Abstract
Conventional multidimensional magic angle spinning (MAS) solid-state NMR (ssNMR) experiments detect the signal arising from the decay of a single coherence transfer pathway (FID), resulting in one spectrum per acquisition time. Recently, we introduced two new strategies, namely DUMAS (DUal acquisition Magic Angle Spinning) and MEIOSIS (Multiple ExperIments via Orphan SpIn operatorS), that enable the simultaneous acquisitions of multidimensional ssNMR experiments using multiple coherence transfer pathways. Here, we combined the main elements of DUMAS and MEIOSIS to harness both orphan spin operators and residual polarization and increase the number of simultaneous acquisitions. We show that it is possible to acquire up to eight two-dimensional experiments using four acquisition periods per each scan. This new suite of pulse sequences, called MAeSTOSO for Multiple Acquisitions via Sequential Transfer of Orphan Spin pOlarization, relies on residual polarization of both 13C and 15N pathways and combines low- and high-sensitivity experiments into a single pulse sequence using one receiver and commercial ssNMR probes. The acquisition of multiple experiments does not affect the sensitivity of the main experiment; rather it recovers the lost coherences that are discarded, resulting in a significant gain in experimental time. Both merits and limitations of this approach are discussed.
Keywords: Magic angle spinning, multiple acquisitions, orphan spin operators, residual polarization
Graphical abstract
INTRODUCTION
Magic angle spinning (MAS) solid-state NMR (ssNMR) spectroscopy is a fundamental technique to study the structure, conformational dynamics, and ligand binding of biomacromolecules that are often recalcitrant to crystallization or not amenable to solution NMR spectroscopy, including fibrils, microcrystalline protein preparations, and membrane proteins [1–7]. However, the inherent low sensitivity of this technique leads to days or weeks of experimental time for routine multidimensional experiments. The classical schemes for signal detection are based on the acquisition of a single coherence pathway, i.e., free-induction decay (FID), which gives rise to one experiment per acquisition time. Under this classical scheme, the NMR spectrometers utilize about 5% of total experimental time for the execution of the pulse sequence and acquisition of the signal, while 95% time is necessary for the recovery of the longitudinal magnetization (T1 relaxation) and/or probe duty cycle (recycle delay).
Recently, we introduced a new class of ssNMR experiments called polarization optimized experiments (POE) that optimize the nuclear spin polarization to acquire multiple experiments utilizing only one receiver and commercially available probes for MAS [8] and static (oriented) experiments [9]. For MAS experiments, POE consists of two different strategies: dual acquisition magic angle spinning (DUMAS) [10, 11] and multiple experiments via orphan spin operators (MEIOSIS) [12]. Both DUMAS and MEIOSIS enable the acquisition of multiple spectra using a single recycle delay per each scan. In the case of the DUMAS experiments, we doubled the capability of the NMR spectrometer, collecting two FIDs per each experiment. The main experiment dictates the total experimental time, while the second acquisition has negligible (less than 5%) contribution to the total experimental time [10]. Although MEIOSIS utilizes principles similar to DUMAS, it exploits 13C and 15N residual polarization for the simultaneous acquisitions of three or four multi-dimensional experiments [12]. The DUMAS and MEIOSIS strategies enabled us to concatenate several pairs of 2D and 3D experiments substantially reducing the experimental time for both crystalline and membrane bound proteins [8].
In this work, we combined the DUMAS and MEIOSIS strategies, showing that it is possible to acquire up to eight two-dimensional experiments using a single pulse sequence with four acquisitions per scan. This new strategy, called MAeSTOSO for Multiple Acquisitions via Sequential Transfer of Orphan Spin pOlarization, combines both the 13C and 15N residual polarization resulting from multiple NC transfer periods. Specifically, the residual 15N/13C polarization originating from each NC (or CN) transfer step is utilized for two different experiments: MAeSTOSO-4 and MAeSTOSO -8, which enable the simultaneous acquisition of four and eight 2D experiments, respectively. We tested the performance of MAeSTOSO-4 and MAeSTOSO-8 on uniformly 13C and 15N labeled microcrystalline ubiquitin.
MATERIAL AND METHODS
All of the NMR experiments were performed on a VNMRS spectrometer operating at a 1H Larmor frequency of 599.61 MHz, equipped with a 3.2 mm scroll coil BioMAS probe [13]. A spinning rate of 12 kHz and a temperature of 2 °C were used in all the experiments. The microcrystalline U-13C,15N-labeled ubiquitin sample was crystallized from 10 mg of powder dissolved in 20 mM citrate buffer at pH 4.1. The protein was then crystallized by adding 2-methyl-2,4-pentanediol (60% final volume) [14]. Approximately 5 mg of microcrystalline 13C,15N-labeled ubiquitin was loaded into the MAS rotor. The 90° pulse length for 1H, 13C, and 15N were set to 2.5, 6, and 6 µs, respectively. For all experiments, the t2 acquisition time was set to 15 ms for each of the four acquisitions with a 10 µs dwell time and a recycle delay of 2 s. The dwell times for 13C and 15N indirect dimensions were set to 30 (t1’) and 240 µs (t1”), respectively. A total of 256 t1’ increments were used for 13C-edited experiments with 128 and 416 scans for MAeSTOSO-4 and MAeSTOSO-8 sequences, respectively; whereas the parallel evolution of 15N was obtained with 32 t1” increments repeated eight times [10, 12]. Hence, the effective number of scans for 15N-edited experiments is eight times the scans used for the 13C-edited experiments.
