Abstract
Natural killer (NK) cells are the primary effectors of the innate immune response against virus-infected cells or cells that have undergone malignant transformation. NK cells recognize their targets through a complex array of activating and inhibitory receptors, which regulate the intensity of the effector response against individual target cells. However, many studies have shown that tumor cells can escape immune cell recognition through a variety of mechanisms, developing resistance to NK cell killing. Using a lentiviral shRNA library, we previously demonstrated that several common signaling pathways modulate susceptibility of tumor cells to NK cell activity. In this study, we focused on one of the genes (PI3KCB), identified in this genetic screen. The PI3KCB gene encodes an isoform of the catalytic subunit of PI3K called P110β. The PI3K pathway has been linked to diverse cellular functions, but has never been associated with susceptibility to NK cell activity. Gene silencing of PI3KCB resulted in increased susceptibility of several tumor cell lines to NK cell lytic activity and induced increased IFN-γ secretion by NK cells. Treatment of primary tumor cells with two different PI3K inhibitors also increased target cell susceptibility to NK cell activity. These effects are due, at least in part, to modulation of several activating and inhibitory ligands and appear to be correlated with PI3K signaling pathway inhibition. These findings identify a new and important role of PI3KCB in modulating tumor cell susceptibility to NK cells and open the way to future combined target immunotherapies.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-016-1804-y) contains supplementary material, which is available to authorized users.
Keywords: NK cells, PI3K, Immunotherapy, shRNA, Tyrosine kinase inhibitors
Introduction
Tumor rejection is a coordinated immune response that involves both adaptive and innate immunity. NK cells represent a major component of the innate immune response, and their lytic activity is determined by a complex balance of signals modulated by the expression of different inhibitory and activating receptors on NK cells and ligands present on the target cell surface [1–3]. Although this wide array of receptors allow NK cells to recognize and eliminate cells showing early signs of tumor transformation, tumor cells can utilize different mechanisms to escape immune recognition [4–6].
Genome-wide shRNA libraries, based on the biological process of RNA interference, have been used to study loss-of-function effects and better understand the mechanisms involved in tumor progression [7–10]. To identify new pathways involved in tumor cell resistance/susceptibility to NK cell lysis, we previously developed a cell–cell interaction screen using a large subset of the TRC1 shRNA library targeting the entire class of protein kinases and phosphatases as well as other genes involved in different cellular functions [8, 11]. Using this approach, we demonstrated that specific downregulation of different proteins, involved in a variety of central pathways, resulted in enhanced tumor cell sensitivity to NK cell-mediated lysis [12].
Phosphatidylinositol 3-kinases (PI3Ks) are a conserved family of lipid kinases divided into three classes (I, II, III). Class I, which is the best characterized, includes two subclasses: IA and IB. Class IA PI3Ks are heterodimers comprised of a catalytic subunit (p110α, p110β and p110δ) and a regulatory subunit (p85α, p85β, p55α, p50α and p55γ) and are activated mainly by receptor tyrosine kinases (RTKs) [13]. Class IA PI3Ks are involved in growth and survival, and a series of mutations, mostly discovered on the p110α (PI3KCA) isoform, have made class IA PI3Ks ideal targets for cancer treatment [14–17]. For these reasons, different PI3K inhibitors have been developed and tested in preclinical as well as phase I and phase II clinical trials in different types of cancer [18–20].
In the present study, we investigated the possible role of one of the 3 PI3K catalytic subunits (PI3KCB, p110β) in modulating tumor cell susceptibility to NK cell lysis. PI3KCB was among the top genes that were found to induce a strong NK interferon-γ (IFN-γ) response when silenced by 2 or more independent shRNAs present in the TRC1 shRNA library. To further characterize this observation, different shRNAs targeting PI3KCB were used to specifically knockdown gene expression in a panel of tumor cell lines. These results show that specific PI3KCB downregulation increased susceptibility to NK-mediated lysis in 3 of 4 tumor cell lines. This effect appears to be associated with upregulation of several activating ligands and downregulation of MHC class I in tumor cells. These findings were confirmed using two different PI3K inhibitors tested on primary tumor cells from patients with multiple myeloma (MM), acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL). Increased susceptibility of primary tumors to NK cell lysis was primarily associated with modulation of MHC class I expression. This study provides new insights into the possible role of PI3KCB in enhancing tumor cell sensitivity to NK cells in hematologic malignancies and suggests that new PI3K inhibitors, being tested as anti-tumor agents, could also have important effects on innate immune responses in vivo.
Materials and methods
Cell culture
shRNAs targeting PI3KCB and controls were obtained from the TRC Consortium at Dana-Farber Cancer Institute (DFCI), and stable transduced PI3KCB-knockdown cell lines were established as previously described [12]. IM-9, K562, U937, Jurkat cell lines as well as primary MM, AML and ALL cells were maintained in RPMI containing 10 % FBS, 2 mmol/L l-glutamine, 10 mmol/L HEPES and 1 mmol/L sodium pyruvate. NKL and NK-92 cell lines [21, 22] were maintained in the same medium added with 50 units/ml for NKL and 200 units/ml of recombinant human IL-2 for NK-92. Primary human NK cells were purified from peripheral blood mononuclear cells (PBMCs) of healthy donors by magnetic cell separation using the NK cell isolation kit (Miltenyi Biotec) and used after overnight activation with 100 units/ml of IL-2. In all experiments, isolated NK cells were tested for purity using CD56 (BD Pharmingen) and CD3 (Beckman Coulter) antibodies, and only when purity was greater than 95 % were cells used for functional assays. All cell lines were purchased from ATCC, rapidly expanded and frozen back in aliquots. Each aliquot was not cultured for more than 2 months. NKL was developed in the laboratory and checked frequently for the NK cell markers by flow cytometry according to the original publication [22].
Western blot analysis
Cell lines were lysed and subject to 7.5 % SDS-PAGE (Ready Gel, Bio-Rad) with Tris–glycine buffer and transferred onto nitrocellulose membranes in 20 % methanol. Membranes were stained with rabbit anti-P110β (Cell Signaling Technology, Inc.) and β-actin (Santa Cruz Biotechnology), followed by a secondary horseradish peroxidase-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories, Inc.) and visualized by chemiluminescence using Pierce® ECL Western Blotting Substrate.
