedigree structure was compatible with autosomal-dominant or X-linked segregation of CAKUT (Figure 1A).
Under the hypothesis that mutation of an autosomal-dominant gene causes CAKUT in this family, we selected three distant cousins (affected individuals A3880-II1, -IV3, and -IV6) for whole exome sequencing (WES). Given the pedigree structure (Figure 1A), the three affected individuals are expected to share about 3.125% of all alleles by descent, allowing us to reduce 32-fold the thousands of variants from normal reference sequence that are expected to result from WES (Table S1).20 We identified a heterozygous truncating mutation (c.1010delG [p.Gly337Valfs∗19]) in T-Box transcription factor 18 (TBX18) (MIM: 604613; GenBank: NM_001080508.1) (Table 1 and Figures 1D and 1E). Segregation analysis revealed that this mutation was shared by all seven available affected family members and was absent from all seven unaffected family members available for study (Figure 1A and Table 1). The LOD score resulting from this segregation pattern (Zmax = 4.2) represents significant linkage (Zmax > 3.3), thereby rendering it extremely likely that the TBX18 truncating mutation causes CAKUT in this family. Furthermore, the mutation was absent from 104 ethnically matched controls, from all available databases (Table 1), as well as from two in-house control cohorts of 480 and 768 individuals with nephrotic syndrome and nephronophthisis, respectively. We also showed by CNV analysis that the affected individuals lacked heterozygous deletions that have previously been associated with CAKUT in the HNF1B locus and the DiGeorge/velocarodiofacial syndrome locus.18
We next performed highly parallel exon sequencing12, 19 of all eight TBX18 exons in 1,295 unrelated individuals with CAKUT. We detected four additional heterozygous missense mutations (c.1570C>T [p.His524Tyr], c.487A>G [p.Lys163Glu], c.490G>A [p.Ala164Thr], and c.1576C>T [p.Pro526Ser]) in five unrelated families with CAKUT (Table 1 and Figures 1D and 1E). All mutations met strict genetic criteria regarding their functional deleteriousness (Table 1). Specifically, all mutated amino acid residues were fully conserved among vertebrates that possess TBX18 orthologs (Table 1; Figure S2). All mutations were absent from >13,000 healthy control alleles in the EVS database (Table 1). Because the two genetic variants p.Ala164Thr and p.Pro526Ser did not exhibit loss of function in any of the functional evaluations described below, we did not consider them as disease causing and will not show the related functional data (data available from the authors).
TBX18 Mutations Interfere with Transcriptional Repression
Tbx18 is a member of the Tbx1 subfamily of T-box transcription factor genes expressed at multiple sites during embryogenesis in the mouse.21 In the urogenital system, expression is confined to the undifferentiated mesenchyme surrounding the distal ureter. In Tbx18−/− mice, descendants of former Tbx18-positive cells do not differentiate into smooth muscle cells of the ureter but mislocalize to the kidney and differentiate into fibroblast-like cells. As a consequence, the renal pelvis becomes dramatically enlarged at the expense of the ureter, and hydronephrosis develops at birth.22, 23 We show by immunohistochemistry that TBX18 localizes to the renal pelvic and ureteral smooth muscle cells (Figure S3).
Previous biochemical analysis showed that the murine TBX18 protein harbors in the N-terminal region a nuclear localization signal (NLS) and an engrailed homology 1 (EH1) domain that mediates transcriptional repression by binding to the WD40 domain of Groucho/TLE transcriptional corepressors (Figure 1D). The downstream T-box domain binds to combinations of cognate DNA binding sites, so-called “T-half-sites.” The T-box also confers protein interaction with TBX18 itself (i.e., homodimerization) and with a number of other tissue-specific transcription factors including NKX2.5, PAX3, GATA4, and SIX1.24, 25 Interaction with SIX1, mutation of which causes branchio-oto-renal syndrome with CAKUT in humans,9 is important for mediating ureteric smooth muscle formation in concert with TBX18.26 No function has yet been assigned to the large C-terminal region of TBX18.
