Abstract
Energy-dependent efflux overexpression and altered outer membrane permeability (influx) can promote multidrug resistance (MDR). The present study clarifies the regulatory pathways that control membrane permeability in the pandemic clone Escherichia coli sequence type 131 (ST131) and evaluates the impact of efflux and influx modulations on biofilm formation, motility, and virulence in the Caenorhabditis elegans model. Mutants of two uropathogenic E. coli (UPEC) strains, MECB5 (ST131; H30-Rx) and CFT073 (ST73), as well as a fecal strain, S250 (ST131; H22), were in vitro selected using continuous subculture in subinhibitory concentrations of ertapenem (ETP), chloramphenicol (CMP), and cefoxitin (FOX). Mutations in genes known to control permeability were shown for the two UPEC strains: MECB5-FOX (deletion of 127 bp in marR; deletion of 1 bp and insertion of an IS1 element in acrR) and CFT073-CMP (a 1-bp deletion causing a premature stop in marR). We also demonstrated that efflux phenotypes in the mutants selected with CMP and FOX were related to the AcrAB-TolC pump, but also to other efflux systems. Alteration of membrane permeability, caused by underexpression of the two major porins, OmpF and OmpC, was shown in MECB5-ETP and mutants selected with FOX. Lastly, our findings suggest that efflux pump-overproducing isolates (CMP mutants) pose a serious threat in terms of virulence (significant reduction in worm median survival) and host colonization. Lack of porins (ETP and FOX mutants) led to a high level of antibiotic resistance in an H30-Rx subclone. Nevertheless, this adaptation created a physiological disadvantage (decreased motility and ability to form biofilm) associated with a low potential for virulence.
INTRODUCTION
Uropathogenic Escherichia coli (UPEC) is the leading causative agent of urinary tract infection (UTI) in both communities and hospitals worldwide (1). The therapeutic management of UTI is particularly problematic because of the increasingly widespread resistance to all classes of antibiotics, including in community-acquired infections (2). With the dissemination of the novel CTX-M family, E. coli has become the species most frequently associated with the production of extended-spectrum β-lactamase (ESBL) (3). In 2008, a globally disseminated and multidrug-resistant (MDR) clone of E. coli O25b:H4-ST131 was identified in clinical samples from three continents (4). Since this first description, sequence type 131 (ST131) was shown to be a successful clone, causing the majority of MDR E. coli infections while exhibiting multiple virulence factors (5, 6). Moreover, it was described as one of the most prevalent clones among phylogenetic group B2 in the digestive carriage of healthy subjects in France (7). Within the ST131 group, various subclonal lineages have been identified. Indeed, a dominant subclone named H30-Rx has extensively expanded. This subgroup harbors allele 30 of fimH, the type 1 fimbrial adhesin gene, and is associated with both blaCTX-M-15 carriage and fluoroquinolone (FQ) resistance by specific topoisomerases mutations (8).
In addition to enzymatic hydrolysis or target modification, energy-dependent efflux and altered outer membrane permeability can promote resistance to antimicrobial agents. In Enterobacteriaceae, overexpression of the chromosomally encoded AcrAB-TolC efflux pump plays a major role in MDR, leading to a cross-resistance phenotype affecting the MICs of quinolones, β-lactams (mainly cefoxitin), chloramphenicol, and cyclines (9). A permeability defect as a result of porin loss or changes in porin structure is involved in β-lactam resistance (10). The two principally affected classes are carbapenems, considered the most powerful agents for the treatment of serious infections caused by ESBL producers (11), and cephamycins, a potential alternative to carbapenems for treating ESBL-producing E. coli isolates (12). In E. coli, the major outer membrane proteins (OMP) allowing the permeation of hydrophilic compounds are OmpC and OmpF (13). Among various genetic regulators, systemic transcriptional activators of the AraC/XylS family, MarA and its homologs SoxS and Rob, control the expression levels of both efflux systems and porins (14, 15). In addition, expression of the acrAB genes is modulated by the local repressor AcrR (16), and the balance of porins is regulated by the two-component system EnvZ-OmpR (17).
Although the impacts of efflux and influx modulation on virulence have been demonstrated in Klebsiella pneumoniae, Salmonella enterica, and Enterobacter aerogenes (18–21), few studies have investigated their impacts in E. coli and none in the pandemic clone ST131. This raises the question of the cost of the successive accumulation of membrane-associated resistance mechanisms to bacterial fitness during antibiotic chemotherapy and its consequences for bacterial colonization ability. Taking into account the role of OMP in various physiological states, the balance of membrane permeability can play a key role in the adaptation of E. coli to host conditions. The aim of this work was to gain new insights into the regulatory pathways that control membrane permeability in E. coli ST131 and to evaluate the impact of efflux and influx modulation on biofilm formation, motility, and virulence in the Caenorhabditis elegans model.
MATERIALS AND METHODS
Bacterial strains.
Experiments were carried out using three strains belonging to phylogenetic group B2: (i) the UPEC strain MECB5, which is a representative isolate of the ST131 H30-Rx subclone and virotype C, isolated from the urine of a patient with cystitis in the south of France (4, 22, 23); (ii) the UPEC reference strain CFT073, isolated from blood and belonging to ST73 (23, 24); and (iii) strain S250, which is a recent (2006) representative of the ST131 H22 subclone and virotype D obtained from the digestive tract of a healthy French individual (7, 23, 25, 26). The main characteristics and virulence traits of these isolates are presented in Table 1. The avirulent E. coli OP50 was used as a food source and a control for the nematode assays (18, 23).
TABLE 1.
Main characteristics and virulence traits of the E. coli clinical isolates used in this study
| Strain | ST (subclone) | ESBL | Phylogroup | Virotypea | Source/yr | Virulence factor-encoding genes | References |
|---|---|---|---|---|---|---|---|
| MECB5 | 131 (H30-Rx) | CTX-M-15 | B2 | C | Urine (cystitis)/2004 | iha, fimH, sat, fyuA, iutA, kpsMII-K5, usp, traT, ompT, iss, malX, irp2 | 4, 22, 23 |
| S250 | 131 (H22) | 0 | B2 | D | Stools of a healthy subject/2006 | fimH, fyuA, iutA, kpsMII-K5, usp, tratT, ompT, iss, malX, irp2, ibeA | 7, 23, M.-H. Nicolas-Chanoine (personal data) |
| CFT073 | 73 | 0 | B2 | NA | Blood (pyelonephritis)/1989 | fimH, sat, fyuA, iutA, kpsMII, ompT, malX, hlyA, papA, papE, sfa/foc, iroN, aer, papC, papG, hly, irp2 | 23, 24 |
NA, not applicable.
In vitro selection of mutants.
Three antibiotics were used for their behavior associated with membrane permeability: (i) ertapenem (ETP), known to select mutants with porin alterations (18, 27); (ii) chloramphenicol (CMP), which selects mutants with efflux pump overexpression (9, 19); and (iii) cefoxitin (FOX), whose resistance has been described with both mechanisms (28, 29). Mutants of each strain were selected using the procedure described by Miller et al. (30) by continuous subculture in Mueller-Hinton (MH) broth containing subinhibitory concentrations (sub-MICs) (0.25× MICs) of ETP, CMP, or FOX. Such growth conditions were repeated for 150 days. The decrease in antibiotic susceptibility was tested weekly.
Clonality analysis.
Parental strains and mutants selected by antibiotic pressure were compared by repetitive sequence-based PCR (rep-PCR) analysis (DiversiLab system; bioMérieux, Marcy l'Étoile, France). Strains with a similarity percentage of ≥95% were considered clonal.
Antibiotic susceptibility tests.
The susceptibilities of the mutants to antibiotics were determined by the agar diffusion method according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the French Antibiogram Committee (CA-SFM) guidelines (http://www.sfm-microbiologie.org). For mutants selected with FOX, MH agar containing cloxacillin at 250 mg/liter (bioMérieux) was used to check the absence of chromosomal AmpC overproduction.
