Abstract
Survivin is a member of the inhibitor-of-apoptosis proteins family. It is overexpressed in many different cancer types but not in the differentiated normal tissue. In addition, overexpression of survivin promotes cancer cell survival and induces chemotherapeutic drug resistance, making it an attractive target for new anticancer interventions. Despite survivin being a promising molecular target for anticancer treatment, it is widely accepted that survivin is only a “semi-druggable” target. Therefore, it is important to develop a new strategy to target survivin for anticancer treatment. In this study, we constructed a novel survivin promoter-driven full-length antisense survivin (pSur/AS-Sur) expression plasmid DNA. Promoter activity assay revealed that the activity of the survivin promoter of pSur/AS-Sur correlated with the endogenous expression of survivin at the transcriptional level in the transfected A549, MDA-MB-231, and PANC-1 cancer cells. Western blot analysis showed that liposomal delivery of pSur/AS-Sur successfully downregulated the expression of survivin in A549, MBA-MB-231, and PANC-1 cells in vitro. In addition, delivery of pSur/AS-Sur induced autophagy, caspase-dependent apoptosis, and caspase-independent apoptosis as indicated by the increased LC3B-II conversion, autophagosome formation, caspase-9/-3 and poly(ADP-ribose) polymerase-1 cleavage, and apoptosis-inducing factor nuclear translocation in A549, MBA-MB-231, and PANC-1 cells. Importantly, liposomal delivery of pSur/AS-Sur was also capable of decreasing the proliferation of the survivin/MDR1 coexpressing multidrug-resistant KB-TAX50 cancer cells and the estrogen receptor-positive tamoxifen-resistant MCF7-TamC3 cancer cells in vitro. In conclusion, the results of this study suggest that delivery of a survivin promoter-driven antisense survivin-expressing plasmid DNA is a promising way to target survivin and to treat survivin-expressing cancers in the future.
Keywords: antisense, apoptosis, cancer, survivin, autophagy, liposomal delivery
Introduction
Survivin is a member of the inhibitor-of-apoptosis proteins family. It is overexpressed in many different cancer types such as breast, colorectal, lung, and pancreatic cancer but not in the differentiated normal tissue.1–5 Survivin expression is positively correlated with poor prognosis and decreased survival rates in oral, breast, and colorectal cancer patients, and its upregulation is related to the promotion of metastasis and the induction of resistance to different chemotherapeutic agents, including etoposide, cisplatin, and paclitaxel.6–14 Therefore, survivin has been proposed as an attractive target for new anticancer interventions.
Despite survivin being a promising molecular target for anticancer treatment, it is widely accepted that survivin is only a “semi-druggable” target. In fact, only a few survivin-specific small-molecule inhibitors and short antisense oligonucleotides (ASOs) have been developed and have successfully reached clinical trials in the past 10 years.15–17 Unfortunately, it has been shown that the most advanced survivin small-molecule inhibitor, YM155, is a substrate of the multidrug-resistant protein (MDR1), which limits its use in clinical settings.16 In addition, it has been shown that the short ASO SPC3042 also targets Bcl-2 with an unknown mechanism, indicating that this drug may not be survivin-specific.17 Hence, it is imperative to develop a new strategy to target survivin for anticancer treatment.
Antisense therapy is the application of ASOs to downregulate the expression of a specific target in cells. Generally, ASOs bind to the targeted messenger RNA (mRNA) complementary based on Watson–Crick base pairing principle.18 The anticancer effect of survivin promoter-driven toxins or p53-expressing plasmid DNAs has already been evaluated in a few studies.19–21 However, the use of survivin promoter-driven antisense survivin-expressing plasmid DNA as an anticancer therapeutic has not been investigated in the past. In this study, we constructed a novel survivin promoter-driven full-length antisense survivin expression plasmid DNA and subsequently evaluated the effectiveness of this plasmid DNA in targeting different survivin-expressing cancer cells, including the MDR1 coexpressing multidrug-resistant KB-TAX50 oral cancer cells and the estrogen receptor-positive (ER+) tamoxifen-resistant MCF7-TamC3 breast cancer cells.
Materials and methods
Cell culture
Human MDA-MB-231 (breast cancer adenocarcinoma), PANC-1 (pancreatic cancer), and A549 (lung cancer) cells were all purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). MDA-MB-231 and PANC-1 cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% fetal bovine serum (FBS) and penicillin–streptomycin–glutamine (PSG). A549 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) medium containing 10% FBS and PSG. The cellular and molecular phenotypes of MCF7-TamC3 and KB-TAX50 have already been characterized in previous studies.22–25 In brief, ER+ tamoxifen-resistant MCF7-TamC3 cancer cells were created by prolonged culture of MCF7 (breast cancer) cells under estrogen-depleted conditions. MCF7-TamC3 cells were cultured in phenol-red-free RPMI containing 5% charcoal-stripped FBS, PSG, and insulin-transferring selenium. KB (oral epidermoid carcinoma)-derived KB-TAX50 cells were generated by paclitaxel-driven selection and displayed overexpression of P-gp170/MDR1. KB-TAX50 cells were cultured in RPMI containing 5% FBS and 50 nM paclitaxel. HUVEC (human umbilical vein endothelial) cells were cultured in EGM2 medium containing 2% FBS, hydrocortisone, human fibroblast growth factor (hFGF), vascular-endothelial growth factor, human epidermal growth factor (hEGF), recombinant human R3 insulin-like growth factor 1 (R3-IGF-1), ascorbic acid, GA-1000, and heparin. All cells were cultured at 37°C in a humidified incubator containing 5% CO2. The use of the aforementioned human cell lines in this study was approved by the review board of Ministry of Science and Technology (Taiwan, ROC) and the biosafety committee of National Cheng Kung University (Taiwan, ROC).
Reverse transcription-polymerase chain reaction to detect the expression of survivin and MDR1 in cancer cells
Total RNA was extracted using the TRIzol® reagent (cat# 15596-026, Thermo Fisher Scientific, Waltham, MA, USA), and complementary DNA (cDNA) was synthesized from RNA using the RevertAid H Minus First Strand cDNA Synthesis Kit (cat# K1631, Thermo Fisher Scientific). Expression of survivin, MDR1, and actin at the transcriptional level was determined by polymerase chain reaction (PCR). The specific primers with the following sequences were used in the study: human β-actin forward primer, 5′-TACCCCATCGAGCACGGCAT-3′; human β-actin reverse primer, 5′-GAAGCAGCCGTGGCCATCTC-3′; human survivin forward primer, 5′-TGCCTGGCAGCCCTTTCTCA-3′; human survivin reverse primer, 5′-GGCGCACTTTCTCCGCAGTT-3′; human MDR1 forward primer, 5′-GCCTGGCAGCTGGAAGACAAATRCACAAAATT-3′; human MDR1 reverse primer, 5′-CAGACAGCAGCTGACAGTCCRAGAACAGGACT-3′. Experiments were repeated three times. The PCR products were resolved in a 1.5% agarose gel.
Promoter activity assay
Cells were seeded at 1×104/well in 96-well plates for 24 hours prior to the transfection of the survivin promoter activity reporter plasmid, pDRIVE-hSurvivin (cat# pdrive-hsurvivin, InvivoGen, San Diego, CA, USA). Cells were washed once with phosphate-buffered saline (PBS) 48 hours posttransfection. Subsequently, 100 µL of β-galactosidase assay reagent was added to each well and incubated at 37°C for 30 minutes with cover. After incubation, 100 µL of β-galactosidase assay stop solution was added to each well to terminate the reaction. Promoter activity was quantified by measuring the absorbance of the solution at 405 nm wavelength using the SpectraMax® M5 microplate reader (Molecular Devices LLC, Sunnyvale, CA, USA).
