Abstract
Low dietary calcium (Ca) intake during growth limits peak bone mass but physiological adaptation can prevent this adverse effect. To assess the genetic control on the physiologic response to dietary Ca restriction (RCR) we conducted a study in 51 BXD lines fed either 0.5% (basal) or 0.25% (low) Ca diets from 4–12 wks of age (n=8/line/diet). Ca absorption (CaAbs), femur bone mineral density (BMD), and bone mineral content (BMC) were examined. ANCOVA with body size as covariate was used to detect significant line and diet main effects, and line-by-diet interactions. Body size-corrected residuals were used for linkage mapping and to estimate heritability (h2). Loci controlling the phenotypes were identified using composite interval mapping on each diet and for the RCR. h2 of basal phenotypes (0.37– 0.43) and their RCR (0.32–0.38) was moderate. For each phenotype we identified multiple QTL on each diet and for the RCR. Several loci affected multiple traits: Chr 1 (88.3–90.6 cM, CaAbs, BMC), Chr 4 (45.8–49.2 cM, CaAbs, BMD, BMC), Chr 8 (28.6–31.6 cM, CaAbs, BMD RCR), and Chr 15 (13.6–24 cM, BMD, BMC), and (32.3–36 cM, CaAbs RCR, BMD). This suggests that gene clusters may regulate interdependent bone-related phenotypes. Using in silico expression QTL (eQTL) mapping and bioinformatic tools we identified novel candidates for the regulation of bone under Ca stress (Ext1, Deptor), and for the first time, we report genes modulating Ca absorption (Inadl, Sc4mol, Sh3rf1 and Dennd3), and both Ca and bone metabolism (Tceanc2, Tll1 and Aadat). Our data reveal gene-by-diet interactions and the existence of novel relationships between bone and Ca metabolism during growth. This article is protected by copyright. All rights reserved
Keywords: Bone, calcium absorption, gene by diet interaction, genetics, QTL, dietary calcium
Introduction
Attaining peak bone mass (PBM) during growth is a critical determinant of osteoporotic fracture in humans.(1) While 60% or more of the total variance in PBM is influenced by genetics,(2) there is growing evidence that environmental factors interact with genetics to define the extent of bone accrual during growth.(3,4) Dietary calcium (Ca) is an essential environmental factor required for skeletal mineralization. Inadequate Ca intake during growth compromises attainment of PBM and increases the risk of fracture later in life.(1) Fortunately, physiological mechanisms are in place to increase intestinal Ca absorption efficiency and renal Ca reabsorption to protect bone in response to dietary Ca stress.(5)
Several investigators have reported that the impact of dietary Ca intake on bone and Ca homeostasis is different across racial groups.(6–8) Similarly, our group has reported that bone parameters, Ca absorption efficiency, and their response to dietary Ca stress were significantly affected by genetic background in a diverse population of 11 inbred mouse lines.(9) Overall, these studies demonstrate that gene-by-environment (GxE) interactions can affect bone and Ca metabolism. Although the impact of genetic variants on bone phenotypes has been examined in many genome-wide association (GWA) and quantitative trait loci (QTL) mapping studies, very few studies have focused on dissecting the GxE interactions influencing bone and none of them have studied Ca absorption. Moreover, despite the importance of dietary Ca intake for attaining and maintaining bone mass, no studies have been conducted to identify the genetic loci regulating bone in different dietary Ca environments. Hence, our understanding of the gene-by diet (GxD) interactions affecting the attainment of PBM is limited.
The purpose of this study was to identify the genetic loci that regulate Ca and bone metabolism on a controlled, optimal Ca diet and to determine how genetics influences the response of mice to dietary Ca stress. For this study we used 51 BXD recombinant inbred (RI) lines(10) and well-defined diets. This is the largest genetic mapping study for bone phenotypes in one RI panel, it is the first mapping study to evaluate intestinal Ca absorption efficiency in any mouse model, and it is the first to evaluate GxD interactions on bone and Ca metabolism.
Materials and Methods
Animal Models
BXD recombinant inbred (RI) mouse lines are each defined by a fixed recombination pattern of alleles from the C57BL/6J (B6) and DBA/2J (DBA) inbred mouse lines.(11) Because each BXD line is inbred, individuals within a line are genetically identical and the panel can be used for experiments that include biological replicates and environmental interventions that test for the existence of gene-by-environment interactions.(11)
Experimental Design
Male mice from 51 BXD RI lines were obtained at 4 wks of age from The Jackson Labs (Bar Harbor, ME) (the lines used are listed in Suppl. Table 1). Four to eight mice per line were obtained at a time and at arrival, an equal number of mice from each line were randomly assigned to either a 0.5% Ca (adequate) or 0.25% Ca (low) diet (AIN93G base with 200 IU vitamin D3/kg diet, Research Diets, New Brunswick, NJ) (n=8/diet/line). Dietary Ca levels were chosen to meet the rodent dietary Ca requirement (0.5% Ca) or to elicit a response to Ca restriction in target organs.(9) Food and water were provided ad libitum. Mice were group-housed (2–4 mice/cage) on TEK-fresh bedding (Harlan Laboratories) in conventional shoebox cages in the Purdue University animal facilities and maintained in an UVB light-free environment on a 12 h-light/dark cycle under standard conditions of temperature and humidity. At 12 wks of age mice were deprived of food 12 h prior to sacrifice. The day of the study, mice were anesthetized with an intraperitoneal injection of ketamine (22 mg/mL) and xylazine (33 mg/mL), and Ca absorption was measured by 45Ca appearance in the serum 10 min after an oral gavage test.(9) Individuals conducting absorption tests and sample collection were blinded to genotype and dietary treatment. Femora were placed in 10% neutral buffered formalin for 48 h and stored in 70% ethanol afterwards. Following removal of muscle from the fixed femora, femur length was recorded using a digital caliper (Mitutoyo America Corporation, Aurora, IL) and BMC (g) and BMD (g/cm2) were determined using a PIXImus II densitometer (Lunar; GE-Healthcare, Madison, WI) at the Indiana University School of Medicine. Scans were conducted in air on 10 bones at a time placed on the company-supplied plastic imaging plate (0.25 × 0.25 mm focal spot size; energy=80 kV, current=400 μA, fixed threshold of 1320). Samples were randomized and positioned uniformly along a grid to minimize potential scan position effects. Regions of interest were manually set for each bone and analysis was done by one investigator (RMR). The coefficient of variation for measurements of femora is 3–5%.(12) All of the experiments were approved by the Purdue University Animal Care and Use Committee.
