Abstract
Emerging strains of influenza, such as avian H5N1 and 2009 pandemic H1N1, are more virulent than seasonal H1N1 influenza, yet the underlying mechanisms for these differences are not well understood. Subtle differences in how a given strain interacts with the immune system are likely a key factor in determining virulence. One aspect of the interaction is the ability of T cells to locate the foci of the infection in time to prevent uncontrolled expansion. Here, we develop an agent based spatial model to focus on T cell migration from lymph nodes through the vascular system to sites of infection. We use our model to investigate whether different strains of influenza modulate this process.
We calibrate the model using viral and chemokine secretion rates we measure in vitro together with values taken from literature. The spatial nature of the model reveals unique challenges for T cell recruitment that are not apparent in standard differential equation models. In this model comparing three influenza viruses, plaque expansion is governed primarily by the replication rate of the virus strain, and the efficiency of the T cell search-and-kill is limited by the density of infected epithelial cells in each plaque. Thus for each virus there is a different threshold of T cell search time above which recruited T cells are unable to control further expansion. Future models could use this relationship to more accurately predict control of the infection.
Keywords: Agent-based model, Systems biology, Immunology, Computational biology, Virology
1. Introduction
Influenza has worldwide epidemic potential with 5 million infections annually and half a million deaths (WHO Influenza Fact Sheet, 2009). Emerging strains of influenza, such as 2003 H5N1 avian and 2009 H1N1 pandemic, are more pathogenic than seasonal strains, yet the mechanisms that control this variability are not well understood. Pathogenicity is a function of both the virus and its interaction with the immune response. Our model explores how various features of the virus and the host immune system interact to produce observed differences between strains.
Identifying the critical components of this complex process, and how they interact, is key to understanding viral pathogenicity and designing therapeutic interventions. We address this challenge through computational modeling, which allows us to study the relative contributions of different aspects of the host immune response and to account for strain-specific viral dynamics in the model. Several earlier theoretical models have been proposed to study specific aspects of influenza infections. Ordinary differential equation (ODE) models that are fit to empirical data (Handel and Antia, 2008; Lee et al., 2009; Miao et al., 2010; Saenz et al., 2010; Murillo et al., 2013; Crauste et al., 2015; Price et al., 2015) have elucidated viral population level dynamics but are unable to perceive localized spatial effects. Spatial models, on the other hand, have been developed to examine interactions between dendritic cells and T cells in the lymph node (Beauchemin et al., 2005; Beltman et al., 2007; Zheng et al., 2008; Mirsky et al., 2011; Celli et al., 2012; Vroomans et al., 2012; Textor et al., 2014), but have not been extended to consider local conditions in the lung. In this paper, we examine how the unique properties of three different strains of influenza, in the presence of chemokines, affect T-cell search in the human lung.
Specifically, we focus on the interactions between activated antigen-specific CD8 T cells, cytokines, and replicating influenza virus. Chemotactic proteins are known to enhance T cell recruitment in both acute infections and chronic inflammatory diseases (Gunn et al., 1998; Medoff et al., 2005; Okada et al., 2005; Castellino et al., 2006; Bromley et al., 2008). Infected epithelial cells secrete chemokines, especially upon contact with CD8 T cells (Zhao et al., 2000; Chan et al., 2005). Previous work showed that efficient recruitment of T cells to the Focus Of Infection (FOI) is crucial for the eventual clearance of the virus (Cerwenka et al., 1999; Kim et al., 2011). As measuring real-time T cell movement in in vivo lung tissue is not currently viable, study these localized processes is aided by spatial modeling of how T cells interact with the FOI and the chemotactic environment. Therefore, instead of developing a comprehensive immune system model, we present a spatially explicit agent-based model (ABM) to describe T cell interactions with chemotactic signals and a dynamically growing plaque. Using the model, we investigate the pathogenic potential of the three influenza strains: seasonal H1N1 (sH1N1), Avian H5N1 (aH5N1), and pandemic H1N1 (pH1N1). These three strains were selected for their differences in in vitro replication rates (Mitchell et al., 2011) as well as differences in in vivo severity of human and animal pneumonia and mortality.
The model is parameterized with values taken from the literature when available. Because chemokine secretion rates are central to our model and appropriate data values are not available, we estimate the parameters using data from physical experiments and our previously published ODE model (Mitchell et al., 2011).
To obtain the physical data, wells of human epithelial cells were infected with one of the three strains and then measured for virus and cytokine concentrations over a 48-hour period. Of the cytokines tested, only the chemokines CXCL10 (IP-10) and CCL5 (RANTES) were expressed differently across the three influenza strains. IP-10 and RANTES are both chemokines that normally attract T cells to the FOI (Hoji and Rinaldo, 2005; Groom and Luster, 2011a). These results suggest that the two chemokines may contribute to the immune system's varying ability to control different strains of influenza, and were thus included in our model.
The analyses of the spatial model reveal several interesting and underappreciated phenomena. The model identified physical constraints on T cells’ ability to clear the rapidly replicating pH1N1 infection. Because T cells in the model use the chemokine gradient to locate the FOI, they tend to cluster in areas of high chemokine concentration. The model suggests that the chemokine response of the host cells can hinder T cells ability to find and clear infected cells in the presence of the rapidly replicating pandemic strain. A sensitivity analysis of the model parameters isolated the effects of each parameter in the model. The analysis shows that most model parameters can vary across a broad range of values without strong effect on predicted outcomes. Further analysis shows model predictions remain unchanged within broad categories of parameter type (e.g., all parameters related to chemokines, parameters related to viral kinetics, etc.). For example, the chemokine-related parameters are stable in that they do not affect the model behavior within biologically plausible ranges. In contrast, parameters related to viral kinetics significantly alter model predictions. Finally we examine the ‘window of control’ which establishes an upper bound on the time it takes T cells to arrive at the FOI while still clearing the infection. We find that this value is consistent with our sensitivity analysis and helps explain the challenge of controlling the highly virulent pH1N1 strain.
2. Methods and models
2.1. Model definition
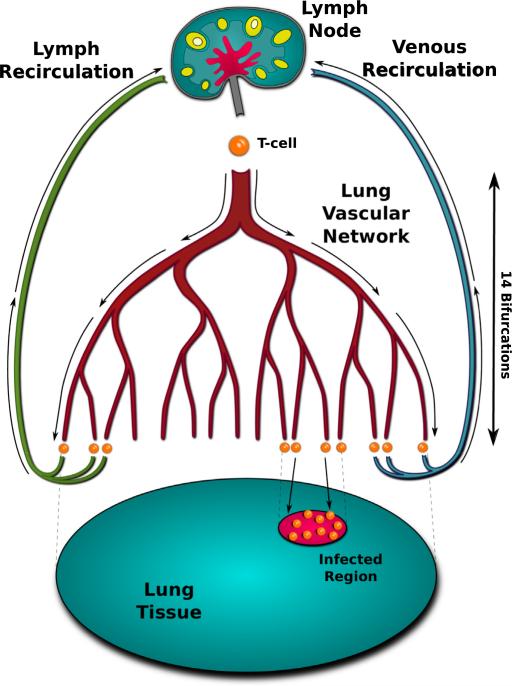
The model focuses on activated CD8 T cells migrating through the vascular and lymph networks, including their movement over tissue after extravasation (Fig. 1). While the volume of the lung consists of spherical alveoli, the spread of influenza throughout the lung occurs through the interconnected bronchiole-alveoli complex, which can be viewed as a connected monolayer. Thus, we represent the lung as a two-dimensional sheet of healthy epithelial cells (S1.3). Because we assume activated T cells descend at random into branches of the vascular network, T cells in the model are introduced uniformly at random across the modeled lung surface. If cytokine signal is detected on the local endothelium the T cell remains in tissue and follows the chemotactic gradient to the FOI. T cells that do not encounter cytokine recirculate to the lymph node where they reenter the bronchial vascular network. Once inside the endothelium, T cells follow the chemokine gradient to localized areas of maximum chemokine concentration. When a T cell contacts an infected epithelial cell it induces apoptosis (S1.2). Otherwise, infected cells continue to secrete virus for a fixed time and then die.
Fig. 1.
Model of T cell search. Activated T cells originate in the lymph node and enter the bloodstream after which they randomly navigate through 14 vascular bifurcations of the bronchial network. Upon reaching a capillary, T cells exit into tissue if cytokine signal is present. In the absence of signal, the T cell recirculates either through the lymph network or through the pulmonary vein back to the top of the network.
