Abstract
Background:
The national mandate for health systems to transition from ICD-9-CM to ICD-10-CM in October 2015 has an impact on research activities. Clinical phenotypes defined by ICD-9-CM codes need to be converted to ICD-10-CM, which has nearly four times more codes and a very different structure than ICD-9-CM.
Methods:
We used the Centers for Medicare & Medicaid Services (CMS) General Equivalent Maps (GEMs) to translate, using four different methods, condition-specific ICD-9-CM code sets used for pragmatic trials (n=32) into ICD-10-CM. We calculated the recall, precision, and F score of each method. We also used the ICD-9-CM and ICD-10-CM value sets defined for electronic quality measure as an additional evaluation of the mapping methods.
Results:
The forward-backward mapping (FBM) method had higher precision, recall and F-score metrics than simple forward mapping (SFM). The more aggressive secondary (SM) and tertiary mapping (TM) methods resulted in higher recall but lower precision. For clinical phenotype definition, FBM was the best (F=0.67), but was close to SM (F=0.62) and TM (F=0.60), judging on the F-scores alone. The overall difference between the four methods was statistically significant (one-way ANOVA, F=5.749, p=0.001). However, pairwise comparisons between FBM, SM, and TM did not reach statistical significance. A similar trend was found for the quality measure value sets.
Discussion:
The optimal method for using the GEMs depends on the relative importance of recall versus precision for a given use case. It appears that for clinically distinct and homogenous conditions, the recall of FBM is sufficient. The performance of all mapping methods was lower for heterogeneous conditions. Since code sets used for phenotype definition and quality measurement can be very similar, there is a possibility of cross-fertilization between the two activities.
Conclusion:
Different mapping approaches yield different collections of ICD-10-CM codes. All methods require some level of human validation.
Keywords: Cohort Identification, Electronic Health Record (EHR), Health Information Technology, Standardized Data Collection
Introduction
Large-scale, multisite observational research studies and pragmatic clinical trials utilize clinical data, including diagnosis data that are encoded with the International Classification of Diseases, 9th Revision, Clinical Modification (ICD-9-CM), collected by health systems as a byproduct of patient care. The national mandate for health systems to migrate to ICD-10-CM in October 2015 has an impact on all research activities that rely on these codes. Further, many current and ongoing investigations will need to manage and analyze data sets that define conditions of interest (i.e., clinical phenotypes) using both ICD-9-CM and ICD-10-CM codes.
The growing availability of electronic health record (EHR) data (encoded in ICD-10-CM) will increasingly be leveraged to support pragmatic clinical trials and quality improvement studies that learning health care comprises. Longitudinal studies in progress and retrospective studies will use ICD-9-CM-based population definitions and will need to understand how those relate to definitions based upon ICD-10-CM. A common challenge for researchers and health administrators moving forward, then, is the need to translate ICD-9-CM-based phenotype definitions, which can include hundreds of codes, into ICD-10-CM and to ensure that the populations retrieved with those codes are clinically equivalent. Although the Centers for Medicare & Medicaid Services (CMS) has produced General Equivalent Maps (GEMs), their use is not straightforward, and different methods for using the GEMs can result in different outcomes.
In the context of pragmatic clinical trials, we explore the use of publicly available mapping files to convert clinical phenotype definitions from ICD-9-CM to ICD-10-CM, compare the outcome of different approaches, and suggest preferred strategies for using the GEMs in automated translation. In addition to the phenotype definitions, we also make use of the value sets defined for electronic quality measurement as an additional way to evaluate the mapping methods. Quality measurement value sets are lists of codes from standard terminologies used to identify sub-populations of patients sharing certain demographic and clinical characteristics, as defined by a clinical quality measure. These value sets are very similar in their function to phenotype definitions. As part of the CMS Meaningful Use of EHR program, certified systems have to demonstrate the electronic submission of data for some selected clinical quality measures. Value sets are published to allow automatic computation of the numerator and denominator of a quality measure.
Pragmatic Clinical Trials and International Classification of Diseases (ICD) Codes
The tremendous costs associated with traditional clinical trials limit their use in addressing the majority of clinical questions and treatment decisions that are based upon insufficient evidence.1–4 Further, the limited generalizability of results inherent in clinical trials has stimulated interest in alternative research models, including observational research and pragmatic trials, to support patient-centered outcomes research.5,6 These alternative research models depend upon access to EHR data collected by health systems as part of the patient care process. The HMO Research Network (HMORN) and other networks have used electronic health care and claims data to advance our understanding of disease.7,8 While electronic claims data have been used in observational research for decades, the growing adoption of EHRs brings the potential to support more sophisticated research activities, such as cohort selection and randomization, to facilitate prospective and interventional research studies.9,10 The routine use of EHR data is a vital component of the envisaged learning health care system, and has become feasible with the widespread adoption and meaningful use of EHRs in health care systems.11
Pragmatic trials are those conducted in actual patient care settings and in cooperation with health care systems.6 The National Institutes of Health (NIH) Health Care Systems Research Collaboratory is funded by the NIH Common Fund to strengthen the national capacity for implementing cost-effective, large-scale research studies that engage health care delivery organizations as research partners, with the assumption that this will make research results more relevant to providers and, ultimately, patients.12 The Collaboratory includes a number of pragmatic trial demonstration projects that are multisite, often cluster-randomized, intervention studies.13 These demonstration projects have developed explicit and reproducible definitions (i.e., clinical phenotypes) using ICD-9-CM and other standardized code systems to identify patients with precise clinical attributes from various organizations and heterogeneous EHRs. These clinical phenotype definitions support a number of research activities, including cohort identification and describing the baseline characteristics (e.g., the proportion of patients with diabetes or hypertension) of different patient populations.
