Abstract
The integration of three-dimensional micropatterning with microfluidics provides a unique opportunity to create perfusable tissue constructs in vitro. Herein, we have used this approach to create a tumor-on-chip with endothelial barrier. Specifically, we photopatterned a mixture of endothelial cells and cancer spheroids within a gelatin methacrylate (GelMA) hydrogel inside of a microfluidic device. The differential motility of endothelial and cancer cells in response to a controlled morphogen gradient across the cell-laden network drove the migration of endothelial cells to the periphery while maintaining the cancer cells within the interior of the hydrogel. The resultant endothelial cell layer forming cell-cell contact via VE-Cadherin junctions was found to encompass the entire the GelMA hydrogel structure. Furthermore, we have also examined the potential of such tumor-on-a-chip system as a drug screening platform using Doxorubicin, a model cancer drug.
Graphical Abstract
Introduction
According to the American Cancer Society, cancer is the second leading cause of death in the United States where one in four deaths in 2015 were cancer-related1. Despite such grim outlooks, survival rate amongst cancer patients has steadily increased from 49% to 68% over the past decade due to the increase in our fundamental understanding of this disease and technological advances1. Some of these technological advances include the development of three-dimensional (3D) in vitro models recapitulating various complexities of the disease. For instance, multicellular tumor spheroids, consisting of cancer cells or multiple cell types, have been developed to study cancer progression and drug efficacy2–7. Other efforts in the area include encapsulation of tumor spheroids within biomaterials to mimic the extracellular matrix environment8–11. 3D in vitro systems incorporating stromal and vascular cells have also been developed12–14. In a recent study, George and colleagues have created vascularized tumor spheroids where the endothelial cells incorporated within the spheroids formed vascular networks in presence of stromal fibroblasts15.
The integration of 3D cell cultures with microfluidics technology can be used to create tumor models with perfusion to facilitate mass transport16, 17. Such microfluidic devices have been used to study various aspects of cancer progression such as tumor growth, presence of high interstitial fluid pressure, and cancer cell extravasation18–24. Such technological platforms have also been used to understand drug-tumor interactions such as drug specificity, penetration into cancer spheroids, and efficacy towards repressing cancer growth20, 24–27. Many of these platforms employ multi-layered or multi-channel devices to create a perfused tumor-a-on-chip system18, 21, 25, 26. Generation of such integrative fluidic system often requires multiple steps and complex fabrication processes.
In this study, we describe a novel yet simple approach to create a tumor-on-a-chip (TOC) device that contains tumor spheroids within an artificial extracellular matrix surrounded by a single-celled endothelial barrier that is assembled through vascular endothelial (VE)-cadherin junctions. Specifically, cancer spheroids along with human umbilical vein endothelial cells (HUVECs) were confined within gelatin methacrylate (GelMA) hydrogel structures through 3D photopatterning and integrated into a microfluidic device28. The differential motility of cancer spheroids and endothelial cells in response to chemotactic gradients generated within the 3D environment was harnessed to drive the migration of endothelial cells to the periphery to form a barrier surrounding the cell-laden GelMA structures. We further validated the potential of this tumor-on-a-chip device as a drug-screening platform by using Doxorubicin, a commonly used anti-cancer drug, as a model compound.
Materials and Methods
Cell culture
MCF7 and HUVECs were obtained from ATCC. MCF7 cells were cultured in growth media (GM) comprised of Dulbecco Modified Eagle’s high glucose media (Hyclone), 10% fetal bovine serum (FBS, Gibco), and 1% Penicillin/Streptomycin (Gibco). HUVECs were cultured in HUVEC Media (HM) containing 79% M199 media (Gibco), 10% FBS (Gibco), 10% endothelial cell growth media (Cell Application, Inc.), and 1% Penicillin/Streptomycin. HUVECs used in this study were limited to cells between passage 6 and 8.
MCF7 Spheroid Formation
MCF7 cells were cultured to about 80% confluency prior to trypsinization. To create spheroids, 1 million MCF7 cells in 4 mL GM were plated in a 60 mm diameter petri dish and cultured on an orbital shaker (VWR, Model No. DS-500E) at 45 rpm in a humidified incubator maintained at 37 °C and 5% CO2. The cultures were maintained for ~20 hours to form spheroids. The average diameter of the spheroids were found to be ~200 μm.
Fabrication of Silicon Mold
Micropatterned silicon molds were fabricated according to the protocol as described previously29. In brief, microfluidic channels were photolithographically defined using NR9-1500PY negative photoresist (Futurrex, Frankling, NJ, USA) on a 4-inch diameter Si wafer. The Si wafer with the photoresist defined was then etched using the deep reactive ion etching (DRIE) process. In the DRIE process, SF6 gas was flowed at 100 sccm throughout the 11 seconds of reaction time, followed by a passivation cycle when C4F8 gas was flowed at 80 sccm for 7 s. A 75 μm of etching depth was achieved under the etching rate of about 0.7 μm per cycle. After the DRIE process, the NR9-1500PY photoresist was removed by immersing in acetone for 4 hours before rinsing with methanol, isopropanol, and deionized water. The Si mold was then dried under compressed nitrogen gas and silanized by vapor deposition of trichlorosilane (TCI Inc, Portland, OR, USA) to facilitate PDMS molding and removal.
Synthesis of lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) as a photoinitiator
First, 2,4,6-trimethylbenzoyl chloride was added drop wise to an equal molar quantity of dimethyl phenylphosphonite under argon while stirring at room temperature30. This mixture was allowed to react for 18 hours. Next, the temperature of the reaction mixture was increased to 50 °C while 4 molar excess of lithium bromide mixed with 2-butanone was added to the reaction mixture. resulting in precipitation to form within 10 minutes. After precipitation, the temperature was cooled to room temperature and allowed to rest for 4 hours. Next, to ensure complete removal of excess lithium bromide, the precipitate was collected by filtration and washed three times using 2-butanone. Finally, the product was dried using a vacuum to remove excess 2-butanone, yielding LAP.
Synthesis of Gelatin Methacrylate (GelMA)
Gelatin was methacrylated in accordance with the protocol described elsewhere31, 32. In brief, 10g of bovine skin gelatin (Sigma Aldrich, St. Louis, MO, USA) was dissolved in 100 mL of PBS and stirred at 60 °C for roughly 1 hour to achieve complete solvation. Next the solution was lowered to 50°C, after which, 8mL of methacrylic anhydride (cat no.: 276685; Sigma Aldrich) was added to the solution drop wise with vigorous stirring. The solution was kept at 50°C with vigorous stirring for an hour after the addition was complete, after which, it is quenched with 2x the volume of PBS (200 mL). The solution was then dialyzed against milliQ water using 12–14 kDa cutoff dialysis tubing (Spectrum Laboratories, Rancho Dominguez, CA, USA) for one week (3 times per day water change) at 40 °C to remove trace contaminants. Next, the GelMA solution was frozen in liquid nitrogen and lyophilized in a freeze dryer for 4 days before being stored at −20 °C until usage.
