Abstract
Vitamin D and intact parathyroid hormone (iPTH) concentrations differ between individuals of African and European descent and may play a role in observed racial differences in bone mineral density (BMD). These findings suggest that mapping by admixture linkage disequilibrium (MALD) may be informative for identifying genetic variants contributing to these ethnic disparities. Admixture mapping was performed for serum 25-hydroxyvitamin D, 1,25-dihydroxyvitamin D, vitamin D-binding protein (VDBP), bioavailable vitamin D, and iPTH concentrations and computed tomography measured thoracic and lumbar vertebral volumetric BMD in 552 unrelated African Americans with type 2 diabetes from the African American-Diabetes Heart Study. Genotyping was performed using a custom Illumina ancestry informative marker (AIM) panel. For each AIM, the probability of inheriting 0, 1, or 2 copies of a European-derived allele was determined. Non-parametric linkage analysis was performed by testing for association between each AIM using these probabilities among phenotypes, accounting for global ancestry, age, and gender. Fine-mapping of MALD peaks was facilitated by genome-wide association study (GWAS) data. VDBP levels were significantly linked in proximity to the protein coding locus (rs7689609, LOD=11.05). Two loci exhibited significant linkage signals for 1,25-dihydroxyvitamin D on 13q21.2 (rs1622710, LOD=3.20) and 12q13.2 (rs11171526, LOD=3.10). iPTH was significantly linked on 9q31.3 (rs7854368, LOD=3.14). Fine-mapping with GWAS data revealed significant known (rs7041 with VDBP, P=1.38×10−82) and novel (rs12741813 and rs10863774 with VDBP, P<6.43×10−5) loci with plausible biological roles. Admixture mapping in combination with fine-mapping has focused efforts to identify loci contributing to ethnic differences in vitamin D-related traits.
Keywords: epidemiology, human association studies, intact parathyroid hormone, Vitamin D, Bone mineral density
1. Introduction
African Americans have lower circulating 25-hydroxyvitamin D and higher intact parathyroid hormone (iPTH) concentrations than European Americans. (11–13, 20, 27) Although these phenomena would be expected to lead to diminished bone synthesis and excessive breakdown, significantly higher bone mineral density (BMD) and lower rates of osteoporosis are seen in African Americans. (2, 10, 21) The recently described lower vitamin D-binding protein (VDBP) levels in African Americans likely counter effects of reduced 25-hydroxyvitamin D concentration. In addition, dietary calcium ingestion is lower in African Americans than European Americans, along with lower risk for calcium containing kidney stones and calcified atherosclerotic plaque. (3, 7, 25, 32) These findings suggest population-specific differences in calcium homeostasis.
Hormones impacting BMD likely have different biologic effects between population groups. For example, higher vitamin D concentrations after supplementation were paradoxically associated with lower BMD and higher fracture rates in African American women in the Women’s Health Initiative. (4) This finding stands in stark contrast to observations in populations of European ancestry. African Americans are also relatively resistant to the catabolic effects of iPTH on bone. (5) The underlying causes of ethnic differences in regulation of vitamin D and iPTH concentrations are poorly understood and likely reflect a complex interplay between genetic and environmental factors.
We performed a mapping by admixture linkage disequilibrium (MALD) analysis in 552 African Americans to determine genomic regions contributing to ethnic differences in serum vitamin D and iPTH concentrations as well as BMD. MALD is a gene-mapping tool used to identify genomic regions associated with differences in prevalence and/or distribution of a trait between two or more ancestral populations. (23) The premise is that if a genetic variant underlies ethnic differences in disease, it will be easier to map the location of that variant in a recently admixed population relative to panmictic ancestral populations. (29) The proportion of alleles at the marker locus that have ancestry from the high-risk population will be increased in affected individuals more than expected by chance, assuming no other influential evolutionary forces (genetic selection or drift). MALD has identified genes and genomic regions implicated in several common diseases. (16, 18, 31, 35)
2. Materials and Methods
2.1. Study participants
Self-reported and unrelated African Americans with type 2 diabetes (T2D) were recruited from internal medicine clinics and community advertising in the African American-Diabetes Heart Study (AA-DHS). Participant examinations were conducted in the Clinical Research Unit of Wake Forest School of Medicine (WFSM) and included interviews for medical history and health behaviors, anthropometric measures, resting blood pressure, 12-lead electrocardiography, fasting blood sampling, and spot urine collection for albumin:creatinine ratio. T2D was defined as a clinical diagnosis after 30 years of age in the absence of diabetic ketoacidosis. The study was approved by the WFSM Institutional Review Board and performed according to the ethical principles defined in the Declaration of Helsinki. All participants provided written informed consent.
2.2. Assays
Measures of 25-hydroxyvitamin D were performed by Quest Diagnostics Nichols Institute (San Juan Capistrano, CA) (8) and LabCorp using liquid chromatography and mass spectrometry and an immunochemiluminometric assay performed on the DiaSorin LIASON® instrument, respectively, with a high paired sample concordance (n=14, R2=0.92). Measures of 1,25-dihydroxyvitamin D were performed at Quest (15) or LabCorp® using liquid chromatography mass spectrometry and column chromatography, radioimmunoassay, respectively. iPTH was measured at LabCorp (Burlington, NC) using an electrochemiluminescence immunoassay.