Construction and production of the survivin promoter-driven antisense survivin (pSur/AS-Sur)-expressing plasmid DNA
Briefly, plasmid encoding the entire coding region of human BIRC5 (coding for survivin) gene (pCMV6-AC-GFP-Survivin, sequence verified, cat# RG205935) was purchased from OriGene (Rockville, MD, USA). Construction of the antisense survivin gene (AS-Sur) with extra BspHI and EcoRI enzymatic cleavage site located on the 5′ and 3′ end, respectively, was carried out by PCR (95°C for 30 seconds, followed up by 68°C for 30 seconds, then 72°C for 30 seconds, for 30 cycles) using the forward primer 5′-AATCATGAATCCATGGCAGCCAG-3′ and the reverse primer 5′-AAGAATTCATGGGTGCCCCGA-3′. The PCR product (AS-Sur) was ligated to the PCR products cloning vector pJET1.2 (cat# K1231, Thermo Fisher Scientific) and then transformed into DH5-α Escherichia coli cells. DNA sequencing was preformed to validate the sequence of the recombinant AS-Sur gene. AS-Sur was excised by digestion with BspHI and EcoRI, and subcloned into the LacZ gene-removed pDRIVE-hSurvivin, which is a mammalian transfectable vector bearing a human survivin promoter. The final product, pSur/AS-Sur, was transformed into DH5-α E. coli cells for long-term storage.
Transfection of pDRIVE-hSurvivin and pSur/AS-Sur plasmid DNA into cancer cells
Lipofectamine® 3000 (cat# L3000015, Thermo Fisher Scientific) was used to transfect different plasmids purified by using the EndoFree® Plasmid Mega Kit (cat# 12381, Qiagen, Hilden, Germany) into the targeted cancer cells. Briefly, cells were seeded onto 96-well plates or 60 mm dishes and allowed to adhere overnight. Appropriate amount of Lipofectamine 3000 reagent was diluted in the Opti-MEM® I medium (cat# 31985, Thermo Fisher Scientific) without serum. Purified DNA was also diluted in the Opti-MEM I medium without serum, and subsequently an appropriate amount of P300 reagent was added to the diluted DNA. Diluted DNA together with P300 reagent was then mixed with the diluted Lipofectamine 3000 reagent (1:1 ratio) and incubated for 5 minutes at room temperature. The transfection mixture was overlaid onto the cells under PSG-free conditions.
Bromodeoxyuridine (BrdU) cell proliferation assay
Incorporation of the thymidine analog, bromodeoxyuridine (BrdU), was measured using the BrdU proliferation assay kit (cat# QIA58, Merck Millipore, Billerica, MA, USA) to determine the effect of pSur/AS-Sur on cell proliferation. Briefly, cells were seeded at 3×103/well in 96-well plates for 24 hours prior to the transfection of pSur/AS-Sur for 4 days. Control (empty plasmid) or pSur/AS-Sur-transfected cells were labeled with BrdU for 5 hours prior to the incubation with anti-BrdU monoclonal antibody for an hour. The immune complex was detected following the incubation with anti-mouse immunoglobulin G, peroxidase conjugate and substrate solution. The reaction was terminated after 30 minutes, and the absorbance of the assay wells was quantified by measuring at 450–540 nm wavelength using the SpectraMax M5 microplate reader (Molecular Devices LLC). The number of proliferating cells is represented by the amount of BrdU incorporation, which directly correlates to the color intensity and the absorbance values. Experiments were performed using duplicate wells and repeated three times.
Lactate dehydrogenase cytotoxicity assay
Lactate dehydrogenase (LDH) assay was performed using the LDH-cytotoxicity assay kit II (cat# cat#ab65393, Abcam, Cambridge, UK). First, PANC-1 cells were seeded at 1×104/well in 96-well plates for 24 hours prior to the transfection of pSur/AS-Sur for 48 hours. Cell cytotoxicity was quantified by measuring the absorbance of the solution at 450 nm wavelength using a SpectraMax M5 microplate reader (Molecular Devices LLC). Duplicate wells were assayed for each condition, and the experiment was repeated three times. Cytotoxicity index for each treatment group was calculated using the equation: (Test sample – Low control)/(High control – Low control), and also by adjusting the untreated control group to 1. Test samples – cells were either transfected with empty plasmid only or with pSur/AS-Sur for 4 days; low control – completely untreated cells (minimal LDH value); high control – completely lysed cells (maximal LDH value).
Ectopic expression of EGFP-LC3B and detection of autophagosome formation
Autophagosome formation in the pSur/AS-Sur-transfected cancer cells was detected by the florescence microscopic analysis. Cells were first seeded onto 60 mm dishes and allowed to adhere overnight. Then, the cells were either transfected with enhanced green fluorescent protein (EGFP)-tagged LC3B (plasmid# 11546, Addgene, Cambridge, MA, USA) alone or cotransfected with EGFP-tagged LC3B and pSur/AS-Sur for 48 hours. LC3B puncta formation in cells was observed under a fluorescence microscope (IX-71, Olympus, Tokyo, Japan). ImageJ software (National Institutes of Health, Bethesda, MA, USA) was used to analyze the number of the green fluorescent puncta present in cells. Experiments were repeated three times.
Monodansylcadaverine staining of acidic vesicular organelles
Monodansylcadaverine (MDC) staining of acidic vesicular organelles (AVOs) was performed for autophagy analysis. First, cells were seeded onto 60 mm dishes and allowed to adhere overnight. Next, the cells were transfected with pSur/AS-Sur using the Lipofectamine 3000 reagent. AVOs were labeled with 0.5 mM MDC in the phenol-red-free RPMI at 37°C for 2 hours. Then, the cells were washed three times with PBS. AVOs in all cells were observed under a fluorescence microscope (IX-71, Olympus). ImageJ software (National Institutes of Health) was used to analyze the number of the green fluorescent puncta present in cells. Experiments were repeated three times.
Western blot analysis
Cells were lysed using the Cellytic™ cell lysis reagent (cat# C2978, Sigma-Aldrich, St Louis, MO, USA) that contained 1 mM phenylmethane sulfonyl fluoride (PMSF), 1 mM NaF, and cocktail protease inhibitors (cat# 04693159001, Hoffmann-La Roche Ltd., Basel, Switzerland). The protein concentration was measured by the method developed by Lowry et al26 using bovine serum albumin as standard. Equal amounts of protein were subjected to SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis on a 6%, 8%, or 10% polyacrylamide gel. The resolved proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane (cat# IPVH00010, Merck Millipore), which was then exposed to 5% nonfat-dried milk or 3% bovine serum albumin in Tris-buffered saline containing 0.1% Tween 20 (TBST) for an hour at room temperature before incubation overnight at 4°C with different primary antibodies ([anti-survivin, anti-XIAP, and anti-AIF; cat# AF886, AF8221, and AF1457, R&D systems, Minneapolis, MN, USA], [anti-actin and anti-γH2AX; cat# MAB1501 and 05636, Merck Millipore], [anti-cleaved caspase-3; cat# 9664, Cell Signaling Technology, Danvers, MA, USA]), ([anti-caspase-9; cat# 05-572, Argonex, Inc., Charlottesville, VA, USA], and [anti-LC3B; cat# TA301543, OriGene]). The PVDF membrane was then washed three times with TBS containing 0.05% Tween 20 before incubation for 1 hour at room temperature with different peroxidase-conjugated secondary antibodies. Immune complexes were finally detected with chemiluminescence reagents, and luminescence protein signals were detected by Luminescence Readers (FUJI LAS-100, Fujifilm, Tokyo, Japan). The intensity of bands was analyzed by the Image Gauge software Version 3.46 (FUJIFILM Science Lab, Tokyo, Japan). Experiments were repeated at least three times.
