Abstract
Members of the Burkholderia cepacia complex (Bcc) cause chronic opportunistic lung infections in people with cystic fibrosis (CF), resulting in a gradual lung function decline and, ultimately, patient death. The Bcc is a complex of 20 species and is rarely eradicated once a patient is colonized; therefore, vaccination may represent a better therapeutic option. We developed a new proteomics approach to identify bacterial proteins that are involved in the attachment of Bcc bacteria to lung epithelial cells. Fourteen proteins were reproducibly identified by two-dimensional gel electrophoresis from four Bcc strains representative of two Bcc species: Burkholderia cenocepacia, the most virulent, and B. multivorans, the most frequently acquired. Seven proteins were identified in both species, but only two were common to all four strains, linocin and OmpW. Both proteins were selected based on previously reported data on these proteins in other species. Escherichia coli strains expressing recombinant linocin and OmpW showed enhanced attachment (4.2- and 3.9-fold) to lung cells compared to the control, confirming that both proteins are involved in host cell attachment. Immunoproteomic analysis using serum from Bcc-colonized CF patients confirmed that both proteins elicit potent humoral responses in vivo. Mice immunized with either recombinant linocin or OmpW were protected from B. cenocepacia and B. multivorans challenge. Both antigens induced potent antigen-specific antibody responses and stimulated strong cytokine responses. In conclusion, our approach identified adhesins that induced excellent protection against two Bcc species and are promising vaccine candidates for a multisubunit vaccine. Furthermore, this study highlights the potential of our proteomics approach to identify potent antigens against other difficult pathogens.
INTRODUCTION
Vaccination is the most effective medical intervention introduced. In the context of the global rise in antimicrobial resistance, vaccines are essential weapons in the fight against bacterial infections. Vaccines do not pose massive selection pressure on the environment, nor do they contribute to antimicrobial resistance (1). However, identification of good vaccine antigens remains challenging. To date, several strategies that identify effective vaccine antigens have been described, including the reverse-vaccinology approach (2). Rappuoli and colleagues pioneered the use of reverse vaccinology to identify novel antigens against Neisseria meningitidis serogroup B. They sequenced the genome, identified 350 surface proteins, and administered these proteins to mice to identify those proteins that were immunogenic (3). This predictive approach assumes that proteins that are able to induce protective immunity are located outside the cell membrane and therefore possess signal sequences (4). Immunoproteomics has also been used to identify novel antigens that elicit an immune response, as recently reviewed (5), but when used in isolation, it has limitations, and no efficacious antigens have yet been identified by using this approach. Indeed, the confirmed prophylactic Bordetella pertussis antigen filamentous hemagglutinin (FHA), a component of most licensed acellular whooping cough vaccines, was undetectable in two immunoproteomic studies (6, 7).
We have developed a novel proteomic-based strategy to identify bacterial adhesins that are involved in host cell attachment and demonstrated that two of these adhesins were protective against the Burkholderia cepacia complex (Bcc). This bacterial pathogen complex comprises a group of 20 species of Gram-negative bacteria (8–11), 2 of which, Burkholderia multivorans and B. cenocepacia, are the most clinically relevant (12, 13). Members of the Bcc are reported to cause infections in up to 5% of cystic fibrosis (CF) patients in the world, which is significant, as CF patients colonized with Bcc experience a more rapid decline than do those colonized with the more commonly acquired pathogen Pseudomonas aeruginosa (14, 15). Once a patient is colonized with Bcc bacteria, these bacteria are rarely eradicated due to the resistance of the Bcc to antibiotics (16) and antimicrobial peptides (17, 18). Strict segregation measures have limited the patient-to-patient spread of the most virulent species, B. cenocepacia (19). Currently, the majority of new acquisitions are from the environment, with B. multivorans being the most frequently acquired (20); therefore, the Bcc still represents a substantial threat to CF patients. B. cenocepacia is subdivided into four clusters by phylogenetic analysis of the recA gene sequence (subgroups IIIA, IIIB, IIIC, and IIID) (21). While all four groups include clinical isolates, subgroup IIIA is associated with more epidemic strains, which have a higher mortality rate than that associated with other B. cenocepacia groups (22). Moreover, Bcc contamination of pharmaceutical formulations, medical devices, and disinfectants has led to a number of outbreaks among both CF and non-CF populations (22). Bcc is also an emerging pathogen in nosocomial infections among chemotherapy patients and other immunosuppressed individuals (23, 24). The high level of antibiotic resistance combined with the continued acquisition of Bcc bacteria from the environment suggests that prevention of infection with a prophylactic vaccine may be a better approach than eradication of existing infections. Only two in vivo mouse vaccination studies have reported protection against the Bcc, both of which involved unpurified outer membrane protein (OMP) preparations (25, 26). No vaccine antigens have been identified for the Bcc to date.
The majority of mucosal pathogens colonize by attaching to host cells and/or host proteins. Previous work in our laboratory has shown that Bcc attaches laterally to the surfaces of epithelial cells, prior to invasion inside the cells (27). Proteins that are involved in bacterial attachment to host cells were previously proven to be excellent vaccine antigens. A classic example is Bordetella pertussis FHA, which is involved in attachment to epithelial cells of the airways (28). FHA has been combined with other proteins with adhesin properties (pertactin, pertussis toxin, and fimbriae 2 and 3) in approved prophylactic vaccines against whooping cough (29).
Little is known about how Bcc attaches to lung epithelial cells. A 22-kDa cable pilus protein was identified as an adhesin; however, it is expressed in only a subset of B. cenocepacia strains, i.e., piliated strains of the subgroup IIIA lineage only (30), and is not expressed in the more frequently acquired species B. multivorans. More recently, Mil-Homens et al. identified two closely related trimeric autotransporters that are involved in B. cenocepacia adhesion to lung epithelial cells (31, 32). We have developed a proteomics approach to identify other bacterial proteins that are involved in the attachment of Bcc to lung epithelial cells in vitro by probing two-dimensional (2D) blots of bacterial proteins with epithelial cells. Several of the identified proteins were subsequently found to be immunoreactive by using sera from Bcc-colonized CF patients. Two of these proteins protected immunized mice from Bcc infection. This approach should also allow the identification of a wide range of adhesins in other bacteria and consequently enable the development of potent vaccine antigens for a range of mucosal pathogens against which there are no protective vaccines to date.
MATERIALS AND METHODS
Ethics statement.
Animal immunizations were performed in compliance with animal protection legislation (Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes) (65). Authorization for immunizations was obtained from the Irish Department of Health and the Health Products Regulation Authority (HRPRA). Animals were maintained and all studies were performed in accordance with regulations of the Irish Department of Health and the HPRA and with the approval of the Maynooth University Ethics Committee. Patient consent was obtained for the use of serum, and ethics approval was obtained from the St. Vincent's University Hospital (SVUH) Ethics and Medical Research Committee and from the ITT Dublin Research and Ethics Committee for the use of stored serum.
Bacterial strains and culture conditions.
To enhance coverage across the two most clinically relevant species, two strains of each species were analyzed, including piliated and nonpiliated B. cenocepacia strains (BC7 and C1394, respectively). The four bacterial strains used, the two B. multivorans strains LMG13010 and C1962 and the two B. cenocepacia strains BC7 (recA IIIA lineage) and C1394 (recA IIIB lineage), are detailed in Table 1 and were obtained from the BCCM/LMG, University of Ghent, Ghent, Belgium, and routinely plated onto B. cepacia-specific agar (BCSA) (33). All bacterial strains were routinely grown in Luria-Bertani (LB) broth at 37°C with orbital shaking (150 rpm). To isolate sufficient quantities of membrane proteins, bacterial cultures were grown in a 10-liter fermentor. Five hundred milliliters of bacterial cultures grown overnight in a 37°C incubator on a shaker at 150 rpm was inoculated into 10 liters of LB medium. The culture was grown at 37°C at 150 rpm without a pH control or antifoam control, with an air supply of 6 liters/min. Bacterial cultures in stationary phase (optical density [OD] of 1.0) were used for OMP extraction. All spent media and the cell pellet were autoclaved after protein extraction.
TABLE 1.
Strains used in this study and their sources
Cell culture.
16HBE14o− and CFBE41o− lung epithelial cells were generous gifts from Dieter Gruenert (UCSF). The human bronchial epithelial cell line 16HBE14o− was routinely grown in Eagle's minimal essential medium (EMEM) with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 1% l-glutamine, and 1% sodium pyruvate and incubated in a 5% CO2 environment at 37°C (34). CFBE41o− cells were routinely grown in minimal essential medium with 10% FBS, 1% penicillin-streptomycin, 1% l-glutamine, 1% sodium pyruvate, and 1% nonessential amino acids and incubated in a 5% CO2 environment at 37°C.
Attachment of Bcc to epithelial cells.
In order to quantitate the attachment of Bcc strains to lung epithelial cells, 16HBE14o− or CFBE41o− cells were seeded into coated chamber slides (4 × 105 cells/chamber) for 24 h prior to incubation individually with B. cenocepacia strain BC7 or C1394 or B. multivorans strain LMG13010 or C1962 for 30 min at a multiplicity of infection (MOI) of 50:1 or 10:1. Cells were gently washed three times with phosphate-buffered saline (PBS) for 5 min each and fixed with 3% (vol/vol) paraformaldehyde for 10 min at room temperature (RT). The cells were then incubated with 5% (wt/vol) bovine serum albumin (BSA) in PBS at room temperature for 1 h to block nonspecific binding. After blocking, the samples were incubated with primary rabbit polyclonal anti-Bcc antibody (1:1,000 dilution) overnight at 4°C (R418, a generous gift from Uma Sajjan) (35), and bound antibodies were detected with a fluorescein isothiocyanate (FITC)-labeled secondary antibody (1:100) for 1 h at room temperature in the dark. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 15 min at room temperature in the dark and visualized under fluorescence and confocal microscopes to count the bacteria attached to the cells.
Preparation of bacterial membrane proteins.
