Abstract
Previous studies showed that sub-MIC levels of β-lactam antibiotics stimulate biofilm formation in most methicillin-resistant Staphylococcus aureus (MRSA) strains. Here, we investigated this process by measuring the effects of sub-MIC amoxicillin on biofilm formation by the epidemic community-associated MRSA strain USA300. We found that sub-MIC amoxicillin increased the ability of USA300 cells to attach to surfaces and form biofilms under both static and flow conditions. We also found that USA300 biofilms cultured in sub-MIC amoxicillin were thicker, contained more pillar and channel structures, and were less porous than biofilms cultured without antibiotic. Biofilm formation in sub-MIC amoxicillin correlated with the production of extracellular DNA (eDNA). However, eDNA released by amoxicillin-induced cell lysis alone was evidently not sufficient to stimulate biofilm. Sub-MIC levels of two other cell wall-active agents with different mechanisms of action—d-cycloserine and fosfomycin—also stimulated eDNA-dependent biofilm, suggesting that biofilm formation may be a mechanistic adaptation to cell wall stress. Screening a USA300 mariner transposon library for mutants deficient in biofilm formation in sub-MIC amoxicillin identified numerous known mediators of S. aureus β-lactam resistance and biofilm formation, as well as novel genes not previously associated with these phenotypes. Our results link cell wall stress and biofilm formation in MRSA and suggest that eDNA-dependent biofilm formation by strain USA300 in low-dose amoxicillin is an inducible phenotype that can be used to identify novel genes impacting MRSA β-lactam resistance and biofilm formation.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) causes numerous infections ranging from mild cellulitis to life-threatening sepsis (1). Although many MRSA infections are health care associated, the increasing incidence of community-acquired MRSA infections represents a growing public health concern (2). Antibiotic resistance and biofilm formation are two virulence factors that contribute to MRSA pathogenesis in both health care and community settings (3). Methicillin resistance is mediated by mecA, a gene that encodes a novel cell wall transpeptidase with low affinity for most β-lactam antibiotics (4). Biofilm formation is mediated by extracellular polymeric substances (EPS), including proteinaceous adhesins, such as fibronectin binding proteins A and B (5, 6); extracellular DNA (eDNA) (7); and polysaccharides, such as poly-N-acetyl-d-glucosamine (PNAG), also known as polysaccharide intercellular adhesin (PIA) (3, 7–9). Methicillin resistance limits treatment options for MRSA, and biofilm formation confers additional antibiotic resistance that limits treatment efficacy.
Several observations suggest a link between MRSA methicillin resistance and biofilm formation. First, most MRSA strains form biofilms that contain eDNA and proteinaceous adhesins in their matrices, whereas methicillin-sensitive S. aureus (MSSA) strains, which lack mecA, usually form biofilms that contain polysaccharides in their matrices (10–13). In addition, deletion of mecA in MRSA strains results in decreased biofilm even in the absence of β-lactam antibiotics (10, 14). Also, MRSA strains are more likely to form biofilm than MSSA strains in the absence of glucose, whereas both groups form biofilms in the presence of glucose (15). Finally, sub-MIC levels of β-lactam antibiotics have been shown to induce eDNA release and biofilm formation in MRSA strains, but not in MSSA strains (16). The latter process may have relevance in hospitals and on farms, where bacteria may be exposed to low levels of antibiotics.
The aim of the present study was to investigate the mechanism of MRSA biofilm formation in the presence of sub-MIC levels of β-lactam antibiotics. Our approach was to measure the effects of low-dose amoxicillin, a clinically relevant antibiotic, on cells and biofilms of the MRSA strain LAC, a well-characterized epidemic community-associated USA300 isolate (16, 17). We cultured biofilms in static and dynamic biofilm reactors, analyzed mutant strains deficient in the production of known biofilm effectors, grew biofilms in EPS-degrading enzymes to study the composition of the biofilm matrix, and used physical and microscopy methods to analyze structural properties of the biofilms. We also investigated the ability of low doses of non-β-lactam cell wall-active agents to stimulate biofilm. Finally, we identified mutants deficient in biofilm induction by screening the Nebraska Transposon Mutant Library (NTML) for mutants that formed decreased biofilm in the presence of sub-MIC amoxicillin. The NTML is a collection of approximately 2,000 mutant derivatives of strain USA300 in which individual genes have been disrupted by insertion of the mariner-based transposon bursa aurealis (18, 19). Our findings suggest that eDNA-dependent biofilm formation by USA300 in low-dose amoxicillin is an inducible mechanistic adaptation to cell wall stress and that this phenotype will be useful for identifying novel genes that impact biofilm formation, the cell wall stress response, and the bactericidal activity of β-lactam antibiotics against MRSA.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are listed in Tables 1 and 2. Unless otherwise indicated, bacteria were cultured in tryptic soy broth (Becton Dickinson and Co.) supplemented with 6 g/liter yeast extract and 8 g/liter glucose. Solid media were further supplemented with 1.2% agar. Antibiotics (amoxicillin, fosfomycin, d-cycloserine, kanamycin, and erythromycin) were purchased from Sigma and added to broth or solid media at the indicated concentrations. Bacterial inocula were prepared in fresh broth from 24-h-old agar colonies, as previously described (7). The inocula were diluted to 105 to 106 CFU/ml unless otherwise indicated. All cultures were incubated at 37°C.
TABLE 1.
Bacterial strains, plasmids, and PCR primers
| Strains, plasmids, and primers | Characteristics or sequencea | Reference(s) or sourceb |
|---|---|---|
| S. aureus | ||
| LAC | CA-MRSA (USA300 clone) | 15, 16 |
| AH1263 | LAC cured of plasmid p03 | 30 |
| JE2 | LAC cured of plasmids p01 and p03 | 17 |
| AH3051 | AH1263 nuc::LtrB nuc2::Erm | 67 |
| AH1919 | AH1263 mutant deficient in extracellular protease production (aur, scpA, splABCDEF, sspAB negative) | 29 |
| NE766 | JE2 icaC::Erm | NARSA |
| KB8766 | JE2 icaC::Km | This study |
| NE619 | JE2 fnbB::Erm | NARSA |
| KB8714 | JE2 fnbB::Km | This study |
| KB8516 | JE2 glcJ::Km | This study |
| SA113 | Biofilm-forming laboratory strain | 68 |
| KM1001 | SA113 glcJ::Erm | This study |
| RN4220 | Restriction-deficient strain | 69 |
| E. coli | ||
| DH5α | Used for cloning and plasmid isolation | Invitrogen |
| Plasmids | ||
| pJB67 | S. aureus expression vector | Jeffrey Bose |
| pJE11 | pJB7 carrying glcJ | This study |
| PCR primers | ||
| 516F | GTGCGTAAAGCATTAAATGCAG | This study |
| 516R | GATGAAATCTCCTGTTGAATC | This study |
| 619F | AATCATGAGGTGATAAGATG | This study |
| 619R | CCTCAGCAGACCTTTTGGC | This study |
| 766F | GATACTTAACTACACGCG | This study |
| 766R | CCATTGACCTAATAGGAC | This study |
| 516C-FNdeI | CCCCCATATGAGTCGAAAAAT | This study |
| 516C-REcoRI | CCCCGAATTCTCATTTCTGTT | This study |
Erm, erythromycin resistance; Km, kanamycin resistance; LtrB, markerless gene deletion. Sequences are shown 5′→3′; restriction sites are underlined.
NARSA, Network on Antimicrobial Resistance in Staphylococcus aureus (http://www.narsa.net).
TABLE 2.
Defined S. aureus Nebraska Transposon Mutant Library strains analyzed in this study
| Phenotype and strain name | Gene no. | Gene namea | Gene product |
|---|---|---|---|
| Biofilm deficiency | |||
| NE460 | SAUSA300_0955 | atl | Autolysin |
| NE1555 | SAUSA300_1148 | codY | Transcriptional repressor |
| NE516 | SAUSA300_1508 | glcJ | Conserved hypothetical protein |
| NE1607 | SAUSA300_2025 | rsbU | σB regulation protein |
| NE1472 | SAUSA300_2023 | rsbW | Anti-σB factor; serine-protein kinase |
| NE1193 | SAUSA300_0605 | sarA | Accessory regulator A |
| Hypersensitivity to amoxicillin | |||
| NE810 | SAUSA300_1642 | cycA (aapA) | d-Serine/d-alanine/glycine transporter |
| NE1022 | SAUSA300_0959 | fmtA | Autolysis and methicillin resistance-related protein |
| NE1360 | SAUSA300_1255 | fmtC (mprF) | Oxacillin resistance-related protein |
| NE1868 | SAUSA300_0032 | mecA | Penicillin-binding protein 2′ |
| NE62 | SAUSA300_0794 | top | Toprim domain protein |
| NE554 | SAUSA300_1865 | vraR | DNA-binding response regulator |
| Hyperresistance to amoxicillin | |||
| NE292 | SAUSA300_0539 | ilvE | Branched-chain amino acid aminotransferase |
| NE1142 | SAUSA300_2264 | rpiRC | Phosphosugar-binding transcriptional regulator |
| NE1714 | SAUSA300_1590 | rsh (relA) | ppGpp hydrolase/synthetase |
| NE914 | SAUSA300_1622 | tig | Trigger factor |
Alternate gene names are in parentheses.
