ABSTRACT
Sleep is essential for health and cognition, but the molecular and neural mechanisms of sleep regulation are not well understood. We recently reported the identification of TARANIS (TARA) as a sleep-promoting factor that acts in a previously unknown arousal center in Drosophila. tara mutants exhibit a dose-dependent reduction in sleep amount of up to ∼60%. TARA and its mammalian homologs, the Trip-Br (Transcriptional Regulators Interacting with PHD zinc fingers and/or Bromodomains) family of proteins, are primarily known as transcriptional coregulators involved in cell cycle progression, and contain a conserved Cyclin-A (CycA) binding homology domain. We found that tara and CycA synergistically promote sleep, and CycA levels are reduced in tara mutants. Additional data demonstrated that Cyclin-dependent kinase 1 (Cdk1) antagonizes tara and CycA to promote wakefulness. Moreover, we identified a subset of CycA expressing neurons in the pars lateralis, a brain region proposed to be analogous to the mammalian hypothalamus, as an arousal center. In this Extra View article, we report further characterization of tara mutants and provide an extended discussion of our findings and future directions within the framework of a working model, in which a network of cell cycle genes, tara, CycA, and Cdk1, interact in an arousal center to regulate sleep.
Keywords: behavior, Cdk1, cell cycle genes, CycA, Drosophila, pars lateralis, sleep, TARANIS
Of all the behaviors required for survival, sleep is one of the most time-consuming, and its loss is linked to many deleterious effects on human health.1,2 It has been demonstrated that people with disrupted sleep schedules, such as shift workers, have an increased risk of cancer, heart disease and diabetes.3,4 Sleep deprivation also impairs cognitive and motor functions.5 Although several theories have been proposed,6-8 the functions of sleep are not yet clear. Identification of genes and neural circuits that control sleep may facilitate elucidation of sleep function.
The Drosophila model for sleep is well suited for discovering sleep regulatory genes through genetic screens. We recently reported the isolation of taranis (tara) from an unbiased genome-wide forward-genetic screen for short-sleeping mutants.9 Mutations in tara resulted in a reduction of total sleep amount due to fewer and shorter sleep bouts, suggesting that loss of tara leads to defects in sleep initiation and maintenance. We found that TARA is expressed widely in neurons and the short-sleeping phenotype of tara mutants can be fully rescued with constitutive and ubiquitous expression of tara. Importantly, adult-specific pan-neuronal expression of tara partially rescued the sleep phenotype, which suggests that TARA has both adult and developmental roles in sleep regulation.
Sleep is controlled mainly by two mechanisms: a circadian mechanism that consolidates sleep to an ecologically relevant time of day and a homeostatic mechanism that ensures an adequate amount of sleep is achieved.10 We examined the free-running locomotor rhythms of tara mutants in constant darkness (DD), and found that most of the severe tara mutants were arrhythmic.9 However, across multiple allelic combinations, the severity of sleep reduction and the degree of arrhythmicity were not highly correlated. Moreover, tara mutants exhibited reduced sleep compared with controls in constant light (LL), which renders both control and mutant flies arrhythmic, demonstrating that the short-sleeping phenotype is not secondary to arrhythmicity. tara mutants also exhibited reduced sleep in DD, suggesting that the role of TARA in sleep is independent of light. In both LL and DD, severe tara mutants lost over 80% of sleep relative to control flies, which is one of the strongest phenotypes documented among sleep mutants. Together, our data suggest that tara regulates sleep amount independently of the circadian mechanism and the light input pathways. These observations leave a defective homeostatic mechanism as the probable cause of reduced sleep in tara mutants. In future studies, we will investigate whether and how TARA controls sleep homeostasis.
To further characterize tara mutant phenotypes, we examined several additional behaviors. First, we found that tara mutants were more likely to wake up in response to brief dim light than control flies (Fig. 1A), which suggests that tara mutants may be more easily aroused, although it is possible that tara mutants are more sensitive to light. Our finding is consistent with previous findings that most short-sleeping flies have lowered arousal threshold,11 and demonstrate that tara mutants can detect dim light. Next, since sleep deprivation can lead to early lethality in flies as well as mammals,12,13 we measured the lifespan of tara mutants. We found that tara mutants had a shorter lifespan compared with control flies (Fig. 1B), suggesting that reduced sleep in tara mutants has consequences for overall fitness, although we cannot rule out the possibility that TARA influences longevity independently of its effect on sleep.14 Like another short-sleeping mutant, sleepless (sss),15 tara mutants could not climb as well as control flies (Fig. 1C). However, despite their climbing defects, tara mutants displayed increased locomotor activity compared with controls, and behaved normally in other behavioral assays. They exhibited neither ether-induced leg shaking nor bang-sensitive paralysis, and performed normally in a taste discrimination assay (Fig. 1D). Altogether, our data suggest that while loss of TARA leads to behavioral deficits often associated with reduced sleep, it has little effect on other behaviors.
Figure 1.
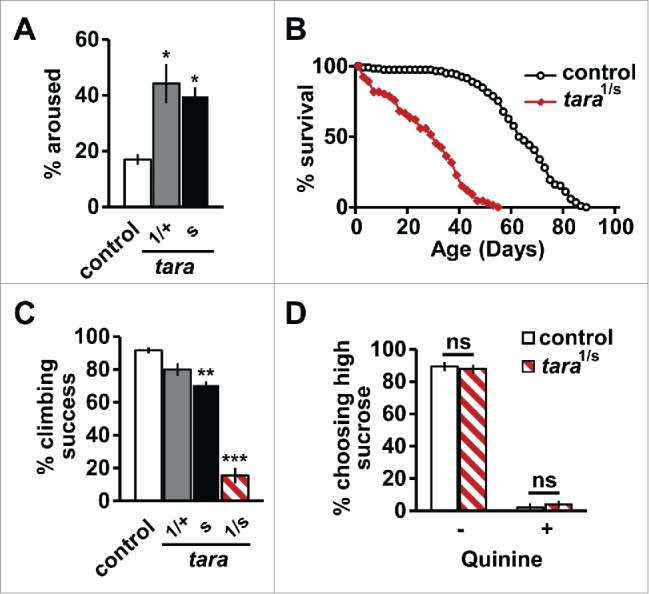
Behavioral phenotypes of tara mutants. (A) Percentage of control and tara flies (n=31−64) that were awakened by a 1 sec pulse of 100 lux light delivered at ZT16. Only flies that were asleep prior to the light pulse are included. (B) Survivorship curves of female control and tara1/s132 flies (n = 66−118). (C) Percentage of control, tara1/+, taras132, and tara1/s132 flies (n = 37−50) that crossed a 10 cm mark within 10 sec against gravity. (D) Percentage of control and tara1/s132 flies (n = 46−68) that chose food with 25 mM sucrose, in the absence or presence of 3 mM quinine, over 5 mM sucrose. Control and tara flies showed an equivalent preference for a higher concentration of sugar and an equivalent avoidance of bitter tasting quinine. Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001, Chi-square test (A, D), log rank test (B), and one-way ANOVA followed by Dunnett post hoc test relative to control flies (C).
