Abstract
Viruses are solely dependent on host cells to propagate; therefore, understanding virus-host interaction is important for antiviral drug development. Since de novo nucleotide biosynthesis is essentially required for both host cell metabolism and viral replication, specific catalytic enzymes of these pathways have been explored as potential antiviral targets. In this study, we investigated the role of different enzymatic cascades of nucleotide biosynthesis in hepatitis E virus (HEV) replication. By profiling various pharmacological inhibitors of nucleotide biosynthesis, we found that targeting the early steps of the purine biosynthesis pathway led to the enhancement of HEV replication, whereas targeting the later step resulted in potent antiviral activity via the depletion of purine nucleotide. Furthermore, the inhibition of the pyrimidine pathway resulted in potent anti-HEV activity. Interestingly, all of these inhibitors with anti-HEV activity concurrently triggered the induction of antiviral interferon-stimulated genes (ISGs). Although ISGs are commonly induced by interferons via the JAK-STAT pathway, their induction by nucleotide synthesis inhibitors is completely independent of this classical mechanism. In conclusion, this study revealed an unconventional novel mechanism of cross talk between nucleotide biosynthesis pathways and cellular antiviral immunity in constraining HEV infection. Targeting particular enzymes in nucleotide biosynthesis represents a viable option for antiviral drug development against HEV. HEV is the most common cause of acute viral hepatitis worldwide and is also associated with chronic hepatitis, especially in immunocompromised patients. Although often an acute and self-limiting infection in the general population, HEV can cause severe morbidity and mortality in certain patients, a problem compounded by the lack of FDA-approved anti-HEV medication available. In this study, we have investigated the role of the nucleotide synthesis pathway in HEV infection and its potential for antiviral drug development. We show that targeting the later but not the early steps of the purine synthesis pathway exerts strong anti-HEV activity. In particular, IMP dehydrogenase (IMPDH) is the most important anti-HEV target of this cascade. Importantly, the clinically used IMPDH inhibitors, including mycophenolic acid and ribavirin, have potent anti-HEV activity. Furthermore, targeting the pyrimidine synthesis pathway also exerts potent antiviral activity against HEV. Interestingly, antiviral effects of nucleotide synthesis pathway inhibitors appear to depend on the medication-induced transcription of antiviral interferon-stimulated genes. Thus, this study reveals an unconventional novel mechanism as to how nucleotide synthesis pathway inhibitors can counteract HEV replication.
INTRODUCTION
Hepatitis E virus (HEV) is a single-stranded positive-sense RNA virus that mainly infects the liver. It is the most common cause of acute viral hepatitis worldwide. In general, HEV infection is a self-limiting disease and is associated with low mortality, but epidemics of hepatitis E occur periodically throughout the developing world, resulting in 70,000 deaths yearly (1). In western countries, HEV primarily affects immunocompromised patients, in particular organ transplant recipients, as well as hematopoietic stem cell transplant recipients (2–5). More than 60% of organ recipients infected with HEV develop chronic hepatitis with rapid progression to cirrhosis (2). Despite being an emerging global health issue, no FDA-approved anti-HEV therapy is currently available. Only alpha interferon (IFN-α), ribavirin, or a combination of these has been used occasionally as an off-label treatment. Thus, further research aimed at understanding its infection biology and developing effective antiviral treatment is urgently required.
Cellular nucleotides, including purines and pyrimidines, are the basic building blocks that form the nucleic acids RNA and DNA. Nucleotides are the fundamental components that are required for cell metabolism, such as genome replication. In vivo, nucleotides can be synthesized de novo through a series of enzymatic reactions or recycled through salvage pathways. Since viral replication heavily relies on the host cells to supply nucleosides, targeting the nucleotide biosynthesis pathway represents an attractive strategy for antiviral drug development. The nucleotide biosynthesis pathways have been well studied for decades (6–8). Numerous compounds have been developed and well characterized to target particular enzymes of this pathway to inhibit viral infections by depletion or causing an imbalance of nucleotide pools (9–18). Among them, inhibitors of IMP dehydrogenase (IMPDH), a key enzyme of the purine synthesis pathway, have been used successfully in the clinic for decades. These drugs, including ribavirin and mycophenolic acid (MPA), used as antiviral or immunosuppressive medication, respectively, have been demonstrated to have broad antiviral activity against a spectrum of viruses, including dengue virus, yellow fever virus (YFV), and hepatitis B, hepatitis C, and hepatitis E viruses (14, 15, 18–21). Likewise, brequinar (BQ) and leflunomide (LFM), inhibitors of dihydroorotate dehydrogenase (DHODH), an essential enzyme of pyrimidine nucleotide synthesis, have been shown to inhibit human polyomavirus type BK virus, YFV, and dengue virus (12, 22).
Besides their function as building blocks of genetic material, free nucleotides also play important roles in cell signaling. We and others have previously reported the potential interaction of nucleotide deprivation and cellular antiviral immune response, such as provoking the expression of interferon-stimulated genes (ISGs) (19, 23). Given that the liver is a major site for nucleotide synthesis, we comprehensively profiled the role of purine and pyrimidine synthesis pathways in HEV cell culture models aimed at identifying potential antiviral drug targets and understanding the cross talk with cellular antiviral immunity against HEV infection.
MATERIALS AND METHODS
Reagents.
Guanosine (CAS 118-00-3), adenosine (CAS 58-61-7), uridine (CAS 58-96-8), 6-thioguanine (6-TG; CAS 154-42-7), lometrexol hydrate (CAS 106400-81-1), methotrexate (MTX) hydrate (CAS 133073-73-1), fludarabine (FA) phosphate (CAS 75607-67-9), brequinar (BQR) sodium salt hydrate (MDL MFCD21363375), leflunomide (LFM) (CAS 75706-12-6), and 6-azauracil (6-AU; CAS 461-89-2) were purchased from Sigma. Twenty-three IMPDH-specific inhibitors were kindly provided by the Center for Drug Design, University of Minnesota. All of the reagents were dissolved in dimethyl sulfoxide (DMSO). The effects of these de novo nucleotide biosynthesis inhibitors on host cell viability were determined by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (see Fig. S7 in the supplemental material). Stocks of JAK inhibitor 1 (CAS 457081-03-7; Santa Cruz Biotech, CA) were dissolved in DMSO at a final concentration of 5 mg/ml. Stocks of CP-690550 (tofacitinib) (Santa Cruz Biotech, CA) were dissolved in DMSO at a final concentration of 10 mg/ml.
Cell culture.
Human hepatoma cell line Huh7 and human embryonic kidney epithelial cell line 293T were cultured in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 IU/ml streptomycin.
Cell culture models.
An HEV replication model with subgenomic HEV sequence coupled with a Gaussia luciferase reporter gene and an HEV infection model containing the full-length HEV genome were used in our study. The construction of two models has been described previously (18). Huh7 cells constitutively expressing the firefly luciferase reporter gene driven by the human PGK promoter were used to represent household luciferase activity for normalizing nonspecific effects on luciferase activity (11). Huh7 cells transduced with a lentiviral transcriptional reporter system expressing the firefly luciferase gene under the control of a promoter containing multiple interferon-stimulated response element (ISRE) promoter elements (SBI Systems Biosciences, Mountain View, CA) was established, and luciferase activity represents ISRE promoter activation.