Stationary-phase (OD of 1.0) cultures were collected by centrifugation for membrane protein extraction as previously described (36). Briefly, after centrifugation at 5,000 × g for 10 min at 4°C, the cell pellets were resuspended in ice-cold PBS containing 5% (vol/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) with a protease inhibitor cocktail (Roche, Penzburg, Germany). Bacterial membranes were collected and washed in a series of five ultracentrifugation steps, and membrane pellets were finally resuspended in 50 mM Tris (pH 8) buffer with a protease inhibitor cocktail. The protein concentration was determined by a bicinchoninic acid assay and Bradford assays (37).
Cell attachment blots.
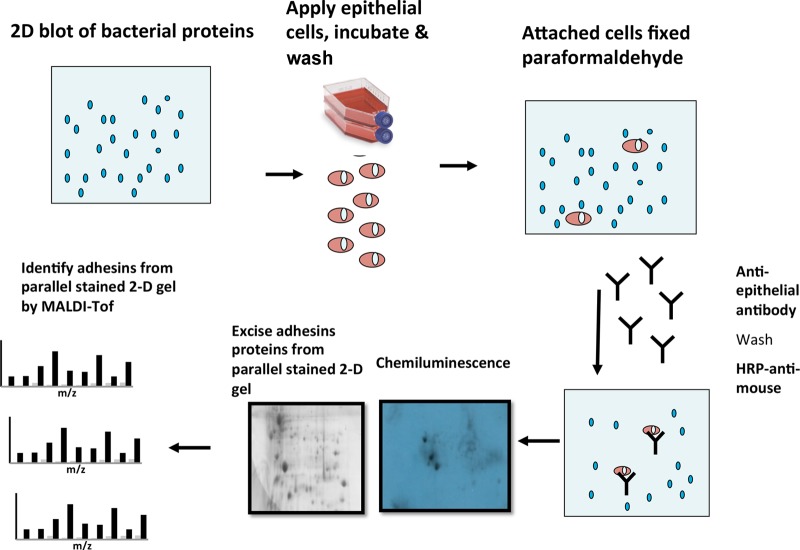
The membrane proteins were solubilized for isoelectric focusing (IEF) in a rehydration solution containing 8 M urea, 2 M thiourea, 4% CHAPS, 1% (vol/vol) Triton, 10 mM Tris base, 65 mM dithiothreitol (DTT), and 0.8% (vol/vol) immobilized pH gradient (IPG) buffer (pH3-11NL) (GE Life Sciences), and duplicate samples were separated on simultaneously run gels by two-dimensional electrophoresis as described previously (36). For each experiment, one gel was stained with Coomassie blue (PageBlue; Fermentas), while the other was transferred onto a polyvinylidene difluoride (PVDF) membrane after equilibration in transfer buffer (25 mM [wt/vol] Tris, 192 mM [wt/vol] glycine, 20% [vol/vol] methanol) (38) for 15 min. Protein transfer to an Immobilon-P membrane was carried out in a Transphor unit (Bio-Rad) at 330 mA for 50 min, and the membrane was blocked overnight with 5% BSA in PBS at 4°C on a shaker at 10 rpm. Lung epithelial cells (either CFBE41o− or 16HBE14o−) were scraped from flasks, washed with PBS, and incubated with the blots for 4 h at 37°C on a shaker at 60 rpm. The blots were rinsed with 20 ml of PBS, and the bound epithelial cells were fixed with 3% paraformaldehyde dissolved in PBS. The cells were detected with an anti-epithelial cell-specific antigen antibody (Millipore) by incubating the blots with the mouse anti-epithelial cell antibody (1:1,000 in 5% [wt/vol] BSA plus 0.04% [wt/vol] Tween 20 in PBS [PBS-T]) overnight at 4°C on a shaker at 10 rpm. The blots were washed three times and incubated with horseradish peroxidase (HRP)-conjugated anti-mouse antibody (1:40,000 in 5% [wt/vol] BSA plus 0.04% [wt/vol] PBS-T) at room temperature. Each blot was then washed five times, and chemiluminescence detection was carried out by using the Santa Cruz luminol detection kit. Each experiment was repeated at least twice with independent bacterial membrane preparations. An overview of this method is shown in Fig. 1.
FIG 1.
Schematic of the workflow used to identify Bcc adhesins from bacterial membrane proteins. The membrane proteins were analyzed by using 2D gels, and pairs of gels were either blotted onto PVDF membranes or stained with Coomassie blue. The blots were probed with lung epithelial cells, followed by detection of bound cells by antibodies. Positive spots on exposed chemiluminescent films were matched to the paired Coomassie blue-stained gels, and the proteins in the corresponding gel-based spots were excised, trypsin digested, and identified by MALDI-TOF MS analysis.
MALDI-TOF MS/MS analysis.
The developed chemiluminescent film was carefully overlaid onto the corresponding Coomassie blue-stained gel. Protein spots that corresponded to positive spots on the film were excised from stained gels and destained with an equal volume of 100 mM ammonium bicarbonate and acetonitrile. Gel digestion was performed with 13 ng/μl of modified porcine trypsin for 2 h on ice, followed by overnight incubation at 37°C. Tubes were cooled to room temperature, and gel pieces were pelleted. Aliquots of 2 to 2.5 μl of the supernatant were withdrawn directly from the digest for matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) tandem mass spectrometry (MS/MS) analysis, without further extraction of the gel pieces (39). MALDI-TOF MS/MS analysis was performed by using a Bruker Ultraflex MALDI-TOF instrument (Bruker Daltonics, Bremen, Germany) with a ground steel target plate as previously described (36).
Immunoblot analysis of membrane proteins from Bcc with CF patient sera.
Two-dimensional blots were prepared from Bcc membrane protein extracts as described above. The blots were blocked with 5% BSA, 2% Marvel dried milk powder overnight at 4°C before probing with pooled serum from seven CF patients who had been positively identified as having a Bcc infection or with sera from six CF patients who had no history of Bcc infection, which were used as negative controls (1:80,000), as previously described (36). The blots were then washed with PBS containing 0.05% Tween 20, before incubation with HRP-conjugated anti-human antibody (1:16,000; Roche), and washed again, and the immunoreactive proteins were detected by chemiluminescence as previously described (36). The data for linocin and OmpW are presented, highlighting the strains in which these two proteins were identified, together with the sequence coverage and Mascot scores obtained.
Cloning and expression of membrane protein.
The presence of each gene in the four strains was confirmed by isolating bacterial genomic DNA using the DNeasy blood and tissue kit (Qiagen, Manchester, England). Specific primers were designed according to linocin and OmpW sequences of the sequenced B. multivorans strain, ATCC 17616, from the NCBI. PCR amplification was carried out with a Hot Start high-fidelity PCR kit (Qiagen) using the following thermal cycle program: initial activation for 5 min at 95°C followed by 35 cycles consisting of denaturation at 94°C for 30 s, annealing at 50 to 68°C for 30 s, and extension at 72°C for 30 s. For the last cycle, the incubation period at 72°C was extended to 5 min and samples were then maintained at 4°C.
Both genes were cloned from B. multivorans LMG13010, as this species is currently the more commonly acquired species of the Bcc. Primers to amplify the genes were designed from the B. multivorans ATCC 17616 sequence, with CACC at the 5′ end to facilitate directional cloning. The forward and reverse primers used were 5′-CACCATGAACAATCTGCACCGCGAACTC and 5′-ATCAGGCGGGCGTGCCGGC for linocin and 5′-CACCATGCATAAACCAATGACA and 5′-CTAGAACTTCATCCCGACACC for OmpW (CACC sites are underlined). Following amplification of the genes by using AccuPrime Taq DNA polymerase, the products were confirmed by agarose gel electrophoresis and cloned into the pET 100 vector. Escherichia coli transformations were carried out with One Shot TOP10 competent cells by heat shock according to the manufacturer's protocol. Transformed cultures were then spread onto LB agar plates with ampicillin (100 μg/ml) and incubated overnight at 37°C, and the presence of the insert was confirmed by restriction digestion and agarose gel electrophoresis. BL21 Star(DE3) One Shot E. coli cells (Life Technologies) were used as the host for expression. Once expression was confirmed in a pilot study using anti-His tag detection in Western blots from 5-ml cultures, larger cultures of BL21 cells (1 liter to 1.5 liters) containing the plasmid of interest were grown in LB medium with ampicillin (100 μg/ml) and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cultures were then centrifuged at 5,000 rpm for 10 min, and the pellets were frozen at −80°C until purification was performed.
Purification of His-tagged recombinant protein.
The expressed bacterial proteins were extracted from bacterial pellets with the B-PER bacterial protein extraction reagent (Thermo Scientific) with an EDTA-free protease inhibitor cocktail (Roche) and incubated for 15 min at room temperature. The lysates were centrifuged at 15,000 × g for 5 min to separate soluble proteins from insoluble proteins. HisPur Ni-nitrilotriacetic acid (NTA) spin columns (Thermo Scientific) were used to purify polyhistidine-tagged proteins from the protein extract. The samples were mixed with an equal volume of equilibration buffer and spun down in the Ni-NTA column. The columns were washed three times with wash buffer containing 25 mM imidazole. After washing the column, the samples were then eluted with 250 mM imidazole, and the fractions were analyzed by SDS-PAGE. The purified recombinant proteins were desalted with a Zeba spin desalting column (Thermo Scientific, Dublin, Ireland). Detoxi gel endotoxin removal columns (Thermo Scientific) were used to remove the endotoxins from the recombinant proteins. The columns were regenerated with 1% sodium deoxycholate and washed with nonpyrogenic water. The protein samples were applied onto the column, and the flowthrough was collected and kept at −80°C. A ToxinSensor Gel Clot endotoxin assay kit (GenScript, Piscataway, NJ) was used to determine the endotoxin levels in the purified proteins. In each case, the endotoxin levels were <0.25 endotoxin units (EU)/ml.
Immunization of recombinant proteins.
