Abstract
This study investigated the effects of HIV-1 infection and antiretroviral therapy (ART) on the expression of intestinal drug efflux transporters, i.e., P-glycoprotein (Pgp), multidrug resistance-associated proteins (MRPs), and breast cancer resistance protein (BCRP), and metabolic enzymes, such as cytochrome P450s (CYPs), in the human upper intestinal tract. Intestinal biopsy specimens were obtained from HIV-negative healthy volunteers, ART-naive HIV-positive (HIV+) subjects, and HIV+ subjects receiving ART (10 in each group). Intestinal tissue expression of drug transporters and metabolic enzymes was examined by microarray, real-time quantitative reverse transcription-PCR (qPCR), and immunohistochemistry analyses. Microarray analysis demonstrated significantly lower expression of CYP3A4 and ABCC2/MRP2 in the HIV+ ART-naive group than in uninfected subjects. qPCR analysis confirmed significantly lower expression of ABCC2/MRP2 in ART-naive subjects than in the control group, while CYP3A4 and ABCG2/BCRP showed a trend toward decreased expression. Protein expression of MRP2 and BCRP was also significantly lower in the HIV+ naive group than in the control group and was partially restored to baseline levels in HIV+ subjects receiving ART. In contrast, gene and protein expression of ABCB1/Pgp was significantly increased in HIV+ subjects on ART relative to HIV+ ART-naive subjects. These data demonstrate that the expression of drug-metabolizing enzymes and efflux transporters is significantly altered in therapy-naive HIV+ subjects and in those receiving ART. Since CYP3A4, Pgp, MRPs, and BCRP metabolize or transport many antiretroviral drugs, their altered expression with HIV infection may negatively impact drug pharmacokinetics in HIV+ subjects. This has clinical implications when using data from healthy volunteers to guide ART.
INTRODUCTION
Despite improved outcomes for persons with HIV, intestinal complications ranging from diarrhea, weight loss, nausea, vomiting, and abdominal pain to gastrointestinal (GI) bleeding, anorectal disease, and GI tumors continue to be common, even in those who have achieved undetectable viral loads and normal CD4+ lymphocyte counts with the use of combination antiretroviral therapy (ART) (1, 2). The persistence of HIV reservoirs in the gut-associated lymphoid tissue (GALT) has been well documented, and detectable levels of HIV replication (RNA, DNA, and p24 protein) have been reported in tissue biopsy specimens from the upper and lower intestinal tracts (3–5). Studies have shown that ART-naive or treated subjects with HIV show decreased intestinal tissue expression of genes involved in mucosal repair and regeneration, while expression of genes regulating inflammation and immune activation is increased (2, 5–7). In addition, the expression of genes involved in intestinal epithelial integrity and barrier function, such as tight junctional proteins, as well as in nutrient and xenobiotic absorptive and digestive functions, such as drug-metabolizing enzymes, is also decreased in HIV-infected subjects compared to HIV-negative healthy volunteers (6). Collectively, these data suggest that HIV infection leads to the disruption of the integrity of the intestinal epithelial barrier and alteration in nutrient and drug absorption. The resulting microbial translocation has been hypothesized to contribute to HIV progression and the development of future comorbidity (8).
Antiretroviral drugs (ARVs) are reported to have highly variable penetration into the ileal and rectal tissues of HIV-infected subjects, and in some cases the tissue concentrations are significantly lower than therapeutic levels required for effective antiviral activity and much lower than the concentrations detected in plasma samples from the same individuals (9–11). Some studies also suggest that poor drug penetration into these intestinal tissues may contribute to the lack of a local therapeutic response and the establishment of an HIV reservoir at this site (10, 11). While drug transporters and metabolic enzymes are known to play a key role in drug disposition at the level of the intestinal epithelium and are suggested to contribute to the persistence of this viral reservoir, experimental evidence to support this rationale is currently lacking (9). Some ARVs (e.g., atazanavir and efavirenz) are also reported to have significantly different plasma pharmacokinetics in HIV-infected subjects and in healthy volunteers. This could potentially be due to the differences in the intestinal drug absorption associated with changes in drug metabolism and/or transport processes (12, 13).
Several efflux transporters belonging to the ATP-binding cassette (ABC) superfamily, such as P-glycoprotein (Pgp) (ABCB1 gene product), multidrug resistance-associated proteins (MRPs) (ABCC1 to -5 gene products), and breast cancer resistance protein (BCRP) (ABCG2 gene product), are expressed at the apical brush border membrane of enterocytes, where they can efflux drugs back into the lumen, lowering their cellular accumulation and transepithelial permeativity and collectively acting as the first biochemical barrier to drug absorption (14, 15). Several drug-metabolizing enzymes, such as oxidative cytochrome P450 (CYP) enzymes and conjugative UDP-glucuronyl transferases (UGTs), are also expressed within enterocytes and play an important role in the first-pass extraction of drugs via metabolic degradation. Lastly, members of the solute carrier (SLC) superfamily, such as various organic cation, organic anion, and nucleotide carriers, facilitate cellular entry of bulky and/or charged drugs, including many ARVs (16).
Systemic absorption and intestinal tissue concentrations of ARVs can be regulated by CYP enzymes and ABC and SLC drug transporters expressed in the intestinal epithelium. For example, CYP3A4 metabolizes HIV protease inhibitors (PIs), nonnucleoside analog reverse transcriptase inhibitors (NNRTIs), and CCR5 receptor antagonists such as maraviroc (17, 18). In addition, CYP2B6 plays an important role in the metabolism of NNRTIs (efavirenz and nevirapine) (19). Among drug transporters, Pgp has a wide range of drug substrates, including all PIs, some nucleoside/nucleotide analog reverse transcriptase inhibitors (NRTIs) such as abacavir and tenofovir disoproxil fumarate, as well as maraviroc and the integrase inhibitors raltegravir and elvitegravir (16, 20). MRP1 to -3 are also capable of transporting many PIs and some NRTIs, while MRP4 and -5 and BCRP are primarily responsible for the efflux of NRTIs (16). Tissue inflammation and cytokine secretion are known to alter functional expression of drug transporters and CYP enzymes in other disease states (e.g., inflammatory bowel syndrome) (21). Furthermore, we and other investigators have previously reported that the functional expression of drug transporters in brain parenchyma, blood-brain and blood-testis barriers, and sigmoid colon epithelium is altered by the HIV antigens, viral infection-associated inflammation, and ARVs (22–28). The main objective of this study was to investigate the impact of HIV-1 infection and suppressive ART on the expression of intestinal drug transporters and metabolic enzymes in the intestinal mucosa of HIV-infected subjects.
MATERIALS AND METHODS
Human subjects and sample collection.
HIV-1-infected subjects were enrolled in the study and divided into two cohorts, ART naive (group B; plasma HIV load of >5,000 copies/ml and no prior exposure to ARVs) and long-term ART treated (group C; plasma HIV load of <50 copies/ml and receiving current ART regimen for >1 year). HIV-1-negative clinically healthy individuals (group A) were enrolled in the study to serve as HIV-negative controls. Jejunal mucosal biopsy samples were collected by gastrointestinal endoscopy under conscious sedation and immediately cryopreserved for transcriptional analysis of HIV RNA and host gene expression by real-time quantitative reverse transcription-PCR (qPCR). Tissue samples were also fixed in paraformaldehyde and embedded in paraffin blocks for immunohistochemical analysis according to a previously published protocol (4). A portion of the jejunal biopsy specimen was collected in RPMI 1640 (Invitrogen, Carlsbad, CA) lymphocyte isolation medium for flow cytometric analysis of the CD4+ T cell subset (4, 29). In addition, peripheral blood samples were collected at the time of gut biopsy for measurement of plasma HIV loads (4, 29). The Institutional Review Board at the University of California, Davis, CA, approved this study protocol, and written informed consent was obtained from all study participants.