Gene silencing by siRNA
Target-validated small interfering RNA (siRNA) oligos were transfected into A549, MDA-MB-231, and PANC-1 cells using the Lipofectamine® RNAiMAX reagent (Thermo Fisher Scientific, cat# 13778150). First, cells were seeded onto 60 mm dishes and cultured in antibiotic-free DMEM and RPMI medium for overnight. Approximately 7.5 µL of survivin siRNA oligomers (stock concentration 10 µM; cat# 6351, Cell signaling Technology) was diluted in 100 µL of Opti-MEM I medium without serum and then mixed with 7.5 µL of Lipofectamine RNAiMAX transfection reagent diluted in 100 µL of Opti-MEM I medium without serum for 20 minutes at room temperature. Cells were overlaid with the transfection mixture and incubated for various durations.
Comet assay
Microscopic slides were gently coated with 100 µL of 1% normal melting point agarose using a coverslip. The slide was placed on ice for 15 minutes to allow the agarose to set. After gelling, the coverslips were removed, 25 µL of the cells suspension (contains 105 cells) was gently mixed with 100 µL of 1.5% low melting point (37°C) agarose and pipetted onto the layer of 1% normal melting point agarose and covered with a coverslip. After 15 minutes on ice, the coverslips were removed and the slides were lowered into freshly made cold lysis buffer (2.5 M NaCl, 100 mM ethylenediaminetetraacetic acid [EDTA], 10 mM Tris, 1% Triton X-100, pH 10) for 30 minutes. To allow DNA unwinding, the slides were placed into an electrophoresis chamber containing cold alkaline electrophoresis buffer (300 mM NaOH, 1 mM EDTA) for 20 minutes. Electrophoresis was performed by setting the power supply to 25 V and adjusting the current to 300 mA for 20 minutes. After electrophoresis, the slides were placed in a freshly made neutralizing buffer (0.4 M Tris, pH 7.5) for 20 minutes. Cell staining was performed with 10 mL per slide of propidium iodide (20 mg/L). The slides were examined with a fluorescence microscope (Optiphot-2, Nikon, Tokyo, Japan) at 20× magnification. Microscopic images of the comets were scored using the TriTek CometScore™ computer software (Sumeduck, VA, USA). From each sample, one slide was prepared, and the images of at least 50 cells from each slide were scored. The tail moment was chosen as our parameter.
Statistical analysis
Each experiment was performed at least three times. Data are presented as mean ± standard deviation. The significance of difference was evaluated with the one-way analysis of variance (one-way ANOVA). A P-value <0.05 was considered statistically significant.
Results
Survivin is overexpressed at the transcriptional level in tumor tissues of patients with different cancer types
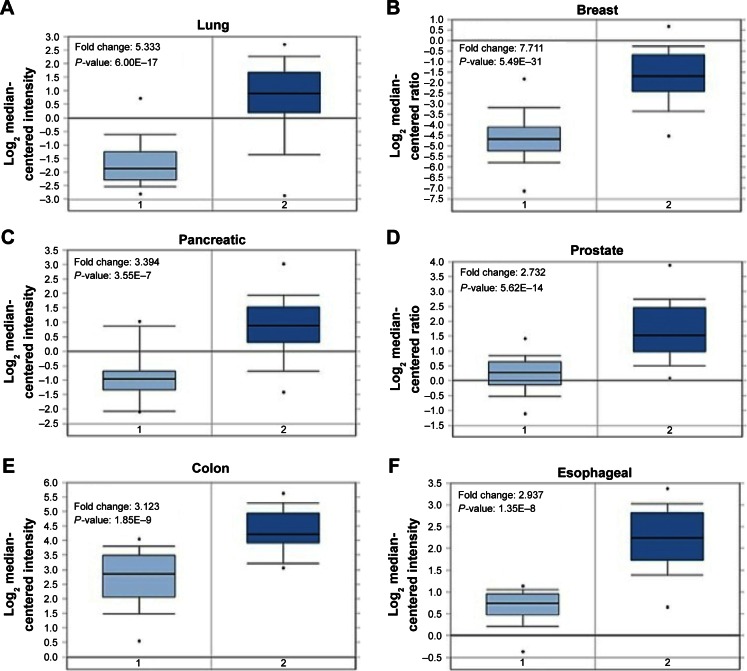
We first started with assessing mRNA expression profiles derived from clinical samples of patients in six different cancer tissues with online database (Oncomine™, www.oncomine.org; The Cancer Genome Atlas, https://tcga-data.nci.nih.gov/tcga/) to determine whether survivin expression is upregulated, probably at the transcriptional level, in tumor tissues.27–31 Retrospective statistical analysis revealed that the amount of survivin RNA transcripts present in the tumor tissues is significantly higher than that in the normal tissues in different cohorts across several cancer types, including lung adenocarcinoma (5.333-fold), invasive breast carcinoma (7.711-fold), pancreatic carcinoma (3.394-fold), prostate carcinoma (2.732-fold), colorectal carcinoma (3.123-fold), and esophageal squamous cell carcinoma (2.937-fold), indicating that survivin gene transcription is upregulated in tumor tissues (Figure 1). These observations suggest that the development of a survivin promoter-driven anticancer gene-expressing plasmid DNA may be beneficial to target survivin-expressing cancer cells.
Figure 1.
Survivin is overexpressed in tumor tissues of patients with different cancer types.
Notes: (A) The mRNA expression level of survivin in normal lung tissue (n=65) and lung adenocarcinoma (n=45) samples (data derived from Hou Lung database). (B) The mRNA expression level of survivin in normal breast tissue (n=61) and invasive breast carcinoma (n=76) samples (data derived from TCGA Breast database). (C) The mRNA expression level of survivin in normal pancreas tissue (n=16) and pancreatic carcinoma (n=36) samples (data derived from Pei Pancreas database). (D) The mRNA expression level of survivin in normal prostate gland (n=28) and prostate carcinoma (n=59) samples (data derived from Grasso Prostate database). (E) The mRNA expression level of survivin in normal colorectal tissue (n=24) and colorectal carcinoma (n=36) samples (data derived from Skrzypczak Colorectal database). (F) The mRNA expression level of survivin in normal esophagus tissue (n=17) and esophageal squamous cell carcinoma (n=17) samples (data derived from Hu Esophagus database). “1” represents normal tissues and “2” represents tumor tissues.
Abbreviation: mRNA, messenger RNA.
