Abstract
Campylobacter jejuni is a prevalent cause of bacterial gastroenteritis in humans worldwide. The mechanisms by which C. jejuni survives stomach acidity remain undefined. In the present study, we demonstrated that the C. jejuni ferric uptake regulator (Fur) plays an important role in C. jejuni acid survival and acid-induced cross-protection against oxidative stress. A C. jejuni Δfur mutant was more sensitive to acid than the wild-type strain. Profiling of the acid stimulon of the C. jejuni Δfur mutant allowed us to uncover Fur-regulated genes under acidic conditions. In particular, Fur was found to upregulate genes involved in flagellar and cell envelope biogenesis upon acid stress, and mutants with deletions of these genes were found to be defective in surviving acid stress. Interestingly, prior acid exposure of C. jejuni cross-protected against oxidative stress in a catalase (KatA)- and Fur-dependent manner. Western blotting and reverse transcription-quantitative PCR revealed increased expression of KatA upon acid stress. Electrophoretic mobility shift assays (EMSAs) demonstrated that the binding affinity between Fur and the katA promoter is reduced in vitro under conditions of low pH, rationalizing the higher levels of expression of katA under acidic conditions. Strikingly, the Δfur mutant exhibited reduced virulence in both human epithelial cells and the Galleria mellonella infection model. Altogether, this is the first study showing that, in addition to its role in iron metabolism, Fur is an important regulator of C. jejuni acid responses and this function cross-protects against oxidative stress. Moreover, our results clearly demonstrate Fur's important role in C. jejuni pathogenesis.
INTRODUCTION
Campylobacter jejuni is a Gram-negative, microaerophilic, curved-rod bacterium that grows over a wide range of temperatures (30 to 47°C), with optimal growth occurring at 42°C (1, 2). C. jejuni is a major cause of foodborne diarrheal illness in humans worldwide (1). It accounts for up to 15% of all diarrheal cases in the world, with an estimated 2.5 million cases occurring per year in the United States alone (1, 3, 4). The main reservoirs for C. jejuni are the avian species and farmed poultry (5). Campylobacteriosis, the infection caused by C. jejuni, is an acute gastroenteritis with symptoms ranging from watery to bloody diarrhea and inflammation of the ileum and jejunum (5). In addition, infection with C. jejuni can lead to other complications, such as the peripheral neuropathies Guillain-Barré syndrome and Miller Fisher syndrome (3).
Enteric pathogens encounter a variety of challenges either within or outside the host (6). In order to establish a successful infection, enteric pathogens must survive the host stomach acidity (6). The stomach pH ranges from 1.5 to 5.5, depending on many variables, including the timing and type of food intake (7, 8). Therefore, enteric pathogens have developed various strategies to sense and combat gastric acidity (6).
Several studies have correlated the ferric uptake regulator (Fur) and acid survival in enteric pathogens (9–11). Fur (17 kDa) is an iron-binding transcriptional repressor (12). Under iron-replete conditions, Fur binds to iron and the Fe2+-Fur complex binds to DNA at a 19-bp inverted repeat sequence known as the Fur box in the promoters of target genes (13). Fur binding to the promoter regions of target genes prevents the recruitment of RNA polymerase and thereby represses gene expression (13). In addition to its role in regulating iron homeostasis, Fur has recently been identified to be the first known transcriptional regulator in Helicobacter pylori that is required for growth under acidic conditions (14). Likewise, a fur mutant of an Escherichia coli strain causing avian septicemia was defective for acid survival and was unable to mount an acid tolerance response (ATR) like that observed in the wild-type (WT) strain (10). Similarly, Fur plays an important role in the acid survival of both Shigella flexneri and Salmonella enterica serovar Typhimurium (9, 11). Fur represses the expression of ryhB, a small regulatory RNA that negatively regulates evgA and ydeP, which are involved in S. flexneri acid survival (11). The fur mutant of S. Typhimurium was defective for acid survival and lacked the inducible pH homeostasis system that is associated with the ATR in wild-type bacteria (9). In addition to its role in acid stress survival, Fur plays an important role in the survival of many bacteria, such as E. coli and H. pylori, in the presence of various environmental stresses (e.g., oxidative stress) (15). Importantly, Fur regulates the expression of many oxidative stress defense genes (e.g., sodB and katA) in enteric pathogens, such as E. coli and C. jejuni (15–18). In addition to oxidative stress genes, Fur also regulates hmp, which encodes a flavohemoglobin that protects S. Typhimurium from nitric oxide stress (19). Therefore, it is suggested that Fur could help bacteria survive harsh conditions and thereby enhance bacterial pathogenesis in the host (15).
As an enteric pathogen, the capacity to survive gastric acidity is a fundamental requirement for C. jejuni to colonize the host and cause disease (20). In C. jejuni, Fur is known to regulate the expression of genes belonging to several functional groups, including energy metabolism, iron acquisition, cell membrane biogenesis, and oxidative stress defense (18, 21, 22). The crystal structure of C. jejuni Fur (CjFur) has recently been characterized, providing a greater understanding of its regulatory function (18). Four different modes of Fur regulation of target genes have been identified in C. jejuni: apo- and holo-CjFur gene repression and activation (18, 23). Moreover, analysis of the CjFur crystal structure revealed the presence of two occupied Zn2+-binding sites (the S1 and S3 sites), in addition to the putative regulatory iron-binding S2 site, per protomer (18). However, the contribution of the CjFur regulatory S2 site to the acid response of C. jejuni has not been characterized previously.
In the present study, we aimed to characterize the role of Fur in C. jejuni acid survival. We studied the transcriptional profile of a C. jejuni Δfur mutant under acidic conditions using a microarray-based approach. We identified C. jejuni genes that were regulated by Fur and acid stress. The contribution of iron sensing to Campylobacter acid survival was also determined. Moreover, we characterized the involvement of Fur in the acid-induced cross-protection of C. jejuni against oxidative stress. Finally, the importance of Fur for C. jejuni virulence in human epithelial cells and host pathogenesis was investigated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni NCTC11168 was acquired from the National Collection of Type Cultures. The bacterial strains used in this study are listed in Table 1. The growth of C. jejuni strains was conducted on Mueller-Hinton (MH) agar plates and MH biphasic cultures as previously described (20). The cultures were incubated at 37°C under microaerophilic conditions (8% O2, 4% H2, 5% CO2, 83% N2) in a MACS-VA500 workstation (Don Whitley, West Yorkshire, England).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | endA1 hsdR17(rK− mK−) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA argF)U169 deoR [ϕ80dlacΔ(lacZΔM15)] | Invitrogen |
| C. jejuni | ||
| AS144 | C. jejuni NCTC11168 | National Collection of Type Cultures |
| AS230 | AS144 Δfur::Camr | Palyada et al. (21) |
| AS960 | AS230 furWT::Camr Kanr | Palyada et al. (17) |
| AS433 | AS144 ΔkatA::Camr | Palyada et al. (21) |
| AS1350 | AS230 furΔS2::Camr Kanr | This study |
| AS1071 | AS144 Δcj0012c::Camr | Flint et al. (31) |
| AS742 | AS144 Δcj1026c::Camr | Flint et al. (31) |
| AS748 | AS144 Δcj0818::Camr | Flint et al. (31) |
| AS751 | AS144 Δcj1024c::Camr | Flint et al. (31) |
| AS757 | AS144 ΔflgG::Camr | Flint et al. (31) |
| AS761 | AS144 ΔflhB::Camr | Flint et al. (31) |
| AS764 | AS144 ΔflgD::Camr | Flint et al. (31) |
| AS765 | AS144 ΔflgK::Camr | Flint et al. (31) |
| AS768 | AS144 ΔflgE2::Camr | Flint et al. (31) |
| AS1336 | AS144 ΔmotA ΔmotB::Kanr | Flint et al. (31) |
| Plasmids | ||
| pRR-Km | Cloning vector used for complementation of mutants, Kanr | Reid et al. (20) |
| pStrepSumofur | Vector used for furΔS2 site-directed mutagenesis | Butcher et al. (18) |
Camr, chloramphenicol resistance gene; Kanr, kanamycin resistance gene.
Site-directed mutagenesis of the furΔS2 mutant.
Site-directed mutagenesis to obtain a fur mutant in which the S2 site was mutated to disrupt iron binding and then inserted into a Δfur background (furΔS2 mutant) was conducted using a QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's recommendations. The furΔS2 mutant was constructed by mutating two histidine residues to alanines (His43Ala-His102Ala) using pStrepSumofur (18) as a template and primers furΔS2H43A(F), furΔS2H43A(R), furΔS2H102A(F), and furΔS2H102A(R) (Table 2). The sequences of the resulting mutants were confirmed by DNA sequencing (Centre de Recherche du CHUL [CHUQ], Québec, Canada).
TABLE 2.
