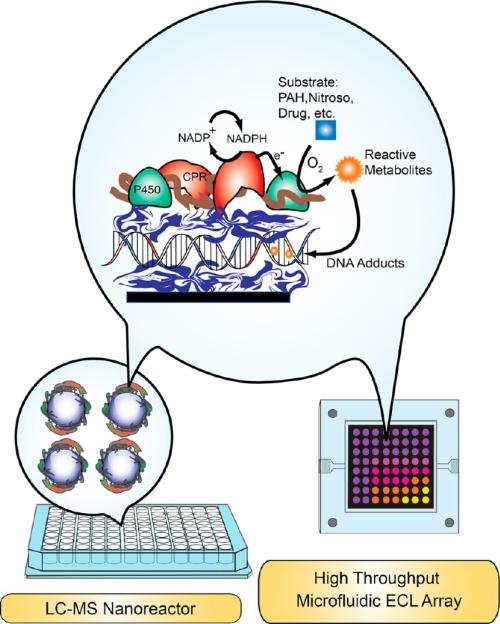
Abstract
Routine in vitro bioassays and animal toxicity studies of drug and environmental chemical candidates fail to reveal toxicity in ~30% of cases. This Feature article addresses research on new approaches to in vitro toxicity testing as well as our own efforts to produce high-throughput genotoxicity arrays and LC–MS/MS approaches to reveal possible chemical pathways of toxicity.
Our modern technological society needs to be continually attuned to potential toxicity issues to ensure public safety. The situation is complicated by the large numbers of industrial, cosmetic, food, and medicinal chemicals that find their way into our bodies,1 either directly or via the food chain, as well as new chemicals and drugs continually entering the market. This issue has engendered numerous methods for toxicity screening, i.e., methods that can reveal toxicity in chemicals by in vitro tests, preferably in high throughput.2–4
During development of new drug candidates, toxicity assessments proceed by using initial panels of toxicity bioassays, then animal toxicity studies and, if the previous studies warrant, human clinical trials.5 Despite extensive in vitro assays and animal toxicity studies, about 30% of developed drug candidates fail at the clinical trial stage due to toxicity issues that had not been discovered in the earlier studies. Drug candidates that are toxic to population subsets may not be identified until they are tested in humans, and a few toxicity-related failures do not manifest until after the drugs are marketed, often with catastrophic consequences.6,7 These facts highlight the need for new approaches to toxicity screening that can identify toxicity with higher accuracy, which is a key objective of research in this area. In addition, the cost of bringing a new drug to market is currently on the order of U.S. $5 billion, so failures represent huge financial losses that drive up general drug costs.8 Development of new chemicals for industries such as agriculture, cosmetics, or food processing often follow similar testing protocols but in the past have traditionally relied on high-dosage animal studies. Human and animal responses to chemicals differ widely, so that animal toxicity studies have questionable significance to humans and can be profoundly misleading for some chemicals.9 Thus, a major challenge is to develop new high-throughput toxicity tests that more accurately predict toxic effects in humans as well as to fully elucidate the complex chemical pathways leading to toxicity. If the latter is achieved for toxic drug candidates, it may provide synthetic chemists guidance to tune out the toxicity while retaining the desired therapeutic effects.
The human biochemistry of chemical toxicity is quite complex and incompletely understood. Although metabolic enzymes have evolved to convert parent chemicals to more soluble metabolites that can be easily cleared from our bodies, in some case the metabolites that form are dangerously reactive to biomolecules in our bodies. In the majority of cases, toxic chemistry is caused by the actions of these reactive metabolites rather than by the parent chemicals.10,11 This complicates toxicity assays since they often need to incorporate representative metabolic enzymes into their protocols to produce the reactive metabolites.4,12
Bioactivation denotes the enzyme-catalyzed generation of reactive metabolites that cause damage to DNA, RNA, proteins, and other biomolecules. Reactive oxygen species (ROS) are byproducts of some metabolic process, and these ROS can oxidize biomolecules and trigger other toxicity pathways.11 When metabolites damage DNA,13,14 they are termed genotoxic. Thus, DNA damage and DNA damage products can be used as end points for genotoxicity assays.2–4 Many chemical pollutants and drugs yield reactive metabolites that form covalent DNA adducts. Examples include styrene, polyaromatic hydrocarbons, nitrosamines, aromatic amines, tamoxifen, and many chemotherapeutic agents.15 Toxic reactions of somewhat obscure origin known as idiosyncratic adverse drug reactions are often linked to reactive metabolites.16
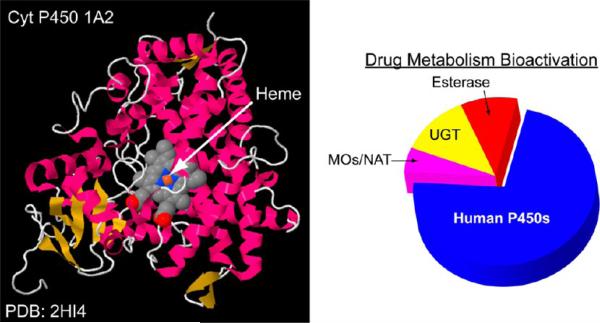
Cytochrome (cyt) P450s are the major enzymes catalyzing oxidations of xenobiotic molecules in humans, and thus are major sources of reactive metabolites. This family of ferric iron heme enzymes are present in all human organs and are involved in 75% of metabolic reactions of known drugs (Figure 1).17 Cyt P450 enzymes use dioxygen and electrons from NADPH delivered by cyt P450 reductase to catalyze oxygen transfers such as hydroxylation of carbon atoms or addition of an oxygen atom to a C–C double bond to make an epoxide. Bioconjugation enzymes that add a water-soluble group to lipophilic parent compounds are also important in metabolism and can lead to deactivation or bioactivation of the parent compound.17c–e Cyt P450-catalyzed reactions are often coupled with bioconjugation enzymes in sequential pathways that can be critical determinants of toxic properties of metabolized chemicals. Different levels of these metabolic enzymes in humans and animals can lead to species differences in metabolism-related toxicity. An important example is the metabolism of Tamoxifen, a therapeutic and preventive breast cancer drug. In humans, an initial metabolic product is the DNA-reactive α-hydroxytamoxifen. In human females, this metabolite is detoxified by a reaction catalyzed by the bioconjugation enzyme glucuronyltransferase yielding a product that is rapidly eliminated from the body.18 Rodent females lack sufficient glucuronyltransferase activity to derivatize all the α-hydroxytamoxifen, and this reactive metabolite can damage DNA in a genotoxic pathway leading to liver cancer.
Figure 1.
Ribbon structure of a metabolic human cytochrome P450 (on left), with pie chart representing the relative importance of various liver enzymes in metablizing known drugs (on right).
Pure single enzymes used for bioactivation in toxicity assays can provide specific metabolic information, but enzyme mixtures are more suitable for general toxicity screening. In some cases, the cells used for bioassays contain subsets of metabolic enzymes including cyt P450s.2,3 Human liver microsomes (HLMs) and rat liver microsomes (RLMs) are also excellent sources of multiple cyt P540 enzymes and contain the cyt P450 reductase (CPR) necessary for the catalytic oxidation cycles. Microsomes are vesicle-like packages of lipids and enzymes that are reconstituted from endoplasmic reticuli of eukaryotic cells. HLMs and RLMs can be obtained commercially and will typically contain a half-dozen or more different cyt P450s in significant amounts for metabolic conversions as well as useful amounts of some bioconjugation enzymes. Many pure bioconjugation enzymes such as sulfotransferases, glutathione transferases, glucuronyltransferases, and N-acetyltransferases are also commercially available. Liver cytosol and S9 liver fractions are additional sources of metabolic enzymes, and similar metabolic enzyme sources from other human organs can be used. In addition, single cyt P450 studies can be readily accomplished using supersomes,19 which are microsome-like materials expressed from E. coli or insect cells to contain a single cyt P450 and its reductase partner.
In this Feature article, we summarize recent advances in high-throughput toxicity screening of chemicals and drugs, focusing largely on chemicals that yield reactive metabolites. These methods may not be in actual use in chemical or drug development activities but represent progressive new systems with clear advantages over traditional approaches. The next section describes recent breakthroughs in high throughput bioassays. Following this, we describe recent research on array-type and liquid chromatography–tandem mass spectrometry (LC–MS/MS) approaches aimed at uncovering possible chemical pathways of metabolite-related toxicity. We wrap up with a brief summary and attempted predictions of what the future may bring.
TOXICITY SCREENING BIOASSAYS
Cell-Based Toxicity Assays
As suggested above, evaluation of vast numbers of potential lead pharmaceutical candidates universally involves initial screening of prospective hit molecules for possible toxicity and other interactions including genotoxicity, hERG (human ether a go-go) channel block gene, drug–drug interactions, and metabolite mediated toxicity.20,21 Some of these events such as genotoxicity and drug–drug interactions are often metabolite related. The relevant assays often include a standard cytotoxicity (cell destruction) assay for result interpretation.21 Cytotoxicity in the form of DNA damage, or genotoxicity, is typically assayed at very early drug development stages to monitor for biologically detrimental effects arising from eventual human exposure to a compound of interest.21,22 Genotoxicity assays include the bacterial reverse mutation mutagenicity assay (Ames test),23 the SOS/umu test,24,25 the micronucleus test (MNT),26 the chromosomal aberration (CA) test, the mouse lymphoma assay (MLA),21 and the Comet assay27,28 for chromosome breakage.
