Abstract
Cell-free mitochondiral DNA (mtDNA) is an immunogenic molecule associated with many inflammatory conditions. We evaluated the relationship between cell-free mtDNA in cerebrospinal fluid (CSF) and neurocognitive performance and inflammation during HIV infection. In a cross-sectional analysis, we evaluated the association of mtDNA levels with clinical assessments, inflammatory markers, and neurocognitive performance in 28 HIV-infected individuals. In CSF, we measured mtDNA levels by droplet digital PCR, and soluble CD14 and CD163, neurofilament light, and neopterin by ELISA. In blood and CSF, we measured soluble IP-10, MCP-1, TNF-α, and IL-6 by ELISA, and intracellular expression of IL-2, IFN-γ, and TNF-α in CD4+ and CD8+ T cells by flow cytometry. We also evaluated the relationship between CSF pleocytosis and mtDNA longitudinally in another set of five individuals participating in an antiretroviral treatment (ART) interruption study. Cell-free CSF mtDNA levels strongly correlated with neurocognitive performance among individuals with neurocognitive impairment (NCI) (r=0.77, p=0.001). CSF mtDNA also correlated with levels of IP-10 in CSF (r=0.70, p=0.007) and MCP-1 in blood plasma (r=0.66, p=0.01) in individuals with NCI. There were no significant associations between inflammatory markers and mtDNA in subjects without NCI, and levels of mtDNA did not differ between subjects with and without NCI. MtDNA levels preceded pleocytosis and HIV RNA following ART interruption. Cell-free mtDNA in CSF was strongly associated with the severity of neurocognitive dysfunction and inflammation only in individuals with NCI. Our findings suggest that within a subset of subjects cell-free CSF mtDNA is associated with inflammation and degree of NCI.
Keywords: Mitochondrial DNA, Inflammation, HAND, Pleocytosis, Droplet digital PCR
Introduction
Mitochondria are near-ubiquitous organelles in human cells (only absent from mature red blood cells) responsible for energy generation, and they are believed to be derived from endosymbiotic bacteria in eukaryotes (Anderson 1981). Eukaryotic cells carry a dynamic network of mitochondria that varies in relation to the energy needs of a cell. This network is made up of hundreds or thousands of mitochondria, each of which carry multiple copies of its own circular mitochondrial DNA (mtDNA) chromosome. Accumulated somatic mutations in mtDNA and changes in mitochondrial fission and fusion dynamics, both of which occur during aging, may play a role in this loss of mitochondrial performance (Biskup and Moore 2006; Chaturvedi and Flint Beal 2013; Itoh et al. 2013). Damaged mitochondria are cleared through mitophagy, during which proteases and endonucleases break down mtDNA and mitochondrial proteins. Unlike nuclear DNA, the covalently closed circular mtDNA is more resistant to degradation by nucleases, and also contains CpG motifs, a Toll-like receptor-9 (TLR-9) ligand (Zhang et al. 2010). Thus, if undigested and released, mtDNA could induce innate immune activation (Zhang et al. 2010).
Cell-free mtDNA is a biomarker that is currently being explored across several disciplines. Recent work in humans and mouse models has demonstrated the importance of cell-free mtDNA in the inflammatory cascade in heart failure and in critically ill patients in the intensive care unit (Konstantinidis and Kitsis 2012; Nakahira et al. 2013; Oka et al. 2012). Free mtDNA has also been detected in the cerebrospinal fluid (CSF) of humans (Kingwell 2013; Podlesniy et al. 2013;Walko et al. 2014; Wang et al. 2013), and levels of cell-free mtDNA in the CSF have been associated with neurologic disease (Podlesniy et al. 2013). Although the source of the cell-free mtDNA found in the CSF remains unclear, cell-free mtDNA levels in CSF were reduced in subjects with Alzheimer’s dementia (Podlesniy et al. 2013), consistent with lower neuronal mtDNA copy numbers, while higher CSF mtDNA levels were associated with a worse prognosis in traumatic brain injury and cases of subarachnoid hemorrhage (Walko et al. 2014; Wang et al. 2013).
Neurocognitive dysfunction remains a common complication of HIV disease despite antiretroviral therapy (ART), with estimates of 18–80 % of individuals demonstrating some evidence of dysfunction on formal testing (Spudich 2014) with imaging (Cardenas et al. 2009) and biomarkers (Jessen Krut et al. 2014). Viral replication or persistence is a possible cause of this neurodegenerative process, possibly through the induction of chronic immune activation in the central nervous system (CNS) (Spudich 2014). Several studies have shown that markers of innate immune activation (soluble CD163 [sCD163] and soluble CD14 [sCD14]) (Burdo et al. 2013; Lyons et al. 2011) and monocyte activation (neopterin) (Brew et al. 1990; Clifford and Ances 2013; Eden et al. 2014; Fuchs et al. 1989) are associated with neurocognitive impairment (NCI). Given the potential role of cell-free mtDNA as a biomarker of innate immune activation and neuronal injury, we explored the relationship between cell-free mtDNA in CSF and inflammation and neurocognitive performance in HIV-infected adults.
