The Astrin/SKAP complex regulates mitotic chromosome alignment and centrosome integrity, but previous work found conflicting results for SKAP function. Here, Kern et al. demonstrate that a previously unappreciated short SKAP isoform mediates mitotic spindle positioning at astral microtubule plus ends.
Abstract
The Astrin/SKAP complex plays important roles in mitotic chromosome alignment and centrosome integrity, but previous work found conflicting results for SKAP function. Here, we demonstrate that SKAP is expressed as two distinct isoforms in mammals: a longer, testis-specific isoform that was used for the previous studies in mitotic cells and a novel, shorter mitotic isoform. Unlike the long isoform, short SKAP rescues SKAP depletion in mitosis and displays robust microtubule plus-end tracking, including localization to astral microtubules. Eliminating SKAP microtubule binding results in severe chromosome segregation defects. In contrast, SKAP mutants specifically defective for plus-end tracking facilitate proper chromosome segregation but display spindle positioning defects. Cells lacking SKAP plus-end tracking have reduced Clasp1 localization at microtubule plus ends and display increased lateral microtubule contacts with the cell cortex, which we propose results in unbalanced dynein-dependent cortical pulling forces. Our work reveals an unappreciated role for the Astrin/SKAP complex as an astral microtubule mediator of mitotic spindle positioning.
Introduction
Mitosis requires assembly of the microtubule-based mitotic spindle to provide the structure and forces for cell division. Multiple molecular players associate with the cell division apparatus to facilitate spindle assembly and chromosome segregation. Previous work from our laboratory and others identified the Astrin/SKAP complex (Schmidt et al., 2010; Dunsch et al., 2011), which comprises Astrin (also referred to as Spag5), the dynein light chain LC8, and the small kinetochore-associated protein SKAP/KNSTRN (Fang et al., 2009; also referred to as C15orf23, Traf4af1, or Kinastrin). The Astrin/SKAP complex is highly expressed in mitosis (Whitfield et al., 2002; Fang et al., 2009; Thiru et al., 2014), where it localizes to aligned kinetochores and the mitotic spindle and plays multiple important roles, including in chromosome alignment and the maintenance of spindle bipolarity (Mack and Compton, 2001; Gruber et al., 2002; Thein et al., 2007; Manning et al., 2010; Schmidt et al., 2010; Dunsch et al., 2011). Although SKAP plays a central role within this complex, previous work found conflicting results for its functions and behavior. Here, we find that the SKAP isoform used in all previous studies of the human protein is exclusively expressed in mammalian testes, whereas mitotic cells instead express a shorter SKAP isoform. Our analysis of the mitotic SKAP isoform reveals a striking localization of this protein along the length of spindle microtubules and to microtubule plus ends, including to astral microtubules, suggesting potential roles for this complex beyond its previously defined functions in chromosome segregation.
Microtubules emanating from the spindle poles interact with two major subcellular sites: kinetochores and the cell cortex. Whereas kinetochores link microtubules to chromosomal DNA to direct chromosome segregation, the cell cortex anchors astral microtubules to the plasma membrane to generate cortical pulling forces that direct spindle positioning and orientation. Spindle positioning is critical for organismal development and cellular viability (Gönczy, 2008; Siller and Doe, 2009; Knoblich, 2010). The position of the mitotic spindle within a dividing cell establishes the cell division plane and the site of the cytokinetic furrow, thereby defining the relative sizes of the two daughter cells. The force to move the spindle within a cell is generated by the interaction of astral microtubule plus ends with the microtubule-based motor cytoplasmic dynein, which is localized to the cell cortex (Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012; McNally, 2013; Kiyomitsu, 2015). Astral microtubules are a unique mitotic population of highly dynamic microtubules that originate from the centrosome and grow toward the cell cortex. When astral microtubules contact the cortex, dynein is thought to establish an “end-on” attachment and generate pulling force to move the spindle toward the cell cortex (Hendricks et al., 2012; Laan et al., 2012). The amount of pulling force on each side of the spindle is regulated through dynamic changes in the relative levels of cortical dynein (Collins et al., 2012; Kiyomitsu and Cheeseman, 2012). As a cell progresses from prometaphase into metaphase, the dynein motors on each side of the cell engage in a brief “tug-of-war” until the spindle is positioned at the cell center. In human cells, mitotic spindle position is controlled by both extrinsic and intrinsic cues (Fink et al., 2011; Kiyomitsu and Cheeseman, 2012). Much of the work on spindle positioning has focused on external or cortical factors, leaving open important questions regarding the function of astral microtubules. Although several microtubule plus-end proteins have been proposed to play roles in spindle positioning, including the end-binding (EB) proteins and Clasp1 (Rogers et al., 2002; Green et al., 2005; Samora et al., 2011; Bird et al., 2013), it remains unclear what protein components and properties of astral microtubule plus ends are required for their proper interactions with cortical dynein.
Our analysis reveals that the Astrin/SKAP complex plays important roles at astral microtubule plus ends for mediating proper spindle positioning. In cells with a plus-end tracking mutant of SKAP, chromosome segregation occurs normally, but metaphase spindles are dramatically mispositioned within the cell. We demonstrate that this spindle mispositioning occurs through an imbalance of forces generated by cortical dynein. SKAP plus-end tracking mutants display an apparent accumulation of lateral interactions between astral microtubules and the cell cortex. We propose that the Astrin/SKAP complex acts to mediate the proper connection between astral microtubule plus ends and cortical dynein.
Results
SKAP has both mitotic and testis/meiosis-specific isoforms
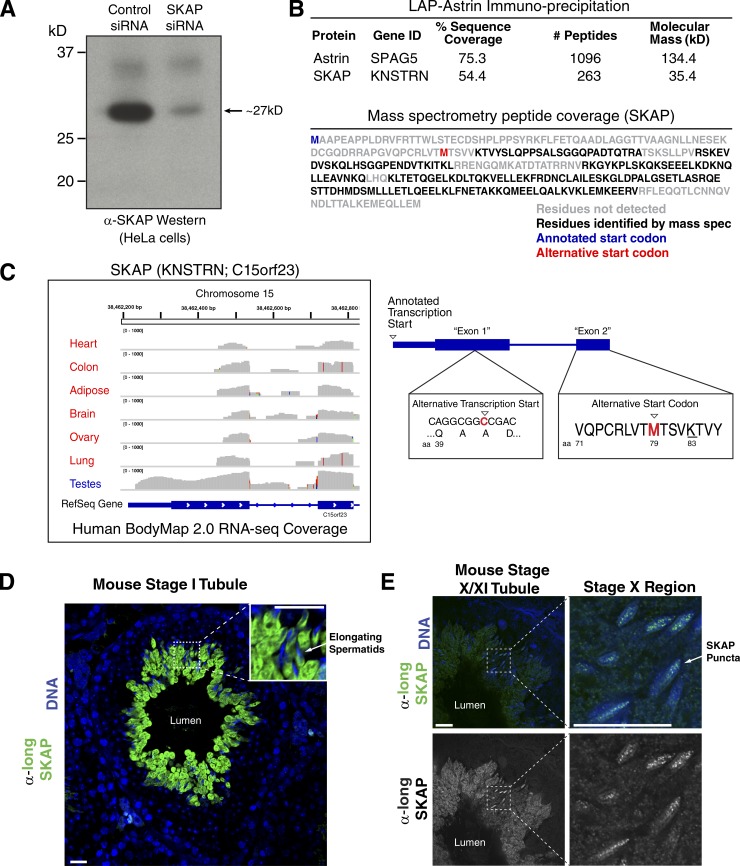
All previous studies on SKAP/KNSTRN have used a consensus annotated database protein sequence (ID: Q9Y448-1) with a predicted molecular mass of 34.5 kD. However, in analyzing the behavior of SKAP in human tissue culture cells, our affinity-purified anti-SKAP antibody detected a protein of ∼27 kD by SDS-PAGE and Western blotting (Fig. 1 A). Based on mass spectrometry analysis of endogenous SKAP isolated from HeLa cells, we were unable to detect peptides from a large region of the N terminus for the annotated SKAP protein (Fig. 1 B). In addition, in RNA-sequencing data from the Human BodyMap 2.0 database, we found that the only tissue with reads spanning the entire annotated SKAP sequence was testis (Fig. 1 C). Indeed, although we were unable to identify peptides corresponding to the annotated SKAP N terminus in mitotic cells based on a mass spectrometry analysis, immunoprecipitation (IP) of SKAP from adult mouse testes identified peptides corresponding to this N-terminal region, as well as copurifying peptides from Astrin (Fig. S1 A). In all other tissues, transcriptional initiation began within the first annotated exon, resulting in an mRNA lacking the previously defined start codon (Fig. 1 C; also see E-MTAB-513 in ArrayExpress: http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-513). Instead, the first in-frame, coding methionine appeared within the previously defined exon 2. The shorter SKAP isoform generated using this downstream start codon has a predicted molecular mass of 26.9 kD, consistent with our mass spectrometry and Western blot analysis (Fig. 1, A and B; and Fig. S1 B). Although SKAP and its binding partner Astrin are conserved throughout vertebrates (Fig. S1 C), the longer, testis-specific SKAP isoform is present only in eutherian mammals (Fig. S1, B and C).
Figure 1.
SKAP has distinct mitotic and testis-specific isoforms. (A) Western blot showing the molecular mass of endogenous SKAP from HeLa cells assessed by protein marker migration. (B, top) Immunoprecipitation mass spectrometry (IP-MS) data pooled from LAP-Astrin IPs analyzed for Astrin and SKAP peptides. (bottom) Identified SKAP peptides mapped against the SKAP amino acid sequence (ID: Q9Y448-1). (C, left) Map of the 5′ end of the SKAP locus with RNA-sequencing reads from Human BodyMap 2.0. (right) Schematic of the transcript for the short (mitotic) SKAP isoform. The underlined lysine indicates the first peptide identifiable in a tryptic digest for short SKAP. (D) Confocal image of long (testis) SKAP IF localization to elongating spermatids in a mouse seminiferous (Stage I) tubule. The seminiferous tubule lumen is indicated. (top right) Zoom-in of elongating spermatids. (E) Testis section showing IF long (testis) SKAP localization to elongating spermatids in a mouse seminiferous tubule transitioning from developmental Stage X to XI. (left) Tubule region imaged with a 40× objective. (right) Boxed region (Stage X) imaged using a 100× objective to indicate DNA localized puncta (∼6× zoom). Bars, 20 µm.
