Abstract
The NLRP3 inflammasome responds to microbes and danger signals by processing and activating proinflammatory cytokines including IL-1β and IL-18. We show that NLRP3 inflammasome activation is restricted to interphase of the cell cycle by NEK7, a serine/threonine kinase previously implicated in mitosis. NLRP3 inflammasome activation requires NEK7, which binds to the NLRP3 leucine-rich repeat domain in a kinase-independent manner downstream from the induction of mitochondrial ROS. This interaction is necessary for NLRP3-ASC complex formation, ASC oligomerization, and caspase-1 activation. NEK7 promotes the NLRP3-dependent cellular inflammatory response to intraperitoneal monosodium urate challenge, and the development of experimental autoimmune encephalitis in mice. Our findings suggest NEK7 serves as a cellular switch that enforces mutual exclusivity between the inflammasome response and cell division.
Inflammasomes are multiprotein complexes that serve as platforms for the activation of caspase-1, leading to the processing and secretion of IL-1β and IL-18, and to the induction of pyroptosis, a form of programmed inflammatory cell death1. The NLRP3 inflammasome is activated in macrophages by a two-step process that involves priming through activation of NF-κB-activating pathways prior to or simultaneously with exposure to a second NLRP3-specific trigger such as extracellular ATP, alum, and the pore-forming toxin nigericin. Upon activation, NLRP3 and the adaptor ASC move from their positions, respectively, at the endoplasmic reticulum (ER) and mitochondria to form a complex at the perinuclear region, an event dependent on microtubule polymerization, acetylation of α-tubulin, and dynein-mediated transport of mitochondria from the periphery2-4. This complex recruits pro-caspase-1, resulting in caspase-1 activation.
Here, forward genetic analysis of inflammasome activation in C57BL/6J mice revealed that NEK7, one of eleven NEK kinases found in vertebrates, is an important component of the NLRP3 inflammasome in macrophages. NEK7 has also been implicated in mitotic spindle formation and separation of centrosomes (with NEK6 and NEK9)5-9, in abscission during cytokinesis8, 10, 11, and in the regulation of interphase centrosomes12. Our findings suggest that NLRP3 inflammasome activation and mitosis cannot occur simultaneously, in part because the quantity of NEK7 present in macrophages is sufficient for only one or the other. Thus, NEK7 acts as a switch between mitosis and inflammasome activation competence, both of which require NEK7.
Results
Impaired NLRP3 inflammasome activation caused by a Nek7 mutation
To identify regulators of NLRP3-mediated inflammation, we carried out a forward genetic screen in which macrophages isolated from C57BL/6J mice carrying homozygous and heterozygous ENU-induced mutations were assayed for IL-1β secretion in response to priming with lipopolysaccharide (LPS) followed by nigericin stimulation (unless otherwise indicated, peritoneal macrophages were used throughout the study). We screened 16,816 G3 mice derived from 811 G1 grandsires bearing 49,590 non-synonymous mutations within the coding regions or splice junctions of 15,927 genes. 16,328 mutations in 9,499 genes were tested three or more times in the homozygous state; these included one or more putative null alleles of 924 genes. Among the phenovariants detected, several mice from a single pedigree displayed diminished IL-1β secretion by macrophages (Fig. 1a). This phenotype was called Cuties.
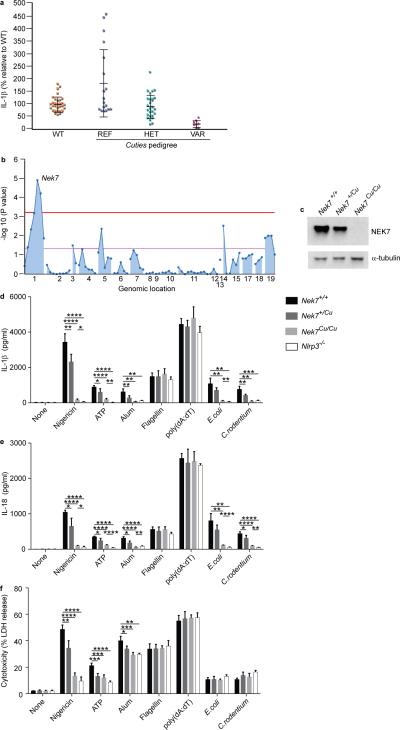
Figure 1.
Impaired NLRP3 inflammasome activation in macrophages from Cuties mice. (a) ELISA analysis of IL-1β secretion by peritoneal macrophages from mice of the Cuties pedigree. Macrophages were primed with LPS and treated with nigericin. Data points represent individual mice; REF, Nek7+/+; HET, Nek7+/Cu; VAR, Nek7Cu/Cu. (b) Manhattan plot showing linkage of a mutation in Nek7 with the Cuties phenotype using a semidominant transmission model (P = 1.153 × 10−5). The −log10 P values are plotted versus the chromosomal positions of 79 mutations identified in the G1 founder of the pedigree. Horizontal red and purple lines represent thresholds of P = 0.05 with or without Bonferroni correction, respectively. (c) Immunoblot showing NEK7 expression in Cuties peritoneal macrophages. ELISA analysis of (d) IL-1β and (e) IL-18 secreted by peritoneal macrophages primed with LPS and treated with the indicated inflammasome stimuli. (f) Peritoneal macrophages were primed with LPS and treated with the indicated inflammasome stimuli. Pyroptosis, as measured by lactate dehydrogenase (LDH) released into the culture medium, is calculated relative to the total LDH activity in lysates of unstimulated cells. In d-f, n = 5 Nek7+/+, 5 Nek7+/Cu, 4 Nek7Cu/Cu, 4 Nlrp3−/− mice. * P≤0.05; ** P≤0.01; *** P≤0.001; **** P≤0.0001 (unpaired, two-tailed Student's t test). Results in d-f are representative of two independent experiments.
To identify the Cuties (Cu) mutation, we sequenced coding exons and flanking splice junctions in genomic DNA from the G1 grandsire of the Cuties pedigree13. 79 mutations were found with 97% coverage of target sequences at ≥ 10 reads. Each of the 79 mutation sites was genotyped in G3 mice of the Cuties pedigree and a mutation in Nek7 showed strongest linkage with the Cuties phenotype using a semidominant model of transmission (−log10[Pnon-linkage] = 4.938) (Fig. 1b). The mutation, a T to A transversion at 138,544,242 bp (GRCm38) on chromosome 1, was predicted to cause a cysteine to premature stop codon substitution at position 53 of the 302 amino acid NEK7 protein. No NEK7 protein was detected in Nek7Cu/Cu macrophages, consistent with nonsense-mediated decay of the transcript (Fig. 1c).
Nek7+/Cu mice appeared normal and showed no internal anatomical abnormalities. At birth, Nek7Cu/Cu mice were similar in size to their littermates, but by two months of age weighed on average 30% less than their wild-type or heterozygous siblings. Nek7Cu/Cu mice had an abnormal gait and slight paresis of the limbs, and were infertile. Consistent with a previous report11, heterozygous mutant crosses (C57BL/6J background) revealed non-Mendelian transmission ratios of the mutation, with wild-type, heterozygous, and homozygous mice representing 34.9%, 60.4%, and 4.7% respectively of offspring at birth.
Further analysis demonstrated reduced IL-1β production by LPS-primed Nek7Cu/Cu macrophages stimulated with ATP and alum (Fig. 1d). Similar results were obtained using bone marrow-derived macrophages (BMDM) and bone marrow-derived dendritic cells (BMDC) (Supplementary Fig. 1a,b). NLRP3-dependent IL-18 production and pyroptosis were also impaired in LPS-primed Nek7Cu/Cu macrophages stimulated with nigericin, ATP, or alum (Fig. 1e,f). The IL-1β and IL-18 responses of Cuties macrophages to E. coli or C. rodentium infection, mediated by non-canonical caspase-11-dependent NLRP3 signaling14, were defective (Fig. 1d,e); however, the programmed death response that is NLRP3-independent but caspase-11-dependent was normal (Fig. 1f). IL-1β and IL-18 production and lactate dehydrogenase (LDH) release by unstimulated versus stimulated Nek7Cu/Cu macrophages were significantly different, indicating that the responses of Nek7Cu/Cu macrophages are severely but not completely impaired. Macrophages from Nek7+/Cu mice exhibited intermediate IL-1β and IL-18 production and pyroptosis relative to Nek7Cu/Cu and wild-type cells, confirming semidominant inheritance of the Cuties phenotype (Fig. 1d-f). Time course analysis of the IL-1β, IL-18, and pyroptosis responses of Cuties macrophages supported that they were inhibited rather than delayed following LPS+nigericin or LPS+ATP stimulation (Supplementary Fig. 1c,d).
