Abstract
Cells that reside within a community can cooperate and also compete with each other for resources. It remains unclear how these opposing interactions are resolved at the population level. Here we investigated such an internal conflict within a microbial biofilm community: Cells in the biofilm periphery not only protect interior cells from external attack, but also starve them through nutrient consumption. We discovered that this conflict between protection and starvation is resolved through emergence of long-range metabolic codependence between peripheral and interior cells. As a result, biofilm growth halts periodically, increasing nutrient availability for the sheltered interior cells. We show that this collective oscillation in biofilm growth benefits the community in the event of a chemical attack. These findings indicate that oscillations support population-level conflict resolution by coordinating competing metabolic demands in space and time, suggesting new strategies to control biofilm growth.
Introduction
Cooperation and competition are complex social interactions that can play critical roles in biological communities. Cooperative behavior often increases the overall fitness of the population through processes such as division of labor and production of common goods1–4. At the same time, individuals in a community compete with each other for limited resources, such as nutrients5–6. Here we investigated bacterial biofilms7–10 to determine how the conflict between the opposing social behaviors of cooperation and competition could be resolved at the community level to increase overall fitness.
Biofilms typically form under environmental stress conditions, such as nutrient limitation11–13. As these bacterial communities grow larger, the supply of nutrients to interior cells becomes limited due to an increase in nutrient consumption associated with the growth of multiple layers of cells in the biofilm periphery. Severe nutrient limitation for interior cells is detrimental to the colony, since the sheltered interior cells are critical to the survival of the biofilm community in the event of an external challenge. This defines a fundamental conflict between the opposing demands for biofilm growth and maintaining the viability of protected (interior) cells (Fig. 1a). The identification of possible mechanisms that ensure the viability of the protected interior cells is fundamental to understanding biofilm development14, 15.
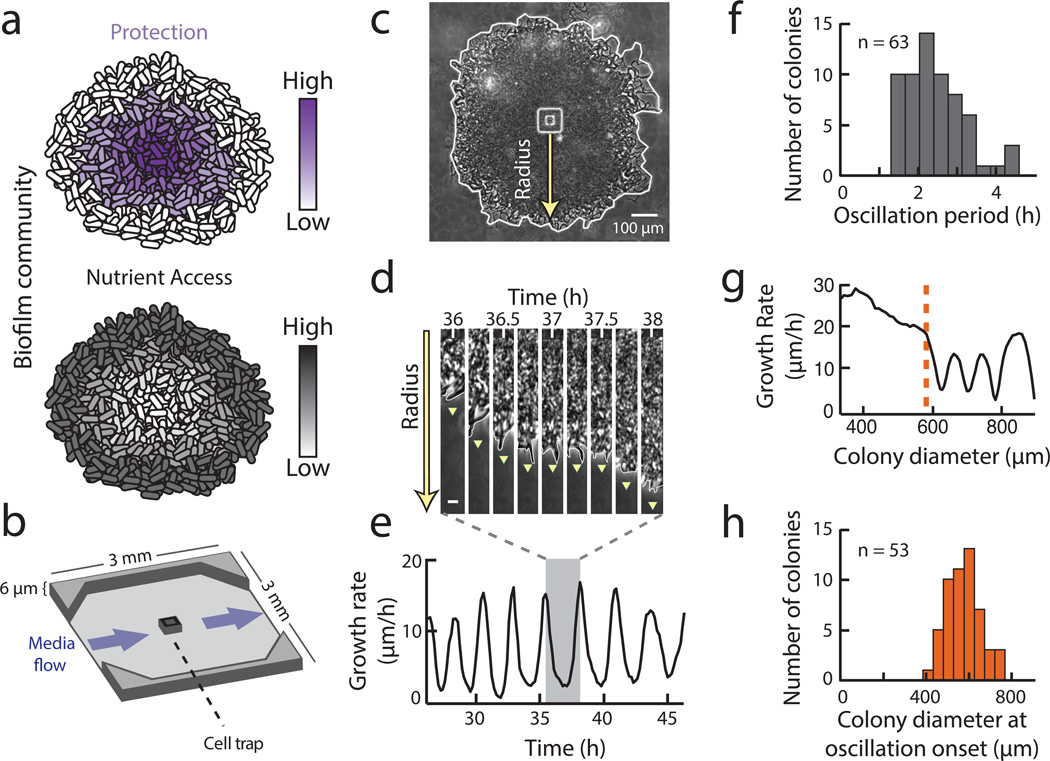
Figure 1.
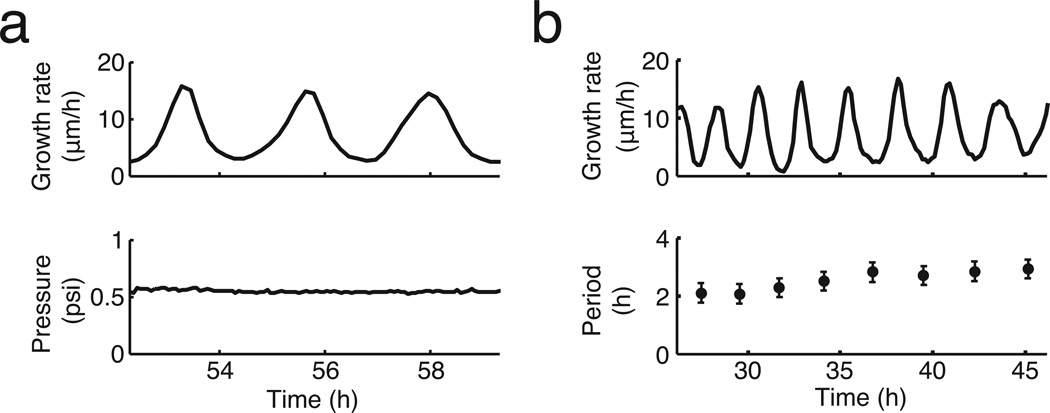
Biofilms grown in microfluidic devices show oscillations in colony expansion. a, Biofilms must reconcile opposing demands for protection from external challenges (gradient indicated in purple) and access to nutrients (gradient indicated in gray). b, Schematic of the microfluidic device used throughout this study. Direction of media flow is indicated by the blue arrows. c, Phase contrast image of a biofilm growing in the microfluidic device. The yellow arrow indicates the region of interest in panel (d). d, Filmstrip of a radius of the biofilm over time shows a pause in colony expansion. This film strip represents one cycle of biofilm oscillations, indicated by the shaded region in panel e. Scale indicates 5 µm. e, Growth rate over time shows persistent oscillations in colony expansion. f, Histogram of the average period of oscillations for each colony (n = 63 colonies, mean = 2.5 hours, s.d. = 0.8 hours). The cell replication time is approximately 3.4 hours under these conditions (Methods: Data Analysis). g. Growth rate as a function of colony diameter (which increases in time) shows that early colony growth does not exhibit oscillations. The orange line indicates the diameter (~600 µm) at which this colony initiates oscillations. h, Histogram of the diameter at which a colony begins to oscillate (n = 53 colonies, mean = 576 µm, s.d. = 85 µm).
In order to directly investigate how Bacillus subtilis biofilms continue expanding while sustaining interior cells, we converted the potentially complex three-dimensional problem to a simpler two-dimensional scenario using microfluidics. Specifically, we used growth chambers that are unconventionally large in the lateral, x-y dimensions (3 × 3 mm), while confining biofilm thickness (z-dimension) to only a few micrometers (Fig. 1b). Therefore, biofilm expansion in this device is predominantly limited to two dimensions, creating a “pancake-like” configuration. In fact, biofilms often form in confined aqueous environments and thus this microfluidic chamber may better mimic those growth conditions11–13. This experimental set-up is thus ideal to interrogate how biofilms can reconcile the opposing benefits of growth and protection during biofilm development.
