Abstract
It is well known that most schizophrenia patients smoke cigarettes. There are different hypotheses postulating the underlying mechanisms of this comorbidity. We used summary statistics from large meta-analyses of plasma cotinine concentration (COT), Fagerström test for nicotine dependence (FTND) and schizophrenia to examine the genetic relationship between these traits. We found that schizophrenia risk scores calculated at P-value thresholds of 5 × 10−3 and larger predicted FTND and cigarettes smoked per day (CPD), suggesting that genes most significantly associated with schizophrenia were not associated with FTND/CPD, consistent with the self-medication hypothesis. The COT risk scores predicted schizophrenia diagnosis at P-values of 5 × 10−3 and smaller, implying that genes most significantly associated with COT were associated with schizophrenia. These results implicated that schizophrenia and FTND/CPD/COT shared some genetic liability. Based on this shared liability, we identified multiple long non-coding RNAs and RNA binding protein genes (DA376252, BX089737, LOC101927273, LINC01029, LOC101928622, HY157071, DA902558, RBFOX1 and TINCR), protein modification genes (MANBA, UBE2D3, and RANGAP1) and energy production genes (XYLB, MTRF1 and ENOX1) that were associated with both conditions. Further analyses revealed that these shared genes were enriched in calcium signaling, long-term potentiation and neuroactive ligand-receptor interaction pathways that played a critical role in cognitive functions and neuronal plasticity.
Large scale genome wide association studies (GWASs) have identified risk genes for many complex human diseases and traits (http://www.genome.gov/gwastudies/), including psychiatric disorders such as schizophrenia and nicotine dependence (ND)1,2,3. These GWASs also show that many human diseases and traits are polygenic in nature and the contribution of individual genes is limited4,5,6. Many of these studies have been deposited in the database of Genotypes and Phenotypes (dbGaP, http://www.ncbi.nlm.nih.gov/gap) and are available for secondary analyses. These datasets provide an opportunity to examine the genetic relationship between correlated traits, and to discover and identify risk genes shared across these traits.
Pleiotropy is a phenomenon in which a single locus affects multiple traits7,8. It accounts for at least a part of the genetic mechanism of many correlated human behaviors and diseases. Pleiotropy can take two forms: either a single process, leading to a cascade of downstream effects (sometimes described as “mediated pleiotropy”), or a single locus influencing multiple traits (sometimes described as “biological pleiotropy”)9. Schizophrenia is highly comorbid with cigarette smoking10. However, the underlying biology of this comorbidity is not well understood11. Several hypotheses have been proposed. The self-medication hypothesis postulates that schizophrenia patients smoke to reduce symptoms and antipsychotics-induced side effects and to improve their attention and working memory12. Alternatively, schizophrenia and ND could have shared some genetic liability (i.e., biological pleiotropy)13, which is supported by recent studies of individual genes14,15,16,17,18,19,20,21. A third possibility is that smoking may be causal to schizophrenia (i.e., mediated pleiotropy)22. To explore the genetic relationship between schizophrenia and ND, we obtained the GWAS summary statistics from the Psychiatric Genomics Consortium (PGC) schizophrenia analyses and ND related traits from our unpublished studies, and conducted polygenic analyses. Under the hypothesis of biological pleiotropy, we expect that genetic risk scores of schizophrenia and ND related traits predict each other; whereas self-medication would anticipate unidirectional (schizophrenia to ND traits) prediction. In this article, we report the findings from these analyses.
Results
Nicotine dependence and schizophrenia share genetic liability
In these analyses, we calculated genetic risk scores for schizophrenia (supplementary Figure 1A) and tested whether the risk scores predicted FTND and CPD. The results were summarized in Table 1. Schizophrenia risk scores predicted FTND score and CPD at the thresholds of P ≤ 5 × 10−3, 5 × 10−2 and 5 × 10−1. The correlation coefficients at these thresholds were all positive, suggesting that a schizophrenia diagnosis was positively associated with cigarette smoking, consistent with the well-known comorbidity between schizophrenia and ND. However, schizophrenia risk scores explained only a very small fraction of the FTND and CPD traits.
Table 1. Schizophrenia risk score prediction of FTND and CPD.
| Threshold (P) | Beta | S.E. | T_value | Pr(>|T|) | R2 |
|---|---|---|---|---|---|
| FTND | |||||
| 5 × 10−5 | 2.41E-04 | 0.000269 | 0.90 | 0.3708 | 0 |
| 5 × 10−4 | 3.30E-04 | 0.000235 | 1.41 | 0.1599 | 1.00E-04 |
| 5 × 10−3 | 3.91E-04 | 0.000194 | 2.01 | 0.0440 | 3.00E-04 |
| 5 × 10−2 | 3.53E-04 | 0.000153 | 2.31 | 0.0211 | 4.00E-04 |
| 5 × 10−1 | 3.22E-04 | 0.000118 | 2.73 | 0.0063 | 7.00E-04 |
| CPD | |||||
| 5 × 10−5 | 1.03E-04 | 8.77E-05 | 1.17 | 0.2424 | 0 |
| 5 × 10−4 | 1.37E-04 | 7.64E-05 | 1.79 | 0.0736 | 1.00E-04 |
| 5 × 10−3 | 1.71E-04 | 6.32E-05 | 2.71 | 0.0067 | 5.00E-04 |
| 5 × 10−2 | 1.56E-04 | 4.99E-05 | 3.13 | 0.0018 | 6.00E-04 |
| 5 × 10−1 | 1.29E-04 | 3.84E-05 | 3.37 | 0.0008 | 8.00E-04 |
FTND and COT risk scores (supplementary Figure 1B,C) were calculated for the subjects of the phase I PGC schizophrenia GWAS samples23 using the summary statistics from the FTND (n = 17,781) and COT (n = 4,548) GWAS meta-analyses. We then evaluated whether the genetic risk scores of COT and FTND could predict the schizophrenia diagnosis using logistic regression. The results were presented in Table 2. The COT risk scores calculated at the P-values of 5 × 10−5, 5 × 10−4 and 5 × 10−3 predicted schizophrenia diagnosis, but FTND risk scores failed to do so. For the P-values thresholds at which the COT risk scores predicted schizophrenia diagnosis, the beta coefficients were also positive, again, confirming the positive phenotypic correlation between ND and schizophrenia.