Statistical Analysis
Statistical analyses were conducted using SAS Enterprise Guide 4.2 (SAS Institute Inc., Cary, NC). Phenotype values with a z-score in the extreme 2.5% of either end of a line/diet group distribution were removed as outliers. Adherence to a normal distribution was assessed by Anderson-Darling tests. For non-normally distributed data, the Box-Cox transformation was performed. Ca absorption values required a natural log transformation to normalize the data. A new phenotype, the response to dietary Ca restriction (RCR), was calculated for each endpoint as we have previously reported.(9) ANCOVA was used to test for significant main effects of genetic background (i.e. line), diet, and line-by-diet interactions; femur length (FL) and body weight (BW) were used as covariates in the model.(13) ANCOVA was also used to determine the effect of genetic background on the RCR parameter. Significant covariate effects were observed on both diets and the RCR for BMD and BMC (BW, FL) and on both diets for Ca absorption (FL). Data were expressed as ANCOVA adjusted least square means (LSmean) ± SEM. Differences were considered significant when p<0.05. The Tukey-Kramer post-hoc test was used to determine differences between LSmeans.
Body size corrected residuals were used for linkage mapping and to calculate heritability estimates. For each phenotype the covariate effect of BW and FL was determined by Pearson’s correlation and removed by linear regression.(13) BMD and BMC were affected by both BW and FL whereas Ca absorption was only affected by FL. Narrow-sense heritability (h2) of bone and Ca absorption phenotypes was calculated for each diet as well as for the RCR parameter using the r2 of a one-way ANOVA (main effect=line). Line means (n=51) for each phenotype on the 0.5 % and 0.25% Ca diets and for their RCR were used for genetic mapping. Using 51 BXD RI strains gave us the power to reliably detect quantitative trait loci (QTL) accounting for 15% of the variance observed in the population with a power of ~ 0.80 and p<0.05.(14)
QTL Mapping
Marker information and BXD genotypes were downloaded from The GeneNetwork (www.genenetwork.org/genotypes/BXD.geno) and the sex-averaged genetic location of each marker was updated using the New Standard Genetic Map for the Laboratory Mouse(15) (http://cgd.jax.org/mousemapconverter). Markers with duplicated genetic locations or perfectly correlated genotypes in our panel of BXD lines were removed. The final genetic map contained 1558 markers. Composite interval mapping (CIM) was conducted on line means of raw (including BW and FL) and co-variate corrected phenotype data using Windows QTL Cartographer v2.5_011 (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). Forward selection identified 5 significant background markers. CIM was carried out using a Haldane map function, 2 cM walking speed, and a 10 cM window. Each diet (0.5% or 0.25% Ca) group and the RCR were mapped separately. Permutations (n=500) were used to determine significance for each analysis. A peak with a logarithm (base 10) of odds (LOD) ≥ 2 was considered putative. QTL plots for raw data are provided in Supplemental Fig. 1.
Bioinformatic Characterization
The QTL candidate region was defined using 1-LOD drop support intervals which approximate 95% confidence intervals.(16) Genetic locations of peaks (cM) were converted to base pairs positions (GRCm38) using the Mouse Map Converter tool. If a QTL was found within ≤ 5cM distance of another, the two loci were reported together and the location expressed as a range. QTL candidate regions were populated with genome features including protein-coding genes, non-coding RNA genes, gene fragments, and unclassified genes from the Mouse Genome Informatics (MGI) database (informatics.jax.org/).(17) Genes were identified as the region from the first to last exon plus 5000 bp upstream of the gene coordinates for exon 1. Regions that were identical by descent (IBD) between B6 and DBA were identified using the Mouse Phylogeny Viewer (http://msub.csbio.unc.edu/)(18) and 100% IBD regions were removed. Genes that were less than 100% IBD and the remaining non-IBD regions were used to query for polymorphisms between B6 and DBA using the SNP/variation query tool at the Mouse Phenome Database (MPD).(19) The Sanger1 and Sanger2 mouse data sets were merged to retrieve variation between the B6 and DBA lines. MPD annotations (dbSNP 138) were used to categorize polymorphisms by gene attribute: intronic, mRNA un-translated region (5′ and 3′ UTR), promoter region (5000 bp upstream from Exon 1), and exon-associated (i.e. synonymous and non-synonymous codons, stop and gain codons, splice sites, or frameshift mutations). Effects of non-synonymous amino acid changes and insertion/deletions were examined for potential functional effects using PROVEAN v1.1 (cutoff = −2.5).(20) Genome-wide DNase1 hypersensitive site (DNase1 HSS) data from the mouse ENCODE project was obtained from the UCSC genome browser and used to identify potential regulatory regions within the QTL regions; DNase1 HSS peak information from 10 mouse adult tissues was merged (fat pad, genital fat pad, heart, kidney, large intestine, liver, lung, skeletal muscle, spleen, and brain) and then overlaid with the polymorphisms in the QTL region.
Expression QTL Mapping
Local (cis) and distal (trans) expression QTL (eQTL) analysis was conducted in silico using microarray databases from the BXD panel at the GeneNetwork (www.genenetwork.org/webqtl/main.py). The GeneNetwork does not contain small intestine microarray data so only bone (GN accession: GN414) and kidney mRNA (GN Accession: GN240) data were used for cis-eQTL detection and only the bone database was used for trans-eQTL. Significance was defined as a Likelihood Ratio Statistic (LRS) score ≥ 15 for cis-eQTL and LRS ≥ 20 for trans-eQTL. The NCBI37/mm9 location of the QTL (Mb) plus an inclusion buffer of 10 Mb was used for the search. Significant associations between transcript abundance of eQTL and our phenotype(s) of interest were determined by Pearson’s Correlation coefficient.
Analysis of Potential Candidate Genes
For candidate genes within QTL regions, we searched for functional annotation using gene ontology (GO) terms at MGI. Information regarding functional association of the genes with skeletal and/or Ca metabolism, and their role in biological pathways was obtained from the Coremine Medical Explorer (www.coremine.com) and the Ingenuity Target Explorer (https://targetexplorer.ingenuity.com). Finally, information of tissue gene expression patterns was obtained using BioGPS (http://biogps.org/).
Results
Phenotype Variability across 51 Lines of the BXD RI Panel
All mice were healthy throughout the study. Body weight (BW) was significantly different among the lines (p<0.0001) but it was not affected by dietary Ca intake. BW variation was observed in the BXD population (mean 25.6 ± 0.7 g, range: 19.7 to 31.5 g) so phenotype data was adjusted for the effect of body size (see methods). Variability in Ca absorption, BMD and BMC was observed among the 51 RI lines for each diet group and for their response to dietary Ca restriction (RCR) (Z scores shown in Suppl. Fig. 2 and Suppl. Fig. 3; LSMeans in Suppl. Table 2). Ca absorption, and femoral BMD and BMC were each significantly affected by line (p<0.0001) and diet (p<0.0001) but only Ca absorption was affected by a significant line-by-diet interaction (p=0.0006). The average effect of dietary Ca restriction in the BXD panel was a 25.5% increase in Ca absorption efficiency (p<0.0001), a 3.5% decline in BMD (p<0.0001), and a 3.9% reduction in BMC (p<0.0001).
