Abstract
We determined the complete nucleotide sequence of a self-transmissible IncL/M plasmid, pKOI-34, from a Klebsiella oxytoca isolate. pKOI-34 possessed the core structure of an IncL/M plasmid found in Erwinia amylovora, pEL60, with two mobile elements inserted, a transposon carrying the arsenic resistance operon and a Tn21-like core module (tnp and mer modules) piggybacking blaIMP-34 as a class 1 integron, In808, where blaIMP-34 confers a resistance to carbapenems in K. oxytoca and Klebsiella pneumoniae.
TEXT
Carbapenem-resistant Enterobacteriaceae (CRE) infections are increasing, which is one of the most important issues in health care facilities around the world (1, 2). In Japan, IMP-6, a variant of IMP-1 with one amino acid substitution (Ser196Gly), has been detected sporadically in clinical isolates of the family Enterobacteriaceae (3). IMP-6 shows a paradoxical effect on carbapenem resistance, i.e., imipenem susceptible but meropenem resistant (4), where a plasmid pKPI-6 harboring blaIMP-6 and blaCTX-M-2 confers this stealth-type carbapenem-resistant phenotype that is not detectable when testing for imipenem resistance (5).
(Preliminary data from this study were presented at the 62nd Annual Meeting of the Japanese Society of Chemotherapy, 2014.)
During a screening for MBL-producing Enterobacteriaceae showing the paradoxical resistance to carbapenems, we found IMP-34 in five MBL-positive isolates of Klebsiella oxytoca (MS5279, MS5280) and Klebsiella pneumoniae (MS5284, MS5285, MS5286) in the Kinki region, which is the geographic center of Japan. The isolates showed intermediate resistance to imipenem but resistance to meropenem (6). IMP-34 has a single amino acid substitution, Glu87Gly, compared to IMP-1. Conjugal transfer experiments showed that blaIMP-34 is located on a self-transmissible plasmid designated pKOI-34.
We sequenced the plasmid DNA of pKOI-34 purified from the E. coli BL21 transconjugant (BL21_pKOI-34) from K. oxytoca strain MS5279. The draft sequences of pKOI-34 were generated using Illumina MiSeq (Nextera paired-end library; 2,354,946 bp), assembled using CLC Genomics Workbench (CLC bio, Cambridge, MA), and sorted using OSLay (7). Gap closing was performed using direct sequencing of PCR products amplified with oligonucleotide primers designed to anneal each end to the neighboring contigs. pKOI-34 is an 87,343-bp plasmid with an average GC content of 53%, and 104 open reading frames (ORFs) (Fig. 1; Table 1). The BLASTP program showed that the amino acid sequence of RepA in pKOI-34 conforms to the IncL/M group. Replicon typing of plasmid preparations from the five strains positive for blaIMP-34 showed that they all belong to the IncL/M group. Sequence comparisons of pKOI-34 with the plasmids registered in the GenBank database show extensive similarity to IncL/M plasmids (see Fig. S1a in the supplemental material).
FIG 1.
Structural features of IncL/M plasmid pKOI-34 (87,343 bp) in comparison with pEL60, which is considered the ancestral strain of the IncL/M plasmid. Two mobile gene elements of 8.8 and 19.5 kb, respectively, are inserted into the IncL/M backbone. Arrows show annotated coding sequences. The 104 identified ORFs are color coded on the basis of function as follows: blue, replication-related genes; green, antibiotic-resistant genes; red, transposases; yellow, conjugation-related genes. The lower schema shows the result of PCR scanning for plasmids carrying blaIMP-34 recovered from K. oxytoca and K. pneumoniae. The 22 sets of PCR primers were designed from the complete sequence of pKOI-34. All plasmids without pKOI-34 showed the same results: set primers 3 and 4 were not detected, and the area corresponding to the mobile gene element containing the arsenic resistance genes is missing. PCR using the forward primer of set 3 and the reverse primer of set 4 showed a DNA fragment of 600 bp (see Table S1 in the supplemental material).
TABLE 1.