The most widely known and well-established Ames assay monitors for frame or base pair shifts that can facilitate bacterial growth upon compound exposure.23 High-throughput modifications to the Ames assay include transition from an agar plate-based to a liquid suspension/microtiter plate based assay (Ames II). This Ames II assay is automated, high throughput, and less costly in terms of test substrate use.29
High-throughput modifications have also been developed for other assays.21,30–33 These established assays may still exhibit drawbacks, however. For instance, hydrophobic compounds can adsorb to microtiter plate plastic, which affects the predicative nature of the assay,34 and the addition of exogenous S9 fraction is necessary to study reactive metabolite mediated genotoxicity.21,24,35 Low specificity can often hamper these assays, which can provide negative outputs, i.e., using aneugens (aneuploidy inducing molecules) in an Ames assay, or false positives from nongenotoxic media.35
Specificity and sensitivity improvements over traditional assays have been reported by Hastwell et al. employing a fusion green fluorescent protein (GFP)-growth arrest and DNA damage (GADD) plasmid in a eukaryotic cellular assay.35,36 This assay, termed GreenScreen (GS) and marketed by Gentronix, utilizes eukaryotic cells, which allows it to potentially identify genotoxins such as certain aneugens and antibiotics that do not produce a response in the prokaryotic Ames assay. The tested genotoxin induces GADD, a downstream cellular reporter of the p53 gene, the presence of which is detected by enhanced fluorescence.35 Compared to a host of traditional genotoxicity assays, GS was shown to be superior to the SOS, and comparable or complementary to the Ames and micronucleus assays (MLA).37
Novel High-Throughput Screening Approaches
Several high content screening (HCS), analysis (HCA), or throughput screening (HTS) approaches have been reported in an effort to more effectively assess drug toxicity. These assays typically monitor phenotypic changes to cells that have been seeded in microtiter plates and typically utilize optical detection strategies. Many of these approaches and their utility have been reported and analyzed elsewhere in the literature.38,39 Significant recent focus has been on image-based HCS, utilizing different fluorescent compounds that incorporate into different areas of a cell and can be imaged to screen for phenotypic changes. Different fluorophore dyes have been used to simultaneously monitor different toxicity end points within the cells via multicolor imaging of different optical fields, providing more accurate toxicity modeling.40 Utilizing this strategy and focusing on both cytotoxicity and mitochondrial toxicity end points, pharmaceuticals known to cause drug-induced liver injury (DILI) were detected in in HepG2 hepatocyte cells with very few false positives.40,41
Liver toxicity is a predominant focus in pharmaceutical toxicity evaluations, and many assays utilize hepatocytes. However, other reports have utilized HCS imaging to improve toxicity prediction in other organs. Kim et al. coupled a previously reported hyperspectral detection system42 to a microfluidic channel plate to develop a high-throughput device designed to detect cardiotoxicity. Progress in this particular area has generally been lacking due to drawbacks of established assays designed to screen for cardiotoxicity.43 For instance, the traditional patch clamp assay typically only screens for a single drug affecting a single ion channel.43 The impact of several drugs affecting the K+ channel, the Na+/K+ pump, and/or initiating apoptosis in cardiac cells (H9-c2(2-1) cells) was assayed simultaneously. Cells exposed to select pharmaceuticals were then exposed to three different fluorophores with nonoverlapping emission wavelengths and excited by an Arion laser. Emission was directed onto an acousto-optical tunable filter (AOTF) that scans from 500 to 630 nm at a certain frequency and directs the diffracted fluorescence emission onto a CCD camera.43 The presence or absence of each fluorophore was indicative of a particular mechanism, i.e., if a particular drug did not impact the K+ channel, thallium ions could enter into the cellular cytosol where they would bind with a Tl+-selective dye that produces emission at 525 nm. Blockage of the K+ channel precluded the Tl+ ions from binding to the dye, resulting in lower fluorescence.43
Beyond small molecule fluorophores and quantum dots, other imaging strategies have been developed with additional specificity in mind. For instance, semiconducting polymer nanoparticles were developed for simultaneous reactive nitrogen species (RNS) and oxygen (ROS) detection in liver cells.44 Figure 2 shows an overview of these particles and the approach. The polymer nanoparticles were formed from poly(2,7-(9,9-dioctylfluorene)-alt-4,7-bis(thiophen-2-yl)benzo-2,1,3-thiadiazole) (PFODBT) and a galactosylated graft copolymer of poly(styrene) and poly(ethylene glycol) (PS-g-PEG-Gal). The PFODBT polymer serves as the chemiluminescence resonance energy transfer (CRET) acceptor and fluorescence resonance energy transfer (FRET) donor while the PS-g-PEG-Gal allowed for localization in liver cells via hepatocyte receptor binding. The particles were further modified with cyanine dye as fluorescence emitter and peroxyoxylate as chemiluminescence donor. Under normal conditions, the polymer donates energy to the cyanine dye resulting in emission at both 680 and 820 nm upon excitation. RNS degrade the cyanine dye, which abolishes the FRET and results in enhanced emission at 680 nm. Likewise, hydrogen peroxide results in chemiluminescence at 680–820 nm due to damage to the peroxyoxylate, which produces a reactive intermediate that excites the PFODBT and cyanine dye. The polymer nanoparticles were used to image acetaminophen and isoniazid induced toxicity in mouse liver. The particles allowed visualization of the threshold hepatatoxicity from acetaminophen ingestion versus dose response damage resulting from isoniazid.44
Figure 2.
Design of CF-SPN for detection of ROS and RNS. (a) Molecular components of CF-SPN are the NIR fluorescent semiconducting polymer PFODBT, a PEG-grafted poly(styrene) copolymer conjugated to galactose for hepatocyte targeting (PS-g-PEG-Gal), the H2O2-specific chemiluminescent substrate CPPO that serves as CRET energy donor, and the FRET acceptor IR775S that degrades after oxidation by ONOO– or –OCl (dark green). PFODBT serves as the CRET energy acceptor and the FRET energy donor. (b) Illustration of the mechanism of simultaneous and differential detection of ONOO– or –OCl and H2O2 by CF-SPN. After drug challenge to the liver, CF-SPN report via the chemiluminescent and fluorescent channels the generation of radical metabolites at safe (left) and toxic (right) drug doses. (c) Hydrodynamic diameter distribution of CF-SPN, determined by dynamic light scattering; A.U., arbitrary units. (d) Transmission electron micrograph of CF-SPNs. Reprinted by permission from Shuhendler, A. J.; Pu, K.; Cui, L. et al. Nat. Biotechnol. 2014, 32, 373–382 (ref 44), copyright Macmillan Publishers Ltd., 2014.
Additional novel high-throughput approaches to detect cellular or metabolite-mediated toxicity have utilized various transduction approaches. Cationic modified amphiphilic periodic mesoporous organosilica (PMO) was utilized as a bioreactor for cyt P450 3A4/CPR adsorption and metabolism studies. The cationic PMO featured ~20 nm diameter pores allowing substrate to access a higher concentration of immobilized enzyme, due to the ionic attraction between microsomes and the PMO surface. On the basis of these considerations, metabolism of model drugs utilizing the modified-PMO was much more efficient versus that in solution, as shown analyzing the reaction medium using LC–MS.45
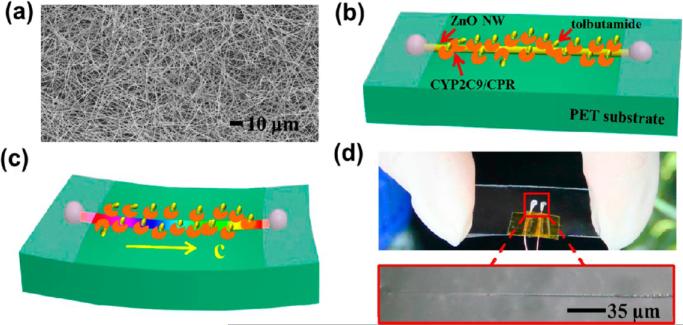
Single zinc oxide nanowire (ZnO NW) semiconductors were fabricated and used to monitor the metabolism properties of cyt P450 2C9/CPR microsomes. An overview of this is shown in Figure 3. In the presence of substrate, currents increased significantly as an external compressive strain was applied to the ZnO NW. This was shown to be due to the piezotronic effect whereby electron transport and transfer from cyt P450 to CPR was enhanced, which facilitated metabolism and increased currents.46 Min et al. developed a 3D-hydrogel electrochemical impedance system by bonding a molded PDMS flow cell onto the top of a deposited interdigitated gold microelectrode array on a glass slide.47 The toxicity of model oncology compounds was tested on HeLa and mouse embryonic fibroblast cells. As the cells were exposed to the compounds, electrochemical resistance in the solution was measured. The charge transfer resistance (Rct) has been shown to be due to changes in cell viability. The anticancer drugs poisoned the cells, which led to altered cellular morphology and lower resistance. While sensitive impedance measurements can be challenging based on competing processes that might occur on the electrode surface,48 this 3-D hydrogel array was validated utilizing a commonly employed 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) cell viability assay. Unlike optical detection methods, the electrochemical impedance system offers real-time, high-throughput cytotoxicity assessment in a label-free manner.47
Figure 3.
(a) Scanning electron microscopy (SEM) image of the as-synthesized ZnO NWs. Schematic of ZnO NW decorated with CYP2C9/CPR-microsomes combined with tolbutamide molecules under (b) no strain and (c) compressive strain. (d) Optical images of a fabricated ZnO NW device. Reprinted from Wang, N.; Gao, C.; Xue, F. et al. ACS Nano 2015, 9, 3159–3168 (ref 46). Copyright American Chemical Society, 2015.
3-D Organ Modeling Approaches
Overall, a set of reliable assays exists for acute pharmaceutical toxicity responses, e.g., multiple assays for genotoxicity that exhibit sufficient accuracy. However, when assaying for metabolism-related processes, in vitro tests are not as clear. The lack of in vitro assays to elucidate organ toxicity is a major challenge for the pharmaceutical industry, and 2D culture systems featuring one cell type do not correlate well with in vivo responses based on how cells behave within different extra-cellular matrixes.20,49 Several reports have recently focused on developing lab or organ-on-a-chip devices that mimic cell environments in an organ or several organs simultaneously for drug development studies.20,49,50 Important issues in developing 3-D arrays include tissue functionality and cell seeding steps, which influence the cellular behavior. Cells utilized in 3-D arrays adhere and aggregate in different manners based on the assay surface or the media agitation conditions, all of which affect their behavior and the approach utilized to assess the toxicity.49
Biocolloid/Spheroid Approaches
Cells grow into 3-D spheroids when they are rocked or rotated as they incubate, and these spheroids can be used as bioreactors to assess drug toxicity.49 The parameters that facilitate spheroid formation are vitally important, as rocking versus rotation incubation conditions can alter important physiologically important hepatocyte spheroid traits such as albumin synthesis, urea cycle, and metabolic enzyme expression.51 Other spheroid formation scaffolds can also be employed, such as bioreactor microcarrier beads or other physical structures onto which cells adhere. Cells can adhere in monolayer or 3-D arrangements on bioreactor beads depending on how they are incubated.49 However, cells grown in a rotating wall vessel forcing a state of continual free fall have been shown to form a desired 3-D structure that physiologically resembles tissue.52 Human vaginal epithelial cells grown on bioreactor beads in this manner were used to detect toxicity of Nonoxynol-9 (N-9), a sexually transmitted infection microbicide that has been shown to have varying degrees of toxicity that have precluded its use. 3-D beads, surgically removed vaginal tissue, and 2-D monolayer cultures were exposed to N-9 and imaged utilizing a fluorescent cell-viability assay.53 N-9 was shown to exhibit comparable toxicity using both the 3-D spheroid and tissue assays, which was presumably due to the similarities in cellular physiology between the bead and tissue. Because cells grown in the monolayer did not express the same cellular functionalities, N-9 toxicity elucidated using the traditional cellular assay was much higher. This work demonstrated the utility of the 3-D rotating wall cellular culture for predictive purposes not only as an upgrade from 2-D cultures but also as an acceptable substitute for tissue or animals.53 Well-plates modified with “nanopillar” structures in the well floors have also been used to control the 3-D size of the spheroids as well as differentiate stem cells into their respective hepatocypte-like cells for toxicity studies.54 Hepatocytes grown in micrometer-scale wells molded on a polystyrene plate formed spheroids of optimal size for toxicity monitoring purposes utilizing acetaminophen as a test compound.55
Chip-Based Devices
Several chip or surface-based devices have been created to mimic organ behavior for drug discovery or metabolism and toxicity elucidation. Many of these have thus far been focused on an attempt to mimic liver behavior as the many of the pharmaceutical issues arise from interactions that occur there.56 Some of these assays have implemented novel and elucidative structure-based detection strategies. Metabolomics-on-a-chip approaches with proton NMR57 and mass spectrometry (MS)58 detection have been utilized to assess the toxicity of model drugs, while also identifying biomarkers related to toxicity. In the latter case, a microflow cell device was constructed consisting of separate chambers for drug metabolism, cytotoxicity, and MS detection (Figure 4). Polyethylene glycol (PEG)-encompassed human liver micro-somes were exposed to acetaminophen in reaction chambers, which then led to either cell culture channels for cytotoxicity assessment or a micro-SPE column feeding directly to electrospray ionization-quadrupole-time-of-flight (ESI-Q-TOF) MS detection. A real time analysis and detection of reactive metabolites coupled with cytotoxicity assessment was then generated.58
Figure 4.