The narrow sense heritability (h2) for BMD and BMC was 43% and 40% in mice on the basal diet and 51% and 48% in mice on the low Ca diet groups, respectively. Heritability of Ca absorption efficiency was 37% in the basal Ca diet group and 40% in the low Ca diet group. The responses to dietary Ca restriction (RCR) for BMD, BMC and Ca absorption were all significantly affected by genetic background (p<0.0001) and had a heritability of 32%, 38% and 36%, respectively. The RCR for each phenotype ranged from −37.3% to 108.9% for Ca absorption, −15 to 11.4% for BMD, and −27.3 to 23.6% for BMC across the BXD panel.
Phenotype Associations
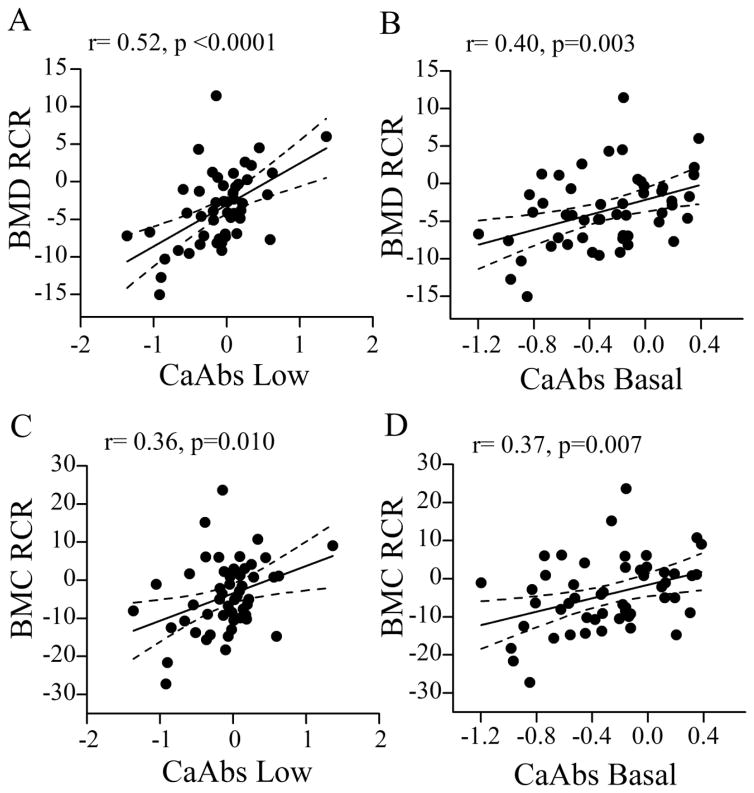
We evaluated associations between bone parameters and Ca absorption efficiency (Suppl. Table 3). BMD was highly correlated to BMC on either diet, and their responses to Ca restriction were also positively and significantly correlated with each other (r > 0.9, p<0.0001). The RCR for Ca absorption was positively correlated to Ca absorption on the low Ca diet (r= 0.49, p<0.001) but negatively correlated to Ca absorption on the basal Ca diet (r = −0.38, p=0.005). This indicates that lines with high basal Ca absorption were less likely to increase Ca absorption efficiency when fed a low Ca diet. The RCR for BMD or BMC were not correlated to their phenotypes on the basal or on the low Ca diets (r = 0.22, p=0.1 and r = 0.32, p = 0.22, respectively). However, the responses of BMD and BMC to dietary Ca restriction were significantly correlated to Ca absorption efficiency on both the low (r=0.52, p < 0.0001 and r=0.36, p=0.010) and basal Ca diets (r=0.40, p = 0.003 and r=0.37, p=0.007) (Fig. 1).
Figure 1. Correlation between bone responses to Ca restriction and Ca absorption efficiency.
(A) BMC response to Ca restriction (RCR) vs. Ca absorption efficiency (CaAbs) on the Low (0.25%) (B) and basal (0.5%) Ca diets. (C) BMD RCR vs. CaAbs efficiency on the Low (D) and Basal Ca diets. ANCOVA line means with 95% confidence intervals are shown (n=51). CaAbs data was log transformed. Pearson’s correlation coefficient (r) and p values are shown.
Genetic Mapping
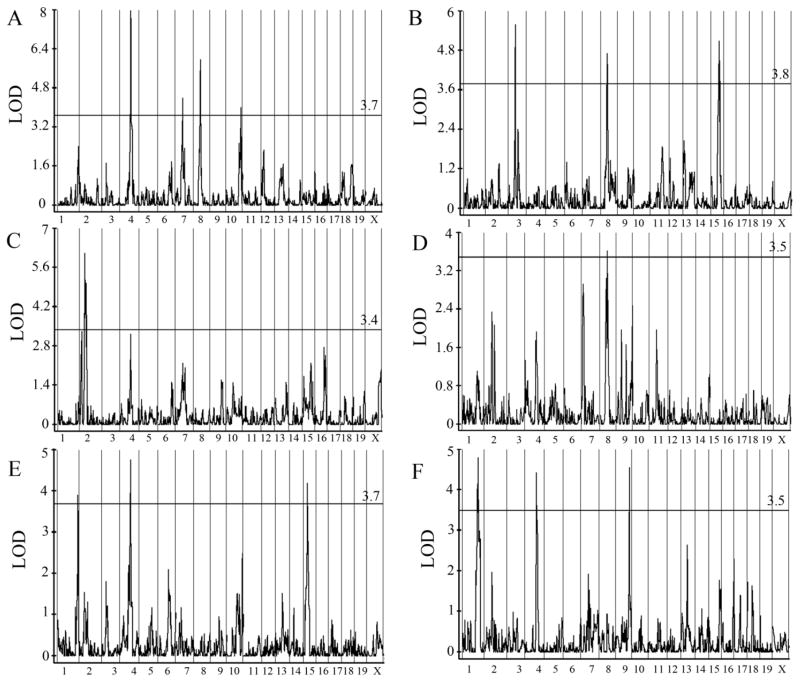
We identified multiple significant QTL for each phenotype in each diet group and for the RCR (co-variate adjusted, Fig. 2; unadjusted raw, Supplmental Fig. 1). When QTL mapping results across analyses were considered together (3 phenotypes, 2 diets, and the RCR), 15 significant QTLs were identified (Table 1, Suppl. Fig. 4). We identified significant QTL controlling the BMD RCR on chromosome (Chr) 8 (QTL8b, 26.9–32.7 cM); the BMC RCR in Chr 1 (QTL1a 66.8–69.03 cM), Chr 4 (QTL4, 44.8–50.7 cM) and Chr 9 (QTL9b, 57–59.74 cM); and for the Ca absorption RCR in Chr 3 (QTL3, 27.9–31.7 cM), Chr 8 (QTL8a, 22.7–26.9 cM), and Chr 15 (QTL15b, 30.87–37.6 cM). Ten of the 15 QTL we identified were significant for more than one phenotype (QTL1b, 3, 4, 7, 8b, 9b, 10, 12, 15a, 15b). Within each phenotype, none of the QTL for the RCR overlapped with those identified at baseline. Loci that modulated traits in both diet groups were observed for BMC on Chr1 and Chr15 and for Ca absorption on Chr12 (Table 1). A list of genome features after IBD filtering as well as the number of polymorphisms associated with non-IBD genes for each these loci are given in Suppl. Table 4.
