Abstract
To determine the distribution and relationship of antimicrobial resistance determinants among extended-spectrum-cephalosporin (ESC)-resistant or carbapenem-resistant Escherichia coli isolates from the aquatic environment in India, water samples were collected from rivers or sewage treatment plants in five Indian states. A total of 446 E. coli isolates were randomly obtained. Resistance to ESC and/or carbapenem was observed in 169 (37.9%) E. coli isolates, which were further analyzed. These isolates showed resistance to numerous antimicrobials; more than half of the isolates exhibited resistance to eight or more antimicrobials. The blaNDM gene was detected in 14/21 carbapenem-resistant E. coli isolates: blaNDM-1 in 2 isolates, blaNDM-5 in 7 isolates, and blaNDM-7 in 5 isolates. The blaCTX-M gene was detected in 112 isolates (66.3%): blaCTX-M-15 in 108 isolates and blaCTX-M-55 in 4 isolates. We extracted 49 plasmids from selected isolates, and their whole-genome sequences were determined. Fifty resistance genes were detected, and 11 different combinations of replicon types were observed among the 49 plasmids. The network analysis results suggested that the plasmids sharing replicon types tended to form a community, which is based on the predicted gene similarity among the plasmids. Four communities each containing from 4 to 17 plasmids were observed. Three of the four communities contained plasmids detected in different Indian states, suggesting that the interstate dissemination of ancestor plasmids has already occurred. Comparison of the DNA sequences of the blaNDM-positive plasmids detected in this study with known sequences of related plasmids suggested that various mutation events facilitated the evolution of the plasmids and that plasmids with similar genetic backgrounds have widely disseminated in India.
INTRODUCTION
The global spread of bacteria showing resistance to a broad spectrum of antimicrobials is universally recognized to be a serious public health concern (1). The Centers for Disease Control and Prevention reported that every year more than 2 million people are infected with antimicrobial-resistant (AMR) pathogens in the United States alone, of which 23,000 die (2). Among the AMR bacteria, extended-spectrum-cephalosporin (ESC)-resistant or carbapenem-resistant members of the family Enterobacteriaceae are recognized to be some of the most serious microbial threats globally because in most cases they also exhibit resistance to other classes of antimicrobials, such as aminoglycosides, fluoroquinolones, macrolides, phenicols, sulfonamides, tetracyclines (TETs), and trimethoprim, leaving few or no therapeutic options (1, 2).
The Indian subcontinent is one of the most important areas for the global risk management of ESC- or carbapenem-resistant Enterobacteriaceae. New Delhi metallo-β-lactamase (NDM) hydrolyzes all β-lactam antimicrobials except monobactam, and most of the NDM-positive isolates of the Enterobacteriaceae exhibit resistance to a broad spectrum of antimicrobials. NDM-1 was first reported in a Klebsiella pneumoniae strain isolated from a Swedish resident who traveled to New Delhi, India (3). Although NDM-1-positive Enterobacteriaceae strains have subsequently been isolated throughout the world, most of the patients have reported a connection with the Indian subcontinent or the Balkan countries (4). The patients had visited and/or were hospitalized there or were potentially linked to other patients who had been hospitalized there. Walsh et al. (5) suggested that the Indian environment is also an important source of infection by NDM-1-positive Enterobacteriaceae, but this is still controversial (6, 7).
Information on environmental contamination by AMR bacteria in India is scanty. Hospital wastewater is reported to be an important source of contamination by AMR Enterobacteriaceae (8, 9). AMR bacteria can also be detected from river water (10, 11). However, in these studies, water samples were collected in a limited area and the molecular characteristics of the antimicrobial resistance determinants were not fully elucidated. More detailed investigations are required to evaluate the importance of the environmental factors that contribute to community-acquired infection by AMR bacteria. The purpose of this study was to manifest the distribution of ESC- and/or carbapenem-resistant Escherichia coli in the Indian aquatic environment as well as sewage treatment plants (STPs), which act as an important anthropogenic source of AMR bacteria in the environment. Further, comparison of the DNA sequences of plasmids conferring AMR was performed to help obtain an understanding of the nationwide pattern of dissemination of AMR determinants.
MATERIALS AND METHODS
Collection of water samples and isolation of E. coli.
A total of 74 water samples were collected from rivers and STPs in the Indian states of Bihar, Goa, Karnataka, Tamil Nadu, and Telangana between February 2013 and May 2014. The water samples were appropriately diluted with sterilized phosphate-buffered saline and spread onto Chromocult coliform agar (Merck KGaA, Darmstadt, Germany) plates to randomly isolate violet colonies (which are positive for both β-galactosidase and β-glucuronidase, which are indicators of the presence of E. coli). Up to 10 violet colonies were obtained from each sample. To check for the production of oxidase and indole, a cytochrome oxidase test strip (Nissui Pharmaceutical, Tokyo, Japan) and the dimethylaminocinnamaldehyde indole reagent (Becton, Dickinson and Company, Sparks, MD) were used. Among the violet colonies, the oxidase-negative and indole-positive isolates were identified to be E. coli and were stored in Luria-Bertani broth (Becton, Dickinson and Company) with 25% glycerol at −80°C until further analyses.
Antimicrobial susceptibility testing.
A Kirby-Bauer disc diffusion test was performed using Mueller-Hinton agar plates (Becton, Dickinson and Company) according to the recommendations of the Clinical and Laboratory Standards Institute (12, 13). The following antimicrobials were tested: ampicillin (AMP; 10 μg), cefazolin (CFZ; 30 μg), cefoxitin (FOX; 30 μg), cefotaxime (CTX; 30 μg), imipenem (IMP; 10 μg), chloramphenicol (CHL; 30 μg), TET (30 μg), streptomycin (STR; 10 μg), kanamycin (KAN; 30 μg), sulfamethoxazole-trimethoprim (SXT; 23.75/1.25 μg), nalidixic acid (NAL; 30 μg), and ciprofloxacin (CIP; 5 μg). The MICs of AMP, CTX, NAL, CIP, and ofloxacin were determined by agar dilution methods according to the recommendations of the Clinical and Laboratory Standards Institute (13).