Features of pKOI-34 ORFs
| ORFs | Position (bp) |
Strand | Gene | Length (no. of aaa) | Source | Description | Identity (%) | Overlap (no. of aa) | Accession no. | |
|---|---|---|---|---|---|---|---|---|---|---|
| Start | Stop | |||||||||
| 1 | 1 | 750 | + | repA | 249 | K. pneumoniae | RepA, IncL/M type replicase protein | 99 | 248/249 | WP_020277896 |
| 2 | 2,120 | 4,153 | − | trbC | 677 | K. pneumoniae (pOXA-48) | Protein involved in plasmid transfer | 99 | 673/677 | YP_006958848 |
| 3 | 4,510 | 4,863 | + | arsR | 117 | K. pneumoniae (plasmid 12) | Arsenic-resistant operon repressor | 100 | 117/117 | YP_002287003 |
| 4 | 4,911 | 5,273 | + | arsD | 120 | K. pneumoniae (plasmid 12) | Arsenic-resistant operon trans-acting repressor | 100 | 120/120 | YP_002287003 |
| 5 | 5,291 | 7,041 | + | arsA | 583 | K. pneumoniae (plasmid 12) | Arsenic pump driving ATPase | 99 | 582/583 | YP_002287002 |
| 6 | 7,091 | 8,380 | + | arsB | 429 | K. pneumoniae (plasmid 12) | Arsenic pump membrane protein | 100 | 429/429 | YP_002287001 |
| 7 | 8,393 | 8,818 | + | arsC | 141 | K. pneumoniae (plasmid 12) | Arsenate reductase | 100 | 141/141 | YP_002287001 |
| 8 | 8,849 | 9,223 | − | arsA | 124 | Enterobacteriaceae | Arsenic transporter ATPase | 100 | 124/124 | WP_004118313 |
| 9 | 9,262 | 9,834 | − | tnpR | 190 | Yersinia enterocolitica | TnpR, site-specific recombinases | 100 | 190/190 | WP_012291338 |
| 10 | 9,998 | 13,006 | + | tnpA | 1,002 | K. pneumoniae (pKP1433) | Transposase, Tn3 family | 100 | 1002/1002 | YP_008003449 |
| 11 | 13,003 | 13,992 | − | trbB | 329 | K. pneumoniae (pCTXM360) | Protein disulfide isomerase | 91 | 300/329 | YP_002333315 |
| 12 | 14,003 | 15,310 | − | trbA | 435 | K. pneumoniae (pCTXM360) | Conjugal transfer protein | 98 | 428/435 | YP_002333316 |
| 13 | 15,310 | 15,705 | − | trbN | 131 | K. pneumoniae (pOXA-48) | Soluble lytic murein transglycosylase and related regulatory proteins | 100 | 131/131 | YP_006958778 |
| 14 | 15,810 | 16,160 | − | 116 | K. pneumoniae | Archaeal fructose-1,6-bisphosphatase | 100 | 116/116 | WP_004187434 | |
| 15 | 16,273 | 16,926 | + | tir | 217 | E. cloacae (pNE1280) | Transfer inhibition protein | 98 | 214/217 | YP_006964687 |
| 16 | 17,019 | 17,276 | + | pemI | 85 | K. pneumoniae (pJEG011) | Growth regulator, antitoxin involved in plasmid maintenance | 100 | 85/85 | YP_007878506 |
| 17 | 17,278 | 17,610 | + | pemK | 110 | K. pneumoniae (pKPoxa-48N1) | Growth inhibitor, toxin involved in plasmid maintenance | 100 | 110/110 | YP_008090835 |
| 18 | 17,703 | 18,137 | + | mucA | 144 | K. pneumoniae (pOXA-48) | SOS response transcriptional repressors | 99 | 143/144 | YP_006958786 |
| 19 | 18,152 | 19,390 | + | mucB | 412 | K. pneumoniae | Nucleotidyltransferase | 99 | 411/412 | AHE47434 |
| 20 | 19,989 | 20,993 | − | tnpA | 334 | K. pneumoniae (pNDM-MAR) | Transposase of IS4321R | 100 | 334/334 | YP_005352261 |
| 21 | 21,072 | 24,044 | − | tnpA | 990 | K. oxytoca (pKOX_R1) | Transposase, Tn3 family protein | 100 | 990/990 | YP_006501624 |
| 22 | 24,047 | 24,604 | − | tnpR | 185 | K. pneumoniae (pIMP-PH114) | Resolvase domain containing protein | 100 | 185/185 | YP_008725240 |
| 23 | 24,910 | 25,923 | − | intl1 | 337 | K. oxytoca (pINCan01) | Integrase | 100 | 337/337 | YP_006961973 |