Microfluidic device for cell culture, metabolite analysis and cytotoxicity assay. (a) The integrated microfluidic device. (b) Microchannels for HLM encapsulation by PEG hydrogels. (c) Design of the on-chip micro-SPE column. (d) Cell culture channel. (e) An image of the microfluidic device filled with a blue dye in the bioreactor part cell culture part. Reproduced from Mao, S.; Gao, D.; Liu, W. et al. Lab Chip, 2012, 12, 219–226 (ref 58), with permission, copyright Royal Society of Chemistry, 2012.
Other innovative 3-D cellular strategies to assess liver toxicity include a perfusion bioreactor devised by Schmelzer et al. that accurately models pharmacokinetics and cyt P450 metabolism issues based on its ability to flow nutrients, gases, and xenobiotics to liver cells utilizing an integrated capillary network.59 Three sets of hollow capillaries, two for counter-current perfusion and one for gas flow, serve hepatic cells grown in the extra-capillary space. A 2 mL volume scaled down version of the device was developed and compared to an earlier larger volume (800 mL) model. It was found that miniaturization did not change the hepatocyte cyt P450 longevity or metabolic properties by measuring standard metabolite production via LC–MS.60
Of course, livers do not operate in isolation in our bodies. Other strategies have been reported to elucidate drug or toxin exposure at alternative organ sites or a multitude of tissue-organ sites simultaneously.50,61 A two-organ tissue microperfusion flow cell device was designed to assess the toxicity of multiple oncology drugs toward a glioblastoma cell line after the drugs were first metabolized in a liver cell compartment.62 HepG2 or glioblastoma cells were immobilized in discs made of porous polylactic acid, which were then positioned into a polycarbonate flow cell. Drugs were introduced into the cell, and were metabolized in the liver compartment before reaching the cancer cells. Cytotoxicity in the glioblastoma cell line was assayed utilizing fluorescence imaging. Cells in the 3-D polymer were much less susceptible to the anticancer drugs compared to those in a 2-D monolayer arrangement owing to the differences in accessibility and system complexity. Also, the expression of cyt P450 levels was varied in the liver compartment and strongly influenced the toxicity assays for ifosfamide.62 Likewise, a liver-kidney two-tissue flow cell constructed of fibronectin-coated PDMS was designed to assess the toxicity of ifosfamide. The drug was metabolized in the liver compartment, and kidney cell toxicity was monitored as the metabolites entered into the adjoining cell. MS was utilized to identify the reactive metabolite structures responsible for the toxicity.63
Dordick et al. reported construction of a 3-D microarray chip designed to assess neural stem cell differentiation and the subsequent neurotoxicity of small molecules.64 The stem cells were mixed with alginate, which was then printed onto a hydrophobic-modified glass slide, clamped in a cell and allowed to proliferate in cell expansion medium. Cytotoxicity was assayed utilizing a fluorescence live/dead cell assay. Despite lower viability and slower proliferation vs monolayer cultures, the stem cells differentiated into glial cells on the chip. Additionally, the chip was used to assess the neurotoxicity (via cytotoxicity) of standard compounds. The assay was able to identify compounds that were toxic to stem cells compared to differentiated cells. It also showed that some compounds, such as acetaminophen, are nontoxic to neural cells at typically utilized concentrations.64
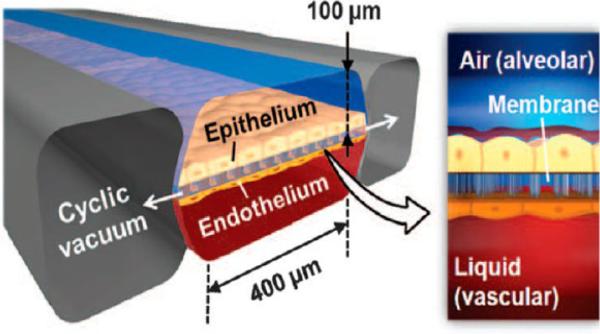
Ingber et al. developed a microfluidic device designed to produce a microengineered state of pulmonary edema (Figure 5).65 The device was fabricated from transparent silicon to house an upper layer of alveolar epithelial cells in close proximity to a bottom layer of vascular endothelial cells separated by a thin membrane. The alveolar layer was exposed to air and medium containing pharmaceuticals flowed through the vascular layer, where the cells were cyclically stretched to mimic the breathing motion. Vascular permeability was assessed by introduction of FITC-inulin into that compartment and periodically measuring the fluorescence of the fluid in the alveolar channel. The device was used to show that IL-2 caused leakage across the vascular layer, resulting in subsequent fibrin development in the alveolar region. Proprietary pharmaceuticals designed to prevent endothelial leakage were also assayed.65 Similarly, a heart cell contraction device was fabricated utilizing neonatal rat myocytes grown on a stretchable silicone membrane coated with polydimethylsiloxane (PDMS) in order to mimic conditions in failing cardiac tissue. This cell was useful in determining the genetic and pathological changes related to heart disease.66
Figure 5.
IL-2-induced pulmonary edema is modeled in a micro-engineered lung-on-a-chip that reproduces the lung microarchitecture and breathing-induced cyclic mechanical distortion of the alveolar-capillary interface. The top “air” portion is the alveolar channel; the bottom “liquid” portion is the vascular channel. From Huh, D.; Leslie, D. C.; Matthews, B. D., et al. Sci. Transl. Med. 2012, 4, 159ra147 (ref 65). Reprinted with permission, copyright AAAS, 2012.
Overall, the landscape of 3-D organ-on-a-chip devices and approaches is constantly changing and improving. These devices continue to become increasingly complex and interconnected and, as a consequence, more predictive of actual toxicity that might be exhibited due to drug exposure. This may be complicated by issues including cellular handling protocols, quality and identity assurances of the cell line used, expensive or complicated equipment, and complexities arising in the data generation.20 Additionally, the usefulness of these devices must be validated with data from animal and clinical studies before their widespread use in pharmaceutical development and testing.50
EXPLORING METABOLITE-RELATED TOXICITY PATHWAYS
For the past 15 years, our research team has been developing sensor arrays and multiplexed LC–MS/MS methods to screen and establish pathways of metabolic toxicity chemistry. Our apporoaches are designed to to reveal the relevant chemical pathways for the effects seen in bioassays. As opposed to bioassays, our cell-free methodologies first employ high-throughput arrays with DNA damage end points to reveal the possibility of toxicity-related chemical reactions. Molecules that produce reactive metabolites are identified by “hits” in these genotoxicity assays. Then, molecular pathways for the “hits” leading to possible toxicity-producing products are investigated in detail by using high-throughput reactions to produce samples for LC–MS/MS. These approaches are designed to complement bioassays, not replace them.
Our approach combines metabolic enzymes, DNA, and polyions in thin films in the screeing arrays,4,67,68 and on magentic particles to produce samples for LC-MS/MS analysis.4,69,70 Enzymes in these thin films produce metabolites in a reaction step, and reactive metabolites form damage DNA in the films. The key to this approach is the close packing of DNA and metabolic enzymes in the films. The concentration of DNA in the films is very high, on the order of 0.1–0.2 M. When reactive metabolites are formed by enzymes in the film in close proximity to DNA, the probablitiy of reaction of these metabolites with DNA bases is very high, as predicted by SN2 second-order kinetics. Many reactive metabolites react with DNA to form nucleobase adducts (Figure 6) that may be stable or could eventually lead to abasic sites.4,10,13 Strand breakage and oxidation of nucleobases is also possible.
Figure 6.
Main nucleophilic reactive atoms of DNA bases for electrophilic reactive metabolites in SN2 reactions. Dark arrows represent the most reactive sites; thinner arrows represent additional reactive sites that may still lead to major nucleobase adducts depending on the properties of the electrophile.
In the genotoxicity arrays, damaged DNA is detected after the enzyme reaction step by subsequent electrochemical or electrochemiluminescent (ECL) detection.71–73 These DNA/enzyme films include an Ru(bipyridyl)-poly(vinylpyridine) polymer as electrochemical catalyst to boost signals in electrochemical detection or to emit light in the ECL process. For LC–MS/MS, similar enzyme/DNA films without the catalytic polymer are grown on silica or magentic beads. Then the enzyme reactions are run in 96 well filter plates, and DNA is hydrolyzed away from the beads to collect nucleobase adducts by filtration into another 96 well plate for LC–MS/MS analysis.70 Both sensor array and LC–MS/MS methods employ layer-by-layer (LbL) film-fabrication to construct stable multi-component films assembled via electrostatic adsorption of alternately charged layers of DNA, polyelectrolytes, and metabolic enzymes.71,73 Films thickness can be controlled on the nanometer scale, and enymes in these films remain stable for a month or more at 4 °C. Our approaches have been built up in stages over the past 15 years, and we summarize the most advanced approaches below.
Microfluidic Electrochemical Arrays
Here we describe fluidic arrays for damage of DNA by reactive metabolites that includes DNA adduct formation and DNA oxidation. As mentioned above, arrays that detect DNA adduct formation and other damage can be based upon thin films of DNA, the ECL-active polymer [Ru(bpy)2(PVP)10](ClO4)2 {RuPVP}] (Scheme 1), and microsomal enzyme sources. Our development of LbL films that combined microsomal cyt P450 reductase (CPR) and cyt P450s accurately reproduced the natural cyt P450 catalytic cycle on electrodes.74 That is, electrons are transferred from electrode to CPR to cyt P450s to mimic the in vivo pathway, i.e., electrode → CPR → cyt P450. This approach was incorporated to activate cyt P450 enzymes in an array format using rat liver microsomes (RLM) as the enzyme source. As explained earlier, RLMs contain the requisite cyt P450s and CPR, and electrons are provided by the electrode array to drive the enzyme-catalyzed catalytic oxidations.
