Abstract
Calcium-dependent protein kinases (CDPKs) are expanded in apicomplexan parasites, especially in Toxoplasma gondii where 14 separate genes encoding these enzymes are found. Although previous studies have shown that several CDPKs play a role in controlling invasion, egress, and cell division in T. gondii, the roles of most of these genes are unexplored. Here we developed a more efficient method for gene disruption using CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein 9) that was modified to completely delete large, multiexonic genes from the genome and to allow serial replacement by recycling of the selectable marker using Cre-loxP. Using this system, we generated a total of 24 mutants in type 1 and 2 genetic backgrounds to ascertain the functions of noncanonical CDPKs. Remarkably, although we were able to confirm the essentiality of CDPK1 and CDPK7, the majority of CDPKs had no discernible phenotype for growth in vitro or infection in the mouse model. The exception to this was CDPK6, loss of which leads to reduced plaquing, fitness defect in a competition assay, and reduced tissue cyst formation in chronically infected mice. Our findings highlight the utility of CRISPR/Cas9 for rapid serial gene deletion and also suggest that additional models are needed to reveal the functions of many genes in T. gondii.
INTRODUCTION
Protein kinases have expanded in eukaryotic genomes to regulate a variety of essential pathways in signaling and development (1, 2). Similar to other eukaryotic cells, apicomplexan parasites contain a diverse array of protein kinases that are important for regulating their complex biology. Apicomplexans contain many members of the major protein kinase families such as AGC (containing protein kinases A, G, and C), CMGC (containing cyclin-dependent, mitogen-activated, and cell division cycle [CDC]-like kinases), calmodulin kinases (CaMK), and casein kinases (CK) (3, 4). Curiously, apicomplexans lack some common kinases like protein kinase C and canonical tyrosine kinases and have reduced number of mitogen-activated protein kinases (3, 4). They also contain several parasite-specific kinases that are expanded, for example, the FIKK (phenylalanine [F]-isoleucine [I]-lysine [K]-lysine [K]) kinases in Plasmodium falciparum (5, 6) and the rhoptry kinases (ROP) implicated in Toxoplasma gondii virulence (7, 8). However, the most novel feature of apicomplexan kinomes is the expansion of plant-like, calcium-dependent serine/threonine (S/T) protein kinases (CDPKs) (9).
CDPKs are found in green algae, higher plants, ciliates, and oomycetes, as well as being in apicomplexan parasites (10). Although they are lacking in animal cells, CDPKs are similar in many ways to calcium-calmodulin-dependent kinases (CaMK). However, instead of the kinase domain being regulated by a separate calmodulin protein, CDPKs have a fused domain structure between an N-terminal serine threonine protein kinase and a C-terminal calcium activation domain (CAD) that consists of a calmodulin-like module linked to the kinase domain by a junctional domain (11, 12). A number of CDPKs have been crystallized, and analysis of their structures has led to an understanding of their unique activation mechanism (11, 12). In the absence of calcium, CDPKs are autoinhibited due to the fact that the CAD occludes the active face of the kinase domain. Peptides from the junctional domain are capable of inhibiting kinase activity of several CDPKs in vitro (13, 14), consistent with the finding that the junctional domain interacts with the kinase domain in the autoinhibited structure (11, 12). Upon binding to calcium, the CAD domain completely reorganizes and flips to the backside of the kinase domain, opening the nucleotide and substrate binding pockets to activate the enzyme (11, 12). CDPKs likely arose by fusion of a calmodulin-like domain containing four EF hands, a structural feature first described in parvalbumin and that is responsible for calcium binding, followed by their diversification in plants and protists (10). The fact that CDPKs are not found in animal cells, combined with the findings that some of them are essential, has made them attractive targets for development of inhibitors (15).
CDPKs in apicomplexans have been broadly classified as having a canonical domain structure consisting of four EF hands in the C-terminal CAD with a single N-terminal S/T kinase domain (9). However, a number of CDPKs contain additional features, including N-terminal extensions, a variable number of EF hands, and additional domains such as pleckstrin homology (PK) domains (9). The canonical CDPKs are among the best studied, and for many of them, their biological roles have been explored in Plasmodium or T. gondii. For example, T. gondii calcium-dependent protein kinase 1 (TgCDPK1) control of microneme secretion and regulated disruption of gene expression revealed a strong phenotype for motility, invasion, and egress (16). Notably, TgCDPK1 is highly unusual in having a small Gly gatekeeper, rendering it sensitive to a class of bulky ATP mimics called pyrazolopyrimidines (16, 17). This fortuitous finding allowed the use of chemical genetics to validate the function of TgCDPK1 (16, 17), to search for potential targets (18), and to expand the chemical series to find more potent inhibitors of parasite growth in vitro (19–21). The orthologue of TgCDPK1, named CDPK4 in Plasmodium berghei (the numbering scheme is different for historical reasons), is not essential during asexual growth in red blood cells but rather is important for controlling microgamete exflagellation (22), suggesting that CDPKs may not play analogous roles across diverse members of the phylum.
Perhaps not surprising given the expansion of the CDPK family, they can also play partially overlapping roles. For example, loss of TgCDPK3 also affects parasite egress, and under some conditions microneme secretion and motility (23). Recent studies indicate that TgCDPK3 acts upstream of TgCDPK1 by controlling calcium homeostasis, and consistent with this, TgCDPK1 overexpression can partially compensate for loss of CDPK3 (24). Although both TgCDPK1 and TgCDPK3 regulate egress in T. gondii, this function is provided by CDPK5 in P. falciparum (25). In both systems, protein kinase G (PKG) is required for egress (23, 25), and invasion (26, 27), demonstrating that cyclic nucleotide- and calcium-dependent kinase pathways are intertwined. Although functional studies have addressed the roles of many canonical CDPKs, much less is known about those with noncanonical domain structures. One exception is CDPK7, which has an unusual domain structure of several incomplete EF hands in an extended N-terminal domain that also has a PK domain (9). Regulated suppression of expression of TgCDPK7 affects cell division in T. gondii (28) and early stages in asexual development in P. falciparum (29), although the precise steps affected by this kinase have not yet been defined.
Herein we sought to identify the functions of eight noncanonical CDPKs in T. gondii using the highly efficient CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein 9) system for genetic disruption (30) or using newly described methods for improved gene deletion. We were easily able to delete all seven noncanonical CDPKs in both the type 1 and type 2 backgrounds and generated several double mutants and even triple mutants. The relatively modest phenotypes of these mutants suggest roles outside of the conventional in vitro culture and mouse models normally used to probe gene function in T. gondii. Nonetheless, the availability of these mutants will foster further studies on their roles in the more complex biology of T. gondii.
MATERIALS AND METHODS
Parasite and host cell cultivation.
The wild-type RH strain (referred to as RH88 in reference 31), previously described mutants that are defective in nonhomologous end joining RHΔku80 (32) and PruΔku80 (33), and genetically modified strains generated here were grown in confluent monolayers of human foreskin fibroblast (HFF) cells in D10 medium composed of Dulbecco's modified Eagle medium (DMEM), 10% fetal bovine serum (FBS) (HyClone) 2 mM glutamine (Sigma), 10 mM HEPES, pH 7.5 (Sigma), and 20 μg/ml gentamicin maintained at 37°C with 5% CO2. Parasites were allowed to egress naturally, harvested by filtration through 3.0-μm polycarbonate membranes, and resuspended in Hanks' balanced salt solution (HBSS) containing 10 mM HEPES and 0.1 mM EGTA (referred to as HHE) as described previously (34). All strains and host cell lines were determined to be mycoplasma negative using the e-Myco plus kit (Intron Biotechnology).
Domain prediction for CDPKs.
A gene ontology (GO) term search of ToxoDB using the terms “calcium binding” and “protein kinase activity” combined with a text word search for “calcium dependent protein kinase” identified 14 protein-coding genes, including previously named CDPKs (15) and two new proteins annotated as calcium-dependent protein kinase (TgME49_240390) and protein kinase domain-containing protein (TgME49_237860). Domain predictions were performed using Prosite (http://prosite.expasy.org/).
Single-gRNA strategy for disruption of CDPK6.
A single guide RNA (gRNA) against the first exon of CDPK6 was generated using Q5 DNA site-directed mutagenesis (New England BioLabs [NEB]) using specific primers (see Table S1 in the supplemental material). A LoxP-DHFR-mCherry (DHFR stands for dihydrofolate reductase) cassette was amplified using primers that contained short regions of homology to CDPK6 using specific primers (Table S1). The amplicon and the single gRNA plasmid were combined and transfected into tachyzoites of the RH strain. Following selection with pyrimethamine, single clones were screened using primers p1 and p2, and reverse transcription-PCR (RT-PCR) was performed using primers R1 and R2, R3 and R4 (Table S1).