To examine how the c.1010delG (p.Gly337Valfs∗19) mutation, which segregated with the CAKUT phenotype in the large family A3880, conveys TBX18 loss of function, we evaluated TBX18 for its known protein domain-related functional features. These include (1) nuclear localization via its NLS, (2) homodimerization, (3) transcriptional repression, (4) DNA interaction via its T-box domain, and (5) interaction with Groucho/TLE and SIX1.
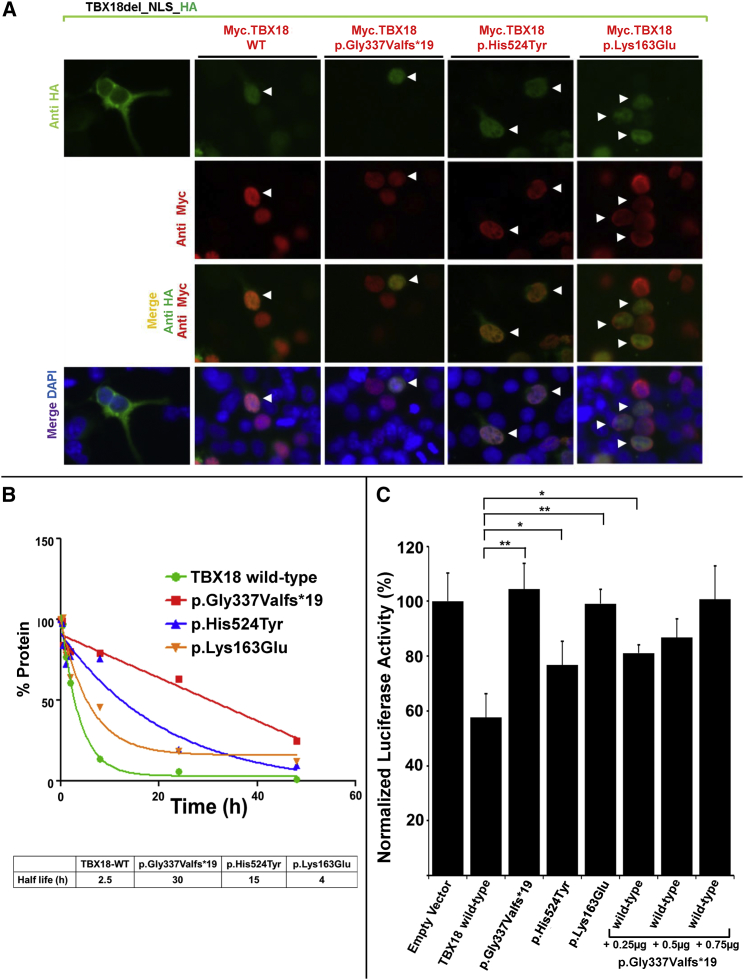
Transfection of expression constructs in HEK293 cells revealed that, whereas a control construct lacking the NLS did not localize to nucleus, the altered forms of TBX18 (p.Gly337Valfs∗19, p.His524Tyr, and p.Lys163Glu) do localize to the nucleus as does the wild-type protein (Figure 2A). Furthermore, all three mutant proteins (that still contained an intact NLS sequence) mediated nuclear localization of a variant of TBX18 lacking the nuclear localization signal (NLS),24 most likely by dimerization with it, indicating that the mutations do not perturb subcellular localization or homodimerization of the TBX18 protein (Figure 2A). We thereby established that the three TBX18 mutants still dimerize and traffic to the nucleus, suggesting a dominant-negative pathomechanism rather than haploinsufficiency. Furthermore, all three mutations affected the half-life of the encoded proteins (Figure 2B). Whereas the half-life of wild-type TBX18 was determined to be 2.5 hr in vitro, the half-lives of the mutant proteins were considerably prolonged to more than 30 hr (p.Gly337Valfs∗19), 15 hr (p.His524Tyr), and 4 hr (p.Lys163Glu) (Figure 2B), consistent with a dominant-negative effect of all three altered proteins.
Figure 2.