Fabrication of Tumor-on-a-Chip Device
The tumor-on-a-chip device was fabricated with slight modifications of a device described previously29. The device includes a trilayer hydrogel system where cell-laden GelMA hydrogels were sandwiched between two polyacrylamide (PAm) hydrogels. The fabrication of the device involves the following steps:
Methacrylation of glass surfaces
To achieve the chemical tethering of PAm hydrogels, glass coverslips were methacrylated as described elsewhere28, 29. Briefly, glass coverslips were cleaned with 1.5 M NaOH for 30 minutes followed by rinsing with DI water and drying with air. The cleaned coverslips were treated with 2% (v/v) 3-(Trimethoxysilyl)propyl methacrylate solution diluted in 0.54% glacial acetic acid and 99.46% ethanol for 5 minutes at room temperature to immobilize methacrylate groups onto the surfaces. Care was taken to aliquot sufficient volume of the reacting solution onto the coverslips to eliminate artifacts associated with their evaporation. The surface modified coverslips were washed with pure ethanol for 10 minutes under gentle stirring to remove excess reactants. The above step was repeated twice, rinsed with DI water, and dried at 50 °C for 30 minutes. The coverslips were used immediately.
Trilayer hydrogel formation
Methacrylated coverslips of 22x60 mm rectangular and 12 mm diameter were used. 3 μL of a polyacrylamide hydrogel precursor solution comprised of 5% (w/v) acrylamide (Am), 0.2% (w/v) Bis-Acrylamide (BisAm), 0.1% (w/v) Ammonium Persulfate (APS), and 0.01% (w/v) N,N,N′,N′-Tetramethylethylenediamine (Sigma-Aldrich) in PBS, was placed in the center of the methacrylated 22x60 mm rectangular coverslip and the droplet was covered with a non-methacrylated 15 mm diameter coverslip. This would result in the bottom layer of the device (Supp. Fig. 1A, B). This process was repeated with a methacrylated 15mm circular coverslip and non-methacrylated square coverslip to fabricate the top layer (Supp. Fig. 1A–B). The precursor solution was left to polymerize for 20 minutes prior to gently removing the non-methacrylated coverslips. The resulting structures containing PAm hydrogels tethered to the circular and rectangular coverslips were allowed to equilibrate in PBS overnight at room temperature to remove trace amounts of unreacted monomers.
Around 5 μL of DI water was placed onto a circular region of the fabricated silicon mold before covering the droplet with PAm hydrogel tethered-15 mm diameter circular coverslip (Supp. Fig. 1C). Polydimethylsiloxane (PDMS, Sylgard 184) base solution was mixed with its curing agent at a weight ratio of 10:1 and degassed to remove air bubbles if any (Supp. Fig. 1D). This mixture was gently poured onto the silicon wafer containing the PAm hydrogel and baked at 60 °C for 2 hours (Supp. Fig. 1E). The PDMS mold containing the hydrogel was detached from the silicon wafer and bonded to the rectangular coverslips containing a PAm hydrogel using UV-Ozone treatment (Supp. Fig. 1F). Care was taken to prevent direct exposure of the PAm hydrogel to deep UV.
The PDMS mold and the glass coverslips were immediately attached to each other while maintaining the alignment between the hydrogels on their respective surfaces. This ensures that the top and bottom of the microfluidics chamber are comprised of PAm hydrogels. The fabrication process was completed by bonding the PDMS mold and glass coverslips at 60 °C. The device was equilibrated in PBS and UV sterilized for 45 minutes prior to using it for cell culture.
Preparation of GelMA solution
Gelatin methacrylate was dissolved in PBS to achieve a 10% wt/v precursor solution. To ensure complete dissolution, the GelMA dispersed PBS was incubated at 60 °C in a water bath for 20 minutes. The GelMA solution was syringe filtered (pore size of 0.22 μm) to remove any insoluble residues and maintained at 37 °C until use.
Encapsulation of cells within GelMA structures in microfluidic device
HUVECs and MCF7 spheroids were encapsulated within GelMA hydrogels. The MCF spheroids were passed through a cell strainer having 100 μm pore size (Corning) to eliminate single cells and small spheroids. Around 50 spheroids with a diameter of ~ 200 μm were dispersed in 5 mL of PBS containing 2 million HUVEC cells. The mixture was centrifuged for 3 minutes at 800 rpm. The supernatant was aspirated and 100 μL of 10% GelMA solution was added to the cell pellet. The cells were resuspended gently using a pipette before the addition of 0.01% ascorbic acid (antioxidant) and 2 μM LAP (photoinitiator) to the solution. This solution was again mixed gently, drawn into a syringe, and injected into the microfluidic device (Supp. Fig. 1G).
This device was placed onto a transparency film photomask containing an ellipse pattern and mounted onto a microscope stage (Supp. Fig. 1H–I). Using the stage controller of the microscope, the position of the fluidic device was moved to locate individual MCF7 spheroids surrounded by HUVECs under brightfield illumination. Each location was exposed to UV light with an excitation and emission wavelengths of approximately 358 and 463 nm, respectively, for 18 seconds. Several locations were photopolymerized before flushing the device with PBS containing 4% Penicillin/Streptomycin. Mixed Media (MM) containing 50% GM and 50% HM was subsequently injected into the device and the GelMA structures containing MCF7 spheroids and HUVECs were cultured at 37 °C and 10% CO2.
