Abstract
GSK2251052 is a broad-spectrum antibacterial inhibitor of leucyl tRNA-synthetase (LeuRS) that has been evaluated in phase II clinical trials. Here, we report the identification of a clinical isolate of Staphylococcus aureus that exhibits reduced susceptibility to GSK2251052 without prior exposure to the compound and demonstrate that this phenotype is attributable to a single amino acid polymorphism (P329) within the editing domain of LeuRS.
TEXT
GSK2251052 (GSK'052) is a novel broad-spectrum antibacterial agent that selectively inhibits bacterial leucyl tRNA-synthetase (LeuRS) (1). Although this compound appears to possess many of the requisite properties of an antibacterial drug for treating infection in humans, it also has the undesirable feature of rapidly selecting resistance in bacteria; in phase II clinical trials involving adult subjects suffering from complicated urinary tract infections, resistance to GSK'052 developed within 2 days of administration in three of 14 patients (1, 2). Here, we report that in addition to arising rapidly in bacteria under selection, reduced susceptibility to GSK'052 is preexisting among clinical isolates of Staphylococcus aureus that have not been exposed to the drug, a phenomenon that is the result of polymorphism in the drug target.
GSK'052 was obtained by chemical synthesis, according to established methodology (3). To evaluate the susceptibility of staphylococcal strains to this compound, a small panel of S. aureus blood culture isolates (n = 52) was tested using the microbroth dilution method, according to CLSI guidelines (4). These isolates were recovered from patients at the Erasmus MC University Medical Center Rotterdam (The Netherlands) between November 2009 and May 2010 and therefore originate from a country in which GSK'052 has never been trialed, and during a period that predates the clinical evaluation of GSK'052 (2). Consequently, it may be stated with some confidence that these isolates have never been exposed to this compound in the clinic. GSK'052 exhibited a MIC of 2 to 4 mg/liter against all isolates, with the exception of one (strain 1372), for which the MIC was 16 mg/liter. This degree of reduced susceptibility to GSK'052 is equivalent to that exhibited by a resistant Escherichia coli strain selected in a patient upon administration of GSK'052 in the phase II clinical trial, and which was associated with microbiological failure (2).
To determine the mechanism for reduced susceptibility to GSK'052 in S. aureus 1372, we proceeded on the basis that this phenotype was likely the result of a polymorphism in the drug target (LeuRS) and subjected the entire leuS gene from this strain to PCR amplification and DNA sequence determination. This revealed that in comparison to the leuS gene of the fully GSK'052-susceptible laboratory strain S. aureus SH1000 (5, 6), leuS1372 encodes a protein containing four amino acid polymorphisms (T311I, S329P, A553G, and Y735F; the SH1000 residue is shown first in each case).
To establish whether one or more of these polymorphisms account for the reduced susceptibility to GSK'052, leuS1372 and leuSSH1000 were PCR amplified using oligonucleotide primers ATCGTTATGTCGACTTTTTTATTGAATAGGAGGA and TGCTTAGTGGATCCATTTCAAAGTCCTCCTTAAA (engineered restriction sites shown underlined) and introduced into the staphylococcal expression vector pLOW (7) for ectopic expression in S. aureus SH1000. Strain SH1000 (pLOW:leuS1372) exhibited a substantial reduction in susceptibility to GSK'052 (MIC, 64 mg/liter) compared to SH1000 (pLOW:leuSSH1000) (MIC, 16 mg/liter), confirming that the reduced susceptibility of S. aureus 1372 to GSK'052 is indeed a consequence of a polymorphism in LeuRS.
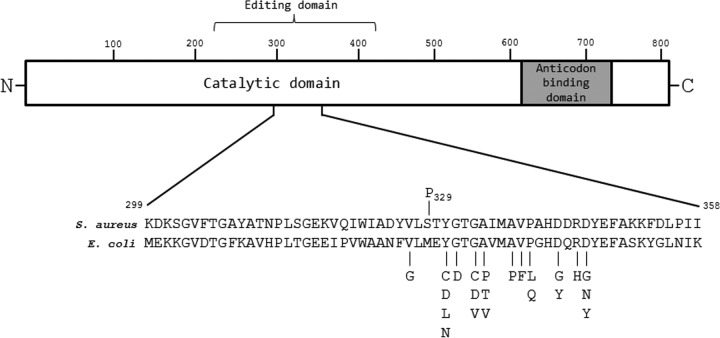
Of the four amino acid polymorphisms in LeuRS1372, two (I311 and G553) are also found encoded in the leuS gene of strains of S. aureus whose genome sequences have been deposited in the public databases (strains MRSA252 [GenBank accession no. BX571856] and Mu50 [GenBank accession no. BA000017]). When we tested these strains, neither was more resistant to GSK'052 than SH1000, implying that neither of these polymorphisms participates in reduced susceptibility to GSK'052. Of the remaining two polymorphisms in LeuRS1372, we considered P329 the most likely candidate for mediating the relative insensitivity of the enzyme to GSK'052, since it resides within the editing domain, a region of the protein containing the majority of amino acid substitutions identified in the LeuRS of clinical isolates of E. coli in which resistance to GSK'052 has evolved (2) (Fig. 1). To test this, the substitution S329P was engineered into pLOW:leuSSH1000 using the Q5 site-directed mutagenesis kit (New England BioLabs, MA, USA) and oligonucleotide primers TTATGTATTACCAACATATGGTACTG (engineered mutation underlined) and TCAGCAATCCAAATTTGTAC. The introduction of this construct into SH1000 resulted in a strain exhibiting the same degree of reduced susceptibility to GSK'052 (MIC, 64 mg/liter) as SH1000 (pLOW:leuS1372), thereby confirming that the polymorphism P329 in LeuRS is responsible for the decreased susceptibility of strain 1372 to GSK'052.