The phenotype definitions of the NIH Collaboratory projects currently include codes from ICD-9-CM, but investigators need to adapt them to ICD-10-CM since health care systems transitioned to it by October 1, 2015. The ICD-10-CM is not an incremental version change from ICD-9-CM. Rather, it is a radical transformation, involving major changes in not only the size of the terminology, but also in the organization, granularity, and semantics (or meaning) of terms.14 The more than 68,000 possible terms in ICD-10-CM more than quadruple the 14,000 terms in ICD-9-CM. Because the Collaboratory demonstration projects are all multiyear studies that span this national ICD-10-CM transition period, investigators need to address both ICD-9-CM and ICD-10-CM in their research data sets.
Automatic Code Translation by the General Equivalent Maps (GEMs)
To ease the burden of researchers who need to translate their cohort or clinical phenotype definitions from ICD-9-CM to ICD-10-CM, we explored the use of published maps between ICD-9-CM and ICD-10-CM for automatic conversion. The General Equivalent Maps (GEMs) are created and maintained by the Centers for Medicare & Medicaid Services (CMS) and the Centers for Disease Control and Prevention (CDC), and serve as a tool for the conversion of data between ICD-9-CM and ICD-10-CM.15 The GEMs are often also referred to as “crosswalks” since they provide important information linking codes from one system with codes in the other system.16 Users are cautioned against using the GEMs for actual coding as they have not been completely validated for clinical use. However, the conversion of data for quality measures and research is specifically listed among the applicable use cases.16 The GEMs are directional and therefore have two types: the forward maps convert ICD-9-CM codes into ICD-10-CM, and the backward maps convert ICD-10-CM codes into ICD-9-CM. Because the relationships between ICD-9-CM and ICD-10-CM codes are often complex and not one-to-one, the use of GEMs is complicated and requires informed consideration.17–20 While the impact of ICD-10-CM transition has been explored in various health care and research settings,21–24 there are few studies on the evaluation of automated translation of codes between ICD-9-CM and ICD-10-CM in the context of phenotype definitions for pragmatic trials.
The forward and backward GEMs are not simple mirror images of each other, as the names may suggest. They are independent maps that differ significantly in scope and coverage (Table 1). The majority of ICD-10-CM codes are not represented in the forward map, and a significant portion of ICD-9-CM codes (25 percent) are not represented in the backward map. The backward map provides 78,034 unique pairs of ICD-9-CM and ICD-10-CM codes (over three times more than the forward map), of which only 18,484 pairs (23.7 percent) are also found in the forward map.
Table 1.
Comparison of the Forward and Backward General Equivalent Maps (GEMs)
| FORWARD GEM | BACKWARD GEM | COMMON TO BOTH GEMS | |
|---|---|---|---|
| Unique ICD-9-CM codes* (% of all ICD-9-CM) | 13,409 (92.0%) | 10,949 (75.0%) | 10,880 (74.7%) |
| Unique ICD-10-CM codes* (% of all ICD-10-CM) | 16,614 (23.8%) | 69,154 (99.0%) | 16,614 (23.8%) |
| Unique ICD-9-CM/ICD-10-CM code pairs | 23,330 | 78,034 | 18,484 |
Note:
not including codes with no maps.
Users of the GEMs often find that they need to apply the forward and backward maps iteratively in order to obtain code maps (or links) that would otherwise be missed. According to Boyd et al.,25 36 percent of the ICD-9-CM codes are involved in so-called convoluted mappings, meaning that they are not simple one-to-one, one-to-many, or many-to-one maps to ICD-10-CM codes. In these complex cases, iterative application of the forward and backward maps will discover more and more links from an ICD-9-CM source code to ICD-10-CM targets (see Methods section). As an example, consider the ICD-9-CM code 648.82 Abnormal glucose tolerance of mother, delivered, with mention of postpartum complication. Using either the forward or the backward GEM alone, one will find the target ICD-10-CM code O99.815 Abnormal glucose complicating the puerperium. With the iterative use of the two GEMs, three additional relevant ICD-10-CM target codes can be found:
O24.430 Gestational diabetes mellitus in the puerperium, diet controlled
O24.434 Gestational diabetes mellitus in the puerperium, insulin controlled
O24.439 Gestational diabetes mellitus in the puerperium, unspecified control.
However, two problems arise when using the forward and backward GEMs iteratively. First, it may take many iterations to exhaust all mapping relationships because some of the convoluted mappings are open-ended. Second, some of the additional codes discovered in this way are not relevant. The aim of this study is to determine the optimal way to use the GEMs in the context of ICD-9-CM code translation in phenotypic definition.
Methods
Generation of the Target ICD-10-CM Codes
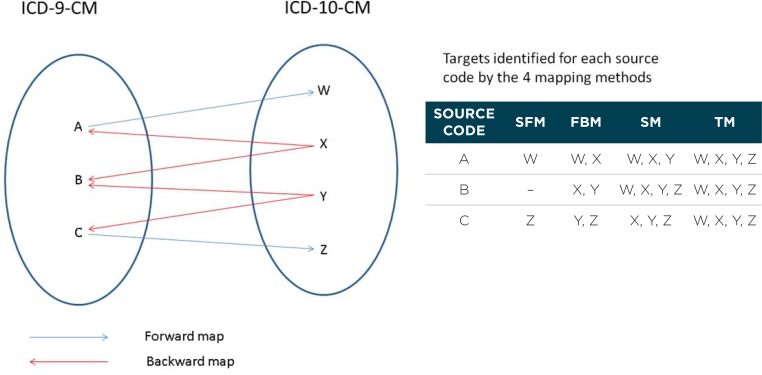
In this study, we compared four progressively more aggressive methods for using the GEMs (Figure 1). The goal of each method was to identify, for each ICD-9-CM code (the source code), one or more corresponding ICD-10-CM codes (the target codes). For all methods, we used a combination of the forward and backward GEMs to discover linkages between ICD-9-CM and ICD-10-CM codes. We treated the linkages in the forward and backward GEMs as the same, ignoring the stated directionality of the maps. In increasing order of aggressiveness, the methods are the following:
Simple forward map (SFM): uses only direct links from the forward GEM. All ICD-10-CM codes linked to an ICD-9-CM code in the forward GEM are used as targets.