For MAeSTOSO-8 the spectra were acquired in interleaved mode with 15N spin lock phase (ϕ) switched between x and −x. For all four acquisitions, the FIDs with phases x and −x are added or subtracted to obtain residual 15N and transferred CN polarization pathways resulting from the first NC transfer [12]. During the simultaneous cross-polarization (SIM-CP) [11], 13C and 15N RF amplitudes were 35 kHz, while the 1H RF amplitude was linearly ramped from 80 to 100% with the center of the ramp corresponding to 59 kHz. The optimal Hartmann–Hahn (HH) contact time for the SIM-CP was estimated to be 500 µs. The 15N RF carrier was centered at 120 ppm, while the 13C RF was centered at 100 ppm. For the specific-CP transfer [15] from 15N to 13Cα (or 13CO), the 13C offset was shifted to 60 (or 177) ppm. During specific-CP, the 15N RF amplitude was set to 5/2· ωr (30 kHz), whereas 13C RF amplitude was 3/2· ωr (18 kHz) and 7/2· ωr (42 kHz) for 13Cα and 13CO specific-CP, respectively. The specific-CP was implemented with an adiabatic ramp and the contact times for NCA and NCO transfers were set to 3.0 and 3.2 ms, respectively. For heteronuclear decoupling (CW or TPPM [16]), the 1H RF amplitude was set to 100 kHz. During the DARR [17] mixing period, the 1H RF amplitude was set to 12 kHz (ωr). The 13C spectra were referenced with respect to the CH2 (methylene) resonance of adamantane at 40.48 ppm and indirectly to 15N, using the relative gyromagnetic ratio of 15N and 13C. All of the spectra were processed and analyzed using the NMRPipe software [18].
RESULTS
Multiple 1D experiments using MAESTOSO-4 and MAESTOSO-8
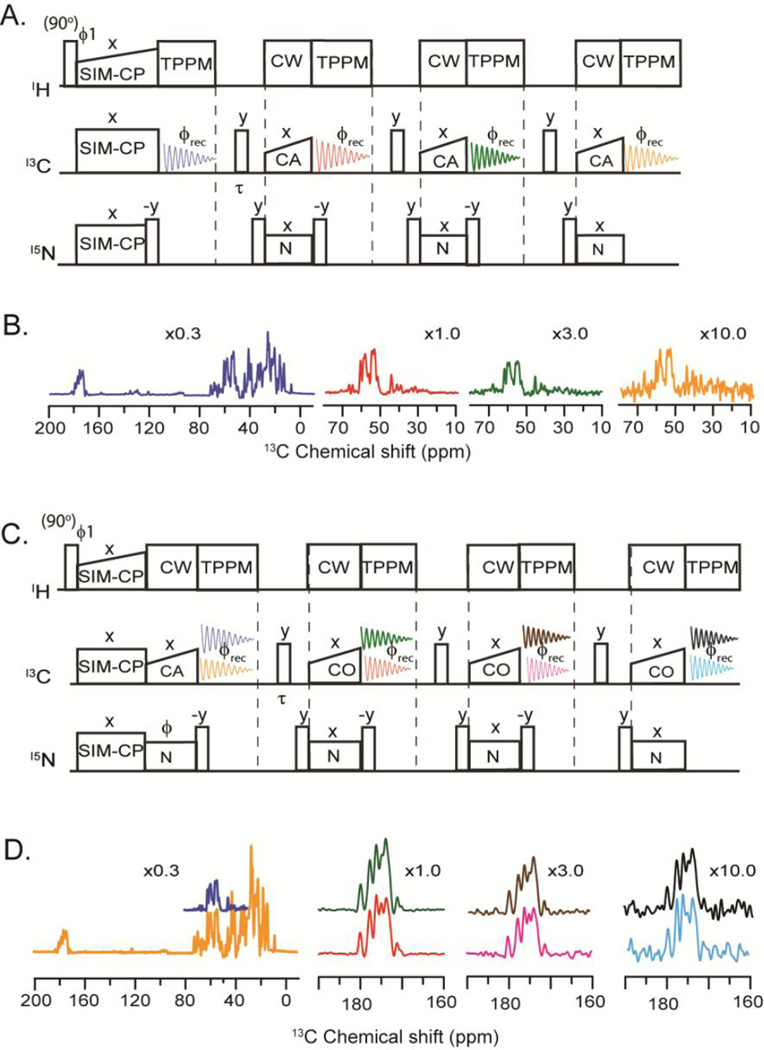
For the 1D MAeSTOSO-4 experiments (Figure 1A), the spin operator formalism can be described by the following equations:
| (1) |
| (2) |
Where Hi, Ci, and Ni (I = x,y,z) represent Pauli spin operators for 1H, 13C, and 15N, respectively. In equation 1, the coefficients ‘a’ and ‘b’ are the amplitudes for the 13C and 15N polarizations. The initial 13C polarization is denoted by ‘aC’, whereas ‘bN’ corresponds to that of 15N. The residual 15N polarization is indicated by ‘bNN’ and ‘bNNN’ and the transferred polarization from 15N to 13Cα is represented by ‘b*N’, ‘b*NN’, and ‘b*NNN’. As shown in Figure 1A, the pulse sequence starts with a 90° excitation pulse on the 1H spin bath followed by SIM-CP [11] that creates initial 13C and 15N polarization. The 13C polarization is acquired in the first acquisition (blue FID), whereas 15N polarization is stored along the z-direction. A 6 ms τ/2−(π/2)− τ/2 is applied at the end of 1st acquisition period to dephase any unwanted polarization that would otherwise interferes with the subsequent acquisition [10]. Note that this spin-echo sequence is applied at the end of the 2nd and 3rd acquisitions as well. After the first FID, a 90° pulse is then applied on 15N and is followed by a specific-CP [15] for the NCA transfer. While the transferred polarization from 15N to 13Cα (bN*·Cx) is acquired during the second acquisition (red FID), the residual 15N polarization (bNN·Nz) is stored along the z-direction. The remainder of the pulse sequence has two additional NCA blocks with a combination of 90° pulses on 15N to transfer the residual 15N polarization to 13Cα, corresponding to the b*NN·Cx and b*NNN·Cx terms (equation 1). These terms are acquired during the third and fourth acquisitions corresponding to the green and orange FIDs, respectively (Figure 1A). The spectra of U-13C,15N ubiquitin acquired with the 1D MAeSTOSO-4 pulse sequence are reported in Figure 1B, where the relative sensitivity for the four acquisitions is obtained by integrating the intensities of the Cα spectral region. The first FID containing the entire 13C spectrum has the highest sensitivity (100%), while the second FID resulting from the transferred polarization from the 15N to Cα has ~32% of the sensitivity of the first one. The third FID has ~30% of the signal of the second, and the remaining 30% of the polarization is further reduced by ~30%, resulting in approximately 10% residual polarization that is transferred via NCA to Cα and detected as a fourth FID. The intensities of four spectra are proportional to the ‘a’ and ‘b*’ coefficients of equation 1. Experimentally, we found that the relative ratios of polarization amplitudes are: 1.00 : 0.32 for aC:b*N and 1.00 : 0.29 : 0.12 for b*N:b*NN:b*NNN.