Static biofilm assay.
Aliquots of inocula (180 μl each) were transferred to the wells of a 96-well microtiter plate (Corning, no. 3599) containing 20 μl of antibiotic dissolved in water at a concentration equal to 10 times the desired final concentration. Control wells were filled with 180 μl of inoculum and 20 μl of water, or 180 of sterile broth and 20 μl of water. The plates were incubated for 18 h. Growth was quantitated by measuring the absorbance of the wells at 450 nm. The amount of biofilm biomass in each well was quantitated by rinsing the wells with water and staining for 1 min with 200 μl of Gram's crystal violet. The wells were then rinsed with water and dried. Bound crystal violet was dissolved in 200 μl of 33% acetic acid and quantitated by measuring the absorbance of the wells at 595 or 620 nm.
Hydrodynamic biofilm assay.
To grow biofilms under various conditions of hydrodynamic flow, 17- by 100-mm polystyrene culture tubes (no. 1485-2810; USA Scientific) were modified as follows. A 1.5-mm hole was drilled in the center of the cap of each tube. A 1.5-mm-diameter polystyrene rod was inserted into the hole so that the rod was suspended in the center of the tube 1 cm from the bottom. The rod was fastened to the cap with epoxy cement, and the rod-cap assembly was sterilized with 70% ethanol. The modified culture tubes were filled with 3 ml of inoculum, or 3 ml of sterile broth as a control, and incubated for 18 h in a gyratory shaker set to 230 rpm. The tubes were incubated with the cap fully closed so the position of the rod was fixed in the center of the tube. Biofilms on the walls of the tube and on the rod were visualized by staining with crystal violet and quantitated by destaining with acetic acid as described above.
Surface attachment assay.
The BioFilm ring test (BioFilm Control, St. Beauzire, France) was used to quantitate attachment of bacterial cells to 96-well polystyrene microtiter plates. The procedure was carried out as described by Chavant et al. (20) with some modifications. Microtiter plates, magnetic beads, a block of 96 magnets in standard microplate format, contrast solution, a plate reader, and image analysis software were obtained from BioFilm Control. Briefly, inocula were supplemented directly with amoxicillin or DNase I at the indicated concentrations, and then aliquots of the inocula (200 μl each) were transferred to the wells of the microtiter plate. Control wells were filled with 200 μl of sterile broth. All the wells were supplemented with 2 μl of magnetic beads (1-μm diameter; no. TON004), and the plate was incubated at 37°C for 3 h. A volume of 100 μl of contrast solution was pipetted into each well, and the plate was then placed on the magnetic block for 1 min. Under these conditions, free beads migrate to the center of the well, forming a spot. Bacterial cells that attach to the bottom of the well block the migration of the beads and reduce the intensity of the spot (21). The plate was imaged using the BioFilm Control plate reader and analyzed with the accompanying image analysis software. The spot intensity was expressed as a biofilm index (BFI) value, which was inversely proportional to the number of attached cells in the well.
Biofilm porosity assay.
The porosity of biofilms was quantitated by measuring the volumetric flow rate of phosphate-buffered saline (PBS) through biofilms cultured in centrifugal filter devices. The procedure was carried out as described by Ganeshnarayan et al. (22) with slight modifications. Briefly, 200-μl aliquots of inoculum supplemented with amoxicillin or DNase I at the indicated concentrations were transferred to the sample reservoirs of Nanosep MF centrifugal filter devices (0.2-μm pore size; no. ODM02C33; Pall Corp.). Control devices were filled with 200 μl of sterile broth. The devices were incubated for 18 h to allow biofilms to form on the filtration membranes. The broth was aspirated, and the reservoirs were filled with 300 μl of PBS. The devices were subjected to low-speed centrifugation (4,300 × g) for 30 s, and flowthrough volumes were measured. The 30-s centrifugation step was repeated five additional times, and the cumulative flowthrough volumes were remeasured after each centrifugation step.
Confocal scanning laser microscopy.
Two-milliliter aliquots of inocula were transferred to 35-mm-diameter glass bottom petri dishes (MatTek). After incubation for 18 h, the broth was aspirated, the dishes were rinsed twice with distilled water, and the biofilms were stained using a LIVE/DEAD BacLight Bacterial Viability kit (Life Technologies) according to the instructions supplied with the kit. Biofilms were imaged using an Olympus FV1000-IX81 inverted confocal microscope equipped with a 60× oil immersion lens. Syto 9 dye was excited by a 488-nm argon laser, and emitted light was collected at 520 nm. Propidium iodide dye was excited by a 559-nm argon laser, and emitted light was collected at 619 nm. Random areas near the center of the dish were selected for viewing. Z-stack images were collected at 1-μm intervals from the base to the top of the biofilm.
EPS-degrading enzymes.
Where indicated, cultures were supplemented with 10 μg/ml DNase I (dornase alfa; Genentech), proteinase K (Sigma), or the PNAG-degrading enzyme dispersin B (7). This was accomplished by adding 0.01 volume of a 1-mg/ml enzyme stock solution to the bacterial inoculum prior to transferring it to the culture vessel.
Isolation and analysis of extracellular DNA.
eDNA was isolated from lawns of bacteria cultured on agar, also known as colony biofilms, using the method described by Karwacki et al. (23). Briefly, 100-μl aliquots of inoculum (>108 CFU) were spread onto agar plates supplemented with 0, 0.1, 0.2, 0.3, 0.4, or 0.5 μg/ml amoxicillin (0× to 0.5× MIC), and the plates were incubated for 24 h. The cell paste from each plate was transferred to a separate, preweighed 1.5-ml microcentrifuge tube; weighed; and resuspended in Tris-EDTA (TE) buffer at a concentration of 1 mg/ml. The tube was mixed by vortex agitation for 10 min, and the cells were pelleted by centrifugation. The supernatant was sterilized by passage through a 0.2-μm-pore-size filter. A 20-μl volume of cell extract was analyzed by agarose gel electrophoresis. eDNA was visualized by staining with ethidium bromide.
Mutant library screen.
The Nebraska Transposon Mutant Library was obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) (http://www.narsa.net). The library was supplied in five 384-well microtiter plates. A volume of 5 μl of each mutant strain was transferred from the well of the 384-well master plate to the well of a 96-well microtiter plate containing 200 μl of fresh broth. The 96-well microtiter plate was incubated at 37°C for 16 h. A 5-μl volume of each culture was then transferred to fresh 96-well microtiter plate wells containing 200 μl of broth supplemented with 0.1, 0.2, or 0.3 μg/ml amoxicillin. The wild-type strain JE2 was used as a positive control, and uninoculated broth served as a negative control. Plates were incubated for 20 ± 2 h. Growth and biofilm were quantitated as described above. The entire mutant library screen was performed on two separate occasions. Approximately 60 mutant strains that exhibited A620 values of <30% that of the control at one or more amoxicillin concentrations on both occasions were considered potential biofilm-deficient mutants. Defined bursa aurealis transposon mutant strains corresponding to each potential mutant were obtained from NARSA and rescreened in the biofilm induction assay described above in 0 to 0.5 μg/ml amoxicillin.
Construction of mutant strains.
Defined bursa aurealis transposon mutant strains NE516 (glcJ), NE619 (fnbB), and NE766 (icaC) were backcrossed to a fresh JE2 background to eliminate possible secondary-site mutations. First, the bursa aurealis transposon, including the erythromycin resistance cassette, was replaced with a kanamycin resistance cassette utilizing the pKAN allelic-exchange plasmid described by Bose et al. (24). Bacteriophage ϕ11 was then propagated on the resulting isolate and used to transduce the mutation into strain JE2 as previously described (25). Transductants were selected on tryptic soy agar plates containing 75 μg/ml kanamycin and then screened for the presence of the kanamycin resistance cassette by PCR utilizing primer pairs 516F/516R, 619F/619R, and 766F/766R (Table 1), which flank the insertion sites of the transposons in NE516, NE619, and NE766, respectively. The reconstructed mutants were named KB8516, KB8619, and KB8766, corresponding to NE516, NE619, and NE766, respectively. Strain KM1001 was constructed by transducing the glcJ::Kan mutation from strain KB8516 to strain SA113 using bacteriophage ϕ11. Transductants were screened for the presence of the kanamycin resistance cassette by PCR using primer pair 516F/516R.
Genetic complementation.
Genetic complementation of KB8516 and KM1001 was achieved by cloning the glcJ open reading frame (ORF) (SAUSA300_1508) into plasmid pJB67, a modified version of plasmid pCN51 (26) that contains an optimized ribosome binding site (27). First, the glcJ ORF from strain JE2 was amplified by PCR using primers 516C-FNdeI and 516C-REcoRI (Table 1). The resulting PCR product (224 bp) was digested with NdeI and EcoRI and ligated into NdeI/EcoRI sites of pJB67. The resulting plasmid (pJE11) was transformed into Escherichia coli DH5α, subjected to DNA sequence analysis to confirm that no errors occurred during PCR amplification, electroporated into the restriction-deficient strain RN4220, and then introduced into strains KB8516 and KM1001 via phage transduction, as described above. All plasmid-harboring strains were cultured in medium supplemented with 10 μg/ml erythromycin to maintain plasmid selection.
