Abstract
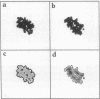
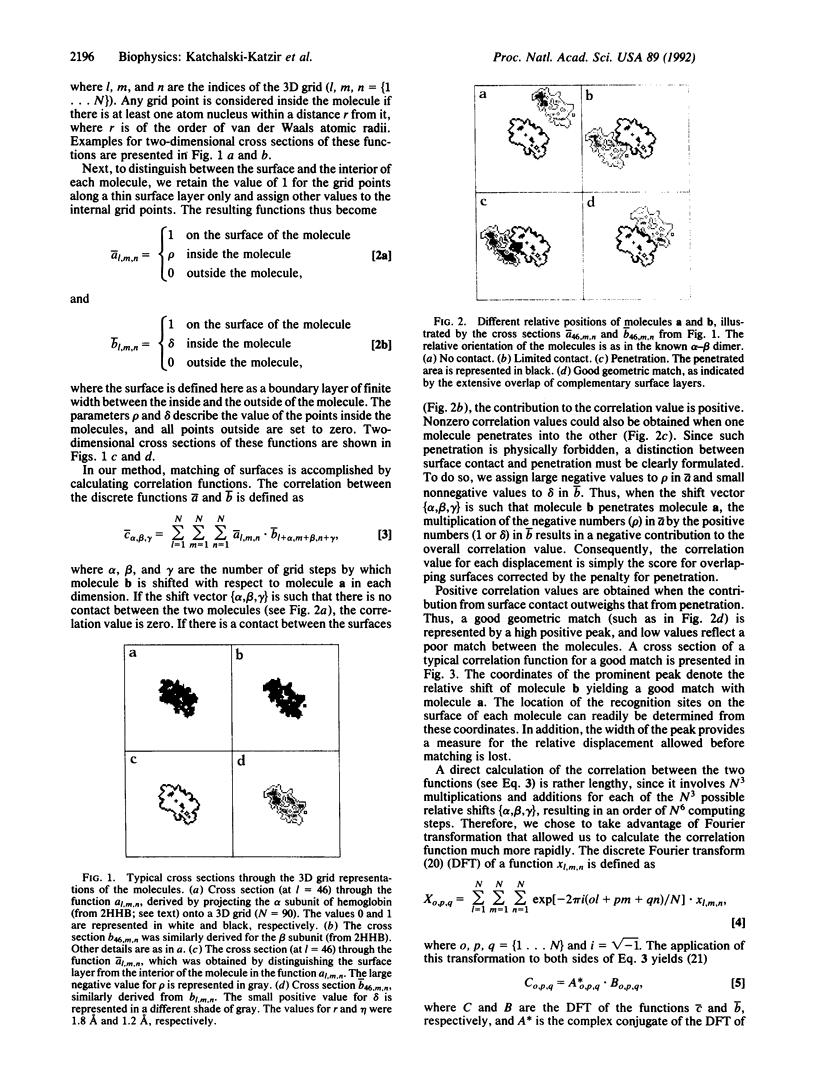
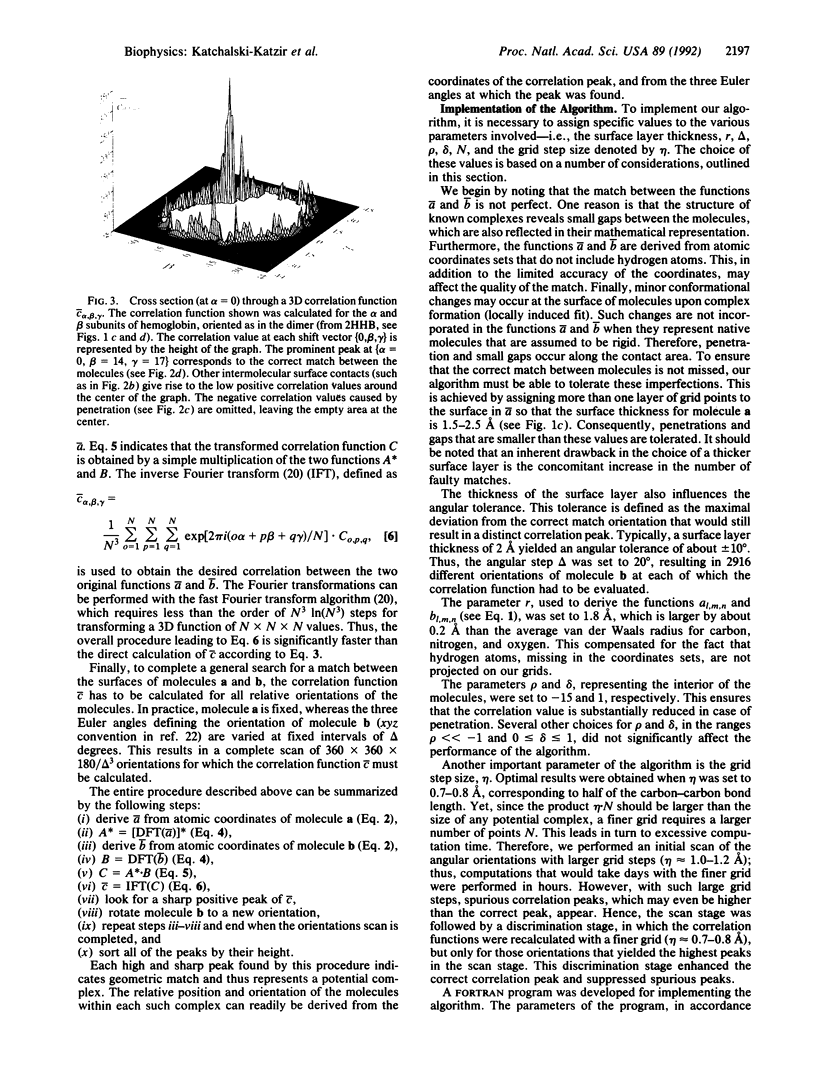
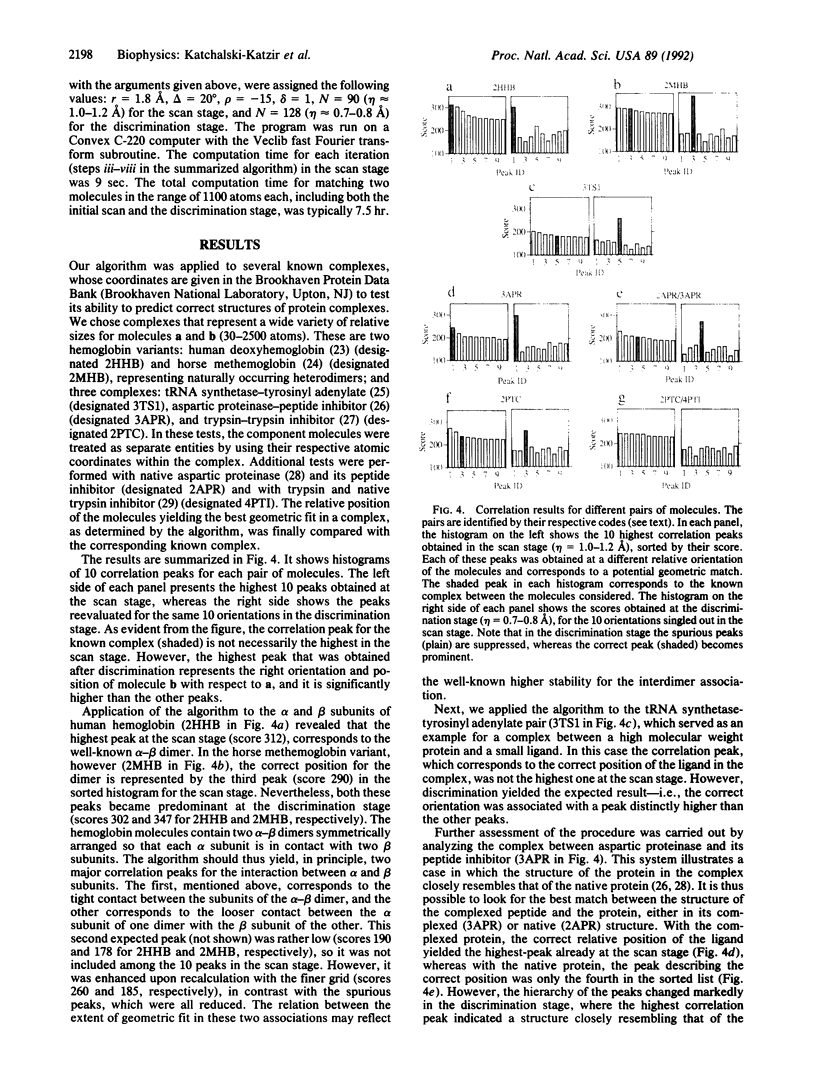
A geometric recognition algorithm was developed to identify molecular surface complementarity. It is based on a purely geometric approach and takes advantage of techniques applied in the field of pattern recognition. The algorithm involves an automated procedure including (i) a digital representation of the molecules (derived from atomic coordinates) by three-dimensional discrete functions that distinguishes between the surface and the interior; (ii) the calculation, using Fourier transformation, of a correlation function that assesses the degree of molecular surface overlap and penetration upon relative shifts of the molecules in three dimensions; and (iii) a scan of the relative orientations of the molecules in three dimensions. The algorithm provides a list of correlation values indicating the extent of geometric match between the surfaces of the molecules; each of these values is associated with six numbers describing the relative position (translation and rotation) of the molecules. The procedure is thus equivalent to a six-dimensional search but much faster by design, and the computation time is only moderately dependent on molecular size. The procedure was tested and validated by using five known complexes for which the correct relative position of the molecules in the respective adducts was successfully predicted. The molecular pairs were deoxyhemoglobin and methemoglobin, tRNA synthetase-tyrosinyl adenylate, aspartic proteinase-peptide inhibitor, and trypsin-trypsin inhibitor. A more realistic test was performed with the last two pairs by using the structures of uncomplexed aspartic proteinase and trypsin inhibitor, respectively. The results are indicative of the extent of conformational changes in the molecules tolerated by the algorithm.