Forward backward map (FBM): uses direct links from both the forward and backward GEMs. This includes all maps in SFM, plus additional map targets identified by the links between ICD-9-CM and ICD-10-CM codes in the backward GEM.
-
Secondary map (SM): uses all maps in FBM, plus additional target codes identified by secondary ICD-9-CM codes.
The following are the steps to generate SM:- Based on FBM, identify secondary ICD-9-CM codes, which are defined as ICD-9-CM codes that share the same target ICD-10-CM code as the primary ICD-9-CM source code. In Figure 1, consider the (primary) ICD-9-CM source code A. It has targets W and X in FBM; while another ICD-9-CM code B has targets X and Y in FBM. Since A and B share the same target X, B is identified as a secondary code of A.
- Add the targets of the secondary codes in FBM to the list of targets for the primary source code. In Figure 1, X and Y are added as targets for source code A.
-
Tertiary map (TM): uses all maps in SM, plus additional target codes identified by tertiary ICD-9-CM codes.
The following are the steps to generate TM:- Based on FBM, identify tertiary ICD-9-CM codes, which are defined as ICD-9-CM codes that share the same ICD-10-CM code as the secondary ICD-9-CM code (identified in the generation of SM). In Figure 1, B has been identified as a secondary code to primary source code A. In FBM, B has targets X and Y, while C has targets Y and Z. Since B and C share the same target Y, C is identified as a tertiary code of A.
- Add the targets of the tertiary codes in FBM to the list of targets for the primary source code. In Figure 1, Y and Z are added as targets for source code A.
Figure 1.
Four Mapping Methods to use the Forward and Backward General Equivalent Maps: Simple Forward Map (SFM), Forward Backward Map (FBM), Secondary Map (SM), Tertiary map (TM)
We chose these four methods for a number of reasons. The SFM and FBM are the most common ways to use the GEMs, and will provide a baseline measure of mapping performance. SM corresponds to the method used in the online transition tool provided by Boyd’s group.26 Given their experience and commentary, we hypothesized that additional iterations will increase the number of ICD-10-CM target codes and may enhance mapping performance. Therefore TM was included to assess whether additional iterations are indeed beneficial.
Evaluation of the Target ICD-10-CM Codes
To evaluate the performance of the four mapping methods, we used a convenience sample of 32 phenotypes (developed to identify research cohorts, characterize risk factors, or define outcomes) from three different pragmatic trials—Collaborative Care for Chronic Pain in Primary Care (PPACT), Strategies and Opportunities to Stop Colorectal Cancer in Priority Populations (STOP CRC), and A Pragmatic Trial of Population-Based Programs to Prevent Suicide Attempt—that were defined by ICD-9-CM codes. The ICD-9-CM codes were translated to ICD-10-CM codes using the four mapping methods based on the 2014 version of the GEMs. The lists of ICD-10-CM codes generated were reviewed by clinical experts. One generalist nurse practitioner (KP) and an MD domain expert for each trial (BG, AP, and MC) reviewed the phenotype name and the ICD-10-CM code sets generated by the maps to determine if each ICD-10-CM code semantically “fit” into the named phenotype condition, based on their understanding of that phenotype and its intent. For example, for the phenotype “active alcohol abuse” the reviewer was asked to look at the ICD-10-CM codes and determine (yes or no) if those codes were appropriate for inclusion in that heading. Reviewers were provided the original phenotype definition (i.e., the set of ICD-9-CM codes that constitute the specified condition) as a reference on the same review sheet.
To limit the scope and time for the evaluation, the reviewers were asked to review only the ICD-10-CM codes generated by the different mapping methods. They were not asked to search for additional ICD-10-CM codes that should have been included. To shorten the list of ICD-10-CM codes for review, we derived an algorithm to “roll up” codes to their parents, as long as the total number of codes in the list was reduced. For example, if the list contained “M47.10, M47.11, M47.12, M47.13, M47.15, M47.16,” which were all children of M47.1, we converted it into “M47.1 exclusion: M47.14” because M47.14 was the only child of M47.1 not included in the list. We did this iteratively until no further reduction in the number of codes was possible. To evaluate the roll up algorithm, one cohort definition from each demonstration project with at least 10 ICD-10-CM codes was manually reviewed to make sure that the final list of codes represented the meaning of the original codes.
To obtain the quality measurement value sets, we used the Value Set Authority Center (VSAC) launched by the United States National Library of Medicine (NLM) in 2012 to provide access to all official versions of value sets.27,28 In the VSAC, we identified all value sets for 2014 Clinical Quality Measures that were dually defined with both ICD-9-CM and ICD-10-CM code lists. We applied the four mapping methods to the ICD-9-CM code lists, and evaluated the resulting ICD-10-CM target codes against the ICD-10-CM codes listed for that measure, using the latter as the gold standard. Since the value sets differed considerably in their sizes, we also analyzed the effect of value set size on the mapping performance.
To evaluate the performance of each mapping method, we calculated the recall, precision and F-score of each method for every phenotype definition and quality measure value set. Note that for the phenotype definitions, we did not measure the true recall because the reviewers were not asked to look for missing ICD-10-CM codes. To give an estimate of recall for the phenotype definitions, we assumed that the most aggressive method (TM) contained all the correct ICD-10-CM codes. We used the F-score (the harmonic mean between recall and precision) as an overall indicator of performance of each mapping method. Based on the distribution of the F-scores, we used the ANOVA test to check the statistical significance of the difference between the four methods. We used the paired samples t-test for pairwise comparison. We used the IBM SPSS for Windows program for statistical computations.