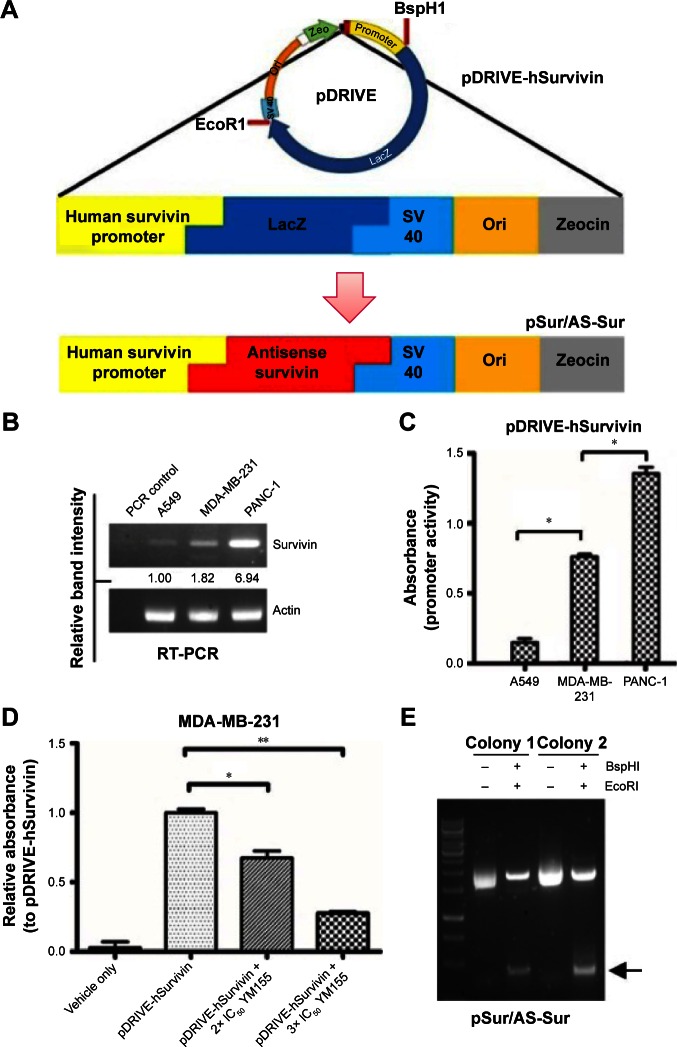
Construction of a human survivin promoter-driven full-length antisense survivin-expressing plasmid DNA (pSur/AS-Sur) for anticancer therapy
Promoter activity assay was first performed to determine whether the selected plasmid, pDRIVE-hSurvivin, which is a DNA vector that harbors a survivin promoter followed up with a luciferase reporter gene and is used for the subsequent construction of the survivin promoter-driven antisense survivin (pSur/AS-Sur)-expressing plasmid DNA, can be activated in cancer cells endogenously expressing survivin at the transcriptional level (Figure 2A). Consistent with the results of the reverse transcription-polymerase chain reaction (RT-PCR) analysis, which showed the differential expression pattern of survivin at the transcriptional level in A549, MDA-MB-231, and PANC-1 cells (Figure 2B), the promoter activity assay revealed a differential activation pattern of the survivin promoter present on pDRIVE-hSurvivin in the tested cancer cells (A549 < MDA-MB-231 < PANC-1) with plasmid transfection (Figure 2C). To confirm that the activation of the luciferase reporter gene observed in the experiment was caused by the activation of the survivin promoter present on pDRIVE-hSurvivin, MDA-MB-231 cells were transfected with pDRIVE-hSurvivin and cotreated with YM155, a pharmacological inhibitor of survivin that inhibits survivin expression by binding onto the promoter of survivin gene, and the expression level of the luciferase reporter gene was determined. The promoter activity assay revealed that YM155 cotreatment significantly attenuated the expression of luciferase in the pDRIVE-hSurvivin-transfected cells, confirming that the activation of the survivin promoter present on pDRIVE-hSurvivin was regulated by the survivin-related transcription factors endogenously expressed in the cancer cells (Figure 2D). These results further indicate that the survivin promoter present on pDRIVE-hSurvivin can be actively and differentially regulated by different cancer cells according to their differential expression pattern of survivin at the transcriptional level.
Figure 2.
Construction of pSur/As-Sur and activity evaluation of the survivin promoter of pDRIVE-hSurvivin.
Notes: (A) Schematic diagram showing the structural difference between pDRIVE-hSurvivin and pSur/As-Sur plasmid DNA. (B) Expression of survivin at the transcriptional level in three different cancer cell lines was examined by RT-PCR. Equal sample loading was verified by actin, and the numbers under each band are the intensities of the band relative to the survivin expression of A549 cells. (C) Cells were transfected with pDRIVE-hSurvivin for 48 hours, and the promoter activity was examined by the β-galactosidase assay. “*” denotes statistical significance with a P-value <0.05 between the testing groups. (D) MDA-MB-231 cells were treated with or without pDRIVE-hSurvivin and cotreated with or without 2× IC50 and 3× IC50 YM155. Promoter activity was examined after 48 hours by the β-galactosidase assay. “*” and “**” denote statistical significance with P-values <0.05 and <0.01, respectively, between the testing groups. (E) The pSur/AS-Sur was incubated with the restriction endonuclease, BspHI and EcoRI, for 10 minutes, and the resulting products were resolved by agarose-gel electrophoresis. Arrow indicates the presence of the AS-Sur gene in pSur/AS-Sur plasmid DNA.
Abbreviations: PCR, polymerase chain reaction; RT-PCR, reverse transcription-polymerase chain reaction; pSur/AS-Sur, survivin promoter-driven antisense survivin.
The luciferase reporter gene then was excised from pDRIVE-hSurvivin and the full-length AS-Sur gene was cloned into the plasmid to make the pSur/AS-Sur plasmid DNA. Results of the restriction enzyme digestion and agarose-gel electrophoresis revealed the presence of a ~430 bp DNA fragment, which is close to the size of the full-length AS-Sur gene (Figure 2E). Results of the DNA sequencing further confirmed the presence of the full-length AS-Sur gene on pSur/AS-Sur (data not shown).
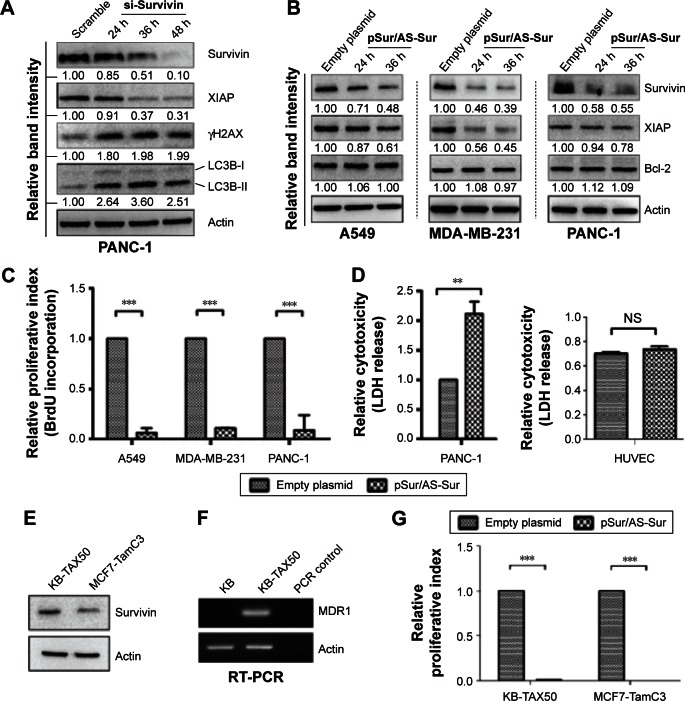
Delivery of pSur/AS-Sur downregulates the expression of survivin and XIAP in cancer cells and inhibits cancer cell proliferation in vitro
Next, we evaluated the specificity of pSur/AS-Sur in targeting survivin in A549, MDA-MB-231, and PANC-1 cells. At the molecular level, survivin stabilizes XIAP through physical interactions with Smad/DIABLO, and the targeting survivin has been shown to downregulate XIAP expression in cancer cells.32,33 Here, Western blot analysis revealed that the downregulation of survivin expression by siRNA decreased XIAP expression as expected (Figure 3A). Similar to the results of cancer cells treated with survivin siRNA, delivery of pSur/AS-Sur concurrently downregulated the expression of survivin and XIAP in A549, MDA-MB-231, and PANC-1 cells in a time-dependent manner (Figure 3B).
Figure 3.
Delivery of pSur/AS-Sur downregulates the expression of survivin and XIAP and inhibits the proliferation of cancer cells.