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Billeter M., Havel T. F., Kuntz I. D. A new approach to the problem of docking two molecules: the ellipsoid algorithm. Biopolymers. 1987 Jun;26(6):777–793. doi: 10.1002/bip.360260602. [DOI] [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Chirgadze Y. u., Kurochkina N., Nikonov S. Molecular cartography of proteins: surface relief analysis of the calf eye lens protein gamma-crystallin. Protein Eng. 1989 Nov;3(2):105–110. doi: 10.1093/protein/3.2.105. [DOI] [PubMed] [Google Scholar]
- Connolly M. L. Shape complementarity at the hemoglobin alpha 1 beta 1 subunit interface. Biopolymers. 1986 Jul;25(7):1229–1247. doi: 10.1002/bip.360250705. [DOI] [PubMed] [Google Scholar]
- DesJarlais R. L., Sheridan R. P., Seibel G. L., Dixon J. S., Kuntz I. D., Venkataraghavan R. Using shape complementarity as an initial screen in designing ligands for a receptor binding site of known three-dimensional structure. J Med Chem. 1988 Apr;31(4):722–729. doi: 10.1021/jm00399a006. [DOI] [PubMed] [Google Scholar]
- Fanning D. W., Smith J. A., Rose G. D. Molecular cartography of globular proteins with application to antigenic sites. Biopolymers. 1986 May;25(5):863–883. doi: 10.1002/bip.360250509. [DOI] [PubMed] [Google Scholar]
- Fermi G., Perutz M. F., Shaanan B., Fourme R. The crystal structure of human deoxyhaemoglobin at 1.74 A resolution. J Mol Biol. 1984 May 15;175(2):159–174. doi: 10.1016/0022-2836(84)90472-8. [DOI] [PubMed] [Google Scholar]