The IL-1β, IL-18, and pyroptosis responses of Cuties macrophages were similar to those of wild-type macrophages when treated with flagellin or poly(dA:dT), inducers of the NLRC4 and AIM2 inflammasomes, respectively (Fig. 1d-f). In addition, tumor necrosis factor (TNF) and IL-6 production by Cuties macrophages in response to Toll-like receptor 4 (TLR4), TLR2/6, or TLR7 stimulation were normal (Supplementary Fig. 1e,f). These findings demonstrate a specific defect in the response of Cuties mice to activators of the NLRP3 inflammasome.
The frequencies of macrophages, neutrophils, and other major lymphocyte populations were normal in the peripheral blood of Cuties mice (Supplementary Fig. 2a). In addition, colony forming unit assays showed normal differentiation and proliferation of Nek7Cu/Cu bone marrow hematopoietic stem cells and myeloid progenitor cells in response to treatment with macrophage colony-stimulating factor (M-CSF) (Supplementary Fig. 2b). Recruitment of exudate cells, macrophages, and neutrophils to the peritoneal cavity in response to i.p. injection with bioactive IL-1β was also normal (Supplementary Fig. 2c). Although RNAi-mediated NEK7 depletion has been reported to result in apoptosis8, we observed a similar frequency of apoptotic cells among wild-type and Nek7Cu/Cu macrophages before or after LPS priming (Supplementary Fig. 2d).
We performed siRNA-mediated knockdown of endogenous NEK7 in the mouse macrophage cell line J774A.1 and observed dramatically reduced IL-1β secretion in response to nigericin or ATP stimulation after LPS priming (Supplementary Fig. 3a). Similarly, in HEK293T cells reconstituted with functional mouse NLRP3 inflammasomes, siRNA-mediated knockdown of endogenous NEK7 inhibited IL-1β secretion in response to nigericin or ATP stimulation (Supplementary Fig. 3b). A null allele of Nek7 generated by CRISPR/Cas9 gene targeting reproduced the Cuties phenotype in homozygous mice (Nek7−/−) (Supplementary Fig. 3c). Importantly, in a cultured human monocyte cell line (THP1) and in primary human monocytes, knockdown of endogenous NEK7 resulted in reduced IL-1β secretion in response to LPS priming plus nigericin or ATP stimulation (Supplementary Fig. 3d,e), whereas LPS-induced IL-6 secretion was unaffected by NEK7 knockdown (Supplementary Fig. 3f). These findings indicated that NEK7 is necessary for NLRP3 inflammasome activation in mouse macrophages and human monocytes.
NEK7 promotes cellular inflammatory responses in vivo
In vivo, intraperitoneal injection of monosodium urate crystals (MSU) induces NLRP3-dependent recruitment of inflammatory cells and IL-1β production in the peritoneal cavity. When challenged with MSU, Cuties mice showed impaired recruitment of total cells, neutrophils, and F4/80-positive monocytes/macrophages to the peritoneal cavity (Fig. 2A). Bone marrow chimeras generated by reconstitution of irradiated wild-type mice with either wild-type or Nek7−/− bone marrow responded similarly to non-chimeric wild-type and Nek7Cu/Cu mice, respectively (Fig. 2B). MSU-induced IL-1β secretion detected in the lavage fluid was also significantly decreased in Nek7−/− chimeras relative to wild-type chimeras (Fig. 2C). Thus, NEK7 functions in the hematopoietic compartment to promote an NLRP3-dependent inflammatory response to intraperitoneal challenge with MSU.
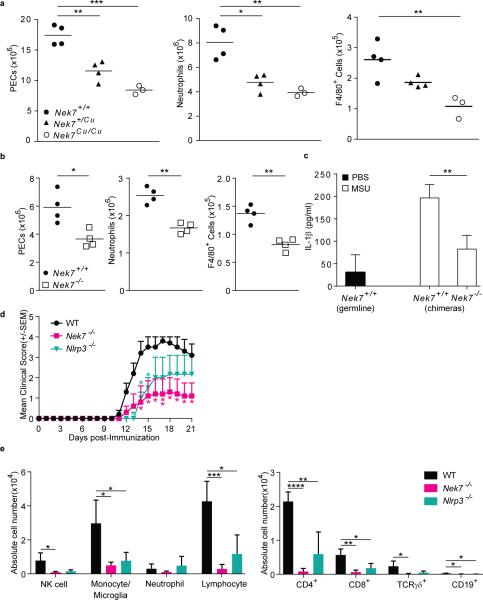
Figure 2.
NEK7 promotes an inflammatory response in vivo. (a,b) Flow cytometric analysis of the number of peritoneal exudate cells (PECs), neutrophils (Ly6G+ F4/80−), and monocytes/macrophages (F4/80+) in the peritoneum of (a) Cuties mutant mice or (b) bone marrow chimeric mice 6 h after intraperitoneal injection of MSU. For bone marrow chimeras, donor genotypes are indicated; recipients were Nek7+/+. Data points represent individual mice; the mean is indicated. (c) ELISA analysis of IL-1β in the lavage fluid of chimeric mice 6 h after MSU injection (n = 4 mice per genotype or chimera type). For bone marrow chimeras, donor genotypes are indicated; recipients were Nek7+/+. (d) Mean clinical score of mice immunized with rhMOG on day 0. P values indicate significance of differences relative to WT. (e) Flow cytometric analysis of the number of the indicated immune cell types in the spinal cords of mice 21 days after immunization with rhMOG (n = 4 mice per genotype). Cell subsets were gated as: NK (NK1.1+CD3ε−), monocytes/microglia (CD11b+Gr1−), neutrophils (CD11b+Gr1+), lymphocytes (CD45hiCD11b−). * P≤0.05; ** P≤0.01; *** P≤0.001; **** P≤0.0001 (unpaired, two-tailed Student's t test). Results in a-c are representative of two independent experiments. Results in d and e are representative of one experiment.
We also investigated whether Cuties mice are protected against IL-1β-driven inflammatory disease using an experimental autoimmune encephalitis (EAE) model15, 16. Recombinant human myelin oligodendrocyte glycoprotein (rhMOG) immunization of wild-type mice induced EAE with paralysis and immune cell infiltration of the CNS. Nek7−/− mice exhibited reduced disease severity relative to wild-type mice (Fig. 2d), as well as reduced recruitment of lymphocytes (CD4, CD8, TCRγδ, CD19+ B cells), monocytes/microglia, and NK cells to the spinal cord (Fig. 2e). Thus, NEK7 is necessary for the development of EAE.
NEK7 is required for inflammasome assembly and activation of caspase-1
Mitochondrial reactive oxygen species (ROS) generation, calcium influx, and reduction of cellular cyclic AMP (cAMP) have been reported to activate the NLRP3 inflammasome2, 172, 17, though their effect(s) have been disputed18. We found that LPS-primed wild-type and Nek7Cu/Cu macrophages induced similar amounts of mitochondrial ROS after nigericin treatment (Fig. 3a). Calcium influx to the cytoplasm induced by ATP was also similar between LPS-primed wild type and Nek7Cu/Cu macrophages, although intracellular calcium levels decreased more rapidly in Nek7Cu/Cu macrophages than in wild-type macrophages over a 30 minute period after ATP administration (Fig. 3b). cAMP, which may bind directly to NLRP3 to inhibit inflammasome assembly17, was more abundant in Nek7Cu/Cu macrophages compared to wild-type macrophages after LPS priming plus nigericin stimulation, and showed a similar trend toward higher amounts in LPS-primed Nek7Cu/Cu macrophages treated with ATP (Fig. 3c). Thus, NLRP3 inflammasome activation may be partially impaired in Nek7Cu/Cu macrophages due to a defective cAMP response in which elevated cAMP concentrations inhibit inflammasome assembly.
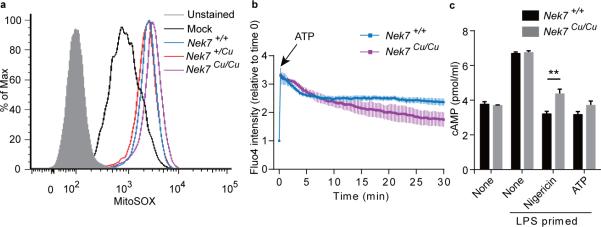
Figure 3.