Oscillations in biofilm growth
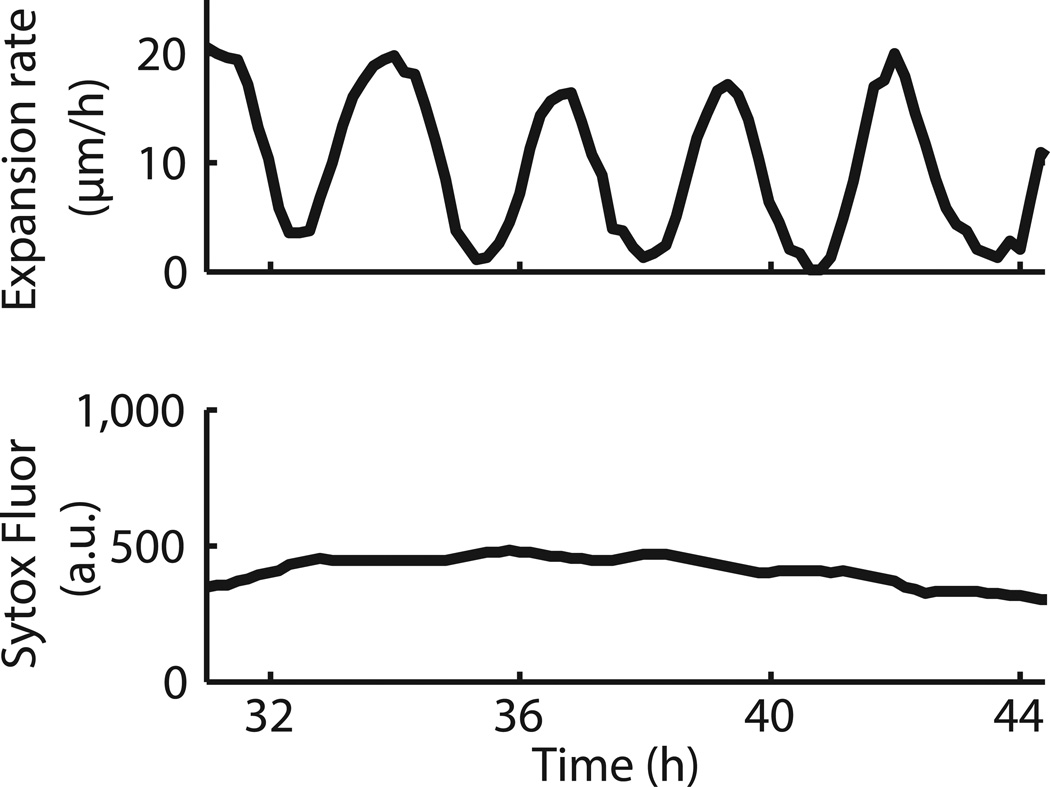
Unexpectedly, we observed oscillations in biofilm expansion despite constant media flow within the microfluidic device (Fig. 1c, d, Supplementary Video 1 and Extended Data Fig. 1a). Specifically, biofilms exhibit periodic reduction in colony expansion that is self-sustained and can last for more than a day (Fig. 1e and Extended Data Fig. 1b). The period of oscillations has a mean of 2.5 ± 0.8 hours (s.d., n = 63 colonies), which is less than the duration of the average cell replication time of 3.4 ± 0.2 hours (s.d., n = 21 cell cycles) under this growth condition (Fig. 1f and Method: Data Analysis). Moreover, oscillations only arise when the biofilm exceeds a certain colony size (Supplementary Video 2). In particular, quantitative measurements obtained from 53 individual biofilms indicate that oscillations emerge in colonies that exceed an average diameter of 580 ± 85 µm (s.d., n = 53 colonies), which corresponds to approximately one million cells (Fig. 1g, h). Together, these data show that oscillations arise during biofilm formation and are self-sustained.
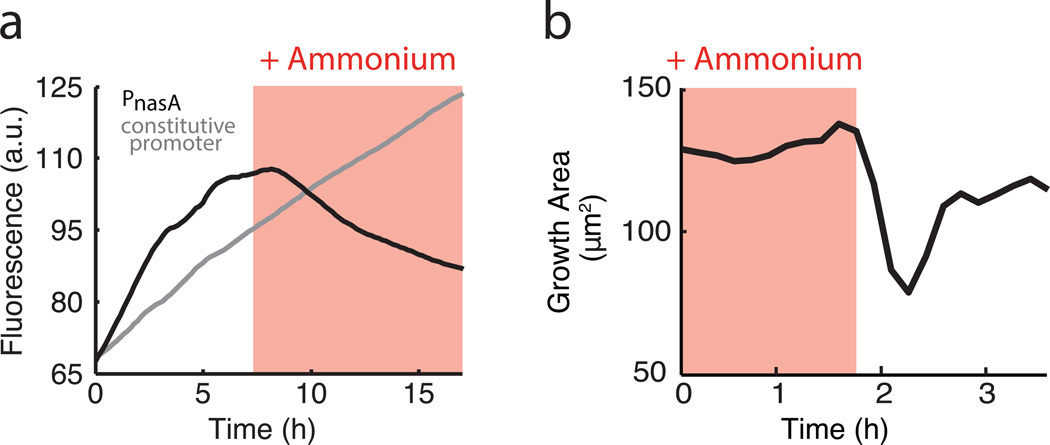
Given that biofilms typically form under nutrient limited conditions and bacterial growth is generally controlled by metabolism, we hypothesized that metabolic limitation plays a key role in the observed periodic halting of biofilm expansion. In particular, after determining that carbon source limitation did not play an essential role in the oscillations (Extended Data Fig. 2), we focused on nitrogen limitation. The standard biofilm growth media (MSgg, see Methods: Growth conditions) used to study B. subtilis biofilm development contains glutamate as the only nitrogen source16. In most organisms including B. subtilis, glutamate is combined with ammonium by glutamine synthetase (GS) to produce glutamine, which is essential for biomass production and growth (Fig. 2a)17. Cells can obtain the necessary ammonium from glutamate through the enzymatic activity of glutamate dehydrogenase (GDH), expressed by the rocG or gudB genes in the undomesticated B. subtilis used in this study (Fig. 2a)18–20. To determine whether biofilms experience glutamine limitation, we measured expression of nasA, one of several genes activated in response to a lack of glutamine21. Results show that biofilms indeed experience glutamine limitation during growth. Specifically, supplementation of growth media directly with glutamine reduced nasA promoter expression, but did not affect expression of a constitutive promoter, confirming glutamine limitation within the biofilm (Fig. 2b). More strikingly, addition of exogenous glutamine eliminated periodic halting of biofilm growth (Fig. 2c and Extended Data Fig. 3a). These findings suggest that glutamine limitation plays a critical role in the observed oscillations during biofilm expansion.
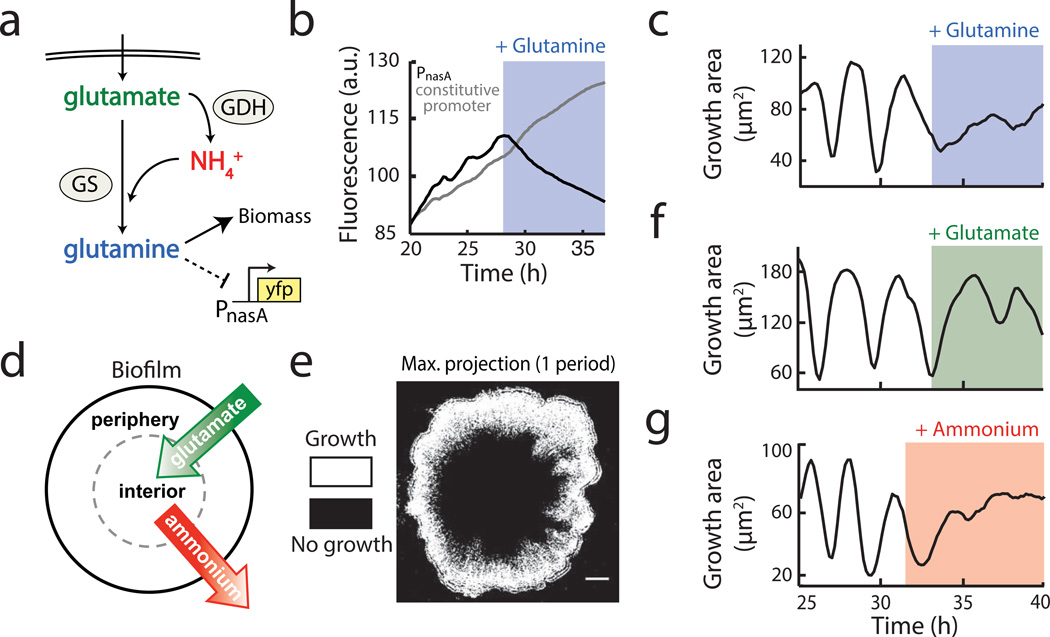
Figure 2.