- Goodford P. J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem. 1985 Jul;28(7):849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- Goodsell D. S., Olson A. J. Automated docking of substrates to proteins by simulated annealing. Proteins. 1990;8(3):195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- Greer J., Bush B. L. Macromolecular shape and surface maps by solvent exclusion. Proc Natl Acad Sci U S A. 1978 Jan;75(1):303–307. doi: 10.1073/pnas.75.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Kim S. H. "Soft docking": matching of molecular surface cubes. J Mol Biol. 1991 May 5;219(1):79–102. doi: 10.1016/0022-2836(91)90859-5. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D., Blaney J. M., Oatley S. J., Langridge R., Ferrin T. E. A geometric approach to macromolecule-ligand interactions. J Mol Biol. 1982 Oct 25;161(2):269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- Ladner R. C., Heidner E. J., Perutz M. F. The structure of horse methaemoglobin at 2-0 A resolution. J Mol Biol. 1977 Aug 15;114(3):385–414. doi: 10.1016/0022-2836(77)90256-x. [DOI] [PubMed] [Google Scholar]
- Lewis R. A., Dean P. M. Automated site-directed drug design: the formation of molecular templates in primary structure generation. Proc R Soc Lond B Biol Sci. 1989 Mar 22;236(1283):141–162. doi: 10.1098/rspb.1989.0018. [DOI] [PubMed] [Google Scholar]
- Novotný J., Handschumacher M., Haber E., Bruccoleri R. E., Carlson W. B., Fanning D. W., Smith J. A., Rose G. D. Antigenic determinants in proteins coincide with surface regions accessible to large probes (antibody domains). Proc Natl Acad Sci U S A. 1986 Jan;83(2):226–230. doi: 10.1073/pnas.83.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet B. K., Kuntz I. D. Protein docking and complementarity. J Mol Biol. 1991 Sep 5;221(1):327–346. doi: 10.1016/0022-2836(91)80222-g. [DOI] [PubMed] [Google Scholar]