Figure 1.
A) 1D MAeSTOSO-4 pulse sequence for acquisition of four spectra. B) Color-coded 1D spectra of U-13C,15N microcrystalline ubiquitin acquired with MAeSTOSO-4 pulse sequence. The relative scaling of the spectra is indicated in the figure. C) 1D MAeSTOSO-8 pulse sequence for the acquisition of eight spectra. D). Color-coded 1D spectra of U-13C,15N microcrystalline ubiquitin acquired using MAeSTOSO-8 pulse sequence. Note that the pulse sequences are implemented with a two-step phase cycle (y, −y) between ϕ1 and ϕrec. For MAeSTOSO-8, the two data sets were recorded with ϕ set to x and −x, respectively. Addition and subtraction of the two data sets prior to Fourier transform deconvolutes them into eight different spectra.
To test how much we can push the residual polarization, we designed MAeSTOSO-8 (Figure 1C) that is able to record eight 1D experiments with four sequential acquisitions in a single scan. The initial 13C and 15N polarization resulting from SIM-CP undergoes bidirectional polarization [11] between 15N and 13CO during NCO specific-CP. At this point, each of the 13C and 15N polarization pathways splits into two components, resulting in four different pathways: CN, CC, NC and NN. While CN and NC represent transferred polarization from Cα to N and vice-versa, CC and NN correspond to residual 13C and 15N polarization, respectively. The spin operator formalism for MAeSTOSO-8 is:
| (3) |
| (4) |
The 13C signals from CC and NC pathways are detected during the first acquisition, while the 15N polarization from NN and CN pathways is stored in the z-direction and detected in the following three acquisition periods. To encode two FIDs per each acquisition period, we utilized the MEIOSIS strategy [12]. Namely, the phase of 15N pulse during the first NCO transfer is switched between 0° and 180° in alternate scans to invert the sign of the NC and CN pathways, while the sign for the CC and NN pathways is left unchanged. The sum and difference of the two scans in the first acquisition give the 13C polarization resulting from the CC and NC pathways, respectively. After the first acquisition, the 15N polarization from the CN and NN pathways is transferred via NCA transfer to Cα followed by a second acquisition and the residual 15N polarization is stored along the z-direction. Unlike MAeSTOSO-4, the residual 15N polarization encodes two different pathways, CN and NN, which are detected during the third and fourth acquisition periods. The density matrix for MAeSTOSO-8 is reported in equation 4, with eight terms corresponding to eight 1D spectra registered. Figure 1D shows the eight MAeSTOSO-8 spectra obtained with the 13C,15N-labeled microcrystalline ubiquitin. The relative sensitivities for CC (first acquisition) and CN (second, third, and fourth acquisitions) pathways are 1.00:0.60:0.16:0.02 (spectra in orange, red, pink and cyan colors). The 15N polarization contributes NC and NN polarization pathways with a relative sensitivity of (spectra in blue, green, brown and black colors) 1.00:0.30:0.09:0.01 recorded in first to fourth acquisitions.