Trafficking of TBX18 to Nuclei via Its Nuclear Localization Signal, Prolonged Half-Life of Mutant Proteins, and Lack of Transcriptional Repression by TBX18 Mutants
(A) Immunofluorescence images in HEK293 cells that were cotransfected with an HA-tagged construct (TBX18del_NLS_HA) that lacks the nuclear localization signal (green), together with either (red) a human Myc-tagged labeled TBX18 wild-type construct (Myc.TBX18_WT) or constructs representing the substitution of family A3880 (Myc.TBX18_Gly337Valfs∗19), the substitution of families A2900 and A2385 (Myc.TBX18_His524Tyr), or the substitution of family ICH_006 (Myc.TBX18_Lys163Glu). Note that the NLS-deficient construct TBX18del_NLS_HA fails to localize to the nucleus (left column). However, in the presence of co-transfection of either Myc.TBX18_WT or either altered protein (red), even the NLS-deficient proteins (green) traffic to the nucleus, most likely by T-box-mediated dimerization with the NLS-containing Myc-tagged construct (red). Double-transfected cells (HA-construct and Myc-construct) are denoted by arrowheads.
(B) Half-life estimation of wild-type and mutant TBX18. Although the half-life of wild-type TBX18 was determined to be 2.5 hr in vitro, the half-lives of the mutant proteins were prolonged to more than 30 hr (p.Gly337Valfs∗19), 15 hr (p.His524Tyr), and 4 hr (p.Lys163Glu).
(C) Luciferase transcriptional reporter assay for TBX18 wild-type and mutant constructs. Compared to empty vector control (set at 100% of luciferase activity), transfection with TBX18 wild-type construct causes transcriptional repression to 58%. All three mutant constructs (p.Gly337Valfs∗19, p.His524Tyr, and p.Lys163Glu) detected in CAKUT-affected families repressed transcriptional activity significantly less than the wild-type construct. Co-transfection of TBX18 wild-type construct and the protein truncating construct p.Gly337Valfs∗19 in increasing amounts yielded a dose-dependent lack of repression for the mutant construct, supporting its dominant-negative effect on transcriptional repression by the wild-type construct. ∗p < 0.05, ∗∗p < 0.01.
We next determined whether the mutations affected the transcriptional activity of TBX18 protein by using cotransfections of TBX18 expression plasmids and a pGL3 reporter plasmid harboring two inverted repeats of T half sites (5′-AGGTGTGAAATCGCACCT-3′). A wild-type TBX18 expression construct mediated transcriptional repression to 58% in this assay, whereas three mutant constructs (p.Gly337Valfs∗19, p.His524Tyr, and p.Lys163Glu) showed significantly less repressive activity (Figure 2C). In addition, co-transfection of TBX18 wild-type construct with the protein truncating construct p.Gly337Valfs∗19 in increasing amounts yielded a dose-dependent lack of repression, supporting a dominant-negative effect of the mutant protein on transcriptional activity of the wild-type protein.
The p.Lys163Glu Substitution Alters TBX18-DNA Interaction
The T-box domain of TBX18 is known to directly interact with cognate sequences on genomic DNA at specific transcription sites. Interestingly, the amino acid residue Lys163, which is altered in individual ICH_006 with CAKUT, resides within the TBX18 T-box domain and is fully conserved in all the H. sapiens TBX paralog proteins (Figures 3A, 3B, and S2). The structures of the T-box domains of TBX1, TBX3, TBX5, and brachury from X. laevis were previously solved.27 Intriguingly, the equivalent of TBX18 Lys163 was shown to interact with the phosphate sugar backbone of the cognate DNA sequence.28 Because reduced transcriptional repression can derive from impaired DNA binding of altered TBX18, we tested by electrophoretic mobility shift assay (EMSA) whether any of the mutations that we found in families affected by CAKUT interfered with DNA binding of the T-box (Figures 3A–3C). As predicted from the structural model, electrophoretic mobility shift assay (EMSA) employing TBX18 binding sites and in vitro translated proteins showed that the TBX18 substitution p.Lys163Glu no longer bound to its cognate site on DNA, whereas p.Gly337Valfs∗19 and p.His524Tyr bound to the site, as did the wild-type TBX18 protein (Figure 3C). Reduced transcriptional repression might also stem from impaired interaction with other transcription factors and/or transcriptional corepressors. However, cotransfection/coimmunoprecipitation experiments revealed that all three altered proteins still bound to SIX1 and Groucho corepressor TLE3 (Figure S4),24 suggesting that other transcription factors might be involved.