To visualize the relative expression of the short and long SKAP isoforms, we used two distinct antibodies: one that detects both isoforms of the human SKAP protein (Schmidt et al., 2010) and one that we generated against the testis-specific N-terminal extension of the mouse SKAP protein (Fig. S1 B). Based on Western blotting, the short SKAP isoform was present in mitotic (nocodazole-arrested) mouse 3T3 cells, juvenile (6-d-old) mouse testis, and adult (70-d-old) mouse testis, which represent cells or tissues with mitotic populations (Fig. S1 D; also see Fig. S1 E). In contrast, we found that the long SKAP isoform was present only in adult mouse testes, which have initiated sperm development (Fig. S1 D). In addition, immunostaining of adult mouse testes with the antibody specific to the long SKAP isoform revealed clear expression in elongating spermatids within the seminiferous epithelium (Fig. 1 D). This localization began as a punctate pattern throughout the DNA (Fig. 1 E) in testis developmental stage X (Russell et al., 1990), then spread to the cytoplasm and increased through stages XI and XII, persisting in stages I and II, before decreasing in stages III and IV (not depicted). In addition to the high levels of expression during late spermatogenesis, immunohistochemistry revealed localization of the testis-specific SKAP isoform to meiosis I spindles (Fig. S1 F) and weaker localization to meiosis II spindles (not depicted). We did not detect spindle localization of the testis-specific SKAP isoform in mitotic cells of the testis, attesting to both the timing of expression and the specificity of the antibody. We also note that recent work found that both the short and long SKAP isoforms detected by Western blotting and by immunostaining using the antibody we generated were abolished in SKAP-knockout mice, and SKAP-knockout mice displayed spermatogenesis defects (Grey et al., 2016). Thus, SKAP is expressed as two isoforms with distinct transcriptional start sites, resulting in a shorter mitotic form (which we will refer to as “short SKAP”) and a longer testis-specific form (which we will refer to as “long SKAP”).
Short SKAP, but not long SKAP, is fully functional to facilitate mitosis
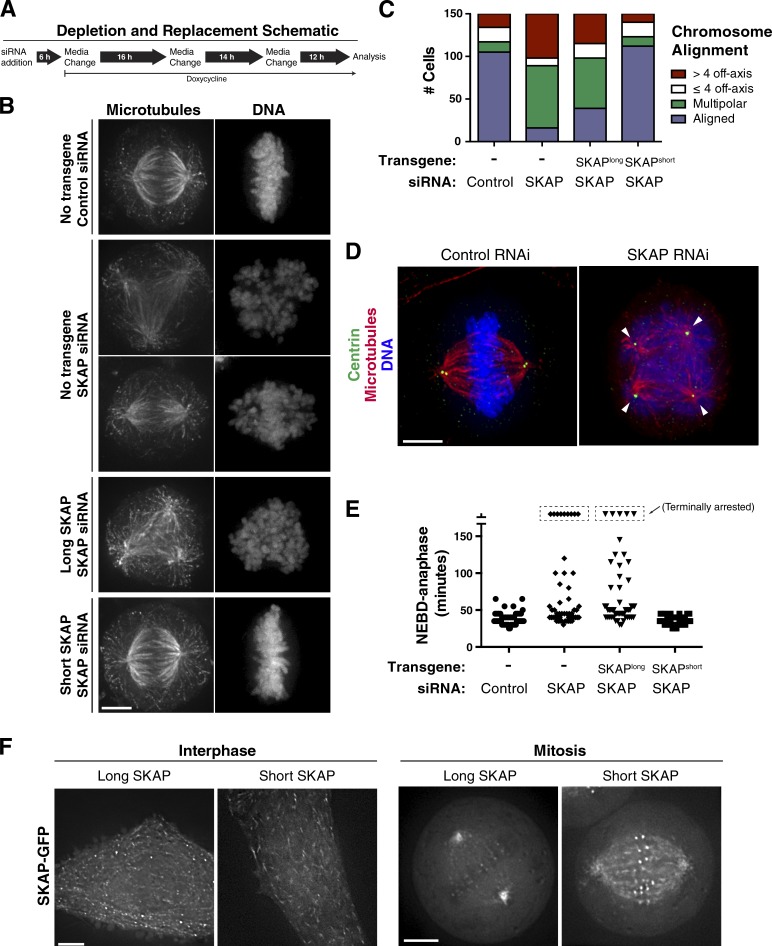
Prior studies on SKAP function in mitosis have relied on unrescued RNAi-based depletions (Schmidt et al., 2010) or ectopic expression of long SKAP (Lee et al., 2014; Tamura et al., 2015). With knowledge of these two SKAP isoforms, we next tested the functionality of each isoform in mitotic human cells by conducting rescue experiments. After SKAP depletion by RNAi (Figs. 2 A and S2 A), we observed dramatic defects in mitosis, including cells with highly misaligned chromosomes, disorganized spindles, and multipolar spindles that are a result of premature centriole splitting based on Centrin localization (Fig. 2, B–E; and Video 1; also see Schmidt et al., 2010; Dunsch et al., 2011). Expression of an RNAi-resistant version of the long (testis-specific) SKAP isoform was unable to rescue these defects (Fig. 2, B, C, and F; and Fig. S2, B and C; also see Materials and methods). In contrast, expression of the short SKAP isoform fully rescued SKAP depletion as assessed by proper SKAP localization (Fig. S2 B), chromosome alignment (Fig. 2, B and C), bipolar spindle assembly, and mitotic timing from nuclear envelope breakdown to anaphase (Figs. 2 E and S2 E). Therefore, the short (mitotic) isoform, but not the long (testis) isoform, rescues SKAP depletion defects in mitosis, demonstrating that these isoforms represent functionally distinct proteins.
Figure 2.
The long and short SKAP isoforms display distinct mitotic functionality. (A) Schematic of SKAP depletion and replacement protocol. Also see Fig. S2 A for a Western blot analyzing the efficiency of SKAP depletion and replacement. (B) IF images showing tubulin and DNA localization for the phenotypes exhibited during SKAP depletion and rescue experiments. From top to bottom: Aligned, multipolar spindles, misaligned (off-axis) chromosomes, multipolar spindles, and aligned chromosomes. Images represent maximal-intensity projections. (C) Quantification of mitotic chromosome alignment for SKAP depletion and rescue experiments from B. Cells were quantified by observing DNA and spindle morphology by IF. Cells were quantified for a phenotype if they displayed GFP reporter expression (if applicable) and displayed chromosome alignment and spindle and cell shape morphology indicative of metaphase or a multipolar mitotic cell morphology. (D) IF images showing a control cell with a bipolar spindle and paired centrioles and a SKAP-depleted multipolar cell with prematurely separated centrioles. (E) Quantification of mitotic timing (nuclear envelope breakdown [NEBD]-anaphase) as assessed from time-lapse videos at 40 h after siRNA addition (see Materials and methods). NEBD and anaphase were scored using DNA and cell morphology. Cells were counted if they entered mitosis during the course of filming. Cells that arrested for longer than 3 h or reached a terminal mitotic phenotype are boxed at the top of the graph. n = 52 cells per condition. Note: this analysis excludes cells in the depletion and long SKAP experiments that were arrested at the start of the video, and thus likely underrepresents the extent of the mitotic arrest. (F) Live cell imaging showing GFP-tagged SKAP cell lines for the two SKAP isoforms in interphase and mitosis. Bars, 5 µm.
To investigate the basis for this functional difference, we analyzed the associations and subcellular localization of the two SKAP isoforms as exogenously expressed GFP fusions (for a complete list of cell lines, see Table S1). We found that both long and short SKAP displayed largely similar interacting partners, including the other Astrin/SKAP complex components (Fig. S2 D). However, despite these similar interactions, the SKAP isoforms displayed distinct localization (Fig. 2 F, Video 2, and Fig. S2 F). In interphase cells, long SKAP-GFP displayed weak microtubule localization, as well as localization to punctate foci throughout the cytoplasm. In contrast, short SKAP-GFP displayed clear localization to microtubule plus ends in interphase cells. In mitosis, a GFP fusion to the long SKAP isoform localized to kinetochores, centrosomes, and spindle microtubules (Fig. 2 F), consistent with previous work (Schmidt et al., 2010). The short isoform of SKAP also localized to aligned kinetochores, spindle microtubules, and centrosomes in mitotic cells. However, the spindle localization of short SKAP was more intense along microtubule lengths and additionally displayed a speckled pattern typical of plus-end tracking proteins (Fig. 2 F), consistent with the interphase data. Therefore, the testes-specific long SKAP isoform is able to associate with the other components of the Astrin/SKAP complex when expressed ectopically in tissue culture cells, but displays distinct localization behavior. As such, long SKAP could act in a dominant manner by replacing endogenous short SKAP within the Astrin/SKAP complex, potentially explaining the defects reported previously for high-level ectopic expression of long SKAP (Lee et al., 2014; Tamura et al., 2015). Together, these analyses demonstrate that the two SKAP isoforms exhibit differential localization in mitotic cells, and that only the shorter SKAP isoform is fully functional to facilitate the multiple roles of SKAP during mitosis.
