Abstract
There is an immediate need to drastically reduce the emissions associated with global fossil fuel consumption in order to limit climate change. However, carbon-based materials, chemicals, and transportation fuels are predominantly made from fossil sources and currently there is no alternative source available to adequately displace them. Gas-fermenting microorganisms that fix carbon dioxide (CO2) and carbon monoxide (CO) can break this dependence as they are capable of converting gaseous carbon to fuels and chemicals. As such, the technology can utilize a wide range of feedstocks including gasified organic matter of any sort (e.g., municipal solid waste, industrial waste, biomass, and agricultural waste residues) or industrial off-gases (e.g., from steel mills or processing plants). Gas fermentation has matured to the point that large-scale production of ethanol from gas has been demonstrated by two companies. This review gives an overview of the gas fermentation process, focusing specifically on anaerobic acetogens. Applications of synthetic biology and coupling gas fermentation to additional processes are discussed in detail. Both of these strategies, demonstrated at bench-scale, have abundant potential to rapidly expand the commercial product spectrum of gas fermentation and further improve efficiencies and yields.
Keywords: gas fermentation, acetogens, Clostridium, syngas, synthetic biology, coupled processes, carbon capture and utilization, low-carbon fuels
Gas fermentation overview
Introduction
In December 2015, 195 countries adopted the Paris Agreement at the end of the 21st Conference of the Parties to the United Nations Framework Convention on Climate Change. The agreement “aims to strengthen the global response to the threat of climate change” and seeks to hold the increase in global average temperature to “well below 2°C above pre-industrial levels” (United Nations, 2015). Though non-binding, this consensus underscores the rising urgency for actions that will limit the amount of greenhouse gasses emitted into the atmosphere. To achieve the goal of staying within the above mentioned 2°C target may require leaving a third of oil reserves, half of gas reserves, and over 80% of current coal reserves unused until 2050 (Friedlingstein et al., 2014; McGlade and Ekins, 2015). However, this time scale is likely too short to switch away from and eliminate the need for carbon-based transportation fuels and chemicals. To meet the demand for these products while simultaneously reducing greenhouse gas emissions, low-carbon fuels (i.e., fuels that emit less CO2 over their life cycle of production and use relative to fossil fuels) and chemicals are needed.
One renewable, non-fossil derived source for low-carbon fuels and chemicals is biomass. Plant biomass is the fourth largest renewable energy resource in the world, following geothermal, solar, and wind (Metz et al., 2007). It accounted for approximately 10% of the global energy supply in 2009 (Vakkilainen et al., 2013). Though the vast majority of this energy was used for inefficient residential heating and cooking, biomass is increasingly being converted to biofuels in efforts to displace fossil fuel-derived transportation fuel. However, only 3.5% of global oil demand for road transport (adjusted for energy content) was met by biofuels in 2013 (International Energy Agency, 2014). In order to successfully displace enough oil and gas reserves to mitigate climate change, significantly more biomass would need to be converted to fuels.
In addition to commercial ethanol production by yeast fermentation of starch/sugar from corn/sugar cane, low-carbon fuels can be produced from lignocellulosic biomass (Naik et al., 2010). There are various proposed process flows but generally multiple pretreatment and hydrolysis steps are necessary to separate cellulose and hemicellulose from the recalcitrant lignin and to break down polysaccharides into fermentable monosaccharides (Geddes et al., 2011). Steps are diverse and include steam explosion, ammonia fiber expansion (AFEX), extrusion, ionic liquid extraction, and dilute acid and enzymatic hydrolysis (Geddes et al., 2011; Brown and Brown, 2013). However, these methods are considered expensive (both in terms of cost and water usage), and no single pretreatment is universally effective (Haghighi Mood et al., 2013). Further challenges include separation of hexose (C6) and pentose (C5) sugars or isolating/designing a robust biocatalyst that can ferment both (Sánchez and Cardona, 2008; Geddes et al., 2011). An alternative to separating the pretreatment/hydrolysis and fermentation is to use cellulolytic microorganisms that are capable of performing both the hydrolysis of lignocellulosic material and sugar fermentation (termed “consolidated bioprocessing process,” or “CBP”). This has been demonstrated in laboratory conditions but not under industrial settings and challenges around conversion rates and productivities remain (Brown and Brown, 2013).
Advantages of gas fermentation
Even with the extensive biomass processing required to produce cellulosic ethanol, the lignin, which can account for up to 40% of plant biomass does not get converted (Sun and Cheng, 2002; Abubackar et al., 2011). The use of the biomass in its entirety as a feedstock is a key advantage inherent to gas fermentation compared to sugar and cellulosic fermentation to produce low-carbon fuels. Biomass can be gasified to a mixture of carbon monoxide (CO), carbon dioxide (CO2), hydrogen (H2), and nitrogen (N2), also called synthesis gas or syngas. Conversion of biomass to syngas allows for utilization of nearly all the available carbon contained within the biomass, including the otherwise inaccessible lignin fraction, and bypasses the expense and inefficiencies of biomass pretreatment.
Once converted into a gas, there are two options for conversion to useful products. Traditionally this has been achieved using the Fischer-Tropsch process (FTP), but the technology has some drawbacks and is very capital intensive. The ability to fix gaseous, inorganic carbon into organic material (autotrophy) is also a prerequisite for life, and routes exist in various forms across all domains of life (Thauer, 2007). Eukaryotes (the most common example being photosynthesis in plants), archaea, and bacteria can all fix carbon by reducing CO2 and/or CO. Anaerobic gas-fermenting bacteria, specifically acetogens, are the focus of this review due to the advantages they possess in low-carbon fuel/chemical production. Advantages of gas fermentation over traditional FTP conversion (Section Fischer Tropsch Process vs. Gas Fermentation), advantages in substrate diversity (Section Substrate Diversity), and advantages inherent in acetogens (Section Acetogens and Wood-Ljungdahl Pathway) are discussed in detail below.
Fischer tropsch process vs. gas fermentation
Converting syngas to low-carbon liquid hydrocarbons with short to medium/long chains has been traditionally achieved via FTP. First developed in 1925, FTP employs high temperature (150–350 °C), elevated pressures (30 bar), and heterogeneous catalysts such as cobalt, ruthenium, and iron (De Klerk et al., 2013). In comparison, gas fermentation takes place at 37 °C and atmospheric pressure, which presents significant energy and cost savings relative to FTP. FTP, unlike gas fermentation, also requires a fixed H2:CO ratio of ideally ~2:1 (De Klerk et al., 2013). However, syngas derived from biomass has typically a lower H2:CO ratio (van der Drift et al., 2001; Datar et al., 2004; Boerrigter and Rauch, 2005; Boateng et al., 2007; Piccolo and Bezzo, 2009; Zheng et al., 2016), often requiring an extra step of water-gas shift reaction (National Energy Technology Laboratory, 2013) at the expense of CO to adjust the H2:CO for FTP. Although chemical processes are generally considered faster than biological approaches, the latter allow near complete conversion efficiencies due to the irreversible nature of biological reactions (Klasson et al., 1991, 1992). Furthermore, the high enzymatic specificities of biological conversions also result in higher product selectivity with the formation of fewer by-products. Crucially, the biocatalysts are also less susceptible to poisoning by sulfur, chlorine, and tars than the inorganic catalysts (Michael et al., 2011; Mohammadi et al., 2011), which reduces the gas pre-treatment costs.
Substrate diversity
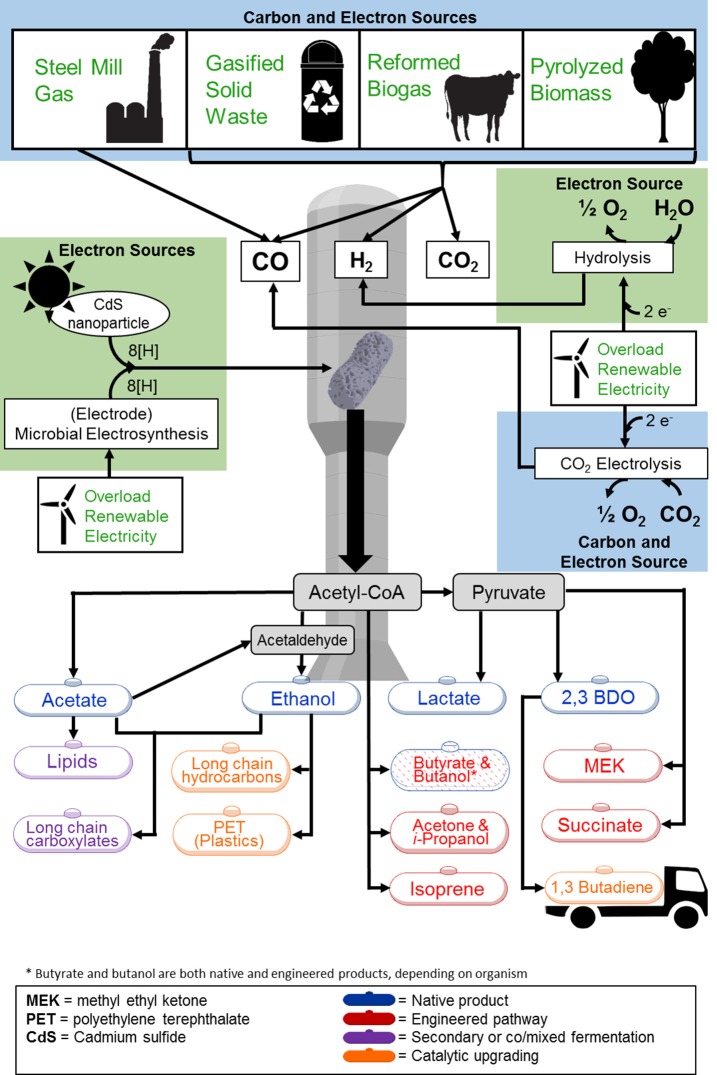
With these capabilities (using gaseous carbon, flexible gas compositions, and tolerance to more contaminants over FTP), gas-fermenting microorganisms can make use of a diverse pool of substrates. Beyond biomass, other organic matter such as municipal solid waste (MSW), or organic industrial waste can be used as input for gasification (National Energy Technology Laboratory, 2014). Furthermore, off-gases from industrial processes, such as steel production (Clarke Energy, 2015) and reformed biogas (Oakley et al., 2011), can serve as direct substrates for gas fermentation (Figure 1). Gas fermentation, therefore, can increase the cyclical carbon emission and fixation in fossil fuel-consuming and carbon-emitting industries. Carbon containing off-gases produced in steel mills, for example, can be sequestered and converted into microbial biomass, fuels, and chemicals. Recycling carbon in this manner can decrease the need for tapping into fossil fuel reserves.
Figure 1.
Overview of feedstock and product options for gas fermentation. Feedstocks to the gas fermentation platform are highlighted in light blue (carbon and electron sources) and green (electron sources). Feedstocks shown are at various stages of commercial deployment. Synthesis of all products shown has been demonstrated including (1) native products (blue text), (2) synthetic products produced through genetic modification (red text), (3) products generated through secondary fermentation of co/mixed cultures (purple text), and (4) products achieved through additional catalytic upgrading (orange text). Acronyms: 2,3-BDO, 2,3-Butanediol; MEK, methyl ethyl ketone.
To fix the relatively oxidized carbon contained in these various syngas sources, acetogens (and other gas-fermenting microorganisms) require reducing equivalents in the form of electrons (such as NAD(P)H or reduced ferredoxin) to reduce the carbon to the central building block acetyl-CoA and further to reduced products such as alcohols. CO and H2 present in syngas themselves can provide these reducing equivalents (see Figure 2 and Section Acetogens and Wood-Ljungdahl Pathway below) by oxidation to CO2 and water (protons), respectively. Reducing equivalents can also be derived from sources other than the syngas sources discussed above.
Figure 2.
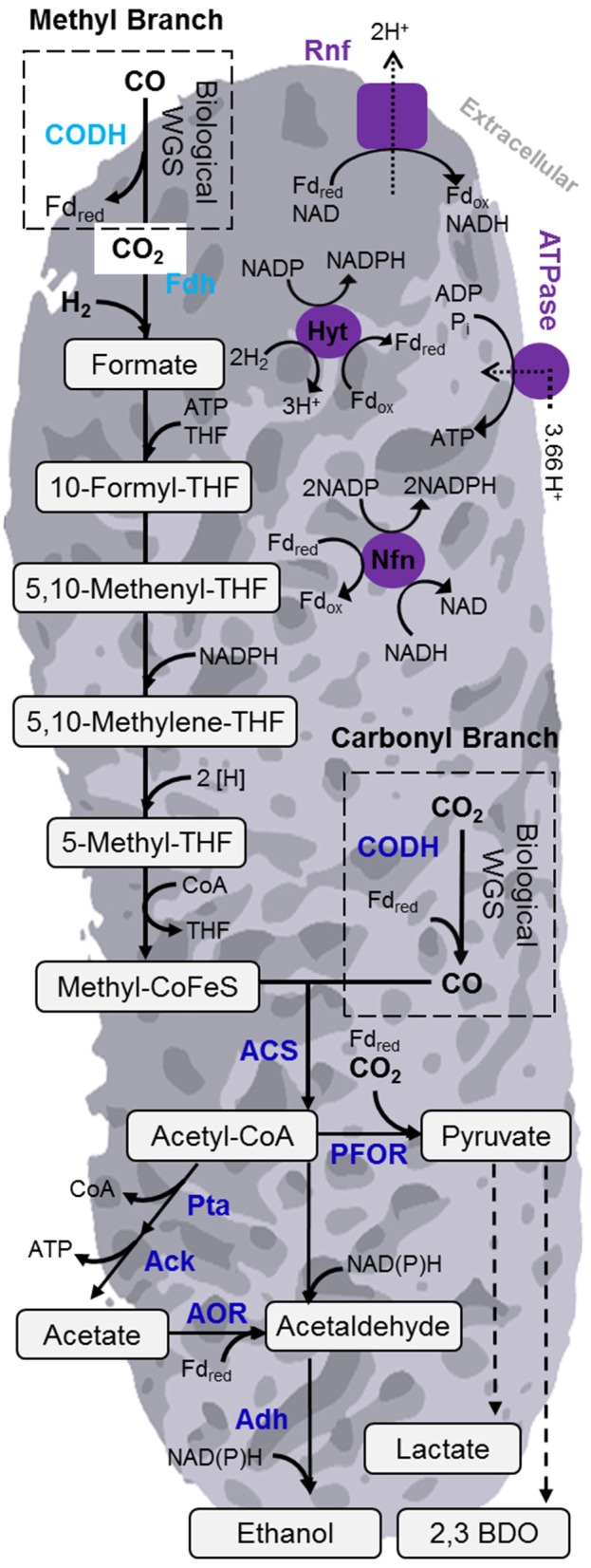
Overview of Wood-Ljungdahl pathway (WLP) and energy conserving mechanisms of acetogen C. autoethanogenum. The WLP is central to the gas fermentation platform for carbon fixation. Noteworthy enzymes are in labeled in blue. The enzymes involved in energy conservation are shown in purple. Acronyms: 2,3-BDO, 2,3-butanediol; AOR, aldehyde:ferredoxin oxidoreductase; ACS, acetyl-CoA synthase; CODH, carbon monoxide dehydrogenase; Nfn, transhydrogenase; PFOR, pyruvate:ferredoxin oxidoreductase; Rnf, Rhodocbacter nitrogen fixation; THF, Tetrahydrofolate; WGS, water-gas shift reaction.
One approach is to use electricity for water electrolysis (produces H2) and/or CO2 electrolysis (produces CO). If the electricity required for water or CO2 electrolysis can be provided by intermittent renewable sources (e.g., wind or solar, see Figure 1), it will not contribute to additional carbon emissions. This approach presents the potential to provide storage of excess electricity during times of low grid demand or surges. Storing excess energy in the form of low-carbon liquid fuels and chemicals is attractive as these chemical are energy dense and no loss of energy over time is observed as for conventional electricity storage.
Several technologies for electrolyzing water using renewable electricity to generate H2 and O2 have already been scaled up and are available commercially. Siemens utilizes a proton-exchange membrane in its Silyzer 200 product with reported overall efficiency of 65–70% (Siemens, 2013). Together with partners Stadtwerke Mainz, Linde and the RheinMain University of Applied Sciences they are operating the world's largest hydrogen electrolysis facility that will be able to process up to 6 mW of power (Aschenbrenner, 2016). Alternatively, Norsk Hydro's subsidiary, NEL Hydrogen, advertises a commercial-scale atmospheric electrolyser (NEL Hydrogen, 2015).
To date, the highest gas fermentation ethanol yields and selectivities have been demonstrated with CO-rich feedstocks (Gaddy et al., 2007), providing additional incentive to develop CO2 electrolysis technology. As mentioned before, it is also of note that during gas fermentation using CO only, a portion of the CO substrate pool must be converted to CO2 to provide necessary reducing equivalents for fixing the gaseous carbon. Some of this CO2 is not fixed and instead emitted from the cell, lowering the yield of carbon substrate fixed. CO2 electrolysis could be used to remedy this situation. Technologies to convert CO2 to CO are still at a pre-commercial stage. Examples include startups such as Opus12 (OPUS 12, 2015) and Dioxide Materials (Ritter, 2015), who report 99% CO2 conversion and greater than 80% efficiency using a proprietary membrane. Alternatively, researchers have found CO2 can be absorbed and reduced to CO using electricity and sponge-like crystals with a efficiency of 90% (Lin et al., 2015).
Another way of providing reducing equivalents is directly from electricity without a gaseous intermediary. Termed microbial electrosynthesis (MES), some acetogenic microorganisms have been shown to use electrical current to reduce CO2 to multi-carbon products (Nevin et al., 2010; Tremblay and Zhang, 2015; Figure 1). It was demonstrated that pure cultures of acetogens such as Clostridium ljungdahlii, Clostridium aceticum, Moorella thermoacetica, and Sporomusa ovata (but not the sodium dependent organism Acetobacterium woodii) form biofilms on the cathode surface of microbial fuels cells and while doing so consume current and generate acetate with 85% electron recovery (Nevin et al., 2011). Photosynthesis, by contrast, only offers between a 2.9 and 4.3% solar-to-biomass conversion efficiency for most crops (Zhang, 2015). MES is an efficient process but biological and engineering challenges remain regarding cultivation and maintenance of a dense biofilm, electron transfer at the cathode, and scale up.
A recent Science paper described an innovative method for direct electron input to the Wood-Ljungdahl pathway (WLP, described in the following section) of an acetogen by photosensitizing the microbes (Sakimoto et al., 2016; Figure 1). When grown with cadmium nitrate and cysteine, M. thermoacetica was able to use biologically generated cadmium sulfide (CdS) semiconducting nanoparticles which absorb light and use the energy to carry out photosynthesis. By feeding reducing equivalents directly into the WJP, solar energy can be converted into acetyl-CoA with 90% selectivity to acetate and 10% selectivity to biomass. This could be an efficient process to convert solar energy into liquid energy (i.e., fuels). However, it is in very early-stage development and requires biological optimization (e.g., production rates, solving issue of photo-oxidation of cell membranes, and selectivity toward other products), and engineering optimization (e.g., scale up and a reactor design where microbes are exposed to sufficient light or light/dark cycles).
Acetogens and wood-ljungdahl pathway
The advantages discussed above can be applied generally to autotrophic microorganisms. Acetogens are particularly attractive for commercialized gas fermentation due to their native ability to synthesize useful products such as ethanol, butanol and 2,3-butanediol and the fact that they are anaerobes. Anaerobic conditions avoid flammability issues working with combustible gases and also makes biological contamination less likely in a sugar and oxygen-free atmosphere. Acetogens are found in over 20 different genera and over 100 different species have been described (Imkamp and Müller, 2007). Table 1 provides an overview of the most noteworthy acetogenic species.
Table 1.
