Abstract
The optic nerve (ON) plays a crucial role in human vision transporting all visual information from the retina to the brain for higher order processing. There are many diseases that affect the ON structure such as optic neuritis, anterior ischemic optic neuropathy and multiple sclerosis. Because the ON is the sole pathway for visual information from the retina to areas of higher level processing, measures of ON damage have been shown to correlate well with visual deficits. Increased intracranial pressure has been shown to correlate with the size of the cerebrospinal fluid (CSF) surrounding the ON. These measures are generally taken at an arbitrary point along the nerve and do not account for changes along the length of the ON. We propose a high contrast and high-resolution 3-D acquired isotropic imaging sequence optimized for ON imaging. We have acquired scan-rescan data using the optimized sequence and a current standard of care protocol for 10 subjects. We show that this sequence has superior contrast-to-noise ratio to the current standard of care while achieving a factor of 11 higher resolution. We apply a previously published automatic pipeline to segment the ON and CSF sheath and measure the size of each individually. We show that these measures of ON size have lower short-term reproducibility than the population variance and the variability along the length of the nerve. We find that the proposed imaging protocol is (1) useful in detecting population differences and local changes and (2) a promising tool for investigating biomarkers related to structural changes of the ON.
Keywords: Magnetic Resonance Imaging, Optic Nerve, Reproducibility
1. Introduction
The human optic nerve (ON) is one of the most challenging structures to image accurately due to the fact that this is a small structure, constantly in motion, surrounded by fatty tissue and adjacent to the maxillary sinuses. However, the ON is integral to visual performance and is clinically indicated in a number of diseases, most notably, multiple sclerosis (MS) [1], but also in several forms of optic neuropathy [2, 3]. Optic neuritis is closely linked with MS, 25% of optic neuritis events eventually develop into MS [4]. Despite this, there is no radiological biomarker of the ON that predicts eventual development of MS. The ON is surrounded by a sheath of cerebrospinal fluid (CSF). The size of this CSF sheath has been shown to correlate with intracranial pressure which, in conjunction with a plethora of diseases, may be associated with increased mortality and less favorable neurological outcomes [5]. These measurements of optic nerve size are often taken at arbitrary points along the length of the ON and they are not investigated as local changes along the length of the nerve due to a lack of contrast.
While important, the ON's size, surrounding fat, position proximal to the maxillary sinuses and mobility combine to make the ON a hostile structure to image with conventional (or advanced) MRI [6]. The ON is typically less than 6mm in diameter; while clinical sequences typically have acceptable resolution (0.6-0.8mm) in-plane, the through-plane resolution is suboptimal (>2mm), which makes characterizing the shape of the ON very difficult. The location of the ON presents challenges in that the surrounding fatty tissue needs to be suppressed to accurately visualize the ON and CSF sheath. Traditional fat suppression techniques can often be insufficient in suppressing olefinic orbital fat [7]. The proximity of the ON to the maxillary sinuses creates magnetic susceptibility distortions which must also be accounted for when designing a sequence to image the ON. The ON's mobility also presents challenges for imaging as subjects make saccadic eye movements during acquisition. These small and fast eye movements subsequently move the ON. Fixation has been used to minimize these effects but does not eliminate eye motion completely and imposes acquisition time constraints. Conventional MR methods for the optic nerve (T1-weighted and T2-weighted) often do not provide contrast between the surrounding CSF and the optic nerve structure (Fig 1A-B) [8]. We developed an alternative MR method, taking advantage of clinical hardware, that can provide excellent resolution, high contrast between optic nerve and surrounding tissues, and offer a method from which advanced morphometrics for identifying biomarkers can be derived.
Figure 1.