Quantification of cell motility within the cell-laden GelMA hydrogel structures
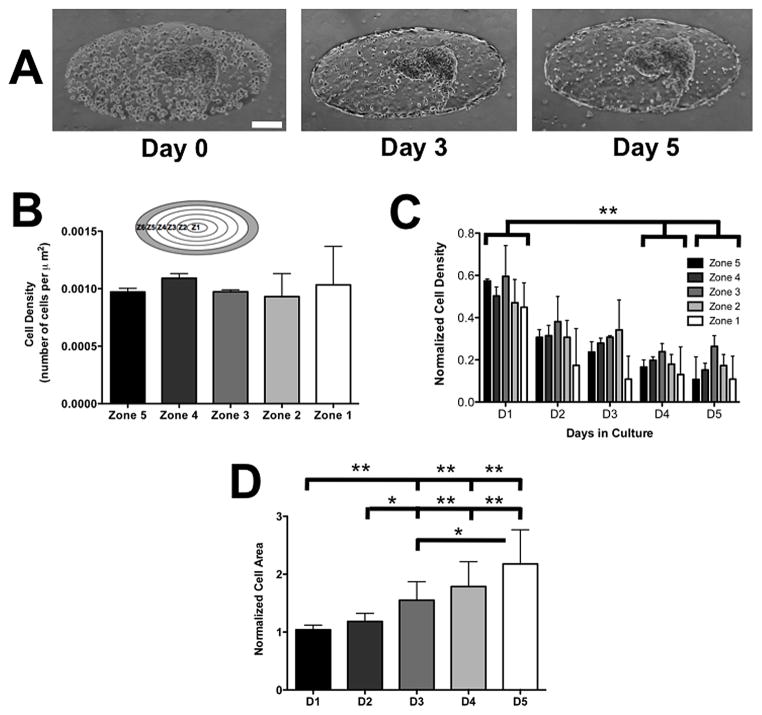
The motility of HUVEC cells and MCF7 spheroids was determined by examining the changes in their local cell density within the GelMA hydrogel as a function of culture time. Brightfield images of the cell-laden GelMA hydrogels were taken for up to 5 days. A custom Matlab software was used to process the brightfield images of the cell-laden GelMA hydrogel by identifying the boundary of the hydrogel structure and partitioning it into smaller zones of Z1 to Z6 as indicated in Figure 4B inset. In order to partition the structure into smaller zones, a Sobel filter was applied to the bright field image of the cell-laden GelMA hydrogel to identify its boundaries. High-pass filter was applied to remove random non-zero values outside the GelMA hydrogel structure. A distance transform was applied to the filtered image resulting in small and large values within the interior and exterior of the ellipse GelMA hydrogel, respectively. The ellipse structure of the cell-laden GelMA hydrogel was identified by applying a low-pass filter onto the distance transform image and converting the filtered results into a binary image. Finally, a built-in Matlab function, Regionprops, was used to identify the centroid, major, and minor axes lengths of the ellipse hydrogel structures. We further created smaller ellipses by reducing the major and minor axes of the outer ellipse structure to create multiple zones as in Fig. 4C.
Figure 4. Co-culture of HUVECs and MCF7 spheroids within GelMA structures.
(A) Brightfield images of HUVECs co-cultured with MCF7 spheroids at immediately after encapsulation (D0), Day 3 (D3), and Day 5 (D5). Scale bar: 200 μm. (B) Cell density within Zones 1 through 5 on D0 within the GelMA hydrogel. Different zones are indicated within the inset. The shaded peripheral region in the diagram, Zone 6, is excluded from the quantification. (C) Changes in the normalized zonal density of HUVECs with culture time. Within each zone, the cell density monitored as a function of culture time was normalized to D0 density and was plotted in the bar graph. (D) Spheroid size, quantified by 2-D area and normalized to D0 size, as a function of culture time. * and ** indicate statistically significant differences of p < 0.05 and 0.01, respectively, as obtained from pair wise t-test. In (C), differences were reported only if the changes in normalized cell density were statistically significant in all zones (Zone 1–5) between different culture days.
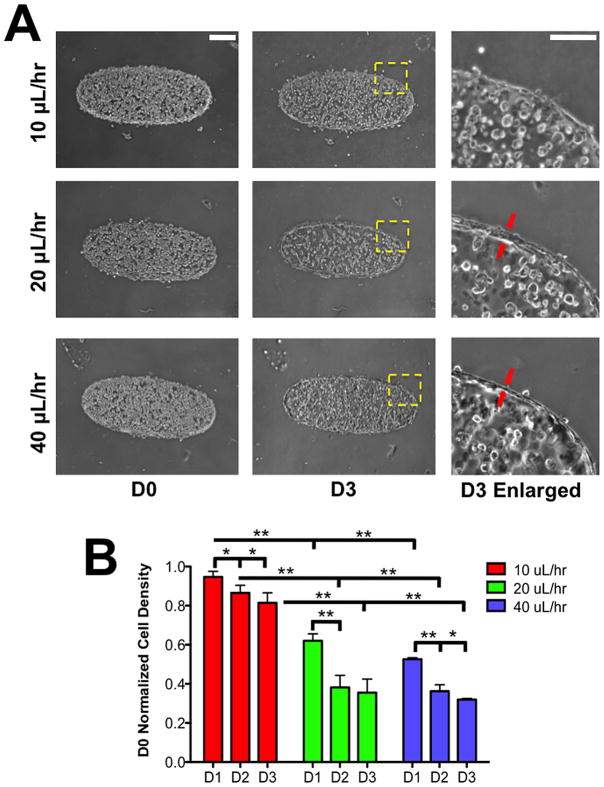
The local cell density was obtained by counting the number of cells divided by the area of each zone. Zone 6 was excluded from all analysis since the number of cells at the perimeter of the ellipse cannot be accurately counted. For quantifying the flow-rate dependent migration of cells in Figure 3B, zones 1 through 5 were merged to form a single area.
Figure 3. Flow-induced migration of encapsulated HUVECs.
(A) Brightfield images of HUVECs within GelMA hydrogels as a function of culture time. Each row represents different flow rates while the 1st and 2nd column represent different culture days —day 0 (D0) indicates the day of encapsulation. Scale bar: 200 μm. The 3rd column represents a magnified image of the region identified by a square window with yellow dashed lines in each row of day 3 (D3) images. The red arrows indicate the HUVECs at the periphery of GelMA structure. Scale bar: 30 μm. (B) The change in local density of HUVECs at different culture times for flow rates of 10, 20, and 40 μL/hr. The y-axis represents the local cell density quantified at specified culture day normalized to D0 cell density. * and ** indicate statistically significant differences of p < 0.05 and 0.01, respectively, as obtained from pair wise t-test.
Quantification of MCF7 spheroids growth within GelMA hydrogel structures
Brightfield images of the GelMA hydrogels containing the spheroids and HUVECs were recorded as a function of culture time (1–5 days) to examine their growth post-encapsulation. The area of the spheroid was quantified by tracing the boundary of the spheroid using the free-hand selection tool on ImageJ.
FITC-Dextran diffusion into cell-laden GelMA hydrogel
GelMA hydrogels containing MCF7 spheroids and endothelial cell layer at the periphery was perfused with PBS containing either 10 μg/mL 10 kDa FITC-conjugated dextran (Sigma-Aldrich) or 100 μg/mL 150 kDa FITC-conjugated dextran. The concentration difference was used to account for the differences in molecular weight. The diffusion of FITC-conjugated dextran molecules into the GelMA hydrogels was monitored by recording the epi-fluoresence images at a time intervals of ~2 min for 30 minutes. Prior to image analysis, the fluorescence intensity within the hydrogel was normalized to the mean intensity outside of the hydrogel. This normalized intensity was used for all analyses. To quantify the diffusion process, the intensity of the normalized fluorescent signal at the central region of the ellipse was used for both the time plots and steady state analysis. The central region consist of a small “zone 1” ellipse as shown in Figure 3B. The region within this small ellipse that overlaps with the cancer spheroid was excluded from the analysis.
