Abstract
Plasmodium parasites employ posttranscriptional regulatory mechanisms as their life cycle transitions between host cell invasion and replication within both the mosquito vector and mammalian host. RNA binding proteins (RBPs) provide one mechanism for modulation of RNA function. To explore the role of Plasmodium RBPs during parasite replication, we searched for RBPs that might play a role during liver stage development, the parasite stage that exhibits the most extensive growth and replication. We identified a parasite ortholog of the Mei2 (Meiosis inhibited 2) RBP that is conserved among Plasmodium species (PlasMei2) and exclusively transcribed in liver stage parasites. Epitope-tagged Plasmodium yoelii PlasMei2 was expressed only during liver stage schizogony and showed an apparent granular cytoplasmic location. Knockout of PlasMei2 (plasmei2−) in P. yoelii only affected late liver stage development. The P. yoelii plasmei2− liver stage size increased progressively until late in development, similar to wild-type parasite development. However, P. yoelii plasmei2− liver stage schizonts exhibited an abnormal DNA segregation phenotype and failed to form exoerythrocytic merozoites. Consequently the cellular integrity of P. yoelii plasmei2− liver stages became increasingly compromised late in development and the majority of P. yoelii plasmei2− underwent cell death by the time wild-type liver stages mature and release merozoites. This resulted in a complete block of P. yoelii plasmei2− transition from liver stage to blood stage infection in mice. Our results show for the first time the importance of a Plasmodium RBP in the coordinated progression of late liver stage schizogony and maturation of new invasive forms.
INTRODUCTION
Malaria is caused by unicellular eukaryotic parasites of the genus Plasmodium. The life cycles of the Plasmodium parasites that cause human disease (including the major human parasite Plasmodium falciparum) and other mammalian parasites such as rodent malaria parasites (including P. berghei and P. yoelii) are similar. The parasites progress through an elaborate life cycle, part within an anopheline mosquito vector and part within the mammalian host. Within the host, the sporozoite stage first infects hepatocytes, within which the parasite then develops as a liver stage (also known as an exoerythrocytic form), an acyclic process that occurs once during a malaria infection. Intrahepatocytic parasite development progresses in a cellular process known as schizogony, an endomitotic process characterized by an enormous increase in cell mass with concomitant DNA replication and organelle replication but without cytokinesis. It is only in the final hours of liver stage schizogony that organelles and DNA completely segregate and are apportioned into tens of thousands of rapidly differentiating exoerythrocytic invasive merozoites. These are released and initiate the infection of erythrocytes. Erythrocytic schizogony is cyclic and generates approximately 20 to 30 new merozoites in each cycle. Parasite schizogony and merozoite formation in both hepatocytes and erythrocytes likely employ shared gene regulatory networks and cell regulatory mechanisms, but it is possible that because exoerythrocytic schizogony exceeds erythrocytic schizogony by more than 3 orders of magnitude and is extremely complex, it has also evolved unique networks and mechanisms that control its progression.
Studies of the asexual intraerythrocytic replication have demonstrated a cyclic pattern of transcript accumulation, with over half of all transcripts achieving maximal expression at only one of the three main developmental stages (1, 2), but the factors governing these waves of expression are poorly understood. Conversely, transcriptome sequencing data sets suggest that the majority of the genome is pervasively transcribed throughout blood stage development (3). Few Plasmodium transcription factors have been identified, with the exception of the 27 members of the Apicomplexan Apetala 2 (ApiAP2) family of transcription factors (4), which are homologous to plant AP2/ethylene response factor DNA-binding proteins, the second largest family of transcription factors in the model plant Arabidopsis thaliana (5). Some of the ApiAP2 family members appear to play very specific roles during the parasite life cycle (as examples, Api-G is critical for gametocyte formation [6, 7], Api-O for ookinetes [8], Api-Sp for sporozoites [9], and Api-L for liver stages [10]). The pervasive transcription of the parasite genome and the paucity of transcription factors thus strongly suggest that there is exquisite posttranscriptional regulation of mRNA that leads to highly organized protein expression during both blood stage and liver stage development. Indeed, comparisons of the P. falciparum blood stage transcriptome and proteomes during development show a delay in translation of a significant number of mRNAs, suggesting that transcribed mRNAs are packaged and repressed, ready for just-in-time translation (11, 12).
Regulation of mRNA translation relies on RNA binding proteins (RBPs), many of which form complexes within the cytoplasm in entities known as processing bodies (P-bodies) or P-granules (13). P-bodies are involved in many aspects of mRNA homeostasis, including both mRNA decay and translational repression, and some studies have begun to reveal their role in the Plasmodium life cycle. For example, the RBP DOZI, a member of the DEAD-box RNA helicase family, is present in P-bodies of gametocytes and is involved in mRNA translational repression (14, 15), whereas in the sporozoite, the P-body resident RBP Puf2 (Pumilio in Drosophila protein and fem-3 binding factor in Caenorhabditis elegans) is involved in mRNA translational repression (16, 17). During the asexual blood stage cycle the RBP Alba1 (Acetylation lowers binding affinity 1) forms P-body-like structures in the trophozoite and schizont (18) and was found to be associated with 1,193 transcripts in the trophozoite, suggesting that Alba1 is involved in the time-dependent assembly of the invasive merozoite (19).
Since no data exist regarding the importance of RBPs during liver stage development, we analyzed transcriptomic and proteomic data sets for liver stage-enriched RBPs, which led us to investigate the role of a Plasmodium Mei2 (Meiosis inhibited 2)-like RBP. Mei2 was initially described in the fission yeast Schizosaccharomyces pombe and plays a critical role in the switch from mitosis to meiosis (20). Mei2 is a member of the largest family of RBPs—those that contain an RNA recognition motif (RRM), a stretch of 70 to 90 amino acids that contain two consensus RNA-interacting motifs, RNP1 and RNP2. RRM-containing proteins are subdivided into 10 separate families (RRM_1 through RRM_10) based on shared amino acid identities between members of each family and Mei2 contains a C-terminal RRM_2, thought to be unique to fungi and plants (21). Here, we show that Plasmodia contain a single Mei2-like gene (here referred to as PlasMei2). In the P. yoelii rodent malaria model, Plasmei2 is only expressed during liver stage development and is localized in distinct cytoplasmic structures reminiscent of P-bodies. To understand the importance of PlasMei2, we created a gene knockout in P. yoelii, which resulted in abnormal liver stage development and failure of the parasite to transition to the blood stage infection.
MATERIALS AND METHODS
Experimental animals.
Six- to eight-week-old female Swiss-Webster (SW) mice from Harlan (Indianapolis, IN) were used for parasite life cycle maintenance and the production of transgenic parasites. Six- to eight-week-old female BALB/cAnN mice from Harlan were used for assessments of parasite infectivity and indirect immunofluorescence assay (IFA). Six- to eight-week-old female BALB/cJ and BALB/cByJ mice from the Jackson Laboratory (Bar Harbor, ME) were used to assess the attenuation and ability of P. yoelii plasmei2– parasites to act as experimental vaccines. P. yoelii 17XNL wild-type and transgenic parasites were cycled between SW mice and Anopheles stephensi mosquitoes for the purposes of sporozoite production. Infected mosquitoes were maintained on sugar water at 24°C and 70% humidity. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Center for Infectious Disease Research has an OLAW Animal Welfare Assurance (A3640-01). The protocol was approved by the Center for Infectious Disease Research Institutional Animal Care and Use Committee.
RT-PCR.