Reproducibility of results and statistics.
All biofilm assays were performed in duplicate to quadruplicate wells that exhibited an average variation of approximately 7% for growth assays and 18% for biofilm assays. All assays were performed 2 to 5 times with similarly significant differences in absorbance values. The significance of differences between mean absorbance values was measured using a 2-tailed Student's t test. Correlation between variables was measured using Spearman's rank correlation coefficient. Growth curves were compared using an analysis of covariance (ANCOVA) assay. P values of <0.05 were considered significant.
RESULTS
Low-dose amoxicillin stimulates USA300 biofilm in a static microtiter plate assay.
Previous studies showed that sub-MIC amoxicillin induces eDNA-dependent biofilm formation in the MRSA strain LAC (26, 28), a well-characterized community-associated USA300 isolate (15, 16). Strain JE2, the parent strain of the Nebraska Transposon Mutant Library, is a derivative of strain LAC that has been cured of two plasmids, p01 and p03 (17). We first measured the ability of sub-MIC amoxicillin to induce biofilm in strain JE2. Figure 1A shows that sub-MIC amoxicillin caused strong biofilm induction, with a peak at approximately 0.2 μg/ml (MIC = 1 μg/ml) after 18 h of growth. This pattern is similar to the pattern of biofilm induction exhibited by strain LAC in the presence of low-dose amoxicillin (26), suggesting that the biofilm induction phenotype in strain LAC is independent of the presence or absence of extrachromosomal elements. When cultured in 0.2 μg/ml amoxicillin, strain JE2 exhibited a small decrease in the growth rate and total biomass compared to cells cultured in antibiotic-free medium (Fig. 1A, inset).
FIG 1.
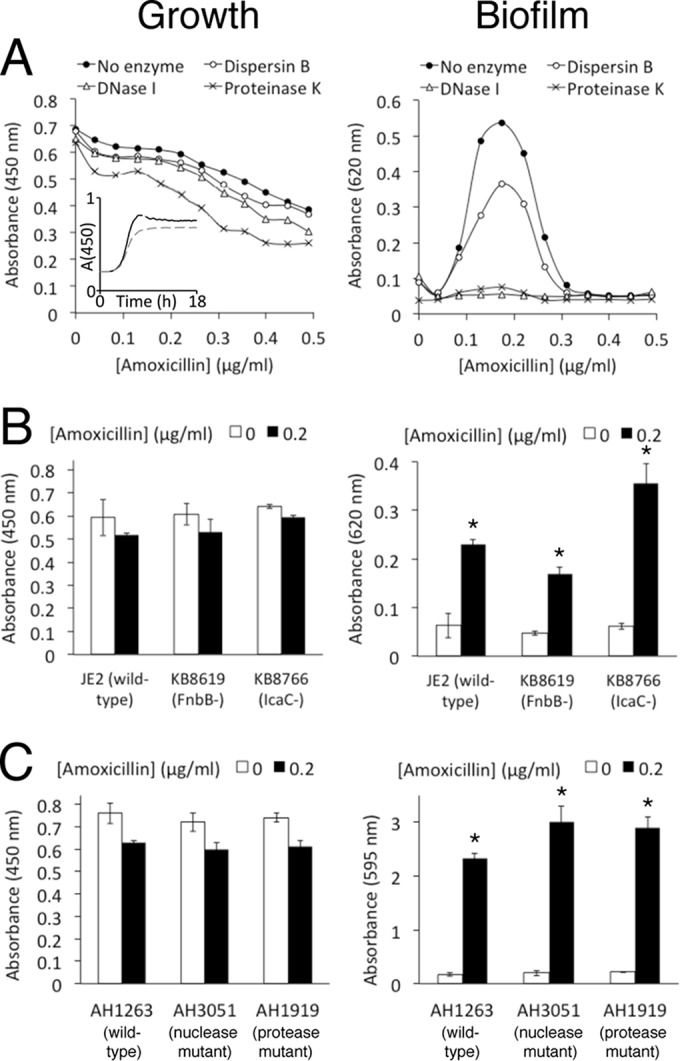
Growth and biofilm formation of wild-type and mutant USA300 strains in static 96-well microtiter plates after 18 h. Broth was supplemented with sub-MIC amoxicillin at the indicated concentrations. (A) Growth and biofilm of strain JE2 in the presence of 10 μg/ml DNase I, dispersin B, or proteinase K. (Inset) Growth of strain JE2 from 0 to 18 h in no antibiotic (solid line) and 0.2 μg/ml amoxicillin (dashed line) in the absence of enzymes. A(450), absorbance at 450 nm. (B) Growth and biofilm of JE2, isogenic fnbB (fibronectin binding protein B) mutant strain KB8619, and isogenic icaC (poly-N-acetyl-d-glucosamine) mutant strain KB8766. (C) Growth and biofilm of wild-type strain AH1263 and isogenic nuclease- and protease-deficient mutant strains AH3051 and AH1919. All the values show means for duplicate wells, and the error bars indicate ranges. Error bars were omitted from panel A for clarity. *, significantly different from no-antibiotic control (P < 0.05).
To investigate the composition of the biofilm matrix formed by strain JE2 in low-dose amoxicillin, we supplemented media with three different EPS-degrading enzymes: DNase I, proteinase K, or the PNAG-degrading enzyme dispersin B (Fig. 1A). None of these enzymes had a major effect on the susceptibility of strain JE2 to amoxicillin killing (Fig. 1A, left). DNase I and proteinase K caused nearly complete inhibition of amoxicillin-induced biofilm, whereas dispersin B caused partial inhibition (Fig. 1A, right). This pattern of inhibition is similar to the pattern of inhibition exhibited by DNase I, proteinase K, and dispersin B against biofilms of strain FPR3757, another USA300 clone, when cultured in low-dose methicillin (16). Figure 1B shows that low-dose amoxicillin also stimulated biofilm in JE2 mutant strains deficient in the production of fibronectin binding protein B (strain KB8619) and PNAG (strain KB8766). Addition of DNase I to the culture medium caused nearly complete inhibition of amoxicillin-induced biofilm in both mutant strains (data not shown). These results suggest that low-dose amoxicillin stimulates predominantly eDNA- and protein-based biofilms in strain JE2.
Previous studies showed that extracellular nucleases and proteases play a role in S. aureus biofilm formation in vitro (29–31). To determine whether extracellular nuclease and protease production contributes to USA300 biofilm formation in low-dose amoxicillin, we cultured strain AH3051, a strain with mutations in extracellular nucleases Nuc1 and Nuc2 (29), and strain AH1919, a strain with mutations in all 10 genes encoding recognized extracellular proteases (31), in broth supplemented with 0.2 μg/ml amoxicillin (Fig. 1C). Both strains were constructed in the background strain AH1263, a derivative of strain LAC that has been cured of plasmid p03 (32) and that exhibits the same pattern of biofilm stimulation in low-dose amoxicillin as strains LAC and JE2 (data not shown). Both mutant strains exhibited biofilm induction in low-dose amoxicillin (Fig. 1C), and addition of DNase I to the culture medium caused nearly complete inhibition of biofilm in both mutant strains (data not shown). These findings suggest that absence of endogenous extracellular nuclease and protease activity does not impact the ability of USA300 to form biofilm in low-dose amoxicillin.
Low-dose amoxicillin stimulates USA300 biofilm under flow conditions.
Figure 2 shows growth and biofilm formation by strain JE2 in polystyrene tubes incubated in a gyratory shaker. Cells cultured in sub-MIC amoxicillin formed more biofilm on the sides of the tubes (Fig. 2A) and on polystyrene rods suspended in the tubes (Fig. 2B) than cells cultured in the absence of antibiotic, as visualized by staining with crystal violet. Addition of DNase I to the medium completely inhibited biofilm formation on the sides of the tubes (Fig. 2C, center graph) and on the polystyrene rods (Fig. 2C, right graph). These findings suggest that low-dose amoxicillin induces physical changes in USA300 cells that cause them to attach to surfaces and form biofilms.
FIG 2.