Overview of acetogens.
| Organism | Substrates | Product(s) | Topt(°C) | pHopt | Genome sequence available | GEM available | References |
|---|---|---|---|---|---|---|---|
| MESOPHILIC MICROORGANISMS | |||||||
| Acetobacterium woodii | H2/CO2, CO | Acetate | 30 | 6.8 | Yes | Genthner and Bryant, 1987; Poehlein et al., 2012 | |
| Acetonema longum | H2/CO2 | Acetate, butyrate | 30–33 | 7.8 | Draft | Kane and Breznak, 1991 | |
| Alkalibaculum bacchi | H2/CO2, CO | Acetate, ethanol | 37 | 8.0–8.5 | Allen et al., 2010; Liu et al., 2012 | ||
| Butyribacterium methylotrophicum | H2/CO2, CO | Acetate, ethanol, butyrate, butanol | 37 | 6 | Zeikus et al., 1980; Lynd et al., 1982; Grethlein et al., 1991 | ||
| Clostridium aceticum | H2/CO2, CO | Acetate | 30 | 8.3 | Yes | Adamse, 1980; Braun et al., 1981; Poehlein et al., 2015 | |
| Clostridium autoethanogenum | H2/CO2, CO | Acetate, ethanol, 2,3-butanediol, lactate | 37 | 5.8–6.0 | Yes | Yes | Abrini et al., 1994; Köpke et al., 2011; Brown et al., 2014; Marcellin et al., 2016 |
| Clostridium carboxidivorans or “P7” | H2/CO2, CO | Acetate, ethanol, butyrate, butanol, lactate | 38 | 6.2 | Draft | Liou et al., 2005; Bruant et al., 2010 | |
| “Clostridium coskatii” | H2/CO2, CO | Acetate, ethanol | 37 | 5.8–6.5 | Zahn and Saxena, 2012 | ||
| Clostridium difficile | H2/CO2, CO | Acetate, ethanol, butyrate | 35–40 | 6.5–7.0 | Yes | Rieu-Lesme et al., 1998; Monot et al., 2011; Köpke et al., 2013b | |
| Clostridium drakei | H2/CO2, CO | Acetate, ethanol, butyrate | 25–30 | 3.6–6.8 | Draft | Küsel et al., 2000; Liou et al., 2005; Gössner et al., 2008; Jeong et al., 2014; Bengelsdorf et al., 2015 | |
| Clostridium formicoaceticum | CO | Acetate, formate | 37 | NR | Andreese et al., 1970; Diekert and Thauer, 1978; Lux and Drake, 1992 | ||
| Clostridium glycolicum | H2/CO2 | Acetate | 37–40 | 7.0–7.5 | Ohwaki and Hungate, 1977; Küsel et al., 2001 | ||
| Clostridium ljungdahlii | H2/CO2, CO | Acetate, ethanol, 2,3-butanediol, lactate | 37 | 6 | Yes | Yes | Tanner et al., 1993; Köpke et al., 2010; Nagarajan et al., 2013 |
| Clostridium magnum | H2/CO2 | Acetate | 30–32 | 7.0 | Schink, 1984; Bomar et al., 1991 | ||
| Clostridium mayombei | H2/CO2 | Acetate | 33 | 7.3 | Kane et al., 1991 | ||
| Clostridium methoxybenzovorans | H2/CO2 | Acetate, formate | 37 | 7.4 | Mechichi et al., 1999 | ||
| “Clostridium ragsdalei” or “P11” | H2/CO2, CO | Acetate, ethanol, 2,3-butanediol, lactate | 37 | 6.3 | Huhnke et al., 2008; Köpke et al., 2011 | ||
| Clostridium scatologenes | H2/CO2, CO | Acetate, ethanol, butyrate | 37–40 | 5.4–7.5 | Küsel et al., 2000; Liou et al., 2005 | ||
| Eubacterium limosum | H2/CO2, CO | Acetate, butyrate | 38–39 | 7.0–7.2 | Yes | Genthner et al., 1981; Genthner and Bryant, 1987; Jeong et al., 2015 | |
| Oxobacter pfennigii | H2/CO2, CO | Acetate, butyrate | 36–38 | 7.3 | Draft | Krumholz and Bryant, 1985 | |
| Blautia productus | H2/CO2, CO | Acetate | 37 | 7 | Lorowitz and Bryant, 1984 | ||
| THERMOPHILIC MICROORGANISMS | |||||||
| Moorella thermoacetica | H2/CO2, CO | Acetate | 55 | 6.5–6.8 | Yes | Yes | Kerby and Zeikus, 1983; Daniel et al., 1990; Pierce et al., 2008 |
| Moorella thermoautotrophica | H2/CO2, CO | Acetate | 58 | 6.1 | Savage et al., 1987 | ||
NR, Not reported; GEM, genome-scale network reconstruction.
What makes the biology of acetogens particularly effective is the WLP for CO2 fixation. The WLP, also known as the reductive acetyl-CoA pathway, is the only linear CO2 fixation pathway to acetyl-CoA (Drake et al., 2008) and considered as the most efficient non-photosynthetic carbon fixation mechanism (Fast and Papoutsakis, 2012). There are several excellent reviews on the detailed mechanism and enzymes of the WLP (Wood, 1991; Drake et al., 2006; Ragsdale and Pierce, 2008; Latif et al., 2014). Briefly, the WLP consists of two branches, a methyl (Eastern) and a carbonyl (Western) branch (Figure 2). In the methyl branch, CO2 is reduced to formate. Next, the formate is activated by condensation with tetrahydrofolate (THF) to form formyl-THF, consuming one molecule of ATP. Over several reactions, formyl-THF is reduced to methyl-THF. In the final step of the methyl branch, the methyl group is transferred to a corrinoid iron-sulfur-containing protein (CoFeSP) and then fused to a molecule of CO from the carbonyl branch to form acetyl-CoA via the bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) complex. When grown autotrophically on CO, the CO2 required for the methyl branch is generated by the CODH-catalyzed water-gas shift reaction. Likewise, during autotrophic growth on CO2, the CO is formed from CO2 by CODH in the carbonyl branch (Figure 2). The WLP is also active during heterotrophic growth, where released CO2 can be re-assimilated (Drake et al., 2006). This ability is also exploited in a concept called acetogenic or anaerobic, non-photosynthetic mixotrophy (ANP) to maximize yield in fermentation of sugar with additional hydrogen (Fast et al., 2015).
Even though the WLP is considered as extremely efficient (Fast and Papoutsakis, 2012), it still requires energy in form of one mol ATP per mol of acetyl-CoA formed. This can be balanced via substrate-level phosphorylation (SLP) during the acetate kinase reaction (Figure 2). However, this would require all acetyl-CoA formed in the WLP to be converted to acetate. Therefore, there must exist another mode of energy production to support cell growth and formation of other products. A membrane based mechanism for energy conservation has been proposed, but up to a few years ago exact details were not known, in particular for acetogens that lack cytochromes (Müller, 2003). This changed with the discovery of an Rnf complex in A. woodii (Müller et al., 2008). The Rnf complex (originally identified in Rhodobacter where it plays a role in nitrogen fixation) is able to build up a transmembrane electrochemical sodium ion gradient via coupling to an exergonic electron-transfer reaction. This gradient can then drive ATP synthesis by a membrane-bound F1FO ATPase (Biegel and Müller, 2010; Biegel et al., 2011; Hess et al., 2016; Figure 2). Subsequently, the same mechanism (although proton rather than sodium ion dependent) was also identified in C. ljungdahlii (Köpke et al., 2010; Tremblay et al., 2012; Hess et al., 2016) and other acetogens such as Clostridium autoethanogenum (Mock et al., 2015) and C. aceticum (Poehlein et al., 2015). Not all acetogens possess such an Rnf complex. M. thermoacetica, for example, was found to contain cytochromes and a membrane-bound Ech (energy converting hydrogenase) complex to produce the necessary ion gradient (Huang et al., 2012).
Around the same time the role of the Rnf complex was elucidated, a novel energy conserving mechanism that involves flavin-based bifurcation of electrons was identified in anaerobes (Buckel and Thauer, 2013; Peters et al., 2016). The initial discovery came for a butyryl-CoA dehydrogenase in a non-acetogenic Clostridium kluyveri (Herrmann et al., 2008). Already, several electron bifurcating enzymes have been identified in acetogens since (Wang et al., 2010, 2013a,b; Schuchmann and Müller, 2012; Bertsch et al., 2013; Weghoff et al., 2014) and it is yet to be determined how prevalent this mechanism is (Peters et al., 2016). Important examples include hydrogenase complexes of A. woodii, M. thermoacetica, and C. autoethanogenum (Schuchmann and Müller, 2012; Wang et al., 2013a,b) or the NADH-dependent reduced ferredoxin:NADP oxidoreductase (Nfn) of C. autoethanogenum (Wang et al., 2010, 2013a; Figure 2).
While these unique mechanisms for energy conservation are widespread in acetogens, they are not found in traditional model organisms like E. coli or yeast. The specific differences in energy metabolism between the various acetogens are described in detail elsewhere (Schuchmann and Müller, 2014; Bertsch and Müller, 2015; Diender et al., 2015; Poehlein et al., 2015). Along with some genetic variations these mechanisms directly dictate the observed product spectrums. As an example, A. woodii, C. aceticum, and M. thermoacetica are only producing acetate under autotrophic conditions, despite having the genetic outfit for ethanol production and being able to produce ethanol under heterotrophic conditions growing on sugars. By contrast, C. autoethanogenum, C. ljungdahlii, “Clostridium ragsdalei,” “Clostridium coskatii” and C. carboxidivorans are able to synthesize more reduced products such as ethanol, butanol or 2,3-butanediol under autotrophic conditions. While the WLP genes and enzymes are highly conserved between all these organisms (Poehlein et al., 2015), they differ in some key enzymes such as presence of an electron-bifurcating hydrogenase, electron bifurcating Nfn, and an aldehyde:ferredoxin oxidoreductase (AOR) that enable formation of these more reduced products (Mock et al., 2015).
For commercial applications, mainly C. ljungdahlii type strain PETC (ATCC 55383 = DSM13528) (Tanner et al., 1993) and later isolates ERI-2 (ATCC 55380) (Gaddy, 1997), C-01 (ATCC 55988) (Gaddy, 2000), and O-52 (ATCC 55989) (Gaddy, 2000); C. autoethanogenum type strain JAI-1 (DSM10061) (Abrini et al., 1994) and evolved versions LBS1560 (DSM19630) (Simpson et al., 2009b) and LBS1561 (DSM23693) (Heijstra et al., 2012); C. carboxidivorans type strain P7 (ATCC BAA-624 = DSM 15234) (Liou et al., 2005); and isolates “C. ragsdalei” (ATCC BAA-622) (Huhnke et al., 2008) and “C. coskatii” (Zahn and Saxena, 2012) are considered. The latter two isolates have only been described in patent literature and not been characterized in a systematic journal. It is worth noting the importance that adaptive laboratory evolution plays in developing the strains being employed for commercial projects. Production strains often need to be adapted for high ethanol production under different fermentation conditions. For example, the C. autoethanogenum type strain JAI-1 has been reported to have low biomass growth, poor ethanol production, and minimal 2,3-butanediol production (Cotter et al., 2009; Guo et al., 2010; Köpke et al., 2011; Abubackar et al., 2015b), but the evolved C. autoethanogenum strain LBS1561 has demonstrated much greater production rates of 195 g/L/d ethanol (Molitor et al., 2016) and 14 g/L/d 2,3-butanediol (Simpson et al., 2014). Highest reported ethanol production rate in the literature was 360 g/L/d with C. ljungdahlii strain C-01, but in this case elevated pressure (1.7-2 bar) rather than atmospheric pressure was used (Gaddy et al., 2007).
Commercialization and life-cycle analysis
The efficiency, diversity of substrates, and product selectivity advantages of gas fermentation have led to scaling up the fermentation process for commercial-scale production of low-carbon fuels using acetogens. Three companies, Coskata, INEOS Bio, and LanzaTech have operated pilot and demonstration plants for extended periods of time. Coskata's technology formed the basis of a new company Synata Bio (Lane, 2016) but has not been scaled up further. INEOS Bio and LanzaTech, on the other hand, are currently scaling up their processes to commercial scale.
INEOS Bio built an 8 million gallon per year (Mgy) semi-commercial facility in Vero Beach, FL as a joint venture with New Planet Energy Holdings, LLC. Commissioned in 2012, the facility uses lignocellulosic biomass and MSW for generating gas fermentation substrates and generates 6 mW of electrical power. In July 2013, the company announced successful production of ethanol in its facility (Schill, 2013). In September 2014, operational changes were imposed to optimize the technology and de-bottleneck the plant to achieve full production capacity (INEOS Bio, 2014).
LanzaTech has successfully operated two 0.1 Mgy pre-commercial plants in different locations in China with two steel companies, BaoSteel and Shougang Steel. Both plants, the first at one of BaoSteel's mills in Shanghai in 2012 and the second at a Shougang steel mill near Beijing in 2013, used steel mill off-gases as substrates for gas fermentation. The Shougang facility was certified by SCS Global Services in 2013 according to Roundtable on Sustainable Biomaterials (RSB) principals (SCS Global Services, 2013). The RSB is a global sustainability standard and certification system for biofuels and biomaterials production. In 2015, both China Steel Corporation of Taiwan and ArcelorMittal of Luxembourg approved commercial projects with LanzaTech. The former will be a 17 Mgy facility with the intention to scale up to 34 Mgy (Lane, 2015b). The latter 9.8 Mgy facility will be built at ArcelorMittal's flagship steel plant in Ghent, Belgium with the intention to construct further plants across ArcelorMittal's operations (Lane, 2015c). If scaled up to its full potential in Europe, the technology could enable the production of around 104 Mgy which would displace 1.6 million barrels of fossil fuel-derived gasoline on a BTU basis. In addition to these two projects, Aemetis, Inc. acquired a license from LanzaTech for the conversion of agricultural waste, forest waste, dairy waste and construction and demolition waste (CDW) to ethanol in California. In a first phase, Aemetis plans to adopt the process by adding an 8 Mgy gas fermentation unit to its existing 60 Mgy first generation biofuel facility in Keyes. This technology enables Aemetis to produce advanced ethanol that is valued up to approximately $3 per gallon more than traditional ethanol (LanzaTech, 2016).
Though these are promising signs that low-carbon fuel production by gas fermentation may be a commercial success in the near future, ongoing analyses in the form of Life Cycle Assessments (LCA) are necessary to ensure that environmental concerns are fully evaluated before commercial deployment. In a recently published cradle-to-grave LCA by Handler et al. (2016), the production of ethanol from gas fermentation in the USA is estimated to result in 67% Greenhouse gas (GHG) reduction (using blast oxygen furnace off-gas from steel manufacturing) and 88–98% GHG reduction (utilizing gasified biomass), when compared to conventional fossil gasoline. In both feedstock scenarios, 20–40% of the carbon in feed-gas is converted into ethanol. Conclusions drawn from LCA are often highly geographically dependent. A separate and older study showed that approximately 50% GHG savings can be attained from a microbial gas-to-ethanol platform based in China, relative to fossil gasoline (Ou et al., 2013).
Process overview
The overall gas fermentation process can be broadly divided into four steps: (1) accumulation or generation of syngas; (2) gas pretreatment; (3) gas fermentation in a bioreactor; and (4) product separation.
Generation of syngas
The versatility of acetogenic bacteria to ferment syngas of diverse compositions means virtually any carbonaceous materials can be gasified to generate the feedstock as discussed in Section Substrate Diversity. When gasification is utilized, the carbonaceous material reacts with steam and air at elevated temperature (600–1000 °C) (Griffin and Schultz, 2012) and high pressure (>30 bar) (Breault, 2010) to form syngas of variable composition (depending on input and process parameters). Although a small amount of energy input is required to heat the incoming feedstock to gasification temperature at the beginning of the process, at steady-state the process is self-sustaining (Handler et al., 2016). Excess heat generated from gasification can be used to generate steam for product distillation and/or electricity (Handler et al., 2016). Depending on the type of gasifier, the starting material may be subjected to drying, commination (size reduction), chipping, pelletization, torrefaction, pyrolysis and/or pulverization prior to gasification (Bronson et al., 2012; Isaksson et al., 2013). Defined by how the reactor brings about contact with the feedstock and reactive gas, there are four main gasifier configurations: fixed/moving bed, fluidized bed, entrained flow, and transport flow (McKendry, 2002; Breault, 2010). Fluidized bed gasifiers are currently the most commonly used biomass gasifiers due to their ease of up-scaling, isothermal operation conditions and high feedstock conversion efficiencies (Swanson et al., 2010; Siedlecki et al., 2011; Griffin and Schultz, 2012).
Gas pretreatment
In addition to the main constituents CO, H2, CO2, input gas streams can also contain impurities such as particulates, tar, aromatics grouped as benzene, toluene, ethylene, xylenes (BTEX) and naphthalene, sulfur compounds such as hydrogen sulfide (H2S), carbonyl sulfide (COS), and carbon disulfide (CS2), halogens such as chlorine and hydrogen fluoride (HF), and other potentially inhibiting gases such as ammonia (NH3), nitric oxide and nitrogen dioxide (NOx), acetylene, oxygen (O2), reactive oxygen species (ROS), and hydrogen cyanide (HCN). These are generated for example during the gasification process, pyrolysis or manufacturing and can be present in fluctuating quantities (Oremland and Taylor, 1975; Abubackar et al., 2011; Griffin and Schultz, 2012; Munasinghe and Khanal, 2012; Zahn, 2015). A complete understanding of impurity species, their concentration fluctuations based on syngas input, process variables as well as installed treatment capacity is critical to maintain optimal productivity. In addition, monitoring impurity accumulation patterns within the fermentation is required to determine biological tolerance levels and the minimal inhibitory concentration (MIC) for designing economical treatment capacity. Understanding the effect of impurities could save treatment costs, however failure to do so can cause delays reaching full scale production capacity as shown in one large-scale syngas fermentation endeavor (Lane, 2014).
Even with gas-fermenting microorganisms' abilities to grow in the presence of low levels of impurities, some impurities necessitate near complete removal from an operational, biological and/or product specificity perspective. Particulates can be removed by cyclone separators and filters. Tars can be condensed and removed by quenching hot syngas, or, alternatively, can be reformed by heating at 800–900°C using olivine, dolomite, and nickel compounds as catalysts, generating additional syngas (McKendry, 2002).
Many contaminants including BTEX are lipophilic compounds that readily dissolve in the cytoplasmic membrane affecting membrane fluidity (Sikkema et al., 1995). Although polycyclic aromatics do not readily dissolve in aqueous phase, they can accumulate and negatively affect operations. Removal technologies for aromatics from gas are commercially available, and techniques to improve efficiency are still being reported (Ye and Ariya, 2015). O2's toxicity above microoxic levels is particularly critical during inoculation of a bioreactor, when little biomass has accumulated to withstand introduction of aerobic conditions. O2 can be tolerated in certain microoxic conditions (Kawasaki et al., 2005). C. ljungdahlii has been shown to detoxify O2 and ROS (likely via rubrerythrin and hydrogen peroxidases) and ethanol formation could actually be stimulated by exposure to O2 (likely due to changes in AOR activity and co-factor metabolism) (Whitham et al., 2015). O2 can be removed by passing the gas over various metal catalysts such as Pt, Pd, and Cu (Yan et al., 2013). Using biological co-culture for O2 removal has also been described (Wu et al., 2016). Sulfur-containing impurities (e.g., H2S and COS) can poison the metal based catalysts (Vega et al., 1990; Rodriguez and Hrbek, 1999) and require prior removal despite the microorganisms' ability to grow in their presence (Griffin and Schultz, 2012).
Acetylene, NOx and HCN are considered particularly troublesome as they are known to inhibit enzymes responsible for initial harvesting of energy from syngas (Anderson et al., 1993; Shima and Ataka, 2011). Cyanide binds to CODH (Ragsdale et al., 1983), a key enzyme of the WLP. NO is a non-competitive inhibitor of hydrogenase activity (Ahmed and Lewis, 2007) while acetylene reversibly inhibits hydrogenases (Krasna and Rittenberg, 1954; He et al., 1989; Maness and Weaver, 2001), which reduce ferredoxin for use in redox reactions (Figure 2). INEOS Bio has identified and reported HCN as a key contaminant that needs treatment from operation of their Vero Beach plant (INEOS Bio, 2014).
Gas fermentation
Treated syngas is next cooled and compressed then sparged into a bioreactor containing the gas-fermenting microorganisms in an aqueous medium. There are a multitude of variables to account for during gas fermentation. Bioreactor design, agitation, gas composition and supply rate, pH, temperature, headspace pressure, oxidation-reduction potential (ORP), nutrients, and amount of foaming in the bioreactor all can contribute to the goal of improving selectivity and yield of the desired product (e.g., ethanol and butanol) as discussed below.