Oligonucleotide primers for amplification of P. yoelii PlasMei2 and 18S rRNA are detailed in Table S1 in the supplemental material. Total RNA from mixed blood stage parasites, midgut sporozoites, salivary gland sporozoites, and liver lobe samples were extracted using TRIzol (Invitrogen) and DNase-treated using Turbo-DNA Free (Ambion). First-strand cDNA was synthesized from RNA by using a Superscript III Platinum RT kit (Invitrogen). The resulting cDNA was used for the amplification of P. yoelii PlasMei2 and 18S rRNA using an Advantage 2 PCR kit (Clontech). cDNA synthesized without reverse transcriptase (RT) was used as a negative control.
Creation of a P. yoelii plasmei2– and P. yoelii PlasMei2-myc.
All oligonucleotide primers used for the creation and analysis of P. yoelii plasmei2– and P. yoelii PlasMei2-myc are detailed in Table S1 in the supplemental material. Deletion of P. yoelii PlasMei2 (PlasmoDB identifier PY17X_1123700) was achieved based on the previously reported gene insertion/marker out (GIMO) strategy (22). In brief, PlasMei2 was deleted using double-crossover homologous recombination. Complementary regions of PlasMei2 upstream and downstream of the open reading frame were ligated into plasmid pL0034 (22) flanking the positive/negative selectable marker, resulting in the creation of a plasmid pL0034_PlasMei2. Linearized pL0034_PlasMei2 was transfected into the blood stage schizonts of P. yoelii line 1971 (22), a marker-free parasite that expresses a green fluorescent protein-luciferase fusion throughout the life cycle under the control of the elongation factor 1α promoter. After transfection and intravenous injection into SW, pyrimethamine was used for the positive selection and downstream cloning of recombinant parasites using standard techniques (23). Gene knockout was confirmed by PCR using methodology we have used on multiple occasions (see reference 24 for a recent example). This led to the creation of the P. yoelii plasmei2– and two separate knockout clones from two independent transfections were initially phenotypically analyzed throughout the life cycle. Creation of P. yoelii PlasMei2-myc also relied upon double crossover homologous recombination using modified plasmid pL0005 (obtained through the MR4 as part of the BEI Resources Repository, National Institute of Allergy and Infectious Diseases, National Institutes of Health: Plasmodium berghei pL0005, MRA-774, deposited by A. P. Waters), which allowed for the addition of a triple-myc epitope tag to the carboxy terminus of PlasMei2. The resultant P. yoelii PlasMei2-myc parasite expresses a single copy of PlasMei2 (with a myc tag) under the control of its endogenous promoter. As for P. yoelii plasmei2–, two individual P. yoelii PlasMei2-myc clones from two independent transfections underwent life cycle phenotypic analysis.
IFA. (i) Liver stage.
BALB/cAnN mice were injected intravenously with approximately 3 × 105 sporozoites, and livers were harvested from euthanized mice at several time points postinfection. Livers were perfused with 1× phosphate-buffered saline (PBS) and fixed in 4% (vol/vol) paraformaldehyde (PFA) in 1× PBS, and lobes were cut into 50-μm sections using a Vibratome apparatus (Ted Pella, Redding, CA). For IFA, sections were permeabilized in 1× Tris-buffered saline (TBS) containing 3% (vol/vol) H2O2 and 0.25% (vol/vol) Triton X-100 for 30 min at room temperature. Sections were then blocked in 1× TBS containing 5% (vol/vol) dried milk (TBS-M) for at least 1 h and incubated with primary antibody in TBS-M at 4°C overnight. After washing in 1× TBS, fluorescent secondary antibodies were added in TBS-M for 2 h at room temperature in a similar manner as above. After further washing, the section was incubated in 0.06% (wt/vol) KMnO4 for 2 min to quench the background fluorescence. Sections were then washed with 1× TBS and stained with DAPI (4′,6′-diamidino-2-phenylindole) at 1 μg/ml in 1× TBS for 5 to 10 min at room temperature to visualize DNA and mounted with FluoroGuard antifade reagent (Bio-Rad, Hercules, CA).
(ii) Blood stage.
Infected red blood cells were processed for IFA using a previously described method (25). Red blood cells were pelleted at 2,000 × g in a microcentrifuge at room temperature for 30 s between manipulations. Cells were washed twice in 1× PBS, fixed in 1× PBS–4% (vol/vol) PFA–0.0075% (vol/vol) glutaraldehyde for 30 min at room temperature, and permeabilized in 1× PBS–0.2% (vol/vol) Triton X-100 for 20 min at room temperature. A 1× PBS–3% (wt/vol) bovine serum albumin (BSA; blocking solution) mixture was applied at 4°C overnight. Primary antibodies were diluted in blocking solution and incubated for 1 h with end-over-end rotation at room temperature. After two washes with 1× PBS, fluorescent secondary antibodies were diluted in blocking solution and incubated with cells for 30 min with end-over-end rotation at room temperature and shielding from light. Nucleic acid was then stained with DAPI in 1× PBS for 5 to 10 min at room temperature. The cells were washed three times with 1× PBS, mixed 1:1 with VectaShield (Vector Laboratories), and applied to a glass slide and coverslip.
Sporozoites.
Sporozoites were isolated and fixed with 4% (vol/vol) PFA in 1× PBS for 20 min at room temperature. Fixed sporozoites were applied to a polylysine-treated microscope slide, which was housed in a wet chamber at 4°C overnight. Sporozoites were washed twice with 1× PBS and then permeabilized and blocked with 2% (wt/vol) BSA–0.2% (vol/vol) Triton X-100 in 1× PBS for 1 h at 37°C. IFA was performed as for the blood stages, but the solutions were applied directly to the slide.
All preparations were analyzed for fluorescence using a fluorescence inverted microscope (Eclipse TE2000-E; Nikon), and images were acquired using Olympus 1×70 DeltaVision deconvolution microscopy.
Sporozoite inoculation and challenge.
P. yoelii plasmei2– sporozoites were isolated from the salivary glands of infected A. stephensi mosquitoes between 14 and 18 days after the infectious blood meal and injected intravenously into the tail vein of recipient mice. To assess the attenuation, sporozoites were injected into highly susceptible BALB/cByJ mice (26). Liver stage-to-blood stage transition (blood stage patency) was assessed by using a Giemsa-stained thin blood smear starting at day 3 after inoculation and ending at day 14, at which time, a negative smear was attributed to complete attenuation. For immunizations, BALB/cJ mice were primed and boosted with P. yoelii sporozoites and subsequently challenged with 10,000 wild-type P. yoelii XNL sporozoites. Breakthrough to blood stage patency was assessed by using a Giemsa-stained thin blood smear starting at day 3 after challenge and ending at day 14, at which time, a negative smear was attributed to complete protection. Mice immunized only with mosquito salivary gland extract were used as controls.
Phenotypic analysis of liverstage P. yoelii plasmei2–.
After IFA, liver stage size was measured by determining the area of the parasite at its greatest circumference. Viability was measured by examining the integrity of the parasitophorous vacuole membrane (PVM) marker Hep17. If Hep17 expression did not completely delineate the PVM, the parasite was considered nonviable. The PVM does break down at the termination of liver stage development, but measurements were made before this event occurs. Liver stage development was also measured using an in vivo imaging system (IVIS) since the parasites used in the present study express luciferase and are thus bioluminescent. Luciferase activity in animals was visualized through imaging of whole bodies using an IVIS Lumina II animal imager (Caliper Life Sciences, USA) as previously described (27–29). Mice were injected with 100 μl of RediJect d-luciferin (PerkinElmer) intraperitoneally prior to being anesthetized using an isoflurane-anesthesia system (XGI-8; Caliper Life Sciences, USA). Measurements were performed within 5 to 10 min after the injection of d-luciferin. Bioluminescence imaging was acquired with a 10-cm field of view, a medium binning factor, and an exposure time of 1 to 5 min. Quantitative analysis of bioluminescence was performed by measuring the luminescence signal intensity using the region-of-interest (ROI) settings of the Living Image 3.0 software. ROIs were placed around the whole animal, and ROI measurements were expressed as the total flux (photons/second).
