Abstract
The selection of resistance-associated variants (RAVs) against single agents administered to patients chronically infected with hepatitis C virus (HCV) necessitates that direct-acting antiviral agents (DAAs) targeting multiple viral proteins be developed to overcome failure resulting from emergence of resistance. The combination of grazoprevir (formerly MK-5172), an NS3/4A protease inhibitor, and elbasvir (formerly MK-8742), an NS5A inhibitor, was therefore studied in genotype 1a (GT1a) replicon cells. Both compounds were independently highly potent in GT1a wild-type replicon cells, with 90% effective concentration (EC90) values of 0.9 nM and 0.006 nM for grazoprevir and elbasvir, respectively. No cross-resistance was observed when clinically relevant NS5A and NS3 RAVs were profiled against grazoprevir and elbasvir, respectively. Kinetic analyses of HCV RNA reduction over 14 days showed that grazoprevir and elbasvir inhibited prototypic NS5A Y93H and NS3 R155K RAVs, respectively, with kinetics comparable to those for the wild-type GT1a replicon. In combination, grazoprevir and elbasvir interacted additively in GT1a replicon cells. Colony formation assays with a 10-fold multiple of the EC90 values of the grazoprevir-elbasvir inhibitor combination suppressed emergence of resistant colonies, compared to a 100-fold multiple for the independent agents. The selected resistant colonies with the combination harbored RAVs that required two or more nucleotide changes in the codons. Mutations in the cognate gene caused greater potency losses for elbasvir than for grazoprevir. Replicons bearing RAVs identified from resistant colonies showed reduced fitness for several cell lines and may contribute to the activity of the combination. These studies demonstrate that the combination of grazoprevir and elbasvir exerts a potent effect on HCV RNA replication and presents a high genetic barrier to resistance. The combination of grazoprevir and elbasvir is currently approved for chronic HCV infection.
INTRODUCTION
Hepatitis C virus (HCV) is a leading cause of chronic liver disease, with an estimated 130 to 170 million people infected globally. WHO estimates that more than 350,000 people die every year from hepatitis C-related liver diseases (1, 2, 3). The introduction of direct-acting antiviral agents (DAAs) as add-ons to the previous standard of care (SOC) consisting of pegylated interferon alpha plus ribavirin (PR) significantly improved sustained virologic response (SVR) rates from 40 to 50% to 65 to 70% in the previously hard-to-cure genotype 1 (GT1) patients after a 24- to 48-week treatment course. Further treatment advancements have been achieved with the introduction of interferon-free all-oral DAAs, with SVR rates now in excess of 90% after 12 weeks of therapy for GT1 patients (4, 5, 6). Recent reports indicate that therapy can be further simplified and likely shortened to <12 weeks in some cases while maintaining high SVR rates. Preexisting baseline resistance-associated variants (RAVs) and resistance selection remain contributory reasons for treatment failure (7). This necessitates that combinations of DAAs targeting multiple viral proteins be developed to overcome treatment failure that may result from emergence of resistance. Thus, despite recent successes, identification of potent antivirals for HCV that can be effectively used in combination is needed.
Two potent DAAs being developed for chronic HCV infection are grazoprevir (formerly known as MK-5172) and elbasvir (formerly known as MK-8742). Grazoprevir is a potent NS3/4A protease inhibitor (PI) (8). The N-terminal third of the NS3 protein (with NS4A as a cofactor) serves as a serine protease required for maturation of other viral nonstructural proteins from a polyprotein precursor (9). Inhibitors of the NS3 protease activity have been validated in the clinic, as exemplified by the first-generation DAAs boceprevir and telaprevir (10). Although efficacious in GT1 patients, first-generation PIs have limited activity against several RAVs, and they also lack pan-genotype activity. Grazoprevir has single-digit nanomolar potency against most HCV genotypes and remains active against common clinical RAVs elicited by the first-generation PIs (8). Elbasvir is a potent HCV NS5A inhibitor with picomolar activities against most HCV genotypes (11). The NS5A protein has become an attractive target for intervention in chronic HCV infection. While it has no enzymatic activity, NS5A is important for RNA synthesis and virus assembly (12, 13, 14). The exact mechanism of NS5A in HCV replication remains unclear; nonetheless, several small-molecule NS5A inhibitors (NS5AIs) have demonstrated clinical activity in patients (15). Viral load reduction is rapid and robust with NS5A inhibitors; however, a low genetic barrier to resistance ensures significant virus escape. Elbasvir shows improved activity against many common NS5A RAVs selected by previous compounds (11). Besides the potential to robustly suppress the HCV load by targeting 2 independent mechanisms, a regimen containing an NS3/4A protease inhibitor and an NS5A inhibitor would also be expected to enhance the suppression of resistant variants if signature RAVs from each inhibitor class do not confer cross-resistance to the other class. Thus, such a combination may enhance sustained virologic response (SVR) rates in patients chronically infected with HCV. In this study, we investigated grazoprevir and elbasvir in genotype 1a replicon cells to evaluate their profile on cross-resistance, HCV RNA reduction, and emergence of resistance.
MATERIALS AND METHODS
Compounds.
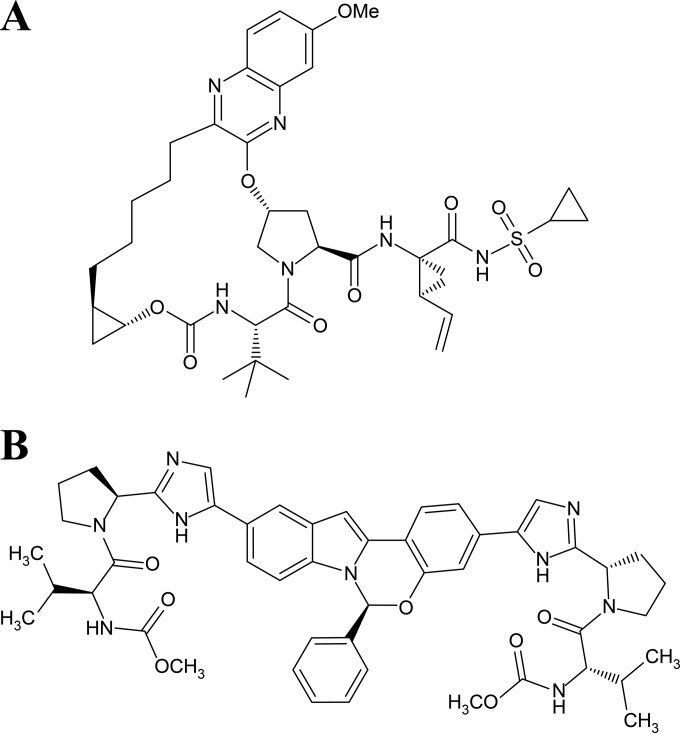
Grazoprevir {N-[[[(1R,2R)-2-[5-(3-hydroxy-6-methoxy-2-quinoxalinyl)pentyl]cyclopropyl]oxy]carbonyl]-3-methyl-l-valyl-(4R)-4-hydroxy-l-prolyl-(1R),2S)-1-amino-N-(cyclopropylsulfonyl)-2-ethenylcyclopropanecarboxamide cyclic (1→2)-ether} (Fig. 1A) was prepared as reported previously (8, 16).
FIG 1.
Chemical structures of the HCV inhibitors used in this study. (A) Chemical structure of the NS3/4A inhibitor grazoprevir. (B) Chemical structure of the NS5A inhibitor elbasvir.
Elbasvir {N,N′-[[(6S)-6-Phenyl-6H-indolo[1,2-c][1,3]benzoxazine-3,10-diyl]bis[1H-imidazole-5,2-diyl-(2S)-2,1-pyrrolidinediyl[(1S)-1-(1-methylethyl)-2-oxo-2,1-ethanediyl]]]bis[carbamic acid] C,C′-dimethyl ester (Fig. 1B) was prepared as reported previously (11).
Stable-replicon assay.
HCV subgenomic replicon cDNAs for GT1a(H77) (17), GT1b(Con1) (18), GT1b(N) (19), GT2a(JFH-1) (20), GT3a(S52) (21), and GT4a(ED43) (21) have been described elsewhere. The GT1b(Con1), GT1b(N), and GT2a(JFH-1) cDNAs were used as model genome backgrounds to receive NS3 and/or NS5A gene sequences from GT2b, GT3a(NZL1), GT5a(SA13), and GT6a(HK6a or GZ52557) to create chimeric replicon genomes; the accession numbers for the chimeric gene segments are listed in Table S1 in the supplemental material. Chimeric cDNAs were designed and made by gene synthesis (Genewiz, South Plainfield, NJ) as cassettes for cloning to replicon background vectors (see Table S1 in the supplemental material). Chimeric replicons with modified NS3 genes included NS3 residues 1 to 181 and the complete NS4A sequences from the genotype/subtype of interest, while replicons bearing new NS5A genes carried the complete NS5A sequence (see Table S1 in the supplemental material). RNA was transcribed from linearized plasmids using T7 Megascript (Ambion/LifeTechnologies) as per the manufacturer's protocol and was used to transfect Huh7 cells for stable cell line generation, as described previously (22).