Immunofluorescence staining of HUVECs
Cells within the device were fixed in 10% Paraformaldehyde (PFA) solution for 10 minutes at room temperature followed by infusing the device with PBS to remove the excess PFA. Blocking buffer, comprised of 0.1% Triton-X100 and 3% Bovine Serum Albumin (BSA), was added and the cells were incubated in this solution for 30 minutes at room temperature. The device was washed with PBS after each step to remove the residual solutions. The fixed cells were treated overnight with rabbit polyclonal VE-Cadherin antibody (Cat. No. D87F2, Cell Signaling) diluted in blocking buffer at 4 °C. The primary antibody solution was removed by washing with PBS. The device was incubated in blocking buffer containing Alexafluor 488 goat anti-rabbit secondary antibody (Cat. No. A-11008, ThermoFisher Scientific) and Rhodamine-conjugated Phalloidin (Cat. No. R415, ThermoFisher Scientific) for 70 minutes at room temperature. The cells were subsequently washed with PBS and stained with 20 μg/mL DAPI solution for 20 minutes to visualize the nuclei. The device was rinsed several times with PBS and imaged using a confocal microscope.
Doxorubicin solution
Doxorubicin (Cat. No. D1515, Sigma-Aldrich) was weighed and dissolved in DMSO to achieve a concentration of 100 mg/mL. This solutions was distributed into small aliquots and stored in −20 °C. The stock solution was thawed and diluted to 100 μg/mL in prewarmed MM (~ 37 °C) and sterilized using a DMSO-resistant syringe filter (Pall Corporations). This 100 μg/mL solution was further diluted in MM to acquire doxorubicin concentrations of 1 and 10 μg/mL used in the study.
Penetration of Doxorubicin into spheroid-laden GelMA structures
The cell-laden GelMA hydrogels were exposed to MM containing 1, 10, and 100 μg/mL Doxorubicin. The presence of Doxorubicin within the spheroids was detected using a Zeiss Observer A1 Microscope with a 10x A-Plan lens after 3 days of incubation on the red fluorescent channel. An exposure time of 700 ms was used for all samples.
Confocal microscopy for imaging immunofluorescently stained cells
Laser scanning confocal microscope was used to obtain Z-stack images of cells stained for nuclei, F-Actin, and VE-Cadherin as well as for the PAm hydrogels embedded with fluorescent beads. A vertical step size of 1 μm was used to acquire the Z-stack images.
Effect of flow rates on mechanical compression of the GelMA hydrogels
To determine whether the GelMA structures are differentially compressed at different flow rates, acellular structures containing 2% (v/v) fluorescent beads of 200 nm diameter (Cat. No. F8782, ThermoFisher Scientific) were used. The fluorescent bead laden-GelMA hydrogels were perfused with MM and allowed to equilibrate for 6 hours at 37 °C. The equilibrated structures were subjected to different flow rates of 10, 20, 40, and 1000 μL/hr. The X-Y images of the hydrogels subjected to different flow rates at 37 °C were acquired by using a spinning disk confocal microscope. The reference state was generated by recording the X-Y section of the ellipse hydrogel structure at a z-position that bisects the top and bottom of the chamber in the absence of any flow. The samples were then exposed to different flow rates and the X-Y images were recorded at the same z-position that was used for the reference state. The 2-D displacement fields, u and v, were obtained by comparing the reference image to the images recorded under different flow rates using Particle Image Velocimetry (PIV). The area strain, Astrain, was calculated by using the following equation:
| (Eq. 7) |
Modeling of mass transfer within the cell-laden GelMA hydrogel structures
COMSOL Version 4.2 was used to solve the 2-D diffusion-reaction equation (Eq. 1, 6) with a convective boundary condition (Eq. 3). The domain of the system was comprised of an ellipse structure with major and minor axes lengths of 1.2 and 0.45 mm, respectively.
Results
Formation and characterization of trilayer hydrogel-based device
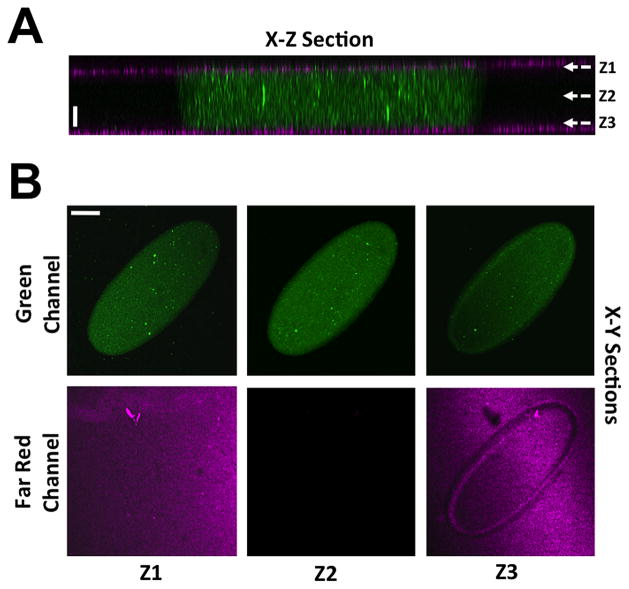
GelMA hydrogels were photopatterned within a microfluidics device to achieve an ellipse structure with major and minor axes lengths of 1.2 mm and 0.45 mm, respectively. A X-Z confocal section of the fluidic chamber depicts a structure embedded with green fluorescent particles sandwiched between two PAm hydrogels containing far red particles (Fig. 1A). The non-adhesive PAm hydrogels were used to eliminate the adhesion (if any) of the encapsulated cells to the surfaces outside GelMA structures. The X-Y confocal sections showed the presence of a tri-layer hydrogel as sections at Z1 and Z3 show both the GelMA and PAm hydrogels while Z2 only shows the GelMA hydrogel (Fig. 1B). The time-lapse recording of the perfused device (visualized by addition of 0.1% green fluorescent beads) shows the robustness of the PAm-GelMA interface, which do not dislodge from shear forces caused by flow rates up to 80 μL/hr (Supplementary Movie 1). Furthermore, visualization of the flow field around various portions of the GelMA structure illustrates the convective mass transport reminiscent of blood flow in vivo (Supplementary Movie 1).
Figure 1. Characterization of the device.
(A) X-Z confocal sections of a photo-patterned GelMA hydrogel sandwiched between two PAm hydrogels. Far red and green fluorescent beads are used to visualize the PAm and GelMA hydrogels, respectively. Scale bar: 50 μm. (B) X-Y confocal sections of the green and far-red channels at Z positions—Z1, Z2, and Z3—listed in (A). Scale bar: 200 μm.