Results
Phenotype Definitions
The selected pragmatic trials used 32 cohort definitions with 3–161 (median 4) ICD-9-CM codes per definition (Table 2). There were altogether 536 unique ICD-9-CM codes, all of which could be mapped by the four different methods. The size of the resulting ICD-10-CM code sets progressively increased as more aggressive mapping methods were used. Overall for SFM, the median size of the ICD-10-CM code sets was comparable to their ICD-9-CM counterparts. There was a sharp increase from SFM to FBM, and also from FBM to SM. The increase from SM to TM was more moderate. Altogether, there were over 7,000 ICD-10-CM targets codes that needed review. Our roll up algorithm reduced the review workload to around 2,000 codes. To ensure that the shortened list of codes represented the same meaning as the original codes, we reviewed three cohort definitions (pelvic and abdominal pain, alcohol abuse, and colon cancer) with 14, 40, and 80 leaf level ICD-10-CM codes respectively. Our roll up algorithm collapsed the lists to 5 codes (1 higher level code, 4 leaf codes), 11 codes (4 high level codes, 2 leaf codes, 5 exclusion codes) and 20 codes (11 high level codes, 9 leaf codes) respectively. By comparing the meaning of the original codes to the shortened lists, we confirmed that the shortened lists were semantically the same as the original lists.
Table 2.
Distribution of ICD-9-CM and Map-generated ICD-10-CM Codes in the Phenotype Definitions
| DEMONSTRATION PROJECT | # OF PHENOTYPE DEFINITIONS | # OF ICD-9-CM CODES/DEFINITION (MEDIAN) | # OF ICD-10-CM CODES/DEFINITION BY MAP METHOD (MEDIAN) | |||
|---|---|---|---|---|---|---|
| SFM | FBM | SM | TM | |||
| Chronic pain | 6 | 14–161 (42) | 18–140 (50) | 36–1,060 (148.5) | 80–1,138 (245) | 80–1,231 (410) |
| Suicide prevention | 23 | 4–41 (4) | 1–130 (2) | 2–323 (9) | 2–340 (48) | 2–372 (292) |
| Colorectal cancer | 3 | 3–14 (13) | 3–14 (11) | 3–77 (14) | 3–89 (14) | 3–115 (14) |
| Overall | 32 | 3–161 (4) | 1–140 (2) | 2–1,060 (14.5) | 2–1,138 (84.5) | 2–1,231 (193.5) |
The performance of the four different mapping methods is summarized in Table 3. The recall, precision, and F score values are the means for the code sets in a demonstration project. FBM was better than SFM in all three metrics (precision, recall, and F score). As expected, the more aggressive methods SM and TM resulted in higher recall at the expense of precision. Using the overall mean F score as a single indicator of performance, FBM was the best (F=0.67), but was close to SM (F=0.62) and TM (F=0.60). Based on the distribution of individual F scores in each method, the overall difference between the four methods was statistically significant (one-way ANOVA, F=5.749, p=0.001). Pairwise comparison between adjacent pairs of methods by paired samples t-test showed that the difference between SFM and FBM was statistically significant (t=−6.184, p<0.0001), while the differences for FBM versus SM and SM versus TM were not.
Table 3.
Performance of the Mapping Methods in Phenotype Definitions Showing Mean Recall, Precision, and F-score Values of the Code Sets Within a Particular Demonstration Project
| DEMONSTRATION PROJECT | SFM | FBM | SM | TM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RECALL* | PREC | F | RECALL* | PREC | F | RECALL* | PREC | F | RECALL* | PREC | F | |
| Chronic pain | 0.39 | 0.78 | 0.40 | 0.76 | 0.68 | 0.65 | 0.96 | 0.63 | 0.70 | 1.0 | 0.55 | 0.64 |
| Suicide prevention | 0.22 | 0.72 | 0.28 | 0.62 | 0.76 | 0.64 | 0.86 | 0.50 | 0.56 | 0.96** | 0.50 | 0.56 |
| Colorectal cancer | 0.71 | 0.89 | 0.68 | 1.0 | 0.89 | 0.93 | 1.0 | 0.84 | 0.91 | 1.0 | 0.78 | 0.87 |
| Overall | 0.30 | 0.75 | 0.34 | 0.68 | 0.76 | 0.67 | 0.89 | 0.56 | 0.62 | 0.97** | 0.53 | 0.60 |
Notes:
Recall was an estimation based on the assumption that all correct codes were included in the most aggressive mapping method TM.
Not 1 as expected because in one definition all ICD-10-CM codes were rated as incorrect so the recall was 0. “Prec” is “precision”; “F” is “F-score.”
Quality Measure Value Sets
A total of 202 quality measure value sets defined by both ICD-9-CM and ICD-10-CM code sets were retrieved from the VSAC. There were altogether 5,545 unique ICD-9-CM codes, of which 2 codes could not be mapped by our selected methods because they were not included in either the forward or backward GEM. The performance of the mapping methods in relation to the size of the ICD-9-CM code sets is summarized in Table 4. The recall, precision, and F-score values shown are the means for the value sets within a particular size range. Based on the overall F-score, the overall best performing mapping method was FBM, followed by SM, TM, and SFM. This trend was the same as the phenotype definition use case. Based on the distribution of F-scores for each value set, the difference in the performance of the four methods was statistically significant (one-way ANOVA, F=40.889, p<0.0005). Pairwise comparisons between adjacent methods (SFM versus FBM, FBM versus SM and SM versus TM) by paired samples t-test were all statistically significant (all with p<0.0001). The number of ICD-9-CM codes in the value sets varied considerably from 1 to 1,212 (mean 58.6, median 6). Smaller value sets generally had better recall, precision, and F-scores, regardless of mapping method. For FBM, value sets with 20 or fewer codes had almost perfect recall (0.97) and precision (0.93).
Table 4.