SKAP microtubule-binding activity is necessary for Astrin/SKAP spindle localization and chromosome segregation
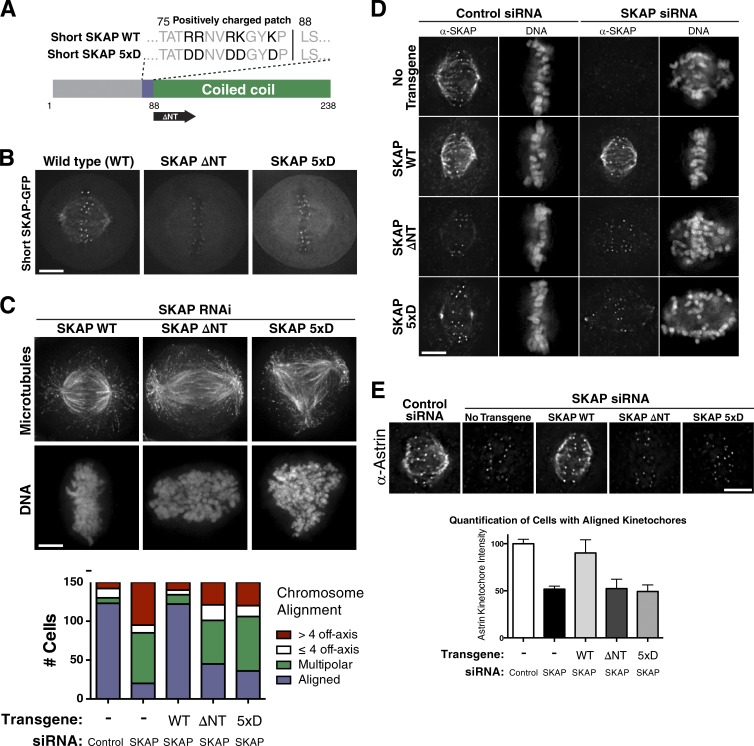
The short, mitosis-specific SKAP isoform displays localization both along the length of microtubules and to microtubule plus ends. To dissect the contributions of this localization to SKAP function, we first sought to completely eliminate the microtubule-binding activity in SKAP. SKAP contains a C-terminal coiled-coil region (Fig. 3 A), which we predicted would be responsible for the association of SKAP with the Astrin/SKAP complex, and an N-terminal region with a patch of positively charged residues immediately preceding the coiled coil that is well suited for associating with negatively charged microtubules. Cells in which endogenous SKAP was replaced by either a SKAP mutant lacking the entire N-terminal region (ΔNT) or a more specific mutant in which a cluster of five positively charged residues was targeted to reverse these charges (SKAP 5xD; Fig. 3 A) resulted in severe defects in chromosome alignment and spindle assembly (Fig. 3, B and C; and Fig. S2 G), similar to those observed after SKAP depletion. In addition, these SKAP ΔNT and 5xD mutants disrupted SKAP microtubule localization but did not prevent SKAP localization to kinetochores (Fig. 3, D and E). However, when quantifying cells with aligned metaphase plates, we did detect a similar relative reduction in kinetochore localization for Astrin in both SKAP-depleted cells and the SKAP microtubule-binding mutants (Fig. 3 E, bottom). Therefore, although SKAP microtubule binding is not strictly necessary for Astrin/SKAP complex kinetochore localization, it contributes to full complex localization at aligned kinetochores. Together, these data indicate that SKAP contains a major microtubule-binding activity of the Astrin/SKAP complex, and that this microtubule binding is critical for the spindle localization and function of this complex.
Figure 3.
SKAP microtubule-binding mutants prevent Astrin/SKAP complex microtubule localization and display defects similar to SKAP depletion. (A) Diagram of SKAP microtubule-binding mutants: a mutant with five positive charged amino acids changed to aspartate (SKAP 5xD) and mutant removing the N terminus to leave only the SKAP coiled coil (SKAP ΔNT). (B) Fluorescence images showing SKAP-GFP localization in cells expressing wild-type (WT) SKAP or the indicated SKAP microtubule-binding mutants. (C, top) IF images showing examples of cells with aligned chromosomes, many off-axis chromosomes, and a multipolar spindle from RNAi-based replacement experiments with SKAP WT, ΔNT, and 5xD, respectively. Images represent maximum-intensity projections. (bottom) Quantification of SKAP mutant cell phenotypes. (D) IF images showing SKAP and DNA localization in the indicated SKAP microtubule-binding mutants. Although most mutant cells display defective chromosome alignment, see Fig. S2 H for cells with aligned chromosomes to highlight the ability of SKAP to localize to kinetochores but not microtubules. DNA is scaled independently for each image. (E, top) IF images showing Astrin localization in the indicated conditions. See Fig. S2 I for cells displaying extreme phenotypes and misaligned plates. (bottom) Kinetochore intensity quantification for Astrin IF in indicated conditions. Each condition represents data from five cells and 50 total kinetochores. SEM is plotted for the averages from each cell in each condition. Using an unpaired two-tailed t test, control and rescue cell kinetochore intensities were statistically indistinguishable (P = 0.2464), and each was significantly different from SKAP-depleted or microtubule-binding mutants (P < 0.0001 for each). The SKAP-depleted and microtubule-binding mutant conditions were statistically indistinguishable from each other (SKAP depleted: ΔNT, P = .9110; SKAP depleted: 5xD, P = .5023; ΔNT: 5xD, P = .6038). Bars, 5 µm.
The short SKAP isoform displays robust plus-end tracking in mitosis
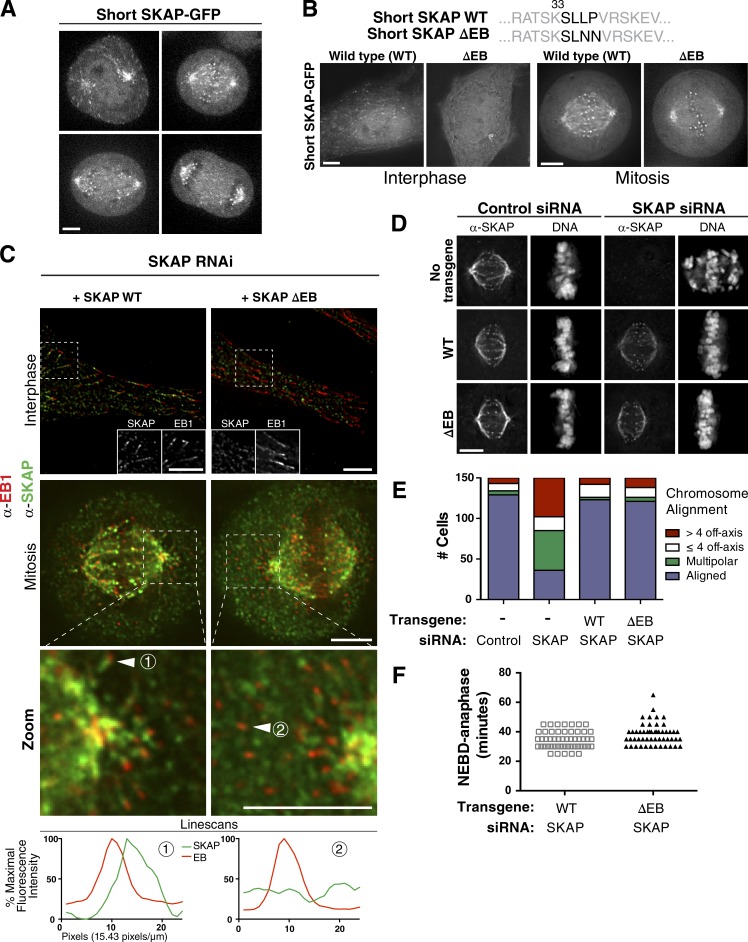
We next sought to precisely analyze the plus-end tracking activity of SKAP. Prior studies on the Astrin/SKAP complex reported plus-end tracking for GFP-Astrin expressed in interphase cells (Dunsch et al., 2011) and identified an interaction between SKAP and EB1 (Wang et al., 2012; Tamura et al., 2015). However, those previous studies did not report a plus-end tracking activity for SKAP in cells. Indeed, we found that the long SKAP isoform did not display robust plus-end tracking activity in interphase or mitosis (Fig. 2 F, Video 2, and Fig. S2 F), possibly because of steric effects or protein instability caused by the N-terminal extension present in long SKAP (Fig. S1 B). In contrast, the short SKAP isoform displayed spindle localization and fluorescent speckles along microtubules indicative of microtubule plus-end tracking (Fig. 2 F, Video 2, and Fig. S2 F). Using live cell imaging, we observed clear localization for short (mitotic) SKAP-GFP to growing microtubule plus-ends (Fig. 4, A and B; and Video 3). In addition, we found that the SKAP and Astrin plus-end signals trailed the extreme tip of the EB1 “comet” in both mitosis and interphase (Figs. 4 C and S3 A), potentially because of the additional microtubule-binding capability of the Astrin/SKAP complex and in agreement with previous studies for Astrin localization (Dunsch et al., 2011).
Figure 4.
Short SKAP displays potent EB motif–dependent microtubule plus-end tracking activity in mitotic cells. (A) Still images from spinning disc confocal videos of short SKAP-GFP (see Video 2). (B, top) Diagram of the SKAP ΔEB mutation. (bottom) Individual fluorescence sections from live cell imaging of mitotic SKAP and SKAP ΔEB mutant GFP-tagged cell lines. (C) Immunofluorescence localization of EB1 and SKAP. (top) SKAP overlaps with the trail of the EB comet, but does not colocalize with the tip. Inset shows a zoomed-in and channel separated view of the boxed region. (middle) Mitotic cells exhibiting wild-type (WT) SKAP plus-end tracking and the ΔEB mutant. (bottom) Maximum-intensity linescans of the microtubule plus ends marked with an arrow above. Intensities are plotted as a percentage of the maximum intensity quantified in this image set. (D) Individual fluorescence sections showing SKAP localization for the depletion and replacement experiments. DNA is scaled independently for each image. (E) Quantification of mitotic chromosome alignment for SKAP depletion and rescue experiments. Cells were quantified by observing DNA and spindle morphology (as in Fig. 2 C). (F) Mitotic timing (nuclear envelope breakdown [NEBD]-anaphase) quantified from videos (as in Fig. 3 E). n = 52 cells/condition. Based on a two-tailed t test, the datasets are statistically different (P = .0065). WT rescue data are duplicated from Fig. 3 E for comparison. Bars, 5 µm.
The plus-end tracking behavior for the Astrin/SKAP complex was dependent on the EB SKAP Sx[I/L]P motif (Honnappa et al., 2009), as mutation of the SKAP EB-interacting motif from SLLP to SLNN (here termed “SKAP ∆EB”) eliminated both SKAP and Astrin plus-end tracking (Fig. 4, B and C; and Fig. S3 A). Importantly, despite eliminating this plus-end tracking activity, the SKAP ΔEB mutant localized to both kinetochores and microtubules, even in the absence of endogenous SKAP (Fig. 4 D). We note that this result for short SKAP localization is different from that reported for long SKAP by others (Tamura et al., 2015), in which a comparable ΔEB mutant also eliminated SKAP microtubule localization. We suspect that long SKAP has reduced capacity to bind and track microtubules, which is consistent with our localization and phenotypic analysis (Fig. 2). Together with our analysis of the SKAP microtubule-binding mutants described earlier, this indicates that SKAP possesses independent microtubule-binding activity in addition to its association with EB family proteins.
To test the role of the SKAP microtubule plus-end tracking behavior in mitotic cells, we next investigated the functional consequences of replacing endogenous SKAP with the short SKAP ΔEB mutant (Fig. S3 B). In contrast to the catastrophic chromosome segregation defects that prevent microtubule localization observed in SKAP mutants (Fig. 3 C), we found that the SKAP ΔEB mutant was able to rescue the depletion of endogenous SKAP for its ability to facilitate proper chromosome alignment and segregation (Fig. 4, D and E) with normal mitotic timing (Fig. 4 F). Therefore, we conclude that Astrin/SKAP plus-end tracking is not necessary for the key chromosome alignment functions of this complex.