TARA contains a conserved Cyclin-A (CycA) binding homology domain, and CycA was previously shown to promote sleep.16 These observations led us to hypothesize that tara interacts with CycA to regulate sleep. Using multiple alleles and RNAi-mediated knockdown, we demonstrated that tara and CycA indeed synergistically interact to promote sleep. Our finding that TARA::GFP fusion protein is enriched in neuronal nuclei 9 is consistent with the previously described role for TARA as a transcriptional co-regulator.17 However, TARA physically binds CycA and regulates its levels at the post-transcriptional level.9 Interestingly, the TRIP-Br1 protein, one of the mammalian homologs of TARA, is enriched in the cytoplasm of mammalian cells.18 Thus, although TARA is expressed mainly in the nucleus, a small pool of TARA may also localize to the cytoplasm. These observations suggest the possibility that TARA regulates sleep through a non-transcriptional mechanism independent of the transcriptional mechanism controlling cell cycle progression.
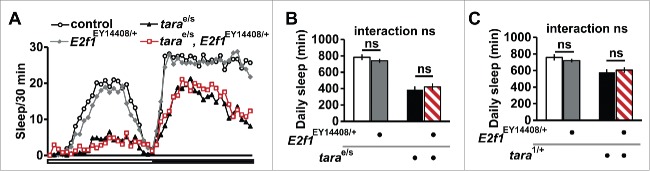
Although TARA may exert its effect on sleep entirely through post-transcriptional mechanisms, it is possible that at least some of the effect of TARA on sleep is through transcriptional regulation. TARA and the Trip-Br family of proteins have been shown to act as coregulators of the E2F1-DP1 transcription complex.19-22 However, we did not find evidence for a genetic interaction between E2f1 and tara for sleep regulation (Fig. 2). TARA may partner with different transcription factors depending on the biological context such as sleep regulation versus cell cycle progression.
Figure 2.
tara and E2f1 appear not to interact for sleep regulation. (A) Sleep profile of background control (white circles), E2f1EY14408/+ (gray diamonds), tarae01264/s132 (black triangles), and E2f1EY14408/+, tarae01264/s132 (open red squares) female flies (n=17−21) in 30 min bins. The white and black bars below the X-axis represent 12 h light and 12 h dark periods, respectively. (B) Total daily sleep amount for the same genotypes indicated in (A). (C) Total daily sleep of the indicated genotypes (n = 16 for all genotypes). Mean ± SEM is shown. ns: not significant, 2-way ANOVA followed by Tukey post hoc test (B, C).
Cyclins regulate cell cycle progression through their modulation of Cyclin-dependent kinases (Cdks). Previous work showed that CycA can physically interact with Cdk1,23 which raises the possibility that Cdk1 may have a role in sleep as well. Indeed our data suggest that CycA regulates sleep through its modulatory action over Cdk1 activity. We found that reduced Cdk1 activity partially rescued the short-sleeping phenotypes of tara and CycA mutants,9 suggesting that Cdk1 is a wake-promoting molecule. Since Cdk1 is regulated through inhibitory phosphorylation of its T14 and Y15 residues, we employed a mutant Cdk1-AF (T14A, Y15F) that cannot be inhibited 24 to show that increased activity of Cdk1 suppresses sleep. Given that CycA and Cdk1 are known to physically interact,23 a direct relationship between CycA and Cdk1 for sleep regulation is likely. Interestingly, we found that both CycA and Cdk1 localize to synaptic regions, which suggests a modulatory role for CycA and Cdk1 over synaptic proteins. Identification of the substrates of the Cdk1 kinase activity relevant for sleep regulation is an important next step we intend to pursue in future experiments.
Recent work in Drosophila has demonstrated that knockdown of Cdk1 significantly reduces seizure duration in both bang sensitive (bas) and bang senseless (bss) mutants,25 which suggests that Cdk1 may modulate ion channel activity and membrane excitability. Several lines of evidence show that ion channels have a dramatic influence over sleep. Shaker, hyperkinetic, ether-à-go-go, redeye, and Rdl genes, which encode a fast delayed rectifier potassium channel,26 cytoplasmic β subunit of Shaker,27 slow delayed rectifier potassium channel,28 nicotinic acetylcholine receptor,29 and GABAA receptor,30,31 respectively, are all implicated in sleep and may be potential phosphorylation targets of Cdk1.
Previous work showed that CycA protein is expressed in a small number of neuronal clusters including ∼14 neurons in the pars lateralis (PL),16 a brain region that together with the pars intercerebralis (PI) is thought to be analogous to the mammalian hypothalamus. In order to manipulate the CycA expressing cells, we made use of a Gal4 driver 32 that labels just the dorsal cluster of CycA expressing cells. Activation of these neurons led to strong sleep suppression, suggesting that they serve as an arousal center.9 Importantly, tara knockdown or Cdk1-AF expression, specifically in PL neurons, also suppressed sleep, suggesting that TARA, CycA, and Cdk1 interact in these neurons to control sleep. Given that increased Cdk1 activity in PL neurons phenocopies activation of those neurons, we propose a model in which TARA upregulates CycA levels to inhibit Cdk1, whose kinase activity increases neuronal excitability of wake-promoting PL neurons (Fig. 3). Whether Cdk1 activity leads to an overall increase in the excitability of PL neurons is an interesting question for future studies.
Figure 3.