Genotyping of the β-lactamase gene by PCR and sequencing.
To determine the genotypes of the β-lactamase gene, PCR was conducted using the primers listed in Table S1 in the supplemental material. PCR amplification was performed using an iCycler apparatus (Bio-Rad Laboratories, Hercules, CA). TaKaRa Ex Taq DNA polymerase (TaKaRa Bio, Shiga, Japan) was used according to the manufacturer's instructions. To determine the whole sequences of the blaCTX-M and blaNDM genes, primer pair seq-CTX-F and seq-CTX-R and primer pair Pre-NDM A and Pre-NDM B, respectively, were used. The nucleotide sequences on both strands were determined using an Applied Biosystems 3130xl genetic analyzer with a BigDye Terminator cycle sequencing kit (version 3.1; Applied Biosystems, Foster City, CA, USA). The sequences were assembled using the Sequencher program (version 4; Hitachi Solutions, Kanagawa, Japan), and DNA alignments and deduced amino acid sequences were examined using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) (14).
Whole-genome sequencing of plasmids and data analysis.
To obtain the draft genome sequence, pulsed-field gel electrophoresis (PFGE) and next-generation sequencing (NGS) were performed. Briefly, plasmid DNA was purified from S1 nuclease-digested genomic DNA that had been separated by PFGE, as previously described (15), and the bands were visualized with SYBR Safe gel stain (Life Technologies Japan, Tokyo, Japan) under a blue-light transilluminator, followed by purification using a ZR-96 Zymoclean gel DNA recovery kit (Zymo Research, Irvine, CA, USA). A DNA sequencing library (insert size, 750 to 1,000 bp) was prepared using a Nextera XT DNA sample preparation kit (Illumina, Inc., San Diego, CA) for sequencing on an Illumina MiSeq sequencer (Illumina) according to the manufacturer's instructions. De novo assembly was performed with the A5-miseq pipeline (16), followed by annotation with the Prodigal program (version 2.60) (17) and a search of the NCBI nucleotide database for homologous sequences by use of the BLASTP program (14). A BLAST Atlases view was generated using a search for homologous sequences by use of the BLASTN program and the GView program (18). A search for the replicon type of the query contigs was performed by use of a search for sequences homologous to the amplicon sequences generated by PCR-based replicon typing (PBRT) and by use of an E value of <1E−10, a cover ratio of ≥90%, and the BLASTN program (19).
Plasmidome network analysis.
These related plasmids were used to perform a network analysis similar to a previously described analysis (20). Briefly, the putative proteins carried by these plasmids were clustered using the UCLUST program (version 6.0.307) and the following parameters after sorting by sequence length, following the instructions accompanying the software: −cluster_smallmem; −id, 1.0; −minsl, 0.9; −minqt, 0.9; −maxqt, 1.1; −query_cov, 0.9; and −target_cov, 0.9. These parameters were chosen to cluster genes that had 100% amino acid sequence identity with at least 90% coverage and a less than 10% length difference. Plasmids sharing at least two homologous genes were connected as a network. Subsequently, a community was detected using the multilevel community method in the igraph library in R and default parameter settings. The network graph was drawn with the Cytoscape program (version 3.2.0) (43).
Nucleotide sequence accession numbers.
The nucleotide sequences of the β-lactamase genes and plasmids were deposited in the DNA Data Bank of Japan under accession numbers LC095449 to LC095574 for β-lactamase genes, AP014876 to AP014877 for the complete nucleotide sequences of two plasmids, and LC056077 to LC056712 and LC069379 to LC069386 for the nucleotide sequences of 644 contigs detected in 47 plasmids.
RESULTS
Selection of cefotaxime- or imipenem-resistant E. coli isolates.
A total of 446 E. coli isolates were obtained from the 74 water samples. Among them, 168 isolates detected in 49 water samples showed resistance to CTX. Twenty of the 168 isolates also exhibited resistance to IMP. One isolate that showed intermediate resistance to CTX and that was detected in a different water sample also exhibited resistance to IMP. The diameter of the inhibition zone of this isolate resistant to IMP was 17 mm. We further examined these 169 CTX- and/or IMP-resistant isolates (37.9%) detected in a total of 50 water samples (67.6%) in this study. These isolates originated from four states, including Bihar, Karnataka, Tamil Nadu, and Telangana, as shown in Table S2 in the supplemental material. We could not detect CTX- and/or IMP-resistant isolates from water samples collected in Goa.
Antimicrobial susceptibility.
The susceptibility data for a few isolates collected from Karnataka have been previously published (8). In the current study, additional isolates from other parts of India were included to understand the nationwide distribution pattern of antimicrobial resistance. Figure 1A shows the prevalence of resistance to the 12 antimicrobials tested among the selected 169 isolates. The prevalence of resistance to FOX, TET, STR, KAN, SXT, NAL, and CIP was 50.0% to 81.1%. These isolates exhibited resistance to more than two antimicrobials, and 58.0% of the isolates showed resistance to eight or more antimicrobials. The most predominant number of antimicrobials to which the isolates showed resistance was 10 (Fig. 1B). The MIC90s of AMP, CTX, and NAL were ≥512 μg/ml, whereas those of CIP and ofloxacin were 256 and 64 μg/ml, respectively (see Table S2 in the supplemental material).
FIG 1.