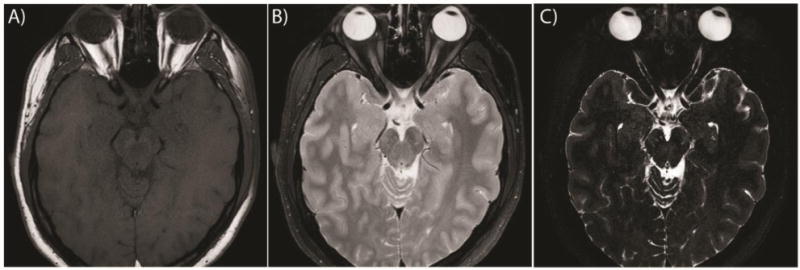
Comparison of the clinical standard of care (A) T1W image, (B) T2W image and (C) our proposed high-resolution sequence for a single subject. Note that the resolution of the clinical standard of care yields one slice containing the ON, which is shown, while a medial slice was chosen for our proposed method (C)
2. Methodology
Sequence Design
We developed and optimized a high-resolution 3-D isotropic turbo spin echo (TSE) with asymmetric k-space sampling (VISTA) with parameters empirically optimized for ON-CSF contrast. This sequence addresses the size of the ON by having sufficiently high resolution in all three planes (≤0.6mm isotropic) and allows for accurate characterization of the ON and CSF. The use of a TSE sequence is chosen due to its inherent robustness to B0 inhomogeneities from continuous refocusing which mitigates the effects of magnetic susceptibility caused by the bone-air interface of the maxillary sinuses. Asymmetrically sampling k-space blurs any ON motion across k-space. The result is a sequence which accurately captures variations in the hostile imaging environment of the ON. This sequence is also optimized for CSF-ON contrast as well as CSF-Fat contrast. We use a SPIR fat suppression technique to minimize fat signal and maximize CSF-Fat contrast. We utilize an extended echo train which leads to a long effective TE and provides good signal in the CSF while suppressing signal within the ON and any remaining fat signal, such as olefinic fat, that has not already been suppressed with the SPIR.
Data Acquisition
10 healthy subjects age 24 to 36 years (average: 28.25, median: 27 years, 6 male/4 female) were enrolled in the imaging study. Imaging was acquired on a 3T Philips Achieva (Philips Medical Systems, Best, The Netherlands) with a 2-channel body coil for transmission and an 8 channel head coil for reception for all sequences. After tri-planar localization, we acquired the all volumes in the axial plane. The VISTA sequence parameters were: 3D TSE TR = 4000ms, TE = 455ms, α = 90°, FOV= 180 × 180 × 20mm3, acquired resolution = 0.55 × 0.55 × 0.55mm3, reconstructed resolution = 0.35 × 0.35 × 0.35mm3, SENSE factor = 2, fat saturation = SPIR, NSA=2 and total scan time = 7:48. For comparison a clinical standard of care T1-weighted (T1W) image was also acquired with parameters: SE TR=400ms, TE=12ms, α = 90°, FOV= 180 × 180 × 33mm3, acquired resolution = 0.70 × 0.88 × 3.0mm3, reconstructed resolution = 0.42 × 0.42 × 3.0mm3, and total scan time = 3:20. A clinical standard of care T2-weighted (T2W) image was also acquired with parameters: TSE TR=3000ms, TE=80ms, α = 90°, FOV= 180 × 180 × 33mm3, acquired resolution = 0.70 × 0.88 × 3.0mm3, reconstructed resolution = 0.42 × 0.42 × 3.0mm3, fat suppression=SPIR and total scan time = 2:48. Subjects were scanned with a baseline scan and again within 30 days of the original scan for short-term reproducibility. Inter-scan time was from 4 to 29 days (average: 19.4 days, median: 23 days). Figure 1 shows the clinical standard of care T1W image (A), T2W image (B) and our optimized imaging method (C). Note the increased contrast between the CSF and ON in our optimized imaging as compared to the standard of care T2W image. The T1W image shows no contrast between CSF and ON, only the ON-Fat boundary is visible.
Data Analysis: Contrast-to-Noise Ratio
To quantitatively compare the three sequences, contrast-to-noise ratio (CNR) was investigated for each of the three sequences for CSF-ON contrast and CSF-Fat contrast. Note that since the CSF-ON boundary is not visible on the T1W images (see Fig. 1a), this comparison was only made for the clinical T2W sequence and our VISTA sequence. CNR is defined as:
| (1) |
where Sa is the mean the signal from tissue a, Sb is the mean signal from tissue b and a is the image noise. Regions of interest (ROIs) were drawn on each scan (T1W, T2W and VISTA) for each of the 10 subjects for both the baseline and short term follow up scans. An ROI was drawn in a homogenous area of white matter in the temporal lobe, and the standard deviation of intensity values within this ROI was taken as an estimate of image noise (σ). ROIs were then drawn such that they encompassed as many pure voxels of each of ON, CSF and orbital fat from a medial ON slice. The mean voxel intensity of each of these ROIs was then taken as the mean signal from that tissue.