Primer used in this study
| Primer purpose and name | Primer sequencea (5′-3′) |
|---|---|
| RT-qPCR | |
| Cj0414F (+) | CTGTTTTAAAGGCAGCAGAACTTAC |
| Cj0414R (−) | CTTCTCCTTGCTCATCTTTAGG |
| Cj0415F (+) | GCTTTAGGTTCTATGGTGGCTTT |
| Cj0415R (−) | AAAAGTGGATCACCCCAAGA |
| Cj0448cF (+) | GGAACATTGCATAGAAGTGTAGATG |
| Cj0448cR (−) | CTAGTTTTCTTACTTCATCGGCAAC |
| clpBF (+) | AAGCCGTACGAAGAAAACCTTATAG |
| clpBR (−) | AATCCACTGTTACACCTTTGCTATC |
| flaBF (−) | CGAACCAATGTCTGCTCTGA |
| flaBF (+) | GCAGGCTCAGGTTTTTCAG |
| grpEF (+) | GCTTTAGAAGCAGCTGTTAATG |
| grpER (−) | CATCTTTGATAAGAGCCACTCC |
| katAF (+) | CTTTAGTCCAAGCAATATCGTTCC |
| katAR (−) | CAGCGACATTGTAAGTATTCACTTC |
| metBF (+) | AAACTTTAGCATTACACGGAGCTT |
| metCF (+) | CTAAACTTATTCATTGTGGCAGAGG |
| metCR (−) | CTCTGTATTTTTCCAAGTTGCGTG |
| metBR (−) | CCCTCCTTCGACATTAGCAA |
| rpsLF (+) | CAAAGAAGGGGAGTTTGCAC |
| rpsLR (−) | TATCAAGAGCACCACGAACG |
| chuA (+) | TGCTTTCAAAATTCATTTTTATCC |
| chuA (−) | TTGGGTGCAAATTTTACTCCTT |
| cfrA (+) | TTGCGCCATTGGTCTCTTA |
| cfrA (−) | GCAACATCTCTATAAGGCTTACTT |
| slyD (+) | TACGATGAAAATGCCGTTCA |
| slyD (−) | TTCGCCAAAAAGCTCCATAC |
| Directed mutagenesis (generation of furΔS2 mutant) | |
| furΔS2H43A(F) | AAAACTCTTTATCACAGTGATACTGCCTACACACCCGAAAGTTTATATATG |
| furΔS2H43A(R) | CATATATAAACTTTCGGGTGTGTAGGCAGTATCACTGTGATAAAGAGTTTT |
| furΔS2H102A(F) | CTTGCCAATAAACCTCACCATGATGCCATGATATGTAAAAATTGCGGAAAA |
| furΔS2H102A(R) | TTTTCCGCAATTTTTACATATCATGGCATCATGGTGAGGTTTATTGGCAAG |
| Transformation of furΔS2 into C. jejuni Δfur | |
| JBCL-Fur4 | GATTTAGATGTCTAGACTCAAAAAGGGGAGTGATATGCTGATAGAAAATGTGGA |
| JBCL-Fur2R | GGGGAAGCTTTCTAGTTATATTTTTACCTTTGCTT |
| AR55 | ATGACATTGCCTTCTGCGT |
| EMSAs | |
| JFC1584 | Cy5-TGCATTTTATTGATAATAAATTTCAAAATAAATTTAGTTT |
| JFC1585 | Cy5-AAACTAAATTTATTTTGAAATTTATTATCAATAAAATGCA |
Underlined sequences represent mutated nucleotides.
Complementation of a C. jejuni Δfur mutant with furΔS2.
The furΔS2 gene construct was introduced into a well-characterized C. jejuni Δfur mutant (21) using the pRR-Km plasmid as previously described (20). The furΔS2 nucleotide sequence was PCR amplified from the pStrepSumofur plasmid containing the furΔS2 mutated insert using Phusion Hot Start II high-fidelity DNA polymerase (Thermo Scientific) and the primers JBCL-Fur4 and JBCL-Fur2R (Table 2). The primers were designed to incorporate a 15-bp region of homology with each end of the XbaI-digested pRR-Km. The furΔS2 PCR product was directionally cloned into the pRR-Km plasmid using an In-Fusion PCR cloning kit (Clontech) following the manufacturer's instructions. The resulting construct was sequenced to confirm the absence of PCR-induced errors in the insert. This final construct was used to transform the C. jejuni Δfur mutant as previously described (20, 21, 24), and transformants were selected on MH agar plates containing both kanamycin (10 μg/ml) and chloramphenicol (20 μg/ml). Finally, the chromosomal insertion of the furΔS2 gene construct into the rRNA locus was confirmed by PCR analysis using the primers AR55 and JBCL-Fur2R (Table 2) as described previously (20).
Acid survival assays.
C. jejuni cells (the wild type and mutants) were grown to the logarithmic phase (optical density at 600 nm [OD600], ∼0.8) in biphasic MH medium. The acid survival assays were performed as previously described by Reid et al. (20) and Le et al. (25). Briefly, bacterial strains were exposed to pH 3 or pH 4 in MH-HCl medium. Aliquots of 2.5 and 5 ml of bacterial culture were added to 10 ml of MH broth adjusted to pH 2.6 or 3 using concentrated HCl. The final pHs of the assay mix were 3 and 4, respectively, and were confirmed using a pH meter. Samples were withdrawn immediately, and at 2-, 4-, 6-, and 8-min intervals after exposure to acid, they were serially diluted into phosphate-buffered saline (PBS; pH 7.4) and plated onto MH agar plates. The plates were incubated at 37°C under microaerophilic conditions for 48 h, and the colonies were counted. The percent bacterial survival (with the percent bacterial survival at time zero representing 100%) was determined as a function of the duration of acid exposure. The results are expressed as the means ± standard errors from three independent experiments. A P value of <0.05 by a two-way analysis of variance (ANOVA) followed by a Bonferroni multiple-comparison test (GraphPad Prism, version 5.03, for Windows; GraphPad Software, San Diego, CA, USA) was considered significant.
Total RNA extraction for real-time reverse transcription-quantitative PCR (RT-qPCR) and microarray analysis.
Wild-type and Δfur mutant C. jejuni strains were grown to mid-log phase in MH broth (pH 7.4) under microaerophilic conditions. The bacteria were exposed to neutral (pH 7) or acidic (pH 3 or 4 for 8 min) conditions in HCl-adjusted MH broth as described above for the acid survival assay. RNA turnover was prevented by adding a 1/10 volume of cold RNA degradation stop solution (10% [vol/vol] buffer-saturated phenol, pH 4.3, in absolute ethanol) (26). Bacterial cells were collected by centrifugation (8,000 × g, 10 min, 4°C), and the cell pellet was resuspended in TE buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA). Total RNA was extracted using a hot phenol-chloroform method (27) and precipitated with absolute ethanol by overnight incubation at −80°C. The RNA was washed three times with 80% cold ethanol to remove any impurities and was resuspended in RNase-free double-distilled H2O (ddH2O). The contaminating genomic DNA was removed from the RNA preparation with an RNase-free DNase I treatment (Epicenter Biotechnologies, Madison, WI). The obtained RNA was further purified using a Qiagen RNeasy minikit (Qiagen, Valencia, CA), and PCR amplification was used to confirm that the preparation was free of genomic DNA. The RNA quality and quantity were ascertained using a Bio-Rad Experion RNA StdSens analysis kit following the manufacturer's protocols. RNA samples were stored at −80°C until further use.
Microarray probe labeling and slide hybridization.
The protocol described by Palyada et al. (21) for probe labeling and slide hybridization was followed in the present study. Briefly, 10 μg of total RNA from each control (C. jejuni wild type) and test sample (Δfur mutant) was converted to cDNA using SuperScript II reverse transcriptase (Invitrogen); 10 μg of random hexamers (Amersham Biosciences); a deoxynucleoside triphosphate mixture of 0.5 mM each dGTP, dATP, and dCTP; 0.16 mM dTTP; and 0.34 mM aminoallyl-dUTP (21). The aminoallyl-labeled cDNA was purified from free amines and unincorporated aminoallyl-dUTP by adding 350 μl of water and spinning the mixture through a Microcon YM-30 filter (Millipore) for 8 min at 8,000 × g, followed by washing with sterile ddH2O. Following concentration and resuspension in NaHCO3, pH 9.0, the aminoallyl-labeled cDNA was coupled to either Cy3 dye (used to label control samples) or Cy5 dye (used to label test samples) by adding 10 μl of Cy3 or Cy5 (GE Healthcare) in dimethyl sulfoxide, followed by incubation in the dark for 1 h at room temperature. Next, the fluorescently labeled cDNA was purified using a QIAquick PCR purification kit according to the manufacturer's instructions (Qiagen). Fluorescent Cy3- and Cy5-labeled cDNAs were combined, and the fluorescently labeled cDNA mix was dried under vacuum with a SpeedVac apparatus and resuspended in 15.14 μl of water, to which the following was added: 2.5 μl of salmon sperm DNA (10 mg/ml), 9 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7), 0.36 μl of 10% SDS, and 9 μl of formamide. The microarray slides used in this study were constructed using PCR-amplified fragments that represent the open reading frames (ORFs) identified in the C. jejuni NCTC11168 genome (2). Prior to hybridization, the arrays were prehybridized at 42°C for 45 min in prehybridization buffer (25% formamide, 5× SSC buffer, 0.1% sodium dodecyl sulfate [SDS], 1% bovine serum albumin [BSA]), rinsed with water, and dried by spinning. The combined probes were denatured for 2 min at 99°C and applied to the microarray slide underneath a coverslip. The slides were placed in a humidified chamber (Arrayit, Sunnyvale, CA) and incubated in the dark overnight at 42°C. Next, the microarray slides were washed once for 5 min at 42°C in 100 ml 2× SSC, 0.1% SDS, once for 10 min in 100 ml 0.1× SSC, 0.1% SDS, and four times for 1 min each time at room temperature in 400 ml 0.1× SSC. The slides were then rinsed with distilled water and dried by centrifugation. Finally, the slides were scanned using a laser-activated confocal scanner (Scan-Array Gx; PerkinElmer) at a 10-μm resolution.
Data collection and analysis.
The signal intensities of each spot were collected using ScanArray software (PerkinElmer). The spot intensities were normalized via locally weighted linear regression (LOWESS) using the MIDAS program (available from the J. Craig Venter Institute [formerly the Institute for Genomic Research]; http://www.jcvi.org/cms/research/software/) as previously described (17, 21, 28). Microarray data were collected from three independent biological replicates with three technical replicates each for both the test and control samples. Finally, the ratio of channel 2 (Cy5) to channel 1 (Cy3) was converted to a log2 value, and the data were statistically analyzed using the empirical Bayes method, as previously described (29). The differentially expressed genes (those with a >1.5-fold differential expression with a P value of <10−4) were subjected to hierarchical clustering using the Genesis program (available from Graz University of Technology, Graz, Austria; http://genome.tugraz.at).
RT-qPCR. (i) Validation of microarray results.