Biofilm growth depends specifically on extracellular ammonium availability. a, Colony growth in MSgg medium depends on the production of glutamine from externally supplied glutamate and self-produced or scavenged ammonium. Glutamine limitation was monitored using YFP expressed from the nasA promoter, which is activated upon glutamine limitation21. b, Addition of 1 mM glutamine (blue shading) represses expression from the PnasA-YFP reporter (black), but does not affect expression from a constitutive reporter (Phyperspank-CFP + 1 mM IPTG, gray). c, Growth area (see Methods: Data Analysis) before and after addition of 1 mM glutamine to an oscillating colony. d, Of the two nutrients required for glutamine production, externally supplied glutamate (green) is most abundant in the biofilm periphery, while biofilm-produced ammonium (red) is most abundant in the biofilm interior. e, Maximum intensity projection over one period of a colony oscillation, made from a difference movie (Methods: Data Analysis), which shows regions of growth (white) and no growth (black). Scale bar represents 100 µm. f, Growth area of an oscillating colony before and after addition of 30 mM glutamate (green shading). g, Growth area of an oscillating colony before and after addition of 1 mM ammonium (red shading).
The synthesis of glutamine requires both glutamate and ammonium, therefore we investigated which of these substrates could be responsible for the observed glutamine limitation. Glutamate is provided in the media and is thus readily available to cells in the periphery of the biofilm. On the other hand, consumption of glutamate by peripheral cells is likely to limit its availability to cells in the biofilm interior (Fig. 2d). One may thus expect that oscillations in biofilm expansion could be due to periodic pausing of cell growth in the biofilm interior. Accordingly, we set out to establish whether interior or peripheral cells exhibited changes in growth. By tracking physical movement within the biofilm, we uncovered that only peripheral cells grow, and that oscillations in biofilm expansion therefore arise exclusively from periodic halting of peripheral cell growth (Fig. 2e, Supplementary Video 3, Extended Data Fig. 4a, and Methods: Data analysis). This finding was further confirmed by single cell resolution analysis that directly showed periodic reduction in the growth of peripheral cells (Extended Data Fig. 4b). This surprising pausing of cell growth in the periphery, despite unrestricted access to glutamate, suggests that glutamate cannot be the limiting substrate for glutamine synthesis. Consistent with this expectation, biofilm oscillations were not quenched by supplementation of the media with glutamate (Fig. 2f). Therefore, it is not glutamate, but ammonium that appears to be the limiting substrate for glutamine synthesis in the biofilm periphery.
Since cells can self-produce ammonium from glutamate, we next sought to determine how peripheral cells could experience periodic ammonium limitation despite a constant supply of glutamate in the media. It is well known that ammonium production is a highly regulated process that is dependent on the metabolic state of the cell and the ambient level of ammonium in the environment22. In particular, since ammonium is in equilibrium with ammonia vapor, which can freely cross the cell membrane and be lost to the extracellular media23, the production of ammonium is known as a “futile cycle”. Cells therefore preferentially use extracellular (ambient) ammonium for growth, rather than producing their own24–26. Since peripheral cells are exposed to media flow, they are particularly susceptible to this futile cycle of ammonia loss. In this sense, since ammonium is not provided in the media, even if all cells produce ammonium, the biofilm interior will be the major source for ambient ammonium (Fig. 2d). Consequently, the simplifying hypothesis is that growth of peripheral cells relies on ammonium produced within the biofilm. To test this conjecture, we supplemented the media with 1 mM ammonium, which eliminated the periodic halting in biofilm expansion (Fig. 2g and Extended Data Fig.3b and 5a). When additional ammonium was suddenly removed from the media, growth in the biofilm periphery halted as expected (Extended Data Fig. 5b). These findings indicate that peripheral cells preferentially rely on extracellular ammonium produced within the biofilm for their growth.
Metabolic codependence between the biofilm periphery and interior
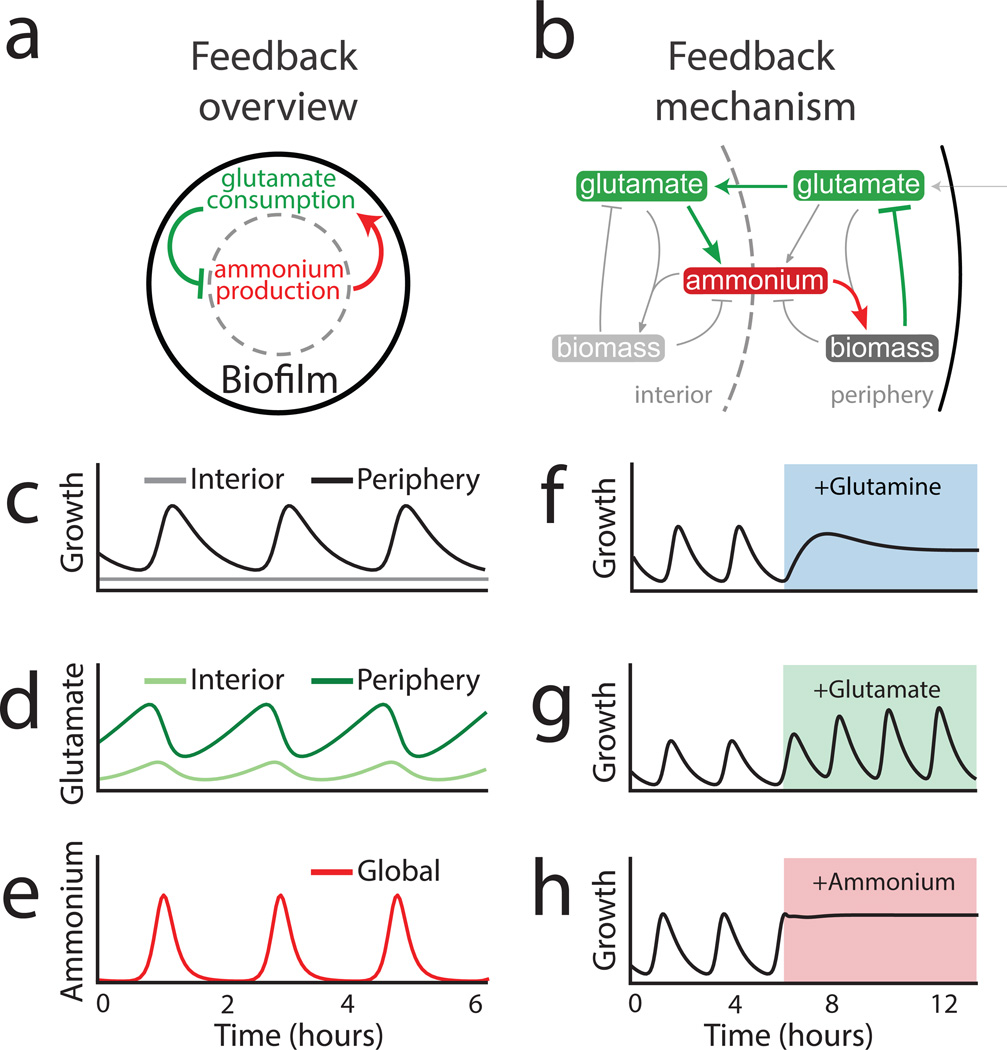
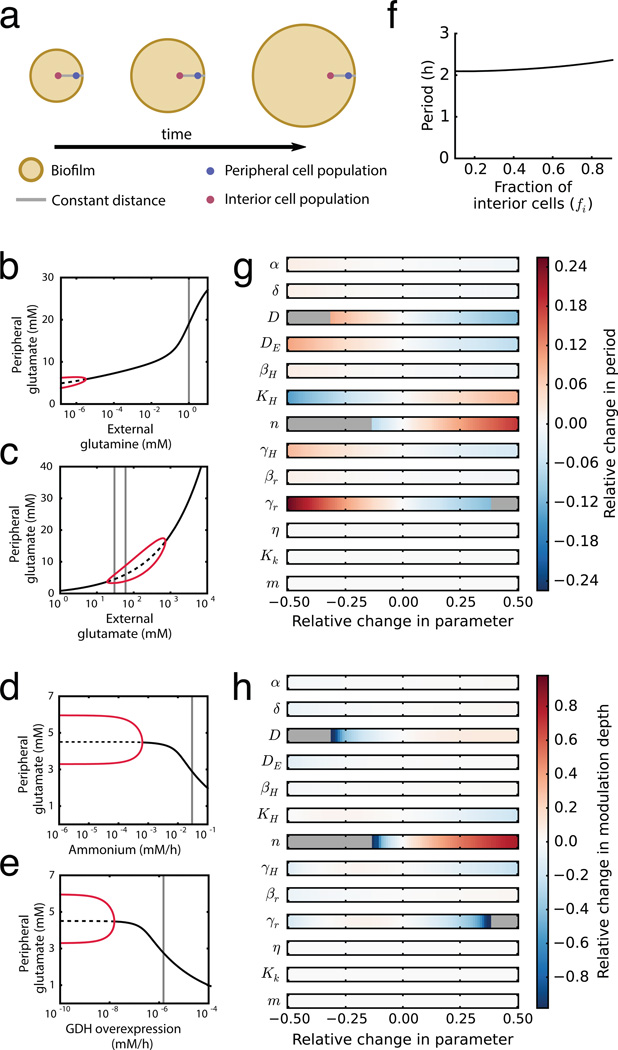
The results described above evoke the intriguing possibility that ammonium limitation for peripheral cells may arise due to glutamate limitation for interior cells. Specifically, persistent consumption of glutamate by peripheral cells can deprive the interior cells of the necessary glutamate for ammonium production. In order to explore this nontrivial hypothesis, we turned to mathematical modeling to develop a conceptual framework and generate experimentally testable predictions. Our model describes separately the metabolic dynamics of interior and peripheral cells and the metabolite exchange between them, where the distinction of the two subpopulations depends on nutrient availability (see Supplementary Information: Mathematical Model). The model thus consists of two main assumptions (Fig. 3a): First, consumption of glutamate during growth of peripheral cells deprives interior cells of this nutrient and thus inhibits ammonium production in the biofilm interior. Second, the growth of peripheral cells depends predominantly on ammonium that is produced by metabolically stressed interior cells. A model based on these two simplifying assumptions (Fig. 3b) generates oscillations consistent with our experimental observations (Fig. 3c–e) and reproduces the effects of supplementing the media with glutamine, glutamate and ammonium (Fig. 3f–h, Extended Data Fig. 6 and Supplementary Information: Mathematical Model). The model also accounts for the observed slight increase of the oscillation period by considering an increase in the ratio of interior to peripheral cells over time (Extended Data Fig.1b and 6f). Therefore, this simple model shows that periodic halting in biofilm growth can result from metabolic codependence between cells in the biofilm periphery and interior that is driven by glutamate consumption and ammonium production, respectively.