VDBP was measured in EDTA plasma samples that had been continuously stored at −80°C without thawing since collection at the baseline visit. Frozen plasma was thawed in a 37°C water bath for 15 minutes, placed on ice, and then centrifuged at 1700 × g (2800 rpm) for 30 minutes at 4°C. VDBP was determined using a Quantikine® Human Vitamin D BP ELISA (cat. no. DDBP0; R&D Systems; Minneapolis, MN) according to manufacturer’s instructions. Serum was pre-diluted 1:2000 or greater in some cases in calibrator diluent before assay. Intra-assay and inter-assay coefficients of variation were <10%. All assays were performed using a single lot of reagents and calibrators at WFSM. Bioavailable vitamin D (BAVD) was computed as described by Powe et al. (27) using dissociation constants for the VDBP genotype variants as described by Arnaud and Constans. (1)
2.3. Bone imaging
Single and multidetector computed tomography (CT) systems incorporating a standardized scanning protocol based on the National Heart Lung and Blood Institute’s Multi-Ethnic Study of Atherosclerosis (MESA) were performed in all participants. Quantitative CT for trabecular vertebral volumetric BMD (mg/cm3) in the thoracic spine (tBMD; T8-T11) and lumbar spine (lBMD; T12-L3) were measured using QCT-5000 volumetric software with a calcium calibration phantom included in each participant’s chest and abdominal CT exam (Image Analysis; Columbia, KY). (30) Coefficients of variation for these measures were <1% for tBMD and lBMD and in sequential studies performed in the same individual the precision error was 2.3%.
2.4. Genotyping
DNA was extracted from peripheral blood using the PureGene system (Gentra Systems, Minneapolis, MN). The MALD study was based on the streamlined panel of 1,509 ancestry informative marker (AIM) single nucleotide polymorphisms (SNPs) genotyped using the Illumina African American admixture panel covering all 22 autosomes and chromosome X. Genome-wide association study (GWAS) data was obtained from the Illumina Omni5 array inclusive of exome chip content. All genotyping was performed in the Center for Genomics and Personalized Medicine Research at WFSM.
2.5. Quality control
Quality control checks were similar for the MALD and GWAS analyses. For MALD, six individuals with call rates <90% and one individual with a heterozygosity score outside of the mean ±4 times the standard error interval were excluded. Twenty-nine SNPs were flagged with Hardy-Weinberg equilibrium p-values <10−3 and two additional SNPs were flagged because their call rate was <98% (none of these SNPs showed a significant result). There was no indication of first-degree familial relationships in the analyzed dataset, i.e. the estimated kinship coefficient ranged between 0 and 0.13. For GWAS, twelve individuals were excluded from the analyses: 6 had call rates <90%, 2 had discordant self-reported and genetically determined sex, 1 had heterozygosity score outside of the mean ±4 times the standard error interval, 2 had the same sample identifier and 1 had 100% European ancestry. We used a classification scheme to rank SNPs and prioritize association results. Similar to the MALD analysis, the scheme was based on the estimated minor allele frequency, the Hardy-Weinberg equilibrium p-value, and the call rate. All SNPs were analyzed; however, higher confidence was placed on common variants with a Hardy-Weinberg equilibrium p-value ≤10−4 and a call rate ≥95%. SNPs with these qualifications also served as the basis for our imputation efforts. SNPs with a minor allele frequency between 1 and 5% that met the Hardy-Weinberg equilibrium and call rate thresholds were also included in imputation.
2.6. Statistical methods
The methodology for the MALD analysis has been described in detail. (6) Briefly, we computed the probability of inheriting 0, 1, or 2 alleles from a specific ancestral population at each AIM. These probabilities are akin to identity by descent probabilities. Therefore, association tests between them and the outcome of interest accounting for global ancestry is a valid test of linkage in the non-parametric linkage framework. (22, 34) This model can be fitted easily using standard statistical packages, facilitating inclusion of covariates and interaction effects. We use logarithm of the odds (LOD) scores to present evidence against the null hypothesis to reinforce the idea that we are performing linkage analysis. Analyses were run using 25-hydroxyvitamin D, 1,25-dihydroxyvitamin D, VDBP, BAVD, iPTH, tBMD, and lBMD as continuous traits. Age, gender, and genome-wide ancestry were included as covariates. For 25-hydroxyvitamin D, 1,25-dihydroxyvitamin D, and BAVD, analyses were conducted by assay site (Quest or LabCorp) and combined using a meta-analysis approach.
GWAS analyses were adjusted for age, gender and principal components. Each SNP was tested for association under the additive genetic model. Imputation was performed using IMPUTE2 (15) using the 1000 genomes integrated reference panel. Statistical analyses were based on the dosage genotype, i.e. the expected count of minor alleles at each marker. The models were fitted with SNPTEST adjusting for the same covariates as in the GWAS.
2.7. Significant effect and correction for multiple testing
We excluded chromosome X from our analyses; therefore, the MALD analyses were run on 1,423 SNPs. A strict Bonferroni correction would place the significance threshold at 3.5×10−5 for a two-sided test, a conservative threshold corresponding to a LOD score of 3.7. We chose to prioritize AIMs that reached a LOD score of 2.5 corresponding to an alpha level of 7.0×10−4. This approach can be viewed as somewhat conservative for identifying suggestive evidence of linkage. Results from the GWAS were evaluated in the context of peaks, i.e. LOD-1 confidence interval based on the next available AIM, lacking significant evidence of linkage. Within this interval, a strict Bonferroni correction for the number of directly genotyped markers examined was used to gauge statistical significance. This approach provides a conservative estimate of significance and no further corrections were made to account for the number of correlated traits examined.