Performance of the Mapping Methods in the Quality Measure Value Sets Showing Mean Recall, Precision, and F-score Values of the Value Sets Within a Particular Size Range
| # ICD-9-CM CODES | # VALUE SETS | SFM | FBM | SM | TM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RECALL | PREC | F | RECALL | PREC | F | RECALL | PREC | F | RECALL | PREC | F | ||
| 1–5 | 99 | 0.60 | 0.96 | 0.67 | 0.97 | 0.94 | 0.95 | 0.97 | 0.87 | 0.89 | 0.98 | 0.86 | 0.88 |
| 6–20 | 56 | 0.65 | 0.96 | 0.73 | 0.98 | 0.93 | 0.95 | 0.98 | 0.86 | 0.91 | 0.98 | 0.85 | 0.89 |
| 21–100 | 31 | 0.62 | 0.94 | 0.70 | 0.90 | 0.88 | 0.87 | 0.94 | 0.78 | 0.82 | 0.94 | 0.72 | 0.77 |
| > 100 | 16 | 0.57 | 0.70 | 0.55 | 0.91 | 0.62 | 0.67 | 0.92 | 0.59 | 0.66 | 0.92 | 0.58 | 0.65 |
| Overall | 202 | 0.62 | 0.94 | 0.68 | 0.96 | 0.90 | 0.92 | 0.97 | 0.83 | 0.87 | 0.97 | 0.81 | 0.85 |
Note: “Prec” is “precision”; “F” is “F-score.”
Discussion
Use of Automatic Code Translation in Phenotype Definitions
After several false starts and delays, the transition from ICD-9-CM to ICD-10-CM finally happened in 2015. Health care providers have adopted the new coding system to ensure continued revenue; researchers and other secondary users of health care data must be prepared to adapt to this change. After October 1, 2015, phenotype definitions that use ICD-9-CM codes to identify cohorts of patients have to shift to ICD-10-CM codes if they are applied to newly-collected data. These ICD-9-CM-based phenotype definitions can include hundreds of codes. Translating them into ICD-10-CM entails significant effort, and automated methods to support these translations reduce this burden. The use of the GEMs is not straightforward because it includes two independent maps in both directions. Different methods for using the GEMs will result in different outcomes, and our findings can inform optimal approaches to using the maps for automated translation.
In this study, we compare four progressively aggressive methods of using the GEMs to translate ICD-9-CM codes used in phenotype definitions to ICD-10-CM codes: (1) simple forward map (SFM); (2) forward backward map (FBM); (3) secondary map (SM); and (4) tertiary map (TM). The papers and online tool from Boyd et al. seem to favor an approach similar to SM, but they did not explain why, nor did they compare the various mapping methods quantitatively.21,25,29 In our study, the different methods are compared quantitatively, and their strengths and weakness are highlighted. The poor results from the simple forward map should caution novice users of the GEMs, who may believe that using the forward map alone will be sufficient to translate ICD-9-CM codes to ICD-10-CM. Since the majority of ICD-10-CM codes (75 percent) are not reachable by the forward map, it is not surprising that the performance of SFM is the worst. The forward backward map combines the forward and backward GEMs regardless of their direction. The two GEMs together include 13,478 (93 percent) of ICD-9-CM codes and 69,154 (99 percent) of ICD-10-CM codes. This is an absolute limitation for any mapping method relying on the GEMs alone, which means that there is a small percentage of ICD-9-CM (7 percent) and ICD-10-CM (1 percent) codes that will not be covered.
Boyd et al. demonstrated that the majority of the ICD-9-CM to ICD-10-CM translations are complex, convoluted, and nonreciprocal.29 This is why one needs to apply the forward and backward maps iteratively to obtain more complete results. In our study, SM (the first iteration) identified several times more ICD-10-CM codes than did FBM. However, not all of the additional ICD-10-CM codes were relevant.
A common source of error related to composite concepts involving more than one medical condition. For example, starting from the ICD-9-CM code 716.80 Other specified arthropathy, site unspecified the FBM found E08.618 Diabetes mellitus due to underlying condition with other diabetic arthropathy, which was a correct target. However, in the SM, E08.618 led to the identification of the secondary ICD-9-CM code 249.80 Secondary diabetes mellitus with other specified manifestations, not stated as uncontrolled, or unspecified. This secondary ICD-9-CM code led to additional ICD-10-CM targets, such as E10.621 Type 1 diabetes mellitus with foot ulcer, which were completely unrelated to the primary ICD-9-CM source code. Such examples highlight the need for thoughtfulness and manual review of mappings generated by aggressive iterative mapping methods.
Based on the F-scores, the FBM was the best performing among all methods (the complete FBM list is available as Appendix A). However, the SM was a close second. For the clinical phenotype use case, SM had a better recall (0.89) over FBM (0.68), but precision dropped considerably (from 0.76 to 0.56). The median number of ICD-10-CM target codes increased six times from FBM to SM, and only one-third of the additional ICD-10-CM codes identified were correct. In practice, the optimal method will depend upon the specific use case, particularly whether higher recall is considered more important than precision or vice versa. In our limited sample of medical conditions, it appears that for clinically distinct and homogenous conditions, such as colorectal cancer, the recall of FBM is already very good, and there is no need to go to more aggressive methods. Conditions with heterogeneous pathology involving multiple organ systems, e.g., chronic pain, might require more aggressive mapping methods. In general, the performance of all mapping methods dropped considerably with diverse and heterogeneous conditions. In such cases, manual review of the map-generated codes and search for missing codes will be essential.
Users of the GEMs should be aware that only billable codes (leaf codes) are included in the GEMs. If their code lists include nonbillable (high level codes), such as some code lists used in quality metrics, they should expand the high level codes to the leaf codes before applying the maps to ensure a more complete translation. The performance of GEM-based mapping also depends on the extent of the changes between ICD-9-CM and ICD-10-CM. Some chapters such as Mental and Behavioral Disorders and Diseases of the Skin and Subcutaneous Tissues have undergone major reorganization. The definitions in the suicide prevention project mostly fall within mental health disorders. As a result, all mapping methods performed more poorly for this group. One example is amphetamine abuse. In ICD-9-CM, this condition has four codes depending on whether it is continuous, episodic, in remission, or unspecified. ICD-10-CM does not distinguish between the time course (which explains why the SFM maps have less ICD-10-CM codes than ICD-9-CM codes), but has added a new axis of classification about the effects of the abuse (e.g., sleep disorder, sexual dysfunction). In the chapters that have changed significantly, more intense scrutiny of GEM-based translations is warranted, and users may need to look outside the GEMs for codes that are not reachable through the GEMs.