Proliferative capacity assay
The viability and proliferation capacity of PI3KCB-knockdown stable cell lines and controls were tested over a 5-day period using Cell Titer-Glo (Promega). The experiments were set up using five plates (one for each day), and each condition was suited with eight replicates. Results were obtained using a Luminoskan Ascent (Thermo Fisher Scientific) and expressed as RLU (Relative Luminescence Units).
IFN-γ release assays and cytotoxicity assays
For IFN-γ assays, stable cell lines expressing individual shRNAs were incubated at 37 °C for 12 h (3:1 E/T ratio) with NKL, NK-92 [21, 22] or with NK cells purified from healthy donors. Supernatants were harvested and incubated with CBA IFN-γ beads according to the manufacturer’s protocol (BD Biosciences) and the level of IFN-γ produced by NK cells was determined by flow cytometry using a BD FACSCanto II (BD Biosciences). Data were analyzed using the FCAP Array™ Analysis Software (Soft Flow, Inc.). For cytotoxicity assays, cells were counted and incubated with NKL, NK-92 or primary NK cells from different healthy donors for 12 h (3:1 E/T ratios). Apoptosis induction of target cells was determined by flow cytometry using an AnnexinV/7AAD assay (BD Pharmingen). PE-conjugated anti-NKG2A antibody (Beckman Coulter) and PE-conjugated anti-CD56 antibody (BD Pharmingen) were used to detect and exclude NK effector cells from the analysis. The level of apoptosis was only calculated for NKG2A-negative or CD56-negative cells. The level of spontaneous apoptosis of target cells without NK cells was subtracted from the final percentage in every experiment and was never more than 6–8 %. For blocking assays, NK cells were plated in 96-well plates and incubated with 1 μg NKG2D purified blocking antibody or 1 μg DNAM-1 purified blocking antibody (R&D system) for 30 min at 37 °C. Specific target cells were then added to the culture (3:1 E/T ratios) and incubated for 12 h at 37 °C. After co-culture, supernatants were harvested and incubated with CBA IFN-γ beads as described above.
Flow cytometry analysis
Modulation of inhibitory and activating ligands was assessed using the following antibodies: mouse anti-MICA and MICB (R&D Systems), mouse anti–MHC class I (clone W6/32), anti-CD155 (R&D Systems) followed by secondary antibodies RPE-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories). PE-conjugated anti-TRAIL-R1, anti-TRAIL-R2, anti-CD95, anti-CD112 and FITC-conjugated anti-CD48 were purchased from Beckman Coulter. PE-conjugated anti-PD-L1 and anti-PD-L2 were purchased from BioLegend. Data were analyzed using the FlowJo software (FlowJo LLC).
PI3KCB inhibitor treatment
Patient tumor samples were obtained under a protocol approved by the Institutional Review Board of the Dana-Farber/Harvard Cancer Center, and informed consent was obtained from each patient. Primary tumor cells from patients with MM (n = 4), AML (n = 4) and ALL (n = 4) containing at least 90 % blasts or CD138+ cells were treated for 4 h with 2 μM of wortmannin (Cell Signaling Technology, Inc) or 5 μM of TGX-221 (Calbiochem, EMD Biosciences). Untreated cells were used as control. Wild-type tumor cell lines were also treated and tested using the same settings as for primary tumor cells. After a wash to remove the inhibitor, tumor cells were stained for ligand expression and the remaining cells were counted and incubated with NKL, NK-92 and primary NK cells for 12 h. Apoptosis induction of target cells was determined by flow cytometry using an AnnexinV/7AAD assay as described above.
Statistics
Student’s t test was used for all 2-sample comparisons, and a p value less than 0.05 was considered significant.
Results
Knockdown of PI3KCB in tumor cells induces increased IFN-γ secretion by NK cells
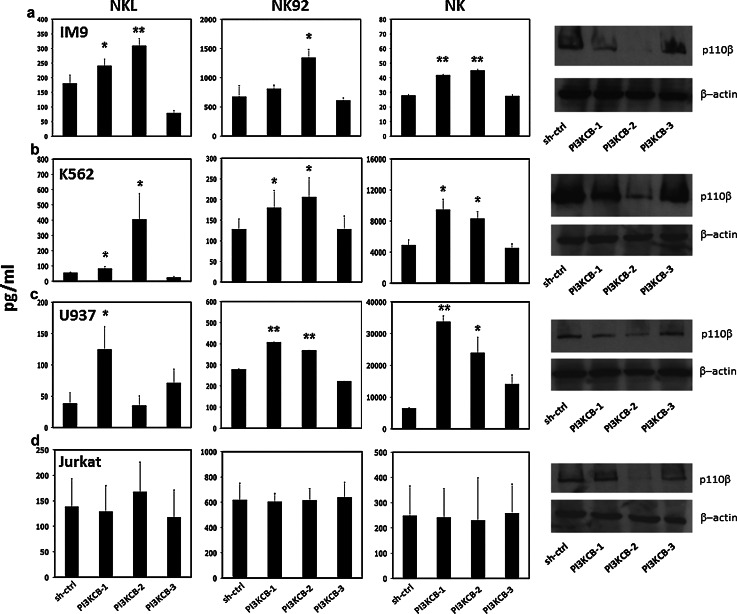
To determine whether specific silencing of the PI3KCB gene would increase sensitivity of tumor cells to NK effector activity, we selected four tumor cell lines representing different hematologic malignancies: IM-9 (multiple myeloma), U937 (acute myeloid leukemia), K562 (chronic myeloid leukemia) and Jurkat (acute T cell leukemia). Using three independent shRNAs (PI3KCB-1, PI3KCB-2, PI3KCB-3) targeting PI3KCB and an irrelevant hairpin as control (shCTRL), we established a series of stable, puromycin-resistant transduced cell lines. Target sequences used to downregulate the expression of PI3KCB gene as well as the sequence of the irrelevant hairpin used as control are shown in Supplementary Tables 1 and 2. To test whether PI3KCB-knockdown affected cell viability, transduced cell lines were first tested for proliferative activity over a 5-day period. As shown in Supplementary Figure 1, silencing a single subunit of PI3K did not affect cell proliferation. Our western blot analysis showed that PI3KCB-2-shRNA was always effective while PI3KCB-3-shRNA did not produce specific silencing in any of the cell lines tested. PI3KCB-1-shRNA, on the contrary, showed differential activity with good protein suppression in IM-9 and U937, moderate in K562 and no effective in Jurkat (Fig. 1). Since the PI3KCB-3-shRNA did not effectively reduce expression of PI3KCB in any cell lines, PI3KCB-3-knockdown cell lines were used as an additional negative control. We next tested each cell line in functional co-culture assays with NKL, NK-92 [21, 22] and primary purified NK cells from different healthy donors to determine NK cell IFN-γ secretion activity in response to specific PI3KCB knockdown. As shown in Fig. 1a, IM-9 cells expressing the 2 shRNAs which led to effective PI3KCB downregulation (PI3KCB-1 and PI3KCB-2) induced 33.3 and 71.1 % higher secretion of IFN-γ by NKL and 20.1 and 99.3 % by NK-92 when compared with shCTRL (p = 0.023, p = 0.006 and p = 0.1, p = 0.01, respectively). IM-9-PI3KCB-knockdown also resulted in increased IFN-γ secretion when tested against primary NK cells. As shown in Fig. 1a, IM-9-PI3KCB-1 and IM-9-PI3KCB-2 induced 49.8 and 61.4 % more IFN-γ secretion when compared with the irrelevant control hairpin (p = 0.0013 and p = 0.0011, respectively). Consistent with these results, K562-PI3KCB-KO (Fig. 1b) and U937-PI3KCB-KO (Fig. 1c) induced significantly higher secretion of IFN-γ by NKL, NK-92 and primary NK cells. In some cases, however, the degree of protein downregulation did not correlate well with the amount of IFN-γ secreted by NK, while in another example (U937-PI3KCB-2 tested against NKL), we did not find any difference compared to the control (p = ns) even if the protein expression was reduced. Conversely, cell lines transduced with PI3KCB-3-shRNA, which was not effective in downregulating PI3KCB, were always comparable to the shCTRL (p = ns). Interestingly, Jurkat-PI3KCB-knockdown cell lines did not show an increased induction of IFN-γ secretion from NKL, NK-92 or primary NK cells (Fig. 1d) suggesting that in this particular tumor cell line, PI3KCB does not modulate NK cell activity.
