Abstract
Polyhydroxyalkanoates (PHAs) are carbon and energy storage polymers produced by a variety of microbial organisms under nutrient-limited conditions. They have been considered as an environmentally friendly alternative to oil-based plastics due to their renewability, versatility and biodegradability. PHA synthase (PhaC) plays a central role in PHA biosynthesis, in which its activity and substrate specificity are major factors in determining the productivity and properties of the produced polymers. However, the effects of modifying the substrate side chain are not well understood because of the difficulty to accessing the desired analogs. In this report, a series of 3-(R)-hydroxyacyl coenzyme A (HACoA) analogs were synthesized and tested with class I synthases from Chromobacterium sp. USM2 (PhaCCs and A479S-PhaCCs) and Caulobacter crescentus (PhaCCc) as well as class III synthase from Allochromatium vinosum (PhaECAv). It was found that, while different PHA synthases displayed distinct preference with regard to the length of the alkyl side chains, they could withstand moderate side chain modifications such as terminal unsaturated bonds and the azide group. Specifically, the specific activity of PhaCCs toward propynyl analog (HHxyCoA) was only 5-fold less than that toward the classical substrate HBCoA. The catalytic efficiency (kcat/Km) of PhaECAv toward azide analog (HABCoA) was determined to be 2.86 × 105 M−1s−1, which was 6.2% of the value of HBCoA (4.62 × 106 M−1s−1) measured in the presence of bovine serum albumin (BSA). These side chain modifications may be employed to introduce new material functions to PHAs as well as to study PHA biogenesis via click-chemistry, in which the latter remains unknown and is important for metabolic engineering to produce PHAs economically.
Keywords: polyhydroxyalkanoate (PHA) synthases, modified side chain, substrate synthesis, enzyme activity, kinetic parameters
TOC
INTRODUCTION
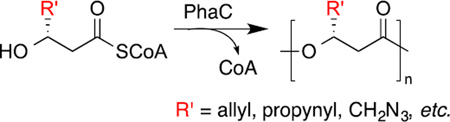
Polyhydroxyalkanoates (PHAs) are polyoxoesters produced by a variety of bacteria under harsh environmental conditions where nutrients other than the carbon source are limited.1–5 As shown in Scheme 1, PHA synthases (PhaCs) catalyze polymerization of 3-(R)-hydroxyacyl coenzyme A (HACoA) to form PHA polymers. Depending on the size of R’ group (side chain), the resulting PHAs will have properties ranging from thermoplastics to elastomers.4, 6 Till now, more than 150 different monomers have been found to incorporate into PHAs by various microbes.7, 8 When the environment becomes hospitable, these polymers will be degraded to release monomers and energy for other biological processes. Due to PHA’s renewability, versatility and biodegradability, it has been considered as an environmentally friendly alternative to petroleum-based plastics.9–12 However, high cost of PHA production has prevented its commercialization and applications.6, 13–15 Thus, there is a growing interest in producing PHAs with defined properties in a cost-effective fashion.16 Understanding PhaC activity toward substrates with various side chains is crucial to achieve this goal as it will identify appropriate synthase candidates that can be selected for protein engineering to lower PHA production cost.
Scheme 1.
Production of PHAs in bacteria
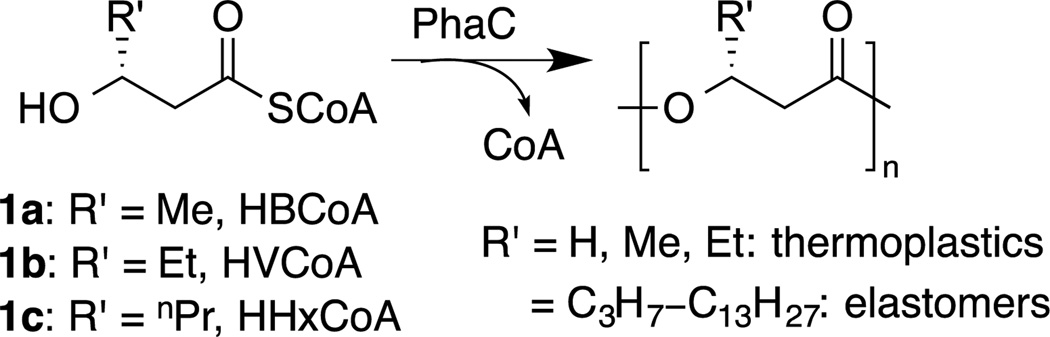
PHA synthases are divided into four classes based on their subunit composition and substrate specificity.17 Class I and class II synthases consist of one subunit (PhaC), whereas class III and class IV synthases require additional subunits PhaE and PhaR, respectively, for their activities. While class II synthases polymerize medium-chain-length (MCL) monomers (C6–C14), all other classes prefer short-chain-length (SCL) monomers (C3–C5) as the substrates.18 Due to limited access to PhaC substrates, studying PHA synthases has been hampered significantly. Thus far, large-scale chemical synthesis of PhaC substrates was reported only for 3-(R)-hydroxybutyryl coenzyme A (HBCoA, R’ = Me in Scheme 1) and 3-(R)-hydroxyvaleryl coenzyme A (HVCoA, R’ = Et).19 Other PhaC substrates with a long side chain, e.g. 3-(R)-hydroxyhexanoate coenzyme A (HHxCoA, R’ = Pr), could be obtained either chemically at extremely low yields or enzymatically in limited quantities.20–23 As far as we know, preparation of PHA substrate analogs with modified side chains has not been reported. However, such analogs are needed for developing biotechnological applications of PhaCs and the produced PHAs.
Here we describe a convenient chemical synthesis of HACoA analogs containing a side chain with various lengths and functional groups in good yields. These analogs were investigated with PHA synthases in order to reveal enzyme substrate specificity. Since class I and class III3 enzymes are the most studied PHA synthases in terms of mechanism and application development,24–32 they were selected for the present study. Specifically, synthases from Chromobacterium sp. USM2 (PhaCCs) and Caulobacter crescentus (PhaCCc) were used as the model enzymes for Class I synthases.33, 34 The synthase from Allochromatium vinosum (PhaECAv) was employed to represent class III enzymes.26
EXPERIMENTAL SECTION
General Information
All chemicals were purchased at the highest purity grade. All solvents were anhydrous. All reactions were performed under argon atmosphere unless otherwise specified. Thin layer chromatography (TLC) was performed using 60 mesh silica gel plates and visualization was performed using short wavelength UV light (254 nm) and basic KMnO4 staining. Absorbance was recorded on an Agilent Cary 100 UV-Vis spectrophotometer or Molecular Devices SpectraMax Plus 384. NMR spectra were recorded on a Varian 400 MHz spectrometer. Chemical shifts of proton (1H NMR) and phosphorus (31P NMR) were reported in ppm relative to the residual solvent peaks and external reference of 85% H3PO4, respectively. High-resolution mass spectrometry (HRMS) was recorded on a Q-Star Elite spectrometer manufactured by Applied Biosystems.
HPLC was performed on a Waters Breeze 2 system equipped with a 1525 pump, a 2998 PDA detector, and a semi-preparative Luna C18-2 column (5 µm, 10 × 250 mm). The column was eluted at 3.0 mL/min using a linear gradient from 5% to 95% methanol in 10 mM ammonium acetate (pH 4.00) over 70 min.