Scheme 1.
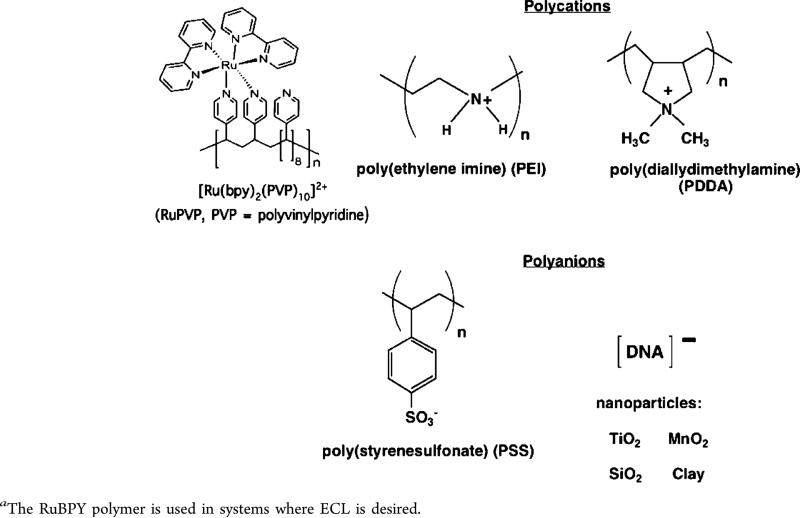
Polyions Used to Make Films by Layer-by-Layer (LbL) Alternate Electrostatic Adsorptiona
All of our methods employ alternate electrostatic layer-by-layer (LbL) film assembly to prepare multicomponent films of enzyme source materials, DNA, using additional polyion layers to provide structural stability.71,73 This approach developed by Lvov and Decher75–78 provides excellent control of thickness for fabricating versatile film architectures on the nanometer scale. In LbL film assembly, an initial layer of charged polyion is adsorbed from a 0.5–3 mg/mL solution onto an oppositely charged array surface, such as the bottom of a well. Loosely bound polyions are removed by washing with water, then a second layer of polyions of opposite charge to the first layer is adsorbed. This process is continued, reversing polyion charge at each step, until the desired architecture, composition, and thickness are achieved. Polyions used in our work are illustrated in Scheme 1. DNA and enzymes are water-soluble polyions that are well suited for this method. We adsorb film layers from single drops of solution in array wells to conserve valuable biomolecules, then wash, and place them in a humidified chamber to dry slowly.73 We use wells printed from hydrophobic ink to locate drops on the arrays, thus restricting the DNA/polyion/enzyme films into specific regions and avoiding cross-contamination during film fabrication.79
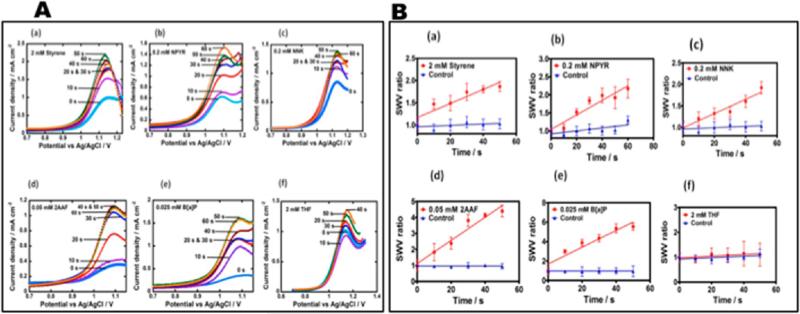
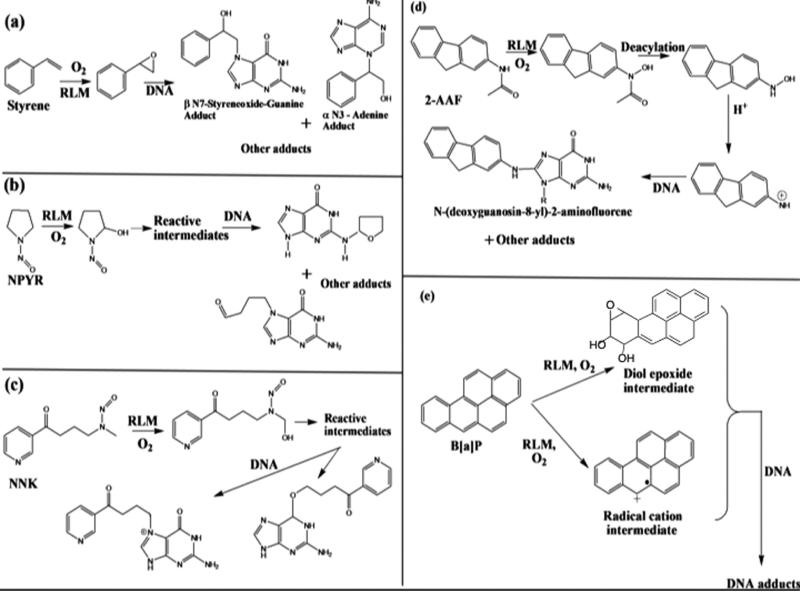
Using LbL films of RLM as enzyme source, RuPVP as electrochemical catalyst, and PDDA and PSS polyions for structural integrity, we designed a novel, rapid microfluidic array using electrochemically driven cyt P450 enzyme catalysis that screens for formation of reactive metabolite from test compounds (Figure 7).80 The device incorporates an eight-electrode screen-printed carbon sensor array coated with the DNA/RuPVP/RLM films. Cyt P450s in RLMs are activated by the electrochemical approach described above. The process features electron donation from the sensors to cyt P450 reductase in the RLMs and subsequent cyt P450 reduction while flowing an oxygenated test reactant solution across the array. Reactive metabolites formed in the films react with DNA, and damaged DNA is detected by catalytic square wave voltammetry (SWV) utilizing the RuPVP polymer (Figure 8). The microfluidic device was tested for a set of pollutant molecules known to form known DNA-reactive metabolites (Scheme 2). In all these reactions, the metabolic process forms reactive metabolites that then form adducts, mainly with guanines and adenines on DNA in the films.
Figure 7.
Microfluidic electrochemical array system used for detection of reactive metabolites formed by liver enzyme cyt P450s.
Figure 8.
Representative data from cyt P450 arrays: (A) SWVs at different enzyme reaction times in anaerobic pH = 7.4 buffer for microfluidic array sensors featuring optimized films of PDDA/PSS/(RuPVP/DNA)2/PDDA/RLM/PDDA/DNA, (SWV ampl. 25 mV; freq 15 Hz; step 4 mV) (a) 2 mM styrene, (b) 0.2 mM NPYR, (c) 0.2 mM NNK, (d) 0.05 mM 2-AAF, (e) 0.025 mM B[a]P, (f) 2 mM THF. (B) Influence of substrate incubation time on SWV peak current ratio (Ip,f/Ip,i) for microfluidic toxicity sensor array using PDDA/PSS/(RuPVP/DNA)2/PDDA/RLM/PDDA/DNA, films: (a) 2 mM styrene, (b) 0.2 mM NPYR, (c) 0.2 mM NNK, (d) 0.05 mM 2-AAF, (e) 0.025 mM B[a]P, (f) 2 mM THF. Controls are incubations without substrate or exposure to the substrate without electrolysis, which gave equivalent results. Reproduced from ref 81. Copyright Royal Society of Chemistry, 2013.
Scheme 2.
DNA Adduct Formation from Metabolic cyt P450 Bioactivation of (a) Styrene, (b) N-Nitrosopyrrolidine (NPYR), (c) 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), (d) N-(9H-Fluoren-2-yl)acetamide (2-AAF), and (e) Benzo[a]pyrene (B[a]P)
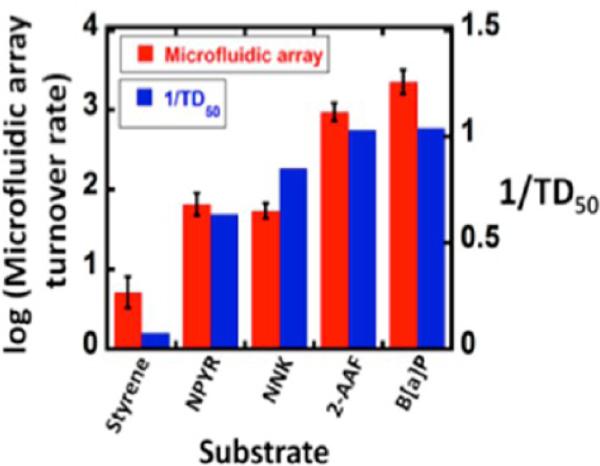
Figure 8 illustrates the type of data obtained from the electrochemical microfluidic cyt P450 array. The SWV peaks increase in height as the DNA damage proceeds with increased time of the enzyme reactions due to partial unraveling of the adducted DNA strands. This unwinding makes the guanines in the DNA more accessible to the catalytic Ru-sites in the film and increases the catalytic reaction rate.4 SWV peak heights are plotted vs reaction time and the slope is proportional to the relative rate of DNA damage,4 as has been confirmed by similar experiments monitored by LC–MS/MS to directly measure the nucleobase adducts. Turnover rates based on SWV responses gave excellent correlation with the reciprocal of rodent liver toxicity metric TD50 (toxic dose 50%) that monitors formation of liver carcinoma resulting from feeding rats these chemicals (Figure 9).80 This type of correlation is supporting evidence for the relevance of the device for toxicity screening. The microfluidic array provides a steady state concentration and flow of reactants that provides much better S/N and reproducibility than single electrode sensors or arrays on which the reaction solutions are deposited and are subject to reactant depletion and evaporation. This approach was later adapted with a multiplexer to accommodate a single micro-fluidic device with four 8-sensor arrays in parallel, and used to investigate reactions of chemicals with cyt P450s coupled with other metabolic enzymes.81
Figure 9.
Correlation of logarithm of turnover rate ({μg protein−1} s−1 mM−1) measured from SWV peaks in the array with reciprocal of reported TD50 values. Reproduced from ref 81. Copyright Royal Society of Chemistry, 2013.