Fig. 1.
Analysis of NK IFN-γ secretion in response to PI3KCB downregulation in IM-9, K562, U937 and Jurkat tumor cell lines NKL, NK-92 and primary NK cells from three different healthy donors incubated with a IM-9-PI3KCB-shRNA, b K562-PI3KCB-shRNA, c U937-PI3KCB-shRNA and d Jurkat-PI3KCB-shRNA tumor cell lines. Bars represent the mean ± SEM of three independent experiments tested in triplicate (*p < 0.05, **p < 0.01 compared to target cells stable transduced with an irrelevant shRNA). The far right columns show expression of PI3KC-p110β measured by western blot for each cell line
PI3KCB-knockdown induces increased susceptibility to NK cell killing
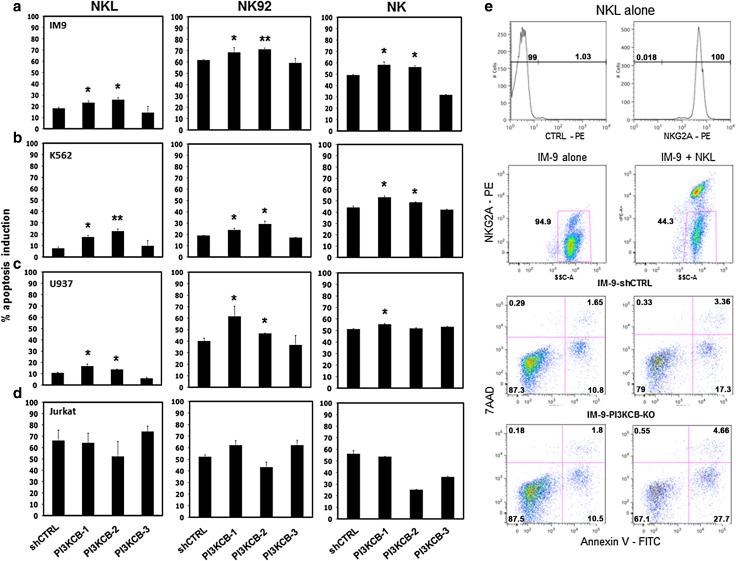
To determine whether increased IFN-γ secretion by NK cells correlated with increased lytic activity, we incubated tumor target cells stable transduced with PI3KCB-shRNAs and irrelevant controls with NKL, NK-92 and primary NK effector cells using an AnnexinV/7AAD flow based assay (Fig. 2e). As shown in Fig. 2a, IM-9-PI3KCB-knockdowns with the two effective PI3KCB-shRNAs (PI3KCB-1 and PI3KCB-2) were significantly more susceptible to NKL cell lysis with 8 and 15 % higher apoptosis induction than the control-shRNA (p = 0.019 and p = 0.023, respectively) and 11 and 15.4 % more than the control-shRNA when incubated with NK-92 (p = 0.043 and p = 0.006, respectively). These results were confirmed in independent experiments using purified NK cells. Primary NK cells induced 17.1 and 13.1 % more apoptosis in IM-9 expressing PI3KCB-1-shRNA and PI3KCB-2-shRNA (p = 0.027 and p = 0.014, respectively) compared to IM-9 cells transduced with the shCTRL (Fig. 2a). Independent experiments performed using K562 and U937 cell lines expressing PI3KCB-1- and PI3KCB-2-specific shRNAs (Fig. 2b, c) also showed significantly increased susceptibility when compared to K562-shCTRL and U937-shCTRL or to the PI3KCB-3-shRNA. These lines were also tested with freshly isolated NK cells, and in this setting, K562 expressing PI3KCB-1-shRNA and PI3KCB-2-shRNA showed significant higher levels of induced apoptosis compared to PI3KCB-shCTRL (p = 0.011 and p = 0.028, respectively), while U937 was more susceptible to purified NK cells when transduced with PI3KCB-1-shRNA (p = 0.02). Overall, the increased NK apoptosis induction upon PI3KCB-knockdown was consistent and significant in almost every condition tested. However, in some cell lines the magnitude of the increased tumor sensitivity was not high and further studies will be necessary to determine the immunologic significance of these changes. Importantly, consistent with the IFN-γ secretion assays, none of the Jurkat-PI3KCB-knockdown cell lines showed different apoptosis induction by NKL, NK-92 or freshly isolated NK cells when compared with the Jurkat-shCTRL cell line (Fig. 2d), confirming that PI3KCB silencing did not influence the sensitivity of this particular tumor cell line to NK effector activity.
Fig. 2.