All procedures involving animals were carried out by licensed personnel according to approved guidelines. Pathogen-free BALB/c mice (females 6 to 8 weeks old) were housed in groups of five under standard pathogen-free conditions, with food and water available. Mice (n = 5 per group) were vaccinated with two administrations of either antigen, 4 weeks apart. Intraperitoneal (i.p.) immunizations were performed with 50 μg of recombinant protein per mouse and an equal volume of Imject alum (Thermo Scientific Pierce, Rockford, IL). Bacterial challenge studies were performed 2 weeks following booster vaccination, with immunosuppression by i.p. injection of 50 μg of cyclophosphamide on days −1 and 4 of challenge (25). The immunized and nonimmunized mice were challenged with 4 × 107 B. cenocepacia BC7 or B. multivorans LMG13010 bacteria suspended in 20 μl of PBS instilled intranasally, and mice were maintained for 5 days before sacrifice by cervical dislocation. The lungs, spleen, and blood were collected under aseptic conditions, and each organ was placed into individual containers containing 1 ml of Ringer's solution. Blood was kept at RT for 4 h to clot and then centrifuged at 1,600 × g for 15 min to collect the serum. Aliquots of serum were then kept at −80°C for further analysis. The lungs and spleen were homogenized, serially diluted in Ringer's solution, and plated onto BCSA plates. All plates were then incubated for 72 h at 37°C prior to manual CFU enumeration.
Determination of antigen-specific antibodies by an ELISA.
An indirect enzyme-linked immunosorbent assay (ELISA) was used to evaluate the specific immunogenic effects of linocin and OmpW. Ninety-six-well Nunc-Immuno MaxiSorp assay plates were coated with 2 μg/well of purified antigen, either linocin or OmpW, in coating buffer (sodium bicarbonate, pH 9.4). After overnight incubation at 4°C, the plates were blocked with 10% BSA in PBS for 1 h at RT. Serial 4-fold dilutions of serum in 1% BSA were added (100 μl/well), and the plates were incubated for 2 h at room temperature. After four washes with PBS–0.05% Tween 20, horseradish peroxidase-conjugated goat anti-mouse IgG1, IgG2a, or IgG2b antibody (Abcam, Cambridge, United Kingdom) was added at a 1:5,000 dilution in 1% BSA. Following 2 h of incubation at RT, the plates were washed four times before tetramethylbenzidine substrate (Pierce, Thermo Scientific, Rockford, IL, USA) was added. Reactions were stopped after 20 min with 2 N H2SO4, and the absorbance was determined at a wavelength of 450 nm. Serum antibody titers were defined as endpoint titers, i.e., the reciprocal of the highest dilution of serum producing an OD above the cutoff value, where the cutoff values were determined as the OD of the corresponding dilution of control sera plus 3 standard deviations.
Visualization of recombinant bacterial cell attachment to lung cells.
CFBE41o− cells were seeded into chamber slides (Lab-Tek; Thermo Scientific) 24 h prior to experiments. E. coli BL21 control and E. coli BL21 recombinant protein-expressing strains at an OD at 600 nm (OD600) of 0.6 were treated by the addition of IPTG (1 mM) to all three cultures for 3 h to induce recombinant protein expression. An aliquot of 500 μl was removed to confirm protein expression by SDS-PAGE analysis. Bacteria were applied to CFBE41o− cells at an MOI of 50:1, and the cells were incubated for 30 min at 37°C in 5% CO2 to allow the bacteria to attach. Cells and bacteria were gently washed four times with PBS (500 μl) and then fixed with 3% paraformaldehyde for 10 min at RT. Cells were washed with PBS and blocked with PBS containing 5% BSA for 1 h at RT before incubation overnight at 4°C with FITC-conjugated anti-E. coli antibody (1:100 dilution in 5% BSA in PBS). The following day, cells were washed three times in PBS before staining with phalloidin-Alexa Fluor 568 (5 U/ml) and DAPI (1 μg/ml) for 1 h in the dark. Samples were then mounted with a drop of Vectashield mounting medium (Vector Laboratories, Peterborough, England) containing DAPI and visualized under a confocal microscope. The cells were counted in 20 randomly selected fields for each strain, and values are expressed as numbers of bacteria per 100 cells.
Cell-mediated immune responses following immunization.
Groups of 5 mice were immunized with linocin or OmpW as outlined above but not treated with cyclophosphamide or challenged. Two weeks after booster immunization, the animals were sacrificed, and spleens from these animals were removed into RPMI 160 medium. Spleen cells from immunized and nonimmunized mice were disrupted, and red blood cells were lysed by using red blood cell lysis buffer (BioLegend, San Diego, CA). Cells were plated at 2 × 105 cells per well into 96-well plates in triplicate and exposed to PBS alone, linocin (50 μg/ml), or OmpW (50 μg/ml). Cells were incubated for 3 days before removal of the supernatant, and the levels of the cytokines gamma interferon (IFN-γ), interleukin-4 (IL-4), IL-6, IL-10, and IL-17A were measured by using a mouse cytometric bead array (Becton Dickinson, United Kingdom) according to the manufacturer's instructions. Flow cytometry standard (FCS) data files were analyzed by FCAP Array software to generate standard curves and to determine the cytokine concentrations for unknown samples.
Statistical analysis.
Attachment of Bcc strains and E. coli strains expressing recombinant linocin or OmpW to lung epithelial cells was analyzed by one-way analysis of variance (ANOVA). Data relating to bacterial CFU in lung and spleen following challenge were analyzed by two-way ANOVA. Data relating to cytokine secretion from splenocytes were analyzed by Student's t tests.
RESULTS
Attachment of Bcc strains to lung epithelial cells.
In order to compare the binding of the bacterial strains to host cells with and without a CF phenotype, fluorescence microscopy was used to quantify the attachment of the two B. multivorans and the two B. cenocepacia strains to lung epithelial cells with (CFBE41o−) and without (16HBE14o−) a CF phenotype. It was evident that all four Bcc strains showed >2-fold more attachment to CFBE41o− cells than to the independently derived cell line 16HBE14o− (P ≤ 0.002) (Fig. 2) at an MOI of 50:1. This 2-fold (or greater) difference was also noted at an MOI of 10:1 for all strains (P < 0.005) except B. cenocepacia C1394 (Fig. 2A). B. multivorans strain LMG13010 showed 4.4- and 6.5-fold more attachment to CFBE41o− cells than to 16HBE14o− cells at MOIs of 10:1 and 50:1, respectively (Fig. 2A and B). The binding of the nonpiliated B. cenocepacia strain C1394 was not significantly different from that of the piliated B. cenocepacia strain BC7 for either cell line (P = 0.128). It was also apparent that, overall, no differences were observed in the attachment of the more virulent B. cenocepacia strain to lung epithelial cells relative to B. multivorans.
FIG 2.
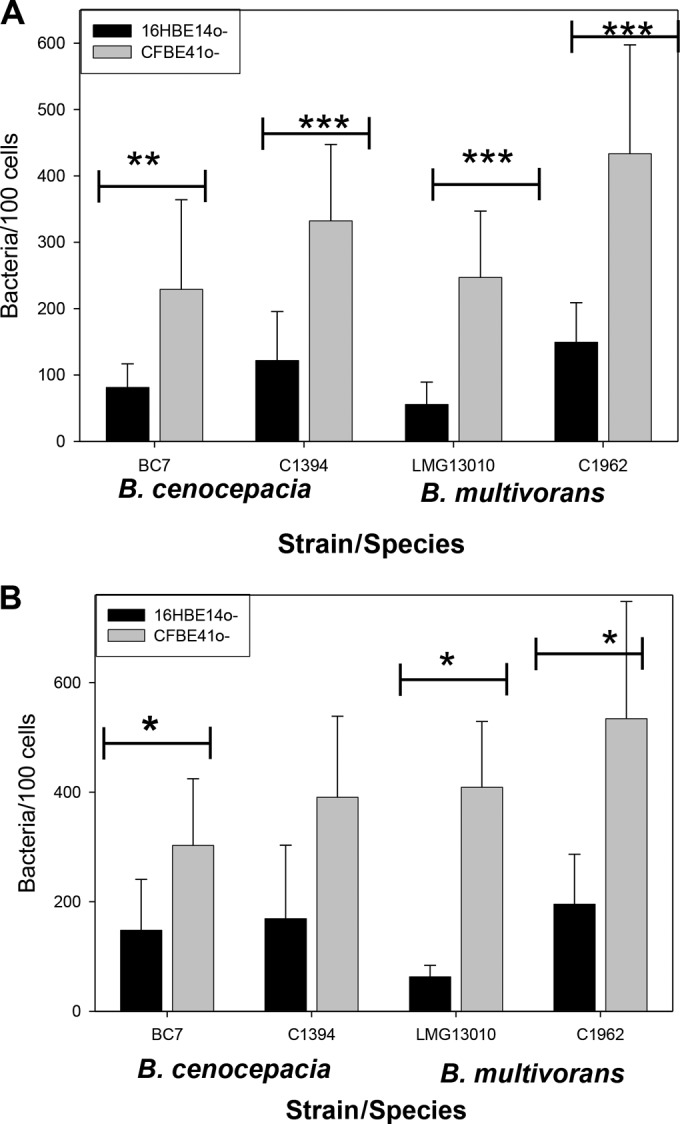
Attachment of Bcc to CFBE41o− and 16HBE14o− cells was examined by confocal microscopy at MOIs of 10:1 (A) and 50:1 (B). Attached bacteria were labeled with FITC-labeled antibody, and cells were stained with DAPI. Each bar represents means (± standard errors of the means) of data from two independent experiments, with 10 fields counted per strain. Statistical significance was determined by one-way ANOVA (***, P < 0.001; **, P < 0.005; *, P < 0.01).
Identification of proteins involved in lung cell attachment.