Gene expression profile analysis.
We measured gene expression changes in the gut biopsy specimens from HIV-infected subjects compared to those from HIV-negative healthy individuals using human genome-specific high-density oligonucleotide microarrays as described previously (6, 30). Total RNA was isolated from jejunal tissue biopsy specimens of five HIV-1 negative healthy volunteers, seven HIV-positive (HIV+) ART-naive subjects, and six HIV+ subjects receiving long-term ART using the RNeasy RNA isolation kit (Qiagen, Valencia, CA). mRNA was amplified, labeled, and hybridized to human whole-genome U95av2 GeneChips (Affymetrix) using standard protocols (Affymetrix, Santa Clara, CA). Cross-sectional analysis of microarray data among all groups, as well as background correction, normalization, and generation of expression values, was performed using dChip analysis software (DNA-Chip analyzer, version 1.3; Harvard University). Samples from HIV-negative participants were used as baseline values for identifying genes differentially expressed in HIV+ naive participants and HIV+ participants on ART. The differential expression was defined as gene expression a minimum of 1.5-fold higher or lower, with statistical confidence (P ≤ 0.05), than the average gene expression observed in healthy control group.
Real-time qPCR analysis.
qPCR analysis was performed to compare expression levels of specific genes between healthy HIV-negative individuals and ARV-naive and -treated HIV+ subjects. Total RNA was reverse transcribed into cDNA using Superscript III (Invitrogen, Carlsbad, CA), and the expression levels of CYP3A4, ABCB1, ABCC2, or ABCG2 or the endogenous control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), were measured using 6-carboxyfluorescein (FAM)-labeled TaqMan gene expression assays and the primer-limited VIC/MGB probe for GAPDH (Life Technologies Inc., Burlington, Ontario, Canada) in the Mastercycler ep realplex 2S thermal cycler (Eppendorf Canada, Mississauga, Ontario, Canada). HIV RNA levels in intestinal tissue and plasma samples were measured by real-time qPCR as described previously (31, 32).
Immunohistochemistry analysis.
Detection and localization of CYP3A4, Pgp, MRP2, or BCRP protein expression in intestinal tissue samples were done by immunohistochemical analysis. Intestinal tissue sections fixed in 4% paraformaldehyde were immunostained with a primary antibody specific to CYP3A4 (1:600 dilution; Sigma-Aldrich, Oakville, Ontario, Canada), Pgp (1:100 dilution; Santa Cruz Biotechnology Inc., Dallas, TX), MRP2 (1:250 dilution; Kamiya Biomedical Company, Seattle, WA), or BCRP (1:100 dilution; Abcam Inc., Toronto, Ontario, Canada). After washing, tissue sections were immunostained with the corresponding Alexa-488-conjugated secondary antibodies (Life Technologies Inc., Burlington, Ontario, Canada). Standard DAPI (4′,6′-diamidino-2-phenylindole) staining was utilized to identify cell nuclei, and the fluorescence images were obtained using a Zeiss LSM700 confocal microscope. See the figure legends for the image quantification procedure.
RESULTS
Study participants.
The gut mucosal gene expression profiling was examined by DNA microarray analysis for five HIV-1-negative healthy volunteers with CD4+ T cell counts in the normal range (632 to 1,203 cells/μl), seven HIV+ naive subjects with plasma viral loads of >5,000 copies/ml and CD4+ T cell depletion with low cell counts (6 to 427 cells/μl), and six HIV+ subjects receiving long-term ART with undetectable viral loads (Table 1).
TABLE 1.
Demographic and clinical characteristics of study subjects used in microarray analysisa
| Group and patient | Age (yr) | Sex | CD4+ cell count (cells/μl) | Yr since HIV-1 diagnosis | ART components | Yr on ART |
|---|---|---|---|---|---|---|
| A (HIV-1-seronegative healthy controls) | ||||||
| P4 | 59 | M | ND | NA | NA | NA |
| P15 | 52 | M | ND | NA | NA | NA |
| P34 | 35 | F | 1,046 | NA | NA | NA |
| P38 | 48 | F | 632 | NA | NA | NA |
| P41 | 43 | F | 1,203 | NA | NA | NA |
| Median (range) | 48 (35–59) | 1,046 (632–1,203) | ||||
| B (HIV-1-seropositive, ART naive) | ||||||
| P13 | 48 | F | 8 | 5 | NA | NA |
| P14 | 36 | M | 6 | 1 | NA | NA |
| P19 | 35 | M | 46 | 1 | NA | NA |
| P20 | 45 | M | 294 | 1 | NA | NA |
| P24 | 34 | M | 34 | 4 | NA | NA |
| P47 | 23 | M | 205 | 1 | NA | NA |
| P48 | 42 | M | 427 | 1 | NA | NA |
| Median (range) | 36 (23–48) | 46 (6–427) | 1 (1–5) | |||
| C (HIV-1-seropositive, receiving ART) | ||||||
| P36 | 35 | M | 250 | 1 | RTV, ABC, 3TC, AZT | 1 |
| P50 | 32 | F | 570 | 1 | EFV, ABC, 3TC, AZT | 1 |
| P57 | 33 | F | 644 | 1 | EFV, ABC, 3TC, AZT | 1 |
| P58 | 36 | M | 300 | 2 | RTV, ABC, 3TC, AZT | 1 |
| P59 | 23 | M | 205 | 2 | EFV, RTV, 3TC | 2 |
| P73 | 37 | M | 468 | 3 | EFV, RTV, ABC, 3TC, AZT | 3 |
| Median (range) | 34 (23–37) | 384 (205–644) | 1.5 (1–3) | 1 (1–3) |
Abbreviations: M, male; F, female; ND, not determined; NA, not applicable; 3TC, lamivudine; ABC, abacavir; AZT, zidovudine; EFV, efavirenz; RTV, ritonavir.
For qPCR and immunohistochemical analyses, 30 participants 25 to 60 years of age were enrolled in this study and divided among three groups (Table 2). Group B consisted of 10 chronically HIV-1-infected ARV-naive subjects who were HIV-1 seropositive for 1 to 10 years (median, 3 years) and had HIV RNA levels of 5,642 to 48,000 copies/ml and CD4+ T cell counts ranging between 10 and 672 cells/μl (median, 265 cells/μl). Group C consisted of 10 HIV-1-infected subjects receiving ART who were HIV seropositive for 1 to 11 years (median, 6 years), were receiving long-term ART (1 to 11 years; median, 6 years), and had undetectable plasma viral loads (<50 copies/ml) and CD4+ T cell counts ranging between 140 and 1107 cells/μl (median, 525 cells/μl). ART regimens contained NRTIs (10/10), PIs (7/10; 2 containing ritonavir-boosted atazanavir, 4 containing ritonavir as the only PI, and 1 containing atazanavir as the only PI), and/or efavirenz (1/10). Group A consisted of 10 HIV-seronegative healthy controls. None of the 30 participants reported being smokers, using any drugs of abuse, or currently using any over-the-counter medications or herbal supplements. Five of the HIV-seronegative participants reported alcohol use.
TABLE 2.