Regardless of the mapping method, our results suggest that automatic translation is not perfect and validation by human review is recommended. However, it is likely that automated translation will save time by reducing the scope of review. The burden of manual review is a real concern, especially in codes sets with hundreds of codes. Very often, all descendants of a subbranch are included in a phenotype definition, so it saves significant time for reviewers if codes are rolled up to their parents. With our roll up algorithm, we managed to reduce the number of codes requiring expert review by 72 percent.
Synergism Between Quality Measurement and Cohort Definition
While clinical quality measurement and pragmatic clinical trials are distinct activities, they both rely on code sets to identify their relevant subpopulations of patients, and there is clear overlap in the function between the phenotype definition code sets and quality measurement value sets. For example, there is a phenotype code set for “colon cancer” in the NIH Collaboratory, and a quality measure value set for “malignant neoplasm of colon,” and both have exactly the same ICD-9-CM codes. Because of this, we have included the quality measure value sets as an additional evaluation of the mapping methods. For the quality measure value sets, the performance of the four mapping methods followed essentially the same trend as in phenotype code sets. Based on the overall F-scores, FBM performed best, followed by SM, TM, and SFM. However, there seemed to be a bigger difference between FBM and the others. The more aggressive methods (SM and TM) resulted in only marginal increase in recall with considerable drop in precision. Therefore, if there is a need to use the GEMs to translate ICD-9-CM code sets for clinical quality measurement, it would seem appropriate to use the FBM mapping method. Note that mapping performance is generally better with smaller value sets. One possible explanation is that smaller value sets may involve more distinct and homogenous conditions (e.g., malignant neoplasm of colon, 13 codes), and that larger value sets tend to be more heterogeneous (e.g., immunocompromised conditions, 149 codes). As we found in cohort definitions, homogenous conditions have better results in automatic mapping. Generally, for small value set (fewer than 20 codes) involving homogeneous conditions that are not in the chapters known to have undergone major reorganization in ICD-10-CM (e.g., mental disorders), the FBM mappings are expected to perform very well, and only minimal manual review will be required.
The existence of code sets used for phenotype definition and quality measurement raises the interesting possibility of “cross-fertilization.” It is conceivable that, in some cases, the same set of codes can serve both functions, as in the colon cancer example above. Indeed, the ICD-10-CM codes in the colon cancer value set are all considered appropriate for phenotype definition by the reviewers. So instead of defining new ICD-10-CM code sets from scratch, the researchers may be able to find quality value sets defined with ICD-10-CM codes that they can reuse. However, to do that one has to search through the thousands of value sets in VSAC. To narrow the search, one can use some similarity measure (e.g., Jaccard coefficient) between the ICD-9-CM phenotype code sets and ICD-9-CM value sets in VSAC.30 In the future, this kind of cross-fertilization between various secondary uses of clinical codes will become more important and perhaps encourage health care organizations to participate in pragmatic trials and nationally coordinated biomedical and health services research, such as HMORN and the Patient Centered Outcomes Research Network (PCORnet). The Phenotype Knowledge Base (PheKB)31 and other repositories of phenotypes should consider partnerships with VSAC and investigate formal linkages between research phenotypes and quality measurement value sets. Some of the phenotype definitions used in this study are posted on PheKB. The use of common value sets for clinical research and quality measurement can enable the generation of evidence from health care organizations and facilitate the vision of learning health care.32,33
Future Research
For future work, we can explore ways to improve the performance of the mapping methods. There is additional information in the GEMs, such as flags for approximate or exact maps, and indicators of combination codes, which can be exploited to refine the mapping algorithms. Another possible strategy is chapter-level refinement. Boyd et al. showed that the mapping relationships for codes from different ICD-9-CM chapters varied considerably.28 This is because the difference between ICD-9-CM and ICD-10-CM is not uniform across all medical specialties. Chapters that do not change radically may require a less aggressive mapping approach. Outside the use of the GEMs, two additional mapping resources may be worthy of consideration. First, the International Health Terminology Standards Development Organisation (IHTSDO) publishes a map from Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) to ICD-9-CM, and the NLM publishes a map from SNOMED CT to ICD-10-CM. Therefore, it is possible to map from ICD-9-CM to ICD-10-CM using SNOMED CT as an intermediary. Second, the Unified Medical Language System (UMLS) has been found to be useful in interterminology mapping.34,35 Mapping relations between ICD-9-CM and ICD-10-CM can be discovered by exploring the synonymy and other relationships within the UMLS. These relationships can then be used to corroborate or supplement the maps derived from the GEMs. In the future, researchers should consider using SNOMED CT codes to define the cohorts. SNOMED CT is a better clinical terminology than ICD because of its coverage, granularity, clinical orientation, and logical underpinning.36 Many quality value sets are already defined in SNOMED CT codes. Although it is true that ICD codes are more commonly found in EHRs at present, SNOMED CT codes will become more ubiquitous with the Meaningful Use initiative.
We note the following limitations in our study. The Collaboratory demonstration projects we used were a convenience sample and are not representative of all pragmatic trials. The phenotype definitions in this study were developed to support a number of purposes for very specific research studies and might not be generalizable or appropriate for other research or quality measurement use cases related to those conditions. Further, the phenotype definitions have not been vetted as national standards. Although we did use two reviewers for each mapping relationship, the reviews by clinical experts have not been independently corroborated.