Double-gRNA CRISPR/Cas9 system.
To generate guide RNAs (gRNAs) to disrupt specific CDPK genes, we modified the previously reported CRISPR Cas9-gRNA plasmid (pSAG1::Cas9-U6::sgUPRT plasmid; Addgene plasmid 54467) (30). The single gRNA in this plasmid was replaced by a short oligonucleotide matching each targeted gene (see Table S2 in the supplemental material) using Q5 DNA site-directed mutagenesis (NEB) with a common reverse primer (5′AACTTGACATCCCCATTTAC3′). CDPK-specific CRISPR-Cas9-gRNA plasmids (Table S2) were sequenced to confirm the correct sequence of the gRNA. To generate clean knockouts in T. gondii, a double-gRNA CRISPR/Cas9 system was designed in which the first gRNA sequence (gRNA1) was placed close to the start codon with the second gRNA (gRNA2) near the stop codon (Table S2) of the same gene. To express two gRNAs from a single plasmid, the gRNA2 expression cassette (U6 promoter-gRNA2-RNA scaffold-TTTT) was amplified using a common set of primers called 2nd gRNA F (F stands for forward) (5′CGAATTGGGTACCCAAGTAAGCAGAAGCACGCTG3′) and 2nd gRNA R (R stands for reverse) (5′TCGACCTCGAGAATTAACCCTCACTAAAGG3′) and cloned into the pCAS9-gRNA1 plasmid using restriction sites KpnI and XhoI. A similar strategy was used to generate a series of pCas9-gRNA1-2 plasmids that were specific for each CDPK gene (Table S2).
A plasmid named pLoxP-DHFR-mCherry containing a pyrimethamine resistance cassette (i.e., LoxP-DHFR 5′UTR-DHFR-mCherry-LoxP-DHFR 3′UTR) (Addgene plasmid 70147) was constructed using Gibson Assembly (New England BioLabs) to combine three PCR amplicons that were amplified using Q5 high-fidelity DNA polymerase (New England BioLabs) with primer set 1, 2, and 3 (see Table S3 in the supplemental material), and a LoxP-DHFR 3′ untranslated region (3′UTR) fragment (set 4) was further cloned to replace the surface antigen 1 (SAG1) 3′UTR. This plasmid was used as a PCR template to generate amplicons containing the DHFR resistance cassette flanked by short homology regions (homology region 1 [HR1] and HR2) to specific CDPK genes that were targeted for disruption by CRISPR/Cas9. HR1 was designed to match the sequence upstream of the Cas9-gRNA1 cleavage site, and HR2 was designed to match the sequence downstream of the Cas9-gRNA2 cleavage site, for each specific CDPK gene (Table S4).
Transfection and subcloning.
Amplicons containing a LoxP-DHFR-mCherry cassette flanked by short homology regions (HR1 and HR2) were purified and combined with their corresponding pCas9-gRNA1-2 plasmids for each CDPK gene. Freshly harvested parasites (3 × 106 to 5 × 106) were electroporated with ∼500 ng of the DHFR amplicon and 3 to 5 μg of a corresponding pCas9-gRNA 1-2 plasmid in 200 μl of Cytomix buffer as described previously (35). Following electroporation, parasites were inoculated onto HFF monolayers in 25-cm2 flasks, grown in D10 medium for 24 h at 5% CO2 and 37°C, followed by addition of 3 μM pyrimethamine. Resistant parasites appeared in 7 to 10 days and were subcloned on HFF monolayers in 96-well plates cultured in D10 medium at 5% CO2 and 37°C.
Single-clone screening and diagnostic PCR.
The genotype of CDPK knockouts was confirmed by diagnostic PCR using lysates of parasite clones grown in 96-well plates (see Table S5 in the supplemental material). For each gene, primer p1 is a forward primer located 300 to 500 bp upstream of a CRISPR Cas9-gRNA1 targeting site; primer p2 is a reverse primer located 100 to 300 bp downstream of a CRISPR Cas9-gRNA2 targeting site; primers p3 and p4 match the endogenous locus of each gene; primer p5 is a reverse primer matching the DHFR 5′UTR of the DHFR-mCherry cassette (5′CGGCCGACAGGACGCTACTG3′); primer p6 is a forward primer from the DHFR 3′UTR of the resistant cassette (5′CGCTCAAGGCTCGATTGTGG3′). Similar diagnostic PCRs were performed for each gene, consisting of the following. PCR1 tested the 5′ integration site using primers p1 and p5. PCR2 tested the 3′ integration site using primer p2 and p6.; PCR3 was performed with primers p3 and p4 to confirm the presence/absence of the endogenous gene. and PCR4 was used to test the insertion of the resistant cassette using primers p1 and p2.
RT-PCR.
First-strand cDNA was generated using a SuperScript III first-strand synthesis kit (Life Technologies). cDNA samples were tested for quality using primers that amplify a full-length cDNA of CDPK1 (F, 5′ATGGGGCAGCAGGAAAGCACTC3′; R, 5′GTTTCCGCAGAGCTTCAAGAGC3′). cDNAs from specific CDPK disruptants and knockouts were tested by RT-PCR using gene-specific primers designed to amplify a central region of each of the targeted genes (see Table S6 in the supplemental material).
Generation of double and triple knockouts.
In order to generate sequential knockouts, the LoxP-DHFR 5′UTR-DHFR-TS-mCherry-LoxP (TS stands for thymidylate synthase) cassette in a single- or double-gene knockout was excised by transfection of Cre recombinase (36). To remove the LoxP-DHFR-mCherry cassette, 50 μg of pmin-Cre-GFP (GFP stands for green fluorescent protein) was electroporated into the knockouts, and parasites were subcloned on day 2 or 3 on HFF monolayers in 96-well plates without drug selection. Clones were screened using PCR4 described above using primers p1 and p2 (see Table S5 in the supplemental material) for each of the targeted genes, in which the excised clones produced a smaller DNA band. Loss of the DHFR cassette in the excised clones was confirmed by sensitivity to 3 μM pyrimethamine. CDPK knockouts that were excised of the DHFR cassette were subsequently used for transfection to generate sequential gene knockouts.
Plaque assay.
Freshly harvested parasites were used to inoculate confluent HFF monolayers in six-well plates with 200 parasites per well for the RHΔku80 background and grown for 6 days in D10 medium at 37°C with 5% CO2, or 250 parasites per well for the PruΔku80 background and grown for 9 days in D10 medium at 37°C with 5% CO2. Two or three independent experiments were performed with three replicate wells each. Monolayers were fixed with ethanol for 5 min, stained with 0.5% crystal violet for 5 min, gently washed with water, and dried at room temperature. Plaque numbers were counted in each well.
Competition assay.
Freshly harvested parasites were seeded on HFF monolayers in a 25-cm2 flask (T25) at a ratio of 1:1 of the wild-type RHΔku80 strain to specific knockout strains and grown in D10 medium at 37°C with 5% CO2. Following replication and natural egress at ∼2-day intervals, parasites were subpassaged by 1:10 dilution to new 25-cm2 flasks containing fresh HFF monolayers. Naturally egressed parasites were also inoculated at ∼2-day intervals onto HFF monolayers grown on glass coverslips in a 24-well plate and cultured in D10 medium at 37°C with 5% CO2. After overnight growth, coverslips were fixed with 4% formalin in phosphate-buffered saline (PBS), permeabilized with 0.05% saponin in PBS containing 10% goat serum, and used for immunofluorescence (IF) staining. Parasites were stained with primary antibodies against mCherry (rat monoclonal antibody [MAb]; Life Technologies) and rabbit anti-SAG1 and then stained with secondary antibodies anti-rat IgG Alexa Fluor 594 (anti-rat IgG labeled with Alexa Fluor 594) and anti-rabbit IgG Alexa Fluor 488 (Life Technologies). Knockout parasites were recognized by staining for mCherry (red), while total parasites were detected with SAG1 (green). Samples were repeated in triplicate, and >250 parasite vacuoles were counted on each slide. The ratio of knockout parasites in the population was calculated at each time point and graphed in Prism. Experiments were repeated three independent times.
Replication assay.