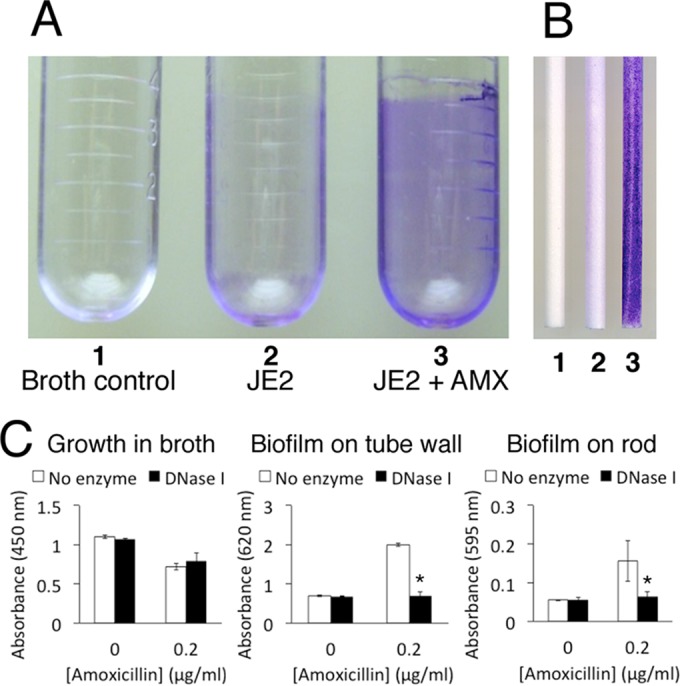
Growth and biofilm formation of MRSA strain JE2 in polystyrene culture tubes incubated in a gyratory shaker. (A) Biofilm formation on the walls of tubes visualized by staining with crystal violet. Tube 1, sterile broth control; tube 2, strain JE2; tube 3, strain JE2 supplemented with 0.2 μg/ml amoxicillin (AMX). (B) Biofilm formation on polystyrene rods suspended in the centers of the tubes in panel A during incubation. Rods 1, 2, and 3 were suspended in tubes 1, 2, and 3, respectively. (C) Quantitation of growth in tubes (in broth), biofilm on the sides of the tubes, and biofilm on the rod in the presence or absence of 10 μg/ml DNase I. The graphs show mean absorbance values from duplicate tubes or rods, and the error bars indicate ranges. *, significantly different from no-enzyme control (P < 0.05).
Low-dose amoxicillin stimulates USA300 surface attachment.
We used the BioFilm ring test (20) to measure the effect of low-dose amoxicillin on the attachment of JE2 cells to polystyrene surfaces. In this assay, inocula were supplemented with magnetic microbeads (1-μm diameter) and incubated in the wells of a 96-well polystyrene microtiter plate. After 3 h, the wells were magnetized using a block of 96 magnets in standard microplate format. Under these conditions, microbeads migrate to the center of the well, forming a spot, but bacterial cells attached to the bottom of the well block the migration of the beads and reduce the intensity of the spot (21). Figure 3A shows photographs of wells that were inoculated with sterile broth, JE2 inoculum, or JE2 inoculum supplemented with low-dose amoxicillin and then incubated for 3 h and magnetized. The spots that formed in wells inoculated with JE2 in the absence of antibiotic were larger than the spots formed in wells inoculated with JE2 in the presence of low-dose amoxicillin. Quantification of spot intensity using image analysis software confirmed that the spot intensity in low-dose amoxicillin was significantly less than the spot intensity in the absence of antibiotic (Fig. 3B). These findings suggest that low-dose amoxicillin increases the ability of JE2 cells to attach to polystyrene.
FIG 3.

Physical characteristics of JE2 biofilms cultured in low-dose amoxicillin. (A and B) Surface attachment assay. (A) Microtiter plate wells were inoculated with sterile broth (Broth control), JE2 inoculum (JE2), or JE2 inoculum supplemented with 0.2 μg/ml amoxicillin (JE2 + AMX). All the wells also contained 1-μm-diameter magnetic microbeads. The microtiter plate was incubated for 3 h, placed on a block of 96 magnets for 1 min, and then imaged using a microplate scanner. Duplicate wells for each condition are shown. (B) Quantitation of the spot intensity in panel A using BioFilm Control software. The spot intensity is expressed in BFI units, which are inversely proportional to the amount of biofilm in the well. *, significantly different (P < 0.05). (C and D) Biofilm porosity assay. (C) Rate of fluid flow through JE2 biofilms (squares), JE2 biofilms cultured in 0.2 μg/ml amoxicillin (triangles), and JE2 biofilms cultured in 0.2 μg/ml amoxicillin plus 10 μg/ml DNase I (diamonds). Fluid flow was measured using centrifugal filter devices. The solid circles show the rate of fluid flow through a control device inoculated with sterile broth. The values show means for duplicate devices. Error bars were omitted for clarity. *, significantly different from strain JE2 (P < 0.05); **, significantly different from strain JE2 plus AMX (P < 0.05). (D) Rate of fluid flow through JE2 biofilms cultured in increasing concentrations of amoxicillin. The values show mean flowthrough volumes and ranges for duplicate centrifugal filter devices after 90 s of centrifugation. *, significantly different from no-antibiotic control (P < 0.05). (E) Confocal scanning laser micrograph of 200-μm2 areas of JE2 biofilms cultured in glass-bottom dishes in the absence or presence of 0.2 μg/ml amoxicillin. The biofilms were stained with LIVE/DEAD stain. Green, live cells; red, dead cells and eDNA; yellow, mixture.
Low-dose amoxicillin decreases USA300 biofilm porosity.
To measure the relative porosity of biofilms cultured in low-dose amoxicillin, we grew JE2 biofilms in 0.2-μm-pore-size spin filters and measured the rate of fluid flow through the biofilms after low-speed centrifugation (Fig. 3C). Biofilms cultured in 0.2 μg/ml amoxicillin (triangles) exhibited a significantly lower rate of fluid flow than biofilms cultured without antibiotic (squares), and DNase I significantly increased the rate of fluid flow in biofilms cultured in low-dose amoxicillin (diamonds). In addition, biofilms cultured in 0.1 to 0.3 μg/ml amoxicillin exhibited significantly lower rates of fluid flow, whereas biofilms cultured in 0.4 to 0.5 μg/ml amoxicillin did not (Fig. 3D). There was a significant negative correlation (ρ = −0.941) between the flow rates exhibited by biofilms cultured in spin filters (Fig. 3D) and the amount of biofilm formed in 96-well microtiter plates (Fig. 1A, right) for amoxicillin concentrations ranging from 0 to 0.5 μg/ml.
USA300 biofilms cultured in low-dose amoxicillin exhibit an altered morphology.
Confocal scanning laser microscopy of JE2 biofilms cultured in glass bottom dishes (Fig. 3E) revealed that biofilms grown in low-dose amoxicillin were thicker than biofilms grown in the absence of antibiotic (approximately 75 μm versus 25 μm, respectively). In addition, biofilms grown in low-dose amoxicillin exhibited more tower- and pillar-shaped structures than biofilms grown in the absence of antibiotic (Fig. 3E).
eDNA is necessary but not sufficient to induce USA300 biofilm.
The maximum amount of biofilm stimulation in strain JE2 occurs at 0.2 μg/ml amoxicillin (Fig. 1A, right). To determine whether the amount of biofilm stimulation correlates with the amount of eDNA released, we isolated eDNA from JE2 colony biofilms cultured in increasing concentrations of amoxicillin (Fig. 4). We also isolated eDNA from colony biofilms of a JE2 mutant strain deficient in the production of Atl autolysin protein (strain NE460). This mutant strain produces significantly less biofilm in sub-MIC amoxicillin than the parental strain, JE2 (see below). Strain JE2 produced more eDNA at 0.2 μg/ml amoxicillin than strain NE460, demonstrating that eDNA release correlates with biofilm. However, both strains produced abundant eDNA at 0.4 to 0.5 μg/ml amoxicillin, although neither strain produced biofilm at these amoxicillin concentrations. These results demonstrate that biofilm stimulation correlates with eDNA release but that eDNA release alone is not sufficient for biofilm formation.
FIG 4.
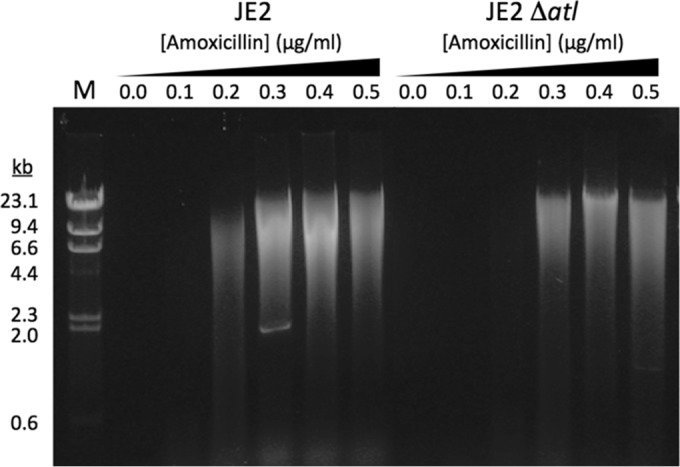
Low-dose amoxicillin induces eDNA release in JE2 and in a JE2 autolysin mutant (strain NE460; JE2 Δatl). Extracellular DNA was isolated from colony biofilms cultured on increasing concentrations of amoxicillin and analyzed by agarose gel electrophoresis. The sizes of molecular size markers (lane M) are indicated on the left.
Low doses of d-cycloserine and fosfomycin also stimulate eDNA-dependent biofilm in strain USA300.
We tested whether low doses of d-cycloserine and fosfomycin, two cell wall-active agents with mechanisms of action different from that of β-lactam antibiotics, could also stimulate JE2 biofilm (Fig. 5). Both agents strongly stimulated JE2 biofilm with a peak of stimulation at approximately 0.1× MIC (Fig. 5A). Biofilm stimulation by both agents was inhibited by DNase I (Fig. 5B).