Stable-replicon cell lines were tested for compound sensitivity. Briefly, replicon cells were seeded in 384-well plates in Dulbecco modified Eagle medium (DMEM) containing 0.5 mg/ml G-418. Concentrations spanning the 50% effective concentration (EC50) for each compound were used independently or in the combination studies for grazoprevir and elbasvir. The compounds were serially diluted individually in 100% dimethyl sulfoxide (DMSO) and then were added to the medium in the presence of 5% fetal calf serum (FCS) with cells in 1:200 dilutions on the day after cells were seeded. The final DMSO concentration was 0.5% (vol/vol). After 72 h of incubation, cells were harvested and subjected to real-time PCR analysis as reported previously (22, 23). The primer/probe sets used for the PCR analysis on an ABI Prism 7900HTS sequence detection system are listed in Table S2 in the supplemental material. The independent and combination studies were tested 3 times with triplicate plates in each experiment (or run).
The threshold cycle numbers (CT) were plotted against the log of compound concentrations and fitted to the sigmoid dose-response model using DataAnalyzer 4.0.30639.0 (Merck Frosst Canada Co.) to obtain the EC90, i.e., the drug concentration needed to achieve 90% inhibition compared to a no-treatment control plate. The EC90 was computed as the drug concentration required for an increase of 3.2 ΔCT over the baseline. The EC50 was the concentration needed to achieve a ΔCT of 1 over the baseline.
Transient-replicon assay.
The rapid transiently expressed HCV replicon system in the full-length GT1a(H77) sequence expressing the Gaussia luciferase (G-luc) gene integrated in frame with the viral polyprotein gene (obtained from Stanley Lemon, University of North Carolina) (24) was used to analyze the phenotypes of several genomes with RAVs. The RAV of interest was introduced by cloning a cassette of the modified target gene (made by gene synthesis; Genewiz, South Plainfield, NJ) into the GT1a–G-luc background. Transfection of this replicon RNA, transcribed from cDNA using T7 Megascript (Ambion/Life Technologies), into Huh 7.5 cells results in replication of the HCV RNA and expression of the G-luc protein. The levels of expressed G-luc protein directly correlate with HCV RNA copy number and viral protein translation (24). The transiently expressed replicon system allows for growth and characterization of viruses bearing mutations that may have detrimental effects on viral fitness and an inability to establish stable replicons. In this protocol, 5 × 106 Huh 7.5 cells were transfected by electroporation with 5 μg replicon RNA on a Bio-Rad Gene Pulser Xcell using the exponential protocol at 270 V, capacitance of 950 μF, and resistance of 100 Ω. Cells were transferred to 75-ml flasks, with 1/50 transferred to one well of a 24-well plate. Culture supernatant was collected and refreshed after 6 h and each day thereafter. All collected culture supernatants were stored at 4°C until luciferase measurement. After 7 days, all culture supernatant samples were assayed for Gaussia luciferase activity using the BioLux Gaussia luciferase assay kit (E3300; New England BioLabs) as per the manufacturer's protocol. Luciferase activity was measured on an Envision plate reader (model 2104) from Perkin-Elmer.
Kinetic analysis of the combination of grazoprevir and elbasvir in GT1a.
The “independent-effects” (25) definition of additive response was used to assess the nature of inhibitor interactions (synergistic, additive, or antagonistic). Inhibitor interactions were analyzed with the aid of MacSynergy software (26), which requires input data in linear scale. To fulfill that requirement the log-scale measurements of threshold cycle number (CT) were converted into relative amounts of RNA using the standard curves from the same assay plate. The standard curves were constructed with total RNA isolated from replicon cells and then serially diluted. The relative amount of replicon RNA from each sample (treated cells in each well) was calculated based on the CT of the sample and the standard curve from the same plate.
MacSynergy calculates an additive response at each combination and defines synergy as a response that exceeds additivity and antagonism as a response that is less than additivity. The application then calculates synergy/antagonism volumes at a ≥95% confidence interval and categorizes the results as synergy, additivity, or antagonism.
Long-term viral kinetic studies.
Stable-replicon cells were seeded in 6-well plates at various cell concentrations for confluence at harvest and then dosed 6 h later at 1× EC90 with grazoprevir and elbasvir independently for a final DMSO concentration of 0.5% (vol/vol) in DMEM containing 10% fetal bovine serum (FBS) without G418. Treated cells were harvested with trypsin-EDTA (0.25%) (Invitrogen, catalog no. 25200-056) at 0, 24, 48, 72, 96, 168, 240, and 336 h (with cell passing at 72, 168, and 240 h), and RNA was isolated utilizing the RNeasy purification kit via the manufacturers protocol (Qiagen, catalog no. 74104). RNA was then subjected to real-time PCR analysis as reported previously (22, 23) using the above-mentioned primers and probes. The threshold cycle numbers (CT) were normalized to the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) housekeeping gene to yield ΔCT, and the log(1/power) (2,ΔCT − average ΔCT of DMSO at day zero) was plotted over time using Prism (GraphPad Software Inc.).
Compound treatment to select for emergence of resistance.
GT1a subgenomic replicon cells (obtained from Stanley Lemon, University of North Carolina) (17) were seeded onto multiple 6-cm tissue culture dishes at a density of 2 × 105 cells/dish (27). Four dishes were prepared for each treatment condition. Twenty-four hours after plating, cells were dosed with various combinations and multiples of the EC90 values of grazoprevir and elbasvir (as indicated in appropriate tables and figures). Media and compounds were refreshed twice a week. In the event that the monolayer reached confluence (e.g., with the DMSO treatment control), those cells were passaged at a 1:10 ratio. At about 3 to 4 weeks postinitiation, when resistant replicon cells formed defined colonies, 3 of the 4 dishes were fixed and stained with a crystal violet solution for colony counting. Cells on the remaining dish were expanded for further analysis (i.e., sequencing of reverse transcription-PCR [RT-PCR] products and phenotypic sensitivity to compounds).
RT-PCR amplification of targeted sequences and cloning.
Total cellular RNA was extracted from resistant replicon cells and processed for RT-PCR. First-strand RT was generated using the Superscript III first-strand synthesis system (Invitrogen), with priming from the 3′ nontranslated region of the HCV genome. PCR amplification was achieved using primers from the subgenomic replicon's encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) and the end of the NS5B gene, allowing for the production of a 6-kb RT-PCR product (23). The desired product was gel purified, used to clone into the pCR2.1-TOPO vector (Invitrogen), and transformed into Escherichia coli TOP10 cells (Invitrogen) for recovery of cDNA. Positive clones were identified by PCR and restriction digest screening. Insert-positive cDNAs were sequenced through the full NS3 and NS5B genes by capillary sequencing (at GenScript, Piscataway, NJ). To evaluate the impact of resistance selection, particular attention was paid to specific variants at amino acid loci prone to resistance selection by HCV inhibitors (“signature” RAVs). The signature NS3 protease loci include positions 36, 54, 55, 56, 80, 107, 122, 132, 155, 156, 158, 168, 170, and 175. Signature NS5A loci include positions 28, 30, 31, 58, and 93.
RESULTS
Potencies of grazoprevir and elbasvir in HCV replicons with NS3/4A or NS5A from different genotypes.
Grazoprevir (Fig. 1A) is a potent, reversibly binding macrocyclic inhibitor of HCV nonstructural protein 3/4A (NS3/4A) in enzyme assays and potently blocks replication in cellular assays (8, 16). The profile of grazoprevir in replicon cells bearing NS3/4A sequences from the major HCV genotypes 1 to 6 is summarized in Table 1. Grazoprevir has low-nanomolar potency against all genotypes tested. It is slightly less potent in the full-length GT3a(S52) replicon. However, when the S52 catalytic domain (residues 1 to 181) replaces the related sequence in JFH-1(GT2a) in a chimeric replicon, the potency of grazoprevir is comparable to that with other chimeric replicons (Table 1). Thus, contextual sequences influence the potency of the inhibitor in the full-length GT3a(S52) replicon. The structural determinant or basis for the reduced potency in the full-length GT3a(S52) is currently unknown.