Phenotypic analysis of blood stage P. yoelii plasmei2–.
To assay the growth of P. yoelii 17XNL and P. yoelii plasmei2– blood stages, blood was removed from infected SW mice when parasitemia was between 0.5 and 1.5%. Blood was diluted in RPMI 1640 medium (HyClone, Logan, UT) so that 100 μl contained 106 parasites. SW mice (five in each group) were then injected intravenously with the infected red blood cells, and the percent parasitemia was monitored every other day until day 11.
RESULTS
The Plasmodium RBP Mei2 (PlasMei2).
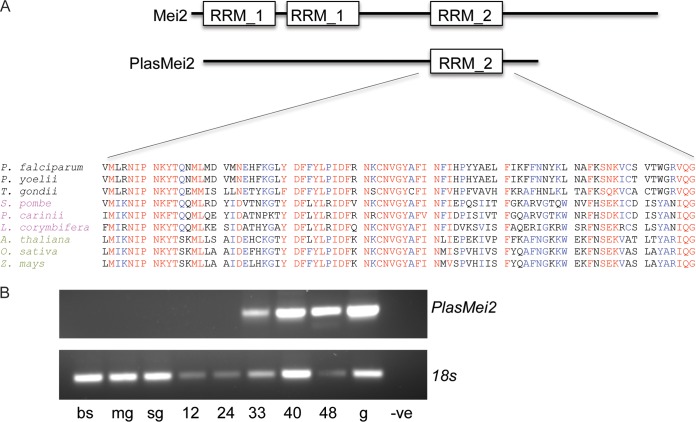
In our efforts to characterize the role of RBPs in preerythrocytic parasite development, we analyzed the available Plasmodium transcriptomic and proteomic data sets through PlasmoDB (http://plasmodb.org/plasmo/) for the presence of proteins that contain RNA binding domains and found a preerythrocytic stage-specific ortholog of Mei2 (PlasMei2) (20). Mei2-like genes are restricted to eukaryotic genomes and fall into three broad clades: the AML and TEL groups of plants, and a fungal clade (21). Interestingly, PlasMei2 cannot be exclusively assigned to any one group. Yeast Mei2 contains three RRMs, two N-terminal RRM_1 domains and a C-terminal RRM_2. Only the C-terminal RRM_2 domain is present in PlasMei2 (Fig. 1A). PlasMei2 is highly conserved among Plasmodium species (see Fig. S1 in the supplemental material) and is the only Plasmodium protein that contains an RRM_2 domain (30). The Plasmodium RRM_2 shows a high degree of similarity to other Mei2 RRM_2 domains, based on a comparison to representative sequences from alveolates, fungi, and plants (Fig. 1A). A recent bioinformatics survey of RBPs in Plasmodium uncovered 72 RRM-containing proteins, 40 of which contain only one RRM domain, among them, PlasMei2, and the predicted three-dimensional structure of PlasMei2 aligned with that of previously a crystalized RRM_2 domain (30). The RRM_2 domain shares amino acid conservation with RRMs of the sex lethal and HuD proteins, both of which bind single-stranded AU-rich RNA, suggesting that PlasMei2 may behave similarly (21).
FIG 1.
PlasMei2: Sequence similarity among the Mei2 family and stage-specific expression. (A) Cartoon of the overall domain structure and alignment of the PlasMei2 RNA recognition motif 2 (RRM_2) with those of other apicomplexa (black), fungi (magenta), and plants (green). Residues in red and blue are either identical or similar and, if red, are shared among all species of the comparison. (B) P. yoelii plasmei2 mRNA expression, as determined by RT-PCR, on mixed blood stages (bs), midgut sporozoites (mg), salivary gland sporozoites (sg), and liver stages at 12, 24, 33, 40, and 48 h after sporozoite infection. Genomic DNA (g) was used as a positive control, and no template was used as a negative control (-ve). RT-PCR of P. yoelii 18S was used as a positive control for each sample.
Analysis of published expression data indicated that PlasMei2 was transcribed during P. yoelii liver stage development (31) and showed no evidence of expression, based on proteomic data sets, in P. falciparum blood stages and sporozoites. This led us to analyze the RNA expression pattern of PlasMei2 in P. yoelii (gene identifier PY17X_1123700) throughout the parasite life cycle. We used RT-PCR to analyze life cycle stages with PlasMei2-specific primers, as well as primers for 18S rRNA as a positive control. Although 18S rRNA transcription was seen in all samples (Fig. 1B), PlasMei2 was transcribed only during mid- to late-liver stage development (Fig. 1B).
PlasMei2 is expressed only during liver stage development.
To further study both the temporal and spatial expression of PlasMei2 protein in liver stages, we used standard transfection procedures to create a transgenic P. yoelii parasite expressing PlasMei2 with a quadruple C-terminal myc epitope tag (P. yoelii PlasmMei2-myc) (see Fig. S2 in the supplemental material). Phenotypic analysis of two individual clones from two transfections suggested that the parasite behaved like the wild type, and thus the tag was not affecting the parasite life cycle. Salivary gland sporozoite numbers were similar to wild type (see Fig. S3A in the supplemental material). Additionally, after the intravenous injection of 10,000 salivary gland sporozoites isolated from wild-type parasites and P. yoelii PlasmMei2-myc clones into groups of five BALB/cJ mice, all mice became blood stage patent with similar kinetics, 3 days after sporozoite injection. We then analyzed Mei2-myc expression throughout the life cycle by IFA using antibodies to myc. As expected, PlasmMei2-myc expression was not observed in blood stages or in salivary gland sporozoites (data not shown). To analyze liver stage expression, infected livers were harvested from BALB/cAnN mice at time points after the intravenous injection of between 0.3 × 106 and 1.0 × 106 P. yoelii PlasmMei2-myc sporozoites. At 20 h of liver stage development, PlasMei2 expression was not observed (Fig. 2A). However, by 32 h of development (Fig. 2A), PlasMei2 expression was observed throughout the liver stage cytoplasm and exhibited a granular localization pattern reminiscent of P-body localization previously seen in Plasmodium asexual blood stages (18), gametocytes (15), and sporozoites (16). Localization continued to be cytoplasmic and granular as the liver stage parasite matured at both 38 and 45 h postinfection (Fig. 2A) before declining to undetectable levels at 52 h (data not shown). Although it cannot be unequivocally stated that PlasMei2 is not additionally localized to the liver stage nuclear region, 45-h liver stage parasites showed a weaker myc signal in areas where parasite DNA was localized (Fig. 2B). Since PlasMei2 was expressed during mid- to late-liver stage development, we next sought to determine the importance of PlasMei2 for liver stage maturation.
FIG 2.