Quantification of HEV replication and infection.
The details for examining HEV replication and infection were described before (18). Briefly, for the HEV replication model (p6-Luc), the activity of secreted Gaussia luciferase in the cell culture medium was measured using a BioLux Gaussia luciferase flex assay kit (New England BioLabs) for the quantification of viral replication, which was normalized by firefly luciferase expression. For the full-length HEV infectious model, SYBR green-based quantitative reverse transcription-PCR (qRT-PCR) was used to quantify the newly formed viral genomic RNA after cell lysis, and the HEV primer sequences are shown in Table S2 in the supplemental material.
Gene knockdown by lentiviral vector-delivered shRNA.
Lentiviral vectors targeting PPAT, GART, ATIC, and DHODH were produced in 293T cells as previously described (11). To generate stable gene knockdown cells, Huh7 cells were transduced with lentiviral vectors. Since the vectors also express a puromycin resistance gene, transduced cells subsequently were selected by adding 2.5 μg/ml puromycin (Sigma) to the cell culture medium. After a pilot study, the short hairpin RNA (shRNA) vectors (see Fig. S1 and Table S3 in the supplemental material) exerting optimal gene knockdown were selected by qPCR with the corresponding primers shown in Table S2 in the supplemental material. Meanwhile, shRNA vector expressing green fluorescent protein (shGFP) was used as a control (shCTR). The amount of HEV was assessed after 3 days of HEV infection in medium containing shGFP cells and knockdown cells. For the experiment comparing the activity of compounds between shGFP and knockdown cells, infectious HEV cells were directly transduced with lentiviral shRNA vectors and selected by puromycin.
Statistical analysis.
Statistical analysis was performed using the nonpaired, nonparametric test (Mann-Whitney test; GraphPad Prism Software). P values of less than 0.05 were considered statistically significant.
RESULTS
Exogenous guanosine, but not uridine, stimulates HEV replication.
Purine and pyrimidine nucleotides are the major cellular energy carriers and constitute the defining subunits of nucleic acids. Two distinct pathways are responsible for the biosynthesis of these two types of nucleotides (Fig. 1A and 2A). Their fundamental role in cellular biochemistry raises the possibility that modifying flux through nucleotide biosynthesis pathways profoundly influences the course of viral infection. Thus, we decided to assess the overall impact of either purine or pyrimidine synthesis on HEV infection. A first indication that such effects exist came from experiments in which we arbitrarily increased the purine and pyrimidine content by supplementation of exogenous guanosine (Fig. 1A) and uridine (Fig. 2A) in human hepatoma cell line (Huh7)-based HEV cell culture models. Guanosine, a purine nucleoside containing guanine attached to a ribose, can be converted to GMP through the purine salvage synthesis pathway and subsequently replenishes the purine nucleotide pool (Fig. 1A). Mechanistically, the cleavage of exogenous guanosine was catalyzed by purine nucleoside phosphorylase (PNP) to form guanine. In the presence of hypoxanthine/guanine phosphoribosyl transferase (HGPRT), guanine was converted to GMP by the addition of ribose 5-phosphate from phosphoribosyl pyrophosphate (PRPP). The supplementation of guanosine dose dependently enhanced HEV replication-related luciferase activity in the subgenomic replicon (p6-Luc) model and increased cellular viral RNA in the full-length (p6) infectious model (Fig. 1B). Likewise, uridine, which is a pyrimidine nucleoside consisting of uracil binding to ribose, commonly presents as UMP to rescue cells from pyrimidine nucleotide depletion (Fig. 2A). In contrast, the supplementation of exogenous uridine had no effect on HEV replication (Fig. 2B). Thus, interaction between at least some of the pathways involved in nucleotide biosynthesis and the HEV infectious process might exist.
FIG 1.

Exogenous guanosine stimulated HEV replication. (A) Schematic overview of de novo biosynthesis of purine nucleotide. PRPP, 5-phosphoribosyl-1-pyrophosphate; PRA, 5-phosphoribosylamine; GAR, glycinamide ribonucleotide; FGAR, formyl-GAR; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide. (B) Huh7 cell-based subgenomic HEV replicons containing the luciferase reporter gene were treated for 24 h, 48 h, and 72 h with a dose range of guanosine (n = 4). Data are presented as means ± standard errors of the means (SEM). Meanwhile, Huh7 cells with the infectious HEV containing the full-length p6 genome were treated for 48 h with a dose range of guanosine (n = 5). Data were normalized to two housekeeping genes (GAPDH and RP2) and are presented relative to the control (CTR) (set as 1). Data represent means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 2.

Exogenous uridine does not affect HEV replication. (A) Schematic overview of de novo biosynthesis of pyrimidine nucleotide. (B) Huh7 cell-based subgenomic HEV replicon containing the luciferase reporter gene was treated for 24 h, 48 h, and 72 h with a dose range of uridine (n = 5). Data are presented as means ± SEM.
Targeting the catalytic steps leading to primary purine nucleotide synthesis (IMP) stimulates HEV replication.
Given the clear proviral effect of exogenous guanosine, we explored potential anti-HEV strategies targeting the different enzymes that are involved in purine nucleotide synthesis. De novo purine is synthesized mainly in the liver and begins with the starting material PRPP. The first fully formed nucleotide, IMP, is catalyzed through 10 reactions by six enzymes (Fig. 1A). We first selectively targeted three key enzymes of this cascade, including amido phosphoribosyltransferase (APRTase), glycinamide ribonucleotide transformylase (GART), and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase (AICARFT), through the use of 6-thioguanine (6-TG), lometrexol, and methotrexate (MTX), respectively. Somewhat counterintuitively, all three compounds increased HEV replication in both cell culture models (Fig. 3). To further clarify the role of their targets, lentivirus-mediated RNA interference (RNAi) was used for the knockdown of the PPAT, GART, and ATIC genes that encode the corresponding enzymes APRTase, GART, and AICARTF, respectively (Fig. 4A). Consistent with the pharmacological results, the downregulation of these enzymes enhanced HEV replication (Fig. 4B). Furthermore, the proviral effects of the pharmacological inhibitors were largely absent in a context in which their targets were silenced, suggesting that pharmacological effects are not due to off-target effects (Fig. 4C).
FIG 3.
Inhibitors of IMP synthesis cascade stimulate HEV replication. The Huh7 cells containing subgenomic HEV replicons with luciferase reporter genes were incubated with increasing doses of 6-TG (A), lometrexol (B), and MTX (C). The luciferase activity was determined at 24 h, 48 h, and 72 h. Accordingly, Huh7 cells infected with full-length HEV were treated with increasing doses of 6-TG (A), lometrexol (B), and MTX (C). The HEV RNA level was quantified by qRT-PCR after 48 h. Data were normalized to two housekeeping genes and are presented relative to the control (CTR) (set as 1). Data represent means ± SEM from five to eight experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 4.