The MICs of a panel of antibiotics, including β-lactams (amoxicillin [AMX], FOX, cefotaxime [CTX], ceftazidime [CAZ], ETP, and imipenem [IMP]), quinolones (nalidixic acid [NAL] and ofloxacin [OFX]), CMP, tetracycline (TET), and tigecycline (TGC), were determined by the broth microdilution method. Tests were carried out in triplicate with or without a fixed concentration (25 μg/ml) of phenylalanine arginine β-naphthylamide (PAβN) (Sigma Chemical Co.), a broad-spectrum efflux pump inhibitor. Efflux pump activity was suspected when PAβN addition induced a 3-fold decrease in the MIC value (31).
SDS-PAGE and immunocharacterization of outer membrane proteins.
Exponential-phase bacteria in Luria-Bertani (LB) broth were pelleted and solubilized as previously described (32). Samples (an amount of bacteria corresponding to 0.02 optical density at 600 nm [OD600] unit) were loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (10% polyacrylamide, 0.1% SDS). After electrophoresis, the proteins were electrotransferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH) in transfer buffer (20 mM Tris, 150 mM glycine, 20% isopropanol, 0.05% sodium dodecyl sulfate). An initial blocking step was performed overnight at 4°C with Tris-buffered sodium (50 mM Tris-HCl, 150 mM NaCl, pH 8) containing skimmed milk powder (10%). The nitrocellulose membranes were then incubated in Tris-buffered sodium containing skimmed milk powder (10%) and Triton X-100 (0.2%) for 2 h at room temperature in the presence of polyclonal antibodies directed against the denatured components of the AcrAB-TolC pump or against the denatured OmpC or OmpF porin. The detection of antigen-antibody complexes was performed with alkaline phosphatase-conjugated AffinitiPure goat anti-rabbit IgG antibodies (Jackson ImmunoResearch, West Grove, PA) (32).
DNA sequencing of the MDR regulator gene.
The presence of mutations in the MDR regulator genes marR, acrR, and soxRS was assessed by PCR and sequencing (33, 34). The sequences and the annealing temperatures (Tm) of the primers used are presented in Table S1 in the supplemental material.
Comparative real-time RT-PCR.
For selected genes involved in MDR regulation, efflux, and permeability (see Table S1 in the supplemental material), transcript level analysis was performed by quantitative reverse transcription (qRT)-PCR. Briefly, MECB5, S250, CFT073, and their respective isogenic mutants selected with ETP, CMP, or FOX were grown in MH broth to an OD600 of ∼0.7. Total RNA from bacterial samples was extracted by using the RNeasy minikit as described by the manufacturer (Qiagen, Courtaboeuf, France). RNA was treated with the RNase-Free DNase set (Qiagen). The purity and concentration were determined using the NanoDrop 2000 spectrophotometer (Fisher Scientific, Pittsburgh, PA, USA). cDNA was synthesized from 1 μg of total RNA for each sample, using the iScript Select cDNA synthesis kit (Bio-Rad, Hercules, CA) with random primers according to the manufacturer's instructions. Real-time PCR assays were performed in a LightCycler480 device using the LightCycler FastStart DNA MasterPlus SYBR Green I kit with 100 ng of cDNA and 10 pmol of target primers (see Table S1 in the supplemental material). The specificity of the PCR products was tested by melting-point analysis. Amplifications were performed in duplicate from three different RNA preparations. The 2−ΔΔCT method was used to analyze transcriptional changes in target genes using gapA as the housekeeping control gene (35, 36). The data were log transformed to obtain a fold change difference between strains.
Nematode-killing assay.
The C. elegans infection assays analyzed survival rates in the presence of the different UPEC strains and their isogenic mutants. Fer-15 mutant worms, which have a temperature-sensitive fertility defect, were maintained and infected as previously described (18, 23). A nematode was considered dead when it no longer responded to touch. E. coli virulence was assessed using the nematode survival curve and by calculating the 50% lethal time (LT50), corresponding to the median time (in days) required to kill 50% of the C. elegans population. At least three replicates per experiment, repeated three times, were performed for each strain.
Motility assays.
The motility of the three parental UPEC strains and their mutants was evaluated using soft-agar LB plates as described previously: swim plates containing 0.25% agar and swarm plates containing 0.5% agar supplemented with 0.5% glucose (37). Briefly, bacteria grown overnight in LB broth were diluted 1,000-fold in LB broth and incubated at 37°C to an OD600 of ∼0.7. Swarm plates were inoculated in the middle of the soft agar by spotting with 5 μl of standardized culture. Swim plates were seeded with the same inoculum below the agar surface using a sterile inoculating needle. The plates were incubated for 16 h at 37°C. The diameters of the migration zones produced by the parental and mutant strains were compared. Swimming and swarming experiments were performed independently three times.
Kinetics of biofilm formation.
The kinetics of early biofilm was explored using the BioFilm ring test (BioFilm Control, Saint Beauzire, France) as described previously (38). Briefly, standardized bacterial cultures were incubated at 37°C in 96-well microtiter plates in the presence of magnetic beads. After different times, the plates were placed onto a magnetic block and put in the reader. The images of each well before and after magnetic attraction were analyzed with the BioFilm Control software, which gives a biofilm index (BFI). A high BFI value (>7) corresponds to high mobility of the beads under magnetic action (no biofilm), while a low value (<2) corresponds to full immobilization of the beads due to the sessile cells. Three experiments with two repeats were performed per strain and per incubation time.
Statistical analysis.
Statistics and graphs were prepared using the software package GraphPad Prism 6.0. The effects of antibiotic selection pressure on the expression of selected genes and on motility were assessed using one-way analysis of variance (ANOVA), followed by Dunnett's multiple-comparison test. Log-transformed data were used for real-time RT-PCR. The kinetics of biofilm formation were compared by two-way ANOVA, followed by Dunnett's multiple-comparison test. For the nematode-killing assays, differences in survival rates between a parental strain and an isogenic mutant were tested by a log-rank (Mantel-Cox) test for statistical significance. A P value of <0.05 was considered to reflect a statistically significant difference.
Nucleotide sequence accession numbers.
Novel sequences of MDR regulators determined in this study have been deposited in GenBank under the following accession numbers: acrR, KT984278 (MECB5), KT984279 (S250), NC_004431.1 (CFT073), and KT984280 (MECB5-FOX); marR, KT984281 (MECB5), KT984282 (S250), NC_004431.1 (CFT073), KT984283 (MECB5-FOX), and KT984284 (CFT073-CMP); and soxR, KT984285 (MECB5), KT984286 (S250), and NC_004431.1 (CFT073).
RESULTS
Selection of resistant mutants.
Continuous subculture of MECB5, S250, and CFT073 with sub-MICs of ETP, CMP, and FOX successfully selected isogenic mutants resistant to these antibiotics, except for ETP resistance, which was obtained only with the CTX-M-15-producing MECB5. No variation of ETP MICs was observed over the weeks for strains S250 and CFT073, and experiments were stopped after 150 days of subculture. Rep-PCR assessed clonal relationships between parental strains and mutants (similarity, >98% [data not shown]). The numbers of days required to obtain mutants are presented in Table 2.
TABLE 2.