Preparation of HACoA Analogs
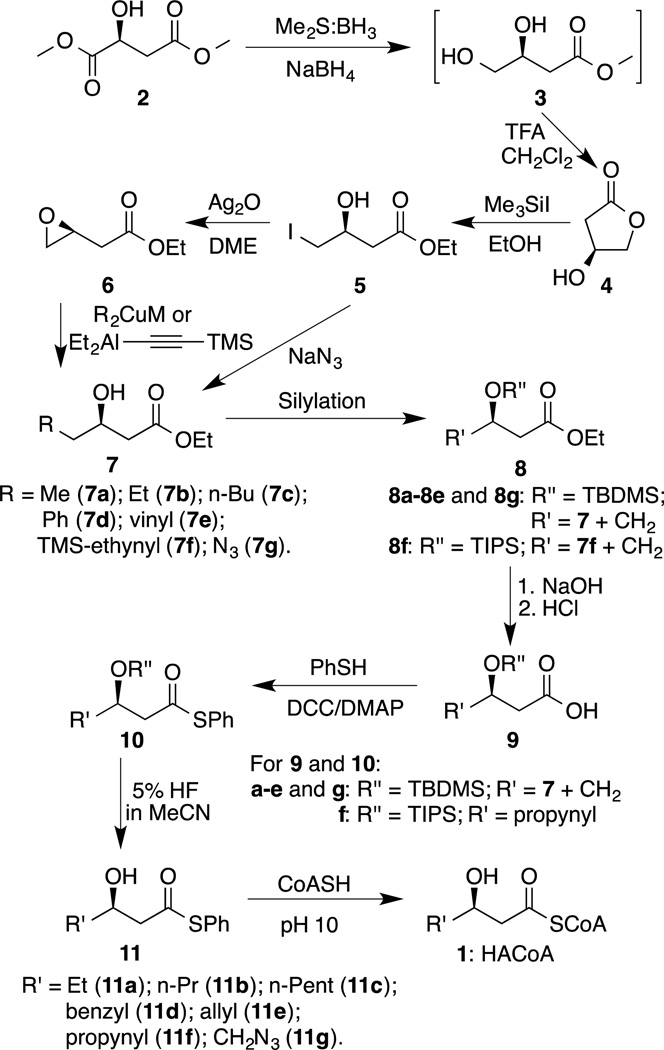
The approach to synthesize HACoA analogs is shown in Schemes 2. The overall yields and HPLC retention times are summarized in Table 1. Spectral data of the end products 1b–h are listed below. Experimental details, spectral data of intermediates 3–11, and copies of NMR spectra are summarized in Supporting Information.
Scheme 2.
Chemical Synthesis of HACoA Analogs
Table 1.
Summary of Synthesized HACoA Analogs
| Num. | R’ | Abbr. | Yield(%)a | tR (min)b |
|---|---|---|---|---|
| 1a | Me | HBCoA | NA | NA |
| 1b | Et | HVCoA | 34 | 22.0 |
| 1c | nPr | HHxCoA | 35 | 25.5 |
| 1d | nPent | HCCoA | 27 | 35.5 |
| 1e | benzyl | HPBCoA | 24 | 33.1 |
| 1f | allyl | HHxeCoA | 30 | 23.2 |
| 1g | propynyl | HHxyCoA | 20 | 20.2 |
| 1h | CH2N3 | HABCoA | 53 | 17.8 |
Total yield from epoxyester 6.
HPLC retention time.
3-(R)-Hydroxyvalerate Coenzyme A (HVCoA) 1b
1H NMR (400 MHz, D2O) δ: 8.64 (s, 1H), 8.40 (s, 1H), 6.18 (d, J = 4.8 Hz, 1H), 4.74 (m, 1H), 4.57 (m, 1H), 4.24 (m, 2H), 4.00 (s, 1H), 3.99–3.92 (m, 1H), 3.83 (d, J = 9.2 Hz, 1H), 3.57 (d, J = 9.2 Hz, 1H), 3.42 (t, J = 6.4 Hz, 2H), 3.31 (t, J = 6.8 Hz, 2H), 2.98 (dt, J = 2.8, 6.4 Hz, 2H), 2.77 (dd, J = 4.0, 15.2 Hz, 1H), 2.69 (dd, J = 8.8, 14.8 Hz, 1H), 2.40 (t, J = 6.8 Hz, 2H), 1.50–1.41 (m, 2H), 0.91 (s, 3H), 0.85 (t, J = 7.2 Hz, 3H), 0.78 (s, 3H); 31P NMR (162 MHz, D2O) δ: 1.54 (s, 1P), −9.77 (d, J = 21.1 Hz, 1P), −10.35 (d, J = 20.9 Hz, 1P); HRMS for [M-H]− (C26H43N7O18P3S−): calcd: 866.1604; found: 866.1577.
3-(R)-Hydroxyhexanoate Coenzyme A (HHxCoA) 1c
1H NMR (400 MHz, D2O) δ: 8.51 (s, 1H), 8.22 (s, 1H), 6.13 (d, 1H, J = 6.0 Hz), 4.55 (m, 1H), 4.20 (m, 1H), 4.05–3.99 (m, 1H), 3.97 (s, 1H), 3.81–3.77 (m, 1H), 3.52 (d, J = 9.2 Hz, 1H), 3.40 (t, J = 6.8 Hz, 2H), 3.29 (t, J = 6.4 Hz, 2H), 2.96 (dt, J = 2.4, 7.2 Hz, 2H), 2.73 (dd, J = 4.4, 15.2 Hz, 1H), 2.67 (dd, J = 8.0, 15.2 Hz, 1H), 2.38 (t, J = 6.4 Hz, 2H), 1.42–1.37 (m, 2H), 1.35–1.26 (m, 2H), 0.85 (s, 3H), 0.83 (t, J = 7.2 Hz, 3H), 0.72 (s, 3H); 31P NMR (162 MHz, D2O) δ: 1.39 (s, 1P), −9.68 (d, J = 25.3 Hz, 1P), −10.14 (d, J = 23.5 Hz, 1P); HRMS for [M-H]− (C27H45N7O18P3S−): calcd: 880.1760; found: 880.1735.
3-(R)-Hydroxycaprylate Coenzyme A (HCCoA) 1d
1H NMR (400 MHz, D2O) δ: 8.50 (s, 1H), 8.21 (s, 1H), 6.13 (d, J = 6.4 Hz, 1H), 4.55 (m, 1H), 4.20 (m, 2H), 4.03− 4.00 (m, 1H), 3.98 (s, 1H), 3.80 (dd, J = 4.4, 10.4 Hz, 1H), 3.53 (dd, J = 4.4, 9.6 Hz, 1H), 3.41 (t, J = 6.4 Hz, 2H), 3.29 (t, 1H, J = 6.4 Hz), 2.98–2.94 (m, 2H), 2.74 (dd, J = 4.4, 15.2 Hz, 1H), 2.67 (dd, J = 8.4, 14.8 Hz, 1H), 2.39 (t, J = 6.4 Hz, 2H), 1.40 (q, J = 7.2 Hz, 2H), 1.35–1.15 (m, 6H), 0.85 (s, 3H), 0.79 (t, J = 6.8 Hz, 3H), 0.72 (s, 3H); 31P NMR (162 MHz, D2O) δ: 1.57 (s, 1P), −9.65 (d, J = 18.9 Hz, 1P), −10.23 (d, J = 14.9 Hz, 1P); HRMS for [M-H]− (C29H49N7O18P3S−): calcd: 908.2073; found: 908.2054.
3-(R)-Hydroxy-4-phenylbutyrate Coenzyme A (HPBCoA) 1e
1H NMR (400 MHz, D2O) δ 8.49 (s, 1H), 8.17 (s, 1H), 7.30–7.18 (m, 5H), 6.11 (d, J = 6.8 Hz, 1H), 4.55 (m, 1H), 4.28 (quin, J = 6.4 Hz, 1H), 4.20 (m, 2H), 3.97 (s, 1H), 3.79 (d, J = 8.8 Hz, 1H), 3.51 (d, J = 9.6 Hz, 1H), 3.39 (t, J = 6.8 Hz, 2H), 3.28 (t, J = 6.4 Hz, 2H), 2.95 (q, J = 6.4 Hz, 2H), 2.75 (d, J = 4.8 Hz, 2H), 2.74 (d, J = 7.2 Hz, 2H), 0.84 (s, 3H), 0.70 (s, 3H); 31P NMR (162 MHz, D2O) δ: 2.65 (s, 1P), −9.62 (m, 1P), −10.14 (m, 1P); HRMS for [M-H]− (C31H45N7O18P3S−): calcd: 928.1760; found: 928.1789.
3-(R)-Hydroxhex-5-enoate Coenzyme A (HHxeCoA) 1f
1H NMR (400 MHz, D2O) δ: 8.51 (s, 1H), 8.23 (s, 1H), 6.14 (d, J = 6.8 Hz, 1H), 5.81–5.74 (m, 1H), 5.12–5.08 (m, 2H), 4.56 (m, 1H), 4.20 (m, 2H), 4.14–4.07 (m, 1H), 3.98 (s, 1H), 3.79 (d, J = 9.6 Hz, 1H), 3.52 (d, J = 9.6 Hz, 1H), 3.41 (t, J = 6.8 Hz, 2H), 3.29 (t, J = 6.4 Hz, 2H), 2.96 (dt, J = 2.4, 6.0 Hz, 2H), 2.77 (dd, J = 4.4, 15.2 Hz, 2H), 2.70 (dd, J = 8.4, 15.2 Hz, 2H), 2.39 (t, J = 6.4 Hz, 2H), 2.25–2.19 (m, 2H), 0.85 (s, 3H), 0.72 (s, 3H); 31P NMR (162 MHz, D2O) δ: 1.82 (s, 1H), −9.73 (m, 1H), −10.12 (m, 1H); HRMS for [M-H]− (C27H43N7O18P3S−): calcd: 878.1604; found: 878.1588.