Astrin/SKAP localization to microtubule plus ends is necessary for proper spindle positioning
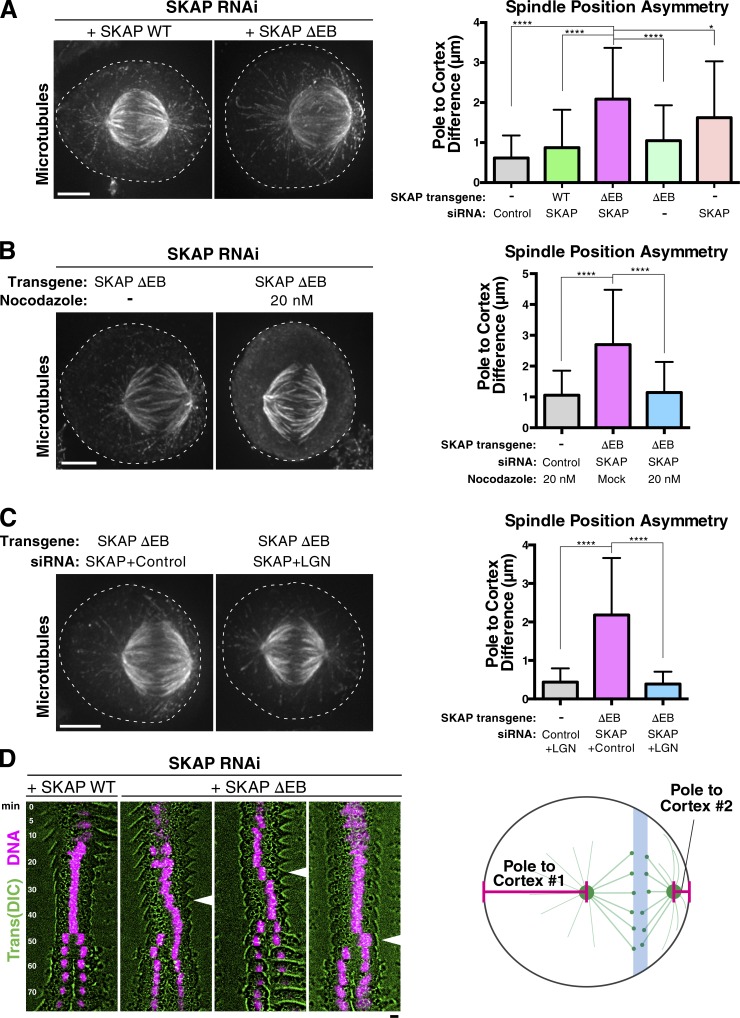
Although the SKAP ΔEB mutant was able to facilitate normal chromosome segregation, during our analysis of chromosome alignment, we unexpectedly observed that many spindles in the SKAP ΔEB mutant were dramatically mispositioned away from the cell midzone, often adjacent to or abutting the cell cortex (Fig. 5 A). In these extreme cases, cells appeared to have centrosomes and their astral microtubules directly in contact with the cortex (Fig. S3 C). In control cells or cells in which wild-type SKAP rescued endogenous SKAP depletion, we found that the spindle was typically positioned symmetrically within the dividing cells with similar distances between each spindle pole and the cell cortex (mean spindle displacement: control, 0.62 µm; rescue, 0.87 µm; Fig. 5 A). In contrast, we found that the spindle was positioned asymmetrically within the cell in SKAP ΔEB mutant cells such that one spindle pole was often much closer to the cell cortex (mean spindle displacement: ΔEB, 2.1 µm; Fig. 5 A), including cells with extremely mispositioned spindles (Fig. 5 A). Expression of the SKAP ΔEB mutant without depletion of the endogenous protein resulted in a modest, but statistically significant, shift in spindle positioning (mean spindle displacement: 1.05 µm; Fig. 5 A), potentially because of the formation of mixed Astrin/SKAP complexes. SKAP-depleted cells display dramatic and pleiotropic defects in chromosome alignment and centrosome stability (Fig. 2), which can influence spindle positioning indirectly (Kiyomitsu and Cheeseman, 2012; Tame et al., 2016), thereby preventing a directed analysis of spindle positioning phenotypes in SKAP-depleted cells with more extreme phenotypes (see Video 1). To test for spindle positioning defects in SKAP-depleted cells without substantial secondary defects, we quantified cells with clearly defined metaphase plates, which likely represent cells with an intermediate SKAP depletion. In these depleted cells, we found substantial spindle mispositioning (mean spindle displacement: 1.62 µm; Fig. 5 A), although slightly less than that observed in the SKAP ΔEB mutant. We conclude that SKAP ΔEB mutant replacement has a potent and specific effect on spindle positioning.
Figure 5.
SKAP ΔEB mutant cells display a spindle mispositioning defect. (A, left) IF images showing microtubules in the SKAP ΔEB mutant spindle. Images represent maximum-intensity projections. The dashed line indicates the cell boundary. (right) Graph showing difference in the distances between each spindle pole and the closest position on the cell cortex (see diagram at bottom right of the figure). A value of 0 represents spindle positioning in the cell center, with equivalent distances to each cell cortex. n = 100 total cells per condition collected from two (ΔEB without depletion) or three independent experiments. Mean and SD are plotted. ****, P < 0.0001, significant difference assessed by an unpaired two-tailed t test. *, difference with P = 0.0153. (B, left) Images showing maximum-intensity projections for microtubule staining to show for spindle mispositioning phenotype and its suppression by LGN depletion. (right) Graph showing the pole–cortex difference for LGN experiment (plotted as in A). n = 100 cells per condition collected from three independent experiments. ****, P < 0.0001, significant difference assessed by an unpaired two-tailed t test. (C, left) Maximum-intensity projections for microtubule staining show the spindle mispositioning phenotype and its suppression by 20 nM nocodazole. (right) Graph showing the pole–cortex difference (as in A and B) for the low-dose nocodazole experiment. n = 100 cells per condition collected from two independent experiments. Mean and SD are plotted. ****, P < 0.0001, significant difference assessed by an unpaired two-tailed t test. (D) Kymographs of videos from cells in which wild-type (WT) SKAP or the SKAP ΔEB mutant replaces endogenous SKAP. Arrows mark the start of the spindle shift in ΔEB cells. Also see Video 4. DIC, differential interference contrast.
To determine the nature of this spindle positioning defect, we next asked whether the mispositioning in the SKAP ΔEB mutants depended on the defined components of the spindle positioning pathway: astral microtubules and cortically localized dynein. Eliminating astral microtubules using low-dose nocodazole treatment (Théry et al., 2005) prevented spindle mispositioning in the SKAP ΔEB mutant (Fig. 5 B). Similarly, depletion of LGN, an upstream cortical recruitment factor for dynein that is required for cortical dynein localization in metaphase (Kiyomitsu and Cheeseman, 2012), eliminated the spindle positioning defects observed for the SKAP ΔEB mutant, resulting instead in a central spindle position (Fig. 5 C). These results demonstrate that aberrant and unbalanced forces generated by astral microtubules and cortical dynein lead to the dramatic spindle positioning defects in the absence of Astrin/SKAP localization to microtubule plus ends.
Based on time-lapse imaging of mitotic cells, SKAP ΔEB mutant cells displayed a directed shift in the position of the metaphase plate toward the cell cortex, usually shortly after chromosome congression (Fig. 5 D and Video 4). These rapid and directed movements are different from the slow spindle oscillations with a narrow amplitude that we have described previously as promoting a central spindle position (Kiyomitsu and Cheeseman, 2012). Such SKAP ΔEB mutant cells entered anaphase with mispositioned spindles but were able to subsequently position the spindle during early anaphase, consistent with an anaphase correction pathway (Kiyomitsu and Cheeseman, 2013). Thus, microtubule plus-end tracking by the Astrin/SKAP complex is necessary for metaphase spindle positioning.
The SKAP plus-end tracking mutant alters astral microtubule behavior at the cell cortex
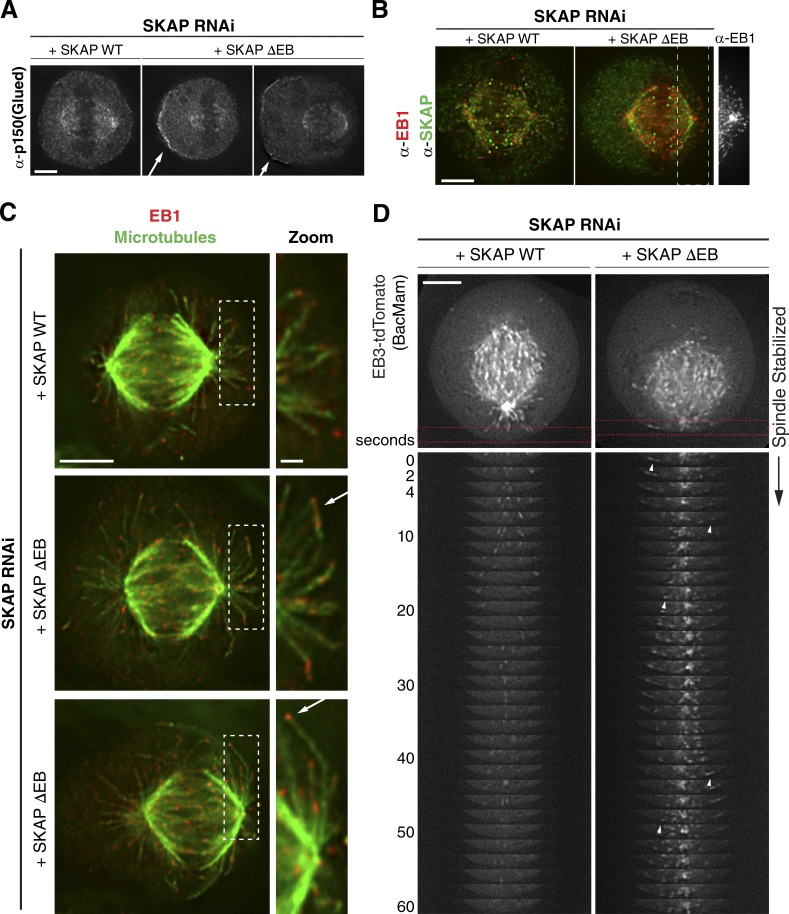
The SKAP ΔEB mutant phenotype could be explained by a failure of dynein to delocalize from the cell cortex as the spindle approaches (Kiyomitsu and Cheeseman, 2012), leading to enhanced spindle forces on one side of the cell. Therefore, we tested whether SKAP ΔEB mutants displayed a misregulation of cortical dynein localization. Using the dynactin subunit p150Glued as a marker of dynein/dynactin localization (Kiyomitsu, 2015), we did not observe a change in cortical localization behavior in the SKAP ΔEB mutant. Similar to what we have reported previously for dynein in control cells (Kiyomitsu and Cheeseman, 2013), p150Glued was able to localize to the cell cortex, but when the spindle was significantly shifted to one side of the cell, p150Glued displayed substantially higher localization to the opposite cell cortex (Fig. 6 A).