Notes: (A) PANC-1 cells were transfected with either scramble siRNA or survivin siRNA for the indicated durations. Expression of different proteins was determined by Western blotting. The numbers under each blot are the intensities of the blot relative to that of the control (scramble). (B) Cancer cells were transfected with either empty plasmid or pSur/AS-Sur for the indicated durations. The expression of survivin, XIAP, and Bcl-2 were examined by Western blotting. Equal protein loading was verified by actin. The numbers under each blot are the intensities of the blot relative to that of the control (empty plasmid). (C) Cancer cells were transfected with pSur/AS-Sur for 4 days. Cell proliferation was evaluated using the BrdU proliferation assay. “***” denotes statistical significance with a P-value <0.001 between the testing groups. (D) PANC-1 and HUVECs were transfected with either an empty plasmid or pSur/AS-Sur for 4 days. Cytotoxicity was evaluated using the LDH assay. “**” denotes statistical significance with a P-value <0.01 between the testing groups. “NS” denotes no statistical significant difference between the testing groups. (E) Expression of survivin in KB-TAX50 and MCF7-TamC3 cells was determined by Western blotting. (F) Expression of MDR1 in KB and KB-TAX50 cells was determined by RT-PCR. (G) KB-TAX50 and MCF7-TamC3 cells were transfected with pSur/AS-Sur for 4 days. Cell proliferation was evaluated using the BrdU proliferation assay. “***” denotes statistical significance with a P-value <0.001 between the testing groups.
Abbreviations: HUVEC, human umbilical vein endothelial cell; RT-PCR, reverse transcription-polymerase chain reaction; siRNA, small interfering RNA; h, hours; BrdU, Bromodeoxyuridine; XIAP, X-linked inhibitor of apoptosis protein; pSur/AS-Sur, survivin promoter-driven antisense survivin.
It has been reported that SPC3042, an antisense inhibitor (a 16-mer locked nucleic acid oligonucleotide) of survivin, which is currently undergoing Phase I/II clinical trials, exhibits an off-target effect in downregulating Bcl-2 in cancer cells. Western blot analysis revealed that delivery of pSur/AS-Sur did not alter the expression of Bcl-2 in A549, MDA-MB-231, and PANC-1 cells, indicating that the full-length AS-Sur may exhibit higher target specificity toward survivin as compared with the short ASOs developed in the past (Figure 3B).
Because delivery of pSur/AS-Sur downregulated the expression of survivin and XIAP in A549, MDA-MB-231, and PANC-1 cells, we sought to determine whether delivery of pSur/AS-Sur can decrease the proliferation of cancer cells. Results from the BrdU cell proliferation assay revealed that delivery of pSur/AS-Sur significantly decreased the proliferation of cancer cells (Figure 3C). Notably, results from the LDH cytotoxicity assay showed that delivery of pSur/AS-Sur induced cell death in PANC-1 cells, suggesting that the decrease of cell proliferation as revealed by the BrdU assay in cells delivered with pSur/AS-Sur are in part caused by the increased cytotoxicity during pSur/AS-Sur activation (Figure 3D). In contrast, the same treatment did not induce cell death in HUVEC cells, which is a noncancerous cell line known for its survivin nonexpressing (or low level expressing) property,34 further suggesting that pSur/AS-Sur exhibits cytotoxicity mostly in survivin-expressing cancer cells but not in normal cells without or with low level of survivin expression (Figure 3D).
Delivery of pSur/AS-Sur inhibits the proliferation of MDR1-expressing cancer cells and ER+/tamoxifen-resistant breast cancer cells in vitro
It has been widely demonstrated that overexpression of MDR1 induces drug resistance to various chemotherapeutic agents, including the survivin inhibitor, YM155, and the microtubule dynamic modulators, vincristine and taxol. In addition, ER+/tamoxifen-resistant breast cancer is frequently found in patients after prolonged tamoxifen treatment. To determine whether delivery of pSur/AS-Sur is also effective in inhibiting cell proliferation in MDR1-expressing multidrug-resistant cancer cells and ER+/tamoxifen-resistant breast cancer cells, the MDR1-expressing KB-TAX50 oral cancer and the ER+/tamoxifen-resistant MCF7-TamC3 breast cancer cells were transfected with pSur/AS-Sur, and cell proliferation was determined by the BrdU assay. Results of the Western blot analysis and the RT-PCR analysis confirmed the expression of survivin in both KB-TAX50 and MCF7-TamC3 cells and the expression of MDR1 in KB-TAX50 cells, respectively (Figure 3E and F). BrdU cell proliferation assay revealed that delivery of pSur/AS-Sur significantly decreased the proliferation of KB-TAX50 and MCF7-TamC3 cancer cells in vitro (Figure 3C). Collectively, these results confirm that delivery of pSur/AS-Sur is capable of decreasing the expression of survivin and XIAP, leading to the inhibition of proliferation and induction of cytotoxicity in survivin-expressing cancer cells regardless of the expression of MDR1.
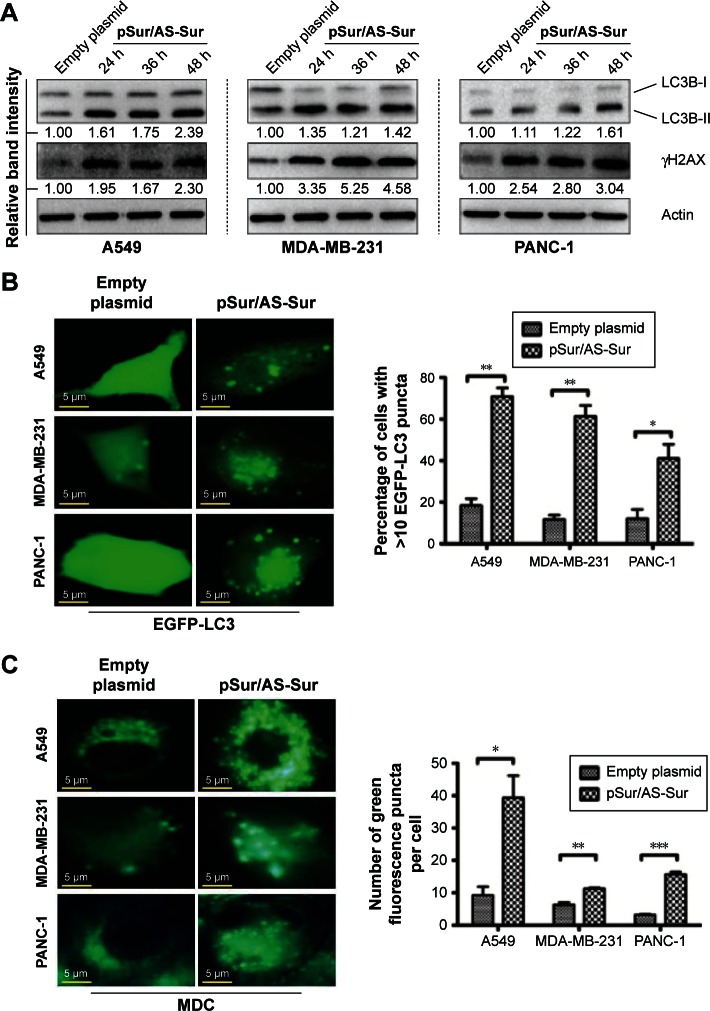
Delivery of pSur/AS-Sur modulates autophagy in cancer cells
Recent studies showed that downregulation of survivin by either pharmacological inhibitor or siRNA induces autophagy in cancer cells.33,35,36 In particular, our previous study revealed that downregulation of survivin by YM155 induced autophagy and autophagy-dependent DNA damage in breast cancer cells.33 To further confirm the target specificity of pSur/AS-Sur, we determined whether delivery of pSur/AS-Sur can also induce autophagy and DNA damage in cancer cells. The Western blot analysis revealed that downregulating survivin expression by siRNA increased LC3B-II conversion and γ-H2AX expression, which is a molecular marker for autophagy and DNA damage, respectively, as expected (Figure 3A). Similar to these results, delivery of pSur/AS-Sur also increased LC3B-II conversion and γH2AX expression in A549, MDA-MB-231, and PANC-1 cells in a time-dependent manner (Figure 4A). To further confirm the effect of pSur/AS-Sur on autophagy modulation, cancer cells were cotransfected with pSur/AS-Sur and a plasmid that overexpresses the EGFP-tagged LC3B and formation of autophagosome/autophagolysosome was determined by fluorescence microscopy. Fluorescence microscopy revealed that EGFP-LC3B was overexpressed and equally distributed inside the cells under pSur/AS-Sur-free conditions (Figure 4B). Delivery of pSur/AS-Sur significantly increased the formation of EGFP-LC3B punctate (green fluorescent punctate) in A549, MDA-MB-231, and PANC-1 cells, indicating the formation of autophagosome and/or autophagolysosome (Figure 4B). Cancer cells were also stained with MDC to further confirm the formation of AVOs, which is also a marker for autophagy, in cells with pSur/AS-Sur delivery. MDC is a fluorescent compound widely used for the detection of AVOs, including lysosome and autophagolysosome. Increased number and/or size of punctate formed in the MDC-stained cells are considered as the formation of AVOs. Fluorescence microscopy again revealed that delivery of pSur/AS-Sur significantly increased the number of AVOs present in cells, indicating the induction of autophagy, further supporting that delivery of pSur/AS-Sur can modulate autophagy in survivin-expressing cancer cells (Figure 4C).