Antibiotic susceptibilities of the three parental E. coli isolates and their isogenic mutants with and without the efflux pump inhibitor PAβN
| Strain | Time to select resistant mutants (days) | MIC (μg/ml) [with PAβN]a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX (≤8, >8) | AMC (≤8, >8) | FOX (≤8, >16) | CTX (≤1, >2) | CAZ (≤1, >4) | ETP (≤0.5, >1) | IMP (≤2, >8) | NAL (≤16, >16) | OFX (≤0.5, >1) | CMP (≤8, >8) | TET (≤4, >8) | TGC (≤1, >2) | ||
| MECB5 | >1,024 [>1,024] | 128 [128] | 4 [4] | >1,024 [>1,024] | >1,024 [>1,024] | 0.16 [0.16] | 0.125 [0.125] | >256 [>256] | 8 [2] | 2 [0.5] | 128 [128] | 1 [0.5] | |
| MECB5-ETP | 80 | >1,024 [>1,024] | >128 [>128] | 16 [8] | >1,024 [>1,024] | >1,024 [>1,024] | 2 [1] | 0.5 [0.25] | >256 [>256] | 16 [4] | 2 [0.5] | 256 [128] | 1 [0.5] |
| MECB5-CMP | 80 | >1,024 [>1,024] | 128 [128] | 16 [4] | >1,024 [>1,024] | >1,024 [>1,024] | 0.16 [0.08] | 0.125 [0.25] | >256 [>256] | 32 [8] | 32 [4] | 256 [128] | 2 [1] |
| MECB5-FOX | 70 | >1,024 [>1,024] | >128 [>128] | 32 [8] | >1,024 [>1,024] | >1,024 [>1,024] | 1 [1] | 0.25 [0.5] | >256 [>256] | 64 [8] | 8 [0.5] | >256 [256] | 2 [1] |
| S250 | 8 [8] | 8 [8] | 2 [2] | 0.125 [0.125] | 0.25 [0.125] | 0.016 [0.008] | 0.25 [0.25] | 8 [1] | 0.063 [0.031] | 2 [0.5] | 2 [1] | 0.25 [0.125] | |
| S250-ETP | Nob | 8 [8] | 8 [8] | 4 [2] | 0.125 [0.125] | 0.25 [0.25] | 0.016 [0.016] | 0.25 [0.25] | 8 [2] | 0.063 [0.063] | 2 [0.5] | 2 [1] | 0.25 [0.25] |
| S250-CMP | 60 | 8 [8] | 8 [8] | 32 [4] | 0.25 [0.125] | 0.5 [0.25] | 0.016 [0.016] | 0.25 [0.25] | 16 [2] | 0.5 [0.063] | 64 [4] | 8 [4] | 1 [0.25] |
| S250-FOX | 70 | 16 [16] | 16 [16] | 64 [8] | 0.5 [0.25] | 0.5 [0.25] | 0.016 [0.016] | 0.25 [0.25] | 16 [1] | 0.063 [0.063] | 4 [1] | 2 [2] | 0.5 [0.25] |
| CFT073 | 8 [8] | 8 [8] | 2 [1] | 0.063 [0.063] | 0.125 [0.125] | 0.008 [0.016] | 0.125 [0.125] | 8 [1] | 0.063 [0.063] | 2 [0.5] | 2 [1] | 0.25 [0.25] | |
| CFT073-ETP | Nob | 8 [8] | 8 [8] | 4 [2] | 0.063 [0.063] | 0.25 [0.25] | 0.016 [0.016] | 0.125 [0.125] | 8 [2] | 0.063 [0.063] | 2 [0.5] | 2 [2] | 0.5 [0.25] |
| CFT073-CMP | 90 | 8 [8] | 8 [8] | 16 [4] | 0.125 [0.063] | 0.25 [0.125] | 0.008 [0.016] | 0.125 [0.125] | 64 [2] | 0.25 [0.063] | 64 [8] | 8 [2] | 1 [0.25] |
| CFT073-FOX | 100 | 16 [16] | 16 [16] | 64 [8] | 0.5 [0.25] | 0.5 [0.25] | 0.008 [0.016] | 0.125 [0.125] | 32 [2] | 0.063 [0.063] | 8 [1] | 2 [2] | 0.25 [0.25] |
AMX, amoxicillin; AMC, amoxicillin associated with clavulanic acid; FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; ETP, ertapenem; IMP. imipenem; NAL, nalidixic acid; OFX, ofloxacin; CMP, chloramphenicol; TET, tetracycline; TGC, tigecycline. The breakpoints according to EUCAST are given in parentheses. The MIC values obtained in the presence of efflux pump inhibitor (PAβN at 25 μg/ml) are shown in brackets.
For strains S250 and CFT073, continuous subculture in a subinhibitory concentration of ETP failed to select resistance to the antibiotic after 150 days of culture.
Antibiotic susceptibility pattern.
The MICs of different antibiotics are detailed in Table 2.
(i) Parental strains.
As described above, MECB5 belonged to the ST131 H30-Rx subclone that produced the CTX-M-15 ESBL responsible for resistance to aminopenicillins and extended-spectrum cephalosporins (ESC) and carried a distinctive gyrA and parC combination (S83L/D87N GyrA and S80I/E84V ParC double substitutions) (22) responsible for quinolone resistance. The S250 and CFT073 parental strains were fully susceptible to all antibiotics tested.
(ii) ETP mutant.
An ETP mutant (Table 2) was obtained only from strain MECB5 (MECB5-ETP). It was resistant to ETP (MIC, 2 μg/ml) but remained susceptible to IMP (MIC, 0.5 μg/ml), according to the revised EUCAST criteria. The mutant was associated with a significant increase in the FOX MIC (4 times higher). Levels of susceptibility to quinolones, CMP, and cyclines were similar to those of the parental strain.
(iii) CMP mutants.
For the CMP mutants (Table 2) obtained from the three strains, CMP resistance (32 to 64 μg/ml) was associated with significantly higher MICs of NAL and OFX (2 to 8 times), TET (2 to 4 times), and TGC (2 to 4 times). Adding PAβN to the medium emphasized the efflux resistance mechanism, leading to a decrease in the MICs of CMP (8- to 16-fold), quinolones (4- to 32-fold), and cyclines (2- to 4-fold).
(iv) FOX mutants.
In the FOX mutants obtained from the three parental strains, an increase in MICs was observed for FOX (8 to 32 times), NAL and OFX (2 to 8 times), CMP (2 to 4 times), and, to a lesser extent, TGC (2 times). For the MECB5-FOX mutant, FOX resistance was associated with ETP resistance (MIC, 1 μg/ml). Antibiotic activity was partially restored by the efflux pump inhibitor, except for ETP, for which PAβN addition had no effect on the MIC. FOX resistance was not attributable to AmpC overproduction, since FOX activity was not restored on MH agar containing cloxacillin (data not shown).
Outer membrane protein profile.
As indicated in Table 3, all the mutants selected with CMP and FOX displayed AcrA and AcrB signals higher than those of the parental strains. Outer membrane proteins (OMP) OmpC and OmpF were detected at the same expression level for CMP mutants. In contrast, no porin was detected in strains MECB5-ETP, MECB5-FOX, and S250-FOX. This lack of porin was observed with antisera that recognized OmpC and OmpF. Interestingly, CFT073-FOX showed OmpC and OmpF production similar to that of the parental strain (Table 3).
TABLE 3.
Analysis of immunodetected membrane proteins in MECB5, S250, CFT073, and their in vitro-selected mutants
| Strain | Membrane protein contenta |
||||
|---|---|---|---|---|---|
| AcrA | AcrB | TolC | OmpC | OmpF | |
| MECB5 | ± | + | + | ++ | ++ |
| MECB5-ETP | + | + | + | − | − |
| MECB5-CMP | ++ | ++ | ++ | + | + |
| MECB5-FOX | ++ | + | ++ | − | − |
| S250 | + | + | ± | ++ | ++ |
| S250-CMP | ++ | ++ | ± | + | + |
| S250-FOX | ++ | ++ | + | − | − |
| CFT073 | + | ± | + | + | + |
| CFT073-CMP | ++ | + | ++ | + | + |
| CFT073-FOX | ++ | ++ | ++ | + | + |
++, very high level of protein production; +, high level of protein production; ±, few protein production; −, lack of protein production.
Sequences of regulator genes.
Alterations in MDR regulators contribute to the MDR phenotype by modulating both efflux and membrane permeability. To gain insight into the regulation of this pathway, genes encoding local (acrR) and global (marR and soxR) repressors were amplified and sequenced.
First, the sequences of the acrR, marR, and soxR genes of MECB5, S250, and CFT073 were compared with those of E. coli strain K-12 substrain MG1655, available in the GenBank database (accession number NC_000913.3). For the three strains studied, the acrR sequences were identical or showed only silent polymorphisms. However, the marR and soxR sequences showed two and one nonsynonymous substitutions, respectively, for the three strains (Fig. 1).