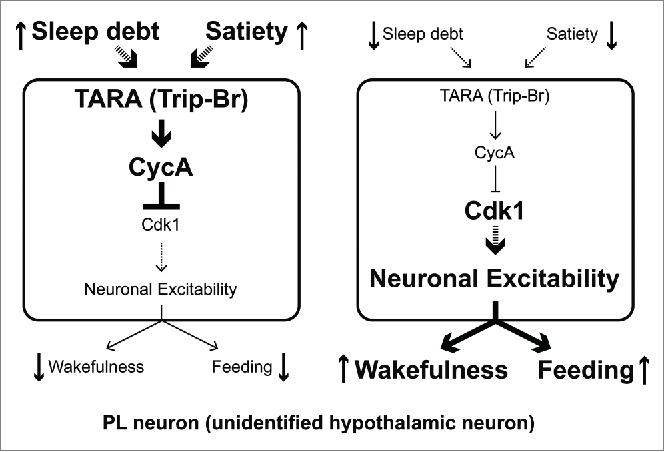
Working model of how TARA promotes sleep. TARA upregulates CycA, which negatively regulates Cdk1 activity in the PL neurons. We propose that increased activity of Cdk1 leads to an increase in the excitability of PL neurons, which promotes wakefulness and feeding. We further speculate that TARA levels and/or activity are modulated by sleep debt and satiety. The two diagrams depict PL neurons when flies have high sleep debt and satiety (left) and when they have low sleep debt and satiety (right), respectively. The mammalian counterparts are indicated in parentheses, and broken lines represent connections that need further investigation.
How TARA is regulated is another interesting question. We did not observe any changes in TARA levels in circadian pacemaker neurons across the day (Fig. 4), but it is possible that TARA levels in PL neurons fluctuate depending on the sleep-wake history. Alternatively, TARA activity rather than its abundance may be under circadian or homeostatic control. Clues to a potential regulator of TARA come from the fact that the PL-Gal4 driver was generated using a fragment of the corazonin (crz) promoter.32 Previous studies found that activation of CRZ neurons using a Gal4 driver that contains the full crz promoter increases food consumption in starved flies,33 and that a subpopulation of PL neurons express Gustatory Receptor 43a (GR43a), which functions as a nutrient sensor.34,35 Although the full crz promoter drives expression in a few neuronal groups outside the PL region, it is plausible that PL neurons themselves are involved in the regulation of starvation response. Starved flies sleep less, presumably to forage for food.36 Moreover, Trip-Br2 is involved in fat metabolism 37 and Trip-Br1 functions in pancreatic β-cells to regulate insulin secretion.38 We speculate that TARA functions in PL neurons to coordinately regulate sleep and feeding in response to metabolic as well as sleep-related signals (Fig. 3). Interestingly, neurons expressing Diuretic Hormone 44 (DH44) in the PI have been implicated in both the regulation of activity-rest rhythms 39 and the detection and consumption of nutritive sugars,40 which suggests that multiple neuronal groups may be involved in the coordination of sleep and metabolism. Both PL and PI regions are proposed to be analogous to the mammalian hypothalamus,41 a major sleep and feeding center.42,43 It may be that an unidentified subpopulation of the hypothalamic neurons function in a manner analogous to PL neurons to integrate sleep and metabolic signals.
Figure 4.
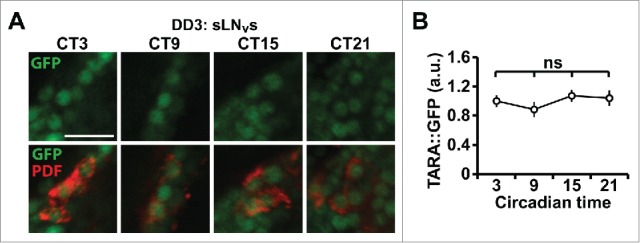
TARA protein levels do not cycle in circadian pacemaker neurons. (A) Immunostaining of TARA::GFP in male fly brains on the 3rd day in DD. We used transgenic flies that carry an artificial exon encoding GFP inserted into an intron of tara in the genome 52 and therefore are expected to produce endogenous levels of TARA protein fused to GFP. Brains were dissected at indicated circadian times (CT) and stained for GFP (green) and PDF (red), which was used to identify small ventral lateral neurons (sLNvs), the pacemaker neurons in DD. Scale bar 10µm. (B) Quantification of TARA::GFP signal in sLNvs. Data from 11−16 brain hemispheres are presented. Mean ± SEM is shown. ns: not significant, 2-way ANOVA followed by Tukey post hoc test (B).
Pan-neuronal knockdown of tara had a stronger effect on sleep than tara knockdown restricted to PL neurons,9 which suggests that TARA also acts in other neuronal groups. A number of neuronal populations have been implicated in the regulation of sleep. These include the mushroom body,44-46 the fan shaped body,47 the PI,48 octopaminergic neurons,48 and the large ventral lateral clock neurons.30,49 Knockdown of tara in these neuronal groups did not result in any significant changes in sleep amount.9 Further investigation of the anatomical loci of TARA function may reveal additional sleep-relevant neuronal populations.
A number of Drosophila sleep factors have been identified in recent years (Table 1), but TARA is particularly interesting because it forms a sleep-regulatory gene network with other cell cycle genes, and functions in an arousal center previously unknown for its role in sleep regulation. Interestingly, several studies have shown that cell cycle regulators have additional functions in adult neurons. For instance, Cyclin E plays a role in memory formation and synaptic plasticity in mice 50; knockdown of several Cyclin/Cdk family members rescues the seizure phenotype of bas and bss mutants in Drosophila 25; and Cyclin-B1 is upregulated in the hypothalamus of patients afflicted with temporal lobe epilepsy.51 It is unknown whether Trip-Br proteins regulate sleep and wakefulness in mammals. Further studies of TARA and its mammalian homologs as well as the PL neurons and the neural circuit they participate in may provide valuable insights into the molecular and neural mechanisms of sleep regulation.
Table 1.
Genes involved in sleep regulation in Drosophila.