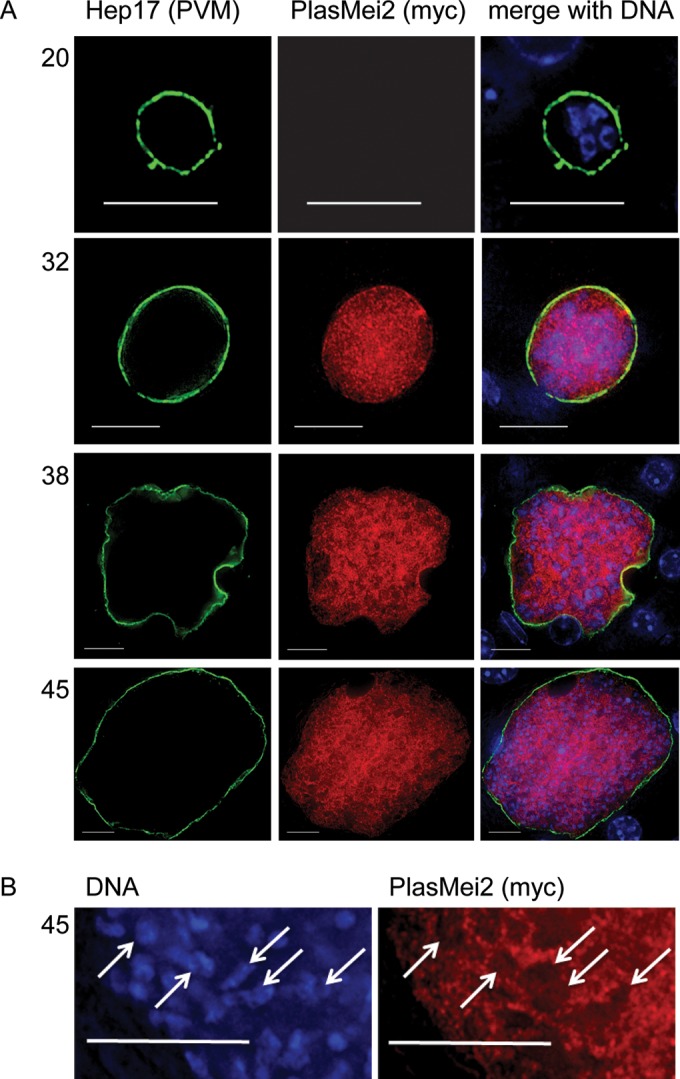
Expression and localization of P. yoelii PlasMei2 during liver stage development. Livers from mice infected with the P. yoelii transgenic parasite PlasMei2-myc were perfused with PBS and fixed in 4% paraformaldehyde at various time points after sporozoite infection. Sections (50 μm) were cut from fixed livers, and IFAs were performed using primary antibodies to the parasite parasitophorous vacuole membrane (PVM) marker Hep17 (green) and to myc (red) for the observation of the spatial expression of PlasMei2-myc. (A) PlasMei2-myc liver stage expression was observed at 20, 32, 38, and 45 h after sporozoite injection. (B) A magnified 45-h liver stage section showing exclusion of PlasMei2-myc expression from nuclear areas as indicated by DNA staining (white arrows). DNA was labeled with DAPI (blue). Images were created from a single deconvolved z-stack. Myc expression is punctate and within the parasite cytoplasm. Scale bar, 10 μm.
PlasMei2 is essential for late liver stage schizogony.
We created a P. yoelii plasmei2– gene knockout parasite (P. yoelii plasmei2–) (see Fig. S4 in the supplemental material) using GIMO technology (22). Individual P. yoelii plasmei2– parasite clones isolated from two individual transfections were analyzed for defects in blood stage replication (see Fig. S3B in the supplemental material) and salivary gland sporozoite production (see Fig. S3A in the supplemental material) and were found to behave like the wild-type parasites. However, P. yoelii plasmei2– sporozoites failed to initiate a blood stage infection when 1 × 103 and 10 × 103 salivary gland sporozoites were injected intravenously into groups of five BALB/cJ mice, whereas mice infected with wild-type sporozoites developed patent blood stage infection within 3 to 4 days (Table 1). This observation suggested that P. yoelii plasmei2– parasites were unable to either infect the liver or complete liver stage development. To assess the severity of the defect, we next injected groups of five highly susceptible BALB/cByJ (26) mice with 5 × 104 P. yoelii plasmei2– sporozoites. Again, no blood stage patency was observed (Table 1). However, sporozoite challenge/blood stage patency assays cannot pinpoint the time point of attenuation during preerythrocytic infection, and thus we examined more closely when the developmental arrest of plasmei2– parasites was taking place.
TABLE 1.
Attenuation of P. yoelii plasmei2− preerythrocytic infection in BALB/c mice
| Mouse strain | Genotypea | No. of sporozoites injected | No. of patent mice/total no. of mice | Day(s) of patency |
|---|---|---|---|---|
| BALB/cJ | WT | 1,000 | 3/3 | 3–4 |
| WT | 10,000 | 7/7 | 3 | |
| plasmei2− | 1,000 | 0/12 | ||
| plasmei2− | 10,000 | 0/56 | ||
| BALB/cByJ | plasmei2− | 50,000 | 0/10 |
WT, wild type.
plasmei2– liver stages grow but fail to form exoerythrocytic merozoites.
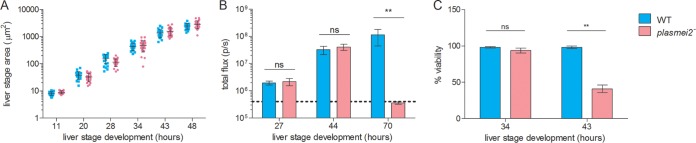
To further explore the P. yoelii plasmei2– phenotype, sporozoites were injected intravenously into BALB/cAnN mice, and the livers were harvested at time different points after infection to assess liver stage development by histology and IFA. Interestingly, P. yoelii plasmei2– liver stages increased in size in a manner similar to that of the wild-type parasites during the course of liver stage development (Fig. 3A), suggesting that PlasMei2 is not required for liver stage growth. Parasite growth was also measured by assessing luciferase activity over time and, in agreement with the IFA analysis, the wild-type and P. yoelii plasmei2– liver stages showed equivalent growth at 27 and 44 h (Fig. 3B; see also Fig. S5 in the supplemental material), whereas only wild-type parasites transitioned to blood stage parasitemia, as measured at 70 h after sporozoite infection (Fig. 3B; see Fig. S5 in the supplemental material). IFA analysis showed no apparent aberrant phenotype of P. yoelii plasmei2– parasites during the first 28 h of development (see Fig. S6 in the supplemental material). However, by 34 h, there was evidence for abnormal development (Fig. 4). Wild-type liver stages showed numerous nuclear centers, but in the majority of P. yoelii plasmei2– liver stages, there was an increase in the size of a proportion of the nuclear centers, suggesting that abnormal DNA segregation was taking place (Fig. 4). This was accompanied by an abnormal expression pattern of the endoplasmic reticulum marker, BiP, which was limited to a perinuclear localization rather than showing scattered expression throughout the liver stage parasite, typically observed in the wild-type liver stages at this time point (Fig. 4). This result suggested that PlasMei2 plays an important role in liver stage endomitosis as early as 34 h after sporozoite invasion.
FIG 3.
Growth of wild-type and P. yoelii plasmei2– liver stages. (A) Livers from wild-type and plasmei2– parasite-infected mice were harvested, perfused with PBS, and fixed in 4% paraformaldehyde at various time points after sporozoite infection. Sections (50 μm) were cut from fixed livers, and IFAs were performed using primary antibody to the parasite parasitophorous vacuole membrane (PVM) marker Hep17 or the parasite plasma membrane marker circumsporozoite protein to delineate parasite size. Determination of approximate liver stage size (based on area at the parasites largest circumference) during development was made in order to compare wild-type and P. yoelii plasmei2– liver stages. At least 20 parasites were assessed at each time point. (B) Groups of five mice were injected with wild-type and plasmei2– sporozoites and luciferase activity (measured flux) was measured at 27, 44, and 70 h after injection. The dotted horizontal line shows background flux. (C) Liver stage viability based on compromised Hep17 expression was analyzed by IFA. If the circumferential expression pattern was incomplete, the parasite was considered nonviable. At 43 h, 42% of P. yoelii plasmei2– liver stage schizonts were nonviable compared to only 2% of wild-type parasites. Comparisons were performed by using an unpaired Student t test; significance is indicated by asterisks (**, P < 0.01). Lack of significance (P > 0.05) was seen in all size comparisons at each time point in panel A and is indicated by “ns” in panels B and C.