One major obstacle immediately present in gas fermentation is the low solubility of the gaseous substrates and combined with an efficient transfer of their masses into the liquid media. CO, H2, and CO2 are soluble to approximately 28 mg/L, 1.6 mg/L, and 1.7 g/L (293 K, 1 atm), respectively, compared to 900 g/L for glucose, a prevalent substrate for traditional fermentations. As gas-fermenting microorganisms consume the gas, substrate availability can become rate-limiting. Increasing flow of the substrate gas can lead to decreased yields of product per mole of carbon fed to the reactor, making reactor design and operation crucial. Continuous stirred tank reactors (CSTR) offer excellent mixing and homogenous distribution of gas substrates to the microorganisms and are most commonly employed at laboratory scale (Ungerman and Heindel, 2007). However, the high power per unit volume required to drive the stirrer renders commercial scale operation economically challenging. Therefore, other less energy-demanding bioreactor designs such as bubble column, loop, and immobilized cell columns and their specific volumetric mass-transfer coefficients (kLa) that describes the efficiency of which a gas can be delivered to a bioreactor have been investigated intensively and reviewed elsewhere (Klasson et al., 1991; Bredwell et al., 1999; Ungerman and Heindel, 2007; Munasinghe and Khanal, 2010; Abubackar et al., 2011; Liew et al., 2013; Orgill et al., 2013).
Next to gas availability determined by reactor kLa, the ratio and partial pressures of CO, H2, and CO2 also influence the product yield, production rate, and selectivity of a gas fermentation (Genthner and Bryant, 1982; Vega et al., 1988; Klasson et al., 1991; Gaddy and Chen, 1998; Hurst and Lewis, 2010; Kantzow et al., 2015). CO and H2 are sources of electrons/reducing equivalents for reducing CO2 in the WLP and generating reduced products over acid products (e.g., ethanol vs. acetate). This product profile reflects the organism's requirement to maintain an internal energy balance that favors growth and is directly influenced by the gas composition and availability. As an example, productivity with C. ljungdahlii was improved from 38.4 g/L/d at 1 atmosphere to 360 g/L/d at 6 atmospheres (Gaddy et al., 2007). Another strategy to control the product profile is lowering the pH of the fermentation culture. This pH change can lead to a (reversible) shift from acidogenesis to solventogenesis allowing an increased production of ethanol and other highly reduced products (Grethlein et al., 1990; Gaddy and Claussen, 1992; Phillips et al., 1993; Guo et al., 2010; Abubackar et al., 2012, 2015b, 2016a,b; Richter et al., 2013b; Martin et al., 2016).
Besides gas, medium composition also affects product yield and selectivity. Nutrient optimization has proven to be a process and species specific requirement. Media optimizations have been conducted for many acetogens including C. autoethanogenum (Cotter et al., 2009; Guo et al., 2010; Simpson et al., 2010; Abubackar et al., 2015a, 2016a), C. ljungdahlii (Phillips et al., 1993), “C. ragsdalei” (Babu et al., 2010; Kundiyana et al., 2010, 2011; Panneerselvam et al., 2010; Maddipati et al., 2011; Phillips et al., 2011; Saxena and Tanner, 2011, 2012), C. aceticum (Sim and Kamaruddin, 2008), and M. thermoacetica (Lundie and Drake, 1984; Savage and Drake, 1986). B vitamins and metals such as zinc, nickel, selenium, and tungsten, required as cofactors for certain enzymes in the central metabolism, are required for bacterial growth and affect product selectivity. For an industrial process it is critically important to keep media cost low, for example by eliminating yeast extract requirements, recycling nutrients and usage of industrial-grade bulk chemicals.
Product separation
Finally, product separation is required to separate the desired metabolic product from the fermentation broth. Distillation systems are common to separate lower boiling point products such as ethanol (Handler et al., 2016) and acetone, but this is considered energy-intensive (and therefore potentially expensive), especially for low concentration products and products with high boiling points (e.g., butanediol). Other technologies to separate fermentation products from broth include liquid-liquid extraction, gas stripping, adsorption, perstraction, pervaporation, and vacuum distillation (Huang et al., 2008; Frolkova and Raeva, 2010; Liew et al., 2013; Molitor et al., 2016) Each of these separation technologies has their own benefits and drawbacks, including potential fouling of membranes (perstraction and pervaporation) and substrate removal (gas stripping and liquid-liquid extraction). Liquid-liquid extraction is also an option for removing acetate from the fermentation broth of gas-fermenting acetogens (Jipa et al., 2009).
Synthetic biology approaches to expand product spectrum of gas fermentation
Overview of synthetic biology for acetogens
Synthetic biology and metabolic engineering approaches offer great promise to improve the efficiency of gas fermentation and to expand the product spectrum beyond native products ethanol, acetate and butanediol to a range of higher-value fuels and commodity chemicals (Latif et al., 2014; National Research Council, 2015; Figure 1). Of particular interest are products that can be separated using the same distillation technology used for ethanol in current commercial endeavors. In these cases, only minimal modifications to the process conditions or fermentation regime would be required but not to the existing plant itself to shift from one product to another. This is a real paradigm shift to typical chemical production plants that cannot react to changing market conditions.
To approach these goals in acetogens, consider the model organisms, E. coli and yeast. These have successfully been reprogrammed to convert sugars at commercially relevant rates and concentrations to new products like farnesene, isobutanol, 1,3-propanediol (1,3-PDO), and 1,4-butanediol (1,4-BDO) (Lan and Liao, 2013; Cho et al., 2014; George et al., 2015; Burk and Van Dien, 2016). Although these advancements cannot be directly transferred to acetogens, similar principles may be applied. Historically, development has been hampered by a lack of available information and tools. E. coli and yeast are well studied and characterized on molecular and systems level with decades of intensive research by thousands of research groups. This has led to the development of sophisticated genetic toolkits that allow precise and rapid design (Temme et al., 2012b) and automated high-throughput strain engineering and prototyping (Gardner, 2013; Burk and Van Dien, 2016). In contrast, acetogens have been viewed as genetically inaccessible until a few years ago and are still considered difficult to work with (Burk and Van Dien, 2016). More specifically, cultivation under an industrial setting, the limited number of genetic tools (Burk and Van Dien, 2016), potential energetic constraints (Bertsch and Müller, 2015), and lack of characterization on genetic and molecular levels are considered key challenges to address for successful development of genetically engineered gas-fermenting platforms.
However, over the last 5 years, great strides have been made that have contributed to the understanding of acetogens on a molecular and systems level. These include elucidation of whole genome sequences for a range of acetogens including C. autoethanogenum (Brown et al., 2014; Humphreys et al., 2015; Utturkar et al., 2015), C. ljungdahlii (Köpke et al., 2010), C. aceticum (Poehlein et al., 2015), M. thermoacetica (Pierce et al., 2008), A. woodii (Poehlein et al., 2012), and E. limosum (Roh et al., 2011); identification of cofactors for key reactions for C. autoethanogenum (Wang et al., 2013a; Mock et al., 2015), A. woodii (Schuchmann and Müller, 2012, 2014), M. thermoacetica (Huang et al., 2012; Mock et al., 2014), and E. limosum (Jeong et al., 2015); and transcriptomics analyses of C. autoethanogenum (Mock et al., 2015; Marcellin et al., 2016) and C. ljungdahlii (Nagarajan et al., 2013; Tan et al., 2013; Whitham et al., 2015). Even more recently, a first complete systems level study comprising of transcriptome, metabolome, and proteome analyses of C. autoethanogenum was published (Marcellin et al., 2016). This study demonstrated that the ATP pool remains constant during autotrophic growth on gas and heterotrophic growth on fructose and dissected the underlying mechanisms. Additionally, a genome-scale model (GEM) was provided to allow more rational strain design. Similar models also exist for C. ljungdahlii (Nagarajan et al., 2013) and M. thermoacetica (Islam et al., 2015). Based on the C. ljungdahlii GEM, spatiotemporal modeling of gas fermentation in a bubble column was performed recently (Chen et al., 2015, 2016). With continued advancement, these GEMs may enable development of efficient strategies and strain designs in a manner similar to that observed for E. coli and yeast.
In parallel, a range of genetic tools have been developed or adapted for gas-fermenting acetogens. Most of the work has focused on acetogenic clostridia, in part due to their properties as commercial production strains for gas-fermentation (see Section Acetogens and Wood-Ljungdahl Pathway) but also due to the presence of pre-existing tools for non-acetogenic clostridia studied for medical purposes (e.g., Clostridium difficile and Clostridium botulinum), or exploited for acetone/isopropanol-butanol-ethanol (ABE/IBE) fermentation (e.g., Clostridium acetobutylicum and Clostridium beijerinckii) and or their ability to degrade cellulosic material (e.g., Clostridium cellulolyticum). An overview of genetic tools is provided in Section Genetic Tools Development. In Section Successful Examples of Metabolic Engineering of Gas-Fermenting Organisms to Expand Product Portfolio, details of several successful examples of metabolic engineering of acetogenic organisms are discussed. These results have been possible due to the enhanced understanding of acetogens and genetic tools development. To accelerate development of new commercially relevant strains and processes, it will be important to further refine and improve the existing tools and models and increase throughput in strain design and prototyping.
Genetic tools development
DNA transfer
The establishment of an efficient protocol to introduce foreign DNA into a microbial host is frequently one of the most challenging steps in any genetic engineering endeavor. This is especially true for Gram-positive bacteria such as clostridia and most other acetogens due to the physical barrier of the thick peptidoglycan cell wall and highly active restriction-modification systems (Pyne et al., 2014). As a result of the inability to transfer DNA, the advancement of genetic tools in clostridia has been severely hindered and lagged behind genetically accessible model organisms such as E. coli or yeast. Some of the well-studied, non-acetogenic clostridia, such as the ABE model organisms C. acetobutylicum (Mermelstein et al., 1992) and C. beijerinckii (Oultram et al., 1988), and important pathogens, such as C. difficile (Ackermann et al., 2001) and C. botulinum (Zhou and Johnson, 1993), have transformation protocols that were established as early as 1988. It is intriguing that C. difficile was recently found to also harbor the WLP and is able to grow autotrophically on H2 and CO2 (Köpke et al., 2013b). Because it is a human pathogen, C. difficile is not considered for industrial use, but studies conducted on the metabolism and development of genetic tools for this pathogen are of interest for acetogens. Methods for genetic manipulation in acetogens other than C. difficile were only beginning to emerge since 2010 (Köpke et al., 2010).
Due to its technical simplicity, higher reproducibility, scalability, greater transformation efficiency, and independence from donor species, electroporation is the most commonly explored method of transformation in microorganisms. Transfer of foreign DNA was successfully demonstrated for C. ljungdahlii (Köpke et al., 2010; Leang et al., 2013), C. autoethanogenum (Köpke and Liew, 2011), C. aceticum (Schiel-Bengelsdorf and Dürre, 2012), A. woodii (Straub et al., 2014), and M. thermoacetica (Kita et al., 2013). With optimizations, a transformation efficiency of up to 1.7 × 104 cfu/μg DNA for C. ljungdahlii was achieved by Leang et al. (2013). Advanced next generation sequencing platforms such as Single Molecule Real-Time (SMRT) sequencing can be employed to reliably detect certain methylation signatures (Flusberg et al., 2010; Fang et al., 2012) as it has been done for C. autoethanogenum (Utturkar et al., 2015). This information can then be used to formulate in vitro or in vivo methylation strategies or disruption of endonucleases to protect foreign DNA from the host's restriction barrier. For instance, genetic disruption of restriction endonuclease in non-acetogen C. acetobutylicum (Dong et al., 2010) and C. cellulolyticum (Cui et al., 2012) resulted in significantly improved transformation efficiency using unmethylated plasmids.
Alternatively, the transfer of foreign DNA into host cells can be accomplished via conjugation. This method requires direct cell-to-cell contact during the transfer of DNA from donor to recipient so this approach is limited by the host range of the donor. However, transfer of DNA via conjugation has been reported to occur in single strand, by which means the incoming DNA can evade the recipient's restriction endonucleases (Jennert et al., 2000; Purdy et al., 2002). Conjugation was reported for C. autoethanogenum (using E. coli strain HB101 as donor) (Mock et al., 2015), and A. woodii (using E. coli strain S17-1 as donor) (Strätz et al., 1994).
The presence of active restriction modification systems (methyltransferases and nucleases) in acetogens, coupled with low transformation efficiencies, means routine molecular biology work such as plasmid purification and ligation cloning have to be performed first in a separate “preparation” host (e.g., E. coli) before introduction into acetogens. This necessitates the development of shuttle plasmids that contain replicon(s) and selection marker(s) that enable the propagation and selection of the plasmids in both acetogens and “preparation” host. The modular Clostridium-E. coli shuttle plasmids developed by Heap et al. (2009) contain customizable components such as different Gram-positive and Gram-negative replicons as well as the traJ gene (for conjugation), and it has been adopted for genetic manipulation work in several acetogens (Köpke and Liew, 2011; Leang et al., 2013; Ueki et al., 2014; Hoffmeister et al., 2016). Future development of high-efficiency transformation protocols, use of non-circularized DNA, direct cloning and intact plasmid purification directly from acetogens will be important to significantly improve the workflow, throughput and automation of recombinant acetogen constructions.
Knock-down of target genes via antisense RNA (asRNA)
When genetic tools development was still in its infancy and stable inactivation mutants were difficult to generate, knock-down of target genes via the actions of plasmid-delivered antisense RNA (asRNA) represented an alternative route for genetic manipulation. In cases where the target gene is essential to survival of the microorganism, knock-down is still an option over a complete knock-out. By applying asRNA, up to 90% reduction in gene expression together with high level of specificity were reported for clostridia (Desai and Papoutsakis, 1999; Tummala et al., 2003; Scotcher et al., 2005).
Intron-based gene inactivation
Some of the first reliable mutagenic tools for clostridia were based on intron-based gene inactivation. The approach utilizes the specificity of mobile group II intron Ll.ltrB from Lactoccocus lactis to propagate into the specified site via a RNA-mediated, retro-homing mechanism (Karberg et al., 2001). Heap et al. (2007) adapted such mobile elements for use in a wide spectrum of clostridia, and the technology is termed “ClosTron.” Without a selection marker, the screening effort necessary for the isolation of the desired integrant can be immense due to the variability in integration frequency between target sites. By incorporating a clostridial retro-transposition activating marker (RAM) based on the ermB gene of the Enterococcus faecalis plasmid pAMβ1, positive selection for the desired integration can be conveniently made by the bacteria's acquisition of erythromycin resistance (Heap et al., 2007, 2010; Kuehne and Minton, 2012). Thanks to these properties, this technique is reproducible and has been widely applied to various clostridia (Heap et al., 2007, 2010; Camiade et al., 2010) including acetogen C. autoethanogenum (Mock et al., 2015; Marcellin et al., 2016). The technique should also be amendable to other non-clostridia mesophilic acetogens such as A. woodii but may not be applicable for some of the thermophilic organisms such as M. thermoacetica. This technique was further optimized to generate multiple insertional mutations in the same strain by recycling of marker through the actions of FLP recombinase, and the delivery of cargo sequence of up to 1 kb was successfully demonstrated in C. sporogenes (Heap et al., 2010).
Stable genome insertion or deletion via homologous recombination
For stable genome insertion, homologous recombination is typically used. To facilitate this method, several counter-selectable markers to allow efficient screening for the rare second recombination event have been developed for clostridia including upp (encoding a uracil-phosphoribosyl-transferase) (Soucaille et al., 2008; Croux et al., 2016), mazF (E. coli toxin) (Al-Hinai et al., 2012a), codA (cytosine deaminase) (Cartman et al., 2012), and pyrE or pyrF (orotate phosphoribosyltransferase) (Ng et al., 2013). PyrF has also been shown to be functional in thermophilic acetogen M. thermoacetica (Kita et al., 2013). For acetogen C. autoethanogenum, two additional counter-selectable marker pheS* (modified phenylalanine tRNA synthase) and thiK (thymidine kinase) have been developed (Walker et al., 2015).
As alternative approach to the use of a counter-selectable marker, it is also possible to rely on single crossover recombination facilitated by an antibiotic marker and then excise the marker. This can either be achieved using above mentioned FLP recombinase (Lee et al., 2016) or using the Cre-lox system (Sauer, 1987). This system utilizes a site-specific recombinase and lox recognition sites, to delete the vector backbone and allowed the recycling of an antibiotic resistance gene. Cre-Lox has been successfully demonstrated in the acetogen C. ljungdahlii (Ueki et al., 2014).
Precise genetic manipulation via allelic-exchange
Recently, a novel genetic tool that allows stable genome insertion via homologous recombination was developed. Termed Allele-Coupled Exchange (ACE), this approach does not employ a counter selective marker to select for the rare second recombination event. Instead, it utilizes the activation or inactivation of gene(s) that result in a selectable phenotype, and asymmetrical homology arms to direct the order of recombination events (Heap et al., 2012). For example, a promoterless primary/secondary adh from C. beijerinckii coupled to the ermB gene was introduced into the downstream region of the thlA gene in the genome of C. acetobutylicum. The native promoter of thlA drives the expression of ermB and adh, allowing selection for the second recombination event using erythromycin or acid-stable analog clarithromycin, and also creation of a recombinant that reduces acetone to the more desirable isopropanol (Heap et al., 2012). Remarkably, the whole genome of phage lambda (48.5 kb minus a 6 kb region) was successfully inserted into the genome of C. acetobutylicum in three successive steps using this genetic tool. This technique was also demonstrated in C. difficile and C. sporogenes, suggesting it is applicable to other Clostridium spp. including acetogens (Heap et al., 2012; Ng et al., 2013).
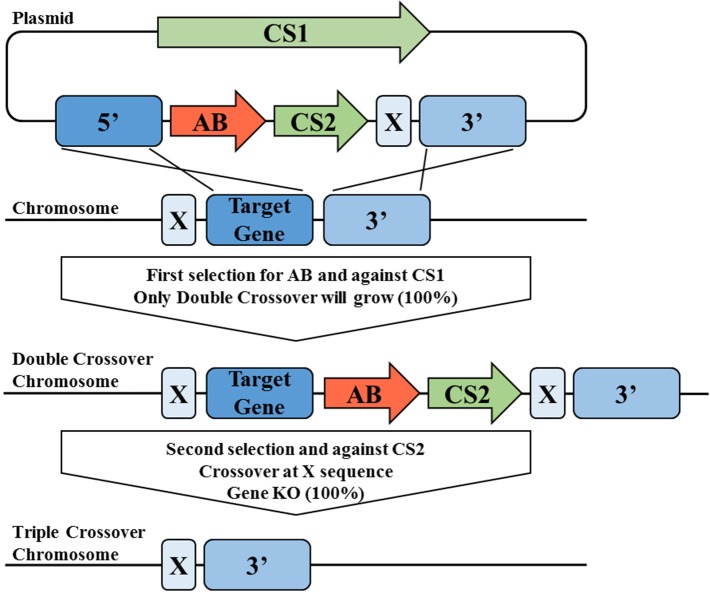
Precise genetic manipulation via triple cross
Recently a novel allelic-exchange based tool termed “Triple Cross” for acetogen C. autoethanogenum was described in a patent application (Walker and Köpke, 2015). Unlike the classical two-step, double-crossover approach which involves one positive and one negative selection markers with two homology arms, the two-step Triple Cross tool utilizes one positive and two negative selection markers with three homology arms. Instead of screening for a first crossover in the first step and a second crossover with a marker recycling in the second step, this invention forces a double crossover directly in the first step using a combination of positive selection marker and a negative selection marker. The optional second step involves the recycling of selection marker through the third homology arms and the second negative selection marker in a third crossover event (Figure 3). Using this technique, a 2,3-butanediol dehydrogenase gene and a secondary alcohol dehydrogenase gene of C. autoethanogenum were successfully in-frame deleted in a “scarless”-manner (Walker and Köpke, 2015). This approach can also be extended to perform other forms of precise genetic manipulation, including insertion and point mutations.
Figure 3.
Overview of the Triple Cross tool for precise genetic manipulations. The triple cross tool relies on two counter-selectable markers (CS1 and CS2) in combination with one antibiotic marker (AB). CS1 and AB1 are located between two homology arms (5′ and 3′) together with a shorter, third homology arm (X). Using selection for AB and against CS1, a direct double crossover event at homology arms 3′ and 5′ is forced in a first step, this is facilitated by having homology arms of different length. In an optional second step, the marker can be recycled using shorter third homology arm X and selection against CS2. Shown is deletion of a target gene, but the same technology can also be used to deliver genes by placing the respective sequence between homology arms X and 3′. Depending how the homology arms are placed, the gene can either be inserted at any given position in the genome or an existing sequence be replaced by a new one.