The relative expression levels of four genes (cj0448c, clpB, grpE, and katA) that were upregulated in the Δfur mutant relative to their regulation in the wild-type strain under acidic conditions and four genes (cj0414, cj0415, flaB, and metB) that were downregulated in the Δfur mutant relative to their regulation in the wild-type strain were analyzed by RT-qPCR. The relative expression level of each gene was normalized to that of rpsL, a gene for which the expression levels remained unchanged in the microarray analysis between the Δfur mutant and wild-type strain. Primers were designed using Primer3Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/) and are listed in Table 2. The analysis was conducted as described previously (2, 21, 28) using a 7300 real-time PCR system (Applied Biosystems) and followed the protocol described in the QuantiTect SYBR green RT-PCR kit (Qiagen). Specific PCR amplification was confirmed by both agarose gel electrophoresis and a melting curve analysis of the products according to the manufacturer's recommendations. The comparative threshold cycle (ΔΔCT) method was used to calculate the relative gene expression (2, 28). The log2 ratio values of gene expression obtained by the microarray analysis were plotted against the log2 ratio values of the relative quantities from the RT-qPCR. Finally, the coefficient of determination (R2) was determined as a measure of the degree of correlation between the microarray data and the real-time RT-qPCR data.
(ii) Quantification of katA.
The relative expression levels of the katA transcripts in both acid-stressed and unstressed C. jejuni NCTC11168 cells were determined as described above. The expression levels of katA were normalized to those of metC (a putative cystathionine beta-lyase), for which the expression levels remained unchanged in acid-stressed and unstressed C. jejuni cells.
(iii) Effect of S2 mutation on iron-dependent Fur regulation.
The relative expression levels of the chuA and cfrA transcripts in the C. jejuni wild-type strain, the Δfur mutant, the Δfur+furΔS2 mutant, and the Δfur mutant complemented with the wild-type fur gene (the Δfur+furWT strain) were determined under iron-replete conditions (18), as described above. RNA was isolated from C. jejuni strains grown to mid-log phase in minimum essential medium alpha (MEMα) supplemented with 10 mM sodium pyruvate and 40 μM FeSO4 under microaerophilic conditions. The relative expression levels of chuA and cfrA were normalized to those of slyD as an endogenous control.
Motility assay.
The motility of the C. jejuni wild type and mutants (the ΔflgD, ΔflgE2, ΔflgH, ΔflgK, ΔflhB, and ΔmotAB mutants) was assayed on 0.4% MH agar plates as previously described (30). Bacterial strains were cultured overnight in biphasic MH medium under microaerophilic conditions. The bacterial cultures were then diluted to an OD600 of 0.02. Ten microliters of bacterial suspension was stabbed into a 0.4% MH agar plate, and the plate was incubated at 37°C under microaerophilic conditions for 24 h. The results are presented as the mean diameter (in millimeters) ± standard error of bacterial migration from the site of inoculation for three biological experiments with three technical replicates each. Statistical analysis was performed using one-way ANOVA followed by a Bonferroni multiple-comparison test, and a P value of <0.05 was considered significant.
Oxidative stress experiments. (i) Disk inhibition assay.
C. jejuni strains were acid stressed by exposing an overnight culture of C. jejuni to acidic conditions (pH 4 for 8 min) in HCl-adjusted MH broth as described above for the acid survival assay. Note that both the C. jejuni Δfur mutant and the wild-type strain remained 100% viable at pH 4 for at least 8 min. After centrifugation at 8,000 × g for 5 min at room temperature, the bacterial pellets were gently washed twice in MH broth (pH 7.4) without resuspending the cells. Next, washed cells were resuspended in MH broth to an OD600 of 1.0. For the control (unstressed bacteria), the same steps except for the exposure to acid were performed. Next, 1 ml of a bacterial suspension in MH broth (acid-stressed or unstressed bacteria) was added to 24 ml of molten MH agar (cooled to 45°C), and the mixture was poured into petri dishes and allowed to solidify. Ten microliters of different molar concentrations of H2O2 (250 to 1,000 mM) were pipetted on top of a 6-mm-diameter paper disk that had been placed on the surface of each MH agar plate, and the plates were then incubated for 24 h at 37°C under microaerophilic conditions (20). The H2O2 sensitivities of both acid-stressed and unstressed C. jejuni strains were determined by measuring the diameters of the growth inhibition zones around the paper disks. The results are expressed as the means ± standard errors from three independent experiments. The data were statistically analyzed using a Student t test. A P value of <0.05 was considered significant.
(ii) Kill curve of C. jejuni exposed to H2O2.
Acid-stressed and unstressed C. jejuni cells (prepared as described above for the disk inhibition assay) were exposed to 10 mM H2O2 in MH broth. The percent bacterial survival for both acid-stressed and unstressed C. jejuni cells was determined immediately and at 4, 8, 15, and 30 min after exposure to H2O2. The percent bacterial survival was determined by counting the viable cells after serially dilution into PBS (pH 7.4), plating of the solution on MH agar plates, and incubation for 48 h at 37°C under microaerophilic conditions. The survival of bacteria exposed to H2O2 was expressed as the percent survival (with the survival at time zero representing 100%) as a function of the duration of exposure to H2O2. The results are expressed as the means ± standard errors from three independent biological experiments. The difference between the capacities of both acid-stressed and unstressed C. jejuni cells to survive exposure to H2O2 was considered significant when the P value was <0.05 using a Student t test.
Western blot analysis.
Five micrograms of protein lysates from acid-stressed and unstressed C. jejuni NCTC11168 cells (prepared as described above for the disk inhibition assay) were separated by SDS-PAGE on a 12% denaturing gel. The proteins were immediately transferred from the gel to a polyvinylidene difluoride (PVDF) membrane (Millipore). The membranes were blocked by overnight incubation in 5% (wt/vol) skim milk and 0.1% Tween 20 in PBS at 4°C. Next, the membranes were incubated with 0.1 μg/ml anti-KatA antiserum (31) for 1 h, followed by three washes with 0.1% Tween 20. Finally, the membranes were incubated for 1 h with a 1:3,000 dilution of anti-rabbit immunoglobulin horseradish peroxidase (HRP)-conjugated antibody (Invitrogen) followed by three washes with 0.1% Tween 20. The immunoblot membrane was developed with a 1:1 mixture of luminol-peroxide solution (Thermo Scientific) for 1 min, and chemiluminescence was detected by the use of X-ray film (Thermo Scientific). Densitometric analysis of the immunoblotting results was performed using ImageJ (version 1.45s) software (http://imagej.nih.gov/ij/).
Electrophoretic mobility shift assays (EMSAs) and calculation of Kds.
The gel shift assays were performed as previously described (18) using the JFC1584 and JFC1585 primers listed in Table 2 and purified recombinant CjFur (18). Forward and reverse Cy5-labeled primers corresponding to 40-bp DNA fragment of the katA promoter region were purchased from Eurofins MWG Operon. Oligonucleotides (10 μM each) were annealed by incubation at 95°C for 10 min and slowly cooled to room temperature in 50 μl of annealing buffer (10 mM Tris, pH 8.0, 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol). For the gel shift assay, 700 nM purified recombinant CjFur (18) was incubated in a final assay volume of 20 μl binding buffer (20 mM bis-Tris borate, 50 mM KCl, 3 mM MgCl2, 5% glycerol, 0.1% Triton X-100, 50 μM MnCl2) for 30 min on ice. The binding buffer was adjusted to the desired pH using a saturated solution of boric acid. Next, the Cy5-katA fragment (1 nM) was added to CjFur and the mixture was incubated for 30 min on ice in the presence of 1 μg poly(dI-dC). The samples were run on a 6% (wt/vol) nondenaturing polyacrylamide gel (19:1) for 50 min at 100 V and 4°C. The gels were freshly prepared with 100 mM bis-Tris borate and 100 μM MnCl2 and were preelectrophoresed at 150 V for 30 min at 4°C. As a control, the binding affinity between C. jejuni PerR (CjPerR) and the katA promoter region was determined under the same conditions. The apparent dissociation constant (Kdapp) of Fur-katA binding under neutral or acidic conditions was determined as described previously (32, 33). The concentration of CjFur (in nanomolar) required to reach half-maximal binding to the katA promoter was calculated using SigmaPlot software. The difference between the dissociation constants (Kds) of CjFur-katA binding under neutral and acidic conditions was considered statistically significant at a P value of <0.05 using the Student t test. For all gel shift assays, the gels were scanned using a Typhoon scanner (Typhoon Trio; GE Healthcare), and the scans were analyzed using ImageQuant TL software (GE Healthcare Life Sciences).
Bacterial interaction with epithelial cells. (i) Epithelial cells.
Human colonic epithelial HCT116 cells were obtained from the American Type Culture Collection and were routinely maintained in MEMα (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% nonessential amino acids. The cells were grown and maintained without antibiotics at 37°C in a 5% CO2 humidified atmosphere. For adherence and invasion assays, confluent HCT116 cells were harvested by trypsinization in 0.01% EDTA. HCT116 cells were adjusted to 106 cells/ml by counting in a hemocytometer, seeded in a 24-well tissue culture plate, and incubated at 37°C in a 5% CO2 humidified atmosphere until a confluent monolayer formed. Prior to the adhesion and invasion assays, the cell monolayers were washed twice with Hanks' balanced salt solution (HBSS) (34) at pH 7.4.
(ii) Adherence and invasion assays.
Assays of the adherence to and invasion of HCT116 cells by the wild-type, fur mutant, and fur-complemented C. jejuni strains were performed as described by Poly et al. (35). Briefly, suspensions of C. jejuni strains were inoculated separately into wells containing confluent monolayers of HCT116 cells at a targeted multiplicity of infection (MOI) of 1,000:1. The actual numbers of bacteria in the inocula added to the monolayers were confirmed retrospectively by serial dilution and plate counting. Bacterium-infected HCT116 cells were incubated for 3 h at 37°C in 5% CO2 to allow bacterial adherence and internalization. To determine the number of bacteria adhering to and internalized in the eukaryotic cells, the epithelial cells were washed twice with HBSS and lysed with 0.1% Triton X-100. The total number of bacteria associated with the eukaryotic cells (extracellular and intracellular bacteria) was determined by serial dilution of the lysates in PBS (pH 7.4), plating of the dilutions on MH agar plates, and counting of the resultant colonies after incubation for 48 h under microaerophilic conditions. To measure bacterial invasion, a gentamicin protection protocol was employed (35). C. jejuni-infected eukaryotic cells were washed twice with HBSS and incubated in fresh 10% FBS–MEMα containing 250 μg/ml gentamicin sulfate for 1 h to kill the remaining viable extracellular bacteria. Intracellular bacteria were determined by washing the infected HCT116 cells twice with HBSS, and the cells were lysed with 0.1% Triton X-100. Following serial dilution in PBS, the released intracellular bacteria were counted as described above by plate counting. The number of bacteria adhering to HCT116 cells was obtained by subtracting the number of intracellular bacteria from the total number of bacteria recovered from eukaryotic cells not subjected to gentamicin treatment. The adhesion and invasion results are expressed as the percentages of adhering and invading bacteria relative to the number of bacteria in the infection dose. The results are expressed as the means ± standard errors from three independent biological experiments with three technical replicates each. Data were statistically analyzed by using one-way ANOVA followed by the Bonferroni multiple-comparison test at a 5% level of significance.