Figure 4.
Delivery of pSur/AS-Sur modulates autophagy in cancer cells.
Notes: (A) Cancer cells were transfected with either empty plasmid or pSur/AS-Sur for the indicated durations. The conversion of LC3B-II and the expression of γH2AX were examined by Western blotting. Equal protein loading was verified by actin. The numbers under each blot are the intensities of the blot relative to that of the control (empty plasmid). (B) Cancer cells were transfected with EGFP-tagged LC3B with or without pSur/AS-Sur cotransfection for 48 hours. LC3 puncta formation in cells (~200 cells) was observed under a fluorescence microscope. The number of puncta present in cells was analyzed using the ImageJ software. (C) Cancer cells were transfected with pSur/AS-Sur for 48 hours and then stained with MDC. The number of puncta present in cells (~200 cells) was analyzed using the ImageJ software. “*,” “**,” and “***” denote statistical significance with P-values <0.05, <0.01, and <0.001, respectively, between the testing groups.
Abbreviations: h, hours; MDC, monodansylcadaverine; pSur/AS-Sur, survivin promoter-driven antisense survivin.
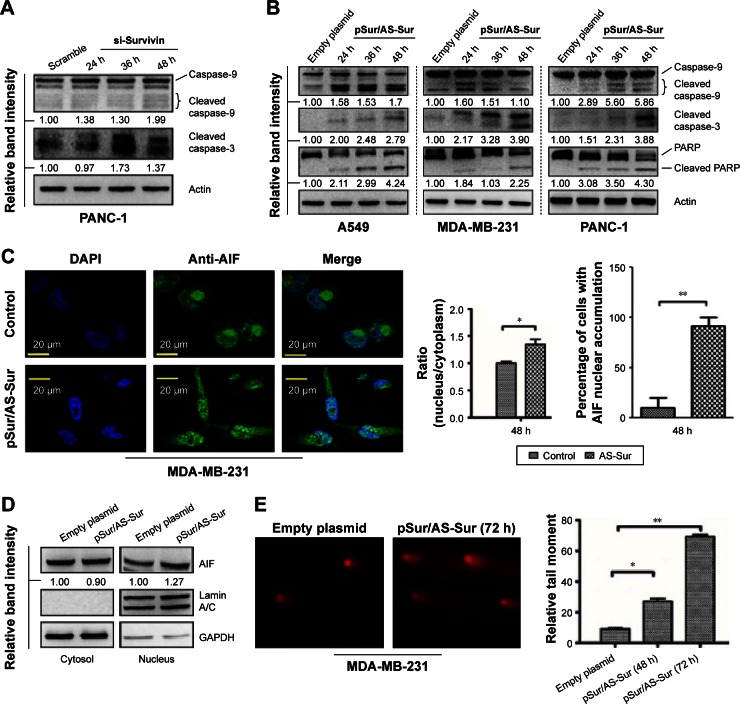
Delivery of pSur/AS-Sur induces caspase-9/-3 activation and apoptosis-inducing factor nuclear translocation in cancer cells
Survivin inhibits apoptosis, and downregulation of survivin triggers caspase-9 and caspase-3 activation, leading to the induction of apoptotic cell death.37 Therefore, to determine whether delivery of pSur/AS-Sur can induce apoptosis in cancer cells, Western blot analysis was performed to detect the expression of the cleaved caspase-3 and caspase-9, which are the molecular markers for the induction of intrinsic apoptosis. Downregulation of survivin by siRNA clearly induced caspase-9 and caspase-3 cleavage in PANC-1 cells, as revealed by the Western blot analysis as expected (Figure 5A). Similar to these results, delivery of pSur/AS-Sur also increased the amount of both the cleaved caspase-9 and caspase-3 present in A549, PANC-1, and MDA-MB-231 cells in a time-dependent manner (Figure 5B). Poly(ADP-ribose) polymerase-1 (PARP-1) plays an important role in DNA repair and apoptosis and serves as one of distinguishing feature of apoptosis and caspase activation.38,39 Western blot analysis further revealed that delivery of pSur/AS-Sur induced PARP cleavage in A549, MDA-MB-231, and PANC-1 cells in a time-dependent manner, supporting that delivery of pSur/AS-Sur can induce caspase-9/-3-involved intrinsic apoptosis in survivin-expressing cancer cells (Figure 5B).
Figure 5.
Delivery of pSur/AS-Sur induces both caspase-dependent and caspase-independent apoptosis in cancer cells.
Notes: (A) PANC-1 cells were transfected with either scramble siRNA or survivin siRNA for the indicated durations. Caspase-9 and caspase-3 cleavages were determined by Western blotting. The numbers under each blot are the intensities of the blot relative to that of the control (scramble). (B) Cancer cells were transfected with either empty plasmid or pSur/AS-Sur for the indicated durations. The expression of the cleaved caspase-9, caspase-3, and PARP was determined by Western blotting. Equal protein loading was verified by actin. The numbers under each blot are the intensities of the blot relative to that of the control (empty plasmid). (C) MDA-MB-231 cells were transfected with pSur/AS-Sur for 48 hours, and AIF (green fluorescent) translocation was determined by immunofluorescence microscopy. Nuclei were countered stained blue with DAPI. The localization of AIF in the nucleus of confocal images was pixel-by-pixel analyzed by the SigmaPlot software (version 10, Systat Software Inc., CA, USA). “*” denotes statistical significance with a P-value <0.05 between the testing groups. “**” denotes statistical significance of P-value <0.01. (D) Nucleic proteins and cytoplasmic proteins were isolated using the cells fractionation assay. Expression of AIF was analyzed by Western blotting. Equal protein loading was verified by either lamin A/C or GAPDH. The numbers under each blot are the intensities of the blot relative to that of the control (empty plasmid). (E) Cells were transfected with either empty plasmid or pSur/AS-Sur for 48–72 hours. DNA damage was detected using the comet assay. A statistically significant difference in the relative tail moment of cells transfected with empty plasmid versus pSur/AS-Sur is denoted by either “*” or “**”. *P<0.05 and **P<0.01.
Abbreviations: siRNA, small interfering RNA; DAPI, 4′,6-diamidino-2-phenylindole; h, hours; PAR, Poly (ADP-ribose) polymerase; AIF, apoptosis-inducing factor; pSur/AS-Sur, survivin promoter-driven antisense survivin.