Figure 3.
Mathematical modeling of a spatial metabolic feedback loop gives rise to oscillations consistent with experimental data. a, The production of ammonium in the interior is limited by and at the same time triggers the consumption of glutamate in the periphery (green and red arrows, respectively), producing a delayed negative feedback loop. b, The excess glutamate not consumed by the biofilm periphery diffuses to the interior, where it can be converted into ammonium (green arrows). The ammonium in turn enhances growth in the periphery (red arrow) and consequently reduces the supply of glutamate to the interior. Model predictions are shown in (c–h): c, Biofilm growth over time. d, Glutamate concentration over time. e, Ammonium concentration over time. f, Colony growth before and after glutamine addition (indicated by blue shading). g, Colony growth before and after addition of glutamate (green shading). (h) h, Colony growth before and after addition of ammonium (red shading).
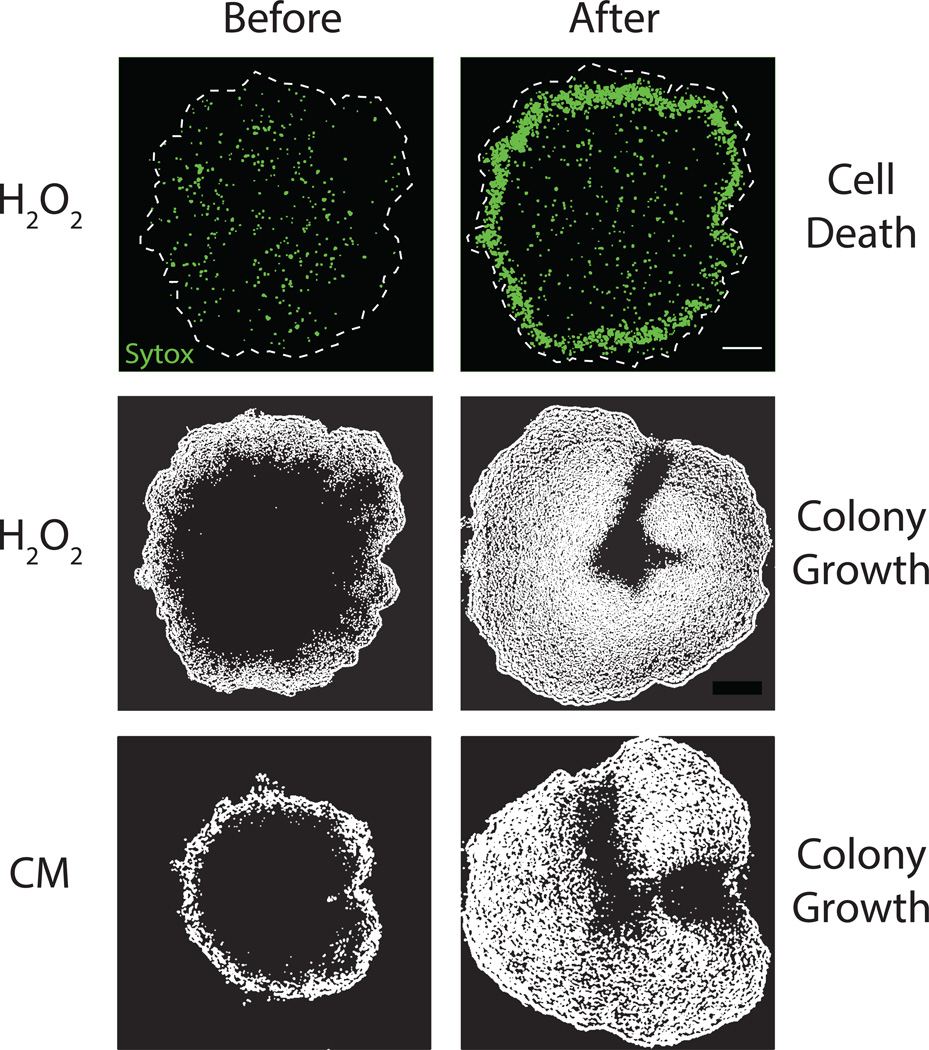
The metabolic codependence between interior and peripheral cells gives rise to the surprising prediction that external attack could promote growth within the biofilm. Specifically, killing of peripheral cells will eliminate their glutamate consumption, which will increase glutamate availability in the biofilm and thereby promote growth of interior cells (Fig. 4a). To test this hypothesis, we measured cell death and growth within oscillating biofilms (Fig. 4b, top and Extended Data Fig. 7). When we exposed the biofilm to media containing hydrogen peroxide (H2O2), we observed increased cell death predominantly in the biofilm periphery (Fig. 4b, bottom and Extended Data Fig. 8). As predicted, death of peripheral cells led to growth of interior cells (Fig. 4c and Extended Data Fig. 8). To verify that this response is not uniquely triggered by H2O2, we exposed biofilms to the antibiotic chloramphenicol and again observed growth of interior cells (Extended Data Fig. 8). These findings further support our hypothesis that glutamate consumption by peripheral cells limits its availability in the biofilm.