FIG 1.
Schematic of the LeuRS protein, with a close up on part of the editing domain, showing the amino acid substitutions that mediate reduced susceptibility to GSK'052 in S. aureus (this study) and E. coli (2). The residue numbering corresponds to the S. aureus sequence, and amino acid substitutions are denoted above and below the sequence alignment for S. aureus and E. coli, respectively.
With a view to understanding how this polymorphism negatively impacts the activity of GSK'052 against LeuRS, we examined the published crystal structure of Thermus thermophilus LeuRS bound to the parent compound (AN2690) of GSK'052 (PDB identification [ID] 2V0C). AN2690 forms an adduct with tRNAleu that becomes trapped in the editing site of the enzyme (8), with residues lying in close proximity to the P329 polymorphism participating in binding the tRNA portion of this adduct. In particular, the preceding residue (L329, T. thermophilus numbering) forms two hydrogen bonds with nucleotide A76 of the tRNAleu (8). The presence of a conformationally rigid proline adjacent to this position would likely serve to constrain the protein backbone, thereby restricting the conformation of this leucine residue and potentially impairing its ability to make these hydrogen bonds; the loss of one or more hydrogen-bonding contacts would reduce the affinity of the enzyme for the tRNAleu drug adduct and thereby lead to reduced susceptibility to the compound.
In conclusion, we have shown that a polymorphism (P329) in the LeuRS enzyme of a clinical isolate of S. aureus mediates reduced susceptibility to GSK'052. While our results do not at this stage enable informed speculation regarding the prevalence of or the underlying reason(s) for this polymorphism, it is clear that its presence is in no part attributable to selection by GSK'052. The identification of a clinical S. aureus isolate that exhibits uniform reduced susceptibility at the level of the drug target to an experimental antibacterial drug with which it has never been challenged, although a phenomenon that has been reported previously (9), is apparently rare or infrequently documented. Our findings raise the possibility that polymorphisms associated with reduced susceptibility to GSK'052 also exist in strains of other bacterial pathogens and underscore the utility of assessing the activities of antibacterial drug candidates against clinical isolates as part of preclinical evaluation to identify any preexisting mechanisms mediating reduced susceptibility.
ACKNOWLEDGMENTS
This work was supported by a grant from the European Union Framework 7 (FP7) program, Health.2013.2.31-1- NABARSI (grant agreement no: 601725). The funders had no role in the study design or the decision to submit the work for publication.
We declare no conflicts of interest.
REFERENCES
- 1.Hernandez V, Crepin T, Palencia A, Cusack S, Akama T, Baker SJ, Bu W, Feng L, Freund YR, Liu L, Meewan M, Mohan M, Mao W, Rock FL, Sexton H, Sheoran A, Zhang Y, Zhang Zhou YKY, Nieman JA, Anugula MR, Keramane EM, Savariraj K, Reddy DS, Sharma R, Subedi R, Singh R, O'Leary A, Simon NL, De Marsh PL, Mushtaq S, Warner M, Livermore DM, Alley MRK, Plattner JJ. 2013. Discovery of a novel class of boron-based antibacterials with activity against Gram-negative bacteria. Antimicrob Agents Chemother 57:1394–1403. doi: 10.1128/AAC.02058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Dwyer K, Spivak AT, Ingraham K, Min S, Holmes DJ, Jakielaszek C, Rittenhouse S, Kwan AL, Livi GP, Sathe G, Thomas E, Van Horn S, Miller LA, Twynholm M, Tomayko J, Dalessandro M, Caltabiano M, Scangarella-Oman NE, Brown JR. 2015. Bacterial resistance to leucyl-tRNA synthetase inhibitor GSK2251052 develops during treatment of complicated urinary tract infections. Antimicrob Agents Chemother 59:289–298. doi: 10.1128/AAC.03774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker SJ, Hernandez VS, Sharma R, Nieman JA, Akama T, Zhang YK, Plattner JJ, Alley MRK, Singh R, Rock F. June 2008. Boron-containing small molecules. U.S. patent 12/142692.
- 4.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed, vol 32 CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 5.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 19:5457–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Neill AJ. 2010. Staphylococcus aureus SH1000 and 8325-4: comparative genome sequences of key laboratory strains in staphylococcal research. Lett Appl Microbiol 51:358–361. doi: 10.1111/j.1472-765X.2010.02885.x. [DOI] [PubMed] [Google Scholar]
- 7.Liew ATF, Theis T, Jensen SO, Garcia-Lara J, Foster SJ, Firth N, Lewis PJ, Harry EJ. 2011. A simple plasmid-based system that allows rapid generation of tightly controlled gene expression in Staphylococcus aureus. Microbiology 157:666–676. doi: 10.1099/mic.0.045146-0. [DOI] [PubMed] [Google Scholar]
- 8.Rock FL, Mao W, Yaremchuk A, Tukalo M, Crepin T, Zhou H, Zhang YK, Hernandez V, Akama T, Baker SJ, Plattner JJ, Shaprio L, Martinis SA, Benkovic SJ, Cusack S, Alley MRK. 2007. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316:1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 9.Watters AA, Jones RN, Leeds JA, Denys G, Sader HS, Fritsche TR. 2006. Antimicrobial activity of a novel peptide deformylase inhibitor, LBM415, tested against respiratory tract and cutaneous infection pathogens: a global surveillance report (2003–2004). J Antimicrob Chemother 57:914–923. doi: 10.1093/jac/dkl093. [DOI] [PubMed] [Google Scholar]