Figure 2. Composite Interval Mapping for Ca Absorption, BMD and BMC.
Composite Interval Mapping (CIM) identified multiple QTL for: Ca Absorption on the (A) Basal diet, (B) and for the response to Ca restriction (RCR); BMD on the (C) Basal diet, (D) and the RCR; and BMC on the (E) Basal diet, (F) and the RCR. Significance was determined separately for each dataset by permutation (n=500), LOD cut off shown as a solid horizontal line (n=51).
Table 1.
QTL Identified for Ca Absorption, BMD, and BMC
| QTL ID | Chr | High Allele | Peak estimate (cM)1 | 1-LOD CI (cM)2 | 1- LOD CI (Mb)2 | CaAbs3 | BMD | BMC | (r2)4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| 0.50% | 0.25% | RCR | 0.50% | 0.25% | RCR | 0.50% | 0.25% | RCR | |||||||
| QTL1a | 1 | DBA | 68.4 | 66.8–69.03 | 155.5–159.4 | 4.8 5 | 0.21 | ||||||||
| QTL1b* | 1 | B6 | 88.3 | 87.9–91.7 | 183.8–187.6 | 3.9 | 4.9 | 0.17; 0.2 | |||||||
| DBA | 90.6 | 2.4 | 0.06 | ||||||||||||
| QTL2* | 2 | B6 | 21.0 | 20.6–21.6 | 29.4–30.4 | 6.1 | 0.28 | ||||||||
| QTL3* | 3 | B6 | 29.6 | 27.9–31.7 | 57.6–68.4 | 5.6 | 0.24 | ||||||||
| DBA | 30.2 | 2.2 | 0.07 | ||||||||||||
| QTL4* | 4 | B6 | 45.8 | 44.8–50.7 | 96.8–108.9 | 3.2 | 4.8 | 0.13; 0.21 | |||||||
| DBA | 49.2 | 8 | 4.4 | 0.27; 0.19 | |||||||||||
| QTL7 | 7 | DBA | 29.9 | 23.9–33.2 | 37.1–54.5 | 2.2 | 0.07 | ||||||||
| B6 | 30.3 | 4.4 | 0.13 | ||||||||||||
| QTL8a | 8 | B6 | 23.7 | 22.7–26.9 | 36.3–47.7 | 4.7 | 0.19 | ||||||||
| QTL8b* | 8 | DBA | 28.6–31.6 | 26.9–32.7 | 47.7–65.3 | 6 | 3.6 | 0.186; 0.16 | |||||||
| QTL9a | 9 | DBA | 43.7 | 43.1–44.7 | 77.4–81.8 | 3.7 | 0.15 | ||||||||
| QTL9b | 9 | DBA | 58.3 | 57–59.74 | 105.7–109.8 | 2.8 | 4.5 | 0.12; 0.19 | |||||||
| QTL10 | 10 | B6 | 62.8–67.8 | 61.7–68.1 | 114.5–120.3 | 4 | 2.5 | 0.11; 0.1 | |||||||
| QTL12 | 12 | DBA | 6.9–8.8 | 5.2–12.2 | 10.4–29.7 | 2 | 3.8 | 0.04; 0.16 | |||||||
| QTL14 | 14 | DBA | 21.1 | 20.7–23.3 | 33.9–45.6 | 4.1 | 0.18 | ||||||||
| QTL15a* | 15 | B6 | 15.1–17.7 | 13.6–24 | 32.4–57.5 | 3.3 | 4.2 | 6.4 | 0.13; 0.18; 0.28 | ||||||
| QTL15b* | 15 | B6 | 32.3–36 | 30.87–37.6 | 69.2–78.8 | 5.1 | 2.2 | 0.22; 0.067 | |||||||
QTL within 5 cM (centimorgans) were reported together.
1-LOD confidence intervals in cM or Mb (Build GRCm38).
Abbreviations: CaAbs (Ca absorption), BMD (Bone Mineral Density), BMC (Bone Mineral Content); 0.50% and 0.25% are dietary Ca levels, RCR (Response to dietary Ca restriction).
(r2) = proportion of the phenotype variance explained by the QTL. If more than one QTL is within a loci the r2 values are reported in order of phenotype and diet,
LOD scores for each loci. LOD > 2 but under the permutation threshold = putative peak (highlighted).
Loci selected for in depth characterization.
Characterization of QTL Regions and Candidate Gene Identification
7 QTL regions were selected for characterization based on: the amount of variation explained by a single QTL (r2), mapping of multiple phenotypes to a loci, and identification of a loci controlling a phenotype in two or more conditions (Table 1). The detailed classification of functional polymorphisms and insertion/deletions (Indels) for each of these loci is in Suppl. Table 5. Genes with stop codon, frameshift, splice site mutations, deleterious non-synonymous polymorphisms, or indels were automatically considered as candidate genes. A frameshift mutation was detected for QTL4 (Chr 4, rs263625036, 3110021N24Rik) and 2 splice site donor (SSD) polymorphisms were identified for QTL8b (Chr8, rs33461690, AW046200, G/C and rs31742488, Tll1, G/A). Potentially deleterious non-synonymous polymorphisms were identified in QTL1b (Chr1, rs32873773, Lyplal1, I58T); QTL4 (Chr 4, rs31915781, Gm12689, S50F, and rs31774248, Gm12689, E 77A); QTL8b (Chr 8, rs33014006, Adam29, S163I, and rs238747469, Gm4975, R18L); and QTL15a (Chr15, rs32502839, Pkhd1l1, N3877T, and rs47743747, Abra, L374V).
Identification of local eQTL coincident with phenotype loci
Cis-eQTL overlapping with our QTL regions whose mRNA levels were significantly correlated with the phenotype and whose gene was not identical by descent between B6 and DBA were considered candidate genes.(21) Cis-eQTL meeting our criterion were detected in: QTL1b (Illumina probe: scl000933.1_43 lacking annotation), QTL4 (Tceanc2, Acot11 and Inadl), QTL8b (Apela, Aadat, Sh3rf1 and Sc4mol), QTL15a (Ext1, Ennp2, Deptor), and QTL15b (Dennd3) (Table 2).