FIG 4.
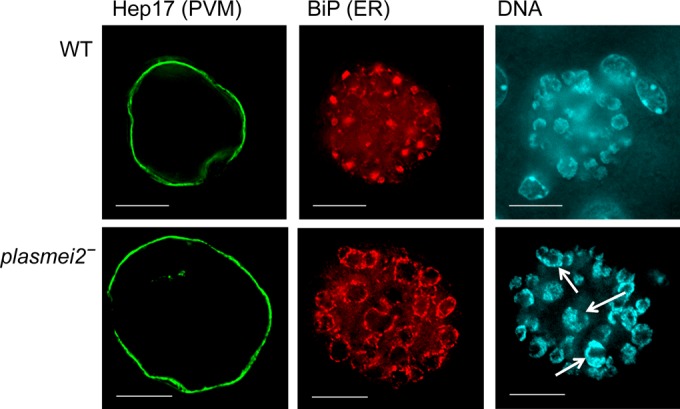
Earliest developmental defects in P. yoelii plasmei2– liver stage parasites. Livers from wild-type (WT) and plasmei2– parasite-infected mice were harvested, perfused with PBS, and fixed in 4% paraformaldehyde at 34 h after sporozoite infection. Sections (50 μm) were cut from fixed livers, and IFAs were performed using primary antibodies to the parasite parasitophorous vacuole membrane (PVM) marker Hep17 (green) to delineate the parasite and BiP (red) to observe the development of the endoplasmic reticulum (ER). DNA was labeled with DAPI (cyan). Compared to wild-type parasites, in which the ER does not enclose the nuclear areas (top panel), the P. yoelii plasmei2– parasites (bottom panel) show an aberrant ER organization that is largely perinuclear and also shows some enlarged nuclear centers (white arrows). The images were created from a single deconvolved z-stack. Scale bar, 10 μm.
The phenotype was far more pronounced at the 45-h time point, and it was clear that normal DNA segregation during endomitosis had ceased, whereas in wild-type parasites, DNA segregation, which precedes exoerythrocytic merozoite formation, had occurred (Fig. 5A). The P. yoelii plasmei2– liver stage mitochondria and apicoplasts also showed signs of unusual development. In the wild-type parasite complex, branched organelle networks were present, as previously described (32), but in the P. yoelii plasmei2– liver stages, the organellar network showed clear signs of abnormal development (Fig. 5B). To determine whether the lack of PlasMei2 also led to liver stage death during development, we estimated the viability of plasmei2– liver stages at 34 and 43 h of development compared to the wild-type liver stages. At 34 h no difference was seen, but at 43 h the percentage of nonviable parasites (based on compromised expression of the parasitophorous vacuole membrane marker Hep17) for P. yoelii plasmei2– was far higher than for the wild type (42% nonviable for P. yoelii plasmei2– versus 2% nonviable for the wild type) (Fig. 3C), and by 52 h no viable P. yoelii plasmei2– parasites were detected (data not shown).
FIG 5.
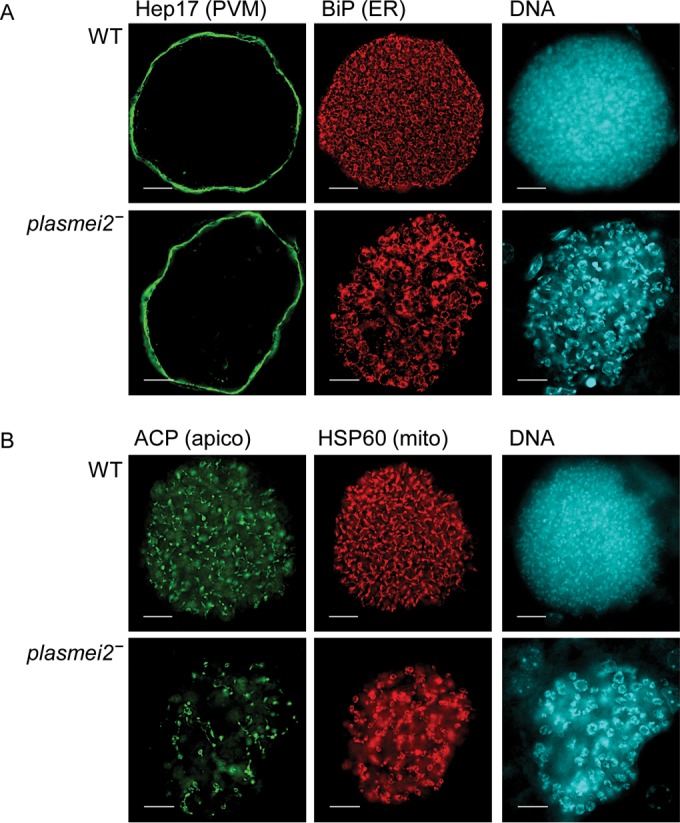
Late developmental defects of P. yoelii plasmei2– liver stage parasites. Livers from wild-type (WT) and plasmei2– parasite-infected mice were harvested, perfused with PBS, and fixed in 4% paraformaldehyde at 45 h after sporozoite infection. Sections (50 μm) were cut from fixed livers, and IFAs were performed using primary antibodies to the parasite parasitophorous vacuole membrane (PVM) marker Hep17 (green) to delineate the parasite, BiP (red) to visualize the endoplasmic reticulum (ER), acyl carrier protein (ACP) to visualize the apicoplast, and HSP60 (red) to visualize the mitochondria. DNA was stained with DAPI (cyan). (A and B) P. yoelii plasmei2– liver stages show anomalous endomitosis with enlarged nuclear centers and lack of normal DNA segregation compared to WT parasites. The ER shows aberrant development (A) and organellar development of apicoplasts and mitochondria appear to be affected (B). The images were created from a single deconvolved z-stack. Scale bar, 10 μm.
Maturation of liver stage parasites is characterized by cytomere formation; cytomeres are invaginations of the parasite plasma membrane that greatly increase the membrane surface area and enable rapid formation of exoerythrocytic merozoites by individuation throughout the exoerythrocytic schizont (33). To determine whether cytomere and merozoite formation were occurring in P. yoelii plasmei2– liver stages, we analyzed expression of merozoite surface protein 1 (MSP1), which is expressed on the parasite plasma membranes of the cytomeres and individual merozoites. P. yoelii plasmei2– liver stages at 43 and 48 h of development did not show cytomere formation and did not contain mature merozoites. MSP1 expression was weak, although some parasites showed strong, aggregated expression of MSP1 within the parasite (Fig. 6). These data show that although the P. yoelii plasmei2– liver stages appears to grow normally in size during development, they do not undergo normal DNA segregation and mitochondrial and apicoplast maturation, and they also do not undergo cytomere formation. Ultimately, lack of PlasMei2 expression abolishes exoerythrocytic merozoite formation.
FIG 6.