In the future, other data types can leveraged for validation of phenotype definitions. For example, medication or laboratory values can be used to identify more records for potentially complete recall sets. A future related activity would be to examine a known subset of patients with chronic diseases before and after the ICD-10-CM transition, and to contrast the assigned ICD-10-CM codes with historical ICD-9-CM codes. Obviously, any practical migration of phenotyping algorithms from ICD-9-CM to ICD-10-CM will ultimately require the use and synthesis of other data types for validation and human review, including a more rigorously defined characterization of a gold standard diagnosis. Even with rigorous GEMs mapping approaches, real-world application would require some level of human review to consider phenotype definitions fully validated.
It is important to mention that although we focused our investigation on the translation of ICD-based phenotype definitions, most phenotyping methods include other data, such as laboratory test results, medications, demographics, and natural language processing, in addition to diagnostic code value sets. While most researchers recognize that ICD code sets by themselves are not sufficient for research phenotyping, these codes are widely used and remain an important component of virtually all of the phenotype definitions posted on PheKB. Given the national impact of implementation to ICD-10-CM in October 2015, specific scrutiny of public GEMs tools is warranted to clarify the research implications of the ICD-9-CM to ICD-10-CM transition.
Conclusion
The transition from ICD-9-CM to ICD-10-CM creates a heavy burden of code translation for clinical researchers using ICD codes in identifying patient cohorts based on clinical criteria. Although national reference mappings and tools exist to support ICD-9-CM to ICD-10-CM conversion, their use is not straightforward. Different approaches yield different sets of ICD-10-CM codes, and users should be aware of the pros and cons of each approach. In most cases, automatic code translation is not accurate enough on its own, and should be used as an auxiliary tool to assist human reviewers. Variation in the migration of phenotype definitions can have an impact on the consistency of definition of cohorts and data collection over time, and can potentially have an impact on study findings if not addressed.
Acknowledgments
This work was supported by the National Library of Medicine intramural research program and the National Institutes of Health (NIH) Common Fund, through a cooperative agreement (U54 AT007748) from the Office of Strategic Coordination within the Office of the NIH Director. The views presented here are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The authors wish to thank researchers from demonstration pragmatic trials of the NIH Collaboratory as well as members of the Phenotype, Data Standards, and Data Quality Core for their support of this work. We are grateful to the following projects for developing their phenotypes and sharing them for this study: Strategies and Opportunities to Stop Colorectal Cancer (STOP CRC, Gloria Coronado, PI; NIH UH3CA188640), Pragmatic Trial of Population-Based Programs to Prevent Suicide Attempt (Gregory Simon, PI; NIH 4UH3MH007755-02), and Collaborative Care for Chronic Pain in Primary Care (PPACT, Lynn DeBar, PI; NIH 4UH3NS088731-02). Thanks also to Steve Emrick of NLM for pulling data from VSAC.
Appendix A
References
- 1.Sung NS, Crowley WF, Jr, Genel M, Salber P, Sandy L, Sherwood LM, et al. Central challenges facing the national clinical research enterprise. JAMA. 2003;289(10):1278–87. doi: 10.1001/jama.289.10.1278. [DOI] [PubMed] [Google Scholar]
- 2.Tricoci P, Allen JM, Kramer JM, Califf RM, Smith SC., Jr Scientific evidence underlying the ACC/AHA clinical practice guidelines. Jama. 2009;301(8):831–41. doi: 10.1001/jama.2009.205. [DOI] [PubMed] [Google Scholar]
- 3.Eisenstein EL, Lemons PW, 2nd, Tardiff BE, Schulman KA, Jolly MK, Califf RM. Reducing the costs of phase III cardiovascular clinical trials. American heart journal. 2005;149(3):482–8. doi: 10.1016/j.ahj.2004.04.049. [DOI] [PubMed] [Google Scholar]
- 4.Eisenstein EL, Collins R, Cracknell BS, Podesta O, Reid ED, Sandercock P, et al. Sensible approaches for reducing clinical trial costs. Clinical trials (London, England) 2008;5(1):75–84. doi: 10.1177/1740774507087551. [DOI] [PubMed] [Google Scholar]
- 5.Glasgow RE, Chambers D. Developing robust, sustainable, implementation systems using rigorous, rapid and relevant science. Clin Transl Sci. 2012;5(1):48–55. doi: 10.1111/j.1752-8062.2011.00383.x. Epub 2012/03/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patsopoulos NA. A pragmatic view on pragmatic trials. Dialogues Clin Neurosci. 2011;13(2):217–24. doi: 10.31887/DCNS.2011.13.2/npatsopoulos. Epub 2011/08/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platt R, Davis R, Finkelstein J, Go AS, Gurwitz JH, Roblin D, et al. Multicenter epidemiologic and health services research on therapeutics in the HMO Research Network Center for Education and Research on Therapeutics. Pharmacoepidemiology and drug safety. 2001;10(5):373–7. doi: 10.1002/pds.607. [DOI] [PubMed] [Google Scholar]
- 8.Moulton G. HMO research network to focus on cancer prevention and control. Journal of the National Cancer Institute. 1999;91(16):1363. doi: 10.1093/jnci/91.16.1363. [DOI] [PubMed] [Google Scholar]
- 9.McGlynn EA, Lieu TA, Durham ML, Bauck A, Laws R, Go AS, et al. Developing a data infrastructure for a learning health system: the PORTAL network. Journal of the American Medical Informatics Association : JAMIA. 2014;21(4):596–601. doi: 10.1136/amiajnl-2014-002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richesson RL, Horvath MM, Rusincovitch SA. Clinical research informatics and electronic health record data. Yearbook of medical informatics. 2014;9(1):215–23. doi: 10.15265/IY-2014-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen L, Aisner D, McGinnis JM, editors. The Learning Healthcare System: Workshop Summary (IOM Roundtable on Evidence-Based Medicine) The National Academies Press; 2007. [PubMed] [Google Scholar]
- 12.Richesson RL, Hammond WE, Nahm M, Wixted D, Simon GE, Robinson JG, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J Am Med Inform Assoc. 2013;20(e2):e226–31. doi: 10.1136/amiajnl-2013-001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collaboratory NHSR Demonstration Projects. 2015. [cited 2015 March 9]; Available from: https://www.nihcollaboratory.org/demonstration-projects/Pages/default.aspx.