FIG 5.
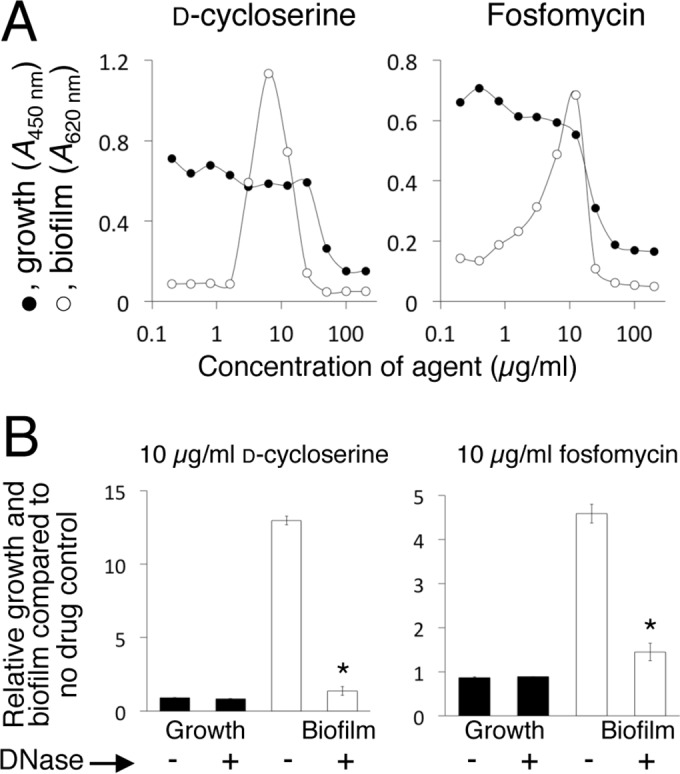
d-Cycloserine and fosfomycin induce eDNA-dependent biofilm in MRSA strain JE2. (A) Growth and biofilm of MRSA strain JE2 in 96-well microtiter plates in the presence of increasing concentrations of d-cycloserine or fosfomycin. The graphs show mean absorbance values from duplicate wells. Error bars were omitted for clarity. (B) Effect of 10 μg/ml DNase I on growth and biofilm of JE2 in the presence of 10 μg/ml d-cycloserine or fosfomycin. The presence (+) or absence (−) of DNase I in the culture is indicated along the bottom. The values show relative growth and biofilm in the presence of the agent compared to growth and biofilm in the no-drug control (absorbance with drug/absorbance without drug × 100). The error bars show the ranges of values from duplicate wells. *, significantly different from no-DNase I control (P < 0.05).
Isolation of biofilm induction mutants.
To identify mutants deficient in the biofilm induction phenotype, we cultured 1,920 NTML mutant strains in low-dose amoxicillin (0.1 to 0.3 μg/ml) and screened for mutants that formed decreased biofilm compared to the parental strain, JE2. Among 60 candidate mutant strains identified in the initial screen, 16 were confirmed to be deficient in biofilm formation in sub-MIC amoxicillin when tested in the biofilm induction assay in 0 to 0.5 μg/ml amoxicillin using defined bursa aurealis transposon mutant strains (Table 2 and Fig. 6). The biofilm-deficient mutants were classified into three groups based on their susceptibility to amoxicillin and biofilm phenotype. The first group, which comprised atl, codY, rsbU, rsbW, sarA (staphylococcal accessory regulator), and a novel gene named glcJ (glucose metabolism; see below), exhibited susceptibility to amoxicillin similar to that exhibited by the wild-type strain JE2 but exhibited decreased biofilm in low-dose amoxicillin (Fig. 6A). The second group, which comprised cycA, fmtA, fmtC, mecA, vraR, and top (Toprim domain protein), exhibited increased susceptibility to amoxicillin killing (hypersensitive) (Fig. 6B). The biofilm induction response in this group of mutants generally exhibited a decreased amplitude and/or a lower stimulatory amoxicillin dose range. The third group, which comprised ilvE, rsh, rpiRC, and tig, exhibited decreased susceptibility to amoxicillin killing and a higher stimulatory amoxicillin dose range (hyperresistant) (Fig. 6C). Several of these genes have previously been shown to play a role in S. aureus β-lactam resistance (fmtA, fmtC, rsh, and vraR), biofilm formation (codY, rsbU, and rsbW), or both processes (atl, mecA, and sarA) (4, 10, 16, 33–39). Two genes (ilvE and fmtA) were previously shown to be induced by cell wall stress (36), and one gene (atl) was previously shown to be required for induction of biofilm by sub-MIC methicillin in strain LAC (16). The identification of numerous known mediators of S. aureus β-lactam resistance and biofilm formation demonstrates that the inducible biofilm phenotype exhibited by USA300 in low-dose amoxicillin is similar to biofilm phenotypes exhibited by other S. aureus strains under various conditions.
FIG 6.
Growth and biofilm of 16 defined NTML mutant strains in the presence of increasing concentrations of sub-MIC amoxicillin. The mutant strains correspond to those listed in Table 2. The name of the inactivated gene in each mutant strain is shown above each set of graphs. The x axes indicate amoxicillin concentrations (in micrograms per milliliter), and the y axes indicate absorbance (450 nm for growth and 620 nm for biofilm). The values show mean absorbance values from 2 to 5 assays. Error bars were omitted for clarity. (A) Biofilm-deficient mutants. (B) Amoxicillin-hypersensitive mutants. (C) Amoxicillin-hyperresistant mutants.
Our mutant library screen also uncovered several novel genes not previously associated with β-lactam resistance, biofilm formation, or cell wall stress in S. aureus. They included glcJ, which exhibited decreased biofilm in the presence of sub-MIC amoxicillin (Fig. 6A); cycA and top, which exhibited increased susceptibility to killing by amoxicillin (Fig. 6B); and ilvE, tig, and rpiRC, which exhibited decreased susceptibility to amoxicillin killing (Fig. 6C). Among these, only tig (trigger factor) has previously been shown to be involved in biofilm in other species of bacteria (40, 41).
glcJ plays a role in glucose metabolism and general biofilm formation.
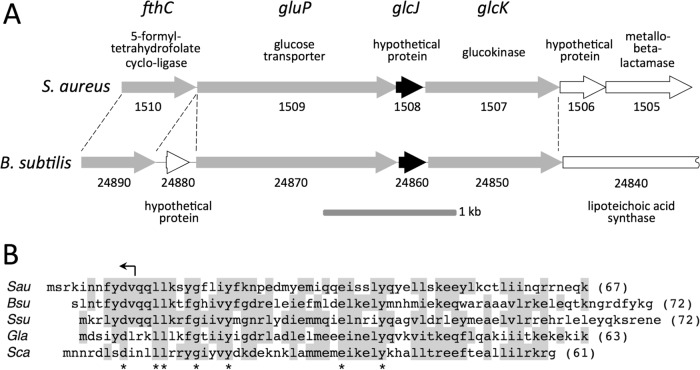
The bursa aurealis transposon in mutant strain NE516 inserted into a novel gene (SAUSA300_1508) that we named glcJ. The glcJ gene is located in a cluster of six genes (SAUSA300_1510 to SAUSA300_1505), which suggests that these genes are translationally coupled and may constitute an operon (Fig. 7A, top). The glcJ gene overlaps the upstream ORF (SAUSA300_1509) by 20 bp and the downstream ORF (SAUSA300_1507) by 4 bp. The glcJ gene and its flanking ORFs (SAUSA300_1509 to SAUSA300_1507) are homologous to the glucokinase operon (gluP-yqgQ-glcK) of Bacillus subtilis (Fig. 7A, bottom), which plays a role in glucose transport and metabolism (42, 43). The glcJ gene encodes a 67-amino-acid-residue protein with homologues present only in Bacilli (Fig. 7B).
FIG 7.
Genetic map and translated amino acid sequence of S. aureus glcJ. (A) Genetic maps of S. aureus glcJ and flanking regions and the homologous B. subtilis glucokinase operon and flanking regions. The arrows indicate ORFs and the direction of transcription. Solid arrows, glcJ; shaded arrows, genes with homologues in both species; open arrows, genes unique to S. aureus or B. subtilis. The numbers below the arrows are gene numbers based on the USA300 numbering system for S. aureus (GenBank accession number NC_007793) and the strain 168 numbering system for B. subtilis (GenBank accession number AL009126). Gene names and putative functions are shown above the S. aureus map. Putative functions of genes unique to B. subtilis are shown below the B. subtilis map. The dashed lines demarcate homologous regions. (B) Amino acid alignment of S. aureus GlcJ (Sau) with homologues from B. subtilis (Bsu), Streptococcus suis (Ssu), Gracilibacillus lacisalsi (Gla), and Salinicoccus carnicancri (Sca). Amino acids present in more than one sequence are shaded. The asterisks indicate amino acids conserved in all five sequences. Protein lengths are in parentheses. The arrow above the S. aureus sequence indicates the location and direction of the transposon insertion in mutant strains NE516, KB8516, and KM1001.