Silencing of enzymes involved in IMP synthesis cascade facilitates HEV replication. (A) Huh7 cells were transduced with lentiviral shRNAs to stably silence the corresponding genes for PPAT, GART, and ATIC (a set of independent shRNA clones targeting each gene was used). Huh7 cells transduced with lentiviral shRNA targeting GFP (shCTR) were used as a control. The efficiency of gene knockdown was analyzed by qRT-PCR. (B) Silencing of PPAT, GART, and ATIC resulted in significant elevation of viral RNA upon inoculation of HEV. HEV RNA levels were determined 72 h after inoculation. (C) Silencing of PPAT, GART, and ATIC abrogated the pro-HEV effects of 6-TG, lometrexol, and MTX. Data were normalized to that for cells without treatment with the three compounds (green bar; set as 1). All data were normalized to two housekeeping genes and are presented relative to the control (CTR) (set as 1) (means ± SEM from four to eight experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
As a bifunctional enzyme, the N-terminal domain of ATIC has AICARFT activity, and the C-terminal domain has IMP cyclohydrolase (IMPCH) activity. FA, an IMPCH inhibitor, also promoted HEV replication but exerted cytotoxicity concurrently (see Fig. S2 in the supplemental material). Thus, these results highlight the interaction of nucleotide biosynthesis and the HEV infection process but also show that the rational design of therapy aimed at exploiting the nucleotide biosynthesis pathway for treatment of HEV is not straightforward.
IMPDH inhibition counteracts HEV replication by depleting the purine nucleotide pool.
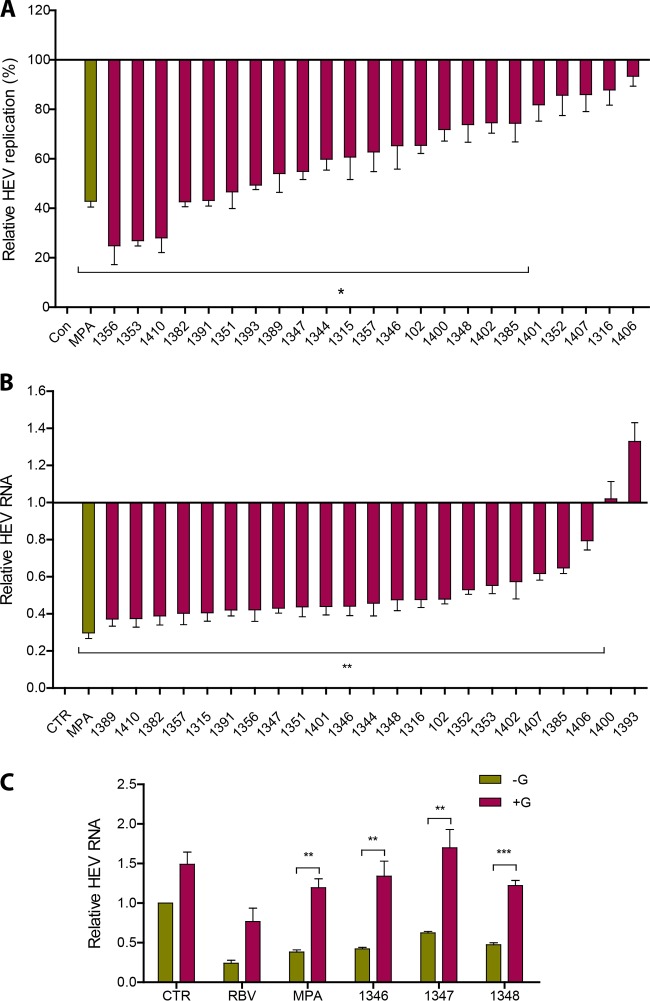
As a branching point in purine synthesis, IMP is converted to either AMP or XMP/GMP (Fig. 1A). IMPDH, an enzyme consisting of two isoforms (IMPDH1 and IMPDH2) in humans, catalyzes the reaction of IMP into XMP for further conversion to GMP. We have previously demonstrated that MPA, a clinically used immunosuppressant that preferentially inhibits IMPDH2, has anti-HEV activity (18). To further explore the potential of targeting this enzyme, a panel of 23 inhibitors was custom designed and synthesized with various affinities in inhibiting IMPDH1 or IMPDH2 (see Table S1 in the supplemental material). As shown in Fig. 5A, HEV replication was inhibited by all 23 IMPDH inhibitors at a concentration of 10 μM as measured by luciferase activity. Accordingly, 21 of the 23 inhibitors also suppressed HEV infection as assessed by full-length HEV genome quantification by qRT-PCR (Fig. 5B). Anti-HEV activity also was observed at 2 μM for 20 of the IMPDH inhibitors (see Fig. S3). To further characterize the inhibition, we selected three representative compounds with anti-HEV activity in both models. Similar to ribavirin and MPA, guanosine supplementation abrogated the anti-HEV activity of these compounds (Fig. 5C), suggesting that the depletion of the purine nucleotide pool is responsible for the antiviral action. Thus, inhibitors with anti-HEV potential exert their action in this respect through targeting nucleotide synthesis.
FIG 5.
IMPDH inhibitors potently inhibit HEV replication by depletion of the purine nucleotide pool. (A) Huh7 HEV replicon luciferase cells were treated with 23 specific IMPDH inhibitors (10 μM) with MPA as a positive control. Luciferase activity was quantified at 24 h after treatment (n = 3). (B) Huh7 cells harboring full-length HEV were treated with 23 specific IMPDH inhibitors with MPA as a positive control. HEV RNA levels were measured by qRT-PCR at 48 h after treatment (n = 5). (C) Supplementation of guanosine abrogated the anti-HEV effects of 3 representative IMPDH inhibitors (1346, 1347, and 1348) (n = 5). Ribavirin (RBV) and MPA served as positive controls. Data were normalized to two housekeeping genes and are presented relative to the control (CTR) (set as 1) (means ± SEM). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Targeting pyrimidine biosynthesis inhibits HEV replication.