(iii) Intraepithelial cell survival assays.
The intracellular survival of C. jejuni strains within HCT116 cells was determined as previously described (24). After bacterial invasion and gentamicin treatment as described above for the binding and invasion assays, infected HCT116 cells were washed three times with HBSS and either lysed with 0.1% Triton X-100 to recover the intracellular bacteria (4 h time point) or further incubated for 24, 48, and 72 h in 10% FBS–MEMα with no antibiotics. Following incubation, the monolayers were washed twice with HBSS and lysed with 0.1% Triton X-100 to determine the numbers of intracellular bacteria at different time points. The numbers of viable intracellular bacteria were determined as described above for the adhesion and invasion assays by serial dilution in PBS and plating on MH agar plates. The results are expressed as the means ± standard errors from three independent biological experiments with at least three technical replicates each. The difference between C. jejuni strains was considered significant at a P value of <0.05 using a two-way ANOVA followed by the Bonferroni multiple-comparison test.
Galleria mellonella larva infection.
The pathogenesis of C. jejuni strains (the NCTC11168 wild-type strain, the Δfur mutant, and the Δfur+furWT strain) was characterized using in vivo G. mellonella killing assays as described previously (36, 37). Larval survival and 50% lethal doses (LD50s) were determined by injection of the C. jejuni strains into G. mellonella. C. jejuni strains were grown to the logarithmic phase in biphasic MH medium at 37°C under microaerophilic conditions. Bacterial pellets were washed in PBS (pH 7.4) and resuspended in the same buffer to the desired bacterial density. The infectious doses (numbers of CFU per milliliter) of both the test and control strains were determined by serial dilution and colony counting on MH agar plates. G. mellonella larvae were obtained from Gecko Gurl (Ottawa, ON, Canada), and only larvae measuring 2.0 to 2.5 cm in length and having a cream-colored cuticle with minimal speckling or discoloration were used. After delivery, the larvae were allowed to acclimate in the lab for at least 24 h by storage in the dark at 10 to 15°C and were used within 10 days. Ten microliters of 10-fold serial dilutions of C. jejuni (107 to 105 CFU in PBS) were injected into the hemocoel of the left hindmost proleg of each larva in cohorts of 10 larvae using a Hamilton 10-μl 901RN syringe (Hamilton Microliter). The infected larvae were maintained in vented petri dishes containing wood chips and were incubated at room temperature under aerobic conditions. Mortality, survival, and the appearance of the G. mellonella larvae were monitored every 24 h for 6 days following inoculation. As negative controls, 10 μl of sterile PBS was injected into each larva in a cohort of 10 larvae, and PBS was not injected into another cohort of 10 larvae. For microscopic examination of C. jejuni-infected larvae, the larvae were chilled on ice, the rear 2 mm of each larva was removed aseptically, and the hemocoel was drained into a sterile microcentrifuge tube. The hemocoel was subjected to Gram staining, and the slides were examined using a BX5 microscope (Olympus Inc., Center Valley, PA). Representative images were photographed with an Olympus DP70 camera. For both acid-stressed and unstressed C. jejuni bacteria, the experiment was performed in at least three independent replicates. Survival curves were plotted using the Kaplan-Meier method, and differences in survival were calculated using the log-rank test (GraphPad Prism). The LD50 was calculated using the Probit method (XLstat 2010 software; Addinsoft, New York, NY, USA) as previously described (37), and differences in bacterial virulence were compared using the Mann-Whitney test. A P value of <0.05 was considered statistically significant.
Microarray data accession number.
Details of the microarray construction and a complete list of the genes represented on the microarrays are available in the Gene Expression Omnibus database under accession number GSE73796.
RESULTS
C. jejuni Δfur is more sensitive to acid than the wild-type strain.
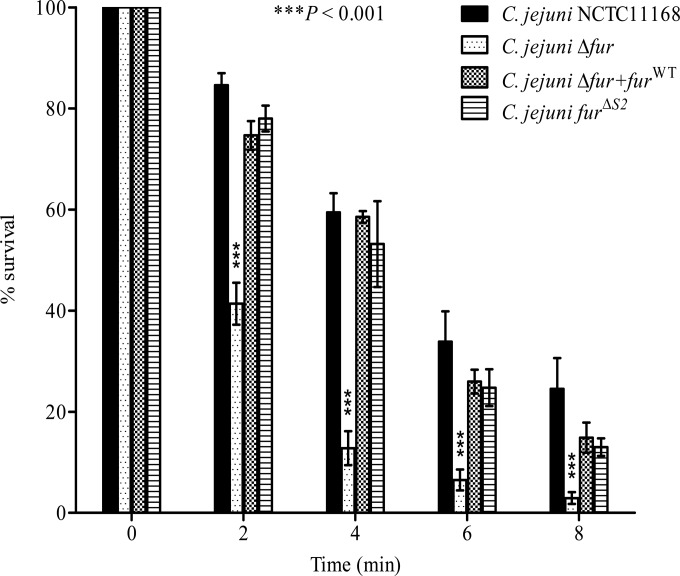
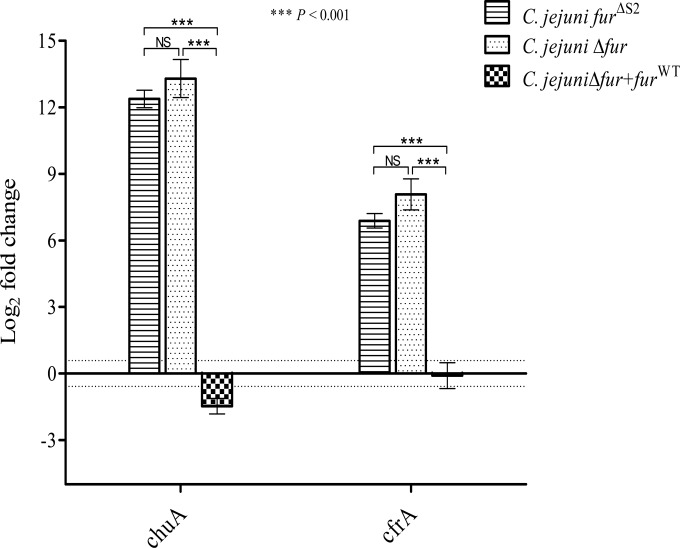
C. jejuni lacks many of the stress regulators (e.g., RpoS) known to be involved in the acid response in other enteropathogens (38–40). Fur participates, directly or indirectly, in the regulation of genes encoding proteins involved in the stress response of many bacteria, such as H. pylori and Salmonella (14, 15). Therefore, we hypothesized that Fur might play a role in the regulation of the acid response genes in C. jejuni. To characterize this role, we compared the capacities of the C. jejuni Δfur mutant and the wild-type strain to survive under acidic conditions. The C. jejuni Δfur mutant was significantly more sensitive to severe acidic conditions (pH 3) than the wild-type strain (Fig. 1). Complementation of the Δfur mutant with the fur gene restored the acid sensitivity phenotype (Fig. 1), confirming the role of Fur in the acid stress response. To test whether this effect was iron dependent or independent, we performed site-directed mutagenesis of the iron-regulatory S2 binding site followed by insertion of this mutant into a Δfur background. The 2 iron-coordinating histidines from the S2 site were replaced by alanines to disrupt iron binding. To confirm the loss of iron sensing, the expression levels of the well-characterized iron-regulated genes chuA and cfrA were monitored in the fur mutant complemented with furΔS2. As shown in Fig. 2, furΔS2 complementation of the fur mutant did not restore chuA and cfrA iron-dependent repression. This suggests that the S2 site is required for iron sensing in C. jejuni Fur. Next, we tested whether the complementation with the furΔS2 gene could restore the acid sensitivity of the Δfur mutant (Fig. 1). As shown in Fig. 1, complementation of the fur mutant with furΔS2 was sufficient to restore acid sensitivity to a level comparable to that for the wild-type strain, demonstrating that the role of Fur in C. jejuni acid stress response is iron independent.
FIG 1.
The C. jejuni Δfur mutant is more acid sensitive than the NCTC11168 wild type. Bacteria were grown to logarithmic phase in a biphasic MH medium culture and were then exposed to acidic conditions at pH 3. Samples were withdrawn immediately and at 2, 4, 6, and 8 min after acid exposure to determine cell viability. Complementation with furWT or furΔS2 restored the acid sensitivity of the C. jejuni Δfur mutant. Data from a minimum of three independent experiments are shown as the mean percent survival ± standard error. P values were determined using a two-way ANOVA followed by the Bonferroni multiple-comparison test.
FIG 2.
RT-qPCR reveals increased expression of chuA and cfrA in C. jejuni Δfur+furΔS2 and Δfur compared to that in the C. jejuni wild-type and Δfur+furWT strains. There was no difference in the expression levels of chuA and cfrA between the C. jejuni Δfur+furΔS2 and Δfur mutants. The data shown are the means ± standard errors from three biological experiments each with three technical replicates. P values were determined using a Student unpaired t test. The dotted lines represent the 1.5-fold cutoff value. NS, not significant.
Transcriptional profile of the C. jejuni Δfur mutant at low pH.