Flow induces concentration gradient within GelMA structures
Fluid flow within the device containing cell-laden hydrogels can impart two effects—(i) compression of the GelMA structures due to increased fluid pressure and (ii) steady state concentration gradient of chemoattractants within the GelMA structures (due to their consumption by the entrapped cells)33. Quantification of flow rate induced changes of the GelMA hydrogels exposed to various flow rates showed no differences in their area strain up to a flow rate of 40 μL/hr (Supp. Fig. 2). However, a significant change in the area strain was observed at 1000 μL/hr, which was used as a positive control (Supp. Fig. 2). Area strain instead of volumetric strain was used to assess the flow induced mechanical compression because the GelMA hydrogels were confined within the fluidics channel that does not allow any vertical displacements.
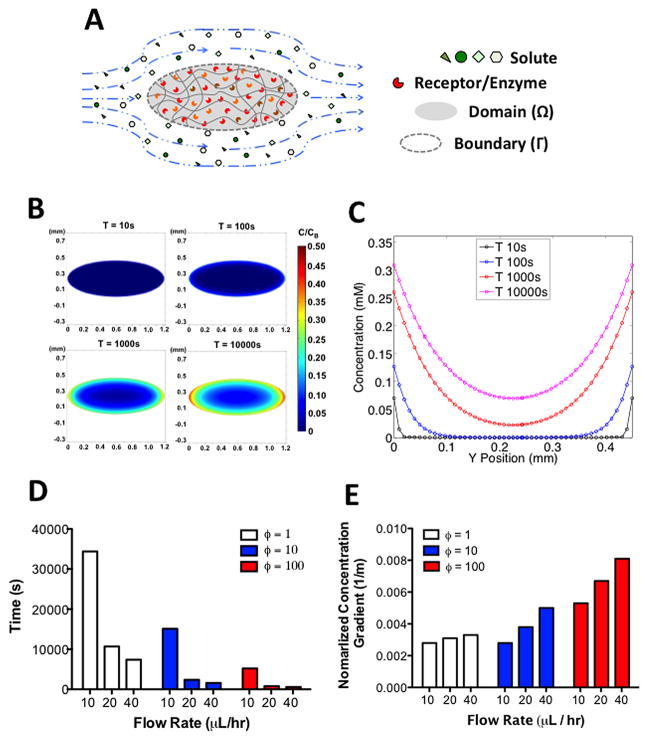
Next we assessed the presence of a chemotactic gradient within the GelMA hydrogel exposed to different flow rates (10, 20, and 40 μL/hr). To model the concentration gradient, we simulated the convective mass transfer of an arbitrary solute from the medium (C) capable of binding to its target enzyme/receptor through a Michaelis-Menten based consumption reaction (RC) (Fig. 2A, Eq. 1).
Figure 2. Concentration gradient within GelMA hydrogels.
(A) Illustration of the diffusion-reaction mass transfer system with convective boundary condition. The interior of the ellipse contains encapsulated cells that consume the soluble factors supplied across the boundary of the ellipse via convection. (B) Heat map of the changes in the normalized concentration (concentration within the GelMA normalized to the bulk concentration in the media) with time. (C) Changes in the normalized concentration profile with time along the minor axis of the ellipse. Time required to reach steady state (D) and maximum normalized concentration gradient at steady state (E) for different flow rates at ϕ= 1, 10, 100.
| (Eq. 1) |
Here, DC is the diffusion coefficient of solute within the GelMA hydrogel. As indicated in Figure 2A, the domain, Ω, of the system is a 2-dimensional ellipse where the consumption of solute occurs via Michaelis-Menten kinetics (Eq. 2) while the perimeter, Γ, of the GelMA hydrogel is governed by convective flux boundary condition (Eq. 3) where the Mass Transfer Coefficient (H) is approximated from the laminar flow over a plate (Eq. 4).
| (Eq. 2) |
| (Eq. 3) |
| (Eq. 4) |
| (Eq. 5) |
In the above equations, kcat is the catalytic coefficient, E0 is the enzyme concentration, KM is the Michaelis constant, L is the characteristic length, and CBulk is the concentration of solute in the bulk solution. Furthermore, we linearized the Michaelis-Menten reaction by assuming that the substrate concentration is much less than KM since high cell density within the hydrogels would increase the consumption rate of solutes such that their concentration remains substantially low. With this assumption, Equation 2 can be simplified as follows:
| (Eq. 6) |
Based on this theoretical framework, we modeled the mass transfer of an arbitrary solute of molecular weight (MW) 75 kDa, more common for serum proteins, whose DC within the GelMA hydrogel was approximated to be 10 μm2/s 34. The concentration and the catalytic efficiency of the enzyme consuming this soluble factor was designated to be 100 nM and 10 mM−1min−1, respectively, based on common enzymes found within the cytosol35, 36. In addition, the resulting concentration profile as a function of time is shown as 2-D heat maps and concentration profiles along the minor axis in Figures 2B and 2C, respectively. These plots suggest that the concentration of solute was substantially higher at the periphery than at the center of the GelMA structure at all time points. This is mainly due to the consumption of the solute by the encapsulated cells. Therefore, at steady state (at 10000s), a large concentration gradient was established within the GelMA structure indicating the presence of a chemotactic gradient (Fig. 2C).
To generalize this model to any proteins or biomolecules, we further examined how the concentration profile changes as a function of ϕ, which is defined as a ratio of DC/AE to (kcat/KM)E0, where AE is the perimeter of the ellipse multiplied by the height of the fluidics chamber. Here, ϕ is a non-dimensionalized parameter that compares the diffusion of a solute to its consumption rate. Our results indicate that increasing flow rates (X-Y) reduces the time required to reach steady state concentration profile at all ϕ values (Fig. 2D and Supp. Fig. 3). The examination of steady state concentration profile indicates increase in concentration gradient throughout the entire ellipse structure with increase in flow rates (Fig. 2E and Supp. Fig. 4). On the other hand, the concentration gradient decreases in the ellipse structure with decreasing ϕ value. Our results indicate that the increase in flow rate exposes the cells within the GelMA hydrogels to greater concentration gradients even at an early time point after encapsulation.
Migration of HUVECs to the periphery of 3D GelMA structures
The above-discussed theoretical analysis predicts the existence of a chemotactic gradient that could drive the migration of HUVECs encapsulated within the GelMA hydrogels. The cell-laden structures were subjected to varying flow rates (10, 20, and 40 μL/hr) for three days post-encapsulation. Brightfield images at 3 days post-encapsulation show that the cell density at the interior of the GelMA structures decreases irrespective of the flow rates with large decrease in cell density with higher flow rates (Fig. 3A 1st and 2nd column). The quantitative analysis of cell density across the GelMA hydrogel as a function of culture time for different flow rates supports the above observation (Fig. 3B). We also observed differences in the spatial distribution of cells within the GelMA hydrogel under different flow rates. At 10 μL/hr, the cells within the GelMA structures were uniformly distributed at day 3 (Fig. 3A 2nd column). On the other hand, flow rates of 20 and 40 μL/hr resulted in the migration of most of the HUVECs to the periphery of the GelMA hydrogel structure (Fig. 3A 1st and 2nd column, 3B). The thickness of the HUVEC layer at the periphery of the GelMA hydrogel was found to be decreased for 40 compared to 20 μL/hr flow rate (Figure 3B 3rd column). For the remainder of the studies, we used cultures exposed to a flow rate of 40 μL/hr.