Figure 6.
Mispositioned metaphase spindles in SKAP ΔEB mutant cells are accompanied by changes in astral microtubule behavior. (A) Individual sections showing IF localization of p150Glued as a marker for dynein/dynactin from cells in which wild-type (WT) SKAP or the SKAP ΔEB mutant replaces endogenous SKAP. Accumulation of cortical dynactin is marked with arrows. We were unable to find an example of aberrantly localized dynactin (i.e., higher on the cortex closer to the shifted spindle) in two independent experiments. (B) IF images (deconvolved sections) of mouse anti-EB1 localization in SKAP WT and SKAP ΔEB cells. (far right) EB1 tracks from the SKAP ΔEB mutant cell showing growth along cortex. Also see Fig. S3 D. (C) IF images (deconvolved sections) showing microtubule and rabbit anti-EB1 localization. Boxed regions are enlarged on right (∼2×) to show plus ends of microtubules growing laterally along the cortex. Images in this panel are scaled individually for clarity. (D) Montages generated from time-lapse videos of EB3-tdTomato in SKAP WT and SKAP ΔEB cells. Top images are the first frame of the video, with the red boxed region used to create the montage. EB3 comets that exhibit clear lateral growth along cortex are marked with arrows. For the SKAP ΔEB cell, Fiji stack registration was used to stabilize the spindle, which exhibited rocking during the course of the video. See Video 5. Bars: 5 µm; (inset/zoom) 1 µm.
In metaphase, the plus ends of astral microtubules contact the cell cortex, but microtubule–cortical contacts typically persist for only a few seconds before growth ceases and the microtubules depolymerize (Samora et al., 2011). Indeed, we only rarely detected microtubule plus ends in proximity to the cell cortex based on EB1 staining in control cells. In contrast, in SKAP ΔEB mutant cells in which we observed significant spindle shifts, we also observed an accumulation of apparent lateral contacts between growing astral microtubules and the cell cortex (Fig. 6 C; also see Fig. 5, A–C; and Fig. S3, C and D). Based on imaging of microtubule plus ends marked by EB3-tdTomato in live cells, we found that in the SKAP ΔEB mutant, astral microtubules often grew along the cell cortex (Fig. 6 D and Videos 5 and 6). This lateral astral microtubule growth increased as the spindle approached the cortex. In cells with substantially shifted spindles, most astral microtubules close to the cortex exhibited lateral cortical growth, whereas astral microtubules on the opposite side generated cortical contacts less frequently (Fig. 5, A–C; Fig. 6 C; Fig. S3 D; and Videos 5 and 6). We conclude that Astrin/SKAP complex localization to microtubule plus ends plays an important role in astral microtubule behavior and is required for proper interactions between astral microtubules and cortical dynein to promote spindle positioning.
The Astrin/SKAP complex binds to and regulates Clasp1 in mitosis
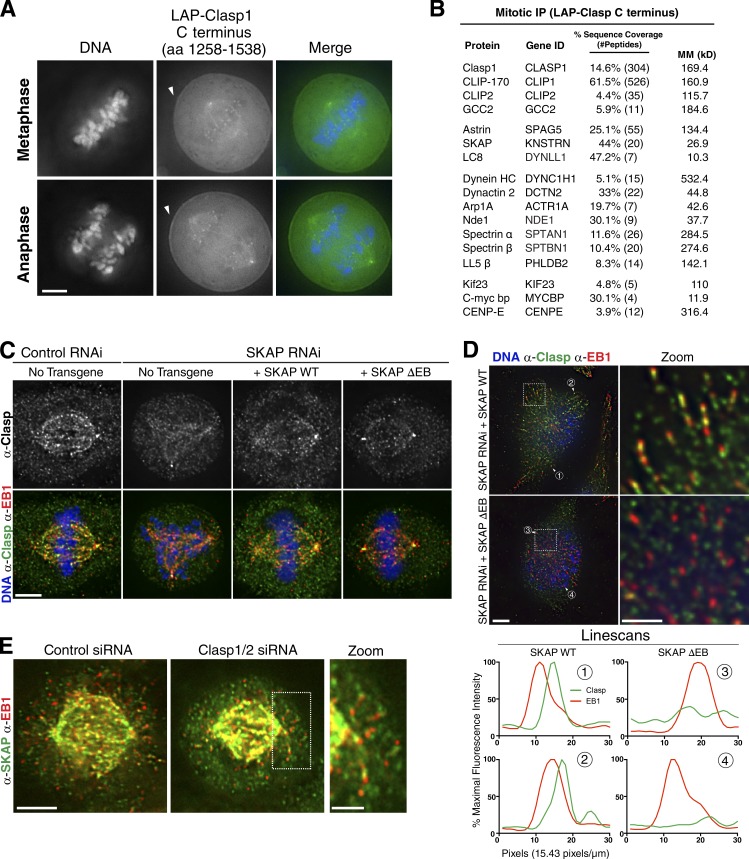
To explore the basis for this spindle positioning phenotype, we next generated an interaction map by conducting multiple immunoprecipitations of Astrin and SKAP from mitotic cells. We found that the Astrin/SKAP complex interacts either directly or indirectly with multiple key proteins that function in microtubule organization and spindle positioning, including Clasp1, Plk1, NuMA, and dynein (Fig. S4 A). Previous work found that Clasp1 plays an important role in microtubule–cortex interactions, spindle positioning, and astral microtubule behavior (Mimori-Kiyosue et al., 2005; Ambrose and Wasteneys, 2008; Samora et al., 2011; Bird et al., 2013). Indeed, Samora et al. (2011) observed spindle mispositioning after Clasp1 depletion. Therefore, we followed up on our results (Schmidt et al., 2010) and previous studies of an interaction between full-length Clasp1 and Astrin (Maffini et al., 2009; Manning et al., 2010). We found that the C-terminal domain of Clasp1 (Maiato et al., 2003), which localizes in mitosis to kinetochores, the spindle, and the cell cortex (Fig. 7 A), was sufficient to coimmunoprecipitate significant amounts of the Astrin/SKAP complex (Fig. 7 B). The Clasp1 C terminus also coimmunoprecipitated previously defined interacting partners including CLIP1/CLIP-170 and CENP-E (Patel et al., 2012), as well as dynein machinery (Fig. 7, A and B). However, affinity purification of the Clasp1 C terminus did not isolate peptides from full-length Clasp1, indicating that this construct is not multimerizing with endogenous Clasp1 and therefore narrowing down the Clasp1 binding region that is sufficient for the Astrin/SKAP complex interaction. To further define the basis for this interaction, we found that ectopically expressed SKAP C-terminal coiled-coil construct was immunoprecipitated by the Clasp1 C terminus (Fig. S4 C), suggesting that Clasp1 interacts with the intact Astrin/SKAP complex but not its microtubule-binding domain.
Figure 7.
The Astrin/SKAP plus-end tracking mutant alters Clasp1 localization behavior during mitosis. (A) Localization of LAP-Clasp1 C terminus in mitosis. Arrows indicate localization to cell cortex. (B) Immunoprecipitation of the LAP-Clasp1 C terminus from mitotic cells displaying identified peptides, with data pooled from two purifications with either 100 or 300 mM KCl in the buffer. (C) IF images of mitotic cells (either normal metaphase spindles or cells with a multipolar phenotype) showing the localization of Clasp1 and EB1. Clasp1 spindle and plus-end localization is reduced in the SKAP ΔEB condition. (D, left) Cocalization of Clasp1 to microtubule plus ends in early prophase cells in which endogenous SKAP is replaced by wild-type (WT) SKAP WT or the SKAP ΔEB mutant. Clasp1 localization to plus ends is eliminated in the SKAP ΔEB mutant. (right) Zoom from boxed region on the left images. (bottom) Maximum-intensity linescans of fluorescence intensity at the microtubule plus ends marked with the indicated numbers above. Intensities are plotted as percentages of the maximum intensity quantified in this image set. (E) IF images showing anti-SKAP and anti-EB1 in control cells or cells codepleted for Clasp1 and Clasp2 (HeLa Flp-In). SKAP localization to plus ends persists in this case. Bars: 5 µm; (inset/zoom) 2 µm.
We next tested the relationship between Clasp1 and SKAP for their localization to microtubules. In control cells, Clasp localized to kinetochores, the spindle, and microtubule plus ends during mitosis (Fig. 7 C). However, in the SKAP ΔEB mutant, we observed a reduction in spindle-bound Clasp and in Clasp localized to microtubule plus ends (Fig. 7, C and D). In contrast, SKAP depletion had no noticeable effect on Clasp plus-end microtubule localization in interphase cells (Fig. S4 B). In reciprocal experiments, we found that SKAP localized normally to microtubules and microtubule plus ends after the individual or simultaneous depletion of Clasp1 and Clasp2 (Fig. 7 E and Fig. S4, C and D). These results indicate that the Astrin/SKAP complex associates with Clasp1 and contributes to its localization in mitosis, including its robust localization to astral microtubule plus ends. In all, we demonstrated that the Astrin/SKAP complex displays multiple activities that may contribute to the connection between astral microtubules and the cell cortex. These include its intrinsic microtubule-binding activities, its association with Clasp1, and interactions with cytoplasmic dynein and dynein-associated proteins (Figs. 7 B and S4 A). The proper targeting of these Astrin/SKAP complex activities to microtubule plus ends is critical for the proper regulation of metaphase spindle position.