Table 2. Genetic risk score to nicotine dependence prediction of schizophrenia diagnosis*.
| Threshold (P) | Beta | S.E. | Wald Z | Pr(>|Z|) | R2 |
|---|---|---|---|---|---|
| Cotinine | |||||
| 5 × 10−5 | 0.0459 | 0.0169 | 2.72 | 0.0066 | 0.0006 |
| 5 × 10−4 | 0.0583 | 0.0169 | 3.45 | 0.0006 | 0.0010 |
| 5 × 10−3 | 0.0435 | 0.0169 | 2.58 | 0.0100 | 0.0005 |
| 5 × 10−2 | 0.0035 | 0.0170 | 0.20 | 0.8384 | 3.4E-06 |
| 5 × 10−1 | −0.0226 | 0.0172 | −1.32 | 0.1886 | 0.0001 |
| FTND | |||||
| 5 × 10−5 | 0.0220 | 0.0242 | 0.91 | 0.3641 | 0.0001 |
| 5 × 10−4 | 0.0227 | 0.0507 | 0.45 | 0.6537 | 2.5E-05 |
| 5 × 10−3 | 0.0331 | 0.0617 | 0.54 | 0.5913 | 3.5E-05 |
| 5 × 10−2 | 0.0033 | 0.0323 | 0.10 | 0.9174 | 1.3E-06 |
| 5 × 10−1 | 0.0281 | 0.0292 | 0.96 | 0.3362 | 0.0001 |
*R2 is the Nagelkerke’s R2 obtained by the R package fmsb.
Identification of shared variants between ND and schizophrenia
Our reciprocal polygenic analyses suggested that there were some shared genetic liability between schizophrenia and ND as defined by the FTND and COT traits. We then proceeded to identify the variants associated with both schizophrenia and ND traits. We computed joint P-values for each marker using the summary statistics from the schizophrenia and COT/FTND/TFC meta-analyses, and assigned a q-value to each of the joint P-values using an FDR method24,25. Table 3 listed the loci identified by the joint analyses with q-values ≤ 0.05. From the joint analyses between schizophrenia and COT, 11 loci reached genome-wide significance for association with both COT and schizophrenia, of which 2 loci had no known genes nearby and 6 were spliced ESTs or long non-coding RNAs. In the analyses between schizophrenia and FTND, 10 loci were identified, and 3 of them were ESTs or non-coding RNAs. The joint analyses between schizophrenia and TFC yielded 15 significant loci. The CHRNA5-CHRNA3-CHRNB4 locus was the only one identified by all three smoking traits. In addition to some genes known to be associated with schizophrenia (HLA-B and MAD1L1), we also identified novel non-coding RNAs and RNA binding protein genes (DA376252, BX089737, LOC101927273, LINC01029, LOC101928622, HY157071, DA902558, RBFOX1 and TINCR), post-translation modification genes (MANBA, UBE2D3 and RANGAP1) and energy production genes (XYLB, MTRF1 and ENOX1).
Table 3. Joint testing of association with schizophrenia and smoking traits.
| SNP | Chr | Location (bp) | Z (ND) | Z (SCZ) | P-value | Q-value | Gene symbol | # markers |
|---|---|---|---|---|---|---|---|---|
| COT | ||||||||
| rs798015 | 1 | 117,320,907 | −3.71 | −3.69 | 5.08E-08 | 0.0020 | DA376252 | 3 |
| rs9850756 | 3 | 18,905,739 | −3.71 | 3.62 | 8.72E-08 | 0.0030 | DA733783 | 1 |
| rs709071 | 3 | 191,426,311 | −3.46 | 3.51 | 2.85E-07 | 0.0081 | – | 1 |
| rs12640124 | 4 | 118,775,172 | 3.61 | −3.50 | 2.12E-07 | 0.0063 | BX089737 | 3 |
| rs2442720 | 6 | 31,320,277 | −3.49 | 4.62 | 2.26E-07 | 0.0067 | HLA-B | 1 |
| rs11779524 | 8 | 8,618,613 | 3.85 | 3.81 | 1.89E-08 | 0.0008 | CF594265 | 4 |
| rs56235824 | 9 | 11,039,320 | 3.44 | 3.55 | 3.38E-07 | 0.0090 | – | 1 |
| rs11788261 | 9 | 138,611,139 | 3.80 | 3.51 | 1.95E-07 | 0.0060 | KCNT1 | 2 |
| rs8042374 | 15 | 78,908,032 | 7.69 | 7.04 | 3.48E-24 | 1.68E-17 | CHRNA5/CHRNA3/CHRNB4 | 270 |
| rs6650723 | 18 | 53,524,269 | 3.76 | 3.51 | 2.07E-07 | 0.0062 | DA696352/LOC101927273 | 5 |
| rs7228837 | 18 | 75,817,606 | 4.23 | 3.65 | 6.64E-08 | 0.0024 | LINC01029 | 19 |
| FTND | ||||||||
| rs56335113 | 1 | 30,427,639 | 3.80 | 5.85 | 2.02E-08 | 0.0021 | – | 10 |
| rs36025078 | 2 | 155,883,716 | −3.90 | −3.59 | 1.13E-07 | 0.0061 | – | 1 |
| rs188499496 | 3 | 38,450,183 | −3.93 | −3.62 | 8.82E-08 | 0.0050 | XYLB | 2 |