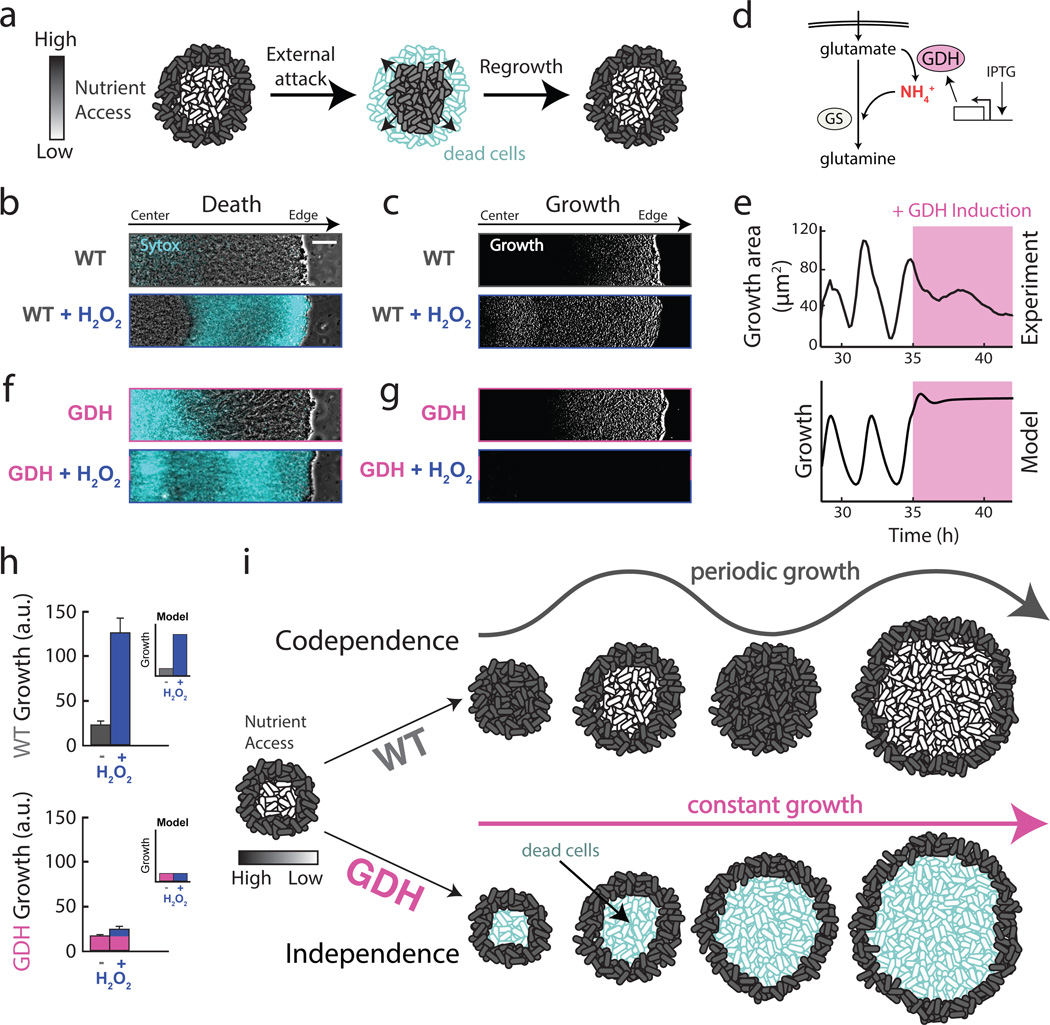
Figure 4.
Metabolic codependence between interior and peripheral cells gives rise to oscillations that make the colony more resilient to external attack. a, Visual representation of the predicted outcome of an external attack on biofilm growth. b, Phase contrast merged with cell death marker (cyan, 1 µM Sytox Green) images of a wild type biofilm region shows cell death with and without challenge by 2% w/w H2O2. Scale bar represents 50 µm. c, In the same biofilm, difference images (white regions indicate cell growth) show wild type growth with and without challenge by H2O2. d, Overexpression of glutamate dehydrogenase (GDH, pink) promotes more production of ammonium from glutamate. e, Experimental (top) and modeling results (bottom) of GDH overexpression (induced with 1 mM IPTG, indicated by pink shading). f, Phase contrast merged with cell death marker (cyan, 1 µM Sytox Green) images of a colony overexpressing GDH with and without challenge by H2O2. g, In the same biofilm, difference images show cell growth during GDH overexpression alone, and with challenge by H2O2. h, Quantification of total biofilm growth rate in wild type (upper, n = 4 colonies) and GDH overexpression (lower, n = 3 colonies) strains upon challenge with H2O2. Error bars represent standard deviations. Modeling data are shown as an inset for each strain. i, Codependence between interior and peripheral cells exhibited in a wild type strain results in a growth strategy that sustains the viability of interior cells, while independence enforced by a GDH overexpression strain results in starvation of interior cells and reduced resilience to external attack.
The benefit of biofilm oscillations
Our model also assumes that glutamate starvation of the biofilm interior reduces the production of ammonium that can support peripheral cell growth. This assumption provokes the question as to why peripheral cells do not simply overcome their dependence on extracellular ammonium by increasing intracellular production27, 28. To address this question, we constructed a strain that contains an inducible copy of the GDH gene rocG (Fig. 4d). We confirmed that GDH overexpression was not toxic to individual cells and did not affect their growth rate (Extended Data Fig. 9). In contrast, the induction of GDH expression in the biofilm quenched growth oscillations (Fig. 4e and Extended Data Fig. 3c) and resulted in high levels of cell death in the colony interior (Fig. 4f, top). This result explains why peripheral cells do not appear to utilize the simple strategy of overcoming their dependence on extracellular ammonium: such a strategy would result in the continuous growth of peripheral cells, starving and ultimately causing the death of sheltered interior cells within the biofilm. Periodic halting of peripheral cell growth due to extracellular ammonium limitation thus promotes the overall viability of the biofilm.
The ability of the biofilm to regenerate itself in the event of an external attack suggested that killing the biofilm interior first would be a more effective strategy for biofilm extermination. Accordingly, we exposed the GDH overexpression strain to hydrogen peroxide and again measured growth and death. As described above, GDH induction causes death of interior cells. Exposing the GDH overexpression strain to hydrogen peroxide resulted in more effective global killing throughout the biofilm (Fig. 4f, g, bottom). While in the wild-type biofilm interior cells begin to grow in response to an external attack, metabolic independence between interior and peripheral cells in the GDH strain interferes with this defense mechanism (Fig. 4h). This outcome is also consistent with modeling predictions (Fig. 4h, inset). Oscillations in biofilm growth that are driven by metabolic codependence thus promote the resilience of the biofilm community by sustaining the viability of the sheltered interior cells that are most likely to survive in the event of an environmental stress (Fig. 4i).
Discussion
The data presented here reveal that intracellular metabolic activity within biofilms is organized in space and time, giving rise to codependence between interior and peripheral cells. Even though bacteria are single-celled organisms, the metabolic dynamics of individual cells can thus be regulated in the context of the community. This metabolic codependence can in turn give rise to collective oscillations that emerge during biofilm formation and promote the resilience of biofilms against chemical attack. The community-level oscillations also support the ability of biofilms to reach large sizes, while retaining a viable population of interior cells. Specifically, periodic halting of peripheral cell growth prevents complete starvation and death of the interior cells. This overcomes the colony size limitation for a viable biofilm interior that would otherwise be imposed by nutrient consumption in the biofilm periphery. Metabolic codependence in biofilms therefore offers an elegant solution that resolves the social conflict between cooperation (protection) and competition (starvation) through oscillations.
The intriguing discovery of biofilm oscillations presented here also provokes new questions. While cellular processes such as swarming or expression of extracellular matrix components are not required for the observed biofilm oscillations (Extended Data Fig. 10), it will be interesting to pursue whether such cellular processes are influenced by oscillatory dynamics29. Another question worth pursuing is whether metabolic codependence can also arise in other biofilm-forming species. Perhaps other metabolic branches where metabolites can be shared among cells could also give rise to oscillations in biofilm growth. It will be exciting to pursue these questions in future studies to obtain a better understanding of biofilm development.
Our observations also suggest future strategies to cope with the intriguing resilience of biofilms in the face of environmental stresses, such as antibiotic exposure. In particular, our findings show that straightforward application of stress (such as H2O2 or chloramphenicol) to the biofilm counterintuitively promotes growth, effectively rejuvenating the biofilm. Death of the colony periphery relieves the repression on the growth of interior cells, allowing them to regenerate a new biofilm periphery and interior. In contrast, manipulation of the metabolic codependence may yield a more effective approach to control biofilm formation. Specifically, promoting continuous growth of peripheral cells can starve the biofilm interior, leaving behind the exposed peripheral cells that can more easily be targeted by external killing factors. Therefore, the metabolically driven collective oscillations in biofilm expansion described here not only reveal fundamental insights into the principles that govern formation of multicellular communities, but also suggest new strategies for manipulating the growth of biofilms.
Methods
Strains and Plasmids
All experiments were done using Bacillus subtilis NCIB 3610. The wild type strain was a gift from Wade Winkler (University of Maryland)30 and all other strains were derived from it (see Supplementary Information: Strains).
Growth conditions
The biofilms were grown using MSgg medium16. It contains 5 mM potassium phosphate buffer (pH 7.0), 100 mM MOPS buffer (pH 7.0, adjusted using NaOH), 2 mM MgCl2, 700 µM CaCl2, 50 µM MnCl2, 100 µM FeCl3, 1 µM ZnCl2, 2 µM thiamine HCl, 0.5% (v/v) glycerol and 0.5% (w/v) monosodium glutamate. The MSgg medium was made from stock solutions on the day of the experiment, and the stock solution for glutamate was made new each week.