3. Results
3.1. Demographic and clinical characteristics
Demographic and clinical characteristics of the 552 participants with relevant phenotype data are presented in Table 1. This subset of individuals was 49% female, and 63.6% were current or former smokers. Participants had a mean (standard deviation) age of 55.5 (9.6) years, diabetes duration 10.0 (7.8) years, hemoglobin A1c 8.1 (2.0) %, fasting serum glucose 149.9 (64.9) mg/dl, HDL-cholesterol 47.9 (13.5) mg/dl, and LDL-cholesterol 106.80 (37.95) mg/dl. The mean (SD) 25-hydroxyvitamin D concentration was 20.3 (11.9) ng/mL and 1,25-dihydroxyvitamin D was 46.2 (17.1) pg/mL. These did not differ by gender (P>0.12). In contrast, VDBP levels were significantly higher in males (P=0.023) while iPTH was significantly higher in females (P=0.0030). tBMD and lBMD were 205.2 (52.1) and 179.5 (46.4) mg/cm3, respectively, and did not differ between genders (P>0.23).
Table 1.
Clinical and laboratory characteristics in the African American-Diabetes Heart Study sample.
| Variable | Men (n=238)
|
Women (n=314)
|
Combined (n=552)
|
p-value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Median | Mean | SD | Median | Mean | SD | Median | ||
| Age (years) | 55.6 | 10.0 | 55.0 | 55.4 | 9.3 | 55.0 | 55.5 | 9.6 | 55.0 | 0.80 |
| BMI (kg/m2) | 32.9 | 7.5 | 31.6 | 37.8 | 9.4 | 36.8 | 35.6 | 8.9 | 34.0 | <0.00010 |
| 25 hydroxyvitamin D (ng/mL) | 19.5 | 10.4 | 18.0 | 20.9 | 12.9 | 17.0 | 20.3 | 11.9 | 18.0 | 0.22 |
| 1,25 dihydroxyvitamin D (pg/mL) | 44.7 | 17.3 | 42.0 | 47.3 | 17.0 | 45.3 | 46.2 | 17.1 | 43.6 | 0.12 |
| Vitamin D binding protein (μg/mL) | 94.3 | 79.1 | 58.8 | 79.7 | 61.7 | 52.5 | 86.2 | 70.3 | 55.5 | 0.023 |
| Intact parathyroid hormone (pg/mL) | 50.8 | 26.6 | 45.0 | 58.9 | 31.8 | 51.0 | 55.4 | 29.9 | 48.0 | 0.0030 |
| Thoracic vBMD (mg/cm3) | 202.0 | 49.1 | 199.0 | 207.8 | 54.3 | 205.3 | 205.2 | 52.1 | 204.5 | 0.23 |
| Lumbar vBMD (mg/cm3) | 178.9 | 43.6 | 178.8 | 179.9 | 48.5 | 176.8 | 179.5 | 46.4 | 178.4 | 0.82 |
| Diabetes duration (years) | 10.0 | 8.1 | 8.0 | 10.0 | 7.6 | 8.0 | 10.0 | 7.8 | 8.0 | 1.00 |
| Hemoglobin A1c (HbA1c, %) | 8.3 | 2.0 | 7.9 | 8.0 | 2.0 | 7.5 | 8.1 | 2.0 | 7.7 | 0.093 |
| High sensitivity C-reactive protein (mg/dL) | 0.76 | 1.13 | 0.31 | 1.30 | 1.94 | 0.69 | 1.06 | 1.65 | 0.50 | 0.00050 |
| Fasting glucose (mg/dL) | 159.7 | 73.0 | 139.0 | 142.1 | 56.5 | 129.0 | 149.9 | 64.9 | 134.0 | 0.0029 |
| LDL-cholesterol (mg/dL) | 104.29 | 38.68 | 101.00 | 108.94 | 37.30 | 100.00 | 106.88 | 37.95 | 100.00 | 0.18 |
| HDL-cholesterol (mg/dL) | 44.7 | 12.2 | 43.0 | 50.4 | 13.9 | 48.0 | 47.9 | 13.5 | 46.0 | <0.00010 |
| Triglycerides (mg/dl) | 139.7 | 170.7 | 108.0 | 118.4 | 88.0 | 97.5 | 127.9 | 131.6 | 102.0 | 0.076 |
| ACE inhibitor use (%) | 43.58% | 38.24% | 40.98% | 0.23 | ||||||
| Current smoker (%) | 27.06% | 20.96% | 24.52% | 0.0045 | ||||||
| Past Smoker (%) | 40.83% | 30.51% | 36.95% | 0.00080 | ||||||
| Hypertension (%) | 83.49% | 84.93% | 84.06% | 0.66 | ||||||
| Lipid-lowering medications (%) | 48.62% | 51.10% | 48.16% | 0.59 | ||||||
Abbreviations: ACE, angiotensin converting enzyme inhibitor; BMI, body mass index; LDL, low density lipoprotein; HDL, high density lipoprotein; CAC, coronary artery calcified plaque; SD, standard deviation; vBMD, volumetric bone mineral density.
3.2. Ancestry proportion distribution
The allele frequency of each AIM estimated in the Yoruba and CEU populations were supplied as prior probabilities in the estimation process. The average proportion of genome-wide African ancestry in this sample was 75 (±15.8) %.
3.3. Regional admixture mapping and fine-mapping results
Admixture mapping was performed using vitamin D, iPTH, and BMD as continuous outcomes. We considered the additive, dominant, and recessive models, allowing us to effectively reduce the models described to 1 degree of freedom tests, with potential improvement in power. (9) Adjusted p-values computed using the maximum of the test statistic observed with the dominant, recessive, and additive models appeared to be one order of magnitude lower than their unadjusted counterparts. Loci with significant (LOD>3.00) and suggestive (LOD>2.5) evidence of linkage as well as fine-mapping results are summarized in Table 2.
Table 2.