| rs11917643 | 3 | 64,171,754 | 3.62 | −4.17 | 8.41E-08 | 0.0048 | PRICKLE2 | 6 |
| rs76923559 | 4 | 34,053,926 | −3.93 | −3.79 | 2.35E-08 | 0.0023 | LOC101928622 | 3 |
| rs147093127 | 5 | 152,086,293 | 3.67 | 4.01 | 5.70E-08 | 0.0037 | LINC01470 | 1 |
| rs9322751 | 6 | 104,015,759 | −3.51 | −3.54 | 2.03E-07 | 0.0089 | HY157071 | 6 |
| rs4994764 | 7 | 1,928,662 | −3.90 | −4.32 | 9.17E-09 | 0.0012 | MAD1L1 | 18 |
| rs3910267 | 11 | 130,810,282 | −3.84 | 5.71 | 1.50E-08 | 0.0018 | SNX19 | 21 |
| rs147144681 | 15 | 78,900,908 | −7.62 | −5.55 | 7.85E-16 | 5.66E-09 | CHRNA5/CHRNA3/CHRNB4 | 106 |
| TFC | ||||||||
| rs4658015 | 1 | 196,053,435 | 3.51 | 3.36 | 5.91E-07 | 0.0094 | – | 1 |
| rs1069267 | 3 | 38,435,023 | −3.63 | −4.43 | 8.02E-08 | 0.0015 | XYLB | 3 |
| rs11722779 | 4 | 103,827,488 | 4.22 | −4.30 | 6.00E-10 | 0.0001 | MANBA/UBE2D3/SLC9B2 | 315 |
| rs2717737 | 8 | 18,459,343 | −3.36 | 3.73 | 6.05E-07 | 0.0096 | PSD3 | 1 |
| rs13264022 | 8 | 21,293,046 | 3.50 | 4.01 | 2.16E-07 | 0.0038 | DA902558 | 15 |
| rs7959287 | 12 | 103,601,638 | 3.84 | 4.49 | 1.54E-08 | 0.0003 | – | 5 |
| rs9594516 | 13 | 41,849,360 | 3.64 | 3.77 | 7.33E-08 | 0.0014 | MTRF1 | 1 |
| rs112531467 | 13 | 43,865,047 | −3.66 | 3.52 | 1.86E-07 | 0.0033 | ENOX1 | 1 |
| rs181676509 | 14 | 104,242,531 | −3.67 | −3.98 | 5.73E-08 | 0.0011 | C14ORF2 | 15 |
| rs147144681 | 15 | 78,900,908 | −5.50 | −5.55 | 1.49E-15 | 1.08E-08 | CHRNA5/CHRNA3/CHRNB4 | 72 |
| rs17665477 | 16 | 6,700,889 | −3.69 | 3.80 | 5.16E-08 | 0.0010 | RBFOX1 | 11 |
| rs12462853 | 19 | 5,538,936 | 3.45 | −3.39 | 4.82E-07 | 0.0078 | TINCR | 2 |
| rs62202174 | 20 | 20,461,125 | −3.38 | −4.24 | 5.26E-07 | 0.0085 | RALGAPA2 | 1 |
| rs5758274 | 22 | 41,664,539 | 3.64 | −4.45 | 7.43E-08 | 0.0014 | RANGAP1 | 1 |
| rs8135804 | 22 | 42,334,660 | 3.40 | 3.57 | 4.54E-07 | 0.0074 | CENPM | 12 |
Pathway enrichment and network interaction analyses
We further explored the pathways shared by schizophrenia and ND by selecting all markers with q-values less than 0.16 from the joint analyses between schizophrenia and smoking traits. After mapping the markers to genes, the genes showing potential association with schizophrenia and COT/FTND/TFC were pooled to search for pathways enriched in both conditions. In these analyses, we selected only the genes identified by at least 2 of the 3 smoking traits, yielding a total of 146 genes. After filtering out the human leukocyte antigen genes (HLA-B, HLA-C, HLA-DOA, HLA-DQA1, HLA-DQB1, HLA-DRB1, and HLA-G) due to their strong linkage disequilibrium26, we used the remaining 139 genes in pathway analyses.
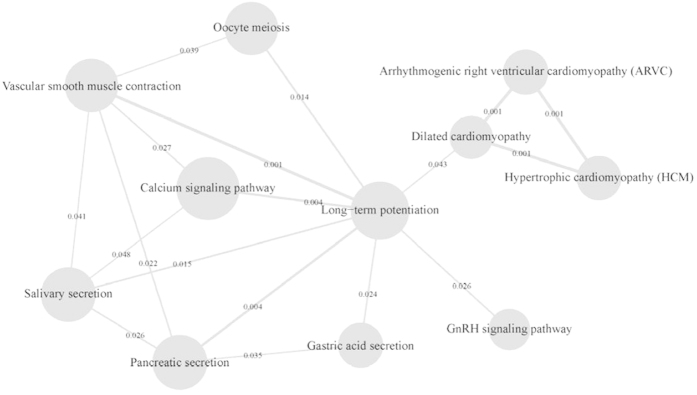
Our analyses identified 16 unique pathways that were shared between schizophrenia and ND (Table 4). The most noticeable pathways were Calcium Signaling, Long-Term Potentiation, Neuroactive Ligand-Receptor Interaction, Phosphatidylinositol Signaling, Cell Adhesion Molecules, and Regulation of Actin Cytoskeleton pathways. Some of these pathways (Calcium Signaling, Long-Term Potentiation, Cell Adhesion Molecules, and Regulation of Actin Cytoskeleton) had been reported to be involved in schizophrenia27,28,29,30,31,32, others (Cell Adhesion Molecules and Neuroactive Ligand-Receptor Interaction) had been implicated in ND33,34. We found that these pathways were enriched in the genes associated with both ND and schizophrenia. Additionally, pathways involved in cardiomyopathy, GnRH signaling, gastric acid secretion and Alzheimer’s disease were also found to be shared between schizophrenia and ND. In the pathway network interaction analyses, we found a network of crosstalk between pathways (Fig. 1), with the Long-Term Potentiation located at the center of these interactions.
Table 4. Pathways enriched in schizophrenia and smoking traits.