ROS, Ca2+ influx, and cAMP in Cuties macrophages in response to NLRP3 inflammasome stimuli. (a) Mitochondrial ROS. Flow cytometric analysis of MitoSOX staining in wild-type peritoneal macrophages primed with LPS (Mock), or macrophages of the indicated genotypes primed with LPS and treated with nigericin. (b) Calcium influx. Peritoneal macrophages were stained with calcium indicator fluo-4/AM. Images of untreated cells were acquired (t=0), then ATP was added to the culture medium and cells were imaged at 15 sec intervals for 30 min. The fold increase in fluorescence intensity of all cells in a field relative to time 0 is shown. (c) Intracellular cAMP in peritoneal macrophages primed with LPS and treated with nigericin or ATP. ** P≤0.01 (unpaired, two-tailed Student's t test). For a-c, n = 3 mice per genotype. Results are representative of two independent experiments.
Upon NLRP3 activation, ASC oligomerizes and forms a complex with NLRP3 at the perinuclear region2, 19-21. NLRP3, ASC, pro-caspase-1, and pro-IL-1β expression were similar in wild-type, Nek7Cu/Cu, and Nek7+/Cu macrophages primed with LPS (Fig. 4a). NEK7 expression was unaffected by LPS priming in wild-type or NLRP3-deficient macrophages (Fig. 4a). However, when primed with LPS and stimulated with either nigericin or ATP, reduced association between NLRP3 and ASC (Fig. 4b), as well as a failure of ASC oligomerization (Fig. 4c) were detected in Nek7Cu/Cu and Nek7+/Cu macrophages compared to wild-type macrophages. Amounts of acetylated α-tubulin, a regulator of NLRP3-ASC binding4, were similar between wild-type, Nek7Cu/Cu, and Nek7−/− macrophages (Fig. 4d,e). Mutant macrophages also secreted reduced amounts of mature IL-1β and active caspase-1 (p10 subunit) relative to wild-type macrophages (Fig. 4a). These findings indicate that NEK7 is dispensable for the induction of core inflammasome components, but necessary for subsequent formation of the NLRP3-ASC complex, and activation of caspase-1.
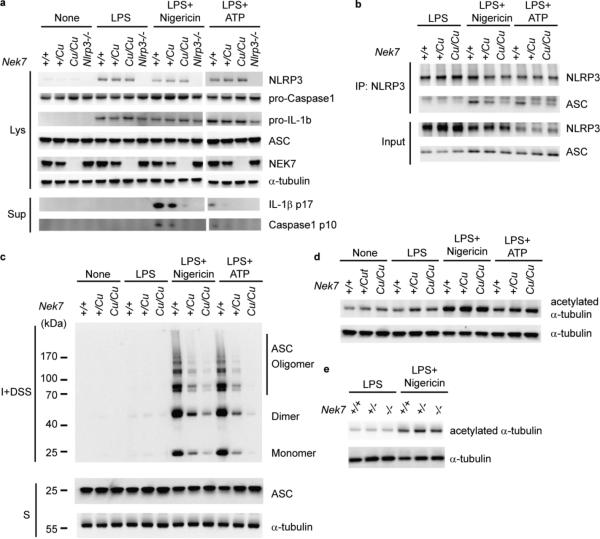
Figure 4.
NEK7 is necessary for NLRP3-ASC complex formation and ASC oligomerization. (a-c) Peritoneal macrophages were primed with LPS and stimulated with nigericin or ATP. Genotypes are for Nek7 except for lanes marked Nlrp3−/−. (a) Immunoblots showing expression of the indicated proteins in cell lysates (Lys) and culture supernatants (Sup). Secreted mature IL-1β p17 and active caspase-1 (p10 subunit) were measured in supernatants. (b) NLRP3-ASC association analyzed by immunoprecipitation and immunoblot. (c) Cell lysates were solubilized with Triton X-100-containing buffer. Insoluble (I) fractions were cross-linked with disuccinimidyl suberate (DSS) to capture ASC oligomers (I+DSS). The soluble (S) and I+DSS fractions were analyzed by immunoblotting with ASC antibodies. (d,e) Immunoblots showing acetylated α-tubulin and total α-tubulin in peritoneal macrophage lysates from (d) Cuties mice or (e) Nek7−/− mice. Results are representative of two independent experiments.
NEK7 directly binds NLRP3 to promote inflammasome assembly
In unstimulated cells, NLRP3 homo-oligomerizes to form inactive preassembled complexes, which undergo conformational changes to form active inflammasome complexes containing ASC upon stimulation22. Consistent with this, using the mouse macrophage cell line RAW264.7 that lacks ASC expression23, we found NLRP3 in complexes ranging from approximately 125 kd to 1000 kd when cells were primed with LPS and extracts subjected to gel filtration chromatography (Fig. 5a). LPS priming plus nigericin treatment resulted in a shift of NLRP3 to higher molecular mass fractions; the recruitment of NLRP3 to these fractions was blocked by siRNA-mediated knockdown of NEK7, suggesting that NLRP3 binds directly to NEK7 to form a complex upon inflammasome stimulation (Fig. 5a). Indeed, when co-expressed in HEK293T cells, NEK7 could be coimmunoprecipitated with NLRP3, but not with NLRC4 or AIM2 (Fig. 5b). The NEK7-NLRP3 interaction was mediated by the intact leucine-rich repeat (LRR) domain of NLRP3 (Fig. 5c,d). Deletions of the N-terminal 20 or 30 amino acids of NEK7 did not affect binding to NLRP3 (Fig. 5e); deletions from the central or C-terminal portion of NEK7 abrogated protein expression and could not be tested. A direct interaction was confirmed via maltose-binding protein (MBP) pull-down of purified recombinant MBP-NEK7 and NLRP3 (Supplementary Fig. 4a). ASC could not interact directly with MBP-NEK7, but was recruited to the NEK7-NLRP3 complex (Supplementary Fig. 4b).
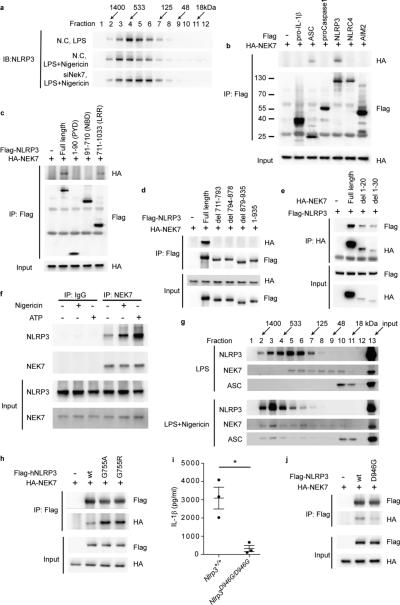
Figure 5.
NEK7 directly interacts with the LRR domain of NLRP3 to promote inflammasome assembly. (a) RAW264.7 cells were transfected with non-targeting control siRNA (N.C.) or Nek7 siRNA (siNek7) and then primed with LPS and stimulated with nigericin. Immunoblot analysis of cell lysates subjected to gel filtration chromatography. (b-e) HEK293T cells were transfected as indicated. The interaction between HA-NEK7 and Flag-NLRP3 proteins was analyzed by immunoprecipitation and immunoblot. (d) Full length NLRP3 or LRR domain mutants lacking the indicated residues were tested. (e) Full length NEK7 or mutants lacking the indicated residues were tested. (f,g) Peritoneal macrophages were primed with LPS and stimulated with nigericin or ATP. (f) Endogenous NEK7-NLRP3 association in wild-type macrophages analyzed by immunoprecipitation and immunoblot. (g) Immunoblot analysis of wild-type macrophage lysates subjected to gel filtration chromatography. (h) HEK293T cells were transfected as indicated. Lysates were analyzed as in b. (i) ELISA analysis of IL-1β secretion by wild type or homozygous Nlrp3D946G mutant peritoneal macrophages primed with LPS and stimulated with nigericin. Data points represent individual mice. * P≤0.05 (unpaired, two-tailed Student's t test). (j) HEK293T cells were transfected as indicated. Lysates were analyzed as in b. Results are representative of two independent experiments.
In LPS-primed macrophages, the interaction between endogenous NEK7 and NLRP3 was significantly enhanced by nigericin or ATP stimulation (Fig. 5f). Moreover, gel filtration chromatography of extracts from macrophages primed with LPS and treated with nigericin demonstrated co-elution of NEK7, NLRP3, and ASC and a shift to higher molecular mass fractions as compared to LPS-stimulated macrophage extracts (Fig. 5g). The NEK7-NLRP3 interaction was partially dependent on microtubule polymerization (Supplementary Fig. 5a). However, ciliobrevin D, an inhibitor of dynein that prevented NLRP3-ASC complex formation at the perinuclear region4 had no effect on the NEK7-NLRP3 interaction (Supplementary Fig. 5a). The NEK7-NLRP3 interaction was also dependent on ROS-induced phosphorylation of NEK7. Treatment of macrophage lysates with the non-specific phosphatase calf intestinal alkaline phosphatase (Supplementary Fig. 5b) or the ROS scavenger N-acetylcysteine (NAC) (Supplementary Fig. 5c) reduced both NEK7 phosphorylation and the interaction between NEK7 and NLRP3. IL-1β production by J774A.1 cells in response to ATP stimulation was dose-dependently diminished by NAC treatment (Supplementary Fig. 5d).