Freshly harvested parasites were seeded on HFF monolayers grown on glass coverslips in 24-well plates and cultured in D10 medium at 37°C with 5% CO2 for 24 h. Monolayers were fixed with 4% formalin in PBS, permeabilized with 0.05% saponin in PBS containing 10% goat serum, and used for IF staining. Monolayers were stained with primary antibodies against mouse anti-IMC1 MAb (1:5,000) or rabbit anti-SAG1 (1:10 000) and followed up with secondary antibodies anti-mouse IgG Alexa Fluor 488 or anti-rabbit IgG Alexa Fluor 488. The number of parasites per vacuole was determined by microscopic examination of >250 parasite-containing vacuoles on each of three separate coverslips, and the percentages of vacuoles with different numbers of parasites were calculated and graphed in Prism. Experiments were repeated three times.
Acute mouse infection with type 1 strain parasites.
Animal studies were conducted at the Washington University School of Medicine and were approved by the Institutional Animal Care and Use Committee in accordance with the Guide for Care and Use of Laboratory Animals (37). Freshly harvested tachyzoites of the wild-type RHΔku80 strain or mutants with this background were tested for acute virulence in the mouse model, as described previously (38). For experiments using the highly virulent type 1 strain background, we used outbred CD1 mice, as they normally succumb to wild-type infection, but provide a sensitive readout for attenuation of virulence. Eight- to ten-week-old female CD1 mice (Jackson Laboratories) were injected intraperitoneally (i.p.) with 100 parasites in a volume of 200 μl of HHE (100 parasites per mouse). Strains were tested independently in groups of five mice, and survival was monitored daily.
Chronic mouse infections with type 2 strain parasites.
Parasites of the PruΔku80 type 2 strain and parasites with gene knockouts in this background were tested for the ability to induce chronic infection in the mouse. In the case of type 2 strains, which show intermediate virulence, the use of more-susceptible inbred mice allows for higher cyst counts, thus potentially revealing differences in chronic infection that might be masked in outbred animals. Freshly harvested tachyzoites grown in culture were inoculated i.p. in a volume of 200 μl of HHE containing 1,000 parasites per animal in 8- to 10-week-old C57BL/6 mice (Jackson Laboratories). Mice were monitored for survival daily, and surviving animals were tested serologically by enzyme-linked immunosorbent assay (ELISA) to establish that they were infected, as described previously (34). Cyst counting was performed as described previously (39). Briefly, at 30 days postinoculation, brains were harvested, and lysed by passing through 16-gauge needles in PBS. The brain homogenate was permeabilized in 0.25% Triton X-100 and blocked with 10% bovine serum albumin (BSA) in PBS for 10 min. The homogenate was centrifuged at 400 × g, resuspended in PBS containing 10% BSA and fluorescein isothiocyanate (FITC)-conjugated Dolichos biflorus lectin (DBL) (1:100 dilution) (Sigma) for 30 min at room temperature. The stained brain homogenate was washed twice with PBS containing 10% BSA and resuspended in PBS. Aliquots of 12.5 μl of the resuspension were examined microscopically using a Zeiss Axioskop microscope, equipped for epifluorescence. The counts from eight coverslips were averaged and used to obtain the total number of cysts in a mouse brain based on adjustment for the volume examined relative to the total brain homogenate.
In vitro differentiation.
Differentiation into bradyzoites was tested using culture in RPMI 1640 medium (pH 8.1) in the absence of CO2 at 37°C as previously described (40). Parasites cultured in D10 medium at 37°C with 5% CO2 for 2 days were processed in parallel as controls for induction. After 2 days of culture, monolayers were fixed with 4% formalin in PBS buffer and permeabilized with 0.5% Triton X-100 in PBS containing 10% goat serum. Monolayers were stained with antibodies against dense granule antigen 7 (GRA7) (rabbit MAb, 1:1,000) (41) followed up with secondary antibodies anti-rabbit Alexa Fluor 594 (1:2,500), and FITC-conjugated DBL (1:1,000). Parasite vacuoles stained fully or partially with DBL were scored as bradyzoite positive, in comparison to the total number of vacuole detected by GRA7 staining. Three independent experiments were performed with triplicate samples for each, and >250 parasite vacuoles were counted on each slide.
Statistics.
Statistical analyses were performed in Prism (GraphPad) using a nonparametric Kruskal-Wallis test with Dunn's multiple comparisons. Linear regression analysis for the competition assay was performed in Prism with a comparison to a theoretical slope of 0. A P value of ≤0.05 was considered significant.
RESULTS
Sequence and expression analyses of noncanonical CDPKs in T. gondii.
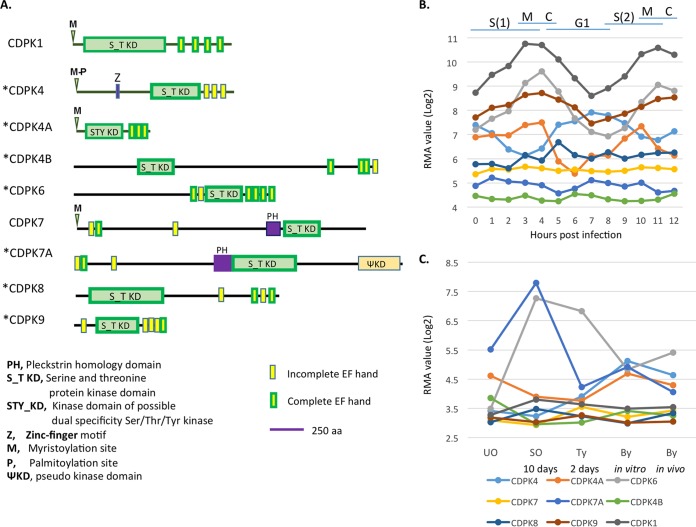
Previous studies have identified 12 CDPKs in T. gondii, many of which have orthologues in other apicomplexans (9). We searched ToxoDB.org using “calcium binding” and “protein kinase activity” as queries for the putative functions and identified two new genes (TgME49_240390 and TgME49_237860) that were candidate CDPKs with a kinase domain and EF hands. Phylogenetic analysis based on the kinase domains suggested that they are close to CDPK4 and CDPK7, and they were named CDPK4B (TgME49_240390) and CDPK7A (TgME49_237860), respectively (see Fig. S1 in the supplemental material), as previously discussed (15). Analyses of domain structures of all CDPKs in T. gondii were performed using an online software PROSITE (http://prosite.expasy.org/), which suggested that these CDPKs can be classed into two groups based on their domain structure. Canonical CDPKs have a serine/threonine (S/T) protein kinase domain immediately followed by three or four EF hands (CDPK1, CDPK2, CDPK2a, CDPK2b, CDPK3, and CDPK5), while noncanonical CDPKs have different arrangements of the kinase domain and EF hands and may also have other domains, such as zinc finger or pleckstrin homology (PH) domains (Fig. 1A). For example, CDPK7 and CDPK7a both contain a PH domain, while CDPK7a also contains a downstream pseudokinase domain. A number of the noncanonical CDPKs also contain EF hands that are N terminal to the kinase domain; these noncanonical CDPKs include CDPK6, CDPK7, CDPK7a, and CDPK9. Finally, all of the noncanonical CDPKs contain one or more incomplete EF hands.
FIG 1.
(A) Domain structures of noncanonical CDPKs in T. gondii. In the diagram of each CDPK, conserved domains are indicated by boxes. Abbreviations and symbols are defined below the domain structures. CDPKs with asterisks were targeted in this study. The domain predictions were performed using the online software Prosite (http://prosite.expasy.org/). aa, amino acids. (B) Gene expression data for CDPK genes in the type 1 RH strain were obtained from http://ToxoDB.org and plotted during two complete cell cycles. S, synthesis, M, mitosis, C, cytokinesis, G1, G1 phase of cell cycle; RMA, robust multiarray average algorithm. (C) Gene expression data from samples of oocysts, tachyzoites, and bradyzoites in the T. gondii type 2 strain M4 were obtained from htpp://ToxoDB.org and plotted for each life cycle stage. The different points indicate unsporulated oocysts (UO), sporulated oocysts on day 10 (SO), tachyzoites in vitro on day 2 (Ty), bradyzoites in vitro on day 8 (By in vitro), and bradyzoites in vivo on day 21 (By in vivo). Colored lines correspond to each CDPK gene in panels B and C.