Summary of SNPs linked (LOD>2.50) in the MALD analysis of vitamin D concentrations, parathyroid hormone concentrations, and bone mineral density with fine-mapping from GWAS data in the AA-DHS sample.
| Allele frequency | MALD | GWAS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP | Gene | Alleles1 | Observed2 | CEU3 | YRI4 | Model | LOD Score | n5 | Beta±SE | P-value6 | R27 |
| 1,25-dihydroxyvitamin D | ||||||||||||
| 13q21.2 | rs1622710 | A/G | 0.73 | 0.05 | 0.83 | Recessive | 3.20 | |||||
| rs183787871 | PCDH20/- | C/T | 0.01 | 34937 | 2.07±0.53 | 9.12E-05 | 0.06 | |||||
| 12q13.2 | rs11171526 | T/A | 0.34 | 0.97 | 0.23 | Additive | 3.10 | |||||
| rs10783612 | CALCOCO1/HOXC13 | C/T | 0.42 | 39571 | −0.34±0.08 | 2.52E-05 | 0.05 | |||||
| 4p15.32 | rs2872795 | C/T | 0.23 | 0.83 | 0.14 | Additive | 2.82 | |||||
| rs144095555 | FAM184B | C/T | 0.01 | 25896 | −0.39±0.11 | 2.18E-04 | 0.001 | |||||
| Vitamin D Binding Protein | ||||||||||||
| 4q13.3 | rs7689609 | A/G | 0.14 | 0.93 | 0.00 | Additive | 11.05 | |||||
| rs7041 | GC | C/A | 0.15 | 4472 | 0.52±0.02 | 1.38E-82 | 0.52 | |||||
| 2q35 | rs16859382 | A/G | 0.38 | 1.00 | 0.19 | Recessive | 2.64 | |||||
| rs719325 | SLC4A3/- | T/C | 0.33 | 145403 | −0.09±0.02 | 2.91E-05 | 0.03 | |||||
| 1q32.2 | rs979698 | A/G | 0.76 | 0.18 | 0.9 | Additive | 2.54 | |||||
| rs1284852 | FLVCR1/VASH2 | A/G | 0.61 | 4930 | −0.079±0.021 | 1.33E-04 | 0.03 | |||||
| rs4951471 | FLVCR1/VASH2 | A/G | 0.61 | −0.079±0.021 | 1.33E-04 | 0.03 | ||||||
| Bioavailable Vitamin D | ||||||||||||
| 17q12 | rs739753 | A/T | 0.25 | 0.81 | 0.15 | Dominant | 2.58 | |||||
| rs4375714 | FAM134C | T/C | 0.46 | 26458 | −0.06±0.01 | 1.03E-04 | 0.03 | |||||
| Intact parathyroid hormone | ||||||||||||
| 9q31.3 | rs7854368 | A/G | 0.29 | 0.74 | 0.17 | Additive | 3.14 | |||||
| rs10125548 | KLF4/ACTL7B | A/G | 0.70 | 7395 | −0.13±0.03 | 7.81E-05 | 0.03 | |||||
| 1q42.2 | rs701185 | A/C | 0.22 | 0.71 | 0.06 | Recessive | 2.69 | |||||
| rs623448 | G/A | 0.07 | 4006 | −0.22±0.06 | 1.46E-04 | 0.02 | ||||||
| Lumbar Volumetric Bone Mineral Density | ||||||||||||
| 15q11.2 | rs2719890 | A/G | 0.73 | 0.03 | 0.88 | Dominant | 2.52 | |||||
| rs8040786 | NDN/PWRN2 | A/G | 0.06 | 927 | 0.61±0.18 | 5.90E-4 | 0.01 | |||||
Reference/other allele,
AA-DHS (nmax=552),
CEPH (Utah Residents with Northern and Western European Ancestry) (n=60),
Yoruba in Ibadan, Nigeria (n=60),
number of variants (directly genotyped and imputed) in the LOD-1 confidence interval,
Additive model,
Variation in phenotype explained by the genetic variant
The most significant linkage signal observed was for VDBP on 4p13-q21.22. This region spanned nine consecutive AIMs with significant LOD scores (LOD>3.0, Figure 1C) under the additive model peaking at a LOD score of 11.05 for AIM rs7689609 (Supplementary Table 1). The group-specific component gene (GC), which encodes the VDBP, resides within the LOD-1 confidence interval and is located 500kb proximal to the peak signal (rs7689609). Fine-mapping of the support interval (>5.24Mb) identified SNP rs7041 as statistically significant (P=1.38×10−82), consistent with previous report. (24) Two additional suggestive linkage signals for VDBP were also detected on at 2q35 and 1q32.2 (Figure 1C). Fine-mapping of these peaks identified only nominally significant association signals, i.e. P>0.05 after strict Bonferroni correction for the number of SNPs contained within the interval (Table 2).
Figure 1.