Methods
Study cohorts
Neurocognitive cohort
Participants were HIV-infected adults enrolled in the University of California San Diego HIV Neurobehavioral Research Program (HNRP) (n=28), of which approximately half (54 %) were on ART. Participants were predominantly male, reflecting the HIV-infected population of San Diego. All participants underwent neurocognitive, neuromedical, and psychiatric evaluations performed by HNRP certified staff, as previously described (Cysique et al. 2011; Heaton et al. 2008). The global deficit score (GDS) was used as a measurement of neurocognitive performance, with NCI defined as GDS≥0.5. The standard GDS cutoff of ≥0.5 is a previously validated and established definition of impairment for HIV-related brain injury (Antinori et al. 2007; Blackstone et al. 2012). CSF (supernatant and pellet), blood, lymphocyte profiles, and demographic and disease characteristics were collected. HIV RNA levels in blood plasma and CSF were measured by Roche Amplicor (version 1.5), with lower limit of quantification of 50 HIV RNA copies/mL.
Interruption cohort
We also evaluated a second set of five HIV-infected individuals, who interrupted ART under the guidance of primary care physicians. Re-initiation of ART was at the discretion of participants and their corresponding medical providers. Biological specimens and clinical data were collected for each subject before, during, and after ART interruption (total specimens n=41). For one participant, there were no specimens available before the interruption of ART. These cases were examined under the rationale that if free mtDNA increased before the onset of pleocytosis, then the pleocytosis might represent a response to elevated mtDNA. However, if the pleocytosis occurred before increases in mtDNA, then the breakdown of these cells might be the source of the mtDNA. This study was approved by the local Institutional Review Board, and all participants provided written informed consent.
DNA extraction, quantification of mtDNA, and genomic DNA
After collection through lumbar puncture, CSF was centrifuged at 250 g and then the supernatant was stored at −80 °C. Total DNA was extracted from CSF supernatant samples using QIAamp DNA Mini Kit (Qiagen, CA) per manufacturer’s protocol. We quantified levels of mtDNA by targeting the NADH dehydrogenase 2 gene (MT-ND2, Applied Biosystems, CA) found on the mitochondrial chromosome and genomic DNA (gDNA) by targeting the Ribonuclease P protein subunit p30 gene (RPP30, Applied Biosystems, CA) by droplet digital PCR (ddPCR, Bio-Rad, CA). First, we digested the extracted DNA using the BamHI HF (New England Biolabs, Ipswich, MA) enzyme and 15 µL of extracted DNA per manufacturer’s protocol. Quantification was carried out in triplicate for a 20 µL reaction, which consisted of 10 µL of 2× Bio-Rad supermix for probes, 1 µL of either 20× Primer/FAM ND2 mix or 20× Primer/VIC RPP30 mix, 4 µL of molecular grade water, and 5 µL of digested DNA. Each 20 µl reaction was loaded into an 8-channel cartridge, and we used 70 µL of oil (Bio-Rad) for droplet generation. The generated droplets were placed and heat-sealed in a 96-well PCR plate for cycling with the following conditions: (1) an initial activation of 95 °C for 10 min, (2) 55 cycles of 94 °C for 30 s and 60 °C for 1 min, (3) enzyme inactivation at 98 °C for 10 min, and 4 °C hold. After amplification, the PCR plate was read and analyzed using the Bio-Rad droplet reader and the QuantaSoft (Bio-Rad, CA) analysis software, respectively (Hindson et al. 2011). Levels of mtDNA were expressed in log10 copies/mL of CSF.
Soluble and inflammatory markers
We measured levels of sCD14 and sCD163 in CSF using Quantikine ELISA Human sCD14 and sCD163 (R&D Systems, Minneapolis, MN) following manufacturer’s protocols. Levels of neurofilament light (NFL) and neopterin in CSF were also measured by ELISA using the NF-Light, (Uman Diagnostics, Umea, Sweden) and BRAHMS (GmbH, Hennigsdorf, Germany) kits, respectively. Additionally, we measured levels of inflammatory cytokines (tumor necrosis factor-α [TNF-α], interlukin-6 [IL-6]) and chemokines (monocyte chemoattractant protein 1 [MCP-1], interferon-γ-induced protein 10 [IP-10]) by the FlexMAP 3D Liquid Bead Suspension Array System (Millipore, MA) in CSF and blood plasma. Results were expressed in picrograms per milliliter (pg/mL), nanograms per milliliter (ng/mL), and nanograms per liter (ng/L) for all soluble inflammatory markers, neopterin, and NFL, respectively.
For the interruption cohort, we measured markers of inflammation and cellular trafficking (IP-10, MCP-1, IL-6, IL-2, IL-8 TNF-α, MIP-1α) in CSF by Meso Scale Discovery platform.