| 24 | 26,174 | 26,506 | + | qacF | 110 | Stenotrophomonas maltophilia | Ethidium bromide resistance protein | 99 | 109/110 | AHN60087 |
| 25 | 26,595 | 27,149 | + | aacA4 | 184 | E. cloacae (pEC-IMP) | Aminoglycoside N(6′)-acetyltransferase | 100 | 184/184 | YP_002791400 |
| 26 | 27,226 | 27,966 | + | blaIMP-34 | 246 | Pseudomonas aeruginosa | Metallo-β-lactamase | 99 | 245/246 | ABF70513 |
| 27 | 28,192 | 28,524 | + | qacE2 | 110 | Uncultured bacterium | Ethidium bromide resistance protein | 99 | 109/110 | ACN22644 |
| 28 | 28,735 | 29,082 | + | qacEΔ1 | 115 | K. pneumoniae (pK29) | Ethidium bromide resistance protein | 100 | 115/115 | YP_001965784 |
| 29 | 29,076 | 29,915 | + | sul1 | 279 | K. pneumoniae (pK29) | Dihydropteroate synthase | 100 | 279/279 | YP_001965785 |
| 30 | 30,043 | 30,543 | + | orf5 | 166 | P. aeruginosa (Rms149) | Acetyltransferase (GNAT) family protein | 100 | 166/166 | YP_245437 |
| 31 | 30,512 | 31,504 | − | tniB | 330 | Salmonella enterica (pOU7519) | NTPb binding protein | 100 | 330/330 | YP_209341 |
| 32 | 31,507 | 33,186 | − | tinA | 559 | K. pneumoniae (pR55) | Transposase and inactivated derivatives | 100 | 559/559 | YP_005352048 |
| 33 | 33,261 | 33,968 | − | urf2 | 235 | S. enterica (pOU7519) | Tn21 protein of unknown function Urf2 | 100 | 235/235 | YP_001598123 |
| 34 | 33,965 | 34,201 | − | merE | 78 | K. pneumoniae (pKpQIL) | Mercuric transport protein | 100 | 78/78 | YP_003560404 |
| 35 | 34,198 | 34,560 | − | merD | 120 | K. pneumoniae (pKpQIL) | Transcriptional regulator | 100 | 120/120 | YP_003560405 |
| 36 | 34,578 | 36,272 | − | merA | 564 | K. pneumoniae (pKpQIL) | Mercuric ion reductase | 100 | 564/564 | YP_003560406 |
| 37 | 36,324 | 36,785 | − | merC | 153 | K. pneumoniae (pKPHS2) | Mercury transport protein | 100 | 153/153 | YP_005229641 |
| 38 | 36,782 | 37,057 | − | merP | 91 | K. pneumoniae (pR55) | Mercury transport protein | 100 | 91/91 | YP_005352054 |
| 39 | 37,071 | 37,421 | − | merT | 116 | K. pneumoniae (pKP048) | Mercury transport protein | 100 | 116/116 | YP_003754062 |
| 40 | 37,493 | 37,927 | + | merR | 144 | E. coli | Mercuric resistance operon regulatory protein | 100 | 144/144 | KKA59047 |
| 41 | 38,006 | 39,010 | + | tnpA | 334 | K. pneumoniae (pNDM-MAR) | Transposase of IS4321R | 100 | 334/334 | YP_005352261 |
| 42 | 39,844 | 40,164 | − | 106 | Enterobacteriaceae | Hypothetical protein | 99 | 106/106 | WP_021561451 | |
| 43 | 40,309 | 40,947 | − | 212 | E. coli | Hypothetical protein | 100 | 212/212 | WP_021561450 | |
| 44 | 41,015 | 41,224 | − | 69 | K. pneumoniae (pCTXM360) | Hypothetical protein | 100 | 69/69 | YP_002333329 | |
| 45 | 41,227 | 41,445 | − | 72 | K. pneumoniae (pCTXM360) | Hypothetical protein | 97 | 71/72 | YP_002333330 | |
| 46 | 41,490 | 42,191 | − | 233 | K. pneumoniae | Hypothetical protein | 92 | 219/233 | WP_023302501 | |
| 47 | 42,208 | 43,539 | − | 443 | K. pneumoniae | Hypothetical protein | 93 | 414/443 | WP_023302502 | |
| 48 | 44,068 | 44,424 | + | 118 | K. pneumoniae | Hypothetical protein | 92 | 113/118 | WP_023302503 | |
| 49 | 44,402 | 44,980 | + | 192 | K. pneumoniae | Hypothetical protein | 93 | 184/192 | WP_023302504 | |
| 50 | 44,982 | 45,389 | + | 135 | K. pneumoniae (pJEG011) | Hypothetical protein | 98 | 134/135 | YP_007878517 | |