Co-Culture of HUVECs and cancer cell spheroids
We examined whether the migration of encapsulated HUVECs to the periphery of the GelMA hydrogel will persist in the presence of cancer spheroids. Brightfield images of the encapsulated cells at days 0, 3, and 5 reveal that the HUVECs indeed migrate to the periphery of the GelMA structures even in the presence of MCF7 spheroids (Fig. 4A).
We investigated the migration of HUVECs at smaller time increments by quantifying the changes in the local cell density as a function of time. To this end, we partitioned the ellipse structure into different zones, and quantified the cell density in each zone as a function of culture time. Immediately after encapsulation, the density of the encapsulated HUVECs across Z1 to Z5 was found to be uniform, suggesting a homogenous distribution of cells within the GelMA hydrogel structures (Fig. 4B). During days 1 to 3, a large number of HUVECs migrated to the periphery of the hydrogel with a decrease of 30–40% in cell density was observed within the GelMA hydrogel. This initial migration of HUVECs to the periphery substantially declined from day 4 and onwards (only 7–10% cell density decrease was observed from day 4 onwards) (Fig. 4C).
On the contrary, encapsulated MCF7 cell clusters remained intact and we did not observe migration of the spheroids or any individual cells migrating away from the spheroid. This observation was validated using labeled MCF7 cells (Supp. Fig. 5). The encapsulated cancer spheroids exhibited a gradual growth as a function of culture time (Fig. 4D). The spheroids were found to grow to approximately twice its size between days 0 and 5.
Characterization of the HUVEC layer at the periphery of the hydrogel structure
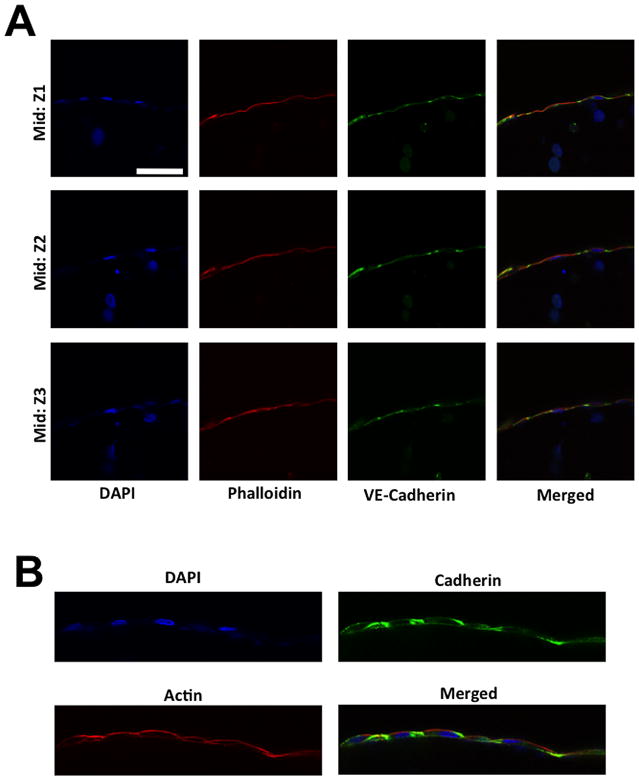
We next investigated whether the assembly of HUVECs around the GelMA structure forms an endothelial barrier. The cell-laden GelMA structures grown for 5 days were used to characterize the HUVEC structure. Confocal images of F-Actin and DAPI stained structures revealed the presence of HUVECs along the entire periphery (observed along the lateral and vertical directions) of the GelMA hydrogel (Fig. 5A). The HUVECs were also observed at both the GelMA-PAm interface (Fig. 5A). Interestingly, the HUVECs were found to be of single cell thickness as illustrated by the nuclei staining of the HUVECs around the ellipse (Fig 5A, DAPI column). In addition, the presence of continuous F-actin indicates that the HUVECs form a continuous monolayer sheet along the periphery (Fig 5A, Phalloidin column). Furthermore, to determine if cell-cell junctions are present within the monolayer sheet, the cell-laden GelMA structures were stained for VE-cadherin. Confocal images of the structures showed the presence of VE-cadherin that connects the neighboring HUVECs to form a single layer along the GelMA structure (Fig. 5B). In addition to the presence of cell-cell junctions, we also assessed the capacity of the endothelial layer to serve as barrier to fluxes of molecules within the media by analyzing the diffusion of FITC-conjugated dextran polymers with molecular weight of 10 and 150 kDa. The diffusion of dextran into the cell-laden GelMA hydrogel can be assessed through fluorescent signal within the ellipse, which increased with time (Supp. Fig. 6A). This diffusion process was quantified by plotting the mean normalized intensity within the central region of the ellipse (Supp. Fig. 6B). As expected, the diffusion of 10 kDa dextran into the hydrogels reached steady state after ~ 15 minutes while that of 150 kDa dextran required ~ 30 minutes. Furthermore, at steady state the fluorescent intensity of high molecular weight dextran was significantly lower than of low molecular weight dextran. Together the findings suggest that the endothelial barrier hinders the diffusion of high molecular weight dextran into the GelMA hydrogel (Supp. Fig. 6C).
Figure 5. Immunostaining of HUVECs cells migrated to the periphery of the GelMA structure.
(A) X-Y confocal sections of HUVECs cells stained for F-Actin (red) and nuclei (blue) at different Z positions: GelMA-PAm interface (labeled as Top and Bottom) and middle of the GelMA hydrogel (labeled as Mid). Green fluorescent beads were embedded within the PAm hydrogels to visualize the presence of the hydrogels. The rows indicate the specified Z positions. The columns 1–3 indicate the specific color channel while column 4 displays the merged image from other channels. Scale bar: 100 μm. (B) High magnification X-Y sections of HUVECs stained for VE-Cadherin (green), F-Actin (red), and nuclei (blue) at the midsection of the GelMA hydrogel. Scale bar: 10 μm. The HUVECs in both images were cultured with MCF7 spheroids for 5 days within the fluidics device prior to staining.