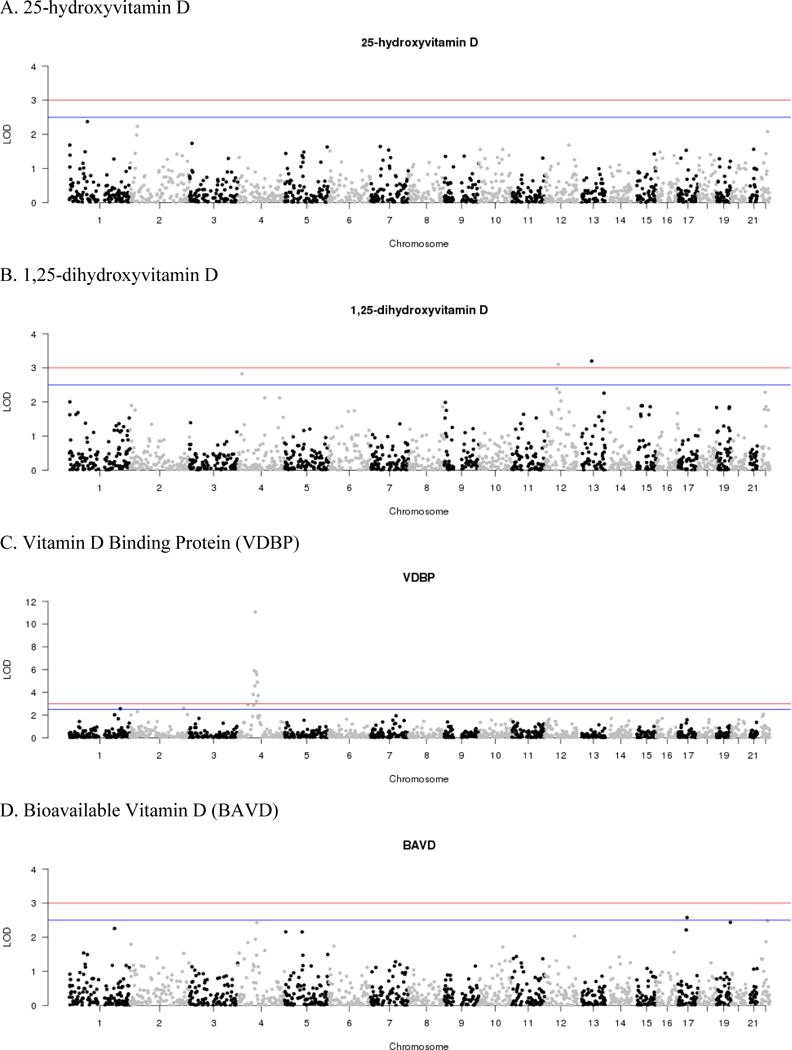
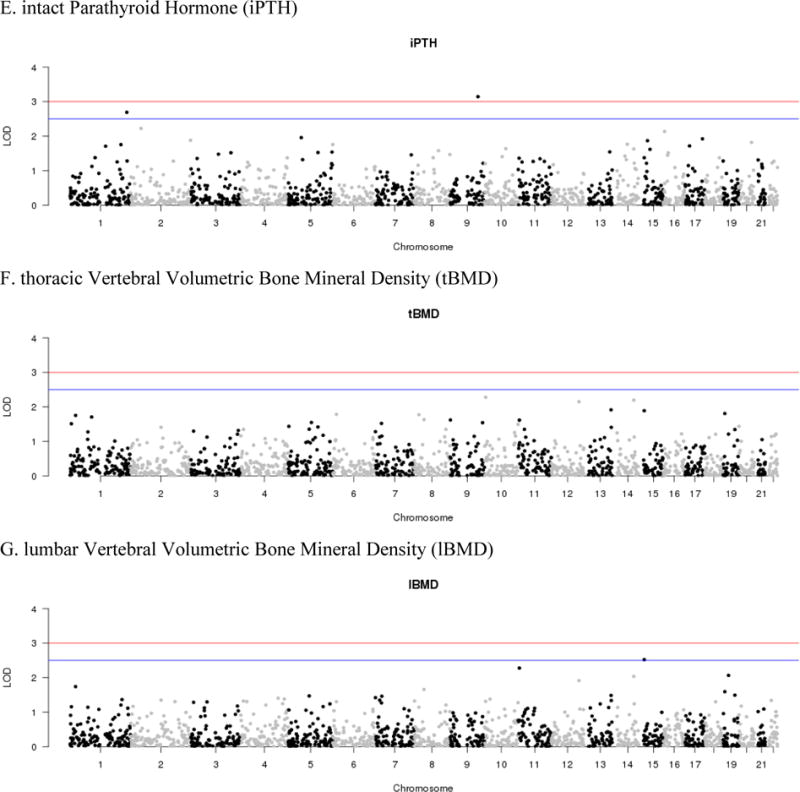
MALD results for the AA-DHS. LOD scores are plotted by chromosome for the seven phenotypes examined: A. 25-hydroxyvitamin D, B. 1,25-dihydroxyvitamin D, C. Vitamin D Binding Protein (VDBP), D. Bioavailable Vitamin D (BAVD), E. intact Parathyroid Hormone (iPTH), F. thoracic vertebral volumetric bone mineral density (tBMD), G. lumbar vertebral volumetric bone mineral density (lBMD)
Two significant linkage signals were observed for 1,25-dihydroxyvitamin D (Figure 1B). The most significant signal was on 13q21.2 (rs1622710, LOD=3.20) along with an additional signal on 12q13.2 (rs11171526, LOD=3.10). Among the confidence interval of these peaks, fine-mapping using GWAS data failed to detect signals of association that survived strict Bonferroni correction. One additional suggestive linkage peaks was observed for 1,25-dihydroxyvitamin D on 4p15.32 (Table 2).
For iPTH, a single significant linkage peak was observed on 9q31.3 (rs7854368, LOD=3.14; Figure 1C). Although not surviving strict Bonferroni correction, rs10125548 was the most associated variant in the region (P=7.81×10−5). Notably, this SNP is located distally to the Kruppel-like factor 4 gene (KLF4) which has been implicated in control of bone homeostasis. (17) A second peak of suggestive significance was observed on 1q42.2 (Table 2). Among the additional phenotypes examined, suggestive evidence of linkage was observed for BAVD and lBMD; however, no signals were observed for 25-hydroxyvitamin D and tBMD (LOD<2.5) (Figure 1).