| Protein function | Gene | Reference |
|---|---|---|
| Neurotransmission | Dopamine transporter | Kume et al., 200553 |
| Dopamine 1-like receptor 1 | Ueno et al., 201254 | |
| 5-hydroxytryptamine (serotonin) receptor 1A | Yuan et al., 200655 | |
| Tyramine β hydroxylase | Crocker and Sehgal, 200856 | |
| Tyrosine decarboxylase 2 | Crocker and Sehgal, 200856 | |
| Octopamine receptor in mushroom bodies | Crocker et al., 201057 | |
| Resistant to dieldrin (GABAAreceptor) | Agosto et al., 2008; Chung et al., 201431,58 | |
| Wide awake | Liu et al., 201459 | |
| GABA transaminase | Maguire et al., 201560 | |
| nicotinic Acetylcholine Receptor α4 | Shi et al., 201429 | |
| nicotinic Acetylcholine Receptor α2 | Wu et al., 201461 | |
| NMDA receptor 1 | Tomita et al., 201562 | |
| Pigment-dispersing factor | Parisky et al., 200830 | |
| Ecdysone receptor | Ishimoto and Kitamoto, 201063 | |
| Sex Peptide | Isaac et al., 200964 | |
| Myoinhibiting peptide precursor | Oh et al., 201465 | |
| Sex peptide receptor | Oh et al., 201465 | |
| short neuropeptide F precursor | Shang et al., 201366 | |
| Diuretic hormone 31 | Kunst et al., 201467 | |
| SIFamide | Park et al., 201468 | |
| SIFamide receptor | Park et al., 201468 | |
| Ion channel signaling | Shaker | Cirelli et al., 200526 |
| Hyperkinetic | Bushey et al., 200769 | |
| quiver (sleepless) | Koh et al., 200815 | |
| Ca2+-channel protein α1 subunit T | Jeong et al., 201570 | |
| Calcineurin B | Nakai et al., 2011; Tomita et al., 201171,72 | |
| Calcineurin A1 | Nakai et al., 2011; Tomita et al., 201171,72 | |
| sarah | Nakai et al., 201172 | |
| Sulfonylurea receptor (ATP-sensitive potassium channel subunit) | Allebrandt et al., 201373 | |
| Transient receptor potential cation channel A1 ortholog | Roessingh et al., 201574 | |
| Cell cycle regulation | Cyclin A | Rogulja and Young, 201216 |
| Regulator of cyclin A1 | Rogulja and Young, 201216 | |
| taranis | Afonso et al., 20159 | |
| Cyclin-dependent kinase 1 | Afonso et al., 20159 | |
| Synaptic development | Fmr1 | Bushey et al., 200975 |
| homer | Naidoo et al., 201276 | |
| Neuroligin 4 | Li et al., 201377 | |
| Neurexin 1 | Larkin et al., 201578 | |
| Cellular signaling | Rolled (ERK) | Foltenyi et al., 2007; Vanderheyden et al., 2013)79,80 |
| Epidermal growth factor receptor | Foltenyi et al., 200780 | |
| spitz | Foltenyi et al., 200780 | |
| Star | Foltenyi et al., 200780 | |
| rhomboid | Foltenyi et al., 200780 | |
| Gold tip | Guo et al., 201181 | |
| Notch | Seugnet et al., 201182 | |
| Delta | Seugnet et al., 201182 | |
| bunched | Seugnet et al., 201182 | |
| basket | Takahama et al., 201283 | |
| foraging | Donlea et al., 201284 | |
| crossveinless c | Donlea et al., 201485 | |
| Metabolism | Insulin-like receptor | Metaxakis et al., 201486 |
| Ribosomal protein S6 kinase | Metaxakis et al., 201486 | |
| forkhead box, sub-group O | Metaxakis et al., 201486 | |
| Lipid storage droplet-2 | Thimgan et al., 201087 | |
| brummer | Thimgan et al., 201087 | |
| fatty acid binding protein | Gerstner et al., 201188 | |
| Circadian | period | Hendricks et al., 200089 |
| cycle | Hendricks et al., 2000; Shaw et al., 200212,90 | |
| Immune/stress response | Heat shock protein 83 | Shaw et al., 200212 |
| Relish | Williams et al., 200791 | |
| bipolar oocyte (bip) | Naidoo et al., 201276 | |
| Anaplastic lymphoma kinase | Bai and Sehgal, 201592 | |
| Protein degradation | Ubiquitin protein ligase E3A | Wu et al., 200893 |
| insomniac | Pfeiffenberger and Allada, 2012; Stavropoulos and Young, 201114,94 | |
| Learning and memory | Cyclic-AMP response element binding protein B | Hendricks et al., 200195 |
| dunce | Hendricks et al., 200195 | |
| rutabaga | Hendricks et al., 200195 | |
| Protein kinase, cAMP-dependent, regulatory subunit type 1 | Crocker and Sehgal, 200856 | |
| amnesiac | Liu et al., 200896 | |
| Other | Catecholamines up | Harbison et al., 200997 |
| Tat interactive protein 60kDa | Pirooznia et al., 201298 | |
| yellow-achaete intergenic RNA | Soshnev et al., 201199 | |
| Activating transcription factor-2 | Shimizu et al., 2008100 | |
| Adar | Robinson et al., 2016101 |
Experimental procedures
E2f1EY14408 was obtained from the Bloomington Stock Center and outcrossed to a w− isogenic background (iso31) for 5 generations. Homozygous E2f1EY14408 are lethal, suggesting that EY14408 is a null or a strong reduction of function allele. All other stocks were described previously.9 The sleep assay and whole-mount immunostaining of adult brains were performed as previously described.9 To assess arousability, flies were subjected to a 1 sec pulse of ∼100 lux light at Zeitgeber Time (ZT) 16. Only flies that were asleep at the time of light pulse were included in the data analysis, and the proportion of flies that started moving within the next 5 min was determined for each genotype. To determine longevity, tara1/s132 mutant and control flies were maintained in a 12 hr: 12 hr LD cycle at 25°C throughout their lifespan. Groups of ∼30 flies (∼15 males and ∼15 females) were collected into food vials within 2 d of eclosion. Males and females were kept together for 2 days, after which they were separated into groups of ∼30 females or males. Flies were transferred to fresh food every 2 days, and the number of dead flies counted. Climbing, leg shaking, bang sensitivity, and taste discrimination assays were performed as described,15 except that flies had to climb 10 cm within 10 sec to be counted as successful climbers and were allowed to feed for 30 min, and 5 or 25 mM sucrose and 3 mM quinine were used.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Bloomington Stock Center for fly stocks; Huihui Pan and Andrea Nam for technical assistance; and Alexandra Kenny and Arzu Ozturk Colak for comments on the manuscript.
Funding
This work was supported by a grant from the National Institutes of Health (R01NS086887 to K.K.) and predoctoral fellowships from the Portuguese Foundation for Science and Technology (SFRH/BD/51726/2011 to D.J.S.A and SFRH/BD/52321/2013 to D.R.M).