Dose dependent response of cells within the GelMA hydrogel to Doxorubicin
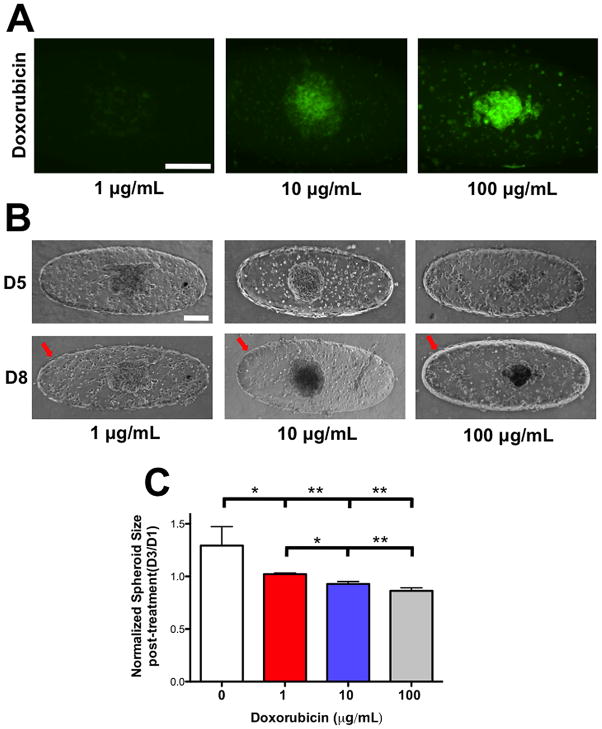
We assessed the potential of our device as a drug screening platform by analyzing the effects of Doxorubicin on HUVEC barrier and the tumor spheroids. Cell laden GelMA structures, cultured for 5 days, were exposed to different concentrations of Doxorubicin (1, 10, and 100 μg/mL) for 3 days. We investigated the penetration of Doxorubicin into the cancer spheroids by using fluorescent imaging. Our results indicate an increased accumulation of Doxorubicin in samples treated with 10 and 100 μg/mL of drugs (Fig. 6A). However, minimal accumulation of Doxorubicin was observed in samples treated with 1 μg/mL concentration (Fig. 6A). Brightfield images of cancer spheroids before and after 3 days of Doxorubicin exposure suggested a decrease in spheroid size and the darkening of the MCF7 spheroids exposed to 10 and 100 μg/mL Doxorubicin (Fig. 6B). To quantify the effects of Doxorubicin, we analyzed the changes in spheroid size before and after treatment with varying concentrations of the drug. MCF7 spheroids exposed to 10 and 100 μg/mL Doxorubicin experienced ~7 and ~15% decrease in spheroid area. On the other hand, the growth of the cancer spheroid was still observed at 1 μg/mL although the growth was ~ 20% less than those cultured in the absence of the drug (Fig. 6C). In addition to cancer spheroids, the drug was also found to be cytotoxic to HUVECs cells as the endothelial barrier was found to have disappeared upon exposure to Doxorubicin. This finding is consistent with other reports, which showed the cytotoxic effect of Doxorubicin on endothelial cells37.
Figure 6. Dose-dependent response of encapsulated tumor spheroids to Doxorubicin.
(A) Fluorescent images to identify Doxorubicin penetration into the cancer spheroids at D8. Increased penetration of Doxorubicin into the MCF7 spheroid is observed at higher dosages. Scale bar: 200 μm. (B) Brightfield images of HUVECs and MCF7 spheroids prior to (D5) and 3 days after Doxorubicin treatment (D8). Red arrow points towards the presence of endothelial barrier and the lack thereof at and above 10 μg/mL of Doxorubicin, respectively. Scale Bar: 200 μm. (C) Change in spheroid size of MCF7 after Doxorubicin treatment for different dosages of Doxorubicin. The spheroid area, obtained from 2-D brightfield images, at D8 is normalized to the area at D5. * and ** indicate statistically significant differences of p < 0.05 and 0.01, respectively, as obtained from pair wise t-test.
Discussion
This study describes the development of a tumor-on-a-chip platform comprised of cancer spheroid encapsulated within a GelMA hydrogel and surrounded by an endothelial barrier. Using a single step process, we simultaneously encapsulated HUVECs and MCF7 spheroids within our device. In addition, we harnessed the differential chemoattractant-induced motility of HUVECs and cancer spheroids to control their confinement and organization within the device.
Our results show the effect of flow rates on the generation of concentration gradient of soluble factors within the cell-laden GelMA structures. Our theoretical analysis does not specify a molecule or protein as a chemoattractant. Instead we used a range of ϕ values, the ratio of the diffusion to the consumption rate of a solute, to generalize the model to encompass the range of biomacromolecules present in the culture media. Specifically, we employed ϕ ranging from 1 to 1000 to account for (i) proteins or molecules with large differences MW which affects the diffusion coefficient and (ii) their specific receptors on the cell surface will vary in their catalytic efficiency38.
Within the GelMA hydrogel containing both HUVECs and MCF7 spheroids, the cancer cells did not migrate in response to the gradient of soluble factors as they were confined to the center of the hydrogel. This lack of motility could be attributed to insufficient strength of the chemotactic gradient to cause the migration of cancer spheroids and/or cancer cells from the spheroids. In addition, the cell-cell contact mediated by the cadherin junctions within the cancer spheroid may also mitigate migration of cancer cells away from the spheres and within the GelMA hydrogel.
The Doxorubicin studies showed the potential of this tumor-on-a-chip platform to assess the response of cells to oncologic drugs. We have assessed the penetration of Doxorubicin into the tumor spheroid and its quantifiable cytotoxic effect on the cancer spheroid. Additionally, the loss of the endothelial barrier in devices exposed to high doses of Doxorubicin (> 10 μg/mL) suggests the lack of target specificity of this oncologic drug. This tumor-on-a-chip device can be translated to assess the efficacy of other cancer therapeutics in a physiologically relevant system that provides a co-culture system to test drug specificity, cancer spheroid in a 3-D environment, as well as an endothelial barrier that can potentially resist drug penetration especially for higher MW compounds18.
Although the platform described here utilizes endothelial and cancer cell co-cultures, our approach provides a versatile framework for establishing systems with increased complexity observed in physiological tumors. In vivo tumor microenvironments are comprised of a variety of resident cells ranging from stromal cells to immune cells39. The incorporation of supporting cells into the device can be accomplished by incorporating these cells into the GelMA structures along with HUVECs and cancer spheroid. The 3D pattern mediated confinement of cells within the device allows the compartmentalization of various cell populations to dissect the interaction and contribution of various cellular populations towards cancer growth individually and in concert. Such an in vitro platform recapitulating various attributes of in vivo tumor microenvironment could not only offers new insights but could also be used as a drug-screening platform. The presence of an encompassing endothelium closely mimics the vasculature present within actual tumors by allowing the circulating cells to attach, roll, and transmigrate. This could provide an additional perspective for analyzing the extravasation of circulating cells into the tumor site, which can be achieved by introducing suspended single cells into the injected media or intravasation of cancer cells into the circulating system.