Besides caspase-dependent apoptosis, survivin can also inhibit caspase-independent apoptosis through interference with the translocation of apoptosis-inducing factor (AIF) from cytoplasm to the nucleus.40,41 Nuclear translocation of AIF has been shown to play an important role in triggering the caspase-independent apoptosis by inducing caspase-independent large-scale DNA fragmentation in cells. To determine whether delivery of pSur/AS-Sur can induce caspase-independent apoptosis, immunofluorescence microscopy and Western blot analysis were performed to detect the intracellular location of AIF in cells. As shown in Figure 5C and D, delivery of pSur/AS-Sur significantly increased the amount of AIF present in the nucleus of MDA-MB-231 cells. Taken together, these results further support that delivery of pSur/AS-Sur can inhibit survivin expression and trigger both caspase-dependent and caspase-independent apoptosis in cancer cells.
Finally, we applied the comet assay to determine whether transfection of pSur/AS-Sur would induce DNA damage, given that caspases activation and AIF nuclear translocation can trigger small-scale DNA fragmentation and large-scale DNA fragmentation, respectively. As shown in Figure 5D, the amount of DNA in comet tail was increased in MDA-MB-231 cells delivered with pSur/AS-Sur, supporting that downregulation of survivin by pSur/AS-Sur can trigger severe DNA damage in survivin-expressing cancer cells.
Discussion
In this study, the ability of pSur/AS-Sur in downregulating survivin expression and inducing cancer cell death was evaluated in A549 (human lung adenocarcinoma), PANC-1 (human pancreatic carcinoma), and MDA-MB-231 (human breast adenocarcinoma) cancer cells in vitro. MDA-MB-231 is a p53-mutant expressing (p53Mut), triple-negative [ER−, human epidermal growth factor receptor 2 (HER2−), progesterone receptor (PR−)], and metastatic aggressive breast cancer cell, which possesses the characteristic and properties of one of the most difficultly treated subtypes of breast cancer in clinical situations. Both the widely used selective estrogen receptor modulator, tamoxifen (Nolvadex®, AstraZeneca, London, UK), and the anti-HER2 monoclonal antibody, trastuzumab (Herceptin®, Genentech, San Francisco, CA, USA), are not applicable for use in treating the triple-negative breast cancers, and traditional chemotherapy remains the mainstay treatment for this type of breast cancers. PANC-1 is also a p53-mutant-expressing cancer cell, whereas A549 is a wild-type p53-expressing cancer cell. Results of this study clearly showed that delivery of pSur/AS-Sur is capable of downregulating survivin expression and inducing autophagy, caspase-involved apoptosis, and caspase-independent apoptosis in A549, MDA-MB-231, and PANC-1 cancer cells in vitro, suggesting that the anticancer activity of pSur/AS-Sur is probably not affected by the p53 status of the targeted cancer cells. Interestingly, delivery of pSur/AS-Sur is also capable of decreasing the proliferation of the MDR1-expressing multidrug-resistant KB-TAX50 oral cancer cells and the ER+ tamoxifen-resistant MCF7-TamC3 breast cancer cells in vitro, further indicating that delivery of pSur/AS-Sur can induce cancer cell death regardless of the tumor types and the MDR1 expression status.
The three-dimension confirmation of the single-stranded RNA oligonucleotides is different from the single-stranded DNA oligonucleotides, and this sometimes creates a thermodynamic challenge for binding of the DNA oligonucleotides onto the targeted mRNA.18 Mechanistically, the production of the single-stranded full-length antisense survivin RNA oligonucleotides by pSur/AS-Sur can potentially overcome the structural issue related to the thermodynamic challenge of using the traditional single-stranded DNA oligonucleotides. In addition, the full-length survivin ASOs expressed by pSur/AS-Sur should exhibit higher target specificity as compared with the previously developed short survivin ASO, which is already in Phase I/II clinical trials. In fact, SPC3042, a short (16 mer) survivin ASO that is currently in clinical trials, was found to induce cancer cells death through nonspecific binding with Bcl-2.17 In contrast, delivery of pSur/AS-Sur did not affect the expression of Bcl-2 in the tested cancer cell lines, suggesting that the full-length antisense survivin oligomers expressed by pSur/AS-Sur may exhibit higher target specificity toward survivin as compared with other short antisense survivin-targeting oligomers that are currently undergoing clinical trials.
Conclusion
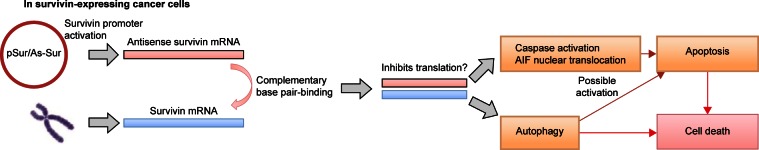
In summary, results of this study confirm that survivin-expressing cancer cells can activate the survivin promoter, which is located on pSur/AS-Sur, to drive the expression of the full-length antisense survivin gene in vitro. Importantly, the results of this study also suggest that delivery of the survivin promoter-driven antisense survivin-expressing plasmid DNA is a promising way to target survivin and to treat survivin-expressing cancers in the future (Figure 6).
Figure 6.
Schematic diagram showing the proposed mechanism of action of pSur/AS-Sur in survivin-expressing cancer cells.
Abbreviations: mRNA, messenger RNA; AIF, apoptosis-inducing factor; pSur/AS-Sur, survivin promoter-driven antisense survivin.