| 51 | 45,542 | 46,087 | − | 181 | K. pneumoniae (pJEG011) | Hypothetical protein | 98 | 178/181 | YP_007878518 | |
| 52 | 46,225 | 46,683 | + | 152 | Erwinia amylovora (pEL60) | Hypothetical protein | 90 | 141/152 | NP_943212 | |
| 53 | 46,680 | 46,928 | + | 82 | K. pneumoniae (pNDM-OM) | Hypothetical protein | 98 | 82/82 | YP_007195536 | |
| 54 | 46,921 | 47,508 | + | 195 | K. pneumoniae (pCTXM360) | Hypothetical protein | 98 | 191/195 | YP_002333340 | |
| 55 | 47,505 | 47,990 | + | 161 | K. pneumoniae (pCTXM360) | Hypothetical protein | 99 | 160/161 | YP_002333341 | |
| 56 | 47,987 | 48,235 | + | 82 | K. pneumoniae (pKPoxa-48N2) | Hypothetical protein | 93 | 81/82 | YP_008110943 | |
| 57 | 48,254 | 48,994 | + | resD | 246 | K. pneumoniae (pKP048) | Resolvase | 99 | 245/246 | YP_006958800 |
| 58 | 49,235 | 50,209 | + | parA | 324 | K. pneumoniae (pCTXM360) | StbA family protein | 100 | 324/324 | YP_002333344 |
| 59 | 50,212 | 50,655 | + | parB | 147 | K. pneumoniae (pCTXM360) | Plasmid stability protein | 100 | 147/147 | YP_002333345 |
| 60 | 50,665 | 51,216 | + | nuc | 183 | K. pneumoniae (pCTXM360) | Catalytic domain of EDTA-resistant nuclease | 100 | 183/183 | YP_002333346 |
| 61 | 51,334 | 51,840 | + | 168 | K. pneumoniae (pCTXM360) | Hypothetical protein | 100 | 168/168 | YP_002333347 | |
| 62 | 51,833 | 52,312 | + | 159 | K. pneumoniae (pCTXM360) | Hypothetical protein | 100 | 159/159 | YP_002333348 | |
| 63 | 52341 | 52,751 | + | 136 | K. pneumoniae (pCTXM360) | Hypothetical protein | 99 | 136/136 | YP_002333349 | |
| 64 | 52,869 | 53,132 | + | 87 | K. pneumoniae (pCTXM360) | Hypothetical protein | 100 | 87/87 | YP_002333350 | |
| 65 | 53,154 | 53,516 | + | 120 | K. pneumoniae (pCTXM360) | Hypothetical protein | 100 | 120/120 | YP_002333351 | |
| 66 | 53,638 | 54,087 | + | 149 | C. freundii (pCTX-M3) | Hypothetical protein | 100 | 149/149 | NP_774994 | |
| 67 | 54,132 | 54,398 | + | korC | 88 | K. pneumoniae (pCTXM360) | Putative transcriptional repressor protein | 100 | 88/88 | YP_002333353 |
| 68 | 54,462 | 55,745 | + | 427 | K. pneumoniae (pCTXM360) | Hypothetical protein | 100 | 427/427 | YP_002333354 | |
| 69 | 56,352 | 56,531 | + | ccgA1 | 59 | E. cloacae (pEI1573) | Hypothetical protein | 100 | 59/59 | YP_006965388 |
| 70 | 56,664 | 57,464 | + | 266 | K. pneumoniae (pCTXM360) | Hypothetical protein | 99 | 265/266 | YP_002333355 | |
| 71 | 57,655 | 57,993 | + | 112 | K. pneumoniae (pOXA-48) | Hypothetical protein | 100 | 112/112 | YP_006958814 | |
| 72 | 58,091 | 58,321 | + | rmoA | 76 | K. pneumoniae (Kp11978) | Hemolysin expression modulating protein | 100 | 76/76 | YP_006958815 |
| 73 | 58,653 | 58,868 | + | 71 | K. pneumoniae (pKPoxa-48N1) | Hypothetical protein | 99 | 71/71 | YP_008090866 | |
| 74 | 58,940 | 59,350 | + | 136 | K. pneumoniae (pOXA-48) | Hypothetical protein | 100 | 136/136 | YP_002333358 | |
| 75 | 59,412 | 59,717 | + | 101 | K. pneumoniae (pNDM-HK) | Hypothetical protein | 100 | 101/101 | YP_006952492 | |
| 76 | 59,918 | 60,358 | + | klcA | 146 | K. pneumoniae (pNDM-OM) | Antirestriction protein | 100 | 146/146 | YP_007195464 |
| 77 | 60,402 | 60,686 | + | 94 | C. freundii (pCTX-M3) | Hypothetical protein | 100 | 94/94 | NP_775004 | |
| 78 | 60,841 | 61,062 | + | 73 | E. coli (pNDM-HK) | hypothetical protein | 100 | 73/73 | YP_006952495 | |