Discussion
We demonstrated that SKAP exists as two distinct transcriptional isoforms and analyzed the novel SKAP isoform in mitosis, which revealed an unexpected role in mitotic spindle positioning. Because the testis SKAP isoform is present only in eutherian mammals (Fig. S1 C) and shows marked sequence divergence even between mammals, it is possible that it has acquired new roles only recently in evolution. For example, it is possible that the N-terminal SKAP extension in testes modifies the structure and behavior of SKAP or facilitates interactions with testis-specific proteins. We note that previous papers from our laboratory and others have conducted studies on the function of SKAP in mitosis using the longer, testis-specific SKAP isoform (Schmidt et al., 2010; Dunsch et al., 2011; Wang et al., 2012; Lee et al., 2014; Tamura et al., 2015). Because this long isoform displays a large reduction or complete loss of microtubule plus-end tracking activity and a reduction in spindle localization and is unable to rescue endogenous SKAP depletion (Fig. 2), previous analyses of SKAP mutations and rescue phenotypes should be reevaluated in the context of this novel mitotic isoform. For example, a prior study identified a recurrent mutation in SKAP in skin cancers based on whole exome sequencing (Lee et al., 2014). However, the major identified mutation, S24F, exists in a portion of SKAP that is not detectably translated in mitotic cells, including in the cultured skin cells used for the study, according to the published Western blots (Lee et al., 2014). It remains possible that some cancers may induce the expression of the long SKAP isoform, that this mutation may instead alter short SKAP expression (because of the mutation’s close proximity to the transcriptional start site), or that this reflects a common background mutation because of a UV-sensitive hot spot occurring in skin cancer.
We find that the depletion of SKAP in human cells results in a range of severe defects in mitosis, including problems in chromosome alignment and the establishment and maintenance of a bipolar spindle. This indicates that SKAP, like its binding partner Astrin, functions in diverse mitotic processes, corresponding to its multiple localizations to kinetochores, microtubules, and centrosomes. Our work demonstrates that the microtubule-binding activity of SKAP is critical for the localization of the Astrin/SKAP complex to microtubules, but not kinetochores, and is required for the core mitotic functions of this complex. In addition to the previously defined roles for the Astrin/SKAP complex during mitosis, our analysis of the SKAP ΔEB mutant revealed an unexpected and unappreciated role for this complex in mitotic spindle positioning. Our work demonstrates that the Astrin/SKAP complex plays an important role at astral microtubule plus ends.
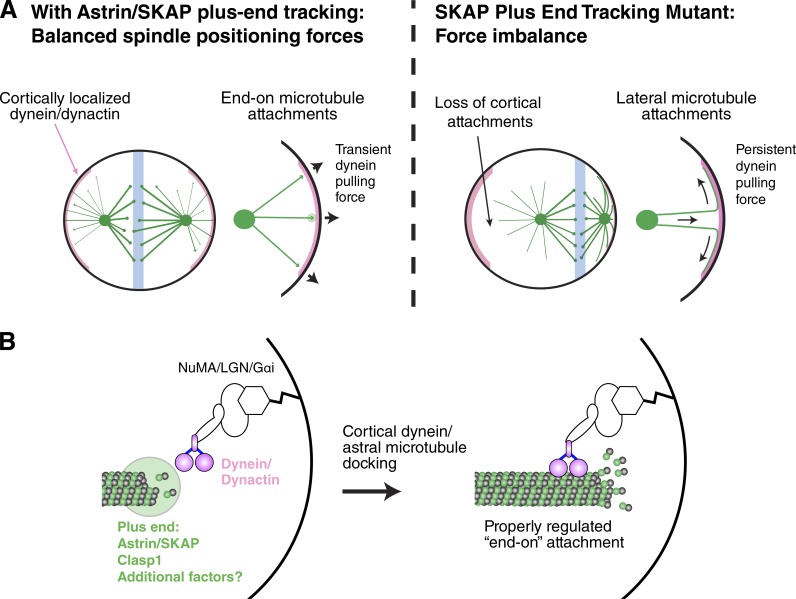
Astral microtubules in metaphase are characterized by transient growth and rapid turnover. Instead of mediating a stable connection with the cell cortex, astral microtubules only occasionally and briefly contact the cortex (Samora et al., 2011). These brief contacts allow cortically localized dynein to use the astral microtubules for transient force generation to drive spindle positioning. This creates a spindle centering system that is rapid and can be quickly regulated by changes in either astral microtubule behavior or cortical dynein recruitment. Interestingly, in the SKAP ΔEB mutant, astral microtubules are altered such that they appear to accumulate lateral, instead of end-on, cortical interactions. We propose that these laterally growing astral microtubules generate more persistent connections with cortical dynein. Because of the persistence of these force-generating interactions, the centrosome closer to the cortex would rapidly obtain a force advantage and the spindle would be pulled to one side of the cell (Fig. 8 A). As the spindle moves toward one side of the cell, the astral microtubules from the distal spindle pole would no longer be able to interact with the far cell cortex, preventing cells from correcting this imbalance, even though cortical dynein localization is still regulated to reduce its localization in the vicinity of the spindle pole (also see Kiyomitsu and Cheeseman, 2012). Thus, we propose that defective astral microtubule interactions with the cell cortex result in the dramatic spindle mispositioning defects that we observed in the SKAP ΔEB mutant. Recent work has described the basis for cortical dynein recruitment and the extrinsic and intrinsic cues that control spindle positioning (Kiyomitsu, 2015). Our work additionally establishes astral microtubule plus-end factors as critical players in spindle positioning.
Figure 8.
Model for Astrin/SKAP complex plus-end tracking activity in promoting proper spindle positioning. (A) Schematic for a model explaining spindle behavior in control cells (left) and SKAP ΔEB mutant cells (right). In wild-type cells, the Astrin/SKAP complex localizes to microtubule plus ends along with Clasp1 and other factors. In these cells, astral microtubules make transient, end-on contacts with the cell cortex and cortical dynein to generate short bursts of force to position the spindle. In SKAP ΔEB cells, the Astrin/SKAP complex is eliminated from microtubule plus ends. In this condition, metaphase astral microtubules make longer-lasting, lateral contacts with cortex and cortical dynein. These persistent connections lead to significant spindle position shifts from stochastic force imbalances and sustained pulling force from cortical dynein. (B) Hypothetical docking model for astral microtubules on cortex. We propose that the Astrin/SKAP complex, along with Clasp1 and other unknown proteins, mediates an engagement between microtubule plus ends and cortical dynein. This docking may be necessary for proper regulation of microtubule dynamics and dynein force production.
Astral microtubules have an array of plus end–associated proteins that modify their behavior (Akhmanova and Steinmetz, 2008, 2015). Our work adds the Astrin/SKAP complex to the list of bona fide astral microtubule plus-end trackers. In addition to the localization of SKAP to microtubule plus ends, we found that the Astrin/SKAP complex associates with diverse proteins that play roles in spindle positioning, including Clasp1 and dynein/dynactin Maiato et al., 2003; Samora et al., 2011; Duellberg et al., 2014). We found that Clasp1 behavior is regulated in mitosis by Astrin/SKAP complex, which may contribute to promoting proper astral microtubule behavior and cortical–microtubule interactions. The combined interactions and activities of these and other proteins may create a modular complex that plays a specific role upon cortical contact (Fig. 8 B). A critical challenge for future work is to define the nature of this astral microtubule–cortical attachment, including the composition and regulation of the astral microtubule plus ends and the activities required for microtubule–dynein engagement and force generation.
Materials and methods
Cell line generation
HeLa Flp-In cells were generated using HeLa LacZeo/TO cells (a gift from S. Taylor, University of Manchester, Manchester, England, UK) and pCDNA5-FRT-TO–based plasmids (Invitrogen). For untagged SKAP constructs with a GFP expression reporter, we generated a vector by fusing the IRES and eGFP sequences from the pIRES2eGFP vector (Clontech) after the open reading frame of the pCDNA5-FRT-TO–based plasmid. Cell lines were made using Flp recombinase–mediated integration based on the manufacturer instructions for the Flp-In TRex System (Invitrogen). Clonal cell lines from retroviral infection were made as described previously (Cheeseman et al., 2004). Cell lines (HeLa, HeLa Flp-In, and 3T3) were maintained in DMEM supplemented with 100 U/ml streptomycin, 100 U/ml penicillin, 2 mM glutamine, and 10% (vol/vol) FBS. Flp-In lines were cultured in HyClone FBS without tetracycline.
Protein depletion, induction, and BacMam expression
For siRNA transfection, we used Lipofectamine RNAiMAX (Invitrogen) and OptiMEM (Gibco). For protein depletion, we used ON-TARGETplus siRNAs (Dharmacon) against SKAP (5′-AGGCUACAAACCACUGAGUAA-3′; Dunsch et al., 2011), LGN (5′-GAACUAACAGCACGACUUA-3′; Kiyomitsu and Cheeseman, 2013), and a nontargeting control. Clasp1 (5′-GGAUGAUUUACAAGACUGG-3′) and Clasp2 (5′-GACAUACAUGGGUCUUAGA-3′; Mimori-Kiyosue et al., 2005) siRNAs were standard siRNAs (Dharmacon). All siRNAs were used at 40 nM. Double RNAi treatments used 40 nM of each siRNA. To rescue SKAP deletion, we developed a system using an inducible, siRNA-resistant, and untagged SKAP (with a GFP expression reporter). Induction of SKAP expression using 2 µg/ml doxycycline replaced at 12- to 16-h intervals throughout the multiday protocols (Figs. 2 A and S2 E) resulted in levels comparable to that of the endogenous protein (Fig. S2, A and G; and Fig. S3 B). We note that the expression of the long SKAP isoform was slightly less than that of short SKAP in these assays (Fig. S2 A), despite identical constructs and expression conditions. The observed expression difference may reflect an inherent instability of the long SKAP protein or the absence of a testis-specific binding partner. For phenotype/mitotic timing videos, imaging took place 40–48 h after siRNA addition. For all other depletion experiments, imaging was conducted at 48 h after siRNA addition. Transient transfections (Video 2) were performed using Effectene with imaging at 24 h after transfection. For EB3-tdTomato expression, we cloned the EB3-tdTomato (Addgene plasmid 50708; gift from E. Dent, University of Wisconsin, Madison, Madison, WI; Merriam et al., 2013) into pACEMAM1 (MultiBacMam; Geneva-Biotech) to generate a BacMam virus. The virus solution was added to cells at 24 h after siRNA addition and removed after 6 h of incubation. Cells were imaged 48 h after siRNA addition.
Antibody generation
The antibody to detect the mouse long SKAP N terminus was raised against GST-mouse long SKAP (aa 1–78), affinity purified using the same protein, and depleted for GST-specific antibodies using a GST column. The affinity-purified human Astrin antibody used for IP-mass spectrometry and immunofluorescence (IF) was raised against Astrin (aa 955–1,193-His) and purified using (aa 965–1,193-His). Antibody affinity purification was performed as described previously (Desai et al., 2003).