Demographic and clinical characteristics of study participantsa
| Group and patient | Age (yr) | Sex | Race | CD4+ cell count (cells/μl) | Plasma viral load (copies/ml) | Yr since HIV-1 diagnosis | ART components | Yr on ART |
|---|---|---|---|---|---|---|---|---|
| A (HIV-1-seronegative healthy controls) | ||||||||
| P110 | 58 | M | AA | ND | NA | NA | NA | NA |
| P112 | 60 | M | C | ND | NA | NA | NA | NA |
| P130 | 35 | F | C | 1,057 | NA | NA | NA | NA |
| P153 | 60 | M | AA | 442 | NA | NA | NA | NA |
| P213 | 43 | F | C | 1,021 | NA | NA | NA | NA |
| P214 | 36 | M | C | 1,559 | NA | NA | NA | NA |
| P229 | 45 | F | AA | 803 | NA | NA | NA | NA |
| P236 | 49 | F | C | 1,835 | NA | NA | NA | NA |
| P238 | 28 | F | C | ND | NA | NA | NA | NA |
| P239 | 36 | F | C | 777 | NA | NA | NA | NA |
| Median (range) | 44 (28–60) | 1,021 (442–1,835) | ||||||
| B (HIV-1-seropositive, ART naive) | ||||||||
| P108 | 38 | M | C | 672 | 5,642 | 5 | NA | NA |
| P115 | 25 | M | C | 175 | 20,000 | 2 | NA | NA |
| P120 | 36 | M | C | 10 | 480,000 | 4 | NA | NA |
| P124 | 43 | M | C | 269 | 246,000 | 1 | NA | NA |
| P152 | 48 | M | C | 184 | 29,200 | 10 | NA | NA |
| P209 | 33 | M | C | 450 | 57,000 | 3 | NA | NA |
| P210 | 30 | M | AA | 618 | 100,000 | 10 | NA | NA |
| P211 | 47 | F | AA | 450 | 20,000 | 2 | NA | NA |
| P223 | 30 | F | C | 192 | 18,000 | 1 | NA | NA |
| P224 | 40 | F | C | 261 | 90,000 | 3 | NA | NA |
| Median (range) | 37 (25–48) | 265 (10–672) | 43,100 (5,642–48,000) | 3 (1–10) | ||||
| C (HIV-1-seropositive, receiving ART) | ||||||||
| P120 | 45 | M | C | 525 | <50 | 11 | RTV, ABC, 3TC | 11 |
| P136 | 25 | M | C | 560 | <50 | 2 | RTV, ABC, 3TC, AZT | 1 |
| P140 | 35 | M | AA | 1,107 | <50 | 1 | ABC, 3TC, AZT | 1 |
| P173 | 31 | M | C | 140 | <50 | 5 | ATV, RTV, TDF, FTC | 5 |
| P189 | 50 | F | C | 585 | <50 | 5 | TDF, FTC, 3TC, AZT | 9 |
| P197 | 45 | F | C | 279 | <50 | 11 | ATV, RTV, TDF | 10 |
| P198 | 50 | F | AA | 508 | <50 | 10 | RTV, TDF, FTC | 10 |
| P208 | 36 | F | C | 322 | <50 | 6 | EFV, TDF, FTC | 5 |
| P219 | 44 | F | AA | ND | <50 | 6 | ATV, ABC, 3TC | 6 |
| P231 | 55 | F | AA | 1,039 | <50 | 10 | RTV, ABC, 3TC | 6 |
| Median (range) | 44.5 (25–55) | 525 (140–1,107) | 6 (1–11) | 6 (1–11) |
Abbreviations: M, male; F, female; AA, African American; C, Caucasian; ND, not determined; NA, not applicable; 3TC, lamivudine; ABC, abacavir; ATV, atazanavir; AZT, zidovudine; EFV, efavirenz; FTC, emtricitabine; RTV, ritonavir; TDF, tenofovir disoproxil fumarate.
Distinct mucosal gene expression profiles in HIV infection with and without ART.
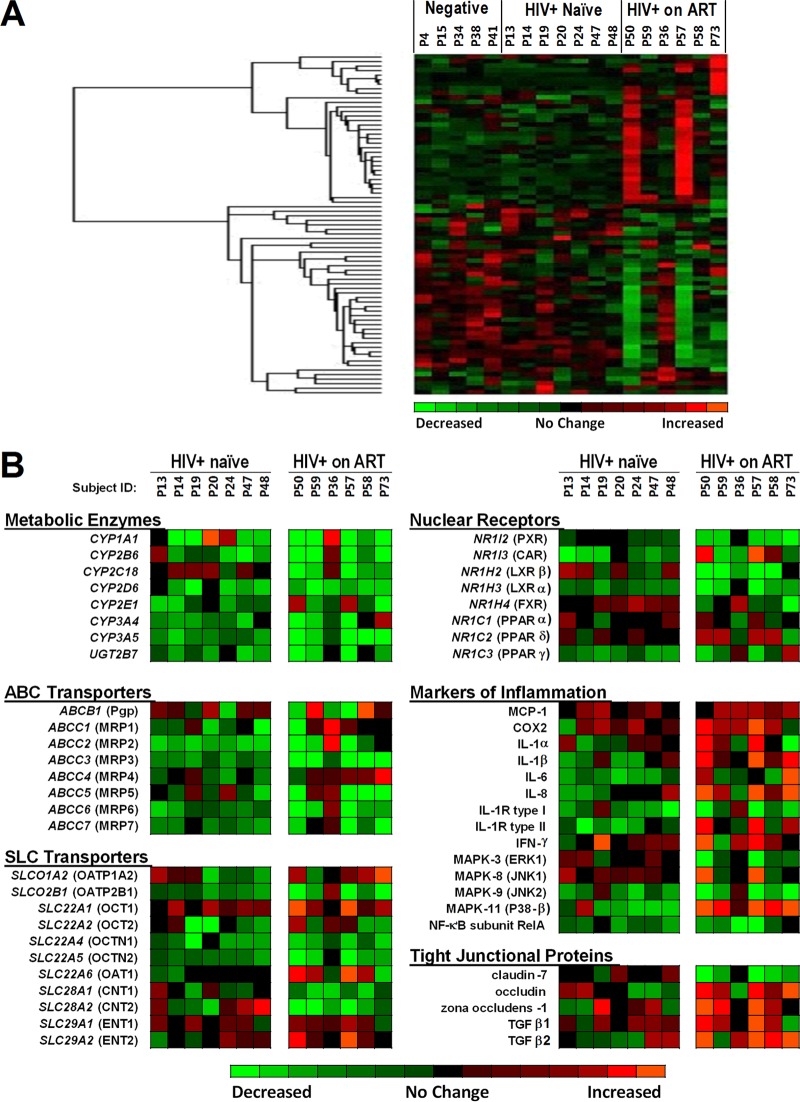
In order to identify differentially expressed gene networks in HIV infection, gene expression profiles were evaluated by high-throughput oligonucleotide microarray analysis in the intestinal biopsy samples from the HIV+ ART-naive subjects and HIV+ subjects on ART compared to those from HIV-negative healthy controls. Hierarchical cluster analysis of the data revealed differential expression of several groups of genes regulating pharmacological and physiological functions, such as those for metabolic enzymes, drug uptake (SLC) and efflux (ABC) transporters, nuclear receptors, and tight junctional proteins, and genes associated with intestinal immune response and inflammation, (Fig. 1A). The gene expression of several CYP enzymes was suppressed in HIV-infected subjects compared to HIV-negative controls. CYP3A4 expression was lower in HIV+ naive subjects than in the healthy controls (P < 0.001). HIV+ subjects receiving ART had significantly lower average expression of CYP3A5 (3.7-fold, P < 0.01), CYP2D6 (2.2-fold, P < 0.01), and UGT2B7 (1.9-fold, P < 0.05) than HIV-negative subjects (Fig. 1B). Among ABC transporters, the expression of ABCC2 (encoding MRP2) was significantly lower in all HIV+ ART-naive subjects (2.1-fold, P < 0.05), and decreased expression of additional MRPs (MRP3, -6, and -7) was observed in some of the subjects. ART-treated HIV+ subjects had significantly lower ABCC3 (encoding MRP3) expression than HIV-negative subjects (3.6-fold, P < 0.01). The expression of ABCB1 (Pgp) was highly variable among the six HIV+ subjects receiving ART, with three subjects showing decreased ABCB1 expression while two subjects had increased ABCB1 expression compared to the HIV-negative controls. Similarly, changes in the gene expression of SLC transporters were variable in HIV-infected subjects and were clearly altered compared to controls (Fig. 1B). The expression of orphan nuclear receptors, i.e., pregnane X receptor, involved in regulating drug transporters and metabolic enzyme transcription, was downregulated in HIV+ ART-treated subjects compared to controls. In contrast, expression of genes associated with the mucosal inflammatory response was elevated in HIV-infected subjects compared to controls. The expression of several genes, including those for interleukin-1α (IL-1α), IL-1β, IL-8, IL-1 receptor (IL-1R) type II, and mitogen-activated protein kinase 11 (MAPK-11) (i.e., p38-β), was upregulated in HIV+ ART-treated subjects. Expression of several other genes involved in inflammatory response such as those for MAPK-9 (i.e., JNK2) and nuclear factor κB (NF-κB) remained upregulated in both cohorts of HIV-infected subjects.