Transposon mutagenesis
The genetic tools described so far constitute “reverse genetics,” in which a defined gene(s) is selectively inactivated or introduced to study the phenotype. In contrast, “forward genetics” do not make any assumptions about the genes involved and aim to determine the genotype resulting in a particular phenotype. To achieve this, a pool of random mutants needs to be generated. This can be accomplished by means of transposon mutagenesis. Earlier efforts of transposon mutagenesis were demonstrated in C. saccharobutylicum (Keis et al., 2001), C. acetobutylicum, and C. beijerinckii, but issues with multiple transposon insertions per mutant, and non-random distribution of insertions were also reported (Bertram and Dürre, 1989; Woolley et al., 1989). Recent developments have seen the successful generation of mono-copy random insertion of transposon Tn1545 into C. cellulolyticum (Blouzard et al., 2010) and mariner transposon Himar1 into C. difficile (Cartman and Minton, 2010; Zhang et al., 2015b). When coupled with next generation sequencing, transposon-directed insertion site sequencing (TraDIS) was performed on C. difficile and identified 404 essential genes for growth and 798 genes that are likely to affect spore production (Dembek et al., 2015). Recently, an inducible transposon system for random mutagenesis has been described for C. acetobutylicum that showed over 90% successful transposition (Zhang et al., 2016).
CRISPR/Cas9
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system is a RNA-guided prokaryotic immune system which can cleave foreign DNA (e.g., phages and plasmids). The type II CRISPR/Cas9 system has been exploited for high-efficiency gene editing in model microorganisms such as E. coli (Jiang et al., 2015), S. cerevisiae (DiCarlo et al., 2013), and Streptococcus pneumoniae (Jiang et al., 2013). Recently, the CRISPR/Cas9 system from Streptococcus pyogenes was successfully applied to various clostridia (Wang et al., 2015; Xu et al., 2015) to deliver one-step, markerless, and highly efficient gene editing. For instance, DNA cargo of sizes 0.7 and 1.7 kbp (but not 3 and 6 kbp) were specifically inserted into the genome of C. cellulolyticum in a single step without marker using CRISPR/Cas9 nickase with single-nick triggered homologous recombination (Xu et al., 2015). This demonstrates potential in making use of this new technology in gas-fermenting microorganisms. Some acetogens as C. autoethanogenum also have a native CRSIPR system, that has been shown to be actively transcribed (Brown et al., 2014).
Genetic parts
In addition to the further development of tools, more validated parts are required to allow efficient metabolic engineering and refinement of the existing tools. This include variable-strength promoters, inducible expression systems, genetic circuits, transcription terminators, and ribosomal bindings sites (RBSs). For promoter library assembly, transcriptomic studies using RNA-seq can be used to identify promoters of various transcription intensities (Sharma and Vogel, 2014; Thomason et al., 2015).
Large libraries of promoters (Temme et al., 2012a), terminators (Chen et al., 2013b), and RBS calculators (Farasat et al., 2014) have been developed and characterized for model organisms like E. coli, dramatically increasing the control of metabolic pathways and allow efficient pathway refactoring and design of genetic circuits (Temme et al., 2012a,b). Though these parts may be transferable to acetogens, proper characterization/confirmation has not been completed.
There has been some progress on parts specifically from and for acetogens, specifically inducible promoter systems. A lactose-inducible system (bgaR-PbgaL) from Clostridium perfringens was successfully adapted for use in C. ljungdahlii (Banerjee et al., 2014) and a tetracycline inducible system (Tet3no) has been demonstrated in C. autoethanogenum (Walker and Köpke, 2015).
To make full use of these and any new genetic parts generated for acetogens in the future, reporter genes will need to be employed to assess the expression level and localization of a gene product. However, commonly used reporters such as green fluorescent protein (GFP) require oxygen for the development of the chromophore responsible for fluorescence. Gene expression reporter systems that function under anaerobic conditions have emerged, including the β-glucuronidase gene (gusA) (Girbal et al., 2003; Dong et al., 2012; Banerjee et al., 2014), chloramphenicol acetyltransferase gene (catP) (Heap et al., 2007; Zhang et al., 2015b), flavin-based fluorescent proteins (Drepper et al., 2007, 2010; Mukherjee and Schroeder, 2015), luxA-luxB reporter system (Phillips-Jones, 2000), and lacZ gene from Thermoanaerobacterium thermosulfurogenes EM1 (Tummala et al., 1999).
Successful examples of metabolic engineering of gas-fermenting organisms to expand product portfolio
Even though genetic tools and parts for acetogens are still underdeveloped, there are already several successful examples of metabolic engineering of acetogens for non-native product synthesis (Figure 1, Table 2). A first proof of concept study that acetogens can be genetically modified was published in 2010 (Köpke et al., 2010). The study successfully showed the production of non-native butanol in C. ljungdahlii. Upon transformation with a plasmid expressing butanol biosynthetic genes (thlA, hbd, crt, bcd, adhE, and bdhA) from ABE-fermenting C. acetobutylicum, the organism was able to synthesize up to 2 mM of butanol from a syngas blend. However, at the end of growth, butanol was assimilated and converted into butyrate. A later study showed improved butanol production from gas in both C. ljungdahlii and C. autoethanogenum (Köpke and Liew, 2011). A similar biosynthetic operon was used, but under control of a different promoter system and including two electron-transferring flavoproteins, EtfAB, while omitting any external alcohol and aldehyde dehydrogenase genes. The recombinant strains produced butanol as the main product from steel mill gas up to a concentration of 25.7 mM (Köpke and Liew, 2011). Ueki et al. (2014) were able to integrate (via single crossover recombination) the butyrate-biosynthetic pathway (thlA-crt-bcd-eftB-etfA-hbd-ptb-buk) from C. acetobutylicum by replacing phosphotransacetylase gene pta, resulting in the production of ~15 mM butyrate under H2/CO2 conditions using C. ljungdahlii.
Table 2.
Summary of genetically modified acetogens.
| Organism | Genetic modifications | Results | References |
|---|---|---|---|
| EXPANDED PRODUCT SPECTRUM | |||
| A. woodii | Plasmid based expression of C. acetobutylicum acetone biosynthetic genes (thlA-ctfAB-adc) using different combinations of promoter (ackA, pta-ack) and plasmid origin (pIP404, pBP1, pCB102, and pCD6) | Continuous production of 26.4 mg/L/h acetone from synthetic syngas in CSTR | Hoffmeister et al., 2016 |
| C. aceticum | Plasmid based expression of C. acetobutylicum acetone biosynthetic genes (thlA-ctfAB-adc) | Production of up to 9 mg/L acetone from synthetic syngas in bottles | Schiel-Bengelsdorf and Dürre, 2012 |
| C. aceticum | Plasmid based expression of synthetic acetone operon of C. acetobutylicum thiolase and acetoacetate decarboxylase genes (thl-adc) and genes for B. subtilis thioesterase (teII) or H. influenzae acyl-CoA thioesterase (ybgC) | Production of up to 59 mg/L acetone a H2/CO2 gas mix | Becker et al., 2012 |
| C. autoethanogenum, C. ljungdahlii | Plasmid based expression of C. acetobutylicum butanol biosynthetic genes (thlA-crt-hbd-bcd-etfAB) | Production of up to 1.93 g/L butanol from steel mill gas and syngas in bottles; Butanol as major product | Köpke and Liew, 2011 |
| C. autoethanogenum, C. ljungdahlii | Plasmid based expression of acetone and isopropanol biosynthetic genes thlA (from C. acetobutylicum) and ctfA-ctfB-adc (from C. beijerinckii) | Production of up to 300 mg/L acetone and 25 mg/L isopropanol from steel mill gas and syngas in bottles; Continuous production of 700 mg/L/d isopropanol from steel mill gas in CSTR | Köpke et al., 2012 |
| C. autoethanogenum | Plasmid based expression of Chloroflexus aurantiacus malonyl-coenzyme A reductase | Production of low levels of 3-hydroxypropionate from steel mill gas and H2/CO2 | Köpke and Chen, 2013 |
| C. autoethanogenum | Plasmid based expression of Klebsiella pneumoniae (S)-specific butanediol dehydrogenase and Klebsiella oxytoca diol dehydratase pddABC in C. autoethanogenum with inactivated native 2,3-butanediol dehydrogenase | Production of up to 370 mg/L meso-2,3-butanediol, MEK and 2-butanol from steel mill gas in bottles | Mueller et al., 2013 |
| C. autoethanogenum | Plasmid base expression of unspecific acyltransferase from Acinetobacter baylyi | Production of low levels of butanoic acid butyl ester from steel mill gas | Liew and Köpke, 2012 |
| C. autoethanogenum | Plasmid based over-expression of DOXP synthase, expression of mevalonate pathway, C. beijerinckii isopentenyl diphosphate isomerase, and either Poplar isoprene synthase or farnesene synthase | Production of low levels of mevalonate, isoprene and franesene from steel mill gas and syngas | Chen et al., 2013a |
| C. ljungdahlii | Plasmid based expression of C. acetobutylicum butanol biosynthetic genes (thlA-crt-hbd-bcd, adhE-bdhA) | Production of up to 148 mg/L n-butanol from synthetic syngas in bottles; Conversion to butyrate at end of growth | Köpke et al., 2010 |
| C. ljungdahlii | Plasmid expression of C. acetobutylicum acetone biosynthetic genes (thlA-ctfAB-adc) under lactose inducible promoter | Production of up to 871 mg/L acetone from CO in bottles | Banerjee et al., 2014 |
| C. ljungdahlii | Chromosomal integration (single crossover recombination) of C. acetobutylicum butyrate biosynthetic genes (thl-crt-bcd-etfB-etfA-hbd-ptb-buk) into pta promoter region | Production of up to 881 mg/L butyrate under H2/CO2 in bottles | Ueki et al., 2014 |
| C. ljungdahlii | Plasmid based expression of mevalonate pathway, E. coli isopentenyl diphosphate isomerase, and Poplar isoprene synthase | Production of up to 68 mg/L mevalonate and low levels of isoprene from syngas in bottles | Beck et al., 2014 |
| C. ljungdahlii | Plasmid based expression of mevalonate pathway, yeast isopentenyl diphosphate isomerase, and Poplar isoprene synthase | Production of up to 68 low levels of isoprene from syngas in bottles | Beck et al., 2014 |
| M. thermoacetica | Genome insertion of ldh from Thermoanaerobacter pseudethanolicus into pyrF locus | Heterotrophic production of up to 613 mg/L lactate from glucose in bottles; Autotrophic production not reported | Kita et al., 2013 |
| ENHANCED PROCESS | |||
| A. woodii | Plasmid based expression of formyl-THF-synthetase, methenyl-THF-cyclohydrolase, methylene-THF-dehydrogenase, and methylene-THF-reductase of C. ljungdahlii | Increase in volumetric acetate production rate by 14% under H2/CO2 conditions in CSTR | Straub et al., 2014 |
| C. autoethanogenum | Inactivation of acetolactate decarboxylase gene budA | Abolishment of 2,3-butanediol production along with enhanced ethanol production by 79%; Small levels of succinate and lactate produced during growth on steel mill gas | Köpke et al., 2013a |
| C. autoethanogenum | Inactivation of lactate dehydrogenase gene ldhA | Abolishment of lactate production | Nagaraju et al., 2015 |
| C. autoethanogenum | Plasmid expression of vitamin biosynthetic genes thiC of “C. ragsdalei” and panBCD of C. beijerinckii | Successful complementation of thiamine and panthothenate biosynthesis pathways; Strains independent of vitamin B1 and B5 supplementation during growth on steel mill gas | Köpke and Al-Sinawi, 2013 |
| C. autoethanogenum | Plasmid expression of native groES and groEL | Increased cell viability when challenged with ethanol during growth on steel mill gas | Simpson et al., 2011 |
| C. difficile | Generation of more than 70,000 unique mutants via transposon mutagenesis, coupled with transposon-directed insertion site sequencing | Identification of 404 essential genes for growth; Identification of 798 genes that are likely to affect sporulation | Dembek et al., 2015 |
| C. ljungdahlii | Chromosomal deletion of adhE1 and/or adhE2 | 6-fold reduction in ethanol concentration of ΔadhE1 mutant (but not ΔadhE2 mutant) under heterotrophic conditions | Leang et al., 2013 |
| C. ljungdahlii | Plasmid expression of formate dehydrogenase from E. coli | 4.3-fold increase in intracellular NADH concentration; 2.3-fold improvement in maximum power density in a sodium formate fed microbial fuel cell | Han et al., 2016 |
Besides butanol and ethanol, production of the other solvents of ABE/IBE fermentation have been demonstrated from gas. By delivering a plasmid that expresses an acetone operon (ctfA, ctfB, adc, and thlA) of C. acetobutylicum under the control of the thlA promoter, C. aceticum was demonstrated to produce up to 0.14 mM acetone using H2/CO2 gas (Schiel-Bengelsdorf and Dürre, 2012). Acetone production increased over 500x from H2/CO2 in C. ljungdahlii using the same genes but under control of an inducible system (Banerjee et al., 2014) and in A. woodii by combinatorial testing of different promoter and plasmid combinations (Hoffmeister et al., 2016). In a different study, it was shown that acetone, depending on the redox state, could be converted to isopropanol in both C. ljungdahlii and C. autoethanogenum (Köpke et al., 2012). If acetone is targeted for production, it is however desirable to either use an organism like A. woodii that is unable to reduce acetone to isopropanol or eliminate this function if the host is C. autoethanogenum or C. ljungdahlii (Hoffmeister et al., 2016). An NADPH dependent primary:secondary alcohol dehydrogenase capable of catalyzing the reduction of acetone to isopropanol was identified to be present in these two organisms (Köpke et al., 2014) and inactivation of this single gene indeed renders the organism unable to reduce acetone further (Köpke et al., 2015). Rather than using genes from ABE fermentation organisms, acetone production was also demonstrated via a thioesterase teII of Bacillus subtilis and an acyl-CoA thioesterase ybgC of Haemophilus influenzae, which improved acetone production in C. aceticum from 0.24 mM to 1 mM on a H2/CO2 gas mix (Becker et al., 2012; Schiel-Bengelsdorf and Dürre, 2012).
Another example is heterologous production of lactate in M. thermoacetica. The gene ldh (encodes lactate dehydrogenase) from Thermoanaerobacter pseudethanolicus was inserted into the genome, resulting in the production of 6.8 mM lactate. Experiments were, however, performed on fructose rather than gas (Kita et al., 2013). Even energy-intense products like isoprene, which require several molecules of ATP, have been successfully produced in acetogens growing on gas. Multiple groups have demonstrated a proof of concept for isoprene production by installation of the mevalonate pathway and an isoprene synthase in either C. autoethanogenum or C. ljungdahlii (Beck et al., 2013, 2014; Chen et al., 2013a; Furutani et al., 2013). In order to optimize these processes, it is important to understand the energy and redox metabolism of acetogens, as this is significantly different to sugar fermenting organisms and governs the product profile (Marcellin et al., 2016).
LanzaTech has demonstrated production of several additional high value molecules via gas fermentation, such as methyl ethyl ketone (MEK) (Mueller et al., 2013), 3-hydroxypropionate (3-HP) (Köpke and Chen, 2013), biodiesel, and jet fuel molecules (Liew and Köpke, 2012). The company also works together with the world's largest nylon producer, U.S.-based Invista, on new processes for production of nylon precursor 1,3-butadiene (Köpke and Havill, 2014; INVISTA, 2015), French start-up Global Bioenergies on direct isobutylene production (Global Bioenergies, 2016), and the German company Evonik Industries on a new process for production of specialty plastics from gas (Evonik, 2013; LanzaTech, 2013).
In addition to expand the product spectrum of acetogens, significant work has also been carried out to enhance current processes (Table 2). Work has focused on improving substrate utilization and ethanol production in the gas fermentation process and eliminating the need to supplement the microbe with vitamins to reduce costs.
Substrate utilization could be improved by overexpressing the WLP. Straub et al. overexpressed the four THF-dependent enzymes of the methyl branch of C. ljungdahlii in A. woodii, resulting in 14% increase in volumetric acetate production rate (Straub et al., 2014). In a similar attempt, formate dehydrogenase (Fdh) of E. coli was heterologously expressed in C. ljungdahlii facilitate electron transfer in electrosynhesis. It was shown that the intracellular NADH concentration increased by 4.3-fold leading to a process improvement (Han et al., 2016).
To improve ethanol production, competing pathways such as the butanediol production pathway were inactivated in C. autoethanogenum. This was achieved by inactivation of the budA gene (encoding an acetolactate decarboxylase) using either allelic exchange or ClosTron mutagenesis (Köpke et al., 2013a). During growth on steel mill gas, the budA deficient mutant synthesized no 2,3-butanediol, but generated 79% more ethanol and 9.9-fold higher lactate than the wild-type control. Interestingly, also significant amount of platform chemical succinate were detected in this engineered strain as an overflow metabolite from the incomplete TCA cycle. A similar effect has also been observed by inactivating the lactate dehydrogenase (ldhA) of the same strain (Nagaraju et al., 2015). To alleviate product inhibition of ethanol, native chaperones GroES and GroEL were overexpressed in C. autoethanogenum with the aim of protecting proteins from misfolding (Simpson et al., 2011). When challenged with various amounts of ethanol [1.5–6% (w/v)], the chaperone overexpression strain consistently outperformed the control strain during growth on steel mill gas. While not resulting in an improvement for ethanol production, important insight came from two studies in C. ljungdahlii, where the role of ethanol biosynthesis genes adhE1 and/or adhE2 was studied and confirmed by generated chromosomal deletions and doing complementation experiments (Leang et al., 2013; Banerjee et al., 2014).
As supplementation of vitamin represent a significant cost factor in a process, work has also been carried out to eliminate the need for vitamin supplementation. An in silico analysis showed that thiamine (vitamin B1) and panthothenate (vitamin B5) biosynthetic pathways in C. autoethanogenum are incomplete but only lack a few genes. A complementation of both pathways was demonstrated by heterologously expressing thiC of “C. ragsdalei” and panBCD of C. beijerinckii in C. autoethanogenum. It was not only shown that the resulting strains no longer need supplementation with these vitamins but thiC and panBCD genes could also be used as selective markers (Köpke and Al-Sinawi, 2013).
Installation of wood-ljungdahl pathway in non-acetogens
Given the challenges working with acetogens (Burk and Van Dien, 2016), it has also been attempted to install the WLP in traditional hosts as E. coli or yeast.
First attempts to demonstrate parts of the WLP in E. coli were already carried out in 1989. Genes encoding methyltransferase (MeTr), CODH/ACS, and CoFeSP from M. thermoacetica were cloned and heterologously expressed in E. coli (Roberts et al., 1989). Although the MeTr was found to be active, activity was not detected for the other two enzymes, and both CODH and CoFeSP were found to be less thermostable than when isolated from the native host. In a later study, it could be shown that the CODH/ACS was functional in both CO oxidation and CO/acetyl-CoA exchange activities after heterologous expression in and purification from E. coli (Loke et al., 2000).
With the advancements in strain engineering, it became possible to express the complete WLP from various source organisms in E. coli (Burk et al., 2010; Trawick et al., 2012). While certain aspects could be demonstrated, growth on CO and H2 could not be shown. One challenge for successful operation of the WLP in non-acetogenic hosts is a lack of intracellular conditions and genetic outfit of traditional hosts as E. coli or yeast promoting co-factor production and assembly of delicate metal centers. Therefore, in an alternative approach, it has also been attempted to implement the pathway into more closely related ABE-fermenting model organism C. acetobutylicum (Al-Hinai et al., 2012a,b). While heterologous gene expression was demonstrated using qRT-PCR and some enzymatic activities were shown, the resultant recombinant C. acetobutylicum was unable to grow autotrophically. The other challenge to make this work may also be yet unknown genes that play a function in operation of the WLP. For methanogens for example it has been predicted that over 200 genes are involved in growth on CO2/H2 (Kaster et al., 2011).
Though a promising approach in theory (in particular in light of the advancements to design and generate synthetic genomes Hutchison et al., 2016), the most efficient use of the WLP in the near future is likely to remain in acetogens.
Coupled processes to expand product spectrum of gas fermentation
While synthetic biology approaches offer great potential to expand the product spectrum of acetogens, coupling gas fermentation with other biological or chemical platforms can allow for generation of products currently impossible to synthesize or economically unviable with gas fermentation alone. The following sections describe specific examples of coupled processes involving gas-fermenting microbes that have the ability to expand the gas fermentation product portfolio and improve product yields and economics. Coupled processes include multi-step bioprocesses (Section Coupling to Other Bioprocesses) that convert metabolites from a primary fermentation in a secondary fermentation (Agler et al., 2011) as well as using heterogeneous catalysts (Section Coupled Catalytic Processes) to chemically convert gas fermentation products.