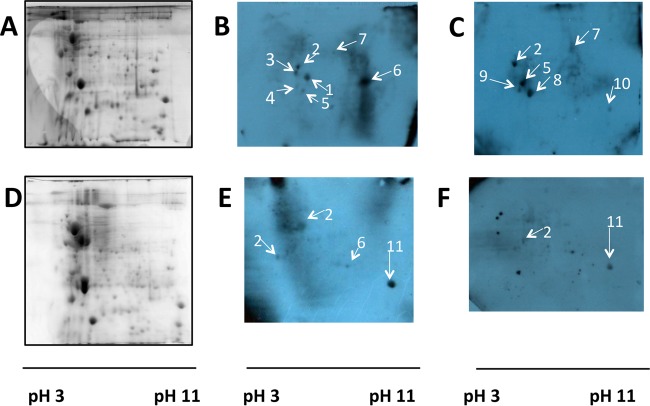
To identify proteins involved in the attachment of Bcc bacteria to host cells, membrane proteins were prepared from two Bcc species, B. multivorans and B. cenocepacia, which represent the most frequently isolated and the most virulent species, respectively. Membrane proteins transferred onto PVDF membranes were probed with either CF or non-CF cells (Fig. 1). Several of the proteins identified were common to both CFBE41o− and 16HBE14o− cell-probed blots. Fourteen distinct proteins were identified across both species (Table 2). There were seven proteins identified from CFBE41o−-probed blots prepared from membrane proteins of the piliated B. cenocepacia strain BC7 (Fig. 3B), three of which were also identified in 16HBE14o− blots (Fig. 3C). The seven proteins were all in the molecular mass range of 15 to 38 kDa and included some proteins with known roles in the attachment of Bcc bacteria, such as the type VI secretion system (T6SS). Other identified proteins had not been shown previously to be involved in host cell attachment of Bcc bacteria, including linocin and OmpW (Fig. 3B and C and Table 2). In each case, the proteins were identified with high sequence coverage (>42%) (Table 2). When blots prepared from the piliated B. cenocepacia strain BC7 were probed with 16HBE14o− cells, six proteins were identified, including a T6SS protein and linocin but not OmpW (Fig. 3C). The proteins identified on the CFBE41o−-probed blots from the nonpiliated B. cenocepacia strain C1394 showed weaker spot intensities but again included OmpW and linocin (Table 2). The C1394 blot probed with 16HBE14o− cells showed only two strong protein spots, corresponding to an Hcp1 family T6SS effector and an OmpA/MotB domain-containing protein (Table 2).
TABLE 2.
Proteins involved in attachment to lung epithelial cells identified from cell blots probed with CFBE41o− or 16HBE14o− cells
| Protein | BCA designation, alias | Functional category | Predicted subcellular locationa (score) | Strain(s) in which protein was identified | Molecular mass (kDa)/pIb | Sequence coverage (%) | Scorec |
|---|---|---|---|---|---|---|---|
| Linocin_M18 bacteriocin protein | Bcen1404 | Encapsulating protein | Cytoplasmic (8.96) | B. cenocepacia BC7 and C1394; B. multivorans C1962 and LMG13010 | 29.0/4.8 | 66–85 | 87–215 |
| OmpW family protein | L1829 | Membrane protein | Outer membrane (9.45) | B. cenocepacia BC7 and C1394; B. multivorans C1962 and LMG13010 | 22.9/7.8 | 73.2–85.9 | 84–143 |
| Alkyl hydroperoxide reductase | L2013 | Oxidoreductase | Cytoplasmic (997) | B. cenocepacia BC7 and C1394; B. multivorans LMG13010 | 20.5/4.9 | 57.1–65.9 | 115–137 |
| Histone family protein DNA-binding protein | M1012 | DNA binding | Unknown (2) | B. cenocepacia BC7 and C1394; B. multivorans LMG13010 | 15.2/11.2 | 46.9–49.7 | 81 |
| Acetoacetyl-CoA reductase | L0833, PhbB | Oxidoreductase | Cytoplasmic (9.26) | B. cenocepacia C1394; B. multivorans LMG13010 | 28.4/7.7 | 53–63 | 90–209 |
| Hcp1 family type VI secretion system effector | L0343, BcsL | Secretion | Extracellular (10) | B. cenocepacia C1394; B. multivorans C1962 | 18.5/6.9 | 52.1–88 | 97–161 |
| Heat shock protein Hsp20 | Bmul5914 | Stress response | Cytoplasmic (8.96) | B. cenocepacia BC7; B. multivorans LMG13010 | 21.4/5.2 | 41.4–52.9 | 96–104 |
| Hypothetical protein Bmul_4930 | M0843 | Lipoproteind | Transmembrane | B. cenocepacia BC7; B. multivorans C1962 | 19.6/10.3 | 58.2–82 | 124 |
| OmpA/MotB domain-containing protein | L2645 | Membrane protein | Outer membrane (9.45) | B. cenocepacia C1394; B. multivorans C1962 | 21.5/10.4 | 42 | |
| Oxidoreductase, aldo/keto reductase family | L0513 | Oxidoreductase | Cytoplasmic (9.97) | B. cenocepacia BC7 | 38.2/5.8 | 69.5 | 246 |
| Putative universal stress protein | BURMUCGD2_RS23075 | Stress response | Cytoplasmic (8.96) | B. cenocepacia C1394; B. multivorans C1962 | 17.6/6.1 | 70.4–95.1 | 196–285 |
| Hypothetical protein Bmul_4929 (also known as protein of unknown function, DUF883, ElaB) | M0844 | Membrane proteind | Membraned | B. multivorans C1962 | 13.9/7.8 | 78.9 | 85 |
| Type VI secretion protein (97% homology between both GIs) | L0341 | Secretion | Cytoplasmic (9.97) | B. cenocepacia BC7 | 19.2/5.1–5.3 | 60.8 | 97–112 |
| Ubiquinone/menaquinone biosynthesis methyltransferase | L0873, UbiE | Methyltransferase | Cytoplasmic (9.96) | B. cenocepacia C1394 | 27.1/9.1 | 67.9–77.4 | 95–210 |
Predicted subcellular localization determined by using PSORTb V3 (http://www.psort.org/) (63).
Theoretical molecular masses (kilodaltons) and isoelectric points were determined by Mascot analysis.
Ranges represent MS/MS ion scores determined by peptide mass fingerprinting (64). Only scores that were deemed to be significant by Mascot analysis (P < 0.05) are reported.
Functional category from UniProt and/or the Burkholderia Genome Database (42).
FIG 3.
Representative Coomassie blue-stained 2D gels (A and D) and cell-probed blots (B, C, E, and F) for piliated B. cenocepacia strain BC7 (A to C) and B. multivorans strain LMG13010 (D to F). Blots in panels B and E were probed with CFBE41o− cells, while blots in panels C and F were probed with 16HBE14o− cells. Spots: 1, Hsp20; 2, linocin; 3, mixture of chorismate mutase and linocin; 4, Ahp reductase; 5, type VI secretion system protein; 6, OmpW family protein; 7, oxidoreductase/aldo-keto reductase; 8, unnamed protein product; 9, unnamed protein product; 10, hypothetical protein Bmul_4390; 11h OmpA/MotB domain protein. Each gel or blot is representative of data from at least two independent experiments.
Membrane proteins from two B. multivorans strains, LMG13010 and C1962, were also prepared and individually analyzed. The 2D gel separation of membrane proteins from either strain showed ∼150 distinct protein spots, which were visible after Coomassie blue staining, where most of the spots were found to be in the pH 4 to 10.5 range. Four spots were reproducibly identified between 20 kDa and 30 kDa as being positive for cellular attachment in the B. multivorans strain LMG13010 blot probed with CFBE41o− cells (Fig. 3E and Table 2). Linocin (Fig. 3E, arrow 2) was identified in two separate regions of the blot and may be involved in binding to other proteins. Four spots were apparent on CFBE41o− cell-probed blots prepared from B. multivorans strain C1962, all of which had been observed in other strains: linocin, OmpW, OmpA/MotB, and the Hcp1 family T6SS effector (Table 2). Two of these four spots were identified on blots probed with non-CF cells in the region of 20 kDa to 30 kDa: linocin and OmpA/MotB domain-containing protein. Consistent with B. cenocepacia, OmpW was not identified on 16HBE14o− cell-probed blots of B. multivorans membrane proteins.
Several proteins that were identified with distinct GenInfo Identifier (GI) sequence identification numbers shared considerable homology and identity with other proteins identified. For example, two hypothetical proteins (GI 134294574 and GI 78065037) were identified, both of which were 96% homologous to Hcp1 family type VI secretion family effectors by BLAST analysis (40). Hcp1 was also identified as being involved in the binding of one strain from each of the two species analyzed, the nonpiliated B. cenocepacia strain C1394 and B. multivorans strain C1962. These proteins were distinct from two other T6SS effector proteins identified (GI 161526099 and GI 493817763). While both of these T6SS effector proteins shared 97% identity to each other across the entire sequence, they shared only 6% identity with Hcp1 within the N-terminal region. Two other closely related proteins were also identified: OmpW (GI 107029064) and OmpW superfamily protein (GI 161524680). These proteins shared 88% identity and are grouped together. The latter protein was more frequently identified, being identified over 15 times across all 4 strains, while OmpW (GI 107029064) was identified in C1394 blots only (n = 3) and is consequently not highlighted in Table 2. Two histone family DNA-binding proteins were also observed (Table 2). Although not identical, they were homologous at the N termini, with 95% identity over 100 amino acids at the N termini. A universal stress family protein was also identified across both species (GI 493452066). This protein shared 94% homology with another identified protein, UspA domain-containing protein (GI 501174111). Overall, six of the proteins are membrane proteins. Although only four proteins were classified as outer membrane proteins on the PSORTb website (http://www.psort.org/), two additional proteins (hypothetical proteins Bmul_4929 and Bmul_4930) were identified as membrane proteins in the UniProt database (41), being classified as a transmembrane protein and a lipoprotein, respectively.
Investigations of immunogenicity of membrane proteins involved in Bcc attachment using CF patient sera.