Table 2.
eQTL in Characterized Regions
| QTL ID | Chr | 1-LOD (Mb)1 (NCBI37/mm9) | Phenotypes associated with QTL | # cis eQTL2 | Correlated with phenotype | # Trans eQTL3 | Correlated with phenotype |
|---|---|---|---|---|---|---|---|
| QTL1b | 1 | 185.6–189.4 | CaAbs basal, BMC basal, BMC LowCa | 1 | scl000933.1_43 (LRS =15.1): BMC Basal, r= −0.56*; BMC Low, r= −0.48* | 0 | |
| QTL2 | 2 | 29.3–30.3 | BMD Basal | 0 | — | 1 | Il8rb (LRS=24): BMCBasal, r=0.4* |
| QTL3 | 3 | 57.4–68.2 | CaAbs RCR, BMD Low Ca | 4 | — | 0 | |
| QTL4 | 4 | 96.4–108.5 | CaAbs basal, BMD Basal, BMC basal, BMC RCR | 7 | Tceanc2 (Kidney) (LRS=50.4): BMD Basal, r=0.41*; CaAbs basal r= −0.38*; Tceanc2 (Bone) (LRS=52.5): CaAbs Basal, r= −0.57*; BMC RCR r= −0.39*, BMD Basal r= 0.4*; Acot11 (LRS=28.2): BMC Basal, r=0.34#; Inadl (LRS=15): CaAbs Basal, r= 0.38* | 3 | Rps18 (LRS=23.5):BMC Basal, r= 0.39*; BMD Basal, r= 0.38*; CaAbs Basal, r= −0.46*, Rps8 (LRS:22.1) BMC Basal, r= 0.36#; CaAbs Basal, r= −0.51* Rbmx (LRS=20.5): BMC Basal, r= 0.39*; BMD Basal, r= 0.37* |
| QTL8b | 8 | 48.8–67.9 | CaAbs basal, BMD RCR | 6 | Apela (LRS=27): CaAbs Basal, r= −0.58*; Aadat (LRS=19.7): CaAbs Basal, r= −0.38#; BMD RCR, r= −0.37#; Sh3rf1(LRS=28.9): CaAbs Basal, r= −0.54*; Sc4mol: (LRS=38.9): CaAbs Basal, r= −0.44* | 0 | |
| QTL15a | 15 | 32.3–57.5 | BMD LowCa, BMC basal, BMC LowCa | 5 | Ext1 (LRS=70.8) : BMC Low, r= 0.4*; BMD Low, r= 0.53*; Enpp2 (LRS=50.3) : BMD Low, r= 0.58*; BMC Basal, r= 0.46*; BMC Low, r= 0.51*; Deptor (LRS=57): BMD Low, r= 0.53* | 1 | Hmga2 (LRS=21.7): BMC Low, r= − 0.34#; BMD Low, r= −0.33# |
| QTL15b | 15 | 69.1–78.6 | CaAbs RCR, BMD Basal | 1 | Dennd3 (LRS=15.3): CaAbs RCR, r= 0.33# | 0 |
1-LOD confidence interval in Mb (Build NCBI37/mm9)
Cis eQTL from the GeneNetwork using the a cut off LRS of 15–999, an inclusion buffer of 10 Mb, and the bone (GN414) and kidney (GN240) databases
Trans eQTL from Gene Network using a cut off LRS of 20–999, an exclusion buffer of 10 Mb, and the bone (GN414) database Significantly correlated with the phenotype of interest
p<0.05,
p<0.1.
The candidate gene list for each QTL included polymorphisms affecting protein coding and cis-eQTL genes. Bioinformatic analysis identified genes whose polymorphisms might affect our phenotypes based on their reported function, their tissue gene expression pattern, and their reported interaction with other genes or proteins. No deleterious mutations or significant cis-eQTL were detected for QTL2 or QTL3. Based on our analysis we identified candidate genes for loci regulating basal Ca absorption (Inadl, Sc4mol, Sh3rf1), the Ca absorption RCR (Dennd3); bone mass on one of the two diets (Acot11, Ext1, Enpp2 and Deptor); and both Ca absorption and basal bone mass (Lyplal1) or bone RCR (Tceanc2, Aadat and Tll1) (Suppl. Table 6).
Discussion
Previous studies have shown that the change in Ca absorption efficiency in response to changes in dietary Ca intake has a strong genetic component.(6,8,9) Consistent with these findings, we reported that basal Ca absorption efficiency and its response to dietary Ca restriction (RCR) are heterogeneous in the BXD RI panel and that they have heritability estimates (37 % and 36% respectively) that are similar to those reported for other Ca and bone metabolism phenotypes in mice and humans.(22,23) We also found that the genetic regulation of the adaptive response (RCR) is independent from the regulation of Ca absorption efficiency or bone mass at baseline indicating that the loci for the RCR contain genes sensitive to the dietary Ca environment.
Several genes have been proposed to contribute to basal and vitamin D-regulated intestinal Ca absorption(24) including candidates from a recent genome-wide study that identified forty-five new 1,25(OH)2D-target genes in the intestine.(25) However, these genes were either located in regions that are identical by descent between B6 and DBA, located outside the QTL we identified, or contained polymorphisms whose variants have no consequence on protein function or mRNA level. Thus, our QTL mapping study has revealed novel variation controlling intestinal Ca absorption and it has the potential to reveal new genes that control Ca absorption efficiency and its response to dietary Ca changes.
Within our list of prioritized loci, several influenced either basal Ca absorption efficiency (QTL4, 8b, 1b) or its RCR (QTL3,15b). Our bioinformatics analysis identified several candidate genes within each loci (Suppl. Table 6). However, the functional link between our candidate genes and Ca metabolism is not straight forward because there is limited information on the role of the genes (Apela) or because their function has not been linked to Ca homeostasis: Lyplal1 (protein depalmitoylation), Sc4mol (lipid/cholesterol metabolism), Sh3rf1 (Rac/JNK signaling), Dennd3 (regulation of GTPase activity). Nevertheless, further studies on these genes could reveal novel roles for them in the regulation of Ca metabolism.
One candidate gene of particular interest is the Inadl gene in QTL4. Inadl helps organize multimeric protein complexes in the plasma membrane(26) and contributes to the formation of tight junctions (TJ).(27) Inadl could influence Ca absorption by affecting the formation of Ca transporter complexes including either TRPV6 (at the apical membrane) or PMCA1b (at the basolateral membrane). It could also affect the TJ proteins that mediate the passive diffusion of small ions across the epithelia (e.g. claudins 2 and 12(28)). Since Inadl expression level is positively correlated to Ca absorption efficiency, we hypothesize that elevated levels of Inadl could enhance the proper formation of transmembrane Ca transport complexes and ultimately increase intestinal Ca absorption.