Mutant NLRP3 proteins containing the LRR domain missense mutations G755A and G755R, associated with neonatal onset multisystem inflammatory disease (NOMID)24-26, showed increased binding to NEK7 in HEK293T cells (Fig. 5h). Conversely, a hypomorphic NLRP3 missense mutant (D946G) that failed to support IL-1β secretion by LPS-primed macrophages treated with nigericin, showed reduced binding to NEK7 (Fig. 5i,j). Together, these data suggest that inflammasome assembly requires the physical association of phosphorylated NEK7 with NLRP3.
NEK7 kinase activity is dispensable for NLRP3 inflammasome activation
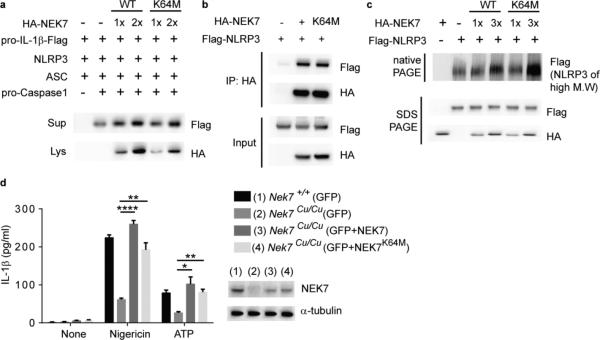
We reconstituted functional NLRP3 inflammasomes in HEK293T cells and showed that overexpression of either wild-type NEK7 or kinase-inactive NEK7 (K64M) (Supplementary Fig. 4c)8 enhanced secretion of mature IL-1β into the culture medium (Fig. 6a). Wild-type NEK7 and NEK7K64M coimmunoprecipitated with similar amounts of NLRP3 in HEK293T cell lysates (Fig. 6b), and a direct interaction between recombinant MBP-NEK7K64M and NLRP3 was also observed (Supplementary Fig. 4a). Moreover, both NEK7 and NEK7K64M promoted recruitment of NLRP3 into high molecular mass protein complexes without significantly changing total NLRP3 expression (Fig. 6c). Finally, when introduced by electroporation into Nek7Cu/Cu macrophages both wild-type NEK7 and NEK7K64M rescued IL-1β secretion in response to LPS priming plus nigericin or ATP (Fig. 6d).
Figure 6.
NEK7 kinase activity is nonessential for NLRP3 inflammasome activation. (a-c) HEK293T cells were transfected as indicated. (a) Immunoblots showing pro-IL-1β-Flag and HA-NEK7 expression in cell lysates (Lys) and culture supernatants (Sup). IL-1β maturation was assessed by p17-Flag immunoblot of the supernatant. (b) The interaction between NEK7 or NEK7K64M and NLRP3 was analyzed by immunoprecipitation and immunoblot. (c) Flag immunoblot of cell lysates subjected to native PAGE to detect NLRP3 in high molecular weight complexes, or SDS-PAGE to detect total NLRP3. (d) Wild type or Nek7Cu/Cu peritoneal macrophages were electroporated with mRNA encoding GFP, GFP + NEK7, or GFP + NEK7K64M. GFP-positive cells were sorted, primed with LPS, and stimulated with nigericin or ATP. Secreted IL-1β was measured by ELISA. n = 3 mice per genotype. (Inset) NEK7 expression was assessed by immunoblot. * P≤0.05; ** P≤0.01; **** P≤0.0001 (unpaired, two-tailed Student's t test). Results are representative of two independent experiments.
We found that NEK6, a related NEK family member 87% identical to NEK7 in its kinase domain27, failed to bind NLRP3 (Supplementary Fig. 6a), and was not required for macrophage production of IL-1β in response to activators of NLRP3, NLRC4, and AIM2 inflammasomes (Supplementary Fig. 6b,c). These results indicate that the kinase activity of NEK7 is nonessential for activation of the NLRP3 inflammasome.
NLRP3 inflammasome activation is blocked in mitotic cells
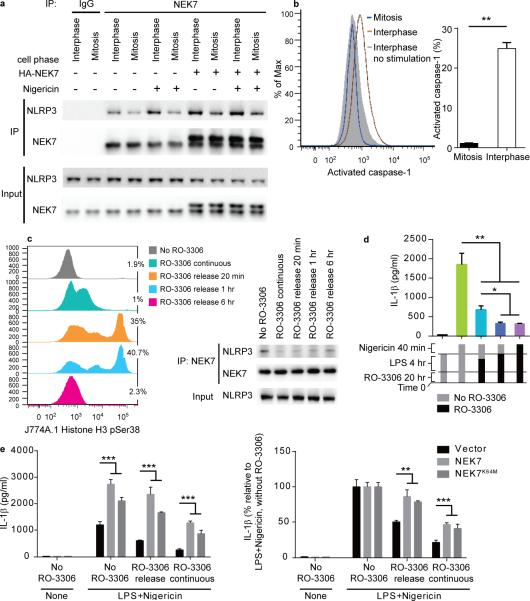
Since NEK7 is required for both NLRP3 inflammasome activation and mitosis, we investigated the interaction between NEK7 and NLRP3 during mitosis versus interphase of the cell cycle. Endogenous NEK7 and NLRP3 coimmunoprecipitated from both mitotic and interphase J774A.1 cells, with a greater amount of NLRP3 bound to NEK7 during interphase than during mitosis (Fig. 7a). LPS priming plus nigericin stimulation of interphase cells, but not mitotic cells, increased the amount of NLRP3 interacting with NEK7 relative to the amount in unstimulated cells at the corresponding phase of the cell cycle (Fig. 7a). Moreover, stimulated cells in interphase displayed increased caspase-1 activation relative to cells in mitosis (Fig. 7b). 25% of stimulated interphase cells expressed activated caspase-1, as compared to only 2% of stimulated mitotic cells (Fig. 7b). Consistent with these data, J774A.1 cells synchronized by chemical arrest at the G2/M phase border28 and released to synchronously enter mitosis (Fig. 7c, left panel) exhibited diminished NLRP3-NEK7 interaction (Fig. 7c, right panel) and produced less IL-1β in response to LPS+nigericin treatment than unsynchronized cells (Fig. 7d), which contained approximately 2% mitotic cells (Fig. 7c, left panel). Note that although they do not maximally express the mitosis marker phosphorylated histone H3, cells with continuous RO-3306 treatment behave as mitotic cells with respect to NEK7-NLRP3 complex formation and IL-1β production, presumably because they are held in a state in which all cellular proteins are poised in their mitosis positions. Similarly, G2/M-synchronized HEK293T cells overexpressing NEK7 and NLRP3 exhibited diminished NLRP3-NEK7 interaction upon release to mitosis, which recovered when cells later entered the G1 phase (Supplementary Fig. 7a).
Figure 7.
NLRP3 inflammasome activation is blocked during mitosis. (a) NEK7-NLRP3 association in LPS-primed J774A.1 mitotic or interphase cells, with or without HA-NEK7 overexpression, analyzed by immunoprecipitation and immunoblot. (b) J774A.1 cells were primed with LPS and stimulated with nigericin together with fluorescent FAM-FLICA-caspase-1 probe specific for activated caspase-1. Flow cytometric analysis of cells containing activated caspase-1 (left); their frequency among mitotic and interphase cells is graphed (right). (c-e) J774A.1 cells were arrested at the G2/M phase border by incubation with RO-3306, then released from arrest for different time periods before analysis. (c) Cells were treated with RO-3306 for 20 h, with LPS added during the last 4 h. Release was in fresh media without RO-3306 and LPS for the indicated times. Flow cytometric analysis of phosphorylated histone H3-positive mitotic cells (left); the percentage of mitotic cells is indicated. The interaction between endogenous NEK7 and NLRP3 was analyzed by immunoprecipitation and immunoblot (right). (d) ELISA analysis of IL-1β in the culture supernatants of J774A.1 cells primed with LPS and stimulated with nigericin. Release from RO-3306 arrest was 1 h before (teal) or concurrent with (blue) nigericin stimulation. (e) ELISA analysis of IL-1β in the culture supernatants of J774A.1 cells stably overexpressing NEK7 or NEK7K64M primed with LPS and stimulated with nigericin. RO-3306 release was at the time nigericin was added. * P≤0.05; ** P≤0.01; *** P≤0.001 (unpaired, two-tailed Student's t test). For b, d, and e, the means of triplicate samples are plotted. Results are representative of two independent experiments.