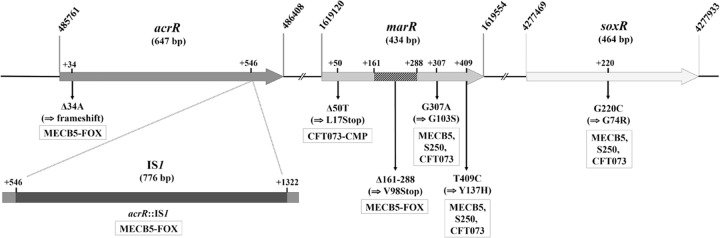
FIG 1.
Schematic diagram of the genomic region comprising the acrR, marR, and soxR genes in E. coli K-12 substrain MG1655 (accession number NC_000913.3). The nucleotide mutations and the deduced protein changes for the three parental strains and their isogenic mutants are shown.
Second, sequences obtained for the in vitro-selected mutants were compared to those of their parental strains. The mutant MECB5-FOX displayed acrR and marR sequences different from those of MECB5 (Fig. 1). acrR showed two alterations: (i) a deletion of 1 bp occurring at nucleotide position +34, which results in a shift in the reading frame throughout the gene, and (ii) a 776-bp insertion occurring at the end of acrR, 546 bp downstream of the ATG start codon. This insertion corresponds to the integration of the insertion sequence IS1 accompanied by a duplication of 8 bp in the target DNA at the site of insertion and flanked by 18-bp left and right inverted repeats at its ends (accession number KT984280) (Fig. 1). marR of MECB5-FOX was disrupted by a 127-bp deletion at positions 161 to 288 downstream of the GTG start codon, resulting in an early frameshift and termination (stop codon at +168; accession number KT984283). Concerning the CFT073-CMP mutant, the marR gene showed a thymine deletion at the +50 nucleotide location, immediately causing a premature stop codon (accession number KT984284) (Fig. 1). The other strains (MECB5-ETP, MECB5-CMP, S250-ETP, S250-CMP, S250-FOX, CFT073-ETP, and CFT073-FOX) harbored acrR and marR sequences identical to those of the parental strains. No mutation was found in the soxR gene of any mutant, except those already present in the parental strains.
Comparative real-time RT-PCR.
To understand behavior changes under antibiotic pressure, the expression levels of the different genes involved in MDR were measured for the isogenic mutants and compared to those of their parental strains. Their log relative transcription levels are shown in Fig. 2 for strain MECB5 and in Fig. S1 and S2 in the supplemental material for S250 and CFT073.
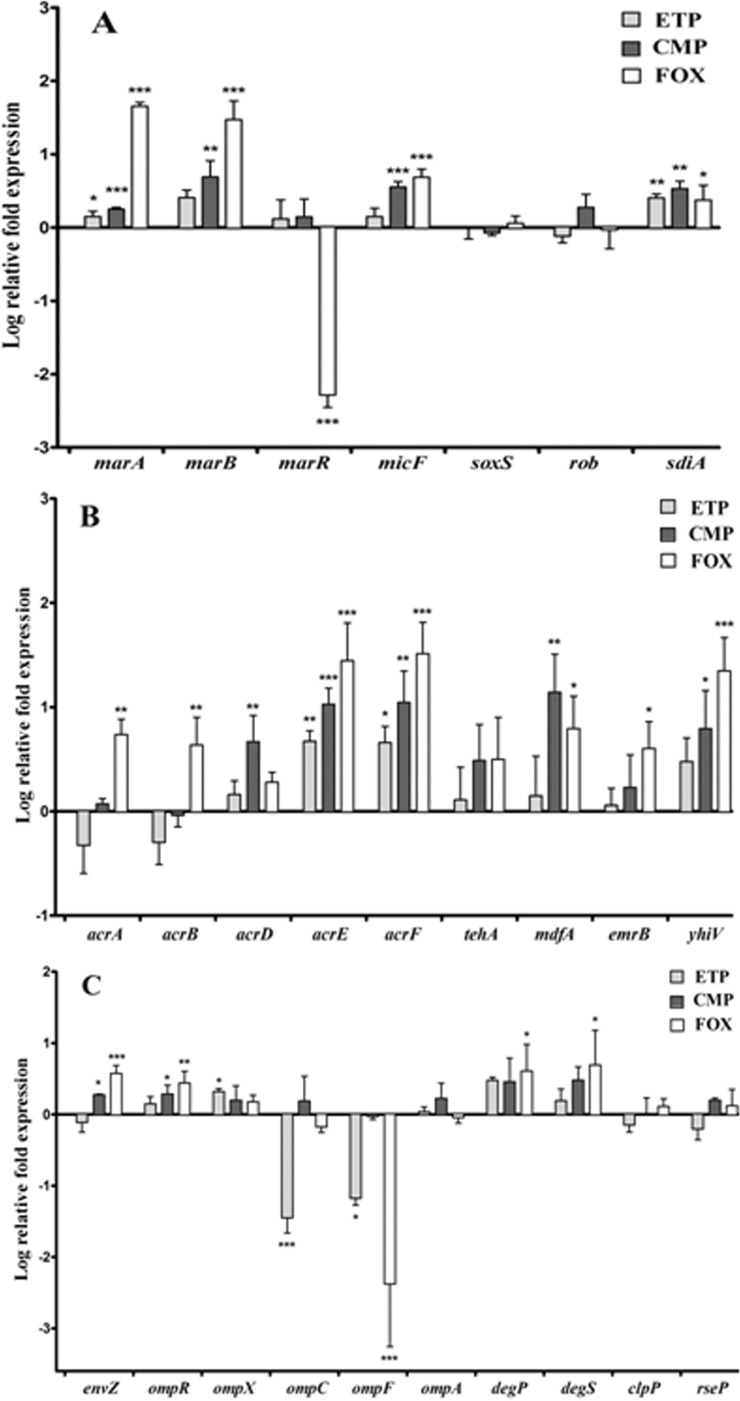
FIG 2.
Relative mRNA expression levels of genes implicated in MDR regulation (A), membrane efflux (B), and permeability (C) for the ST131 H30-Rx E. coli strain MECB5 and its mutants in vitro selected with ETP, CMP, and FOX. The log-transformed averages of relative fold changes of mutants compared to the parental strain for each antibiotic are presented. The error bars represent the standard deviations from three different RNA preparations. Significant differences from the parental MECB5 using Dunnett's test are indicated; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Genes responding to stress. (i) The marRAB operon and micF.
The marA gene, which encodes a transcriptional activator targeting many genes involved in resistance to stress (14), was significantly overexpressed in mutants MECB5-ETP (P < 0.05) and MECB5-CMP and MECB5-FOX (P < 0.001) compared to their parental strains (Fig. 2A). marA was also significantly upregulated in mutants S250-CMP (P < 0.01) and CFT073-CMP and FOX (P < 0.001) (see Fig. S1A and S2A in the supplemental material). Similar upregulations were observed with marB, which encodes a periplasmic protein that modulates marA expression (39), and the noncoding RNA stress response gene micF (40) (P < 0.01 to 0.001). MarR is an autorepressor of the operon marRAB. Comparative RT-PCR showed a significant decrease in marR expression in MECB5-FOX (P < 0.001), consistent with the sequencing results (the primer used, marR_R, targets a region deleted from the gene). In contrast, disrupted marR of mutant CFT073-CMP (stop codon at +51) was overexpressed (P < 0.001) (see Fig. S2A in the supplemental material).
(ii) soxS, rob, and sdiA genes.
Expression of soxS and rob, encoding two other positive transcriptional regulators belonging to the AraC/XylS family (15), appeared to be less impacted than marA (Fig. 2A). rob was upregulated only in strain S250-CMP (P < 0.05) (see Fig. S1A in the supplemental material). The expression of sdiA, encoding a quorum-sensing transcription factor (41), was significantly increased in the ST131 mutants regardless of the antibiotic pressure (P < 0.05 to < 0.001) and in CFT073-FOX (P < 0.01).
Genes encoding efflux pump systems. (i) acrAB.
The expression of genes encoding the major resistance-nodulation-division (RND) efflux pump in Enterobacteriaceae was principally increased in mutants harboring MDR regulator alterations, MECB5-FOX and CFT073-CMP (P < 0.05 to < 0.001) (Fig. 2B; see Fig. S2B in the supplemental material).