- 14.Steindel SJ. International classification of diseases, 10th edition, clinical modification and procedure coding system: descriptive overview of the next generation HIPAA code sets. Journal of the American Medical Informatics Association : JAMIA. 2010;17(3):274–82. doi: 10.1136/jamia.2009.001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CMS . ICD-10. Centers for Medicare and Medicaid Services; 2015. [cited 2015 March 9]; Available from: http://www.cms.gov/Medicare/Coding/ICD10/. [PubMed] [Google Scholar]
- 16.CMS . General Equivalence Mappings Frequently Asked Questions. Centers for Medicare & Medicaid Services; 2014. [Google Scholar]
- 17.Jones LM, Nachimson S. Use Caution When Entering the Crosswalk: A Warning About Relying on GEMs as Your ICD-10 Solution. 2014 [Google Scholar]
- 18.Wollman J. ICD -10: A master data challenge. Health Manag Technol. 2011;32(7):16, 20–1. [PubMed] [Google Scholar]
- 19.Bhuttar VK. Crosswalk options for legacy systems. Implementing near-term tactical solutions for ICD-10. J Ahima. 2011;82(6):34–7. [PubMed] [Google Scholar]
- 20.Butler R, Bonazelli J. Converting MS-DRGs to ICD-10-CM/PCS. Methods used, lessons learned. J Ahima. 2009;80(11):40–3. [PubMed] [Google Scholar]
- 21.Boyd AD, Yang YM, Li J, Kenost C, Burton MD, Becker B, et al. Challenges and remediation for Patient Safety Indicators in the transition to ICD-10-CM. Journal of the American Medical Informatics Association : JAMIA. 2015;22(1):19–28. doi: 10.1136/amiajnl-2013-002491. Epub 2014/09/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caskey R, Zaman J, Nam H, Chae SR, Williams L, Mathew G, et al. The transition to ICD-10-CM: challenges for pediatric practice. Pediatrics. 2014;134(1):31–6. doi: 10.1542/peds.2013-4147. Epub 2014/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venepalli NK, Qamruzzaman Y, Li JJ, Lussier YA, Boyd AD. Identifying clinically disruptive International Classification of Diseases 10th Revision Clinical Modification conversions to mitigate financial costs using an online tool. Journal of oncology practice / American Society of Clinical Oncology. 2014;10(2):97–103. doi: 10.1200/JOP.2013.001156. Epub 2014/02/13. [DOI] [PubMed] [Google Scholar]
- 24.Venepalli NK, Shergill A, Dorestani P, Boyd AD. Conducting Retrospective Ontological Clinical Trials in ICD-9-CM in the Age of ICD-10-CM. Cancer informatics. 2014;13(Suppl 3):81–8. doi: 10.4137/CIN.S14032. Epub 2014/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyd AD, Li JJ, Burton MD, Jonen M, Gardeux V, Achour I, et al. The discriminatory cost of ICD-10-CM transition between clinical specialties: metrics, case study, and mitigating tools. Journal of the American Medical Informatics Association : JAMIA. 2013;20(4):708–17. doi: 10.1136/amiajnl-2012-001358. Epub 2013/05/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lussier Research Group: The University of Arizona On-line ICD-10-CM Conversion Tool. Available from: http://www.lussierlab.org/transition-to-ICD9CM/.
- 27.Bodenreider O, Nguyen D, Chiang P, Chuang P, Madden M, Winnenburg R, et al. The NLM value set authority center. Studies in health technology and informatics. 2013;192:1224. [PMC free article] [PubMed] [Google Scholar]
- 28.NLM . LM Value Set Authority Center (VSAC) Bethesda, MD: National Library of Medicine; 2015. [updated Feb 11, 2015; cited 2015 March 11]; Available from: https://vsac.nlm.nih.gov/. [Google Scholar]
- 29.Boyd AD, John’ Li J, Kenost C, Joese B, Min Yang Y, Kalagidis OA, et al. Metrics and tools for consistent cohort discovery and financial analyses post-transition to ICD-10-CM. Journal of the American Medical Informatics Association : JAMIA. 2015 doi: 10.1093/jamia/ocu003. Epub 2015/02/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winnenburg R, Bodenreider O. Metrics for assessing the quality of value sets in clinical quality measures. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2013;2013:1497–505. Epub 2014/02/20. [PMC free article] [PubMed] [Google Scholar]
- 31.University V PheKB 2012. [cited 2013 May 24]; Available from: http://www.phekb.org/.
- 32.Richesson RL, Hammond WE, Nahm M, Wixted D, Simon GE, Robinson JG, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. Journal of the American Medical Informatics Association : JAMIA. 2013;20(e2):e226–31. doi: 10.1136/amiajnl-2013-001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Office of the National Coordinator for Health Information Technology Health IT Enabled Quality Improvement: A Vision to Achieve Better Health and Health Care. Available from: https://www.healthit.gov/sites/default/files/HITEnabledQualityImprovement-111214.pdf.
- 34.Fung KW, Bodenreider O. Utilizing the UMLS for semantic mapping between terminologies. AMIA Annu Symp Proc. 2005:266–70. [PMC free article] [PubMed] [Google Scholar]
- 35.Fung KW, Bodenreider O, Aronson AR, Hole WT, Srinivasan S. Combining lexical and semantic methods of inter-terminology mapping using the UMLS. Studies in health technology and informatics. 2007;129(Pt 1):605–9. Epub 2007/10/04. [PMC free article] [PubMed] [Google Scholar]
- 36.Fung KW, Xu J. An exploration of the properties of the CORE problem list subset and how it facilitates the implementation of SNOMED CT. Journal of the American Medical Informatics Association. 2015. [DOI] [PMC free article] [PubMed]