To examine the expression of CDPK genes during intracellular development, we analyzed previously collected microarray data collected from a synchronized time course of intracellular development (42). Expression data for CDPK genes were obtained from ToxoDB and plotted over the course of two complete cell cycles (Fig. 1B). Comparison of these profiles indicated that several of the noncanonical CDPKs (i.e., CDPK4, CDPK4A, CDPK6, and CDPK9) varied their expression levels during the cell cycle, and most of these showed a pattern similar to that of CDPK1 (Fig. 1B). In contrast, the remaining genes (i.e., CDPK4B, CDPK7, CDPK7A, and CDPK8) were constitutively expressed and also showed overall lower expression levels (Fig. 1B). We also examined the expression levels of CDPK genes during different developmental life stages based on previous transcriptomic data (43). Expression data for CDPK genes were obtained from ToxoDB and plotted across different life cycle stages of development (Fig. 1C). Several of the genes, including CDPK4, CDPK4A, CDPK6, and CDPK7A, were differentially regulated during parasite development, while the remaining genes (i.e., CDPK4B, CDPK7, CDPK8, and CDPK9) were constitutively expressed (Fig. 1C).
Generation of single CDPK knockouts in the type 1 RHΔku80 strain.
We initially used CRISPR/Cas9 and a single gRNA placed downstream of the ATG to target the eight noncanonical CDPKs in the RH strain (Fig. 1A). Based on PCR screening of gDNA, disruptants were obtained for all of the noncanonical CDPKs with the exception of CDPK1 and CDPK7 (data not shown). However, mRNA from downstream of the targeted sites was still detected by RT-PCR in the disruptants for each of the genes. One example of this is illustrated for the single gRNA CDPK6 disruptant (see Fig. S2 in the supplemental material). These residual transcripts could be due to alterative splicing, internal sites for transcription initiation, or read through the DHFR-mCherry cassette. Rather than decipher among these potential alternatives, we redesigned the CRISPR strategy to delete the majority of the coding sequences (CDS) for each of the noncanonical CDPKs.
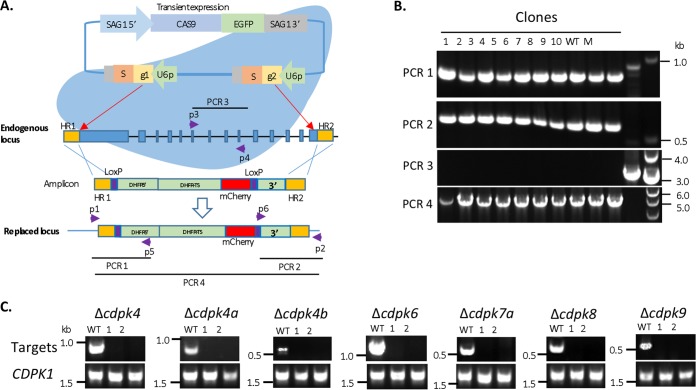
Because double-strand DNA breaks (DSBs) generated by CRISPR/Cas9 are efficiently repaired by the homology-directed DNA repair (HDR) mechanism in the RHΔku80 background (44), we took advantage of this information to develop a system to generate clean knockouts. We developed a system in which two CRISPR gRNAs (gRNA1 and gRNA2) were expressed from a single CRISPR/Cas9 plasmid that generates two DSBs separately located at the 5′ and 3′ regions of the CDS. This plasmid was cotransfected with a resistance marker containing a LoxP-DHFR 5′UTR-DHFR-mCherry-LoxP-DHFR 3′UTR cassette flanked by short homology regions (HR1 and HR2, each of 42 bp) that were designed to repair the DSBs (Fig. 2A). We tested the efficiency of this system for deletion of cdpk6. A pyrimethamine-resistant (Pyrr) pool was generated in about 8 days, and diagnostic PCR indicated the presence of both flanking regions for the correct insertion of the DHFR cassette into the endogenous locus. In contrast, transfections performed with the CRISPR/Cas9 gRNA1-2 plasmid and a resistance cassette without short homology regions did not produce a Pyrr pool. Although transfections using only the resistance cassette containing the short homology regions (HR1 and HR2) produced a Pyrr pool after 2 weeks, they remained negative by diagnostic PCRs designed to detect the knockout (Fig. 2A and data not shown). The Pyrr pool generated from transfection of the double-CRISPR/Cas9 plasmid plus the resistance cassette with flanking HRs was cloned by limiting dilutions, and single-cell clones were screened by diagnostic PCRs (Fig. 2A). Consistent with the high targeting efficiency in Δku80 strains, all clones screened were positive (Fig. 2B). The trial with the Δcdpk6 generation clearly suggested that the strategy using double-CRISPR gRNAs and the resistance cassette flanked by short homology regions was highly efficient in generating gene knockouts.
FIG 2.
Generation of knockout strains for noncanonical CDPKs in T. gondii using a double CRISPR gRNA system. (A) Schematic of the double CRISPR gRNA system used to delete a noncanonical CDPK gene. The strategy uses two short homology regions (HRs) flanking a LoxP-DHFR-mCherry cassette. Abbreviations: EGFP, enhanced green fluorescent protein; S, the scaffold for gRNA; g1, gRNA1; g2, gRNA2; U6p, T. gondii U6 promoter; HR1 and HR2, short homology regions 1 and 2 (42 bp). The red box in the amplicon is mCherry fused with DHFR-TS. The purple boxes are loxP sites. PCR1 and PCR2 detected the correct insertion sites using the primer pairs p1-p5 and p2-p6, respectively. PCR3 detected the endogenous locus with primers p3 and p4. PCR4 detected the correct insertion of the DHFR cassette using primers p1 and p2. (B) PCR screening for Δcdpk6 knockouts demonstrated the efficiency of the process. Single clones were screened with the PCRs shown in panel A. M, molecular size markers. (C) RT-PCR screening for CDPK gene knockouts in the RHΔku80 background. Two clones for each knockout strain were tested by RT-PCR. The CDPK1 gene served as a positive control for cDNA template quality. The wild-type parental strain RHΔku80 (WT) is shown for comparison.
The double-gRNA strategy was then applied to all noncanonical CDPKs in this study. With the exception of CDPK7, we were able to obtain Pyrr pools for each of the targeted CDPKs. The Pyrr pools and single clones for the noncanonical CDPKs knockouts were efficiently generated and tested by the diagnostic PCRs as illustrated in Fig. 2A. Several independent single clones for each strain were picked up for screening, and the majority were confirmed to be knockouts (see Fig. S3A in the supplemental material). Total RNAs from two clones of each strain were extracted and used for the first cDNA strand synthesis and RT-PCR analyses. As expected, mRNAs of the targeted genes in the knockouts were not detected (Fig. 2C). These studies confirm that it is possible to delete the CDS for all of the noncanonical CDPKs, with the exception of CDPK7, which has previously been reported to be essential (28).
Phenotypic analyses for single CDPK knockouts in the type 1 RHΔku80 strain.
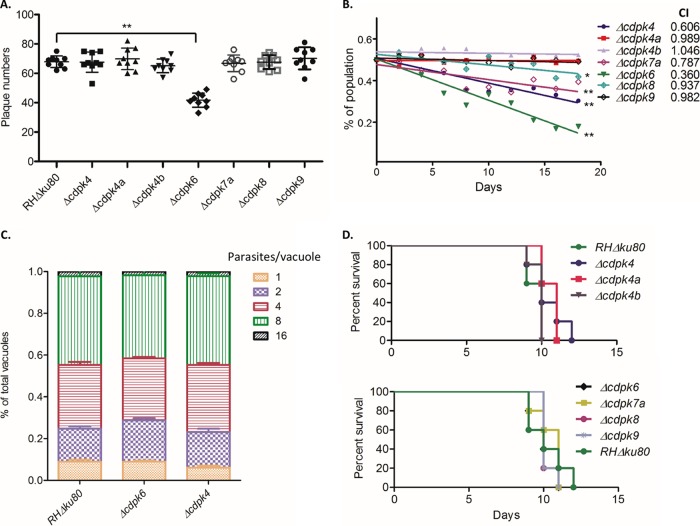
To test the phenotype during the lytic cycle, CDPK knockouts were compared for plaque formation on HFF monolayers, an assay that captures invasion, replication, egress, and spread. All of the knockouts produced plaques with equal efficiency except for the Δcdpk6 strain, which had a mild but significant defect (Fig. 3A). The fitness of the knockouts was further tested by a competition assay in which they were cocultured with the background strain RHΔku80. The competition assay was initiated with equal number of wild-type and knockout parasites and the parasites were passaged for 18 days, during which time the proportion of knockouts was determined at 2-day intervals. Under the null hypothesis, it is expected that the knockout and the wild type would grow the same and the competition index (CI), defined by the proportion of mutant versus wild-type parasites at day 18 divided by proportion of at day 1 (the initial ratio was 1:1 of mutant to wild type) (45), is expected to equal 1. Most of the knockout strains did not have obvious growth defects, although Δcdpk6, Δcdpk4, and Δcdpk7a strains showed mild fitness defects that were significant (i.e., the slope was significantly different from 0 and the CI is lower than 1) (Fig. 3B). However, the growth defects of Δcdpk6 and Δcdpk4 strains were not attributed to differences in parasite replication in HFF monolayers (Fig. 3C), consistent with the fact there was no obvious difference in the size of plaques formed by the knockouts (data not shown). We also tested the survival of mice challenged with wild-type and knockout strains. Mice infected with the wild type or the various CDPK knockouts all succumbed to challenge and died with very similar kinetics, indicating there is no overt in vivo defect (Fig. 3D).