References
- 1.Crocker A, Sehgal A. Genetic analysis of sleep. Gen Dev 2010; 24:1220-35; PMID:20551171; http://dx.doi.org/ 10.1101/gad.1913110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palma J-A, Urrestarazu E, Iriarte J. Sleep loss as risk factor for neurologic disorders: A review. Sleep Med 2013; 14:229-36; PMID:23352029; http://dx.doi.org/ 10.1016/j.sleep.2012.11.019 [DOI] [PubMed] [Google Scholar]
- 3.Wang XS, Armstrong ME, Cairns BJ, Key TJ, Travis RC. Shift work and chronic disease: the epidemiological evidence. Occup Med 2011; 61:78-89; PMID:21355031; http://dx.doi.org/ 10.1093/occmed/kqr001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buxton OM, Cain SW, O'Connor SP, Porter JH, Duffy JF, Wang W, Czeisler CA, Shea SA. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med 2012; 4:129ra43; PMID:22496545; http://dx.doi.org/ 10.1126/scitranslmed.3003200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tefft BC. Prevalence of motor vehicle crashes involving drowsy drivers, United States, 1999−2008. Accid Anal Prev 2012; 45:180-6; PMID:22269499; http://dx.doi.org/ 10.1016/j.aap.2011.05.028 [DOI] [PubMed] [Google Scholar]
- 6.Tononi G, Cirelli C. Sleep and the price of plasticity: from synaptic and cellular homeostasis to memory consolidation and integration. Neuron 2014; 81:12-34; PMID:24411729; http://dx.doi.org/ 10.1016/j.neuron.2013.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O'Donnell J, Christensen DJ, Nicholson C, Iliff JJ, et al.. Sleep drives metabolite clearance from the adult brain. Science 2013; 342:373-7; PMID:24136970; http://dx.doi.org/ 10.1126/science.1241224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt MH. The energy allocation function of sleep: a unifying theory of sleep, torpor, and continuous wakefulness. Neuroscience and biobehavioral reviews 2014; 47:122-53; PMID:25117535; http://dx.doi.org/ 10.1016/j.neubiorev.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 9.Afonso DJ, Liu D, Machado DR, Pan H, Jepson JE, Rogulja D, Koh K. TARANIS Functions with Cyclin A and Cdk1 in a Novel Arousal Center to Control Sleep in Drosophila. Curr Biol 2015; 25:1717-26; PMID:26096977; http://dx.doi.org/ 10.1016/j.cub.2015.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms 1999; 14:557-68; PMID:10643753 [DOI] [PubMed] [Google Scholar]
- 11.Wu MN, Koh K, Yue Z, Joiner WJ, Sehgal A. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep 2008; 31:465-72; PMID:18457233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 2002; 417:287-91; PMID:12015603; http://dx.doi.org/ 10.1038/417287a [DOI] [PubMed] [Google Scholar]
- 13.Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. Physiological correlates of prolonged sleep deprivation in rats. Science 1983; 221:182-4; PMID:6857280; http://dx.doi.org/ 10.1126/science.6857280 [DOI] [PubMed] [Google Scholar]
- 14.Stavropoulos N, Young MW. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 2011; 72:964-76; PMID:22196332; http://dx.doi.org/ 10.1016/j.neuron.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science 2008; 321:372-6; PMID:18635795; http://dx.doi.org/ 10.1126/science.1155942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogulja D, Young MW. Control of sleep by cyclin A and its regulator. Science 2012; 335:1617-21; PMID:22461610; http://dx.doi.org/ 10.1126/science.1212476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calgaro S, Boube M, Cribbs DL, Bourbon HM. The Drosophila gene taranis encodes a novel trithorax group member potentially linked to the cell cycle regulatory apparatus. Genetics 2002; 160:547-60; PMID:11861561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zang ZJ, Gunaratnam L, Cheong JK, Lai LY, Hsiao L-L, O'Leary E, Sun X, Salto-Tellez M, Bonventre JV, Hsu SIH. Identification of PP2A as a novel interactor and regulator of TRIP-Br1. Cellular signalling 2009; 21:34-42; PMID:18940248; http://dx.doi.org/ 10.1016/j.cellsig.2008.09.018 [DOI] [PubMed] [Google Scholar]
- 19.Hsu SI, Yang CM, Sim KG, Hentschel DM, O'Leary E, Bonventre JV. TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. EMBO J 2001; 20:2273-85; PMID:11331592; http://dx.doi.org/ 10.1093/emboj/20.9.2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manansala MC, Min S, Cleary MD. The Drosophila SERTAD protein Taranis determines lineage-specific neural progenitor proliferation patterns. Dev Biol 2013; 376:150-62; PMID:23376107; http://dx.doi.org/ 10.1016/j.ydbio.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 21.Darwish H, Cho JM, Loignon M, Alaoui-Jamali MA. Overexpression of SERTAD3, a putative oncogene located within the 19q13 amplicon, induces E2F activity and promotes tumor growth. Oncogene 2007; 26:4319-28; PMID:17260023; http://dx.doi.org/ 10.1038/sj.onc.1210195 [DOI] [PubMed] [Google Scholar]
- 22.Hayashi R, Goto Y, Ikeda R, Yokoyama KK, Yoshida K. CDCA4 is an E2F transcription factor family-induced nuclear factor that regulates E2F-dependent transcriptional activation and cell proliferation. J Biol Chem 2006; 281:35633-48; PMID:16984923; http://dx.doi.org/ 10.1074/jbc.M603800200 [DOI] [PubMed] [Google Scholar]
- 23.Meyer CA, Jacobs HW, Datar SA, Du W, Edgar BA, Lehner CF. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J 2000; 19:4533-42; PMID:10970847; http://dx.doi.org/ 10.1093/emboj/19.17.4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayeni JO, Varadarajan R, Mukherjee O, Stuart DT, Sprenger F, Srayko M, Campbell SD. Dual phosphorylation of cdk1 coordinates cell proliferation with key developmental processes in Drosophila. Genetics 2014; 196:197-210; PMID:24214341; http://dx.doi.org/ 10.1534/genetics.113.156281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin W-H, He M, Baines RA. Seizure suppression through manipulating splicing of a voltage-gated sodium channel. Brain 2015; 138:891-901; PMID:25681415; http://dx.doi.org/ 10.1093/brain/awv012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature 2005; 434:1087-92; PMID:15858564; http://dx.doi.org/ 10.1038/nature03486 [DOI] [PubMed] [Google Scholar]