FIG 1.
DNA microarray analysis of mucosal gene expression in treated and untreated HIV-1 infection. (A) Gene expression changes in the gut biopsy specimens of HIV-negative participants (n = 5), HIV+ ART-naive participants (n = 7), and HIV+ participants on ART (n = 6). The patients were arranged based on when the microarray sample was analyzed (see Materials and Methods for details). (B) Differentially expressed genes with important pharmacological and physiological functions, such as genes for metabolic enzymes, drug efflux and uptake transporters (i.e., ABC and SLC drug transporters), nuclear receptors, and tight junctional proteins, and genes associated with the intestinal immune response and inflammation, were identified through hierarchical cluster analysis. For clarity, the sequence of patients was organized by patient identification number matching the arrangement that was used in panel A. Abbreviations: CYP, cytochrome P450 enzyme; UGT, UDP-glucuronyl transferase; Pgp, P-glycoprotein; MRP, multidrug resistance-associated protein; BCRP, breast cancer resistance protein; OATP, organic anion transporting polypeptide; OAT, organic anion transporter; OCT, organic cation transporter; CNT, concentrative nucleoside transporter; ENT, equilibrative nucleoside transporter; PXR, pregnane X receptor; CAR, constitutive androstane receptor; FXR, farnesoid X receptor; LXR, liver X receptor; PPAR, peroxisome proliferator-activated receptor; IL, interleukin; IFN, interferon; MCP, monocyte chemoattractant protein; COX, cyclooxygenase; MAPK, mitogen-activated protein kinase; NF, nuclear factor; TGF, transforming growth factor.
Effect of HIV-1 infection on expression of drug metabolic enzymes and transporters.
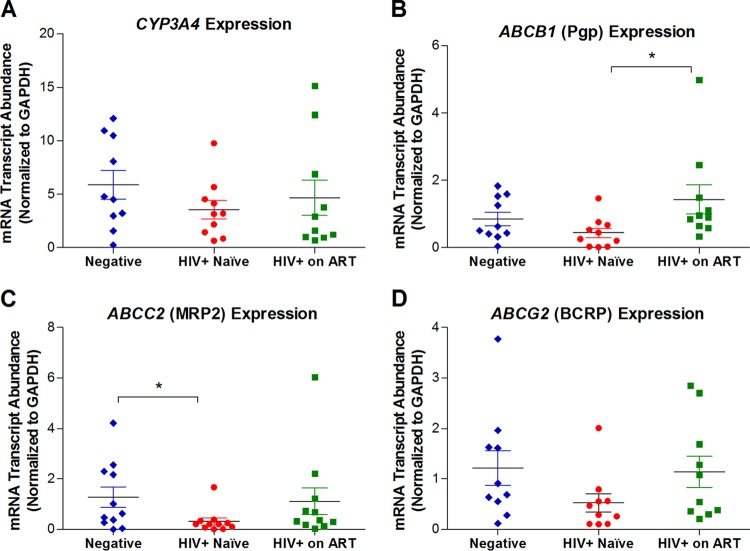
We investigated the impact of HIV infection on the mucosal expression of genes regulating the transport and metabolism of ARVs commonly used in HIV therapy. Gene and protein expression of selected drug transporters and metabolic enzymes (CYP3A4, ABCB1, ABCC2, and ABCG2) in intestinal tissue biopsy specimens from HIV-infected and uninfected subjects (Table 2) were examined by real-time qPCR and immunohistochemistry, respectively. The qPCR data (Fig. 2) showed that the gene expression of CYP3A4, ABCB1, ABCC2, and ABCG2 in the intestinal biopsy specimens was consistently decreased in HIV-infected subjects (Fig. 2). These results were in agreement with the findings for the gene expression changes based on the DNA microarray analysis. Long-term ART resulted in the restoration of expression of all of these genes. However, a certain degree of variability in the expression levels was observed among the six HIV+ subjects receiving ART. ART-treated subjects had increased ABCB1 expression compared to ARV-naive HIV+ subjects (3.2-fold, P < 0.05). The microarray data set did not include ABCG2 (i.e., BCRP), an apical efflux carrier that can transport NRTIs and hence has an important role in restricting drug tissue concentrations and transepithelial permeativity (16). ABCG2 mRNA levels were decreased in HIV-infected subjects compared to HIV-negative healthy individuals but showed a trend of increased expression in HIV+ subjects receiving ART (Fig. 2D).
FIG 2.
Real-time qPCR analysis of gene expression of selected intestinal metabolic enzymes and drug efflux transporters. Total RNA extracted from jejunal tissue biopsy specimen obtained from each subject was used for analysis of mRNA expression levels of each target gene using TaqMan gene expression assays specific for human CYP3A4 (A), ABCB1 (B), ABCC2 (C), or ABCG2 (D), obtained from Life Technologies Inc., Burlington, Ontario, Canada (assay Hs00604506_m1, Hs00184500_m1, Hs00166123_m1, or Hs01053790_m1, respectively). In each sample, the expression of the target gene was normalized to an internal control, GAPDH, by the ΔCT method. Statistically significant differences in mRNA expression between groups (negative versus HIV+ naïve, negative versus HIV+ on ART, and HIV+ naive versus HIV+ on ART) were determined using GraphPad Prism (version 5.01; Graph Pad Software, San Diego, CA) by applying the nonparametric two-tailed Mann-Whitney test with significance defined by a P value of <0.05.