Intracellular cytokine assay
Peripheral blood mononuclear cells (PBMCs) were collected in Cell Preparation Tubes (BD Biosciences), washed in phosphate-buffered saline, and resuspended in culture media (X VIVO 15 media with 5 % human serum [Lonza, Germany]). CSF cells were collected from 25–40 cc CSF, spun down, resuspended in culture media, and counted in trypan blue to determine viability. Intracellular cytokine staining without antigen stimulation in PBMCs and CSF cells was performed as directed for BD FastImmune kits (BD Biosciences, San Jose, CA) in Lamoreaux et al. (2006). Briefly, PBMCs were plated at 100,000 cells/well and CSF cells at 5000–20,000 cells/well in V-bottom 96-well plates. After 2 h of culture, brefelden A and Golgi Stop (both from BD Biosciences, San Jose, CA) were added to the plates and cells were incubated overnight (18 h) at 37 °C and 5 % CO2. Cells were then washed using PBS with 1 % BSA and 0.1 % NaN3, fixed using BD Perm 2 (BD Biosciences, San Jose, CA), and stained using CD4-PerCP-Cy5.5 and CD8-APC antibodies in combination with either TNF-α-PE and CD107a-FITC or IL-2-FITC and interferon-γ (IFN-γ)-PE. Isotype controls were included to detect background fluorescence. All antibodies were from BD Biosciences. Samples were acquired on a FACS Calibur II (BD Biosciences, San Jose, CA) within 24 h. Data were analyzed using Flow-Jo software (Tree Star) version 9. Gates for cytokine expression were set on PBMC files and then applied to CSF file data.
Statistical analyses
All statistical analyses were performed using R statistical software (Team 2013). Variables were either log- or square root-transformed to approximate a normal distribution assessed by a Shapiro test with a significance of p<0.05. A Mann- Whitney or t test was used to assess statistical difference between study groups. We evaluated the association of mtDNA with clinical, immunological, and sociodemographical variables using fixed effects linear models. Additionally, longitudinal associations of mtDNA with other variables were assessed using mixed-effects regression analysis to adjust for repeated measurements. We reported the Akaike information criterion (AIC) as our measurement of model selection. Association of categorical variables was assessed using a Fisher exact test. Given the small sample size, we reported the Cohen’s d as a measure of effect size in addition to the p value for our t test statistics.
Results
Study population
Neurocognitive assessments were performed on all 28 subjects and 14 had NCI. The median age of the participants was 46.5 years and 97 % were male. A summary of the demographic and disease characteristics of the subjects is provided in Table 1(A). Table 1(B) describes the clinical and demographic characteristics of the additional five subjects who underwent treatment interruption and were analyzed in a longitudinal fashion.
Table 1.
Characteristics of study participants
| A. HNRP cohort | ||||
| Clinical variable | All samples (n=28) | NCI (n=14) | Normal (n=14) | p value* |
| Gender (% males) | 96 % | 100 % | 0.93 | 1 |
| Ethnicity (% Caucasian) | 80 % | 79 % | 64 % | 0.68 |
| Age | 46.5 (37.75–49.25) | 47 (39–49) | 45.5 (35.5–52) | 0.65 |
| ART naive | 46 % | 50 % | 43 % | 1 |
| CSF HIV RNA (log10 copies/mL) | 2.58 (1.68–341) | 2.25 (1.68–3.71) | 2.58 (1.68–3.26) | 0.94 |
| CSF VL >50 HIV RNA copies/mL (%) | 62 % | 57 % | 67 % | 0.70 |
| CD4+ T cell counts (cells/µL) | 386 (155.5–347) | 391 (356.25–481.25) | 369 (347–500) | 0.77 |
| CD4 percentage (%) | 22 (17.2–25.7) | 20 (17.15–23.98) | 23.1 (21.7–28.3) | 0.3 |
| CD8+ T cell counts (cells/µL) | 1006 (767.5–1260.5) | 1112 (908.5–1470.5) | 945 (655–1126) | 0.08 |
| CD8 percentage (%) | 54.6 (47.2–59.15) | 54.05 (50.35–58.68) | 54.6 (40.1–59,6) | 0.65 |
| CD4/CD8 ratio | 0.41 (0.28–0.49) | 0.4 (0.28–0.45) | 0.42 (0.36–0.76) | 0.34 |
| CD4 nadir (cells/µL) | 244 (124.5-330.75) | 240 (99–303.25) | 252 (145.5-341.75) | 0.67 |
| Global deficit score | 0.49 (0.17–0.68) | 0.72 (0.58–1.26) | 0.17 (0.11–0.22) | 7.32E−06 |
| B. ART interruption cohort | ||||
| Clinical variable | All samples (n=5) | |||
| Gender (% males) | 100 % | |||
| Ethnicity (% Caucasian) | 80 % | |||
| Age | 43 (34–51) | |||
| CSF VL <50 HIV RNA copies/mL (%) | 100 % | |||
| CD4+ T cell counts (cells/µL) | 863.5 (700.25–1040) | |||