TABLE 1.
Replicon profiles of grazoprevir and elbasvir in HCV genotypes 1 to 6
| Sequence identificationa | Grazoprevir |
Elbasvir |
||
|---|---|---|---|---|
| EC50 (nM)b | EC90 (nM) | EC50 (nM) | EC90 (nM) | |
| GT1a (NC004102, H77) | 0.4 ± 0.2 | 0.9 ± 0.5 | 0.004 ± 0.002 | 0.006 ± 0.002 |
| GT1b (AJ238799, Con1) | 0.5 ± 0.3 | 1.1 ± 0.6 | 0.003 ± 0.001 | 0.006 ± 0.004 |
| GT2a (AB047639, JFH1) | 2.3 ± 1.2 | 7.1 ± 3.1 | 0.003 ± 0.001 | 0.019 ± 0.01 |
| GT2b (AY232740) | 3.7 ± 1.1 | 7.8 ± 2.1 | NA | NA |
| GT2b (AB030907) | NA | NA | 3.4 ± 2.6 | 11 ± 4.8 |
| GT2b (AB030907) M31L | NA | NA | 0.32 ± 0.38 | 1.0 ± 1.2 |
| GT3a (GU945445, GU945457.1) | 7.6 ± 3.2 | 20.5 ± 9.0 | NA | NA |
| GT3a (GU814263, S52), catalytic | 2.1 ± 1 | 10.2 ± 1.0 | NA | NA |
| GT3a (S52), full-length | 35 ± 15 | 153 ± 35 | 0.14 ± 0.09 | 0.49 ± 0.19 |
| GT3a (NC009824) | NA | NA | 0.03 ± 0.01 | 0.12 ± 0.06 |
| GT4a (GU814265, ED43) | 0.3 ± 0.2 | 0.8 ± 0.4 | 0.0003 ± 0.0001 | 0.0005 ± 0.0001 |
| GT5a (AF064490, SA13) | 6.6 ± 0.6 | 12.8 ± 2.2 | 0.001 ± 0.001 | 0.002 ± 0.002 |
| GT6a_(JN180455.1) | 0.9 ± 0.1 | 2.3 ± 0.4 | NA | NA |
| GT6 (DQ278892) | 0.2 ± 0.04 | 0.3 ± 0.1 | 0.009 ± 0.006 | 0.017 ± 0.009 |
Numbers in parentheses are GenBank accession numbers and/or strain designations. Additional details about the replicons are in Table S1 in the supplemental material. Parental replicons of Con1 (GT1b) and JFH1 (GT2a) were used as controls.
Values are means ± standard deviations (n ≥ 3). NA, not available.
Elbasvir (Fig. 1B) is a potent NS5A inhibitor with broad genotype activity (11). It is highly potent against HCV replicons bearing NS5A sequences from GT1a, -1b, -2a(31L), -3a, -4a, -5a, and -6, with EC50s in the low-picomolar range. It is less active against the GT2b replicon, with an EC50 of 3.4 nM. This loss of potency is in part due to the amino acid substitution L31M within the GT2 NS5A sequence (Table 1), which occurs naturally and is prevalent among natural GT2 infections in public databases.
Absence of cross-resistance from NS3 and NS5A signature RAVs on elbasvir and grazoprevir, respectively.
As a prelude to investigating their combined activity on HCV RNA replication, the potencies of grazoprevir and elbasvir were assessed in GT1a(H77) replicons bearing key RAVs from the other inhibitor class for cross-resistance. A summary of the measured fold shifts in potency (relative to the activity of the inhibitors in the wild type [WT]) against signature RAVs from the other DAA class is presented in Table 2. Replicons with RAVs associated with resistance to elbasvir remain susceptible to grazoprevir. Conversely, replicons resistant to grazoprevir are susceptible to elbasvir. Thus, RAVs from one inhibitor class are susceptible to inhibition by the other DAA, with no evidence of cross-resistance.
TABLE 2.
Potency fold shifts for grazoprevir and elbasvir on common RAVs elicited by NS5A and NS3/4A protease inhibitors, respectively
| Target | Replicona | Fold shiftb |
|
|---|---|---|---|
| Grazoprevir | Elbasvir | ||
| Common NS3 RAVs | WT | 1 | 1 |
| V36A | 1.2 | 1.3 | |
| T54S | 1.1 | 1.3 | |
| Y56H | 46 | 1 | |
| Q80K | 1.1 | 1 | |
| S122R | 1.8 | 1 | |
| R155K | 3.0 | 0.4 | |
| A156S | 2.5 | 1 | |
| D168A | 114 | 0.2 | |
| D168Y | 27 | 1 | |
| V170T | 2.0 | 0.3 | |
| Common NS5A RAVs | M28T | 0.8 | 15 |
| M28V | 0.9 | 1 | |
| Q30E | 0.9 | 56 | |
| Q30R | 1.0 | 16 | |
| L31 M | 1.1 | 10 | |
| L31V | 1.2 | 61 | |
| H58D | 1.1 | 6 | |
| Y93H | 1.0 | 220 | |
| Y93N | 1.1 | 929 | |
GT1a(H77) replicons bearing the designated change in NS3 or NS5A.
EC90 values were used, relative to that for WT GT1a(H77).
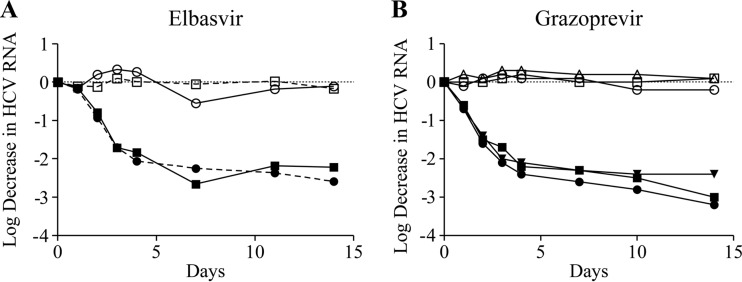
To provide further support for a lack of cross-resistance, the activity of the inhibitors on HCV RNA was monitored over a 14-day period on GT1a(H77) replicons bearing prototypic RAVs and compared to that on the WT GT1a(H77) replicon. Figure 2 demonstrates that grazoprevir inhibits the NS5A RAVs Y93H and Q30D with kinetics comparable to those for WT replicon. Similarly, elbasvir inhibits NS3 RAV R155K with kinetics comparable to that for the WT replicon. Hence, a signature RAV from the other mechanism affects neither the potency nor the rate of viral RNA decline for the inhibitor of the other mechanism.
FIG 2.
Kinetics of HCV RNA reduction in GT1a(H77) replicons bearing NS3 and NS5A RAVs treated with elbasvir and grazoprevir. (A) Inhibition of GT1a_R155K (□, ■) and wild-type GT1a (○, ●) with DMSO (open symbols) and 6 pM elbasvir (closed symbols) over 14 days. (B) Inhibition of Q30D (□, ■), Y93H (▽, ▼), and wild-type GT1a (○, ●) with DMSO (open symbols) and 15 nM grazoprevir (closed symbols) over 14 days.
Interaction between grazoprevir and elbasvir in GT1a replicon cells.
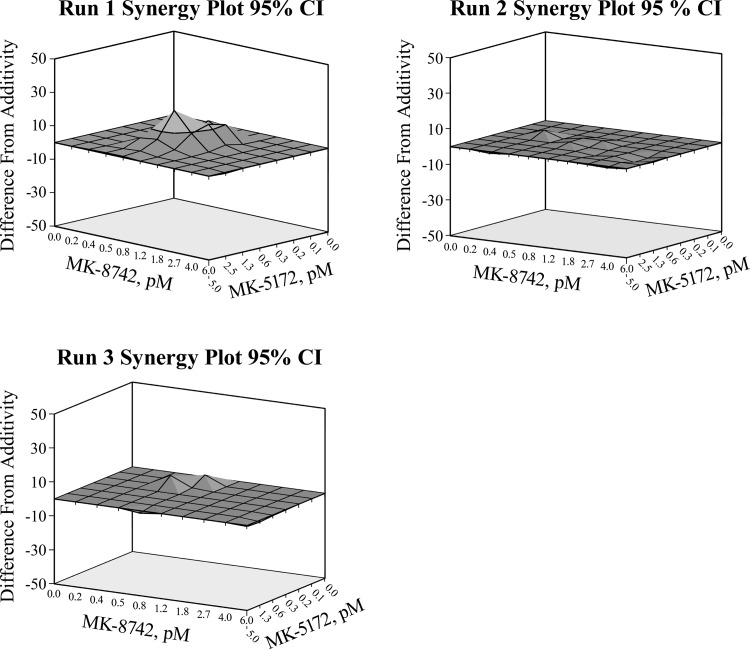
Having demonstrated that grazoprevir and elbasvir can inhibit RAVs elicited by the other inhibitor with no evidence of cross-resistance, the nature of their interactions as components of a combination to reduce HCV RNA was investigated. Concentrations of each compound spanning up to 10 times the EC50 for inhibiting HCV RNA synthesis in GT1a(H77) replicon cells over 72 h were studied in a matrix and quantified. Figure 3 shows synergy plots demonstrating additivity over most of the dose ranges studied in 3 runs (each performed in triplicate) for the grazoprevir-elbasvir combination in GT1a(H77) replicons. Table 3 summarizes the computed synergy volumes for the 3 independent runs using MacSynergy (26). The data demonstrate that grazoprevir and elbasvir interact additively to inhibit HCV RNA synthesis, with no evidence of antagonism.