Many proteins expressed under in vitro culture conditions may not be expressed during in vivo infection. Therefore, the expression of the antigens during infection was confirmed by immunoblotting of Bcc membrane proteins with pooled sera from Bcc-colonized CF patients. All immunoblots probed with Bcc-positive sera showed strong antibody responses to membrane proteins from all four strains. The corresponding membrane proteins on parallel Coomassie blue-stained gels were analyzed by MALDI-TOF MS. Over 50 proteins were identified as being immunogenic (36). Linocin and OmpW were identified as being immunoreactive on 2D blots prepared from membrane fractions of all four strains examined when serum from Bcc-colonized patients was used (Table 3). Neither protein was identified in control blots from patients who had no history of Bcc colonization. This shows that these two proteins are both expressed by the bacteria during human infection and exposed to, and targeted by, the immune system.
TABLE 3.
Identification of linocin and OmpW from 2D blots probed with sera from Bcc-colonized CF patients compared with control blots using sera from CF patients with no history of Bcc infection
| GI sequence identifier | Protein (species) | Molecular mass (kDa)/pIa | Strain | Sequence coverage (%) | Scoreb |
|---|---|---|---|---|---|
| GI 161524680 | OmpW family protein | 22.9/9 | BC7 | 27–85 | 54–142 |
| C1394 | 10 | 74–78 | |||
| LMG10303 | 29–85 | 108–253 | |||
| C1962 | 36–84 | 43–222 | |||
| GI 221198593 | Linocin (B. multivorans) | 28.8/4.8 | BC7 | 73.8–78.2 | 155–572 |
| GI 170735569 | Linocin (B. cenocepacia)c | 28.8/4.8 | C1394 | 41.7–50.9 | 76–89 |
| GI 221198593 | Linocin (B. multivorans) | 28.8/4.8 | LMG13010 | 34–81 | 193–488 |
| GI 221198593 | Linocin (B. multivorans) | 28.8/4.8 | C1962 | 48.3–90 | 105–141 |
Theoretical molecular masses (kilodaltons) and isoelectric points were determined by Mascot analysis.
Ranges represent MS/MS ion scores determined by mass fingerprinting. Only scores that were deemed to be significant by Mascot analysis (P < 0.05) are reported.
Query coverage of 96%; 92% identity.
Confirmation of the role of linocin and OmpW in lung cell attachment.
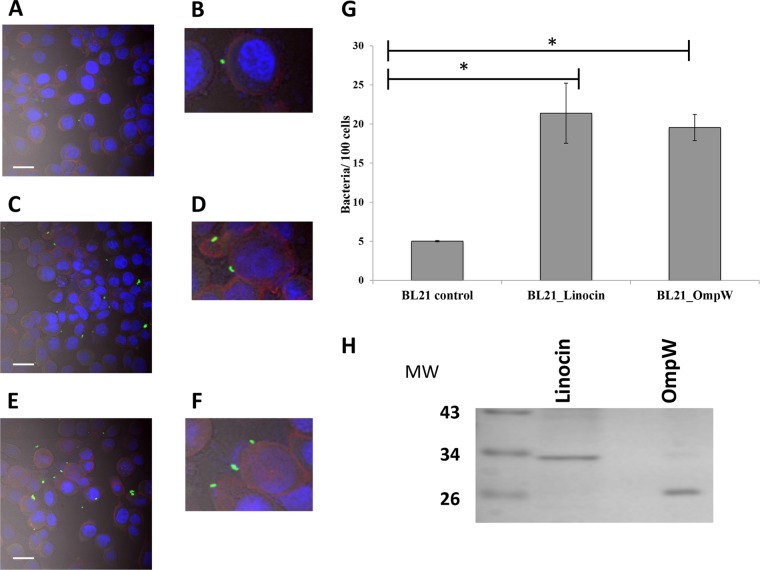
Due to the facts that both linocin and OmpW were identified in all four strains examined as being involved in attachment to CFBE41o− cells, were expressed during human infection, and were immunogenic, these two proteins were selected for investigation as protective antigens. Both proteins were cloned and individually expressed in E. coli BL21 Star cells. Transformation was confirmed by restriction digestion of extracted plasmid DNA, and recombinant protein expression was identified in both the membrane (insoluble) and soluble fractions of whole-cell lysates for both antigens. In order to confirm that the individual proteins were involved in host cell attachment, the affinity of the recombinant E. coli clones (E. coli-Lin and E. coli-OmpW) for lung epithelial cells was investigated in order to examine whether the expression of these proteins enhanced attachment to lung epithelial cells. As expected, the E. coli BL21 Star strain showed a low level of attachment to CFBE41o− cells, with only 5.02 ± 0.08 cells of the expression strain BL21 Star attaching per 100 CFBE41o− cells within 30 min. When recombinant linocin-expressing E. coli cells were applied, attachment increased by 4.2-fold (P = 0.026), confirming the role of linocin in attachment (Fig. 4). The OmpW-expressing recombinant strain showed a 3.9-fold (P = 0.006) increase in attachment to CFBE41o− cells over control BL21 cells.
FIG 4.
(A to F) Attachment of recombinant E. coli cells expressing either linocin (C and D) or OmpW (E and F) to CFBE41o− cells by confocal microscopy compared with E. coli BL21 controls (A). Bacteria (induced with 1 mM IPTG) were applied to CFBE41o− cells (MOI of 50:1) for 30 min and stained with FITC-conjugated anti-E. coli antibody, and epithelial cells were counterstained with phalloidin and DAPI and superimposed with differential interference contrast imaging. Bars = 20 μm. Panels B, D, and F show zoomed images to highlight bacteria interacting with epithelial cells. (G) Bacterial cell counts in 20 randomly selected fields for each strain, expressed as the number of bacteria attached per 100 epithelial cells, from two independent experiments. *, P < 0.05 as determined by one-way ANOVA. (H) Purified recombinant linocin and OmpW on a Coomassie blue-stained SDS-PAGE gel. MW, molecular weight (in thousands).
Protection of immunized mice against challenge.
Both antigens were purified by using Ni-NTA affinity columns followed by desalting and subsequent purification on endotoxin affinity columns to remove lipopolysaccharide (LPS). The purified proteins were confirmed to have endotoxin levels of <0.25 EU/ml. The identity of the recombinant proteins was confirmed by MALDI-TOF MS/MS analysis of the individual protein bands on SDS-PAGE gels following purification (Fig. 4E).
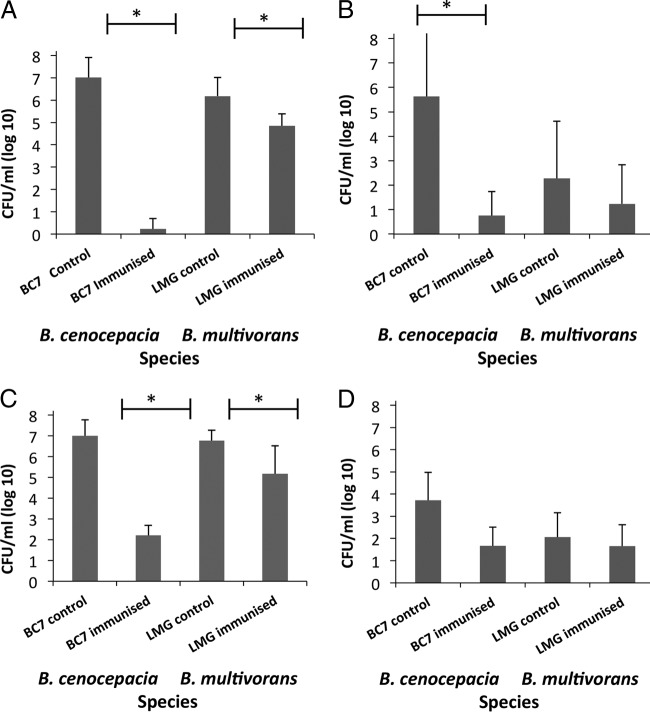
It was clear that linocin immunization protected mice from challenge with B. cenocepacia and B. multivorans (Fig. 5). Immunization with linocin reduced the B. cenocepacia burden to barely detectable levels (mean value of 3 CFU/ml/lung; P = 0.000002) (Fig. 5A). Although the effect on B. multivorans was not as potent using this protocol, the CFU in the lung were reduced by 4,000-fold compared to those in unimmunized controls (P = 0.015). Bcc bacteria are invasive pathogens, and they escape the lung and colonize the spleen. Immunization with linocin protected mice from invasion of B. cenocepacia into the spleen to 0.001% (P = 0.0004) (Fig. 5B). B. multivorans is generally less invasive than B. cenocepacia, and this was confirmed in this study, where the unimmunized controls showed less splenic invasion than the B. cenocepacia controls. Splenic invasion of B. multivorans was reduced by linocin immunization to 10% of that in unimmunized LMG13010-challenged mice (P = 0.018).
FIG 5.
Protection of mice following immunization with linocin (A and B) and OmpW (C and D). Bacterial CFU were counted in lung (A and C) and spleen (B and D) homogenates from triplicate plates prepared individually from groups of 5 mice. Each study was done two independent times, with comparable results. Statistical significance was determined by two-way ANOVA (*, P < 0.05).
OmpW-immunized mice were also protected against B. cenocepacia, with a reduction in lung bacterial counts of >4 logs (60,000-fold reduction in bacterial counts; P = 0.022) (Fig. 5C). Immunization with OmpW protected against B. multivorans with a 1-log reduction in bacterial counts (P = 0.013) (the lung bacterial count was reduced to 8% of that in controls). Although splenic invasion was reduced, this did not reach statistical significance following OmpW immunization using the protocols described above (Fig. 5D).
Antibody responses following immunization.
The levels of antigen-specific antibodies were measured in sera of immunized mice relative to controls. Pooled sera from mice in the immunized groups showed high endpoint titers of antigen-specific IgG1, IgG2a, and IgG2b. The IgG1 and IgG2a titers in response to linocin immunization were 0.79 × 106 and 1.05 × 106, respectively. The ratio of IgG2a to IgG1 antibodies in response to linocin (1.32) was indicative of a bias toward a Th1 response. IgG responses to OmpW immunization were comparable to those to linocin. The IgG2a titers (1.05 × 106) were comparable to the IgG1 titer (0.95 × 106), such that the ratio of IgG2a to IgG1 in response to OmpW was suggestive of a mixed Th1/Th2 response.