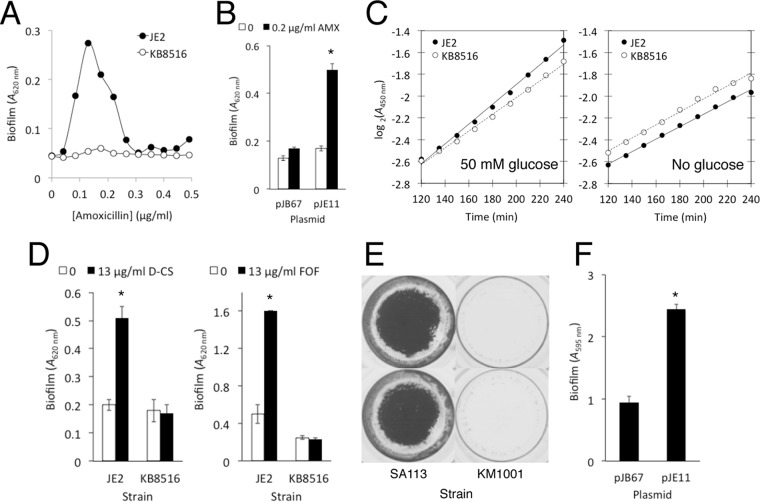
To confirm that glcJ contributes to amoxicillin-induced biofilm in strain JE2, we backcrossed the mutation from the defined bursa aurealis transposon mutant strain NE516 to a clean JE2 background in order to eliminate possible secondary-site mutations. The resulting reconstructed mutant, designated KB8516, exhibited the same biofilm-deficient phenotype in low-dose amoxicillin as strain NE516 (Fig. 6A and 8A.). A plasmid harboring the wild-type glcJ gene restored the ability of strain KB8516 to form biofilm in low-dose amoxicillin (Fig. 8B). The mutant strain KB8516 exhibited a lower growth rate than strain JE2 in medium supplemented with 50 mM glucose (Fig. 8C, left), but not in medium lacking glucose (Fig. 8C, right), suggesting that glcJ plays a role in glucose metabolism. In addition, the glcJ mutant strain KB8516 was deficient in biofilm formation in sub-MIC d-cycloserine and fosfomycin (Fig. 8D).
FIG 8.
Biofilm formation by S. aureus strains JE2 and SA113 and their isogenic glcJ mutants, KB8516 and KM1001, respectively, in 96-well microtiter plates. (A) Biofilm formation by strains JE2 and KB8516 in increasing concentrations of amoxicillin. (B) Biofilm formation by KB8516 harboring plasmid pJB67 (vector control) or pJE11 (glcJ) in no antibiotic (0) or 0.2 μg/ml amoxicillin. The values show means and ranges for duplicate wells. *, significantly different from the pJB67 control (P < 0.05). (C) Growth curves for strains JE2 and KB8516 in static microtiter plates. Bacteria were cultured in 17 g/liter tryptone, 3 g/liter Soytone (BD Biosciences), 5 g/liter NaCl, 2.5 g/liter K2HPO4 supplemented with 9 g/liter (50 mM) glucose or no glucose. The graphs show mean absorbance values from 3 or 4 wells. (D) Biofilm formation by JE2 and KB8515 in 0 μg/ml or 13 μg/ml d-cycloserine (D-CS) or fosfomycin (FOF). The values show means and ranges from duplicate wells. *, significantly different from the no-drug control (P < 0.05). (E) Biofilm formation by S. aureus strains SA113 (wild type) and KM1001 (SA113 ΔglcJ) in antibiotic-free medium. The biofilm was visualized by staining with crystal violet. Duplicate wells of each strain are shown. (F) Biofilm formation by strain KM1001 harboring plasmid pJB67 (vector control) or pJE11 (glcJ) in antibiotic-free medium. The values show means and ranges for duplicate wells. *, significantly different from the pJB67 control (P < 0.05).
To determine whether the glcJ mutant strain KB8516 is specifically impaired in its ability to form biofilm in low-dose amoxicillin, we transduced the glcJ mutation from strain KB8516 to S. aureus strain SA113, a laboratory strain that forms robust biofilms in the absence of low-dose antibiotics. The resulting mutant strain, named KM1001, exhibited significantly less biofilm than the wild-type strain SA113 in drug-free medium (Fig. 8E), and a plasmid harboring the wild-type glcJ gene restored the ability of strain KM1001 to form biofilm in drug-free medium (Fig. 8F). These results suggest that glcJ is a novel gene involved in glucose metabolism and general biofilm formation.
DISCUSSION
Previous studies showed that sub-MIC levels of β-lactam antibiotics induce biofilm formation in genetically diverse MRSA strains in vitro (16, 28, 44–46). This process is of interest because it can provide information about biofilm regulation and the signaling pathways involved in global gene regulation in response to cell wall stress (16). It may also be relevant in environments where bacteria are exposed to low levels of antibiotics, such as hospitals and farms. In the present study, we investigated this process by measuring the effects of low doses of amoxicillin on cells and biofilms of MRSA strain LAC. This strain was chosen because it represents a well-characterized epidemic community-associated USA300 isolate (17, 18), and it is the parent strain of the Nebraska Transposon Mutant Library, which is an arrayed collection of 1,920 transposon mutants that is useful for phenotypic screening and the identification of candidate genes for future research (19).
Using EPS-degrading enzymes, we found that biofilms formed by strain LAC in sub-MIC amoxicillin contained eDNA and proteinaceous adhesins in their matrices, with PNAG polysaccharide playing only a minor role in biofilm cohesion (Fig. 1A). These results are consistent with those of previous studies showing that most MRSA strains form eDNA- and protein-based biofilms in the absence of antibiotics (10). Biofilms formed by MRSA strains FPR3757 (USA300) and 11490 (USA 500) in low-dose methicillin were also shown to contain primarily eDNA and proteinaceous adhesins in their matrices (16). We also found that LAC strains deficient in the production of extracellular nucleases and proteases formed strong biofilms in low-dose amoxicillin (Fig. 1C), suggesting that the weak biofilm phenotype exhibited by strain LAC in the absence of antibiotics and the strong biofilm phenotype exhibited in the presence of low-dose amoxicillin are not mediated by up- or downregulation of nuclease and protease activity (29–31).
The fact that strain LAC formed biofilms in low-dose amoxicillin under flow conditions (Fig. 2) suggests that biofilm formation in low-dose amoxicillin is an inducible phenotype resulting from physical changes in LAC cells that promote intercellular adhesion and surface attachment and not simply an artifact of the static microtiter plate biofilm assay. This is important because amoxicillin can cause cell lysis and eDNA release. It is possible that at certain amoxicillin concentrations in the static biofilm assay, there is an optimal balance between cell lysis and cell growth. Under these conditions, the cells may simply become trapped in an adhesive eDNA matrix that may not be representative of a true biofilm. The formation of eDNA-dependent biofilm under flow conditions is inconsistent with this model. In addition, we found that LAC cells exposed to low-dose amoxicillin became more adherent to surfaces, and LAC biofilms cultured in low-dose amoxicillin were thicker and less porous and contained more three-dimensional (3D) mushroom- and pillar-like structures with signs of water channel development than biofilms cultured in the absence of drug (Fig. 3). Taken together, these data suggest that the USA300 strain LAC biofilm phenotype can be induced by low-dose amoxicillin.
The S. aureus major autolysin protein, Atl, is a murein hydrolase involved in peptidoglycan turnover and daughter cell separation (47–49). Atl has also been shown to play a role in eDNA release and biofilm formation in clinical MRSA strains (13, 50). In fact, an atl mutant of strain LAC was deficient in biofilm formation in low-dose methicillin (16). Recent evidence suggests that the function of AtlA in biofilm formation involves lysis of the cells and release of genomic DNA into the biofilm matrix during early biofilm formation (51). To study the role of Atl-mediated eDNA release in LAC biofilm formation in low-dose amoxicillin, we analyzed eDNA produced by strain LAC, and by an isogenic atl mutant, in the presence of increasing concentrations of sub-MIC amoxicillin (0 to 0.5× MIC) (Fig. 4). The production of eDNA increased with increasing concentrations of sub-MIC amoxicillin in both strains, presumably due to amoxicillin-mediated cell lysis. In strain LAC, however, eDNA production correlated with biofilm formation at amoxicillin concentrations ranging from 0 to 0.2 μg/ml, but not at amoxicillin concentrations ranging from 0.3 to 0.5 μg/ml. In addition, the atl mutant strain produced abundant eDNA, but little biofilm, at amoxicillin concentrations ranging from 0.3 to 0.5 μg/ml (Fig. 4 and 6). These findings suggest that eDNA release is necessary but not sufficient for biofilm formation by strain LAC in sub-MIC amoxicillin and that other adhesins, such as eDNA binding proteins or eDNA binding polysaccharides, may also be involved in biofilm cohesion (52).