The microfluidic system in Figure 7 was also adapted to measure oxidized DNA dissolved in solution.82 Relative to the DNA damage toxicity end point, some metabolites contribute to oxidative stress in humans by generating reactive oxygen species (ROS) that can oxidize DNA in a complex redox pathway. We used the microfluidic electrochemical array to rapidly detect oxidation in DNA samples using sensor arrays fabricated by wet-chemistry patterning of gold compact discs. These eight-sensor Au arrays were incorporated into the 60 μL microfluidic channel (Figure 7). Array sensors were coated with 7 nm thick osmium bipyridyl poly(vinylpyridine) chloride [Os(bpy)2(PVP)10Cl]+ films. This Os complex was used for surface electrochemical catalysis to selectively oxidize 7,8-dihydro-8-oxoguanine (8-oxodG), a major product of DNA oxidation, at a much lower potential than the intact DNA bases. Oxidized ds-DNA was measured by catalytic square wave voltammograms (SWV) at levels as low as one 8-oxodG per 6600 nucleobases. Fast assays (<1 min) and moderate sample consumption (15 pmol DNA) suggest potential for research and clinical applications. Practical use was illustrated by detecting DNA oxidation from quinones in cigarette smoke and ash extracts in dispersions with NADPH and Cu2+, initiating a complex redox pathway producing ROS that oxidize dG in DNA to 8-oxodG.82
ECL Arrays
Electrochemical generation of light with a luminescent dye and coreactant is known as electrochemiluminescence (ECL) and is a very sensitive detection method for bioanalysis. In many applications, the dye Ru(bpy)32+ produces visible ECL light through a complex redox process involving a sacrificial reductant, such as tripropylamine (TPrA), at oxidizing electrode potentials to yield excited-state [Ru-(bpy)32+]* that emits light at 610 nm.83 The reaction pathway involves oxidized and reduced forms of TprA and Ru(bpy)32+ and depends on concentrations of both.84 In our systems, we used the RuPVP catalyst polycation (Scheme 1) which is readily incorporated in LbL films and generates ECL light when electrochemically activated using guanines in DNA as coreactants.85 A representation of the LbL films (Figure 10) shows the polyanionic DNA, enzyme sources such as microsomes with net anionic charge, and RuPVP, and/or polyions such as PDDA or PSS to improve film stability.4,73
Figure 10.
Films of DNA, polyions and metabolic enzyme sources used in arrays and on magnetic beads for screening, and pathway studies of metabolite-related toxicity pathways. Blue layers on nonreactive polyions or RuPVP ECL polymer.
Liver Enzyme Array
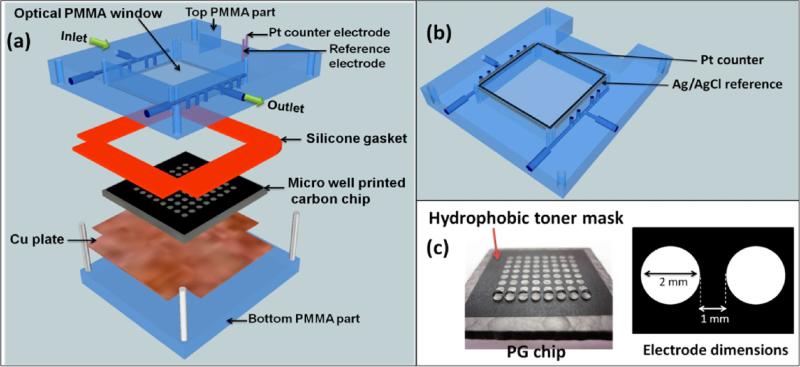
Our most advanced ECL metabolic toxicity screening array features a 64-microwell reactor chip in a microfluidic chamber (Figure 11).79 The chip is a conductive pyrolytic graphite (PG) sheet onto which are printed 64 microwells using a computer printed pattern that is heat-transferred onto the PG. The computer ink is hydrophobic enough so that the 10–20 nm deep wells readily hold 1 μL droplets in place for the sequential adsorption of LbL film layers (Figure 11c).
Figure 11.
64-microwell ECL chip and the fluidic reaction chamber: (a) assembly of the flow cell, (b) underside view of reference and counter electrode wires in the top poly(methyl methacrylate) (PMMA) plate, (c) pyrolytic graphite (PG) chip with computer-printed microwells. The first row shows 1 μL water droplets on each of the wells. The microwell chip is used in a fluidic reactor system similar to Figure 7. Reproduced from ref 79. Copyright Royal Society of Chemistry, 2013.
The enzyme/RuPVP/DNA films are fabricated on the array by placing 1 μL droplets of relevant adsorbate solutions in the microwells at 4 °C to make each layer. The well holds the droplet in place during layer adsorption in a humidified chamber. Enzyme sources for the films can include liver microsomes, pure human bioconjugation enzymes, human liver S9 enzyme fractions (HS9), and supersomes of cyt P450s.
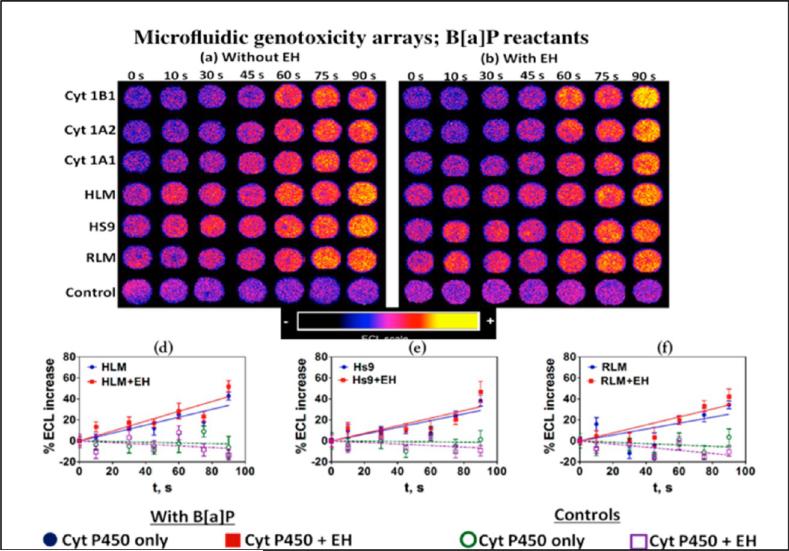
The 64-microwell ECL array reactor (Figure 11) chip was used to replace the voltammetric sensor array in the fluidic system in Figure 7. This new system was first evaluated by investigating metabolic DNA damage and cyt P450 inhibition using benzopyrene (B[a]P) as a test molecule. The B[a]P metabolism pathway involves two oxidations by cyt P450 with hydrolysis of an intermediate by epoxide hydrolase (EH) to give the ultimate probable carcinogen B[a]P-7,8-dihydrodiol-9,10-epoxide (BPDE).86 BPDE reacts directly with DNA to form adducts with guanines and adenines (Scheme 2e). The ECL chip wells were loaded with replicate thin films including DNA, RuPVP, and the various enzyme preparations listed above and using wells with and without EH to provide the key epoxide hydrolysis step to give BPDE. Enzyme cofactors, oxygen, and reactants necessary to drive all the enzyme reactions on the chip are pumped into the array in a constant flow. As in the electrochemical toxicity arrays, microsomal cyt P450s are activated electrochemically. Once the reactions proceed for a desired time, the electrode potential is turned off and wash buffer is passed through the array. Then, 1.25 V vs Ag/AgCl is applied for 180 s in a dark box to generate ECL that is measured with a charge coupled-device CCD camera positioned above the array.4,79 Electrochemical oxidation of RuII in RuPVP initiates a multistep redox pathway featuring guanines in DNA as coreactants to generate electronically excited RuII* sites that decay to emit visible ECL light.85
ECL for wells containing cyt P450s in supersomes or other enzyme source materials increased with enzyme reaction time (Figure 12a,b). To analyze the data, a software package evaluates ECL intensity and provides graphs of % ECL increase vs enzyme reaction time (Figure 12d–f). The slopes of these linear plots directly monitor relative rates of DNA damage, as confirmed in several LC–MS/MS studies.4 The increase in ECL output results from the formation of covalent nucleobase adducts of metabolites and possibly strand breaks that disorder the DNA double helix and make guanines in the films more accessible to the RuIII sites in the LbL films. Slightly larger relative DNA damage rates were found when EH was added to the cyt P450 enzyme wells, suggesting formation of more BPDE when both cyt P450 and EH are present to provide more DNA damage. Interpretation is clouded a bit since HLM and RLM may contain small amounts of EH.
Figure 12.
ECL array data obtained with microwells containing RuIIPVP/enzyme/DNA film assemblies reacted with oxygenated 25 μM of B[a]P + pH = 7.4 buffer with bioelectronic activation of cyt P450s at −0.65 V vs Ag/AgCl (0.14 M KCl) for 0–90 s. Enzyme sources are Cyt P450 supersomes, human and rat liver microsomes (HLM and RLM), epoxy hydrolase (EH), and human S9 fractions (HS9). Controls contain cyt P450 1B1 with or without EH and were subjected to the same reaction conditions without bioelectronic activation of cyt P450s. Reconstructed, recolorized array images are shown (a) without EH and (b) with EH. Graphs show examples of influence of enzyme reaction time on ECL increase for (d) HLM, (e) Hs9, (f) RLM. Reproduced from ref 79. Copyright Royal Society of Chemistry, 2013.
LC–MS/MS analysis of products of magnetic bead bioreactors coated with the same enzyme/DNA films as in the above arrays was also done. Reactions with B[a]P and analysis of the resulting nucleobase adducts after hydrolysis of the DNA gave product ion spectra that confirmed formation of adducts dG-BPDE and dA-BPDE when cyt P450 and EH were on the beads.79
Organ-Specific Enzyme Array
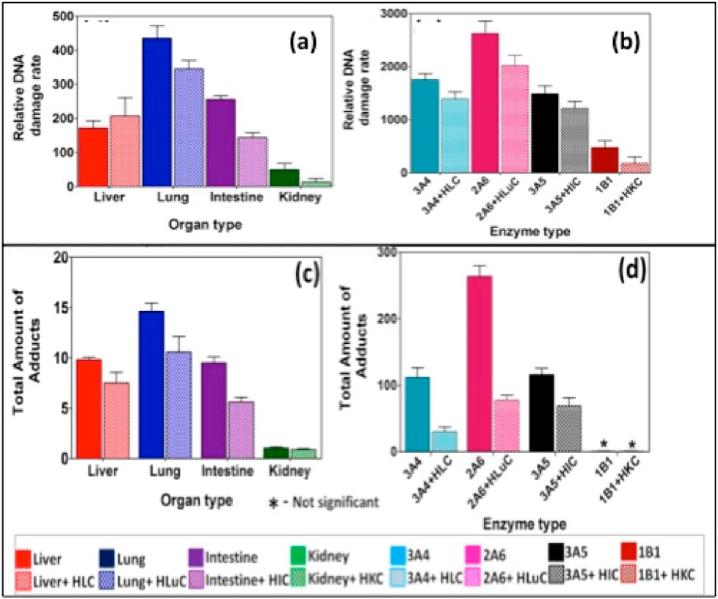
Liver metabolism has been the traditional target of drug and pollutant toxicity studies. However, other organs also produce reactive metabolites and should be considered in a complete toxicity assessment. We used the enzyme/DNA ECL arrays described above to evaluate chemical genotoxicity pathways from reactive metabolites formed on the array by a representative range of liver, lung, kidney, and intestinal enzymes.87 DNA damage rates were measured on the 64-microwell chip loaded with films of DNA, RuPVP, and a wide range of metabolic enzymes. Multiple enzyme reactions were run on test compounds using the array to measure relative DNA damage rates. A companion LC–MS/MS method4,70 was used to react target compounds with magnetic beads coated with DNA and the same organ enzymes in 96 well plates, after which DNA was hydrolyzed and nucleobase-metabolite adducts identified and measured. Results revealed nucleobase adducts from DNA damage, specific enzymes that produce the reactive metabolites, influence of bioconjugation enzymes, relative dynamics of enzymes from different organs, and pathways of possible genotoxic chemistry.