FIG 3.
The combination of grazoprevir and elbasvir additively inhibits HCV RNA replication in GT1a(H77) replicon cells. Synergy plots of three independent runs (performed in triplicate) were analyzed by MacSynergy. The difference from additivity occurs above and below the additivity surface. The dark gray area is within −10 to 10 from the additive surface, and the light gray area is 10 to 30 above the additive surface.
TABLE 3.
Synergy volume from MacSynergy analysis of the grazoprevir-elbasvir combination
| Log vol, synergy/antagonism, 95% confidence intervala | Effect |
|---|---|
| 4.42/−0.03 | Minor but significant synergy |
| 0.5/−0.45 | Additive |
| 0.66/−0.24 | Additive |
Synergy volumes (21): log volume of <2, additive; log volume of >2 and <5, minor but significant synergy; log volume of >5 and <9, moderate synergy; log volume of >9, strong synergy.
Colony formation assay to assess emergence of resistance with the combination of grazoprevir and elbasvir.
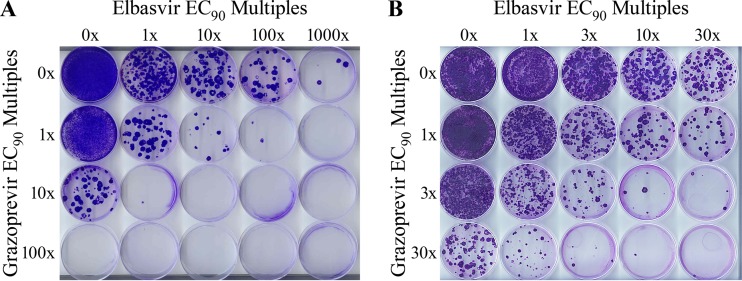
Next, the concurrent inhibition of NS3/4A and NS5A by grazoprevir and elbasvir, respectively, on emergence of resistance was studied. GT1a(H77) replicon cells were placed under various levels of compound selective pressure to assess the relative emergence of resistance from the combination. Inhibitor concentrations at equivalent multiples of each compound's intrinsic potency (i.e., the EC90 in genotype 1a replicons) were evaluated. Resistant cell colonies appeared within 3 to 4 weeks of selection; details of the selection procedure are described in Materials and Methods. Representative images of the inhibitor matrices are shown in Fig. 4A and B. Control cells, treated with DMSO only, as well as cells treated with low concentrations of each inhibitor independently (e.g., at 1× EC90), grew into near monolayers as expected. Dose-dependent reductions of resistant colonies were seen from treatments with either grazoprevir or elbasvir as a single agent. Further inspection of Fig. 4A shows that the numbers of resistant colonies are further reduced when both grazoprevir and elbasvir are used in combination. For example, the addition of 1× EC90 of grazoprevir further decreases the number of emergent colonies with increasing concentrations of elbasvir (Fig. 4A, row 1 versus row 2). As single agents, 100× and 1,000× EC90 of grazoprevir and elbasvir, respectively, were required to suppress substantially the emergence of resistant colonies (<10 colonies) (Fig. 4A). In combination, 10× EC90 of each compound practically blocked the emergence of resistant colonies. The dose dependence of suppression of resistant colonies was finely mapped with lower concentrations of the inhibitor combination to enable a broader accounting and analysis of potential resistance pathways. A finer titration with 3-fold increases (rather than the original 10-fold) in inhibitor concentration was therefore conducted (Fig. 4B). As observed with the original study (Fig. 4A), a dose-dependent decrease in the number of selected resistant colonies was similarly observed with the more gradual titration of the inhibitor combination.
FIG 4.
Representative images of a colony formation assay for the combination of grazoprevir and elbasvir in GT1a(H77) replicon cells. Multiples of the EC90 values of both inhibitors were titrated in a matrix and scored for the emergence of resistant colonies. Higher concentrations of the combination were evaluated in panel A than in panel B to finely map the combinatorial effect.
The average numbers of resistant colonies (n = 3) for the independent agents and the various combinations (Fig. 4B) are summarized in Table 4. The data showed a greater reduction in the number of resistant colonies when the inhibitors are used in combination than when either inhibitor is used as a single agent. Resistant colonies emerged at low multiples (<10-fold) of the EC90 values for the combination. At >10× EC90, the emergence of resistant colonies was suppressed. The data demonstrate that the grazoprevir-elbasvir combination is more effective in suppressing the emergence of resistant colonies than either inhibitor acting alone.
TABLE 4.
Summary of colony counts in genotype 1a(H77) replicons treated with combination of grazoprevir and elbasvira
| Grazoprevir EC90 multiple | No. of resistant colonies counted (% of input) with elbasvir EC90 multipleb: |
||||
|---|---|---|---|---|---|
| 0 | 1× | 3× | 10× | 30× | |
| 0 | (100c) | TMTCd | 293 (0.15) | 181 (0.09) | 144 (0.072) |
| 1× | TMTC | 435 (0.22) | 179 (0.09) | 82 (0.04) | 39 (0.02) |
| 3× | ∼1,000 (∼0.5) | 255 (0.12) | 83 (0.042) | 9 (0.0045) | 1 (0.0020) |
| 10× | 120 (0.06) | 38 (0.019) | 9 (0.0045) | 1 (0.0005) | 0 |
For GT1a(H77), the grazoprevir EC90 is 1.5 nM and the elbasvir EC90 is 6 pM.
The values represent the average from n = 3 to 4 dishes for each treatment condition, with an input of 2 × 105 cells.
No drug treatment yielded a lawn of cells, which was set at 100%.
TMTC, too many to count; splitting of the cell culture precludes quantitation.
Resistance phenotypes of pooled surviving colonies.
In order to define the impact of resistance-associated variants in surviving colonies to inhibition, GT1a(H77) replicon-cell populations were recovered from a number of inhibitor combinations and expanded for further study. The extent of resistance in these populations was estimated by measuring the susceptibility of the pooled resistant colonies for that condition. The potencies of grazoprevir and elbasvir were tested on representative cell populations using the standard replicon assay (described in Materials and Methods). The data are summarized in Table 5. There was a direct correlation between susceptibility and the concentration of inhibitor used for selection. In cell lines selected with elbasvir alone, susceptibility decreased with increasing concentration of the NS5A inhibitor used for selection. These cells remained susceptible to grazoprevir with no shift in potency. Similarly, cells selected with grazoprevir alone showed a dose-dependent reduction in susceptibility with increasing inhibitor concentration used for selection; the cells remained susceptible to elbasvir. This absence of cross-resistance was consistent with data presented in Table 2 and Fig. 2. As expected, the shift in potency in the selected colonies was much higher with cells selected with elbasvir than with those selected with grazoprevir at equivalent multiples of the EC90. Only cell lines selected with lower concentrations (<10× EC90) of the grazoprevir-elbasvir combination were successfully established for phenotypic characterization. There was a greater reduction in the potency of elbasvir (20- to 1,500-fold) than in that of grazoprevir (2- to 4-fold). The fold shift in potency with the inhibitor combination was fairly comparable (within 3-fold) for each inhibitor to that when the resistant colonies were selected with the comparable concentrations with either agent independently (Table 5). Cell lines from the higher-concentration inhibitor combinations did not survive continued passages. However, sequencing results could be obtained from initial passages in some cases; those results are described below.
TABLE 5.