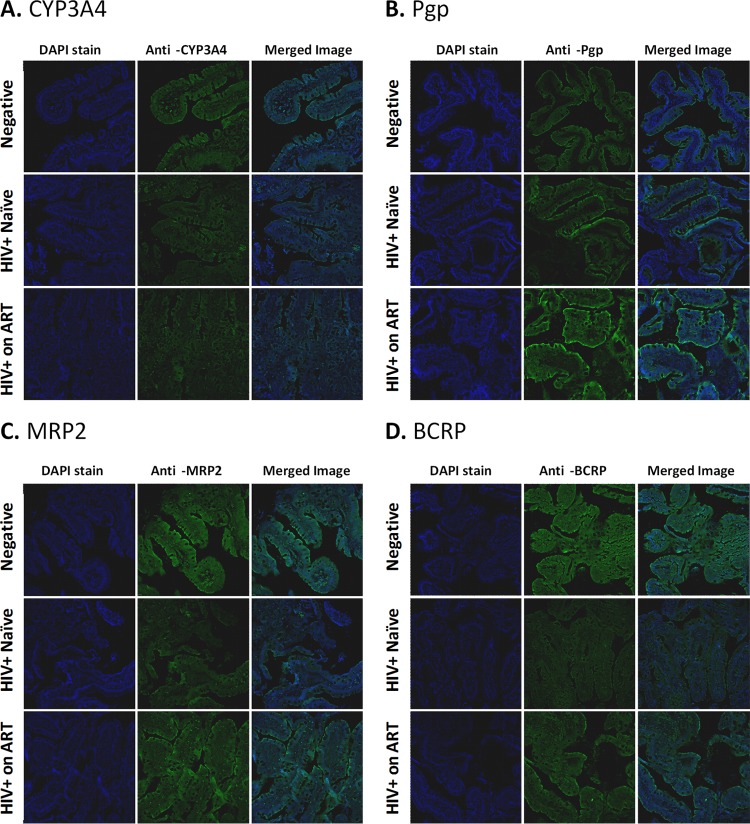
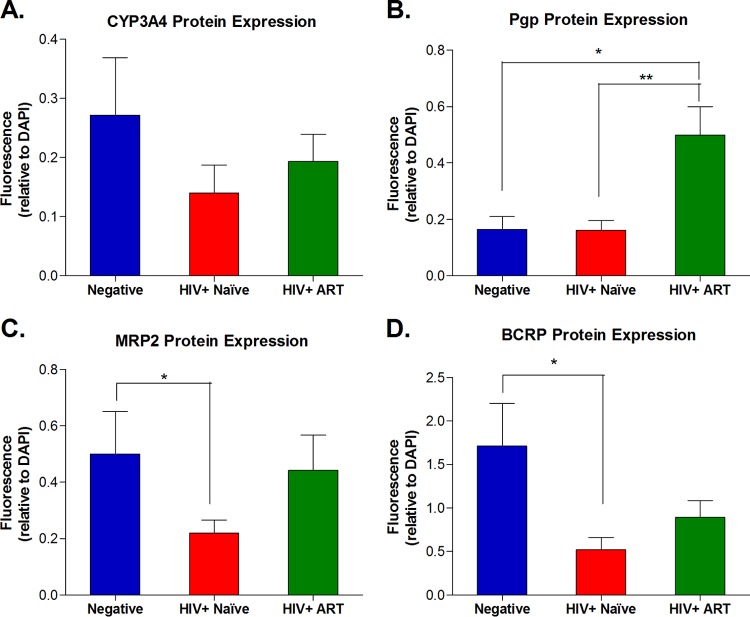
For the selected four genes involved in the pharmacokinetics of ARVs (i.e., CYP3A4, ABCB1, ABCC2, and ABCG2), we evaluated their protein expression (i.e., CYP3A4, Pgp, MRP2, and BCRP, respectively) in intestinal biopsy specimens by immunohistochemical analysis (Fig. 3 and 4). Overall, the changes in protein expression of these genes reflected the differences observed in the mRNA transcript levels. CYP3A4 expression showed a trend toward downregulation in HIV+ therapy-naive subjects compared to healthy controls (1.9-fold decrease), and this effect was partially reversed in ART-treated HIV+ subjects. Pgp expression was significantly upregulated in HIV+ subjects receiving ART compared to the HIV+ naive group (3.4-fold, P < 0.01) and was also higher than that in the healthy control group (3.3-fold, P < 0.05). Protein expression of both MRP2 and BCRP was significantly lower in HIV-infected ARV-naive subjects than in healthy controls (2.3- and 3.3-fold, respectively; P < 0.05). This downregulation appeared to be partially reversed in ART-treated subjects, with 2- and 1.7-fold-higher protein expression of MRP2 and BCRP, respectively, than in ARV-naive subjects.
FIG 3.
Immunohistochemistry analysis of protein expression of selected intestinal metabolic enzymes and drug efflux transporters. To detect protein expression and localization, paraffin-embedded jejunal tissue biopsy specimens from each subject, fixed in 4% paraformaldehyde and mounted onto glass slides, were immunostained with anti-CYP3A4 mouse polyclonal antibody from Sigma-Aldrich (Oakville, Ontario, Canada) at a 1:600 dilution (A), anti-Pgp mouse monoclonal D-11 antibody from Santa Cruz Biotechnology, Inc. (Dallas, TX) at a 1:100 dilution (B), anti-MRP2 mouse monoclonal M2III-6 antibody from Kamiya Biomedical Company (Seattle, WA) at a 1:250 dilution (C), or anti-BCRP rat monoclonal BXP-21 antibody from Abcam Inc. (Toronto, Ontario, Canada) at a 1:100 dilution (D). After washing, each tissue slice was immunostained with the corresponding Alexa-488-conjugated secondary antibodies, i.e., donkey anti-rat IgG for BCRP or donkey anti-mouse IgG for all other proteins (Life Technologies Inc., Burlington, Ontario, Canada). Standard DAPI staining was used to identify cell nuclei. For each gene of interest, the entire set of tissue slices was immunostained in a single experiment with all steps performed simultaneously, and images were obtained at constant exposure, zoom, and background settings using a Zeiss LSM700 confocal microscope.
FIG 4.
Group comparison of protein expression levels estimated from relative fluorescence intensities of immunostained jejunal tissue slices. For each protein of interest, all fluorescent images obtained from tissue slices from each subject were analyzed using the Image Pro Premier software (Media Cybernetic, Rockville, MD). The fluorescence intensity (integrated optical density [OD], lumens/pixel2) for the protein of interest, i.e., CYP3A4 (A), Pgp (B), MRP2 (C), or BCRP (D), was normalized to the intensity of nuclear staining (i.e., the integrated OD measured for DAPI) within the same area. This analysis was performed in triplicate on three different areas within each tissue slice obtained from each study subject. The average normalized intensity ratio (gene of interest/DAPI) for each subject was used to estimate the mean ± standard error of the mean (SEM) for the study group. Statistically significant differences in protein expression between groups (negative versus HIV+ naïve, negative versus HIV+ on ART, and HIV+ naive versus HIV+ on ART) were determined using GraphPad Prism software, version 5.01 (Graph Pad Software, San Diego, CA) by applying the nonparametric two-tailed Mann-Whitney test with significance defined by a P value of <0.05.
DISCUSSION
In this study, we demonstrated differential expression of metabolic enzymes and drug transporters between healthy individuals and HIV-1-infected subjects. Chronically HIV-1-infected subjects without prior exposure to ART demonstrated a trend toward lower expression of CYP3A4 enzyme, which metabolizes all PIs and NNRTIs and many other drugs. Furthermore, drug efflux transporters that are localized to the brush border membrane, such as MRP2, which mediates efflux of PIs and NRTIs, and BCRP, which is capable of transporting many NRTIs and rilpivirine, were also downregulated in HIV-infected ARV-naive subjects (16). Given the localization of MRP2 and BCRP at the apical brush border membrane of enterocytes, downregulation in their protein expression is likely to lead to increased drug accumulation within enterocytes and increased drug permeability across the intestinal epithelium in the absorptive direction (33). These findings are in agreement with previously reported downregulation of drug efflux transporters in the sigmoid colon epithelia of ART-naive HIV-infected men, who had significantly lower Pgp and MRP2 protein expression than healthy individuals (27). Due to limited sample availability, we were unable to compare the concentrations of ARVs achieved in the intestinal tissue or blood plasma of HIV-infected and uninfected subjects. However, our data on differential protein expression of these enzymes/transporters caution against the use of pharmacokinetic data obtained in healthy volunteers to guide therapy in chronically HIV-1-infected ART-naive subjects due to the potential for treatment-associated intestinal tissue toxicity. Further studies are needed to evaluate the differences in tissue concentrations and plasma pharmacokinetics of ARVs between healthy volunteers and HIV+ patients and to determine needed modifications to ART dosing regimens to help optimize ART efficacy and safety in HIV+ patients.