- 27.Bushey D, Huber R, Tononi G, Cirelli C. Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci 2007; 27:5384-93; PMID:17507560; http://dx.doi.org/ 10.1523/JNEUROSCI.0108-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, et al.. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 2010; 327:348-51; PMID:20075256; http://dx.doi.org/ 10.1126/science.1183090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi M, Yue Z, Kuryatov A, Lindstrom JM, Sehgal A. Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, et al.. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 2008; 60:672-82; PMID:19038223; http://dx.doi.org/ 10.1016/j.neuron.2008.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol 2009; 19:386-90; PMID:19230663; http://dx.doi.org/ 10.1016/j.cub.2009.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi S-H, Lee G, Monahan P, Park JH. Spatial regulation of Corazonin neuropeptide expression requires multiple cis-acting elements in Drosophila melanogaster. J Comparative Neurol 2008; 507:1184-95; PMID:18181151; http://dx.doi.org/ 10.1002/cne.21594 [DOI] [PubMed] [Google Scholar]
- 33.Hergarden AC, Tayler TD, Anderson DJ. Allatostatin-A neurons inhibit feeding behavior in adult Drosophila. Proc Natl Acad Sci 2012; 109:3967-72; http://dx.doi.org/ 10.1073/pnas.1200778109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyamoto T, Slone J, Song X, Amrein H. A Fructose Receptor Functions as a Nutrient Sensor in the Drosophila Brain. Cell 2012; 151:1113-25; PMID:23178127; http://dx.doi.org/ 10.1016/j.cell.2012.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto T, Amrein H. Diverse roles for the Drosophila fructose sensor Gr43a. Fly 2014; 8:19-25; PMID:24406333; http://dx.doi.org/ 10.4161/fly.27241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keene AC, Duboué ER, McDonald DM, Dus M, Suh GSB, Waddell S, Blau J. Clock and cycle Limit Starvation-Induced Sleep Loss in Drosophila. Curr Biol 2010; 20:1209-15; PMID:20541409; http://dx.doi.org/ 10.1016/j.cub.2010.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liew CW, Boucher J, Cheong JK, Vernochet C, Koh HJ, Mallol C, Townsend K, Langin D, Kawamori D, Hu J, et al.. Ablation of TRIP-Br2, a regulator of fat lipolysis, thermogenesis and oxidative metabolism, prevents diet-induced obesity and insulin resistance. Nat Med 2013; 19:217-26; PMID:23291629; http://dx.doi.org/ 10.1038/nm.3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Marcos PJ, Pantoja C, Gonzalez-Rodriguez A, Martin N, Flores JM, Valverde AM, Hara E, Serrano M. Normal Proliferation and Tumorigenesis but Impaired Pancreatic Function in Mice Lacking the Cell Cycle Regulator Sei1. PloS one 2010; 5:e8744; PMID:20090907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavanaugh Daniel J, Geratowski Jill D, Wooltorton Julian RA, Spaethling Jennifer M, Hector Clare E, Zheng X, Johnson Erik C, Eberwine James H, Sehgal A. Identification of a Circadian Output Circuit for Rest:Activity Rhythms in Drosophila. Cell 2014; 157:689-701; PMID:24766812; http://dx.doi.org/ 10.1016/j.cell.2014.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dus M, Lai Jason S-Y, Gunapala Keith M, Min S, Tayler Timothy D, Hergarden Anne C, Geraud E, Joseph Christina M, Suh Greg SB. Nutrient Sensor in the Brain Directs the Action of the Brain-Gut Axis in Drosophila. Neuron 2015; 87:139-51; PMID:26074004; http://dx.doi.org/ 10.1016/j.neuron.2015.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol 2007; 302:309-23; PMID:17070515; http://dx.doi.org/ 10.1016/j.ydbio.2006.09.035 [DOI] [PubMed] [Google Scholar]
- 42.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature 2005; 437:1257-63; PMID:16251950; http://dx.doi.org/ 10.1038/nature04284 [DOI] [PubMed] [Google Scholar]
- 43.Waterson Michael J, Horvath Tamas L. Neuronal Regulation of Energy Homeostasis: Beyond the Hypothalamus and Feeding. Cell metabolism 2015; 22:962-70; PMID:26603190; http://dx.doi.org/ 10.1016/j.cmet.2015.09.026 [DOI] [PubMed] [Google Scholar]
- 44.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature 2006; 441:757-60; PMID:16760980; http://dx.doi.org/ 10.1038/nature04811 [DOI] [PubMed] [Google Scholar]
- 45.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 2006; 441:753-6; PMID:16760979; http://dx.doi.org/ 10.1038/nature04739 [DOI] [PubMed] [Google Scholar]
- 46.Sitaraman D, Aso Y, Jin X, Chen N, Felix M, Rubin GM, Nitabach MN. Propagation of Homeostatic Sleep Signals by Segregated Synaptic Microcircuits of the Drosophila Mushroom Body. Curr Biol 2015; 25(22):2915-27; PMID:26455303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 2011; 332:1571-6; PMID:21700877; http://dx.doi.org/ 10.1126/science.1202249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron 2010; 65:670-81; PMID:20223202; http://dx.doi.org/ 10.1016/j.neuron.2010.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 2008; 18:1537-45; PMID:18771923; http://dx.doi.org/ 10.1016/j.cub.2008.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Odajima J, Wills ZP, Ndassa YM, Terunuma M, Kretschmannova K, Deeb TZ, Geng Y, Gawrzak S, Quadros IM, Newman J, et al.. Cyclin E constrains Cdk5 activity to regulate synaptic plasticity and memory formation. Dev Cell 2011; 21:655-68; PMID:21944720; http://dx.doi.org/ 10.1016/j.devcel.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagy Z, Esiri MM. Neuronal cyclin expression in the hippocampus in temporal lobe epilepsy. Exp Neurol 1998; 150:240-7; PMID:9527893; http://dx.doi.org/ 10.1006/exnr.1997.6753 [DOI] [PubMed] [Google Scholar]
- 52.Quinones-Coello AT, Petrella LN, Ayers K, Melillo A, Mazzalupo S, Hudson AM, Wang S, Castiblanco C, Buszczak M, Hoskins RA, et al.. Exploring strategies for protein trapping in Drosophila. Genetics 2007; 175:1089-104; PMID:17179094; http://dx.doi.org/ 10.1534/genetics.106.065995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci 2005; 25:7377-84; PMID:16093388; http://dx.doi.org/ 10.1523/JNEUROSCI.2048-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, Kume K. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci 2012; 15:1516-23; PMID:23064381; http://dx.doi.org/ 10.1038/nn.3238 [DOI] [PubMed] [Google Scholar]