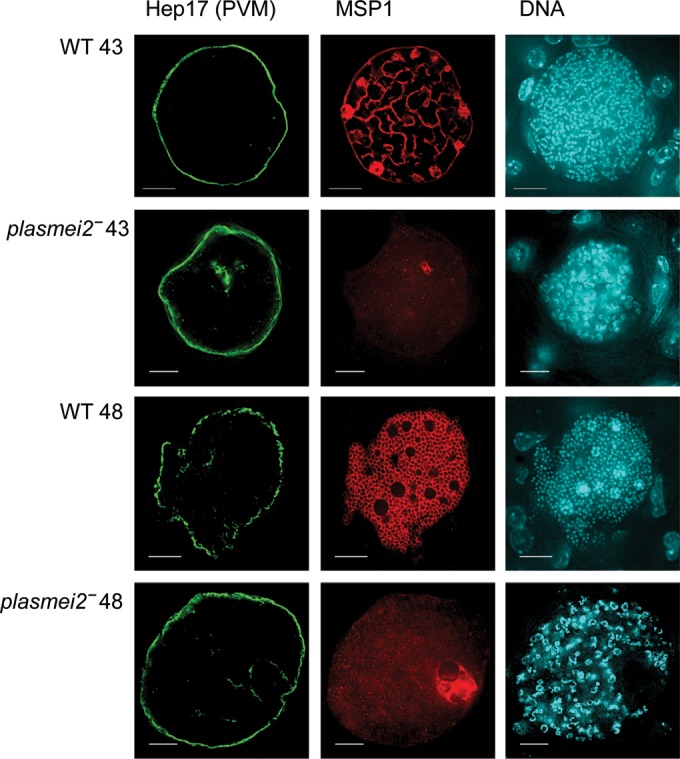
Disruption of cytomere formation and merozoite morphogenesis in P. yoelii plasmei2– liver stage parasites. Livers from wild-type (WT) and plasmei2– parasite-infected mice were harvested, perfused with PBS, and fixed in 4% paraformaldehyde at 43 and 48 h after sporozoite infection. Sections (50 μm) were cut from fixed livers, and IFAs were performed using primary antibodies to merozoite surface protein 1 (MSP1; red) to visualize the liver stage plasma membrane for analysis of cytomere formation and merozoite formation. Hep17 (green) visualizes the parasitophorous vacuole membrane (PVM). DNA was stained with DAPI (cyan). Cytomere formation is occurring in the WT parasites at 43 h (first panel), and individual exoerythrocytic merozoites are clearly differentiating at 48 h (third panel). P. yoelii plasmei2– liver stages show strongly reduced and abnormal MSP1 expression and lack of cytomere formation and exoerythrocytic merozoite differentiation at 43 and 48 h, respectively (second and fourth panels). Note the aberrant DNA segregation in plasmei2– liver stages. The images were created from a single deconvolved z-stack. Scale bar, 10 μm.
Protection by immunization with plasmei2– sporozoites.
Attenuated parasites that arrest their development in the liver engender potent protective immune responses that completely protect against wild-type sporozoite challenge (34). To determine whether P. yoelii plasmei2– could confer protection, groups of BALB/cJ mice were immunized three times with 10,000 P. yoelii plasmei2– sporozoites (Table 2). All immunized mice remained blood stage negative after immunization, demonstrating again the lack of breakthrough blood stage infection. Immunized mice and naive controls were then challenged with 10,000 wild-type sporozoites; while all control mice developed blood stage parasitemia, all immunized mice were completely protected, and a subset of mice, after a rechallenge with wild-type sporozoites, remained protected (Table 2).
TABLE 2.
Immunization with P. yoelii plasmei2− sporozoites protects against infectiona
| Prime | Boost |
Challengeb (days after last immunization) |
Protectionc
(after challenge) |
Rechallenge (days after challenge) |
Protection (after rechallenge) |
|
|---|---|---|---|---|---|---|
| 1 (days after prime) | 2 (days after boost 1) | |||||
| 10,000 | 10,000 (35) | 10,000 (45) | 46 | 9/9 | 56 | 9/9 |
| 10,000 | 10,000 (31) | 10,000 (45) | 39 | 12/12 | ||
| Naive | 46 | 0/10 | ||||
Primes and boosts were performed via intravenous injection of sporozoites. Naive mice were immunized with salivary gland extract from uninfected mosquitoes and were not rechallenged since they all became patent after the first challenge.
Challenges were performed by intravenous injection of 10,000 wild-type sporozoites.
Protection was evaluated by Giemsa-stained thin smear for blood stage patency from days 3 through 14 after challenge. A mouse was considered protected if it remained nonpatent for the duration of the 14-day time course. Protection is expressed as the number of mice protected/total number of mice tested.
DISCUSSION
In the present study, we show that a liver stage-specific RBP, PlasMei2, has an essential role in the completion of endomitosis during liver stage schizogony. However, PlasMei2 is not required for blood stage schizogony, highlighting that liver stage schizogony is regulated by unique factors. Although the lack of PlasMei2 had no effect on asexual or sexual blood stage development in P. yoelii, evidence exists for PlasMei2 transcription in the mature P. falciparum gametocyte (35), suggesting that PlasMei2 may be required for P. falciparum gametocyte maturation, although this remains to be confirmed. The essentiality of PlasMei2 expression in the liver is likely necessitated by the far greater magnitude of liver stage parasite replication, compared to the blood stage, requiring an even more tightly regulated process to ensure proper endomitosis and the coordinated maturation of tens of thousands of exoerythrocytic merozoites. Also, there could be qualitative differences between merozoite maturation in the erythrocyte host cell versus the hepatocyte that require further checkpoints of developmental control. Cyclins are a family of proteins important for cell cycle progression and recently, a Plasmodium P-type cyclin, CYC3 was shown to be critical for normal endomitosis during oocyst development but not necessary for liver stage development (36). Since PlasMei2 also appears to play a role in endomitosis, it is tempting to hypothesize that liver stage-specific expression of a cyclin other than CYC3 could work alongside PlasMei2 to coordinate liver stage development.
The difficulty in obtaining large numbers of liver stage parasites has thus far precluded us from performing immunoprecipitation analyses to determine the RNA and proteins that potentially interact with PlasMei2. Such analyses have been extremely helpful in identifying both RNA transcripts that bind to Plasmodium RBPs in the case of P. falciparum Alba1 (19), and proteins that are part of the P-body nucleoprotein complex, in the case of the P. berghei DOZI (15), which interestingly contains Alba1. Further studies might uncover both the RNAs and the proteins that interact with PlasMei2, and this information should further inform the molecular mechanisms by which PlasMei2 exerts its essential role in liver stage development.
The canonical Mei2 family proteins contain two N-terminal RRM_1 motifs and a C-terminal RRM_2, whereas PlasMei2 contains only the RRM_2. In the fission yeast Schizosaccharomyces pombe, Mei2 forms a dot structure in the nucleus, together with a noncoding meiRNA, resulting in the stabilization of meiosis-specific transcripts (37), and its expression regulates a mitosis-meiosis switch. Although the PlasMei2 contains just an RRM_2, it is tempting to hypothesize that an evolutionarily conserved function links the mitotic-meiotic switch in S. pombe to the endomitotic replication-DNA segregation switch in P. yoelii. In plants, however, the family of Mei2 proteins, known as AML and TEL, act in a different biological context to regulate various aspects of developmental pattern formation (21). Thus, since Mei2-like proteins have such divergent functions, it is not possible to reasonably predict the function of PlasMei2. However, PlasMei2 does have orthologs in other apicomplexans, including Toxoplasma gondii, Neospora caninum, Hammondia hammondi, and Gregarina niphandrodes, as well as other alveolates, including Vitrella brassicaformis and Perkinsus marinus. Therefore, conservation of Mei2 has been maintained throughout alveolate evolution, although the function of Mei2 in any alveolate has yet to be determined.
The disparity between mRNA profiles and protein expression during asexual blood stage development in Plasmodium (11, 12) suggests a role of RNA regulation by RBPs in controlling translation. Recently, it was elegantly shown that the RBP P. falciparum Alba1 plays a critical role in regulating the translation of RNA transcripts necessary for the formation of the invasive organelles of merozoites, demonstrating the importance of RBPs in the coordinated maturation of the asexual blood stage schizont (19). Although P. falciparum Alba1 appears to be specifically involved in the maturation of the merozoite, it is expressed throughout the parasite's erythrocytic cycle (19), suggesting that it plays a role in RNA homeostasis throughout blood stage development. Alba1 is also expressed in sporozoites (38) and liver stages (31), suggesting that it has multiple roles in the parasite life cycle. It is possible that Alba1 function in liver stages is linked to PlasMei2. PlasMei2 is not expressed early in liver stage development but well expressed at 30 h into liver stage development. This corresponds well with the occurrence of the plasmei2– phenotype, which is first observable at 34 h of development. Interestingly, we did not see evidence for PlasMei2 expression at the end of liver stage maturation, suggesting that it is necessary for the proper orchestration of endomitosis but not the completion of merozoite formation or for merozoite egress.