Many studies have examined the independent effects of genetics and diet on bone but very few of these studies are focused on GXD interactions. Our work is the first carefully controlled study to identify loci underlying the bone responses to dietary Ca stress. We found that genetic factors contribute up to 43% of the variation observed in BMD and BMC at baseline and up to 38% of the variation in RCR. Several of our bone loci were located within the 95% confidence intervals of loci from other studies: i.e. QTL1b (87.6 cM, whole body aBMD), QTL10 (62.5 cM, femur aBMD), and QTL15b (33.8 cM, femur aBMD; and 38.7 cM, whole-body aBMD).(29) Within our loci, we found a number of genes known to regulate bone metabolism in humans: osteonectin (Spock 3/SPARC)(30) within QTL8b, osteoprotegerin (TNFRSF11B)(31) in QTL15a, and the leptin receptor (Lepr) gene (32) in QTL4. However, the Lepr gene is 100% IBD between B6 and DBA mice and neither of the other two genes contain deleterious polymorphisms affecting protein function or gene expression. Hence, it is unlikely that these three genes are contributors to the phenotype variation observed in BMD and BMC in the BXD population.
Wang et al.(33) recently used a panel of male mice from 46 BXD RI lines and identified a significant loci controlling femur and tibia BMD in the same genomic region where our QTL15a is located. They identified a non-synonymous coding polymorphism (V813L) in the Trichorhino-phalangeal Syndrome Type I Protein (TRPS1) gene (rs32398060, B6=C, DBA=G) as the variant controlling the phenotype. However, we found that the polymorphism was classified as neutral using the PROVEAN software (score = −0.01), suggesting that the sequence variation does not affect protein function. In addition, while TRPS1 mRNA is a cis-eQTL at the QTL15a locus, TRPS1 mRNA levels were not correlated with either BMD or BMC in our BXD population. Therefore, we do not consider TRPS1 to be a strong candidate gene for QTL15a.
The most promising candidate gene in QTL15a is exostoses multiple 1 (Ext1). Ext1 has glycosyltransferase activity required for heparan sulfate (HS) biosynthesis,(34) a component of the extracellular matrix (ECM) critical for endochondral development.(35) Ext1 expression is very high in osteoblasts and osteoclasts, and mice with a hypomorphic mutation in Ext1 have impaired chondrocyte and bone differentiation, and defects in HS production.(35) In humans, a splice variant of Ext1 has been reported as a causative locus for hereditary multiple exostosis (HME), a disease characterized by a low PBM and osteoporosis,(36) and a polymorphism near the Ext1 gene was recently identified as responsible for height variation in a population of Korean adults.(37) In our study, bone Ext1 mRNA level was positively correlated with BMD and BMC in mice on the low Ca diet. We hypothesize that reduced Ext1 transcript levels will reduce production of HS, reduce signals for endochondral ossification, and lower BMD and BMC, especially under dietary Ca stress.
In addition to the loci regulating basal BMD and BMC, we identified loci for their RCR in QTL8b, QTL1a, QTL4, and QTL9b. These loci did not co-localize with loci controlling basal bone mass. This indicates that there are independent genetic controls on basal bone mass and its RCR. As a result, future bone genetics studies in humans should control for habitual dietary Ca intake. QTL4 and QTL8b also controlled basal Ca absorption efficiency. Others have shown that higher Ca absorption efficiency during growth is associated with higher bone deposition in children(38) and mice(9) while reduced Ca absorption efficiency in adulthood is associated with increased hip fracture risk.(39,40) Consistent with a functional relationship between bone and Ca absorption efficiency, we found that the RCR for BMC and BMD were significantly, positively correlated to intestinal Ca absorption efficiency (Fig. 1, Suppl. Table 3). These findings suggest that genetic factors leading to high Ca absorption efficiency protect bone under dietary Ca stress. In addition, the co-localization of these complementary phenotypes within a locus suggests the existence of novel relationships between bone and Ca metabolism during growth.
QTL4 is a particularly interesting locus controlling BMC and BMD on the basal Ca diet, the BMC RCR, and basal Ca absorption efficiency (Table 1). Others have mapped a number of other bone-related traits to this locus including femur mechanical properties,(41) spine and femur BMD, and spine and femur μCT phenotypes.(42,43) Several potential candidate genes are in this region but there is not an obvious link between their biological roles and Ca/bone metabolism. However, one gene, Tceanc2, encodes a protein whose GO annotation identifies it as involved in DNA binding, regulation of mRNA processing, and transcription elongation by RNA polymerase II. Tceanc2 mRNA expression was positively correlated with basal BMD and negatively correlated with basal Ca absorption and the BMC RCR. Although, very little is known regarding the role of this gene in mice or humans, its fundamental biological function in regulatory events suggests it could impact many physiologic processes, including bone and Ca metabolism.
QTL8b controls both basal Ca absorption efficiency and the BMD RCR. Two interesting candidate genes, Tll1 and Aadat, are within this locus. Tll1 is highly expressed in osteoblasts and is a member of the mammalian bone morphogenetic protein-1 (BMP1)-like proteinases that regulate the formation, organization, and mineralization of extracellular matrix (ECM).(44) In BXD mice, a G/A polymorphism affects a splice donor site in Tll1. Since the polymorphism is associated with a positive impact on the BMD RCR and basal Ca absorption, we hypothesize that alternative splicing results in a more active isoform of the Tll1 protein that increases the formation of ECM matrix, increases bone mass, and protects bone from dietary Ca restriction. However, while this hypothesis explains the role of Tll1 in the regulation of bone mass, the role of this gene in the modulation of Ca absorption is less clear and may be indirect.
Aadat mRNA levels were negatively associated with both basal Ca absorption and the RCR for BMD and this suggests Aadat is a promising co-regulator of Ca and bone metabolism. Aadat catalyzes the production of kynurenic acid (KYNA), a non-competitive glutamate receptor antagonist.(45) Glutamate-mediated excitation of neurotransmitters can stimulate thyroid function.(46) In rats, glutamate receptor agonists can increase TSH and thyroid hormone concentrations whereas glutamate receptor antagonists decrease them.(47) A recent human GWAS identified Aadat as a novel locus associated with the variation in free T4 levels.(48) We hypothesize that decreased Aadat levels increase thyroid hormone levels which others have shown can enhance intestinal Ca absorption,(49) chondrocytes maturation, osteoblast differentiation, and osteoclasts activation.(50)
Our study has several strengths and weaknesses that should be noted. One strength of our approach was that by using the BXD RILs, with numerous recombination events during inbreeding, we had higher resolution mapping than is possible with F2 populations. In addition, since the RI have genotypes that are fixed as homozygotes they can be used for the study of GXE interactions. As a result, our study is unique in that it has determined the genetic architecture of the response of bone and Ca metabolism to the stress environment of low dietary Ca intake. Some limitations in our study also exist. First, the BXD panel was built with two founders so captures just a portion of the genetic diversity available in the mouse genome. Second, although we used more lines than any other bone RI mapping study, the relatively small size of our population (51 lines) limits our power to detect small effect size QTLs. Third, we studied only male mice, and so we are likely missing important loci that are influenced by the female sex hormones. Finally, the assignment of arbitrary gene boundaries for our QTL characterization may have missed other plausible candidate genes.