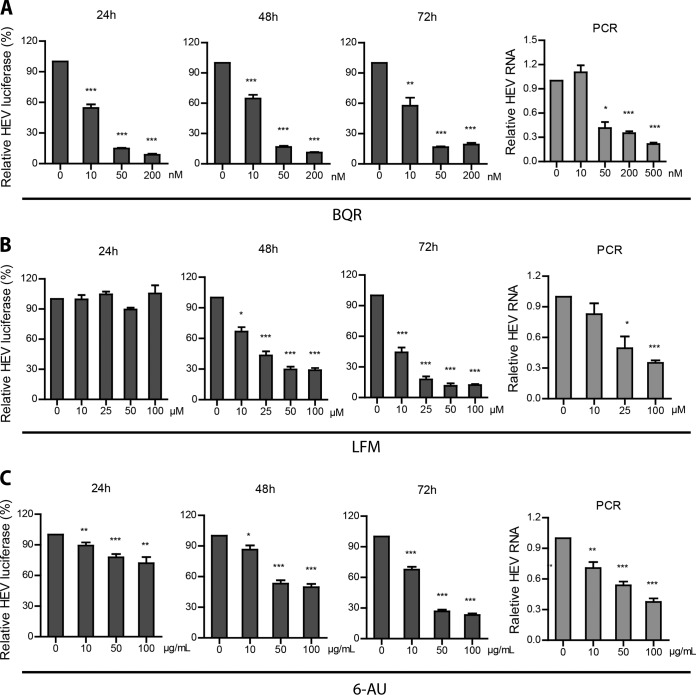
Even though the supplementation of exogenous uridine has no effect on HEV, inhibitors of pyrimidine synthesis have been widely reported to inhibit the infection of a broad spectrum of other viruses, prompting further exploration of the role of pyrimidine biosynthesis in HEV replication. Thus, we selected two catalytic enzymes involved in de novo pyrimidine synthesis for further study. DHODH, which localizes to the mitochondria, is a critical enzyme that converts dihydroorotate to orotate. Brequinar (BQR) and leflunomide (LFM) are well-known clinically tested DHODH inhibitors. Treatment with BQR (10 to 500 nM) results in a significant reduction of HEV replication-related luciferase activity in the subgenomic replicon assay system (Fig. 6A). Concordantly, BQR also dose dependently inhibits cellular viral RNA in our infectious HEV model. Treatment with 500 nM BQR for 48 h resulted in 78% ± 17% (means ± standard deviations [SD]; n = 7; P < 0.001) inhibition of HEV genomic RNA level (determined by qRT-PCR) compared with that of the control (Fig. 6A). Similar results were observed with treatment of LMF (Fig. 6B). The specificity of these effects was confirmed in experiments in which we examined the cognate target of these inhibitors, DHODH, by lentiviral RNAi-mediated silencing. Consistent with previous results, the knockdown of DHODH inhibited HEV replication and abrogated the anti-HEV effect of BQR (Fig. 7), and this enzyme emerged as a relevant target in anti-HEV therapy.
FIG 6.
Inhibition of pyrimidine nucleotide synthesis suppresses HEV replication. Huh7 cells containing subgenomic HEV replicons with luciferase report genes were treated with increasing doses of BQR (A), LFM (B), and 6-AU (C). The luciferase activity was determined after 24 h, 48 h, and 72 h. Accordingly, Huh7 cells harboring infectious HEV also were treated with increasing doses of BQR (A), LFM (B), and 6-AU (C). HEV RNA was quantified by qRT-PCR after 48 h of treatment. Data were normalized to two housekeeping genes and are presented relative to the control (CTR) (set as 1). Data represent means ± SEM from four to seven experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 7.

Anti-HEV activity by BQR can be attributed to the inhibition of its target, DHODH. (A) Huh7 cells were transduced with lentiviral shRNA to stably silent DHODH (DHODH shRNA positive). Huh7 cells transduced with lentiviral shRNA targeting GFP were used as a control (DHODH shRNA negative). DHODH knockdown was assessed by qRT-PCR (n = 3). DHODH knockdown resulted in the significant inhibition of HEV replication. HEV RNA levels were determined 72 h after HEV inoculation (n = 6). (B) DHODH knockdown abrogated the anti-HEV effect of BQR (n = 7). Data were normalized to cells without BQR treatment (green bar; set as 1). All data were normalized to two housekeeping genes and are presented relative to the control (shCTR) (set as 1) (means ± SEM). **, P < 0.01; ***, P < 0.001.
To further identify potential anti-HEV targets, we also examined orotidine-5′-monophosphate decarboxylase (ODCase), the downstream enzyme of DHODH that catalyzes the decarboxylation of OMP to UMP. To this end, we employed 6-azauracil (6-AU), a potent inhibitor of ODCase. As shown in Fig. 6C, HEV replication was dose dependently inhibited by 6-AU. Conversely, supplementation with uridine fully restored the HEV infectious potential despite the presence of BQR, LMF, or 6-AU (Fig. 8). In conjunction, these results show that the depletion of the pyrimidine nucleotide pool is a powerful anti-HEV strategy.
FIG 8.
Uridine reverses the anti-HEV activity mediated by pyrimidine inhibition. The Huh7 subgenomic HEV replicon was incubated with BQR (A), LFM (B), and 6-AU (C) and supplemented with increasing doses of uridine. After 72 h, luciferase activity was determined. Accordingly, Huh7 cells harboring full-length HEV RNA were treated with BQR (A), LFM (B), and 6-AU (C) and supplemented with 200 μM uridine. HEV viral RNA was assessed by qRT-PCR 48 h after treatment. Data were normalized to two housekeeping genes and are presented relative to the control (CTR) (set as 1). Data represent means ± SEM from four to seven experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Inhibitors of purine and pyrimidine synthesis provoke cellular antiviral immune responses through nucleotide depletion.
We previously demonstrated that the IMPDH inhibitor MPA can induce the expression of ISGs to combat hepatitis C virus (HCV) infection, although the underlying mechanism remained unclear (19). ISGs are the ultimate antiviral effectors and generally are assumed to be induced solely through the action of antiviral cytokines, especially interferons. In HEV infection models, we observed that MPA as well as three other selected IMPDH inhibitors were able to induce the expression of a panel of antiviral ISGs (Fig. 9A), challenging this dogma. The induction of ISGs by IMPDH inhibitors was associated with purine nucleotide depletion, since the supplementation of guanosine at least partly abrogated the induction of ISGs (Fig. 9B).
FIG 9.
Inhibition of IMPDH stimulates ISG expression through purine nucleotide deprivation. (A) Huh7 cells infected with HEV were treated with MPA or 3 other IMPDH inhibitors (1346, 1347, and 1348). The expression of a panel of ISGs was determined by qRT-PCR after 48 h of treatment. Data were normalized to basal ISG expression without treatment (gray bar; set as 1). (B) Supplementation of guanosine abrogated the induction of ISGs by IMPDH inhibitors. The expression of ISGs was determined by qRT-PCR 48 h after treatment. Data were normalized to basal ISG expression without treatment (purple bar; set as 1). All data were normalized to two housekeeping genes and represent means ± SEM from four experiments.
In parallel, we also investigated the effects of pyrimidine synthesis inhibitors. We employed an interferon response reporter in which Huh7 cells are stably integrated with an interferon-stimulated response element (ISRE)-driven luciferase gene that measures ISG transcription upon interferon stimulation. BQR potently induces luciferase activity in this reporter assay and triggers the expression of a panel of ISGs (Fig. 10A; also see Fig. S4 in the supplemental material). The supplementation of uridine completely abrogated these effects on ISG transcription (Fig. 10B; also see Fig. S4). Similar results also were observed with another pyrimidine synthesis inhibitor, 6-AU, targeting ODCase (Fig. 10C). Thus, both purine and pyrimidine synthesis pathways can interact with cellular antiviral immune responses, providing a rational explanation for their antiviral effects.
FIG 10.