Potencies of grazoprevir and elbasvir in genotype 1a(H77) cell lines selected with the independent agents or combinations of inhibitors at different concentrations
| EC90 multiple used for selectiona |
Inhibitor potency |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Elbasvir |
Grazoprevir |
||||||||
| Grazoprevir | Elbasvir | EC50 |
EC90 |
EC50 |
EC90 |
||||
| Value, nM | Fold shiftb | Value, nM | Fold shift | Value, nM | Fold shift | Value, nM | Fold shift | ||
| 0 | 0 | 0.002 | 1 | 0.003 | 1 | 1 | 1 | 2 | 1 |
| 0 | 1× | 0.3 | 150 | 2.4 | 800 | 1 | 1 | 2 | 1 |
| 0 | 10× | 0.5 | 250 | 2.3 | 767 | 1 | 1 | 1.2 | 0.6 |
| 0 | 100× | 5 | 2,500 | 22 | 7,333 | 1 | 1 | 2 | 1 |
| 0 | 1,000× | 36 | 18,000 | 63 | 21,000 | 1 | 1 | 2 | 1 |
| 1× | 0 | 0.003 | 1.5 | 0.004 | 1 | 2 | 2 | 10 | 5 |
| 10× | 0 | NTc | NT | NT | NT | 48 | 48 | 92 | 46 |
| 1× | 1× | 0.04 | 20 | 1 | 333 | 4 | 4 | 7 | 3.5 |
| 1× | 10× | 1.3 | 650 | 4.6 | 1,533 | 2 | 2 | 3 | 1.5 |
EC90 multiples used for GT1a(H77) selection: grazoprevir EC90, 1.5 nM; elbasvir EC90, 6 pM.
Change in potency relative to the DMSO (0×/0×) control.
NT, not tested.
Identification of RAVs in surviving colonies.
In an effort to identify mutations potentially responsible for the emergence of resistance, total RNA was extracted from each of the available surviving GT1a(H77) replicon-cell lines and used as the template for RT-PCR amplification of viral genes for sequencing. Of particular interest were samples from cells selected with both grazoprevir and elbasvir. Given the potential for mutations to occur on the same genome, a clonal sequencing strategy that amplified a 6-kb product, spanning the entire NS3-NS5B segment of the viral genome, was employed to monitor concurrently both cognate targets of the inhibitors (details are described in Materials and Methods) to facilitate analysis of linkage. In order to obtain sequencing data from higher-concentration inhibitor combinations that failed to survive continuous passaging, very early passages were employed. A summary of key positions and variants identified by sequencing of viral genomes selected with the indicated inhibitor concentrations is in Table 6. There were a few amino acid substitutions that were observed in control replicon cells (treated with DMSO only; see Table S3 in the supplemental material); these were attributed to genetic drifts in the replicons. The changes observed included substitutions at amino acid positions 41 and 174 in NS3; these changes were not studied further (see Table S3 in the supplemental material).
TABLE 6.
Changes at key RAV positions from clonal sequencing of genotype 1a(H77) replicons selected with grazoprevir and elbasvir in combination or independently
| Elbasvir EC90 multiple (concn) | Change(s) (%)a with grazoprevir EC90 multiple (concn): |
|||
|---|---|---|---|---|
| 0 | 1× (1.5 nM) | 3× (4.5 nM) | 10× (15 nM) | |
| 0 | WT [20] | R155K (20), D168E (10) [10] | NA | D168A [1] |
| 1× (6 pM) | Q30R (100) [5] | R109K (93), Q30R (93), L31V (7) [14] | NA | D168V (86), D168S (7), D168E (7), Q30H (100) [14] |
| 3× (18 pM) | M28T (33), Q30H (7) Q30R (33), Q30E (7), Y93C (33), Y93H (7) [15] | R155K (11), M28K (11) Q30R (22), L31 M (78), Y93C (11) [9] | R155K (31), D168E (54), D168V (15), M28T (23), Q30D (46), Q30R (38) [13] | |
| 10× (60 pM) | M28I (6), Q30R (44), L31V (28), L31 M (6), Y93H (17), Y93C (6) [18] | R109K (32), D168A (32), M28T (16), M28K (5), Q30R (42), Q30D (5), Q30E (5), Y93C (5) [19] | ||
| 30× (180 pM) | M28T (18), Q30R (64), Q30E (9), L31V (9), Y93H (18) [11] | NA | ||
| 100× (600 pM) | Y93N (100) [4] | R109K (100), Q30E (67), Q30R (33) [3] | ||
| 1,000× (6 nM) | M28K (40), Q30R (60), L31V (60) [5] | |||
Boldface, NS3 protease RAVs; lightface, NS5A RAVs. The number in brackets is the number of clones sequenced for that sample. NA, not available.
Resistance selection with grazoprevir and elbasvir as independent agents.
Few RAVs were identified when grazoprevir was independently applied to the GT1a(H77) replicon cells (Table 7). At a concentration of 1× EC90, only two NS3 RAVs were identified: R155K and D168E. At 10× EC90, the only surviving clone had a change at position 168, D168A (Table 6; see Table S4 in the supplemental material). Clones from higher-concentration grazoprevir treatment conditions did not survive.
TABLE 7.
Potencies of grazoprevir in transiently expressed genotype 1a(H77) replicons bearing NS3 RAVs from resistant colonies
| Variant | EC50, nMa | Fold shift vs WTb | Relative fitnessc |
|---|---|---|---|
| WT | 0.3 ± 0.1 | 1 | 1 |
| R109K | 0.6 ± 0.11 | 2 | 2.4 |
| D168A | 82 ± 12 | 273 | 0.5 |
| D168E | 11 ± 1.3 | 37 | 0.4 |
| D168V | 46 ± 20 | 153 | 0.3 |
| R155K+D168A | 1.9 ± 0.7 | 6 | 0.7 |
Average ± standard deviation; n ≥ 3.
Change in potency relative to the WT GT1a(H77) control, rounded up to whole numbers for easy discussion in the text.
Fitness range: ≤0.02, unfit; 0.03 to 0.5, moderately fit; >0.5, fit.
Increasing concentrations of elbasvir resulted in substantial changes in the GT1a(H77) NS5A gene. At 1× EC90 elbasvir, the only consequential change observed was Q30R (see Table S4 in the supplemental material). While amino acid changes at positions 20, 24, and 81 were observed, phenotypic characterization of the independent changes K24E/G/N/R/T and R81S/M showed they do not reduce the potency of elbasvir (Table 8). At 3× EC90, changes at positions 28, 30, and 93 of NS5A were observed. In addition to Q30R, Q30E and Q30H were observed but at a lower frequency (single clones only). Amino acid changes at Y93 involved mostly Y93C, with a single clone bearing the Y93H substitution. There were three examples of linked GT1a(H77) NS5A RAVs at this concentration; the clones harbored M28T+Q30H, Q30R+Y93H, and K24T+M28T+Q30R linked substitutions (see Table S4 in the supplemental material). Additional linked substitutions were observed at 10× EC90 elbasvir, including changes at K24. These changes involved mostly doubly and triply substituted NS5A amino acids, including K24E/T/R+Q30R, K24E+L31V, K24T+Y93H, Q30R+R81W, K24R+R81M+Y93C, K24Q+M28I+L31M, and K24Q+R81S+Y93H. In all cases, one of the amino acids substituted involved either position 30, 31, or 93 (see Table S4 in the supplemental material). At 30× EC90, the most common change was Q30R, which was linked in some instances with K24A/T. There were very few examples of Y93H and L31V. Only 4 clones survived at 100× EC90 elbasvir; all 4 clones bore a Y93N amino acid change. At the highest dose tested, 1,000× EC90, the 5 surviving replicons harbored only 2 types of linked changes, comprising K24I+M28K or Q30R+L31V (see Table S4 in the supplemental material).
TABLE 8.