4. Discussion
Four genomic regions were identified using admixture mapping that were significantly linked (LOD>3.0) with 1,25-dihydoroxyvitamin D (n=2), VDBP (n=1) and iPTH (n=1) concentrations in African Americans, as well as six regions with suggestive linkage (LOD>2.5) among the seven traits examined. Nearly all published reports demonstrate that individuals of African ancestry have lower vitamin D and VDBP and higher iPTH levels than individuals of European ancestry. Rates of osteoporosis, calcified atherosclerotic plaque, and nephrolithiasis also differ markedly between these ancestral groups, possibly contributing to these findings.
The most significant signal observed from the MALD analysis was linkage of VDBP to 4q13.3 (rs7689609, LOD=11.05). Fine-mapping of this peak, which spanned nine consecutive markers, using GWAS data confirmed the association of a previously identified missense (threonine to glycine) variant (24), rs7041 (P=1.38×10−82) with comparatively nominal evidence of association with 25-hydroxyvitamin D. Among the additional two peaks with suggestive evidence of linkage to VDBP, fine-mapping of the peak on 1q32.2 (rs979698, LOD=2.54) revealed two nominally significant correlated (r2=1.00) association signals (P>0.05 after Bonferroni correction for 4930 tests). SNPs rs1284852 and rs4951471 (P=1.33×10−4) are located distally to FLVCR1, a protein involved in heme transportation. (28) Notably, the peak observed for VDBP is in close proximity to a peak for coronary artery calcification (rs7530895, LOD=2.9) that was previously observed in this population. (6)
Among the seven phenotypes examined, only 1,25-dihydroxyvitamin D exhibited multiple significant linkage signals: 13q21.2 (LOD=3.20) and 12q13.2 (LOD=3.10). However, fine-mapping using data from the GWAS failed to reveal significant associations that survived multiple correction testing (P>2.52×10−5). This can partially be attributed to broad genomic interval encompasses by these linkage peaks, i.e. 6.7–7.8Mb. Interestingly, the linkage interval observed on 12q13.2 did encompass the calcium binding and coiled-coil domain 1 gene (CALCOCO1) with a distal SNP showing nominal evidence of association (P=2.52×10−5). This observation is interesting in that the gene product CoCoA is a secondary activator of β-catenin signaling (33), for which 1,25-dihydroxyvitamin D has been observed to function as a repressor. (19) The REGARDS Study (14), a population-based investigation of stroke incidence in African American and European American, does have 1,25-dihydroxyvitamin D and genetic data from the Illumina Human Exome array; however, the variants associated or suitable proxies from AA-DHS were not available. Further efforts to replicate these findings are limited by the fact that few studies outside of the AA-DHS have large scale genetic data and comparable vitamin D-related phenotypes on African Americans with or without diabetes.
Admixture mapping is not without limitations. The AIMs selected for this study are not completely informative as the power of the approach is maximized when the average ancestry proportion is close to 50%; our proportions were estimated to be approximately 75% and 25% in this sample for African and Europeans, respectively. In addition this method depends on a probabilistically calculation of the European ancestry at each locus. Nonetheless, admixture mapping remains a valuable and proven gene-mapping technique and combining admixture mapping and association tests can increase statistical power.(26)
In conclusion, admixture mapping identified novel genomic regions which contribute to ethnic differences in iPTH and 1,25-dihyroxyvitamin D concentrations between African Americans and European Americans. Fine-mapping of both significant and suggestive linkage intervals implicated known genes with biology relevant to vitamin D metabolism, e.g. β-catenin signaling and bone homeostasis, as well as novel loci that have the potential to improve our understanding of ethnic-specific differences observed among related vitamin D traits.
Highlights.
Admixture mapping can be used to determine genomic regions contributing to observed ethnic differences in serum vitamin D, iPTH, and BMD.
Novel loci linked to 1,25-dihydroxyvitamin D were identified on 13q21.2 (rs1622710, LOD=3.20) and 12q13.2 (rs11171526, LOD=3.10).
iPTH was significantly linked to 9q31.3 (rs7854368, LOD=3.14) proximal to a regulatory protein of sodium-phosphate cotransport (AKAP).
Fine-mapping of admixture mapping regions has focused efforts to identify loci contributing to ethnic differences in vitamin D-related traits.
Acknowledgments
The investigators acknowledge the cooperation of our Diabetes Heart Study (DHS) and AA-DHS participants and study recruiter Cassandra Bethea. This work was supported by NIH RO1 DK071891 (BIF), RO1 HL092301 (DWB) and the General Clinical Research Center of Wake Forest School of Medicine MO1-RR-07122.
Abbreviations
- iPTH
intact parathyroid hormone
- BMD
bone mineral density
- VDBP
vitamin D-binding protein
- MALD
mapping by admixture linkage disequilibrium
- T2D
type 2 diabetes
- AA-DHS
African American-Diabetes Heart Study
- WFSM
Wake Forest School of Medicine
- BAVD
bioavailable vitamin D
- CT
computed tomography
- AIM
ancestry informative marker
- GWAS
genome-wide association study
- tBMD
thoracic volumetric bone mineral density
- lBMD
lumbar volumetric bone mineral density
- LOD
logarithm of the odds
- HHAT
hedgehog acyltransferase gene
- CAMK1G
calcium/calmodulin-dependent protein kinase 1G gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nicholette D. Palmer, Email: nallred@wakehealth.edu.
Jasmin Divers, Email: jdivers@wakehealth.edu.
Lingyi Lu, Email: llu@wakehealth.edu.
Thomas C. Register, Email: register@wakehealth.edu.
J. Jeffrey Carr, Email: j.jeffrey.carr@vanderbilt.edu.
Pamela J. Hicks, Email: panderso@wakehealth.edu.
S. Carrie Smith, Email: suscsmit@wakehealth.edu.
Jianzhao Xu, Email: jixu@wakehealth.edu.
Suzanne E. Judd, Email: sejudd@uab.edu.