Genetically attenuated Plasmodium parasites that arrest during liver stage development are powerful immunogens and in experimental vaccination provide complete protection from infectious sporozoite challenge (34). We have shown here that immunization with plasmei2– parasites also confers complete protection against wild-type sporozoite challenge. Interestingly, liver stage parasites that arrest late in development, such as FASII mutants, provide superior protection compared to parasites that arrest early (39). Late liver stage arresting parasites, owing to their expression of cross-stage-specific antigens in late-liver stage development, also engender protection against blood stage parasite challenge (40). Thus, it is possible that the later the attenuated liver stage arrests, the better the overall immune response will be at protecting from both liver- and blood stage parasites. In the present study, we have generated a late liver stage arresting parasite fundamentally different from the FASII mutants; P. yoelii plasmei2– liver stages are the same size as their wild-type counterparts but are cleared from the liver by 52 h, whereas FASII mutants are significantly smaller (41). Further studies will determine whether P. yoelii plasmei2– liver stages are superior immunogens and, more importantly, whether P. falciparum plasmei2– has a similar phenotype to P. yoelii plasmei2– since the creation of a late liver stage arresting P. falciparum parasite has been elusive (42). Indeed, since a late liver stage arresting P. falciparum parasite has the potential to be a more potent immunogen than an early liver stage arresting parasite, P. falciparum plasmei2– could play a significant role in the development of novel vaccines for malaria.
The phenotype of the P. yoelii plasmei2– parasites is unique; these parasites do not appear to suffer defects in liver stage growth but show a clear perturbation of endomitosis and differentiation into exoerythrocytic merozoites. This is in contrast to another parasite protein thought to be involved in mRNA homeostasis, SAP1, whose deletion results in a very early liver stage growth arrest (43, 44) caused by the premature destruction of a range of transcripts necessary for the initial phase of liver stage development (45). Conversely, the deletion of genes encoding enzymes of the type II fatty acid biosynthesis (FASII) pathway, like PlasMei2, causes a late arrest during liver stage development and a failure to produce exoerythrocytic merozoites but with an observable impact on liver stage growth (41, 46).
In summary, we have shown that the RBP PlasMei2 has an essential role in the endomitotic process that culminates in maturation of liver stage exoerythrocytic merozoites, and further studies will help to uncover the precise molecular mechanism by which Mei2 exerts its function through interaction with liver stage RNAs. Of equal importance is the pursuit of Mei2 as a target for late-liver stage attenuation, which could critically contribute to the development of a highly protective, whole-organism malaria vaccine.
Supplementary Material
ACKNOWLEDGMENTS
We thank Center for Infectious Disease Research insectary employees Will Betz and Heather Kain for help in maintaining parasites in the mosquito vector. We also thank Center for Infectious Disease Research vivarium staff for help with rodent maintenance.
This study was funded through charitable donations to the Center for Infectious Disease Research.
S.H.I.K. and A.M.V. conceived the experimental plan. D.A.D., M.J.D., and A.M.V. carried out the experiments. S.H.I.K., A.M.V., and D.A.D. wrote the paper.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01417-15.
REFERENCES
- 1.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozdech Z, Zhu J, Joachimiak MP, Cohen FE, Pulliam B, DeRisi JL. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol 4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel TN, Hon CC, Zhang Q, Lopez-Rubio JJ, Scheidig-Benatar C, Martins RM, Sismeiro O, Coppee JY, Scherf A. 2014. Strand-specific RNA-seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum. BMC Genomics 15:150. doi: 10.1186/1471-2164-15-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Painter HJ, Campbell TL, Llinas M. 2011. The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol Biochem Parasitol 176:1–7. doi: 10.1016/j.molbiopara.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riechmann JL, Meyerowitz EM. 1998. The AP2/EREBP family of plant transcription factors. Biol Chem 379:633–646. [DOI] [PubMed] [Google Scholar]
- 6.Kafsack BF, Rovira-Graells N, Clark TG, Bancells C, Crowley VM, Campino SG, Williams AE, Drought LG, Kwiatkowski DP, Baker DA, Cortes A, Llinas M. 2014. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507:248–252. doi: 10.1038/nature12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinha A, Hughes KR, Modrzynska KK, Otto TD, Pfander C, Dickens NJ, Religa AA, Bushell E, Graham AL, Cameron R, Kafsack BF, Williams AE, Llinas M, Berriman M, Billker O, Waters AP. 2014. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature 507:253–257. doi: 10.1038/nature12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuda M, Iwanaga S, Shigenobu S, Mair GR, Janse CJ, Waters AP, Kato T, Kaneko I. 2009. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol Microbiol 71:1402–1414. doi: 10.1111/j.1365-2958.2009.06609.x. [DOI] [PubMed] [Google Scholar]
- 9.Yuda M, Iwanaga S, Shigenobu S, Kato T, Kaneko I. 2010. Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol Microbiol 75:854–863. doi: 10.1111/j.1365-2958.2009.07005.x. [DOI] [PubMed] [Google Scholar]
- 10.Iwanaga S, Kaneko I, Kato T, Yuda M. 2012. Identification of an AP2-family protein that is critical for malaria liver-stage development. PLoS One 7:e47557. doi: 10.1371/journal.pone.0047557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foth BJ, Zhang N, Mok S, Preiser PR, Bozdech Z. 2008. Quantitative protein expression profiling reveals extensive posttranscriptional regulation and posttranslational modifications in schizont-stage malaria parasites. Genome Biol 9:R177. doi: 10.1186/gb-2008-9-12-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, Yates JR III, Winzeler EA. 2004. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res 14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Chua NH. 2011. Processing bodies and plant development. Curr Opin Plant Biol 14:88–93. doi: 10.1016/j.pbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mair GR, Braks JA, Garver LS, Wiegant JC, Hall N, Dirks RW, Khan SM, Dimopoulos G, Janse CJ, Waters AP. 2006. Regulation of sexual development of Plasmodium by translational repression. Science 313:667–669. doi: 10.1126/science.1125129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mair GR, Lasonder E, Garver LS, Franke-Fayard BM, Carret CK, Wiegant JC, Dirks RW, Dimopoulos G, Janse CJ, Waters AP. 2010. Universal features of posttranscriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog 6:e1000767. doi: 10.1371/journal.ppat.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindner SE, Mikolajczak SA, Vaughan AM, Moon W, Joyce BR, Sullivan WJ Jr, Kappe SH. 2013. Perturbations of Plasmodium Puf2 expression and RNA-seq of Puf2-deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell Microbiol 15:1266–1283. doi: 10.1111/cmi.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller K, Matuschewski K, Silvie O. 2011. The Puf-family RNA-binding protein Puf2 controls sporozoite conversion to liver stages in the malaria parasite. PLoS One 6:e19860. doi: 10.1371/journal.pone.0019860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chene A, Vembar SS, Riviere L, Lopez-Rubio JJ, Claes A, Siegel TN, Sakamoto H, Scheidig-Benatar C, Hernandez-Rivas R, Scherf A. 2012. PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites. Nucleic Acids Res 40:3066–3077. doi: 10.1093/nar/gkr1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vembar SS, Macpherson CR, Sismeiro O, Coppee JY, Scherf A. 2015. The PfAlba1 RNA-binding protein is an important regulator of translational timing in Plasmodium falciparum blood stages. Genome Biol 16:212. doi: 10.1186/s13059-015-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egel R, Nielsen O, Weilguny D. 1990. Sexual differentiation in fission yeast. Trends Genet 6:369–373. doi: 10.1016/0168-9525(90)90279-F. [DOI] [PubMed] [Google Scholar]