Potencies of elbasvir in transiently expressed genotype 1a(H77) replicons bearing NS5A RAVs from resistant colonies
| Variant | EC50, nMa | Fold shift vs WTb | Relative fitnessc |
|---|---|---|---|
| WT | 0.007 ± 0.004 | 1 | 1 |
| K24E | NTd | NT | Unfit |
| K24N | 0.003 ± 0.002 | 0.4 | 0.1 |
| K24R | 0.007 ± 0.004 | 1 | 0.8 |
| K24T | 0.003 ± 0.002 | 0.4 | 0.9 |
| M28T | 0.11 ± 0.03 | 15 | 0.8 |
| M28V | 0.01 ± 0.01 | 1 | 1.0 |
| Q30D | NT | NT | Unfit |
| Q30E | 0.39 ± 0.02 | 56 | 0.9 |
| Q30G | 0.59 ± 0.31 | 84 | 0.7 |
| Q30H | 0.044 ± 0.03 | 6 | 1.3 |
| Q30R | 0.11 ± 0.009 | 16 | 0.8 |
| L31 M | 0.07 ± 0.03 | 10 | 1.5 |
| L31V | 0.43 ± 0.13 | 61 | 0.9 |
| R81 M | 0.005 ± 0.005 | 1 | 1.3 |
| R81S | 0.005 ± 0.004 | 1 | 0.4 |
| R81W | 0.008 ± 0.005 | 1 | 0.7 |
| Y93C | 0.08 ± 0.004 | 11 | 0.3 |
| Y93H | 1.54 ± 0.78 | 220 | 0.2 |
| Y93N | 6.5 ± 1.25 | 929 | 0.5 |
| M28T+Q30H | 16 ± 1 | 2,286 | 0.7 |
| Q30H+L31V | 10 ± 1 | 1,429 | 1.1 |
| Q30R+L31V | 50 ± 10 | 7,143 | 1.1 |
| L31V+Y93H | 272 ± 115 | 38,857 | 0.5 |
| L31V+Y93N | 375 ± 145 | 53,571 | 0.5 |
| K24T+M28T+Q30R | 49 | 7,000 | 1.2 |
| K24Q+M28T+L31 M | 0.06 | 9 | 0.4 |
| K24 M+Q30R+R81 M | 0.48 ± 0.01 | 69 | 1.0 |
| M28T+Q30H+L31V | 392 ± 34 | 56,000 | 0.2 |
| M28T+Q30R+L31V | 624 ± 97 | 89,143 | 0.7 |
| M28V+Q30H+L31V | 17 ± 5 | 2,429 | 0.8 |
| M28T+Q30R+L31V+Y93H | 5,000 | 714,286 | 0.2 |
Average ± standard deviation; n ≥ 3.
Change in potency relative to the WT GT1a(H77) control.
Fitness range: ≤0.02, unfit; 0.03 to 0.5, moderately fit; >0.5, fit.
NT, not tested.
Resistance selection with the combination of grazoprevir and elbasvir.
Findings from sequence analyses of GT1a(H77) HCV replicon genomes that emerged from the treatment of cells with the combination of grazoprevir and elbasvir are summarized in Table 6. With a combination of 1× EC90 for both agents, here referred to as 1×/1× EC90 (where the first number refers to the fold multiple of the grazoprevir EC90 and the second the fold multiple of the elbasvir EC90), no variant at positions usually associated with NS3 RAVs was identified. Most clones from this group had changes at codons 109 and 131 in NS3; changes in codon 109 of NS3 were mostly R109K, which is considered a compensatory mutation. In NS5A, there were changes primarily at position 30. The majority of the NS5A changes (12/14 clones) harbored a Q30R substitution. The Q30R substitution and the only L31V variant observed in NS5A were linked to changes in NS3 at positions 41, 109, and 131 on the same genome.
Resistance selection with the 1×/3× combination yielded clones with primarily L31M substitutions in NS5A. Only one out of 9 clones established harbored an R155K change in NS3; this variant was linked with L31M and Y93C NS5A RAVs in a triple substitution. An NS5A-K24T change linked with NS5A-Q30R was observed in 2 additional clones. A further increase in the concentration of elbasvir to 1×/10× for the combination resulted in more changes not only in NS5A but also in NS3; 32% of the clones retained an NS3-D168A substitution. Single NS5A RAVs, Q30R and M28T (to a lesser extent), were observed at these inhibitor concentrations. Both RAVs were linked with an NS3-Q41R change and in some cases NS3-R109K on the same replicon genome (see Table S5 in the supplemental material). Other, less frequent changes included Q30D/E and M28K in NS5A and D168E in NS3; these were observed as single clones only. At a higher-concentration inhibitor combination, 1×/100× EC90, only three clones were successfully recovered, bearing NS3 R109K linked to NS5A Q30E/R. After a few passages, these cells stopped growing and were not characterized further.
Increasing the concentrations of grazoprevir beyond 1× EC90 generally resulted in fewer resistant colonies that harbored increased multiply linked substitutions (Table 6; see Table S6 in the supplemental material). At 10×/1× EC90, changes in NS3 were found only at position 168. The three types of variants observed included NS3-D168E/S/V substitutions, with the valine variant being the most prominent, occurring in 12 out of 14 clones established. Each of the 14 clones had a Q30H substitution in NS5A, with 2 of the clones also bearing an NS5A-K24R change. At a 3×/3× EC90, multiple-substitution changes, including R155K and D168E/S in NS3 coupled with M28T, Q30R, and R81W in NS5A, were observed. While NS3-R155K and NS3-D168E/V were never found on the same genome, NS5A-Q30R was frequently found on the same genome with NS5A-K24T, M28T, or R81W. A Q41R substitution in NS3 was found in all clones with this inhibitor combination. Colonies isolated at inhibitor combination concentrations greater than 3×/3× EC90 failed to grow and died off readily.
Phenotypic characterization of variants.
The substitutions observed following sequencing of HCV RNA from the surviving stable GT1a(H77) replicon colonies were introduced into and transiently expressed in G-luc-bearing GT1a(H77) HCV replicons and tested for susceptibility to grazoprevir and elbasvir. The potencies of grazoprevir and elbasvir in the GT1a(H77) transient replicons bearing NS3 and NS5A RAVs, respectively, are summarized in Tables 7 and 8. Generally, the results from the transient replicons were comparable to those for the stable replicons, except for a few outliers. Of note, elbasvir was 10-fold less active in the transient replicon bearing L31M but remained equipotent in the stable replicon relative to the WT replicon (data not shown). In the GT1a(H77) NS3 gene, the most resistant RAV was identified at position 168: D168E, D168A, and D168V caused potency losses of 37-, 273-, and 153-fold, respectively, relative to the wild type (Table 7), and were less fit than the wild type. The activity of grazoprevir against R109K was within 2-fold of that of the wild type. For GT1a(H77) NS5A, Y93N and Y93H were the most resistant single RAVs identified, causing potency losses of 929- and 220-fold, respectively, to elbasvir (Table 8); M28K, tested in the stable-replicon system, caused a 4,598-fold potency loss. Changes at K24 and R81 did not cause potency losses to elbasvir. The majority of multiple mutations in the NS5A gene caused substantial potency losses to elbasvir, especially if linked with an L31 or Y93 amino acid substitution. Generally, the higher the number of amino acid substitutions in NS5A, the greater the potency loss to elbasvir. One exception observed is for the triple mutant K24Q+M28T+L31M, which was found to confer only a 9-fold shift to elbasvir, fairly comparable to the losses caused by M28T and L31M.
DISCUSSION
The ability of HCV to readily select RAVs against single agents administered to patients necessitates that DAAs targeting multiple viral proteins be developed to overcome failure resulting from emergence of resistance. To this end, the combination of grazoprevir and elbasvir was evaluated in GT1a(H77) replicon cells. The discovery and initial characterization of grazoprevir (formerly known as MK-5172), a potent inhibitor of HCV NS3/4A protease, were reported previously (8, 16). In this study, the activity of the compound in HCV replicons with NS3/4A from additional viral genotypes was evaluated. The data demonstrate that grazoprevir maintains potent activity across all major HCV genotypes. It maintains a subnanomolar to nanomolar EC50 in all genotypes tested. There is a modest shift in potency in the GT3a(S52) subgenomic replicon. This shift in potency appears to be related to sequences beyond the catalytic domain, as the intrinsic activity of grazoprevir on the GT3a(S52) catalytic domain is comparable to its activity in other genotypes (Table 1). The contribution of an adaptive mutation(s) in the course of establishing the replicon cell line cannot be discounted. As with grazoprevir, the discovery of elbasvir (formerly known as MK-8742), an inhibitor of HCV NS5A, was also reported previously (11). A more complete profile of the inhibitor is presented in this report (Table 1). Elbasvir has broad HCV genotype activity, with EC50 and EC90 values in the low picomolar range. Elbasvir is less active in GT2b, which is attributable to a methionine polymorphism at position 31 in lieu of a valine residue [as found in GT2a(JFH1)]. In general, both DAAs are potent inhibitors of HCV replication in all major HCV genotypes.