FIG 3.
Phenotypic analyses of CDPK gene knockouts in the RHΔku80 strain. (A) Plaque formation performed in six-well plates. Data for the mutants were compared to the wild-type RHΔku80 strain using a Kruskal-Wallis test with Dunn's multiple comparisons (P ≤ 0.001). The value for the Δcdpk6 mutant was significantly different (P ≤ 0.001) from the value for the wild-type RHΔku80 strain as indicated by the bar and two asterisks. The data are from a combination of three independent experiments with three wells each. Each symbol represents the value for one well. Bars indicate means ± standard deviations. (B) Competition assay for the knockouts based on coculture with the wild-type RHΔku80 strain. The ratio of parasites was tested every 2 days (x axis). The number of knockout vacuoles (mCherry positive) divided by the total number of vacuoles (SAG1 staining) × 100 = percentage of the population (y axis). The competition index (CI) (to the right of the knockout strain symbols) is defined by the proportion of mutant versus wild-type parasites on day 18 divided by the proportion present on day 1 (the initial ratio was 1:1 of mutant and wild type) (45). The average values for three independent experiments are shown plotted as a linear regression. The slope of the line was tested under the null hypothesis that for mutants that showed no growth defect versus the wild type, the plot of the line should have a slope of zero; for several mutants, this hypothesis was rejected as shown by the significance levels (**, P ≤ 0.0001; *, P ≤ 0.001). (C) Replication assay for Δcdpk6 and Δcdpk4 strains compared to the wild-type RHΔku80 strain. Parasites grown for 24 h were stained with antibodies against IMC1 and SAG1, followed by fluorescently conjugated secondary antibodies. The proportion of vacuoles with different numbers of parasites was calculated based on microscopic examination (y axis). The data are from three independent experiments, each with three internal replicates. (D) Survival of mice inoculated with the wild-type RHΔku80 strain or CDPK knockouts. CD1 mice were inoculated i.p. with 100 parasites. There were five mice in each group.
Generation and phenotypic testing of CDPK knockouts in the type 2 PruΔku80 strain.
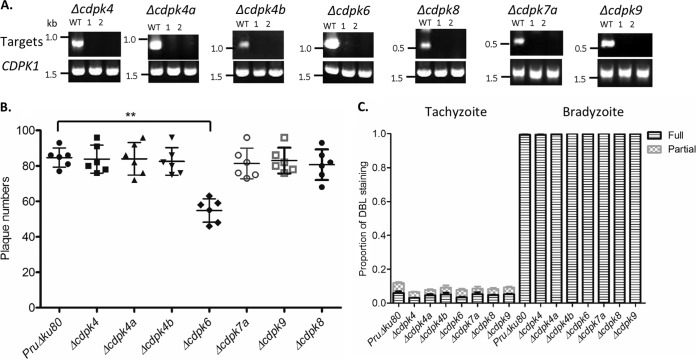
Based on transcriptomic and microarray data found in ToxoDB, it is likely that the noncanonical CDPKs are expressed at similar levels in tachyzoites and bradyzoites during development. However, given the absence of phenotypes in the tachyzoite stage, we wondered whether these noncanonical CDPKs might have roles in the bradyzoite stage. As the RHΔ80ku strain used here does not efficiently differentiate to other life cycle stages, we undertook the generation of knockouts for all of the nonessential CDPK knockouts in the type 2 PruΔku80 strain (33) using a similar double-CRISPR/Cas9 strategy. The same plasmids and PCR amplicons were transfected into the PruΔku80 strain. Knockout clones were isolated and tested by diagnostic PCR to confirm the loss of the gene and insertion of the resistance cassette (see Fig. S3B in the supplemental material) and loss of expression by RT-PCR (Fig. 4A). Plaque assays performed for the various CDPK knockouts in the PruΔku80 strain showed results similar to those of the knockouts in the RHΔku80 strain with only the Δcdpk6 strain showing a mild yet significant defect in growth (Fig. 4B). When assayed using an in vitro differentiation protocol based on induction by culture at pH 8.1, all of the knockouts efficiently switched to bradyzoites, as revealed by induction of cyst wall staining detected with Dolichos bifloris lectin staining (Fig. 4C).
FIG 4.
Generation of CDPK knockouts in the type 2 strain PruΔku80 strain. (A) Confirmation of gene knockouts using RT-PCR for specific genes. Two clones for each knockout strain and the wild-type PruΔku80 strain (WT) are shown. The CDPK1 gene served as a positive control for cDNA template quality. (B) Plaque formation performed in six-well plates. Data for the mutants were compared to the RHΔku80 strain using a Kruskal-Wallis test with Dunn's multiple comparisons. The value for the Δcdpk6 mutant was significantly different (P ≤ 0.01) from the value for the wild-type RHΔku80 strain as indicated by the bar and two asterisks. The data are from two independent experiments with three replicates each. (C) In vitro differentiation into bradyzoites based on staining with FITC-conjugated Dolichos biflorus lectin (DBL). Parasites were induced at pH 8.1 for 2 days (Bradyzoite) or cultured under standard D10 medium conditions (Tachyzoite). Samples were visualized by staining with GRA7 antibodies and fluorescent secondary antibodies in combination with FITC-labeled DBL. Vacuoles were scored as partially or fully DBL positive. The data are from two independent experiments, each with three internal replicates.
Generation of multiple CDPK knockouts.
Previous studies with other CDPKs have shown that in some cases they can compensate for each other or that they work in common pathways (23, 24). Hence, we considered that the lack of phenotypes for noncanonical CDPKs might reflect redundant functions such that single-gene knockouts remain normal. Therefore, we set out to generate multiple gene knockouts both in the RHΔku80 and PruΔku80 backgrounds. We chose pairwise double knockouts based on phylogenetic similarity, although many of the CDPKs lie on long branch lengths and hence are quite dissimilar from each other (15).
Because the DHFR resistance cassette is flanked by loxP sites, we excised this marker from the single knockouts by introducing Cre recombinase, as described previously (36). In the RHΔku80 background, we excised the DHFR cassette in the Δcdpk4a::DHFR-mCherry and Δcdpk6::DHFR-mCherry mutants by transfection of a plasmid pCre-GFP, obtaining ∼20% of clones that lacked the resistance cassette, as confirmed by PCR (data not shown). The cleaned-up Δcdpk6 mutant was then transfected with the CRISPR/Cas9 gRNA1-2/CDPK4 plasmid and the DHFR-mCherry cassette flanking short homology regions from CDPK4 to generate the double deletion Δcdpk6 Δcdpk4::DHFR-mCherry as confirmed by diagnostic PCRs and RT-PCRs (see Fig. S4A in the supplemental material). The cleaned-up knockout of Δcdpk4a was separately transfected with the CRISPR/Cas9 gRNA1-2 plasmids targeting CDPK4, CDPK6, and CDPK4b, together with the DHFR-mCherry cassette and corresponding gene-specific short homology regions. The double knockouts of Δcdpk4a Δcdpk4::DHFR-mCherry, Δcdpk4a Δcdpk6::DHFR-mCherry, and Δcdpk4a Δcdpk4b::DHFR-mCherry were efficiently generated as confirmed by diagnostic PCRs and RT-PCRs (Fig. S4A). Furthermore, two of the double knockouts (Δcdpk4a Δcdpk6::DHFR-mCherry and Δcdpk4a Δcdpk4b::DHFR-mCherry) were transfected with Cre to remove the DHFR selection cassette and then transfected with the CRISPR/Cas9 gRNA1-2/CDPK4 plasmid and the DHFR-mCherry cassette with short homology regions from CDPK4 to generate triple-gene knockouts as confirmed by RT-PCR (Fig. S4A). Four double-gene knockouts in the PruΔku80 strain were generated in the same way, including Δcdpk4a Δcdpk4::DHFR-mCherry, Δcdpk4a Δcdpk6::DHFR-mCherry, Δcdpk4a Δcdpk4b::DHFR-mCherry, and Δcdpk8 Δcdpk9::DHFR-mCherry, as confirmed by RT-PCR (Fig. S4B).