| 79 | 61,134 | 61,568 | + | ssb | 144 | K. pneumoniae (pOXA-48) | Single-stranded DNA binding protein | 100 | 144/144 | YP_006958822 |
| 80 | 61,624 | 61,935 | + | 103 | K. pneumoniae (pOXA-48) | Hypothetical protein | 99 | 103/103 | YP_006958823 | |
| 81 | 62,068 | 62,604 | + | 178 | Serratia marcescens (R830b) | Hypothetical protein | 100 | 178/178 | YP_006964602 | |
| 82 | 63,106 | 63,471 | − | mobC | 121 | K. pneumoniae (pCTXM360) | MobC protein involved in plasmid mobilization | 98 | 119/121 | YP_002333286 |
| 83 | 63,746 | 64,063 | + | mobB | 105 | K. pneumoniae (pCTXM360) | MobB protein involved in plasmid mobilization | 99 | 105/105 | YP_002333287 |
| 84 | 64,050 | 66,029 | + | mobA | 659 | K. pneumoniae (pKPoxa-48N1) | MobA protein involved in plasmid mobilization | 99 | 659/659 | YP_008090802 |
| 85 | 66,043 | 66,543 | + | traH | 166 | K. pneumoniae (pCTXM360) | Conjugative transfer protein | 99 | 165/166 | YP_002333289 |
| 86 | 66,540 | 67,319 | + | traI | 259 | K. pneumoniae (pCTXM360) | Conjugative transfer protein | 99 | 259/259 | YP_002333290 |
| 87 | 67,330 | 68,493 | + | traJ | 387 | K. pneumoniae | Tfp pilus assembly protein | 100 | 387/387 | WP_020805653 |
| 88 | 68,483 | 68,743 | + | traK | 86 | K. pneumoniae (pCTXM360) | Conjugative transfer protein | 100 | 86/86 | YP_002333292 |
| 89 | 68,768 | 72,118 | + | pri | 1,116 | K. oxytoca | DNA primase | 96 | 1077/1116 | WP_031942492 |
| 90 | 72,060 | 72,596 | + | traL | 178 | K. pneumoniae (pKPoxa-48N1) | Conjugative transfer protein | 100 | 178/178 | YP_008090807 |
| 91 | 72,562 | 73,212 | + | 216 | K. pneumoniae (pKPoxa-48N1) | Hypothetical protein | 97 | 213/216 | YP_008090808 | |
| 92 | 73,190 | 73,972 | + | traM | 260 | K. pneumoniae (pCTXM360) | Conjugative transfer protein | 98 | 257/260 | YP_002333295 |
| 93 | 73,981 | 75,132 | + | traN | 383 | K. pneumoniae (pCTXM360) | Conjugative transfer protein | 98 | 380/383 | YP_002333296 |
| 94 | 75,144 | 76,493 | + | traO | 449 | K. pneumoniae (pCTXM360) | Conjugative transfer protein | 100 | 449/449 | YP_002333297 |
| 95 | 76,505 | 77,209 | + | traP | 234 | K. pneumoniae (pNDM-OM) | Conjugative transfer protein | 98 | 232/234 | YP_007195483 |
| 96 | 77,233 | 77,763 | + | traQ | 176 | K. pneumoniae (pNDM-OM) | Conjugative transfer protein | 100 | 176/176 | YP_007195484 |
| 97 | 77,780 | 78,169 | + | traR | 129 | K. pneumoniae (pNDM-OM) | Conjugative transfer protein | 100 | 129/129 | YP_006964622 |
| 98 | 78,215 | 78,709 | + | 164 | K. pneumoniae (pNDM-OM) | Hypothetical protein | 100 | 164/164 | YP_007195486 | |
| 99 | 78,706 | 81,756 | + | traU | 1,016 | K. pneumoniae (pCTXM360) | Conjugative transfer protein | 99 | 1014/1016 | YP_002333302 |
| 100 | 81,753 | 82,961 | + | traW | 402 | E. cloacae (pNE1280) | Conjugative transfer protein | 100 | 402/402 | YP_006964668 |
| 101 | 82,958 | 83,608 | + | traX | 216 | Enterobacteriaceae | Conjugative transfer protein | 100 | 216/216 | WP_004187488 |
| 102 | 83,508 | 85,781 | + | traY | 757 | E. cloacae (pNE1280) | Conjugative transfer protein | 100 | 757/757 | YP_006964669 |
| 103 | 85,784 | 86,437 | + | excA | 217 | K. pneumoniae (pCTXM360) | Phosphoglycerol transferase | 100 | 216/217 | YP_002333306 |
| 104 | 86,511 | 86,741 | + | repC | 76 | E. cloacae | Replication regulatory protein | 99 | 76/76 | YP_006964671 |
aa, amino acids.