| CD4 percentage (%) | 35 (33.75–37.25) | |||
| CD8+ T cell counts (cells/µL) | 881.5 (810–958.25) | |||
| CD8 percentage (%) | 39.5 (34.75–43.25) | |||
| CD4/CD8 ratio | 0.94 (0.80–1.18) | |||
| CD4 nadir (cells/µL) | 281 (215–394.75) |
Median and interquantile range values are shown for the both cohort (A, B) and by groups defined by neurocognitive performance (NCI vs NCN) in the HNRP cohort (A)
Represent the p value of a double-tailed Mann-Whitney or a Fisher test
mtDNA, neuropsychological performance, and markers of inflammation
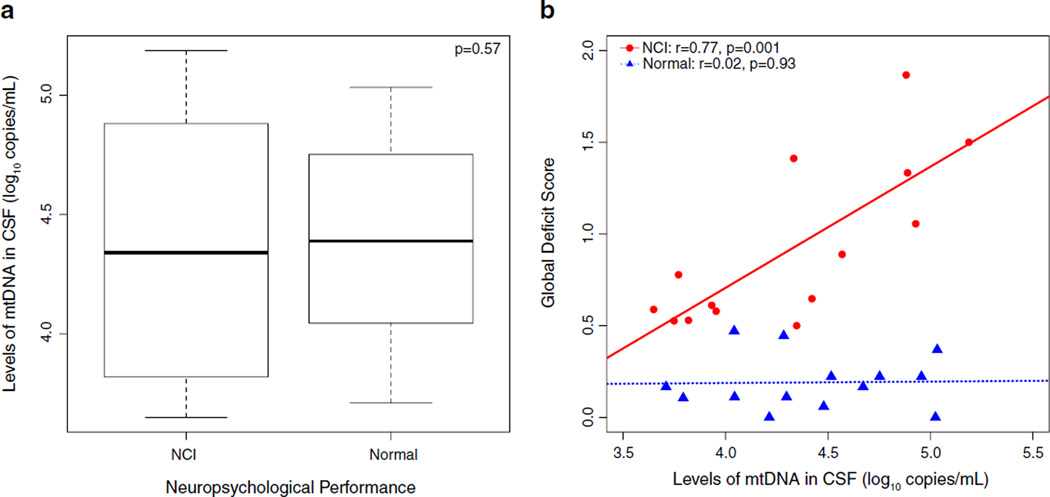
There was no difference in the level of CSF cell-free mtDNA between subjects with NCI and those without NCI (Fig. 1a, p= 0.57). However, higher levels of mtDNA were associated with a higher GDS only in individuals with NCI (Fig. 1b, r=0.77, p= 0.001). These positive associations remained significant when accounting for the effects blood plasma and CSF HIV RNA levels, CD4+ T cell counts, protein level in CSF, and age. Furthermore, cell-free mtDNA was more strongly correlated with NCI severity than CSF HIV RNA levels, CD4+ T cell counts, CD8+ T cell counts, or nadir CD4+ T cell counts (Table 2). In particular, among the seven domains tested by the neurocognitive battery, higher cell-free mtDNA levels correlated with worse learning (r=0.62, p=0.02) and motor (r=0.57, p=0.03) functioning with a trend toward worse speed of information processing (r=0.47, p=0.08, Supplementary Table 1). Furthermore, since we noticed that individuals with a borderline GDS (GDS >0.4) seemed to group along the regression line with the NCI group, we included them in a sub-analysis and found that the individuals with borderline NCI also demonstrated a correlation between NCI and mtDNA levels (r=0.75, p=0.0008).
Fig. 1.
Mitochondrial DNA in CSF and neuropsychological performance. a There was no statistical difference in levels of mtDNA between the groups with and without neurocognitive impairment (NCI vs normal). b While there was no association of mtDNA in CSF in the normal group with, higher levels of mtDNA in CSF in individuals with NCI were correlated with worse neurocognitive deficits
Table 2.
Associations between clinical markers with NCI
| All participants | NCI | Normal | ||||
|---|---|---|---|---|---|---|
| Correlation | p value | Correlation | p value | Correlation | p value | |
| mtDNA | 0.29 | 0.14 | 0.77 | 0.001 | 0.02 | 0.94 |
| CSF HIV RNA | 0.09 | 0.65 | 0.20 | 0.48 | −0.55 | 0.06 |
| CD4 Nadir | −0.06 | 0.77 | −0.19 | 0.54 | 0.40 | 0.19 |
| CD4 T cell counts | 0.08 | 0.67 | −0.07 | 0.79 | 0.34 | 0.25 |
| CD8+ T cell counts | 0.17 | 0.39 | 0.001 | 0.99 | −0.44 | 0.13 |
| CD4/CD8 ratio | −0.10 | 0.59 | −0.07 | 0.8 | 0.57 | 0.04 |
Associations were tested with all participants and also by groups defined by neuropsychological performance (NCI vs normal).MtDNA correlated strongly with the severity of NCI. There were no association between GDS and CSF VL or lymphocyte profiles
To determine if the cell-free mtDNA within the CSF was coming from the death or degeneration of CNS cells, we quantified the cell-free levels of the genomic target RPP30within CSF supernatants as a marker of general cell death. We also evaluated the presence of NFL in the CSF as a specific indicator of neuronal damage. There were no significant differences in the levels of RPP30 or NFL between the groups with and without NCI (Supplementary Fig. 1, p=0.79 and p=0.90, respectively), and there were no significant associations between mtDNA levels with RPP30 and NFL in either group of individuals with or without NCI (data not shown). This suggested that the major source of mtDNA in the CSF may not be death of neurons or other cell types present in the CSF. We also evaluated if cell-free mtDNA correlated with markers of inflammation (sCD14, sCD163, neopterin, IP-10,MCP-1, TNF-α, and IL-6) and found no such correlations (data not shown). Similarly, we found no statistical difference in any of the soluble inflammatory biomarkers between those with or without NCI (only CSF sCD163 levels trended toward being higher in the NCI group, Cohen’s d=0.69, p=0.08). Individuals with NCI did have a significantly higher percentage of CD8+ T cells that were producing IFN-γ (Cohen’s d=1.33, p=0.02, two-tailed t test) and TNF-α (Cohen’s d=1.11, p=0.01, two-tailed t test) in CSF.