3-(R)-Hydroxyhex5-ynoate Coenzyme A (HHxyCoA) 1g
1H NMR (400 MHz, D2O) δ: 8.52 (s, 1H), 8.24 (s, 1H), 6.15 (d, J = 6.4 Hz, 1H), 4.57 (m, 1H), 4.26–4.19 (m, 3H), 3.99 (s, 1H), 3.80 (d, J = 6.8 Hz, 1H), 3.54 (dd, J = 7.2, 14.4 Hz, 1H), 3.42 (t, J = 6.8 Hz, 2H), 3.31 (t, J = 6.4 Hz, 2H), 2.98 (t, J = 6.4 Hz, 2H), 2.89–2.84 (m, 2H), 2.49–2.32 (m, 5H), 0.86 (s, 3H), 0.73 (s, 3H); 31P NMR (162 MHz, D2O) δ: 1.96 (s, 1P), −9.66 (d, J = 22.7 Hz, 1P), −10.04 (d, J = 22.7 Hz, 1P); HRMS for [M-H]− (C27H41N7O18P3S−): calcd: 876.1447; found: 876.1467.
3-(S)-Hydroxy-4-azidobutyrate Coenzyme A (HABCoA) 1h
1H NMR (400 MHz, D2O) δ: 8.52 (s, 1H), 8.24 (s, 1H), 6.15 (d, J = 6.4 Hz, 1H), 4.56 (m, 1H), 4.21 (m, 3H), 3.98 (s, 1H), 3.83–3.76 (m, 1H), 3.55–3.51 (m, 1H), 3.44–3.37 (m, 3H), 3.32–3.27 (m, 3H), 2.98 (t, J = 6.0 Hz, 2H), 2.81–2.79 (m, 2H), 2.40 (t, J = 6.8 Hz, 2H), 0.86 (s, 3H), 0.73 (s, 3H); 31P NMR (162 MHz, D2O) δ: 1.70 (s, 1P), −9.66 (d, J = 19.8 Hz, 1P), −10.21 (d, J = 19.3 Hz, 1P); HRMS for [M-H]− (C25H40N10O18P3S−): calcd: 893.1461; found: 893.1484
Construction of wild-type (wt-) and A479S-PhaCCs Expression Plasmids
The wt-PhaCCs gene on the plasmid pET-PhaCCs33 was amplified by PCR using the forward primer 5’−TAGCGCATATGCAGCAGTTTGTCAATTCCCTG–3’ and reverse primer 5’−TTACTAAGCTTCAGTTCAAGGCGGCGGCGACGGGAG–3’ to introduce the unique restriction sites for NdeI and HindIII (underlined in the sequence) into the N- and C-terminus of the PhaCCs gene. The amplified gene was digested with NdeI and HindIII and ligated into pET28-MHL (Addgene plasmid # 26096) digested with the same enzymes to give plasmid pET28-MHL-PhaCCs. To generate the A479S-PhaCCs mutant, a QuickChange kit from Agilent Technologies was employed using 5’−CATCCGGCCACATCTCCGGCTCGATCAAC–3’ and 5’−GTTGATCGAGCCGGAGATGTGGCCGGATG–3’ as the forward and reverse primers to introduce the mutation (underlined in the sequence). The genes were sequenced to verify their correctness.
Expression and Purification of PHA Synthases
Synthase PhaCCc and PhaECAv were expressed and purified according to the previous published procedures.26, 33
The wt- and A479S-PhaCcs were overexpressed in E. coli BL21 (DE3) cells (Lucigen). A single colony was inoculated into 80 mL Luria Broth (LB) medium at 37°C overnight in the presence of 50 µg/mL kanamycin. This seed culture was used to inoculate 4 L LB medium at 37°C. When OD600 reached 0.6, the culture was cooled to 25 °C and protein expression was induced by addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM. After growth for an additional 4 h at 25 °C, 2 g/L cells were harvested by centrifugation at 5000 × g for 10 min at 4 °C and stored at −80 °C.
All of the following steps were carried out at 4 °C. Purification buffers consisted of buffer A [400 mM NaCl and 50 mM potassium phosphate (KPi), pH 7.8] and increasing concentrations of imidazole. Buffers B (lysis), C (wash), and D (elution) contained 10, 30, and 250 mM imidazole, respectively. The cell pellets were re-suspended in buffer B and lysed by sonication (25 × 30-s pulsed cycle). The cell debris was removed by centrifugation at 18,000 × g for 45 min and the supernatant was incubated with 15 mL of Ni-NTA resin that had been pre-equilibrated with buffer B. The resin was washed with ten column volumes of buffer C and then eluted with five column volumes of buffer D. The fractions containing PhaCCs were collected, concentrated, and exchanged into buffer E (100 mM NaCl and 50 mM KPi, pH 7.8) using an Amicon Ultra-15 (30K, EMD Millipore). The purified protein was stored in aliquots at −80 °C until further use. Protein purity was assessed by 12% acrylamide SDS-PAGE. The protein concentration was determined by the bicinchoninic acid assay using a BSA calibration curve.35
Enzyme Activity Assays
Enzyme assays were performed in either a continuous or discontinuous approach.28, 36 A continuous enzyme assay was employed for wt-PhaCCs, A479S-PhaCCs, and PhaECAv. Reactions were carried out at 30 °C in a final volume of 160 µL consisting of KPi (pH 7.8, 100 mM), 5,5'-dithiobis-(2-nitrobenzoic acid) (DTNB, 0.3 mM), BSA (0.2 mg/mL), TritonX-100 (50 mg/L, only for PhaCCs), HBCoA 1a or its analogs 1b–1h (0.8 or 1.6 mM), and the synthase at different concentrations. Reactions were initiated by the addition of enzyme. Formation of 3-thio-6-nitrobenzoate (TNB) dianion was determined by the absorbance at 412 nm using an extinct coefficient of 13.7 mM −1 cm−1. The reaction rate was determined by the slope of the initial fast phase and represented in the enzyme specific activity (SA, U/mg). One unit is defined as 1 µmol of substrate consumed per minute. The measurement was done in triplicate.
Enzyme activity of PhaCCc was determined by a discontinuous DTNB assay.34 Briefly, reactions were carried out at 30 °C in a final volume of 200 µL consisting of KPi (100mM, pH 7.8), BSA (0.2 mg/mL), HBCoA 1a or its analogs 1b–1h (1.6 mM), and PhaCCc at different concentrations. Reactions were initiated by the addition of enzyme. At various times, 20 µL aliquots were removed from the reaction mixtures and quenched with 20 µL of 10% trichloroacetic acid. Samples were centrifuged at 13,000 × g for 1 min to remove the precipitated proteins, and then 35 µL was added to 125 µL of 2 mM DTNB in 0.5 M KPi (pH 7.8). Formation of TNB dianion and the reaction rate (done in triplicate) were determined as described above.
Determination of Kinetic Parameters of PhaCCc and PhaECAv with Selected HACoA Analogs
Assays were conducted in the same way as described above. Concentrations of substrates and synthases are 0.1–6.4 mM HVCoA with 2.2 µM PhaCCc, 0.4–30.0 mM HHxeCoA with 32.6 µM PhaCCc, 0.1–3.2mM HHxyCoA with 32.6 µM PhaCCc, 0.2–40.0 mM HABCoA with 32.6 µM PhaCCc, 0.025–1.6 mM HBCoA with 5.8 nM PhaECAv, 0.02–1.6 mM HVCoA with 12.7 nM PhaECAv, 0.01–1.6 mM HHxeCoA with 63.6 nM PhaECAv, 0.01–1.6 mM HHxyCoA with 254 nM PhaECAv, and 0.01–1.6 mM HABCoA with 50.9 nM PhaECAv. Each substrate concentration was run in triplicate. The kinetic parameters were determined by fitting the data to Michaelis-Menten equation 1 using SigmaPlot.