Supplementary Material
Acknowledgments
The authors acknowledge that this study was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01 AR063184-02. The authors also acknowledge the University of California San Diego Neuroscience Microscopy Shared Facility funded through NS047101. AA acknowledges the support from ARCS foundation and the Ruth L. Kirschstein National Research Service Award NIH/NHLBI T32 HL 105373.
References
- 1.American Cancer Society. 2016 [Google Scholar]
- 2.Gong X, Lin C, Cheng J, Su JS, Zhao H, Liu TL, Wen XJ, Zhao P. Plos One. 2015;10(6):e0130348. doi: 10.1371/journal.pone.0130348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta G, Hsiao AY, Ingram M, Luker GD, Takayama S. J Control Release. 2012;164:192–204. doi: 10.1016/j.jconrel.2012.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S. Analyst. 2011;136:473–478. doi: 10.1039/c0an00609b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minchinton AI, Tannock IF. Nat Rev Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 6.Doublier S, Belisario DC, Polimeni M, Annaratone L, Riganti C, Allia E, Ghigo D, Bosia A, Sapino A. Bmc Cancer. 2012;12(4):1471–2407. 12–4. doi: 10.1186/1471-2407-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauhan VP, Jain RK. Nat Mater. 2013;12:958–962. doi: 10.1038/nmat3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debnath J, Muthuswamy SK, Brugge JS. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 9.Wei SC, Fattet L, Tsai JH, Guo YR, Pai VH, Majeski HE, Chen AC, Sah RL, Taylor SS, Engler AJ, Yang J. Nat Cell Biol. 2015;17:678–U306. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, Gurski LA, Zhang C, Harrington DA, Farach-Carson MC, Jia XQ. Biomaterials. 2012;33:9049–9060. doi: 10.1016/j.biomaterials.2012.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Nat Methods. 2006;3:369–375. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 12.Lovitt CJS, TB, Avery VM. Biology. 2014;3:345–367. doi: 10.3390/biology3020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo BR, Bhardwaj P, Choi S, Gonzalez J, Eguiluz RCA, Wang K, Mohanan S, Morris PG, Du BH, Zhou XK, Vahdat LT, Verma A, Elemento O, Hudis CA, Williams RM, Gourdon D, Dannenberg AJ, Fischbach C. Sci Transl Med. 2015;7(301):301ra130. doi: 10.1126/scitranslmed.3010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Upreti M, Jamshidi-Parsian A, Koonce NA, Webber JS, Sharma SK, Asea AAA, Mader MJ, Griffin RJ. Transl Oncol. 2011;4:365–U138. doi: 10.1593/tlo.11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehsan SM, Welch-Reardon KM, Waterman ML, Hughes CCW, George SC. Integr Biol-Uk. 2014;6:603–610. doi: 10.1039/c3ib40170g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim T, Doh I, Cho YH. Biomicrofluidics. 2012;6(3):034107. doi: 10.1063/1.4739460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moya M, Tran D, George SC. Stem Cell Res Ther. 2013;4(Suppl 1):S15. doi: 10.1186/scrt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. P Natl Acad Sci USA. 2012;109:13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung JH, Shuler ML. Lab Chip. 2009;9:1385–1394. doi: 10.1039/b901377f. [DOI] [PubMed] [Google Scholar]
- 20.Walsh CL, Babin BM, Kasinskas RW, Foster JA, McGarry MJ, Forbes NS. Lab Chip. 2009;9:545–554. doi: 10.1039/b810571e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwak B, Ozcelikkale A, Shin CS, Park K, Han B. J Control Release. 2014;194:157–167. doi: 10.1016/j.jconrel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaw KC, Manimaran M, Tay EH, Swaminathan S. Lab Chip. 2007;7:1041–1047. doi: 10.1039/b707399m. [DOI] [PubMed] [Google Scholar]
- 23.Hsiao AY, Torisawa YS, Tung YC, Sud S, Taichman RS, Pienta KJ, Takayama S. Biomaterials. 2009;30:3020–3027. doi: 10.1016/j.biomaterials.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albanese A, Lam AK, Sykes EA, Rocheleau JV, Chan WCW. Nat Commun. 2013;4:2718. doi: 10.1038/ncomms3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bersini S, Jeon J, Dubini G, Arrigoni C, Moretti M, Kamm RD. J Tissue Eng Regen M. 2014;8:203–203. [Google Scholar]
- 26.Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, Moretti M, Kamm RD. Biomaterials. 2014;35:2454–2461. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagley AF, Scherz-Shouval R, Galie PA, Zhang AQ, Wyckoff J, Whitesell L, Chen CS, Lindquist S, Bhatia SN. Cancer Res. 2015;75:3255–3267. doi: 10.1158/0008-5472.CAN-15-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davey SK, Aung A, Agrawal G, Lim HL, Kar M, Varghese S. Tissue Eng Part C-Me. 2015;21:1188–1196. doi: 10.1089/ten.tec.2015.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aung A, Bhullar IS, Theprungsirikul J, Davey SK, Lim HL, Chiu YJ, Ma XY, Dewan S, Lo YH, McCulloch A, Varghese S. Lab Chip. 2016;16:153–162. doi: 10.1039/c5lc00820d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Biomaterials. 2009;30:6702–6707. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang HM, Shih YRV, Hwang Y, Wen C, Rao V, Seo T, Varghese S. Acta Biomater. 2014;10:4961–4970. doi: 10.1016/j.actbio.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aung A, Gupta G, Majid G, Varghese S. Arthritis Rheum-Us. 2011;63:148–158. doi: 10.1002/art.30086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaemmerer E, Melchels FPW, Holzapfel BM, Meckel T, Hutmacher DW, Loessner D. Acta Biomater. 2014;10:2551–2562. doi: 10.1016/j.actbio.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 35.Sun ZJ, Murry DJ, Sanghani SP, Davis WI, Kedishvili NY, Zou Q, Hurley TD, Bosron WF. J Pharmacol Exp Ther. 2004;310:469–476. doi: 10.1124/jpet.104.067116. [DOI] [PubMed] [Google Scholar]
- 36.Di Carlo D, Aghdam N, Lee LP. Anal Chem. 2006;78:4925–4930. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- 37.Chen YL, Tsai YT, Lee CY, Lee CH, Chen CY, Liu CM, Chen JJ, Loh SH, Tsai CS. Plos One. 2014;9(9):e106812. doi: 10.1371/journal.pone.0106812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. Biochemistry-Us. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 39.Liu YA, Zeng G. J Immunother. 2012;35:299–308. doi: 10.1097/CJI.0b013e3182518e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.