Figure 3.
Three-Dimensional Model of Crystal Structure of Human TBX18 Paralog TBX1 in Complex with Cognate DNA
(A) Ribbon diagram of the TBX1-DNA complex in orthogonal views. Depicted are residues of two TBX18 monomers. β-pleated sheets are marked as yellow arrows, α helices in red, and DNA is marked in white as space-filling model. Residues marked in blue are those found at the protein-DNA interface (see B). Arrows point to the position of the amino acid residue Lys163 (red) found altered (p.Lys163Glu) in family ICH_006 with CAKUT. Inset on the right shows that Lys163 (arrow pointing to red Lys163 side chain) is in direct vicinity to the DNA interaction interface (white) (modeled in TBX1, PDB: 4A04).
(B) Partial sequence alignment of TBX18 T-box domain and its paralogs, for which protein 3D crystal structure data are available (H. sapiens TBX1, TBX3, and TBX5 and X. laevis brachyury). Amino acid residue numbers are shown per alignment row. Residues in blue are in the vicinity of the TBX1 protein-DNA interface (see A). Residue Lys163 found altered (p.Lys163Glu) in family ICH_006 with CAKUT is shown in red.
(C) DNA binding assay of wild-type TBX18 and TBX18 mutants detected in individuals with CAKUT show that p.Lys163Glu substitution fails to bind to DNA. Electrophoretic mobility shift assay (EMSA) was performed with equal amounts of in vitro synthesized myc-tagged constructs Myc.TBX18_wild-type (wild-type), Myc.TBX18_Gly337Valfs∗19, Myc.TBX18_His524Tyr, or Myc.TBX18_Lys163Glu. All alleles except for p.Lys163Glu bind to the TBX18 DNA target site as indicated by retardation of the labeled probe (red arrowheads in lanes 3). Specificity of the binding is confirmed by the addition of anti-myc antibody that results in formation of a super shifted complex (black arrowheads in lanes 5).
CAKUT Phenotype in Tbx18+/− Mice
Previous work reported that mice homozygous for a null allele of Tbx18 show hydroureter and hydronephrosis. To describe the phenotypic quality and penetrance of these changes in more detail, we re-inspected a large number of both heterozygous and homozygous Tbx18-deficient mice that we published previously.23 In 74% of newborn embryos homozygous for a null allele of Tbx18 (n = 23), we detected UPJO; the remaining 26% featured a bilateral hydroureter. Interestingly, we found that only 7% of Tbx18+/− (n = 41) heterozygous mice had a mild proximal hydroureter, confirming the recessive nature of the null mutation and demonstrating only a very minor role for haploinsufficiency. This further confirms that the mutations identified in humans with CAKUT exert a dominant-negative effect rather than haploinsufficiency of these alleles. These findings might also explain why in humans with CAKUT dominant TBX18 mutations are so infrequent, because a dominant-negative effect usually results from only a few very specific amino acid changes, and because the mice with recessive mutations were perinatal lethal and our cohort did not include such severe phenotypes.23
Discussion
We here identify dominant-negative mutations of TBX18 as causing CAKUT. By employing TBX18 half-life estimation, luciferase assay, and DNA interaction test by EMSA, we demonstrate loss of function for three different mutations in four families with CAKUT. Because TBX18 is essential for developmental specification of the ureteric mesenchyme and ureteric smooth muscle cells, we hereby implicate ureter smooth muscle cell development in the pathogenesis of human CAKUT.
TBX18 belongs to the T-box gene family, a large group of transcription factors involved in many aspects of embryonic development.29 In mice, Tbx18 has been shown to be selectively expressed in ureteral mesenchyme and to regulate ureteral smooth muscle differentiation.23 Mouse models lacking Tbx18 show malformed ureters resulting in various types of obstructive uropathy phenotypes as well as kidney parenchymal damage. The latter was suggested to be secondary to urine outflow obstruction,23 but TBX18 might also play a role during kidney development.30 Similar to the heterogeneity of kidney and urinary tract phenotypes previously seen in Tbx18-deficient mice, the phenotypes we observed in individuals with TBX18 mutations were broad and heterogeneous with predominant obstructive uropathy (Table 1).