Acknowledgments
This work is kindly supported by the following grants: MOST 104-2320-B-006-029 (Ministry of Science and Technology, Taiwan ROC) and NHRI-EX105-10237SC (National Health Research Institutes, Taiwan ROC). We also thank the technical services provided by the Bio-image Core Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology, Taiwan.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 2.Ryan BM, Konecny GE, Kahlert S, et al. Survivin expression in breast cancer predicts clinical outcome and is associated with HER2, VEGF, urokinase plasminogen activator and PAI-1. Ann Oncol. 2006;17(4):597–604. doi: 10.1093/annonc/mdj121. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki H, Toyoda M, Shinohara H, et al. Expression of survivin correlates with apoptosis, proliferation, and angiogenesis during human colorectal tumorigenesis. Cancer. 2001;91(11):2026–2032. doi: 10.1002/1097-0142(20010601)91:11<2026::aid-cncr1228>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 4.Olie RA, Simoes-Wust AP, Baumann B, et al. A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res. 2000;60(11):2805–2809. [PubMed] [Google Scholar]
- 5.Satoh K, Kaneko K, Hirota M, Masamune A, Satoh A, Shimosegawa T. Expression of survivin is correlated with cancer cell apoptosis and is involved in the development of human pancreatic duct cell tumors. Cancer. 2001;92(2):271–278. doi: 10.1002/1097-0142(20010715)92:2<271::aid-cncr1319>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Gu X, Lin HL. Analysis of survivin expression in subtypes of lymphoma. Ai Zheng. 2004;23(6):655–661. Chinese. [PubMed] [Google Scholar]
- 7.Sarela AI, Macadam RC, Farmery SM, Markham AF, Guillou PJ. Expression of the antiapoptosis gene, survivin, predicts death from recurrent colorectal carcinoma. Gut. 2000;46(5):645–650. doi: 10.1136/gut.46.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaffaroni N, Daidone MG. Survivin expression and resistance to anticancer treatments: perspectives for new therapeutic interventions. Drug Resist Updat. 2002;5(2):65–72. doi: 10.1016/s1368-7646(02)00049-3. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Ma X, Wu X, Liu X, Liu L. Prognostic significance of survivin in breast cancer: meta-analysis. Breast J. 2014;20(5):514–524. doi: 10.1111/tbj.12303. [DOI] [PubMed] [Google Scholar]
- 10.Goossens-Beumer IJ, Zeestraten EC, Benard A, et al. Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. Br J Cancer. 2014;110(12):2935–2944. doi: 10.1038/bjc.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Li T, Liu Q, et al. Clinical and prognostic significance of HIF-1alpha, PTEN, CD44v6, and survivin for gastric cancer: a meta-analysis. PLoS One. 2014;9(3):e91842. doi: 10.1371/journal.pone.0091842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selemetjev S, Dencic TI, Marecko I, et al. Evaluation of survivin expression and its prognostic value in papillary thyroid carcinoma. Pathol Res Pract. 2014;210(1):30–34. doi: 10.1016/j.prp.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Cao M, Yie SM, Wu SM, et al. Detection of survivin-expressing circulating cancer cells in the peripheral blood of patients with esophageal squamous cell carcinoma and its clinical significance. Clin Exp Metastasis. 2009;26(7):751–758. doi: 10.1007/s10585-009-9274-7. [DOI] [PubMed] [Google Scholar]
- 14.Yie SM, Lou B, Ye SR, et al. Clinical significance of detecting survivin-expressing circulating cancer cells in patients with non-small cell lung cancer. Lung Cancer. 2009;63(2):284–290. doi: 10.1016/j.lungcan.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 15.Carrasco RA, Stamm NB, Marcusson E, Sandusky G, Iversen P, Patel BK. Antisense inhibition of survivin expression as a cancer therapeutic. Mol Cancer Ther. 2011;10(2):221–232. doi: 10.1158/1535-7163.MCT-10-0756. [DOI] [PubMed] [Google Scholar]
- 16.Iwai M, Minematsu T, Li Q, Iwatsubo T, Usui T. Utility of P-glycoprotein and organic cation transporter 1 double-transfected LLC-PK1 cells for studying the interaction of YM155 monobromide, novel small-molecule survivin suppressant, with P-glycoprotein. Drug Metab Dispos. 2011;39(12):2314–2320. doi: 10.1124/dmd.111.040733. [DOI] [PubMed] [Google Scholar]
- 17.Hansen JB, Fisker N, Westergaard M, et al. SPC3042: a proapoptotic survivin inhibitor. Mol Cancer Ther. 2008;7(9):2736–2745. doi: 10.1158/1535-7163.MCT-08-0161. [DOI] [PubMed] [Google Scholar]
- 18.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1(5):347–355. [PubMed] [Google Scholar]
- 19.Liu C, Sun B, An N, et al. Inhibitory effect of Survivin promoter-regulated oncolytic adenovirus carrying P53 gene against gallbladder cancer. Mol Oncol. 2011;5(6):545–554. doi: 10.1016/j.molonc.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang R, Lin F, Wang X, et al. The therapeutic potential of survivin promoter-driven siRNA on suppressing tumor growth and enhancing radiosensitivity of human cervical carcinoma cells via downregulating hTERT gene expression. Cancer Biol Ther. 2007;6(8):1301–1307. doi: 10.4161/cbt.6.8.4505. [DOI] [PubMed] [Google Scholar]
- 21.Zhang HZ, Wang Y, Gao P, et al. Silencing stathmin gene expression by survivin promoter-driven siRNA vector to reverse malignant phenotype of tumor cells. Cancer Biol Ther. 2006;5(11):1457–1461. doi: 10.4161/cbt.5.11.3272. [DOI] [PubMed] [Google Scholar]
- 22.Leung E, Kannan N, Krissansen GW, Findlay MP, Baguley BC. MCF-7 breast cancer cells selected for tamoxifen resistance acquire new phenotypes differing in DNA content, phospho-HER2 and PAX2 expression, and rapamycin sensitivity. Cancer Biol Ther. 2010;9(9):717–724. doi: 10.4161/cbt.9.9.11432. [DOI] [PubMed] [Google Scholar]
- 23.Kuo CC, Hsieh HP, Pan WY, et al. BPR0L075, a novel synthetic indole compound with antimitotic activity in human cancer cells, exerts effective antitumoral activity in vivo. Cancer Res. 2004;64(13):4621–4628. doi: 10.1158/0008-5472.CAN-03-3474. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh CC, Kuo YH, Kuo CC, et al. Chamaecypanone C, a novel skeleton microtubule inhibitor, with anticancer activity by trigger caspase 8-Fas/FasL dependent apoptotic pathway in human cancer cells. Biochem Pharmacol. 2010;79(9):1261–1271. doi: 10.1016/j.bcp.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 25.Leung EY, Kim JE, Askarian-Amiri M, Joseph WR, McKeage MJ, Baguley BC. Hormone resistance in two MCF-7 breast cancer cell lines is associated with reduced mTOR signaling, decreased glycolysis, and increased sensitivity to cytotoxic drugs. Front Oncol. 2014;4:221. doi: 10.3389/fonc.2014.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- 27.Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5(4):e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pei H, Li L, Fridley BL, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16(3):259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skrzypczak M, Goryca K, Rubel T, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu N, Clifford RJ, Yang HH, et al. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576. doi: 10.1186/1471-2164-11-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dohi T, Okada K, Xia F, et al. An IAP-IAP complex inhibits apoptosis. J Biol Chem. 2004;279(33):34087–34090. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- 33.Cheng SM, Chang YC, Liu CY, et al. YM155 down-regulates survivin and XIAP, modulates autophagy and induces autophagy-dependent DNA damage in breast cancer cells. Br J Pharmacol. 2015;172(1):214–234. doi: 10.1111/bph.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahboubi K, Li F, Plescia J, et al. Interleukin-11 up-regulates survivin expression in endothelial cells through a signal transducer and activator of transcription-3 pathway. Lab Invest. 2001;81(3):327–334. doi: 10.1038/labinvest.3780241. [DOI] [PubMed] [Google Scholar]
- 35.Vequaud E, Seveno C, Loussouarn D, et al. YM155 potently triggers cell death in breast cancer cells through an autophagy-NF-kB network. Oncotarget. 2015;6(15):13476–13486. doi: 10.18632/oncotarget.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JYC, Kuo CW, Tsai SL, et al. Inhibition of HDAC3- and HDAC6-Promoted survivin expression plays an important role in SAHA-induced autophagy and viability reduction in breast cancer cells. Front Pharmacol. 2016;7:81. doi: 10.3389/fphar.2016.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dohi T, Beltrami E, Wall NR, Plescia J, Altieri DC. Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J Clin Invest. 2004;114(8):1117–1127. doi: 10.1172/JCI22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Zhao S, Song J. Caspase-dependent apoptosis and -independent poly(ADP-ribose) polymerase cleavage induced by transforming growth factor beta1. Int J Biochem Cell Biol. 2004;36(2):223–234. doi: 10.1016/s1357-2725(03)00215-2. [DOI] [PubMed] [Google Scholar]
- 39.Soldani C, Lazze MC, Bottone MG, et al. Poly(ADP-ribose) polymerase cleavage during apoptosis: when and where? Exp Cell Res. 2001;269(2):193–201. doi: 10.1006/excr.2001.5293. [DOI] [PubMed] [Google Scholar]
- 40.Cande C, Cohen I, Daugas E, et al. Apoptosis-inducing factor (AIF): a novel caspase-independent death effector released from mitochondria. Biochimie. 2002;84(2–3):215–222. doi: 10.1016/s0300-9084(02)01374-3. [DOI] [PubMed] [Google Scholar]
- 41.Croci DO, Cogno IS, Vittar NB, et al. Silencing survivin gene expression promotes apoptosis of human breast cancer cells through a caspase-independent pathway. J Cell Biochem. 2008;105(2):381–390. doi: 10.1002/jcb.21836. [DOI] [PubMed] [Google Scholar]