| (1) |
RESULTS AND DISCUSSION
Chemical Synthesis of HACoA Analogs 1b–h
A series of analogs 1b–h were synthesized according to the strategy developed by Stubbe et al. except for some modifications in order to improve yields.19 As shown in Scheme 2, commercially available dimethyl (S)-malate 2 was regioselectively reduced to 1, 2-diol 3 by borane-dimethyl sulfide (Me2S:BH3) and catalytic amount of sodium borohydride (NaBH4).37 It has been reported that addition of NaBH4 greatly increases the reduction rate though Me2S:BH3 itself also affords the same product.37 After the reduction reaction was quenched with methanol and solvents were removed, crude product 3 was directly converted to 3-(S)-hydroxy-4-butanolide 4 in the presence of catalytic trifluoroacetic acid (TFA) at a temperature between 24 and 28 °C. It has to be noted that the temperature control for this step was crucial as the yield dramatically decreased when the reaction temperature reached 35 °C or higher. We suspect that the butanolide 4 may undergo polymerization in an acidic condition at elevated temperatures. Our TLC result also showed that the diol 3 was still present when the lactonization step was carried out at 20 °C even for 48 hrs. Thus, by carefully monitoring temperatures, the butanolide 4 was obtained with a total yield of 87% for two steps. Treatment of 4 with absolute ethanol and Me3SiI gave iodohydrin in a high yield at 95% after silica gel chromatography.38 It has to be pointed out that, although Me3SiI could silylate a hydroxyl group, no trimethylsilyl (TMS)-protected product was observed in the ring opening of butanolide 4. The iodohydrin 5 was converted to epoxyester 6 in a 92% yield by refluxing with Ag2O at 80 °C for 4 hrs.38 Interestingly, the epoxidation took place only in dimethoxyethane (DME), instead of acetonitrile, although both solvents have similar physical properties.
To obtain an enantiomerically pure 3-(R)-hydroxyester 7, a nucleophile would react with either the iodohydrin 5 or epoxyester 6.19, 38 Since the ester group in 6 is reactive toward organolithium and Grignard reagents, organocuprates were employed as the nucleophiles to open the epoxide ring. Generation of the organocuperate was accomplished by mixing Me2S:CuBr complex and organolithium (or Grignard reagent) at −60 °C followed by incubation with stirring at −25 °C for 1 h. Epoxyester 6 was then added into the organocuperate mixture and the ring-opening reaction was complete within 4 h at −25 °C. It has to be noted that the ethynylcopper generated from lithium trimethylsilylacetylide failed to give the expected product 7f. Thus, the ethynylaluminum prepared from the corresponding lithium reagent and diethylaluminum chloride at 0 °C was used as the nucleophile.
Once the 3-(R)-hydroxyester 7 was purified, it was reacted with a silylating agent under basic conditions to yield ester 8 by protecting the hydroxyl group. While tert-butyldimethylsilyl chloride (TBDMSCl) would efficiently silylate 7a–7e and 7g, the yield was extremely low for 7f. The low yield could be attributed to the presence of large trimethylsilyl (TMS) group in 7f, which might partially shield the −OH and lower its nucleophilicity to react with TBDMSCl through a SN2 mechanism. Formation of 8f was achieved in the presence of triisopropylsilyl trifluoromethanesulfonate (TIPS-OTf) because the OTf− is a much better leaving group than the Cl−.
Alkaline hydrolysis of ester 8 followed by acidification gave carboxylic acid 9, which was subsequently coupled with benzenethiol to yield thioester 10 in the presence of N,N’-dicyclohexylcarbodiimide (DCC) and catalytic 4-(dimethylamino)pyridine (DMAP). Diluted hydrofluoric acid (5% HF in acetonitrile) was utilized to quickly remove the TBDMS- or TIPS-protecting group to afford 3-(R)-hydroxythioester 11. To synthesize the target compounds, 5–10 equiv. 11a–11g were dissolved in acetonitrile and mixed with 1 equiv. CoASH in 50 mM phosphate buffer at pH 10. The reaction was monitored by HPLC and usually complete within 48 to 72 h. Excess 11 was removed by multiple ether extractions, resulting in more than 90% purity for HACoA analogs 1b−1h, which would be directly used for enzyme assays. Samples for analytical analysis were further purified by semi-preparative HPLC and their structures were confirmed by 1H NMR, 31P NMR, and HRMS. The overall yields and HPLC retention times are summarized in Table 1.
Selection and Purification of PHA Synthases
Since class I and class III enzymes are the most studied PHA synthases,2, 19, 24–32, 34, 39–42 they were also selected as the focus of the present investigation. Specifically, PhaCCs, PhaCCc, and PhaECAv were used to study their reactivity toward HACoA analogs with various side chains. Both the PhaCCs and PhaCCc belong to class I synthases. The PhaCCs is known as the most active PHA synthases so far, which can polymerize monomers such as 3-(R)-hydroxybutyrate (HB), 3-(R)-hydroxyvalerate (HV), and 3-(R)-hydroxyhexanoate (HHx).33 Since not many PHA synthases have the ability to use both the SCL and MCL monomers, the PhaCCs’s capacity to produce copolymers with mixed monomeric chain lengths highlights its great potential for various applications. In addition, the mutant A479S- PhaCCs was reported to display higher preference to MCL monomers than the wt,20 suggesting that the mutation may facilitate the enzyme to adapt to side chain modifications. Thus, it was also included for our study.
The PhaCCc was selected as a second representative of class I synthases for the current study due to its uniqueness. First, it is the only known class I PHA synthase that lacks of a lag phase in its polymerization kinetics.34 Second, priming of PhaCCc with saturated trimeric HBCoA (named sTCoA)24 demonstrated that approximately one equivalent CoA per PhaC was released during enzyme assay.34 This was quite different from other reported class I synthases such as the one from Ralstonia eutropha (PhaCRe), in which the labeling stoichiometry was determined to be 0.5 equivalent CoA per PhaC.24 All of these suggest that PhaCCc represents an unusual example of class I PHA synthases and is worth further investigation.
The PhaECAv is a prototypical class III synthase, which has been extensively studied as a model to understand the mechanism of polymerization catalyzed by PHA synthases.24, 26, 27, 29–32, 43 However, its substrate specificity in terms of R’ group in HACoA 1 (Scheme 2) still remains to be explored. Understanding the broadness of substrate specificity will permit the synthesis of hybrids of SCL- and MCL-PHAs as well as introduction of novel monomeric units that are rarely present in natural polyesters.
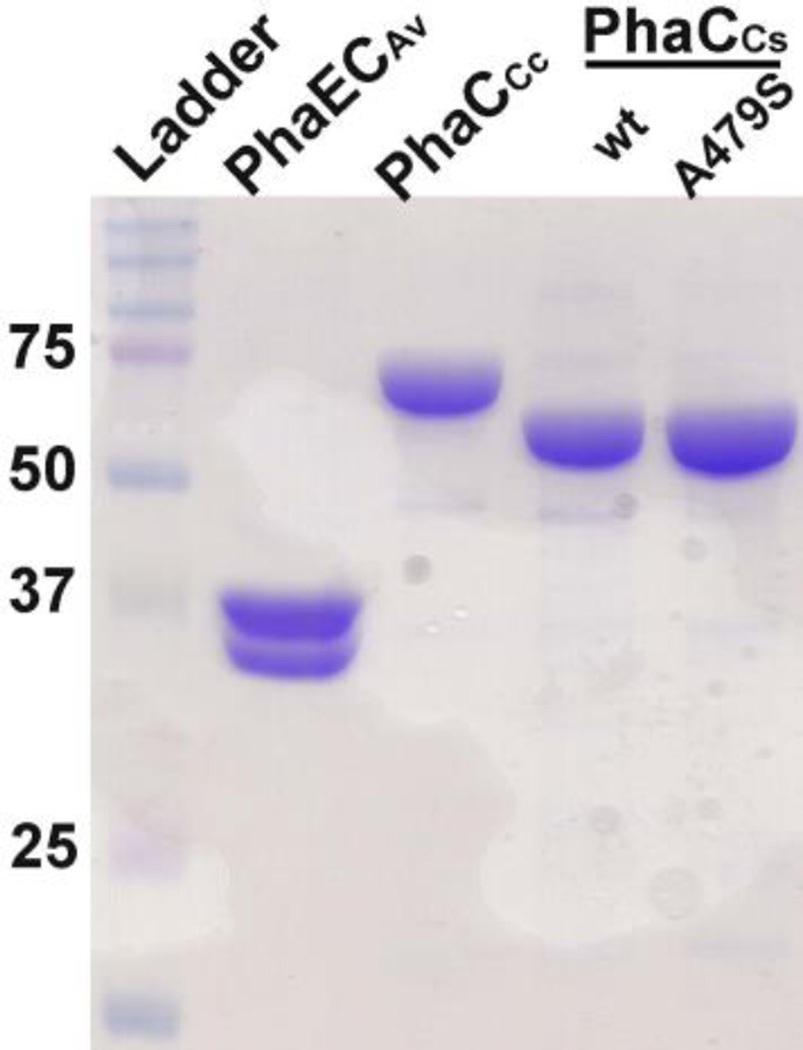
The purifications of PhaCCc and PhaECAv were performed according to the previously reported protocols.26, 33 The wt and A479S-PhaCCs were purified as N-terminal His-tagged proteins containing a tobacco etch virus (TEV) protease cleavage site. Unlike purification of PhaCRe, a prototypical class I synthase, in which detergent Hecamag at 0.05% is required in order to prevent its nonspecific surface binding,40 the synthase PhaCCs could be obtained in the absence of any detergent. Moreover, removal of the His-tag did not affect its enzyme activity (data not shown). Therefore, the assays were carried out in the presence of His-tag. All proteins were purified to more than 95% homogeneous judged by SDS-PAGE shown in Figure 1. The molecular weights (MWs) of PhaCCs, PhaCCc and PhaECAv were calculated to be 66, 73, and 39/39 KDa, respectively, corresponding to their bands shown in Figure 1.