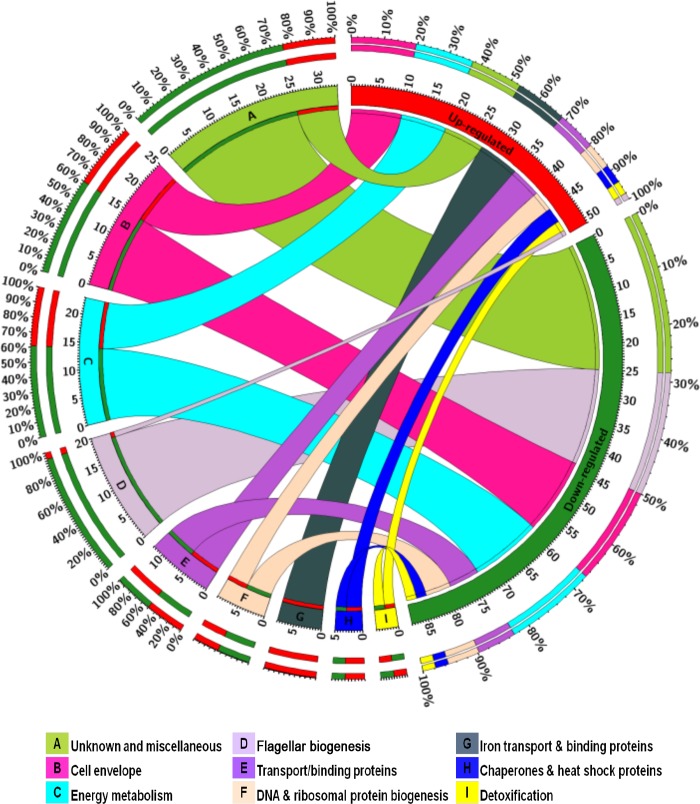
The acid survival assays showed that the Δfur mutant is defective in acid survival compared to the C. jejuni wild type. As a first step to better understand the mechanism of acid survival in C. jejuni, we investigated the transcriptional profiles of both the C. jejuni wild-type strain and the Δfur mutant under low-pH conditions. The main objective of our microarray experiment was to identify C. jejuni genes that are both Fur and acid regulated. A total of 141 genes were identified. These differentially expressed genes were subjected to hierarchical clustering analysis (see Fig. S1 in the supplemental material) and were grouped into three major clusters (designated groups A, B, and C; see Table S1 in the supplemental material). Also, up- and downregulated genes were grouped by functional category (Fig. 3). The microarray data were validated using RT-qPCR for a subset of genes that was either up- or downregulated by transcriptome profiling. A strong correlation (R2 = 0.85) between the data obtained from the microarray experiment and those obtained by RT-qPCR was identified.
FIG 3.
Functional categorization of Fur- and acid-responsive genes. The total numbers of up- and downregulated genes are represented in red and green segments, respectively. Colored segments labeled A to I represent each functional category, and the ribbon size indicates the number of genes within each category that are either up- or downregulated. The outermost colored segments represent the relative contribution of each functional category to the total number of up- and downregulated genes. The figure was constructed using Circos Table Viewer (46).
The role of differentially expressed genes in Campylobacter acid survival.
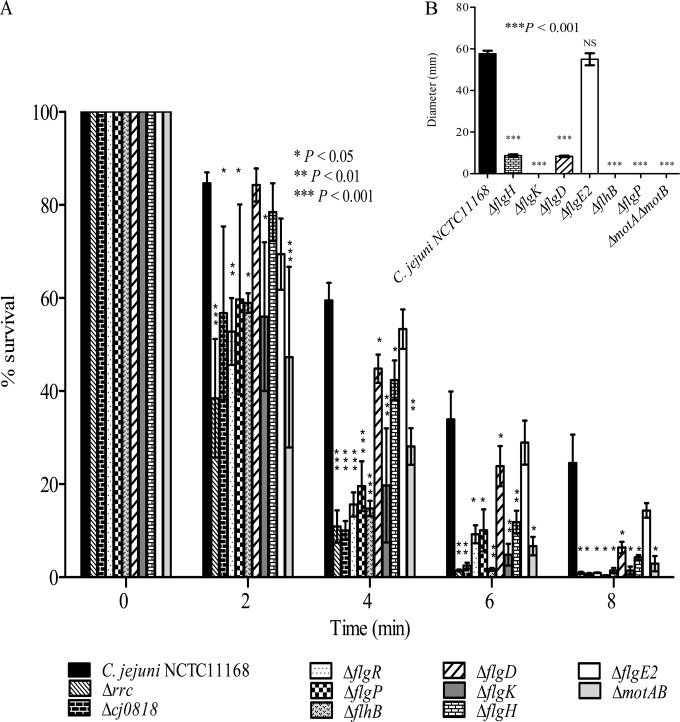
The contribution of differentially expressed genes identified by microarray analysis to C. jejuni acid survival was further characterized by testing the capacity of mutants with deletion mutations of various highly differentially expressed genes to survive acid stress. The tested mutants included those with deletion mutations of genes encoding proteins involved in flagellar biogenesis (the ΔflgD, ΔflgE2, ΔflgH, ΔflgK, ΔflhB, and ΔflgP mutants), cell membrane structure (the Δcj0818 mutant), the oxidative stress response (the Δrrc mutant), and signal transduction (the ΔflgR mutant). Moreover, the acid sensitivity of C. jejuni ΔmotAB was compared to that of the wild-type strain to further determine the importance of motility for C. jejuni acid survival. Both the motA and motB genes, which encode the flagellar stator proteins, are required for C. jejuni motility but not for flagellar biogenesis (41). In contrast to the ΔflgE2 mutant, which was not sensitive to acid, deletion mutations of the other genes (rrc, cj0818, flgD, flgH, flgK, flgP, flgR, flhB, and motAB) significantly increased the acid sensitivity of C. jejuni (Fig. 4A). Interestingly, the acid-sensitive mutants with deletion mutations of flagellar genes, namely, C. jejuni ΔflgD, ΔflgH, ΔflgK, ΔflgP, ΔflgR, ΔflhB, and ΔmotAB, were defective for motility compared to the wild-type strain, while C. jejuni ΔflgE2 remained motile (Fig. 4B). Altogether, these results suggest a link between C. jejuni motility and the bacterial capacity to survive acid stress.
FIG 4.
(A) Acid survival of C. jejuni mutants relative to the wild-type strain. The C. jejuni Δrrc, Δcj0818, ΔflgR, ΔflgP, ΔflhB, ΔflgD, ΔflgK, ΔflgH, and ΔmotAB mutants but not the ΔflgE2 mutant were more sensitive to acid than the wild-type strain. (B) Acid-sensitive C. jejuni mutants were defective in motility on soft MH agar compared to the motility of the wild-type strain. Results are represented as the mean diameters of bacterial migration from the site of inoculation ± standard errors for three biological experiments each with three technical replicates. Data from a minimum of three independent experiments are shown as the mean ± standard error. P values were determined using ANOVA followed by a Bonferroni multiple-comparison test.
Fur protects C. jejuni against oxidative stress upon acid exposure.
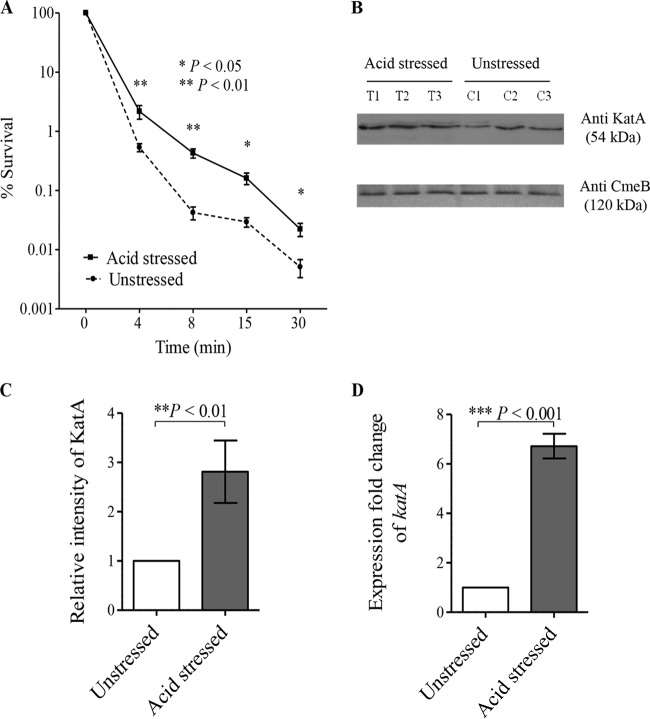
Interestingly, the expression of the catalase katA gene was found to be responsive to acid stress by our microarray analysis (see Fig. S1 and Table S1 in the supplemental material), in agreement with the findings of previous studies, which revealed the upregulation of katA in response to acid exposure (20, 25). The increased expression of katA might in turn enhance C. jejuni survival upon exposure to H2O2. Based on these observations, we hypothesized that acid stress might cross-protect C. jejuni against oxidative stress. To test this hypothesis, we compared the capacities of acid-stressed and unstressed C. jejuni cells to survive exposure to H2O2 stress. Interestingly, acid-stressed C. jejuni bacteria were more resistant to oxidative stress than unstressed bacteria, as indicated by the results of a disk inhibition assay (Table 3) and kill curves (Fig. 5A). The catalase KatA is the main protein responsible for the detoxification of H2O2 in C. jejuni (31), and its expression is known to be Fur regulated (18). To test the contribution of Fur in the acid stress-induced cross-protection of C. jejuni against H2O2 stress, we compared the capacities of acid-stressed and unstressed C. jejuni mutants with fur and katA deletion mutations to survive the oxidative stress induced by H2O2 exposure. In contrast to the wild-type strain, no differences in the capacity to survive oxidative stress were identified between the acid-stressed and unstressed C. jejuni Δfur and ΔkatA mutants (Table 3). These results clearly indicate that both Fur and KatA are essential for C. jejuni H2O2 cross-protection in response to acidic conditions. Importantly, the protein and transcript expression levels of KatA were significantly increased in response to acid stress, as revealed by Western blotting (Fig. 5B and C) and RT-qPCR, respectively (Fig. 5D). The induced expression of KatA in acid-stressed C. jejuni bacteria could account for the enhanced survival of C. jejuni in response to oxidative stress upon acid exposure.
TABLE 3.