Together, these data indicate that a higher basal quantity of the NEK7-NLRP3 complex exists in interphase cells than in mitotic cells, and that inflammasome stimulation increases the amount of NLRP3 bound to NEK7 and the amount of activated caspase-1 specifically in interphase cells, resulting in greater IL-1β production from interphase than from mitotic cells. We also found that overexpression of NEK7 in J774A.1 cells enhanced NEK7-NLRP3 binding in mitotic cells stimulated with LPS+nigericin compared to mitotic cells without NEK7 overexpression (Fig. 7a, compare lanes 6 and 10). In addition, IL-1β production by mitotic J774A.1 cells was enhanced by overexpression of NEK7 or NEK7K64M (Fig. 7e). Our findings suggest that a limiting amount of cellular NEK7 may be available for either mitosis or inflammasome activation, but not both simultaneously.
We tested whether component(s) of mitotic cell extracts activate the NLRP3 inflammasome in a NEK7-dependent manner. We found that IL-1β production induced by mitotic cell extracts was similar between LPS-primed Nek7−/− and wild-type macrophages and dependent on the AIM2 inflammasome (Supplementary Fig. 7b and data not shown). Thus, the function of NEK7 in inhibiting simultaneous mitosis and inflammasome activation is not likely to be the prevention of aberrant inflammasome activation induced by nuclear activators exposed during breakdown of the nuclear membrane in mitosis.
Discussion
A member of the NIMA-related serine/threonine kinase family, NEK7 has been implicated in mitotic progression downstream of NEK9 (refs. 5-9). Depletion of NEK7 by RNAi arrests cells in mitosis and induces apoptosis; these defects stem from impaired mitotic spindle formation that may be secondary to deficiencies in microtubule organization7, 8, 10. If allowed to progress past the spindle assembly checkpoint, NEK7-depleted cells then arrest in cytokinesis8, 10, 11. Mouse embryo fibroblasts derived from homozygous NEK7 null embryos displayed aneuploidy, polyploidy, and increased frequencies of binuclear cells, confirming the role of NEK7 in mitosis and cytokinesis11. Our findings that only 18.8% of the expected number of Nek7Cu/Cu offspring were born from heterozygous crosses, and these mice displayed infertility, abnormal gait, and slight paralysis of the limbs, support an important but redundant role for NEK7 in mitosis. Here, we established that NEK7, by direct interaction with NLRP3, is also an important component of the NLRP3 inflammasome, contributing to its assembly in macrophages in response to priming with LPS and stimulation with nigericin, ATP, or alum. The observation that binding of NEK7 to NLRP3 requires ROS, a mediator for multiple activators of NLRP329, supports a broad role for NEK7 in NLRP3 inflammasome activation.
Our findings strongly suggest that NEK7 serves non-redundant functions in NLRP3 inflammasome activation and mitosis that cannot occur simultaneously. In support of this hypothesis, caspase-1 activation and IL-1β production induced by LPS+nigericin were robust during interphase but greatly reduced during mitosis, and NEK7 overexpression partially rescued IL-1β production by mitotic cells. This latter point, and the fact that LPS+nigericin stimulation failed to increase endogenous NEK7-NLRP3 interaction in mitotic cells, suggest that cellular NEK7 amounts are limiting in the choice between mitosis and inflammasome activation. The cellular significance of precluding NLRP3 inflammasome activation during mitosis is unknown. We hypothesized that this regulation may prevent aberrant activation of the NLRP3 inflammasome by putative nuclear activators exposed to the cytosol upon breakdown of the nuclear envelope during mitosis. However, mitotic cell extracts induced similar IL-1β production by LPS-primed Nek7−/− and wild-type macrophages, which was dependent on the AIM2 inflammasome, suggesting that NLRP3 is not activated by nuclear components exposed during mitosis.
During mitosis, the nuclear membrane dismantles, vesicle trafficking halts, Golgi and ER membranes reorganize, actin and microtubule cytoskeletons dramatically restructure, chromosomes condense, and transcription and translation slow or stop30. These cellular changes may prohibit an NLRP3-dependent inflammatory response, which requires microtubule polymerization4, localization of NLRP3 and ASC at the perinuclear region2, 3, secretion of IL-1β and IL-18 (ref. 31), and the induction of IL-1β- and IL-18-dependent transcriptional programs. IL-1β is known to induce the production of H2O2 and oxygen radicals capable of damaging DNA32, 33, which is not readily repaired during mitosis34. The phagocytic response also involves the generation of abundant ROS during the respiratory burst. Thus, NEK7 dependence may have evolved as one of several proximal switches to avoid a futile or potentially damaging inflammatory response during cell division. In principle, if NEK7 is a switch between mitosis and the NLRP3 inflammasome response, the converse situation in which mitosis cannot be initiated in cells with activated NLRP3 inflammasomes should be true, possibly for similar reasons, but this has not been formally tested.
NEK7 binds directly to NLRP3, an interaction dependent on the LRR domain of NLRP3 and required for inflammasome assembly. Notably, several mutations within the LRR domain of NLRP3 have been linked with the autoinflammatory disease NOMID24-26. Our findings suggest that the aberrant activation of NLRP3 inflammasomes in myeloid cells of patients with such mutations may stem from an increased association between NLRP3 and NEK7. Targeting this interaction may represent an alternative to neutralizing IL-1β for the treatment of NLRP3-mediated autoinflammatory diseases.
Online Methods
Mice
Eight- to ten-week old male and female mice (Mus musculus) on a pure C57BL/6J background were used in experiments. Male C57BL/6J mice purchased from The Jackson Laboratory were mutagenized with ENU as described35. Mutagenized G0 males were bred to C57BL/6J females, and the resulting G1 males were crossed to C57BL/6J females to produce G2 mice. G2 females were backcrossed to their G1 sires to yield G3 mice, which were screened for phenotype. Whole exome sequencing and mapping were performed as described13. Nek7Cuties, Nlrp3D946G (Nlrp3Park3) and Nek6 mutant strains were generated by ENU mutagenesis and are described at http://mutagenetix.utsouthwestern.edu.
All experimental procedures using mice were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Texas Southwestern Medical Center, and were conducted in accordance with institutionally approved protocols and guidelines for animal care and use. All mice were maintained at the University of Texas Southwestern Medical Center in accordance with institutionally approved protocols. Animals were to be excluded from analysis only if they displayed obvious illness or death; these conditions were not observed and no animals were excluded. No randomization of the allocation of samples or animals to experimental groups was performed.
Bone marrow chimeras
Recipient wild-type (CD45.1) mice were lethally irradiated with 950 rads using a 137Cs source and injected intravenously 2-3 h later with 5 × 106 bone marrow cells derived from the tibia and femurs of the respective donors (CD45.2). Seven weeks after engrafting, the chimeras were assessed by in vivo MSU challenge.
Generation of Nek7−/− mice using CRISPR/Cas9 targeting
Female C57BL/6J mice were superovulated by injection with 6.5 U pregnant mare serum gonadotropin (PMSG; Millipore), then 6.5 U human chorionic gonadotropin (hCG; Sigma-Aldrich) 48 h later. The superovulated mice were subsequently mated with C57BL/6J male mice overnight. The following day, fertilized eggs were collected from the oviducts and in vitro transcribed Cas9 mRNA (50 ng/μl) and Nek7 small base-pairing guide RNA (50 ng/μl; 5’-CTGCTTAATTAAATTACCTG-3’) were injected into the cytoplasm or pronucleus of the embryos. The injected embryos were cultured in M16 medium (Sigma-Aldrich) at 37°C and 95% air/5% CO2. For the production of mutant mice, 2-cell stage embryos were transferred into the ampulla of the oviduct (10-20 embryos per oviduct) of pseudopregnant Hsd:ICR (CD-1) (Harlan Laboratories) females. Chimeric mutant mice were first crossed with C57BL/6J mice and their offspring were intercrossed to generate the Nek7−/− mice. Nek7−/− mice have 4 bp net deletion in Nek7 exon 2 (5 bp deletion [italic] and 1 bp insertion [underlined]: ‘ttccagc a cacag gtaatttaatta’). The Nek7−/− mice were genotyped by capillary sequencing using 5’-CCGGAGAAGTGGAAATGGTGT-3’ and 5’-CCAGACTATCAGTAACCCTCAAAGCC-3’ as PCR primers, and 5’-TTATGTGAACTAAACAGAGCTTGG-3’ as the sequencing primer.