Microfluidics
We used the CellASIC ONIX Microfluidic Platform and the Y04D microfluidic plate (EMD Millipore). It provides unconventionally large chambers, allowing the formation of colonies containing millions of cells, yet still leaves room for media flow. Media flow in the microfluidic chamber was driven by a pneumatic pump from the CellASIC ONIX Microfluidic Platform, and the pressure from the pump was kept stable during the course of the oscillation. In most of the experiments, we used a pump pressure of 1 psi with only one media inlet open (there are 6 media inlets in the Y04D plate), which corresponds to a flow speed of ~16 µm/s in the growth chamber.
On the day before the experiment, cells from −80°C glycerol stock were streaked onto LB agar plate and incubated at 37°C for overnight. The next day morning, a single colony was picked from the plate and inoculated into 3 ml of LB broth in a 50 ml conical tube, and then incubated in 37°C shaker. After 2.5 hours of incubation, the cell culture was centrifuged at 2100 rcf for 1 min, and then the cell pellet was re-suspended in MSgg and then immediately loaded into microfluidics. After the loading, cells in the microfluidic chamber were incubated at 37°C for 90 min, and then the temperature was kept at 30°C for the rest of the experiment.
Time-Lapse Microscopy
The growth of the biofilms was recorded using phase contrast microscopy. The microscopes used were Olympus IX81 and IX83, and DeltaVision PersonalDV. To image entire biofilms, 10× lens objectives were used in most of the experiments. Images were taken every 10 min. Whenever fluorescence images were recorded, we used the minimum exposure time that still provided a good signal-to-noise ratio.
Data analysis
ImageJ (National Institutes of Health) and MATLAB (MathWorks) were used for image analysis. In house software was also developed to perform colony detection and quantification of colony expansion. Multiple methods of colony detection were used to ensure the accuracy of the analysis. To detect regions of expansion in a biofilm, we performed image differencing on snapshots of the biofilm from time-lapse microscopy videos. Specifically, we calculated the difference between two consecutive phase contrast images (taken 10 min apart) by finding the absolute difference between each pixel in each image. We then generated an image stack based on these results. The intensity values from the stack correlate with the expansion inside the biofilm. The growth area was determined by converting difference images to binary images and then measuring the area of the colony growth region (white pixels). To measure cell replication time, we tracked the length and division of individual cells in the biofilm periphery (Extended Data Fig. 4b).
Extended Data
Extended Data Figure 1.
Characterization of biofilm growth oscillations. a, (Top) Growth rate over time of an oscillating colony. (Bottom) The pressure that drives media flow in the microfluidic chamber is constant over time (see Methods: Microfluidics). b, (Top) Growth rate of an oscillating colony. (Bottom) Period of each oscillation cycle, measured peak to peak. The error bars (±20 min) are determined by the imaging frequency (1 frame/10 min). The period slightly increases over time (see also Extended Data Fig. 6f and Supplementary Information: Mathematical Model).
Extended Data Figure 2.
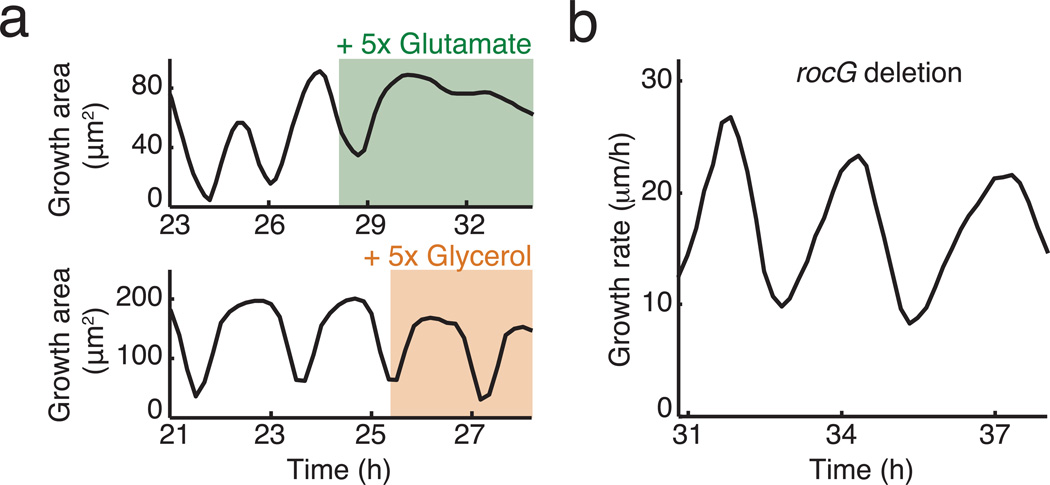
Roles of carbon and nitrogen in biofilm growth oscillations. a, Effect of increasing carbon (glycerol) or nitrogen (glutamate) availability on the oscillations. While increasing glutamate by 5 times of the normal MSgg levels leads to quenching of the oscillation, increasing glycerol by 5 times does not. b, Colony growth of mutant strain with rocG deletion. B. subtilis NCIB 3610 has two glutamate dehydrogenases (GDH), rocG and gudB. While gudB is constitutively expressed, rocG expression is subject to carbon catabolite repression18. The oscillatory growth of the rocG deletion strain indicates that carbon-source dependent regulation of rocG expression is not required for biofilm oscillations.
Extended Data Figure 3.
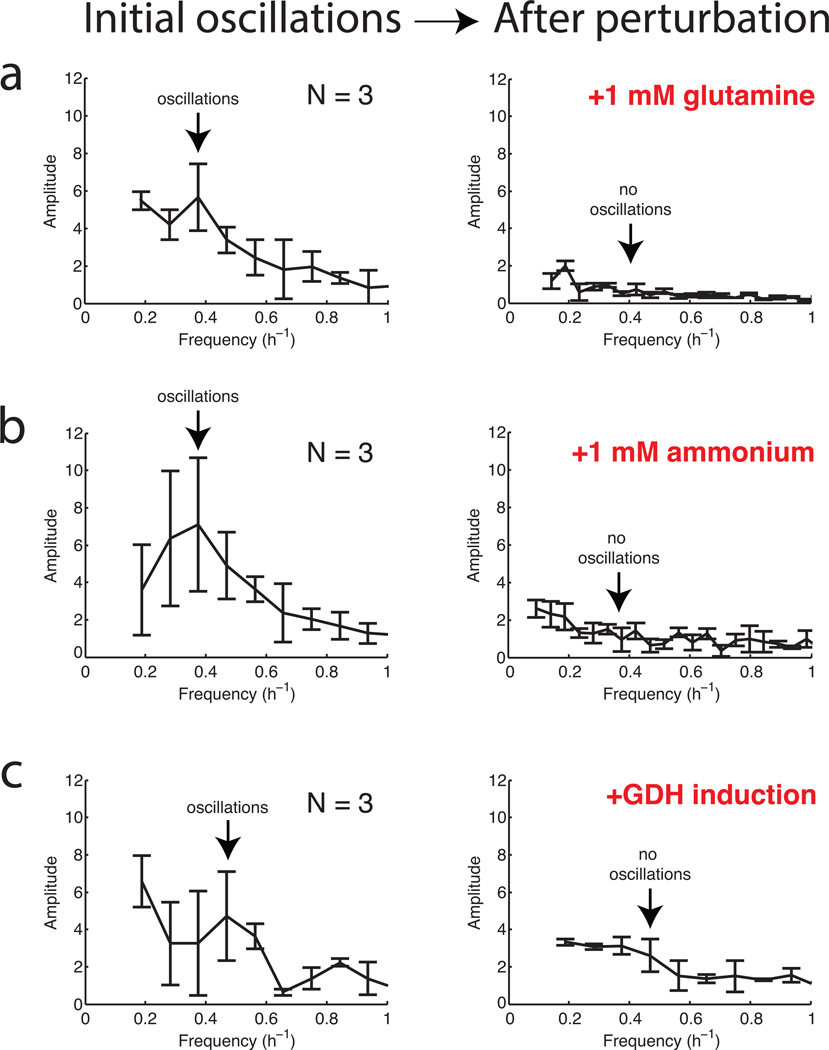
Fourier transform of biofilm growth rates before and after addition of a, 1 mM glutamine, b, 1 mM ammonium, and c, 1 mM IPTG to induce Phyperspank-RocG. The error bars show standard deviations (n = 3 colonies for each condition). The arrows indicate the frequency of oscillations for each condition before perturbation (left) and the lack of oscillations after perturbation (right).
Extended Data Figure 4.