Inhibition of pyrimidine synthesis stimulates ISG expression through pyrimidine nucleotide depletion. (A) Huh7 cells infected with HEV were treated with BQR or 6-AU. After 48 h, the expression of a panel of ISGs was determined by qRT-PCR. Data were normalized to basal ISG expression without treatment (gray bar; set as 1). (B) Supplementation of uridine completely abrogated the induction of ISGs by BQR (B) or 6-AU (C).The expression of ISGs was determined by qRT-PCR at 48 h after treatment. Data were normalized to basal ISG expression without treatment (purple bar; set as 1). All data were normalized to two housekeeping genes and represent means ± SEM from five experiments. *, P < 0.05; **, P < 0.01.
The induction of ISGs by nucleotide synthesis inhibitors is independent of the JAK-STAT machinery.
Classically, ISGs are thought to be induced only by interferons through the activation of the JAK-STAT pathway. Briefly, the binding of interferons to their receptors leads to the activation of Janus activated kinase 1 (JAK1), resulting in the tyrosine phosphorylation of downstream substrates, including signal transducer and activator of transcription 1 (STAT1) and STAT2. The complex of STAT1-STAT2-IRF9 (IFN-regulatory factor 9) enters the nucleus and binds to the ISRE motifs in the target gene, subsequently regulating ISG transcription and mediating the innate antiviral immune response.
To assess whether the induction of ISGs by nucleotide synthesis inhibitors also occurs via this classical pathway, we blocked the JAK-STAT cascade by employing the pharmacological JAK inhibitors JAK inhibitor 1 and CP-690550, which conceivably were identified to conceivably impair the expression of ISGs triggered by IFN-α (see Fig. S5 in the supplemental material). Surprisingly, the induction of ISGs as well as the anti-HEV effects of these inhibitors were not affected (Fig. 11). These results revealed that targeting nucleotide synthesis provokes ISG induction via a noncanonical mechanism that is independent of classical interferon signaling.
FIG 11.
ISG induction and the anti-HEV activity triggered by nucleotide synthesis inhibitors are independent of the JAK-STAT signaling pathway. The induction of ISGs (A and C) and the anti-HEV effects (B and D) by MPA were quantified in the presence or absence of JAK inhibitor 1 (A and B) or CP-690550 (CP) (C and D). The induction of ISGs was normalized to basal ISG expression without MPA treatment (purple bar; set as 1). The relative HEV RNA levels were normalized to cells without treatment of MPA (set as 1). Similarly, the induction of ISGs (E and G) and the anti-HEV effects (F and H) mediated by BQR were quantified in the presence or absence of JAK inhibitor 1 (E and F) or CP (G and H). The induction of ISGs was normalized to basal ISG expression without BQR treatment (purple bar; set as 1). The relative HEV RNA levels were normalized to cells without BQR treatment (set as 1). Data were normalized to two housekeeping genes and represent means ± SEM from three or four experiments.
DISCUSSION
Nucleotides are key components involved in host cell metabolism and virus infection. Most of the inhibitors targeting de novo nucleotide biosynthesis have been well characterized by many studies, and their efficacy in inhibiting nucleotide synthesis has been thoroughly demonstrated (16, 17, 24–31). Based on these findings, we profiled and established the effects and mechanism of action of inhibiting de novo nucleotide biosynthesis on HEV replication. Unexpectedly, targeting the early steps of the purine nucleotide synthesis pathway (before the primary purine IMP formed) leads to the enhancement of HEV replication, whereas targeting a later step (IMPDH enzyme) results in potent antiviral activity against HEV, an effect apparently relating to purine nucleotide depletion. The inhibition of the pyrimidine nucleotide synthesis pathway also inhibits HEV replication. Mechanistically, these effects are related to an unconventional interaction with cell-autonomous antiviral immunity dependent on the very strong induction of antiviral ISGs.
It is counterintuitive that targeting the upstream enzymes of the purine pathway (before IMP formed) by pharmacological inhibitors facilitates HEV replication, but the specificity became evident from silencing genes encoding the enzymes involved. Supplementation with exogenous purine nucleotides (adenosine or guanosine) in culture medium in the presence of these purine synthesis inhibitors was not capable of abrogating the stimulation of HEV replication, suggesting these proviral effects only partly relate to the nucleotide synthesis pathway (see Fig. S6A to C in the supplemental material). It is worth noting that targeting the early stage of purine synthesis results in the depletion of the ATP and/or GTP pool. Cellular energy metabolism mediated by ATP might be important for the host cells to defend against virus infection (32, 33). Therefore, insufficient ATP levels might facilitate HEV infection by escaping from host cellular immunity. However, how the ATP levels regulate virus infection deserves further investigation. Similarly, a previous study reported proviral activity by nucleotide biosynthesis inhibitors LFM and FK778 in the hepatitis B virus model, although these two compounds generally are antiviral against other viruses (17). Thus, the question of whether the pro-HEV effects of targeting the early steps of the purine pathway are specific to this virus or a general phenomenon in virus biology remains unanswered.
IMPDH, as a target for antiviral drug development for a broad spectrum of viruses, has been widely investigated. We previously demonstrated that the IMPDH inhibitors ribavirin and MPA inhibit HEV replication in vitro (18, 20). This study further validated this notion by showing the anti-HEV potential of 23 specifically designed IMPDH inhibitors. The levels of efficacy of 23 IMPDH inhibitors on HEV infection were consistent to various degrees, which might be due to their different abilities and variable affinities in inhibiting IMPDH1 and IMPDH2. As a competitive IMPDH inhibitor, ribavirin has been used in the clinic to treat chronic hepatitis C for decades. However, ribavirin monotherapy has a barely detectable effect on HCV viral load reduction (34), but when combined with IFN-α it doubles the response rate compared to that with IFN-α alone (35). In contrast, ribavirin monotherapy as an off-label treatment appears very effective for treating chronic HEV infection, in that viral clearance was observed in the majority of the patients as reported by a recent large retrospective multicenter study (36), although prospective randomized trials still are required to confirm the findings. In addition to IMPDH inhibition, ribavirin also possesses pleiotropic biological properties, including immunomodulation, inhibition of gene translation, interaction with viral RNA-dependent RNA polymerase (RdRp), and mutation of virus (37–39). Thus, the exact anti-HEV mechanism used by ribavirin remains to be further elucidated, but the present study provides evidence that the answers lie in its relation to nucleotide biosynthesis.
As a noncompetitive IMPDH inhibitor, MPA has been used as an immunosuppressant to prevent allograft rejection following organ transplantation (40). Despite the opposing effects on HEV of inhibitors targeting early or later steps of the purine synthesis cascade, we demonstrated that the anti-HEV action of MPA was independent of those early-step enzymes (see Fig. S6D in the supplemental material). Interestingly, clinical evidence appears to support our experimental observation that the use of immunosuppressive treatments containing mycophenolate mofetil (the prodrug of MPA) leads to more frequent HEV clearance in heart transplant recipients (41). Nevertheless, because of limited patient numbers, we still do not have sufficient evidence to draw a solid conclusion regarding the in vivo effect of MPA. A recent cohort study reported the anti-HEV activity by ribavirin was not affected by MPA in patients, but they did not analyze the direct effect of MPA on HEV infection (42).