(ii) Other RND transporters.
Expression of acrD, encoding an RND transporter able to expel highly hydrophilic molecules, like aminoglycosides; acrE and acrF, encoding close homologs of AcrA and AcrB; and yhiV (mdtF) was significantly upregulated in the three mutants selected with CMP (P < 0.05 to < 0.001). acrEF and yhiV were also overexpressed in MECB5 and CFT073 mutants selected with FOX (P < 0.05 to < 0.001).
(iii) Major facilitator superfamily (MFS) and small multidrug resistance (SMR) transporters.
The increase in MdfA activity was very significant among all CMP mutants (P < 0.01 to < 0.001) and, to a lesser extent, MECB5-FOX and S250-FOX (P < 0.05). The increased level of activity of tehA and emrB genes was much less evident: tehA was significantly upregulated for S250-CMP and CFT073-CMP mutants, while emrB showed the highest level of activity only for the mutant S250-CMP.
Genes encoding proteins involved in OM permeability. (i) envZ, ompR, and ompX regulators.
The expression of envZ, which encodes an osmoregulatory sensor protein (17), was upregulated for MECB5-FOX (P < 0.001) and the three CMP mutants (P < 0.05) (Fig. 2C; see Fig. S1C and S2C in the supplemental material). The expression level of ompR, which encodes the cytosolic response regulator, followed this trend. ompX, encoding another porin expression downregulator (42), was overexpressed for the MECB5-ETP mutant (P < 0.05) and strains S250 and CFT073 selected with CMP and FOX (P < 0.05 to < 0.001).
(ii) ompC, ompF, and ompA porins.
The upregulation of ompX observed for MECB5-ETP was associated with a significant decrease in both ompC and ompF expression (P < 0.05 to < 0.001). For S250-ETP and CFT073-ETP, which remained susceptible to ETP, no modification of ompC and ompF expression could be noted. A significant decrease in ompF expression without modification of that of ompC was assessed both for mutants selected with FOX and for S250-CMP and CFT073-CMP (P < 0.05 to < 0.001). These data were similar in MECB5-FOX, S250-CMP, CFT073-CMP, and CFT073-FOX with increased expression levels of ompR and envZ, encoding the two-component system that allows ompC transcription and ompF repression (43). Unlike that of ompC and ompF, transcription of ompA, which encodes an architectural protein, was conserved after antibiotic pressure.
Proteases.
Increased expression of genes that encode proteases DegP and DegS, responsible for posttranslational control of OMP expression (44), was noted for MECB5 and CFT073 mutants selected with FOX (P < 0.05). The expression of other proteases, ClpP and RseP, remained identical for all strains.
Strain virulence.
The C. elegans model was used to explore the possible relationship between in vitro acquisition of resistance to ETP, CMP, or FOX and modifications of virulence behavior.
First, the life span of C. elegans feeding on the parental strains was compared to that obtained when nonpathogenic E. coli OP50 was used as food (Table 4). The LT50, i.e., the median survival in days, was significantly reduced when the worms fed on MECB5, S250, and CFT073 (LT50, 5 ± 0.5, 5 ± 0.5, and 4 ± 0.5 days, respectively) in comparison with OP50 (LT50, 7 ± 0.5 days) indicating that the three parental strains studied are virulent for nematodes (P < 0.0001 for both strains). The ST131 strains showed similar virulence profiles (P value, nonsignificant [NS]). However, CFT073 was significantly more virulent than MECB5 and S250 (P < 0.05).
TABLE 4.
LT50s of C. elegans infected with MECB5, S250, and CFT073 parental strains and their respective mutants
| Strain | LT50 (days) (mean ± SE) |
P valuea |
|
|---|---|---|---|
| Studied strain vs. OP50 | Mutant vs. parental strain | ||
| OP50 | 7 ± 0.5 | NA | NA |
| MECB5 | 5 ± 0.5 | <0.0001 | NA |
| MECB5-ETP | 6 ± 0.5 | 0.0009 | 0.0007 |
| MECB5-CMP | 4 ± 0.5 | <0.0001 | 0.0006 |
| MECB5-FOX | 6 ± 0.5 | 0.0064 | <0.0001 |
| S250 | 5 ± 0.5 | <0.0001 | NA |
| S250-CMP | 4 ± 0.5 | <0.0001 | 0.0019 |
| S250-FOX | 6 ± 0.5 | 0.0016 | 0.0003 |
| CFT073 | 4 ± 0.5 | <0.0001 | NA |
| CFT073-CMP | 4.5 ± 0.5 | <0.0001 | 0.5842 (NS) |
| CFT073-FOX | 6 ± 0.5 | 0.0069 | <0.0001 |
Differences in survival rates were tested by a log-rank (Mantel-Cox) test for statistical significance. A P value of <0.05 was considered to reflect a statistically significant difference. NA, not applicable; NS, nonsignificant.
As shown in Table 4, CMP pressure selected more virulent strains (P < 0.01), except in the case of CFT073-CMP, which killed worms as fast as the parental strain (P value, NS). Conversely, the nematodes died more slowly when fed on FOX mutants and MECB5-ETP (P < 0.001).
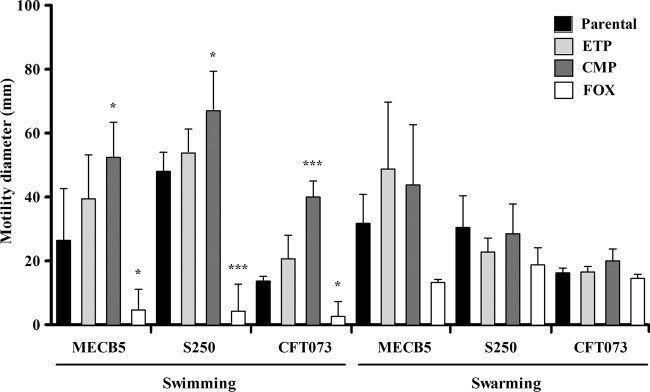
Motility assays.
To determine if the changes in virulence were linked to a motility variation, bacterial swimming and swarming motilities were quantified on soft agar. As shown in Fig. 3, all the CMP mutants showed a significant increase in swimming area (P < 0.05 to < 0.001). In contrast, swimming behavior was considerably affected for the three mutants selected with FOX (P < 0.05 to < 0.001). ETP pressure did not seem to influence swimming motility (P value, NS). Swarming behavior was not significantly different between parental and mutant strains.
FIG 3.
Comparative results of swimming and swarming assays for MECB5, S250, and CFT073 parental strains and their isogenic mutants selected with ETP, CMP, and FOX. The error bars represent the standard deviations from at least three independent assays. Significant differences from the parental strains using Dunnett's test are indicated; *, P < 0.05; ***, P < 0.001.
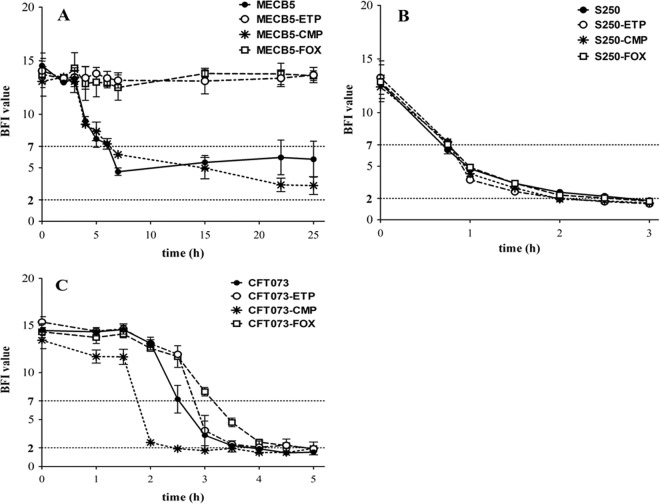
Kinetics of biofilm formation.
The BioFilm ring test was performed to examine the effect of antibiotic pressure on the capacity of E. coli to form biofilm and to determine whether this effect might influence virulence in worms. The results revealed different profiles among parental strains and among their mutants (Fig. 4).
FIG 4.