NTP, nucleoside triphosphate.
The plasmid pEL60 (60,145 bp) from the plant pathogen Erwinia amylovora is considered the ancestral IncL/M plasmid. It does not possess any resistance genes and shares its entire sequence with other IncL/M plasmid backbones (8, 9) (see Fig. S1a in the supplemental material). The nucleotide sequence identity of pKOI-34 with pEL60 is 53% (46,561 bp), and the common backbone sequence possesses genes for replication (rep), a toxin-antitoxin system (pemIK) that elevates mutation frequency conferring a UV-resistant phenotype (mucAB) (10), plasmid stability (parAB), endonuclease (nuc), primase (pri), and mobility/conjugal transfer genes (trbCBAN, tir, mobAB, traHIJK, and traLMNOPQUWXY) (Fig. 1). The pKOI-34 repA gene shows 93% nucleotide identity with pEL60 (53 nucleotide substitutions resulting in 13 amino acid variants) and 99% nucleotide identity with the majority of the other IncL/M plasmids (5 to 8 nucleotide substitutions, resulting in 1 or 2 amino acid substitutions).
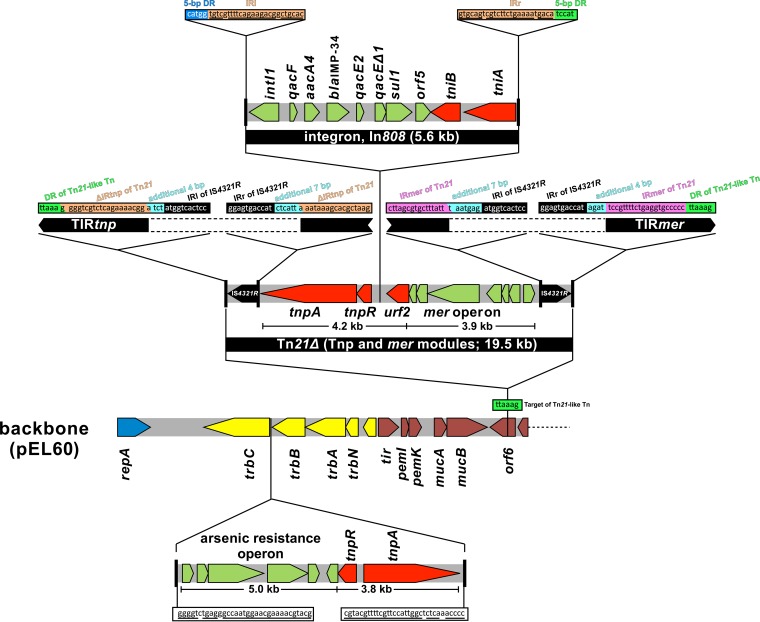
pKOI-34 possesses two mobile genetic elements. One is an 8.8-kb element consisting of tnpA, tnpR, and the arsenic resistance operon (10); and the other is a Tn21-like transposon, including two IS4321R (19.5 kb) (Fig. 2).
FIG 2.
The schema for the insertion region of pKOI-34 was based on the IncL/M backbone of pEL60. pKOI-34 possesses two mobile gene elements. One is an 8.8-kb region containing transposase (tnpA), resolvase (tnpR), and arsenic resistance genes. Another is aTn21-like transposon, consisting of tnpAR, an integron cassette structure of the class I integron (intl1, qacF, aacA4, blaIMP-34, qacE2, qacEΔ1, sul1, orf5), tniAB, urf2, and the mercury resistance operon. This transferable structure (Tn21-like transposon and IR4321R) was inserted into orf6, and the direct repeat sequence TTAAA (green) was generated. The Tn21-like transposon has an incomplete inverted repeat site, IRtnp and IRmer, on the left and right sides, respectively. The IRtnp of the Tn21-like transposon (orange) is truncated on the left side of IS4321R, and the IRmer of the Tn21-like transposon (pink) is truncated on the right side of IS4321R. Black, inverted repeat of IS4321R; green, direct repeat sequence of Tn21-like transposon; orange, IRtnp of the Tn21-like transposon; light blue, additional sequence; black, inverted repeat of IS4321R; pink, IRmer of Tn21-like transposon. The arsenic resistance operon has an incomplete inverted repeat (underlined).