| Pathway | Genes found in pathway | # gene in pathway | # gene observed | # gene expected | Observed/expected ratio | Raw P-value | Adjusted P-value |
|---|---|---|---|---|---|---|---|
| Calcium signaling pathway | ATP2B2, CACNA1C, CACNA1I, CHRM3, ITPR1, ITPR2 | 177 | 6 | 0.57 | 10.59 | 2.41E-05 | 0.0004 |
| Long-term potentiation | CACNA1C, ITPR1, ITPR2, PPP1CB | 70 | 4 | 0.22 | 17.86 | 7.81E-05 | 0.0007 |
| Neuroactive ligand-receptor interaction | CHRM3, CHRNA3, CHRNA5, CHRNB4, NR3C1, THRB | 272 | 6 | 0.87 | 6.89 | 0.0003 | 0.0011 |
| Salivary secretion | ATP2B2, CHRM3, ITPR1, ITPR2 | 89 | 4 | 0.28 | 14.05 | 0.0002 | 0.0011 |
| Pancreatic secretion | ATP2B2, CHRM3, ITPR1, ITPR2 | 101 | 4 | 0.32 | 12.38 | 0.0003 | 0.0011 |
| Vascular smooth muscle contraction | CACNA1C, ITPR1, ITPR2, PPP1CB | 116 | 4 | 0.37 | 10.78 | 0.0005 | 0.0013 |
| Oocyte meiosis | ITPR1, ITPR2, PPP1CB, SPDYA | 112 | 4 | 0.36 | 11.16 | 0.0005 | 0.0013 |
| Phosphatidylinositol signaling system | INPP5K, ITPR1, ITPR2 | 78 | 3 | 0.25 | 12.02 | 0.0020 | 0.0036 |
| Gastric acid secretion | CHRM3, ITPR1, ITPR2 | 74 | 3 | 0.24 | 12.67 | 0.0018 | 0.0036 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | CACNA1C, ITGAV, SGCD | 74 | 3 | 0.24 | 12.67 | 0.0018 | 0.0036 |
| Hypertrophic cardiomyopathy (HCM) | CACNA1C, ITGAV, SGCD | 83 | 3 | 0.27 | 11.30 | 0.0024 | 0.0039 |
| Dilated cardiomyopathy | CACNA1C, ITGAV, SGCD | 90 | 3 | 0.29 | 10.42 | 0.0031 | 0.0046 |
| GnRH signaling pathway | CACNA1C, ITPR1, ITPR2 | 101 | 3 | 0.32 | 9.28 | 0.0042 | 0.0058 |
| Cell adhesion molecules (CAMs) | ITGAV, NCAM2, PTPRM | 133 | 3 | 0.43 | 7.05 | 0.0091 | 0.0117 |
| Alzheimer’s disease | CACNA1C, ITPR1, ITPR2 | 167 | 3 | 0.53 | 5.61 | 0.0167 | 0.0188 |
| Regulation of actin cytoskeleton | CHRM3, ITGAV, PPP1CB | 213 | 3 | 0.68 | 4.40 | 0.0313 | 0.0331 |
Figure 1. Pathway crosstalk network.
The size of the node is proportional to the P-values of pathway enrichment test. The thickness of the edge is proportional to the P-values of pathway crosstalk.
Discussion
It is well known in psychiatric clinics that a large proportion of schizophrenia patients smoke cigarettes and smoke heavily13. The dominant hypothesis to explain the comorbidity is self-medication12, i.e., that schizophrenia patients smoke to ameliorate impairments in cognitive function and suppress psychotic symptoms. Another hypothesis contends that schizophrenia and ND share some genetic liability, and the high prevalence rate of cigarette smoking in schizophrenia patients is a manifestation that is partially due to the common liability13. A third possibility is that smoking may be a risk factor for the development of schizophrenia, given that smoking initiation typically predates the onset of schizophrenia22. These three hypotheses are not mutually exclusive, and all three may contribute to the observed co-occurrence of schizophrenia and smoking.
Previous studies examining this issue have largely focused on individual functions/symptoms or genes using relatively small sample sizes. Here we took a systematic approach, and examined the entire genome using large GWAS datasets and multiple traits. We observed different patterns between the reciprocal polygenic analyses (comparing Tables 1 and 2). When we used the genetic risk scores of schizophrenia to predict ND traits, the association was evident at P-values ≥ 5 × 10−3, with the association strength increased as the P-value threshold became larger (Table 1). Given that the PGC schizophrenia GWAS did not control for smoking status and quantity, and there was a large difference of smoking prevalence between schizophrenia patients and controls (on average, 65% or more schizophrenia patients smoke, and about 20% people smoke in the general population), we would expect that the PGC schizophrenia GWAS identify top candidates for ND related traits. But what we found was not the case. These top ranked candidates (i.e. those with P-values ≤ 5 × 10−5) from the PGC schizophrenia meta-analysis1 were not predict ND related traits. A likely explanation for these results is that genes most strongly associated with schizophrenia do not directly contribute to the smoking behaviors in schizophrenia patients. In other words, the reason why schizophrenia patients smoke is that they want to improve their cognitive functions and to suppress psychotic symptoms, not because that they are addicted to nicotine as regular smokers in the general population do. These results are consistent with the self-medication hypothesis.
In contrast, when we used COT risk scores to predict schizophrenia diagnosis, we found that smaller P-values produced stronger signals (Table 2), indicating that genes most strongly associated with ND were associated with schizophrenia. The results imply that either ND and schizophrenia share some genetic liability, or ND is a risk factor of schizophrenia. These fit the predictions of the shared liability hypothesis and that smoking is a causal risk for schizophrenia. Of note, these two explanations are not mutually exclusive. But without data on smoking of the patients we are unable to test the latter possibility (e.g., by stratifying our sample on smoking status).
Assuming biological pleiotropy to be the underlying mechanism, we devised a test to discover the variants shared between ND and schizophrenia. Using this approach, we identified multiple genes associated with both conditions (Table 3). Of these genes, the CHRNA5-CHRNA3-CHRNB4 cluster had been found to be associated with CPD2,3 and other ND related traits, and it was reported to be associated with schizophrenia in the latest schizophrenia GWAS meta-analysis from PGC1. Several of the genes had been reported to be associated with schizophrenia (HLA-B and MAD1L1)28 and epilepsy (KCNT1, PRICKLE2 and RBFOX1)35,36,37, suggesting that they might play a role in smoking behaviors as well. Our analyses also identified some novel genes shared between schizophrenia and ND, including a group of long non-coding RNAs and RNA binding protein genes (DA376252, BX089737, LOC101927273, LINC01029, LOC101928622, HY157071, DA902558, RBFOX1 and TINCR), a group of post-translation modification genes (MANBA, UBE2D3, and RANGAP1) and a group of energy production genes (XYLB, MTRF1 and ENOX1). Long non-coding RNAs were suggested to play a role in schizophrenia38,39, the identification of multiple long non-coding RNAs was intriguing.