Figure 1.
SDS-PAGE of purified PHA synthases.
Selection of Assay Methods
PHA synthases can be assayed by DTNB using either a continuous or discontinuous approach.28, 36 Formation of the released CoA (Scheme 1) is determined by the breakage of disulfide bond in DTNB to form TNB.44 While the continuous approach is more convenient than the discontinuous one, the latter is more accurate as it eliminates the interference caused by surface (solvent-exposed) cysteines in certain PHA synthases. To verify whether a continuous DTNB assay could be used for the synthases of interest, their SA were determined by both methods. It was found that all of them except for PhaCCc had similar SA values (data not shown). Thus, we decided to assay PhaCCc and other synthases in this report using discontinuous and continuous methods, respectively.
Activation Factors of Enzyme Specific Activity
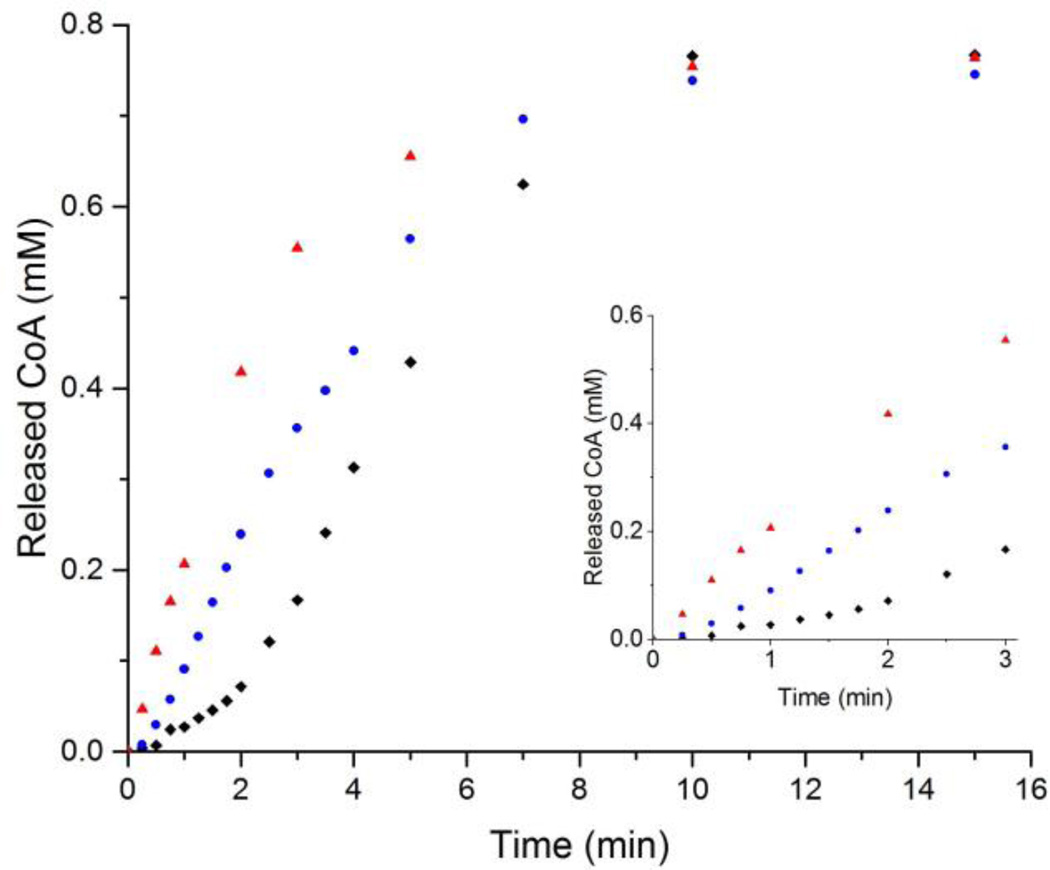
Before the PhaCCc was reported in 2015,34 kinetics of polymerization reactions catalyzed by recombinant class I PHA synthases was found to contain a lag phase.28, 33 Although the exact cause of the lag phase remains an enigma, it is thought to relate to protein dimerization.28 For example, equilibrium of class I PhaCRe shifts from monomers to dimers in the presence of synthetic PHA primers, resulting in significant reduction of the lag phase and increase of enzyme activity.28 Same phenomena were also observed with addition of non-ionic detergents like Hecameg and TritonX-100 at concentration of 0.2–0.4 CMC.45 As depicted in Figure 2, polymerization of HBCoA catalyzed by PhaCCs displays a lag phase for ~3 min (black square), which is reduced to ~ 45 s in the presence of 50 mg/L TritonX-100 (blue dot). The activity (slope of the linear part) was also increased slightly in the presence of detergent. Since molecular mechanisms of PhaC activation and lag phase elimination are complicated,45 kinetic parameters of any class I PHA synthase have not been determined. However, for the first time, the lag phase was absent from PhaCCc (a class I synthase, red triangle). Thus, its kinetic parameters could be derived from the Michaelis-Menten equation and used to make quantitative comparisons between different substrates in terms of catalytic efficiency (kcat/KM).
Figure 2.
Polymerization of HBCoA (0.8 mM) catalyzed by PhaCCc (79.5 nM, red triangle), PhaCCs (3.5 nM) in the presence of 50 mg/L TritonX-100 (blue dot), and PhaCCs only (3.8 nM, black square). The inset is the expansion of the progress curves during 0–3 min.
The PhaECAv activity was reported to increase 1.4-fold when BSA was added to a concentration of 0.5 mg/mL (7.5 µM) in the assay mixture.46 Since the BSA was shown to bind PHB granules in vitro,47 it was proposed that the observed increase resulted from the BSA interacting with hydrophobic polymers to prevent the growing chain from blocking the active site.48 Therefore, the proposed interaction would allow for great substrate access to PhaC.48 However, other PHB granule binding proteins such as PhaP1 did not exhibit the same effect.46 In addition, BSA is often used to eliminate nonspecific binding of enzymes to apparatus surface.49 This nonspecific binding will lower the effective concentration of the active enzyme. The effect becomes much more pronounced when the enzyme concentration is in low nanomolar ranges. Indeed, the SA of PhaECAv in the present study was determined to be 373 U/mg when the assay was carried out in the presence of BSA at 0.2 mg/mL between 5 nM enzyme and 1.6 mM HBCoA. The SA dropped 2.5-fold to 150 U/mg when the BSA was absent. The activation effect became marginal when the assay mixture contained 0.25 µM PhaECAv and 1.6 mM HHxyCoA.
The BSA activation effect was also investigated with two class I synthases. It was found that, while addition of BSA would increase SA of PhaCCs with HBCoA (574 U/mg vs. 280 U/mg in the absence of BSA), it did not have an extensive effect on PhaCCc with HBCoA (58 U/mg vs. 50 U/mg in the absence of BSA). Since presence of BSA did not inhibit activities of the synthases, for simplicity, all assays were performed with BSA at 0.2 mg/mL.
Enzyme Activity Toward HACoA Analogs
To the best of our knowledge, no one has systematically investigated the reactivity of class I and III PhaCs toward HACoA analogs in terms of side chain length and its modification. Yet the information is crucial because identity of R not only determines the material property of PHA polymers, but also allows for modification of the resulting PHAs to introduce new functionalities.