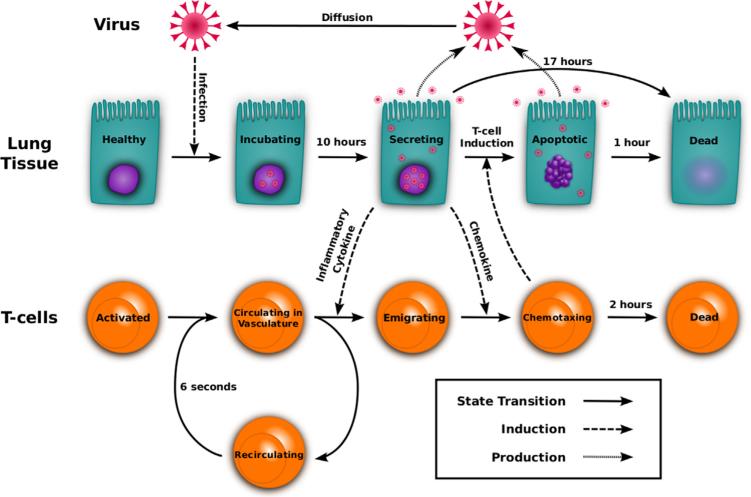
The model is initialized with a single infected cell. After the incubation period, the infected cell begins secreting virus and chemokine at rates determined by the virus strain and chemokine type (Table 1). Virus from expressing cells diffuses locally, infecting neighboring cells. Chemokine diffuses from secreting cells, creating a spreading region of stimulation around the FOI. After four simulated days, Immunoglobulin M (IgM) is introduced into the model by increasing the viral decay rate. After five simulated days, representing lymph node stimulation and T cell proliferation, activated T cells exit the lymph node at a constant rate and travel through the vasculature to the tissue as described above. These cell and molecular interactions and state transitions are depicted in Fig. 2.
Table 1.
Strain-specific parameters. Small text values show 95% confidence intervals resulting from 1000 bootstrapping runs for each parameter (Wu and Jackknife, 1986). Bootstrapping for the chemokine values was performed using the original fit of Eq. S1 to the data in Fig. 3 to produce new data sets. Viral production values and confidence intervals are taken from Mitchell et al. (2011).
| Strain | IP-10 production (pg/s · cell) | RANTES production (pg/s · cell) | Viral production (PFU/s · cell) |
|---|---|---|---|
| Avian H5N1 | 2.0e – 4 | 1.3e – 5 | 5.4e – 5 |
| 8.4e – 5–4.2e – 4 | 7.9e – 6–1.9e – 5 | 4.4e – 5–3.7e – 4 | |
| Seasonal H1N1 | 1.8e – 4 | 8.9e – 7 | 3.8e – 4 |
| 1.2e – 4–3.0e – 4 | 4.8e – 7–1.6e – 6 | 2.8e – 4–1.5e – 3 | |
| Pandemic H1N1 | 8.7e – 5 | 4.3e – 6 | 5.1e – 3 |
| 1.7e – 5–7.1e – 4 | 5.0e – 7–3.5e – 5 | 2.8e – 3–5.3e – 3 |
Fig. 2.
Visual representation of the model. Healthy epithelial cells infected by virus begin secreting virus after the incubation delay. Activated T cells traverse the bronchial vascular network and may be recruited by inflammatory cytokine. Chemotaxing T cells climb the chemokine gradient and induce apoptosis in infected cells. Solid arrows represent a cell state transition from one behavior to another. Dashed arrows display the mechanism used to induce a transition. Dotted arrows indicate the production of new virus.
2.2. Model parameters
We include only parameters that directly address the role of T cells and T cell migration because our study focuses on the role of T cells and the chemokine effects of T cells in influenza virus infection. Thus, we included parameters that affected virus, T cells, and chemokines. With regards to the influenza, we chose parameters involving viral replication, infectivity in epithelial cells, and virus decay and diffusion rates based on our earlier study. The chemokine decay rate, diffusion rate, and secretion rate parameters are relevant to dynamic chemokine gradients required for T cell chemotaxis. Because we wanted to use our model to test the role of T cells and T cell migration in influenza clearance, we included multiple T cell parameters, such as T cell production rate, T cell death rate, T cell kill time, and T cell migration rate. We added IgM because many studies have argued for the importance of antibody mediated virus clearance. However, because our study is not addressing the role of antibody in clearance of influenza, we did not test the full range of B cell responses directly by including them in our parameters.
Model parameters are listed in Tables 1 and 2. These values were taken from the literature when available. Other parameters were determined by matching empirical data to ODE models, and some parameters were estimated using biologically plausible ranges. All estimated parameters were studied in a sensitivity analysis to test their impact on model behavior (Table 3).
Table 2.
Default values used for the model in bold. Min and max represent the extreme values tested in the sensitivity analysis (Table 3 and Figs. S3–S5).
| Referenced parameters | Units | Min | Value | Max | Source |
|---|---|---|---|---|---|
| aViral diffusion in airway | μm2/s | 3.18e – 4 | 3.18e – 2 | 3.18 | Beauchemin et al. (2006) |
| aViral decay in airway | day–1 | 0.01 | 1 | 100 | Lee et al. (2009) |
| Chemokine diffusion rate | μm2/s | 3.18e – 3 | 0.318 | 318 | Beauchemin et al. (2006) |
| Incubation time | hours | 5 | 10 | 20 | Mitchell et al. (2011) |
| Epithelial cell radius | μm | – | 5 | – | Elbert et al. (1999) |
| T Cell radius | μm | – | 5 | – | Abbas et al. (2011) |
| T Cell production rate | cells/h | 125 | 1257 | 3750 | Miao et al. (2010) |
| T Cell speed | μm/min | 6e – 2 | 6 | 600 | Egen et al. (2011) |
| Blood circulation time | seconds | 1 | 6 | 3600 | Banerjee and Moses (2010) |
| T Cell sensitivity to chemokine | ng/mL | – | 100 | – | Nandagopal et al. (2011) |
| Onset of T Cell lymph node exit | days | – | 5 | – | Banerjee et al. (2011) |
| aIgM viral decay factor | – | 1 | 10 | 1000 | Diamond et al. (2003) |
| IgM onset | days | – | 4 | – | Diamond et al. (2003) |
| Estimated parametersb | Units | Min | Value | Max | Footnote |
|---|---|---|---|---|---|
| Chemokine decay rate | Hz | 3.8e – 6 | 3.8e – 4 | 3.8e – 2 | 1 |
| aInfectivity | min/virion | 12 | 120 | 1200 | 2 |
| Expression time | min | 100 | 1000 | 3000 | Mitchell et al. (2011) 3 |
| T Cell expected kill time | min | 0 | 10 | 100 | 4 |
| Apoptosis time | hours | 0 | 1 | 2 | Ganusov and De Boer (2008) 5 |
| T Cell age (at FOI) | min | 12 | 120 | 1200 | 6 |
| T Cell age (in blood) | days | 0.04 | 4 | 400 | 6 |
Denotes parameters determined to be sensitive by the one-factor-at-a-time sensitivity analysis. Values were taken from experimental literature if possible and from earlier modeling papers if not. Parameters not found in the literature were estimated as follows: (1) Corresponds to a 30 min half-life. (2) Epithelial cells are infected at a probabilistic rate such that the expected time for infection in the presence of a single virion is 2 h. This scales linearly with the number of virions in the cell's vicinity. (3) Chosen as a plausible median time (1000 min) between 6 h and 24 h. (4) T cells induce apoptosis in nearby virus-secreting epithelial cells at a probabilistic rate such that the expected time to induce apoptosis is 10 min. This rate does not scale with T cell numbers. (5) Calculated for low T cell densities. (6) Chosen to be at the lower end of biologically plausible values because increased T cell counts are shown not to affect the model behavior.
All estimated parameter values are examined in the sensitivity analysis (Table 3, S2.3).
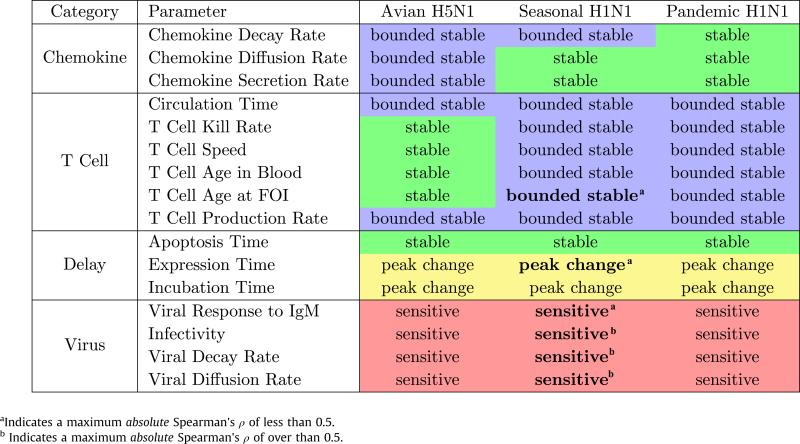
Table 3.
Sensitivity results: The above parameters were varied over predetermined ranges in isolation, resulting in new model runs for every new value tested (Figs. S3–S5). The results of the sensitivity analysis were then qualitatively evaluated for each individual parameter. A model run's behavior was determined by examining the height of the peak of the infection at day 5 post-infection and the number of infected cells at day 10 post-infection. Each combination of influenza strain and free parameter was classified as belonging to one of four categories. Parameters were classified as stable if all runs follow the same behavior, bounded stable if intermediate parameter adjustments did not affect the model's behavior, even if the more extreme adjustments did, peak change if the peak of the infection differs but the result at day 10 is the same, and sensitive if any level of change in the parameter affects the resulting model behavior. PRCC analysis was also performed for the sH1N1 strain over these parameters (Figs. S6–S8). Bold text in the seasonal column denotes significant Spearman rank correlation (p < 0.01) over the time period where the parameter was active.