Multidimensional CC and NC correlation experiments using MAeSTOSO-4
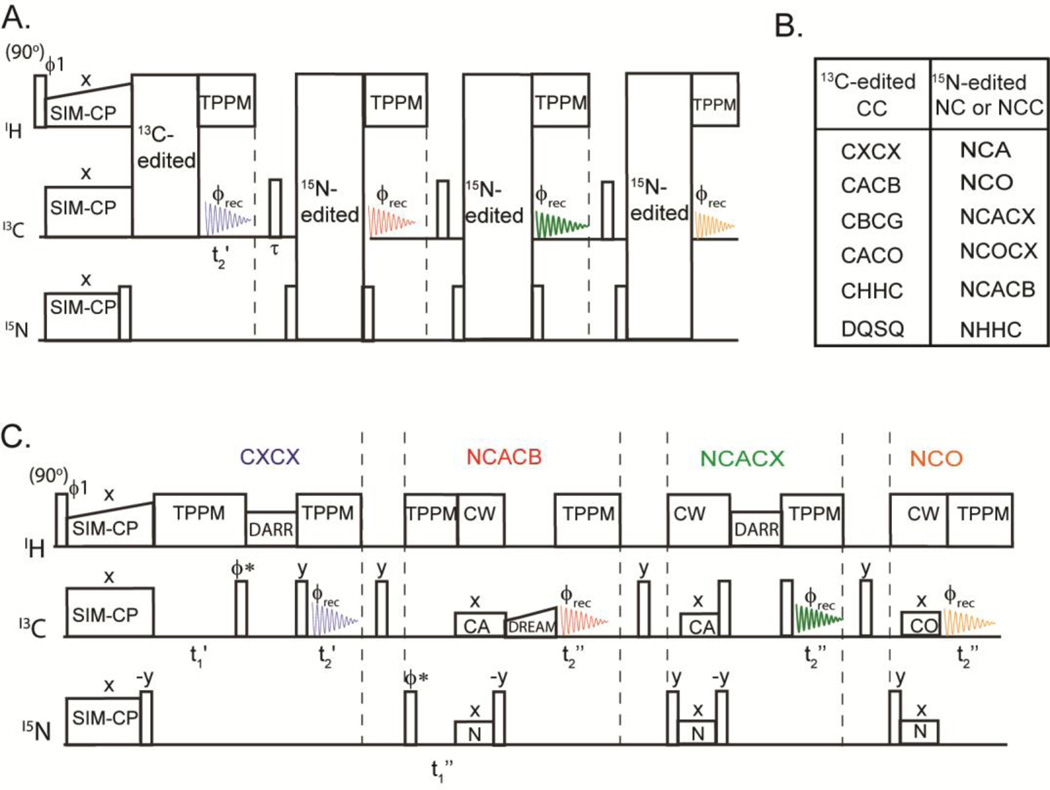
The general scheme for the 2D MAeSTOSO-4 is shown in Figure 2A. In this pulse sequence, the 13C polarization from SIM-CP is used to acquire one 2D CC correlation experiment during the first acquisition, whereas the 15N polarization is exploited for 15N-edited NC correlation experiments in the subsequent three acquisitions. Figure 2B shows different combinations of CC and NC correlation experiments that can be concatenated into the 2D MAeSTOSO-4. An example of this scheme is shown in Figure 2C, where CXCX and three NC correlation experiments (NCACB, NCACX, and NCO) are combined. For the CC experiment, the t1 evolution period is coded right after the SIM-CP block followed by a CC mixing and the t2’ acquisition time. Note that the t1” evolution for 15N-edited experiments is implemented after the first acquisition and all three 15N-edited experiments share the same t1‘’ evolution period. In this scheme, a DARR sequence [17] is used for homonuclear 13C-13C transfer for both CXCX and NCACX experiments [19], while for the CACB transfer we implemented a DREAM sequence [20]. The heteronuclear polarization transfer for NCA or NCO is obtained using the specific-CP sequence [15]. Note that other combinations of CC and NC experiments can be achieved by permutating the hetero- and homonuclear mixing periods.
Figure 2.
A) General scheme of the 2D MAeSTOSO-4 pulse sequence for acquisition of CC and NC correlation experiments. B) Possible combinations of experiments that can be concatenated with the 2D MAeSTOSO-4 strategy. The 13C-edited CC correlation experiment is acquired in the 1st acquisition, whereas 15N edited NC or NCC are acquired in 2nd, 3rd and 4th acquisitions. C) Example of 2D MAeSTOSO-4, the CXCX and NCACX use DARR mixing periods for homonuclear 13C-13C transfer, and DREAM mixing for CACB transfer during NCACB. The NCA and NCO polarization transfer steps are achieved using specific-CP. The phase ϕ* is switched between y and x for phase sensitive t1 acquisition.
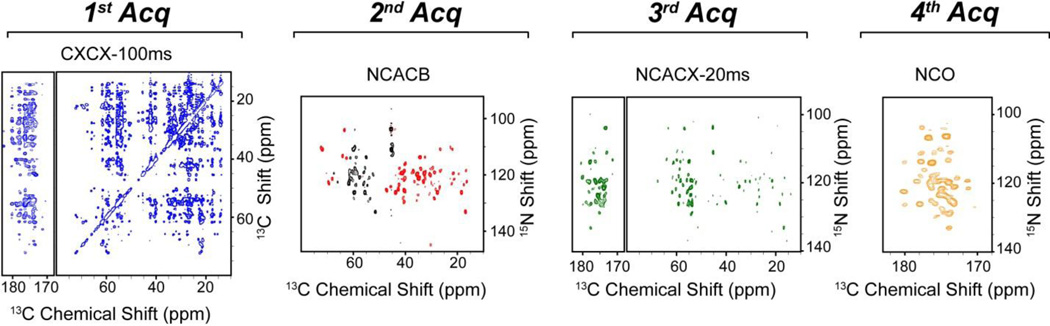
The four 2D CC and NC correlation spectra of U-13C,15N ubiquitin acquired with the MAeSTOSO-4 pulse sequence are reported in Figure 3. For the CXCX and NCACX experiments, we utilized the DARR mixing times of 100 and 20 ms, respectively; whereas a 2.5 ms DREAM [20] mixing period was used for the CACB transfer. The total experimental time for 2D MAeSTOSO-4 spectra was 38 hrs. To give an estimate of the total acquisition time for 2D MAeSTOSO-4 versus the conventional single-acquisition experiments, we compared the intensity of the first increments of the pulse sequences. Specifically, for the MAeSTOSO-4 spectra we considered the first increment of the 2D data in Figure 3, whereas for the conventional experiments we set the dwell time in the indirect dimension to zero. To acquire these four spectra with identical signal-to-noise ratios, MAeSTOSO-4 required 35 min, whereas 75.2 min was necessary for the individual CXCX, NCACB, NCACX, and NCO. Therefore, the total estimated time required to acquire these 2D experiments using conventional pulse sequences is ~2.2 times longer than the corresponding MAeSTOSO-4 experiment.