Given that cell-free mtDNA levels in CSF showed a correlation with degree of impairment in the NCI group, we investigated subjects with and without NCI separately. We found no associations between mtDNA and any soluble or cellular inflammatory measures in those without NCI. In the group with NCI, there were significant associations between mtDNA and the inflammatory markers IP-10 in CSF (Fig. 2a, r=0.7, p= 0.007) andMCP-1 in blood plasma (Fig. 2b, r=0.66, p=0.01). Compared to all of the measured soluble biomarkers of inflammation, mtDNA levels had the strongest correlation with the severity of NCI (Supplementary Table 2). Further, higher levels of mtDNA were associated with a higher percentage of CD8+ T cells producing IFN-γ (Fig. 2c, r=0.8, p=0.01) and IL-2 (Fig. 2d, r=0.69, p=0.04) in the CSF. There was also a significant association between higher levels of CSF cell-free mtDNA and higher lymphocyte counts in the CSF (Fig. 2e, r=0.9, p=0.03).
Fig. 2.
Associations of mtDNA and inflammation in individuals with NCI. Higher levels of mtDNA in CSF were associated with higher levels of inflammatory biomarkers in CSF and blood (a, b), higher proportion of CD8+ T cells that produce IFN-γ and IL-2 (c, d), and more lymphocyte counts in CSF (e). These associations were only seen in individuals with NCI
mtDNA, HIV RNA, CD4+ T cell count, and markers of inflammation
Active HIV RNA replication can initiate inflammatory responses and therefore may lead to the release of mtDNA into the CSF via death of inflammatory cells. To evaluate whether the presence of HIV replication was associated with CSF cell-free mtDNA levels, we compared the levels of cell-free mtDNA between the 16 subjects with a detectable (>50 copies/mL) CSF HIV RNA and the 12 subjects with an undetectable (<50 copies/mL) HIV RNA.
We found no statistical difference (Supplementary Fig. 2a, p=0.98); however, when we evaluated mtDNA in relationship with the other clinical and immunological measurements in individuals with a detectable CSF HIV RNA, higher levels of mtDNA were associated with higher levels of IP-10 in CSF (r=0.70, p=0.002),MCP-1 in blood plasma (r=0.53, p=0.04) and the levels of RPP30 in CSF (r=0.83, p=7.1 × 10−5) (Supplementary Fig. 2b–d). Similar findings were seen in individuals with detectable blood plasma HIV RNA (data not shown). No significant associations were present in subjects who had undetectable CSF HIV RNA or plasma HIV RNA.
Stratifying individuals by CD4+ T cell count, we found that participants with CD4+ T cell counts ≥350 cells/µl had significantly higher levels of CSF cell-free mtDNA (Supplementary Fig. 3a) than individuals with CD4+ T cell counts <350 cells/µl (p=0.02).We also found that when we segregated individuals by CD4 counts, we found that higher levels of CSF cell-free mtDNA were associated with higher CSF HIV RNA levels (r=0.78, p=0.007) and higher CSF IP-10 levels, (r=0.68, p= 0.03) and showed a trend for higher levels of neopterin in CSF (r=0.57, p=0.08) (Supplementary Fig. 3b–d) only in individuals with CD4+ T cell counts <350 cells/µl. No significant associations were among individuals with CD4 ≥350 cells/µl.
mtDNA, HIV RNA, and pleocytosis
Since we observed associations between mtDNA and CSF lymphocyte counts, we analyzed the temporal association between these two measures in order to determine whether mtDNA appeared to precede or follow CSF pleocytosis. To do this, we utilized CSF samples from a treatment interruption study to determine if the CSF cell-free mtDNA preceded or followed the development of CSF pleocytosis and viral rebound. We performed an investigation of the dynamics of mtDNA, pleocytosis, and HIV RNA levels in the CSF of five individuals, who underwent a structured ART interruption. In all cases, mtDNA levels appeared to increase prior to development of pleocytosis as well as before HIV RNA levels in both CSF and blood plasma (Fig. 3a–e). Furthermore, we also investigated the dynamics of mtDNA and several inflammation markers in CSF after ART interruption. In all cases, levels of mtDNA and inflammatory markers also rose after ART interruption. Using mixed-effects regression analyses in a univariate analysis, mtDNA was proportionally associated with HIV RNA levels in CSF (p=0.05) and plasma (p=0.03), IP- 10 (0.02), IL-2 (p=0.01), MIP-1α (0.04), TNF-α, (0.04) sCD14 (0.04), and pleocytosis (0.03). In a multivariate mixed-effect analysis, pleocytosis could be predicted better using levels of mtDNA and HIV RNA levels in CSF than HIV RNA levels in blood plasma (AIC 371.6 vs. 391, Supplementary Table 3).