Because we are interested in the interplay between the virus and the induced immune response, we infected human epithelial cells with the three different strains of influenza in vitro and measured the resulting cytokine and chemokine responses. We extended our previously published differential equation model (Mitchell et al., 2011) to obtain values for per-cell chemokine production rates. T cell production rates were derived from measured replication rates in vitro (Miao et al., 2010) using another differential equation model. T cell production is assumed to be constant after day 5 (Martin-Fontecha et al., 2003).
2.3. Model implementation
The model is implemented using an updated version of the CyCells software (Warrender et al., 2006), (github.com/drew-levin/cycells), a modeling platform for two- or three-dimensional agent-based simulations of the immune response (S1.1). Due to computational constrains our model is scaled to the size of a mouse lung (approximately 100 cm2), a well-established practice in spatially explicit influenza models (Miller et al., 2003; Allan et al., 2006; Ingulli et al., 2009). Parameters affected by this choice include the T cell production rate, T cell circulation time, and the total size of the lung (Table 2).
Parameters that are independent of the chosen influenza strain are shown in Table 2. Strain-specific values are shown in Table 1. Further details regarding the model definition are included in S1.2.
In the model, epithelial cells are stationary and are described by one of five sequential states: healthy, virus-incubating, virus-expressing, apoptotic, and dead (Bachem et al., 1996; Beauchemin et al., 2005; Mitchell et al., 2011). Healthy cells remain unchanged unless infected by virus. Once infected, the cell transitions from incubating to expressing after a 10 h incubation delay (Table 2). Expressing cells secrete virus and chemokine for a fixed 16.7 h and then die (Table 2). Expressing cells initiate apoptosis sooner if they are contacted by activated T cells. Apoptoic cells continue to secrete virus and chemokine and then die after one hour (Table 2). Dead cells take up space and do not regenerate over the course of an infection. Cell regrowth is not implemented as the rate of regrowth is unknown.
T cells are described by two states: circulating and chemotaxing. T cells emerge from the lymph node at five days post-infection (p.i.) at a rate of 1257 cells per hour (Models for Parameter Estimation, Table 2). T cell travel time from the lymph node to a random location on the lung's surface is six seconds. If chemokine is not encountered, the circulating T cell returns to a new location in the lung after another six seconds. If a circulating cell encounters chemokine, it changes state to chemotaxing and follows the chemotactic gradient to the FOI. Circulating T cells decay exponentially with an average lifespan of four days. Chemotaxing T cells follow the gradient through the two-dimensional lung endothelium, inducing apoptosis when they encounter expressing epithelial cells. Chemotaxing T cells decay exponentially with an average lifespan of two hours. Justifications for these parameter values are listed in Table 2.
In addition to T cells, the model contains two types of particles: virus and chemokine. Both are produced at constant rates by expressing epithelial cells. Virus diffuses through the lung tissue, infecting healthy cells at a rate proportional to the virus concentration at the location of the cell. Chemokine diffuses across the tissue but has no direct effect beyond recruiting T cells. Both particle types decay exponentially. IgM is modeled by increasing the viral decay rate by a factor of 10 after the fourth day (Table 2).
2.4. Models for parameter estimation
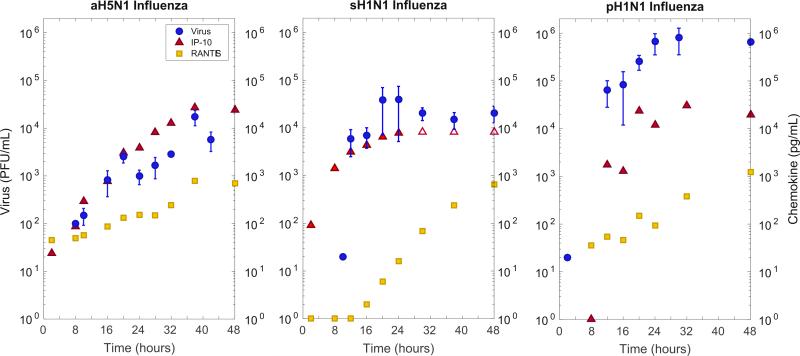
To provide estimates of chemokine concentrations and secretion rates in lung tissue, chemokine levels were measured at 4–6 h intervals during the first 48 h of infection in wells containing approximately one million human bronchial epithelial cells (Fig. 3, Table S1). The dynamic viral loads at these intervals have been reported previously by us for cultures infected with seasonal H1N1 virus, pandemic H1N1 virus, and avian H5N1 virus (Mitchell et al., 2011). IP-10 concentration increases were observed by 8 h post-infection (p.i.), and RANTES by 16 h p.i.
Fig. 3.
Empirical viral and cytokine titers for three strains of influenza: Avian H5N1, Seasonal sH1N1, and Pandemic pH1N1. Viral titer (blue circles) is in PFU/mL, and IP-10 (red triangle) and RANTES (yellow square) are shown in pg/mL. sH1N1 IP-10 secretion exceeded measurement accuracy above 8500 pg/mL and these three values (empty red triangles) were not included in the model fitting. An extended differential equation model from Mitchell et al. (2011) was fit to IP-10 and RANTES data (Eq. (1)). These fits were used to obtain chemokine production values for use in the spatial CyCells model. Human bronchial epithelial cells were infected at an MOI of 0.01 (10,000 virions) with one of the three strains of influenza. Apical fluid for viral secretion and basal media for chemokine secretion were collected at the given time intervals post-infection. Viral culture was performed by a standard plaque assay and chemokine levels were measured using 30 μl aliots for a panel of 17 chemokines and cytokines (not shown). (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
We estimate chemokine production rates, r, by adapting the delay differential equation model of influenza infection described in Mitchell et al. (2011) Eq. (1) by adding one new equation (Ċ = rI1τ3 – dC) to model chemokine production. Strain-specific values for r were found by fitting the equations to the experimental data in Table S1 using a genetic algorithm to minimize the log squared error between the model and the data while holding the rest of the parameter values constant (Fig. S1, Table S3). Next, 1000 bootstrapping runs using resampled residuals were performed on each modeled strain to generate confidence intervals over r. The results of these fits are shown in Table 1 (viral secretion rates are from previous fits in Mitchell et al., 2011). IP-10 and RANTES secretion rates are aggregated in the spatial model (S2.2):
| (1) |
τ subscript variables denote delay terms, signifying the value is the population quantity in existence at time t – τ. Table S2 summarizes population and parameter values and descriptions.
We calculate the rate of production, σ, of CD8 T cells using a differential equation model from Miao et al. (2010). The equations model T cell production and subsequent search over an area of infected lung tissue:
| (2) |
Nc is the number of circulating activated antigen-specific CD8 T cells, N′f is the number of circulating T cells that have found and exit into a region of lung tissue expressing chemokines, Nf is the number of circulating T cells that have found an infected region. T is the number of uninfected target cells, I is the number of productively infected cells, and V is the viral titer in serum. The infected region is assumed to be of radius r and is within a region expressing chemokines of radius r′ (r < r′). The lung is modeled as a circular region with radius R. The area of the infected region is equal to the area of an infected cell (of radius rcell) multiplied by the number of infected cells. See Fig. 1 for more details about the model. The area of the region expressing chemokine was found to be related non-linearly to the number of infected cells where a and b are constants that depend on the viral strain. a and b were fit to 5 experimental runs of a spatial model implemented in CyCells (R2 = 0.997, P < 0.01).
Circulating CD8 T cells (Nc) are assumed to be released from lymph nodes at a constant rate σ and circulate to the N′f population over a time defined by trc. We assume these circulating cells transition into N′f and Nf at rates proportional to the areas of the chemokine expressing and infected regions relative to the whole lung area.
The average time an exiting CD8 T cell takes to migrate to the infected region is equal to the difference in the radii between the two regions (r′ – r) times the T cell speed, vtcell. Circulating cells that are in the chemokine expressing region (N′f) move into the infected region after performing chemotaxis for that time. Target cells (T) become infected by virus at rate βTV, where β is the rate constant characterizing infection. Infected cells (I) die at rate δ in addition to being lysed by T cells (Nf) at a rate ke. Finally the viral titers (V) increase due to production of virus at rate p by infected cells. Virus is also cleared due to uptake by infected cells (at a rate −βTV) and due to antibody (at a rate γ(t) that changes after 5 days post-infection). The initial viral titer was initialized to 10,000 PFUs and the initial number of target cells was one million. The initial number of infected cells is assumed to be zero. Parameter values are listed in Table S4. This ODE was fit to data taken from Miao et al. (2010) using Matlab's nlinfit function in order to obtain a value for σ. The final value was found to be 1257 per hour.