Induction of target cell apoptosis by NK cells after PI3KCB silencing a IM-9, b K562, c U937 and d Jurkat tumor co-incubated with NKL, NK-92 and primary NK cells. Bars represent the mean ± SEM percent apoptosis induction in three independent experiments tested in triplicate. (*p < 0.05, **p < 0.01 compared to target cells stable transduced with an irrelevant shRNA). e Representative example of IM-9-PI3KCB-knockdown or IM-9-shRNA-CTRL co-incubated with NKL. NKL are 100 % NKG2A+ and the analysis was performed on gated target cells (NKG2A negative). Target cells alone (left scatter plots) were analyzed for spontaneous apoptosis and the values were subtracted from the level of apoptosis induced by NKL
PI3KCB knockdown modulates expression of NK cell ligands
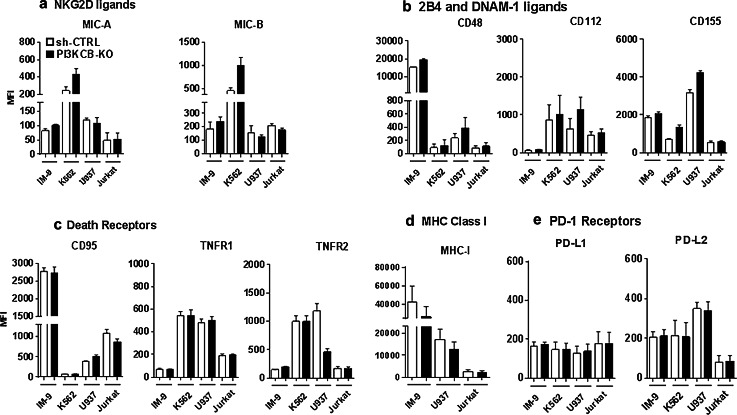
NK cells recognize their targets using a complex balance of activating/inhibitory ligands/receptors expressed on the membrane surface of target/NK cells [23]. To define the mechanism responsible for increased susceptibility of tumor cells to NK cell killing following PI3KCB silencing, we examined expression of different activating/inhibitory ligands on tumor cell lines after PI3KCB knockdown. Our panel included MICA, MICB (NKG2D ligands), CD48 (2B4 ligand), CD112, CD155 (DNAM-1 ligands), TNF-R1, TNF-R2 (TRAIL ligands), CD95 (FAS ligand) PD-L1, PD-L2 (PD-1 ligands) as well as MHC class I. These ligands are known to be key molecules that can trigger the cytolytic activity of innate effector cells and are often modulated in tumor cells [24–26]. As shown in Fig. 3a, following PI3KCB silencing, the expression of NKG2D ligands MICA and MICB was upregulated in IM-9 and K562 cell lines but not in U937 and Jurkat. Specifically, MICB expression increased 30.4 % in IM-9-PI3KCB-knockdown and 121 % in K562-PI3KCB-knockdown while MICA upregulation was less pronounced and in the order of 22.3 % for IM-9-PI3KCB-knockdown and 77.5 % for K562-PI3KCB-knockdown when compared to the same cell lines transduced with an irrelevant shRNA. Conversely, U937-PI3KCB-knockdown showed increased expression of DNAM-1 ligands with an increase of 81.9, 32.7 and 64.5 % in CD112, CD115 and CD48, respectively, when compared with U937-shRNA control. In addition to the upregulation of several activating ligands, IM-9-PI3KCB-knockdown and U937-PI3KCB-knockdown showed a decreased expression of MHC class I (Fig. 3d) while none of the death receptors such as TRAIL-R1, TRAIL-R2 or FAS ligand (CD95) and PD-1 receptors (PD-L1 and PD-L2) were modulated upon PI3KCB knockdown (Fig. 3c, e). Consistent with the lack of increased susceptibility of Jurkat-PI3KCB-knockdown lines, we did not find any modulation of NK ligands in Jurkat cell lines. Although other mechanisms could be involved in Jurkat resistance to NK after PI3KCB-knockdown observed in our previous experiments, the absence of modulation of any of the NK ligands tested likely explains why Jurkat-PI3KCB-knockdown cells did not become more susceptible to NK effector cells. Furthermore, as shown in Supplementary Figure 2, treatment with two different PI3K inhibitors (wortmannin and TGX-221) confirmed an enhanced sensitivity of IM-9, K562 and U937 to NK cell activity, but also in this setting there was no effect on Jurkat cells. These inhibitors induced an upregulation of MICA and MICB on IM-9 and K562 and consistent with the shRNAs studies, MHC-I was downregulated.
Fig. 3.
Modulation of activating/inhibitory ligands in PI3KCB knockdown tumor cells IM-9-PI3KCB-2, K562-PI3KCB-2, U937-PI3KCB-2 and Jurkat-PI3KCB-2 tumor cell lines were analyzed for a panel of activating and inhibitory ligands. Bars represent the mean values ± SEM of MFI in three different experiments of a NKG2D ligands, b 2B4 and DNAM-1 ligands, c death receptors, d MHC-I expression and e PD-1 ligands in PI3KCB-knockdown compared with the same lines transduced with an irrelevant shRNA
Overall, these data show that inhibition of PI3KCB induces expression of activating ligands. In some tumor cell lines such as IM-9 and K562, this effect was primarily observed in the NKG2D ligand family (MICA and MICB). In other tumor cell lines such as U937, this effect was more pronounced in the DNAM-1 or 2B4 ligand families (CD112, CD155 and CD48), likely reflecting different patterns of expression of these ligands in different tumors.