Our findings support a dominant-negative effect of defective TBX18 proteins on the wild-type protein. In particular, we show that TBX18 altered proteins dimerize with the wild-type TBX18, were nonfunctional by themselves, and when co-expressed with wild-type TBX18 showed a dominant-negative effect. Our findings are consistent with the general consensus that proteins that dimerize are particularly prone, when altered, to exert dominant-negative effects by sequestering functioning molecules into inactive dimers.
Tbx18-null mice die shortly after birth due to severe malformations of the axial skeleton.31 In addition, TBX18 was suggested to regulate the normal development of the sinoatrial node.32, 33 Consequently, Tbx18 gene therapy was recently used for biological pacing in porcine model of heart block.34 None of the individuals in the current study presented with bone or heart malformations. Overall the phenotypes of the individuals with dominant TBX18 mutations were milder when compared to the severe Tbx18 recessive null mice phenotype. Nonetheless, the dominant human phenotype was more severe when compared to the phenotype of the heterozygous mice that appear normal. This apparent discrepancy might again be explained by a dominant-negative effect in which specific heterozygous mutations cause more severe effects than simple heterozygous null alleles of the same gene.
When considering the anatomical location and developmental stage of TBX18 expression, we postulate that the following mechanisms play a role in the pathogenesis of TBX18 loss of function. Because our immunohistochemistry data (Figure S3) as well as expression data from the LacZ-Tbx18−/+ mouse model23 demonstrate that TBX18 is localized in the urinary system only in the ureter and only during development of the urogenital tract, the relevant pathogenic effect must act during ureter development. Furthermore, because it was shown that Tbx18 is expressed in the ureteric mesenchyme and is required to prepattern the ureteric mesenchyme sub-lineage, loss of Tbx18 function will interfere with proper differentiation of the histologic layering of the ureter.22 It is therefore likely that the CAKUT phenotypes that we observed in addition to obstructive uropathy in individuals with TBX18 mutations, including renal hypo/dysplasia, occur by loss of essential TBX18 function during nephrogenesis.30 Alternatively, hypo/dysplasia might develop in way of a “sequence,” in which lack of urine transport in a maldeveloped ureter and the resultant intrarenal pressure interferes with proper renal development.13
Our discovery of dominant TBX18 mutations as causing human CAKUT via lack of repression of TBX18 transcriptional activity for the first time implicates ureter smooth muscle cell development in the pathogenesis of human CAKUT.
Acknowledgments
We are grateful the families who contributed to this study. We thank Seymour Rosen, Heather J. Cordell, William G. Newman, Stanley Swierzewski III, Frank Croke, John Blanchete, Dicken Ko, Ghaleb Daouk, John Herrin, and the UK VUR Study Group for contributing renal pathology, radiology, and clinical materials. This research was supported by grants from the NIH to R.S.L. (DK096238), to R.P.L. (5U54HG006504), to H.J. (DK 46718), to W.L. (DK078226), and to F.H. (DK088767) and from the March of Dimes to W.L. and F.H. This work was supported by grants from the German Research Foundation (DFG) to S.S.L. within SFB1140 and the Emmy Noether programme and to A.K. by an individual grant (Ki728/7-1) and within the Cluster of Excellence REBIRTH (From Regenerative Biology to Reconstructive Therapy) at Hannover Medical School. A.S.W. received project grants from Kidney Research UK, the Medical Research Council (G0600040 and MR/L002744/1), and the Wellcome Trust (066647). H.M.S. was funded by fellowships from NIHR UK and the Wellcome Trust. A.S.W.’s and H.M.S.’s research was facilitated by the Manchester Academic Health Sciences Centre. A.V. is a recipient of a Fulbright postdoctoral scholar award and of a Manton Center fellowship award from Boston Children’s Hospital. F.H. and R.P.L. are Investigators of the Howard Hughes Medical Institute. F.H. is the Warren E. Grupe Professor of Pediatrics at Harvard Medical School.