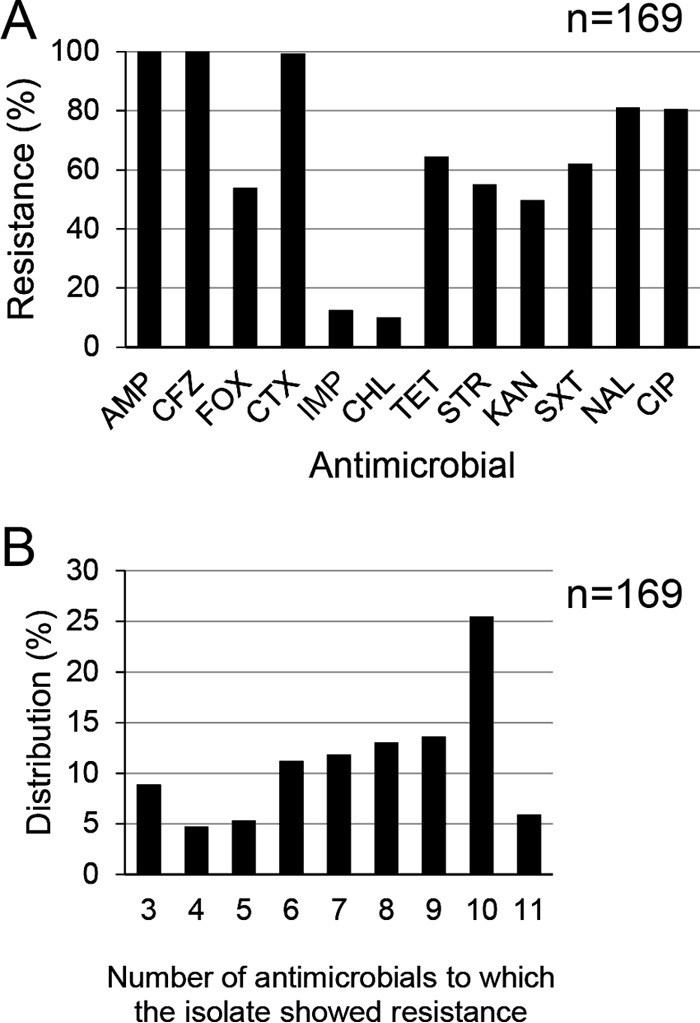
(A) Distribution of resistance to 12 antimicrobials among the extended-spectrum-cephalosporin-resistant and/or carbapenem-resistant E. coli isolates. The x axis indicates the antimicrobials used in this study: ampicillin (AMP), cefazolin (CFZ), cefoxitin (FOX), cefotaxime (CTX), imipenem (IMP), chloramphenicol (CHL), tetracycline (TET), streptomycin (STR), kanamycin (KAN), sulfamethoxazole-trimethoprim (SXT), nalidixic acid (NAL), and ciprofloxacin (CIP). The y axis indicates the prevalence of antimicrobial-resistant isolates. (B) The numbers of antimicrobials to which the same E. coli isolates for which the results are shown in panel A are resistant.
Genotype distribution of β-lactamase genes.
As shown in Fig. 2, the genes for the TEM, SHV, CTX-M, OXA, NDM, CMY, and ACC β-lactamases were detected in this study. The CTX-M β-lactamase gene was the most predominant and was detected in 112 isolates (66.3%), comprising 108 isolates carrying blaCTX-M-15 and 4 isolates carrying blaCTX-M-55. The distributions of TEM, OXA, and CMY were 43.8%, 39.6%, and 40.2%, respectively. In addition, 1 to 4 β-lactamase genes were detected in each isolate. Among the 21 isolates with resistance to IMP, NDM was detected in 14, comprising 2 isolates carrying blaNDM-1, 7 isolates carrying blaNDM-5, and 5 isolates carrying blaNDM-7. The diameters of the inhibition zones for IMP (≥23 mm for susceptible, 20 to 22 mm for intermediate, ≤19 mm for resistant) were 6 to 9 mm for these isolates, whereas those for the remaining seven isolates without blaNDM genes were 12 to 19 mm. In addition, 1 to 3 different β-lactamase genes (other than NDM) were detected in each of the seven isolates.
FIG 2.
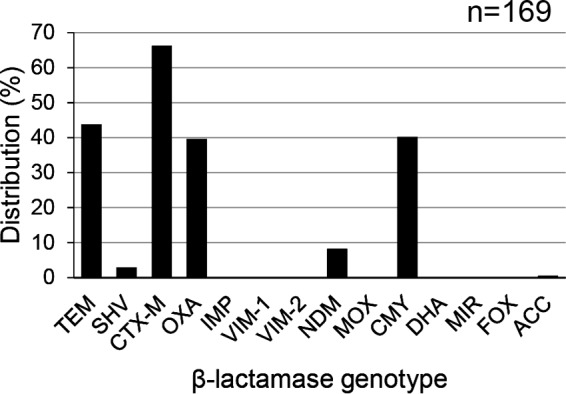
Distribution of β-lactamase genes among the extended-spectrum-cephalosporin-resistant and/or carbapenem-resistant E. coli isolates.
Identification of antimicrobial resistance genes and replicon types in the whole-genome sequences of the plasmids.