Marguerite R. Irvin, Email: irvinr@uab.edu.
Orlando M. Gutierrez, Email: ogutierr@uab.edu.
Donald W. Bowden, Email: dbowden@wakehealth.edu.
Lynne E. Wagenknecht, Email: lwgnkcht@wakehealth.edu.
Carl D. Langefeld, Email: clangefe@wakehealth.edu.
Barry I. Freedman, Email: bfreedma@wakehealth.edu.
References
- 1.Arnaud J, Constans J. Affinity differences for vitamin D metabolites associated with the genetic isoforms of the human serum carrier protein (DBP) Hum Genet. 1993;92:183–8. doi: 10.1007/BF00219689. [DOI] [PubMed] [Google Scholar]
- 2.Barrett-Connor E, Siris ES, Wehren LE, Miller PD, Abbott TA, Berger ML, Santora AC, Sherwood LM. Osteoporosis and fracture risk in women of different ethnic groups. J Bone Miner Res. 2005;20:185–94. doi: 10.1359/JBMR.041007. [DOI] [PubMed] [Google Scholar]
- 3.Bild DE, Detrano R, Peterson D, Guerci A, Liu K, Shahar E, Ouyang P, Jackson S, Saad MF. Ethnic differences in coronary calcification: the Multi-Ethnic Study of Atherosclerosis (MESA) Circulation. 2005;111:1313–20. doi: 10.1161/01.CIR.0000157730.94423.4B. [DOI] [PubMed] [Google Scholar]
- 4.Cauley JA, Danielson ME, Boudreau R, Barbour KE, Horwitz MJ, Bauer DC, Ensrud KE, Manson JE, Wactawski-Wende J, Shikany JM, Jackson RD. Serum 25-hydroxyvitamin D and clinical fracture risk in a multiethnic cohort of women: the Women’s Health Initiative (WHI) J Bone Miner Res. 2011;26:2378–88. doi: 10.1002/jbmr.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosman F, Morgan DC, Nieves JW, Shen V, Luckey MM, Dempster DW, Lindsay R, Parisien M. Resistance to bone resorbing effects of PTH in black women. J Bone Miner Res. 1997;12:958–66. doi: 10.1359/jbmr.1997.12.6.958. [DOI] [PubMed] [Google Scholar]
- 6.Divers J, Palmer ND, Lu L, Register TC, Carr JJ, Hicks PJ, Hightower RC, Smith SC, Xu J, Cox AJ, Hruska KA, Bowden DW, Lewis CE, Heiss G, Province MA, Borecki IB, Kerr KF, Chen YD, Palmas W, Rotter JI, Wassel CL, Bertoni AG, Herrington DM, Wagenknecht LE, Langefeld CD, Freedman BI. Admixture mapping of coronary artery calcified plaque in African Americans with type 2 diabetes mellitus. Circ Cardiovasc Genet. 2013;6:97–105. doi: 10.1161/CIRCGENETICS.112.964114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman BI, Hsu FC, Langefeld CD, Rich SS, Herrington DM, Carr JJ, Xu J, Bowden DW, Wagenknecht LE. The impact of ethnicity and sex on subclinical cardiovascular disease: the Diabetes Heart Study. Diabetologia. 2005;48:2511–8. doi: 10.1007/s00125-005-0017-2. [DOI] [PubMed] [Google Scholar]
- 8.Freedman BI, Wagenknecht LE, Hairston KG, Bowden DW, Carr JJ, Hightower RC, Gordon EJ, Xu J, Langefeld CD, Divers J. Vitamin d, adiposity, and calcified atherosclerotic plaque in african-americans. J Clin Endocrinol Metab. 2010;95:1076–83. doi: 10.1210/jc.2009-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum Hered. 2002;53:146–52. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 10.George A, Tracy JK, Meyer WA, Flores RH, Wilson PD, Hochberg MC. Racial differences in bone mineral density in older men. J Bone Miner Res. 2003;18:2238–44. doi: 10.1359/jbmr.2003.18.12.2238. [DOI] [PubMed] [Google Scholar]
- 11.Gutierrez OM, Farwell WR, Kermah D, Taylor EN. Racial differences in the relationship between vitamin D, bone mineral density, and parathyroid hormone in the National Health and Nutrition Examination Survey. Osteoporos Int. 2011;22:1745–53. doi: 10.1007/s00198-010-1383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris SS, Dawson-Hughes B. Seasonal changes in plasma 25-hydroxyvitamin D concentrations of young American black and white women. Am J Clin Nutr. 1998;67:1232–6. doi: 10.1093/ajcn/67.6.1232. [DOI] [PubMed] [Google Scholar]
- 13.Harris SS, Soteriades E, Coolidge JA, Mudgal S, Dawson-Hughes B. Vitamin D insufficiency and hyperparathyroidism in a low income, multiracial, elderly population. J Clin Endocrinol Metab. 2000;85:4125–30. doi: 10.1210/jcem.85.11.6962. [DOI] [PubMed] [Google Scholar]
- 14.Howard VJ, Cushman M, Pulley L, Gomez CR, Go RC, Prineas RJ, Graham A, Moy CS, Howard G. The reasons for geographic and racial differences in stroke study: objectives and design. Neuroepidemiology. 2005;25:135–43. doi: 10.1159/000086678. [DOI] [PubMed] [Google Scholar]