Phenotype comorbidity is common in complex diseases and traits7,8. Pleiotropy, or shared genetic liability, may be an underlying mechanism of these comorbidities. Under this condition, different approaches have been developed to identify genes shared by the comorbid conditions40,41, and these approaches seem more powerful than standard GWAS8,42. Another advantage of these methods is that they can use the large number of GWAS datasets produced by a single phenotype/trait analyses. The approach we used to identify these shared loci is conservative. In our analyses, we excluded all markers reaching genome-wide significance from both schizophrenia and smoking traits and required a balanced contribution from both traits. Under this condition, if a marker reached genome wide significance for schizophrenia but had a modest association with ND traits (say P-values between 10−4 to 5 × 10−6), it was excluded from our joint testing. Similarly, some markers would be excluded if they reached genome wide significance in ND traits. Because the GWASs used have different sample sizes, and therefore varied in their statistical power, it is inevitable that we would miss some markers from the more powerful GWAS when we required balanced summary statistics in the joint testing.
Our pathway analyses identified multiple pathways shared by schizophrenia and ND. The most significant pathways were Calcium Signaling, Long-Term Potentiation and Neuroactive Ligand-Receptor Interaction. These pathways are involved in neurotransmitter transduction and communication between neurons, and they are essential for cognitive functions. These pathways have been shown to be involved in schizophrenia28,43,44 and ND45,46. The Cell Adhesion Molecules and Regulation of Actin Cytoskeleton pathways have also been reported in schizophrenia31,47,48,49 and ND45,50,51. Thus, our results are consistent with these studies. It is worth noting that the cardiomyopathy pathways were identified in our analyses and that, in a previous study, we found that CMYA5 was associated with schizophrenia52. Another gene, NDUFV2, causative to hypertrophic cardiomyopathy53,54, the genetic form of cardiomyopathy, was also found to be associated with schizophrenia55,56,57. Pathway crosstalk analyses showed that many of these pathways interact with each other and together they form an interlinked network with the Long-Term Potentiation pathway at the center of these interactions. In animal studies, nicotine alters long-term potentiation58,59,60 and learning and memory61. In humans, smoking may alleviate cognitive impairment62, and both nicotine withdrawal and schizophrenia are associated with cognitive impairments63,64. Thus, compensating cognitive impairments may be a common motivational factor between regular smokers and schizophrenia patients.
In summary, our results supported the self-medication hypothesis. We also found evidence that schizophrenia and ND share some genetic liability and these results did not contradict the hypothesis that smoking was a causal risk factor for schizophrenia. Assuming shared liability and a balanced contribution, we identified novel candidate genes associated with both schizophrenia and ND. Analyses of the shared genes revealed multiple pathways and an interacting network centered on long-term potentiation. These results provided some new insights for our understanding of smoking behaviors in both schizophrenia patients and the general population.
Methods
Phenotypes and GWAS datasets
For schizophrenia, we obtained the summary statistics from the PGC GWAS of schizophrenia1. This study used 52 independent samples, of them 46 were case control samples of European ancestry, 3 were Asian case control samples and 3 were European family samples. Since the samples were collected from different countries, both the criteria for Diagnostic and Statistical Manual of Mental Disorders (DSM) and International Classification of Diseases (ICD) were used in the diagnosis of the patients. Please see original paper1 for details. We selected to use the summary statistics of the 46 European case control samples (32,405 cases and 46,839 controls). For ND-related traits, we used the summary statistics of our cotinine study65 and 2 unpublished datasets (manuscripts in preparation). One data used the sum scores of the Fagerström Test for Nicotine Dependence (FTND)66 as a trait, which is a commonly used phenotype for ND based on self-reported smoking behaviors. The second data used a single item of the FTND questionnaire, “How soon after you wake up do you smoke your first cigarette”, or time to smoke the first cigarette (TFC) as a trait. This question can be seen as a measure of nicotine withdrawal since the half-life of nicotine in the human body is about 2 hours67. Smokers often experience nicotine withdrawal in the morning after not smoking overnight. The third data65 used the plasma cotinine concentration (COT) as a trait. Cotinine is the major metabolite of nicotine, and its half-life is much longer than that of nicotine. Therefore, its concentration in plasma can be considered an index of nicotine intake in recent days68,69. Because the quantity of nicotine intake is one of the most important measures of ND, COT may be considered a measure of ND as well. In these studies, FTND, TFC and COT were treated as quantitative traits. The sample size for FTND was 16,237, excluding the Netherlands Twin Registry sample because some of its subjects were also used in COT GWAS. The sample sizes for TFC and COT were 15,705 and 4,575 respectively. The FTND and TFC measures were derived from the same subjects, therefore, only FTND was used in polygenic analyses. TFC were used only for the identification of shared genes between schizophrenia and ND related phenotypes. The samples used in these 3 ND related GWASs were listed in Supplementary Table S1. All subjects used in this study were of European ancestry.
Polygenic analyses
Schizophrenia risk scores were calculated for 9 independent smoking related studies (Table S1, n = 10,794) with FTND and CPD measures using the summary statistics from the PGC schizophrenia meta-analysis. The control subjects from the Molecular Genetics of Schizophrenia (MGS) were included in the GWASs of both FTND and PGC schizophrenia, therefore they were excluded from this analysis. Risk scores for COT and FTND were calculated for 13,326 individuals from the NIMH genetics consortium repository (https://www.nimhgenetics.org/). We estimated the risk scores for each trait using the algorithms implemented in the PLINK software70. Specifically, the risk score for an individual was the sum of the number of risk alleles multiplied by the logarithm of odds ratio (OR, for schizophrenia) or beta coefficient (for FTND and COT), which was then normalized subsequently by the product of maximal number of risk alleles and log(OR)s/beta coefficients. For each trait, we calculated risk scores at 5 P-value thresholds: 5 × 10−5, 5 × 10−4, 5 × 10−3, 5 × 10−2 and 5 × 10−1. The numbers of markers used to calculate schizophrenia risk scores at these thresholds were 6,014, 94,804, 268,070, 1,021,476 and 5,370,899. The numbers of markers used for FTND and COT were 731, 6,312, 55,378, 500,542 and 4,752,196; and 1,621, 6,357, 48,575, 473,100, and 4,737,313 respectively. We then tested whether schizophrenia risk scores predicted FTND scores and vice versa using logistic (schizophrenia) and linear regression (FTND scores). Since the number of cigarettes smoked per day (CPD) was available from the FTND datasets, we also tested whether the genetic risk scores for schizophrenia predicted the CPD phenotype. Because we did not have individual genotypes for all datasets used in the COT meta-analyses, we used only the COT risk score to predict schizophrenia diagnosis. Sex, age and study were included as covariates in regression analyses.