Blocking the target-effector conjugate using NKG2D and DNAM-1 antibodies
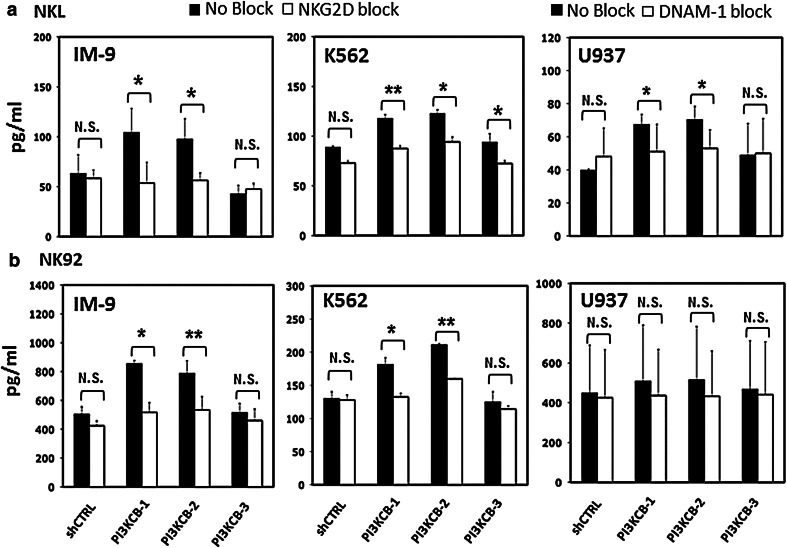
To further define the involvement of NKG2D and DNAM-1 ligands in the increased susceptibility of tumor cells following PI3KCB-knockdown, we undertook a series of experiments using blocking antibodies against NKG2D and DNAM-1 receptors. NKL and NK-92 were pre-incubated with NKG2D and DNAM-1 receptor blocking antibodies and then incubated with either shCTRL or PI3KCB-knockdown cell lines. PI3KCB-3-shRNA, which was not effective in downregulating PI3KCB protein, was also used as a further negative control. As shown in Fig. 4, the activity of NKL and NK-92 was decreased after NKG2D or DNAM-1 was blocked. In IM-9, the difference of IFN-γ secretion between no-block versus block conditions was significant only in PI3KCB-knockdown lines (52 % decrease; p = 0.02 and 42 % decrease; p = 0.01 for IM-9-PI3KCB-1-shRNA and IM-9-PI3KCB-2-shRNA, respectively; Fig. 4a). When we used NK-92 effector cells blocked with NKG2D antibody, we found 39.4 and 32 % decreases in IM-9-PI3KCB-1-shRNA and IM-9-PI3KCB-2-shRNA susceptibility compared with NK-92 unblocked (p = 0.02 and p = 0.002, respectively; Fig. 4b). NKG2D blocking also decreased activity of NKL and NK-92 against IM-9-shCTRL or K562-shCTRL (Fig. 4a, b), but these differences were not significant. The reduction in reactivity of NKL and NK-92 was also specifically related to effective PI3KCB knockdown when K562-PI3KCB-knockdown were tested with NK-92 pre-incubated with NKG2D blocking antibodies (Fig. 4b) and U937-PI3KCB-knockdown were tested with NKL blocked with DNAM-1 antibodies (Fig. 4a). Conversely, when we tested K562-PI3KCB-knockdown against NKG2D blocked NKL and U937-PI3KCB-knockdown against DNAM-1 blocked NK-92, we found a consistent reduction in reactivity between PI3KCB-knockdown lines and shCTRLs.
Fig. 4.
NK IFN-γ secretion after NKG2D or DNAM-1 ligand blocking in PI3KCB-knockdown tumor cell lines a IM-9, K562 and U937-PI3KCB-knockdown or shCTRL control were incubated with NKL and b NK-92 with or without NKG2D or DNAM-1 blocking antibodies. Bars represent the mean values ± SEM of IFN-γ secreted by NKL or NK-92 in three independent experiments tested in triplicate. (*p < 0.05, **p < 0.01 compared NK IFN-γ secretion against target cells-PI3KCB-knockdown treated with or without blocking antibody)
Although other ligands might also be involved, these experiments confirmed that the increased susceptibility of PI3KCB-knockdown cells could be specifically related to overexpression of several activating ligands.
Effects of PI3KCB inhibitors on susceptibility of tumor cells to NK cell-mediated lysis
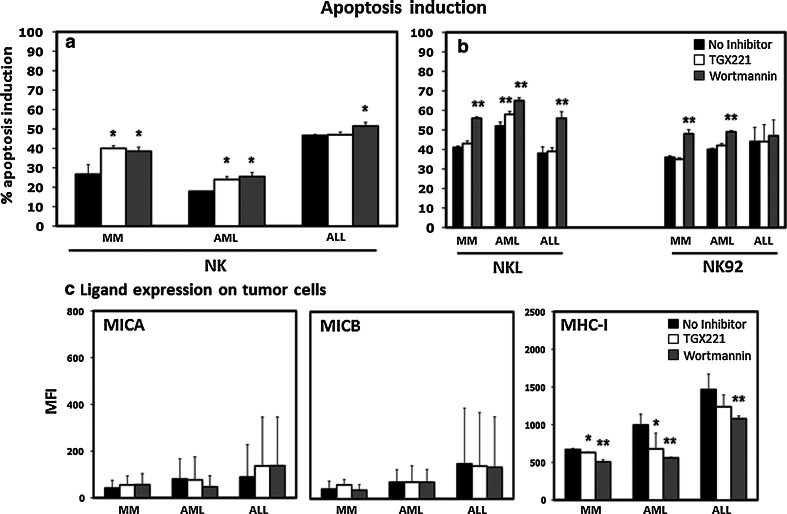
Because of their importance in many cellular functions [15, 27, 28], the PI3K pathway has been targeted in various settings, and several inhibitors are being tested in clinical trials [29, 30]. Our genetic screen and the subsequent validation studies showed that reduced expression of PI3KCB led to increased tumor susceptibility to NK cells. To determine whether available inhibitors targeting PI3K would also sensitize tumor cells to NK cell activity, we treated primary tumor cells from three different hematologic malignancies (MM, AML and ALL) with two PI3K inhibitors (TGX-221 and wortmannin). TGX-221 is a reversible inhibitor specific for PI3KCB, while wortmannin acts irreversibly on the whole PI3K complex and consequently also on PI3KCB. In preliminary experiments, we established a suitable time treatment of 4 h and a concentration of inhibitor of 5 μM for TGX-221 and 2 μM for wortmannin (data not shown). Pre-treated primary tumor cells were subsequently incubated with primary NK effectors for 12 h. As shown in Fig. 5a, MM cells treated with TGX-221 and wortmannin were more susceptible to primary NK cell lysis when compared with untreated cells (13.3 and 11.8 % higher than untreated condition, p = 0.033 and p = 0.045, respectively). TGX-221 and wortmannin-treated AML cells were also more susceptible than untreated cells 6.1 % (p = 0.013) and 7.6 % (p = 0.018), respectively. ALL cells showed an increased susceptibility to NK cell killing only when treated with wortmannin (p = 0.02), but there was no difference when treated with TGX-221 (p = ns). The effects of TGX-221 and wortmannin on primary tumor cells using primary NK cells were also confirmed using NKL and NK-92. However, using these two established NK cell lines, the increased primary tumor cell susceptibility was stronger with wortmannin than TGX-221, probably due to its irreversible activity or to the different receptors present on these NK cell lines. As shown in Fig. 5b, after treatment with wortmannin, MM cell susceptibility was increased by 15 % (p = 0.001) and 12 % (p = 0.001) when co-incubated with NKL and NK-92, respectively. We did not detect any effect when MM cells were pre-treated with TGX-221 (p = ns). TGX-221 was more efficient in inducing AML susceptibility to NK cell lines; however, it was still less effective than wortmannin while ALL cells were more susceptible when pre-treated with wortmannin and co-incubated with NKL effector cells (p = 0.001). These results show that PI3K inhibitors can also induce increased susceptibility to NK cell lysis, but they also suggest that similarly to what we observed in tumor cell lines, some tumors may be less affected by PI3K targeting.