Coupling to other bioprocesses
Multi-step bioprocesses can consist of combinations of pure cultures, co-cultures, and/or open (undefined) mixed cultures with natural and/or genetically modified strains. Potential advantages of co-cultures and mixed cultures include microbial symbiotic relationships, increased substrate utilization, and improved product yields. For example, undesirable inhibitory or toxic byproducts of a metabolic pathway can be consumed by a second microbe, or two host cells can work together to ensure the optimal environment for all pathway enzymes by effectively dividing the pathway (Zhang et al., 2015a; Zhou et al., 2015). Other advantages of mixed cultures include protection against contamination by foreign bacteria, resistance to bacteriophages, oxygen removal and production of growth factors by facultative anaerobes in an anaerobic reactor (Angenent and Wrenn, 2008), improved stability, and obligately mutual metabolic activity (syntrophy) that enables production or degradation of complex organics (Morris et al., 2013). As compared to pure cultures, disadvantages include potentially longer period to reach steady-state, difficulty understanding the interaction between microbes in the fermentation, reduced reproducibility due to challenges in maintaining population dynamics, and inability to optimize parameters for each strain.
H2/CO2 to lipids
Most gas fermentation work to date has been carried out on CO-containing gas streams. CO acts as both a carbon and energy source, providing sufficient energy to synthesize reduced products such as ethanol and butanol. Using CO2 as the carbon source for ethanol and other reduced product formation is highly desirable but difficult to achieve. The more oxidized CO2 can only act as a carbon source and typically requires H2 for its fixation. H2/CO2 gas fermentations mostly produce acetate, though production of ethanol (Mock et al., 2015) and other products such as acetone (Schiel-Bengelsdorf and Dürre, 2012; Hoffmeister et al., 2016) or butyrate (Ueki et al., 2014) have been described, albeit at lower productivity compared to CO. Additionally, ethanol production from CO is thermodynamically favorable over CO2 (Reactions 1-4; the free energy (ΔG°') of Reactions 1–4 was calculated from data given in Thauer et al. (1977)).
| (1) |
| (2) |
| (3) |
| (4) |
Using CO as the sole carbon and energy source to form ethanol results in CO2 formation (Reaction 1), lowering the amount of fixed carbon. The presence of H2 can reduce or eliminate CO2 (Reactions 2–4), but in practice, the H2 uptake is often limited in the presence of CO through hydrogenase inhibition by CO (Purec et al., 1962; Bennett et al., 2000; Pandelia et al., 2010). To obtain the desired H2 uptake to limit CO2 production, the dissolved CO concentration must therefore be low. Unfortunately, under such conditions, organic acid (e.g., acetate and butyrate) production becomes the favored fermentation product over more reduced alcohols (e.g., ethanol, butanol) (Hurst and Lewis, 2010; Devarapalli and Atiyeh, 2015). Using CO2 as the carbon source does not inhibit H2 uptake as seen with CO but again results in lower productivity and is thermodynamically less favorable than using CO due to the lower energy content of CO2.
If acetate, on the other hand, is the target of the fermentation, using H2/CO2 is an attractive option. It has been demonstrated that A. woodii, for example, can be used for productive fermentations, with a space time yield of 148 g/L/d of acetate in a bioreactor with a submersed filtration unit and a final acetate concentration of 44 g/L after 11 days (Kantzow et al., 2015). Even though acetate can be produced at high volumetric production rates, the process has some challenges that need to be overcome to make it economically viable. The fermentation process requires the costly constant addition of base to compensate for the acid produced in order to keep the pH constant. Post fermentation recovery of the acetate then requires the addition of another strong acid to protonate the acetate.
To overcome economic drawbacks of acetate fermentation, a secondary fermentation process can be employed wherein acetate is upgraded to more valuable products. For instance, Gong et al. demonstrated lipid synthesis from acetate by multiple oleaginous yeast strains with one, Cryptococcus curvatus ATCC 20509, yielding a lipid concentration of 4.2 g/L and 73.4% of the cell dry weight (Gong et al., 2015). The acetate in this work was not sourced from gas fermentation, however. Recent work from Hu et al., presented an integrated bioprocess where acetate made from CO2 and H2 by M. thermoacetica was converted to lipids by Yarrowia lipolytica (Hu et al., 2016). The two-stage system produced 18 g/L of triacylglycerides at 36% lipid content of cell weight. This work not only validates this coupled bioprocess approach but also provides a baseline for further experiments. One potential improvement, for example, could be made in the efficiency of carbon utilization by feeding any CO2 produced by the yeast back to the acetogen (Stephanopoulos, 2011).
Gas to elongated carboxylic acids and conversion to alcohols
Longer-chain linear alcohols than ethanol are more valuable as drop-in fuels due to their energy density, compatibility with current transportation infrastructure, and greater ease of separation from aqueous fermentation broth. They are also valuable commodity chemicals, as production of linear alcohols through petrochemical means can be challenging since existing technologies such as linear alpha oligomerization are not selective. Gas-fermentation products of acetogens can be converted to longer carboxylic acids by other microorganisms, and a number of microorganisms, including acetogens themselves, can reduce carboxylic acids of varying lengths to their corresponding alcohols. Coupling these processes could then allow for synthesis of low-carbon drop-in fuels or valuable chemicals from gas feedstocks.
Chain elongation of short-chain fatty acids and alcohols such as acetate and ethanol to longer carboxylic acids can occur via the reverse β-oxidation pathway (Barker et al., 1945; Spirito et al., 2014; Angenent et al., 2016). Two molecules of acetyl-CoA (sourced from ethanol) condense to form acetoacetyl-CoA, which is then reduced to butyryl-CoA. The energy required for elongation and subsequent reduction can be provided indirectly by the oxidation of ethanol to acetyl-CoA (Spirito et al., 2014), which provides reducing equivalents such as NADH and ATP through SLP and the Rnf complex as described above for acetogens. As this is a cyclical pathway, multiple rounds of elongation can take place with the addition of acetyl-CoA with each cycle. Use of this approach has resulted in production of caproate (C6) and caprylate (C8) from acetate with either ethanol or H2 as the electron donor source (Steinbusch et al., 2011; Angenent et al., 2016). Chain extension for longer odd-numbered carboxylic acids can also occur if propionate (C3) is provided as the substrate, which is converted to valerate (C5) and eventually heptanoate (C7) (Grootscholten et al., 2013).
This approach has already shown to successfully couple to acetogenic gas fermentation. A dilute ethanol and acetate product stream from syngas fermentation with C. ljungdahlii was used as the substrate for a separate anaerobic reactor, which produced butyrate and caproate (Vasudevan et al., 2014). Because mixed cultures used for chain elongation may also convert substrates such as acetate to methane by acetoclastic methanogens, care must be taken to ensure culture conditions do not favor methanogen growth, which would limit the yield of longer chain carboxylic acids. Techniques to inhibit acetoclastic methanogens include pH control (Agler et al., 2014) and high H2 partial pressure, which is estimated to confer greater growth advantage to homoacetogens over hydrogenotrophic methanogens (Spirito et al., 2014).
Despite the prevalent use of mixed cultures, these works and others allow for speculation that the non-acetogenic C. kluyveri may be responsible for the chain elongation based on previous characterizations of that microorganism (Barker et al., 1945; Seedorf et al., 2008). C. kluyveri was the only anaerobic, clostridial bacterium isolated to pure culture known to utilize ethanol and acetate as sole energy sources until recently with the isolation of “Clostridium pharus” (Levinson, 2014). Because C. kluyveri is considered the dominant chain-elongating microorganism in some of these cases, it could be possible to couple a pure culture of this bacteria to a gas fermentation platform to achieve longer chain carboxylic acids from syngas.
Gas-fermenting acetogens can be used to reduce carboxylic acids into their respective alcohols with CO or H2 present in syngas as a source of electron donors (Steinbusch et al., 2008; Simpson et al., 2009a). The carboxylic acid is reduced to its corresponding aldehyde via the reduced ferredoxin-dependent AOR (Köpke et al., 2010; Isom et al., 2015; Mock et al., 2015). The reduction of the aldehyde to the corresponding alcohol is completed by an alcohol dehydrogenase. Pure cultures of C. ljungdahlii and “C. ragsdalei” have been reported to convert externally added carboxylic acids (acetate, propionate, n-butyrate, iso-butyrate, n-valerate, and n-caproate) into their corresponding alcohols using syngas as the source of electrons (Perez et al., 2013; Isom et al., 2015). Isom et al. for example, showed “C. ragsdalei” can convert caproate at 62%, valerate at 82%, and butyrate at 100% efficiency to their respective alcohols.
This capability possessed by acetogens allows for interesting coupling scenarios to produce alcohols that were chain extended from ethanol and acetate. Coskata, for example, had described using a symbiotic co-culture of C1-fixing and C3-producing microorganisms or, alternatively, a multi-zone process containing these microorganisms in separate zones. Syngas would be consumed and converted to ethanol and acetate by acetogens in one fermentation zone (Datta et al., 2014; Enzien et al., 2014). These products would transfer to the second fermentation zone where they could be converted to propionate by separate microorganisms. This could then be reduced to propanol using acetogens and syngas as an electron donor back in the first fermentation zone. A similar syntrophic co-culture of C. kluyveri, an acetogenic clostridium, and/or a butyrogenic microorganism for the production of butanol was also described by Coskata (Datta and Reeves, 2014). Here the acetogen would produce ethanol and acetate, which could be converted to butyric acid and butanol by whichever species contain the genes for NADPH dependent acetyl-CoA reductase and either a butyryl-CoA acetate transferase or a butyrate kinase. Diender et al. recently demonstrated production of butyrate, caproate, butanol and hexanol by co-culture of C. kluyveri and C. autoethanogenum fed with CO and H2 (Diender et al., 2016).
Richter et al. even integrated three bioprocesses to produce alcohols, with the final step of carboxylic acid conversion to alcohols completed by syngas fermentation (Richter et al., 2013a). First, the conventional yeast fermentation product corn beer was fed to an undefined anaerobic culture where butyrate and caproate were produced. The caproate product was selectively removed and the effluent from this process was then fed to the acetogen C. ljungdahlii, which reduced the residual short and medium chain carboxylic acids (n-butyric acid and unremoved n-caproic acid) into their respective alcohols using syngas as an electron source.
Coupled catalytic processes
Another option for expanding the products formed from gas fermentation is coupling gas fermentation to chemical synthesis platforms. Combining biologically synthesized products from gas fermentation with conversion processes employing heterogeneous catalysts may allow for conversion of syngas substrates to products that may be difficult or currently impossible to produce solely via enzymatic catalysis (e.g., diesel/jet fuels and thermoplastic polymers). These chemical processes are generally faster than biological syntheses, and, if performed carefully, may still offer an overall reduction in GHG emissions for these products compared to traditional production from fossil fuels. Two reviews published recently focus on coupling biological fermentation products with chemical processes and explore more in depth the scope of work accomplished to date (Goulas and Toste, 2016; Schwartz et al., 2016). Detailed here are processes more immediately relevant to commercial gas fermentation efforts and/or not covered in those reviews.
Alcohol-to-jet fuel
Although ethanol (and potentially other short-chain alcohols) produced by gas fermentation can serve as an additive or potential replacement to gasoline, it is not considered to be an energy dense enough molecule to serve as a jet fuel. Jet fuels are typically aliphatic and contain 8–16 carbon atoms (Atsonios et al., 2015). Molecules this large can be made from syngas substrates via coupled biological processes (see above), but it may be more efficient to convert the small alcohols to jet fuel chemically by employing a process called alcohol-to-jet (ATJ).
To obtain jet fuel from these small alcohols, ethanol and butanol (separated from the fermentation broth) are first dehydrated to corresponding alkenes ethylene and butylene (Harvey and Meylemans, 2011; Zhang and Yu, 2013). The alkenes are then oligomerized to a desired range of sizes (i.e., number of carbon atoms) that fit the end application (Heveling et al., 1998; Harvey and Meylemans, 2011). Reaction conditions are tuned to avoid extensive polymerization to thermoplastics like poly(ethylene) when fuels are the desired product (Atsonios et al., 2015).
There are currently multiple industrial ATJ projects for both ethanol and butanol conversions to jet fuel (Lane, 2015a; LanzaTech, 2015). The ATJ fuels have undergone and continue to undergo rigorous testing to establish their use as drop-in fuels for commercial aircraft (Luning Prak et al., 2015). Successfully meeting these standards will expand the volume of fuels that may be sourced from renewable resources, such as biomass, and aid in keeping fossil fuels typically needed for aviation fuel in the ground.
Another promising approach to convert ethanol (and other short-chain alcohols) to jet (or biodiesel) fuels is via extractive catalytic upgrading directly from the fermentation broth (Anbarasan et al., 2012; Bormann et al., 2014; Sreekumar et al., 2014, 2015; Baer et al., 2016). This technology requires strains that co-produce acetone (Becker et al., 2012; Köpke et al., 2012, 2015; Schiel-Bengelsdorf and Dürre, 2012) which harbors a nucleophilic α-carbon amenable to C–C bond formation with the electrophilic alcohols.
Ethanol and butanediol to butadiene
As mentioned, alkenes derived from gas fermentation alcohols are sources for ubiquitous thermoplastic and rubber products. Ethylene made from dehydration of ethanol can be more fully polymerized beyond oligomers for ATJ fuel and converted to poly(ethylene). Poly(butadiene) is another polymer used in mass production of products such as nylon or automobile tires. The monomer of this synthetic rubber, 1,3-butadiene, can be formed from gas fermentation products ethanol (Makshina et al., 2012) and 2,3-butanediol (Duan et al., 2015). These processes have been reviewed more in depth elsewhere (Makshina et al., 2014). 1,3-butadiene is also a precursor for nylon production (Carraher, 2010), and LanzaTech is currently a part of multiple collaborations to develop gas fermentation production of 2,3-butanediol for the purpose of chemical conversion to 1,3-butadiene (Köpke and Havill, 2014).
Summary and outlook
Gas fermentation has developed incredibly over the past years, leading to multiple endeavors for commercial production of ethanol from syngas. Significant strides in genetic manipulation of gas-fermenting bacteria (acetogens specifically) has granted access to new products and routes for enhancing production rates and yields. A focus for the future is continuing to develop these genetic tools and techniques and move toward high-throughput genetic engineering and screening. Given the challenges inherent in heterologous pathway expression (e.g., balancing of fluxes of each enzyme reaction to reduce accumulation of toxic intermediates, redox imbalance, and metabolic burden from overproduction of enzymes), an automated platform would allow for larger-scale and greater efficiency in testing modifications.
While these tools and platforms develop, coupled processes, both biological and chemical, presently allow for synthesis of products currently unattainable in single gas-fermenting microorganisms. However, the urgency of global climate change hangs over this work. Even if these advances in gas fermentation are able to expand to produce aviation fuels and thermoplastics at commercially-relevant rates, it is unlikely to develop quickly enough to completely replace the consumption of fossil fuels for their production. The combination of gas fermentation with established chemical processes, however, could more quickly bring renewable aviation fuels and synthetic rubber to relevancy. Sourcing fuels and chemicals for polymer synthesis from renewable resources like biomass may be attainable with gas fermentation.
Author contributions
FL, MM, RT, CM, and MK drafted the manuscript and all authors edited and wrote the manuscript. FL, MM, RT contributed equally. All authors gave approval for publication of the manuscript.
Funding
We thank the following investors in LanzaTech's technology: Sir Stephen Tindall, Khosla Ventures, Qiming Venture Partners, Softbank China, the Malaysian Life Sciences Capital Fund, Mitsui, Primetals, CICC Growth Capital Fund I, L.P. and the New Zealand Superannuation Fund.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. LanzaTech, Inc. has commercial interest in gas fermentation.
References
- Abrini J., Naveau H., Nyns E. J. (1994). Clostridium autoethanogenum, sp. nov., an anaerobic bacterium that produces ethanol from carbon monoxide. Arch. Microbiol. 161, 345–351. 10.1007/BF00303591 [DOI] [Google Scholar]
- Abubackar H. N., Bengelsdorf F. R., Dürre P., Veiga M. C., Kennes C. (2016a). Improved operating strategy for continuous fermentation of carbon monoxide to fuel-ethanol by clostridia. Appl. Energy 169, 210–217. 10.1016/j.apenergy.2016.02.021 [DOI] [Google Scholar]
- Abubackar H. N., Fernández-Naveira Á., Veiga M. C., Kennes C. (2016b). Impact of cyclic pH shifts on carbon monoxide fermentation to ethanol by Clostridium autoethanogenum. Fuel 178, 56–62. 10.1016/j.fuel.2016.03.048 [DOI] [Google Scholar]
- Abubackar H. N., Veiga M. C., Kennes C. (2012). Biological conversion of carbon monoxide to ethanol: Effect of pH, gas pressure, reducing agent and yeast extract. Bioresour. Technol. 114, 518–522. 10.1016/j.biortech.2012.03.027 [DOI] [PubMed] [Google Scholar]
- Abubackar H. N., Veiga M. C., Kennes C. (2015a). Carbon monoxide fermentation to ethanol by Clostridium autoethanogenum in a bioreactor with no accumulation of acetic acid. Bioresour. Technol. 186, 122–127. 10.1016/j.biortech.2015.02.113 [DOI] [PubMed] [Google Scholar]
- Abubackar H. N., Veiga M. C., Kennes C. (2015b). Ethanol and acetic acid production from carbon monoxide in a Clostridium strain in batch and continuous gas-fed bioreactors. Int. J. Environ. Res. Public Heal. 12, 1029–1043. 10.3390/ijerph120101029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abubackar H. N., Veiga M. C., Kennes C., Coruña L. (2011). Biological conversion of carbon monoxide: rich syngas or waste gases to bioethanol. Biofuels Bioprod. Biorefining 5, 93–114. 10.1002/bbb.25621924896 [DOI] [Google Scholar]
- Ackermann G., Tang Y. J., Henderson J. P., Rodloff A. C., Silva J., Cohen S. H. (2001). Electroporation of DNA sequences from the pathogenicity locus (PaLoc) of toxigenic Clostridium difficile into a non-toxigenic strain. Mol. Cell. Probes 15, 301–306. 10.1006/mcpr.2001.0373 [DOI] [PubMed] [Google Scholar]
- Adamse A. D. (1980). New isolation of Clostridium aceticum (Wieringa). Antonie Van Leeuwenhoek 46, 523–531. 10.1007/BF00394009 [DOI] [PubMed] [Google Scholar]
- Agler M. T., Spirito C. M., Usack J. G., Werner J. J., Angenent L. T. (2014). Development of a highly specific and productive process for n-caproic acid production: applying lessons from methanogenic microbiomes. Water Sci. Technol. 69, 62–68. 10.2166/wst.2013.549 [DOI] [PubMed] [Google Scholar]
- Agler M. T., Wrenn B. A., Zinder S. H., Angenent L. T. (2011). Waste to bioproduct conversion with undefined mixed cultures: the carboxylate platform. Trends Biotechnol. 29, 70–78. 10.1016/j.tibtech.2010.11.006 [DOI] [PubMed] [Google Scholar]
- Ahmed A., Lewis R. S. (2007). Fermentation of biomass-generated synthesis gas: effects of nitric oxide. Biotechnol. Bioeng. 97, 1080–1086. 10.1002/bit.21305 [DOI] [PubMed] [Google Scholar]
- Al-Hinai M. A., Fast A. G., Indurthi D., Papoutsakis E. T. (2012b). Installing the wood-ljungdahl (WL) pathway in Clostridium acetobutylicum. Clostridium XII Conf. [Google Scholar]
- Al-Hinai M. A., Fast A. G., Papoutsakis E. T. (2012a). Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration. Appl. Environ. Microbiol. 78, 8112–8121. 10.1128/AEM.02214-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T. D., Caldwell M. E., Lawson P. A., Huhnke R. L., Tanner R. S. (2010). Alkalibaculum bacchi gen. nov., sp. nov., a CO-oxidizing, ethanol-producing acetogen isolated from livestock-impacted soil. Int. J. Syst. Evol. Microbiol. 60, 2483–2489. 10.1099/ijs.0.018507-0 [DOI] [PubMed] [Google Scholar]
- Anbarasan P., Baer Z. C., Sreekumar S., Gross E., Binder J. B., Blanch H. W., et al. (2012). Integration of chemical catalysis with extractive fermentation to produce fuels. Nature 491, 235–239. 10.1038/nature11594 [DOI] [PubMed] [Google Scholar]
- Anderson M. E., DeRose V. J., Hoffman B. M., Lindahl P. A. (1993). Identification of a cyanide binding site in CO dehydrogenase from Clostridium thermoaceticum using EPR and ENDOR spectroscopies. J. Am. Chem. Soc. 115, 12204–12205. 10.1021/ja00078a085 [DOI] [Google Scholar]
- Andreese J. R., Gottscha G., Schlegel H. G. (1970). Clostridium formicoaceticum nov spec - Isolation, description and distinction from Clostridium aceticum and Clostridium thermoaceticum. Arch. Microbiol. 72, 154. [DOI] [PubMed] [Google Scholar]
- Angenent L. T., Richter H., Buckel W., Spirito C. M., Steinbusch K. J., Plugge C., et al. (2016). Chain elongation with reactor microbiomes: open-culture biotechnology to produce biochemicals. Environ. Sci. Technol. 50, 2796–2810. 10.1021/acs.est.5b04847 [DOI] [PubMed] [Google Scholar]
- Angenent L. T., Wrenn B. A. (2008). Optimising mixed-culture bioprocessing to convert wastes into bioenergy, in Bioenergy, eds Demain A., Harwood C. S., Wall J. D. (Washington, DC: American Society of Microbiology; ), 179–194. [Google Scholar]
- Aschenbrenner N. (2016). World's Largest Hydrogen Electrolysis Facility. SIEMENS. Available online at: http://www.siemens.com/innovation/en/home/pictures-of-the-future/energy-and-efficiency/smart-grids-and-energy-storage-largest-hydrogen-electrolysis-facility.html [Accessed March 23, 2016].