Sensitivity of C. jejuni NCTC11168, a Δfur mutant, and a ΔkatA mutant to H2O2 before and after exposure to acid stress
| Strain and stress condition | Inhibition zone diama (mm) after exposure to H2O2 at concn of: |
||
|---|---|---|---|
| 250 mM | 500 mM | 1,000 mM | |
| C. jejuni NCTC11168 | |||
| Acid stressed | 12.11 ± 0.44* | 18.22 ± 0.44* | 22.44 ± 0.44* |
| Unstressed | 15.00 ± 0.19 | 20.22 ± 0.11 | 24.00 ± 0.19 |
| C. jejuni Δfur mutant | |||
| Acid stressed | 12.22 ± 0.22 | 14.88 ± 0.11 | 17.99 ± 0.19 |
| Unstressed | 12.10 ± 0.29 | 14.55 ± 0.29 | 17.88 ± 0.22 |
| C. jejuni ΔkatA mutant | |||
| Acid stressed | 20.77 ± 0.22 | 26.44 ± 0.29 | 32.22 ± 0.11 |
| Unstressed | 20.66 ± 0.33 | 26.44 ± 0.22 | 32.00 ± 0.19 |
The diameter of the inhibition zone is represented as the mean clear zone ± standard error from three independent experiments for C. jejuni strains after exposure to different molar concentrations (250 to 1,000 mM) of H2O2. The differences in H2O2 sensitivities between acid-stressed and unstressed C. jejuni strains (C. jejuni NCTC11168 and the Δfur and ΔkatA mutants) were analyzed for statistical significance using a Student t test. *, P < 0.05.
FIG 5.
(A) Prior acid exposure enhances C. jejuni survival in response to H2O2. Both acid-stressed and unstressed C. jejuni bacteria were exposed to 10 mM H2O2, and the percent bacterial survival was determined immediately and at 4, 8, 15, and 30 min after H2O2 exposure by counting the number of viable cells. (B) KatA expression levels were higher in acid-stressed C. jejuni bacteria than unstressed bacteria, as determined by Western blotting. KatA levels from three biological replicates of acid-stressed C. jejuni bacteria (T1, T2, and T3) and unstressed C. jejuni bacteria (C1, C2, and C3) were quantified by immunoblotting using anti-KatA antiserum, and anti-CmeB antibodies were used as a loading control for the total protein content. (C) There was a significant increase in KatA levels in acid-stressed C. jejuni bacteria relative to those in unstressed bacteria, as determined by the densitometric quantification of KatA levels. (D) RT-qPCR revealed that katA expression increases in acid-stressed C. jejuni bacteria compared with that in unstressed bacteria. The data shown are the means ± standard errors from three biological experiments with three technical replicates each. P values were determined by Student's unpaired t test.
Fur binding to the katA promoter region is alleviated under acidic conditions in vitro.
The induction of katA expression by acid stress suggests that its expression is modulated by acid exposure. Given that Fur directly binds the promoter region of katA (18), we tested this hypothesis by assessing the impact of acidic pH on Fur DNA binding. The effect of acid on Fur binding to the promoter region of katA was determined using EMSAs. As shown in Fig. 6A, the affinity of Fur binding to the katA promoter was higher at neutral pH 7 than under acidic conditions (pH 6.5 and pH 6). Importantly, no difference in binding affinity between C. jejuni PerR (CjPerR) and the katA promoter was detected under the same conditions (Fig. 6B). Moreover, the apparent dissociation constant (Kdapp) of Fur binding to the katA promoter was determined at both pH 7 and pH 6.5. Interestingly, under neutral conditions, Fur bound to the katA promoter with a Kdapp of 293.73 ± 38.66 nM, which is significantly lower (P < 0.05, t test) than the Kdapp value obtained under acidic conditions (584.03 ± 33.82 nM). These results indicate that the binding between Fur and the katA promoter is reduced under acidic conditions, explaining the observed increase in katA expression (Fig. 5B to D).
FIG 6.
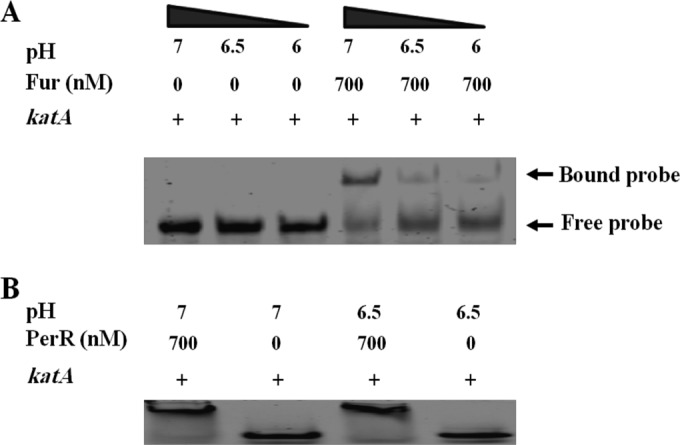
EMSAs of CjFur binding to Cy5-katA at different pHs. (A) The binding of CjFur (700 nM) to the promoter region of katA (1 nM) was determined under both neutral and acidic conditions. The capacity of CjFur to retard the electrophoretic mobility of Cy5-katA in a 6% nondenaturing polyacrylamide gel was characterized at different pH values (pH 7, 6.5, and 6). CjFur binding to the katA promoter was higher at neutral pH 7 than under acidic conditions (pH 6.5 and pH 6). (B) The binding between CjPerR (700 nM) and the katA promoter (1 nM) was examined under the same conditions described in the legend to panel A for CjFur-katA binding, with no differences being observed.
Acid stress enhances C. jejuni virulence in a Fur-dependent manner.
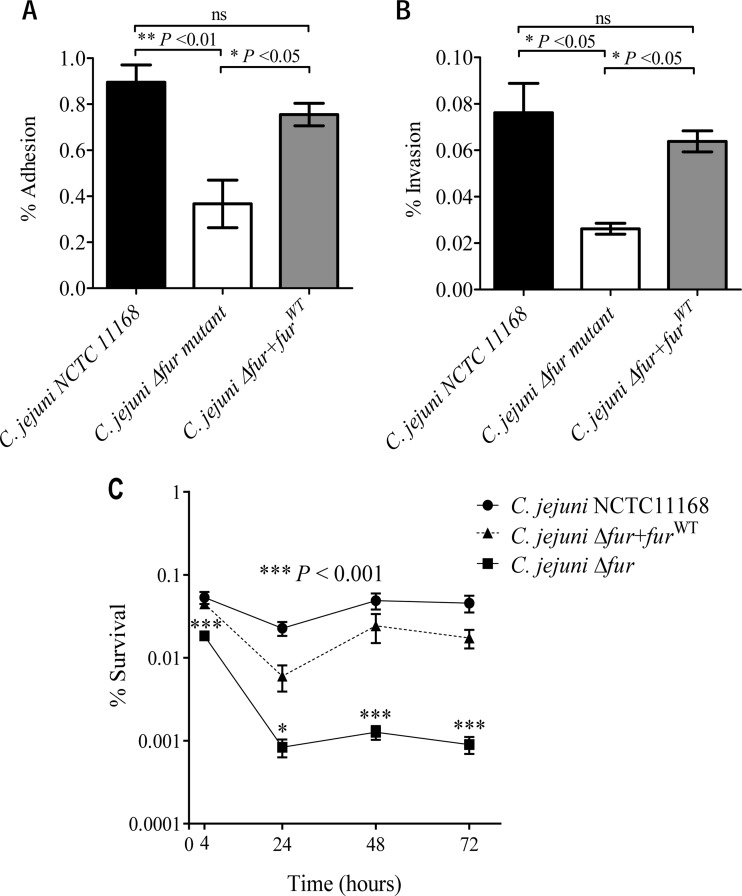
The microarray data revealed the differential expression of numerous genes involved in C. jejuni pathogenesis, such as genes involved in flagellar biogenesis, in addition to the classical Fur-regulated iron acquisition genes, upon acid exposure in a Fur-dependent manner. This observation suggests that acid exposure could enhance C. jejuni virulence and that Fur might play a significant role in this process that extends beyond iron acquisition. The capacity of the C. jejuni Δfur mutant to adhere to and invade eukaryotic cells was examined and compared to that of the wild-type strain. As shown in Fig. 7A and B, the C. jejuni Δfur mutant exhibited a significantly reduced capacity to adhere to and invade HCT116 cells (0.37% ± 0.1% and 0.02% ± 0.002%, respectively) relative to the wild-type strain (0.89% ± 0.07% and 0.08% ± 0.01%, respectively) (P < 0.05). Moreover, the wild-type strain exhibited a significant increase in its capacity to survive intracellularly within HCT116 cells compared to that of the fur mutant (Fig. 7C). Complementation of the Δfur mutant with the fur gene (resulting in the C. jejuni Δfur+furWT strain) restored the wild-type adhesion, invasion, and intracellular survival phenotypes. These results indicate that Fur is important for C. jejuni virulence traits related to human epithelial cell adhesion and invasion.
FIG 7.
The C. jejuni Δfur mutant is affected in adhesion (A), invasion (B), and intracellular survival (C) in HCT116 cells relative to the wild-type strain. (A and B) Monolayers of HCT116 cells were infected with the wild-type, Δfur, or Δfur+furWT C. jejuni strain, and the degrees of bacterial adherence (A) and invasion (B) are shown. (C) Percent intracellular bacterial survival over a 72-h period for each strain. The data shown are the means ± standard errors from three independent experiments. P values were determined using ANOVA followed by the Bonferroni multiple-comparison test. *, P < 0.05; ns, not significant.
Fur is required for C. jejuni pathogenesis in Galleria mellonella.