Figure 3.
Color-coded 2D CC and NC correlation spectra of U-13C,15N ubiquitin obtained using the 2D MAeSTOSO-4 pulse sequence reported in Figure 2C.
Multidimensional CC, NC, and C(N)C experiments using MAeSTOSO-8
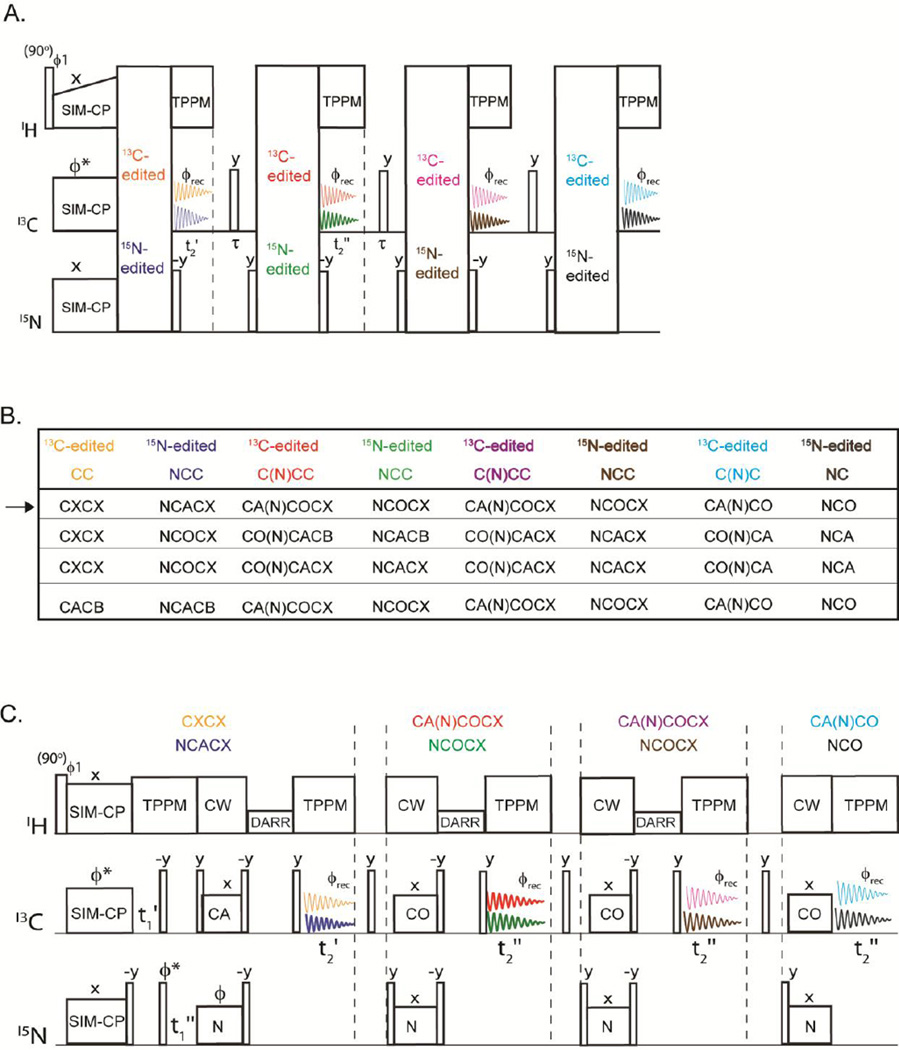
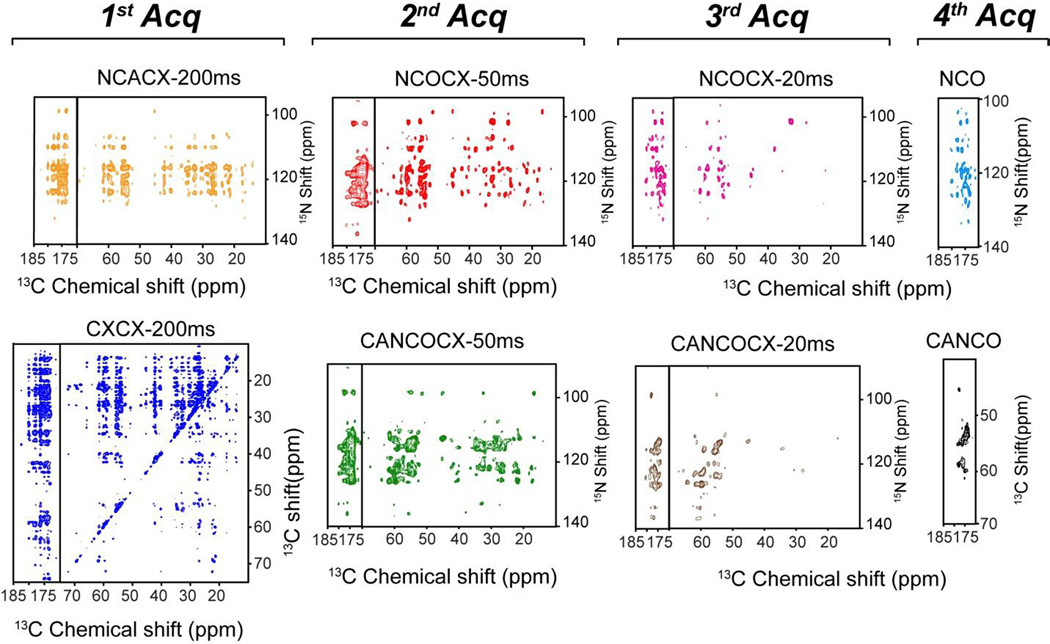
The 2D MAeSTOSO-8 pulse sequence enables the acquisition of eight 2D spectra simultaneously using four acquisitions per scan (Figure 4A). Generated from the SIM-CP sequence, the initial 13C and 15N polarization evolves for a t1’ evolution period and is followed by an NCO transfer, creating the CC, CN, NC and NN pathways. The 13C polarization of CC and NC pathways utilizes DARR mixing period for homonuclear 13C-13C homonuclear transfer followed by t2’ acquisition, while the 15N polarization from CN and NN pathways is stored along the z-direction. Therefore, the first acquisition generates 2D CXCX and NCACX correlation experiments, respectively. The 15N polarization from CON and NN pathways is then transferred via NCA to Cα followed by a DARR mixing period and a second acquisition period, t2”, to obtain 2D CO(N)CACX and NCACX spectra. The residual 15N polarization from CON and NN pathways remaining after the NCA transfer is exploited for recording two more sets of 2D C(N)C and NC correlation experiments in third and fourth acquisitions, respectively. A shorter DARR mixing period (20 ms) is used prior to the third acquisition period. This gives 2D CA(N)COCX and NCOCX in the third acquisition, whereas CA(N)CO and NCO were collected in the fourth acquisition. To deconvolute the eight experiments in 2D MAeSTOSO-8, we employed a phase switching scheme previously used for the 2D MEIOSIS experiments, where the phase of 15N spin-lock during first NC period was altered between 0 and 180°. Although we tested only one combination, the MAeSTOSO-8 strategy can be used to concatenate several different combinations of experiments (Figure 4B). As shown in Figures 1C and 1D the relative sensitivity of the 3rd and 4th acquisitions in MAeSTOSO-8 is weaker. Therefore, to optimize the experiments in the 2D MAeSTOSO-8, one must use relatively higher sensitive experiments that contain shorter 13C-13C mixing times (or no mixing times) prior to the acquisition of the 3rd and 4th FIDs. The sensitivity of 1D MAeSTOSO-8 spectra should be used as a reference to optimize the combination of experiments in Figure 4B. The performance of 2D MAeSTOSO-8 is shown in Figure 5, where eight 2D spectra were collected on U-13C,15N-labeled microcrystalline ubiquitin for a total experimental time of 5.2 days. The NCACX-200 ms and CA(N)COCX-50 ms experiments were obtained from transferred polarization via SIM-CP and bidirectional specific-CP. Therefore, they used the polarization generated by SIM-CP and their sensitivity is similar to that of the conventional single acquisition experiments. The remaining six experiments utilized either the 13C or 15N residual polarizations and are less sensitive than the corresponding 2D experiments acquired separately. Figure 2S compares the sensitivity of MAeSTOSO-8 and conventional single- acquisition experiments as for the MAeSTOSO-4 experiments. The total experimental time for eight spectra using conventional methods is ~2.5 times longer than the corresponding experiments acquired with the MAeSTOSO-8 pulse sequence.
Figure 4.
A) General scheme for 2D MAeSTOSO-8 strategy for acquisition of the CC, NC, NCC, C(N)C and C(N)CC experiments. B) Table of experiments that can be concatenated using the MAeSTOSO-8 strategy. C) Example of a pulse sequence of 2D MAeSTOSO-8 that combines eight experiments shown by the arrow in the table reported in B.