Extensive data sets were obtained for styrene, NNK, and 2-AAF, but for brevity we illustrate here only the final results for NNK (Figure 13). Values presented in the figure are normalized for the relative amounts of enzyme in each experiment. In nearly all cases, inclusion of cytosolic bioconjugation enzymes decreased the relative rate of DNA damage and the amount of nucleobase adducts found, consistent with the roles these enzymes play in detoxication.87 The lone exception was relative damage rates for liver enzymes that were within experimental error of one another (Figure 13a). Furthermore, the lung enzymes are clearly the most active, again consistent with previous literature on NNK metabolism.87
Figure 13.
Results from genotoxicity study of oxygenated 25 μM NNK in pH 7.4 buffer with electrochemical activation of cyt P450s at −0.65 V vs Ag/AgCl (0.14 M KCl): relative DNA damage rates ({μg of protein}−1 s−1 mM−1) from ECL arrays (a and b) and without human cytosols from the same organ (e.g., HLuC = human lung cytosol) for (a) human organ tissue enzymes, (b) cyt P450 supersomes. Controls for cyt P450s (green) and cyt P450s + EH (purple) behaved equivalently without substrate or with substrate but no activation of cyt P450s. Bottom panels c and d are total DNA adducts found (pmol {μg of protein}−1 {mM of NNK}−1) in hydrolyzed samples by LC–MS/MS after reactions in 96 well plates with 150 μM NNK at pH = 7.4 for 18 h using magnetic beads coated with DNA and (c) human organ microsomes and (d) cyt P450 supersomes. Color codes are the same for all panels. Reproduced from ref 87. Copyright Royal Society of Chemistry, 2015.
Correlations between relative DNA damage rates from the cell-free ECL array and organ-specific Comet assays that monitor DNA damage in cells were found. These results illustrate the power of the combined DNA/enzyme microarray/LC–MS/MS approach to efficiently explore a broad spectrum of organ-specific metabolic genotoxic pathways for drugs and environmental chemicals. Experimental details of the 96-well plate LC–MS/MS methodology have been summarized elsewhere.4,70
Damage to Tumor Suppressor Genes
The LC–MS/MS methods mentioned above were redesigned to detect and identify adducted DNA bases from reactive metabolite reactions. We have begun extending this approach to identify sites of codon damage on important genes related to toxicity responses. Tumor suppressor genes provide protection against cancer by coding for proteins that carry out relevant protective tasks.88 TP53 (or p53) was one of the first tumor suppressor genes discovered89 and is found to be damaged in about half of all human cancers. Extensive databases bringing together research on tumors and cell cultures reveal that mutations of specific p53 codons are well correlated with specific types of cancers.90 If frequently damaged p53 codons in cancers correlate with in vitro chemical damage sites, predictions of organ-specific cancer risks might be enabled. We recently described LC–MS/MS methodology to identify the most frequently adducted nucleobases within codons in a 32 base pair (bp) oligonucleotide representing part of p53 exon 7 with 5 possible reactive codons.91 The methodology is based on LC–MS polynucleotide sequencing and enables nucleobase reactivity studies of double stranded oligonucleotides with more than 20 base pairs and multiple reactive sites. BPDE undergoes nucleophilic substitution by DNA bases, and we reacted it with the p53 fragment in solution. LC–MS/MS sequencing of products from the reaction of BPDE with 32 bp oligonucleotide was done after cleavage by a restriction endonuclease to produce strands of less than 20 bases that are amenable to direct LC–MS/MS sequencing.
Analysis of the chemically adducted p53 fragment revealed most frequently reacted guanines in codons 248 and 244. These codons are also mutated with high frequency in human tumors.90 Codon 248 is mutated in lung, head and neck, colorectal, and skin cancers, while codon 244 is mutated in small cell lung cancer. All involve possible initiation related to BPDE exposure. This approach, once developed into a high-throughput format, should be useful for screening drugs and environmental chemicals to predict risks for organ specific cancers based on their p53 codon damage patterns.
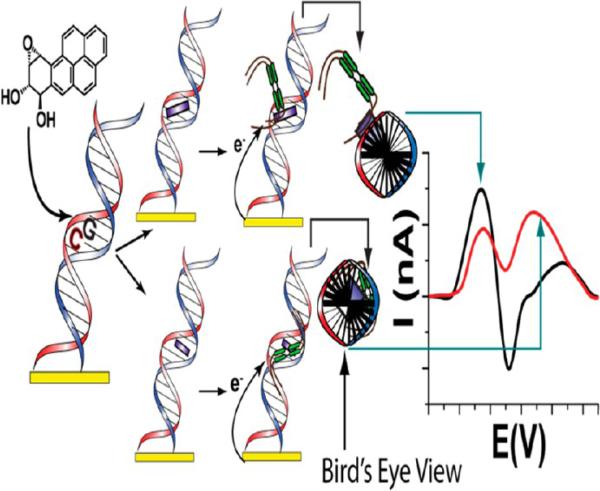
We have also detected sequence specific BPDE-related TP53 damage at codon 273 electrochemically utilizing DNA hybridization sensors.92,93 Unmodified DNA and 5′-methyl cytosine modified DNA oligomers were employed. Specially synthesized viologen molecules associate with DNA based on its morphology and produce signals based on this association upon electrochemical reduction to detect the changes to DNA after exposure to BPDE. An overview of the sensor and the origin of the electrochemical signals derived from BPDE exposure is shown in Figure 14.
Figure 14.
Overview of the sequence specific DNA damage electrochemical sensor. DNA oligomers spanning codons 270–276 of the TP53 gene were exposed to anti-BPDE. The codon 273 section is highlighted and DNA oligomers may have featured cytosine methylation. BPDE adducts DNA (purple) and can either be located in the minor groove (top) or intercalate (bottom). Viologen-containing molecules of the general form C12H25V2+C6H12V2+C12H25 (V2+ = 4,4′-bipyridyl or viologen) are associated with the adducted helices. Resulting voltammetry is dependent on the resulting location of the viologen on the DNA oligomer. Minor groove BPDE (top) forces viologen outside of the DNA helix, producing positive shifted voltammetry upon reduction. Intercalated BPDE (bottom) allows violgen to bind within the minor groove, resulting in reduction waves that are negative shifted. Reproduced from ref 93. Copyright American Chemical Society, 2013.
When exposed to unmodified DNA, (±)-anti-BPDE enantiomers are known to adduct hotspot guanines and eventually orient within the DNA minor groove.94 Epigenetics also influences the adduct orientation. For instance, hotspot codons often feature the –CGx– sequence, where x = any base. The cytosine in this sequence (5′ to the guanine) is often methylated at the 5-C location within the pyrimidine ring, which may force the BP-adduct to an intercalated state within the DNA base stack due to favorable hydrophobic interactions. However, this process ultimately depends on BPDE stereo-chemistry.95 The electrochemical sensor utilized oligomers featuring different combinations of cytosine methylation exposed to either racemic (+/−)-anti-BPDE or enantiomerically pure (+)-anti-BPDE to show how this complicated BPDE-DNA adduct interplay unfolds. First, current signals acquired when DNA was exposed to anti-BPDE showed preferential binding at the codon 273 sequence as compared to control sequences missing the hotspot guanine.92 However, these signals were also dependent on both the BPDE stereochemistry and DNA methylation status, which both dictate where the BPDE will eventually bind. BPDE bound in the minor groove caused viologen to be reduced at positive potentials compared to its standard formal potential, while intercalated BPDE caused viologen to be reduced at negatively shifted potentials.93 Essentially, minor groove bound BPDE caused instability in oxidized viologen by causing it to bind outside of the protective DNA helix, whereas intercalated BPDE caused viologen to bind within the stabilizing minor groove location.
Validation of the electrochemical results was performed utilizing UV-spectroscopy and nano-ESI MS, which confirmed that the guanine within the codon 273 site was preferentially adducted.93 Specifically, the coding, nontranscribed strand containing the 5′-CGT-3′ sequence was preferentially adducted as compared to the complementary sequence guanine. BPDE preferentially bound to this site compared to the complementary sequence independent of cytosine methylation status. However, methylation of the complementary strand cytosine caused a significant increase in BPDE binding at the guanine site. Directing of BPDE binding is thought to be due to the change in hydrophobicity in the DNA introduced by methylation, which makes the initial BPDE association with DNA more favorable.96
OUTLOOK FOR THE FUTURE
This Feature article describes emerging high-throughput technologies that take advantage of modern robotics, micro-fluidics, and materials-printing approaches that can have a significant effect on improving the future success of toxicity prediction in the future. As mentioned above, huge drug failure costs and human safety are the main driving forces, whereas regulatory factors are mostly adequate to the task. However, it is important to realize limitations and roadblocks in our current approaches. The first limitation for drug and environmental chemical development is that it is essential to combine a suite of high-throughput bioassays including modern toxicity tests, genotoxicity assessment, metabolite related toxicity, computational approaches, and establish pathways for any toxic reactions. An important issue here is what exact tests should be included to minimize effort and cost, while maximizing success in toxicity prediction. We believe that genotoxicity chemistry arrays are very important to help reveal toxicity pathways that are active in the other tests and possibly in humans. When serious toxicity is suspected from the results of these tests, the magnetic bead bioreactor approach with LC–MS/MS can reveal detailed chemical pathways of metabolite-related toxicity.4,18,97–99 In this respect, elucidation of specific pathways of toxicity may facilitate synthetic strategies that can design toxicity out of desired chemical products while retaining the beneficial pharmacological effects.
A related roadblock in toxicity prediction is the lack of fundamental understanding of all the complex toxic responses and interactions in the human body, including those collected under the heading of idiosyncratic toxicity. Innovative 3-D cellular culture assays may eventually be able to shed light on this type of toxicity based on more accurate modeling of the in vivo responses. Currently, idiosyncratic toxicity appears in clinical testing much more often than is desirable. Additionally, most of these assays are still in the developmental stage and must be extensively tested and validated before regulatory agencies will accept data generated by their use.20 Efforts are underway to bring industry and regulatory agencies together so that the implementation of any useful drug development and toxicology screening technology will not be unnecessarily delayed.100 Of course, we need to be aware that bioassays, toxic pathway analyses and animal model studies will almost always, at least for the foreseeable future, fall short of 100% human toxicity prediction.