Effects of ETP, CMP, and FOX resistance on E. coli biofilm formation. The kinetics of the early phase of biofilm formation for strains MECB5 (A), S250 (B), and CFT073 (C) and their respective mutants were determined by the BioFilm ring test (BioFilm Control). Means ± standard errors of the mean of BFIs for at least three independent replicates are presented. Statistical differences between the strains and their mutants at each time were obtained by using two-way ANOVA, followed by Dunnett's multiple-comparison test.
(i) MECB5 strains.
For the MECB5 parental strain, biofilm was being formed after 7 h of incubation (BFI = 4.63 ± 0.20), but longer incubation (up to 24 h) failed to completely immobilize the beads (Fig. 4A). The same trend could be noted for the MECB5-CMP mutant during the first hours, but then, the number of sessile bacteria was significantly increased after 15 h (BFI = 5.47 ± 0.45; P < 0.0001) and 22 h (BFI = 3.40 ± 0.36; P < 0.0001). In contrast, no biofilm formation was detected for MECB5-ETP and MECB5-FOX mutants after 24 h, showing a significant difference between these strains and the parental strain (P < 0.0001).
(ii) S250 strains.
The kinetics of biofilm formation for the S250 strain was very fast, with a biofilm constituted after 2 h (BFI = 2.56 ± 0.09) (Fig. 4B). Antibiotic pressure had no significant impact on these kinetics of formation: the BFI values for each mutant were similar to those for the parental strain.
(iii) CFT073 strains.
For strain CFT073 (Fig. 4C), biofilm formation was obtained after 4 h (BFI = 1.9 ± 0.06). The CFT073-CMP mutant showed significantly faster biofilm formation than the parental strain, with the beads immobilized after 2.5 h (BFI = 1.90 ± 0.06) (P < 0.0001). As observed for strain MECB5, growth with sub-MICs of ETP or FOX altered the capacity of CFT073 to form a biofilm. Biofilm formation was significantly delayed for CFT073-FOX at 2.5, 3, and 3.5 h (P < 0.0001). For CFT073-ETP, the difference was significant only after 2.5 h of incubation (P < 0.0001).
DISCUSSION
Since early 2000, the spread of E. coli ST131, particularly the H30-Rx subclone, has led to a worldwide increase of ESBL-producing and FQ-resistant E. coli isolates in both community and hospital settings (45). The present study describes a link between the antibiotic pressure and the adaptive strategy used by bacteria involving the modulation of influx/efflux balance in the inner and outer membrane. As a consequence, this modification of membrane permeability observed in our mutants might have some side effects on the virulence capacity and the fitness of E. coli.
Continued growth in the presence of nonlethal antibiotic levels is a crucial aspect of the current antibiotic resistance crisis. Sub-MICs of antibiotics are present in many natural environments (46). They also occur in patients during therapy and are likely to accelerate the evolution of resistance by triggering some modification in the regulation of protein expression, by increasing mutagenesis in targeted genes, and/or by favoring recombination. We evaluated the behavior of two representative strains of E. coli ST131, MECB5 (H30-Rx) and S250 (H22), and the CFT073 UPEC reference strain grown under antimicrobial pressure with antibiotics known to interact with membrane influx and/or efflux: ETP, CMP, and FOX (17, 24–26, 44). Mutants resistant to these antibiotics were obtained for all the strains with the exception of the ESC-susceptible strains S250 and CFT073, for which no increase in ETP MICs was noted after 5 months of subculture. These results were in agreement with those of Tangden and Girlich, who also observed that carbapenem resistance in E. coli was selected only among ESBL-producing strains (27, 47).
Concerning the MECB5-ETP mutant, Western blot analysis indicated that resistance to ETP is associated with a lack of expression of the major OmpC and OmpF porins. Interestingly, qRT-PCR showed that the loss of transcription was particularly important for ompC, as previously observed by Girlich et al. (27), and was related to overexpression of ompX rather than to that of the envZ-ompR control system. This role of OmpX in porin expression (at transcriptional and translational regulation levels) has been previously reported during short exposure to chemicals (42).
In the CMP mutants, the important efflux activity was confirmed by overexpression of the main pump AcrAB-TolC, leading to higher MICs of CMP, FOX, quinolones, and cyclines. Interestingly, genes encoding other efflux systems less frequently described in clinical resistance (36)—acrEF, mdfA, yhiV, and, to a lesser extent, acrD and tehA—were highly overexpressed, suggesting that the increase in E. coli efflux is a global phenomenon that involves several pumps of different families.
Finally, FOX pressure resulted in increasing efflux and reduced influx. The efflux activity significantly increased in the MECB5-FOX strain, for which the repressor AcrR was no longer functional. This mutant was obtained fairly quickly (70 days of subculture), and this raises the question of the use of cephamycins in the treatment of infections with ESBL producers even if the subculture time is long and not observed in real life (12). Inactivating mutations in the acrR gene were previously reported in MDR clinical isolates of E. coli, S. enterica, E. aerogenes, and K. pneumoniae (48–52). To our knowledge, this is the first description of a frameshift loss associated with an IS1 insertion in the acrR gene. acrR::IS1 has been described only once in an E. coli veterinary strain (DE28) (53). In this isolate, insertion occurred earlier in the sequence of acrR, at nucleotide +394 (+546 in our study) (accession number JN652619.1). Moreover, for the three FOX mutants, a lack of OmpF expression could be noted, while that of OmpC was less affected, or even increased in the case of CFT073-FOX. Such differences of expression can be explained by the increased activity of MicF, a small antisense RNA affecting ompF expression at a posttranscriptional level (40), and the activation of the two-component system EnvZ-OmpR. After phosphorylation by EnvZ, OmpR promotes expression of ompC, which encodes a porin that exhibits narrower channels than the OmpF porin (17).
The most important modulation of influx/efflux regulation in the bacterial membrane was observed for MECB5-FOX and CFT073-CMP. These two mutants harbored alterations in the global MDR regulator marR, highlighting the key role of the activator MarA in E. coli, which is devoid of the activator RamA. The other AraC/XylS activators, SoxS and Rob, for which gene expression varied only slightly in selected mutants, appeared less involved in the regulation of MDR. In contrast, overexpression of sdiA was observed in all the ST131 mutants studied. SdiA is homologous to the LuxR family of quorum-sensing transcription factors, which has been previously described as involved in acrAB, acrD, and acrEF overexpression in ESC- or quinolone- resistant isolates (41, 54, 55). In this study, SdiA appeared to be involved in the adaptive resistance of the ST131 clone, independently of the antibiotic pressure used. Nevertheless, its intrinsic role is difficult to assess, due to the concomitant upregulation of marA in most ST131 mutants.
A link between the modulation of the influx/efflux balance at the membrane level and virulence has been previously described in different enterobacteria (18–21, 56). In E. coli, lack of porins was associated with low invasion and virulence profiles (57, 58), while the ABC-type pump MacAB-TolC seemed to increase the pathogenicity of enterotoxigenic E. coli (ETEC) by the secretion of enterotoxin II (59). Some publications have demonstrated the usefulness and interest of the C. elegans nematode to study E. coli virulence (23, 60). The present study highlights, in this model, the direct impact of strong modifications in the membrane organization and physiology on bacterial virulence. Indeed, the lack of both OmpC and OmpF in the ST131 H30-Rx strain selected with ETP was associated with reduced virulence. This finding suggests that the alteration of outer membrane porins might be a serious disadvantage for bacterial fitness compared to β-lactamase production (21, 23). On the other hand, the overexpression of efflux systems was correlated with increased virulence of the ST131 mutants selected with CMP. These bacterial efflux systems export toxic compounds, such as antibiotics, as well as virulence factors important for the colonization of human and animal cells (adhesins, toxins, and an iron capture system) (9). The expression of AcrAB-TolC has been described as a prerequisite in the early bacterial colonization step, for instance, to pass the bile salt barrier or host defensin activity (19, 20). The unchanged virulence potential observed for the CFT073-CMP derivative can be explained by the decreased expression of ompF following micF overexpression in this strain harboring a prematurely interrupted marR gene. Another possible explanation is that the C. elegans model has some limitations when the worms are fed with highly virulent strains, such as CFT073. As described in K. pneumoniae (18), increased efflux systems did not appear to counterbalance the lack of porins in the FOX mutants that showed attenuated virulence.