Data Analysis: ON Structural Reproducibility
Our segmentation begins with a previously described multi-atlas segmentation method [9], which automatically segments the orbits, optic chiasm and ON. This method uses 35 manually labeled atlas images, which include both healthy controls as well as ON drusen and MS patients. The target image to be segmented is registered to each of the 35 atlas images using an affine registration and non-rigid registration [10]. The manual labels of the atlas images are then transformed to the target space using these registrations and are fused using non-local spatial STAPLE[11, 12]. The segmentation of the ON includes both the ON and CSF sheath and so we must refine our segmentation to separate the two structures and measure them independently.
We utilize a previously described model [13] which can be seen in Equation (2) to fit the ON and CSF sheath in the coronal plane and extract the radii of both. The model is a difference of two Gaussian distributions which matches the intensity profile of the ON in the coronal plane. The second Gaussian is scaled by an exponential term and has a scaling factor on the covariance matrix in the range (0,1) such that the second Gaussian is always smaller than the first Gaussian. The covariance matrix is formulated with the correlation term as a sigmoid function to improve stability later on, during the optimization process.
| (2) |
| (3) |
| (4) |
We define the error as the sum of squared difference between the model and the image, which can be seen in Equation (5). The derivative can be seen in Equation (6) where the last term is the partial derivative with respect to each of the eight model parameters. These partial derivatives are computed analytically and can be seen in Equations (7)(8)(9)(10)(11)(12). Note that the derivatives for σy and μy are omitted as they are a direct substitutions into equations (7) and (10), respectively. In summary, the complete model is composed of eight terms: Θ = [σx, σy, σ2, I0, μx, μy, β, ρ]. We then fit the model to the ON in the coronal plane using an iterative conjugate gradient descent optimization method on all eight parameters [13].
The model parameters are correlated with the radii of the ON and CSF sheath through a random forest regression[14] using 1.2 million synthetic training images. Six of the eight model parameters are used for the regression, the centroids are omitted as they are dependent solely on field of view. These training images were generated by simulating partial volume effects of imaging two concentric tubular structures with 0.6 mm isotropic voxels using a Monte Carlo simulation. This model is then tilted at varying angles relative to the imaging plane and the size of each of the concentric tubes is varied to generate the training set. The regression is validated using tenfold cross validation which shows the predicted radii to correlate with the true underlying radii with an explanatory R-squared greater than 0.95 for both ON and sheath radii.
The shape of the ON often results in a variable number of coronal slices between individuals. For population comparison each ON was interpolated to be the same length as the longest ON in the population, we refer to this as posterior normalized slice. A three-element moving window median filter is also applied across slices to reduce noise in the measurements.
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
3. Results
Contrast-to-Noise Ratio
Figure 2 shows the results of the CNR analysis for the CSF-Fat contrast (left) and the ON-CSF contrast (right). We can see that the T1W sequence has significantly higher CNR for the CSF-Fat boundary than the T2W or VISTA sequences (Wilcoxon rank-sum test; p<0.01). The T2W and VISTA both have similar but sufficient CNR to distinguish CSF and orbital fat, due to the SPIR fat suppression. The ON-CSF boundary is indistinguishable in the T1W imaging and is therefore excluded from comparison. The VISTA sequence has significantly superior CNR to the clinical T2W sequence (Wilcoxon rank-sum test; p<0.01). The Rose criterion states that if the CNR is below 3-5, two structures become difficult to distinguish [16]. These results suggest the ON-CSF contrast is often difficult to distinguish on clinical T2W while the proposed VISTA CNR is well above this criterion.
Figure 2.
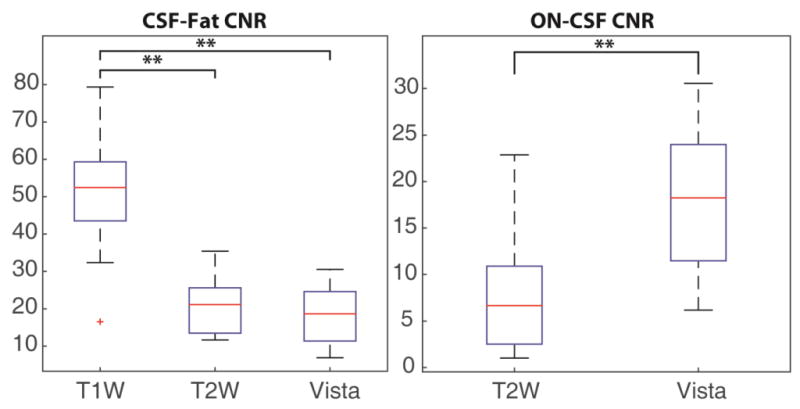
Contrast-to-noise ratio (CNR) comparison for contrast between CSF-Fat (left) and ON-CSF (right) for the clinical standard of care T1W, T2W and the optimized VISTA sequence. ** indicates the results are significantly different by Wilcoxon rank-sum test at p<0.01.