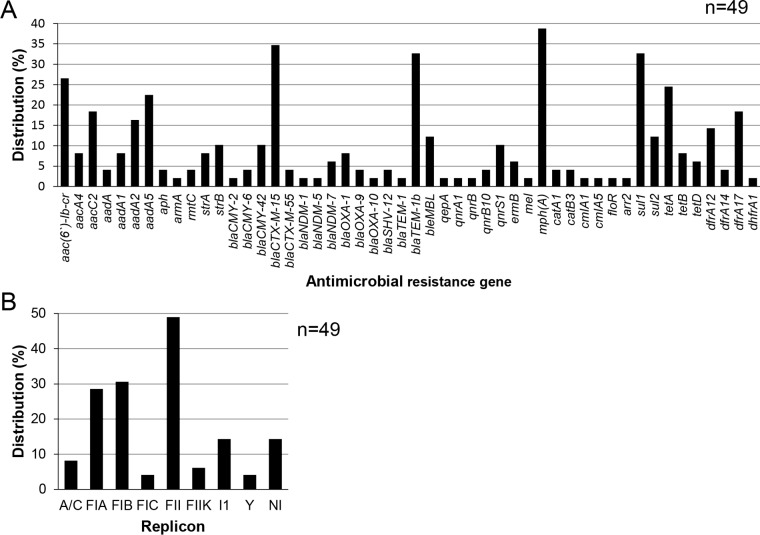
We selected 36 isolates based on their AMR patterns and sample origins (see Table S2 in the supplemental material). Some of the 36 isolates may have originated from the same sample but were selected as they showed resistance to different antimicrobials. Whole-genome sequencing of the plasmids from these isolates yielded a total of 49 plasmid genome sequences (2 complete sequences and 47 draft sequences) (see Table S3 in the supplemental material). Fifty antimicrobial resistance genes which contribute to resistance to aminoglycosides, β-lactams, bleomycin, fluoroquinolones, macrolides, phenicols, rifampin, sulfonamides, TETs, and trimethoprim were detected in these sequences (Fig. 3A; see also Fig. S1 in the supplemental material). The mph(A) gene, which contributes to macrolide resistance, was the most prevalent among the 50 genes, and its prevalence was 38.8%. blaCTX-M-15 and blaTEM-1b were the most prevalent β-lactamase genes, and their prevalences were 34.7% and 32.7%, respectively. The prevalence of aac(6′)-Ib-cr, which contributes to resistance to both aminoglycosides and fluoroquinolones, was 26.5%.
FIG 3.
(A) Distribution of genes conferring antimicrobial resistance detected in the sequences of 49 selected plasmids obtained from extended-spectrum-cephalosporin-resistant and/or carbapenem-resistant E. coli isolates; (B) distribution of replicon types detected in the 49 selected plasmids.
A total of eight PBRT amplicon sequences (replicon types) were detected in 42/49 plasmids. We could not identify any replicon type in the remaining seven plasmids (Fig. 3B). Up to three replicon types were detected in 1 plasmid, and 5 different combinations of the replicon types were observed among 17 plasmids. The most prevalent combination of replicon types was FIA, FIB, and FII (see Fig. S1 and Table S3 in the supplemental material). This type of plasmid is subsequently described to be FIA+FIB+FII in this article. A solitary replicon type was detected in the remaining 25 plasmids.
Network analysis of the plasmid sequences.
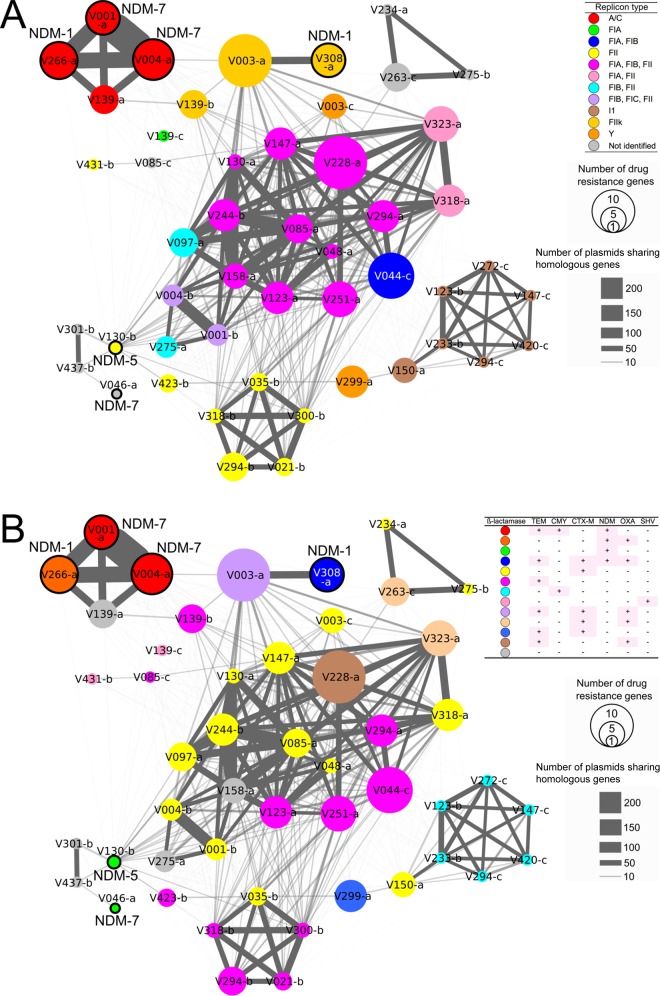
Network analysis was used to determine the number of genes in each plasmid shared by other plasmids. These numbers reflect the width of the lines between each plasmid in Fig. 4. Network analysis of the 49 plasmids revealed that plasmids belonging to the same replicon type shared more genes (Fig. 4A). Seventeen plasmids belonging to replicon types FIA+FIB, FIA+FIB+FII, FIA+FII, FIB+FII, and FIB+FIC+FII formed the largest community and shared 7 to 124 genes with each other. Fifteen of the 17 plasmids (pV044-c, pV048-a, pV085-a, pV097-a, pV123-a, pV130-a, pV147-a, pV158-a, pV228-a, pV244-b, pV251-a, pV275-a, pV294-a, pV318-a, and pV323-a) were detected in 12 water samples collected from three STPs in Karnataka. The remaining two plasmids (pV001-b and pV004-b) were detected in two river water samples collected in Bihar. Five replicon type FII plasmids (pV021-b, pV035-b, pV294-b, pV300-b, and pV318-b), which were detected in four water samples collected from two STPs in Karnataka, formed a community and shared 37 to 64 genes with each other. Each plasmid in this community also shared <24 genes between plasmids in the largest community. Four A/C plasmids (pV001-a, pV004-a, pV139-a, and pV266-a), which were detected in four water samples collected from an STP in Karnataka and a river in Bihar, formed a community and shared 72 to 289 genes with each other. Six I1 plasmids (pV123-b, pV147-c, pV233-b, pV272-c, pV294-c, and pV420-c), which were detected in six water samples collected from three STPs in Karnataka and a river in Tamil Nadu, formed a community and shared 22 to 54 genes with each other.