Published: July 30, 2015
Footnotes
Supplemental Data include four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.07.001.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
Ensembl Genome Browser, http://www.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
GenitoUrinary Molecular Anatomy Project (GUDMAP), http://www.gudmap.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Renal Genes, http://www.renalgenes.org
SeattleSeq Annotation 137, http://snp.gs.washington.edu/SeattleSeqAnnotation137/
UCSC Human Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway
Supplemental Data
References
- 1.Smith J.M., Stablein D.M., Munoz R., Hebert D., McDonald R.A. Contributions of the Transplant Registry: The 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS) Pediatr. Transplant. 2007;11:366–373. doi: 10.1111/j.1399-3046.2007.00704.x. [DOI] [PubMed] [Google Scholar]
- 2.Chen F. Genetic and developmental basis for urinary tract obstruction. Pediatr. Nephrol. 2009;24:1621–1632. doi: 10.1007/s00467-008-1072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woolf A.S. A molecular and genetic view of human renal and urinary tract malformations. Kidney Int. 2000;58:500–512. doi: 10.1046/j.1523-1755.2000.00196.x. [DOI] [PubMed] [Google Scholar]
- 4.Pohl M., Bhatnagar V., Mendoza S.A., Nigam S.K. Toward an etiological classification of developmental disorders of the kidney and upper urinary tract. Kidney Int. 2002;61:10–19. doi: 10.1046/j.1523-1755.2002.00086.x. [DOI] [PubMed] [Google Scholar]
- 5.Sanna-Cherchi S., Sampogna R.V., Papeta N., Burgess K.E., Nees S.N., Perry B.J., Choi M., Bodria M., Liu Y., Weng P.L. Mutations in DSTYK and dominant urinary tract malformations. N. Engl. J. Med. 2013;369:621–629. doi: 10.1056/NEJMoa1214479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saisawat P., Kohl S., Hilger A.C., Hwang D.Y., Yung Gee H., Dworschak G.C., Tasic V., Pennimpede T., Natarajan S., Sperry E. Whole-exome resequencing reveals recessive mutations in TRAP1 in individuals with CAKUT and VACTERL association. Kidney Int. 2014;85:1310–1317. doi: 10.1038/ki.2013.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang D.Y., Dworschak G.C., Kohl S., Saisawat P., Vivante A., Hilger A.C., Reutter H.M., Soliman N.A., Bogdanovic R., Kehinde E.O. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 2014;85:1429–1433. doi: 10.1038/ki.2013.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohl S., Hwang D.Y., Dworschak G.C., Hilger A.C., Saisawat P., Vivante A., Stajic N., Bogdanovic R., Reutter H.M., Kehinde E.O. Mild recessive mutations in six Fraser syndrome-related genes cause isolated congenital anomalies of the kidney and urinary tract. J. Am. Soc. Nephrol. 2014;25:1917–1922. doi: 10.1681/ASN.2013101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruf R.G., Xu P.X., Silvius D., Otto E.A., Beekmann F., Muerb U.T., Kumar S., Neuhaus T.J., Kemper M.J., Raymond R.M., Jr. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc. Natl. Acad. Sci. USA. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vivante A., Kohl S., Hwang D.Y., Dworschak G.C., Hildebrandt F. Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr. Nephrol. 2014;29:695–704. doi: 10.1007/s00467-013-2684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu W., van Eerde A.M., Fan X., Quintero-Rivera F., Kulkarni S., Ferguson H., Kim H.G., Fan Y., Xi Q., Li Q.G. Disruption of ROBO2 is associated with urinary tract anomalies and confers risk of vesicoureteral reflux. Am. J. Hum. Genet. 2007;80:616–632. doi: 10.1086/512735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halbritter J., Porath J.D., Diaz K.A., Braun D.A., Kohl S., Chaki M., Allen S.J., Soliman N.A., Hildebrandt F., Otto E.A., GPN Study Group Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum. Genet. 2013;132:865–884. doi: 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichikawa I., Kuwayama F., Pope J.C., 4th, Stephens F.D., Miyazaki Y. Paradigm shift from classic anatomic theories to contemporary cell biological views of CAKUT. Kidney Int. 2002;61:889–898. doi: 10.1046/j.1523-1755.2002.00188.x. [DOI] [PubMed] [Google Scholar]