IF and Western blotting
IF and Western blotting (WB) were conducted using antibodies against human SKAP (Schmidt et al., 2010; 1:2,000 for IF, 1:4,000 for WB), rabbit anti-mouse SKAP N terminus (this study; 1:400 IF, 1:1,000 WB), tubulin (for human cell experiments: DM1α; Sigma-Aldrich; 1:1,000 for IF, 1:2,000 for WB; for mouse cell experiments: T5186; Sigma-Aldrich; 1:1,000 for IF, 1:2,000 for WB), Centrin2 (Backer et al., 2012; 1:12,000 for IF), rabbit anti-Astrin (this study; 1:2,000 for IF), mouse anti-Astrin (Yang et al., 2006; diluted from media for IF), mouse anti-EB1 (5/EB1; BD Transduction Laboratories; 1:250), rabbit anti-EB1 (H-70; Santa Cruz Biotechnology; 1:200), Clasp (Xorbit; 1:2,000 for IF; a gift from R. Heald, University of California, Berkeley, Berkeley, CA; Hannak and Heald, 2006), p150Glued (1/p150Glued; BD Transduction Laboratories; 1:250), goat anti-GFP (a gift of A. Ladurner, Ludwig Maximilians University of Munich, Munich, Germany; 1:2,000 for WB), and centromeres (ACA Antibodies; 1:100). For immunofluorescence, cells were plated on glass coverslips coated with poly-l-lysine (Sigma-Aldrich). For nocodazole experiments, the indicated concentration of nocodazole (Sigma-Aldrich) or vehicle control was added to cells for 1 h before fixation (47 h after siRNA addition). Fixation conditions were as follows: for IF with SKAP or Astrin visualized at kinetochores (Figs. 2 A, 3 C, 4 D, and S3 B), cells were first preextracted for 5 min in 0.1% Triton X-100 in PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 4 mM MgSO4, pH 7), followed by fixation for 10 min in 4% formaldehyde in PHEM buffer. For IF with microtubules visualized (including all experiments with spindle shifts quantified in Fig. 5), cells were fixed for 10 min in 4% formaldehyde in PHEM buffer. For IF with p150Glued, cells were fixed in PBS with 2% sucrose and 3% formaldehyde. For IF with EB1 or Centrin2 antibodies, cells were fixed for 5 min in methanol at −20°C. All other fixation steps were performed at RT immediately after removal from 37°C incubator. Blocking and primary antibody dilutions were performed using AbDil (20 mM Tris, 150 mM NaCl, 0.1% Triton X-100, 3% BSA, and 0.1% NaN3, pH 7.5). Washing and secondary antibody dilution steps were performed in PBS plus 0.1% Triton X-100 (PBS-TX). Cy2-, Cy3-, and Alexa Fluor 647–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were used at 1:300. DNA was visualized by incubating cells in 1 µg/ml Hoechst-33342 (Sigma-Aldrich) in PBS-TX for 2 min followed by PBS-TX rinsing. Coverslips were mounted using PPDM (0.5% p-phenylenediamine and 20 mM Tris-Cl, pH 8.8, in 90% glycerol).
SKAP models and phylogeny
The SKAP gene model (Fig. 2 B) was generated using the Integrated Genomics Viewer (Robinson et al., 2011) with data from Human BodyMap 2.0 (Illumina). SKAP proteins were aligned and visualized using Jalview software (Robinson et al., 2011). Phylogenetic tree was made using data from TimeTree (Hedges et al., 2006).
Live and fixed-cell imaging
Fixed-cell and live cell images were acquired on a DeltaVision Core microscope (Applied Precision) equipped with a CoolSnap HQ2 CCD camera (Photometrics). Images were deconvolved and planes projected (maximum intensity) as noted in the figure legends. Paired images are scaled equivalently unless otherwise noted. Fixed cells were imaged using a 100×, 1.4-NA U-PlanApo objective (Olympus). For live-cell imaging, cells were plated on 35-mm glass-bottom microwell dishes (MatTek) and imaged in CO2-independent media (Invitrogen) with 100 ng/ml Hoechst-33342 (Sigma-Aldrich) at 37°C. For mitotic timing/phenotype videos, to assess the IRES GFP reporter, points were selected and imaged before video start. For video imaging, points were imaged with three Z-sections 5 µm apart using fluorescent and transmitted light at the lowest level usable for data collection. Images were collected at 5-min intervals for 8 h using a using a 40×, 1.35-NA U-PlanApo objective (Olympus). For EB3-tdTomato videos, GFP-positive (transgene expressing) cells were selected and the 100× objective was used to collect images every 2 s for 60 s with a 0.5-s exposure time. Spinning disc confocal videos were taken with a Ti-E inverted microscope (Nikon) using a Revolution laser system (Andor) using an Apo 100× TIRF objective (NA 1.49; Nikon) and an iXon 897E EMCCD camera (Andor) in a 37°C chamber. Images were collected every 0.5 s for 30 s with a 240-ms exposure time. Images and videos were processed using Softworx (Applied Precision), Metamorph (Molecular Devices), ImageJ/Fiji, and Photoshop/Illustrator software (Adobe). Distance measurements for quantifications were made using Softworx and then plotted and analyzed using Prism (GraphPad). Kinetochore intensity measurements were made using Metamorph. Projected image stacks (maximal intensity) were quantified by selecting regions with kinetochores and nearby regions for background subtraction. Kymographs and line scans were generated using Metamorph. Statistical analyses (two-tailed t tests) were performed using Prism software where indicated.
Mouse immunostaining
Wild-type 129S2/SvPasCrl mice were purchased from Charles River Laboratories. All animals were housed at the Whitehead Institute for Biomedical Research and maintained according to protocols approved by the Massachusetts Institute of Technology Committee on Animal Care. Adult male mice (60–70 d old) were killed in CO2 and the testes were removed. For histologic analysis, testes were fixed in Bouin’s solution or 4% paraformaldehyde overnight and processed into paraffin wax, and 10-µm sections were cut and mounted on Superfrost slides. For immunohistology, tissues were rehydrated and blocked in 5% normal donkey serum. Primary antibodies (mouse SKAP N terminus) were applied in PBS with 5% normal donkey serum overnight at 4°C. Immunolocalization was performed by fluorescence (using FITC-conjugated donkey anti–rabbit [Jackson ImmunoResearch Laboratories] and mounted using Prolong mounting media with DAPI [Molecular Probes]) or colorimetrically (using ImmPRESS HRP anti-Rabbit detection and developed with a DAB substrate kit [Vector Laboratories], counterstained with Harris’s hematoxylin, and mounted using Permount [Fisher Scientific]).
IP and mass spectrometry
For direct antibody IP (Figs. S2 D and S4 A), the indicated antibodies were coupled to beads as described previously (Cheeseman and Desai, 2005). For mitotic IP, unless otherwise noted, cells were arrested with nocodazole (Sigma-Aldrich) at 330 nM for 14 h and harvested by shake-off. For interphase IPs, mitotic cells were removed by shake-off and the remaining cells were harvested. GFPLAP tagged proteins or endogenous proteins were isolated from HeLa cells as described previously (Cheeseman and Desai, 2005). Salt concentrations for bead binding and washing are indicated in the figure legends. Purified proteins were identified by mass LTQ XL Ion trap mass spectrometer (Thermo Fisher Scientific) using MudPIT and SEQUEST software as described previously (Washburn et al., 2001). For the mouse testis IP, testes were collected as described earlier from one 3-mo-old mouse. Testes were then detunicated and resuspended in HeLa lysis buffer (Cheeseman and Desai, 2005) to a final volume of ∼2 ml. For protease inhibition, one half of a Complete Mini, EDTA-free tablet (Roche) and 1 mM PMSF were added. This mix was subjected to Dounce homogenizer strokes until a suspension was achieved (as determined by light microscope). This mix was used with our standard localization and affinity purification (LAP) protocol (Cheeseman and Desai, 2005) using wash buffer with 150 mM KCl.
For IP followed by Western blot, 10-cm plates containing the LAP-Clasp1 C terminus stable cell line were incubated for 12 h with BacMam virus containing myc-SKAP (tag = MEQKLISEEDLGSS-) constructs. At 12 h, virus-containing media was replaced, and cells were incubated for 8 h before addition of 330 nM nocodazole. Cells were arrested in nocodazole for 15 h before harvest by shake-off. Each condition had ∼2 million recovered cells. Cells were lysed by sonication in 0.5 ml IP-Western buffer (50 mM Hepes, pH 7.4, 150 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 0.05% NP-40 substitute) with added 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, and 1 mM PMSF. Lysed cells were pelleted at 22,000 g for 10 min, and then 200 µl supernatant was bound for 2 h to 25 µl GFP-coupled beads (see LAP protocol) or 25 µl anti-myc (9E10)–coupled Sepharose beads (Covance). Cells were rinsed three times in 500 µl IP-Western buffer and then washed twice for 5 min. Bound protein was eluted with 100 µl of 100 mM glycine at pH 2.6. Elutions and input samples were prepared with gel loading buffer and loaded for Western blotting. For Western blotting, the SKAP full-length and goat anti-GFP antibodies were used.
Online supplemental material
Fig. S1 shows SKAP alignment, phylogeny, and further SKAP isoform characterization in testes. Fig. S2 shows interphase interactions of SKAP and validation of the SKAP depletion and replacement protocols. Fig. S3 shows immunofluorescence of Astrin on mitotic plus ends, validation of the SKAP ΔEB mutant replacement experiment, and additional characterization of the astral microtubule phenotype. Fig. S4 shows data for Astrin and SKAP interactions in mitotic cells, validation of Clasp reagents, and further characterization of the Clasp interaction with the Astrin/SKAP complex. Video 1 depicts SKAP depleted cells going through mitosis. Video 2 compares interphase localization of the short and long SKAP isoforms. Video 3 shows localization of SKAP on mitotic plus ends by spinning disc microscopy. Video 4 shows examples of mitotic cells undergoing a spindle shift in the SKAP ΔEB condition. Video 5 shows EB3-marked plus ends in control, depletion, and replacement conditions. Video 6 shows additional EB3-marked plus ends in examples of SKAP ΔEB replacement cells. Table S1 lists cell lines used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201510117/DC1.