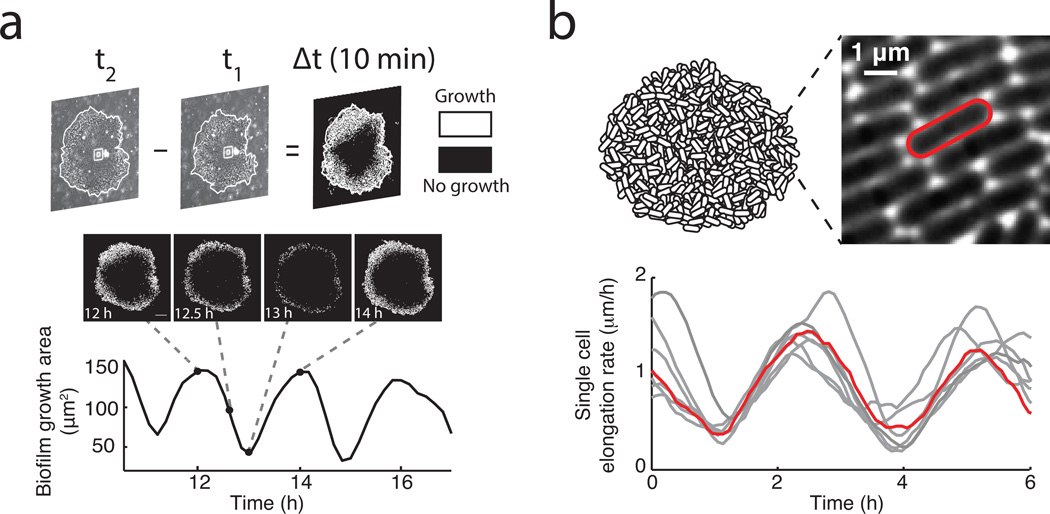
Measurements of cell growth within oscillating biofilms. a, (Top) Visual representation of the method through which difference movies are generated (Methods: Data Analysis). Growth is represented by white pixels, and lack of growth is indicated by black pixels. (Middle) Film strip and (bottom) growth area over time of an oscillating colony. Dashed lines show the position of each image on the time trace. Scale bar represents 100 µm. b, (Top left) schematic of a biofilm. (Top right) high magnification phase contrast image of biofilm periphery focused at the bottom layer of cells. (Bottom panel) time traces depicting elongation rates of single cells in gray. Highlighted in red is the single cell time trace for the cell outlined in red in the top right panel. The periodic slowdown of the growth of individual peripheral cells is responsible for the observed periodic reduction in biofilm expansion.
Extended Data Figure 5.
Effects of external ammonium on biofilm development. a, Addition of external ammonium (red shading, 1 mM) represses expression from the PnasA-YFP reporter (black), but does not affect expression from a constitutive reporter (Phyperspank-CFP + 1 mM IPTG, gray). b, Removal of external ammonium (red shading, 13 mM) causes halting of colony growth.
Extended Data Figure 6.
Mathematical model of biofilm growth. a, The model describes the dynamics of two cell populations in a biofilm, interior and peripheral. As the biofilm grows, there is a constant distance between the interior population and the biofilm edge. b–e, Bifurcation diagrams showing systematic analysis on the effects of external glutamine, external glutamate, ammonium uptake, and GDH overexpression respectively. The red lines correspond to the extrema of oscillations in peripheral glutamate (stable limit cycle). The solid black line denotes stable fixed point. The dashed black line corresponds to an unstable fixed point. The vertical gray lines highlight the state of the system for each nutrient addition experiment shown in Fig. 3 of the main text. f, Model prediction of oscillation period as function of interior cell fraction in the whole biofilm. g–h, Sensitivity analysis of oscillation period and modulation depth to changes in model parameters. Modulation depth is defined as the amplitude of the oscillations divided by the mean value. Gray color denotes parameter regions where the system does not oscillate.
Extended Data Figure 7.
Temporal profile of cell death within an oscillating biofilm. a, Colony growth rate. b, Average fluorescence intensity of a cell death marker (Sytox Green, 1 µM, Life Technologies) from the same colony shown in (a).
Extended Data Figure 8.
Effect of external attack with hydrogen peroxide (H2O2, 0.15% v/v) or chloramphenicol (CM, 5 µg/ml). (Top) cell death shown by Sytox Green (1 µM). (Middle and bottom) colony growth shown by image differencing (see Extended Data Fig. 4a and Methods: Data Analysis). Scale bar represents 100 µm. The white dashed lines indicate colony edge.
Extended Data Figure 9.
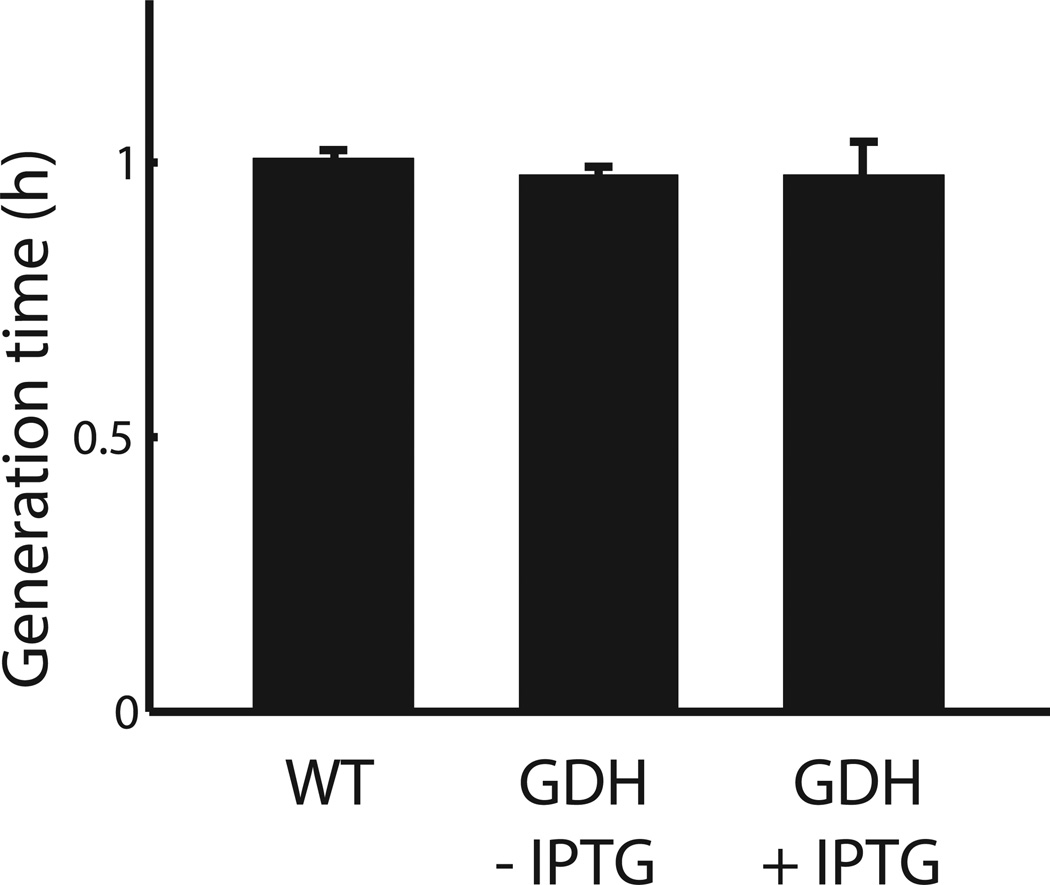
Effect of GDH induction on cell growth. Wild type and Phyperspank-RocG (uninduced or induced with 10 mM IPTG) strains were grown in liquid culture (MSgg medium, 30°C). Cell generation times were measured using OD600. Error bars show standard deviations (n = 3 replicates).
Extended Data Figure 10.