- 15.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, Fink NE, Sadler JH, Weir MR, Abboud HE, Adler SG, Divers J, Iyengar SK, Freedman BI, Kimmel PL, Knowler WC, Kohn OF, Kramp K, Leehey DJ, Nicholas SB, Pahl MV, Schelling JR, Sedor JR, Thornley-Brown D, Winkler CA, Smith MW, Parekh RS. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–92. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JH, Kim K, Youn BU, Lee J, Kim I, Shin HI, Akiyama H, Choi Y, Kim N. Kruppel-like factor 4 attenuates osteoblast formation, function, and cross talk with osteoclasts. J Cell Biol. 2014;204:1063–74. doi: 10.1083/jcb.201308102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS, Berns JS, Briggs W, Cho ME, Dart RA, Kimmel PL, Korbet SM, Michel DM, Mokrzycki MH, Schelling JR, Simon E, Trachtman H, Vlahov D, Winkler CA. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–84. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larriba MJ, Gonzalez-Sancho JM, Barbachano A, Niell N, Ferrer-Mayorga G, Munoz A. Vitamin D Is a Multilevel Repressor of Wnt/b-Catenin Signaling in Cancer Cells. Cancers (Basel) 2013;5:1242–60. doi: 10.3390/cancers5041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Looker AC, Dawson-Hughes B, Calvo MS, Gunter EW, Sahyoun NR. Serum 25-hydroxyvitamin D status of adolescents and adults in two seasonal subpopulations from NHANES III. Bone. 2002;30:771–7. doi: 10.1016/s8756-3282(02)00692-0. [DOI] [PubMed] [Google Scholar]
- 21.Looker AC, Melton LJ, 3rd, Borrud LG, Shepherd JA. Lumbar spine bone mineral density in US adults: demographic patterns and relationship with femur neck skeletal status. Osteoporos Int. 2012;23:1351–60. doi: 10.1007/s00198-011-1693-z. [DOI] [PubMed] [Google Scholar]
- 22.McKeigue PM. Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am J Hum Genet. 1998;63:241–51. doi: 10.1086/301908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKeigue PM. Prospects for admixture mapping of complex traits. Am J Hum Genet. 2005;76:1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moy KA, Mondul AM, Zhang H, Weinstein SJ, Wheeler W, Chung CC, Mannisto S, Yu K, Chanock SJ, Albanes D. Genome-wide association study of circulating vitamin D-binding protein. Am J Clin Nutr. 2014;99:1424–1431. doi: 10.3945/ajcn.113.080309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman AB, Naydeck BL, Whittle J, Sutton-Tyrrell K, Edmundowicz D, Kuller LH. Racial differences in coronary artery calcification in older adults. Arterioscler Thromb Vasc Biol. 2002;22:424–30. doi: 10.1161/hq0302.105357. [DOI] [PubMed] [Google Scholar]
- 26.Pasaniuc B, Zaitlen N, Lettre G, Chen GK, Tandon A, Kao WH, Ruczinski I, Fornage M, Siscovick DS, Zhu X, Larkin E, Lange LA, Cupples LA, Yang Q, Akylbekova EL, Musani SK, Divers J, Mychaleckyj J, Li M, Papanicolaou GJ, Millikan RC, Ambrosone CB, John EM, Bernstein L, Zheng W, Hu JJ, Ziegler RG, Nyante SJ, Bandera EV, Ingles SA, Press MF, Chanock SJ, Deming SL, Rodriguez-Gil JL, Palmer CD, Buxbaum S, Ekunwe L, Hirschhorn JN, Henderson BE, Myers S, Haiman CA, Reich D, Patterson N, Wilson JG, Price AL. Enhanced statistical tests for GWAS in admixed populations: assessment using African Americans from CARe and a Breast Cancer Consortium. PLoS Genet. 2011;7:e1001371. doi: 10.1371/journal.pgen.1001371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powe CE, Evans MK, Wenger J, Zonderman AB, Berg AH, Nalls M, Tamez H, Zhang D, Bhan I, Karumanchi SA, Powe NR, Thadhani R. Vitamin D-binding protein and vitamin D status of black Americans and white Americans. N Engl J Med. 2013;369:1991–2000. doi: 10.1056/NEJMoa1306357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, Sassa S, Wood BL, Abkowitz JL. Identification of a human heme exporter that is essential for erythropoiesis. Cell. 2004;118:757–66. doi: 10.1016/j.cell.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 29.Redden DT, Divers J, Vaughan LK, Tiwari HK, Beasley TM, Fernandez JR, Kimberly RP, Feng R, Padilla MA, Liu N, Miller MB, Allison DB. Regional admixture mapping and structured association testing: conceptual unification and an extensible general linear model. PLoS Genet. 2006;2:e137. doi: 10.1371/journal.pgen.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Register TC, Divers J, Bowden DW, Carr JJ, Lenchik L, Wagenknecht LE, Hightower RC, Xu J, Smith SC, Hruska KA, Langefeld CD, Freedman BI. Relationships between serum adiponectin and bone density, adiposity and calcified atherosclerotic plaque in the African American-Diabetes Heart Study. J Clin Endocrinol Metab. 2013;98:1916–22. doi: 10.1210/jc.2012-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, Bera O, Semana G, Kelly MA, Francis DA, Ardlie K, Khan O, Cree BA, Hauser SL, Oksenberg JR, Hafler DA. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–8. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 32.Scales CD, Jr, Smith AC, Hanley JM, Saigal CS. Prevalence of kidney stones in the United States. Eur Urol. 2012;62:160–5. doi: 10.1016/j.eururo.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang CK, Kim JH, Li H, Stallcup MR. Differential use of functional domains by coiled-coil coactivator in its synergistic coactivator function with beta-catenin or GRIP1. J Biol Chem. 2006;281:3389–97. doi: 10.1074/jbc.M510403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X, Cooper RS, Elston RC. Linkage analysis of a complex disease through use of admixed populations. Am J Hum Genet. 2004;74:1136–53. doi: 10.1086/421329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37:177–81. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]