- Suguna K., Bott R. R., Padlan E. A., Subramanian E., Sheriff S., Cohen G. H., Davies D. R. Structure and refinement at 1.8 A resolution of the aspartic proteinase from Rhizopus chinensis. J Mol Biol. 1987 Aug 20;196(4):877–900. doi: 10.1016/0022-2836(87)90411-6. [DOI] [PubMed] [Google Scholar]
- Suguna K., Padlan E. A., Smith C. W., Carlson W. D., Davies D. R. Binding of a reduced peptide inhibitor to the aspartic proteinase from Rhizopus chinensis: implications for a mechanism of action. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7009–7013. doi: 10.1073/pnas.84.20.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwicker J. Investigating protein-protein interaction surfaces using a reduced stereochemical and electrostatic model. J Mol Biol. 1989 Mar 20;206(2):381–395. doi: 10.1016/0022-2836(89)90487-7. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Deisenhofer J., Huber R. Comparison of two highly refined structures of bovine pancreatic trypsin inhibitor. J Mol Biol. 1987 Jan 5;193(1):145–156. doi: 10.1016/0022-2836(87)90633-4. [DOI] [PubMed] [Google Scholar]
- Wodak S. J., Janin J. Computer analysis of protein-protein interaction. J Mol Biol. 1978 Sep 15;124(2):323–342. doi: 10.1016/0022-2836(78)90302-9. [DOI] [PubMed] [Google Scholar]
- Yue S. Y. Distance-constrained molecular docking by simulated annealing. Protein Eng. 1990 Dec;4(2):177–184. doi: 10.1093/protein/4.2.177. [DOI] [PubMed] [Google Scholar]
- Zielenkiewicz P., Rabczenko A. Protein-protein recognition: method for finding complementary surfaces of interacting proteins. J Theor Biol. 1984 Nov 7;111(1):17–30. doi: 10.1016/s0022-5193(84)80193-9. [DOI] [PubMed] [Google Scholar]
- Zielenkiewicz P., Rabczenko A. Searching for interacting surfaces of proteins--the improved method. J Theor Biol. 1985 Oct 21;116(4):607–612. doi: 10.1016/s0022-5193(85)80090-4. [DOI] [PubMed] [Google Scholar]