LETTER
The OXA-23, OXA24/40, and OXA-58 class D carbapenemases are typically found in Acinetobacter species; OXA-48-like enzymes are generally found in Enterobacteriaceae (1). There have been a few reports of Acinetobacter-type OXA genes in Enterobacteriaceae (2–4), but the real prevalence of such isolates may be underreported, since most surveillance efforts go into identifying the most common “big five” carbapenemases KPC, NDM, OXA-48, VIM, and IMP (5) among Enterobacteriaceae. Indeed, the development of fast commercial molecular diagnostic kits further feeds this bias.
Here we report the unexpected finding of a blaOXA-23-positive isolate during routine surveillance. A Proteus mirabilis isolate was collected in September 2014 from the urine of a 74-year-old female patient with inoperable pancreatic cancer and with an unknown travel history. The patient died 20 days later; the role of the infection in her death was uncertain. The unusual phenotype of the isolate in our disk testing panel caught the eye and, combined with a borderline UV-spectrophotometric imipenem hydrolysis assay result, prompted more-detailed tests (as previously described [6], with additional MIC testing using a custom Sensititre broth microdilution panel [Thermo Fisher Scientific, Vantaa, Finland]). The isolate was susceptible to all beta-lactams, but the zone around the carbapenem disks was outside the EUCAST wild-type cutoff, although still on the susceptible side even of the EUCAST screening breakpoints (Table 1). The isolate had no extended-spectrum β-lactamase (ESBL) or transferable AmpC β-lactamases that could have caused this (PCR negative for CTX-M, TEM, SHV, CIT, DHA, MOX, FOX, ACC, and EBC), and wild-type susceptibility to third-generation cephalosporins is not typically seen in strains with porin mutations or upregulated efflux. A negative result from an imipenem-plus-EDTA inhibitor test (Rosco, Taastrup, Denmark) showed that no metallo-β-lactamase was involved. The isolate was PCR negative for KPC, NDM, OXA-48, VIM, IMP, and GES. PCR screening for OXA-23, OXA-24/40, OXA-51, and OXA-58 gave a positive band for OXA-23. Sanger sequencing confirmed the result. The isolate was later subjected to whole-genome sequencing, using an Illumina MiSeq sequencer (Illumina Inc.) with an Illumina Nextera XT DNA sample preparation kit and a Nextera XT index kit with 24 indices for library preparation and MiSeq reagent kit V2 (300 cycles) utilizing 150-bp paired-end sequencing. The reads were compiled using Velvet assembler version 1.1.04 (7), which is included in Ridom SeqSphere+ software (Ridom SeqSphere+ version 2.4.0; Ridom GmbH, Münster, Germany). Species identification and the presence of the OXA-23 were confirmed with the corresponding SpeciesFinder and ResFinder databases on the Center for Genomic Epidemiology (CGE) server (https://cge.cbs.dtu.dk/services/). In addition to blaOXA-23, ResFinder found 9 other resistance genes [aadA1, aac(3)-IIa, strB, strA, aph(3′)-Ic, floR, sul2, tet(J), and dfrA1]. The CGE Plasmid Finder found no plasmids; this result was not further confirmed.
TABLE 1.
Susceptibility test results for the OXA-23-positive P. mirabilis ESBL4969 isolate
| Compound | Disk assay |
MICa |
|||
|---|---|---|---|---|---|
| Content (μg) | Diam (mm) | Interpretation | mg/liter | Interpretation | |
| Meropenem | 10 | 26 | S, close to EUCAST screening breakpointb | 2 | S, outside ECOFFc |
| Ertapenem | 10 | 25 | S, close to breakpoint | ||
| Imipenem | 10 | 26 | No MBLd | ||
| Imipenem/EDTA | 10/750 | 26 | No MBL | ||
| Cefoxitin | 30 | 25 | S | ||
| Cefepime | 30 | 30 | S | ≤1 | S |
| Cefalexin | 30 | 19 | S | ||
| Ceftazidime | 30 (diagnostic) | 30 | S, no ESBL | ≤1 | S |
| Ceftazidime-clavulanic acid | 30/10 | 30 | S, no ESBL | ||
| Cefotaxime | 30 (diagnostic) | 34 | S, no ESBL | ||
| Cefotaxime-clavulanic acid | 30/10 | 35 | S, no ESBL | ||
| Temocillin | 30 | 22 | No OXA-48-like enzymeb | ||
| Piperacillin-tazobactam | 30/6 | 21 | S, close to ECOFF | 8 | S, outside ECOFF |
| Ciprofloxacin | ≤0.12 | S | |||
| Fosfomycin | ≤4 | S | |||
| Gentamicin | >16 | R | |||
| Amikacin | 8 | S | |||
| Tobramycin | 16 | R | |||
| Trimethoprim-sulfamethoxazole | >8 | R | |||
Broth microdilution, Sensititre (ThermoFisher).
EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. V 1.0, 2013. http://www.eucast.org/resistance_mechanisms/. S, sensitive.
ECOFF, epidemiological cutoff, EUCAST. http://www.eucast.org/mic_distributions_and_ecoffs/.
MBL, metallo-β-lactamase.