Prior to studying the two DAAs in combination, their abilities to inhibit GT1a(H77) RAVs elicited by inhibitors from the other class were evaluated. RAVs elicited by NS5A inhibitors were potently inhibited by grazoprevir, an NS3/4A protease inhibitor, with potencies comparable to that for the wild-type reference strain. Similarly, elbasvir, an NS5A inhibitor, also potently inhibited RAVs elicited in NS3/4A by inhibitors of that class. Hence, RAVs from either inhibitor class do not confer resistance to an inhibitor from the other class. To confirm this finding, the kinetics of inhibition was monitored over a longer period (14 days) using prototypical RAVs from each class. Grazoprevir inhibited GT1a-Q30R and GT1a-Y93H, key clinically relevant NS5A RAVs, with kinetics comparable to that of the wild-type GT1a(H77) replicon. Conversely, R155K, a key clinically relevant RAV elicited by first-generation NS3/4A protease inhibitors, was inhibited by elbasvir with kinetics comparable to those for the WT GT1a(H77) replicon. These studies provided the impetus to study the compounds in combination. Grazoprevir and elbasvir interacted additively in combination to inhibit HCV RNA synthesis at concentrations spanning their EC50s. More importantly, the inhibitor combination did not exhibit antagonistic behavior. The profile of the inhibitor combination suggested that they may complement each other to block the emergence of resistance. Grazoprevir and elbasvir independently exhibited a dose-dependent suppression of resistant colonies in GT1a(H77) replicon cells. There was a reduction in the number of resistant colonies with increasing multiples of EC90 for each compound. However, the combination of grazoprevir and elbasvir was more effective at blocking the emergence of resistance. As independent agents, 100× EC90 and 1,000× EC90 of grazoprevir and elbasvir, respectively, were required to effectively suppress the emergence of resistant colonies. In combination, only 10× EC90 of each inhibitor practically blocked the emergence of resistant colonies. This suggests the grazoprevir-elbasvir combination presents a potent inhibitory activity with a high genetic barrier to resistance. Thus, apart from being highly efficacious, lower concentrations of the inhibitor combination may also provide a larger therapeutic window.
To obtain insights into the pathways of resistance, RNA was isolated from resistant replicon cells and sequenced. Variants that had not been previously established as stable replicons were tested by introducing the changes into the cDNA of the GT1a(H77)-Gluc virus and transiently expressing transcribed RNA in Huh7.5 cells, and the potencies of grazoprevir and elbasvir were evaluated (Tables 7 and 8) as discussed in Materials and Methods. The major amino acid substitutions identified in NS3 were R155K and D168A/E/S/V. The potency of grazoprevir in GT1a(H77) NS3-R155K is shifted only modestly (3-fold) relative to that in the wild-type replicon. The loss in potency of grazoprevir against the GT1a(H77) NS3-D168 variants was more substantial, ranging from 2- to 273-fold (Tables 2 and 7). With increasing concentration of grazoprevir, changes at D168 became more prominent, suggesting that it is the dominant pathway for resistance against the inhibitor. The structural basis for the potency of grazoprevir in NS3 has been reported previously (28). Substitutions at D168 negatively influence ionic interactions within the active site and consequently grazoprevir/protein interactions, resulting in a loss of inhibitor potency in GT1a(H77) replicons. In NS5A, RAVs primarily involved changes at positions 30, 31, and 93 that have also been observed for other inhibitors of this class (29). These positions remained the dominant loci with elbasvir. The absence of a three-dimensional structure of an inhibitor-NS5A complex makes it difficult to explain the structural basis for the loss in elbasvir potency. The two alternate structures reported for an NS5A homodimer (30, 31) further cloud the potential modes of interaction of elbasvir with the protein. Nonetheless, the side chains of the substituted proteins at positions 30, 31, and 93 are very different in size, charge, and aromaticity, such that natural interactions between the protein and inhibitor will be expected to be altered. As reported in Table 8, substitutions at these positions caused substantial potency losses for elbasvir, with the single Y93N amino acid change resulting in a >900-fold reduction in potency. Secondary changes in NS5A included K24E/I/M/Q/T located at end of the N-terminal amphipathic helix and R81M/S/W, which did not confer any potency losses to elbasvir. The highest concentrations (100 to 1,000× EC90) of elbasvir selected variants that had mostly linked substitutions that required two or more nucleotide changes, underscoring the higher genetic barrier to resistance. Only 4 to 5 resistant replicon clones were successfully characterized at the highest elbasvir dose (1,000× EC90) presumably as a result of low replication fitness of the variants. An assessment of the pattern of resistance suggests the existence of three main NS5A pathways involving M28, Q30/L31, and Y93. The M28 pathway more frequently emerged together with the Q30/L31 pathway but not with the Y93 pathway.
In combination with grazoprevir, increasing elbasvir concentrations elicited primarily the Q30/L31 pathway. This may be a result of a low replicative fitness for the Y93 variants in replicon cells (13). The data for the grazoprevir-elbasvir combination indicate cross talk between NS3 and NS5A. An increase in the concentration of elbasvir, while maintaining the concentration of grazoprevir constant, resulted in the selection of variants not only in NS5A but also in NS3. The selective pressure from NS5A inhibition as a component of the replicase complex caused the NS3 gene to acquire substitutions that promoted fitness. The Q41R substitution in NS3, which is known to promote replicative fitness in replicons (24), was frequently observed under these conditions. Additionally, these higher elbasvir concentrations promoted the selection of R109K in NS3. While R109K does not substantially affect the potency of grazoprevir in GT1a replicons, it markedly increases fitness (Table 7). As NS3 is not the target of elbasvir, selection of a variant(s) to reduce the intrinsic potency would be inconsequential. However, generating relevant substitutions in the NS3 gene to increase the overall fitness of surviving NS5A variants is a strategy that may attenuate or overcome the effectiveness of the NS5A inhibitor within the replicase complex; this has also been suggested for GT1a replicons selected with Asunaprevir and daclatasvir (32). In contrast, increasing the concentration of grazoprevir in the combination resulted in fewer selections of NS3-Q41R and R109K substitutions and more variants at D168 to directly counter the effect on the cognate target. As a corollary, increasing selective pressure from higher grazoprevir concentrations in the combination resulted in preferential selection of Q30H and L31M over Q30R and L31V in NS5A, respectively. While they cause less potency loss to elbasvir, GT1a(H77) replicons with NS5A-Q30H and L31M have higher relative fitness. Most of the changes observed in NS3 and NS5A described here occurred at ≤10-fold EC90 values. At higher concentrations, comparable to levels routinely observed in the clinic (33), most replicons do not survive; this enhanced potency may contribute to the efficacy of the combination in the clinic.
Collectively, these results demonstrate that the combination of grazoprevir, an NS3/4A inhibitor, and elbasvir, an NS5A inhibitor, potently inhibits HCV RNA synthesis, with no evidence of antagonism, and present a high genetic barrier to resistance. The inhibitor combination presents an attractive alternative as an oral, interferon-free DAA for patients chronically infected with HCV. In recent clinical studies, the in vitro activities translated to robust clinical efficacy for the combination. In treatment-naive GT1a patients given a once-daily dose of 50 mg/100 mg elbasvir-grazoprevir, an SVR rate of 95% was achieved in patients (34). The RAVs and dominant resistance pathways observed in clinical studies of GT1a-infected subjects largely mirrored what was observed in vitro. The most common treatment-emergent (TE) NS5A RAV in virologic failures was at Q30. Similarly, D168 amino acid substitutions accounted for most TE NS3 RAVs in virologic failures. The combination has been approved (January 2016) for the treatment of chronic HCV infections.
Supplementary Material
ACKNOWLEDGMENTS
Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., is developing grazoprevir (MK-5172) and elbasvir (MK-8742) as components of a combination therapy for chronic HCV infections. The design, execution, and interpretation of this study were performed by the authors, who are or were employees of Merck and Co. As present or former employees of Merck, authors may own stock and/or stock options in the company. All authors had full access to any pertinent data upon request. Each coauthor approved an essentially final version of the manuscript.
The opinions expressed in this report represent the consensus of the authors and do not necessarily reflect the formal position of Merck.
We thank Richard Raubertas and Stuart Black for helpful discussions. We also thank Michele McColgan for assistance with the figure files.
Funding Statement
The work described in this paper was funded and supported by Merck & Co.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00051-16.