Identification of shared risk genes
While there are papers looking at pleiotropy from a conditional FDR point of view71, we arrive to qualitatively similar conclusions using a somewhat simpler approach of family-wise error rate. Our test attempts to discover shared risk genes between schizophrenia and ND using summary statistics from their respective GWASs. To ensure that such a test is not overly influenced by a strong signal in just one trait, we implemented a “weakest link” approach (i.e., choosing the larger P-value of the pair of trait tests at the SNP under investigation)72. In more detail, let Xj and Pj be the χ2 distributed statistics and their associated (background enrichment adjusted) P-values, j = 1, …, m, for association tests between the m traits and a SNP. As the overlap statistic of all traits we use 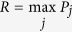 (or, alternatively,
(or, alternatively,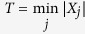 ). Under the assumption that the trait tests are independent, the P-value (also denoted as overlap P-value) for a given overlap statistic, r, at a SNP is
). Under the assumption that the trait tests are independent, the P-value (also denoted as overlap P-value) for a given overlap statistic, r, at a SNP is 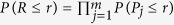 . If we further assume that (under the null hypothesis - H0) none of the traits is associated with the genetic variant, the overlap P-value simplifies to P(R ≤ r) = rm (1). Otherwise, P(Pj ≤ r) can be computed based on the distribution of the j-th trait P-values. For instance, for two phenotype configuration and a putative threshold of 5 × 10−8, the parametric version of our method requires that, for a significant pleiotropic signal, the p-values for both phenotypes to be <2.2 × 10−4 (
. If we further assume that (under the null hypothesis - H0) none of the traits is associated with the genetic variant, the overlap P-value simplifies to P(R ≤ r) = rm (1). Otherwise, P(Pj ≤ r) can be computed based on the distribution of the j-th trait P-values. For instance, for two phenotype configuration and a putative threshold of 5 × 10−8, the parametric version of our method requires that, for a significant pleiotropic signal, the p-values for both phenotypes to be <2.2 × 10−4 ( ). This substantially less than 5 × 10−8 p-value threshold is similar in spirit to the one from Andeassen et al.71 While the overlap p-value (1) does eliminate most of the influence of an extreme signal for one phenotype, it does not eliminate it completely. However, for a putative threshold of 5 × 10−8 in (1), under the worst case scenario of an extreme signal in one phenotype, the false positive rate per SNP is still rather small, i.e. 2.2 × 10−4. Even more, as seen in Andreassen et al., the false positive rate is likely to be substantially lower. Moreover, a worst-case-scenario 2.2 × 10−4 false positive rate is adequate for the pathway analyses73. We used FDR24 to evaluate the approximate significance of the genetic overlap (described by relation (1)) between schizophrenia and smoking phenotypes. To select promising markers for pathway and network analyses we applied a threshold of q-value ≤ 0.16, corresponding to a factor of 2 in Akaike Information Criterion penalty in a likelihood ratio χ2 test with 1 degree of freedom.
). This substantially less than 5 × 10−8 p-value threshold is similar in spirit to the one from Andeassen et al.71 While the overlap p-value (1) does eliminate most of the influence of an extreme signal for one phenotype, it does not eliminate it completely. However, for a putative threshold of 5 × 10−8 in (1), under the worst case scenario of an extreme signal in one phenotype, the false positive rate per SNP is still rather small, i.e. 2.2 × 10−4. Even more, as seen in Andreassen et al., the false positive rate is likely to be substantially lower. Moreover, a worst-case-scenario 2.2 × 10−4 false positive rate is adequate for the pathway analyses73. We used FDR24 to evaluate the approximate significance of the genetic overlap (described by relation (1)) between schizophrenia and smoking phenotypes. To select promising markers for pathway and network analyses we applied a threshold of q-value ≤ 0.16, corresponding to a factor of 2 in Akaike Information Criterion penalty in a likelihood ratio χ2 test with 1 degree of freedom.
Pathway and network analyses
We conducted pathway enrichment analysis of genes with at least one marker with q-values lower than 0.16 from the joint testing of schizophrenia and COT/FTND/TFC traits. If a marker was within a gene region, it was assigned to the gene; otherwise, it was mapped to its most proximate gene using the 50-kb flanking regions (both 5′ and 3′ sides). Genes identified using SNPs associated with COT, FTND, or TFC were merged for the pathway enrichment analysis, for which we used the hypergeometric test implemented in the tool WebGestalt (2013 update)74 and the canonical pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. We required each pathway to have at least three genes from our gene list and no more than 300 genes from the reference genome. The P-values from hypergeometric tests were further adjusted by the Benjamini-Hochberg method23. Only pathways with adjusted P-values < 0.05 were considered statistically significantly enriched.
We further examined pathways interaction using the Characteristic Sub-Pathway Network (CSPN) algorithm31,75 the human protein-protein interaction (PPI) network76. We restricted the analysis specifically to the aforementioned merged gene set and their enriched pathways. In the final step, we selected the significant pathway interaction pairs based on permutation P-values less than 0.05.
Additional Information
How to cite this article: Chen, J. et al. Genetic relationship between schizophrenia and nicotine dependence. Sci. Rep. 6, 25671; doi: 10.1038/srep25671 (2016).
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants ML011177, MH101054 and DA032246. JK has been supported by the Academy of Finland (grants # 265240 and 263278). We are grateful to the PGC schizophrenia analysis group for providing the summary statistics of its GWAS meta-analysis. Although unrelated to this research, H. Kranzler has been a consultant or advisory board member for Alkermes, Lilly, Lundbeck, Otsuka, Pfizer, and Roche. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which is supported by AbbVie, Alkermes, Ethypharm, Lilly, Lundbeck, and Pfizer.
Footnotes
Author Contributions J.C. and S.-A.B. performed polygenic and joined analyses, and wrote the paper; H.Y., Z.Z. and P.J. performed pathway and network analyses; K.S.K., H.R.K., J.G., L.F., C.M., R.P., Y.M., D.I.B., B.W.J.H.P., R.F.T., J.J.W., J.V., J.K. and M.M. contributed to the samples used this manuscript and commented on the text; cotinine meta-analysis group and FTND meta-analysis group contributed samples to this paper; and XC conceived, designed and wrote the paper. All authors have read and agreed on the content of this manuscript.