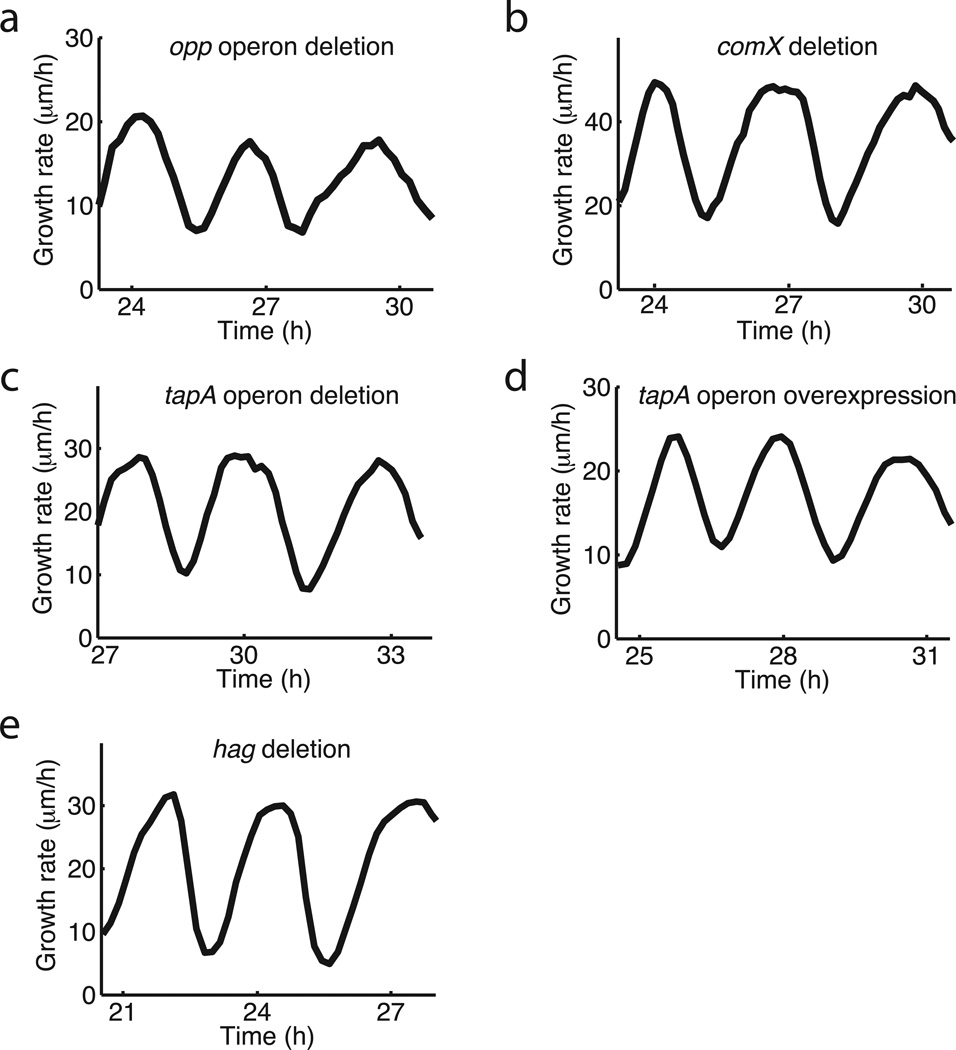
Growth rate oscillations persist in various mutant strains. a, opp operon deletion (deficient in quorum sensing). b, comX deletion (deficient in quorum sensing). c, tapA operon deletion (extracellular matrix component deletion). d, tapA operon overexpression (Phyperspank-tapA operon, 1mM IPTG). e, hag deletion (deficient in swimming and swarming). These results show that the corresponding genes and processes are not required for biofilm oscillations.
Supplementary Material
Acknowledgments
We thank the anonymous referees for their constructive comments during the review process. We would also like to thank Katherine Süel, Tolga Çağatay, Roy Wollman, Terry Hwa and Michael Elowitz for comments during the writing of the manuscript. A.P. is a Simons Foundation Fellow of the Helen Hay Whitney Foundation. J.H. is supported by the UCSD Cell and Molecular Genetics Training Grant. J.G.O. is supported by the Ministerio de Economia y Competitividad (Spain) and FEDER, under project FIS2012-37655-C02-01, and by the ICREA Academia Programme. This research was funded by the National Institutes of Health, National Institute of General Medical Sciences Grant R01 GM088428 and the National Science Foundation Grant MCB-1450867 (both to G.M.S.).
G.M.S. and J.L. have a pending patent through the University of California San Diego based on this work (SD2015-219).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions G.M.S., J.L., M.A., and J.G.O. designed the research, J.L. performed the experiments, J.L., J.H., and M.A. performed the data analysis, M.G.S. and J.G.O. performed the mathematical modeling, D.D.L., S.L., and M.A. made the bacteria strains, G.M.S., A.P., J.L., J.H., M.G.S, and J.G.O. wrote the manuscript. All authors discussed the manuscript.
The authors declare no competing financial interest.
References
- 1.Ben-Jacob E, Cohen I, Levine H. Cooperative self-organization of microorganisms. Adv Phys. 2000;49:395–554. [Google Scholar]
- 2.Eldar A. Social conflict drives the evolutionary divergence of quorum sensing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13635–13640. doi: 10.1073/pnas.1102923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregor T, Fujimoto K, Masaki N, Sawai S. The Onset of Collective Behavior in Social Amoebae. Science. 2010;328:1021–1025. doi: 10.1126/science.1183415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingreen NS, Levin SA. Cooperation among microorganisms. Plos Biol. 2006;4:1486–1488. doi: 10.1371/journal.pbio.0040299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nature reviews. Microbiology. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira NM, Niehus R, Foster KR. Evolutionary limits to cooperation in microbial communities. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:17941–17946. doi: 10.1073/pnas.1412673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies D. Understanding biofilm resistance to antibacterial agents. Nature reviews. Drug discovery. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 8.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical microbiology reviews. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes & development. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends in microbiology. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 13.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nature reviews. Microbiology. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 14.Asally M, Kittisopikul M, Rue P, Du Y, Hu Z, Cagatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Suel GM. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilking JN, Zaburdaev V, De Volder M, Losick R, Brenner MP, Weitz DA. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunka K, Commichau FM. Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. Molecular microbiology. 2012;85:213–224. doi: 10.1111/j.1365-2958.2012.08105.x. [DOI] [PubMed] [Google Scholar]
- 18.Stannek L, Thiele MJ, Ischebeck T, Gunka K, Hammer E, Volker U, Commichau FM. Evidence for synergistic control of glutamate biosynthesis by glutamate dehydrogenases and glutamate in Bacillus subtilis. Environmental microbiology. 2015 doi: 10.1111/1462-2920.12813. [DOI] [PubMed] [Google Scholar]
- 19.Belitsky BR, Sonenshein AL. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. Journal of bacteriology. 1998;180:6298–6305. doi: 10.1128/jb.180.23.6298-6305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. The origins of 168, W23, and other Bacillus subtilis legacy strains. Journal of bacteriology. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano MM, Yang F, Hardin P, Zuber P. Nitrogen regulation of nasA and the nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. Journal of bacteriology. 1995;177:573–579. doi: 10.1128/jb.177.3.573-579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleiner D. Bacterial Ammonium Transport. Fems Microbiol Lett. 1985;32:87–100. [Google Scholar]
- 23.Castorph H, Kleiner D. Some properties of a Klebsiella pneumoniae ammonium transport negative mutant (Amt-) Archives of microbiology. 1984;139:245–247. doi: 10.1007/BF00402008. [DOI] [PubMed] [Google Scholar]
- 24.Boogerd FC, Ma HW, Bruggeman FJ, van Heeswijk WC, Garcia-Contreras R, Molenaar D, Krab K, Westerhoff HV. AmtB-mediated NH3 transport in prokaryotes must be active and as a consequence regulation of transport by GlnK is mandatory to limit futile cycling of NH4+/NH3. Febs Lett. 2011;585:23–28. doi: 10.1016/j.febslet.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 25.Jayakumar A, Schulman I, MacNeil D, Barnes EM., Jr Role of the Escherichia coli glnALG operon in regulation of ammonium transport. Journal of bacteriology. 1986;166:281–284. doi: 10.1128/jb.166.1.281-284.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim M, Zhang Z, Okano H, Yan D, Groisman A, Hwa T. Need-based activation of ammonium uptake in Escherichia coli. Molecular systems biology. 2012;8:616. doi: 10.1038/msb.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Commichau FM, Gunka K, Landmann JJ, Stulke J. Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. Journal of bacteriology. 2008;190:3557–3564. doi: 10.1128/JB.00099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Detsch C, Stulke J. Ammonium utilization in Bacillus subtilis: transport and regulatory functions of NrgA and NrgB. Microbiology. 2003;149:3289–3297. doi: 10.1099/mic.0.26512-0. [DOI] [PubMed] [Google Scholar]
- 29.Anyan ME, Amiri A, Harvey CW, Tierra G, Morales-Soto N, Driscoll CM, Alber MS, Shrout JD. Type IV pili interactions promote intercellular association and moderate swarming of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18013–18018. doi: 10.1073/pnas.1414661111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irnov I, Winkler WC. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Molecular microbiology. 2010;76:559–575. doi: 10.1111/j.1365-2958.2010.07131.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.