2.5. Materials
In an effort to obtain per-cell chemokine and cytokine production rates, human epithelial cells were infected with influenza in vitro. Epithelial cell culture and supernatant collection was performed as described in Mitchell et al. (2011). Briefly, undiffer-entiated human tracheal epithelial cells (University of Miami) were cultured for 4 weeks to achieve fully differentiated confluent monolayers on collagen-coated transwell inserts, or commercial differentiated human bronchial epithelial cells (EpiAirway Tissue, MatTek Corp., Ashland, MA) used immediately upon receipt were infected at an MOI of 0.01 with either seasonal H1N1 virus A/New Caledonia/20/99 (sH1N1), the 2009 H1N1 pandemic strain A/California/04/09 (pH1N1), or avian H5N1 virus A/Hong Kong/483/97 (aH5N1) derived from a fatal human infection. Basal media was collected from previously undisturbed triplicate or quadruplicate wells at 0, 6, 10, 12, 16, 20, 24, 30, 36, 42, 48, and 72 h after infection, and stored at −80 °C until assay. Subsequently, apical fluid for virus secretion was collected before and after treatment of the monolayer with protease (Pronase, Sigma) to optimize the collection of infectious virus (Mitchell et al., 2011). Quantitative viral culture was performed by standard plaque assay. Quantitative chemokine levels were performed in 30 μL aliquots for a panel of chemokines (IL-8, MCP-1, MIP-1α, MIP-1β, RANTES, IP-10, eotaxin) and cytokines (interferon-gamma, IL-1α, IL-1β, IL-1RA, IL-2, IL-3, IL-4, IL-6, IL-10, IL-12p40, IL-15, IL-17, TNFα) (Luminex Assay, Luminex Corp.) and reported as ng/mL basal media sampled from a total volume of 4 mL. Only IP-10, RANTES, and TNFα showed increases in production (Table S1). TNF activity is not incorporated in the model. Data for other chemokines and cytokines is not shown.
3. Results
3.1. Model results
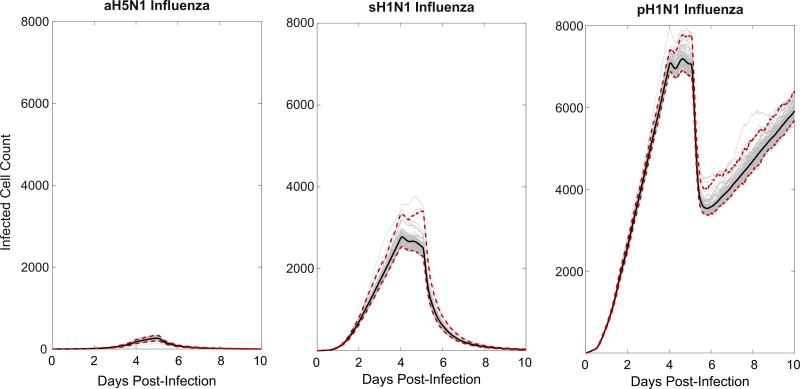
Because the CyCells model is stochastic, we conducted 100 runs of the model with the same parameter set for each of the three influenza strains, each run initialized with a unique random seed (Fig. 4). We count the number of infected cells in the simulated plaque at every time point. For the avian and seasonal strains, all model runs cleared the infection by day 10. Conversely, each simulation of the 2009 pandemic influenza led to uncontrolled infection.
Fig. 4.
Model results: Time series plots of 100 runs of aH5N1 (A), sH1N1 (B), and pH1N1 (C) infections (gray). Each run took the calculated viral production and chemokine production rates for the three different strains of influenza as input (Table 1) and reported the total number of infected cells, including incubating, virus secreting and apoptotic, but not including dead cells. Therefore the figures approximate the rate of plaque growth over time. IP-10 and RANTES were simulated in each run, except for aH5N1, which produced only RANTES. Each run was initialized identically for each strain save for the random seed. The middle line shows the mean while the red dashed lines show the 96% credible confidence interval. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
We estimated variance across runs by calculating the standard deviation at each time point. For aH5N1, the maximum standard deviation was 35, which occurred at day 4.8 where the mean value was 257. For sH1N1, the maximum standard deviation was 212, which occurred at day 5.1 where the mean value was 2528. For pH1N1, the maximum standard deviation was 231, which occurred at day 5.1 where the mean value was 6988 (Fig. 4).
In all studied strains, virus population growth slows at four days p.i. (Fig. 4), due to IgM appearance. Subsequently, the simulated CD8 T cell response causes the number of infected cells to decline quickly after day five p.i. aH5N1 is cleared completely, sH1N1 is cleared more slowly. In contrast, at day six p.i., pH1N1 replication overwhelms the IgM and T cell response. These results are consistent with previously reported in vitro differences among the three different strains (Mitchell et al., 2011).
3.2. Spatial effects
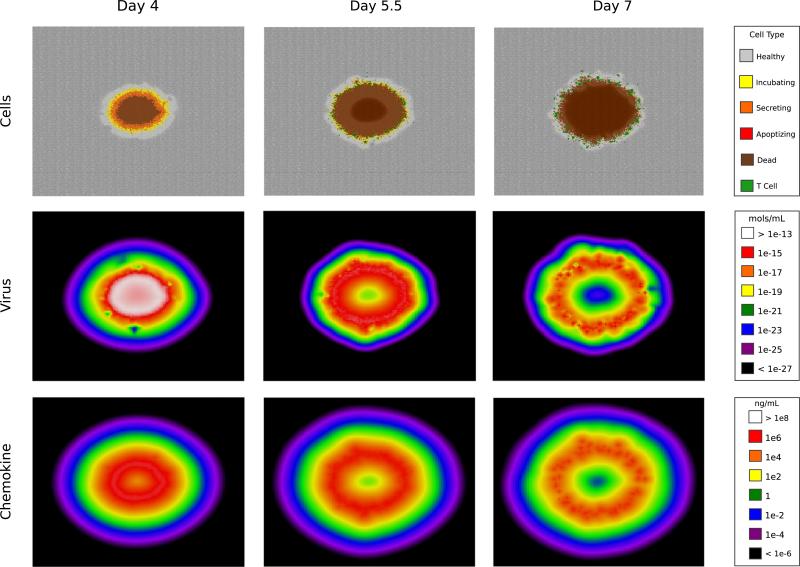
Plaque growth in the model is illustrated in Fig. 5 and Videos S1–S3. Because the virus particles are an order of magnitude larger than the chemokine molecules, they diffuse more slowly according to the Stokes–Einstein equation. However, chemokine molecules decay more quickly than the virus (Table 2). In the model these countervailing characteristics produce similar spatio-temporal profiles for the two particle types (Fig. 5). Before day 4 the plaque is populated primarily by virus-incubating and virus-secreting cells, with a small percentage of dead cells (Panel A). Over time, cells in the plaque's interior die, and active cells comprise a decreasing proportion of the plaque. T cells arrive at day 5 and begin killing the virus-secreting cells (Panel B). By day 6 many expressing cells have been eliminated and the plaque is populated by dead cells (Panel C).
Fig. 5.
Simulated sH1N1 infection. Screenshots from day 4, day 5.5, and day 7. The top row shows the spreading focus of infection through the color coding of individual cells: healthy cells in uninfected tissue (gray), virus-incubating cells (yellow), virus-secreting cells (orange), apoptotic cells (red), dead cells (brown), and T cells arriving at day 5 (green). Free virus and chemokine particles are represented by compartmentalized concentrations of mols/mL and ng/mL. Chemokine shown is an aggregate of total IP-10 and RANTES concentrations. See Videos S1–S3 for an animated visualization of each row. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.jtbi.2016.02.022.
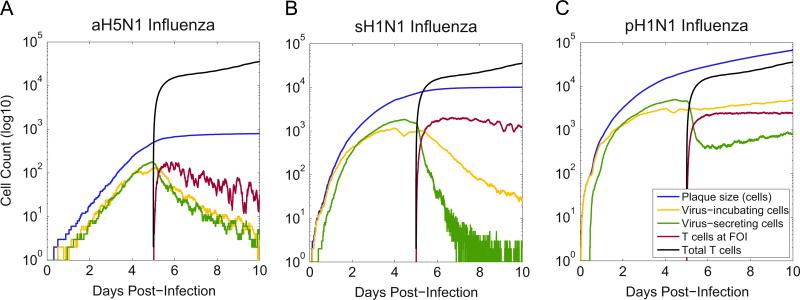
In the aH5N1 infection, the plaque is densely populated with infected cells, allowing T cells to find secreting cells easily and clear the infection (Fig. 6A). However, secreting cells were not cleared in either the sH1N1 or pH1N1 simulations (Fig. 4). In these runs, virus-secreting cells accounted for at most 10% of the active cell population and less than 1% of the total plaque at 6 days p.i. (Fig. 6). T cells continue to accumulate, but they arrive at a slower rate than the rate at which new cells become infected. Further, the newly arriving T cells are less efficient because the infected epithelial cells are more sparsely distributed in the plaque.
Fig. 6.