The three inhibitors used in our study that interfere with pyrimidine synthesis have been described in many previous studies (16, 29–31). Adding to the previous knowledge that pyrimidine synthesis inhibitors, such as BQR and LFM, have broad antiviral activity against a spectrum of viruses (16, 23, 43), we now report their potent anti-HEV activity. Both BQR and LFM are immunosuppressive agents, although whether the mechanism of action is solely via pyrimidine inhibition remains controversial and unclear (44–46). The efficacy of BQR against graft rejection has been investigated extensively in preclinical models (47–49), and LFM has been proposed as an off-label immunosuppressive therapy in bone marrow (11) and renal (50) transplantation. In addition, DHODH inhibitors have been explored as treatments of other diseases, including malaria, autoimmune and inflammatory diseases, cancer, rheumatoid arthritis, and psoriasis (51–55). Given the bifunctional antiviral and immunosuppressive effects of BQR and LFM, these regimens may hold the potential to treat HEV-infected organ recipients.
Interestingly, nucleotide synthesis interacts with cellular antiviral immune responses. Here, we demonstrated a direct effect of the depletion of nucleotide pools on the transcription of antiviral ISGs. ISGs are antiviral effectors that are thought to be induced by interferons only. Although hundreds of ISGS have been identified, recent functional studies of individual ISGs surprisingly have found that only a small subset of ISGs actually have potent or broad antiviral activities; these ISGs include IRF1, DDX58, and IRF7 (56, 57). It is these antiviral ISGs that are induced in our HEV models upon treatment with nucleotide synthesis inhibitors. Consistent with this, previous studies in HCV models reported that the induction of IRF1 or IRF7 was associated with the antiviral activity of MPA (19) or ribavirin (58), respectively. Furthermore, the antiviral activity of inhibitors of pyrimidine biosynthesis against measles virus, chikungunya virus, and West Nile virus also was associated with the induction of ISGs (23).
For now, the mechanistic details of inhibitors of nucleotide biosynthesis that can induce ISGs remain obscure. Classically, the transcription of ISGs is initiated from the binding of interferons to their receptors, which subsequently drives the activation of the JAK-STAT cascade (56). The inhibition of JAK1 in the phosphorylation STAT1, the key event of interferon signaling transduction, often results in the complete blockage of antiviral interferon responses (59). However, exceptions also exist, in that ISGs can be induced in the absence of JAK1 or STAT1 activation (60, 61). Here, we found that the induction of ISGs and the anti-HEV effects by nucleotide synthesis inhibitors are independent of the classical JAK-STAT cascade, suggesting the involvement of a noncanonical mechanism that is independent of interferons, and the identification of these mechanisms should have substantial value for our understanding of antiviral immunity.
In conclusion, selectively targeting host enzymes involved in de novo nucleotide biosynthesis potently inhibits HEV replication. Furthermore, nucleotide biosynthesis pathways interact with cellular immune responses, and all of the pharmacological inhibitors exerting anti-HEV activity are capable of triggering antiviral ISG transcription. Thus, targeting nucleotide biosynthesis represents a viable option for antiviral drug development against HEV.
Supplementary Material
ACKNOWLEDGMENTS
We thank Suzanne U. Emerson (National Institute of Allergy and Infectious Diseases, NIH, USA) for generously providing the plasmids to generate subgenomic and full-length HEV genomic RNA.
We thank the Center for Drug Design, University of Minnesota, for generous financial support of developing new IMPDH inhibitors.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02700-15.
REFERENCES
- 1.Kamar N, Bendall R, Legrand-Abravanel F, Xia NS, Ijaz S, Izopet J, Dalton HR. 2012. Hepatitis E. Lancet 379:2477–2488. doi: 10.1016/S0140-6736(11)61849-7. [DOI] [PubMed] [Google Scholar]
- 2.Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel JP, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358:811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, de Man RA, de Knegt RJ, Metselaar HJ, Peppelenbosch MP, Pan Q. 2013. Epidemiology and management of chronic hepatitis E infection in solid organ transplantation: a comprehensive literature review. Rev Med Virol 23:295–304. doi: 10.1002/rmv.1751. [DOI] [PubMed] [Google Scholar]
- 4.Kamar N, Garrouste C, Haagsma EB, Garrigue V, Pischke S, Chauvet C, Dumortier J, Cannesson A, Cassuto-Viguier E, Thervet E, Conti F, Lebray P, Dalton HR, Santella R, Kanaan N, Essig M, Mousson C, Radenne S, Roque-Afonso AM, Izopet J, Rostaing L. 2011. Factors associated with chronic hepatitis in patients with hepatitis E virus infection who have received solid organ transplants. Gastroenterology 140:1481–1489. doi: 10.1053/j.gastro.2011.02.050. [DOI] [PubMed] [Google Scholar]
- 5.van der Eijk AA, Pas SD, Cornelissen JJ, de Man RA. 2014. Hepatitis E virus infection in hematopoietic stem cell transplant recipients. Curr Opin Infect Dis 27:309–315. doi: 10.1097/QCO.0000000000000076. [DOI] [PubMed] [Google Scholar]
- 6.Lane AN, Fan TW. 2015. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res 43:2466–2485. doi: 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffatt BA, Ashihara H. 2002. Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis Book 1:e0018. doi: 10.1199/tab.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blakley RL, Vitols E. 1968. The control of nucleotide biosynthesis. Annu Rev Biochem 37:201–224. doi: 10.1146/annurev.bi.37.070168.001221. [DOI] [PubMed] [Google Scholar]
- 9.Christopherson RI, Lyons SD, Wilson PK. 2002. Inhibitors of de novo nucleotide biosynthesis as drugs. Acc Chem Res 35:961–971. doi: 10.1021/ar0000509. [DOI] [PubMed] [Google Scholar]
- 10.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL Jr, Haussinger D, Diago M, Carosi G, Dhumeaux D, Craxi A, Lin A, Hoffman J, Yu J. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med 347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 11.Avery RK, Bolwell BJ, Yen-Lieberman B, Lurain N, Waldman WJ, Longworth DL, Taege AJ, Mossad SB, Kohn D, Long JR, Curtis J, Kalaycio M, Pohlman B, Williams JW. 2004. Use of leflunomide in an allogeneic bone marrow transplant recipient with refractory cytomegalovirus infection. Bone Marrow Transplant 34:1071–1075. doi: 10.1038/sj.bmt.1704694. [DOI] [PubMed] [Google Scholar]