Fig. 3.
Dynamics of mtDNA when ART in interrupted. In all cases, when ART is interrupted, mtDNA levels rise before the CSF white blood cell (WBC) count; therefore, levels of mtDNA precede pleocytosis (a–e). Also, levels of mtDNA rose before CSF and plasma HIV RNA levels (viral load (VL)) (a–e). Furthermore, levels of mtDNA also rose before inflammation markers in all cases (f–j). *Represent cytokines that we square root transformed for visual purposes
Discussion
This study found that cell-free mtDNA in the CSF is strongly correlated with the severity of NCI among impaired HIV-infected individuals and that CSF mtDNA levels were more strongly correlated with the severity of NCI than other factors previously associated with HIV associated neurocognitive disease (e.g., CD4 nadir, CD4+ T cell count, or CSF HIV RNA) (Clifford and Ances 2013).Although the associations between mtDNA and both worse impairment and inflammation were only seen within the 14 individuals with NCI, they were independent of CD4+ T cell count and CSF HIV RNA in these subjects. Furthermore, the association between mtDNA and NCI did not seem to be due to neuronal damage, since levels of NFL, a marker of axonal damage, and RPP30, a housekeeping gene, were not associated with mtDNA. If neuronal damage differed between groups, we would have expected the levels of NFL to be higher and the ratio of RPP30 to mtDNA lower in the NCI group, since neurons have more mtDNA than any other cell type.
Markers of inflammation, such as neopterin in CSF and MCP-1 in blood, have also been associated with NCI during HIV infection (Burdo et al. 2013; Clifford and Ances 2013; Kamat et al. 2012; Lyons et al. 2011). For the first time, here, we demonstrated a strong association between CSF cell-free mtDNA and both soluble and cellular biomarkers of inflammation in CSF and blood of HIV-infected individuals with NCI. This is interesting since mtDNA is an immunogenic molecule containing unmethylated CpG motifs, which are ligands for TLR-9 (Zhang et al. 2010). Given that mtDNA is a pro-inflammatory molecule, we hypothesize that individuals with NCI may have inflammatory responses that differ from individuals without NCI, and thus also respond differently to the presence of mtDNA.
The source of the cell-free mtDNA in the CSF remains unclear, but possibilities include inflammatory cells, neurons, or glia. We evaluated the possibility that the cell-free mtDNA in CSF was due to release of mtDNA from dying inflammatory cells, but found that mtDNA levels rose prior to the onset of pleocytosis and rise in inflammatory markers in subjects undergoing treatment interruption, suggesting that a rise in mtDNA may predict an increase in inflammation. The lack of association between NFL and mtDNA in the CSF also suggested that the source of the mtDNA may not be axonal injury. However, we do not discard the possibility that inflammatory cells not present in the CSF, i.e., attached to the meninges, could also be the source of the mtDNA observed. In addition to cell death, other proposed mechanisms by which mtDNA could be released include dysregulated autophagy (Oka et al. 2012) and incomplete transmitophagy (Davis et al. 2014). The process of transmitophagy was recently described by Davis et al. (2014), where mitochondria were released from neurons and degraded by adjacent astrocytes, suggesting that degradation of mitochondria may not always take place within the cell of origin and that mitochondrial products, like mtDNA, could be released extracellularly as part of a transcellular process. Such a mechanism could lead to increased levels of mtDNA and pro-inflammatory mitochondrial proteins released into the CSF.
Currently, investigators interested in HIV cure research in studying the effects of interventions on time to viral rebound in individuals undergoing a structured treatment interruption (Hurst et al. 2015; Li et al. 2015). To enhance the safety of such studies, biomarkers predicting viral rebound prior could be used to reinitiate ART prior to detectable viral replication while still allowing researchers to obtain the information necessary for the study. Here, we demonstrate that levels of mtDNA rose prior to both CSF and plasma HIV viral rebound, making mtDNA a potential candidate biomarker for predicting viral rebound following ART interruption.
As any evaluation of an observational cohort, this study has several limitations. First, our cohort was relatively small and diverse with regard to disease stage, ART use, and other factors. This limits the power to detect differences and to adjust for influential covariates. Despite this, several of our hypotheses were supported by results with medium to large effect sizes. Second, as a cross-sectional study, we cannot determine whether mtDNA levels can predict NCI. Third, it is statistically difficult to interpret correlations with GDS in the individuals without NCI since the score has a limited dynamic range with a cutoff using population norms among individuals without NCI; therefore, a lack of correlation here may not be correct. In other words, there could be an association between mtDNA levels and NCI but the effect is most pronounced among those people who have frank and observable NCI. Further variations in GDS between individuals make it more difficult to interpret borderline scores. This is highlighted in our subanalysis of subjects with borderline GDS scores, i.e., not meeting the >0.5 cutoff for NCI, where we found a correlation between mtDNA level and GDS, like that found among those with frank NCI. Fourth, we do not have a HIV negative group, therefore it is difficult to understand completely the dynamics of mtDNA in relation to HIV infection. Lastly, our larger analyses of neurocognitive functioning cannot determine causality because of their cross-sectional design, and our longitudinal analyses were small and descriptive and may be prone to sampling bias.