Simulated infections of aH5N1, sH1N1, and pH1N1. Plotted values: total plaque size (blue), number of virus incubating cells (yellow), number of virus secreting cells (green), total number of T cells (black), and T cells at the focus of infection (FOI) (red). T cells clear secreting and incubating cells in aH5N1, fail to clear incubating cells in sH1N1, and fail to clear either type of infected cell in pH1N1. The number of incubating cells (yellow) after day 5 differs markedly among the three strains indicating that the T cells have differing success at controlling the infection. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
Finally, the spatial nature of our model reveals that the locations of peak chemokine concentration can move more slowly than the rate infected cells and virus expand, thus misdirecting the T cells. It takes time for infected cells to begin producing chemokine, while the preexisting areas of high chemokine density are slow to decay. Thus, T cells whose movements respond to the spatial layout of the chemokine gradient can become trapped, failing to locate new regions of infected cells in the growing plaque. The differences in the infection outcomes between the three strains illustrate the effect of the infection spreading faster than the chemokine maxima can move in the case of pH1N1. The velocity of the chemokine maxima positions depends on a combination of many factors, including the chemokine diffusion, decay, delay, and secretion rates (Table 2).
3.3. Sensitivity analysis
We performed two sensitivity analyses of 16 model parameters to identify those that have an effect on determining model outcomes (S2.3–S2.4). We studied all but five of the parameters listed in Tables 1 and 2, excluding T cell radius, T cell sensitivity to chemokine, T cell onset time, IgM onset time, and the viral production rate. T cell radius is known (Abbas et al., 2011) and does not factor strongly into the spatial model's implementation. T cell sensitivity was set to mimic a realistic threshold of response to cytokine (see S2.1 for a more complete explanation of how T cell sensitivity was determined). The two onset parameters are known (Diamond et al., 2003). Viral production is an independent variable, as discussed above, and determined by each of the three strains (Mitchell et al., 2011).
Each of the remaining 16 parameters was varied independently (over multiple orders of magnitude when appropriate) for each viral strain using a one-factor-at-a-time (OFAT) approach, and the model was rerun once for each parameter setting (S2.3). Results were compared for the quantitative size of the peak infection and the number of uncleared infected cells at day 10 p.i. Based on these runs, we classified each parameter for each strain as follows (Table 3): stable parameters are those that do not affect the model behavior over any of the different values tested; bounded stable parameters are stable across a biologically plausible range of values but model behavior diverges at some threshold; peak change parameters affect the peak size of the infection, but do not change the ultimate number of infected cells at the end of the run; and sensitive parameters substantively change the outcome of the simulation within biologically plausible ranges.
Table 3 shows that four parameters are sensitive: viral response to IgM, infectivity, viral decay rate, and viral diffusion. In addition, Fig. 4 shows that viral production rate is a sensitive parameter, varying among the three panels. The four parameters (infectivity, viral decay rate, viral diffusion rate, and viral production rate) are specific viral characteristics. Further, viral response to IgM, as coded in the model, directly affects viral decay rate and can be considered a viral parameter. None of the T cell parameters nor the chemokine parameters has a strong effect on the model's prediction of control or lack of control of viral replication. This result supports the hypothesis that virus dynamics dominate the course of an influenza infection.
Partial rank correlation coefficient (PRCC) analysis was performed on the sH1N1 strain as described in Marino et al. (2008) using the same 16 parameters plus the viral secretion rate to determine the strength and significance of the parameters’ partial correlation with the model output (S2.4). The five viral parameters are confirmed by the PRCC analysis to be significantly correlated with model output (Table 3). Consistent results between the two approaches suggests the less computationally intensive OFAT analysis is also accurate for aH5N1 and pH1N1.
3.4. Window of control
Our sensitivity analysis suggests that viral parameters are the most sensitive and have the largest effect on the course of the infection (Table 3). Here we examine the role of T cells by assessing multiple factors that control the overall T cell response. We seek to identify the maximum amount of time virus-secreting cells may produce virus while still allowing the infection to be cleared. We name this threshold the ‘window of control’. This window can be defined as the combination of the time of T cell arrival at a virus-secreting cell (Tarr), the time it takes for the T cell to induce apoptosis (Tkill) and the time for the infected cell to apoptose (Tapop). We examine how much virus a virus-secreting cell can produce inside this window. Values above one suggest a growing infection, while values below one suggest infection control.
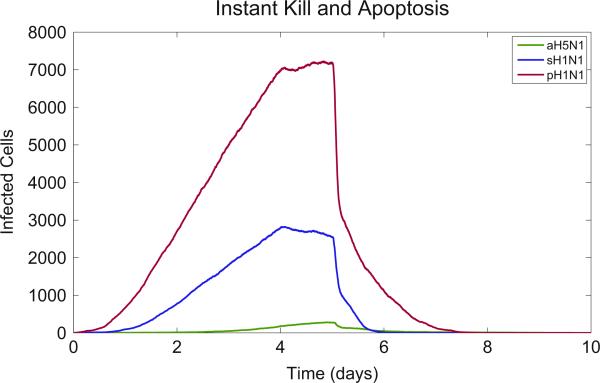
An epithelial cell infected with pH1N1 produces new virus at the rate of 5.1e – PFU/s (Table 1). Assuming instantaneous T cell arrival at a new virus-secreting cell, T cell-induced apoptosis occurs within 10 min (Tkill) and viral secretion continues for 60 min after that (Tapop) according to our model (Table 2). Under these circumstances the cell produces 21 new viral particles that can infect new cells in this 70 min window that occurs after T cell arrival. In contrast, with the sH1N1 strain, a single infected cell produces 1.6 viral particles, and for the aH5N1 strain this number is 0.2.
Even if T cell arrival and apoptosis time were instantaneous, a cell would still secrete virus for 10 min (Tkill), producing 3 new virions in the case of pH1N1. Only in the case where both Tkill and Tapop are simultaneously reduced can the pandemic infection be limited to less than one new virion per infected cell. We confirm this effect in the model by first setting both relevant parameters (apoptosis time and T cell kill time) to zero, and as expected all three strains were cleared (Fig. 7). We now ask how much T cell arrival delay the model can tolerate and still clear a given viral strain. To answer this, we formulate the basic equations of the model in terms of R0, the viral replication rate, and solve for the case where R0 < 1:
| (3) |
where p is the secretion rate of the virus and E is viral effectiveness, i.e. the proportion of virus that infects cells. Eq. (1) assumes each free virus particle can infect no more than one healthy epithelial cell.
Fig. 7.
Simulated infections of aH5N1, sH1N1, and pH1N1 with instant cell death. The model results show that the combined delay of the T cell kill time and apoptosis time form a barrier to infection clearance. Removing both delays results in infection clearance for all strains.
p, Tkill, and Tapop are model parameters (Table 2). E incorporates both spatial effects of viral diffusion and temporal effects of viral decay. As the infected plaque expands, some secreted virus diffuses to cells that are already infected or dead (the existing plaque) and do not contribute to new infections. This effect is minimal in the beginning when the edge of the plaque is small and tightly curved. As the plaque expands, its radius grows and the edge approaches the limiting case of a straight line when 50% of the virus could be expected to diffuse over healthy cells and 50% over infected or dead cells. In the presence of healthy cells, virions infect the cells at a rate of 0.5/h and decay at a rate of 0.42/h in the presence of IgM after day 4 p.i. (Table 2). This suggests that 50% of free virus contacts healthy cells and 54% of that virus succeeds in infecting a healthy cell, implying that E ≈ 0.27 (see S2.5 for a more complete explanation of how E was determined). Solving for Tarr where R0 < 1 gives:
| (4) |
Using the model parameters, this equation suggests that Tarr < 18 h for aH5N1 and Tarr < 93 min for sH1N1. pH1N1, however, cannot be cleared for any value of Tarr in this scenario.
The above calculation suggests a combined transition point for sH1N1 at 163 min (R0 = 1 for Tarr + Tkill + Tapop = 163 min), which is consistent with the sensitivity analysis (Fig. S5: Apoptosis Time: 90–120 min) and suggests that the modeled Tarr is between 33 and 63 min for sH1N1. These data show that the balance between viral production and T cell response is a key factor in clearance of infection: pH1N1 can theoretically be cleared with instantaneous T cell effectiveness (Fig. 7), but in actuality viral production exceeds T cell response time and this leads to uncontrolled infection.
4. Discussion
We used a spatial model to study how T cell search in the lung affects the host's ability to clear virus. CD8 T cells conduct two ‘searches’ in two different tissue environments, first to encounter antigen-loaded dendritic cells in the lymph node, and second to encounter local inflammatory signals in the infected lung. Search problems in the lymph node have been simulated using live cell imaging data to provide reliable parameters of cell movement (Mirsky et al., 2011; Vroomans et al., 2012). Our model focused on the second search, i.e. the process of recruiting activated CD8 effector cells to infected sites in the lung, which is not as well understood. In contrast to the dense 3D lymph node volume, our model approximates a mouse lung as a 2D surface where the epithelial cell monolayer is 100 cm2 (only the area containing the FOI is modeled explicitly). T cells must somehow rapidly locate the infection, which at day 5 p.i. is approximately 0.5 mm in diameter. Our model revealed four main insights regarding chemokine aided T cell search in the lung.
First, the model consistently clears the aH5N1 strain by day 10 p.i., contains sH1N1 by day 10 p.i., and fails to clear the pH1N1 infection. Second, the model revealed spatial constraints on T cell search when the infection spreads more quickly than the chemokine gradient can diffuse. In these cases, T cells become trapped in areas of high chemokine concentration which lag behind the expanding infection. High concentrations of chemokine also attract and trap arriving T cells, thus limiting the direct benefit of increasing numbers of T cells. Third, the sensitivity analysis tested the individual effects of each parameter on the model's behavior. The analysis shows that infection outcomes are highly sensitive to the properties of the specific virus. Finally, we examined the window of control which suggests that pandemic influenza could not be cleared even if T cell arrival was instantaneous.