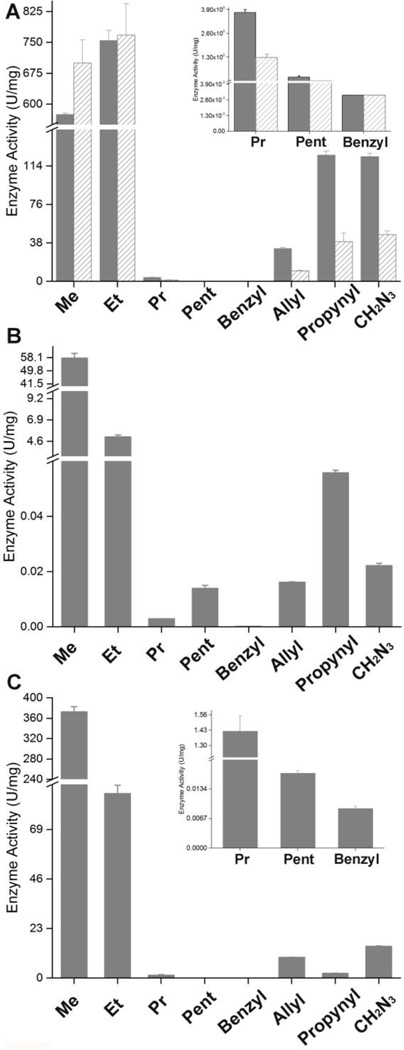
The synthase substrate specificity with a series of HACoA analogs is summarized in Figure 3. Consistent with synthase classification, both class I and class III enzymes have higher activities with SCL monomers (C4–C5) than with MCL monomers (≥C6). However, the trends are different between synthases. Activity of wt-PhaCCs increased in the order of C8〈〈C6〈〈C4〈C5, in which the PhaCCs displayed 131% and 0.65% HBCoA (R’ = Me) activities toward HVCoA (R’ = Et) and HHxCoA (R’ = Pr), respectively. While A479S-PhaCCs displayed the same order as the wt with regard to chain length, it exhibited higher activity for both HBCoA and HVCoA and lower activity for HHxCoA than wt. Our results on A479S-PhaCCs were exactly opposite to those previously reported, in which the mutant had decreased activity for both HBCoA and HVCoA and increased activity for HHxCoA relative to the wt.20 While the exact cause of the discrepancy is unknown, it may arguably be attributed to the presence of BSA as well as different epitope tags and purification methods.50, 51 The order of PhaCCc reactivity was determined to be C6〈C8〈〈C5〈〈C4, implying that the active site of PhaCCc prefers HBCoA and may be smaller than that of PhaCCs. A slightly higher activity with HCCoA (R’ = Pent) than with HHxCoA may result from increased interactions between the side chain of HCCoA and hydrophobic residues in the substrate entrance channel. The activity of PhaECAv dropped significantly with increasing length of the side chain in substrates. For example, the polymerization rates of HVCoA, HHxCoA, and HCCoA catalyzed by PhaECAv were measured at 23%, 0.38%, and 0.005% rate of HBCoA, respectively.
Figure 3.
Enzyme SA toward HACoA analogs 1a–1h at 1.6 mM. (A) wt- (solid bar) and A479S-PhaCCs (patterned bar). The enzyme concentrations are (wt/A479S) 2.0/2.4 nM, 2.0/2.4 nM, 234/285 nM, 703/854 nM, 703/854 nM, 47/57 nM, 7.8/9.5 nM, and 7.8/9.5 nM for HBCoA (Me), HVCoA (Et), HHxCoA (Pr), HCCoA (pent), HPBCoA (benzyl), HHxeCoA (allyl), HHxyCoA (propynyl), and HABCoA (CH2N3), respectively. (B) PhaCCc. The enzyme concentrations are 81.5 nM, 2.17 µM, 65.21 µM, 32.60 µM, 32.60 µM, 32.60 µM, 32.60 µM, and 32.60 µM for HBCoA, HVCoA, HHxCoA, HCCoA, HPBCoA, HHxeCoA, HHxyCoA, and HABCoA, respectively. (C) PhaECAv. The enzyme concentrations are 5.1 nM, 12.7 nM, 509 nM, 10.17 µM, 10.17 µM, 63.6 nM, 254 nM, 50.9 nM for HBCoA, HVCoA, HHxCoA, HCCoA, HPBCoA, HHxeCoA, HHxyCoA, and HABCoA, respectively.
To quantitatively compare the effects of chain length on the activities of class I and class III PHA synthases, their kinetic parameters were determined by fitting the data to Michaelis-Menten equation and the results are summarized in Table 2. Only PhaCCc was selected to represent class I synthases because it is the only known class I synthase that does not contain a lag phase. Kinetic parameters were not measured with HHxCoA and HCCoA due to their marginal activities.
Table 2.
Kinetic Parameters of PhaCs with HACoA Analogs
| Substrate | Class I synthase: PhaCCc | Class III synthase: PhaECAv | ||||
|---|---|---|---|---|---|---|
| Km (mM) | kcat (s−1) | kcat/Km (M−1s−1) | Km (mM) | kcat (s−1) | kcat/Km (M−1s−1) | |
| HBCoA | 0.29a | 75.0a | 2.59 × 105 | 0.11 | 508 | 4.62 × 106 |
| HVCoA | 2.07 | 14.4 | 6.96 × 103 | 0.11 | 98.1 | 8.92 × 105 |
| HHxeCoA | 3.22 | 0.06 | 1.75 × 101 | 0.05 | 12.7 | 2.54 × 105 |
| HHxyCoA | 1.54 | 0.13 | 8.25 × 101 | 0.05 | 2.99 | 5.98 × 104 |
| HABCoA | 9.46 | 0.19 | 2.01 × 101 | 0.07 | 20.0 | 2.86 × 105 |
From reference 34
While the catalytic efficiency (kcat/Km) of PhaCCc toward HBCoA was reported to be 2.59 × 105 M−1s−1,34 it dropped 37-fold to 6.96 × 103 M−1s−1 when HVCoA was employed as the substrate. The reduction in catalytic efficiency results from the low binding affinity and slowed turnover number when an additional methylene group is added onto the side chain. This is in contrast with the other class I synthase in the present report, PhaCCs that displays higher activity with HVCoA than HBCoA.
Although addition of BSA enhances in vitro activity of PhaECAv,46 its kinetic parameters have never been determined in the presence of BSA. Our study indicates that BSA could significantly boost the turnover number of PhaECAv toward HBCoA (from 65 s−1 to 508 s−1) without affecting the Km (0.11 mM with BSA vs 0.13 mM without BSA).19 Thus, the catalytic efficiency is improved by more than 8-fold from 5.03 × 105 M−1s−1 in the absence of BSA to 4.62 × 106 M−1s−1 in the presence of BSA. Similar to PhaCCc, the catalytic efficiency of PhaECAv toward HVCoA decreased 5.2-fold compared with HBCoA. However, unlike PhaCCc, the decrease is exclusively caused by the lowered reaction rate (kcat), which indicates that the additional methylene group in the side chain may disrupt optimal alignment of the substrate and catalytic residues for polymerization reactions.
We also investigated the effects of side chain modifications on the reactivity of PHA synthases. It was discovered that introduction of aromatic ring nearly abolished the enzyme activity. This is not surprising because the size of the aromatic ring may prevent HPBCoA 1e from entering the active site. But both class I and class III PHA synthases can sustain minor side chain modification to different degrees depending on the individual enzyme.
As shown in Figure 3 and for all synthases, incorporation of a terminal double bond in the side chain results in the increased activity relative to the saturated side chain with the same length. The enzyme activities of HHxeCoA with wt-PhaCCs, PhaCCc, and PhaECAv are 8.6-, 5.4-, and 6.8-fold higher than HHxCoA, respectively. Presence of a terminal triple bond in the side chain would further improve activities of class I synthases, in which wt-PhaCCs and PhaCCc exhibited 3.9- and 3.5-fold higher activity with HHxyCoA than with HHxeCoA, respectively. The observed increasing trend suggest that removal of hydrogen atoms in the side chain may facilitate substrate binding, which is consistent with the Km values summarized in Table 2. However, the trend is reversed in class III PhaECAv. The enzyme SA decreased 4.2-fold when the terminal double bond in HHxeCoA was replaced with a triple bond in HHxyCoA (Figure 3). The replacement did not affect the substrate binding, but significantly lowered the reaction turnover number, implying that the optimal alignment for polymerization reactions may be further disrupted in HHxyCoA.