- 12.Farasati NA, Shapiro R, Vats A, Randhawa P. 2005. Effect of leflunomide and cidofovir on replication of BK virus in an in vitro culture system. Transplantation 79:116–118. doi: 10.1097/01.TP.0000149338.97084.5F. [DOI] [PubMed] [Google Scholar]
- 13.Chong AS, Zeng H, Knight DA, Shen J, Meister GT, Williams JW, Waldman WJ. 2006. Concurrent antiviral and immunosuppressive activities of leflunomide in vivo. Am J Transplant 6:69–75. doi: 10.1111/j.1600-6143.2005.01152.x. [DOI] [PubMed] [Google Scholar]
- 14.Takhampunya R, Ubol S, Houng HS, Cameron CE, Padmanabhan R. 2006. Inhibition of dengue virus replication by mycophenolic acid and ribavirin. J Gen Virol 87:1947–1952. doi: 10.1099/vir.0.81655-0. [DOI] [PubMed] [Google Scholar]
- 15.Ying C, Colonno R, De Clercq E, Neyts J. 2007. Ribavirin and mycophenolic acid markedly potentiate the anti-hepatitis B virus activity of entecavir. Antiviral Res 73:192–196. doi: 10.1016/j.antiviral.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Wang QY, Bushell S, Qing M, Xu HY, Bonavia A, Nunes S, Zhou J, Poh MK, Florez de Sessions P, Niyomrattanakit P, Dong H, Hoffmaster K, Goh A, Nilar S, Schul W, Jones S, Kramer L, Compton T, Shi PY. 2011. Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J Virol 85:6548–6556. doi: 10.1128/JVI.02510-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoppe-Seyler K, Sauer P, Lohrey C, Hoppe-Seyler F. 2012. The inhibitors of nucleotide biosynthesis leflunomide, FK778, and mycophenolic acid activate hepatitis B virus replication in vitro. Hepatology 56:9–16. doi: 10.1002/hep.25602. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Zhou X, Debing Y, Chen K, Van Der Laan LJ, Neyts J, Janssen HL, Metselaar HJ, Peppelenbosch MP, Pan Q. 2014. Calcineurin inhibitors stimulate and mycophenolic acid inhibits replication of hepatitis E virus. Gastroenterology 146:1775–1783. doi: 10.1053/j.gastro.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 19.Pan Q, de Ruiter PE, Metselaar HJ, Kwekkeboom J, de Jonge J, Tilanus HW, Janssen HL, van der Laan LJ. 2012. Mycophenolic acid augments interferon-stimulated gene expression and inhibits hepatitis C virus infection in vitro and in vivo. Hepatology 55:1673–1683. doi: 10.1002/hep.25562. [DOI] [PubMed] [Google Scholar]
- 20.Debing Y, Emerson SU, Wang Y, Pan Q, Balzarini J, Dallmeier K, Neyts J. 2014. Ribavirin inhibits in vitro hepatitis E virus replication through depletion of cellular GTP pools and is moderately synergistic with alpha interferon. Antimicrob Agents Chemother 58:267–273. doi: 10.1128/AAC.01795-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leyssen P, De Clercq E, Neyts J. 2006. The anti-yellow fever virus activity of ribavirin is independent of error-prone replication. Mol Pharmacol 69:1461–1467. doi: 10.1124/mol.105.020057. [DOI] [PubMed] [Google Scholar]
- 22.Qing M, Zou G, Wang QY, Xu HY, Dong H, Yuan Z, Shi PY. 2010. Characterization of dengue virus resistance to brequinar in cell culture. Antimicrob Agents Chemother 54:3686–3695. doi: 10.1128/AAC.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucas-Hourani M, Dauzonne D, Jorda P, Cousin G, Lupan A, Helynck O, Caignard G, Janvier G, Andre-Leroux G, Khiar S, Escriou N, Despres P, Jacob Y, Munier-Lehmann H, Tangy F, Vidalain PO. 2013. Inhibition of pyrimidine biosynthesis pathway suppresses viral growth through innate immunity. PLoS Pathog 9:e1003678. doi: 10.1371/journal.ppat.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson JA, Carpenter JW, Rose LM, Adamson DJ. 1975. Mechanisms of action of 6-thioguanine, 6-mercaptopurine, and 8-azaguanine. Cancer Res 35:2872–2878. [PubMed] [Google Scholar]
- 25.Beardsley GP, Moroson BA, Taylor EC, Moran RG. 1989. A new folate antimetabolite, 5,10-dideaza-5,6,7,8-tetrahydrofolate is a potent inhibitor of de novo purine synthesis. J Biol Chem 264:328–333. [PubMed] [Google Scholar]
- 26.Pizzorno G, Moroson BA, Cashmore AR, Beardsley GP. 1991. (6R)-5,10-Dideaza-5,6,7,8-tetrahydrofolic acid effects on nucleotide metabolism in CCRF-CEM human T-lymphoblast leukemia cells. Cancer Res 51:2291–2295. [PubMed] [Google Scholar]
- 27.Budzik GP, Colletti LM, Faltynek CR. 2000. Effects of methotrexate on nucleotide pools in normal human T cells and the CEM T cell line. Life Sci 66:2297–2307. doi: 10.1016/S0024-3205(00)00559-2. [DOI] [PubMed] [Google Scholar]
- 28.Carr SF, Papp E, Wu JC, Natsumeda Y. 1993. Characterization of human type I and type II IMP dehydrogenases. J Biol Chem 268:27286–27290. [PubMed] [Google Scholar]
- 29.Silva HT Jr, Cao W, Shorthouse RA, Loffler M, Morris RE. 1997. In vitro and in vivo effects of leflunomide, brequinar, and cyclosporine on pyrimidine biosynthesis. Transplant Proc 29:1292–1293. doi: 10.1016/S0041-1345(96)00523-4. [DOI] [PubMed] [Google Scholar]
- 30.Greene S, Watanabe K, Braatz-Trulson J, Lou L. 1995. Inhibition of dihydroorotate dehydrogenase by the immunosuppressive agent leflunomide. Biochem Pharmacol 50:861–867. doi: 10.1016/0006-2952(95)00255-X. [DOI] [PubMed] [Google Scholar]
- 31.Lopez JM, Marks CL, Freese E. 1979. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim Biophys Acta 587:238–252. doi: 10.1016/0304-4165(79)90357-X. [DOI] [PubMed] [Google Scholar]
- 32.Seo JY, Yaneva R, Hinson ER, Cresswell P. 2011. Human mtt megalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 332:1093–1097. doi: 10.1126/science.1202007. [DOI] [PubMed] [Google Scholar]
- 33.Rawling DC, Fitzgerald ME, Pyle AM. 2015. Establishing the role of ATP for the function of the RIG-I innate immune sensor. eLife 4:e09391. doi: 10.7554/eLife.09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pawlotsky JM, Dahari H, Neumann AU, Hezode C, Germanidis G, Lonjon I, Castera L, Dhumeaux D. 2004. Antiviral action of ribavirin in chronic hepatitis C. Gastroenterology 126:703–714. doi: 10.1053/j.gastro.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Poynard T, Marcellin P, Lee SS, Niederau C, Minuk GS, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C, Albrecht J. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT). Lancet 352:1426–1432. [DOI] [PubMed] [Google Scholar]