Reagents
Ultra-pure LPS, MALP-2 and R848 were obtained from Enzo Life Sciences; for stimulation of human monocytes, LPS (O111:B4) was from List Biologicals. Recombinant murine IL-1β (rIL-1β) was from R&D Systems. The inflammasome agonists Alum, MSU and poly(dA:dT) were from InvivoGen; nigericin was from Sigma-Aldrich; ATP was from GE Healthcare; and flagellin was from AdipoGen. RO-3306, N-acetylcysteine, colchicine, nocodazole, and cytochalasin D were from Sigma-Aldrich. Ciliobrevin D was from Xcess Biosciences. cDNAs encoding NEK7, pro-IL-1β, NLRP3 (human and mouse), ASC, pro-caspase-1 and AIM2 (GE Healthcare) were amplified using standard PCR techniques and subsequently inserted into mammalian expression vectors. All point mutations were introduced using QuickChange II XL site-directed mutagenesis kit (Agilent Technologies). All constructs were confirmed by sequencing. Recombinant NLRP3 and ASC protein were obtained from Abnova. Recombinant MBP-tagged NEK7 and NEK7K64M were expressed and purified from Escherichia coli. The following antibodies were used: mouse IL-1β (R&D Systems, AF-401-NA); NLRP3 (cryo-2) and ASC (AL177) (AdipoGen); HA (F7), mouse caspase-1 p10 (sc-514), and NEK7 (N-20) (Santa Cruz Biotechnology); NEK7 (EPR4900) (GeneTex); NEK6 (EPR5283) (Abcam); Flag (M2), β-actin (A2228) and α-tubulin (T6199) (Sigma-Aldrich); GAPDH (Cell Signaling Technology, 8884); CD45.2 (109835), CD11b (101237), and histone H3-phosphorylated (Ser28) (641006) (BioLegend); CD16/32 (553142), Ly6G (560602), CD3 (553062), CD4 (563727), CD8α (553035), B220 (557957), NK1.1 (564143) and F4/80 (565410) (BD Biosciences).
None of the cell lines used (J774A.1, THP1, RAW264.7, HEK293T) are listed in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample.
Flow cytometry
Blood was collected in Minicollect Tubes (Mercedes Medical), centrifuged at 700g to separate serum, and red blood cells remaining in the serum lysed (eBioscience) before immune cell staining and flow cytometric analysis. Cells were incubated with mAb to CD16/CD32 and labeled for 1 h at 4°C using fluorochrome-conjugated mAb against mouse CD3, CD4, CD8α, B220, NK1.1, F4/80, CD11b. Data were acquired using an LSRFortessa Cell Analyzer (BD Biosciences).
Isolation and culture of peritoneal macrophages, BMDM, BMDC
Thioglycollate-elicited macrophages were recovered 4 days after i.p. injection of 2 ml BBL thioglycollate medium, brewer modified (4%; BD Biosciences) by peritoneal lavage with 5 ml phosphate buffered saline (PBS). The peritoneal macrophages were cultured in DMEM cell culture medium (DMEM containing 10% FBS (Gemini Bio Products), 1% penicillin and streptomycin (Life Technologies) at 37°C and 95% air/5% CO2. For electroporation, 2×106 macrophages were electroporated with GFP mRNA only (2 μg), GFP mRNA (0.5 μg) + Nek7 mRNA (2 μg), or GFP mRNA (0.5 μg) + Nek7K64M mRNA (2 μg). Cell Line Nucleofector Kit V (Lonza) and program Y-001 were applied. Murine BMDMs were collected by flushing bone marrow cells from femurs and tibiae of mice. These cells were cultured for 7 days in DMEM cell culture medium containing 10% conditioned medium from L929 cells. For BMDCs, bone morrow cells were cultured in Petri dishes in 10 ml DMEM cell culture medium containing 10 ng/ml of murine GM-CSF (R&D Systems). On day 3 of culture, this was replaced with fresh GM-CSF medium. Loosely adherent cells were transferred to a fresh Petri dish and cultured for an additional 4 days.
Measurement of cytokine production
Cells were seeded onto 96-well plates at 1×105 cells per well and stimulated as follows: LPS (10 ng/ml, 4 h); MALP-2 (400 pg/ml, 4 h); R848 (20 ng/ml, 4 h); nigericin (10 μg/ml, 1 h); ATP (5 mM, 1 h); alum (400 μg/ml, 8 h), Escherichia coli, (ATCC 11775, 16 h) and Citrobacter rodentium (ATCC 51459, 16 h). Poly(dA:dT) (4 μg/ml, 8 h) and flagellin (500 ng/ml, 2 h) were complexed with Lipofectamine 2000 (Life Technologies) and transfected according to the manufacturer's instructions.
HEK293T cells were washed with cold PBS and lysed in Homogenization buffer (10 mM Tris-HCl pH 7.5, 2 mM MgCl2, 250 mM sucrose) by 20 strokes in a Dounce homogenizer. The homogenate was centrifuged at 5,000g for 10 min, and the supernatant was used as cytosol extract. Mitotic cell extract was prepared from HEK293T cells treated overnight with RO-3306 in the culture medium followed by 1 h release in fresh medium. The cells were washed with cold PBS and lysed in Homogenization buffer with gentle sonication. Following priming with LPS, 1.5 μg extract was transfected per 105 cells using Lipofectamine 2000 for 14 h.
Primary human monocytes were stimulated with LPS (1 ng/ml, 4 h) and nigericin (10 μg/ml, 1 h). Cytokine concentrations in the supernatants were measured using enzyme-linked immunosorbent assay (ELISA) kits for human IL-1β, IL-6, mouse IL-1β, IL-18, IL-6, and TNF (eBioscience).
Lactate dehydrogenase assay
The release of lactate dehydrogenase (LDH) into the culture medium was determined by Pierce LDH cytotoxicity assay kit according to the manufacturer's instructions.
Measurement of mitochondrial ROS
Peritoneal macrophages were stained with MitoSOX for 30 min in accordance with the manufacturer's instructions (Life Technologies). The cells were suspended in 1% Fetal Bovine Serum (FBS) in PBS and then analyzed by flow cytometry using an LSRFortessa Cell Analyzer.
Measurement of intracellular cAMP
1×106 peritoneal macrophages were treated as indicated. The cells were resuspended in cold 0.1 N HCl/Cell Lysis Buffer (provided by the kit). The cAMP in the cell lysates was assayed using the Mouse/Rat cAMP Parameter Assay Kit (R&D Systems) following the manufacturer's instructions.
Calcium measurement
Peritoneal macrophages were plated on 4-chambered coverglass dishes (Fisher Scientific) at a density of 4×104 cells per well and incubated with fluo-4/AM (Life Technologies). Images of untreated cells were acquired (t = 0), then cells were treated with 5 mM ATP in DMEM cell culture medium and imaged every 15 sec for 30 min. Images were acquired using an Andor Confocal Spinning Disk microscope using the 488-nm laser and emission in the range of 500-560 nm. Images were analyzed using ImageJ 1.47v software. Absolute intensity for all cells in a field at different time points was obtained and normalized to time 0 to obtain the fold increase in intensity. Data are displayed as the mean fluorescence intensity of all cells in a field (about 20 cells per field) relative to time 0 and are representative of four independent fields.
ASC oligomerization assay
Peritoneal macrophages were lysed with TBS buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl) containing 0.5% Triton X-100, EDTA-free protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche). The lysates were centrifuged at 6,000g at 4°C for 15 min, and the pellets and supernatants were used as the Triton-insoluble and Triton-soluble fractions, respectively. For the detection of ASC oligomerization, the Triton-insoluble pellets were washed twice with TBS buffer and then resuspended in 300 μl TBS buffer. The resuspended pellets were crosslinked for 30 min at 37°C with 2 mM disuccinimidyl suberate (DSS) (Pierce) and then centrifuged for 15 min at 6,000g. The pellets were dissolved in SDS sample buffer.
siRNA-mediated interference
J774A.1 or THP1 cells (ATCC) in 12-well cell culture plates were transfected with 40 pM small interfering RNA using Lipofectamine RNAiMAX (Life Technologies). After incubation for 48 h, the cells were washed and used in experiments. The mouse and human NEK7 siRNA (SMART pool) and the non-targeting siRNA were from Dharmacon (GE Healthcare).