Figure 5.
Color-coded 2D spectra of ubiquitin acquired using the MAeSTOSO-8 combination of experiments shown in Figure 4C.
DISCUSSION
The inefficiency of the polarization transfer schemes together with undetected (orphan) spin operators contribute to the intrinsic insensitivity of the ssNMR experiments [21]. To overcome these limitations, we have been developing a class of experiments (POE) that optimize the sensitivity by recovering residual polarization as well as orphan spin operators to obtain multiple NMR experiments [8, 10, 11]. These experiments do not require special experimental settings or additional hardware for the spectrometer. Spectrometers equipped with one receiver and commercial probes are sufficient to execute them. However, the architecture of the pulse sequences needs to be optimized to be able to generate multiple polarization pathways via SIM-CP [11] and bidirectional polarization transfer [12], with the concatenation of multi-dimensional schemes and phase cycling. The first POE were the 2D DUMAS and MEIOSIS suite of experiments that are now routinely used in our laboratory for both crystalline and membrane proteins [8, 10–12, 22]. In the case of 2D DUMAS, we were able to combine CC and NC correlation experiments. On the other hand, with MEIOSIS we obtained simultaneous acquisition of CC, NC, NCC, and C(N)C experiments. The MAeSTOSO strategy (MAeSTOSO-4 and -8) combines both DUMAS and MEIOSIS schemes to push the residual polarization even further. Of course, the sensitivity of the 15N residual polarization is lowered by approximately 30–35 % for each NC transfer, resulting in about 10 and 4% residual polarization after two and three NC transfer periods. As the number of building blocks in MAeSTOSO increases, the signal decreases due to the lower amount of residual polarization transferrable and the intrinsic T1ρ of the protein. Nonetheless, achieving up to eight different transfers and corresponding experiments is quite remarkable. A key to achieving the successful application of MAeSTOSO is the use of a larger number of scans to accumulate sufficient residual polarization to drive the third and fourth acquisitions. To this extent, it is important to code the low-sensitivity experiments, which require a larger number of scans, during the first two acquisitions of the pulse scheme and the more sensitive experiments for the other acquisitions. In this way, it is possible to accumulate in the main experiment sufficient residual polarization to drive the acquisition of the other experiments. Also, it should be noted that for more sensitive experiments, e.g., 2D CXCX (20 ms), NCACX (20 ms), NCA, NCO, CA(N)CO etc., DUMAS and MEIOSIS strategies are preferable over the MAeSTOSO. On the other hand, for low sensitivity experiments, e.g., 2D CXCX (100–200 ms mixing time) or NCOCX (100–200 ms mixing times) that require a significantly higher number of scans, the MAESTOSO strategy is preferable as the accumulation of residual polarization is such that one can acquire simultaneously a greater number of experiments and optimize the total experimental time. Typically, longer mixing times are necessary to obtain distance restraints for protein structure determination. These experiments require several days for insensitive samples such as those of membrane proteins. In these cases, MAeSTOSO can represent a cost-effective solution to acquire multiple experiments using one pulse sequence.