Cellular immune responses.
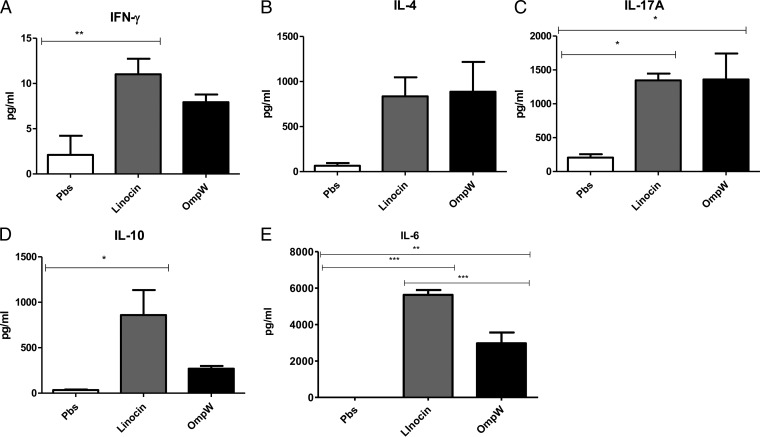
The cytokine responses of antigen-stimulated spleen cells were measured in order to determine whether one of the antigens resulted in immunological memory and to investigate the T-cell response elicited with this antigen. When previously vaccinated spleen cells were restimulated with linocin, they produced higher levels of IL-6, IL-17A, and IL-10 than did spleen cells from mice that had not been vaccinated (P < 0.05, as determined by Student's t test) (Fig. 6). OmpW had a lower ability to stimulate cytokine responses, with only IL-17A and IL-6 showing statistically significant secretion (P < 0.05) upon reexposure to the antigen. This suggests that the mice immunized with either linocin or OmpW developed immunological memory and were able to initiate a greater immune response upon reexposure than that in nonvaccinated mice. IL-17 secretion is indicative of a typical murine Th17 response, while IL-6 and IL-10 are indicative of a Th2 cell response. Although the level of IFN-γ secretion was low in response to both antigens, overall, the cytokine profile is suggestive of a mixed Th1/Th17/Th2 response. Overall, the potential for both antigens is confirmed in these experiments due to the induction of immunological memory and potent recall cytokine production.
FIG 6.
Cellular response of mouse spleen cells upon reexposure to linocin or OmpW (50 μg/ml). Cytokine levels were measured in restimulated spleen cells from either five linocin-vaccinated mice (gray bars), five OmpW-vaccinated mice (black bars), or five sham-vaccinated mice (PBS controls) (clear bars) following exposure to linocin, OmpW, or PBS, respectively. All mice were cyclophosphamide free. (A) IFN-γ; (B) IL-4; (C) IL-17A; (D) IL-10; (E) IL-6. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (as determined by Student's t test).
DISCUSSION
This study describes a method to identify potentially potent vaccine antigens by identifying bacterial proteins that mediate high-affinity adherence to host cells. While this method may not identify all adhesins, it allows the selection of avidly binding adhesins. The effectiveness of this method was demonstrated by the identification of two novel potent antigens, linocin and OmpW, which protected mice against two different species of the cystic fibrosis-associated pathogens in the Bcc.
Broad identification methods such as reverse vaccinology have been excellent in identifying previously unidentified antigens for a host of bacterial infections (2). The very high number of identifiable antigens can be a limiting factor, as considerable “weeding out” of the less immunogenic antigens is required. In contrast, our novel “cell blot”-based method does not aim to identify all adhesins involved in host cell attachment but selects for strongly binding adhesins, which consequently have a high potential as potent vaccine antigens. Neither linocin nor OmpW was previously shown to be involved in the virulence or host cell attachment of any Burkholderia species, highlighting the potential for the cell blot method to identify novel adhesins and potential virulence factors. The role of these antigens as adhesins was confirmed by an increase in the attachment of E. coli cells recombinantly expressing each of these proteins. Furthermore, immunoproteomic analysis showed that CF patients colonized with Bcc bacteria produce antibodies that specifically recognize these proteins, demonstrating that these proteins are expressed during human infection and that they are immunoreactive. Moreover, neither protein was identified when blots of Bcc membrane proteins were probed with serum from CF patients with no history of Bcc infection.
This is the first study that involves the use of recombinant subunit antigens to protect mice against Bcc bacteria. Previously, mice were protected from B. cenocepacia by using enriched OMPs that were administered intranasally as a nanoemulsion preparation (25, 26). Both of those studies demonstrated the potential for OMPs as protective antigens but were carried out with unpurified OMP preparations, which are ill defined, rather than pure specific identified antigens. A 17-kDa OMP was identified as an immunodominant antigen in the latter study (26). Those previous studies were also carried out with non-CF mice, as both BALB/c and CD-1 mice are well-established models for Bcc infection.
Linocin is expressed by the two most clinically relevant species of the Bcc, B. multivorans and B. cenocepacia, and has homologs in at least seven Bcc species, including B. ambifaria, B. pyrrocinia, B. vietnamiensis, B. dolosa, and B. lata. The gene is also present in non-Bcc species, including B. gladioli, B. phymatum, B. xenovorans, and Burkholderia sp., but not in any of the sequenced B. pseudomallei strains. It is well conserved, with identities of 81 to 100% across Bcc species (40), and has been identified as being involved in attachment to lung epithelial cells in both individual strains of both Bcc species examined. It is predicted to be an encapsulating protein for peroxidase (42). Linocin was previously described as having antimicrobial activity in Brevibacterium linens (43) and was subsequently identified as having a role in T-cell stimulation in Mycobacterium tuberculosis but showed no bacteriocin activity (44). It is widely distributed in mycobacterial species (44). Sutter et al. showed that linocin-like proteins form large assemblies and are involved in the encapsulation of enzymes (45). This protein was not shown to be involved in host cell attachment or in the pathogenesis of Bcc bacteria prior to this study. The dual roles of linocin in the attachment of Bcc bacteria to lung epithelial cells and in immunogenicity during human Bcc infection, together with its previously reported role in eliciting an immune response in an unrelated respiratory pathogen, suggested that it would make a potent protective antigen.
OmpW is an outer membrane protein that is also expressed by both Bcc species examined and is involved in the attachment of both Bcc species to CFBE41o− cells, and its role in attachment was confirmed by using the recombinant Bcc OmpW-expressing E. coli strain. BLAST analysis of a Bcc OmpW family protein (GI 161524680) indicated that the most closely related proteins are all expressed by members of the Bcc. Other strains that express homologous OmpW proteins include strains of another CF-associated pathogen, Pandoraea pnomenusa, and another proteobacterium, Sphingomonas (86% identity) (40). Its homolog in P. aeruginosa is OprG (GI 610414171; 32% identity) (40), which is involved in host cell toxicity associated with P. aeruginosa (46). More recently, OprG expression was downregulated from early to late isolates colonizing adults with CF, which correlated with a reduction of virulence in the later-colonizing isolates (47). OmpW proteins have been identified in bacteria that colonize the gastrointestinal tract, e.g., Vibrio cholerae (48). Its role in the attachment of, and immune response to, a number of intestinal pathogens was recently reviewed (49). OmpW proteins have been associated with the attachment of Vibrio alginolyticus; OmpW-deficient mutants showed a 10-fold reduction in mouse colonization (50). OmpW was recently identified as being related to the immune response in inflammatory bowel disease associated with an oral pathogen, Porphyromonas gingivalis (51), and is immunogenic in Salmonella-induced reactive arthritis (52). We have recently shown that immunization of mice with OmpW also protects against B. pseudomallei infection (W. T. Casey, N. Spink, F. Cia, C. Collins, M. Romano, R. Berisio, G. J. Bancroft, and S. McClean, submitted for publication). Furthermore, it was demonstrated to have protective efficacy as a vaccine antigen against the fish pathogen Aeromonas hydrophila (53).
OmpW family proteins are found throughout Gram-negative proteins, which could potentially raise concerns regarding commensal bacteria. The similarity with E. coli OmpW is weak, showing only 36% identity across 88% of the sequence. Other Gram-positive commensals with OmpW-like domains include Bacteroides fragilis; however, the homology with Bcc OmpW is minimal, with only 38% identity over only 3% of the sequence being observed (40). Furthermore, only four pathogens were found to share >37% identity across 90% of the sequence; the remainder shared <30% homology.
Although we did not target proteins that bound specifically to CF cells, we were interested in comparing the binding of Bcc bacteria to epithelial cells of CF and non-CF origins and found that Bcc bacteria bound at least 2-fold more to CFBE41o− cells than to 14HBE14o− cells. This may be due, at least in part, to the different proteins identified on membrane protein blots probed with CFBE41o− cells relative to 16HBE14o− cells, and more proteins were identified on CFBE41o− cell blots than on 16HBE14o− cell blots. Indeed, both OmpW and alkyl hydroperoxide (Ahp) reductase were absent in all 16HBE14o− cell-probed blots. Further analysis of any specificity in the binding of OmpW or Ahp to cells with a CF phenotype will need to be addressed by using primary lung epithelial cells from people with and without CF.
Several other proteins identified as being involved in host cell attachment in this study were also found to be immunogenic in our previously reported immunoproteomic analysis (36). These proteins include Ahp reductase, which was immunogenic across all 4 of the strains used; Hcp1; OmpA/MotB domain-containing protein; oxidoreductase/aldo-keto reductase; and acetyl coenzyme A (acetyl-CoA) acetyltransferase. This suggests that their exposure to, and stimulation of, the host humoral system is also a result of their role in attachment to host epithelial cells and will be the focus of future work. It is worth highlighting that although we used a membrane preparation for our blots, we identified eight proteins that are predicted to be cytosolic proteins. We and others have reported this previously (36, 54). In particular, Riedel et al. showed that of the 304 intracellular proteins identified in their full proteomic analysis of B. cenocepacia, 46 were located in extracellular or surface fractions (54). These proteins that are predicted to be cytosolic may be expressed in outer membrane vesicles, resulting in their consequent inclusion in membrane preparations; for example, elongation factor Tu is predicted to be cytosolic but has been shown to be secreted in outer membrane vesicles and was immunoprotective against B. thailandensis in mice (55).