Phenotypic analysis of multiple CDPK knockouts.
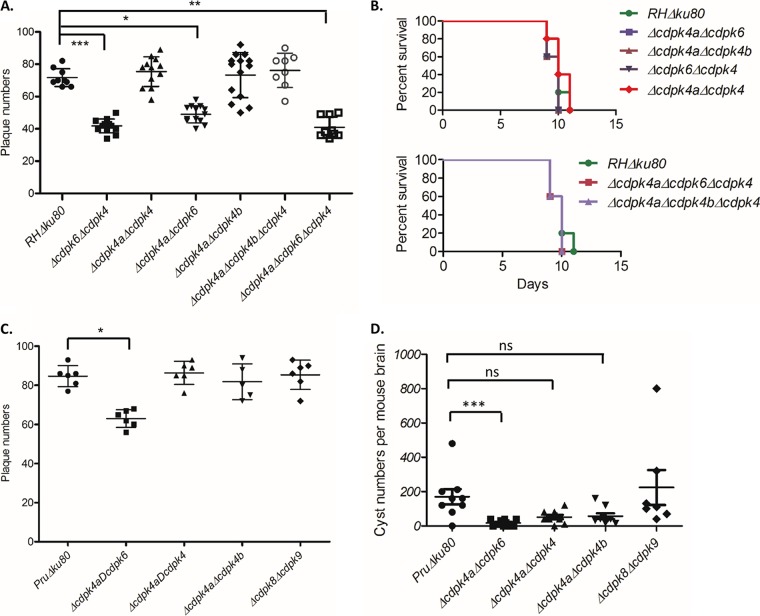
The growth of four double and two triple knockouts in the RHΔku80 strain was assayed by plaque formation on HFF monolayers as described above. All of the knockouts containing Δcdpk6 had mild yet significant reductions in plaque formation, similar to the single Δcdpk6 mutant (Fig. 5A). The double- and triple-gene knockouts in the RHΔku80 strain were also examined for acute virulence in CD1 mice. All of the mice inoculated with the wild type or multiple CDPK knockouts succumbed to infection, with approximately equal time to death (Fig. 5B). The growth of four double-gene knockouts in the PruΔku80 strain was also tested using plaque formation on HFF monolayers. We obtained similar plaque results with only the Δcdpk6 knockout showing a mild, yet significant defect (Fig. 5C). Four of the double-gene knockouts in the PruΔku80 strain were used to infect C57BL/6 mice by i.p. injection. Interestingly, we found that the double-gene knockout Δcdpk4a Δcdpk6::DHFR-mCherry formed significantly fewer cysts in mouse brains (Fig. 5D). Although the cyst burden was also lower with the two other double-gene knockouts Δcdpk4a Δcdpk4::DHFR-mCherry and Δcdpk4a Δcdpk4b::DHFR-mCherry, this difference was not significant (Fig. 5D).
FIG 5.
Phenotype analyses of multiple gene knockouts of noncanonical CDPKs in the RHΔku80 and PruΔku80 strains. (A) Plaque formation performed in six-well plates. Data for the mutants were compared to the RHΔku80 strain using a Kruskal-Wallis test with Dunn's multiple comparisons. Values that are significantly different from the value for the RHΔku80 strain are indicated by bars and asterisks as follows: *, P ≤ 0.01; **, P ≤ 0.001; ***, P ≤ 0.0001. The data are from three independent experiments with three wells each. (B) Survival of mice inoculated with the wild-type RHΔku80 strain or double or triple CDPK knockouts. CD1 mice were inoculated i.p. with 100 parasites. There were five mice in each group. (C) Plaque formation performed in six-well plates. Data for the mutants were compared to the PruΔku80 strain using a Kruskal-Wallis test with Dunn's multiple comparisons. The value that is significantly different (P ≤ 0.01) from the value for the RHΔku80 strain is indicated by a bar and asterisk. Data are from two independent experiments with three replicates each. (D) Cyst formation of double knockouts in the PruΔku80 strain tested on C57BL/6 mice. Each mouse was i.p. infected with 1,000 parasites (five mice in each group). Data from two independent experiments were combined and tested by a Kruskal-Wallis test with Dunn's multiple comparisons. The value that was significantly different (P < 0.0005) from the value for the PruΔku80 strain is indicated by the bar and three asterisks. Values that were not significantly different (ns) from the value for the PruΔku80 strain are also indicated.
DISCUSSION
The CDPK family is amplified and diversified in T. gondii, which contains more members of this group of kinases than either Plasmodium or Cryptosporidium (9). Although previous studies have revealed that TgCDPK1 and TgCDPK3 are important in controlling egress and invasion and CDPK2 is essential for bradyzoite development (46) in T. gondii, little is known about the noncanonical CDPKs in T. gondii. Here we disrupted seven noncanonical CDPK genes in both the type 1 and type 2 strains of T. gondii. Remarkably, none of these genes was essential for growth and only Δcdpk6 mutants had a mild plaquing defect, which resulted in decreased fitness and lower chronic tissue cyst burden. To interrogate the possibility of redundancy, we generated a rapid system for generating serial mutants using CRISPR to recycle selection cassettes. The lack of an overt phenotype for most mutants was not simply due to redundancy as pairwise double and triple mutants also failed to reveal additional phenotypes over that of the single Δcdpk6 mutant. Thus, while our findings provide a more efficient process for generating serial gene deletions in T. gondii, they also highlight the limitations of existing models for studying the roles of individual genes in the biology of infection.
The initial development of CRISPR/Cas9 technology in T. gondii relied on gene disruptions caused by nicking of the DNA with a single gRNA and insertion/deletion of a few base pairs or insertion of a selectable marker to disrupt the gene coding function (30, 44). However, for multiple exonic genes where the gene models are often in question, this strategy may not lead to loss of function. For example, in the present study, when we attempted to disrupt CDPK genes with a single gRNA, the presence of residual mRNAs confounded the interpretation of these disruptant mutants. When employed in a wild-type background, the single-gRNA strategy might also result in nonhomologous insertion of the drug resistance cassette, further complicating the analysis of mutants. As such, we modified the original CRISPR/Cas9 method to incorporate two gRNAs that are placed near the 5′ and 3′ ends of the CDS, thus removing the entire coding sequence and replacing it with a selectable marker. This process is extremely efficient and when combined with loxP sites flanking the selectable marker, it can easily be used to recycle the marker for subsequent rounds of selection. This recycling strategy also potentially alleviates the concern of using mutant forms of DHFR for selection, which impart resistance to the clinically important drug pyrimethamine: after generation of the mutant, the mutant DHFR gene can be completely removed from the genome. Alternatively, other markers such as HXGPRT can be used for both positive and negative selection, and this strategy has the advantage of not employing pyrimethamine selection. Collectively, we have developed 24 independent mutants in T. gondii using an improved method for gene deletion by CRISPR. Among the set of mutants were seven mutants with individual gene deletions, four double mutants, and two triple mutants in the RHΔku80 background and seven mutants with individual gene deletions and four double mutants in the PruΔku80 background. The use of CRISPR/Cas9 for serial gene deletions is thus extremely efficient for interrogating the members of multigene families including screening for redundant functions.
Despite efficiently generating 24 separate mutants, we were able to discern only a mild phenotype for one of the genes studied here: Δcdpk6 mutants showed a decrease in the number of PFU in both the type 1 RHΔku80 and type 2 PruΔku80 backgrounds. While our manuscript was being reviewed, another study describing disruption of the noncanonical CDPKs using a single-gRNA strategy was published (47). This study also failed to find a growth defect in the disruptants; however, they also did not report a defect for disruption of CDPK6. This difference from our findings may be due to the limitations described above for the use of single-gRNA disruptants or other technical differences in how the phenotypes were tested. The decrease in plaque number observed with our Δcdpk6 mutant was not the result of a replication defect or of a defect in entry or egress. Rather, it is likely that loss of CDPK6 results in decreased survival when the parasites are extracellular, as is the case when they are inoculated in the plaque assay. In the type 1 background, this did not result in a decrease in virulence as would be expected, since infection with a single viable organism of type 1 strains causes lethal infection (48). In contrast, the lack of CDPK6 in the type 2 background results in a significant decrease in the number of tissue cysts in the brain. This defect is likely due to a decrease in expansion and dissemination rather than an intrinsic defect in differentiation, as all mutants showed the same capacity to differentiate into bradyzoites in vitro, indicating that bradyzoite genes are upregulated normally. Finally, although we did not complement the defect in the Δcdpk6 mutants by restoring expression of the protein, the fact that we observed this same phenotype for four separately derived mutants in the type 1 and type 2 strains supports the idea that the decreased fitness of this mutant is due to loss of CDPK6.