Optic Nerve Structural Reproducibility
The radius of the ON (lower line) and CSF sheath (upper line) can be seen in Figure 3 for 8 subjects in scan 1 (blue) and 8 subjects in scan 2 (red). Two scans, from two separate subjects, were segmentation failures and are excluded, along with the corresponding rescan, from this analysis. The error bars are standard deviation and illustrate the population variability. The lower line is the radius of the ON along the length of the nerve and the upper line is the radius of the CSF sheath along the nerve. Note the labels indicating which direction is proximal to the globes and optic chiasm. The computed reproducibility (green) shows the standard deviation of the difference of each scan-rescan measurement divided by This factor is divided out to account for the summation of two random variables. These error bars are plotted on the overall mean distribution for comparison. We here note that the computed reproducibility is less than the population variability and the variability along the length of the nerve. This data supports that this tool is useful in detecting local population differences.
Figure 3.
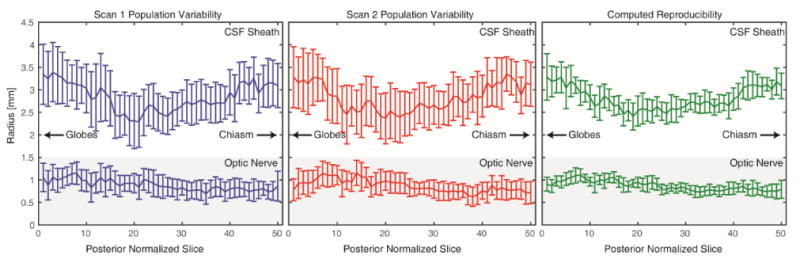
Comparison of population variability for scan 1 (blue) and scan 2 (red) as well as the computed reproducibility as the standard deviation of the difference of each scan-rescan measurement divided by . The computed reproducibility error bars are shown on the overall population mean distribution for comparison.
4. Discussion
We have demonstrated the superiority of our proposed VISTA sequence in characterizing the ON and in generating contrast between ON and CSF. This sequence has ON-CSF CNR that is consistently above the Rose criterion and therefore more consistent in accurately visualizing the ON as compared to a current clinical standard of care with 11.1 fold higher volumetric resolution than the clinical sequence. This high contrast in conjunction with the isotropic high resolution of this sequence allows for characterization of the size and shape of the ON along the length of the ON, which was previously not possible. The improved imaging facilitates automated processing and algorithm development.
We have also applied a fully automated pipeline to measure the radius of the ON and CSF sheath along the length of the nerve. We have demonstrated that this pipeline is sufficiently accurate to detect local population changes in both ON and CSF sheath size. The results are consistent with previous results that population variability should not vary across individuals based on age or sex [17]. Previously ON biomarkers have been investigated using manual or semi-automated methods which measured the ON at arbitrary points along the ON [4, 18, 19]. This proposed pipeline could be applied to disease populations to identify relevant disease biomarkers for disease onset. By measuring along the entire length of the nerve, we can identify meaningful areas of local changes which can lead to a more informed characterization of the ON by the clinical community.
All tools used and developed in this work are available in open source from their respective authors. The tools that implement the ON specific components of analysis are based on the Java Image Science Toolkit (JIST)[20] and Non-Local STAPLE [12]. The ON-CSF measurement code is primarily written in MATLAB (The MathWorks, Inc., Natick, Massachusetts, United States) and bundled into an automated program (i.e., “spider”[21]) that combines these tools using PyXNAT[22] for XNAT [23] and is available through the NITRC project MASIMATLAB (http://www.nitrc.org/projects/masimatlab).
Acknowledgments
Research reported in this publication was supported in part by the National Institutes of Health R21EY024036, R01EY023240 and 5T32EY007135. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This project was supported in part by ViSE/VICTR VR3029. The project described was supported by the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN.