- Atsonios K., Kougioumtzis M.-A. D., Panopoulos K., Kakaras E. (2015). Alternative thermochemical routes for aviation biofuels via alcohols synthesis: Process modeling, techno-economic assessment and comparison. Appl. Energy 138, 346–366. 10.1016/j.apenergy.2014.10.056 [DOI] [Google Scholar]
- Babu B. K., Atiyeh H. K., Wilkins M. R., Huhnke R. L. (2010). Effect of the reducing agent dithiothreitol on ethanol and acetic acid production by. ASABE 3, 19–35. 10.13031/2013.35924 [DOI] [Google Scholar]
- Baer Z. C., Bormann S., Sreekumar S., Grippo A., Toste F. D., Blanch H. W., et al. (2016). Co-production of acetone and ethanol with molar ratio control enables production of improved gasoline or jet fuel blends. Biotechnol. Bioeng. [Epub ahead of print]. 10.1002/bit.25978. [DOI] [PubMed] [Google Scholar]
- Banerjee A., Leang C., Ueki T., Nevin K. P., Lovley D. R. (2014). Lactose-inducible system for metabolic engineering of Clostridium ljungdahlii. Appl. Environ. Microbiol. 80, 2410–2416. 10.1128/AEM.03666-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker H. A., Kamen M. D., Bornstein B. T. (1945). The synthesis of butyric and caproic acids from ethanol and acetic acid by Clostridium kluyveri. Proc. Natl. Acad. Sci. U.S.A. 31, 373–381. 10.1073/pnas.31.12.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck Z. Q., Cervin M., Chotani G., Diner B. A., Fan J., Peres C. M., et al. (2014). Recombinant Anaerobic Acetogenic Bacteria for Production of Isoprene and/or Industrial Bio-Products Using Synthesis Gas. US2014/0234926. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Beck Z. Q., Cervin M., Chotani G. K., Caroline P., Sanford K. J., Scotcher M., et al. (2013). Compositions and Methods of Producing Isoprene and/or Industrial Bio-Products Using Anaerobic Microorganisms. WO2013/181647. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Becker F. U., Grund G., Orschel M., Doderer K., Löhden G., Brand G., et al. (2012). Cells and Method for Producing Acetone. US20120101304 A1. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Bengelsdorf F. R., Poehlein A., Schiel-Bengelsdorf B., Daniel R., Dürre P. (2015). Genome sequence of the acetogenic bacterium oxobacter pfennigii DSM 3222T. Genome Announc. 3:e01408–15. 10.1128/genomeA.01408-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B., Lemon B. J., Peters J. W. (2000). Reversible carbon monoxide binding and inhibition at the active site of the Fe-only hydrogenase. Biochemistry 39, 7455–7460. 10.1021/bi992583z [DOI] [PubMed] [Google Scholar]
- Bertram J., Dürre P. (1989). Conjugal transfer and expression of streptococcal transposons in Clostridium acetobutylicum. Arch. Microbiol. 151, 551–557. 10.1007/BF00454874 [DOI] [Google Scholar]
- Bertsch J., Müller V. (2015). Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria. Biotechnol. Biofuels 8:210. 10.1186/s13068-015-0393-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsch J., Parthasarathy A., Buckel W., Müller V. (2013). An electron-bifurcating caffeyl-CoA reductase. J. Biol. Chem. 288, 11304–11311. 10.1074/jbc.M112.444919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel E., Müller V. (2010). Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 107, 18138–18142. 10.1073/pnas.1010318107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel E., Schmidt S., González J. M., Müller V. (2011). Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 68, 613–634. 10.1007/s00018-010-0555-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouzard J.-C., Valette O., Tardif C., de Philip P. (2010). Random mutagenesis of Clostridium cellulolyticum by using a Tn1545 derivative. Appl. Environ. Microbiol. 76, 4546–4549. 10.1128/AEM.02417-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boateng A., Banowetz G., Steiner J., Barton T., Taylor D., Hicks K., et al. (2007). Gasification of Kentucky bluegrass (Poa pratensis l.) straw in a farm-scale reactor. Biomass Bioenergy 31, 153–161. 10.1016/j.biombioe.2006.08.001 [DOI] [Google Scholar]
- Boerrigter H., Rauch R. (2005). Review of applications of gases from biomass gasification, in Handbook Biomass Gasification, ed Knoef H. A. M. (Enschede: Biomass Technology Group; ), 211–230. [Google Scholar]
- Bomar M., Hippe H., Schink B. (1991). Lithotrophic growth and hydrogen metabolism by Clostridium magnum. Fems Microbiol. Lett. 83, 347–350. 10.1111/j.1574-6968.1991.tb04488.x [DOI] [PubMed] [Google Scholar]
- Bormann S., Baer Z. C., Sreekumar S., Kuchenreuther J. M., Dean Toste F., Blanch H. W., et al. (2014). Engineering Clostridium acetobutylicum for production of kerosene and diesel blendstock precursors. Metab. Eng. 25, 124–130. 10.1016/j.ymben.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Braun M., Mayer F., Gottschalk G. (1981). Clostridium aceticum (Wieringa), a microorganism producing acetic acid from molecular hydrogen and carbon dioxide. Arch. Microbiol. 128, 288–293. 10.1007/BF00422532 [DOI] [PubMed] [Google Scholar]
- Breault R. W. (2010). Gasification processes old and new: a basic review of the major technologies. Energies 3, 216–240. 10.3390/en3020216 [DOI] [Google Scholar]
- Bredwell M. D., Srivastava P., Wordon R. M. (1999). Reactor design issues for synthesis-gas fermentations. Biotechnol. Prog. 15, 834–844. 10.1021/bp990108m [DOI] [PubMed] [Google Scholar]
- Bronson B., Preto F., Mehrani P. (2012). Effect of pretreatment on the physical properties of biomass and its relation to fluidized bed gasification. Environ. Prog. Sustain. Energy 31, 335–339. 10.1002/ep.11659 [DOI] [Google Scholar]
- Brown S. D., Nagaraju S., Utturkar S., De Tissera S., Segovia S., Mitchell W., et al. (2014). Comparison of single-molecule sequencing and hybrid approaches for finishing the genome of Clostridium autoethanogenum and analysis of CRISPR systems in industrial relevant clostridia. Biotechnol. Biofuels 7, 1–18. 10.1186/1754-6834-7-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. R., Brown R. C. (2013). A review of cellulosic biofuel commercial-scale projects in the United States. Biofuels, Bioprod. Biorefining 7, 235–245. 10.1002/bbb.1387 [DOI] [Google Scholar]
- Bruant G., Lévesque M.-J., Peter C., Guiot S. R., Masson L. (2010). Genomic analysis of carbon monoxide utilization and butanol production by Clostridium carboxidivorans strain P7. PLoS ONE 5:e13033. 10.1371/journal.pone.0013033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel W., Thauer R. K. (2013). Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113. 10.1016/j.bbabio.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Burk M. J., Schilling C. H., Burgard A. P., Trawick J. D. (2010). Methods and Organisms for Utilizing Synthesis Gas or Other Gaseous Carbon Sources and Methanol. US7803589. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Burk M. J., Van Dien S. (2016). Biotechnology for chemical production: challenges and opportunities. Trends Biotechnol. 34, 187–190. 10.1016/j.tibtech.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Camiade E., Peltier J., Bourgeois I., Couture-Tosi E., Courtin P., Antunes A., et al. (2010). Characterization of Acp, a peptidoglycan hydrolase of Clostridium perfringens with N-acetylglucosaminidase activity that is implicated in cell separation and stress-induced autolysis. J. Bacteriol. 192, 2373–2384. 10.1128/JB.01546-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraher C. E., Jr. (2010). Introduction to Polymer Chemistry, 2nd Edn. Boca Raton: CRC Press. [Google Scholar]
- Cartman S. T., Kelly M. L., Heeg D., Heap J. T., Minton N. P. (2012). Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl. Environ. Microbiol. 78, 4683–4690. 10.1128/AEM.00249-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartman S. T., Minton N. P. (2010). A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 76, 1103–1109. 10.1128/AEM.02525-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Gomez J. A., Höffner K., Phalak P., Barton P. I., Henson M. A. (2016). Spatiotemporal modeling of microbial metabolism. BMC Syst. Biol. 10, 21. 10.1186/s12918-016-0259-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Gomez J., Höffner K., Barton P., Henson M. (2015). Metabolic modeling of synthesis gas fermentation in bubble column reactors. Biotechnol. Biofuels 8, 89. 10.1186/s13068-015-0272-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Liew F., Koepke M. (2013a). Recombinant Microorganisms and Uses Therefor. US20130323820. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Chen Y.-J., Liu P., Nielsen A. A. K., Brophy J. A. N., Clancy K., Peterson T., et al. (2013b). Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 10, 659–664. 10.1038/nmeth.2515 [DOI] [PubMed] [Google Scholar]
- Cho C., Choi S. Y., Luo Z. W., Lee S. Y. (2014). Recent advances in microbial production of fuels and chemicals using tools and strategies of systems metabolic engineering. Biotechnol. Adv. 33, 1455–1466. 10.1016/j.biotechadv.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Clarke Energy (2015). Steel Production Gas for Power Production. Available online at: https://www.clarke-energy.com/steel-production-gas/ [Accessed March 29, 2016].
- Cotter J. L., Chinn M. S., Grunden A. M. (2009). Influence of process parameters on growth of Clostridium ljungdahlii and Clostridium autoethanogenum on synthesis gas. Enzyme Microb. Technol. 44, 281–288. 10.1016/j.enzmictec.2008.11.002 [DOI] [Google Scholar]
- Croux C., Nguyen N.-P.-T., Lee J., Raynaud C., Saint-Prix F., Gonzalez-Pajuelo M., et al. (2016). Construction of a restriction-less, marker-less mutant useful for functional genomic and metabolic engineering of the biofuel producer Clostridium acetobutylicum. Biotechnol. Biofuels 9, 23. 10.1186/s13068-016-0432-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G. Z., Hong W., Zhang J., Li W. L., Feng Y., Liu Y. J., et al. (2012). Targeted gene engineering in Clostridium cellulolyticum H10 without methylation. J. Microbiol. Methods 89, 201–208. 10.1016/j.mimet.2012.02.015 [DOI] [PubMed] [Google Scholar]
- Daniel S. L., Hsu T., Dean S. I., Drake H. L. (1990). Characterization of the H2-dependent and CO-dependent chemolithotrophic potentials of the acetogens Clostridium thermoaceticum and Acetogenium kivui. J. Bacteriol. 172, 4464–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar R. P., Shenkman R. M., Cateni B. G., Huhnke R. L., Lewis R. S. (2004). Fermentation of biomass-generated producer gas to ethanol. Biotechnol. Bioeng. 86, 587–594. 10.1002/bit.20071 [DOI] [PubMed] [Google Scholar]
- Datta R., Enzien M., Hickey R., Levinson W. (2014). Method for Production of n-Propanol and Other c3-containing Products from Syngas by Symbiotic Co-cultures of Anaerobic Microorganisms. WO2014140335. [Google Scholar]
- Datta R., Reeves A. (2014). Syntrophic Co-culture of Anaerobic Microorganism for Production of n-Butanol from Syngas. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- De Klerk A., Li Y. W., Zennaro R. (2013). Greener Fischer-Tropsch Processes for Fuels and Feedstocks. Weinheim: John Wiley & Sons, Inc. [Google Scholar]
- Dembek M., Barquist L., Boinett C. J., Cain A. K., Mayho M., Lawley T. D., et al. (2015). High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. MBio 6, e02383–e02314. 10.1128/mBio.02383-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R. P., Papoutsakis E. T. (1999). Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum. Appl. Environ. Microbiol. 65, 936–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarapalli M., Atiyeh H. K. (2015). A review of conversion processes for bioethanol production with a focus on syngas fermentation. Biofuel Res. J. 2, 268–280. 10.18331/BRJ2015.2.3.5 [DOI] [Google Scholar]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., Church G. M. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343. 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert G. B., Thauer R. K. (1978). Carbon monoxide oxidation by Clostridium thermoaceticum and Clostridium formicoaceticum. J. Bacteriol. 136, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diender M., Stams A. J. M., Sousa D. Z. (2015). Pathway and bioenergetics of anaerobic carbon monoxide fermentation. Front. Microbiol. 6:1275 10.3389/fmicb.2015.01275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diender M., Stams A. J. M., Sousa D. Z. (2016). Production of medium-chain fatty acids and higher alcohols by a synthetic co-culture grown on carbon monoxide or syngas. Biotechnol. Biofuels 9:82. 10.1186/s13068-016-0495-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Tao W., Zhang Y., Li Y. (2012). Development of an anhydrotetracycline-inducible gene expression system for solvent-producing Clostridium acetobutylicum: a useful tool for strain engineering. Metab. Eng. 14, 59–67. 10.1016/j.ymben.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Dong H., Zhang Y., Dai Z., Li Y. (2010). Engineering Clostridium strain to accept unmethylated DNA. PLoS ONE 5:e9038. 10.1371/journal.pone.0009038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake H. L., Gössner A. S., Daniel S. L. (2008). Old acetogens, new light. Ann. N.Y. Acad. Sci. 1125, 100–128. 10.1196/annals.1419.016 [DOI] [PubMed] [Google Scholar]
- Drake H. L., Küsel K., Matthies C. (2006). Acetogenic prokaryotes, in The Prokaryotes, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (New York, NY: Springer; ), 354–420. 10.1007/0-387-30742-7_13 [DOI] [Google Scholar]
- Drepper T., Eggert T., Circolone F., Heck A., Krauss U., Guterl J.-K., et al. (2007). Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25, 443–445. 10.1038/nbt1293 [DOI] [PubMed] [Google Scholar]
- Drepper T., Huber R., Heck A., Circolone F., Hillmer A.-K., Büchs J., et al. (2010). Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl. Environ. Microbiol. 76, 5990–5994. 10.1128/AEM.00701-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H., Yamada Y., Sato S. (2015). Efficient production of 1,3-butadiene in the catalytic dehydration of 2,3-butanediol. Appl. Catal. A Gen. 491, 163–169. 10.1016/j.apcata.2014.12.006 [DOI] [Google Scholar]
- Enzien M., Hickey R., Levinson W., Vermeiren W. (2014). Method for Production of n-Propanol and/or Ethanol by Fermentation of Multiple Substrates in A Symbiotic Manner. US20140273124. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Evonik (2013). Bacteria Like the Taste of Syngas. Available online at: http://corporate.evonik.com/en/media/search/pages/news-details.aspx?newsid=40331 [Accessed March 30, 2016].
- Fang G., Munera D., Friedman D. I., Mandlik A., Chao M. C., Banerjee O., et al. (2012). Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 30, 1232–1239. 10.1038/nbt.2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farasat I., Kushwaha M., Collens J., Easterbrook M., Guido M., Salis H. M. (2014). Efficient search, mapping, and optimization of multi-protein genetic systems in diverse bacteria. Mol. Syst. Biol. 10, 731. 10.15252/msb.20134955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast A. G., Papoutsakis E. T. (2012). Stoichiometric and energetic analyses of non-photosynthetic CO2-fixation pathways to support synthetic biology strategies for production of fuels and chemicals. Curr. Opin. Chem. Eng. 1, 380–395. 10.1016/j.coche.2012.07.005 [DOI] [Google Scholar]
- Fast A. G., Schmidt E. D., Jones S. W., Tracy B. P. (2015). Acetogenic mixotrophy: novel options for yield improvement in biofuels and biochemicals production. Curr. Opin. Biotechnol. 33, 60–72. 10.1016/j.copbio.2014.11.014 [DOI] [PubMed] [Google Scholar]
- Flusberg B. A., Webster D. R., Lee J. H., Travers K. J., Olivares E. C., Clark T. A., et al. (2010). Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–472. 10.1038/nmeth.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlingstein P., Andrew R. M., Rogelj J., Peters G. P., Canadell J. G., Knutti R., et al. (2014). Persistent growth of CO2 emissions and implications for reaching climate targets. Nat. Geosci. 7, 709–715. 10.1038/ngeo2248 [DOI] [Google Scholar]
- Frolkova A. K., Raeva V. M. (2010). Bioethanol dehydration: state of the art. Theor. Found. Chem. Eng. 44, 545–556. 10.1134/S0040579510040342 [DOI] [Google Scholar]
- Furutani M., Uenishi A., Iwasa K., Jennewein S., Fischer R. (2013). Recombinant Cell, and Method for Producing Isoprene. US20150284742. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Gaddy G. L. (1997). Clostridium Strain Which Produces Acetic Acid from Waste Gases. US5593886. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Gaddy J. L. (2000). Clostridium Strains Which Produce Ethanol from Substrate-Containing Gases. WO2000068407. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Gaddy J. L., Chen G. (1998). Bioconversion of Waste Biomass to Useful Products. US5821111. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Gaddy J. L., Claussen E. C. (1992). Clostridiumm ljungdahlii, an Anaerobic Ethanol and Acetate Producing Microorganism. US5173429. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Gaddy J. L., Dinesh K. A., Ko C. W., Phillips J. R., Basu R., Wikstrom C. V., et al. (2007). Methods for Increasing the Production of Ethanol from Microbial Fermentation. US7285402. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Gardner T. S. (2013). Synthetic biology: from hype to impact. Trends Biotechnol. 31, 123–125. 10.1016/j.tibtech.2013.01.018 [DOI] [PubMed] [Google Scholar]
- Geddes C. C., Nieves I. U., Ingram L. O. (2011). Advances in ethanol production. Curr. Opin. Biotechnol. 22, 312–319. 10.1016/j.copbio.2011.04.012 [DOI] [PubMed] [Google Scholar]
- Genthner B. R., Bryant M. P. (1982). Growth of Eubacterium limosum with carbon monoxide as the energy source. Appl. Environ. Microbiol. 43, 70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genthner B. R. S., Bryant M. P. (1987). Additional characteristics of one-carbon-compound utilization by Eubacterium limosum and Acetobacterium woodi. Appl. Environ. Microbiol. 53, 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genthner B. R. S., Davis C. L., Bryant M. P. (1981). Features of rumen and sewage sludge strains of Eubacterium limosum, a methanol-utilizing and H2-CO2-utilizing species. Appl. Environ. Microbiol. 42, 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George K. W., Alonso-Gutierrez J., Keasling J. D., Lee T. S. (2015). Isoprenoid drugs, biofuels, and Chemicals–Artemisinin, farnesene, and beyond, in Biotechnology of Isoprenoids., eds Schrader J., Bohlmann J. (Cham: Springer International Publishing; ), 355–389. [Google Scholar]
- Girbal L., Mortier-barrière I., Rouanet C., Croux C., Mortier-barrie I., Soucaille P. (2003). Development of a sensitive gene expression reporter system and an inducible promoter-repressor system for Clostridium acetobutylicum. Appl. Environ. Microbiol. 69, 4985–4988. 10.1128/AEM.69.8.4985-4988.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Bioenergies (2016). Global Bioenergies and LANZATECH Strengthen Cooperation. Available online at: http://www.global-bioenergies.com/global-bioenergies-and-lanzatech-strengthen-cooperation/?lang=en [Accessed March 30, 2016].