The 8.8-kb element was inserted at the 5′ end of trbC that was related to recombination, bacterial conjugation, and DNA transfer. This region showed 99% homology to the plasmid pZA1001, R46, and pCC416 (accession nos. CP001723, AY046276, and AJ704863, respectively).
The 19.5-kb element has a Tn21-like structure that divides orf6 into two truncated sequences (Fig. 2). The sites of insertion, defined by a 6-bp duplication of the target DNA TTAAAG, is AT rich, similar to Tn21 (11) (Fig. 2, green box). The core element of the Tn21-like structure is a composite of the Tn21 tnp and mer modules forming Tn21Δ. Both sides of the 38-bp Tn21Δ terminal inverted repeat (TIR) are interrupted by the 1,327-bp IS4321R. IS4321 insertion creates 11-bp inverted repeats (IRl and IRr of IS4321) and additional flanking bases on both sides. IS4321R is a member of the IS1111 family that targets a specific position in the 38-bp complete TIR in the family of Tn21 transposons (12). A similar IS insertion into a 38-bp TIR is observed in many plasmids carrying TIR from the family of Tn21 transposons, Tn21/Tn501 (12). In Tn21, In2 is integrated between tnpR and urf2 in Tn21Δ, generating an imperfect inverted repeat of 25 bp and a 5-bp direct repeat TCCAT. However, in this Tn21-like structure, In808 carrying blaIMP-34 is present instead of In2 (Fig. 2). Besides blaIMP-34, In808 possesses other resistance genes, i.e., qacF, qacE2, and aacA4 (4). In808 is flanked by an imperfect inverted repeat of 25 bp (IRl, IRr). The TCCAT sequence was found on the right side but not on the left side, suggesting a later swapping event of an integron module between In2 and In808.
The other four plasmids recovered from MS5280, MS5284, MS5285, and MS5286 carrying blaIMP-34 were examined using the PCR scanning method (13). Twenty-two primer sets covering the total DNA sequence with the PCR products of pKOI-34 were used (Fig. 1; see Table S1 in the supplemental material). PCR scanning data showed that primer sets 3 and 4 generated no amplicon, but the other 20 sets of the primer pairs yielded PCR products of the expected sizes in all four plasmids. The data suggest that all four plasmids were identical to pKOI-34 but lacked the mobile element carried by the transposon containing the arsenic resistance operon inserted between trbC and trbB. To examine the junction between the sequence corresponding to the plasmid backbone and the transposon, an ∼600-bp amplicon was generated with PCR using the set 3 forward primer and set 4 reverse primer (see Table S1). The nucleotide sequence of the ∼600-bp amplicon was identical to the trbC to trbB region of pEL60. This indicates that the four plasmids recovered from MS5280, MS5284, MS5285, and MS5286 are pKOI-34 variants lacking the transposon carrying the arsenic resistance operon. MS5279 and MS5280 were isolated in 2004, MS5285 and MS5286 in 2006, and MS5284 in 2007. Therefore, it may be reasonable to assume that the arsenic operon was lost in MS5280, MS5284, MS5285, and MS5286. In any case, this fact is not strong enough support that this element was lost in these strains.
Multilocus sequence typing (MLST) was performed using the protocol published by Larsen et al. (14). MLST showed that K. pneumoniae MS5284, MS5285, and MS5286 belong to ST334, and K. oxytoca MS5279 and MS5280 belong to ST171 (gapA, 3; infB, 4; mdh, 15; pgi, 4; phoE, 18; rpoB, 3; tonB, 4).
The number of members of the IncL/M multidrug resistance (MDR) plasmids harboring broad-spectrum β-lactam resistance is increasing worldwide. pKOI-34 is the newest member of this group and the first IncL/M drug resistance plasmid found in Japan. Bonnin et al. (15) suggest that the evolution of the IncL/M MDR plasmids was through the acquisition of resistance genes and insertion sequences. There are two integration hot spots in the IncL/M plasmids; one is located between the rep locus and trbC, and the other is near pemIK (15) (see Fig. S1b in the supplemental material) (16, 17).