Our results demonstrate that sub-MIC levels of two non-β-lactam cell wall-active antibiotics, d-cycloserine and fosfomycin, also induced eDNA-dependent biofilm formation in strain LAC (Fig. 5). d-Cycloserine and fosfomycin inhibit different steps in the intracellular pathway leading to the biosynthesis of lipid II, the bacterial cell wall precursor (53). In addition, sublethal doses of the glycopeptide antibiotic vancomycin have previously been shown to induce eDNA-dependent biofilm in several vancomycin-susceptible and vancomycin-resistant MRSA strains (44, 54–56). Vancomycin binds to the terminal d-Ala-d-Ala residues of NAG/NAM peptides, thereby blocking proper cell wall assembly (54). Taken together, these findings suggest that biofilm formation may be a general response to cell wall stress. In one study, however, sublethal doses of several non-cell wall-active antibiotics, including linezolid, daptomycin, tigecycline, trimethoprim-sulfamethoxazole, and rifampin, failed to induce biofilm in strain LAC (28), which suggests that biofilm formation may not be a response to all cell stressors.
A screen of the Nebraska Transposon Mutant Library identified 16 mutants deficient in biofilm formation in low-dose amoxicillin (Table 2 and Fig. 6). Three of these genes encode well-studied mediators of S. aureus biofilm formation and β-lactam resistance: atl (discussed above), sarA, and mecA. Mutations in sarA decrease S. aureus biofilm and increase antibiotic susceptibility both in vitro and in vivo (31, 57), whereas mutations in mecA, which encodes penicillin binding protein 2a, impede resistance to β-lactam antibiotics (58). Several other genes identified in our screen have also been associated with biofilm formation (codY), β-lactam resistance (fmtC and vraR), or both processes (rsbU, rsbW, fmtA, and rsh) (4, 10, 16, 33–39). In addition, two genes (ilvE and fmtA) were previously shown to be induced by cell wall stress (36), and four genes (sarA, atl, codY, and rsbU) were specifically shown to limit biofilm in strain USA300 (38). The identification of numerous known mediators of S. aureus β-lactam resistance and biofilm formation validate the Nebraska Transposon Mutant Library screen and highlight the complex and highly interactive circuits that regulate these processes.
Genome-wide transcriptional profiling of S. aureus in response to challenge by a variety of cell wall-active antibiotics has revealed a cell wall stress stimulon consisting of 17 genes that exhibit altered expression in response to these agents (36, 59, 60). Several of the genes encode enzymes involved in cell wall metabolism, and their induction may constitute a bacterial response that protects and repairs the damaged cell wall. Only two of the genes that we identified in our screen for biofilm induction mutants (atl and fmtA) are members of the cell wall stress stimulon. In addition, the cell wall stress stimulon has been shown to be regulated by the vraSR two-component system (36), yet the vraR mutant characterized in our screen exhibited a level of biofilm induction similar to that exhibited by the wild-type strain, albeit at a lower stimulatory-dose range (Fig. 6B). Taken together, these findings suggest that the cell wall stress-induced biofilm phenotype may be regulated by a pathway different than that regulating the cell wall stress stimulon. However, it is difficult to directly compare the results of previous studies on the cell wall stress stimulon with those of the present study because previous studies tested antibiotics at concentrations greater than or equal to the MIC (36, 59, 60), whereas the antibiotic concentrations that induced biofilm in the present study were less than the MIC. Previous studies also showed that β-lactam-induced biofilm in strain USA300 is not affected by mutations in the agr quorum-sensing system (16). Other potential regulators of amoxicillin-induced biofilm that were identified in our screen include CodY, a stationary-phase regulatory protein that targets more than 200 S. aureus genes (61); rpiRC and rsh, which may regulate biofilm formation by affecting the biosynthesis of the alarmone (p)ppGpp (62); and SAUSA300_0794, a putative topoisomerase that may play a role in the transcription of stress response genes (63). More experiments are needed to identify the regulators of amoxicillin-induced biofilm in strain USA300.
Besides the vraR mutant, five other mutants (cycA, fmtA, fmtC, mecA, and top) exhibited increased susceptibility to amoxicillin killing and a concomitant decreased biofilm induction amplitude and/or a lower stimulatory amoxicillin dose range compared to those exhibited by the wild-type strain LAC (Fig. 6B). Conversely, a higher stimulatory amoxicillin dose range was characteristic of all four mutants that exhibited decreased susceptibility to amoxicillin killing (ilvE, rpiRC, rsh, and tig) (Fig. 6C). These findings are consistent with those of Ng et al. (28), who showed that the methicillin concentration that induced maximum biofilm formation in 39 different MRSA clinical isolates was inversely proportional to the susceptibility of each strain to methicillin. These observations help further validate the biofilm induction phenotype and the Nebraska Transposon Mutant Library screen.
Our results identified the novel gene glcJ as a potential effector of S. aureus biofilm formation. Homologues of glcJ are present in the genomes of most Bacilli, but not in the genomes of other classes of Firmicutes (Mollicutes and Clostridia) or other Bacteria. In most Bacilli, the glcJ homologue is translationally coupled to glucokinase (glcK). In S. pneumoniae, glcK is located 3 kb upstream from the glcJ homologue. In addition, gluP, which encodes a glucose exporter (43), is translationally coupled to glcJ in S. aureus and B. subtilis (Fig. 7). The three-dimensional structure of the B. subtilis GlcJ homologue, YqgQ, comprises three α-helical bundles with inferred single-stranded nucleic acid binding activity based on sequence and structural homology (64). The role of glcJ is unknown, but its proximity to gluP and glcK suggests that it may play a role in glucose metabolism. Consistent with this hypothesis, a glcJ mutant of strain LAC exhibited a doubling time in medium supplemented with glucose, but not in medium lacking glucose (Fig. 8C), longer than the doubling time exhibited by the wild-type strain. Previous studies showed that glucose stimulates MRSA cell death (65) and biofilm formation (12, 66).
In summary, our results link cell wall stress and biofilm formation in MRSA. Our assay provides a simple model for studying an antibiotic-induced biofilm phenotype and a screen to identify novel effectors of drug resistance and biofilm formation in MRSA. Since cell wall synthesis and biofilm formation are both viable targets for new antimicrobial drugs, understanding cell wall stress-induced biofilm could facilitate target discovery and the identification of novel cell wall-active agents that enhance the activity of currently used antimicrobial drugs. More experiments are needed to determine whether sub-MIC levels of antibiotics enhance biofilm formation in clinical settings or contribute to the evolution of antibiotic-resistant bacteria in agricultural settings.
ACKNOWLEDGMENTS
We thank Jeffrey Bose (University of Kansas Medical Center) for providing plasmid pJB67, Karen LoVetri (Kane Biotech) for providing dispersin B, Thierry Bernardi and Christian Provost (BioFilm Control) for providing equipment and supplies for the BioFilm ring test, Donald A. Campbell and Ben Kussin-Shoptaw (American University) for technical assistance, and three anonymous reviewers for helpful comments.
The work was funded by NIH grants AI097182 (to J.B.K.) and AI083211 (to K.W.B.) and by American University.