The in vivo quantitative chemokine parameters in the infected lung are difficult to estimate. There is clear evidence from multiple models that chemokines and chemokine receptors are required for effector T cell localization to infected tissues (Christensen et al., 2004, 2006; Dawson et al., 2000; De Lemos et al., 2005; Fadel et al., 2008; Gadhamsetty et al., 2014; Groom and Luster, 2011a, 2011b; Hsieh et al., 2006; Klein et al., 2005; Kohlmeier et al., 2009, 2011; Thapa et al., 2008; Wareing et al., 2004; Wuest and Carr, 2008; Pawelek et al., 2012). However, the actual effect of chemokines on effector T cells in tissues is still largely unknown. Blood levels documented in virulent influenza infections (De Jong et al., 2006) may not reflect lung tissue concentrations. Dynamic chemokine concentrations secreted by bronchial epithelial cells in vitro depend on infection intensity and cell maturation state (Mitchell et al., 2011; Chan et al., 2010, 2005; Zeng et al., 2011) and may better approximate chemokine levels in tissue. We have determined the level of chemokine released by infected bronchial epithelial cells (Table 1) and used these as the best approximation for chemokine levels in tissues. Our model did not incorporate the potential contributions from other cytokines that did not show different levels in the physical experiment. This includes chemokines such as CXCL8/IL-8 detected in bronchial cell cultures (Matsukura et al., 1996; Arndt et al., 2002) as well as chemokines secreted by immigrant macrophages (Julkunen et al., 2000), neutrophils (Lim et al., 2015), and amplification of epithelial cell secretion by CD8 T cells (Zhao et al., 2000). The sensitivity analysis classifies the chemokine secretion rate as stable, suggesting the total amount of chemokine is not a significant factor in determining viral clearance in our model.
Although leukocytes exhibit directional behavior to chemokines (Li Jeon et al., 2002; McDonald et al., 2010), CD8 T cells have not yet been shown to climb chemokine gradients. While T cell chemotaxis may not be proven, our previous work suggests severe limitations to viral clearance in the absence of this ability (Banerjee et al., 2011). Our sensitivity analysis shows that slowing T cell speed (and consequentially, the ability of T cells to climb gradients) reduces clearance of sH1N1 and pH1N1 strains. These results are consistent with the hypothesis that T cells are guided by chemokine in the lung epithelium.
Previous ODE models have studied how viral dynamics affect the course of infection (Handel and Antia, 2008; Lee et al., 2009; Miao et al., 2010; Mitchell et al., 2011; Saenz et al., 2010; Crauste et al., 2015; Price et al., 2015). The use of an ABM can complement spatially homogeneous differential equation models (Beltman et al., 2007; Textor et al., 2014; Vroomans et al., 2012; Zheng et al., 2008). In this study, we tested how chemokine-directed T cell search contributes to infection clearance. Explicitly modeling T cell chemotaxis revealed unique challenges to viral clearance brought about by infections that spread more rapidly than the chemokine gradient could decay and diffuse. Furthermore, our model was also able to account for spatial constraints of viral diffusion and infection. Fig. 5 illustrates how new virus diffuses over cells that are no longer able to be infected. ODE models account for this discrepancy by lowering rates of infection by a constant amount, but this assumes that the proportion of unsuitable virions will always be the same. Our model visualizes how the early infection has a higher proportion of virus overlapping healthy cells (Fig. 5).
In our spatial model, CD8 T cells climb a chemokine gradient to find infected epithelial cells and cluster at local maxima of chemokine concentration. Because T cells are clustered, they cannot cover the expanding plaque effectively, where infected cells on the periphery become more highly dispersed as the plaque grows. Thus, T cells in the model become redundant at a relatively low threshold, beyond which additional T cells do not improve clearance rates. These observations provide an explanation for pH1N1 dynamics that would be obscured without the visualizations provided by spatial modeling.
The window of control describes the limit on the amount of time an infected cell may secrete virus while still allowing the infection to be cleared. Studying the time of T cell arrival at virus-secreting cells in the model revealed one reason why T cells fail to clear pH1N1 infections. We formalized this effect in an equation which calculates an upper bound on the search time for T cells to find new virus-secreting cells, and we showed that this value is consistent with the results of our sensitivity analysis.
Our model captures important quantitative and spatial aspects of T cell response to influenza infection which have not been addressed by earlier T cell models. The results of our sensitivity analysis are important for future biological modeling and experimental design. Empirical verification of the model's sensitive parameters (viral response to IgM, infectivity, viral decay rate, viral diffusion rate, and viral production rate) will be valuable to future studies. Finally, knowledge of the spatial nature of the interaction between T cells and chemokines can aid in the design of future immune system models.
Supplementary Material
HIGHLIGHTS.
A spatial agent-based model examines T cell search in an influenza infected lung.
Experimental data show that chemokines CXCL10 and CCL5 are stimulated by infection.
Rapidly expanding plaques of infection create challenges for chemotaxing T cells.
Sensitivity analysis isolates the effects of each parameter in the model.
Model parameters are used to determine an upper bound on T cell search time.
Acknowledgments
We thank F. Asperti-Boursin (UNM Pathology and Computer Science), S. Jordan (UNM Computer Science and Biology), and G. Bezerra, B. Edwards, ThanhVu Nguyen, and G. Stelle (UNM Computer Science) for suggestions concerning the manuscript, cellular and chemokine behavior, and agent-based modeling.
This publication was funded by NIH Grants 1R21-AI-73607 (to F.K.), U01-AI-074561 (to F.K.), DARPA Grant P-1070-113237 (to S.F.), and NSF Grant EF 1038682 (to M.M.) and a James S. McDonnell Foundation Complex Systems Scholar Award (to M.M.).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.jtbi.2016.02.022.
References
- Abbas AK, Lichtman AHH, Pillai S. Cellular and Molecular Immunology. Saunders; 2011. [Google Scholar]
- Allan RS, Waithman J, Bedoui S, Jones CM, Villadangos JA, Zhan Y, Lew AM, Shortman K, Heath WR, Carbone FR. Migratory dendritic cells transfer antigen to a lymph node-resident dendritic cell population for efficient CTL priming. Immunity. 2006;25(1):153–162. doi: 10.1016/j.immuni.2006.04.017. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/16860764〉. [DOI] [PubMed] [Google Scholar]
- Arndt U, Wennemuth G, Barth P, Nain M, Al-Abed Y, Meinhardt A, Gemsa D, Bacher M. Release of macrophage migration inhibitory factor and CXCL8/interleukin-8 from lung epithelial cells rendered necrotic by influenza A virus infection. J. Virol. 2002;76(18):9298–9306. doi: 10.1128/JVI.76.18.9298-9306.2002. URL 〈 http://view.ncbi.nlm.nih.gov/pubmed/12186913〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem A, Hochstättler W, Malich M. The simulated trading heuristic for solving vehicle routing problems. Discret. Appl. Math. 1996;65(1):47–72. [Google Scholar]
- Banerjee S, Moses M. Scale invariance of immune system response rates and times: perspectives on immune system architecture and implications for artificial immune systems. Swarm Intell. 2010;4(4):301–318. http://dx.doi.org/10.1007/s11721-010-0048-2, ISSN 1935-3812, URL 〈 http://www.springerlink.com/index/10.1007/s11721-010-0048-2〉. [Google Scholar]
- Banerjee S, Levin D, Moses M, Koster F, Forrest S. The value of inflammatory signals in adaptive immune responses. ICARIS. 2011;6825:1–14. [Google Scholar]
- Beauchemin C, Samuel J, Tuszynski J. A simple cellular automaton model for influenza A viral infections. J. Theor. Biol. 2005;232(2):223–234. doi: 10.1016/j.jtbi.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Beauchemin C, Forrest S, Koster FT. Modeling influenza viral dynamics in tissue. LNCS Artif. Immune Syst. 2006;4163:23–36. ISSN 03029743, URL 〈 http://www.springerlink.com/content/9754174j200113jv/〉. [Google Scholar]
- Beltman JB, Marée AFM, De Boer RJ. Spatial modelling of brief and long interactions between T cells and dendritic cells. Immunol. Cell Biol. 2007;85(4):306–314. doi: 10.1038/sj.icb.7100054. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/17420768〉. [DOI] [PubMed] [Google Scholar]
- Bromley SK, Mempel TR, Luster AD. Orchestrating the orchestrators: chemokines in control of T cell traffic. Nat. Immunol. 2008;9(9):970–980. doi: 10.1038/ni.f.213. http://dx. doi.org/10.1038/ni.f.213, ISSN 1529-2916, URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/18711434〉. [DOI] [PubMed] [Google Scholar]
- Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440(7086):890–895. doi: 10.1038/nature04651. http://dx.doi.org/10.1038/nature04651, ISSN 14764687, URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/16612374〉. [DOI] [PubMed] [Google Scholar]