ACKNOWLEDGMENTS
J.F.R. thanks the National Institute of Environmental Health Sciences (NIEHS), NIH, USA, Grant No. ES03154 for financial support. E.G.H. is grateful to the E. Carolina Univ. Office of Undergraduate Research and North Carolina Biotechnology Center for financial support. The authors thank co-workers and collaborators named in joint publications for their many excellent contributions, without which research progress would not have been possible.
Biography
Eli Hvastkovs is an Associate Professor in the East Carolina University Department of Chemistry. He received his Ph.D. in chemistry from the University of Wyoming under Dan Buttry and was a postdoctoral researcher at the University of Connecticut under the guidance of James Rusling and John Schenkman. His research focuses on developing electrochemical methods to assess a variety of biological processes.
James F. Rusling was awarded a Ph.D. from Clarkson University in 1979. He is a Professor of Chemistry at University of Connecticut, Professor of Surgery and member of the Neag Cancer Center at the University of Connecticut Health Center, and adjunct Professor of Chemistry at National University of Ireland, Galway. Current research includes new cancer diagnostic arrays measuring biomarker proteins, ECL, and mass spectrometric arrays for toxicity screening and fundamental bioelectrochemistry.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.U.S. Environmental Protection Agency [July 21, 2015];Toxicology Testing in the 21st Century (Tox21) [Google Scholar]
- 2.Steinberg P, editor. High-Throughput Screening Methods in Toxicity Testing. John Wiley & Sons; Hoboken, NJ: 2013. pp. 433–452. [Google Scholar]
- 3.Lynch AM, Sasaki JC, Elespuru R, Jacobson-Kram D, Thybaud V, et al. Environ. Mol. Mutagen. 2011;52:205–223. doi: 10.1002/em.20614. [DOI] [PubMed] [Google Scholar]
- 4.Hvastkovs EG, Schenkman JB, Rusling JF. Annu. Rev. Anal. Chem. 2012;5:79–105. doi: 10.1146/annurev.anchem.111808.073659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayne JT, Ku WW, Kennedy SP. Curr. Opin. Drug Discovery Dev. 2006;9:75–83. [PubMed] [Google Scholar]
- 6.Nassar AEF, Kamel AM, Clarimont C. Drug Discovery Today. 2004;9:1055–1064. doi: 10.1016/S1359-6446(04)03297-0. [DOI] [PubMed] [Google Scholar]
- 7.Kramer JA, Sagartz JE, Morris DL. Nat. Rev. Drug Discovery. 2007;6:636–649. doi: 10.1038/nrd2378. [DOI] [PubMed] [Google Scholar]
- 8. [March 31, 2016]; http://www.forbes.com/sites/matthewherper/2013/08/11/howthe-staggering-cost-of-inventing-new-drugs-is-shaping-the-future-of-medicine/, Forbes.com.
- 9.Shanks N, Greek R, Greek J. Philos. Ethics Humanit. Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. DOI: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby WB, editor. Enzymatic Basis of Detoxification. I/II. Academic; New York: 1980. [Google Scholar]
- 11.Davis CD, Hanumegowda UM. In: Drug Metabolism Handbook. Nasser AF, Hollenberg PF, Scatina J, editors. John Wiley & Sons; Hoboken, NJ: 2009. pp. 561–628. [Google Scholar]
- 12.Caldwell GW, Yan L. Curr. Opin. Drug Discov. Devel. 2006;9:47–60. [PubMed] [Google Scholar]
- 13.Friedberg EC. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 14.Phillips DH, Farmer PB, Beland FA, Nath RG, Poirier MC, et al. Environ. Mol. Mutagen. 2000;35:222–233. doi: 10.1002/(sici)1098-2280(2000)35:3<222::aid-em9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 15.a Bond JA. Crit. Rev. Toxicol. 1989;19:227–249. doi: 10.3109/10408448909037472. [DOI] [PubMed] [Google Scholar]; b Umemoto A, Komaki K, Monden Y, Suwa M, Kanno Y, et al. Chem. Res. Toxicol. 2001;14:1006–1013. doi: 10.1021/tx010012d. [DOI] [PubMed] [Google Scholar]; c Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, et al. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 16.Claesson A, Spjuth O. Mini-Rev. Med. Chem. 2013;13:720–729. doi: 10.2174/1389557511313050009. [DOI] [PubMed] [Google Scholar]
- 17.a Schenkman JB, Greim H. Cytochrome P450. 1st ed. Springer; Berlin, Germany: 1993. [Google Scholar]; b Ortiz de Montellano PR, editor. Cytochrome P450. 3rd ed. Kluwer/Plenum; New York: 2005. [Google Scholar]; c Liebler DC, Guengerich FP. Nat. Rev. Drug Discovery. 2005;4:410–420. doi: 10.1038/nrd1720. [DOI] [PubMed] [Google Scholar]; d Ioannides C, Lewis DFV. Curr. Top. Med. Chem. 2004;4:1767–1788. doi: 10.2174/1568026043387188. [DOI] [PubMed] [Google Scholar]; e Guengerich FP. Chem. Res. Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L, Krishnan S, Zhang Y, Schenkman JB, Rusling JF. Chem. Res. Toxicol. 2009;22:341–347. doi: 10.1021/tx8004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a [December 9, 2015];Cytochrome P450 Enzyme Mapping in Drug Discovery using Corning Supersomes Enzymes. Application Note 467, http://csmedia2.corning.com/LifeSciences/media/pdf/an_DL_GT_048_Cytochrome_P450_Enzyme_Mapping_in_DD_using_Corning_Supersomes_Enzymes.pdf.; b [December 9, 2015];Reaction Phenotyping Methods using Recombinant Enzymes and HLM, BD Biosciences. http://www.bdbiosciences.com/documents/webinar_2012_07_HLM.pdf.
- 20.Roth A, Singer T. Adv. Drug Delivery Rev. 2014:69–70. 179–189. doi: 10.1016/j.addr.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Kramer JA, Sagartz JE, Morris DL. Nat. Rev. Drug Discovery. 2007;6:636–649. doi: 10.1038/nrd2378. [DOI] [PubMed] [Google Scholar]
- 22.Zeiger E. Environ. Mol. Mutagen. 2010;51:781–791. doi: 10.1002/em.20602. [DOI] [PubMed] [Google Scholar]
- 23.Ames BN, McCann J, Yamasaki E. Mutat. Res. 1975;31:347–363. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- 24.Oda Y, Nakamura S, Oki I, Kato T, Shinagawa H. Mutat. Res. 1985;147:219–229. doi: 10.1016/0165-1161(85)90062-7. [DOI] [PubMed] [Google Scholar]
- 25.Liebler DC, Guengerich FP. Nat. Rev. Drug Discovery. 2005;4:410–420. doi: 10.1038/nrd1720. [DOI] [PubMed] [Google Scholar]
- 26.Galloway SM, Aardema MJ, Ishidate M, Jr., Ivett JL, Kirkland DJ, Morita T, Mosesso P, Sofuni T. Mutat. Res. 1994;312:241–261. doi: 10.1016/0165-1161(94)00012-3. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki YF, Sekihashi K, Izumiyama F, Nishidate E, Saga A, Ishida K, Tsuda S. Crit. Rev. Toxicol. 2000;30:629–799. doi: 10.1080/10408440008951123. [DOI] [PubMed] [Google Scholar]
- 28.Singh NP, McCoy MT, Tice RR, Schneider EL. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 29.Flückiger-Isler S, Kamber M. In: Genotoxicity and DNA Repair: A Practical Approach; Methods in Pharmacology and Toxicology. Sierra LM, Gaivão I, editors. Springer; New York: 2014. pp. 23–41. [Google Scholar]
- 30.Kiskinis E, Suter W, Hartmann A. Mutagenesis. 2002;17:37–43. doi: 10.1093/mutage/17.1.37. [DOI] [PubMed] [Google Scholar]
- 31.Flückiger-Isler SM, Baumeister K, Braun V, Gervais N, Hasler-Nguyen R, Reimann J, Van Gompel H-G, Wunderlich G, Engelhardt G. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2004;558:181–197. doi: 10.1016/j.mrgentox.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Styles JA, Clark H, Festing MFW, Rew DA. Cytometry. 2001;44:153–155. doi: 10.1002/1097-0320(20010601)44:2<153::aid-cyto1095>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 33.Shuga J, Zhang J, Samson LD, Lodish HF, Griffith LG. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8737–8742. doi: 10.1073/pnas.0701829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith KEC, Heringa MB, Uytewaal M, Mayer P. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2013;750:12–18. doi: 10.1016/j.mrgentox.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Hastwell PW, Chai L-L, Roberts KJ, Webster TW, Harvey JS, Rees RW, Walmsley RM. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2006;607:160–175. doi: 10.1016/j.mrgentox.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Van Gompel J, Woestenborghs F, Beerens D, Mackie C, Cahill PA, Knight AW, Billinton N, Tweats DJ, Walmsley RM. Mutagenesis. 2005;20:449–454. doi: 10.1093/mutage/gei062. [DOI] [PubMed] [Google Scholar]
- 37.Luzy AP, Orsini N, Linget J-M, Bouvier GJ. Appl. Toxicol. 2013;33:1303–1315. doi: 10.1002/jat.2793. [DOI] [PubMed] [Google Scholar]
- 38.O'Brien PJ. Basic Clin. Pharmacol. Toxicol. 2014;115:4–17. doi: 10.1111/bcpt.12227. [DOI] [PubMed] [Google Scholar]
- 39.Judson R, Kavlock R, Martin M, Reif D, Houck K, et al. ALTEX. 2013;30:51–56. doi: 10.14573/altex.2013.1.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garside H, Marcoe KF, Chesnut-Speelman J, Foster AJ, Muthas D, Gerry Kenna J, Warrior U, Bowes J, Baumgartner J. Toxicol. In Vitro. 2014;28:171–181. doi: 10.1016/j.tiv.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Persson M, Løye AF, Mow T, Hornberg JJ. J. Pharmacol. Toxicol. Methods. 2013;68:302–313. doi: 10.1016/j.vascn.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Naoghare PK, Kim MJ, Song JM. Anal. Chem. 2008;80:5407–5417. doi: 10.1021/ac800452x. [DOI] [PubMed] [Google Scholar]
- 43.Kim MJ, Lee SC, Pal S, Han E, Song JM. Lab Chip. 2011;11:104–114. doi: 10.1039/c0lc00110d. [DOI] [PubMed] [Google Scholar]