In pKOI-34, two target sites for inserted sequences are different from those previously reported (see Fig. S1a,b in the supplemental material). The arsenic resistance operon was located between trbC and trbB, and the other Tn21-like transposon was located within the orf6 of pEL60.
In conclusion, we report the complete sequence of pKOI-34, an IncL/M type conjugal plasmid carrying blaIMP-34. pKOI-34 possesses a pEL60 backbone with two inserted mobile elements, a Tn21-like architecture with a class 1 integron, In808 instead of In2, and a transposon carrying the arsenic resistance operon.
We show here that the spread of the blaIMP-34 gene in K. oxytoca and K. pneumoniae is linked to the spread of pKOI-34 or its derivatives.
Nucleotide sequence accession number.
The complete sequence of pKOI-34 has been deposited in GenBank (accession number AB715422).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jim Nelson for editorial assistance.
N.S. received the 62nd presentation award in the category of basic research conferred by the Director of the West Japan Branch of the Japanese Society of Chemotherapy.
This research was supported by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02507-15.
REFERENCES
- 1.McKenna M. 2013. Antibiotic resistance: the last resort. Nature 499:394–396. doi: 10.1038/499394a. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M. 2001. Plasmid-encoded metallo-beta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother 45:1343–1348. doi: 10.1128/AAC.45.5.1343-1348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shigemoto N, Kuwahara R, Kayama S, Shimizu W, Onodera M, Yokozaki M, Hisatsune J, Kato F, Ohge H, Sugai M. 2012. Emergence in Japan of an imipenem-susceptible, meropenem-resistant Klebsiella pneumoniae carrying blaIMP-6. Diagn Microbiol Infect Dis 72:109–112. doi: 10.1016/j.diagmicrobio.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Kayama S, Shigemoto N, Kuwahara R, Oshima K, Hirakawa H, Hisatsune J, Jové T, Nishio H, Yamasaki K, Wada Y, Ueshimo T, Miura T, Sueda T, Onodera M, Yokozaki M, Hattori M, Ohge H, Sugai M. 2015. Complete nucleotide sequence of the IncN plasmid encoding IMP-6 and CTX-M-2 from emerging carbapenem-resistant Enterobacteriaceae in Japan. Antimicrob Agents Chemother 59:1356–1359. doi: 10.1128/AAC.04759-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shigemoto N, Kayama S, Kuwahara R, Hisatsune J, Kato F, Nishio H, Yamasaki K, Wada Y, Sueda T, Ohge H, Sugai M. 2013. A novel metallo-β-lactamase, IMP-34, in Klebsiella isolates with decreased resistance to imipenem. Diagn Microbiol Infect Dis 76:119–121. doi: 10.1016/j.diagmicrobio.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 7.Richter DC, Schuster SC, Huson DH. 2007. OSLay: optimal syntenic layout of unfinished assemblies. Bioinformatics 23:1573–1579. doi: 10.1093/bioinformatics/btm153. [DOI] [PubMed] [Google Scholar]
- 8.Partridge SR, Ginn AN, Paulsen IT. 2012. pEl1573 carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother 56:6029–6032. doi: 10.1128/AAC.01189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster GC, McGhee GC, Jones AL, Sundin GW. 2004. Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl Environ Microbiol 70:7539–7544. doi: 10.1128/AEM.70.12.7539-7544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AHY, Bao JY-J, Lok S, Lo JYC. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. doi: 10.1371/journal.pone.0017989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 63:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge SR, Hall RM. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J Bacteriol 185:6371–6384. doi: 10.1128/JB.185.21.6371-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi T, Hayashi T, Takami H, Ohnishi M, Murata T, Nakayama K, Asakawa K, Ohara M, Komatsuzawa H, Sugai M. 2001. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect Immun 69:7760–7771. doi: 10.1128/IAI.69.12.7760-7771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnin RA, Nordmann P, Carattoli A, Poirel L. 2013. Comparative genomics of IncL/M-type plasmids: evolution by acquisition of resistance genes and insertion sequences. Antimicrob Agents Chemother 57:674–676. doi: 10.1128/AAC.01086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gołebiewski M, Kern-Zdanowicz I, Zienkiewicz M, Adamczyk M, Zylinska J, Baraniak A, Gniadkowski M, Bardowski J, Cegłowski P. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum beta-lactamase gene blaCTX-M-3. Antimicrob Agents Chemother 51:3789–3795. doi: 10.1128/AAC.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.