- Celli S, Day M, Müller AJ, Molina-Paris C, Lythe G, Bousso P. How many dendritic cells are required to initiate a T-cell response? Blood. 2012;120:3945–3948. doi: 10.1182/blood-2012-01-408260. http://dx.doi.org/10.1182/blood-2012-01-408260, ISSN 00064971. [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Morgan TM, Harmsen AG, Dutton RW. Migration kinetics and final destination of Type 1 and Type 2 CD8 effector cells predict protection against pulmonary virus infection. J. Exp. Med. 1999;189(2):423–434. doi: 10.1084/jem.189.2.423. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2192982&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MCW, Cheung CY, Chui WH, Tsao SW, Nicholls JM, Chan YO, Chan RWY, Long HT, Poon LLM, Guan Y, Peiris JSM. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 2005;6(1):135. doi: 10.1186/1465-9921-6-135. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/16283933〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RWY, Yuen KM, Yu WCL, Ho CCC, Nicholls JM, Peiris JSM, Chan MCW. Influenza H5N1 and H1N1 virus replication and innate immune responses in bronchial epithelial cells are influenced by the state of differentiation. PLoS ONE. 2010;5(1):9. doi: 10.1371/journal.pone.0008713. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2806912&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JE, Nansen A, Moos T, Lu B, Gerard C, Christensen JP, Thomsen AR. Efficient T-cell surveillance of the CNS requires expression of the CXC chemokine receptor 3. J. Neurosci. 2004;24(20):4849–4858. doi: 10.1523/JNEUROSCI.0123-04.2004. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/15152045〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JE, De Lemos C, Moos T, Christensen JP, Thomsen AR. CXCL10 is the key ligand for CXCR3 on CD8+ effector T cells involved in immune surveillance of the lymphocytic choriomeningitis virus-infected central nervous system. J. Immunol. 2006;176(7):4235–4243. doi: 10.4049/jimmunol.176.7.4235. URL 〈 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16547260〉. [DOI] [PubMed] [Google Scholar]
- Crauste F, Terry E, Mercier IL, Mafille J, Djebali S, Andrieu T, Mercier B, Kaneko G, Arpin C, Marvel J, Gandrillon O. Predicting pathogen-specific CD8 T cell immune responses from a modeling approach. J. Theor. Biol. 2015;374:66–82. doi: 10.1016/j.jtbi.2015.03.033. http://dx.doi.org/10.1016/j.jtbi.2015.03.033, ISSN 00225193, URL 〈 http://www.sciencedirect.com/science/article/pii/S0022519315001484〉. [DOI] [PubMed] [Google Scholar]
- Dawson TC, Beck M.a., Kuziel W.a., Henderson F, Maeda N. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am. J. Pathol. 2000;156(6):1951–1959. doi: 10.1016/S0002-9440(10)65068-7. http://dx.doi.org/10.1016/S0002-9440(10)65068-7, ISSN 0002-9440, URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1850091&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJD, Chau TNB, Hoang DM, Chau NVV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha DQ, Guan Y, Peiris JSM, Chinh NT, Hien TT, Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006;12(10):1203–1207. doi: 10.1038/nm1477. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/16964257〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lemos C, Christensen JE, Nansen A, Moos T, Lu B, Gerard C, Christensen JP, Thomsen AR. Opposing effects of CXCR3 and CCR5 deficiency on CD8+ T cell-mediated inflammation in the central nervous system of virus-infected mice. J. Immunol. 2005;175(3):1767–1775. doi: 10.4049/jimmunol.175.3.1767. URL 〈 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16034118〉. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Sitati EM, Friend LD, Higgs S, Shrestha B, Engle M. A critical role for induced IgM in the protection against West Nile virus infection. J. Exp. Med. 2003;198(12):1853–1862. doi: 10.1084/jem.20031223. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2194144&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egen JG, Rothfuchs AG, Feng CG, Horwitz MA, Sher A, Germain RN. Intravital imaging reveals limited antigen presentation and T cell effector function in mycobacterial granulomas. Immunity. 2011;34(5):807–819. doi: 10.1016/j.immuni.2011.03.022. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/21596592〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbert KJ, Schäfer UF, Schäfers HJ, Kim KJ, Lee VH, Lehr CM. Monolayers of human alveolar epithelial cells in primary culture for pulmonary absorption and transport studies. Pharm. Res. 1999;16(5):601–608. doi: 10.1023/a:1018887501927. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/10349999〉. [DOI] [PubMed] [Google Scholar]
- Fadel S, Bromley S, Medoff B, Luster A. CXCR3-deficiency protects influenza-infected CCR5-deficient mice from mortality. Eur. J. Immunol. 2008;38(12):3376–3387. doi: 10.1002/eji.200838628. http://dx.doi.org/10.1002/eji.200838628.CXCR3-deficiency, URL 〈 http://onlinelibrary.wiley.com/doi/10.1002/eji.200838628/full〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadhamsetty S, Marée AFM, Beltman JB, De Boer RJ. A general functional response of cytotoxic T lymphocyte-mediated killing of target cells. Biophys. J. 2014;106(8):1780–1791. doi: 10.1016/j.bpj.2014.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusov VV, De Boer RJ. Estimating in vivo death rates of targets due to CD8 T-cell-mediated killing. J. Virol. 2008;82(23):11749–11757. doi: 10.1128/JVI.01128-08. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2583656&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 2011a;89(2):207–215. doi: 10.1038/icb.2010.158. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/21221121〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom JR, Luster AD. CXCR3 in T cell function. Exp. Cell Res. 2011b;317(5):620–631. doi: 10.1016/j.yexcr.2010.12.017. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3065205&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn MD, Tangemann K, Tam C, Cyster JG, Rosen SD, Williams LT. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 1998;95(1):258–263. doi: 10.1073/pnas.95.1.258. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=18193&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel A, Antia R. A simple mathematical model helps to explain the immunodominance of CD8 T cells in influenza a virus infections. J. Virol. 2008;82(16):7768–7772. doi: 10.1128/JVI.00653-08. 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2519595&tool=pmcentrez&rendertype=abstract 〈 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoji A, Rinaldo CR. Human CD8+ T cells specific for influenza A virus M1 display broad expression of maturation-associated phenotypic markers and chemokine receptors. Immunology. 2005;115(2):239–245. doi: 10.1111/j.1365-2567.2005.02135.x. URL 〈 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15885130〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M-F, Lai S-L, Chen J-P, Sung J-M, Lin Y-L, Wu-Hsieh BA, Gerard C, Luster A, Liao F. Both CXCR3 and CXCL10/IFN-inducible protein 10 are required for resistance to primary infection by dengue virus. J. Immunol. 2006;177(3):1855–1863. doi: 10.4049/jimmunol.177.3.1855. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/16849497〉. [DOI] [PubMed] [Google Scholar]
- Ingulli E, Funatake C, Jacovetty EL, Zanetti M. Cutting edge: antigen presentation to CD8 T cells after influenza A virus infection. J. Immunol. 2009;182(1):29–33. doi: 10.4049/jimmunol.182.1.29. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/19109130〉. [DOI] [PubMed] [Google Scholar]
- Julkunen I, Melén K, Nyqvist M, Pirhonen J, Sareneva T, Matikainen S. Inflammatory responses in influenza A virus infection. Vaccine. 2000;19(Supplement 1):S32–7. doi: 10.1016/s0264-410x(00)00275-9. http://dx.doi.org/10.1016/S0264-410X(00)00275-9, ISSN 0264410X, URL 〈 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11163460〉. [DOI] [PubMed] [Google Scholar]
- Kim TS, Sun J, Braciale TJ. T cell responses during influenza infection: getting and keeping control. Trends Immunol. 2011;32(5):225–231. doi: 10.1016/j.it.2011.02.006. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/21435950〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RS, Lin E, Zhang B, Luster AD, Tollett J, Samuel MA, Engle M, Diamond MS. Neuronal CXCL10 directs CD8+ T-cell recruitment and control of West Nile virus encephalitis. J. Virol. 2005;79(17):11457–11466. doi: 10.1128/JVI.79.17.11457-11466.2005. URL 〈 http://jvi.asm.org/cgi/content/abstract/79/17/11457〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeier JE, Cookenham T, Miller SC, Roberts AD, Christensen JP, Thomsen AR, Woodland DL. CXCR3 directs antigen-specific effector CD4+ T cell migration to the lung during parainfluenza virus infection. J. Immunol. 2009;183(7):4378–4384. doi: 10.4049/jimmunol.0902022. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2757292&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeier JE, Reiley WW, Perona-Wright G, Freeman ML, Yager EJ, Connor LM, Brincks EL, Cookenham T, Roberts AD, Burkum CE, Sell S, Winslow GM, Blackman MA, Mohrs M, Woodland DL. Inflammatory chemokine receptors regulate CD8(+) T cell contraction and memory generation following infection. J. Exp. Med. 2011;208(8):1621–1634. doi: 10.1084/jem.20102110. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/21788409〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Topham DJ, Park SY, Hollenbaugh J, Treanor J, Mosmann TR, Jin X, Ward BM, Miao H, Holden-Wiltse J, Perelson AS, Zand M, Wu H. Simulation and prediction of the adaptive immune response to influenza A virus infection. J. Virol. 2009;83(14):7151–7165. doi: 10.1128/JVI.00098-09. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2704765&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Jeon N, Baskaran H, Dertinger SKW, Whitesides GM, Van De Water L, Toner M. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat. Biotechnol. 2002;20(8):826–830. doi: 10.1038/nbt712. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/12091913〉. [DOI] [PubMed] [Google Scholar]