To understand whether a modification of fitness could contribute to the difference in virulence phenotypes observed, we examined the motility of mutants and their capacity to form biofilm. CMP pressure greatly improved swimming motility for all the mutants. FOX pressure led to the opposite effect. These different behaviors could contribute to the virulence variations observed in C. elegans, as motility has been reported to enhance bacterial spreading during infection (61).
Biofilm production is an important pathogenicity factor, as it can protect bacteria from the immune system and antimicrobial drugs (62). Very different behaviors were observed in the two ST131 parental isolates. The H22 strain formed early biofilm very quickly (within 2 h). This ability was stable, independently of the antibiotic pressure used. On the other hand, the H30-Rx strain was not able to form a full early biofilm within 24 h, showing that the ability to form a biofilm is not the key factor that accounts for the success of the subclone. These results are consistent with those of Olesen et al. and Shin and Ko, who showed that among ST131 strains, biofilm production was strongly associated with non-H30 subclones and non-CTX-M isolates (63, 64). A previous study had shown that sub-MICs of antibiotics (aminoglycosides, tetracycline, and fluoroquinolones) triggered biofilm formation (65). In our study, CMP pressure indeed induced this effect, speeding up biofilm formation in CFT073-CMP and increasing the number of sessile bacteria in ST131 MECB5-CMP. Nevertheless, the kinetics was found to be slowed down in ETP or FOX derivatives, emphasizing the lower colonization capacity of the porin-deficient strains. Interestingly, sub-MIC ETP also influenced the behavior of CFT073, which remained susceptible to the antibiotic, slowing down biofilm formation. The delay in biofilm formation observed in FOX mutants (MECB5-FOX and CFT073-FOX) could be explained by the decreased swimming motility, which is an essential parameter in biofilm organization (66).
The main limitation of the study is the need to cultivate the parental strains over a long period, which is not under normal physiological conditions. This could lead to the emergence of some mutations in the bacterial genome that could explain the modification of virulence. However, the technique used in our study has been previously published (18, 29, 30) and was demonstrated to be a good approach to evaluate the impacts of antibiotics on bacterial fitness.
In conclusion, the modification of outer membrane permeability due to the loss of porins observed in the E. coli ST131 mutants leads to a high level of resistance to all β-lactams in isolates belonging to the ST131 subclone H30-Rx and producing CTX-M-15. Nevertheless, because of the role of porins in the uptake of nutrients required for bacterial survival and growth, their loss creates a physiological disadvantage. FOX and ETP pressure promotes the selection of isolates exhibiting alterations of membrane permeability and consequently imposes an important cost for bacterial fitness involving a low potential of virulence. Conversely, our findings suggest that efflux pump-overproducing isolates pose a serious threat in terms of host colonization and virulence compared to membrane-altered isolates that exhibit a restricted capability to survive under stress conditions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by INSERM.
We thank Mariella Lomma for her editing assistance.
We have no conflict of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02872-15.
REFERENCES
- 1.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen YH, Ko WC, Hsueh PR. 2013. Emerging resistance problems and future perspectives in pharmacotherapy for complicated urinary tract infections. Expert Opin Pharmacother 14:587–596. doi: 10.1517/14656566.2013.778827. [DOI] [PubMed] [Google Scholar]
- 3.Cantón R, Coque TM. 2006. The CTX-M β-lactamase pandemic. Curr Opin Microbiol 9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. [DOI] [PubMed] [Google Scholar]
- 5.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 6.Brisse S, Diancourt L, Laouénan C, Vigan M, Caro V, Arlet G, Drieux L, Leflon-Guibout V, Mentré F, Jarlier V, Nicolas-Chanoine MH. 2012. Phylogenetic distribution of CTX-M- and non-extended-spectrum-β-lactamase-producing Escherichia coli isolates: group B2 isolates, except clone ST131, rarely produce CTX-M enzymes. J Clin Microbiol 50:2974–2981. doi: 10.1128/JCM.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leflon-Guibout V, Blanco J, Amaqdouf K, Mora A, Guize L, Nicolas-Chanoine MH. 2008. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J Clin Microbiol 46:3900–3905. doi: 10.1128/JCM.00734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee R, Johnson JR. 2014. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li XZ, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagès JM, James CE, Winterhalter M. 2008. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 11.Vardakas KZ, Tansarli GS, Rafailidis PI, Falagas ME. 2012. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum β-lactamases: a systematic review and meta-analysis. J Antimicrob Chemother 67:2793–2803. doi: 10.1093/jac/dks301. [DOI] [PubMed] [Google Scholar]
- 12.Kernéis S, Valade S, Geri G, Compain F, Lavollay M, Rostane H, Carbonnelle E, Mainardi JL. 2015. Cefoxitin as a carbapenem-sparing antibiotic for infections caused by extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Dis 47:789–795. doi: 10.3109/23744235.2015.1062133. [DOI] [PubMed] [Google Scholar]
- 13.Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 14.Alekshun MN, Levy SB. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother 41:2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin RG, Gillette WK, Rhee S, Rosner JL. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol 34:431–441. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 16.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol 19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 17.Mizuno T, Mizushima S. 1990. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol Microbiol 4:1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 18.Bialek S, Lavigne JP, Chevalier J, Marcon E, Leflon-Guibout V, Davin A, Moreau R, Pagès JM, Nicolas-Chanoine MH. 2010. Membrane efflux and influx modulate both multidrug resistance and virulence of Klebsiella pneumoniae in a Caenorhabditis elegans model. Antimicrob Agents Chemother 54:4373–4378. doi: 10.1128/AAC.01607-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJV. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol 8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 21.Lavigne JP, Sotto A, Nicolas-Chanoine MH, Bouziges N, Bourg G, Davin-Regli A, Pagès JM. 2012. Membrane permeability, a pivotal function involved in antibiotic resistance and virulence in Enterobacter aerogenes clinical isolates. Clin Microbiol Infect 18:539–545. doi: 10.1111/j.1469-0691.2011.03607.x. [DOI] [PubMed] [Google Scholar]
- 22.Lavigne JP, Marchandin H, Delmas J, Moreau J, Bouziges N, Lecaillon E, Cavalie L, Jean-Pierre H, Bonnet R, Sotto A. 2007. CTX-M β-lactamase-producing Escherichia coli in French hospitals: prevalence, molecular epidemiology, and risk factors. J Clin Microbiol 45:620–626. doi: 10.1128/JCM.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavigne JP, Vergunst AC, Goret L, Sotto A, Combescure C, Blanco J, O'Callaghan D, Nicolas-Chanoine MH. 2012. Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One 7:e34294. doi: 10.1371/journal.pone.0034294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 207:919–928. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Marzoa J, Fernández V, De La Cruz F, Martínez-Martínez L, Alonso MP, Nicolas-Chanoine MH, Johnson JR, Johnston B, López-Cerero L, Pascual Á, Rodríguez-Baño J. 2013. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 51:3358–3367. doi: 10.1128/JCM.01555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girlich D, Poirel L, Nordmann P. 2009. CTX-M expression and selection of ertapenem resistance in Klebsiella pneumoniae and Escherichia coli. Antimicrob Agents Chemother 53:832–834. doi: 10.1128/AAC.01007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Martínez L, Hernández-Allés S, Albertí S, Tomás J, Benedi V, Jacoby G. 1996. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob Agents Chemother 40:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bialek-Davenet S, Marcon E, Leflon-Guibout V, Lavigne JP, Bert F, Moreau R, Nicolas-Chanoine MH. 2011. In vitro selection of ramR and soxR mutants overexpressing efflux systems by fluoroquinolones as well as cefoxitin in Klebsiella pneumoniae. Antimicrob Agents Chemother 55:2795–2802. doi: 10.1128/AAC.00156-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller K, O'Neill AJ, Chopra I. 2002. Response of Escherichia coli hypermutators to selection pressure with antimicrobial agents from different classes. J Antimicrob Chemother 49:925–934. doi: 10.1093/jac/dkf044. [DOI] [PubMed] [Google Scholar]