FIG 4.
Network community analysis of the 49 selected plasmids based on the whole or draft plasmid genome sequences. Each circle represents a plasmid. The circle diameters correlate with the number of antimicrobial resistance genes in the plasmid. blaNDM-positive plasmids are highlighted by black borders. Plasmids sharing at least 2 homologous genes (at least 100% identity at the amino acid level, 90% ORF coverage, and a length difference of less than 10%) are connected by gray lines. The widths of the gray lines correlate with the quantity of homologous genes shared. Each plasmid is colored by replicon type (A) or the pattern of possession of β-lactamase genes (B). The prefix “p” was removed from the plasmid names.
Twelve different patterns of detection of β-lactamase genes in one plasmid were observed. Plasmids with the same replicon types did not exclusively show the same patterns of β-lactamase genes in most cases (Fig. 4B; see also Fig. S1 in the supplemental material). NDM was detected in six plasmids (pV001-a, pV004-a, pV046-a, pV130-b, pV266-a, and pV308-a) which belonged to replicon types A/C, FII, and FIIk and one not identifiable replicon type. These plasmids were detected in five water samples collected from a river in Bihar and two STPs in Karnataka (see Table S3 in the supplemental material).
Comparison of DNA sequences of blaNDM-positive plasmids.
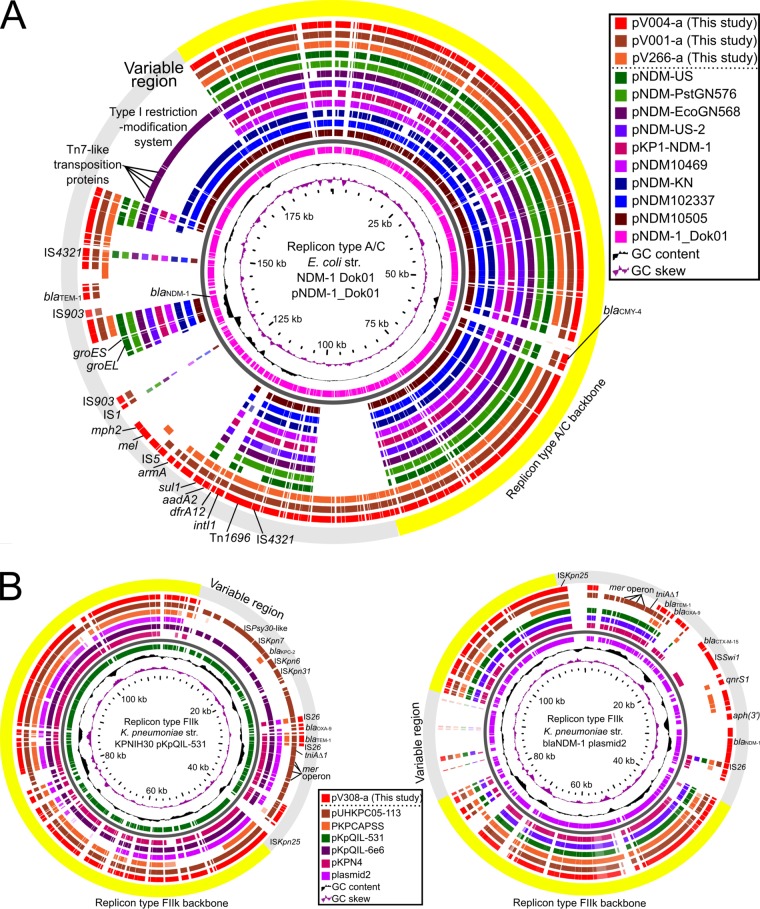
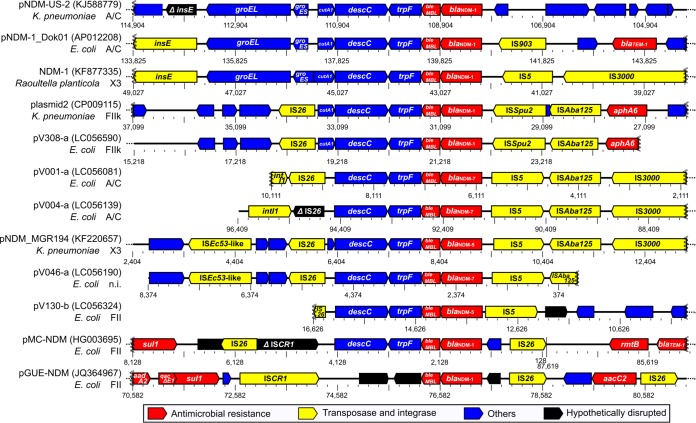
To assess the evolution of blaNDM-positive plasmids, we compared the whole-genome sequences of the six blaNDM-positive plasmids detected in this study with known sequences of related plasmids. The replicon type A/C backbone of plasmids pV001-a, pV004-a, and pV266-a showed high degrees of sequence similarity with the backbone of known replicon type A/C plasmids containing blaNDM genes. Most of the antimicrobial resistance genes were located in the variable region; the exception was blaCMY-4 (Fig. 5A). The blaNDM genes of pV001-a, pV004-a, and pV266-a were located in the variable regions of these plasmids. The replicon type FIIk backbone of pV308-a showed a high degree of sequence similarity with that of several known blaKPC-2-positive plasmids (Fig. 5B, left). Although the blaKPC-2 gene was not detected in pV308-a, a genetic region containing antimicrobial resistance genes blaNDM-1, aph(3′), qnrS1, and blaCTX-M-15 was located in the variable region in this plasmid (Fig. 5B, right). Three open reading frames (ORFs) adjacent to the blaNDM gene of the six plasmids, including bleMBL, trpF, and descC, were common with those found in other related plasmid sequences. Multiple transposase genes were located in the flanking regions (Fig. 6).