When an azide group was installed in the side chain, the corresponding analog, HABCoA, had higher reactivity with all synthases than MCL substrates (HHxCoA and HCCoA) and HHxeCoA. However, HABCoA showed different effects on the activity of PHA synthases compared to HHxyCoA. While the wt-PhaCCs had similar activities toward HHxyCoA and HABCoA, the other class I synthase, PhaCCc, displayed a 2.5-fold lower activity with HABCoA than with HHxyCoA. The class III PhaECAv showed a different pattern, in which it could polymerize HABCoA 6.5-fold faster than HHxyCoA. Although PhaC structures are still unknown, our results suggest that the active site of class III synthases may be more polar than that of class I synthases, which would facilitate binding of the HACoA analog containing a polar azide group.
Importance of Side Chain Modifications and Their Potential Applications
Introduction of unsaturated side chains into PHAs is of great interest as it offers a new avenue to modify the polymers to exhibit functionalities en route towards new PHA-based biomaterials.6 For example, the side chains containing a terminal double bond can be oxidized to produce hydroxyl pendants for attaching new functional moieties.52
Additionally, HACoA analogs with a modified side chain could be also employed to study biogenesis of the PHA granules, which is still a challenge at this time. PHA biosynthesis represents a widespread phenomenon in nature named template-independent polymerizations (TIPs).3 Such examples include generation of natural rubber from isopentenyl pyrophosphates,53 formation of starch from ADP-glucose,54 and many more.55–58 While both TIP and template-dependent polymerization (e.g. formation of nucleic acids and polypeptides) reactions use soluble substrates, the polymers produced in TIP reactions undergo phase transition to generate insoluble granules inside the cell.3 However, mechanism of granule formation remains unknown. Thus, PHA biosynthesis has been selected as a model system to study this challenging process. Three models have been proposed for biogenesis and structures of PHA granules.59 Differentiation of these models is important because PHA granules are increasingly recognized as potential functionalized beads for use in biotechnological and biomedical applications.60 Therefore, introduction of reactive handles that may enable direct imaging of PHA granule formation and structure is of great interest in general.
Incorporation of terminal alkyne and azide into the side chain may create an opportunity to study PHA granule formation using click-chemistry. The copper-catalyzed alkyne-azide cycloadditions (CuAAC) has emerged as one of the most useful reactions for bioconjugation.61 When these reactive handles are incorporated by the synthases, the produced PHAs will be visible if fluorescence is introduced using CuAAC. This gives a unique opportunity to image granule formation in vivo and provides a potential way to understand the biogenesis of PHA granules. The information is important for metabolic engineering such that the PHAs with defined properties can be generated economically.
Our study has demonstrated that both class I and class III synthases are sensitive to the modifications in the alkyl side chain. As shown in Table 2, the substrate binding affinities are weakened significantly for PhaCCc in addition to the dramatically slowed reaction rates, resulting in 104-fold decrease in catalytic efficiency relative to the classical substrate HBCoA. The detrimental effects are much smaller on PhaCCs, in which the activities only dropped 5–18 folds compared with HBCoA (Figure 3). While the class III PhaECAv displayed higher affinities than HBCoA toward analogs containing a terminal unsaturated bond or azide group, its reaction rates with the modified analogs were much slower than that of HBCoA, resulting in 16–80-fold decrease in catalytic efficiency. These results showed that both the HHxyCoA and HABCoA could be potentially employed to image biogenesis of PHA granules as they could be efficiently incorporated into the polymers produced by PhaCCs and PhaECAv. Additionally, during the review process of this paper, Nomura et al reported an in vivo synthesis of clickable PHA polymers utilizing ω-azido fatty acids as the feedstocks.62 Since HHxCoA could be metabolically generated from 3-(R)-hydroxy-5-hexynoic acid63 and exhibits high reactivity toward PhaCCs (22% activity of HBCoA), it is currently used as a probe to investigate the formation of PHA granules.
CONCLUSIONS
In this study, a series of HACoA analogs were conveniently synthesized in high yields from an enantiomerically pure chemical that was commercially available. They were used to study the substrate specificity of class I and class III PHA synthases in terms of side chain length and chain modifications. Our results have shown that inclusion of BSA in the reaction mixture significantly enhances in vitro activities of PhaCCs and PhaECAv. Generally, activities of class I and class III PhaCs decrease with the increasing carbon chain length. However, the wt-PhaCCs displayed 1.3-fold higher activity toward HVCoA than HBCoA. While the mutant A479S-PhaCCs exhibited improved activity with SCL relative to the wt, its activity with MCL substrate was lower than that of the wt. Although introduction of an aromatic ring abolished the enzyme activity, all synthases could sustain minor modifications such as the unsaturated bonds and azide group. The SA of PhaCCs toward HHxyCoA and HABCoA were 125 and 123 U/mg, respectively, which were ~22% activity of the classical substrate HBCoA. The catalytic efficiency of PhaECAv toward HABCoA and HHxyCoA were determined to be 2.86 × 105 and 5.98 × 104 M−1s−1, respectively, which corresponded to 1.3–6.2% activity of HBCoA (4.62 × 106 M−1s−1). Thus, these side chain modifications may be used to introduce new material functions to PHAs as well as to study the challenging PHA biogenesis via click-chemistry.
Supplementary Material
Acknowledgments
The authors would like to thank Profs. JoAnne Stubbe (MIT) and Kumar Sudesh (USM) for providing plasmids pRBPhaCCc and pET-PhaCCs, respectively. We also wish to thank Prof. John Tomich for reading the manuscript.
Funding Sources
This work was supported in part by funds from Johnson Cancer Research Center and NIH P30GM110761.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Synthesis and characterization of compounds, copies of NMR spectra, and kinetic analysis of the compounds with PHA synthases. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.Wang Y, Yin J, Chen GQ. Curr. Opin. Biotech. 2014;30:59–65. doi: 10.1016/j.copbio.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Stubbe J, Tian J. Nat. Prod. Rep. 2003;20:445–457. doi: 10.1039/b209687k. [DOI] [PubMed] [Google Scholar]
- 3.Stubbe J, Tian JM, He AM, Sinskey AJ, Lawrence AG, Liu PH. Annu. Rev. Biochem. 2005;74:433–480. doi: 10.1146/annurev.biochem.74.082803.133013. [DOI] [PubMed] [Google Scholar]
- 4.Steinbüchel A, Hein S. Adv. Biochem. Eng. Biot. 2001;71:81–123. doi: 10.1007/3-540-40021-4_3. [DOI] [PubMed] [Google Scholar]
- 5.Sudesh K, Abe H, Doi Y. Prog. Polym. Sci. 2000;25:1503–1555. [Google Scholar]
- 6.Chen GQ. Chem. Soc. Rev. 2009;38:2434–2446. doi: 10.1039/b812677c. [DOI] [PubMed] [Google Scholar]
- 7.Kessler B, Weusthuis R, Witholt B, Eggink G. Adv. Biochem. Eng. Biot. 2001;71:159–182. doi: 10.1007/3-540-40021-4_5. [DOI] [PubMed] [Google Scholar]