- Lim K, Hyun Y-M, Lambert-Emo K, Capece T, Bae S, Miller R, Topham DJ, Kim M. Neutrophil trails guide influenza-specific CD+ 8 T cells in the airways. Science. 2015;349(6252):aaa4352. doi: 10.1126/science.aaa4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Hogue IB, Ray CJ, Kirschner DE. A methodology for performing global uncertainty and sensitivity analysis in systems biology. 2008 doi: 10.1016/j.jtbi.2008.04.011. URL 〈 http://dx.doi.org/10.1016/j.jtbi.2008.04.011〉. [DOI] [PMC free article] [PubMed]
- Martin-Fontecha A, Sebastiani S, Höpken UE, Uguccioni M, Lipp M, Lanzavecchia A, Sallusto F. Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J. Exp. Med. 2003;198(4):615–621. doi: 10.1084/jem.20030448. ISSN 0022-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura S, Kokubu F, Noda H, Tokunaga H, Adachi M. Expression of IL-6, IL-8, and RANTES on human bronchial epithelial cells, NCI-H292, induced by influenza virus A. J. Allergy Clin. Immunol. 1996;98(6 Pt 1):1080–1087. doi: 10.1016/s0091-6749(96)80195-3. URL 〈 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8977509〉. [DOI] [PubMed] [Google Scholar]
- McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CCM, Beck PL, Muruve DA, Kubes P. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 2010;330(6002):362–366. doi: 10.1126/science.1195491. URL 〈 http://www.sciencemag.org/cgi/doi/10.1126/science.1195491〉. [DOI] [PubMed] [Google Scholar]
- Medoff BD, Thomas SY, Banerji A, Wain JC, Zhang H, Lilly CM, Ginns LC, Luster AD. Pathogenic T-cell recruitment into the airway in human disease. Ann. N. Y. Acad. Sci. 2005;1062:220–241. doi: 10.1196/annals.1358.026. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/16461804〉. [DOI] [PubMed] [Google Scholar]
- Miao H, Hollenbaugh JA, Zand MS, Holden-Wiltse J, Mosmann TR, Perelson AS, Wu H, Topham DJ. Quantifying the early immune response and adaptive immune response kinetics in mice infected with influenza A virus. J. Virol. 2010;84(13):6687–6698. doi: 10.1128/JVI.00266-10. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2903284&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Wei SH, Cahalan MD, Parker I. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proc. Natl. Acad. Sci. U. S. A. 2003;100(5):2604–2609. doi: 10.1073/pnas.2628040100. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/18431356〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky HP, Miller MJ, Linderman JJ, Kirschner DE. Systems biology approaches for understanding cellular mechanisms of immunity in lymph nodes during infection. 2011 doi: 10.1016/j.jtbi.2011.06.037. http://dx.doi.org/10.1016/j.jtbi.2011.06.037. [DOI] [PMC free article] [PubMed]
- Mitchell H, Levin D, Forrest S, Beauchemin C.a.a., Tipper J, Knight J, Donart N, Layton RC, Pyles J, Gao P, Harrod KS, Perelson AS, Koster F. Higher level of replication efficiency of 2009 (H1N1) pandemic influenza virus than those of seasonal and avian strains: kinetics from epithelial cell culture and computational modeling. J. Virol. 2011;85(2):1125–1135. doi: 10.1128/JVI.01722-10. http://dx.doi.org/10.1128/JVI.01722-10, ISSN 1098-5514 URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3019989&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo LN, Murillo MS, Perelson AS. Towards multiscale modeling of influenza infection. Journal of theoretical biology. 2013;332:267–290. doi: 10.1016/j.jtbi.2013.03.024. http://dx.doi.org/10.1016/j.jtbi.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandagopal S, Wu D, Lin F. Combinatorial guidance by CCR7 ligands for T lymphocytes migration in Co-existing chemokine fields. PLoS ONE. 2011;6(3):11. doi: 10.1371/journal.pone.0018183. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3064588&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Miller MJ, Parker I, Krummel MF, Neighbors M, Hartley SB, O'Garra A, Cahalan MD, Cyster JG. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3(6):e150. doi: 10.1371/journal.pbio.0030150. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1088276&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek KA, Huynh GT, Quinlivan M, Cullinane A, Rong L, Perelson AS. Modeling within-host dynamics of influenza virus infection including immune responses. PLoS Comput. Biol. 2012;8(6) doi: 10.1371/journal.pcbi.1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price I, Mochan-Keef ED, Swigon D, Ermentrout GB, Lukens S, Toapanta FR, Ross TM, Clermont G. The inflammatory response to influenza A virus (H1N1): an experimental and mathematical study. J. Theor. Biol. 2015;374:83–93. doi: 10.1016/j.jtbi.2015.03.017. http://dx.doi.org/10.1016/j.jtbi.2015.03.017, ISSN 1095-8541, URL 〈 http://www.sciencedirect.com/science/article/pii/S0022519315001265〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz RA, Quinlivan M, Elton D, MacRae S, Blunden AS, Mumford JA, Daly JM, Digard P, Cullinane A, Grenfell BT, McCauley JW, Wood JLN, Gog JR. Dynamics of influenza virus infection and pathology. J. Virol. 2010;84(8):3974–3983. doi: 10.1128/JVI.02078-09. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/20130053〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor J, Henrickson SE, Mandl JN, von Andrian UH, Westermann J, de Boer RJ, Beltman JB. Random migration and signal integration promote rapid and robust T cell recruitment. PLoS Comput. Biol. 2014;10(8):e1003752. doi: 10.1371/journal.pcbi.1003752. ISSN 1553-7358, URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/25102014〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa M, Welner RS, Pelayo R, Carr DJJ. CXCL9 and CXCL10 expression are critical for control of genital herpes simplex virus type 2 infection through mobilization of HSV-specific CTL and NK cells to the nervous system. J. Immunol. 2008;180(2):1098–1106. doi: 10.4049/jimmunol.180.2.1098. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2185792&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroomans RMA, Marée AFM, De Boer RJ, Beltman JB. Chemotactic migration of T cells towards dendritic cells promotes the detection of rare antigens. PLoS Comput. Biol. 2012;8(11):e1002763. doi: 10.1371/journal.pcbi.1002763. http://dx.doi.org/10.1371/journal.pcbi.1002763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareing MD, Lyon AB, Lu B, Gerard C, Sarawar SR. Chemokine expression during the development and resolution of a pulmonary leukocyte response to influenza A virus infection in mice. J. Leukoc. Biol. 2004;76(4):886–895. doi: 10.1189/jlb.1203644. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/15240757〉. [DOI] [PubMed] [Google Scholar]
- Warrender C, Forrest S, Koster F. Modeling intercellular interactions in early Mycobacterium infection. Bull. Math. Biol. 2006;68(8):2233–2261. doi: 10.1007/s11538-006-9103-y. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/17086496〉. [DOI] [PubMed] [Google Scholar]
- WHO Influenza Fact Sheet. Technical Report. World Health Organisation; 2009. [Google Scholar]
- Wu CFJ, Jackknife Bootstrap and other resampling methods in regression analysis. Ann. Stat. 1986;14(4):1261–1295. http://dx.doi.org/10.1214/aos/1176350142, ISSN 00905364, URL 〈 http://projecteuclid.org/euclid.aos/1176350142〉. [Google Scholar]
- Wuest TR, Carr DJJ. Dysregulation of CXCR3 signaling due to CXCL10 deficiency impairs the antiviral response to herpes simplex virus 1 infection. J. Immunol. 2008;181(11):7985–7993. doi: 10.4049/jimmunol.181.11.7985. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2596651&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Pappas C, Katz JM, Tumpey TM. The 2009 pandemic H1N1 and triple-reassortant swine H1N1 influenza viruses replicate efficiently but elicit an attenuated inflammatory response in polarized human bronchial epithelial cells. J. Virol. 2011;85(2):686–696. doi: 10.1128/JVI.01568-10. URL 〈 http://www.ncbi.nlm.nih.gov/pubmed/21047961〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MQ, Stoler MH, Liu AN, Wei B, Soguero C, Hahn YS, Enelow RI. Alveolar epithelial cell chemokine expression triggered by antigen-specific cytolytic CD8(+) T cell recognition. J. Clin. Investig. 2000;106(6):R49–R58. doi: 10.1172/JCI9786. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=381394&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Jin B, Henrickson SE, Perelson AS, Von Andrian UH, Chakraborty AK. How antigen quantity and quality determine T-cell decisions in lymphoid tissue. Mol. Cell. Biol. 2008;28(12):4040–4051. doi: 10.1128/MCB.00136-08. URL 〈 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2423119&tool=pmcentrez&rendertype=abstract〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.