Our main objective in these studies was to identify potential vaccine antigens. Both individual antigens tested protected mice from both B. cenocepacia and B. multivorans challenges following two immunizations. Protection was greatest against B. cenocepacia over B. multivorans, and lung CFU were reduced in mice immunized with OmpW and, to a greater extent, linocin. Both antigens also elicited potent serological responses. Of particular interest was the finding that both antigens were able to overcome the polarizing Th2 effect of the adjuvant alum, inducing mixed Th1/Th2/Th17 responses, indicating that both antigens in particular have some adjuvant properties despite the undetectable levels of endotoxin in these preparations. Linocin proved to be effective as a stimulant of the cellular host response, with stimulation of IL-17, IL-6, and IL-10 production from antigen-restimulated splenocytes. Restimulation of spleen cells from OmpW-vaccinated mice with OmpW also stimulated IL-6, IL-10, and IL-17A comparably to the linocin profile albeit at reduced levels. The role of IL-17 in protection against extracellular bacterial pathogens has been well described. While the cell-mediated responses required to protect against Bcc have not been studied to date, the stimulation of IL-17A by both antigens is likely to contribute to protection against Bcc infection by these antigens, which have both an extracellular and an intracellular component to their lifestyle (27, 56–60). IL-17-stimulating antigens were recently shown to confer protection against P. aeruginosa in immunization studies (61). Although further studies, such as immunization and protection of another animal species, toxicity studies, and clinical trials, on the potential of these two antigens as vaccine candidates to protect or treat CF patients and other susceptible populations against Bcc infection need to be carried out, the current data indicate that the two antigens OmpW and linocin show promise as protective antigens. Indeed, the recent finding that linocin-like proteins form large assemblies (45) further highlights the potential of linocin as a vaccine antigen.
In summary, the cell blot method identified several proteins that are involved in attachment to human lung epithelial cells. Although many of these proteins have no previously known role in attachment to date, several were recently found to be immunogenic in CF patients (36). Furthermore, two of the adhesins were protective antigens in a mouse challenge study. The method that we have developed to identify bacterial adhesins that bind avidly to host cells could be applied to any bacterial pathogen that colonizes mucosal surfaces. By probing 2D membranes with relevant host epithelial cells, with rigorous wash steps, bacterial adhesins can be identified by using MALDI-TOF MS analysis. We have validated this method for the identification of novel adhesins that were expressed in two Bcc species. This highlights the potential of our cell blot method to identify novel and potent antigens against highly multidrug-resistant bacteria.
ACKNOWLEDGMENTS
The Centre of Applied Science for Health is funded by Programme for Research in Third Level Institutions (PRTLI) Cycle 4, supported by the European Union Regional Development Plan and the Irish Government National Development Plan 2007-2013 and administered by the Higher Education Authority in Ireland. S.M. and M.C. are members of European COST Action BM1003: Microbial Cell Surface Determinants of Virulence as Targets for New Therapeutics in Cystic Fibrosis.
We are grateful to Dieter Gruenert (UCSF) for the gift of 16HBE14o− and CFBE41o− cells and to Umadevi Sajjan, University of Michigan, for the gift of the anti-Bcc antibody.
Funding Statement
This work was funded in part by Technological Sector Research (Strand III) support for M.S. Cassandra Collins was funded by Enterprise Ireland Commercialisation Fund, cofunded by the European Regional Development Fund (CF0133015). Ruth Dennehy was supported by Science Foundation Ireland under grant number SFI 11/RFP.1/BMT/3307.
REFERENCES
- 1.Mishra RP, Oviedo-Orta E, Prachi P, Rappuoli R, Bagnoli F. 2012. Vaccines and antibiotic resistance. Curr Opin Microbiol 15:596–602. doi: 10.1016/j.mib.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Masignani V, Rappuoli R, Pizza M. 2002. Reverse vaccinology: a genome-based approach for vaccine development. Expert Opin Biol Ther 2:895–905. doi: 10.1517/14712598.2.8.895. [DOI] [PubMed] [Google Scholar]
- 3.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 4.Felgner PL, Kayala MA, Vigil A, Burk C, Nakajima-Sasaki R, Pablo J, Molina DM, Hirst S, Chew JS, Wang D, Tan G, Duffield M, Yang R, Neel J, Chantratita N, Bancroft G, Lertmemongkolchai G, Davies DH, Baldi P, Peacock S, Titball RW. 2009. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc Natl Acad Sci U S A 106:13499–13504. doi: 10.1073/pnas.0812080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dennehy R, McClean S. 2012. Immunoproteomics: the key to discovery of new vaccine antigens against bacterial respiratory infections. Curr Protein Pept Sci 13:807–815. doi: 10.2174/138920312804871184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altindis E, Tefon BE, Yildirim V, Ozcengiz E, Becher D, Hecker M, Ozcengiz G. 2009. Immunoproteomic analysis of Bordetella pertussis and identification of new immunogenic proteins. Vaccine 27:542–548. doi: 10.1016/j.vaccine.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Tefon BE, Maass S, Ozcengiz E, Becher D, Hecker M, Ozcengiz G. 2011. A comprehensive analysis of Bordetella pertussis surface proteome and identification of new immunogenic proteins. Vaccine 29:3583–3595. doi: 10.1016/j.vaccine.2011.02.086. [DOI] [PubMed] [Google Scholar]
- 8.Coenye T, Vandamme P, Govan JR, LiPuma JJ. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J Clin Microbiol 39:3427–3436. doi: 10.1128/JCM.39.10.3427-3436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenye T, Vandamme P, LiPuma JJ, Govan JR, Mahenthiralingam E. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J Clin Microbiol 41:2797–2798. doi: 10.1128/JCM.41.6.2797-2798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahenthiralingam E, Baldwin A, Drevinek P, Vanlaere E, Vandamme P, Lipuma JJ, Dowson CG. 2006. Multilocus sequence typing breathes life into a microbial metagenome. PLoS One 1:e17. doi: 10.1371/journal.pone.0000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peeters C, Zlosnik JE, Spilker T, Hird TJ, LiPuma JJ, Vandamme P. 2013. Burkholderia pseudomultivorans sp. nov., a novel Burkholderia cepacia complex species from human respiratory samples and the rhizosphere. Syst Appl Microbiol 36:483–489. doi: 10.1016/j.syapm.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 12.McClean S, Callaghan M. 2009. Burkholderia cepacia complex: epithelial cell-pathogen confrontations and potential for therapeutic intervention. J Med Microbiol 58:1–12. doi: 10.1099/jmm.0.47788-0. [DOI] [PubMed] [Google Scholar]
- 13.Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 14.Courtney JM, Dunbar KE, McDowell A, Moore JE, Warke TJ, Stevenson M, Elborn JS. 2004. Clinical outcome of Burkholderia cepacia complex infection in cystic fibrosis adults. J Cyst Fibros 3:93–98. doi: 10.1016/j.jcf.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Callaghan M, McClean S. 2012. Bacterial host interactions in cystic fibrosis. Curr Opin Microbiol 15:71–77. doi: 10.1016/j.mib.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Chernish RN, Aaron SD. 2003. Approach to resistant gram-negative bacterial pulmonary infections in patients with cystic fibrosis. Curr Opin Pulm Med 9:509–515. doi: 10.1097/00063198-200311000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Saiman L, Tabibi S, Starner TD, San Gabriel P, Winokur PL, Jia HP, McCray PB Jr, Tack BF. 2001. Cathelicidin peptides inhibit multiply antibiotic-resistant pathogens from patients with cystic fibrosis. Antimicrob Agents Chemother 45:2838–2844. doi: 10.1128/AAC.45.10.2838-2844.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird RM, Brown H, Smith AW, Watson ML. 1999. Burkholderia cepacia is resistant to the antimicrobial activity of airway epithelial cells. Immunopharmacology 44:267–272. doi: 10.1016/S0162-3109(99)00122-8. [DOI] [PubMed] [Google Scholar]
- 19.McDowell A, Mahenthiralingam E, Dunbar KE, Moore JE, Crowe M, Elborn JS. 2004. Epidemiology of Burkholderia cepacia complex species recovered from cystic fibrosis patients: issues related to patient segregation. J Med Microbiol 53:663–668. doi: 10.1099/jmm.0.45557-0. [DOI] [PubMed] [Google Scholar]
- 20.Baldwin A, Mahenthiralingam E, Drevinek P, Pope C, Waine DJ, Henry DA, Speert DP, Carter P, Vandamme P, Lipuma JJ, Dowson CG. 2008. Elucidating global epidemiology of Burkholderia multivorans in cases of cystic fibrosis by multilocus sequence typing. J Clin Microbiol 46:290–295. doi: 10.1128/JCM.01818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, Vandamme P. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol 38:3165–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahenthiralingam E, Baldwin A, Dowson CG. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol 104:1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- 23.Vardi A, Sirigou A, Lalayanni C, Kachrimanidou M, Kaloyannidis P, Saloum R, Anagnostopoulos A, Sotiropoulos D. 4 October 2012. An outbreak of Burkholderia cepacia bacteremia in hospitalized hematology patients selectively affecting those with acute myeloid leukemia. Am J Infect Control doi: 10.1016/j.ajic.2012.04.325. [DOI] [PubMed] [Google Scholar]
- 24.Fishman JA. 2011. Infections in immunocompromised hosts and organ transplant recipients: essentials. Liver Transpl 17(Suppl 3):S34–S37. doi: 10.1002/lt.22378. [DOI] [PubMed] [Google Scholar]