In summary, our study is novel in that it is the first to examine either the genetic architecture of intestinal Ca absorption efficiency or the response of bone mass and Ca absorption to dietary Ca restriction. Our data revealed strong genetic effects on basal Ca and bone metabolism and their responses to Ca stress. Importantly, the genetic variation controlling the response to low Ca intake is independent of the genetic variation controlling baseline levels of these phenotypes. This demonstrates that there are independent genetic effects that should be respected in the design of future bone genetic studies. Moreover, our study confirms the existence of GxD interactions influencing bone accrual during growth and shows that accrual of bone during growth is dependent on high Ca absorption efficiency. A critical finding from our study was the evidence for novel regulation of Ca and bone homeostasis under conditions of Ca stress, and the possibility of common loci controlling Ca and bone phenotypes during growth.
Supplementary Material
Acknowledgments
This work was supported by NIH grant ES019103 to JCF. CONACyT, Mexico provided a partial graduate scholarship to PRF.
Footnotes
This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: [10.1002/jbmr.2760]
Disclosures
JCF is on the Scientific Advisory Board for Innophos, Inc. All other authors have no conflicts of interest to report.
Authors’ roles: Study design: JCF. Study execution: RAR, LW and PRF. Data collection: RAR. Data analysis: PRF, LW, MZ. Data interpretation: JCF, PRF, RAR. Drafting manuscript: PRF and JCF. Approval of final manuscript: JCF, PRF, RAR, LW and MZ. JCF takes responsibility for the integrity of the data analysis.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Heaney RP, Abrams S, Dawson-Hughes B, et al. Peak bone mass. Osteoporos Int. 2000;11(12):985–1009. doi: 10.1007/s001980070020. [DOI] [PubMed] [Google Scholar]
- 2.Bonjour JP, Chevalley T, Rizzoli R, Ferrari S. Gene-environment interactions in the skeletal response to nutrition and exercise during growth. Med Sport Sci. 2007;51:64–80. doi: 10.1159/000103005. [DOI] [PubMed] [Google Scholar]
- 3.Kelly PJ, Eisman JA, Sambrook PN. Interaction of genetic and environmental influences on peak bone density. Osteoporosis International. 1990;1(1):56–60. doi: 10.1007/BF01880417. [DOI] [PubMed] [Google Scholar]
- 4.Ackert-Bicknell CL, Karasik D. Impact of the environment on the skeleton: is it modulated by genetic factors? Curr Osteoporos Rep. 2013;11(3):219–28. doi: 10.1007/s11914-013-0151-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Y, Peng X, Porta A, et al. Calcium transporter 1 and epithelial calcium channel messenger ribonucleic acid are differentially regulated by 1,25 dihydroxyvitamin D3 in the intestine and kidney of mice. Endocrinology. 2003;144(9):3885–94. doi: 10.1210/en.2003-0314. [DOI] [PubMed] [Google Scholar]
- 6.Weaver CM, McCabe LD, McCabe GP, et al. Vitamin D status and calcium metabolism in adolescent black and white girls on a range of controlled calcium intakes. J Clin Endocrinol Metab. 2008;93(10):3907–14. doi: 10.1210/jc.2008-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu K, Greenfield H, Zhang Q, et al. Growth and bone mineral accretion during puberty in Chinese girls: a five-year longitudinal study. J Bone Miner Res. 2008;23(2):167–72. doi: 10.1359/jbmr.071006. [DOI] [PubMed] [Google Scholar]
- 8.Braun M, Palacios C, Wigertz K, et al. Racial differences in skeletal calcium retention in adolescent girls with varied controlled calcium intakes. Am J Clin Nutr. 2007;85(6):1657–63. doi: 10.1093/ajcn/85.6.1657. [DOI] [PubMed] [Google Scholar]
- 9.Replogle RA, Li Q, Wang L, Zhang M, Fleet JC. Gene-by-Diet Interactions Influence Calcium Absorption and Bone Density in Mice. J Bone Miner Res. 2014;29(3):657–65. doi: 10.1002/jbmr.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey DW. Recombinant-inbred strains. An aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation. 1971;11(3):325–7. doi: 10.1097/00007890-197103000-00013. [DOI] [PubMed] [Google Scholar]
- 11.Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez Franco GE, O’Neil TK, Litscher SJ, Urban-Piette M, Blank RD. Accuracy and precision of PIXImus densitometry for ex vivo mouse long bones: comparison of technique and software version. J Clin Densitom. 2004;7(3):326–33. doi: 10.1385/jcd:7:3:326. [DOI] [PubMed] [Google Scholar]
- 13.Lang DH, Sharkey NA, Lionikas A, et al. Adjusting data to body size: a comparison of methods as applied to quantitative trait loci analysis of musculoskeletal phenotypes. J Bone Miner Res. 2005;20(5):748–57. doi: 10.1359/JBMR.041224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belknap JK, Mitchell SR, O’Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behav Genet. 1996;26(2):149–60. doi: 10.1007/BF02359892. [DOI] [PubMed] [Google Scholar]
- 15.Cox A, Ackert-Bicknell CL, Dumont BL, et al. A new standard genetic map for the laboratory mouse. Genetics. 2009;182(4):1335–44. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupuis J, Siegmund D. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics. 1999;151:373–86. doi: 10.1093/genetics/151.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–6. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang JR, de Villena FP, McMillan L. Comparative analysis and visualization of multiple collinear genomes. BMC Bioinformatics. 2012;13(Suppl 3):S13. doi: 10.1186/1471-2105-13-S3-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grubb SC, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2014;42:D825–34. doi: 10.1093/nar/gkt1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verdugo RA, Farber CR, Warden CH, Medrano JF. Serious limitations of the QTL/microarray approach for QTL gene discovery. BMC Biol. 2010;8:96. doi: 10.1186/1741-7007-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein RF, Mitchell SR, Phillips TJ, Belknap JK, Orwoll ES. Quantitative trait loci affecting peak bone mineral density in mice. J Bone Miner Res. 1998;13(11):1648–56. doi: 10.1359/jbmr.1998.13.11.1648. [DOI] [PubMed] [Google Scholar]
- 23.Hunter DJ, Lange M, Snieder H, et al. Genetic contribution to renal function and electrolyte balance: a twin study. ClinSci (Lond) 2002;103(3):259–65. doi: 10.1042/cs1030259. [DOI] [PubMed] [Google Scholar]