The idea of multiple acquisitions using discarded coherences was introduced by Takegoshi and co-workers [23]. Recently, a number of research groups have also been developing multiple acquisition MAS experiments with one or multiple receivers for solution and solid-state NMR under fast spinning conditions [24–32] and several of these experiments can be further developed using the MAESTOSO strategy.
Finally, one should take into account that the POE schemes (DUMAS, MEIOSIS, and MAeSTOSO) may challenge the hardware of the NMR spectrometer due to the increase of RF duty cycles. Usually, for microcrystalline protein samples, the acquisition times are ~15–20 ms, under moderate spinning speeds (10–15 kHz). In our case, we used 15 ms to avoid possible RF heating. While shorter acquisition times might represent a shortcoming of the method, for membrane proteins embedded into lipids 15 ms are sufficient, as the relaxation rates are faster than microcrystalline proteins. Nonetheless, we should point out that under fast-MAS conditions MAeSTOSO can be implemented with longer acquisition times, as low decoupling powers are usually employed. Finally, we anticipate that the utilization of longer acquisition times will be possible with advancements in probe design. The pulse sequences described here have been extensively tested with both BioMAS and E-free ssNMR probes and we found they are well within the RF capabilities of the commercial spectrometers and probes.
CONCLUSIONS
In conclusion, we added a new strategy (MAeSTOSO) to the class of POE. MAeSTOSO enables the acquisition of up to eight 2D experiments simultaneously using commercial ssNMR probe and one receiver. Depending on the sensitivity of the experiments planned, one can choose DUMAS, MEIOSIS or MAeSTOSO schemes for concatenating multi-dimensional experiments. For low-sensitivity experiments, where a significant number of scans are required for high sensitivity, the accumulation of residual polarization justifies the use of the MAeSTOSO strategy. Although demonstrated for 2D experiments, the development of 3D MAeSTOSO pulse sequences is an ongoing effort in our laboratory. We anticipate that the combination of these methods with other sensitivity enhancement methods such as 1H detection and DNP will further enhance the application of ssNMR to biomacromolecules.
Supplementary Material
Highlights.
New solid-state NMR strategy for the acquisition of up to 8 spectra simultaneously.
Orphan spin operators and residual polarization are combined for multiple acquisitions.
Reduction of the experimental time up to 40%.
Acknowledgments
The experiments were carried out at the Minnesota NMR Center with the support of the National Institute of Health (GM64742 and GM72701).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ardenkjaer-Larsen JH, Boebinger GS, Comment A, Duckett S, Edison AS, Engelke F, Griesinger C, Griffin RG, Hilty C, Maeda H, Parigi G, Prisner T, Ravera E, van Bentum J, Vega S, Webb A, Luchinat C, Schwalbe H, Frydman L. Facing and Overcoming Sensitivity Challenges in Biomolecular NMR Spectroscopy. Angewandte Chemie. 2015;54:9162–9185. doi: 10.1002/anie.201410653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker LA, Baldus M. Characterization of membrane protein function by solid-state NMR spectroscopy. Current opinion in structural biology. 2014;27C:48–55. doi: 10.1016/j.sbi.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Hong M, Zhang Y, Hu F. Membrane protein structure and dynamics from NMR spectroscopy. Annual review of physical chemistry. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDermott A, Polenova T. Solid state NMR: new tools for insight into enzyme function. Current opinion in structural biology. 2007;17:617–622. doi: 10.1016/j.sbi.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Renault M, Cukkemane A, Baldus M. Solid-State NMR Spectroscopy on Complex Biomolecules. Angewandte Chemie-International Edition. 2010;49:8346–8357. doi: 10.1002/anie.201002823. [DOI] [PubMed] [Google Scholar]
- 6.Tang M, Comellas G, Rienstra CM. Advanced solid-state NMR approaches for structure determination of membrane proteins and amyloid fibrils. Accounts of chemical research. 2013;46:2080–2088. doi: 10.1021/ar4000168. [DOI] [PubMed] [Google Scholar]
- 7.Tycko R, Wickner RB. Molecular structures of amyloid and prion fibrils: consensus versus controversy. Accounts of chemical research. 2013;46:1487–1496. doi: 10.1021/ar300282r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopinath T, Veglia G. Multiple acquisition of magic angle spinning solid-state NMR experiments using one receiver: application to microcrystalline and membrane protein preparations. Journal of magnetic resonance. 2015;253:143–153. doi: 10.1016/j.jmr.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gopinath T, Mote KR, Veglia G. Simultaneous acquisition of 2D and 3D solid-state NMR experiments for sequential assignment of oriented membrane protein samples. Journal of biomolecular NMR. 2015;62:53–61. doi: 10.1007/s10858-015-9916-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gopinath T, Veglia G. 3D DUMAS: simultaneous acquisition of three-dimensional magic angle spinning solid-state NMR experiments of proteins. Journal of magnetic resonance. 2012;220:79–84. doi: 10.1016/j.jmr.