- 21.Jeffares DC, Phillips MJ, Moore S, Veit B. 2004. A description of the Mei2-like protein family; structure, phylogenetic distribution and biological context. Dev Genes Evol 214:149–158. doi: 10.1007/s00427-004-0384-6. [DOI] [PubMed] [Google Scholar]
- 22.Lin JW, Annoura T, Sajid M, Chevalley-Maurel S, Ramesar J, Klop O, Franke-Fayard BM, Janse CJ, Khan SM. 2011. A novel “gene insertion/marker out” (GIMO) method for transgene expression and gene complementation in rodent malaria parasites. PLoS One 6:e29289. doi: 10.1371/journal.pone.0029289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janse CJ, Ramesar J, Waters AP. 2006. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat Protoc 1:346–356. doi: 10.1038/nprot.2006.53. [DOI] [PubMed] [Google Scholar]
- 24.Lindner SE, Sartain MJ, Hayes K, Harupa A, Moritz RL, Kappe SH, Vaughan AM. 2014. Enzymes involved in plastid-targeted phosphatidic acid synthesis are essential for Plasmodium yoelii liver-stage development. Mol Microbiol 91:679–693. doi: 10.1111/mmi.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, Cowman AF, McFadden GI. 2004. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol 137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Kaushansky A, Austin LS, Mikolajczak SA, Lo FY, Miller JL, Douglass AN, Arang N, Vaughan AM, Gardner MJ, Kappe SH. 2015. Susceptibility to Plasmodium yoelii preerythrocytic infection in BALB/c substrains is determined at the point of hepatocyte invasion. Infect Immun 83:39–47. doi: 10.1128/IAI.02230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franke-Fayard B, Waters AP, Janse CJ. 2006. Real-time in vivo imaging of transgenic bioluminescent blood stages of rodent malaria parasites in mice. Nat Protoc 1:476–485. doi: 10.1038/nprot.2006.69. [DOI] [PubMed] [Google Scholar]
- 28.Mwakingwe A, Ting LM, Hochman S, Chen J, Sinnis P, Kim K. 2009. Noninvasive real-time monitoring of liver-stage development of bioluminescent Plasmodium parasites. J Infect Dis 200:1470–1478. doi: 10.1086/606115. [DOI] [PubMed] [Google Scholar]
- 29.Ploemen IH, Prudencio M, Douradinha BG, Ramesar J, Fonager J, van Gemert GJ, Luty AJ, Hermsen CC, Sauerwein RW, Baptista FG, Mota MM, Waters AP, Que I, Lowik CW, Khan SM, Janse CJ, Franke-Fayard BM. 2009. Visualization and quantitative analysis of the rodent malaria liver stage by real-time imaging. PLoS One 4:e7881. doi: 10.1371/journal.pone.0007881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy BP, Shrestha S, Hart KJ, Liang X, Kemirembe K, Cui L, Lindner SE. 2015. A bioinformatic survey of RNA-binding proteins in Plasmodium. BMC Genomics 16:890. doi: 10.1186/s12864-015-2092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, Daly TM, Bergman LW, Kappe SH. 2008. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci U S A 105:305–310. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanway RR, Mueller N, Zobiak B, Graewe S, Froehlke U, Zessin PJ, Aepfelbacher M, Heussler VT. 2011. Organelle segregation into Plasmodium liver-stage merozoites. Cell Microbiol 13:1768–1782. doi: 10.1111/j.1462-5822.2011.01657.x. [DOI] [PubMed] [Google Scholar]
- 33.Graewe S, Stanway RR, Rennenberg A, Heussler VT. 2012. Chronicle of a death foretold: Plasmodium liver-stage parasites decide on the fate of the host cell. FEMS Microbiol Rev 36:111–130. doi: 10.1111/j.1574-6976.2011.00297.x. [DOI] [PubMed] [Google Scholar]
- 34.Butler NS, Vaughan AM, Harty JT, Kappe SH. 2012. Whole parasite vaccination approaches for prevention of malaria infection. Trends Immunol 33:247–254. doi: 10.1016/j.it.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Barragan MJ, Lemieux J, Quinones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, Su XZ. 2011. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12:587. doi: 10.1186/1471-2164-12-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roques M, Wall RJ, Douglass AP, Ramaprasad A, Ferguson DJ, Kaindama ML, Brusini L, Joshi N, Rchiad Z, Brady D, Guttery DS, Wheatley SP, Yamano H, Holder AA, Pain A, Wickstead B, Tewari R. 2015. Plasmodium P-type cyclin CYC3 modulates endomitotic growth during oocyst development in mosquitoes. PLoS Pathog 11:e1005273. doi: 10.1371/journal.ppat.1005273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harigaya Y, Yamamoto M. 2007. Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res 15:523–537. doi: 10.1007/s10577-007-1151-0. [DOI] [PubMed] [Google Scholar]
- 38.Lindner SE, Swearingen KE, Harupa A, Vaughan AM, Sinnis P, Moritz RL, Kappe SH. 2013. Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol Cell Proteomics 12:1127–1143. doi: 10.1074/mcp.M112.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler NS, Schmidt NW, Vaughan AM, Aly AS, Kappe SH, Harty JT. 2011. Superior antimalarial immunity after vaccination with late liver stage-arresting genetically attenuated parasites. Cell Host Microbe 9:451–462. doi: 10.1016/j.chom.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sack BK, Keitany GJ, Vaughan AM, Miller JL, Wang R, Kappe SH. 2015. Mechanisms of stage-transcending protection following immunization of mice with late liver stage-arresting genetically attenuated malaria parasites. PLoS Pathog 11:e1004855. doi: 10.1371/journal.ppat.1004855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaughan AM, O'Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, Cowman AF, Kappe SH. 2009. Type II fatty acid synthesis is essential only for malaria parasite late liver-stage development. Cell Microbiol 11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan SM, Janse CJ, Kappe SH, Mikolajczak SA. 2012. Genetic engineering of attenuated malaria parasites for vaccination. Curr Opin Biotechnol 23:908–916. doi: 10.1016/j.copbio.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Aly AS, Mikolajczak SA, Rivera HS, Camargo N, Jacobs-Lorena V, Labaied M, Coppens I, Kappe SH. 2008. Targeted deletion of SAP1 abolishes the expression of infectivity factors necessary for successful malaria parasite liver infection. Mol Microbiol 69:152–163. doi: 10.1111/j.1365-2958.2008.06271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silvie O, Goetz K, Matuschewski K. 2008. A sporozoite asparagine-rich protein controls initiation of Plasmodium liver-stage development. PLoS Pathog 4:e1000086. doi: 10.1371/journal.ppat.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aly AS, Lindner SE, MacKellar DC, Peng X, Kappe SH. 2011. SAP1 is a critical posttranscriptional regulator of infectivity in malaria parasite sporozoite stages. Mol Microbiol 79:929–939. doi: 10.1111/j.1365-2958.2010.07497.x. [DOI] [PubMed] [Google Scholar]
- 46.Pei Y, Tarun AS, Vaughan AM, Herman RW, Soliman JM, Erickson-Wayman A, Kappe SH. 2010. Plasmodium pyruvate dehydrogenase activity is only essential for the parasite's progression from liver infection to blood infection. Mol Microbiol 75:957–971. doi: 10.1111/j.1365-2958.2009.07034.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.