References
- 1.O.N.S. Group. Multiple sclerosis risk after optic neuritis: final optic neuritis treatment trial follow-up. Archives of neurology. 2008;65(6):727. doi: 10.1001/archneur.65.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O.N.S. Group. Visual function 15 years after optic neuritis: a final follow-up report from the Optic Neuritis Treatment Trial. Ophthalmology. 2008;115(6):1079. doi: 10.1016/j.ophtha.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Rizzo JF, III, Andreoli CM, Rabinov JD. Use of magnetic resonance imaging to differentiate optic neuritis and nonarteritic anterior ischemic optic neuropathy. Ophthalmology. 2002;109(9):1679–1684. doi: 10.1016/s0161-6420(02)01148-x. [DOI] [PubMed] [Google Scholar]
- 4.Toosy AT, Mason DF, Miller DH. Optic neuritis. The Lancet Neurology. 2014;13(1):83–99. doi: 10.1016/S1474-4422(13)70259-X. [DOI] [PubMed] [Google Scholar]
- 5.Geeraerts T, Newcombe VF, Coles JP, et al. Use of T2-weighted magnetic resonance imaging of the optic nerve sheath to detect raised intracranial pressure. Critical Care. 2008;12(5):R114. doi: 10.1186/cc7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker G. Technical issues for the study of the optic nerve with MRI. Journal of the neurological sciences. 2000;172:S13–S16. doi: 10.1016/s0022-510x(99)00271-3. [DOI] [PubMed] [Google Scholar]
- 7.Hernando D, Karampinos D, King K, et al. Removal of olefinic fat chemical shift artifact in diffusion MRI. Magnetic Resonance in Medicine. 2011;65(3):692–701. doi: 10.1002/mrm.22670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hickman SJ. Optic nerve imaging in multiple sclerosis. Journal of Neuroimaging. 2007;17(s1):42S–45S. doi: 10.1111/j.1552-6569.2007.00136.x. [DOI] [PubMed] [Google Scholar]
- 9.Harrigan RL, Panda S, Asman AJ, et al. Robust optic nerve segmentation on clinically acquired computed tomography. Journal of Medical Imaging. 2014;1(3):034006–034006. doi: 10.1117/1.JMI.1.3.034006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avants BB, Tustison NJ, Song G, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asman AJ, Landman BA. Formulating Spatially Varying Performance in the Statistical Fusion Framework. Medical Imaging, IEEE Transactions on. 2012;31(6):1326–1336. doi: 10.1109/TMI.2012.2190992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asman AJ, Landman BA. Non-local statistical label fusion for multi-atlas segmentation. Med Image Anal. 2013;17(2):194–208. doi: 10.1016/j.media.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrigan RL, Plassard AJ, Bryan FW, et al. Disambiguating the Optic Nerve from the Surrounding Cerebrospinal Fluid Sheath in MS-related Atrophy. Magnetic Resonance in Medicine. 2014 doi: 10.1002/mrm.25613. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nocedal J, Wright SJ. [Conjugate gradient methods] Springer. 2006 [Google Scholar]
- 15.Breiman L, Friedman J, Stone CJ, et al. [Classification and regression trees] CRC press. 1984 [Google Scholar]
- 16.Cherry SR, Sorenson JA, Phelps ME. [Physics in nuclear medicine] Elsevier Health Sciences. 2012 [Google Scholar]
- 17.Harrigan RL, Plassard AJ, Mawn LA, et al. Constructing a statistical atlas of the radii of the optic nerve and cerebrospinal fluid sheath in young healthy adults. :941303, 941303–7. doi: 10.1117/12.2081887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hickman S, Miszkiel K, Plant G, et al. The optic nerve sheath on MRI in acute optic neuritis. Neuroradiology. 2005;47(1):51–55. doi: 10.1007/s00234-004-1308-x. [DOI] [PubMed] [Google Scholar]
- 19.Yiannakas MC, Wheeler-Kingshott CA, Berry AM, et al. A method for measuring the cross sectional area of the anterior portion of the optic nerve in vivo using a fast 3D MRI sequence. Journal of Magnetic Resonance Imaging. 2010;31(6):1486–1491. doi: 10.1002/jmri.22202. [DOI] [PubMed] [Google Scholar]
- 20.Lucas BC, Bogovic JA, Carass A, et al. The Java Image Science Toolkit (JIST) for rapid prototyping and publishing of neuroimaging software. Neuroinformatics. 2010;8(1):5–17. doi: 10.1007/s12021-009-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, Burns SS, Lauzon CB, et al. Integration of XNAT/PACS, DICOM, and Research Software for Automated Multi-modal Image Analysis. Proc SPIE. 2013:8674. doi: 10.1117/12.2007621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz Y, Barbot A, Thyreau B, et al. PyXNAT: XNAT in Python. Front Neuroinform. 2012;6:12. doi: 10.3389/fninf.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcus DS, Olsen TR, Ramaratnam M, et al. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]