- 55.Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol 2006; 16:1051-62; PMID:16753559; http://dx.doi.org/ 10.1016/j.cub.2006.04.032 [DOI] [PubMed] [Google Scholar]
- 56.Crocker A, Sehgal A. Octopamine regulates sleep in Drosophila through protein kinase A-dependent mechanisms. J Neurosci 2008; 28:9377-85; PMID:18799671; http://dx.doi.org/ 10.1523/JNEUROSCI.3072-08a.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a Neural Circuit that Underlies the Effects of Octopamine on Sleep:Wake Behavior. Neuron 2010; 65:670-81; PMID:20223202; http://dx.doi.org/ 10.1016/j.neuron.2010.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci 2008; 11:354-9; PMID:18223647; http://dx.doi.org/ 10.1038/nn2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S, Lamaze A, Liu Q, Tabuchi M, Yang Y, Fowler M, Bharadwaj R, Zhang J, Bedont J, Blackshaw S, et al.. WIDE AWAKE Mediates the Circadian Timing of Sleep Onset. Neuron 2014; 82:151-66; PMID:24631345; http://dx.doi.org/ 10.1016/j.neuron.2014.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maguire SE, Rhoades S, Chen W-F, Sengupta A, Yue Z, Lim JC, Mitchell CH, Weljie AM, Sehgal A. Independent Effects of γ-Aminobutyric Acid Transaminase (GABAT) on Metabolic and Sleep Homeostasis. J Biol Chem 2015; 290:20407-16; PMID:26124278; http://dx.doi.org/ 10.1074/jbc.M114.602276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu M, Robinson James E, Joiner William J. SLEEPLESS Is a Bifunctional Regulator of Excitability and Cholinergic Synaptic Transmission. Curr Biol 2014; 24:621-9; PMID:24613312; http://dx.doi.org/ 10.1016/j.cub.2014.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomita J, Ueno T, Mitsuyoshi M, Kume S, Kume K. The NMDA Receptor Promotes Sleep in the Fruit Fly, Drosophila melanogaster. PloS one 2015; 10:e0128101; PMID:26023770; http://dx.doi.org/ 10.1371/journal.pone.0128101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishimoto H, Kitamoto T. The Steroid Molting Hormone Ecdysone Regulates Sleep in Adult Drosophila melanogaster. Genetics 2010; 185:269-81; PMID:20215472; http://dx.doi.org/ 10.1534/genetics.110.114587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Isaac RE, Li C, Leedale AE, Shirras AD. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc Biol Sci 2009; 277:65-70; http://dx.doi.org/ 10.1098/rspb.2009.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh Y, Yoon SE, Zhang Q, Chae HS, Daubnerova I, Shafer OT, Choe J, Kim YJ. A homeostatic sleep-stabilizing pathway in Drosophila composed of the sex peptide receptor and its ligand, the myoinhibitory peptide. PLoS Biol 2014; 12:e1001974; PMID:25333796; http://dx.doi.org/ 10.1371/journal.pbio.1001974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shang Y, Donelson Nathan C, Vecsey Christopher G, Guo F, Rosbash M, Griffith Leslie C. Short Neuropeptide F Is a Sleep-Promoting Inhibitory Modulator. Neuron 2013; 80:171-83; PMID:24094110; http://dx.doi.org/ 10.1016/j.neuron.2013.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, Nitabach MN. Calcitonin gene-related Peptide neurons mediate sleep-specific circadian output in Drosophila. Curr Biol 2014; 24:2652-64; PMID:25455031; http://dx.doi.org/ 10.1016/j.cub.2014.09.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park S, Sonn JY, Oh Y, Lim C, and Choe J. SIFamide and SIFamide Receptor Define a Novel Neuropeptide Signaling to Promote Sleep in Drosophila. Mol Cells 2014; 37:295-301; PMID:24658384; http://dx.doi.org/ 10.14348/molcells.2014.2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bushey D, Huber R, Tononi G, Cirelli C. Drosophila Hyperkinetic Mutants Have Reduced Sleep and Impaired Memory. J Neurosci 2007; 27:5384-93; PMID:17507560; http://dx.doi.org/ 10.1523/JNEUROSCI.0108-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeong K, Lee S, Seo H, Oh Y, Jang D, Choe J, Kim D, Lee J-H, Jones WD. Ca-α1T, a fly T-type Ca2+ channel, negatively modulates sleep. Scientific Reports 2015; 5:17893; PMID:26647714; http://dx.doi.org/ 10.1038/srep17893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomita J, Mitsuyoshi M, Ueno T, Aso Y, Tanimoto H, Nakai Y, Aigaki T, Kume S, Kume K. Pan-Neuronal Knockdown of Calcineurin Reduces Sleep in the Fruit Fly, Drosophila melanogaster. J Neurosci 2011; 31:13137-46; PMID:21917797; http://dx.doi.org/ 10.1523/JNEUROSCI.5860-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakai Y, Horiuchi J, Tsuda M, Takeo S, Akahori S, Matsuo T, Kume K, Aigaki T. Calcineurin and Its Regulator Sra/DSCR1 Are Essential for Sleep in Drosophila. J Neurosci 2011; 31:12759-66; PMID:21900555; http://dx.doi.org/ 10.1523/JNEUROSCI.1337-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allebrandt KV, Amin N, Muller-Myhsok B, Esko T, Teder-Laving M, Azevedo RVDM, Hayward C, van Mill J, Vogelzangs N, Green EW, et al.. A KATP channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Molecular psychiatry 2013; 18:122-32; PMID:22105623; http://dx.doi.org/ 10.1038/mp.2011.142 [DOI] [PubMed] [Google Scholar]
- 74.Roessingh S, Wolfgang W, Stanewsky R. Loss of Drosophila melanogaster TRPA1 Function Affects “Siesta” Behavior but Not Synchronization to Temperature Cycles. J Biol Rhythms 2015; 30:492-505; PMID:26459465; http://dx.doi.org/ 10.1177/0748730415605633 [DOI] [PubMed] [Google Scholar]
- 75.Bushey D, Tononi G, Cirelli C. The Drosophila fragile X mental retardation gene regulates sleep need. J Neurosci 2009; 29:1948-61; PMID:19228950; http://dx.doi.org/ 10.1523/JNEUROSCI.4830-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Naidoo N, Ferber M, Galante RJ, McShane B, Hu JH, Zimmerman J, Maislin G, Cater J, Wyner A, Worley P, et al.. Role of Homer Proteins in the Maintenance of Sleep-Wake States. PloS one 2012; 7:e35174; PMID:22532843; http://dx.doi.org/ 10.1371/journal.pone.0035174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Zhou Z, Zhang X, Tong H, Li P, Zhang ZC, Jia Z, Xie W, Han J. Drosophila Neuroligin 4 Regulates Sleep through Modulating GABA Transmission. J Neurosci 2013; 33:15545-54; PMID:24068821; http://dx.doi.org/ 10.1523/JNEUROSCI.0819-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Larkin A, Chen MY, Kirszenblat L, Reinhard J, van Swinderen B, Claudianos C. Neurexin-1 regulates sleep and synaptic plasticity in Drosophila melanogaster. Eur J Neurosci 2015; 42(7):2455-66 [DOI] [PubMed] [Google Scholar]