In conclusion, cell-free mtDNA in the CSF of HIV-infected subjects correlates with the severity of NCI and the degree of inflammation in subjects with NCI. While mtDNA levels did not differ between impaired and unimpaired subjects, this biomarker may have prognostic value in determining response to therapeutic interventions among impaired individuals, but such uses would need to be evaluated in separate studies. Further, the source and mechanisms of release of mtDNA in the CSF during HIV infection remain important unanswered questions, and future studies to address these questions could provide crucial insight into the causal mechanisms underlying NCI in HIV-infected individuals.
Supplementary Material
Acknowledgments
This work was supported by the Department of Veterans Affairs and grants from the National Institutes of Health: AI093163, AI100665, AI036214, AI007384, MH062512, MH081482, MH101012, DA026306, 1S10RR031646-01, PST5TP2, MH097673; the James B. Pendleton Charitable Trust; and the CNPq-Brazil [MFO]. Additionally, the authors would like to acknowledge the contribution of the research volunteers, the CFAR Genomics and Translational Virology Cores, and the HNRP. Particularly, we would like to thank Parris S. Jordan and Michael Potter for helping with the measurements of soluble inflammatory markers.
DMS has received grant support from ViiV Pharmaceuticals and has served as consultant for Gen-Probe, Hologic, and Testing Talent Services. SLL has served as a consultant for GlaxoSmithKline and Merck & Co.
Footnotes
Meetings Some of these data were presented at the Conference of Retroviruses and Opportunistic Infections in Boston, MA, from March 3 to 6, 2014, and in Seattle, WA, from February 23 to 26, 2015.
Electronic supplementary material The online version of this article (doi:10.1007/s13365-015-0384-5) contains supplementary material, which is available to authorized users.
Conflict of interest JPS, RC,MFO,SG, SRV, TRCD,MRG, JDS, BM, MM, MC, RJE, and SRM do not have any commercial or other associations that might pose a conflict of interest.
References
- Anderson L. Identification of mitochondrial proteins and some of their precursors in two-dimensional electrophoretic maps of human cells. Proc Natl Acad Sci U S A. 1981;78:2407–2411. doi: 10.1073/pnas.78.4.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, Wojna VE. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69:1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup S, Moore DJ. Detrimental deletions: mitochondria, aging and Parkinson’s disease. Bioessays. 2006;28:963–967. doi: 10.1002/bies.20471. [DOI] [PubMed] [Google Scholar]
- Blackstone K, Moore DJ, Franklin DR, Clifford DB, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, Ellis RJ, Atkinson JH, Grant I, Heaton RK. Defining neurocognitive impairment in HIV: deficit scores versus clinical ratings. Clin Neuropsychol. 2012;26:894–908. doi: 10.1080/13854046.2012.694479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brew BJ, Bhalla RB, Paul M, Gallardo H, McArthur JC, Schwartz MK, Price RW. Cerebrospinal fluid neopterin in human immunodeficiency virus type 1 infection. Ann Neurol. 1990;28:556–560. doi: 10.1002/ana.410280413. [DOI] [PubMed] [Google Scholar]
- Burdo TH, Weiffenbach A, Woods SP, Letendre S, Ellis RJ, Williams KC. Elevated sCD163 in plasma but not cerebrospinal fluid is a marker of neurocognitive impairment in HIV infection. AIDS. 2013;27:1387–1395. doi: 10.1097/QAD.0b013e32836010bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas VA, Meyerhoff DJ, Studholme C, Kornak J, Rothlind J, Lampiris H, Neuhaus J, Grant RM, Chao LL, Truran D, Weiner MW. Evidence for ongoing brain injury in human immunodeficiency virus-positive patients treated with antiretroviral therapy. J Neurovirol. 2009;15:324–333. doi: 10.1080/13550280902973960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi RK, Flint Beal M. Mitochondrial diseases of the brain. Free Radic Biol Med. 2013;63:1–29. doi: 10.1016/j.freeradbiomed.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Clifford DB, Ances BM. HIV-associated neurocognitive disorder. Lancet Infect Dis. 2013;13:976–986. doi: 10.1016/S1473-3099(13)70269-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cysique LA, Franklin D, Jr, Abramson I, Ellis RJ, Letendre S, Collier A, Clifford D, Gelman B, McArthur J, Morgello S, Simpson D, McCutchan JA, Grant I, Heaton RK, Group C, Group H. Normative data and validation of a regression based summary score for assessing meaningful neuropsychological change. J Clin Exp Neuropsychol. 2011;33:505–522. doi: 10.1080/13803395.2010.535504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CH, Kim KY, Bushong EA, Mills EA, Boassa D, Shih T, Kinebuchi M, Phan S, Zhou Y, Bihlmeyer NA, Nguyen JV, Jin Y, Ellisman MH, Marsh-Armstrong N. Transcellular degradation of axonal mitochondria. Proc Natl Acad Sci U S A. 2014;111:9633–9638. doi: 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden A, Franklin DR, Fuchs D, Grant I, Letendre S, Marcotte TD, Nilsson S, Price RW, Zetterberg H, Gisslen M. CNS Immunoactivation in HIV Patients on ART With HIV-Associated Mild Neurocognitive Impairment; Conference on Retroviruses and Opportunistic Infections; 2014. Abstract 490. [Google Scholar]