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gopinath T, Veglia G. Dual acquisition magic-angle spinning solid-state NMR-spectroscopy: simultaneous acquisition of multidimensional spectra of biomacromolecules. Angewandte Chemie. 2012;51:2731–2735. doi: 10.1002/anie.201108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopinath T, Veglia G. Orphan spin operators enable the acquisition of multiple 2D and 3D magic angle spinning solid-state NMR spectra. The Journal of chemical physics. 2013;138:184201. doi: 10.1063/1.4803126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stringer JA, Bronnimann CE, Mullen CG, Zhou DH, Stellfox SA, Li Y, Williams EH, Rienstra CM. Reduction of RF-induced sample heating with a scroll coil resonator structure for solid-state NMR probes. Journal of magnetic resonance. 2005;173:40–48. doi: 10.1016/j.jmr.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Igumenova TI, McDermott AE, Zilm KW, Martin RW, Paulson EK, Wand AJ. Assignments of carbon NMR resonances for microcrystalline ubiquitin. Journal of the American Chemical Society. 2004;126:6720–6727. doi: 10.1021/ja030547o. [DOI] [PubMed] [Google Scholar]
- 15.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Molecular Physics. 1998;95:1197–1207. [Google Scholar]
- 16.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decouplings in rotating solids. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 17.Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chemical physics letters. 2001;344:631–637. doi: 10.1063/1.2364503. [DOI] [PubMed] [Google Scholar]
- 18.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 19.Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. Backbone and side-chain 13C and 15N signal assignments of the alpha-spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 Tesla. Chembiochem : a European journal of chemical biology. 2001;2:272–281. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Verel R, Ernst M, Meier BH. Adiabatic dipolar recoupling in solid-state NMR: the DREAM scheme. Journal of magnetic resonance. 2001;150:81–99. doi: 10.1006/jmre.2001.2310. [DOI] [PubMed] [Google Scholar]
- 21.Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. The Journal of chemical physics. 1973;59:569–590. [Google Scholar]
- 22.Mote KR, Gopinath T, Traaseth NJ, Kitchen J, Gor'kov PL, Brey WW, Veglia G. Multidimensional oriented solid-state NMR experiments enable the sequential assignment of uniformly 15N labeled integral membrane proteins in magnetically aligned lipid bilayers. Journal of biomolecular NMR. 2011;51:339–346. doi: 10.1007/s10858-011-9571-8. [DOI] [PubMed] [Google Scholar]
- 23.Fukuchi M, Inukai M, Takeda K, Takegoshi K. Double-acquisition: Utilization of discarded coherences in a 2D separation experiment using the States method. Journal of magnetic resonance. 2008;194:300–302. doi: 10.1016/j.jmr.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Herbst C, Riedel K, Ihle Y, Leppert J, Ohlenschlager O, Gorlach M, Ramachandran R. MAS solid state NMR of RNAs with multiple receivers. Journal of biomolecular NMR. 2008;41:121–125. doi: 10.1007/s10858-008-9247-1. [DOI] [PubMed] [Google Scholar]
- 25.Kupce E, Kay LE, Freeman R. Detecting the "afterglow" of 13C NMR in proteins using multiple receivers. Journal of the American Chemical Society. 2010;132:18008–18011. doi: 10.1021/ja1080025. [DOI] [PubMed] [Google Scholar]
- 26.Kupce E. NMR with multiple receivers. Topics in current chemistry. 2013;335:71–96. doi: 10.1007/128_2011_226. [DOI] [PubMed] [Google Scholar]
- 27.Banigan JR, Gayen A, Traaseth NJ. Combination of (1) (5)N reverse labeling and afterglow spectroscopy for assigning membrane protein spectra by magic-angle-spinning solid-state NMR: application to the multidrug resistance protein EmrE. Journal of biomolecular NMR. 2013;55:391–399. doi: 10.1007/s10858-013-9724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellstedt P, Herbst C, Hafner S, Leppert J, Gorlach M, Ramachandran R. Solid state NMR of proteins at high MAS frequencies: symmetry-based mixing and simultaneous acquisition of chemical shift correlation spectra. Journal of biomolecular NMR. 2012;54:325–335. doi: 10.1007/s10858-012-9680-z. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen AB, Szekely K, Gath J, Ernst M, Nielsen NC, Meier BH. Simultaneous acquisition of PAR and PAIN spectra. Journal of biomolecular NMR. 2012;52:283–288. doi: 10.1007/s10858-012-9616-7. [DOI] [PubMed] [Google Scholar]
- 30.Demers JP, Vijayan V, Lange A. Recovery of bulk proton magnetization and sensitivity enhancement in ultrafast magic-angle spinning solid-state NMR. The journal of physical chemistry. B. 2015;119:2908–2920. doi: 10.1021/jp511987y. [DOI] [PubMed] [Google Scholar]
- 31.Martineau C, Decker F, Engelke F, Taulelle F. Parallelizing acquisitions of solid-state NMR spectra with multi-channel probe and multi-receivers: applications to nanoporous solids. Solid state nuclear magnetic resonance. 2013;55–56:48–53. doi: 10.1016/j.ssnmr.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Das BB, Opella SJ. Multiple acquisition/multiple observation separated local field/chemical shift correlation solid-state magic angle spinning NMR spectroscopy. Journal of magnetic resonance. 2014;245:98–104. doi: 10.1016/j.jmr.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.