REFERENCES
- 1.Stryjewski ME, Corey GR. 2014. Methicillin-resistant Staphylococcus aureus: an evolving pathogen. Clin Infect Dis 58:S10–S19. doi: 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- 2.Otto M. 2013. Community-associated MRSA: what makes them special? Int J Med Microbiol 303:324–330. doi: 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lencastre H, Oliveira D, Tomasz A. 2007. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiol 10:428–435. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O'Gara JP. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol 190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCourt J, O'Halloran DP, McCarthy H, O'Gara JP, Geoghegan JA. 2014. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol Lett 353:157–164. doi: 10.1111/1574-6968.12424. [DOI] [PubMed] [Google Scholar]
- 7.Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol 74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol 20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 9.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol 178:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, O'Gara JP. 2012. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog 8:e1002626. doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, O'Gara JP. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol 45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzpatrick F, Humphreys H, O'Gara JP. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol 43:1973–1976. doi: 10.1128/JCM.43.4.1973-1976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP. 2011. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun 79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudkin JK, Edwards AM, Bowden MG, Brown EL, Pozzi C, Waters EM, Chan WC, Williams P, O'Gara JP, Massey RC. 2012. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J Infect Dis 205:798–806. doi: 10.1093/infdis/jir845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzpatrick F, Humphreys H, O'Gara JP. 2006. Environmental regulation of biofilm development in methicillin-resistant and methicillin-susceptible Staphylococcus aureus clinical isolates. J Hosp Infect 62:120–122. doi: 10.1016/j.jhin.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan JB, Izano EA, Gopal P, Karwacki MT, Kim S, Bose JL, Bayles KW, Horswill AR. 2012. Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 3:e00198-12. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller LG, Perdreau-Remington F, Rieg G, Mehdi S, Perlroth J, Bayer AS, Tang AW, Phung TO, Spellberg B. 2005. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med 352:1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 18.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 19.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of non-essential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavant P, Gaillard-Martinie B, Talon R, Hébraud M, Bernardi T. 2007. A new device for rapid evaluation of biofilm formation potential by bacteria. J Microbiol Methods 68:605–612. doi: 10.1016/j.mimet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Puig C, Domenech A, Garmendia J, Langereis JD, Mayer P, Calatayud L, Liñares J, Ardanuy C, Marti S. 2014. Increased biofilm formation by nontypeable Haemophilus influenzae isolates from patients with invasive disease or otitis media versus strains recovered from cases of respiratory infections. Appl Environ Microbiol 80:7088–7095. doi: 10.1128/AEM.02544-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganeshnarayan K, Shah SM, Libera MR, Santostefano A, Kaplan JB. 2009. Poly-N-acetylglucosamine matrix polysaccharide impedes fluid convection and transport of the cationic surfactant cetylpyridinium chloride through bacterial biofilms. Appl Environ Microbiol 75:1308–1314. doi: 10.1128/AEM.01900-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karwacki MT, Kadouri DE, Bendaoud M, Izano EA, Sampathkumar V, Inzana TJ, Kaplan JB. 2013. Antibiofilm activity of Actinobacillus pleuropneumoniae serotype 5 capsular polysaccharide. PLoS One 8:e63844. doi: 10.1371/journal.pone.0063844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol 204:587–636. doi: 10.1016/0076-6879(91)04029-N. [DOI] [PubMed] [Google Scholar]
- 26.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol 70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller WG, Lindow SE. 1997. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191:149–153. doi: 10.1016/S0378-1119(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 28.Ng M, Epstein SB, Callahan MT, Piotrowski BO, Simon GL, Roberts AD, Keiser JF, Kaplan JB. 2014. Induction of MRSA biofilm by low-dose β-lactam antibiotics: specificity, prevalence and dose-response effects. Dose Response 12:152–161. doi: 10.2203/dose-response.13-021.Kaplan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714. doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beenken KE, Spencer H, Griffin LM, Smeltzer MS. 2012. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect Immun 80:1634–1638. doi: 10.1128/IAI.06134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, Skinner RA, Tackett AJ, Horswill AR, Smeltzer MS. 2012. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol 86:1183–1196. doi: 10.1111/mmi.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiger T, Goerke C, Fritz M, Schäfer T, Ohlsen K, Liebeke M, Lalk M, Wolz C. 2010. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun 78:1873–1883. doi: 10.1128/IAI.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JH, Kim CH, Hacker J, Ziebuhr W, Lee BK, Cho SH. 2008. Molecular characterization of regulatory genes associated with biofilm variation in a Staphylococcus aureus strain. J Microbiol Biotechnol 18:28–34. [PubMed] [Google Scholar]
- 35.Komatsuzawa H, Ohta K, Fujiwara T, Choi GH, Labischinski H, Sugai M. 2001. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 203:49–54. doi: 10.1111/j.1574-6968.2001.tb10819.x. [DOI] [PubMed] [Google Scholar]
- 36.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Verma V, Belcheva A, Singh A, Fridman M, Golemi-Kotra D. 2012. Staphylococcus aureus methicillin-resistance factor fmtA is regulated by the global regulator SarA. PLoS One 7:e43998. doi: 10.1371/journal.pone.0043998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atwood DN, Loughran AJ, Courtney AP, Anthony AC, Meeker DG, Spencer HJ, Gupta RK, Lee CY, Beenken KE, Smeltzer MS. 2015. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiologyopen 4:436–451. doi: 10.1002/mbo3.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Götz F. 2002. Staphylococcus and biofilms. Mol Microbiol 43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 40.Wen ZT, Suntharaligham P, Cvitkovitch DG, Burne RA. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect Immun 73:219–225. doi: 10.1128/IAI.73.1.219-225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wickström C, Chávez de Paz L, Davies JR, Svensäter G. 2013. Surface-associated MUC5B mucins promote protease activity in Lactobacillus fermentum biofilms. BMC Oral Health 13:43. doi: 10.1186/1472-6831-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesak LR, Mesak FM, Dahl MK. 2004. Bacillus subtilis GlcK activity requires cysteines within a motif that discriminates microbial glucokinases into two lineages. BMC Microbiol 4:6. doi: 10.1186/1471-2180-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mesak LR, Mesak FM, Dahl MK. 2004. Expression of a novel gene, gluP, is essential for normal Bacillus subtilis cell division and contributes to glucose export. BMC Microbiol 4:13. doi: 10.1186/1471-2180-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirani ZA, Jamil N. 2011. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J Basic Microbiol 51:191–195. doi: 10.1002/jobm.201000221. [DOI] [PubMed] [Google Scholar]
- 45.Haddadin RN, Saleh S, Al-Adham IS, Buultjens TE, Collier PJ. 2010. The effect of subminimal inhibitory concentrations of antibiotics on virulence factors expressed by Staphylococcus aureus biofilms. J Appl Microbiol 108:1281–1291. doi: 10.1111/j.1365-2672.2009.04529.x. [DOI] [PubMed] [Google Scholar]
- 46.Subrt N, Mesak LR, Davies J. 2011. Modulation of virulence gene expression by cell wall active antibiotics in Staphylococcus aureus. J Antimicrob Chemother 66:979–984. doi: 10.1093/jac/dkr043. [DOI] [PubMed] [Google Scholar]
- 47.Yamada S, Sugai M, Komatsuzawa H, Nakashima S, Oshida T, Matsumoto A, Suginaka H. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J Bacteriol 178:1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Götz F. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett 259:260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 49.Götz F, Heilmann C, Stehle T. 2014. Functional and structural analysis of the major amidase (Atl) in Staphylococcus. Int J Med Microbiol 304:156–163. doi: 10.1016/j.ijmm.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 50.McCarthy H, Rudkin JK, Black NS, Gallagher L, O'Neill E, O'Gara JP. 2015. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front Cell Infect Microbiol 5:1. doi: 10.3389/fcimb.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bose JL, Lehman MK, Fey PD, Bayles KW. 2012. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One 7:e42244. doi: 10.1371/journal.pone.0042244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okshevsky M, Regina VR, Meyer RL. 2015. Extracellular DNA as a target for biofilm control. Curr Opin Biotechnol 33:73–80. doi: 10.1016/j.copbio.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Dengler V, Meier PS, Heusser R, Berger-Bächi B, McCallum N. 2011. Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics. BMC Microbiol 11:16. doi: 10.1186/1471-2180-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdelhady W, Bayer AS, Seidl K, Moormeier DE, Bayles KW, Cheung A, Yeaman MR, Xiong YQ. 2014. Impact of vancomycin on sarA-mediated biofilm formation: role in persistent endovascular infections due to methicillin-resistant Staphylococcus aureus. J Infect Dis 209:1231–1240. doi: 10.1093/infdis/jiu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdelhady W, Bayer AS, Seidl K, Nast CC, Kiedrowski MR, Horswill AR, Yeaman MR, Xiong YQ. 2013. Reduced vancomycin susceptibility in an in vitro catheter-related biofilm model correlates with poor therapeutic outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57:1447–1454. doi: 10.1128/AAC.02073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu CY, Lin MH, Chen CC, Chien SC, Cheng YH, Su IN, Shu JC. 2011. Vancomycin promotes the bacterial autolysis, release of extracellular DNA, and biofilm formation in vancomycin-non-susceptible Staphylococcus aureus. FEMS Immunol Med Microbiol 63:236–247. doi: 10.1111/j.1574-695X.2011.00846.x. [DOI] [PubMed] [Google Scholar]
- 57.Weiss EC, Zielinska A, Beenken KE, Spencer HJ, Daily SJ, Smeltzer MS. 2009. Impact of sarA on daptomycin susceptibility of Staphylococcus aureus biofilms in vivo. Antimicrob Agents Chemother 53:4096–4102. doi: 10.1128/AAC.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peacock SJ, Paterson GK. 2015. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem 84:577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 59.McAleese F, Wu SW, Sieradzki K, Dunman P, Murphy E, Projan S, Tomasz A. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J Bacteriol 188:1120–1133. doi: 10.1128/JB.188.3.1120-1133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, Singh VK, Jayaswal RK, Wilkinson BJ. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 61.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jolivet-Gougeon A, Bonnaure-Mallet M. 2014. Biofilms as a mechanism of bacterial resistance. Drug Discov Today Technol 11:49–56. doi: 10.1016/j.ddtec.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 63.Tse-Dinh YC. 2009. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res 37:731–737. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lakshminarasimhan D, Eswaramoorthy S, Burley SK, Swaminathay S. 2010. Structure of YqgQ protein from Bacillus subtilis, a conserved hypothetical protein. Acta Crystallogr F Struct Biol Cryst Commun 66:8–11. doi: 10.1107/S1744309109047009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas VC, Sadykov MR, Chaudhari SS, Jones J, Endres JL, Widhelm TJ, Ahn JS, Jawa RS, Zimmerman MC, Bayles KW. 2014. A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog 10:e1004205. doi: 10.1371/journal.ppat.1004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knobloch JK-M, Horstkotte MA, Rohde H, Mack D. 2002. Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med Microbiol Immunol 191:101–106. doi: 10.1007/s00430-002-0124-3. [DOI] [PubMed] [Google Scholar]
- 67.Kiedrowski MR, Crosby HA, Hernandez FJ, Malone CL, McNamara JO II, Horswill AR. 2014. Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PLoS One 9:e95574. doi: 10.1371/journal.pone.0095574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iordanescu S, Surdeanu M. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J Gen Microbiol 96:277–281. doi: 10.1099/00221287-96-2-277. [DOI] [PubMed] [Google Scholar]
- 69.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]