FIG 5.
Circular alignments of the DNA sequences of four blaNDM-positive plasmids obtained in this study and known related plasmid sequences. The visualized area shows that the percent identity of similar genes between the reference plasmid and other plasmids was at least 80%. The known sequences of the following plasmids (GenBank accession numbers) were included: pNDM-US (NZ_CP006661), pNDM-PstGN576 (KJ802405), pNDM-EcoGN568 (KJ802404), pNDM-US-2 (KJ588779), pKP1-NDM-1 (KF992018), pNDM10469 (JN861072), pNDM-KN (JN157804), pNDM102337 (JF714412), pNDM10505 (JF503991), pNDM-1_Dok01 (AP012208), pKPCAPSS (KP008371), pKpQIL-531 (CP008833), pKpQIL-6e6 (CP007730), pKPN4 (CP000649), and plasmid2 (CP009115). (A) Alignment of replicon type A/C plasmids. Draft genome sequence data for plasmids pV001-a, pV004-a, and pV266-a were obtained in this study. Ten known sequences of blaNDM-positive plasmids were included, and pNDM-1_Dok01 was used as a reference. (B) Alignments of replicon type FIIk plasmid sequences. Draft genome sequence data for plasmid pV308-a were obtained in this study. Six known sequences of blaKPC-2- or blaNDM-1-positive plasmids were included, and pKpQIL-531 (left) and plasmid2 (right) were used as references.
FIG 6.
Linear alignment of DNA sequences of blaNDM genes and the flanking regions of various plasmids. Draft genome sequence data for five blaNDM-positive plasmids obtained in this study were used. Data for pV266-a were omitted because the blaNDM-1-positive contig originating from this plasmid does not contain flanking regions. Seven known sequences of blaNDM-positive plasmids, as indicated, were included. GenBank accession numbers are given in parentheses.
DISCUSSION
In this study, more than half of the 169 ESC- and/or carbapenem-resistant E. coli isolates exhibited resistance to eight or more antimicrobials (Fig. 1B). Eighty percent of the 169 E. coli isolates also exhibited a high level of resistance to fluoroquinolones (see Table S2 in the supplemental material). Fluoroquinolone resistance is mainly mediated by the accumulation of point mutations in the chromosomal genes encoding DNA gyrase and/or DNA topoisomerase IV (21). Although we did not check the DNA sequences of the quinolone resistance-determining regions of these genes in this study, most of the fluoroquinolone-resistant isolates should have these point mutations because wild-type E. coli isolates are highly susceptible to this antibiotic (21). Plasmid-mediated quinolone resistance genes, including aac(6′)-Ib-cr, qepA, qnrA1, qnrB, qnrB10, and qnrS1, that contribute to the low level of resistance to fluoroquinolones (22) were detected among the 49 plasmids analyzed in this study (Fig. 3A). To some extent, these genes contribute to the fluoroquinolone resistance of these isolates.
Among the 12 antimicrobials to which the susceptibility of the isolates was tested, the rate of resistance to CHL was the lowest (10.1%) (Fig. 1A). Historically, CHL, AMP, and SXT were used for the treatment of typhoid fever. Since the global emergence of multidrug-resistant Salmonella enterica serovar Typhi isolates, CIP has become the first-line drug of choice for the treatment of typhoid fever in India (23). The lower level of CHL consumption compared with the level of consumption of the other antimicrobials could be reflected by the lower prevalence of resistance to this antibiotic among the isolates (24).
We detected seven different β-lactamase genes, of which CTX-M was predominant (Fig. 2). Most of the CTX-M-positive E. coli isolates had the blaCTX-M-15 gene, which is quite common in India (11, 25, 26). The blaCMY-2, blaCMY-6, blaCMY-42, blaOXA-1, blaOXA-9, and blaSHV-12 β-lactamase genes, in addition to the blaCTX-M gene, were detected in the plasmid sequences (Fig. 3A) and can confer resistance to ESCs (27–29). In this study, we found 21 IMP-resistant E. coli isolates. blaNDM genes were detected in 14 of these 21 isolates. The diameters of the IMP inhibition zone for the remaining 7 isolates were larger than those for these 14 isolates with blaNDM genes. Because we detected 1 to 3 different β-lactamase genes other than the blaNDM gene, expression of an extended-spectrum β-lactamase and/or a AmpC β-lactamase combined with the decreased permeability of the cell membrane may have contributed to the lower level of resistance to IMP among the seven isolates (30).
Limited reports on the prevalence of β-lactamase genes among bacteria isolated from the environment are available. Bajaj et al. (11) reported that blaTEM was the most widespread (100%) β-lactamase gene, followed by blaCTX-M (16%), among 61 E. coli isolates which originated from a river in the northern part of India. They did not select E. coli isolates according to their antimicrobial resistance, while we selected E. coli isolates showing resistance to ESC and/or carbapenem for the detailed analyses. The difference in β-lactamase gene prevalence may be due to the difference in the methodology of selection of E. coli isolates. blaCTX-M-15 is also the most prevalent β-lactamase gene conferring ESC resistance among bacteria in the environment in Bangladesh (31). blaCTX-M genes seem to be commonly detected from the environment in East Asia, Europe, and Australia (32–35). blaNDM genes have been detected in the environment in Bangladesh and China (31, 36). Novovic et al. (37) could not detect blaNDM-positive bacteria from environmental waters in Serbia, which is recognized to be an area where NDM-1-producing bacteria are the most prevalent.