- 36.Kamar N, Izopet J, Tripon S, Bismuth M, Hillaire S, Dumortier J, Radenne S, Coilly A, Garrigue V, D'Alteroche L, Buchler M, Couzi L, Lebray P, Dharancy S, Minello A, Hourmant M, Roque-Afonso AM, Abravanel F, Pol S, Rostaing L, Mallet V. 2014. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N Engl J Med 370:1111–1120. doi: 10.1056/NEJMoa1215246. [DOI] [PubMed] [Google Scholar]
- 37.Tam RC, Lau JY, Hong Z. 2001. Mechanisms of action of ribavirin in antiviral therapies. Antivir Chem Chemother 12:261–272. doi: 10.1177/095632020101200501. [DOI] [PubMed] [Google Scholar]
- 38.Paeshuyse J, Dallmeier K, Neyts J. 2011. Ribavirin for the treatment of chronic hepatitis C virus infection: a review of the proposed mechanisms of action. Curr Opin Virol 1:590–598. doi: 10.1016/j.coviro.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 39.Feld JJ, Hoofnagle JH. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 40.Manzia TM, De Liguori Carino N, Orlando G, Toti L, De Luca L, D'Andria D, Cardillo A, Anselmo A, Casciani CU, Tisone G. 2005. Use of mycophenolate mofetil in liver transplantation: a literature review. Transplant Proc 37:2616–2617. doi: 10.1016/j.transproceed.2005.06.073. [DOI] [PubMed] [Google Scholar]
- 41.Pischke S, Stiefel P, Franz B, Bremer B, Suneetha PV, Heim A, Ganzenmueller T, Schlue J, Horn-Wichmann R, Raupach R, Darnedde M, Scheibner Y, Taubert R, Haverich A, Manns MP, Wedemeyer H, Bara CL. 2012. Chronic hepatitis E in heart transplant recipients. Am J Transplant 12:3128–3133. doi: 10.1111/j.1600-6143.2012.04200.x. [DOI] [PubMed] [Google Scholar]
- 42.Kamar N, Lhomme S, Abravanel F, Cointault O, Esposito L, Cardeau-Desangles I, Del Bello A, Dorr G, Lavayssiere L, Nogier MB, Guitard J, Ribes D, Goin AL, Broue P, Metsu D, Saune K, Rostaing L, Izopet J. 2015. An early viral response predicts the virological response to ribavirin in hepatitis E virus organ transplant patients. Transplantation 99:2124–2131. doi: 10.1097/TP.0000000000000850. [DOI] [PubMed] [Google Scholar]
- 43.Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML. 2011. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci U S A 108:5777–5782. doi: 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chong AS, Rezai K, Gebel HM, Finnegan A, Foster P, Xu X, Williams JW. 1996. Effects of leflunomide and other immunosuppressive agents on T cell proliferation in vitro. Transplantation 61:140–145. doi: 10.1097/00007890-199601150-00026. [DOI] [PubMed] [Google Scholar]
- 45.Xu X, Williams JW, Shen J, Gong H, Yin DP, Blinder L, Elder RT, Sankary H, Finnegan A, Chong AS. 1998. In vitro and in vivo mechanisms of action of the antiproliferative and immunosuppressive agent, brequinar sodium. J Immunol 160:846–853. [PubMed] [Google Scholar]
- 46.Cherwinski HM, Cohn RG, Cheung P, Webster DJ, Xu YZ, Caulfield JP, Young JM, Nakano G, Ransom JT. 1995. The immunosuppressant leflunomide inhibits lymphocyte proliferation by inhibiting pyrimidine biosynthesis. J Pharmacol Exp Ther 275:1043–1049. [PubMed] [Google Scholar]
- 47.Makowka L, Sher LS, Cramer DV. 1993. The development of brequinar as an immunosuppressive drug for transplantation. Immunol Rev 136:51–70. doi: 10.1111/j.1600-065X.1993.tb00654.x. [DOI] [PubMed] [Google Scholar]
- 48.Cramer DV, Chapman FA, Makowka L. 1993. The use of brequinar sodium for transplantation. Ann N Y Acad Sci 696:216–226. [DOI] [PubMed] [Google Scholar]
- 49.Cramer DV, Chapman FA, Jaffee BD, Jones EA, Knoop M, Hreha-Eiras G, Makowka L. 1992. The effect of a new immunosuppressive drug, brequinar sodium, on heart, liver, and kidney allograft rejection in the rat. Transplantation 53:303–308. doi: 10.1097/00007890-199202010-00009. [DOI] [PubMed] [Google Scholar]
- 50.Chon WJ, Josephson MA. 2011. Leflunomide in renal transplantation. Expert Rev Clin Immunol 7:273–281. doi: 10.1586/eci.11.20. [DOI] [PubMed] [Google Scholar]
- 51.Boa AN, Canavan SP, Hirst PR, Ramsey C, Stead AM, McConkey GA. 2005. Synthesis of brequinar analogue inhibitors of malaria parasite dihydroorotate dehydrogenase. Bioorg Med Chem 13:1945–1967. doi: 10.1016/j.bmc.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 52.Leban J, Vitt D. 2011. Human dihydroorotate dehydrogenase inhibitors, a novel approach for the treatment of autoimmune and inflammatory diseases. Arzneimittelforschung 61:66–72. doi: 10.1055/s-0031-1296169. [DOI] [PubMed] [Google Scholar]
- 53.Baumann P, Mandl-Weber S, Volkl A, Adam C, Bumeder I, Oduncu F, Schmidmaier R. 2009. Dihydroorotate dehydrogenase inhibitor A771726 (leflunomide) induces apoptosis and diminishes proliferation of multiple myeloma cells. Mol Cancer Ther 8:366–375. doi: 10.1158/1535-7163.MCT-08-0664. [DOI] [PubMed] [Google Scholar]
- 54.Herrmann ML, Schleyerbach R, Kirschbaum BJ. 2000. Leflunomide: an immunomodulatory drug for the treatment of rheumatoid arthritis and other autoimmune diseases. Immunopharmacology 47:273–289. doi: 10.1016/S0162-3109(00)00191-0. [DOI] [PubMed] [Google Scholar]
- 55.Norman P. 2013. Evaluation of WO2013076170: the use of a dihydroorotate dehydrogenase inhibitor for the treatment of psoriasis. Expert Opin Ther Pat 23:1391–1394. doi: 10.1517/13543776.2013.831075. [DOI] [PubMed] [Google Scholar]
- 56.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas E, Feld JJ, Li Q, Hu Z, Fried MW, Liang TJ. 2011. Ribavirin potentiates interferon action by augmenting interferon-stimulated gene induction in hepatitis C virus cell culture models. Hepatology 53:32–41. doi: 10.1002/hep.23985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart CE, Randall RE, Adamson CS. 2014. Inhibitors of the interferon response enhance virus replication in vitro. PLoS One 9:e112014. doi: 10.1371/journal.pone.0112014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leaman DW, Pisharody S, Flickinger TW, Commane MA, Schlessinger J, Kerr IM, Levy DE, Stark GR. 1996. Roles of JAKs in activation of STATs and stimulation of c-fos gene expression by epidermal growth factor. Mol Cell Biol 16:369–375. doi: 10.1128/MCB.16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shresta S, Sharar KL, Prigozhin DM, Snider HM, Beatty PR, Harris E. 2005. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J Immunol 175:3946–3954. doi: 10.4049/jimmunol.175.6.3946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.