Supplementary Material
Acknowledgments
We thank Tomomi Kiyomitsu, Terry Orr-Weaver, and members of Cheeseman, Page, and Burge Laboratories for their support and constructive comments; the Bioinformatics and Research Computing group at Whitehead for assistance with the analysis of SKAP isoforms; Wendy Salmon and the W.M. Keck Microscopy Facility at Whitehead for help with spinning disc microscopy; Dirk de Rooij for help with testis staging and identifying meiotic spindles; Corinne Grey and Bernard de Massy for discussions and sharing results before publication; and Rebecca Heald for suggesting and providing the Xorbit/Clasp1 antibody.
P.K. Nicholls is a Hope Funds for Cancer Research Fellow supported by the Hope Funds for Cancer Research (HFCR-15-06-06) and the recipient of an Early Career Fellowship (National Health and Medical Research Council GNT1053776). This work was supported by a Scholar award to I.M. Cheeseman from the Leukemia and Lymphoma Society, a grant from the National Institutes of Health/National Institute of General Medical Sciences to I.M. Cheeseman (GM088313), and a Research Scholar grant to I.M. Cheeseman from the American Cancer Society (121776).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- EB
- end binding
- IF
- immunofluorescence
- IP
- immunoprecipitation
- LAP
- localization and affinity purification
References
- Akhmanova A., and Steinmetz M.O.. 2008. Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., and Steinmetz M.O.. 2015. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16:711–726. 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- Ambrose J.C., and Wasteneys G.O.. 2008. CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Mol. Biol. Cell. 19:4730–4737. 10.1091/mbc.E08-06-0665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer C.B., Gutzman J.H., Pearson C.G., and Cheeseman I.M.. 2012. CSAP localizes to polyglutamylated microtubules and promotes proper cilia function and zebrafish development. Mol. Biol. Cell. 23:2122–2130. 10.1091/mbc.E11-11-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird S.L., Heald R., and Weis K.. 2013. RanGTP and CLASP1 cooperate to position the mitotic spindle. Mol. Biol. Cell. 24:2506–2514. 10.1091/mbc.E13-03-0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1. [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Niessen S., Anderson S., Hyndman F., Yates J.R. III, Oegema K., and Desai A.. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. 10.1101/gad.1234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins E.S., Balchand S.K., Faraci J.L., Wadsworth P., and Lee W.L.. 2012. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol. Biol. Cell. 23:3380–3390. 10.1091/mbc.E12-02-0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., and Oegema K.. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duellberg C., Trokter M., Jha R., Sen I., Steinmetz M.O., and Surrey T.. 2014. Reconstitution of a hierarchical +TIP interaction network controlling microtubule end tracking of dynein. Nat. Cell Biol. 16:804–811. 10.1038/ncb2999 [DOI] [PubMed] [Google Scholar]
- Dunsch A.K., Linnane E., Barr F.A., and Gruneberg U.. 2011. The astrin-kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J. Cell Biol. 192:959–968. 10.1083/jcb.201008023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Seki A., and Fang G.. 2009. SKAP associates with kinetochores and promotes the metaphase-to-anaphase transition. Cell Cycle. 8:2819–2827. 10.4161/cc.8.17.9514 [DOI] [PubMed] [Google Scholar]
- Fink J., Carpi N., Betz T., Bétard A., Chebah M., Azioune A., Bornens M., Sykes C., Fetler L., Cuvelier D., and Piel M.. 2011. External forces control mitotic spindle positioning. Nat. Cell Biol. 13:771–778. 10.1038/ncb2269 [DOI] [PubMed] [Google Scholar]
- Gönczy P. 2008. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9:355–366. 10.1038/nrm2388 [DOI] [PubMed] [Google Scholar]
- Green R.A., Wollman R., and Kaplan K.B.. 2005. APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol. Biol. Cell. 16:4609–4622. 10.1091/mbc.E05-03-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C., Espeut J., Ametsitsi R., Kumar R., Luksza M., Brun C., Verlhac M.H., Suja J.A., and de Massy B.. 2016. SKAP, an outer kinetochore protein, is required for mouse germ cell development. Reproduction. 151:239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber J., Harborth J., Schnabel J., Weber K., and Hatzfeld M.. 2002. The mitotic-spindle-associated protein astrin is essential for progression through mitosis. J. Cell Sci. 115:4053–4059. 10.1242/jcs.00088 [DOI] [PubMed] [Google Scholar]
- Hannak E., and Heald R.. 2006. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J. Cell Biol. 172:19–25. 10.1083/jcb.200508180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S.B., Dudley J., and Kumar S.. 2006. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics. 22:2971–2972. 10.1093/bioinformatics/btl505 [DOI] [PubMed] [Google Scholar]
- Hendricks A.G., Lazarus J.E., Perlson E., Gardner M.K., Odde D.J., Goldman Y.E., and Holzbaur E.L.. 2012. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr. Biol. 22:632–637. 10.1016/j.cub.2012.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J., Buey R.M., Lawera A., et al. . 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 138:366–376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T. 2015. Mechanisms of daughter cell-size control during cell division. Trends Cell Biol. 25:286–295. 10.1016/j.tcb.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., and Cheeseman I.M.. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317. 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., and Cheeseman I.M.. 2013. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 154:391–402. 10.1016/j.cell.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A. 2010. Asymmetric cell division: Recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11:849–860. 10.1038/nrm3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Busso C., and Gönczy P.. 2012. Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199:97–110. 10.1083/jcb.201203166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., López M.P., Vale R.D., Jülicher F., Reck-Peterson S.L., and Dogterom M.. 2012. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 148:502–514. 10.1016/j.cell.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Bhaduri A., Mah A., Johnson W.L., Ungewickell A., Aros C.J., Nguyen C.B., Rios E.J., Siprashvili Z., Straight A., et al. . 2014. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat. Genet. 46:1060–1062. 10.1038/ng.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack G.J., and Compton D.A.. 2001. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc. Natl. Acad. Sci. USA. 98:14434–14439. 10.1073/pnas.261371298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffini S., Maia A.R., Manning A.L., Maliga Z., Pereira A.L., Junqueira M., Shevchenko A., Hyman A., Yates J.R. III, Galjart N., et al. . 2009. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr. Biol. 19:1566–1572. 10.1016/j.cub.2009.07.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Fairley E.A., Rieder C.L., Swedlow J.R., Sunkel C.E., and Earnshaw W.C.. 2003. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 113:891–904. 10.1016/S0092-8674(03)00465-3 [DOI] [PubMed] [Google Scholar]
- Manning A.L., Bakhoum S.F., Maffini S., Correia-Melo C., Maiato H., and Compton D.A.. 2010. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 29:3531–3543. 10.1038/emboj.2010.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J. 2013. Mechanisms of spindle positioning. J. Cell Biol. 200:131–140. 10.1083/jcb.201210007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriam E.B., Millette M., Lumbard D.C., Saengsawang W., Fothergill T., Hu X., Ferhat L., and Dent E.W.. 2013. Synaptic regulation of microtubule dynamics in dendritic spines by calcium, F-actin, and drebrin. J. Neurosci. 33:16471–16482. 10.1523/JNEUROSCI.0661-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., and Akhmanova A.. 2005. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168:141–153. 10.1083/jcb.200405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K., Nogales E., and Heald R.. 2012. Multiple domains of human CLASP contribute to microtubule dynamics and organization in vitro and in Xenopus egg extracts. Cytoskeleton (Hoboken). 69:155–165. 10.1002/cm.21005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., Rogers G.C., Sharp D.J., and Vale R.D.. 2002. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158:873–884. 10.1083/jcb.200202032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L.D., Ettlin R.A., Sinha Hikim A.P., and Clegg E.D.. 1990. Histological and Histopathological Evaluation of the Testis. Cache River Press, Clearwater, FL: 286 pp. [Google Scholar]
- Samora C.P., Mogessie B., Conway L., Ross J.L., Straube A., and McAinsh A.D.. 2011. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat. Cell Biol. 13:1040–1050. 10.1038/ncb2297 [DOI] [PubMed] [Google Scholar]
- Schmidt J.C., Kiyomitsu T., Hori T., Backer C.B., Fukagawa T., and Cheeseman I.M.. 2010. Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J. Cell Biol. 191:269–280. 10.1083/jcb.201006129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K.H., and Doe C.Q.. 2009. Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11:365–374. 10.1038/ncb0409-365 [DOI] [PubMed] [Google Scholar]
- Tame M.A., Raaijmakers J.A., Afanasyev P., and Medema R.H.. 2016. Chromosome misalignments induce spindle-positioning defects. EMBO Rep. 17:317–325. 10.15252/embr.201541143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N., Simon J.E., Nayak A., Shenoy R., Hiroi N., Boilot V., Funahashi A., and Draviam V.M.. 2015. A proteomic study of mitotic phase-specific interactors of EB1 reveals a role for SXIP-mediated protein interactions in anaphase onset. Biol. Open. 4:155–169. 10.1242/bio.201410413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein K.H., Kleylein-Sohn J., Nigg E.A., and Gruneberg U.. 2007. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J. Cell Biol. 178:345–354. 10.1083/jcb.200701163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M., Racine V., Pépin A., Piel M., Chen Y., Sibarita J.B., and Bornens M.. 2005. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 7:947–953. 10.1038/ncb1307 [DOI] [PubMed] [Google Scholar]
- Thiru P., Kern D.M., McKinley K.L., Monda J.K., Rago F., Su K.C., Tsinman T., Yarar D., Bell G.W., and Cheeseman I.M.. 2014. Kinetochore genes are coordinately up-regulated in human tumors as part of a FoxM1-related cell division program. Mol. Biol. Cell. 25:1983–1994. 10.1091/mbc.E14-03-0837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhuang X., Cao D., Chu Y., Yao P., Liu W., Liu L., Adams G., Fang G., Dou Z., et al. . 2012. Mitotic regulator SKAP forms a link between kinetochore core complex KMN and dynamic spindle microtubules. J. Biol. Chem. 287:39380–39390. 10.1074/jbc.M112.406652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M.P., Wolters D., and Yates J.R. III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242–247. 10.1038/85686 [DOI] [PubMed] [Google Scholar]
- Whitfield M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., and Botstein D.. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 13:1977–2000. 10.1091/mbc.02-02-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.C., Hsu Y.T., Wu C.C., Chen H.T., and Chang M.S.. 2006. Silencing of astrin induces the p53-dependent apoptosis by suppression of HPV18 E6 expression and sensitizes cells to paclitaxel treatment in HeLa cells. Biochem. Biophys. Res. Commun. 343:428–434. 10.1016/j.bbrc.2006.02.166 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.