The results of network analysis suggest that plasmids sharing replicon types tend to form a community, which is based on the predicted gene similarity among the plasmids. The largest community consisted of different combinations of replicon type FIA, FIB, and/or FII (Fig. 4A). In this community, the integration of plasmids with different replicon types seemed to facilitate the evolution of these plasmids. In contrast, the remaining three communities consisted of plasmids with a single replicon type, A/C, FII, or I1 (Fig. 4A). Two different β-lactamase gene sets were observed among the communities of replicon types A/C and FII (Fig. 4B), suggesting that evolution had occurred within the variable regions of these plasmids. The plasmids within three of the four communities were detected in two different states, suggesting that dissemination of the ancestor plasmids to multiple states has occurred in India.
Three different blaNDM genes, blaNDM-1, blaNDM-5, and blaNDM-7, from six plasmids with replicon types A/C, FII, FIIk, and untypeable were detected in this study (Fig. 4A). blaNDM-5 and blaNDM-7 were reported to be variants of blaNDM-1, and each one has two amino acid substitutions compared with the sequence of blaNDM-1: V87L and M154L for blaNDM-5 and D130N and M154L for blaNDM-7 (38–40). Among these substitutions, M154L was reported to increase the hydrolytic activity of the enzymes (41). The results of comparisons of the DNA sequences of blaNDM-positive plasmids of replicon types A/C and FIIk showed that the backbone regions among these plasmids were highly conserved (Fig. 5A and B). The variable region contained multiple antimicrobial resistance genes, including blaNDM. The region adjacent to the blaNDM gene was highly conserved and was flanked by several insertion sequences (Fig. 6). These observations suggest that the point mutation, recombination, and transposition of the variable region facilitated the evolution of these plasmids. blaNDM genes were detected in plasmids belonging to replicon types A/C, L/M, FII, FIIk, FIB-M, and HI1, which have been found to be harbored by more than 30 species of bacteria thus far (4, 20).
In summary, most of the ESC- and/or carbapenem-resistant E. coli isolates from rivers and STPs in India evaluated in this study exhibited resistance to multiple antimicrobials, in addition to β-lactams. Fifty different resistance genes were detected in plasmids with various genetic backgrounds. Among the β-lactamase genes, blaCTX-M was predominant, and blaNDM was also detected. The results of network analysis and comparison of the DNA sequences suggest that various mutation events facilitated the evolution of the plasmids and that plasmids with similar genetic backgrounds have widely disseminated in India. Eighty percent of these isolates also exhibited resistance to fluoroquinolones. As ESCs and fluoroquinolones are often used for the treatment of serious infectious diseases in humans (42), environmental contamination with Enterobacteriaceae with this type of resistance is a serious threat to public health. The prevention of environmental contamination by anthropogenic sources is required to reduce community-acquired infection in India and to prevent the worldwide dissemination of ESC- or carbapenem-resistant Enterobacteriaceae.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nobuyoshi Yamashita and Sachi Taniyasu of the National Institute of Advanced Industrial Science and Technology for their technical assistance on environmental water sampling.
The Dr. T. M. A. Pai Endowment Chair of Earth Sciences at Manipal University is thanked for supporting this study at Manipal.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01950-15.
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland: http://www.who.int/drugresistance/documents/surveillancereport/en/. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013/. [Google Scholar]
- 3.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrazeg M, Diene S, Medjahed L, Parola P, Drissi M, Raoult D, Rolain J. 2014. New Delhi metallo-β-lactamase around the world: an eReview using Google maps. Euro Surveill 19(20):pii=20809. doi: 10.2807/1560-7917.ES2014.19.20.20809. [DOI] [PubMed] [Google Scholar]
- 5.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 6.Deshpande P, Shetty A, Kapadia F, Hedge A, Soman R, Rodrigues C. 2010. New Delhi metallo 1: have carbapenems met their doom? Clin Infect Dis 51:1222. doi: 10.1086/656921. [DOI] [PubMed] [Google Scholar]
- 7.Shahid M, Khan F, Shah MS, Shukla I, Shujatullah F, Khan HM, Malik A, Khan IM. 2012. NDM-1 in the Indian environment: hitherto the problem is not disquieting. Asian Pac J Trop Med 5:335–336. doi: 10.1016/S1995-7645(12)60053-4. [DOI] [PubMed] [Google Scholar]
- 8.Akiba M, Senba H, Otagiri H, Prabhasankar VP, Taniyasu S, Yamashita N, Lee K, Yamamoto T, Tsutsui T, Joshua DI, Balakrishna K, Bairy I, Iwata T, Kusumoto M, Kannan K, Guruge KS. 2015. Impact of wastewater from different sources on the prevalence of antimicrobial-resistant Escherichia coli in sewage treatment plants in South India. Ecotoxicol Environ Saf 115:203–208. doi: 10.1016/j.ecoenv.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Chandran SP, Diwan V, Tamhankar AJ, Joseph BV, Rosales-Klintz S, Mundayoor S, Lundborg CS, Macaden R. 2014. Detection of carbapenem resistance genes and cephalosporin, and quinolone resistance genes along with oqxAB gene in Escherichia coli in hospital wastewater: a matter of concern. J Appl Microbiol 117:984–995. doi: 10.1111/jam.12591. [DOI] [PubMed] [Google Scholar]