- 25.Bertot GM, Restelli MA, Galanternik L, Aranibar Urey RC, Valvano MA, Grinstein S. 2007. Nasal immunization with Burkholderia multivorans outer membrane proteins and the mucosal adjuvant adamantylamide dipeptide confers efficient protection against experimental lung infections with B. multivorans and B. cenocepacia. Infect Immun 75:2740–2752. doi: 10.1128/IAI.01668-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makidon PE, Knowlton J, Groom JV II, Blanco LP, LiPuma JJ, Bielinska AU, Baker JR Jr. 2010. Induction of immune response to the 17 kDa OMPA Burkholderia cenocepacia polypeptide and protection against pulmonary infection in mice after nasal vaccination with an OMP nanoemulsion-based vaccine. Med Microbiol Immunol 199:81–92. doi: 10.1007/s00430-009-0137-2. [DOI] [PubMed] [Google Scholar]
- 27.Caraher E, Duff C, Mullen T, Mc Keon S, Murphy P, Callaghan M, McClean S. 2007. Invasion and biofilm formation of Burkholderia dolosa is comparable with Burkholderia cenocepacia and Burkholderia multivorans. J Cyst Fibros 6:49–56. doi: 10.1016/j.jcf.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Liu DF, Phillips E, Wizemann TM, Siegel MM, Tabei K, Cowell JL, Tuomanen E. 1997. Characterization of a recombinant fragment that contains a carbohydrate recognition domain of the filamentous hemagglutinin. Infect Immun 65:3465–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marzouqi I, Richmond P, Fry S, Wetherall J, Mukkur T. 2010. Development of improved vaccines against whooping cough: current status. Hum Vaccin 6:543–553. doi: 10.4161/hv.6.7.11413. [DOI] [PubMed] [Google Scholar]
- 30.Sajjan U, Wu Y, Kent G, Forstner J. 2000. Preferential adherence of cable-piliated Burkholderia cepacia to respiratory epithelia of CF knockout mice and human cystic fibrosis lung explants. J Med Microbiol 49:875–885. doi: 10.1099/0022-1317-49-10-875. [DOI] [PubMed] [Google Scholar]
- 31.Mil-Homens D, Leca MI, Fernandes F, Pinto SN, Fialho AM. 2014. Characterization of BCAM0224, a multifunctional trimeric autotransporter from the human pathogen Burkholderia cenocepacia. J Bacteriol 196:1968–1979. doi: 10.1128/JB.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mil-Homens D, Fialho AM. 2012. A BCAM0223 mutant of Burkholderia cenocepacia is deficient in hemagglutination, serum resistance, adhesion to epithelial cells and virulence. PLoS One 7:e41747. doi: 10.1371/journal.pone.0041747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry DA, Campbell ME, LiPuma JJ, Speert DP. 1997. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J Clin Microbiol 35:614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goncz KK, Feeney L, Gruenert DC. 1999. Differential sensitivity of normal and cystic fibrosis airway epithelial cells to epinephrine. Br J Pharmacol 128:227–233. doi: 10.1038/sj.bjp.0702772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sylvester FA, Sajjan US, Forstner JF. 1996. Burkholderia (basonym Pseudomonas) cepacia binding to lipid receptors. Infect Immun 64:1420–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinoy M, Dennehy R, Coleman L, Carberry S, Schaffer K, Callaghan M, Doyle S, McClean S. 2013. Immunoproteomic analysis of proteins expressed by two related pathogens, Burkholderia multivorans and Burkholderia cenocepacia, during human infection. PLoS One 8:e80796. doi: 10.1371/journal.pone.0080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Biochemistry 76:4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2007. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.UniProt Consortium. 2014. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res 42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FS. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valdes-Stauber N, Scherer S. 1994. Isolation and characterization of linocin M18, a bacteriocin produced by Brevibacterium linens. Appl Environ Microbiol 60:3809–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenkrands I, Rasmussen PB, Carnio M, Jacobsen S, Theisen M, Andersen P. 1998. Identification and characterization of a 29-kilodalton protein from Mycobacterium tuberculosis culture filtrate recognized by mouse memory effector cells. Infect Immun 66:2728–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. 2008. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol 15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 46.McPhee JB, Tamber S, Bains M, Maier E, Gellatly S, Lo A, Benz R, Hancock RE. 2009. The major outer membrane protein OprG of Pseudomonas aeruginosa contributes to cytotoxicity and forms an anaerobically regulated, cation-selective channel. FEMS Microbiol Lett 296:241–247. doi: 10.1111/j.1574-6968.2009.01651.x. [DOI] [PubMed] [Google Scholar]
- 47.Harmer C, Alnassafi K, Hu H, Elkins M, Bye P, Rose B, Cordwell S, Triccas JA, Harbour C, Manos J. 2013. Modulation of gene expression by Pseudomonas aeruginosa during chronic infection in the adult cystic fibrosis lung. Microbiology 159:2354–2363. doi: 10.1099/mic.0.066985-0. [DOI] [PubMed] [Google Scholar]
- 48.Nandi B, Nandy RK, Sarkar A, Ghose AC. 2005. Structural features, properties and regulation of the outer-membrane protein W (OmpW) of Vibrio cholerae. Microbiology 151:2975–2986. doi: 10.1099/mic.0.27995-0. [DOI] [PubMed] [Google Scholar]
- 49.McClean S. 2012. Eight stranded beta-barrel and related outer membrane proteins: role in bacterial pathogenesis. Protein Pept Lett 19:1013–1025. doi: 10.2174/092986612802762688. [DOI] [PubMed] [Google Scholar]
- 50.Qian R, Chu W, Mao Z, Zhang C, Wei Y, Yu L. 2007. Expression, characterization and immunogenicity of a major outer membrane protein from Vibrio alginolyticus. Acta Biochim Biophys Sin (Shanghai) 39:194–200. doi: 10.1111/j.1745-7270.2007.00268.x. [DOI] [PubMed] [Google Scholar]
- 51.Wei B, Dalwadi H, Gordon LK, Landers C, Bruckner D, Targan SR, Braun J. 2001. Molecular cloning of a Bacteroides caccae TonB-linked outer membrane protein identified by an inflammatory bowel disease marker antibody. Infect Immun 69:6044–6054. doi: 10.1128/IAI.69.10.6044-6054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh R, Shasany AK, Aggarwal A, Sinha S, Sisodia BS, Khanuja SP, Misra R. 2007. Low molecular weight proteins of outer membrane of Salmonella Typhimurium are immunogenic in Salmonella induced reactive arthritis revealed by proteomics. Clin Exp Immunol 148:486–493. doi: 10.1111/j.1365-2249.2007.03362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maiti B, Shetty M, Shekar M, Karunasagar I, Karunasagar I. 2012. Evaluation of two outer membrane proteins, Aha1 and OmpW of Aeromonas hydrophila as vaccine candidate for common carp. Vet Immunol Immunopathol 149:298–301. doi: 10.1016/j.vetimm.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 54.Riedel K, Carranza P, Gehrig P, Potthast F, Eberl L. 2006. Towards the proteome of Burkholderia cenocepacia H111: setting up a 2-DE reference map. Proteomics 6:207–216. doi: 10.1002/pmic.200500097. [DOI] [PubMed] [Google Scholar]
- 55.Nieves W, Heang J, Asakrah S, Honer zu Bentrup K, Roy CJ, Morici LA. 2010. Immunospecific responses to bacterial elongation factor Tu during Burkholderia infection and immunization. PLoS One 5:e14361. doi: 10.1371/journal.pone.0014361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bevivino A, Pirone L, Pilkington R, Cifani N, Dalmastri C, Callaghan M, Ascenzioni F, McClean S. 2012. Interaction of environmental Burkholderia cenocepacia strains with cystic fibrosis and non-cystic fibrosis bronchial epithelial cells in vitro. Microbiology 158:1325–1333. doi: 10.1099/mic.0.056986-0. [DOI] [PubMed] [Google Scholar]
- 57.Chiu CH, Ostry A, Speert DP. 2001. Invasion of murine respiratory epithelial cells in vivo by Burkholderia cepacia. J Med Microbiol 50:594–601. doi: 10.1099/0022-1317-50-7-594. [DOI] [PubMed] [Google Scholar]
- 58.Duff C, Murphy PG, Callaghan M, McClean S. 2006. Differences in invasion and translocation of Burkholderia cepacia complex species in polarised lung epithelial cells in vitro. Microb Pathog 41:183–192. doi: 10.1016/j.micpath.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 59.Sajjan U, Moreira J, Liu M, Humar A, Chaparro C, Forstner J, Keshavjee S. 2004. A novel model to study bacterial adherence to the transplanted airway: inhibition of Burkholderia cepacia adherence to human airway by dextran and xylitol. J Heart Lung Transplant 23:1382–1391. doi: 10.1016/j.healun.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 60.Schwab U, Leigh M, Ribeiro C, Yankaskas J, Burns K, Gilligan P, Sokol P, Boucher R. 2002. Patterns of epithelial cell invasion by different species of the Burkholderia cepacia complex in well-differentiated human airway epithelia. Infect Immun 70:4547–4555. doi: 10.1128/IAI.70.8.4547-4555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu W, Huang J, Duan B, Traficante DC, Hong H, Risech M, Lory S, Priebe GP. 2012. Th17-stimulating protein vaccines confer protection against Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 186:420–427. doi: 10.1164/rccm.201202-0182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mahenthiralingam E, Coenye T, Chung JW, Speert DP, Govan JR, Taylor P, Vandamme P. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol 38:910–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pappin DJ, Hojrup P, Bleasby AJ. 1993. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol 3:327–332. doi: 10.1016/0960-9822(93)90195-T. [DOI] [PubMed] [Google Scholar]
- 65.European Parliament and the Council of the European Union. 2010. Directive 2010/63/Eu on the protection of animals used for scientific purposes, p b7 Institute for Health and Consumer Protection, Ispra, Italy. [Google Scholar]