Previous studies on the roles of CDPKs in the biology of T. gondii have focused primarily on TgCDPK1 and TgCDPK3, which are important in controlling invasion and egress. TgCDPK1 regulates microneme secretion and is essential for this process as shown using molecular and chemical genetic approaches (16, 17). In contrast, while TgCDPK3 can be disrupted, it contributes to the control of microneme secretion and plays an important role in response to stimulated egress (23, 49, 50). Consistent with this, TgCDPK3 mutants have reduced virulence in vivo (51). Among the other CDPKs, only TgCDPK7 has been reported to be essential, and genetic knockdown studies indicate that it plays a role in cell division (28). Our attempts to disrupt these genes using CRISPR/Cas9 confirmed that they are essential for in vitro growth. Remarkably, based on the current findings, the remaining noncanonical CDPKs appear to be nonessential for growth in vitro or infection in the mouse model. This raises the intriguing question of why this gene family is amplified and diversified when it appears to play a nonessential role in the biology. In particular, it is unclear why the number and placement of EF hands varies so much among the noncanonical members. For example, TgCDPK4a and -4b contain only three intact EF hands, while TgCPK4 has three incomplete EF hands and none that are complete (Fig. 1). In Arabidopsis, there are also some CDPKs containing degenerated EF hands with unknown functions (52). However, there are no reports on CDPKs in plants containing N-terminal EF hands or a PH domain. Given the importance of the EF hand domains in regulating the activity of canonical CDPKs (11, 12), it is unclear why they have been modified so extensively in other members. Two possibilities might account for this pattern. First, it is possible that CDPKs are gradually accumulating mutations and becoming degenerate due to the fact that many of them are no longer essential. This might be inferred from the domain architecture of TgCDPK7a, which has a duplicated pseudokinase domain downstream of the main kinase domain and only a single intact EF hand located at the N terminus (Fig. 1). However, even if such degenerate CDPKs lack regulation, they might still serve important roles in substrate binding, scaffolding, etc., as is seen in the case of ROP5, a pseudokinase that lacks catalytic activity and yet is critical for virulence in T. gondii (53, 54). An alternative possibility is that CDPKs play roles in other aspects of biology not tested here, for example in other hosts or during other life cycle stages. CDPKs are expressed widely throughout the life cycle, so it remains possible that they are important during the sexual phase that occurs in the cat intestine or during oocyst development. For example, CDPK4A is slightly elevated in unsporulated oocysts, while CDPK6 and CDPK7A are elevated in sporulated oocysts, which suggests that they may play roles during sporozoite formation or transmission via oocysts. Unfortunately, we cannot test this possibility in the genetic backgrounds used here due to loss of the development of the sexual stages in these lines.
In summary, we have developed an efficient system for serial gene deletion using CRISPR/Cas9 combined with loxP to efficiently recycle selectable markers. This system allowed us to rapidly generate two dozen mutants in the CDPK family in T. gondii. Although the majority of these lack discernible phenotypes in standard assays, this approach should be useful in testing the function of other multigene families in T. gondii. Our findings also suggest that additional models are needed to ascertain gene function on T. gondii, as the currently available models may not adequately assess biological function.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Sibley laboratory, including Kevin Brown, Bang Shen, and Drew Etheridge, for their advice and sharing plasmids, Nathaniel Jones and Zi Teng Wang for their advice on monitoring bradyzoite differentiation, and Jennifer Barks for technical assistance.
Funding Statement
Partial funding was provided by the NIH. The funders did not have any role in the study design, data collection and interpretation, or decision to publish the work.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01173-15.
For a commentary on this article, see doi:10.1128/IAI.00175-16.
REFERENCES
- 1.Hanks SK, Hunter T. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9:576–596. [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Miranda-Saavedra D, Gabaldon T, Barton GJ, Langsley G, Doerig C. 2012. The kinomes of apicomplexan parasites. Microbes Infect 14:796–810. doi: 10.1016/j.micinf.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Talevich E, Tobin AB, Kannan N, Doerig C. 2012. An evolutionary perspective on the kinome of malaria parasites. Philos Trans R Soc Lond B Biol Sci 367:2607–2618. doi: 10.1098/rstb.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunes M, Goldring JPD, Doerig C, Scherf A. 2007. A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Mol Microbiol 63:391–403. [DOI] [PubMed] [Google Scholar]
- 6.Schneider AG, Mercereau-Puijalon O. 2005. A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics 6:30. doi: 10.1186/1471-2164-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter CA, Sibley LD. 2012. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat Rev Microbiol 10:766–778. doi: 10.1038/nrmicro2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talevich E, Kannan N. 2013. Structural and evolutionary adaptation of rhoptry kinases and pseudokinases, a family of coccidian virulence factors. BMC Evol Biol 13:117. doi: 10.1186/1471-2148-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billker O, Lourido S, Sibley LD. 2009. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe 5:612–622. doi: 10.1016/j.chom.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valmonte GR, Arthur K, Higgins CM, MacDiarmid RM. 2014. Calcium-dependent protein kinases in plants: evolution, expression and function. Plant Cell Physiol 55:551–569. doi: 10.1093/pcp/pct200. [DOI] [PubMed] [Google Scholar]
- 11.Wernimont AK, Amani M, Qiu W, Pizarro JC, Artz JD, Lin YH, Lew J, Hutchinson A, Hui R. 2011. Structures of parasitic CDPK domains point to a common mechanism of activation. Proteins 79:803–820. doi: 10.1002/prot.22919. [DOI] [PubMed] [Google Scholar]
- 12.Wernimont AK, Artz JD, Finerty P, Lin Y, Amani M, Allali-Hassani A, Senisterra G, Vedadi M, Tempel W, Mackenzie F, Chau I, Lourido S, Sibley LD, Hui R. 2010. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol 17:596–601. doi: 10.1038/nsmb.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranjan R, Ahmed A, Gourinath S, Sharma P. 2009. Dissection of mechanisms involved in the regulation of Plasmodium falciparum calcium dependent protein kinase 4 (PfCDPK4). J Biol Chem 284:15267–15276. doi: 10.1074/jbc.M900656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azevedo MF, Sanders PR, Krejany E, Nie CQ, Fu P, Bach LA, Wunderlich G, Crabb BS, Gilson PR. 2013. Inhibition of Plasmodium falciparum CDPK1 by conditional expression of its J-domain demonstrates a key role in schizont development. Biochem J 452:433–441. doi: 10.1042/BJ20130124. [DOI] [PubMed] [Google Scholar]
- 15.Hui R, El Bakkouri M, Sibley LD. 2015. Designing selective inhibitors for calcium-dependent protein kinases in apicomplexans. Trends Pharmacol Sci 36:452–460. doi: 10.1016/j.tips.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lourido S, Shuman J, Zhang C, Shokat KM, Hui R, Sibley LD. 2010. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature 465:359–362. doi: 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ojo KK, Larson ET, Keyloun KR, Castaneda LJ, DeRoucher AE, Inampudi KK, Kim JE, Arakaki TL, Murphy RC, Zhang L, Napuli AJ, Maly DJ, Verlinde CLMJ, Buckner FS, Parsons M, Hol WGJ, Merritt EA, Van Voorhis WC. 2010. Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors. Nat Struct Mol Biol 17:602–607. doi: 10.1038/nsmb.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lourido S, Jeschke GR, Turk BE, Sibley LD. 2013. Exploiting the unique ATP-binding pocket of Toxoplasma calcium-dependent protein kinase 1 to identify its substrates. ACS Chem Biol 8:1155–1162. doi: 10.1021/cb400115y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lourido S, Zhang C, Lopez MS, Tang K, Barks J, Wang Q, Wildman SA, Shokat KM, Sibley LD. 2013. Optimizing small molecule inhibitors of calcium-dependent protein kinase 1 to prevent infection by Toxoplasma gondii. J Med Chem 56:3068–3077. doi: 10.1021/jm4001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SM, Murphy RC, Geiger JA, DeRocher AE, Zhang Z, Ojo KK, Larson ET, Perera BG, Dale EJ, He P, Reid MC, Fox AM, Mueller NR, Merritt EA, Fan E, Parsons M, Van Voorhis WC, Maly DJ. 2012. Development of Toxoplasma gondii calcium-dependent protein kinase 1 (TgCDPK1) inhibitors with potent anti-toxoplasma activity. J Med Chem 55:2416–2426. doi: 10.1021/jm201713h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy RC, Ojo KK, Larson ET, Castellanos-Gonzalez A, Perera BG, Keyloun KR, Kim JE, Bhandari JG, Muller NR, Verlinde CL, White AC, Merritt EA, Van Voorhis WC, Maly DJ. 2010. Discovery of potent and selective inhibitors of calcium-dependent protein kinase 1 (CDPK1) from C. parvum and T. gondii. ACS Med Chem Lett 1:331–335. doi: 10.1021/ml100096t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, Brinkmann V. 2004. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117:503–514. doi: 10.1016/S0092-8674(04)00449-0. [DOI] [PubMed] [Google Scholar]