- Gong Z., Shen H., Zhou W., Wang Y., Yang X., Zhao Z. K. (2015). Efficient conversion of acetate into lipids by the oleaginous yeast Cryptococcus curvatus. Biotechnol. Biofuels 8, 1–9. 10.1186/s13068-015-0371-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gössner A. S., Picardal F., Tanner R. S., Drake H. L. (2008). Carbon metabolism of the moderately acid-tolerant acetogen Clostridium drakei isolated from peat. FEMS Microbiol. Lett. 287, 236–242. 10.1111/j.1574-6968.2008.01313.x [DOI] [PubMed] [Google Scholar]
- Goulas K. A., Toste F. D. (2016). Combining microbial production with chemical upgrading. Curr. Opin. Biotechnol. 38, 47–53. 10.1016/j.copbio.2015.12.019 [DOI] [PubMed] [Google Scholar]
- Grethlein A. J., Worden R. M., Jain M. K., Datta R. (1990). Continuous production of mixed alcohols and acids from carbon monoxide. Appl. Biochem. Biotechnol. 24–25, 875–884. 10.1007/BF02920301 [DOI] [Google Scholar]
- Grethlein A. J., Worden R. M., Jain M. K., Datta R. (1991). Evidence for production of n-butanol from carbon monoxide by Butyribacterium methylotrophicum. J. Ferment. Bioeng. 72, 58–60. 10.1016/0922-338X(91)90147-9 [DOI] [Google Scholar]
- Griffin D. W., Schultz M. A. (2012). Fuel and chemical products from biomass syngas: A comparison of gas fermentation to thermochemical conversion routes. Environ. Prog. Sustain. Energy 31, 219–224. 10.1002/ep.11613 [DOI] [Google Scholar]
- Grootscholten T. I. M., Steinbusch K. J. J., Hamelers H. V. M., Buisman C. J. N. (2013). High rate heptanoate production from propionate and ethanol using chain elongation. Bioresour. Technol. 136, 715–718. 10.1016/j.biortech.2013.02.085 [DOI] [PubMed] [Google Scholar]
- Guo Y., Xu J., Zhang Y., Xu H., Yuan Z., Li D. (2010). Medium optimization for ethanol production with Clostridium autoethanogenum with carbon monoxide as sole carbon source. Bioresour. Technol. 101, 8784–8789. 10.1016/j.biortech.2010.06.072 [DOI] [PubMed] [Google Scholar]
- Haghighi Mood S., Hossein Golfeshan A., Tabatabaei M., Salehi Jouzani G., Najafi G. H., Gholami M., et al. (2013). Lignocellulosic biomass to bioethanol, a comprehensive review with a focus on pretreatment. Renew. Sustain. Energy Rev. 27, 77–93. 10.1016/j.rser.2013.06.033 [DOI] [Google Scholar]
- Han S., Gao X., Ying H., Zhou C. C. (2016). NADH gene manipulation for advancing bioelectricity in Clostridium ljungdahlii microbial fuel cells. Green Chem. 18, 2473–2478. 10.1039/C5GC02696B [DOI] [Google Scholar]
- Handler R. M., Shonnard D. R., Griffing E. M., Lai A., Palou-Rivera I. (2016). Life Cycle Assessments of ethanol production via gas fermentation: anticipated greenhouse gas emissions for cellulosic and waste gas feedstocks. Ind. Eng. Chem. Res. 55, 3253–3261. 10.1021/acs.iecr.5b03215 [DOI] [Google Scholar]
- Harvey B. G., Meylemans H. A. (2011). The role of butanol in the development of sustainable fuel technologies. J. Chem. Technol. Biotechnol. 86, 2–9. 10.1002/jctb.2540 [DOI] [Google Scholar]
- He S. H., Woo S. B., DerVartanian D. V., Le Gall J., Peck H. D. (1989). Effects of acetylene on hydrogenases from the sulfate reducing and methanogenic bacteria. Biochem. Biophys. Res. Commun. 161, 127–133. 10.1016/0006-291X(89)91570-2 [DOI] [PubMed] [Google Scholar]
- Heap J. T., Ehsaan M., Cooksley C. M., Ng Y. K., Cartman S. T., Winzer K., et al. (2012). Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res. 40, e59. 10.1093/nar/gkr1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap J. T., Kuehne S. A., Ehsaan M., Cartman S. T., Cooksley C. M., Scott J. C., et al. (2010). The ClosTron: mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 80, 49–55. 10.1016/j.mimet.2009.10.018 [DOI] [PubMed] [Google Scholar]
- Heap J. T., Pennington O. J., Cartman S. T., Carter G. P., Minton N. P. (2007). The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70, 452–464. 10.1016/j.mimet.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Heap J. T., Pennington O. J., Cartman S. T., Minton N. P. (2009). A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78, 79–85. 10.1016/j.mimet.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Heijstra B. D., Kern E., Köpke M., Segovia S., Liew F. (2012). Novel Bacteria and Methods of Use Thereof. US20130217096. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Herrmann G., Jayamani E., Mai G., Buckel W. (2008). Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190, 784–791. 10.1128/JB.01422-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess V., Gallegos R., Jones J. A., Barquera B., Malamy M. H., Müller V. (2016). Occurrence of ferredoxin:NAD + oxidoreductase activity and its ion specificity in several Gram-positive and Gram-negative bacteria. PeerJ 4, e1515. 10.7717/peerj.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heveling J., Nicolaides C. P., Scurrell M. S. (1998). Catalysts and conditions for the highly efficient, selective and stable heterogeneous oligomerisation of ethylene. Appl. Catal. A Gen. 173, 1–9. 10.1016/S0926-860X(98)00147-1 [DOI] [Google Scholar]
- Hoffmeister S., Gerdom M., Bengelsdorf F. R., Linder S., Fluchter S., Ozturk H., et al. (2016). Acetone production with metabolically engineered strains of Acetobacterium woodii. Metab. Eng. 36, 37–47. 10.1016/j.ymben.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Hu P., Chakraborty S., Kumar A., Woolston B., Liu H., Emerson D., et al. (2016). Integrated bioprocess for conversion of gaseous substrates to liquids. Proc. Natl. Acad. Sci. U.S.A. 113, 3773–3778. 10.1073/pnas.1516867113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. J., Ramaswamy S., Tschirner U. W., Ramarao B. V. (2008). A review of separation technologies in current and future biorefineries. Sep. Purif. Technol. 62, 1–21. 10.1016/j.seppur.2007.12.011 [DOI] [Google Scholar]
- Huang H., Wang S., Moll J., Thauer R. K. (2012). Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2. J. Bacteriol. 194, 3689–3699. 10.1128/JB.00385-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhnke R. L., Lewis R. S., Tanner R. S. (2008). Isolation and Characterization of Novel Clostridial Species. US7704723. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Humphreys C. M., McLean S., Schatschneider S., Millat T., Henstra A. M., Annan F. J., et al. (2015). Whole genome sequence and manual annotation of Clostridium autoethanogenum, an industrially relevant bacterium. BMC Genomics 16, 1–10. 10.1186/s12864-015-2287-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K. M., Lewis R. S. (2010). Carbon monoxide partial pressure effects on the metabolic process of syngas fermentation. Biochem. Eng. J. 48, 159–165. 10.1016/j.bej.2009.09.004 [DOI] [Google Scholar]
- Hutchison C. A., Chuang R.-Y., Noskov V. N., Assad-Garcia N., Deerinck T. J., Ellisman M. H., et al. (2016). Design and synthesis of a minimal bacterial genome. Science (80-.). 351, 1371–1494. 10.1126/science.aad6253 [DOI] [PubMed] [Google Scholar]
- Imkamp F., Müller V. (2007). Acetogenic bacteria, in Encyclopedia of Life Sciences (Chichester, UK: John Wiley & Sons, Ltd; ). 10.1002/9780470015902.a0020086 [DOI] [Google Scholar]
- INEOS Bio (2014). INEOS Bio Provides Operational Update. Available online at: http://www.ineos.com/businesses/ineos-bio/news/ineos-bio-provides-operational-update [Accessed January 27, 2016].
- INVISTA (2015). INVISTA and LanzaTech Make Breakthrough for Bio-derived Butadiene Production. Available online at: http://www.invista.com/en/news/pr-invista-and-lanzatech-make-breakthrough-for-bio-derived-butadiene-production.html [Accessed March 30, 2016].
- International Energy Agency (2014). Medium-Term Renewable Energy Market Report 2014. Weinheim: International Energy Agency. [Google Scholar]
- Isaksson J., Asblad A., Berntsson T. (2013). Influence of different pretreatment methods on biomass gasification and production of fischer-tropsch crude integrated with a pulp and paper mill. Chem. Eng. Trans. 35, 559–564. 10.3303/CET1335093 [DOI] [Google Scholar]
- Islam M. A., Zengler K., Edwards E. A., Mahadevan R., Stephanopoulos G. (2015). Investigating Moorella thermoacetica metabolism with a genome-scale constraint-based metabolic model. Integr. Biol. 7, 869–882. 10.1039/c5ib00095e [DOI] [PubMed] [Google Scholar]
- Isom C. E., Nanny M. A., Tanner R. S. (2015). Improved conversion efficiencies for n-fatty acid reduction to primary alcohols by the solventogenic acetogen “Clostridium ragsdalei.” J. Ind. Microbiol. Biotechnol. 42, 29–38. 10.1007/s10295-014-1543-z [DOI] [PubMed] [Google Scholar]
- Jennert K. C. B., Tardif C., Young D. I., Young M. (2000). Gene transfer to Clostridium cellulolyticum ATCC 35319. Microbiology 146, 3071–3080. 10.1099/00221287-146-12-3071 [DOI] [PubMed] [Google Scholar]
- Jeong J., Bertsch J., Hess V., Choi S., Choi I.-G., Chang I. S., et al. (2015). A model for energy conservation based on genomic and experimental analyses in a carbon monoxide-utilizing, butyrate-forming acetogen, Eubacterium limosum KIST612. Appl. Environ. Microbiol. 84, 4782–4790. 10.1128/aem.00675-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y., Song Y., Shin H. S., Cho B.-K. (2014). Draft genome sequence of acid-tolerant clostridium drakei SL1T, a potential chemical producer through syngas fermentation. Genome Announc. 2:e00387–14. 10.1128/genomeA.00387-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A. (2013). CRISPR-assisted editing of bacterial genomes. Nat. Biotechnol. 31, 233–239. 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S. (2015). Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514. 10.1128/AEM.04023-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jipa I., Dobre T., Stroescu M., Stoica A. (2009). Acetic acid extraction from fermentation broth experimental and modelling studies. Rev. Chim. Bucharest Orig. Ed. 60, 1084–1089. [Google Scholar]
- Kane M. D., Brauman A., Breznak J. A. (1991). Clostridium mayombei sp. nov, an H2/CO2 acetogenic bacterium from the gut of the African soil-feeding termite, Cubitermes speciosus. Arch. Microbiol. 156, 99–104. 10.1007/BF00290980 [DOI] [Google Scholar]
- Kane M. D., Breznak J. A. (1991). Acetonema longum gen nov SP-nov, an H2/CO2 acetogenic bacterium from the termite, Pterotermes occidentis. Arch. Microbiol. 156, 91–98. 10.1007/BF00290979 [DOI] [PubMed] [Google Scholar]
- Kantzow C., Mayer A., Weuster-Botz D. (2015). Continuous gas fermentation by Acetobacterium woodii in a submerged membrane reactor with full cell retention. J. Biotechnol. 212, 11–18. 10.1016/j.jbiotec.2015.07.020 [DOI] [PubMed] [Google Scholar]
- Karberg M., Guo H. T., Zhong J., Coon R., Perutka J., Lambowitz A. M. (2001). Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol. 19, 1162–1167. 10.1038/nbt1201-1162 [DOI] [PubMed] [Google Scholar]
- Kaster A. K., Goenrich M., Seedorf H., Liesegang H., Wollherr A., Gottschalk G., et al. (2011). More than 200 genes required for methane formation from H2 and CO2 and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus. Archaea 2011, 973848. 10.1155/2011/973848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S., Watamura Y., Ono M., Watanabe T., Takeda K., Niimura Y. (2005). Adaptive responses to oxygen stress in obligatory anaerobes Clostridium acetobutylicum and Clostridium aminovalericum. Appl. Environ. Microbiol. 71, 8442–8450. 10.1128/AEM.71.12.8442-8450.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keis S., Shaheen R., Jones D. T. (2001). Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov and Clostridium saccharobutylicum sp. nov. Int. J. Syst. Evol. Microbiol. 51, 2095–2103. 10.1099/00207713-51-6-2095 [DOI] [PubMed] [Google Scholar]
- Kerby R., Zeikus J. G. (1983). Growth of Clostridium thermoaceticum on H2/CO2 or CO as energy source. Curr. Microbiol. 8, 27–30. 10.1007/BF0156731018636591 [DOI] [Google Scholar]
- Kita A., Iwasaki Y., Sakai S., Okuto S., Takaoka K., Suzuki T., et al. (2013). Development of genetic transformation and heterologous expression system in carboxydotrophic thermophilic acetogen Moorella thermoacetica. J. Biosci. Bioeng. 115, 347–352. 10.1016/j.jbiosc.2012.10.013 [DOI] [PubMed] [Google Scholar]
- Klasson K. T., Ackerson M. D., Clausen E. C., Gaddy J. L. (1991). Bioreactor design for synthesis gas fermentations. Fuel 70, 605–614. 10.1016/0016-2361(91)90174-9 [DOI] [Google Scholar]
- Klasson K. T., Ackerson M. D., Clausen E. C., Gaddy J. L. (1992). Bioconversion of synthesis gas into liquid or gasesous fuels. Enzyme Microb. Technol. 14, 602–608. 10.1016/0141-0229(92)90033-K27125997 [DOI] [Google Scholar]
- Köpke M., Al-Sinawi B. (2013). Selection Method and Recombinant Microorganisms and Uses Therefor. US20130316364. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Köpke M., Al-Sinawi B., Jensen R. O., Mueller A. P., Hill R. E. (2015). Microorganisms and Methods for the Production of Ketones. US20150152445. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Köpke M., Chen W. Y. (2013). Recombinant Microorganisms and Uses Therefor. US20130323806. Available online at: https://www.google.com/patents/US20130323806. [Google Scholar]
- Köpke M., Gerth M. L., Maddock D. J., Mueller A. P., Liew F., Simpson S. D., et al. (2014). Reconstruction of an acetogenic 2,3-butanediol pathway involving a novel NADPH-dependent primary-secondary alcohol dehydrogenase. Appl. Environ. Microbiol. 80, 3394–3403. 10.1128/AEM.00301-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köpke M., Havill A. (2014). LanzaTech's route to bio-butadiene. Catal. Rev. 27, 7–12. [Google Scholar]
- Köpke M., Held C., Hujer S., Liesegang H., Wiezer A., Wollherr A., et al. (2010). Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. U.S.A. 107, 13087–13092. 10.1073/pnas.1004716107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köpke M., Liew F. M. (2011). Recombinant Microorganism and Methods of Production Thereof. US 2011/0236941 A1. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Köpke M., Mihalcea C., Liew F., Tizard J. H., Ali M. S., Conolly J. J., et al. (2011). 2,3-Butanediol production by acetogenic bacteria, an alternative route to chemical synthesis, using industrial waste gas. Appl. Environ. Microbiol. 77, 5467–5475. 10.1128/AEM.00355-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köpke M., Nagaraju S., Chen W. (2013a). Recombinant Microorganisms and Methods of Use Thereof. US20140206901. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Köpke M., Simpson S., Liew F. M., Chen W. (2012). Fermentation Process for Producing Isopropanol Using a Recombinant Microorganism. US20120252083 A1. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Köpke M., Straub M., Dürre P. (2013b). Clostridium difficile is an autotrophic bacterial pathogen. PLoS ONE 8:e62157. 10.1371/journal.pone.0062157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasna A. I., Rittenberg D. (1954). The inhibition of hydrogenase by nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 40, 225–227. 10.1073/pnas.40.4.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumholz L. R., Bryant M. P. (1985). Clostridium pfennigii sp. nov uses methoxyl groups of monobenzenoids and produces butyrate. Int. J. Syst. Bacteriol. 35, 454–456. 10.1099/00207713-35-4-454 [DOI] [Google Scholar]
- Kuehne S. A., Minton N. P. (2012). ClosTron-mediated engineering of Clostridium. Bioengineered 3, 247–254. 10.4161/bioe.21004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundiyana D. K., Huhnke R. L., Wilkins M. R. (2010). Syngas fermentation in a 100-L pilot scale fermentor: design and process considerations. J. Biosci. Bioeng. 109, 492–498. 10.1016/j.jbiosc.2009.10.022 [DOI] [PubMed] [Google Scholar]
- Kundiyana D. K., Wilkins M. R., Maddipati P., Huhnke R. L. (2011). Effect of temperature, pH and buffer presence on ethanol production from synthesis gas by “Clostridium ragsdalei”. Bioresour. Technol. 102, 5794–5799. 10.1016/j.biortech.2011.02.032 [DOI] [PubMed] [Google Scholar]
- Küsel K., Dorsch T., Acker G., Stackebrandt E., Drake H. L. (2000). Clostridium scatologenes strain SL1 isolated as an acetogenic bacterium from acidic sediments. Int. J. Syst. Evol. Microbiol. 50(Pt 2), 537–546. 10.1099/00207713-50-2-537 [DOI] [PubMed] [Google Scholar]
- Küsel K., Karnholz A., Trinkwalter T., Devereux R., Acker G., Drake H. L. (2001). Physiological ecology of Clostridium glycolicum RD-1, an aerotolerant acetogen isolated from sea grass roots. Appl. Environ. Microbiol. 67, 4734–4741. 10.1128/AEM.67.10.4734-4741.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan E. I., Liao J. C. (2013). Microbial synthesis of n-butanol, isobutanol, and other higher alcohols from diverse resources. Bioresour. Technol. 135, 339–349. 10.1016/j.biortech.2012.09.104 [DOI] [PubMed] [Google Scholar]
- Lane J. (2014). On the Mend: Why INEOS Bio Isn't Producing Ethanol in Florida. Biofuels Dig. Available online at: http://www.biofuelsdigest.com/bdigest/2014/09/05/on-the-mend-why-ineos-bio-isnt-reporting-much-ethanol-production/ [Accessed January 28, 2016].
- Lane J. (2015a). Alcohol to renewable jet fuel: the Digest's 2015 8-Slide Guide to Gevo's ATJ, in Biofuels Dig. Available online at: http://www.biofuelsdigest.com/bdigest/2015/12/16/alcohol-to-renewable-jet-fuel-the-digests-2015-8-slide-guide-to-gevos-atj/ [Accessed January 29, 2016].
- Lane J. (2015b). China steel green-lights commercial-scale LanzaTech advanced biofuels project, in Biofuels Dig. Available online at: http://www.biofuelsdigest.com/bdigest/2015/04/22/china-steel-green-lights-46m-for-commercial-scale-lanzatech-advanced-biofuels-project/ [Accessed January 27, 2016].
- Lane J. (2015c). Steel's Big Dog jumps into low carbon fuels: ArcelorMittal, LanzaTech, Primetals Technologies to construct $96M biofuel production facility, in Biofuels Dig. Available online at: http://www.biofuelsdigest.com/bdigest/2015/07/13/steels-big-dog-jumps-into-low-carbon-fuels-arcelormittal-lanzatech-primetals-technologies-to-construct-96m-biofuel-production-facility/ [Accessed January 27, 2016].
- Lane J. (2016). Coskata's Technology Re-emerges as Synata Bio. Available online at: http://www.biofuelsdigest.com/bdigest/2016/01/24/coskatas-technology-re-emerges-as-synata-bio/ [Accessed March 30, 2016].
- LanzaTech (2013). Evonik and LanzaTech Working on Bio-Processed Precursors for Specialty Plastics. Available online at: http://www.lanzatech.com/evonik-and-lanzatech-working-on-bio-processed-precursors-for-specialty-plastics/ [Accessed July 26, 2015]
- LanzaTech (2015). Fuels. Available online at: http://www.lanzatech.com/innovation/markets/fuels/ [Accessed January 29, 2016].
- LanzaTech (2016). Aemetis Acquires License from LanzaTech with California Exclusive Rights for Advanced Ethanol from Biomass. Available online at: http://www.lanzatech.com/aemetis-acquires-license-lanzatech-california-exclusive-rights-advanced-ethanol-biomass-including-forest-ag-wastes/ [Accessed March 25, 2016].