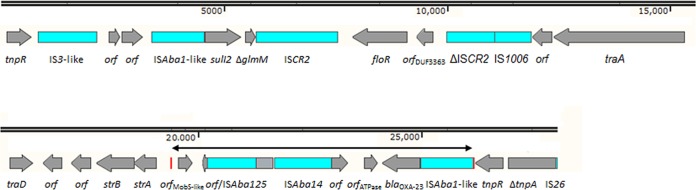
Using primers designed in house, a number of PCR assays and Sanger sequencing were used to confirm and extend the genetic environment of blaOXA-23 and to fill in gaps between the involved contigs (8) (see Table S1 in the supplemental material). The blaOXA-23-containing region was analyzed using Geneious 7.1 software (Biomatters Ltd., Auckland, New Zealand) and annotated with Prokka version 1.4.0 (9), which is included in Galaxy/CRS4 (Orione) (10). The OXA-23 gene was located downstream of an ISAba1-like insertion sequence element, resembling the structure of transposon Tn2008, although not surrounded by a classic 9-bp target site duplication (Fig. 1). Instead, a flanking duplication of 22 bp (GATGAAGCGCGGAGGTGGCTCA) was detected, which could define the site of insertion in several potential backgrounds such as uncultured bacterium plasmid pHHV216, Pseudomonas aeruginosa plasmid pMRVIM0713, and Bordetella bronchiseptica plasmid R906 (GenBank accession numbers FJ012880, KP975076, and KF743818, respectively). The neighboring region also carried sul2, floR, strB, and strA and a variety of insertion elements.
FIG 1.
Genetic structure of the blaOXA-23 region. Genes are shown as gray-labeled arrows, with the arrowhead indicating the direction of transcription. Insertion sequence elements are shown as labeled blue boxes. The red vertical rectangles represent two 22-bp direct repeats, which might be the result of a target site duplication. GenBank accession number KU302354.
There is only one previous report of OXA-23 in P. mirabilis, in isolates collected in the 1990s in France (2). Considering the transferable elements surrounding the gene in our isolate and that La et al. found OXA-23 in an Escherichia coli plasmid (3), as well as the phenotype of relative susceptibility of our isolate, OXA genes may well be spreading silently “under the radar.” It is doubtful whether active screening to find these genes in relatively susceptible strains would be cost-effective, but researchers in reference laboratories should at least keep this possibility in mind.
Nucleotide sequence accession number.
The sequence of the OXA-23-containing region has been deposited in GenBank with accession number KU302354.
Supplementary Material
ACKNOWLEDGMENTS
We express our thanks to Toni Huovinen, Tuula Rantasalo, Minna Lamppu, and Heli Laaksonen for expert technical assistance and to Ville Mäkelä, Turku Centre for Biotechnology, for help with the sequencing.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03119-15.
REFERENCES
- 1.Evans BA, Amyes SA. 2014. OXA β-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet R, Marchandin H, Chanal C, Sirot D, Labia R, De Champs C, Jumas-Bilak E, Sirot J. 2002. Chromosome-encoded class D β-lactamase OXA-23 in Proteus mirabilis. Antimicrob Agents Chemother 46:2004–2006. doi: 10.1128/AAC.46.6.2004-2006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La M-V, Jureen R, Lin RTP, Teo JWP. 2014. Unusual detection of an Acinetobacter class D carbapenemase gene, blaOXA-23, in a clinical Escherichia coli isolate. J Clin Microbiol 52:3822–3823. doi: 10.1128/JCM.01566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leski TA, Bangura U, Jimmy DH, Ansumana R, Lizewski SE, Li RW, Stenger DA, Taitt CR, Vora GJ. 2013. Identification of blaOXA-51-like, blaOXA-58, blaDIM-1, and blaVIM carbapenemase genes in hospital Enterobacteriaceae isolates from Sierra Leone. J Clin Microbiol 51:2435–2438. doi: 10.1128/JCM.00832-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Findlay J, Hopkins KL, Meunier D, Woodford N. 2015. Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J Antimicrob Chemother 70:1338–1342. doi: 10.1093/jac/dku571. [DOI] [PubMed] [Google Scholar]
- 6.Österblad M, Kirveskari J, Hakanen AJ, Tissari P, Vaara M, Jalava J. 2012. Carbapenemase-producing Enterobacteriaceae in Finland: the first years (2008–11). J Antimicrob Chemother 67:2860–2864. doi: 10.1093/jac/dks299. [DOI] [PubMed] [Google Scholar]
- 7.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nigro SJ, Farrugia DN, Paulsen IT, Hall RM. 2013. A novel family of genomic resistance islands, AbGRI2, contributing to aminoglycoside resistance in Acinetobacter baumannii isolates belonging to global clone 2. J Antimicrob Chemother 68:554–557. doi: 10.1093/jac/dks459. [DOI] [PubMed] [Google Scholar]
- 9.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 10.Cuccuru G, Orsini M, Pinna A, Sbardellati A, Soranzo N, Travaglione A, Uva P, Zanetti G, Fotia G. 2014. Orione, a web-based framework for NGS analysis in microbiology. Bioinformatics 30:1928–1929. doi: 10.1093/bioinformatics/btu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.