- 23.Lourido S, Tang K, Sibley LD. 2012. Distinct signalling pathways control Toxoplasma egress and host-cell invasion. EMBO J 31:4524–4534. doi: 10.1038/emboj.2012.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treeck M, Sanders JL, Gaji RY, LaFavers KA, Child MA, Arrizabalaga G, Elias JE, Boothroyd JC. 2014. The calcium-dependent protein kinase 3 of Toxoplasma influences basal calcium levels and functions beyond egress as revealed by quantitative phosphoproteome analysis. PLoS Pathog 10:e1004197. doi: 10.1371/journal.ppat.1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, Hopp CS, Bright AT, Westenberger S, Winzeler E, Blackman MJ, Baker DA, Wandless TJ, Duraisingh MT. 2010. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science 328:910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alam MM, Solyakov L, Bottrill AR, Flueck C, Siddiqui FA, Singh S, Mistry S, Viskaduraki M, Lee K, Hopp CS, Chitnis CE, Doerig C, Moon RW, Green JL, Holder AA, Baker DA, Tobin AB. 2015. Phosphoproteomics reveals malaria parasite protein kinase G as a signalling hub regulating egress and invasion. Nat Commun 6:7285. doi: 10.1038/ncomms8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiersma HI, Galuska SE, Tomley FM, Sibley LD, Liberator PA, Donald RGK. 2004. A role for coccidian cGMP-dependent protein kinase in motility and invasion. Int J Parasitol 34:369–380. doi: 10.1016/j.ijpara.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Morlon-Guyot J, Berry L, Chen CT, Gubbels MJ, Lebrun M, Daher W. 2014. The Toxoplasma gondii calcium-dependent protein kinase 7 is involved in early steps of parasite division and is crucial for parasite survival. Cell Microbiol 16:95–114. doi: 10.1111/cmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar P, Tripathi A, Ranjan R, Halbert J, Gilberger T, Doerig C, Sharma P. 2014. Regulation of Plasmodium falciparum development by calcium-dependent protein kinase 7 (PfCDPK7). J Biol Chem 289:20386–20396. doi: 10.1074/jbc.M114.561670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen B, Brown KM, Lee TD, Sibley LD. 2014. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio 5(3):e01114-14. doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan A, Behnke MS, Dunay IR, White MW, Sibley LD. 2009. Phenotypic and gene expression changes among clonal type I strains of Toxoplasma gondii. Eukaryot Cell 8:1828–1836. doi: 10.1128/EC.00150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. 2009. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot Cell 8:520–529. doi: 10.1128/EC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fox BA, Falla A, Rommereim LM, Tomita T, Gigley JP, Mercier C, Cesbron-Delauw MF, Weiss LM, Bzik DJ. 2011. Type II Toxoplasma gondii KU80 knockout strains enable functional analysis of genes required for cyst development and latent infection. Eukaryot Cell 10:1193–1206. doi: 10.1128/EC.00297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan A, Ajzenberg D, Mercier A, Demar M, Simon S, Darde ML, Wang Q, Verma SK, Rosenthal BM, Dubey JP, Sibley LD. 2014. Geographic separation of domestic and wild strains of Toxoplasma gondii in French Guiana correlates with a monomorphic version of chromosome 1a. PLoS Negl Trop Dis 8:e3182. doi: 10.1371/journal.pntd.0003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Etheridge RD, Alagan A, Tang K, Turk BE, Sibley LD. 2014. ROP18 and ROP17 kinase complexes synergize to control acute virulence of Toxoplasma in the mouse. Cell Host Microbe 15:537–550. doi: 10.1016/j.chom.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heaslip AT, Nishi M, Stein B, Hu K. 2011. The motility of a human parasite, Toxoplasma gondii, is regulated by a novel lysine methyltransferase. PLoS Pathog 7:e1002201. doi: 10.1371/journal.ppat.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 38.Howe DK, Summers BC, Sibley LD. 1996. Acute virulence in mice is associated with markers on chromosome VIII in Toxoplasma gondii. Infect Immun 64:5193–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang ZT, Harmon S, O'Malley KL, Sibley LD. 2015. Reassessment of the role of aromatic amino acid hydroxylases and the effect of infection by Toxoplasma gondii on host dopamine. Infect Immun 83:1039–1047. doi: 10.1128/IAI.02465-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fux B, Nawas J, Khan A, Gill DB, Su C, Sibley LD. 2007. Toxoplasma gondii strains defective in oral transmission are also defective in developmental stage differentiation. Infect Immun 75:2580–2590. doi: 10.1128/IAI.00085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alaganan A, Fentress SJ, Tang K, Wang Q, Sibley LD. 2014. Toxoplasma GRA7 effector increases turnover of immunity-related GTPases and contributes to acute virulence in the mouse. Proc Natl Acad Sci U S A 111:1126–1131. doi: 10.1073/pnas.1313501111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behnke M, Wooten JC, Lehmann M, Radke J, Lucas O, Nawas J, Sibley LD, White M. 2010. Coordinated progression through two subtranscriptions underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One 5:e12354. doi: 10.1371/journal.pone.0012354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fritz HM, Buchholz KR, Chen X, Durbin-Johnson B, Rocke DM, Conrad PA, Boothroyd JC. 2012. Transcriptomic analysis of toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PLoS One 7:e29998. doi: 10.1371/journal.pone.0029998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S. 2014. Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One 9:e100450. doi: 10.1371/journal.pone.0100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uboldi AD, McCoy JM, Blume M, Gerlic M, Ferguson DJ, Dagley LF, Beahan CT, Stapleton DI, Gooley PR, Bacic A, Masters SL, Webb AI, McConville MJ, Tonkin CJ. 2015. Regulation of starch stores by a Ca2+-dependent protein kinase is essential for viable cyst development in Toxoplasma gondii. Cell Host Microbe 18:670–681. doi: 10.1016/j.chom.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 47.Wang JL, Huang SY, Li TT, Chen K, Ning HR, Zhu XQ. 26 October 2015. Evaluation of the basic functions of six calcium-dependent protein kinases in Toxoplasma gondii using CRISPR-Cas9 system. Parasitol Res Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 48.Su C, Howe DK, Dubey JP, Ajioka JW, Sibley LD. 2002. Identification of quantitative trait loci controlling acute virulence in Toxoplasma gondii. Proc Natl Acad Sci U S A 99:10753–10758. doi: 10.1073/pnas.172117099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garrison E, Treeck M, Ehret E, Butz H, Garbuz T, Oswald BP, Settles M, Boothroyd J, Arrizabalaga G. 2012. A forward genetic screen reveals that calcium-dependent protein kinase 3 regulates egress in Toxoplasma. PLoS Pathog 8:e1003049. doi: 10.1371/journal.ppat.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy JM, Whitehead L, van Dooren GG, Tonkin CJ. 2012. TgCDPK3 regulates calcium-dependent egress of Toxoplasma gondii from host cells. PLoS Pathog 8:e1003066. doi: 10.1371/journal.ppat.1003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lavine MD, Knoll LJ, Rooney PJ, Arrizabalaga G. 2007. A Toxoplasma gondii mutant defective in responding to calcium fluxes shows reduced in vivo pathogenicity. Mol Biochem Parasitol 155:113–122. doi: 10.1016/j.molbiopara.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klimecka M, Muszynska G. 2007. Structure and functions of plant calcium-dependent protein kinases. Acta Biochim Pol 54:219–233. [PubMed] [Google Scholar]
- 53.Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, Sibley LD. 2011. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc Natl Acad Sci U S A 108:9631–9636. doi: 10.1073/pnas.1015338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reese ML, Zeiner GM, Saeij JP, Boothroyd JC, Boyle JP. 2011. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc Natl Acad Sci U S A 108:9625–9630. doi: 10.1073/pnas.1015980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.