- 79.Vanderheyden WM, Gerstner JR, Tanenhaus A, Yin JC, Shaw PJ. ERK Phosphorylation Regulates Sleep and Plasticity in Drosophila. PloS one 2013; 8:e81554; PMID:24244744; http://dx.doi.org/ 10.1371/journal.pone.0081554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci 2007; 10:1160-7; PMID:17694052; http://dx.doi.org/ 10.1038/nn1957 [DOI] [PubMed] [Google Scholar]
- 81.Guo F, Yi W, Zhou M, Guo A. Go signaling in mushroom bodies regulates sleep in Drosophila. Sleep 2011; 34:273-81; PMID:21358844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seugnet L, Suzuki Y, Merlin G, Gottschalk L, Duntley Stephen P, Shaw Paul J. Notch Signaling Modulates Sleep Homeostasis and Learning after Sleep Deprivation in Drosophila. Curr Biol 2011; 21:835-40; PMID:21549599; http://dx.doi.org/ 10.1016/j.cub.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takahama K, Tomita J, Ueno T, Yamazaki M, Kume S, Kume K. Pan-neuronal knockdown of the c-Jun N-terminal Kinase (JNK) results in a reduction in sleep and longevity in Drosophila. Biochem Biophy Res Commun 2012; 417:807-11; PMID:22197814; http://dx.doi.org/ 10.1016/j.bbrc.2011.12.040 [DOI] [PubMed] [Google Scholar]
- 84.Donlea J, Leahy A, Thimgan MS, Suzuki Y, Hughson BN, Sokolowski MB, Shaw PJ. foraging alters resilience/vulnerability to sleep disruption and starvation in Drosophila. Proc Natl Acad Sci 2012; 109:2613-8; http://dx.doi.org/ 10.1073/pnas.1112623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Donlea JM, Pimentel D, Miesenbock G. Neuronal machinery of sleep homeostasis in Drosophila. Neuron 2014; 81:860-72; PMID:24559676; http://dx.doi.org/ 10.1016/j.neuron.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Metaxakis A, Tain LS, Grönke S, Hendrich O, Hinze Y, Birras U, Partridge L. Lowered Insulin Signalling Ameliorates Age-Related Sleep Fragmentation in Drosophila. PLoS Biol 2014; 12:e1001824; PMID:24690889; http://dx.doi.org/ 10.1371/journal.pbio.1001824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, Shaw PJ. The Perilipin Homologue, Lipid Storage Droplet 2, Regulates Sleep Homeostasis and Prevents Learning Impairments Following Sleep Loss. PLoS Biol 2010; 8:e1000466; PMID:20824166; http://dx.doi.org/ 10.1371/journal.pbio.1000466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gerstner JR, Vanderheyden WM, Shaw PJ, Landry CF, Yin JC. Fatty-acid binding proteins modulate sleep and enhance long-term memory consolidation in Drosophila. PloS one 2011; 6:e15890; PMID:21298037; http://dx.doi.org/ 10.1371/journal.pone.0015890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron 2000; 25:129-38; PMID:10707978; http://dx.doi.org/ 10.1016/S0896-6273(00)80877-6 [DOI] [PubMed] [Google Scholar]
- 90.Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms 2003; 18:12-25; PMID:12568241; http://dx.doi.org/ 10.1177/0748730402239673 [DOI] [PubMed] [Google Scholar]
- 91.Williams JA, Sathyanarayanan S, Hendricks JC, Sehgal A. Interaction Between Sleep and the Immune Response in Drosophila: A Role for the NFκB Relish. Sleep 2007; 30:389-400; PMID:17520783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bai L, Sehgal A. Anaplastic Lymphoma Kinase Acts in the Drosophila Mushroom Body to Negatively Regulate Sleep. PLoS Genet 2015; 11:e1005611; PMID:26536237; http://dx.doi.org/ 10.1371/journal.pgen.1005611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu Y, Bolduc FV, Bell K, Tully T, Fang Y, Sehgal A, Fischer JA. A Drosophila model for Angelman syndrome. Proc Natl Acad Sci 2008; 105:12399-404; http://dx.doi.org/ 10.1073/pnas.0805291105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pfeiffenberger C, Allada R. Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila. PLoS Genet 2012; 8:e1003003; PMID:23055946; http://dx.doi.org/ 10.1371/journal.pgen.1003003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci 2001; 4:1108-15; PMID:11687816; http://dx.doi.org/ 10.1038/nn743 [DOI] [PubMed] [Google Scholar]
- 96.Liu W, Guo F, Lu B, Guo A. amnesiac regulates sleep onset and maintenance in Drosophila melanogaster. Biochem Biophy Res Commun 2008; 372:798-803; PMID:18514063; http://dx.doi.org/ 10.1016/j.bbrc.2008.05.119 [DOI] [PubMed] [Google Scholar]
- 97.Harbison ST, Carbone MA, Ayroles JF, Stone EA, Lyman RF, Mackay TFC. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat Genet 2009; 41:371-5; PMID:19234472; http://dx.doi.org/ 10.1038/ng.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pirooznia SK, Chiu K, Chan MT, Zimmerman JE, Elefant F. Epigenetic regulation of axonal growth of Drosophila pacemaker cells by histone acetyltransferase tip60 controls sleep. Genetics 2012; 192:1327-45; PMID:22982579; http://dx.doi.org/ 10.1534/genetics.112.144667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Soshnev AA, Ishimoto H, McAllister BF, Li X, Wehling MD, Kitamoto T, Geyer PK. A Conserved Long Noncoding RNA Affects Sleep Behavior in Drosophila. Genetics 2011; 189:455-68; PMID:21775470; http://dx.doi.org/ 10.1534/genetics.111.131706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shimizu H, Shimoda M, Yamaguchi T, Seong K-H, Okamura T, Ishii S. Drosophila ATF-2 Regulates Sleep and Locomotor Activity in Pacemaker Neurons. Mol Cell Biol 2008; 28:6278-89; PMID:18694958; http://dx.doi.org/ 10.1128/MCB.02242-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Robinson JE, Paluch J, Dickman DK, Joiner WJ. ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat Commun 2016; 7:10512. [DOI] [PMC free article] [PubMed] [Google Scholar]