- 44.Shuhendler AJ, Pu K, Cui L, Uetrecht JP, Rao J. Nat. Biotechnol. 2014;32:373–382. doi: 10.1038/nbt.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang X, Zhang P, Qiao L, Feng X, Zhang X, Girault XX, Liu B. Anal. Chem. 2014;86:10870–10876. doi: 10.1021/ac503024h. [DOI] [PubMed] [Google Scholar]
- 46.Wang N, Gao C, Xue F, Han Y, Li T, Cao X, Zhang X, Zhang Y, Wang ZL. ACS Nano. 2015;9:3159–3168. doi: 10.1021/acsnano.5b00142. [DOI] [PubMed] [Google Scholar]
- 47.Tran TB, Cho S, Min J. Biosens. Bioelectron. 2013;50:453–459. doi: 10.1016/j.bios.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 48.Bogomolova A, Komarova E, Reber K, Gerasimov T, Yavuz O, Bhatt S, Aldissi M. Anal. Chem. 2009;81:3944–3949. doi: 10.1021/ac9002358. [DOI] [PubMed] [Google Scholar]
- 49.Astashkina A, Grainger DW. Adv. Drug Delivery Rev. 2014:69–70. 1–18. doi: 10.1016/j.addr.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Esch EW, Bahinski A, Huh D. Nat. Rev. Drug Discovery. 2015;14:248–260. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brophy CM, Luebke-Wheeler JL, Amiot BP, Khan H, Remmel RP, Rinaldo P, Nyberg SL. Hepatology. 2009;49:578–586. doi: 10.1002/hep.22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radtke AL, Herbst-Kralovetz MM. J. Visualized Exp. 2012;62:e3868. doi: 10.3791/3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hjelm BE, Berta AN, Nickerson CA, Arntzen CJ, Herbst-Kralovetz MM. Biol. Reprod. 2010;82:617–627. doi: 10.1095/biolreprod.109.080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takayama K, Kawabata K, Nagamoto Y, Kishimoto K, Tashiro K, Sakurai F, Tachibana M, Kanda K, Hayakawa T, Furue MK, Mizuguchi H. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 55.Nakamura K, Mizutani R, Sanbe A, Enosawa S, Kasahara M, Nakagawa A, Ejiri Y, Murayama N, Miyamoto Y, Torii T, Kusakawa S, Yamauchi J, Fukuda M, Yamazaki H, Tanoue A. J. Biosci. Bioeng. 2011;111:78–84. doi: 10.1016/j.jbiosc.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 56.Polini A, Prodanov L, Bhise NS, Manoharan V, Dokmeci MR, Khademhosseini A. Expert Opin. Drug Discovery. 2014;9:335–352. doi: 10.1517/17460441.2014.886562. [DOI] [PubMed] [Google Scholar]
- 57.Choucha-Snouber L, Aninat C, Grsicom L, Madalinski G, Brochot C, Poleni PE, Razan F, Guillouzo CG, Legallais C, Corlu A, Leclerc E. Biotechnol. Bioeng. 2013;110:597–608. doi: 10.1002/bit.24707. [DOI] [PubMed] [Google Scholar]
- 58.Mao S, Gao D, Liu W, Wei H, Lin J-M. Lab Chip. 2012;12:219–226. doi: 10.1039/c1lc20678h. [DOI] [PubMed] [Google Scholar]
- 59.Schmelzer E, Mutig K, Schrade P, Bachmann S, Gerlach JC, Zeilinger K. Biotechnol. Bioeng. 2009;103:817–827. doi: 10.1002/bit.22283. [DOI] [PubMed] [Google Scholar]
- 60.Zeilinger K, Schreiter T, Darnell M, Soderdahl T, Lubberstedt M, Dillner B, Knobeloch D, Nussler AK, Gerlach JC, Andersson TB. Tissue Eng., Part C. 2011;17:549–556. doi: 10.1089/ten.TEC.2010.0580. [DOI] [PubMed] [Google Scholar]
- 61.Bhatia SN, Ingber DE. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 62.Ma L, Barker J, Zhou C, Li W, Zhang J, Lin B, Foltz G, Küblbeck J, Honkakoski P. Biomaterials. 2012;33:4353–4361. doi: 10.1016/j.biomaterials.2012.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choucha-Snouber L, Aninat C, Grsicom L, Madalinski G, Brochot C, Poleni PE, Razan F, Guillouzo CG, Legallais C, Corlu A, Leclerc E. Biotechnol. Bioeng. 2013;110:597–608. doi: 10.1002/bit.24707. [DOI] [PubMed] [Google Scholar]
- 64.Meli L, Barbosa HSC, Hickey AM, Gasimli L, Nierode G, Diogo MM, Linhardt RJ, Cabral JMS, Dordick JS. Stem Cell Res. 2014;13:36–47. doi: 10.1016/j.scr.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, McAlexander MA, Ingber DE. Sci. Transl. Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCain ML, Sheehy SP, Grosberg A, Goss JA, Parker KK. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9770–9775. doi: 10.1073/pnas.1304913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rusling JF. Biosens. Bioelectron. 2004;20:1022–1228. doi: 10.1016/j.bios.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 68.Rusling JF. In: Electrochemistry of Nucleic Acids and Proteins. Palecek E, Scheller F, Wang J, editors. Elsevier; Amsterdam, The Netherlands: 2005. pp. 433–450. [Google Scholar]
- 69.Zhao L, Schenkman JB, Rusling JF. Anal. Chem. 2010;82:10172–10178. doi: 10.1021/ac102317a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao L, Bajrami B, Rusling JF. In: Cytochrome P450 Protocols. 3rd ed. Phillips IR, Shephard EA, Ortiz de Montellano PR, editors. Humana Press, Springer; New York: 2013. pp. 129–134. [Google Scholar]
- 71.Rusling JF, Hvastkovs EG, Hull DO, Schenkman JB. Chem. Commun. 2008:141–154. doi: 10.1039/b709121b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hvastkovs EG, So M, Krishnan S, Bajrami B, Tarun T, et al. Anal. Chem. 2007;79:1897–906. doi: 10.1021/ac061975q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rusling JF, Wasalathanthri D, Zhao L, Schenkman JB. Soft Matter. 2014;10:8145–8156. doi: 10.1039/c4sm01679c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krishnan S, Wasalathanthri D, Zhao L, Schenkman JB, Rusling JF. J. Am. Chem. Soc. 2011;133:1459–1465. doi: 10.1021/ja108637s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lvov Y. In: Protein Architecture: Interfacing Molecular Assemblies and Immobilization Biotechnology. Lvov Y, Möhwald H, editors. Marcel Dekker; New York: 2000. pp. 125–167. [Google Scholar]
- 76.Lvov Y. Nanostructured Materials, Micelles and Colloids. In: Nalwa RW, editor. Handbook of Surfaces and Interfaces of Materials. Vol. 3. Academic Press; San Diego, CA: 2001. pp. 170–189. [Google Scholar]
- 77.Zhang X, Chen H, Zhang H. Chem. Commun. 2007:1395–1405. doi: 10.1039/b615590a. [DOI] [PubMed] [Google Scholar]
- 78.Ariga K, Hill JP, Ji Q. Phys. Chem. Chem. Phys. 2007;9:2319–2340. doi: 10.1039/b700410a. [DOI] [PubMed] [Google Scholar]
- 79.Wasalathanthri DP, Malla S, Bist I, Tang CK, Faria RC, Rusling JF. Lab Chip. 2013;13:4554–4562. doi: 10.1039/c3lc50698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wasalathanthri DP, Mani V, Tang CK, Rusling JF. Anal. Chem. 2011;83:9499–9506. doi: 10.1021/ac202269t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wasalathanthri DP, Faria RC, Malla S, Joshi AA, Schenkman JB, Rusling JF. Analyst. 2013;138:171–178. doi: 10.1039/c2an35993f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song B, Pan S, Tang C, Li D, Rusling JF. Anal. Chem. 2013;85:11061–11067. doi: 10.1021/ac402736q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Forster RJ, Bertoncello P, Keyes TE. Annu. Rev. Anal. Chem. 2009;2:359–85. doi: 10.1146/annurev-anchem-060908-155305. [DOI] [PubMed] [Google Scholar]
- 84.Miao W, Choi JP, Bard AJ. J. Am. Chem. Soc. 2002;124:14478–14485. doi: 10.1021/ja027532v. [DOI] [PubMed] [Google Scholar]
- 85.Dennany L, Forster RJ, Rusling JF. J. Am. Chem. Soc. 2003;125:5213–5218. doi: 10.1021/ja0296529. [DOI] [PubMed] [Google Scholar]
- 86.Benigni R, Bossa C. Chem. Rev. 2011;111:2507–2536. doi: 10.1021/cr100222q. [DOI] [PubMed] [Google Scholar]
- 87.Wasalathanthri DP, Li D, Song D, Zheng Z, Choudhary D, Jansson I, Lu X, Schenkman JB, Rusling JF. Chem. Science. 2015;6:2457–2468. doi: 10.1039/c4sc03401e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haber D, Harlow E. Nat. Genet. 1997;16:320–322. doi: 10.1038/ng0897-320. [DOI] [PubMed] [Google Scholar]
- 89.May P, May E. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 90.Hjortsberg L, Rubio-Nevado JM, Hamroun D, Beroud C, Claustre M, Soussi T. [April 16, 2015];The p53 Mutation handbook 2.0. http://p53.free.fr/Database/p53_cancer_db.html.
- 91.Malla S, Kadimisetty K, Fu Y-J, Choudhary D, Jansson I, Lu X, Schenkman JB, Rusling JF. Chem. Science. 2015;6:5554–5563. doi: 10.1039/c5sc01403d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Satterwhite JE, Pugh AM, Danell AS, Hvastkovs EG. Anal. Chem. 2011;83:3327–3335. doi: 10.1021/ac103091v. [DOI] [PubMed] [Google Scholar]
- 93.Satterwhite JE, Trumbo CM, Danell AS, Hvastkovs EG. Anal. Chem. 2013;85:1183–1191. doi: 10.1021/ac303077h. [DOI] [PubMed] [Google Scholar]
- 94.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 95.Broyde S, Wang L, Zhang L, Rechkoblit O, Geacintov NE, Patel DJ. Chem. Res. Toxicol. 2008;21:45–52. doi: 10.1021/tx700193x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pradhan P, Graeslund A, Seidel A, Jernstroem B. Chem. Res. Toxicol. 1999;12:816–821. doi: 10.1021/tx980230f. [DOI] [PubMed] [Google Scholar]
- 97.Pan S, Li D, Zhao L, Schenkman JB, Rusling JF. Chem. Res. Toxicol. 2013;26:1229–1239. doi: 10.1021/tx400147c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bajrami B, Zhao L, Schenkman JB, Rusling JF. Anal. Chem. 2009;81:9921–9929. doi: 10.1021/ac9015853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bajrami B, Krishnan S, Rusling JF. Drug Metab. Lett. 2008;2:158–162. doi: 10.2174/187231208785425854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sittampalam S, Eglen R, Ferguson S, Maynes JT, Olden K, Schrader L, Shelper T, Ferrer M. Assay Drug Dev. Technol. 2015;13:254–261. doi: 10.1089/adt.2015.29001.rtd. [DOI] [PMC free article] [PubMed] [Google Scholar]