- 31.Malléa M, Chevalier J, Eyraud A, Pagès JM. 2002. Inhibitors of antibiotic efflux pump in resistant Enterobacter aerogenes strains. Biochem Biophys Res Commun 293:1370–1373. doi: 10.1016/S0006-291X(02)00404-7. [DOI] [PubMed] [Google Scholar]
- 32.Mallea M, Chevalier J, Bornet C, Eyraud A, Davin-Regli A, Bollet C, Pagès JM. 1998. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology 144:3003–3009. doi: 10.1099/00221287-144-11-3003. [DOI] [PubMed] [Google Scholar]
- 33.Nicoloff H, Perreten V, Levy SB. 2007. Increased genome instability in Escherichia coli lon mutants: relation to emergence of multiple-antibiotic-resistant (Mar) mutants caused by insertion sequence elements and large tandem genomic amplifications. Antimicrob Agents Chemother 51:1293–1303. doi: 10.1128/AAC.01128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koutsolioutsou A, Peña-Llopis S, Demple B. 2005. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob Agents Chemother 49:2746–2752. doi: 10.1128/AAC.49.7.2746-2752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Viveiros M, Dupont M, Rodrigues L, Couto I, Davin-Regli A, Martins M, Pagès JM, Amaral L. 2007. Antibiotic stress, genetic response and altered permeability of Escherichia coli. PLoS One 2:e365. doi: 10.1371/journal.pone.0000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dusane DH, Hosseinidoust Z, Asadishad B, Tufenkji N. 2014. Alkaloids modulate motility, biofilm formation and antibiotic susceptibility of uropathogenic Escherichia coli. PLoS One 9:e112093. doi: 10.1371/journal.pone.0112093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chavant P, Gaillard-Martinie B, Talon R, Hébraud M, Bernardi T. 2007. A new device for rapid evaluation of biofilm formation potential by bacteria. J Microbiol Methods 68:605–612. doi: 10.1016/j.mimet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Vinué L, McMurry L, Levy S. 2013. The 216 bp marB gene of the marRAB operon in Escherichia coli encodes a periplasmic protein which reduces the transcription rate of marA. FEMS Microbiol Lett 345:49–55. doi: 10.1111/1574-6968.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogel J, Papenfort K. 2006. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol 9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Rahmati S, Yang S, Davidson AL, Zechiedrich EL. 2002. Control of the AcrAB multidrug efflux pump by quorum sensing regulator SdiA. Mol Microbiol 43:677–685. doi: 10.1046/j.1365-2958.2002.02773.x. [DOI] [PubMed] [Google Scholar]
- 42.Dupont M, James CE, Chevalier J, Pagès JM. 2007. An early response to environmental stress involves regulation of OmpX and OmpF, two enterobacterial outer membrane pore-forming proteins. Antimicrob Agents Chemother 51:3190–3198. doi: 10.1128/AAC.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai SJ, Inouye M. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem 277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- 44.Castillo-Keller M, Vuong P, Misra R. 2006. Novel mechanism of Escherichia coli porin regulation. J Bacteriol 188:576–586. doi: 10.1128/JB.188.2.576-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 47.Tangden T, Adler M, Cars O, Sandegren L, Lowdin E. 2013. Frequent emergence of porin-deficient subpopulations with reduced carbapenem susceptibility in ESBL-producing Escherichia coli during exposure to ertapenem in an in vitro pharmacokinetic model. J Antimicrob Chemother 68:1319–1326. doi: 10.1093/jac/dkt044. [DOI] [PubMed] [Google Scholar]
- 48.Sato T, Yokota SI, Uchida I, Okubo T, Usui M, Kusumoto M, Akiba M, Fujii N, Tamura Y. 2013. Fluoroquinolone resistance mechanisms in an Escherichia coli isolate, HUE1, without quinolone resistance-determining region mutations. Front Microbiol 4:125. doi: 10.3389/fmicb.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Dzink-Fox J, Chen M, Levy SB. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother 45:1515–1521. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olliver A, Vallé M, Chaslus-Dancla E, Cloeckaert A. 2004. Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett 238:267–272. doi: 10.1111/j.1574-6968.2004.tb09766.x. [DOI] [PubMed] [Google Scholar]
- 51.Pradel E, Pagès JM. 2002. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob Agents Chemother 46:2640–2643. doi: 10.1128/AAC.46.8.2640-2643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneiders T, Amyes SGB, Levy SB. 2003. Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob Agents Chemother 47:2831–2837. doi: 10.1128/AAC.47.9.2831-2837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren Y, Li J, Zhang X, Zeng Z, Zhen P, Zeng D, Liu Y, Jiang H. 2012. Contribution of insertion within acrR to acrAB expression and ciprofloxacin resistance in a veterinary Escherichia coli isolate. J Anim Vet Adv 11:2436–2439. doi: 10.3923/javaa.2012.2436.2439. [DOI] [Google Scholar]
- 54.Wei Y, Lee JM, Smulski DR, Larossa RA. 2001. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J Bacteriol 183:2265–2272. doi: 10.1128/JB.183.7.2265-2272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavío MM, Aquili VD, Poveda JB, Antunes NT, Sánchez-Céspedes J, Vila J. 2010. Quorum-sensing regulator sdiA and marA overexpression is involved in in vitro-selected multidrug resistance of Escherichia coli. J Antimicrob Chemother 65:1178–1186. doi: 10.1093/jac/dkq112. [DOI] [PubMed] [Google Scholar]
- 56.Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother 55:1485–1493. doi: 10.1128/AAC.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rolhion N, Carvalho FA, Darfeuille-Michaud A. 2007. OmpC and the σE regulatory pathway are involved in adhesion and invasion of the Crohn's disease-associated Escherichia coli strain LF82. Mol Microbiol 63:1684–1700. doi: 10.1111/j.1365-2958.2007.05638.x. [DOI] [PubMed] [Google Scholar]
- 58.Turner AK, Terry TD, Sack DA, Londoño-Arcila P, Darsley MJ. 2001. Construction and characterization of genetically defined aro omp mutants of enterotoxigenic Escherichia coli and preliminary studies of safety and immunogenicity in humans. Infect Immun 69:4969–4979. doi: 10.1128/IAI.69.8.4969-4979.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. 2008. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol 190:7693–7698. doi: 10.1128/JB.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diard M, Baeriswyl S, Clermont O, Gouriou S, Picard B, Taddei F, Denamur E, Matic I. 2007. Caenorhabditis elegans as a simple model to study phenotypic and genetic virulence determinants of extraintestinal pathogenic Escherichia coli. Microbes Infect 9:214–223. doi: 10.1016/j.micinf.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 61.Lane MC, Alteri CJ, Smith SN, Mobley HLT. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hernández-Jiménez E, del Campo R, Toledano V, Vallejo-Cremades MT, Muñoz A, Largo C, Arnalich F, García-Rio F, Cubillos-Zapata C, López-Collazo E. 2013. Biofilm vs. planktonic bacterial mode of growth: which do human macrophages prefer? Biochem Biophys Res Commun 441:947–952. doi: 10.1016/j.bbrc.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 63.Olesen B, Frimodt-Møller J, Leihof RF, Struve C, Johnston B, Hansen DS, Scheutz F, Krogfelt KA, Kuskowski MA, Clabots C, Johnson JR. 2014. Temporal trends in antimicrobial resistance and virulence-associated traits within the Escherichia coli sequence type 131 clonal group and its H30 and H30-Rx subclones, 1968 to 2012. Antimicrob Agents Chemother 58:6886–6895. doi: 10.1128/AAC.03679-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin J, Ko KS. 2015. Effect of plasmids harbouring blaCTX-M on the virulence and fitness of Escherichia coli ST131 isolates. Int J Antimicrob Agents 46:214–218. doi: 10.1016/j.ijantimicag.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 65.Linares JF, Gustafsson I, Baquero F, Martinez JL. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A 103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wood TK, González Barrios AF, Herzberg M, Lee J. 2006. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol 72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.