In contrast to the observed downregulation of transporters and metabolic enzymes in ART-naive subjects, in ART-treated subjects, we observed a trend toward an increase in expression of several transporters and metabolic enzymes. In particular, Pgp expression was significantly higher in the ART-treated group than in ARV-naive subjects (3.4-fold, P < 0.01) or controls (3.3-fold, P < 0.05). These findings are in agreement with our previous study demonstrating 1.9- and 1.5-fold-higher Pgp and MRP2 protein expression, respectively, in the sigmoid colon biopsy specimens from HIV+ subjects receiving ART than in the HIV+ treatment-naive group and/or the control group (27). Pgp is expressed at the apical membrane of enterocytes, where it mediates active efflux of drug substrates, including all PIs, some NRTIs (e.g., abacavir and tenofovir disoproxil fumarate [TDF]), and other ARVs (e.g., raltegravir, elvitegravir, and maraviroc) back into the intestinal lumen (16, 33). Hence, increased Pgp expression can lead to increased clearance of these ARVs from intestinal tissue in ART-treated HIV+ patients and will likely decrease their intracellular accumulation in enterocytes and permeativity across the intestinal epithelium. In addition, we observed that some ART-treated subjects had higher transcriptional expression of several drug uptake transporters (OCT1, OAT1, OATP1A2, ENT1, and ENT2), which could potentially compensate for the adverse effect of increased drug efflux and/or metabolism of common drug substrates such as PIs (substrates of organic anion transporting polypeptides [OATPs]), NRTIs (substrates of organic anion transporters [OATs], organic cation transporters [OCTs], concentrative nucleoside transporters [CNTs], and equilibrative nucleoside transporters [ENTs]), raltegravir (substrate of OAT1 and peptide transporter 1), and rilpivirine (substrate of OCT1) (16, 34–36). Due to limited tissue availability, in this study we did not evaluate protein expression of these drug uptake transporters. Interestingly, ritonavir is a known substrate of Pgp, so its accumulation within enterocytes and intestinal permeativity may be adversely impacted by increased Pgp-mediated efflux. Since most PI-based ART regimens include ritonavir as a boosting agent to inhibit CYP3A4-mediated metabolism of other PIs, its inhibitory effect on CYP3A4 may be diminished in patients with induced Pgp expression, leading to higher metabolism and lower concentrations of other PIs and ARVs. Indeed, some ARVs (e.g., atazanavir and efavirenz) are reported to have lower plasma concentrations and bioavailability in HIV-infected subjects than in healthy volunteers (12, 13). Other factors may also contribute to the observed pharmacokinetic differences, such as race, ethnicity, gender, age, diet, altered composition of the gastrointestinal lumen (e.g., pH differences), modified barrier integrity (e.g., tight junctional proteins), and/or interplay between intestinal drug uptake, efflux, and metabolism of ARVs (12, 13). Furthermore, expression of drug transporters and enzymes may change over time, leading to progressive changes in tissue and plasma levels of ARVs in long-term-treated patients and resulting in late failure of a previously suppressive regimen. Additional studies investigating differences in ARV pharmacokinetics between HIV-1-infected ARV-naive subjects and well-matched healthy volunteers using a parallel-group design are needed to confirm the reported differences, further explore the mechanisms behind these changes, and determine appropriate dosing of ARVs in HIV-infected patients at different stages of treatment to help optimize the efficacy and safety of ART.
Microarray analysis performed on intestinal biopsy samples demonstrated that other genes with critical pharmacological functions, such as genes for nuclear receptors, inflammatory cytokines, and other mediators of the cellular inflammatory response, are also differentially expressed in HIV-1-infected compared to HIV-negative subjects. Our group and others have previously demonstrated that long-term ARV treatment can upregulate Pgp expression at the blood-brain barrier by directly interacting with orphan nuclear receptors (e.g., pregnane X receptor and constitutive androstane receptor), transcription factors that when activated can upregulate the expression of metabolic enzymes and drug transporters (28, 37, 38). In particular, atazanavir, efavirenz, ritonavir, and other PIs were found to activate human pregnane X receptor, while abacavir, efavirenz, and nevirapine were found to activate human constitutive androstane receptor (28). Since all HIV+ ART-treated subjects enrolled in this study were receiving one or more of these drugs in combination with other ARVs, we propose that the observed upregulation of Pgp expression could be due to the interactions of these ARVs with nuclear receptors. In addition, several previous studies have reported regulation of drug transporters by HIV-1-associated pathogenesis and inflammation at other blood-tissue barriers and in the brain parenchyma. In cultured rat and human fetal astrocytes, HIV-1 envelope protein gp120 was demonstrated to trigger an inflammatory response via secretion of cytokines (IL-1β, IL-6, and tumor necrosis factor alpha [TNF-α]) which were found to regulate the expression of Pgp and MRP1 through NF-κB and JNK signaling pathways (22, 24, 39). Pgp expression was upregulated by IL-1β and TNF-α but profoundly downregulated by IL-6, with an overall decrease in expression with gp120 treatment or HIV-1 isolates exposure (24, 39). In contrast, MRP1 expression was upregulated by TNF-α and unaltered by IL-1β or IL-6, with an overall increase in expression following gp120 treatment (22). These effects were further confirmed in vivo. For example, intracerebral administration of gp120 to rodents was also shown to trigger the release of IL-1β, IL-6, TNF-α, and inducible nitric oxide as well as the activation of ERK1/2 and JNK pathways in different brain regions (40). Involvement of similar signaling pathways was also reported for the gp120-mediated inflammatory response in cardiac myocytes (41, 42). Our study provides initial evidence that these nuclear receptors and inflammatory pathways may be involved in differential regulation of intestinal expression of drug transporters and metabolic enzymes in the context of HIV infection and ART; however, further investigation is needed.
Persistent HIV replication in the GALT of HIV-infected subjects receiving long-term suppressive ART, despite undetectable plasma viral loads, continues to be one of the primary obstacles to HIV eradication and may be a source of persistent immune activation and inflammation that could contribute to HIV progression and comorbidity (2–6). Many studies have documented that the gut HIV reservoir may be the source for new HIV infection in circulating immune cells, leading to viral rebound and treatment failure in long-term-treated subjects (43–45). Although several factors may contribute to the formation of the viral reservoir in the GALT, subtherapeutic concentrations of ARVs in the intestinal tissue and the potential for emergence of drug resistance may play a role in the observed local HIV replication. Indeed, previous studies have demonstrated that ARVs can have low and often subtherapeutic concentrations in lymphocytes isolated from the ileum and rectum-associated lymphoid tissue even in subjects receiving long-term ART who have undetectable plasma viral loads and that these low drug concentrations are associated with high viral counts detected in these tissues (10, 11, 43). Although tissue drug concentrations in the upper intestinal tract have not been investigated to date, formation of a viral reservoir is observed in GALT tissue isolated from the upper jejunum of HIV-infected subjects (6, 31). Similarly to intestinal enterocytes, gut-associated lymphocytes also express several drug efflux (e.g., Pgp, BCRP, and MRP1, -2, -4, and -5) and uptake (e.g., OATP1A2, OATP3A1, and ENT1 and -2) transporters, which may be differentially expressed in HIV-1-infected and uninfected individuals and could affect the intracellular levels of ARVs achieved in this cellular compartment (16, 46, 47).
Several limitations are associated with the design of this study, such as a small number of study participants, the range of ARVs used, the variability in ART duration, and other confounding factors that may have impacted the expression of these enzymes and transporters, such as concurrent unreported use of natural compounds, illicit drugs, and/or other prescribed and nonprescribed medications. We also did not control for other potential confounding factors such as age, gender, and ethnicity. A larger study could be designed to account for some of these confounders and compare subjects with more versus less advanced HIV or those with good versus poor CD4+ T cell recovery.