- 24.Fleet JC, Schoch RD. Molecular mechanisms for regulation of intestinal calcium absorption by vitamin D and other factors. Crit Rev Clin Lab Sci. 2010;47(4):181–95. doi: 10.3109/10408363.2010.536429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SM, Riley EM, Meyer MB, et al. 1,25-Dihydroxyvitamin D3 Controls a Cohort of Vitamin D Receptor Target Genes in the Proximal Intestine That Is Enriched for Calcium-regulating Components. J Biol Chem. 2015;290(29):18199–215. doi: 10.1074/jbc.M115.665794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philipp S, Flockerzi V. Molecular characterization of a novel human PDZ domain protein with homology to INAD from Drosophila melanogaster. FEBS Lett. 1997;413(2):243–8. doi: 10.1016/s0014-5793(97)00877-6. [DOI] [PubMed] [Google Scholar]
- 27.Michel D, Arsanto JP, Massey-Harroche D, Beclin C, Wijnholds J, Le Bivic A. PATJ connects and stabilizes apical and lateral components of tight junctions in human intestinal cells. J Cell Sci. 2005;118(Pt 17):4049–57. doi: 10.1242/jcs.02528. [DOI] [PubMed] [Google Scholar]
- 28.Fujita H, Sugimoto K, Inatomi S, et al. Tight junction proteins claudin-2 and -12 are critical for vitamin D-dependent Ca2+ absorption between enterocytes. Mol Biol Cell. 2008;19(5):1912–21. doi: 10.1091/mbc.E07-09-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ackert-Bicknell CL, Karasik D, Li Q, et al. Mouse BMD quantitative trait loci show improved concordance with human genome-wide association loci when recalculated on a new, common mouse genetic map. J Bone Miner Res. 2010;25(8):1808–20. doi: 10.1002/jbmr.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Termine JD, Kleinman HK, Whitson SW, Conn KM, McGarvey ML, Martin GR. Osteonectin, a bone-specific protein linking mineral to collagen. Cell. 1981;26(1 Pt 1):99–105. doi: 10.1016/0092-8674(81)90037-4. [DOI] [PubMed] [Google Scholar]
- 31.Simonet WS, Lacey DL, Dunstan CR, et al. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–19. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 32.Fairbrother UL, Tanko LB, Walley AJ, Christiansen C, Froguel P, Blakemore AI. Leptin receptor genotype at Gln223Arg is associated with body composition, BMD, and vertebral fracture in postmenopausal Danish women. J Bone Miner Res. 2007;22(4):544–50. doi: 10.1359/jbmr.070114. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Lu W, Zhang L, et al. Trps1 differentially modulates the bone mineral density between male and female mice and its polymorphism associates with BMD differently between women and men. PLoS One. 2014;9(1):e84485. doi: 10.1371/journal.pone.0084485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J Biol Chem. 1998;273(41):26265–8. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 35.Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev Cell. 2004;6(6):801–13. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Lemos MC, Kotanko P, Christie PT, et al. A novel EXT1 splice site mutation in a kindred with hereditary multiple exostosis and osteoporosis. J Clin Endocrinol Metab. 2005;90(9):5386–92. doi: 10.1210/jc.2004-2520. [DOI] [PubMed] [Google Scholar]
- 37.Kim JJ, Lee HI, Park T, et al. Identification of 15 loci influencing height in a Korean population. J Hum Genet. 2010;55(1):27–31. doi: 10.1038/jhg.2009.116. [DOI] [PubMed] [Google Scholar]
- 38.Abrams SA, Copeland KC, Gunn SK, Gundberg CM, Klein KO, Ellis KJ. Calcium absorption, bone mass accumulation, and kinetics increase during early pubertal development in girls. J Clin Endocrinol Metab. 2000;85(5):1805–9. doi: 10.1210/jcem.85.5.6508. [DOI] [PubMed] [Google Scholar]
- 39.Bryant RJ, Wastney ME, Martin BR, et al. Racial differences in bone turnover and calcium metabolism in adolescent females. J Clin Endocrinol Metab. 2003;88(3):1043–7. doi: 10.1210/jc.2002-021367. [DOI] [PubMed] [Google Scholar]
- 40.Ensrud KE, Duong T, Cauley JA, et al. Low fractional calcium absorption increases the risk for hip fracture in women with low calcium intake. Study of Osteoporotic Fractures Research Group. Ann Intern Med. 2000;132(5):345–53. doi: 10.7326/0003-4819-132-5-200003070-00003. [DOI] [PubMed] [Google Scholar]
- 41.Volkman SK, Galecki AT, Burke DT, Miller RA, Goldstein SA. Quantitative trait loci that modulate femoral mechanical properties in a genetically heterogeneous mouse population. J Bone Miner Res. 2004;19(9):1497–505. doi: 10.1359/JBMR.040506. [DOI] [PubMed] [Google Scholar]
- 42.Beamer WG, Shultz KL, Donahue LR, et al. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res. 2001;16(7):1195–206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- 43.Beamer WG, Shultz KL, Coombs HF, 3rd, Horton LG, Donahue LR, Rosen CJ. Multiple quantitative trait loci for cortical and trabecular bone regulation map to mid-distal mouse chromosome 4 that shares linkage homology to human chromosome 1p36. J Bone Miner Res. 2012;27(1):47–57. doi: 10.1002/jbmr.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muir AM, Ren Y, Butz DH, et al. Induced ablation of Bmp1 and Tll1 produces osteogenesis imperfecta in mice. Hum Mol Genet. 2014;23(12):3085–101. doi: 10.1093/hmg/ddu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu HQ, Pereira EF, Bruno JP, Pellicciari R, Albuquerque EX, Schwarcz R. The astrocyte-derived alpha7 nicotinic receptor antagonist kynurenic acid controls extracellular glutamate levels in the prefrontal cortex. J Mol Neurosci. 2010;40(1–2):204–10. doi: 10.1007/s12031-009-9235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alfonso M, Duran R, Arufe MC. Effect of excitatory amino acids on serum TSH and thyroid hormone levels in freely moving rats. Horm Res. 2000;54(2):78–83. doi: 10.1159/000053236. [DOI] [PubMed] [Google Scholar]
- 47.Arufe MC, Duran R, Perez-Vences D, Alfonso M. Endogenous excitatory amino acid neurotransmission regulates thyroid-stimulating hormone and thyroid hormone secretion in conscious freely moving male rats. Endocrine. 2002;17(3):193–7. doi: 10.1385/ENDO:17:3:193. [DOI] [PubMed] [Google Scholar]
- 48.Porcu E, Medici M, Pistis G, et al. A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet. 2013;9(2):e1003266. doi: 10.1371/journal.pgen.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar V, Prasad R. Thyroid hormones stimulate calcium transport systems in rat intestine. Biochimica et Biophysica Acta-Molecular Basis of Disease. 2003;1639(3):185–94. doi: 10.1016/j.bbadis.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 50.Wojcicka A, Bassett JH, Williams GR. Mechanisms of action of thyroid hormones in the skeleton. Biochim Biophys Acta. 2013;1830(7):3979–86. doi: 10.1016/j.bbagen.2012.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.