Isolation and culture of human monocytes and shRNA-mediated interference
shRNA lentiviruses for human NEK7 (sequence 1: ACATTAGCCAACTTTCGAA; sequence 2: GGAACACATGCATTCTCGA; sequence 3: GAGGCTAATTCCTGAAAGA) and non-targeting control were obtained from Dharmacon (GE Healthcare). Monocytes were isolated from heparinized blood of normal human donors by Ficoll Paque centrifugation followed by negative isolation using Miltenyi Pan Monocyte Isolation reagents. The cells were plated at a density of 1×105 cells per square cm in 24-well plates in IMDM containing 10% heated FBS (Hyclone Defined) and 30 ng/ml GM-CSF (R&D Systems). After 7 days in culture, the cells were infected at a multiplicity of infection of 5 particles per cell. Two days later, the efficiency of infection was estimated by assessing red fluorescent protein (RFP)-positive cells. In some cases, RFP-expressing cells were selected using puromycin (4 μg/ml) for 7 days or sorted for RFP-positive cells using flow cytometry. RFP-enriched populations of infected cells (>90% RFP positive) were stimulated with LPS (1 ng/ml, 4 h) and then treated with nigericin (10 μg/ml, 1 h). Cell culture medium (sup) was harvested for ELISA to measure IL-1β and IL-6 (BioLegend). In addition, cells were collected and NEK7 gene expression was measured using MagMax Total RNA Isolation (Ambion) followed by reverse transcription (RT)-PCR using SuperScript VILO Master Mix (Invitrogen) and TaqMan assays and gene expression master mix (Applied Biosystems) (not shown). Expression of NEK7 protein was estimated by immunoblot.
Reconstruction of the inflammasome system in HEK293T cells
HEK293T cells were plated in 24-well microplates at a density of 2×105 cells per well and incubated overnight. The cells were transfected with plasmids expressing mouse pro-IL-1β-Flag (200 ng/well) and mouse inflammasome components ASC (20 ng/well), pro-caspase-1 (100 ng/well) and NLRP3 (200 ng/well) using Lipofectamine 2000. These modified cells were then co-transfected with plasmids encoding mouse NEK7 or specific siRNA to knockdown the endogenous NEK7. The total amount of DNA was adjusted to a concentration of 1 μg per well with empty vector. The cells were washed with culture medium 36 h after transfection and were further incubated for 12 h. Cell culture medium was collected and analyzed for IL-1β maturation by immunoblot analysis.
Immunoprecipitation
24 h after transient transfections, HEK293T cells were lysed in cold NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl at pH 7.4, 150 mM NaCl) supplemented with Roche Complete Protease Inhibitor. Flag-tagged proteins were immunoprecipitated with Anti-FLAG M2 Magnetic Beads (Sigma-Aldrich). For the endogenous interaction assay, macrophages or J774A.1 cells were lysed with NP-40 lysis buffer with Complete Protease Inhibitor. The cell lysates were incubated with the indicated antibodies and Protein G Mag Sepharose (GE Healthcare) overnight at 4°C. For the NLRP3-ASC interaction assay, macrophages or J774A.1 cells were lysed with NP-40 lysis buffer and gentle sonication.
Gel filtration chromatography
Fresh lysates from mouse macrophages were prepared using NP-40 lysis buffer and gentle sonication. Soluble lysate (0.6 mg of total protein) was run on a gel filtration chromatography column (Superdex 200 Increase 10/300 GL, GE Healthcare) in NP-40 lysis buffer with 1mM dithiothreitol (DTT). 1 ml fractions were collected and 1.5% of each fraction subjected to immunoblot analysis.
In vitro kinase assay
The kinase assay was performed with 10 μg recombinant MBP-NEK or MBP-NEK7K64M and 15 μg β-casein (Sigma-Aldrich) as a substrate using the Universal Kinase Activity Kit (R&D Systems) according to the manufacturer's instructions. MBP-NEK and MBP-NEK7K64M were expressed in E.coli and purified with amylose beads.
Bone marrow progenitor colony forming unit assay
Bone marrow cells were collected from tibiae and femurs and treated with red blood cell lysis buffer (Sigma-Aldrich). 105 cells and 50 ng/ml mouse M-CSF (eBioscience) were added to Mouse Methylcellulose Base Media (R&D Systems) according to the manufacturer's instructions. Individual colonies were counted 8 days later.
Apoptosis analysis
Peritoneal macrophages were plated in 96-well clear flat bottom ultra-low attachment microplates (Corning). The cells were treated as indicated and then stained with Annexin V and 7-AAD (BD Biosciences) following the manufacturer's instructions.
In vivo MSU and IL-1β challenge
Mice were challenged by i.p. injection with MSU (1 mg) or one dose of active recombinant murine IL-1β in sterile PBS (50 ng/100 μl). After 6 h, the mice were euthanized and peritoneal cavities were lavaged with 5 ml PBS. PECs were collected and analyzed by flow cytometry using an LSRFortessa Cell Analyzer. For IL-1β ELISA analysis, the peritoneal fluid was concentrated using Amicon Ultra 10K filtration units (Millipore) and then processed.
Induction of EAE and flow cytometric analysis of leukocyte infiltration in CNS
rhMOG (1-125aa) was generated as previously described36. EAE was induced by subcutaneous immunization at four sites on the back with 100 μg of rhMOG emulsified in complete Freund's adjuvant (Difco) containing 5 mg/ml of mycobacteria (BD Biosciences). On days 0 and 2, mice were injected i.p. with 300 ng pertussis toxin (List Biological Laboratories). Clinical disease was assessed as follows: 0, no disease; 1, loss of tail tone; 2, weakness of hind limbs; 3, partial hind limb paralysis; 4, total hind limb paralysis with or without front limb paralysis; 5, moribund or death.
On day 21 after rhMOG immunization, mice were euthanized and perfused via the left ventricle with cold PBS supplemented with 10 U/ml heparin (Fisher Scientific). Spinal cords were harvested from the perfused animals. Tissues were pressed through a 70 μm cell strainer into RPMI 1640 (Corning). Spinal cord cells were pelleted by centrifugation at 390g for 10 min at 4°C. Cell pellets were resuspended in 30% Percoll (GE Healthcare) and centrifuged at 390g for 20 min at room temperature with brake off. The cell pellets were collected, washed with RPMI 1640, and counted. For staining with the 10-color survey panel, the following anti-mouse monoclonal antibodies were used: CD3ε (145-2C11), TCRβ (H57-597), CD4 (RM4-5), B220 (RA3-6B2), CD19 (1D3), CD11b (ICRF44), Gr1 (RB6-8C5), NK1.1 (PK136) (eBioscience or BD Biosciences); CD45 (clone 30-F11, BioLegend). The cell viability dye (Tonbo Biosciences) was used to differentiate viable cells from dead cells.
Infection and treatments of J774A.1 cells
J774A.1 cells were infected with retroviruses encoding mouse NEK7, NEK7K64M and empty vector (pMXs-IRES-Puro, Cell Biolabs). 48 h after the infection, the cells were selected and cultured with 2 μg/ml puromycin (InvivoGen).
LPS-primed J774A.1 cells were treated with colchicine (10 μM), nocodazole (10 μM), cytochalasin D (5 μM) or ciliobrevin D (10 μM) for 2 h, followed by nigericin stimulation for 1 h. LPS-primed J774A.1 cells were treated with ATP and NAC for 1 h. For calf intestinal alkaline phosphatase (CIP) treatment, 100 mg cell lysate was incubated with 50 units of CIP (New England Biolabs) at 37°C for 1 h. Immunoprecipitation was performed as described above. For CIP treatment, the immunoprecipitated complex was further incubated with 50 units of CIP for 1 h at 37°C in a 50 μl reaction. Immunoprecipitates were separated by standard SDS-PAGE, or by Phos-tag SDS-PAGE (Wako Chemicals) to separate phosphorylated proteins according to their degree of phosphorylation.
Intracellular activated caspase-1 staining
To measure intracellular activated caspase-1, a fluorescent labeled inhibitor of caspases (FLICA) probe specific for active caspase-1 (ImmunoChemistry Technologies) was used. The probe is cell-permeable and covalently binds to the active form of caspase-1, which is retained within the cell while unbound probe diffuses out of the cell and is washed away. After LPS priming, the FAM-FLICA-caspase-1 probe was added directly to the cell culture medium together with nigericin, incubated for 1 h at 37°C, and washed twice with the supplied wash buffer. Cells were fixed and stained to label mitotic cells before analysis by flow cytometry using an LSRFortessa Cell Analyzer.