In summary, this study demonstrated that the expression of drug efflux transporters and metabolic enzymes can be significantly altered by both HIV-1 infection and ART. The data imply a high complexity of regulation of drug transporters and metabolic enzymes in the context of HIV-1 infection and its pharmacotherapy and caution against the use of pharmacokinetic studies in healthy volunteers to guide therapy or drug interaction issues in those with HIV.
ACKNOWLEDGMENTS
This study was supported by an operating grant from the Canadian Foundation for AIDS Research (grant 20023), awarded to R. Bendayan, and by grants from the National Institutes of Health (NIH) R01 (DK61297 and AI43274), awarded to S. Dandekar. R. Bendayan and S. Walmsley are recipients of Ontario HIV Treatment Network (OHTN) Career Scientist awards. S. Sankaran is supported by a Building Interdisciplinary Research Careers in Women's Health award (K12 HD051958) funded by the National Institute of Child Health and Human Development (NICHD), Office of Research on Women's Health (ORWH), and the National Institute on Aging (NIA). O. Kis was supported by Ph.D. studentships from the OHTN, Ministry of Health of Ontario, and the Canadian Institutes of Health Research (CIHR) Frederick Banting and Charles Best Canada Graduate Scholarship.
O. Kis performed qPCR analysis and immunohistochemistry imaging experiments evaluating gene and protein expression of the drug transporters and metabolic enzymes in gut tissue biopsy specimens, statistical data analysis, and preparation of figures and the first draft of the manuscript. The isolation of gut tissue biopsy specimens, analysis of HIV load and CD4+ cell counts, assessment of other patient characteristics, and DNA microarray analysis were performed by S. Sankaran-Walters in the laboratory of S. Dandekar. All authors contributed to data analysis, preparation of figures, and statistical analysis. R. Bendayan provided expertise on the experimental design and preparation of the manuscript for submission. S. Dandekar provided guidance and supervision for the experimental design and analysis of the gut biopsy specimen collection and immunologic, molecular, and virologic analyses and contributed to the preparation of the manuscript.
REFERENCES
- 1.Coodley GO, Loveless MO, Merrill TM. 1994. The HIV wasting syndrome: a review. J Acquir Immune DeficSyndr 7:681–694. [PubMed] [Google Scholar]
- 2.McGowan I, Elliott J, Fuerst M, Taing P, Boscardin J, Poles M, Anton P. 2004. Increased HIV-1 mucosal replication is associated with generalized mucosal cytokine activation. J Acquir Immune Defic Syndr 37:1228–1236. doi: 10.1097/01.qai.0000131846.12453.29. [DOI] [PubMed] [Google Scholar]
- 3.Anton PA, Mitsuyasu RT, Deeks SG, Scadden DT, Wagner B, Huang C, Macken C, Richman DD, Christopherson C, Borellini F, Lazar R, Hege KM. 2003. Multiple measures of HIV burden in blood and tissue are correlated with each other but not with clinical parameters in aviremic subjects. AIDS 17:53–63. doi: 10.1097/00002030-200301030-00008. [DOI] [PubMed] [Google Scholar]
- 4.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol 77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guadalupe M, Sankaran S, George MD, Reay E, Verhoeven D, Shacklett BL, Flamm J, Wegelin J, Prindiville T, Dandekar S. 2006. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. J Virol 80:8236–8247. doi: 10.1128/JVI.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sankaran S, Guadalupe M, Reay E, George MD, Flamm J, Prindiville T, Dandekar S. 2005. Gut mucosal T cell responses and gene expression correlate with protection against disease in long-term HIV-1-infected nonprogressors. Proc Natl Acad Sci U S A 102:9860–9865. doi: 10.1073/pnas.0503463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Schacker T, Carlis J, Beilman G, Nguyen P, Haase AT. 2004. Functional genomic analysis of the response of HIV-1-infected lymphatic tissue to antiretroviral therapy. J Infect Dis 189:572–582. doi: 10.1086/381396. [DOI] [PubMed] [Google Scholar]
- 8.Sandler NG, Douek DC. 2012. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol 10:655–666. doi: 10.1038/nrmicro2848. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J. 2011. HIV/AIDS research. Tissue says blood is misleading, confusing HIV cure efforts. Science 334:1614. doi: 10.1126/science.334.6063.1614. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW. 2014. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 111:2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cory TJ, Schacker TW, Stevenson M, Fletcher CV. 2013. Overcoming pharmacologic sanctuaries. Curr Opin HIV AIDS 8:190–195. doi: 10.1097/COH.0b013e32835fc68a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukonzo JK, Nanzigu S, Rekic D, Waako P, Roshammar D, Ashton M, Ogwal-Okeng J, Gustafsson LL, Aklillu E. 2011. HIV/AIDS patients display lower relative bioavailability of efavirenz than healthy subjects. Clin Pharmacokinet 50:531–540. doi: 10.2165/11592660-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 13.Busti AJ, Hall RG, Margolis DM. 2004. Atazanavir for the treatment of human immunodeficiency virus infection. Pharmacotherapy 24:1732–1747. doi: 10.1592/phco.24.17.1732.52347. [DOI] [PubMed] [Google Scholar]
- 14.Oude Elferink RP, de Waart R. 2007. Transporters in the intestine limiting drug and toxin absorption. J Physiol Biochem 63:75–81. doi: 10.1007/BF03174087. [DOI] [PubMed] [Google Scholar]
- 15.Takano M, Yumoto R, Murakami T. 2006. Expression and function of efflux drug transporters in the intestine. Pharmacol Ther 109:137–161. doi: 10.1016/j.pharmthera.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Kis O, Robillard K, Chan GN, Bendayan R. 2010. The complexities of antiretroviral drug-drug interactions: role of ABC and SLC transporters. Trends Pharmacol Sci 31:22–35. doi: 10.1016/j.tips.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Wynn GH, Zapor MJ, Smith BH, Wortmann G, Oesterheld JR, Armstrong SC, Cozza KL. 2004. Antiretrovirals. I. Overview, history, and focus on protease inhibitors. Psychosomatics 45:262–270. [DOI] [PubMed] [Google Scholar]
- 18.Zapor MJ, Cozza KL, Wynn GH, Wortmann GW, Armstrong SC. 2004. Antiretrovirals. II. Focus on non-protease inhibitor antiretrovirals (NRTIs, NNRTIs, and fusion inhibitors). Psychosomatics 45:524–535. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Tompkins LM. 2008. CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9:598–610. doi: 10.2174/138920008785821710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sankatsing SU, Beijnen JH, Schinkel AH, Lange JM, Prins JM. 2004. P glycoprotein in human immunodeficiency virus type 1 infection and therapy. Antimicrob Agents Chemother 48:1073–1081. doi: 10.1128/AAC.48.4.1073-1081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cressman AM, Petrovic V, Piquette-Miller M. 2012. Inflammation-mediated changes in drug transporter expression/activity: implications for therapeutic drug response. Expert Rev Clin Pharmacol 5:69–89. doi: 10.1586/ecp.11.66. [DOI] [PubMed] [Google Scholar]