REFERENCES
- 1.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 2.WHO. 2014. Guidelines for the screening, care and treatment of persons with hepatitis C infection. WHO, Geneva, Switzerland: http://www.who.int/hiv/pub/hepatitis/hepatitis-c-guidelines/en/ Accessed 3 September 2015. [Google Scholar]
- 3.Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. 2014. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol 61:S45–S57. doi: 10.1016/j.jhep.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 4.Herbst DA, Reddy K. 2013. Sofosbuvir, a nucleotide polymerase inhibitor, for the treatment of chronic hepatitis C virus infection. Expert Opin Invest Drugs 22:527–536. doi: 10.1517/13543784.2013.775246. [DOI] [PubMed] [Google Scholar]
- 5.Younossi ZM, Stepanova M, Marcellin P, Afdhal N, Kowdley KV, Zeuzem S, Hunt SL. 2015. Treatment with ledipasvir and sofosbuvir improves patient-reported outcomes: results from the ION-1, -2, and -3 clinical trials. Hepatology 61:1798–1808. doi: 10.1002/hep.27724. [DOI] [PubMed] [Google Scholar]
- 6.Deeks ED. 2015. Ombitasvir/paritaprevir/ritonavir plus dasabuvir: a review in chronic HCV genotype 1 infection. Drugs 75:1027–1038. doi: 10.1007/s40265-015-0412-z. [DOI] [PubMed] [Google Scholar]
- 7.Cento V, Chevaliez S, Perno CF. 2015. Resistance to direct-acting antiviral agents: clinical utility and significance. Curr Opin HIV AIDS 10:381–389. doi: 10.1097/COH.0000000000000177. [DOI] [PubMed] [Google Scholar]
- 8.Summa V, Ludmerer S, McCauley JA, Fandozzi C, Burlein C, Claudio G, Coleman PJ, Dimuzio JM, Ferrara M, Di Filippo M, Gates AT, Graham DJ, Harper S, Hazuda DJ, Huang Q, McHale C, Monteagudo E, Pucci V, Rowley M, Rudd MT, Soriano A, Stahlhut MW, Vacca JP, Olsen DB, Liverton NJ, Carroll SS. 2012. MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob Agents Chemother 56:4161–4167. doi: 10.1128/AAC.00324-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolykhalov A, Mihalik K, Feinstone SM, Rice CM. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol 74:2046–2051. doi: 10.1128/JVI.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soriano V, Vispo E, Poveda E, Labarga P, Martin-Carbonero L, Fernandez-Montero JV, Barreiro P. 2011. Directly acting antivirals against hepatitis C virus. J Antimicrob Chemother 66:1673–1686. doi: 10.1093/jac/dkr215. [DOI] [PubMed] [Google Scholar]
- 11.Coburn CA, Meinke P, Chang W, Fandozzi CM, Graham DJ, Hu B, Huang Q, Kargman S, Kozlowski J, Liu R, McCauley JA, Nomeir AA, Soll RM, Vacca JP, Wang D, Wu H, Zhong B, Olsen DB, Ludmerer SW. 2013. Discovery of MK-8742: an HCV NS5A inhibitor with broad genotype activity. Chem Med Chem 8:1930–1940. doi: 10.1002/cmdc.201300343. [DOI] [PubMed] [Google Scholar]
- 12.Macdonald A, Harris M. 2004. Hepatitis C virus NS5A: tales of promiscuous protein. J Gen Virol 85:2485–2502. doi: 10.1099/vir.0.80204-0. [DOI] [PubMed] [Google Scholar]
- 13.McGivern DR, Masaki T, Williford S, Ingravallo P, Feng Z, Lahser F, Asante-Appiah E, Neddermann P, De Francesco R, Howe AY, Lemon SM. 2014. Kinetic analyses reveal potent and early blockade of hepatitis C virus assembly by NS5A inhibitors. Gastroenterology 147:453–462. doi: 10.1053/j.gastro.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross-Thriepland D, Harris M. 2015. Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J Gen Virol 96:727–738. doi: 10.1099/jgv.0.000009. [DOI] [PubMed] [Google Scholar]
- 15.Rai DWL, Jiang X, Zhan P, Jia H, De Clercq E, Liu X. 2016. The changing face of hepatitis C: recent advances on HCV inhibitors targeting NS5A. Curr Med Chem 22:1860–1879. doi: 10.2174/0929867322666150209150920#sthash.6keCr4Wi.dpuf. [DOI] [PubMed] [Google Scholar]
- 16.Harper S, McCauley J, Rudd MT, Ferrara M, DiFilippo M, Crescenzi B, Koch U, Petrocchi A, Holloway MK, Butcher JW, Romano JJ, Bush KJ, Gilbert KF, McIntyre CJ, Nguyen KT, Nizi E, Carroll SS, Ludmerer SW, Burlein C, DiMuzio JM, Graham DJ, McHale CM, Stahlhut MW, Olsen DB, Monteagudo E, Cianetti S, Giuliano C, Pucci V, Trainor N, Fandozzi CM, Rowley M, Coleman PJ, Vacca JP, Summa V, Liverton NJ. 2012. Discovery of MK-5172, a macrocyclic hepatitis C virus NS3/4a protease inhibitor. ACS Med Chem Lett 3:332–336. doi: 10.1021/ml300017p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi M, Lemon S. 2004. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J Virol 78:7904–7915. doi: 10.1128/JVI.78.15.7904-7915.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blight KJ, Kolykhalov A, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda M, Yi M, Lemon SM. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficently in cultured Huh7 cells. J Virol 76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Saeed M, Scheel T, Gottwein J, Marukian S, Dustin L, Bukh J, Rice CM. 2012. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob Agents Chemother 56:5365–5373. doi: 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong X, Bogen S, Chase R, Girijavallabhan V, Guo Z, Njoroge FG, Prongay A, Saksena A, Skelton A, Xia E, Ralston R. 2008. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res 77:177–185. doi: 10.1016/j.antiviral.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Gu Z, Graci J, Lahser F, Breslin JJ, Jung SP, Crona JH, McMonagle P, Xia E, Liu S, Karp G, Zhu J, Huang S, Nomeir A, Weetall M, Almstead NG, Peltz SW, Tong X, Ralston R, Colacino JM. 2013. Identification of PTC725, an orally bioavailable small molecule that selectively targets the hepatitis C virus NS4B protein. Antimicrob Agents Chemother 57:3250–3261. doi: 10.1128/AAC.00527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimakami T, Welsch C, Yamane D, McGivern DR, Yi M, Zeuzem S, Lemon SM. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prichard MN, Prichard L, Shipman C Jr. 1993. Strategic design and three-dimensional analysis of antiviral drug combinations. Antimicrob Agents Chemother 37:540–545. doi: 10.1128/AAC.37.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prichard MN, Shipman C. 1990. A three-dimentional model to analyze drug-drug interactions. Antiviral Res 14:181–205. doi: 10.1016/0166-3542(90)90001-N. [DOI] [PubMed] [Google Scholar]
- 27.Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res 70:28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Guan Y, Sun H, Pan P, Li Y, Li D, Hou T. 2015. Exploring resistance mechanisms of HCV NS3/4A protease mutations to MK5172: insight from molecular dynamics simulations and free energy calculations. Mol Biosyst 11:2568–2578. doi: 10.1039/C5MB00394F. [DOI] [PubMed] [Google Scholar]
- 29.Fridell RA, Wang C, Sun JH, O'Boyle DR II, Nower P, Valera L, Qiu D, Roberts S, Huang X, Kienzle B, Bifano M, Nettles RE, Gao M. 2011. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935. doi: 10.1002/hep.24594. [DOI] [PubMed] [Google Scholar]
- 30.Love RA, Brodsky O, Hickey MJ, Wells PA, Cronin CN. 2009. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J Virol 83:4395–4403. doi: 10.1128/JVI.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tellinghuisen TL, Marcotrigiano J, Rice CM. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelosi LA, Voss S, Liu M, Gao M, Lemm JA. 2012. Effect on hepatitis C virus replication of combinations of direct-acting antivirals, including NS5A inhibitor Daclatasvir. Antimicrob Agents Chemother 56:5230–5239. doi: 10.1128/AAC.01209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merck and Co., Inc. 2016. ZEPATIERTM (elbasvir and grazoprevir) tablets, for oral use. Prescribing information. Merck and Co., Inc., Kenilworth, NJ. https://www.merck.com/product/usa/pi_circulars/z/zepatier/zepatier_pi.pdf.
- 34.Zeuzem S, Ghalib R, Reddy KR, Pockros PJ, Ben Ari Z, Zhao Y, Brown DD, Wan S, DiNubile MJ, Nguyen BY, Robertson MN, Wahl J, Barr E, Butterton JR. 2015. Grazoprevir-elbasvir combination therapy for treatment-naive cirrhotic and noncirrhotic patients with chronic hepatitis C virus genotype 1, 4, or 6 infection: a randomized trial. Ann Intern Med 163:1–13. doi: 10.7326/M15-0785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.