- 14.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E., Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 16.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 17.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanna-Cherchi S., Kiryluk K., Burgess K.E., Bodria M., Sampson M.G., Hadley D., Nees S.N., Verbitsky M., Perry B.J., Sterken R. Copy-number disorders are a common cause of congenital kidney malformations. Am. J. Hum. Genet. 2012;91:987–997. doi: 10.1016/j.ajhg.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halbritter J., Diaz K., Chaki M., Porath J.D., Tarrier B., Fu C., Innis J.L., Allen S.J., Lyons R.H., Stefanidis C.J. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J. Med. Genet. 2012;49:756–767. doi: 10.1136/jmedgenet-2012-100973. [DOI] [PubMed] [Google Scholar]
- 20.Otto E.A., Hurd T.W., Airik R., Chaki M., Zhou W., Stoetzel C., Patil S.B., Levy S., Ghosh A.K., Murga-Zamalloa C.A. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat. Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraus F., Haenig B., Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech. Dev. 2001;100:83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- 22.Bohnenpoll T., Bettenhausen E., Weiss A.C., Foik A.B., Trowe M.O., Blank P., Airik R., Kispert A. Tbx18 expression demarcates multipotent precursor populations in the developing urogenital system but is exclusively required within the ureteric mesenchymal lineage to suppress a renal stromal fate. Dev. Biol. 2013;380:25–36. doi: 10.1016/j.ydbio.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 23.Airik R., Bussen M., Singh M.K., Petry M., Kispert A. Tbx18 regulates the development of the ureteral mesenchyme. J. Clin. Invest. 2006;116:663–674. doi: 10.1172/JCI26027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farin H.F., Bussen M., Schmidt M.K., Singh M.K., Schuster-Gossler K., Kispert A. Transcriptional repression by the T-box proteins Tbx18 and Tbx15 depends on Groucho corepressors. J. Biol. Chem. 2007;282:25748–25759. doi: 10.1074/jbc.M703724200. [DOI] [PubMed] [Google Scholar]
- 25.Farin H.F., Mansouri A., Petry M., Kispert A. T-box protein Tbx18 interacts with the paired box protein Pax3 in the development of the paraxial mesoderm. J. Biol. Chem. 2008;283:25372–25380. doi: 10.1074/jbc.M802723200. [DOI] [PubMed] [Google Scholar]
- 26.Nie X., Sun J., Gordon R.E., Cai C.L., Xu P.X. SIX1 acts synergistically with TBX18 in mediating ureteral smooth muscle formation. Development. 2010;137:755–765. doi: 10.1242/dev.045757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Omari K., De Mesmaeker J., Karia D., Ginn H., Bhattacharya S., Mancini E.J. Structure of the DNA-bound T-box domain of human TBX1, a transcription factor associated with the DiGeorge syndrome. Proteins. 2012;80:655–660. doi: 10.1002/prot.23208. [DOI] [PubMed] [Google Scholar]
- 28.Müller C.W., Herrmann B.G. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature. 1997;389:884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- 29.Naiche L.A., Harrelson Z., Kelly R.G., Papaioannou V.E. T-box genes in vertebrate development. Annu. Rev. Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- 30.Xu J., Nie X., Cai X., Cai C.L., Xu P.X. Tbx18 is essential for normal development of vasculature network and glomerular mesangium in the mammalian kidney. Dev. Biol. 2014;391:17–31. doi: 10.1016/j.ydbio.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bussen M., Petry M., Schuster-Gossler K., Leitges M., Gossler A., Kispert A. The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes Dev. 2004;18:1209–1221. doi: 10.1101/gad.300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapoor N., Liang W., Marbán E., Cho H.C. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiese C., Grieskamp T., Airik R., Mommersteeg M.T., Gardiwal A., de Gier-de Vries C., Schuster-Gossler K., Moorman A.F., Kispert A., Christoffels V.M. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 34.Hu Y.F., Dawkins J.F., Cho H.C., Marbán E., Cingolani E. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci. Transl. Med. 2014;6:245ra94. doi: 10.1126/scitranslmed.3008681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.