- 22.Ronaldson PT, Ashraf T, Bendayan R. 2010. Regulation of multidrug resistance protein 1 by tumor necrosis factor alpha in cultured glial cells: involvement of nuclear factor-kappaB and c-Jun N-terminal kinase signaling pathways. Mol Pharmacol 77:644–659. doi: 10.1124/mol.109.059410. [DOI] [PubMed] [Google Scholar]
- 23.Robillard KR, Hoque MT, Bendayan R. 2014. Expression of ATP-binding cassette membrane transporters in a HIV-1 transgenic rat model. Biochem Biophys Res Commun 444:531–536. doi: 10.1016/j.bbrc.2014.01.092. [DOI] [PubMed] [Google Scholar]
- 24.Ashraf T, Ronaldson PT, Persidsky Y, Bendayan R. 2011. Regulation of P-glycoprotein by human immunodeficiency virus-1 in primary cultures of human fetal astrocytes. J Neurosci Res 89:1773–1782. doi: 10.1002/jnr.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy U, Bulot C, Honer zu BK, Mondal D. 2013. Specific increase in MDR1 mediated drug-efflux in human brain endothelial cells following co-exposure to HIV-1 and saquinavir. PLoS One 8:e75374. doi: 10.1371/journal.pone.0075374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zembruski NC, Buchel G, Jodicke L, Herzog M, Haefeli WE, Weiss J. 2011. Potential of novel antiretrovirals to modulate expression and function of drug transporters in vitro. J Antimicrob Chemother 66:802–812. doi: 10.1093/jac/dkq501. [DOI] [PubMed] [Google Scholar]
- 27.De Rosa MF, Robillard KR, Kim CJ, Hoque MT, Kandel G, Kovacs C, Kaul R, Bendayan R. 2013. Expression of membrane drug efflux transporters in the sigmoid colon of HIV-infected and uninfected men. J Clin Pharmacol 53:934–945. doi: 10.1002/jcph.132. [DOI] [PubMed] [Google Scholar]
- 28.Chan GN, Patel R, Cummins CL, Bendayan R. 2013. Induction of P-glycoprotein by antiretroviral drugs in human brain microvessel endothelial cells. Antimicrob Agents Chemother 57:4481–4488. doi: 10.1128/AAC.00486-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sankaran-Walters S, Macal M, Grishina I, Nagy L, Goulart L, Coolidge K, Li J, Fenton A, Williams T, Miller MK, Flamm J, Prindiville T, George M, Dandekar S. 2013. Sex differences matter in the gut: effect on mucosal immune activation and inflammation. Biol Sex Differ 4:10. doi: 10.1186/2042-6410-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sankaran S, George MD, Reay E, Guadalupe M, Flamm J, Prindiville T, Dandekar S. 2008. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J Virol 82:538–545. doi: 10.1128/JVI.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macal M, Sankaran S, Chun TW, Reay E, Flamm J, Prindiville TJ, Dandekar S. 2008. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol 1:475–488. doi: 10.1038/mi.2008.35. [DOI] [PubMed] [Google Scholar]
- 32.Dewar RL, Highbarger HC, Sarmiento MD, Todd JA, Vasudevachari MB, Davey RT Jr, Kovacs JA, Salzman NP, Lane HC, Urdea MS. 1994. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J Infect Dis 170:1172–1179. doi: 10.1093/infdis/170.5.1172. [DOI] [PubMed] [Google Scholar]
- 33.Konig J, Muller F, Fromm MF. 2013. Transporters and drug-drug interactions: important determinants of drug disposition and effects. Pharmacol Rev 65:944–966. doi: 10.1124/pr.113.007518. [DOI] [PubMed] [Google Scholar]
- 34.Estudante M, Morais JG, Soveral G, Benet LZ. 2013. Intestinal drug transporters: an overview. Adv Drug Deliv Rev 65:1340–1356. doi: 10.1016/j.addr.2012.09.042. [DOI] [PubMed] [Google Scholar]
- 35.Weiss J, Haefeli WE. 2013. Potential of the novel antiretroviral drug rilpivirine to modulate the expression and function of drug transporters and drug-metabolising enzymes in vitro. Int J Antimicrob Agents 41:484–487. doi: 10.1016/j.ijantimicag.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Moss DM, Kwan WS, Liptrott NJ, Smith DL, Siccardi M, Khoo SH, Back DJ, Owen A. 2011. Raltegravir is a substrate for SLC22A6: a putative mechanism for the interaction between raltegravir and tenofovir. Antimicrob Agents Chemother 55:879–887. doi: 10.1128/AAC.00623-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan GN, Hoque MT, Cummins CL, Bendayan R. 2011. Regulation of P-glycoprotein by orphan nuclear receptors in human brain microvessel endothelial cells. J Neurochem 118:163–175. doi: 10.1111/j.1471-4159.2011.07288.x. [DOI] [PubMed] [Google Scholar]
- 38.Zastre JA, Chan GN, Ronaldson PT, Ramaswamy M, Couraud PO, Romero IA, Weksler B, Bendayan M, Bendayan R. 2009. Up-regulation of P-glycoprotein by HIV protease inhibitors in a human brain microvessel endothelial cell line. J Neurosci Res 87:1023–1036. doi: 10.1002/jnr.21898. [DOI] [PubMed] [Google Scholar]
- 39.Ronaldson PT, Bendayan R. 2006. HIV-1 viral envelope glycoprotein gp120 triggers an inflammatory response in cultured rat astrocytes and regulates the functional expression of P-glycoprotein. Mol Pharmacol 70:1087–1098. doi: 10.1124/mol.106.025973. [DOI] [PubMed] [Google Scholar]
- 40.Ashraf T, Jiang W, Hoque MT, Henderson J, Wu C, Bendayan R. 2014. Role of anti-inflammatory compounds in human immunodeficiency virus-1 glycoprotein120-mediated brain inflammation. J Neuroinflammation 11:91. doi: 10.1186/1742-2094-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kan H, Xie Z, Finkel MS. 2000. HIV gp120 enhances NO production by cardiac myocytes through p38 MAP kinase-mediated NF-kappaB activation. Am J Physiol Heart Circ Physiol 279:H3138–H3143. [DOI] [PubMed] [Google Scholar]
- 42.Kan H, Xie Z, Finkel MS. 2004. p38 MAP kinase-mediated negative inotropic effect of HIV gp120 on cardiac myocytes. Am J Physiol Cell Physiol 286:C1–C7. [DOI] [PubMed] [Google Scholar]
- 43.Chun TW, Nickle DC, Justement JS, Meyers JH, Roby G, Hallahan CW, Kottilil S, Moir S, Mican JM, Mullins JI, Ward DJ, Kovacs JA, Mannon PJ, Fauci AS. 2008. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis 197:714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 44.Imamichi H, Degray G, Dewar RL, Mannon P, Yao M, Chairez C, Sereti I, Kovacs JA. 2011. Lack of compartmentalization of HIV-1 quasispecies between the gut and peripheral blood compartments. J Infect Dis 204:309–314. doi: 10.1093/infdis/jir259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zalar A, Figueroa MI, Ruibal-Ares B, Bare P, Cahn P, de Bracco MM, Belmonte L. 2010. Macrophage HIV-1 infection in duodenal tissue of patients on long term HAART. Antiviral Res 87:269–271. doi: 10.1016/j.antiviral.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Janneh O, Owen A, Chandler B, Hartkoorn RC, Hart CA, Bray PG, Ward SA, Back DJ, Khoo SH. 2005. Modulation of the intracellular accumulation of saquinavir in peripheral blood mononuclear cells by inhibitors of MRP1, MRP2, P-gp and BCRP. AIDS 19:2097–2102. doi: 10.1097/01.aids.0000194793.36175.40. [DOI] [PubMed] [Google Scholar]
- 47.Janneh O, Hartkoorn RC, Jones E, Owen A, Ward SA, Davey R, Back DJ, Khoo SH. 2008. Cultured CD4T cells and primary human lymphocytes express hOATPs: intracellular accumulation of saquinavir and lopinavir. Br J Pharmacol 155:875–883. doi: 10.1038/bjp.2008.320. [DOI] [PMC free article] [PubMed] [Google Scholar]