- 10.Ahammad ZS, Sreekrishnan TR, Hands CL, Knapp CW, Graham DW. 2014. Increased waterborne blaNDM-1 resistance gene abundances associated with seasonal human pilgrimages to the Upper Ganges River. Environ Sci Technol 48:3014–3020. doi: 10.1021/es405348h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajaj P, Singh NS, Kanaujia PK, Virdi JS. 2015. Distribution and molecular characterization of genes encoding CTX-M and AmpC β-lactamases in Escherichia coli isolated from an Indian urban aquatic environment. Sci Total Environ 505:350–356. doi: 10.1016/j.scitotenv.2014.09.084. [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial disk susceptibility tests; approved standard, 11th ed CLSI document M02-A11. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Shahada F, Sekizuka T, Kuroda M, Kusumoto M, Ohishi D, Matsumoto A, Okazaki H, Tanaka K, Uchida I, Izumiya H, Watanabe H, Tamamura Y, Iwata T, Akiba M. 2011. Characterization of Salmonella enterica serovar Typhimurium isolates harboring a chromosomally encoded CMY-2 β-lactamase gene located on a multidrug resistance genomic island. Antimicrob Agents Chemother 55:4114–4121. doi: 10.1128/AAC.00560-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 17.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. 2010. Interactive microbial genome visualization with GView. Bioinformatics 26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Yamashita A, Sekizuka T, Kuroda M. 2014. Characterization of antimicrobial resistance dissemination across plasmid communities classified by network analysis. Pathogens 3:356–376. doi: 10.3390/pathogens3020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopkins KL, Davies RH, Threlfall EJ. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents 25:358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev 22:664–689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harish BN, Menezes GA. 2011. Antimicrobial resistance in typhoidal salmonellae. Indian J Med Microbiol 29:223–229. doi: 10.4103/0255-0857.83904. [DOI] [PubMed] [Google Scholar]
- 24.Ganguly NK, Arora NK, Chandy SJ, Fairoze MN, Gill JP, Gupta U, Hossain S, Joglekar S, Joshi PC, Kakkar M, Kotwani A, Rattan A, Sudarshan H, Thomas K, Wattal C, Easton A, Laxminarayan R, Global Antibiotic Resistance Partnership—India Working Group. 2011. Rationalizing antibiotic use to limit antibiotic resistance in India. Indian J Med Res 134:281–294. [PMC free article] [PubMed] [Google Scholar]
- 25.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob Agents Chemother 55:1274–1278. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diwan V, Chandran SP, Tamhankar AJ, Stalsby Lundborg C, Macaden R. 2012. Identification of extended-spectrum beta-lactamase and quinolone resistance genes in Escherichia coli isolated from hospital wastewater from central India. J Antimicrob Chemother 67:857–859. doi: 10.1093/jac/dkr564. [DOI] [PubMed] [Google Scholar]
- 27.Hentschke M, Kotsakis SD, Wolters M, Heisig P, Miriagou V, Aepfelbacher M. 2011. CMY-42, a novel plasmid-mediated CMY-2 variant AmpC β-lactamase. Microb Drug Resist 17:165–169. doi: 10.1089/mdr.2010.0137. [DOI] [PubMed] [Google Scholar]
- 28.Jacoby GA. 2009. AmpC β-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeifer Y, Cullik A, Witte W. 2010. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol 300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Patel JB, Rasheed JK, Kitchel B. 2009. Carbapenemases in Enterobacteriaceae: activity, epidemiology, and laboratory detection. Clin Microbiol Newsl 31:55–62. doi: 10.1016/j.clinmicnews.2009.03.005. [DOI] [Google Scholar]
- 31.Toleman MA, Bugert JJ, Nizam SA. 2015. Extensively drug-resistant New Delhi metallo-β-lactamase-encoding bacteria in the environment, Dhaka, Bangladesh, 2012. Emerg Infect Dis 21:1027–1030. doi: 10.3201/eid2106.141578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amos GCA, Hawkey PM, Gaze WH, Wellington EM. 2014. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J Antimicrob Chemother 69:1785–1791. doi: 10.1093/jac/dku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gundogdu A, Jennison AV, Smith HV, Stratton H, Katouli M. 2013. Extended-spectrum β-lactamase producing Escherichia coli in hospital wastewaters and sewage treatment plants in Queensland, Australia. Can J Microbiol 59:737–745. doi: 10.1139/cjm-2013-0515. [DOI] [PubMed] [Google Scholar]
- 34.Korzeniewska E, Harnisz M. 2013. Extended-spectrum beta-lactamase (ESBL)-positive Enterobacteriaceae in municipal sewage and their emission to the environment. J Environ Manage 128:904–911. doi: 10.1016/j.jenvman.2013.06.051. [DOI] [PubMed] [Google Scholar]
- 35.Zou LK, Li LW, Pan X, Tian GB, Luo Y, Wu Q, Li B, Cheng L, Xiao JJ, Hu S, Zhou Y, Pang YJ. 2012. Molecular characterization of β-lactamase-resistant Escherichia coli isolated from Fu River, China. World J Microbiol Biotechnol 28:1891–1899. doi: 10.1007/s11274-011-0987-9. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Qiu S, Wang Y, Qi L, Hao R, Liu X, Shi Y, Hu X, An D, Li Z, Li P, Wang L, Cui J, Wang P, Huang L, Klena JD, Song H. 2013. Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLoS One 8:e64857. doi: 10.1371/journal.pone.0064857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novovic K, Filipic B, Veljovic K, Begovic J, Mirkovic N, Jovcic B. 2015. Environmental waters and blaNDM-1 in Belgrade, Serbia: endemicity questioned. Sci Total Environ 511:393–398. doi: 10.1016/j.scitotenv.2014.12.072. [DOI] [PubMed] [Google Scholar]
- 38.Cuzon G, Bonnin RA, Nordmann P. 2013. First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS One 8:e61322. doi: 10.1371/journal.pone.0061322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. 2013. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-β-lactamase with increased carbapenemase activity. J Antimicrob Chemother 68:1737–1740. doi: 10.1093/jac/dkt088. [DOI] [PubMed] [Google Scholar]
- 40.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordmann P, Boulanger AE, Poirel L. 2012. NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother 56:2184–2186. doi: 10.1128/AAC.05961-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization. 2012. Critically important antimicrobials for human medicine, 3rd revision World Health Organization, Geneva, Switzerland: http://www.who.int/foodsafety/publications/antimicrobials-third/en/. [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. http://genome.cshlp.org/content/13/11/2498.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.