Contributor Information
Cotinine meta-analysis group:
Jennifer J. Ware, Xiangning Chen, Jacqueline M. Vink, Anu Loukola, Camelia Minica, Rene Pool, Yuri Milaneschi, Massimo Mangino, Cristina Menni, Jingchun Chen, Roseann Peterson, Kirsi Auro, Leo-Pekka Lyytikäinen, Juho Wedenoja, Alex I. Stiby, Gibran Hemani, Gonneke Willemsen, Jouke Jan Hottenga, Tellervo Korhonen, Markku Heliövaara, Markus Perola, Richard Rose, Lavinia Paternoster, Nic Timpson, Catherine A. Wassenaar, Andy Z. X. Zhu, George Davey Smith, Olli Raitakari, Terho Lehtimäki, Mika Kähönen, Seppo Koskinen, Timothy Spector, Brenda W. J. H. Penninx, Veikko Salomaa, Dorret I. Boomsma, Rachel F. Tyndale, Jaakko Kaprio, and Marcus Munafò
FTND meta-analysis group:
Jennifer J. Ware, Xiangning Chen, Jacqueline M. Vink, Anu Loukola, Camelia Minica, Jingchun Chen, Roseann Peterson, Nic Timpson, Michelle Taylor, Dorret I. Boomsma, Jaakko Kaprio, Marcus Munafò, Hermine Maes, Brien Riley, Kenneth S. Kendler, Joel Gelernter, Richard Sherva, Lindsay Farrer, Henry R. Kranzler, Brion Maher, and Michael Vanyukov
References
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson T. E. et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 42, 448–453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 42, 441–447 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G. et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol. Psychiatry 16, 996–1005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S. et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41, 56–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran S. et al. Abundant pleiotropy in human complex diseases and traits. Am. J. Hum. Genet. 89, 607–618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N., Cotsapas C., Lee P. H., Purcell S. M. & Smoller J. W. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 14, 483–495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G. & Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23, R89–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziedonis D. et al. Tobacco use and cessation in psychiatric disorders: National Institute of Mental Health report. Nicotine Tob. Res. Off. J. Soc. Res. Nicotine Tob. 10, 1691–1715 (2008). [DOI] [PubMed] [Google Scholar]
- de Leon J. & Diaz F. J. Genetics of schizophrenia and smoking: an approach to studying their comorbidity based on epidemiological findings. Hum. Genet. 131, 877–901 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari V. & Postma P. Nicotine use in schizophrenia: the self medication hypotheses. Neurosci. Biobehav. Rev. 29, 1021–1034 (2005). [DOI] [PubMed] [Google Scholar]
- de Leon J. & Diaz F. J. A meta-analysis of worldwide studies demonstrates an association between schizophrenia and tobacco smoking behaviors. Schizophr. Res. 76, 135–157 (2005). [DOI] [PubMed] [Google Scholar]
- De Luca V. et al. Evidence of association between smoking and alpha7 nicotinic receptor subunit gene in schizophrenia patients. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 29, 1522–1526 (2004). [DOI] [PubMed] [Google Scholar]
- Hong L. E. et al. A CHRNA5 allele related to nicotine addiction and schizophrenia. Genes Brain Behav. 10, 530–535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak G. et al. Association of polymorphisms in the BDNF, DRD1 and DRD3 genes with tobacco smoking in schizophrenia. Ann. Hum. Genet. 74, 291–298 (2010). [DOI] [PubMed] [Google Scholar]
- Novak G., Boukhadra J., Shaikh S. A., Kennedy J. L. & Le Foll B. Association of a polymorphism in the NRXN3 gene with the degree of smoking in schizophrenia: a preliminary study. World J. Biol. Psychiatry Off. J. World Fed. Soc. Biol. Psychiatry 10, 929–935 (2009). [DOI] [PubMed] [Google Scholar]
- Stephens S. H. et al. Association of the 5′-upstream regulatory region of the alpha7 nicotinic acetylcholine receptor subunit gene (CHRNA7) with schizophrenia. Schizophr. Res. 109, 102–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y. et al. Association of functional dopamine-beta-hydroxylase (DBH) 19 bp insertion/deletion polymorphism with smoking severity in male schizophrenic smokers. Schizophr. Res. 141, 48–53 (2012). [DOI] [PubMed] [Google Scholar]
- Jackson K. J., Fanous A. H., Chen J., Kendler K. S. & Chen X. Variants in the 15q25 gene cluster are associated with risk for schizophrenia and bipolar disorder. Psychiatr. Genet. 23, 20–28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukola A. et al. Genome-wide association study on detailed profiles of smoking behavior and nicotine dependence in a twin sample. Mol. Psychiatry 19, 615–624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser M. et al. Higher rates of cigarette smoking in male adolescents before the onset of schizophrenia: a historical-prospective cohort study. Am. J. Psychiatry 161, 1219–1223 (2004). [DOI] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 43, 969–976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995). [Google Scholar]
- Benjamini Y. & Yekutieli D. Quantitative trait Loci analysis using the false discovery rate. Genetics 171, 783–790 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P. et al. A bias-reducing pathway enrichment analysis of genome-wide association data confirmed association of the MHC region with schizophrenia. J. Med. Genet. 49, 96–103 (2012). [DOI] [PubMed] [Google Scholar]
- Jia P., Wang L., Meltzer H. Y. & Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophr. Res. 122, 38–42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Föcking M. et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol. Psychiatry 20, 424–432 (2015). [DOI] [PubMed] [Google Scholar]
- Ripke S. et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45, 1150–1159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. et al. Focal adhesion dynamics are altered in schizophrenia. Biol. Psychiatry 74, 418–426 (2013). [DOI] [PubMed] [Google Scholar]
- Zhao Z. et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol. Psychiatry 20, 563–572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salavati B. et al. Imaging-based neurochemistry in schizophrenia: a systematic review and implications for dysfunctional long-term potentiation. Schizophr. Bull. 41, 44–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishimoto A. et al. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Hum. Mol. Genet. 16, 2880–2891 (2007). [DOI] [PubMed] [Google Scholar]
- Thorgeirsson T. E. et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 452, 638–642 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal D. et al. RBFOX1 and RBFOX3 mutations in rolandic epilepsy. PloS One 8, e73323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcia G. et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat. Genet. 44, 1255–1259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H. et al. Mutations in prickle orthologs cause seizures in flies, mice, and humans. Am. J. Hum. Genet. 88, 138–149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol. Psychiatry 19, 410–416 (2014). [DOI] [PubMed] [Google Scholar]