- Fuchs D, Chiodi F, Albert J, Asjo B, Hagberg L, Hausen A, Norkrans G, Reibnegger G, Werner ER, Wachter H. Neopterin concentrations in cerebrospinal fluid and serum of individuals infected with HIV-1. AIDS. 1989;3:285–288. doi: 10.1097/00002030-198905000-00006. [DOI] [PubMed] [Google Scholar]
- Heaton RK, Cysique LA, Jin H, Shi C, Yu X, Letendre S, Franklin DR, Ake C, Vigil O, Atkinson JH, Marcotte TD, Grant I, Wu Z San Diego HIVNRCG. Neurobehavioral effects of human immunodeficiency virus infection among former plasma donors in rural China. J Neurovirol. 2008;14:536–549. doi: 10.1080/13550280802378880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst J, Williams J, Pace M, Willberg C, Phillips R, Frater J, Thornhill J, Fidler S, Hamlyn E, Babiker A. Conference on retroviruses and opportunistic infections. Seattle, Washington: 2015. Biomarkers to predict viral rebound at antiretroviral therapy interruption in SPARTAC. [Google Scholar]
- Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen Krut J, Mellberg T, Price RW, Hagberg L, Fuchs D, Rosengren L, Nilsson S, Zetterberg H, Gisslen M. Biomarker evidence of axonal injury in neuroasymptomatic HIV-1 patients. PLoS One. 2014;9:e88591. doi: 10.1371/journal.pone.0088591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat A, Lyons JL, Misra V, Uno H, Morgello S, Singer EJ, Gabuzda D. Monocyte activation markers in cerebrospinal fluid associated with impaired neurocognitive testing in advanced HIV infection. J Acquir Immune Defic Syndr. 2012;60:234–243. doi: 10.1097/QAI.0b013e318256f3bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingwell K. Alzheimer disease: CSF levels of mitochondrial DNA—a new biomarker for preclinical Alzheimer disease? Nat Rev Neurol. 2013;9:420. doi: 10.1038/nrneurol.2013.134. [DOI] [PubMed] [Google Scholar]
- Konstantinidis K, Kitsis RN. Cardiovascular biology: escaped DNA inflames the heart. Nature. 2012;485:179–180. doi: 10.1038/485179a. [DOI] [PubMed] [Google Scholar]
- Lamoreaux L, Roederer M, Koup R. Intracellular cytokine optimization and standard operating procedure. Nat Protoc. 2006;1:1507–1516. doi: 10.1038/nprot.2006.268. [DOI] [PubMed] [Google Scholar]
- Li J, Kearny M, Shao W, Keele B, Gandhi R. Conference on retroviruses and opportunistic infections. Seattle, Washington: 2015. Identifying HIV variants that rebound after treatment interruption. [Google Scholar]
- Lyons JL, Uno H, Ancuta P, Kamat A, Moore DJ, Singer EJ, Morgello S, Gabuzda D. Plasma sCD14 is a biomarker associated with impaired neurocognitive test performance in attention and learning domains in HIV infection. J Acquir Immune Defic Syndr. 2011;57:371–379. doi: 10.1097/QAI.0b013e3182237e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, Quintana C, Osorio JC, Wang Z, Zhao Y, Lawler LA, Christie JD, Meyer NJ, Mc Causland FR, Waikar SS, Waxman AB, Chung RT, Bueno R, Rosas IO, Fredenburgh LE, Baron RM, Christiani DC, Hunninghake GM, Choi AM. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10:e1001577. doi: 10.1371/journal.pmed.1001577. discussion e1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Akira S, Yamamoto A, Komuro I, Otsu K. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlesniy P, Figueiro-Silva J, Llado A, Antonell A, Sanchez-Valle R, Alcolea D, Lleo A, Molinuevo JL, Serra N, Trullas R. Low cerebrospinal fluid concentration of mitochondrial DNA in preclinical Alzheimer disease. Ann Neurol. 2013;74:655–668. doi: 10.1002/ana.23955. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. 2013 URL: http://www.r-project.org/ [Google Scholar]
- Spudich SS. CROI 2014: neurologic complications of HIV infection. Top Antivir Med. 2014;22:594–601. [PMC free article] [PubMed] [Google Scholar]
- Walko TD, 3rd, Bola RA, Hong JD, Au AK, Bell MJ, Kochanek PM, Clark RS, Aneja RK. Cerebrospinal Fluid Mitochondrial DNA: A Novel DAMP in Pediatric Traumatic Brain Injury. Shock. 2014;41:499–503. doi: 10.1097/SHK.0000000000000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Yang TM, Lin WC, Lin YJ, Tsai NW, Liou CW, Kwan AL, Lu CH. The value of serial plasma and cerebrospinal fluid nuclear and mitochondrial deoxyribonucleic acid levels in aneurysmal subarachnoid hemorrhage. J Neurosurg. 2013;118:13–19. doi: 10.3171/2012.8.JNS112093. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.