Fig. 5.
Induction of apoptosis in primary tumor cells by NK effector cells and ligand modulation after treatment with PI3KCB inhibitors a percent of apoptosis induction by primary purified NK or b by NKL and NK-92 after pre-treatment of primary MM (n = 4), AML (n = 4) and ALL (n = 4) tumor cells with PI3K inhibitors. c Modulations of MICA, MICB and MHC-I in primary tumor cells after PI3K inhibitor treatment. Bars represent the mean ± SEM percent of three independent experiments. (*p < 0.05, **p < 0.01 compared to target cells treated without inhibitors)
PI3K inhibitors modulate MHC class I expression in primary tumor cells
Our previous experiments showed that the increased susceptibility of different tumor cell lines to NK cell killing after PI3KCB knockdown was probably due to specific modulation of different NK activating ligands. In particular, MICA, MICB (NKG2D ligands) and CD112, CD155 (DNAM-1 ligands) were upregulated when PI3KCB was specifically silenced, while MHC-I expression was downregulated in IM-9 and U937. When expression of NK activating ligands was examined in primary tumor cells (Fig. 5c), treatment with PI3K inhibitor resulted in only a minor upregulation of MICA in ALL cells and no difference in MICB expression in MM, AML and ALL for MICB (p = ns). However, MHC-I expression decreased following PI3K inhibitor treatment and this effect appeared to be consistent in the three types of tumor cells tested (Fig. 5c). Although both inhibitors induced a clear effect, wortmannin appeared to be more effective than TGX-221 in MM, AML and ALL (p = 0.008, p = 0.022 and p = 0.016, respectively). None of the other markers tested showed significant change in expression. Although more experiments will be necessary to completely clarify the mechanisms, these findings suggest that depending on the tumor cell type tested, PI3K silencing can affect the expression of different activating ligands on the tumor cells or acting on MHC class I expression. The expression of these ligands profoundly affects the recognition of tumor cells by NK cells making the tumor cells more susceptible to recognition and elimination by the innate immune system.
Discussion
PI3KCB (p110β) is one of the four isoforms of class I PI3K, an important class of kinases involved in many cellular functions and often deregulated in different tumors. Although mutations have only been found in the α isoform, several studies have demonstrated the involvement of the other members of class I PI3K in cancer progression and development [13, 17, 31]. Here, we show that various tumor cell lines as well as primary tumor cells become more sensitive to NK cell killing when PI3KCB is silenced or targeted by inhibitors. Our initial analysis was conducted in four different tumor cell lines stably transduced with three independent shRNAs targeting PI3KCB. Our experiments with different NK effector cells showed that in three out of four tumor cell lines tested, reduced expression of PI3KCB in the tumor cells was associated with increased secretion of IFN-γ by NK cells. Interestingly, while these experiments confirmed the specificity of our previous evidence obtained in a larger unbiased shRNA screening, PI3KCB silencing was not always effective in sensitizing tumor cells to NK cell activation, and despite successful PI3KCB-knockdown, susceptibility of Jurkat cells was not affected in any of our assays.
When NK cells recognize cellular targets, they become an important source of immunomodulatory cytokines such as IFN-γ or TNF-α, which in turn can activate and attract immune cells as well as increasing tumor susceptibility [1, 32]. Increased INF-γ secretion in response to PI3KCB-knockdown prompted us to investigate whether PI3KCB silencing also induced increased NK killing. Our data show that the three cell lines found to increase NK IFN-γ secretion were also more susceptible to NK cell killing in almost every condition tested. Consistent with previous results, killing of Jurkat did not change, indicating that PI3KCB might not play the same role in every tumor cell and that tumors can employ different mechanisms to escape immune cell recognition. Since NK cell recognition and activation is driven by the level of expression of different activating/inhibitory ligands expressed on the target cell surface, we investigated whether PI3KCB downregulation could have an effect on several NK ligands expressed on tumor cells. These experiments showed that expression of various NK ligands was modulated by PI3KCB downregulation, but this varied among the cell lines tested reflecting the different patterns of expression of these ligands in different tumors. Thus, activating ligands such as MICA and MICB showed a consistent pattern of upregulation on K562-PI3KCB-knockdown, while MHC class I whose absence on target cells leads to enhanced NK cell recognition, was found to be significantly downregulated on IM-9-PI3KCB-knockdown and U937-PI3KCB-knockdown. These variations in the expression of several ligands when PI3KCB is downregulated could explain the increased susceptibility that the stable PI3KCB-knockdown targets showed when incubated with different NK effector cells. Importantly, NK ligand expression was not modulated on Jurkat cells suggesting a possible explanation on the lack of increased susceptibility that Jurkat cells showed in our IFN-γ and AnnexinV/7AAD experiments.
PI3K has been studied primarily for its important critical contribution to tumor development [31, 33]; for these reasons, several molecule inhibitors have been developed to target pan, isoform-specific and dual PI3K/mTOR or PI3K/AKT pathways [17, 18, 20]. Although recent clinical trials have suggested that better outcomes can be obtained with combined PI3K/mTOR or PI3K/AKT inhibition rather than targeting PI3K alone, new pan inhibitors are being tested and these studies will eventually define the efficacy of these promising drugs [29]. In addition to their anti-tumor effects, studies have shown that many kinase inhibitors are also able to modulate immune functions. Immunomodulatory drugs such as thalidomide, lenalidomide, pomalidomide or tyrosine kinase inhibitors like imatinib have also shown potent effects on T and NK cell activation [34, 35], while other drugs have immune suppressive activity [36]. In many cases, targeting a protein kinase directly modulates ligand expression on tumor cells resulting in increased tumor susceptibility to immune recognition. Conversely, as we previously showed for JAK1/JAK2 inhibition, increased tumor susceptibility to immune effector cells was only induced when NK effector cells were engaging with the tumor cells preventing IFN-γ secreted by the effector cells to upregulate PD-L1 on the tumor cell surface [37].