- Barry G. et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494 (2014). [DOI] [PubMed] [Google Scholar]
- Zhernakova A., van Diemen C. C. & Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat. Rev. Genet. 10, 43–55 (2009). [DOI] [PubMed] [Google Scholar]
- Andreassen O. A. et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am. J. Hum. Genet. 92, 197–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen O. A., Thompson W. K. & Dale A. M. Boosting the power of schizophrenia genetics by leveraging new statistical tools. Schizophr. Bull. 40, 13–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Trent S., Thomas K. L., O’Donovan M. C. & Owen M. J. Genetic risk for schizophrenia: convergence on synaptic pathways involved in plasticity. Biol. Psychiatry 77, 52–58 (2015). [DOI] [PubMed] [Google Scholar]
- Purcell S. M. et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. & Li M. D. Common and Unique Biological Pathways Associated with Smoking Initiation/Progression, Nicotine Dependence, and Smoking Cessation. Neuropsychopharmacology 35, 702–719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S. F. et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 16, 36–49 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins A. K. et al. Neurexin 1 (NRXN1) splice isoform expression during human neocortical development and aging. Mol. Psychiatry, 10.1038/mp.2015.107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionel A. C. et al. Rare exonic deletions implicate the synaptic organizer Gephyrin (GPHN) in risk for autism, schizophrenia and seizures. Hum. Mol. Genet. 22, 2055–2066 (2013). [DOI] [PubMed] [Google Scholar]
- Todarello G. et al. Incomplete penetrance of NRXN1 deletions in families with schizophrenia. Schizophr. Res. 155, 1–7 (2014). [DOI] [PubMed] [Google Scholar]
- Nussbaum J. et al. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum. Mol. Genet. 17, 1569–1577 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut L. J. et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 16, 24–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol. Psychiatry 16, 1117–1129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénit P. et al. Mutant NDUFV2 subunit of mitochondrial complex I causes early onset hypertrophic cardiomyopathy and encephalopathy. Hum. Mutat. 21, 582–586 (2003). [DOI] [PubMed] [Google Scholar]
- Liu H.-Y., Liao P.-C., Chuang K.-T. & Kao M.-C. Mitochondrial targeting of human NADH dehydrogenase (ubiquinone) flavoprotein 2 (NDUFV2) and its association with early-onset hypertrophic cardiomyopathy and encephalopathy. J. Biomed. Sci. 18, 29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M. et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol. Psychiatry 17, 887–905 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akarsu S. et al. Mitochondrial complex I and III gene mRNA levels in schizophrenia, and their relationship with clinical features. J. Mol. Psychiatry 2, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washizuka S. et al. Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with schizophrenia in the Japanese population. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 141B, 301–304 (2006). [DOI] [PubMed] [Google Scholar]
- Jia Y., Yamazaki Y., Nakauchi S., Ito K.-I. & Sumikawa K. Nicotine facilitates long-term potentiation induction in oriens-lacunosum moleculare cells via Ca2+ entry through non-alpha7 nicotinic acetylcholine receptors. Eur. J. Neurosci. 31, 463–476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., An L., Yang B., Si L. & Zhang T. Nicotine-induced impairments of spatial cognition and long-term potentiation in adolescent male rats. Hum. Exp. Toxicol. 33, 203–213 (2014). [DOI] [PubMed] [Google Scholar]
- Reisiger A.-R. et al. Nicotine self-administration induces CB1-dependent LTP in the bed nucleus of the stria terminalis. J. Neurosci. Off. J. Soc. Neurosci. 34, 4285–4292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutlu M. G. & Gould T. J. Nicotinic modulation of hippocampal cell signaling and associated effects on learning and memory. Physiol. Behav. 155, 162–171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall F. S. et al. Negative affective states and cognitive impairments in nicotine dependence. Neurosci. Biobehav. Rev. 58, 168–185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashare R. L., Falcone M. & Lerman C. Cognitive function during nicotine withdrawal: Implications for nicotine dependence treatment. Neuropharmacology 76 Pt B, 581–591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva O. A. & Levin E. D. Glutamate and nicotinic receptor interactions in working memory: importance for the cognitive impairment of schizophrenia. Neuroscience 195, 21–36 (2011). [DOI] [PubMed] [Google Scholar]
- Ware J. J. et al. Genome-Wide Meta-Analysis of Cotinine Levels in Cigarette Smokers Identifies Locus at 4q13.2. Sci. Rep. 6, 20092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatherton T. F., Kozlowski L. T., Frecker R. C. & Fagerström K. O. The Fagerström Test for Nicotine Dependence: a revision of the Fagerström Tolerance Questionnaire. Br. J. Addict. 86, 1119–1127 (1991). [DOI] [PubMed] [Google Scholar]
- Luck W. & Nau H. Nicotine and cotinine concentrations in serum and milk of nursing smokers. Br. J. Clin. Pharmacol. 18, 9–15 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaiyandi V., Goodz S. D., Sellers E. M. & Tyndale R. F. CYP2A6 genotype, phenotype, and the use of nicotine metabolites as biomarkers during ad libitum smoking. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 15, 1812–1819 (2006). [DOI] [PubMed] [Google Scholar]
- Benowitz N. L., Dains K. M., Dempsey D., Yu L. & Jacob P. Estimation of nicotine dose after low-level exposure using plasma and urine nicotine metabolites. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 19, 1160–1166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen O. A. et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 9, e1003455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdeli T. B. et al. On schizophrenia as a ‘disease of humanity’. Schizophr. Res. 143, 223–224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P. et al. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am. J. Hum. Genet. 85, 13–24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Kirov S. & Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–748 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. & Li S. Detection of characteristic sub pathway network for angiogenesis based on the comprehensive pathway network. BMC Bioinformatics 11 Suppl 1, S32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. et al. Integrated network analysis platform for protein-protein interactions. Nat. Methods 6, 75–77 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.