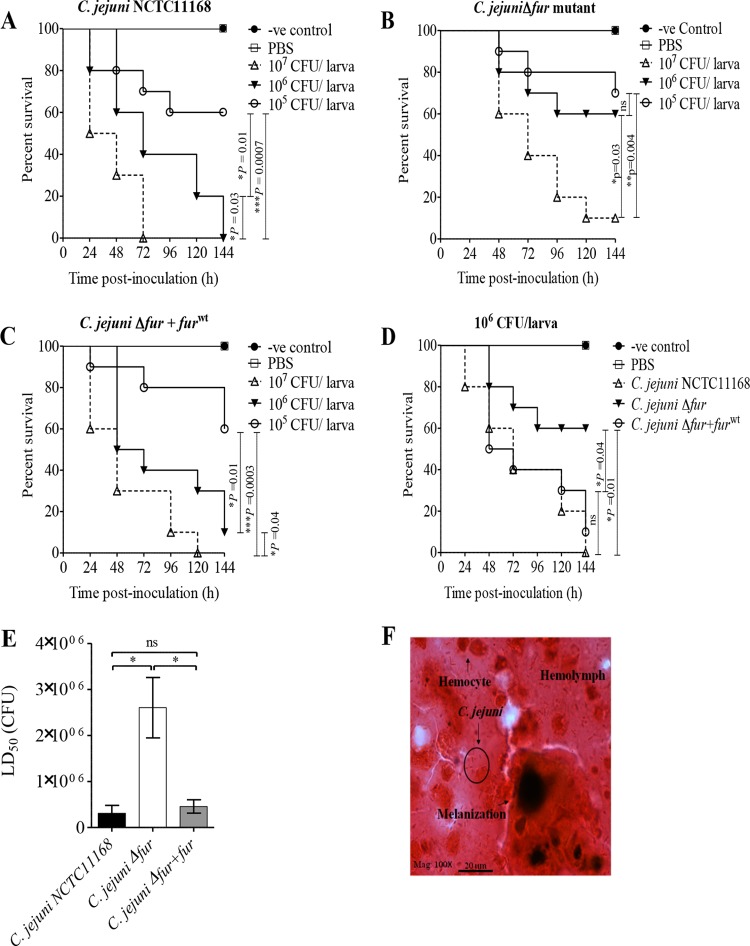
The contribution of Fur to C. jejuni pathogenesis was examined using G. mellonella larvae as an infection model (36) and by inoculating the larvae with the wild-type strain, the fur mutant, and the Δfur+furWT strain. All C. jejuni strains caused dose-dependent killing in Galleria larvae (Fig. 8A to C), and in contrast to the findings for larvae infected with one of the C. jejuni strains (the wild type, the fur mutant, or the Δfur+furWT strain), no death was recorded for the control groups, either uninoculated or PBS-inoculated larvae. At the 106-CFU infection dose, wild-type C. jejuni killed significantly more wax worms than the fur mutant (P = 0.01) (Fig. 8D). Importantly, this defect in the capacity of the C. jejuni Δfur mutant to kill larvae was restored by complementation with the furWT gene (Fig. 8D). Moreover, as shown in Fig. 8E, both the C. jejuni wild-type and Δfur+furWT strains had significantly lower LD50 values (3.1 × 105 and 4.6 × 105 CFU, respectively) than the Δfur mutant (2.6 × 106 CFU). Finally, histopathological examination of C. jejuni-infected larvae revealed bacterial cells surrounded by larval hemocytes, coagulated hemolymph, and melanin pigments (Fig. 8F), demonstrating that Fur plays an important role in C. jejuni pathogenesis.
FIG 8.
Fur is important for C. jejuni pathogenesis in G. mellonella larvae. G. mellonella larvae (n = 10 larvae/group) were inoculated with the serially diluted (107, 106, and 105 CFU) C. jejuni wild-type, Δfur, or Δfur+furWT strain. Larval survival was monitored every 24 h for 6 days and was plotted using Kaplan-Meier survival curves. (A to C) Survival curves of G. mellonella larvae inoculated with different C. jejuni strains: the wild type (A), the Δfur mutant (B), or the Δfur+furWT strain (C). (D) The C. jejuni wild-type and Δfur+furWT strains killed more larvae than the Δfur mutant at the 106-CFU dose (P = 0.01 and 0.04, respectively; log-rank test). (E) The C. jejuni Δfur mutant has a significantly higher LD50 than either the wild-type or Δfur+furWT strain; thus, this mutant is less virulent. *, P < 0.05, which was considered significant using a Mann-Whitney rank sum test; ns, not significant. (F) Microscopic examination of C. jejuni-infected larvae revealed C. jejuni cells surrounded by larval hemocytes, hemolymph, and melanin pigment. Gram stain was used. Mag, magnification; -ve, negative.
DISCUSSION
Many regulators (e.g., RpoS) which help enteric pathogens survive in the presence of various stresses are absent in C. jejuni (38–40). While C. jejuni harbors a ferric iron uptake regulator (Fur) that has been shown to be involved in the regulation of iron metabolism and oxidative stress response genes (18–20, 30, 31), its potential role in the acid response in C. jejuni has not yet been characterized. Our results demonstrate that a C. jejuni Δfur mutant is affected in its ability to survive acid exposure, suggesting a role for Fur in the acid stress response in C. jejuni. This observation was further confirmed by the restoration of the acid sensitivity of the Δfur mutant by complementation with the furWT gene. Interestingly, our results also demonstrate that the CjFur S2 metal binding site is required for iron sensing and that the CjFur-mediated acid response is iron independent, similar to what has been observed in S. Typhimurium (43, 44).
In other enteric bacteria, Fur is a known regulator of acid shock proteins (ASPs), which protect and repair the damages that occur in cellular components following acid exposure (12, 45, 46). Therefore, it is tempting to speculate that the acid survival defect of the C. jejuni Δfur mutant results from the absence of ASP expression. To test this hypothesis and identify these ASPs, we compared the transcriptomes of the Δfur mutant and the wild-type C. jejuni strain under acidic conditions using genome-wide microarrays. Notably, the changes in gene expression in both the Δfur mutant and wild-type C. jejuni were not a result of cells dying at pH 4; rather, they reflected the bacterial response to acid stress. However, it should be noted that a significant decrease in cell viability was observed in both the Δfur mutant and the wild-type strain at pH 3 and could result from bacteria entering into a viable but nonculturable (VBNC) state upon exposure to strong acidic conditions (47). Nevertheless, the upregulation of many genes at pH 3 and the similarities in gene expression between that at pH 3 and that at pH 4 suggest that C. jejuni remains transcriptionally active at pH 3, despite a decrease in cell viability.
The transcriptomic results indicate that flagellar biogenesis genes (flaABDG, flgBDEGHIKMG2K2E2, and fliDKS) were repressed in the C. jejuni Δfur mutant at low pH compared to their levels of expression in the wild-type strain. Flagella are believed to partially confer protection against acid through their ability to rapidly move bacteria from an acidic environment to one with a more suitable pH (48, 49). Reid et al. (20) demonstrated that a decrease in pH signals entry into the host, causing C. jejuni to highly express flagellar genes that are required to escape the stomach acidity and rapidly localize the bacterium to the protective mucus layer. Similarly, many flagellar genes were upregulated in the closely related bacterium H. pylori upon acid exposure (48). The role of the flagellar genes in C. jejuni acid resistance was clearly demonstrated by assessing the acid stress phenotype of mutants with deletion mutations in flagellar gene flgD, flgH, flgK, flgR, or flhB. These mutants were found to be significantly impaired in their capacity to survive under acidic conditions. Interestingly, the flagellar mutants defective in bacterial motility (the ΔflgD, ΔflgH, ΔflgK, and ΔflhB mutants) were all found to be acid sensitive, while the ΔflgE2 mutant, which was still motile, was not affected in its survival in response to acid stress. These observations suggest an important role for bacterial motility in C. jejuni acid survival. To further investigate this finding, we characterized the capacity of the ΔflgP and ΔmotAB mutants to survive under acidic conditions. FlgP and MotAB have been shown to be required for C. jejuni motility but not for flagellar biogenesis (41, 50). The flgP gene encodes an outer membrane lipoprotein, which is important for Campylobacter motility (41, 50), while the motA and motB genes code for the flagellar stator proteins, which utilize the transmembrane proton motive force (PMF) to drive flagellar rotation (41, 51). Interestingly, both the ΔflgP and ΔmotAB mutants displayed significantly increased sensitivity to acid stress compared to that of the C. jejuni wild-type strain. Flagellar mutation could disrupt the proton potential of the inner cell membrane and thereby interfere with the electron transport chain (ETC) (30). The electron transport chain might play an important role in Campylobacter acid survival by decreasing the intracellular proton concentration and thereby reversing the cytoplasmic acidity.
Interestingly, KatA expression was significantly higher in acid-stressed C. jejuni bacteria than unstressed bacteria, as revealed by Western blotting and RT-qPCR analyses. The upregulation of KatA in acid-stressed C. jejuni bacteria suggests that acid exposure induces catalase expression, in agreement with the findings of previous studies (17, 25). Moreover, the enhanced survival of the C. jejuni wild-type strain in the presence of H2O2 following acid exposure, a finding that was not seen in either the Δfur or the ΔkatA mutant, suggests that both Fur and KatA are essential for the acid-enhanced C. jejuni H2O2 resistance. These results indicate that Fur plays an important role in the acid-induced cross-protection of C. jejuni against oxidative stress. It is possible that the increased iron solubility under acidic conditions might negatively affect bacterial growth by promoting the generation of damaging hydroxyl radicals through the Fenton reaction (52). Thereby, it is tempting to speculate that the acid induction of KatA expression might enable C. jejuni to cope with iron-mediated oxidative stress at low pH. Given the known role of Fur in katA gene expression, we tested the hypothesis that Fur repression might be released in response to acid stress. In agreement with this hypothesis, the affinity of CjFur binding to the katA promoter was significantly reduced at acidic pH, with the dissociation constant (Kd) being higher than that under neutral conditions. These results indicate that the Fur-mediated repression of katA is released in acidic environments, enabling catalase expression and protection against oxidative stress. In addition to Fur, the peroxide stress regulator PerR from C. jejuni has also been shown to directly regulate katA (16), and of particular interest in the context of the current study, perR expression has previously been reported to be upregulated in response to acid shock (20). However, we have shown here that CjPerR binding to the katA promoter region is not affected by acidic conditions, suggesting that the acid induction of katA expression is mainly CjFur mediated and that the upregulation of CjPerR minimally affects katA expression.
Fur is important for C. jejuni host pathogenesis.