- 8.Steinbüchel A, Valentin HE. FEMS Microbiol. Lett. 1995;128:219–228. [Google Scholar]
- 9.Chen GQ. Plastics from bacteria: Natural functions and applications. Heidelberg ; New York: Springer; 2010. [Google Scholar]
- 10.Sudesh K, Iwata T. Clean-Soil Air Water. 2008;36:433–442. [Google Scholar]
- 11.Keshavarz T, Roy I. Curr. Opin. Microbiol. 2010;13:321–326. doi: 10.1016/j.mib.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Snell KD, Peoples OP. Biofuel Bioprod. Bior. 2009;3:456–467. [Google Scholar]
- 13.van Geemen D, Driessen-Mol A, Grootzwagers LGM, Soekhradj-Soechit RS, Vis PWR, Baaijens FPT, Bouten CVC. Regen. Med. 2012;7:59–70. doi: 10.2217/rme.11.100. [DOI] [PubMed] [Google Scholar]
- 14.Brigham CJS, A J. Int. J. Biotech. Well. Indus. 2012;1:53–60. [Google Scholar]
- 15.Lau NS, Ch'ng DHE, Chia KH, Wong YM, Sudesh K. J. Biobased Mater. Bio. 2014;8:118–129. (2014) [Google Scholar]
- 16.Koller M, Bona R, Braunegg G, Hermann C, Horvat P, Kroutil M, Martinz J, Neto J, Pereira L, Varila P. Biomacromolecules. 2005;6:561–565. doi: 10.1021/bm049478b. [DOI] [PubMed] [Google Scholar]
- 17.Rehm BH. Biochem. J. 2003;376:15–33. doi: 10.1042/BJ20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuge T, Hyakutake M, Mizuno K. Appl. Microbiol. Biot. 2015;99:6231–6240. doi: 10.1007/s00253-015-6777-9. [DOI] [PubMed] [Google Scholar]
- 19.Yuan W, Jia Y, Tian JM, Snell KD, Muh U, Sinskey AJ, Lambalot RH, Walsh CT, Stubbe J. Arch. Biochem. Biophys. 2001;394:87–98. doi: 10.1006/abbi.2001.2522. [DOI] [PubMed] [Google Scholar]
- 20.Chuah JA, Tomizawa S, Yamada M, Tsuge T, Doi Y, Sudesh K, Numata K. Appl. Environ. Microbiol. 2013;79:3813–3821. doi: 10.1128/AEM.00564-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su L, Lenz RW, Takagi Y, Zhang SM, Goodwin S, Zhong LH, Martin DP. Macromolecules. 2000;33:229–231. [Google Scholar]
- 22.Gerngross TU, Martin DP. P. Natl. Acad. Sci. USA. 1995;92:6279–6283. doi: 10.1073/pnas.92.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukui T, Chou K, Harada K, Orita I, Nakayama Y, Bamba T, Nakamura S, Fukusaki E. Metabolomics. 2014;10:190–202. [Google Scholar]
- 24.Jia Y, Kappock TJ, Frick T, Sinskey AJ, Stubbe J. Biochemistry. 2000;39:3927–3936. doi: 10.1021/bi9928086. [DOI] [PubMed] [Google Scholar]
- 25.Jia Y, Yuan W, Wodzinska J, Park C, Sinskey AJ, Stubbe J. Biochemistry. 2001;40:1011–1019. doi: 10.1021/bi002219w. [DOI] [PubMed] [Google Scholar]
- 26.Muh U, Sinskey AJ, Kirby DP, Lane WS, Stubbe J. Biochemistry. 1999;38:826–837. doi: 10.1021/bi9818319. [DOI] [PubMed] [Google Scholar]
- 27.Tian JM, Sinskey AJ, Stubbe J. Biochemistry. 2005;44:8369–8377. doi: 10.1021/bi050331u. [DOI] [PubMed] [Google Scholar]
- 28.Wodzinska J, Snell KD, Rhomberg A, Sinskey AJ, Biemann K, Stubbe J. J. Am. Chem. Soc. 1996;118:6319–6320. [Google Scholar]
- 29.Chen C, Cao RK, Shrestha R, Ward C, Katz BB, Fischer CJ, Tomich JM, Li P. ACS Chem. Biol. 2015;10:1330–1339. doi: 10.1021/cb5009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, Chakraborty S, Stubbe J. Biochemistry. 2009;48:9202–9211. doi: 10.1021/bi901329b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Chen C, Cao R, Maurmann L, Li P. ChemBioChem. 2015;16:156–166. doi: 10.1002/cbic.201402380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Shrestha R, Buckey RM, Jewell J, Bossmann SH, Stubbe J, Li P. ACS Chem. Biol. 2014;9:1773–1779. doi: 10.1021/cb5002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhubalan K, Chuah JA, Shozui F, Brigham CJ, Taguchi S, Sinskey AJ, Rha C, Sudesh K. Appl. Environ. Microb. 2011;77:2926–2933. doi: 10.1128/AEM.01997-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckley RM, Stubbe J. Biochemistry. 2015;54:2117–2125. doi: 10.1021/bi501405b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 36.Zhang SM, Yasuo T, Lenz RW, Goodwin S. Biomacromolecules. 2000;1:244–251. doi: 10.1021/bm005513c. [DOI] [PubMed] [Google Scholar]
- 37.Saito S, Hasegawa T, Inaba M, Nishida R, Fujii T, Nomizu S, Moriwake T. Chem. Lett. 1984:1389–1392. [Google Scholar]
- 38.Larcheveque M, Henrot S. Tetrahedron. 1990;46:4277–4282. [Google Scholar]
- 39.Cho M, Brigham CJ, Sinskey AJ, Stubbe J. Biochemistry. 2012;51:2276–2288. doi: 10.1021/bi2013596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerngross TU, Snell KD, Peoples OP, Sinskey AJ, Csuhai E, Masamune S, Stubbe J. Biochemistry. 1994;33:9311–9320. doi: 10.1021/bi00197a035. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence AG, Choi J, Rha C, Stubbe J, Sinskey AJ. Biomacromolecules. 2005;6:2113–2119. doi: 10.1021/bm0501048. [DOI] [PubMed] [Google Scholar]
- 42.Sim SJ, Snell KD, Hogan SA, Stubbe J, Rha CK, Sinskey AJ. Nat. Biotechnol. 1997;15:63–67. doi: 10.1038/nbt0197-63. [DOI] [PubMed] [Google Scholar]
- 43.Tian J, Sinskey AJ, Stubbe J. Biochemistry. 2005;44:1495–1503. doi: 10.1021/bi047734z. [DOI] [PubMed] [Google Scholar]
- 44.Eyer P, Worek F, Kiderlen D, Sinko G, Stuglin A, Simeon-Rudolf V, Reiner E. Anal. Biochem. 2003;312:224–227. doi: 10.1016/s0003-2697(02)00506-7. [DOI] [PubMed] [Google Scholar]
- 45.Ushimaru K, Sangiambut S, Thomson N, Sivaniah E, Tsuge T. Appl. Microbiol. Biot. 2013;97:1175–1182. doi: 10.1007/s00253-012-4089-x. [DOI] [PubMed] [Google Scholar]
- 46.Jossek R, Reichelt R, Steinbuchel A. Appl. Microbiol. Biot. 1998;49:258–266. doi: 10.1007/s002530051166. [DOI] [PubMed] [Google Scholar]
- 47.Horowitz DM, Sanders JKM. Can. J. Microbiol. 1995;41:115–123. [Google Scholar]
- 48.Niamsiri N, Bergkvist M, Delamarre SC, Cady NC, Coates GW, Ober CK, Batt CA. Colloid Surface B. 2007;60:68–79. doi: 10.1016/j.colsurfb.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 49.Acker MGA, D S. Perspect. Sci. 2014;1:56–73. [Google Scholar]
- 50.Horchani H, Ouertani S, Gargouri Y, Sayari A. J. Mol. Catal. B–Enzym. 2009;61:194–201. [Google Scholar]
- 51.Smith MA, Gonzalez J, Hussain A, Oldfield RN, Johnston KA, Lopez KM. Enzyme Res. 2016 doi: 10.1155/2016/5098985. Article ID 5098985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee MY, Park WH, Lenz RW. Polymer. 2000;41:1703–1709. [Google Scholar]
- 53.Steinbüchel A, Koyama T. Biopolymers. Vol. 2, Polyisoprenoids. Weinheim ; New York: Wiley-VCH; 2001. [Google Scholar]
- 54.Ball SG, Morell MK. Annu. Rev. Plant Biol. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- 55.McNamara JT, Morgan JLW, Zimmer J. Annu. Rev. Biochem. 2015;84:895–921. doi: 10.1146/annurev-biochem-060614-033930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roach PJ. Curr. Mol. Med. 2002;2:101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- 57.Shiba T, Tsutsumi K, Ishige K, Noguchi T. Biochemistry-Moscow+ 2000;65:315–323. [PubMed] [Google Scholar]
- 58.Yother J. Annu. Rev. Microbiol. 2011;65:563–581. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 59.Jendrossek D, Pfeiffer D. Environ. Microbiol. 2014;16:2357–2373. doi: 10.1111/1462-2920.12356. [DOI] [PubMed] [Google Scholar]
- 60.Draper JL, Rehm BH. Bioengineered. 2012;3:203–208. doi: 10.4161/bioe.19567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinto A, Ciesla JH, Palucci A, Sutliff BP, Nomura CT. ACS Macro Lett. 2016;5:215–219. doi: 10.1021/acsmacrolett.5b00823. [DOI] [PubMed] [Google Scholar]
- 63.Gong YM, Wan X, Jiang ML, Hu CJ, Hu HH, Huang FH. Prog. Lipid. Res. 2014;56:19–35. doi: 10.1016/j.plipres.2014.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.