To confirm studies with tumor cell lines, we performed additional experiments on primary tumor cells using two small molecule inhibitors targeting the PI3K complex in primary leukemia cells. In our hands, the pan PI3K inhibitor wortmannin was more effective than the isoform-specific TGX-221 in inducing tumor susceptibility to NK cell lysis. The differential activity shown by the two inhibitors tested in this study could be in part due to their different mechanisms of action and their different specificities for targets in the PI3K pathway [38, 39]. This emphasized that different inhibitors could have different effects on tumor cells even though they target the same protein complex. This effect has already been described in other studies where sorafenib and sunitinib, two approved tyrosine kinase inhibitors for renal cell carcinoma treatment [40, 41], had comparable anti-tumor efficacy but differential activity in sensitizing tumor cells to immune cell responses [42–44]. Moreover, the sensitivity to NK cells lysis, after PI3K inhibitor treatment, was different in the three primary leukemia cells tested. While MM and AML cells became more susceptible to NK cells in most of our tested conditions, ALL tumor cells showed increased susceptibility only when tested against primary purified NK cells. Our subsequent analysis of PI3K inhibitor effect on NK ligand expression showed a marked reduction of MHC-class I expression but contrary to what we saw in tumor cell lines, no modulation was noted on NKG2D and DNAM-1 ligands. MHC-I expression was significantly reduced in MM and AML while in ALL it was only reduced when cells were treated with wortmannin. These results could explain the possible differential efficacy that the two PI3K inhibitors had on ALL compared to MM and AML cells. Moreover, our previous study [12] showed that more than one shRNA targeting another subunit of PI3K (PI3KCA) also induced increased IFN-γ secretion by NK cells. Increased susceptibility of these tumors to NK cell lysis after pre-treatment with the pan PI3K inhibitor wortmannin supports the hypothesis that the other PI3K sub-units may also play a role in modulating tumor cell susceptibility to these effector cells. It is also important to note that while MHC class I downregulation on tumor cells treated with PI3K inhibitors or silenced by PI3K-shRNAs can enhance NK cell killing, it may also affect T cell recognition leading tumors to be resistant to cytotoxic T cells. Further studies considering other selective inhibitors or specific shRNAs will be necessary to determine and confirm whether these phenomena can be extended to other subunits of the PI3K complex and whether other compartments of the immune system are affected.
Taken together, these studies have identified a new potential role for PI3KCB in modulating tumor susceptibility to NK cells in different hematological malignancies and show that this effect is mostly due to modulation of several activating/inhibitory ligands expressed on tumor cells. Although it is important to note that several studies have shown how inhibition of PI3K on immune cells may impair some immune-functional activity [45], our findings suggest that targeting the PI3K complex in tumor cells can also have important immunological aspects that may enhance future immunotherapeutic approaches.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Funding
R21AI088521 (NIH), Multiple Myeloma Research Foundation (MMRF) and Claudia Adams Barr Research Program (Roberto Bellucci), P01CA078378 (NIH), PO1CA142106 (NIH), CA183560 (NIH) (Jerome Ritz).
Abbreviations
- ALL
Acute lymphocytic leukemia
- AML
Acute myeloid leukemia
- CD
Cluster of differentiation
- ET ratio
Effector to target ratio
- IFN
Interferon
- MACS
Magnetic-activated cell sorting
- MFI
Mean intensity fluorescence
- MHC
Major histocompatibility complex
- MICA
MHC class I polypeptide-related sequence A
- MICB
MHC class I polypeptide-related sequence B
- MM
Multiple myeloma
- NK cells
Natural killer cells
- NKL
Natural killer line
- PBMC
Peripheral blood mononuclear cell
- PD-1
Programmed death 1 receptor
- PD-L1
Programmed death ligand 1
- PE
Phycoerythrin
- PI3Ks
Phosphatidylinositol 3-kinases
- RPMI
Roswell Park Memorial Institute (culture medium)
- SDS-PAGE
Sodium dodecylsulfate polyacrylamide gel electrophoresis
- TRAIL
TNF-related apoptosis-inducing ligand
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Jerome Ritz, Phone: 617-632-3465, Email: Jerome_ritz@dfci.Harvard.edu.
Roberto Bellucci, Phone: 617-632-3536, Email: Roberto_Bellucci@dfci.harvard.edu.
References
- 1.Raulet DH, Guerra N. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nat Rev Immunol. 2009;9(8):568–580. doi: 10.1038/nri2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smyth MJ, Hayakawa Y, Takeda K, Yagita H. New aspects of natural-killer-cell surveillance and therapy of cancer. Nat Rev Cancer. 2002;2(11):850–861. doi: 10.1038/nrc928. [DOI] [PubMed] [Google Scholar]
- 3.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 5.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell. 2010;142(6):847–856. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smyth MJ, Dunn GP, Schreiber RD. Cancer immunosurveillance and immunoediting: the roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv Immunol. 2006;90:1–50. doi: 10.1016/S0065-2776(06)90001-7. [DOI] [PubMed] [Google Scholar]
- 7.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428(6981):431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 8.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3(9):715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 9.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37(11):1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 10.Tiedemann RE, Zhu YX, Schmidt J, Yin H, Shi CX, Que Q, Basu G, Azorsa D, Perkins LM, Braggio E, Fonseca R, Bergsagel PL, Mousses S, Stewart AK. Kinome-wide RNAi studies in human multiple myeloma identify vulnerable kinase targets, including a lymphoid-restricted kinase, GRK6. Blood. 2010;115(8):1594–1604. doi: 10.1182/blood-2009-09-243980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 12.Bellucci R, Nguyen HN, Martin A, Heinrichs S, Schinzel AC, Hahn WC, Ritz J. Tyrosine kinase pathways modulate tumor susceptibility to natural killer cells. J Clin Invest. 2012;122(7):2369–2383. doi: 10.1172/JCI58457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 14.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 15.Matheny RW, Jr, Adamo ML. PI3K p110 alpha and p110 beta have differential effects on Akt activation and protection against oxidative stress-induced apoptosis in myoblasts. Cell Death Differ. 2010;17(4):677–688. doi: 10.1038/cdd.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMullen JR, Jay PY. PI3K(p110alpha) inhibitors as anti-cancer agents: minding the heart. Cell Cycle. 2007;6(8):910–913. doi: 10.4161/cc.6.8.4124. [DOI] [PubMed] [Google Scholar]