Mitotic cell staining and sorting
Where indicated, cells were treated for at least 20 h with RO-3306 (10 μM) in the culture medium to synchronize cells at the G2/M phase border, and then released from the RO-3306-imposed cell cycle block for different time periods by replacing with fresh culture medium.
Cells were collected and fixed in Fixation/Permeabilization solution (BD Biosciences) for 20 min at 4°C, followed by staining with Alexa Fluor 647-conjugated phosphorylated (Ser28) histone H3 antibody (BioLegend) in 50 μl of BD Perm/Wash buffer (BD Biosciences). Cells were analyzed by flow cytometry or sorted before immunoprecipitation and immunoblot analysis.
Statistical analysis
Comparisons of differences were between two unpaired experimental groups in all cases. An unpaired t-test (Student's t test) is appropriate and was used for such comparisons. The phenotypic performance of mice (C57BL/6J) and primary cells of these mice is expected to follow a normal distribution, as has been observed in large datasets from numerous phenotypic screens conducted by our group. Variation within each dataset obtained by measurements from mice or primary cells was assumed to be similar between genotypes since all strains were generated and maintained on the same pure inbred background (C57BL/6J); experimental assessment of variance was not performed.
Data represent means ± s.d. in all graphs depicting error bars unless indicated. The statistical significance of differences between experimental groups was determined using GraphPad Prism 6 and the Student's t test (unpaired, two-tailed). P<0.05 was considered statistically significant. No pre-specified effect size was assumed, and in general three animals or replicates for each genotype or condition were used in experiments; this sample size was sufficient to demonstrate statistically significant differences in comparisons between two unpaired experimental groups by unpaired t-test. The investigator was not blinded to genotypes or group allocations during any experiment.
Supplementary Material
Acknowledgements
This work was supported by NIH grant U19 AI100627. Nlrp3−/− mice (B6.129S6-Nlrp3tm1Bhk/J) were a gift from Dr. Hasan Zaki (University of Texas Southwestern Medical Center, USA). The NLRC4 plasmid was a gift from Dr. Feng Shao (National Institute of Biological Sciences, Beijing, China).
Footnotes
Author contributions
H.S. and B.B. designed the study and analyzed data with suggestions from N.L.M. and R.J.U.; H.S., Y.W., L.S., D.C., V.V.K. and J.C.M. performed experiments; H.S. and Y.W. identified the Cuties phenotype; X.L., X.Z., M.T. and M.F. generated the Nek7 knockout mice; D.P., C.H.B., S.H., S.L. L.S. J.Q. and Q.S. performed genome mapping and genotyping; H.S., J.R. and S.A. maintained the Cuties mice; H.S., P.J., E.M.Y.M., and B.B. edited the figures; H.S., E.M.Y.M., and B.B. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Gross O, Thomas CJ, Guarda G, Tschopp J. The inflammasome: an integrated view. Immunol. Rev. 2011;243:136–151. doi: 10.1111/j.1600-065X.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 3.Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misawa T, et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 5.Roig J, Mikhailov A, Belham C, Avruch J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002;16:1640–1658. doi: 10.1101/gad.972202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belham C, et al. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 2003;278:34897–34909. doi: 10.1074/jbc.M303663200. [DOI] [PubMed] [Google Scholar]
- 7.Yissachar N, Salem H, Tennenbaum T, Motro B. Nek7 kinase is enriched at the centrosome, and is required for proper spindle assembly and mitotic progression. FEBS Lett. 2006;580:6489–6495. doi: 10.1016/j.febslet.2006.10.069. [DOI] [PubMed] [Google Scholar]
- 8.O'Regan L, Fry AM. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 2009;29:3975–3990. doi: 10.1128/MCB.01867-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertran MT, et al. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 2011;30:2634–2647. doi: 10.1038/emboj.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Lee K, Rhee K. NEK7 is a centrosomal kinase critical for microtubule nucleation. Biochem. Biophys. Res. Commun. 2007;360:56–62. doi: 10.1016/j.bbrc.2007.05.206. [DOI] [PubMed] [Google Scholar]
- 11.Salem H, et al. Nek7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene. 2010;29:4046–4057. doi: 10.1038/onc.2010.162. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Kim S, Rhee K. NEK7 is essential for centriole duplication and centrosomal accumulation of pericentriolar material proteins in interphase cells. J. Cell. Sci. 2011;124:3760–3770. doi: 10.1242/jcs.078089. [DOI] [PubMed] [Google Scholar]
- 13.Wang T, et al. Real-time resolution of point mutations that cause phenovariance in mice. Proc. Natl. Acad. Sci. U. S. A. 2015 doi: 10.1073/pnas.1423216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs CA, et al. Experimental autoimmune encephalomyelitis is exacerbated by IL-1 alpha and suppressed by soluble IL-1 receptor. J. Immunol. 1991;146:2983–2989. [PubMed] [Google Scholar]
- 16.Martin D, Near SL. Protective effect of the interleukin-1 receptor antagonist (IL-1ra) on experimental allergic encephalomyelitis in rats. J. Neuroimmunol. 1995;61:241–245. doi: 10.1016/0165-5728(95)00108-e. [DOI] [PubMed] [Google Scholar]
- 17.Lee GS, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz-Planillo R, et al. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell. Host Microbe. 2010;8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Compan V, et al. Cell volume regulation modulates NLRP3 inflammasome activation. Immunity. 2012;37:487–500. doi: 10.1016/j.immuni.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Pelegrin P, Barroso-Gutierrez C, Surprenant A. P2X7 receptor differentially couples to distinct release pathways for IL-1beta in mouse macrophage. J. Immunol. 2008;180:7147–7157. doi: 10.4049/jimmunol.180.11.7147. [DOI] [PubMed] [Google Scholar]
- 24.Matsubayashi T, Sugiura H, Arai T, Oh-Ishi T, Inamo Y. Anakinra therapy for CINCA syndrome with a novel mutation in exon 4 of the CIAS1 gene. Acta Paediatr. 2006;95:246–249. doi: 10.1080/08035250500341451. [DOI] [PubMed] [Google Scholar]
- 25.Aksentijevich I, Remmers EF, Goldbach-Mansky R, Reiff A, Kastner DL. Mutational analysis in neonatal-onset multisystem inflammatory disease: comment on the articles by Frenkel et al and Saito et al. Arthritis Rheum. 2006;54:2703–4. doi: 10.1002/art.22022. author reply 2704-5. [DOI] [PubMed] [Google Scholar]
- 26.Jesus AA, et al. Phenotype-genotype analysis of cryopyrin-associated periodic syndromes (CAPS): description of a rare non-exon 3 and a novel CIAS1 missense mutation. J. Clin. Immunol. 2008;28:134–138. doi: 10.1007/s10875-007-9150-7. [DOI] [PubMed] [Google Scholar]
- 27.Kandli M, Feige E, Chen A, Kilfin G, Motro B. Isolation and characterization of two evolutionarily conserved murine kinases (Nek6 and nek7) related to the fungal mitotic regulator, NIMA. Genomics. 2000;68:187–196. doi: 10.1006/geno.2000.6293. [DOI] [PubMed] [Google Scholar]
- 28.Vassilev LT, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper GM. The Cell: A Molecular Approach. Sinauer Associates; Sunderland, Massachusetts: 2000. [Google Scholar]
- 31.Eder C. Mechanisms of interleukin-1beta release. Immunobiology. 2009;214:543–553. doi: 10.1016/j.imbio.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Li Q, Engelhardt JF. Interleukin-1beta induction of NFkappaB is partially regulated by H2O2-mediated activation of NFkappaB-inducing kinase. J. Biol. Chem. 2006;281:1495–1505. doi: 10.1074/jbc.M511153200. [DOI] [PubMed] [Google Scholar]
- 33.Bertram C, Hass R. Cellular responses to reactive oxygen species-induced DNA damage and aging. Biol. Chem. 2008;389:211–220. doi: 10.1515/BC.2008.031. [DOI] [PubMed] [Google Scholar]
- 34.Orthwein A, et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–193. doi: 10.1126/science.1248024. [DOI] [PubMed] [Google Scholar]
- 35.Georgel P, Du X, Hoebe K, Beutler B. ENU mutagenesis in mice. Methods Mol. Biol. 2008;415:1–16. doi: 10.1007/978-1-59745-570-1_1. [DOI] [PubMed] [Google Scholar]
- 36.Lyons JA, Ramsbottom MJ, Cross AH. Critical role of antigen-specific antibody in experimental autoimmune encephalomyelitis induced by recombinant myelin oligodendrocyte glycoprotein. Eur. J. Immunol. 2002;32:1905–1913. doi: 10.1002/1521-4141(200207)32:7<1905::AID-IMMU1905>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.