The transcriptional profile of the Δfur mutant under acidic conditions revealed the differential expression of several genes involved in C. jejuni virulence. In addition to the iron acquisition genes, many genes involved in flagellar biogenesis were differentially expressed in response to acid conditions in a Fur-dependent manner. These data indicate that Fur might play a key role in C. jejuni pathogenesis, especially upon exposure to acidic conditions, like those encountered in the host gastrointestinal tract. Indeed, deletion of the fur gene significantly reduced the capacity of C. jejuni to adhere to, invade, and survive intracellularly within HCT116 cells, which are important virulence traits of C. jejuni (25, 53). Moreover, in wax worm larvae, the C. jejuni fur mutant had a significantly higher LD50 than the wild-type strain. The importance of Fur for C. jejuni virulence is most likely multifactorial and could be related to its role in regulating the expression of genes responsible for several biological functions known to contribute to Campylobacter pathogenesis, such as iron acquisition, energy metabolism, and flagellar biogenesis (18, 21, 22). Deregulation of these genes might significantly affect the ability of C. jejuni to kill wax worms. The role of Fur in flagellar gene regulation and the importance of flagella for C. jejuni pathogenesis (42, 45) could account for the virulence defect of the Δfur mutant in the G. mellonella larvae relative to the virulence of the wild-type strain. From all the information mentioned above, it is clear that Fur plays a critical role in C. jejuni host pathogenesis.
In summary, the results of our study indicate that, in addition to its role in iron metabolism, Fur is an important regulator of the C. jejuni acid response. Moreover, Fur plays a fundamental role in the acid-induced cross-protection of C. jejuni against oxidative stress. The contribution of Fur to C. jejuni host pathogenesis was also highlighted herein.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from CIHR (MOP grant 84224) to A.S. and a Ph.D. scholarship to M.A. from the Egyptian Government through the Egyptian Bureau of Cultural and Educational Affairs.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01377-15.
REFERENCES
- 1.Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg Infect Dis 5:28–35. doi: 10.3201/eid0501.990104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stintzi A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J Bacteriol 185:2009–2016. doi: 10.1128/JB.185.6.2009-2016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allos BM. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis 32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 4.Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bender J, Shiferaw B, Reddy S, Ahuja SD, Helfrick DL, Hardnett F, Carter M, Anderson B, Tauxe RV. 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin Infect Dis 38(Suppl 3):S285–S296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- 5.Young KT, Davis LM, Dirita VJ. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 6.Bearson S, Bearson B, Foster JW. 1997. Acid stress responses in enterobacteria. FEMS Microbiol Lett 147:173–180. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 7.Dressman JB, Berardi RR, Dermentzoglou LC, Russell TL, Schmaltz SP, Barnett JL, Jarvenpaa KM. 1990. Upper gastrointestinal (GI) pH in young, healthy men and women. Pharm Res 7:756–761. doi: 10.1023/A:1015827908309. [DOI] [PubMed] [Google Scholar]
- 8.Foster JW. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol 2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 9.Foster JW, Hall HK. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J Bacteriol 174:4317–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu C, Ngeleka M, Potter AA, Allan BJ. 2002. Effect of fur mutation on acid-tolerance response and in vivo virulence of avian septicemic Escherichia coli. Can J Microbiol 48:458–462. doi: 10.1139/w02-042. [DOI] [PubMed] [Google Scholar]
- 11.Oglesby AG, Murphy ER, Iyer VR, Payne SM. 2005. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Mol Microbiol 58:1354–1367. doi: 10.1111/j.1365-2958.2005.04920.x. [DOI] [PubMed] [Google Scholar]
- 12.Hantke K. 2001. Iron and metal regulation in bacteria. Curr Opin Microbiol 4:172–177. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 13.Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol 184:5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bijlsma JJ, Waidner B, van Vliet AHM, Hughes NJ, Hag S, Bereswill S, Kelly DJ, Vandenbroucke-Grauls CM, Kist M, Kusters JG. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect Immun 70:606–611. doi: 10.1128/IAI.70.2.606-611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect Immun 77:2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Vliet AH, Baillon ML, Penn CW, Ketley JM. 1999. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J Bacteriol 181:6371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palyada K, Sun YQ, Flint A, Butcher J, Naikare H, Stintzi A. 2009. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. doi: 10.1186/1471-2164-10-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butcher J, Sarvan S, Brunzelle JS, Couture JF, Stintzi A. 2012. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci U S A 109:10047–10052. doi: 10.1073/pnas.1118321109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford MJ, Goldberg DE. 1998. Regulation of the Salmonella typhimurium flavohemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J Biol Chem 273:34028–34032. [DOI] [PubMed] [Google Scholar]
- 20.Reid AN, Pandey R, Palyada K, Naikare H, Stintzi A. 2008. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl Environ Microbiol 74:1583–1597. doi: 10.1128/AEM.01507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palyada K, Threadgill D, Stintzi A. 2004. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol 186:4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes K, Mulholland F, Pearson BM, Pin C, McNicholl-Kennedy J, Ketley JM, Wells JM. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151(Pt 1):243–257. doi: 10.1099/mic.0.27412-0. [DOI] [PubMed] [Google Scholar]
- 23.Butcher J, Handley RA, van Vliet AH, Stintzi A. 2015. Refined analysis of the Campylobacter jejuni iron-dependent/independent Fur- and PerR-transcriptomes. BMC Genomics 16:498. doi: 10.1186/s12864-015-1661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naikare H, Palyada K, Panciera R, Marlow D, Stintzi A. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect Immun 74:5433–5444. doi: 10.1128/IAI.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le MT, Porcelli I, Weight CM, Gaskin DJH, Carding SR, van Vliet AHM. 2012. Acid-shock of Campylobacter jejuni induces flagellar gene expression and host cell invasion. Eur J Microbiol Immunol 1:12–19. doi: 10.1556/EuJMI.2.2012.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci U S A 99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao H, Bausch C, Richmond C, Blattner FR, Conway T. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol 181:6425–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stintzi A, Marlow D, Palyada K, Naikare H, Panciera R, Whitworth L, Clarke C. 2005. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect Immun 73:1797–1810. doi: 10.1128/IAI.73.3.1797-1810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldi P, Long AD. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 30.Flint A, Sun YQ, Butcher J, Stahl M, Huang H, Stintzi A. 2014. Phenotypic screening of a targeted mutant library reveals Campylobacter jejuni defenses against oxidative stress. Infect Immun 82:2266–2275. doi: 10.1128/IAI.01528-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flint AY, Sun Q, Stintzi A. 2012. Cj1386 is an ankyrin-containing protein involved in heme trafficking to catalase in Campylobacter jejuni. J Bacteriol 194:334–345. doi: 10.1128/JB.05740-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Nandi S, Martel A, Antoun A, Ioshikhes I, Blais A. 2012. Discovery, optimization and validation of an optimal DNA-binding sequence for the Six1 homeodomain transcription factor. Nucleic Acids Res 40:8227–8239. doi: 10.1093/nar/gks587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carey M, Smale ST. 2000. Transcriptional regulation in eukaryotes: concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 34.Hanks JH, Wallace RE. 1949. Relation of oxygen and temperature in the preservation of tissues by refrigeration. Proc Soc Exp Biol Med 71:196–200. doi: 10.3181/00379727-71-17131. [DOI] [PubMed] [Google Scholar]
- 35.Poly F, Threadgill D, Stintzi A. 2004. Identification of Campylobacter jejuni ATCC 43431-specific genes by whole microbial genome comparisons. J Bacteriol 186:4781–4795. doi: 10.1128/JB.186.14.4781-4795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Champion OL, Karlyshev AV, Senior NJ, Woodward M, La Ragione R, Howard SL, Wren BW, Titball RW. 2010. Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J Infect Dis 201:776–782. doi: 10.1086/650494. [DOI] [PubMed] [Google Scholar]
- 37.Olsen RJ, Watkins ME, Cantu CC, Beres SB, Musser JM. 2011. Virulence of serotype M3 group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence 2:111–119. doi: 10.4161/viru.2.2.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 39.Birk TT, Takamiya Wik MM, Lametsch RR, Knochel SS. 2012. Acid stress response and protein induction in Campylobacter jejuni isolates with different acid tolerance. BMC Microbiol 12:174. doi: 10.1186/1471-2180-12-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park SF. 2002. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int J Food Microbiol 74:177–188. doi: 10.1016/S0168-1605(01)00678-X. [DOI] [PubMed] [Google Scholar]
- 41.Mertins S, Allan BJ, Townsend HG, Koster W, Potter AA. 2013. Role of motAB in adherence and internalization in polarized Caco-2 cells and in cecal colonization of Campylobacter jejuni. Avian Dis 57:116–122. doi: 10.1637/10235-050412-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 42.Grant CC, Konkel ME, Cieplak W Jr, Tompkins LS. 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun 61:1764–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall HK, Foster JW. 1996. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J Bacteriol 178:5683–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foster JW. 1991. Salmonella acid shock proteins are required for the adaptive acid tolerance response. J Bacteriol 173:6896–6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernando U, Biswas D, Allan B, Willson P, Potter AA. 2007. Influence of Campylobacter jejuni fliA, rpoN and flgK genes on colonization of the chicken gut. Int J Food Microbiol 118:194–200. doi: 10.1016/j.ijfoodmicro.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 46.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tangwatcharin P, Chanthachum S, Khopaibool P, Griffiths MW. 2006. Morphological and physiological responses of Campylobacter jejuni to stress. J Food Prot 69:2747–2753. [DOI] [PubMed] [Google Scholar]
- 48.Merrell DS, Goodrich ML, Otto G, Tompkins LS, Falkow S. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect Immun 71:3529–3539. doi: 10.1128/IAI.71.6.3529-3539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suerbaum S. 1995. The complex flagella of gastric Helicobacter species. Trends Microbiol 3:168–170. doi: 10.1016/S0966-842X(00)88913-1. [DOI] [PubMed] [Google Scholar]
- 50.Sommerlad SM, Hendrixson DR. 2007. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J Bacteriol 189:179–186. doi: 10.1128/JB.01199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol 270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 52.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konkel ME, Joens LA. 1989. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect Immun 57:2984–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.