- 17.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4(4):257–262. doi: 10.1016/S1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 19.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chene P, De Pover A, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, Garcia-Echeverria C. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7(7):1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 20.Maira SM, Stauffer F, Schnell C, Garcia-Echeverria C. PI3K inhibitors for cancer treatment: where do we stand? Biochem Soc Trans. 2009;37(Pt 1):265–272. doi: 10.1042/BST0370265. [DOI] [PubMed] [Google Scholar]
- 21.Gong JH, Maki G, Klingemann HG. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia. 1994;8(4):652–658. [PubMed] [Google Scholar]
- 22.Robertson MJ, Cochran KJ, Cameron C, Le JM, Tantravahi R, Ritz J. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp Hematol. 1996;24(3):406–415. [PubMed] [Google Scholar]
- 23.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 24.El-Sherbiny YM, Meade JL, Holmes TD, McGonagle D, Mackie SL, Morgan AW, Cook G, Feyler S, Richards SJ, Davies FE, Morgan GJ, Cook GP. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res. 2007;67(18):8444–8449. doi: 10.1158/0008-5472.CAN-06-4230. [DOI] [PubMed] [Google Scholar]
- 25.Farag SS, Fehniger TA, Ruggeri L, Velardi A, Caligiuri MA. Natural killer cell receptors: new biology and insights into the graft-versus-leukemia effect. Blood. 2002;100(6):1935–1947. doi: 10.1182/blood-2002-02-0350. [DOI] [PubMed] [Google Scholar]
- 26.Pietra G, Vitale M, Manzini C, Balsamo M, Moretta L, Mingari MC (2012) Melanoma cells inhibit NK cell functions. Cancer Res 72(20):5430; author reply on comment on “Melanoma cells inhibit natural killer cell function by modulating the expression of activating receptors and cytolytic activity [Cancer Res. 2012]. doi:10.1158/0008-5472 [DOI] [PubMed]
- 27.Bunney TD, Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer. 2010;10(5):342–352. doi: 10.1038/nrc2842. [DOI] [PubMed] [Google Scholar]
- 28.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20(1):87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maira SM. PI3K inhibitors for cancer treatment: five years of preclinical and clinical research after BEZ235. Mol Cancer Ther. 2011;10(11):2016. doi: 10.1158/1535-7163.MCT-11-0792. [DOI] [PubMed] [Google Scholar]
- 30.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10(3):143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 31.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 32.Caligiuri MA. Human natural killer cells. Blood. 2008;112(3):461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 34.Borg C, Terme M, Taieb J, Menard C, Flament C, Robert C, Maruyama K, Wakasugi H, Angevin E, Thielemans K, Le Cesne A, Chung-Scott V, Lazar V, Tchou I, Crepineau F, Lemoine F, Bernard J, Fletcher JA, Turhan A, Blay JY, Spatz A, Emile JF, Heinrich MC, Mecheri S, Tursz T, Zitvogel L. Novel mode of action of c-kit tyrosine kinase inhibitors leading to NK cell-dependent antitumor effects. J Clin Invest. 2004;114(3):379–388. doi: 10.1172/JCI21102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayashi T, Hideshima T, Akiyama M, Podar K, Yasui H, Raje N, Kumar S, Chauhan D, Treon SP, Richardson P, Anderson KC. Molecular mechanisms whereby immunomodulatory drugs activate natural killer cells: clinical application. Br J Haematol. 2005;128(2):192–203. doi: 10.1111/j.1365-2141.2004.05286.x. [DOI] [PubMed] [Google Scholar]
- 36.Salih J, Hilpert J, Placke T, Grunebach F, Steinle A, Salih HR, Krusch M. The BCR/ABL-inhibitors imatinib, nilotinib and dasatinib differentially affect NK cell reactivity. Int J Cancer. 2010;127(9):2119–2128. doi: 10.1002/ijc.25233. [DOI] [PubMed] [Google Scholar]
- 37.Bellucci R, Martin A, Bommarito D, Wang K, Hansen SH, Freeman GJ, Ritz J. Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. OncoImmunology. 2015;4(6):e1008824. doi: 10.1080/2162402X.2015.1008824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351(Pt 1):95–105. doi: 10.1042/bj3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26(1):127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 40.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, Rolland F, Demkow T, Hutson TE, Gore M, Freeman S, Schwartz B, Shan M, Simantov R, Bukowski RM. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356(2):125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 41.Goodman VL, Rock EP, Dagher R, Ramchandani RP, Abraham S, Gobburu JV, Booth BP, Verbois SL, Morse DE, Liang CY, Chidambaram N, Jiang JX, Tang S, Mahjoob K, Justice R, Pazdur R. Approval summary: sunitinib for the treatment of imatinib refractory or intolerant gastrointestinal stromal tumors and advanced renal cell carcinoma. Clin Cancer Res. 2007;13(5):1367–1373. doi: 10.1158/1078-0432.CCR-06-2328. [DOI] [PubMed] [Google Scholar]
- 42.Hipp MM, Hilf N, Walter S, Werth D, Brauer KM, Radsak MP, Weinschenk T, Singh-Jasuja H, Brossart P. Sorafenib, but not sunitinib, affects function of dendritic cells and induction of primary immune responses. Blood. 2008;111(12):5610–5620. doi: 10.1182/blood-2007-02-075945. [DOI] [PubMed] [Google Scholar]
- 43.Krusch M, Salih J, Schlicke M, Baessler T, Kampa KM, Mayer F, Salih HR. The kinase inhibitors sunitinib and sorafenib differentially affect NK cell antitumor reactivity in vitro. J Immunol. 2009;183(12):8286–8294. doi: 10.4049/jimmunol.0902404. [DOI] [PubMed] [Google Scholar]
- 44.Ozao-Choy J, Ma G, Kao J, Wang GX, Meseck M, Sung M, Schwartz M, Divino CM, Pan PY, Chen SH. The novel role of tyrosine kinase inhibitor in the reversal of immune suppression and modulation of tumor microenvironment for immune-based cancer therapies. Cancer Res. 2009;69(6):2514–2522. doi: 10.1158/0008-5472.CAN-08-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang K, Zhong B, Gilvary DL, Corliss BC, Hong-Geller E, Wei S, Djeu JY. Pivotal role of phosphoinositide-3 kinase in regulation of cytotoxicity in natural killer cells. Nat Immunol. 2000;1(5):419–425. doi: 10.1038/80859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.