- Latif H., Zeidan A. A., Nielsen A. T., Zengler K. (2014). Trash to treasure: production of biofuels and commodity chemicals via syngas fermenting microorganisms. Curr. Opin. Biotechnol. 27, 79–87. 10.1016/j.copbio.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Leang C., Ueki T., Nevin K. P., Lovley D. R. (2013). A genetic system for Clostridium ljungdahlii: a chassis for autotrophic production of biocommodities and a model homoacetogen. Appl. Environ. Microbiol. 79, 1102–1109. 10.1128/AEM.02891-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-H., Kim H. J., Shin Y.-A., Kim K. H., Lee S. J. (2016). Single crossover-mediated markerless genome engineering in Clostridium acetobutylicum. J. Microbiol. Biotechnol. 26, 725–729. 10.4014/jmb.1512.12012 [DOI] [PubMed] [Google Scholar]
- Levinson W. E. (2014). A Butyrate Producing Clostridium Species, Clostridium pharus. US20140242654. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Liew F., Köpke M. (2012). Recombinant Microorganisms Make Biodiesel. US20120252083. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Liew F. M., Köpke M., Simpson S. D. (2013). Gas fermentation for commercial biofuels production, in Biofuel Production-Recent Developments and Prospects, ed Fang Z. (Rijeka, Croatia: InTech; ), 125–174. [Google Scholar]
- Lin S., Diercks C. S., Zhang Y.-B., Kornienko N., Nichols E. M., Zhao Y., et al. (2015). Covalent organic frameworks comprising cobalt porphyrins for catalytic CO2 reduction in water. Science 349, 1208–1213. 10.1126/science.aac8343 [DOI] [PubMed] [Google Scholar]
- Liou J. S.-C., Balkwill D. L., Drake G. R., Tanner R. S. (2005). Clostridium carboxidivorans sp. nov., a solvent-producing clostridium isolated from an agricultural settling lagoon, and reclassification of the acetogen Clostridium scatologenes strain SL1 as Clostridium drakei sp. nov. Int. J. Syst. Evol. Microbiol. 55, 2085–2091. 10.1099/ijs.0.63482-0 [DOI] [PubMed] [Google Scholar]
- Liu K., Atiyeh H. K., Tanner R. S., Wilkins M. R., Huhnke R. L. (2012). Fermentative production of ethanol from syngas using novel moderately alkaliphilic strains of Alkalibaculum bacchi. Bioresour. Technol. 104, 336–341. 10.1016/j.biortech.2011.10.054 [DOI] [PubMed] [Google Scholar]
- Loke H. K., Bennett G. N., Lindahl P. A. (2000). Active acetyl-CoA synthase from Clostridium thermoaceticum obtained by cloning and heterologous expression of acsAB in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 12530–12535. 10.1073/pnas.220404397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorowitz W. H., Bryant M. P. (1984). Peptostreptococcus productus strain that grows rapidly with CO as the energy source. Appl. Environ. Microbiol. 47, 961–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundie L. L., Drake H. L. (1984). Development of a minimally defined medium for the acetogen Clostridium thermoaceticum. J. Bacteriol. 159, 700–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luning Prak D. J., Jones M. H., Trulove P., McDaniel A. M., Dickerson T., Cowart J. S. (2015). Physical and chemical analysis of alcohol-to-jet (ATJ) fuel and development of surrogate fuel mixtures. Energy and Fuels 29, 3760–3769. 10.1021/acs.energyfuels.5b00668 [DOI] [Google Scholar]
- Lux M. F., Drake H. L. (1992). Reexamination of the metabolic potentials of the acetogens Clostridium aceticum and Clostridium formicoaceticum - chemolithoautotrophic and aromatic-dependent growth. Fems Microbiol. Lett. 95, 49–56. 10.1111/j.1574-6968.1992.tb05341.x [DOI] [PubMed] [Google Scholar]
- Lynd L., Kerby R., Zeikus J. G. (1982). Carbon monoxide metabolism of the methylotrophic acidogen Butyribacterium methylotrophicum. J. Bacteriol. 149, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddipati P., Atiyeh H. K., Bellmer D. D., Huhnke R. L. (2011). Ethanol production from syngas by Clostridium strain P11 using corn steep liquor as a nutrient replacement to yeast extract. Bioresour. Technol. 102, 6494–6501. 10.1016/j.biortech.2011.03.047 [DOI] [PubMed] [Google Scholar]
- Makshina E. V., Dusselier M., Janssens W., Degrève J., Jacobs P. A., Sels B. F. (2014). Review of old chemistry and new catalytic advances in the on-purpose synthesis of butadiene. Chem. Soc. Rev. 43, 7917–7953. 10.1039/C4CS00105B [DOI] [PubMed] [Google Scholar]
- Makshina E. V., Janssens W., Sels B. F., Jacobs P. A. (2012). Catalytic study of the conversion of ethanol into 1,3-butadiene. Catal. Today 198, 338–344. 10.1016/j.cattod.2012.05.031 [DOI] [Google Scholar]
- Maness P.-C., Weaver P. (2001). Evidence for three distinct hydrogenase activities in Rhodospirillum rubrum. Appl. Microbiol. Biotechnol. 57, 751–756. 10.1007/s00253-001-0828-0 [DOI] [PubMed] [Google Scholar]
- Marcellin E., Behrendorff J. B., Nagaraju S., DeTissera S., Segovia S., Palfreyman R. W., et al. (2016). Low carbon fuels and commodity chemicals from waste gases – systematic approach to understand energy metabolism in a model acetogen. Green Chem. [Epub ahead of print]. 10.1039/C5GC02708J. [DOI] [Google Scholar]
- Martin M. E., Richter H., Saha S., Angenent L. T. (2016). Traits of selected Clostridium strains for syngas fermentation to ethanol. Biotechnol. Bioeng. 113, 531–539. 10.1002/bit.25827 [DOI] [PubMed] [Google Scholar]
- McGlade C., Ekins P. (2015). The geographical distribution of fossil fuels unused when limiting global warming to 2°C. Nature 517, 187–190. 10.1038/nature14016 [DOI] [PubMed] [Google Scholar]
- McKendry P. (2002). Energy production from biomass (Part 3): gasification technologies. Bioresour. Technol. 83, 55–63. 10.1016/S0960-8524(01)00120-1 [DOI] [PubMed] [Google Scholar]
- Mechichi T., Labat M., Patel B. K., Woo T. H., Thomas P., Garcia J. L. (1999). Clostridium methoxybenzovorans sp. nov., a new aromatic o-demethylating homoacetogen from an olive mill wastewater treatment digester. Int. J. Syst. Bacteriol. 49(Pt 3), 1201–1209. 10.1099/00207713-49-3-1201 [DOI] [PubMed] [Google Scholar]
- Mermelstein L., Welker N., Bennet G., Papoutsakis E. (1992). Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Biotechnology 10, 190–195. 10.1038/nbt0292-190 [DOI] [PubMed] [Google Scholar]
- Metz B., Davidson O. R., Bosch P. R., Dave R., Meyer L. A. (eds.). (2007). Contribution of Working Group III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, 2007. Cambridge: Cambridge University Press. [Google Scholar]
- Michael K., Steffi N., Peter D. (2011). The past, present, and future of biofuels – biobutanol as promising alternative, in Biofuel Production-Recent Developments and Prospects, ed dos Santos Bernades M. A. (Rijeka: InTech; ), 451–486. [Google Scholar]
- Mock J., Wang S., Huang H., Kahnt J., Thauer R. K. (2014). Evidence for a hexaheteromeric methylenetetrahydrofolate reductase in Moorella thermoacetica. J. Bacteriol. 196, 3303–3314. 10.1128/JB.01839-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock J., Zheng Y., Mueller A. P., Ly S., Tran L., Segovia S., et al. (2015). Energy conservation associated with ethanol formation from H2 and CO2 in Clostridium autoethanogenum involving electron bifurcation. J. Bacteriol. 197, 2965–2980. 10.1128/JB.00399-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M., Najafpour G. D., Younesi H., Lahijani P., Uzir M. H., Mohamed A. R. (2011). Bioconversion of synthesis gas to second generation biofuels: a review. Renew. Sustain. Energy Rev. 15, 4255–4273. 10.1016/j.rser.2011.07.124 [DOI] [Google Scholar]
- Molitor B., Richter H., Martin M. E., Jensen R. O., Juminaga A., Mihalcea C., et al. (2016). Carbon recovery by fermentation of CO-rich off gases – turning steel mills into biorefineries. Bioresour. Technol. [Epub ahead of print]. 10.1016/j.biortech.2016.03.094. [DOI] [PubMed] [Google Scholar]
- Monot M., Boursaux-Eude C., Thibonnier M., Vallenet D., Moszer I., Medigue C., et al. (2011). Reannotation of the genome sequence of Clostridium difficile strain 630. J. Med. Microbiol. 60, 1193–1199. 10.1099/jmm.0.030452-0 [DOI] [PubMed] [Google Scholar]
- Morris B. E. L., Henneberger R., Huber H., Moissl-Eichinger C. (2013). Microbial syntrophy: Interaction for the common good. FEMS Microbiol. Rev. 37, 384–406. 10.1111/1574-6976.12019 [DOI] [PubMed] [Google Scholar]
- Mueller A. P., Koepke M., Nagaraju S. (2013). Recombinant Microorganisms and Uses Therefor. US20130330809. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Mukherjee A., Schroeder C. M. (2015). Flavin-based fluorescent proteins: emerging paradigms in biological imaging. Curr. Opin. Biotechnol. 31, 16–23. 10.1016/j.copbio.2014.07.010 [DOI] [PubMed] [Google Scholar]
- Müller V. (2003). Energy conservation in acetogenic bacteria. Appl. Environ. Microbiol. 69, 6345–6353. 10.1128/AEM.69.11.6345-6353.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller V., Imkamp F., Biegel E., Schmidt S., Dilling S. (2008). Discovery of a ferredoxin:NAD+-oxidoreductase (Rnf) in Acetobacterium woodii: a novel potential coupling site in acetogens. Ann. N.Y. Acad. Sci. 1125, 137–146. 10.1196/annals.1419.011 [DOI] [PubMed] [Google Scholar]
- Munasinghe P. C., Khanal S. K. (2010). Syngas fermentation to biofuel: evaluation of carbon monoxide mass transfer coefficient (kLa) in different reactor configurations. Biotechnol. Prog. 26, 1616–1621. 10.1002/btpr.473 [DOI] [PubMed] [Google Scholar]
- Munasinghe P. C., Khanal S. K. (2012). Syngas fermentation to biofuel: Evaluation of carbon monoxide mass transfer and analytical modeling using a composite hollow fiber (CHF) membrane bioreactor. Bioresour. Technol. 122, 1–7. 10.1016/j.biortech.2012.03.053 [DOI] [PubMed] [Google Scholar]
- Nagarajan H., Sahin M., Nogales J., Latif H., Lovley D. R., Ebrahim A., et al. (2013). Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii. Microb. Cell Fact. 12, 118. 10.1186/1475-2859-12-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraju S., Al-Sinawi B., DeTissera S., Koepke M. (2015). Recombinant Microorganisms and Methods of Use Thereof. US20150210987. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Naik S. N., Goud V. V., Rout P. K., Dalai A. K. (2010). Production of first and second generation biofuels: a comprehensive review. Renew. Sustain. Energy Rev. 14, 578–597. 10.1016/j.rser.2009.10.003 [DOI] [Google Scholar]
- National Energy Technology Laboratory (2013). Water Gas Shift & Hydrogen Production. Available online at: http://www.netl.doe.gov/research/coal/energy-systems/gasification/gasifipedia/water-gas-shift [Accessed March 30, 2016].
- National Energy Technology Laboratory (2014). Alternatives/Supplements to Coal - Feedstock Flexibility. Available online at: http://www.netl.doe.gov/research/coal/energy-systems/gasification/gasifipedia/feedstock [Accessed March 29, 2016].
- National Research Council (2015). Industrialization of Biology: A Roadmap to Accelerate the Advanced Manufacturing of Chemicals. Washington, DC: National Academies Press. [PubMed] [Google Scholar]
- NEL Hydrogen (2015). Efficient Electrolysers for Hydrogen Production. Available online at: http://www.nel-hydrogen.com/brochures-and-downloads/ [Accessed March 30, 2016].
- Nevin K. P., Hensley S. A., Franks A. E., Summers Z. M., Ou J., Woodard T. L., et al. (2011). Electrosynthesis of organic compounds from carbon dioxide is catalyzed by a diversity of acetogenic microorganisms. Appl. Environ. Microbiol. 77, 2882–2886. 10.1128/AEM.02642-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevin K. P., Woodard T. L., Franks A. E., Summers Z. M., Lovley D. R. (2010). Microbial electrosynthesis: feeding microbes electricity to convert carbon dioxide and water to multicarbon extracellular organic compounds. MBio 1, e00103–e00110. 10.1128/mBio.00103-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Y. K., Ehsaan M., Philip S., Collery M. M., Janoir C., Collignon A., et al. (2013). Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyre alleles. PLoS ONE 8:e56051. 10.1371/journal.pone.0056051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley S. D., Coombes J. A., Simpson S. D., Heijstra B. D., Schultz M. A., Molloy S. (2011). Improved Fermentation of Waste Gases. US20130045517. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Ohwaki K., Hungate R. E. (1977). Hydrogen utilization by clostridia in sewage sludge. Appl. Environ. Microbiol. 33, 1270–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OPUS 12 (2015). A Profitable Solution to CO2. Available online at: http://www.opus-12.com/ [Accessed March 30, 2016].
- Oremland R. S., Taylor B. F. (1975). Inhibition of methanogenesis in marine sediments by acetylene and ethylene: validity of the acetylene reduction assay for anaerobic microcosms. Appl. Microbiol. 30, 707–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgill J. J., Atiyeh H. K., Devarapalli M., Phillips J. R., Lewis R. S., Huhnke R. L. (2013). A comparison of mass transfer coefficients between trickle-bed, hollow fiber membrane and stirred tank reactors. Bioresour. Technol. 133, 340–346. 10.1016/j.biortech.2013.01.124 [DOI] [PubMed] [Google Scholar]
- Ou X., Zhang X., Zhang Q., Zhang X. (2013). Life-cycle analysis of energy use and greenhouse gas emissions of gas-to-liquid fuel pathway from steel mill off-gas in China by the LanzaTech process. Front. Energy 7, 263–270. 10.1007/s11708-013-0263-9 [DOI] [Google Scholar]
- Oultram J. D., Loughlin M., Swinfield T. J., Brehm J. K., Thompson D. E., Minton N. P. (1988). Introduction of plasmids into whole cells of Clostridium acetobutylicum by electroporation. FEMS Microbiol. Lett. 56, 83–88. 10.1111/j.1574-6968.1988.tb03154.x [DOI] [Google Scholar]
- Pandelia M. E., Ogata H., Currell L. J., Flores M., Lubitz W. (2010). Inhibition of the [NiFe] hydrogenase from Desulfovibrio vulgaris Miyazaki F by carbon monoxide: an FTIR and EPR spectroscopic study. Biochim. Biophys. Acta 1797, 304–313. 10.1016/j.bbabio.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Panneerselvam A., Wilkins M. R., Delorme M. J. M., Atiyeh H. K., Huhnke R. L. (2010). Effects of various reducing agents on syngas fermentation by “Clostridium ragsdalei.” ASABE 2, 135–144. 10.13031/2013.34831 [DOI] [Google Scholar]
- Perez J. M., Richter H., Loftus S. E., Angenent L. T. (2013). Biocatalytic reduction of short-chain carboxylic acids into their corresponding alcohols with syngas fermentation. Biotechnol. Bioeng. 110, 1066–1077. 10.1002/bit.24786 [DOI] [PubMed] [Google Scholar]
- Peters J. W., Miller A. F., Jones A. K., King P. W., Adams M. W. W. (2016). Electron bifurcation. Curr. Opin. Chem. Biol. 31, 146–152. 10.1016/j.cbpa.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Phillips J. R., Hall A., Remondet N. M., Wilkins M. R., Huhnke R. L. (2011). Designing syngas fermentation medium for fuels and bulk chemicals production. ASABE. 7004 10.13031/2013.37400 [DOI] [Google Scholar]
- Phillips J. R., Klasson K. T., Claussen E. C., Gaddy J. L. (1993). Biological production of ethanol from coal synthesis gas - medium development studies. Appl. Biochem. Biotechnol. 39, 559–571. 10.1007/BF02919018 [DOI] [Google Scholar]
- Phillips-Jones M. K. (2000). Use of a lux reporter system for monitoring rapid changes in α-toxin gene expression in Clostridium perfringens during growth. Fed. Eur. Microbiol. Soc. Microbiol. Lett. 188, 29–33. 10.1016/s0378-1097(00)00208-1 [DOI] [PubMed] [Google Scholar]
- Piccolo C., Bezzo F. (2009). A techno-economic comparison between two technologies for bioethanol production from lignocellulose. Biomass Bioenergy 33, 478–491. 10.1016/j.biombioe.2008.08.008 [DOI] [Google Scholar]
- Pierce E., Xie G., Barabote R. D., Saunders E., Han C. S., Detter J. C., et al. (2008). The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10, 2550–2573. 10.1111/j.1462-2920.2008.01679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlein A., Cebulla M., Ilg M. M., Bengelsdorf F. R., Schiel-Bengelsdorf B., Whited G., et al. (2015). The complete genome sequence of Clostridium aceticum: a missing link between Rnf- and cytochrome-containing autotrophic acetogens. MBio 6:e01168–15. 10.1128/mBio.01168-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlein A., Schmidt S., Kaster A.-K., Goenrich M., Vollmers J., Thürmer A., et al. (2012). An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS ONE 7:e33439. 10.1371/journal.pone.0033439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy D., O'Keeffe T. A. T., Elmore M., Herbert M., McLeod A., Bokori-Brown M., et al. (2002). Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46, 439–452. 10.1046/j.1365-2958.2002.03134.x [DOI] [PubMed] [Google Scholar]
- Purec L., Krasna A. I., Rittenberg D. (1962). The inhibition of hydrogenase by carbon monoxide and the reversal of this inhibition by light. Biochemistry 1, 270–275. 10.1021/bi00908a013 [DOI] [PubMed] [Google Scholar]
- Pyne M. E., Bruder M., Moo-Young M., Chung D. A., Chou C. P., Perry Chou C. (2014). Technical guide for genetic advancement of underdeveloped and intractable Clostridium. Biotechnol. Adv. 32, 623–641. 10.1016/j.biotechadv.2014.04.003 [DOI] [PubMed] [Google Scholar]
- Ragsdale S. W., Ljungdahl L. G., DerVartanian D. V. (1983). Isolation of carbon monoxide dehydrogenase from Acetobacterium woodii and comparison of its properties with those of the Clostridium thermoaceticum enzyme. J. Bacteriol. 155, 1224–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale S. W., Pierce E. (2008). Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784, 1873–1898. 10.1016/j.bbapap.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter H., Loftus S. E., Angenent L. T. (2013a). Integrating syngas fermentation with the carboxylate platform and yeast fermentation to reduce medium cost and improve biofuel productivity. Environ. Technol. 34, 1983–1994. 10.1080/09593330.2013.826255 [DOI] [PubMed] [Google Scholar]
- Richter H., Martin M., Angenent L. (2013b). A two-stage continuous fermentation system for conversion of syngas into ethanol. Energies 6, 3987–4000. 10.3390/en6083987 [DOI] [Google Scholar]
- Rieu-Lesme F., Dauga C., Fonty G., Dore J. (1998). Isolation from the rumen of a new acetogenic bacterium phylogenetically closely related to Clostridium difficile. Anaerobe 4, 89–94. 10.1006/anae.1998.0153 [DOI] [PubMed] [Google Scholar]
- Ritter S. K. (2015). CO2 Electrolyzer Nears Commercialization. Available online at: http://cen.acs.org/articles/93/i13/CO2-Electrolyzer-Nears-Commercialization.html [Accessed March 30, 2016].
- Roberts D. L., James-Hagstrom J. E., Garvin D. K., Gorst C. M., Runquist J. A., Baur J. R., et al. (1989). Cloning and expression of the gene cluster encoding key proteins involved in acetyl-CoA synthesis in Clostridium thermoaceticum: CO dehydrogenase, the corrinoid/Fe-S protein, and methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 86, 32–36. 10.1073/pnas.86.1.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. A., Hrbek J. (1999). Interaction of sulfur with well-defined metal and oxide surfaces: unraveling the mysteries behind catalyst poisoning and desulfurization. Acc. Chem. Res. 32, 719–728. 10.1021/ar9801191 [DOI] [Google Scholar]
- Roh H., Ko H.-J., Kim D., Choi D. G., Park S., Kim S., et al. (2011). Complete genome sequence of a carbon monoxide-utilizing acetogen, Eubacterium limosum KIST612. J. Bacteriol. 193, 307–308. 10.1128/JB.01217-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakimoto K. K., Wong A. B., Yang P. (2016). Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production. Science 351, 74–77. 10.1126/science.aad3317 [DOI] [PubMed] [Google Scholar]
- Sánchez O. J., Cardona C. A. (2008). Trends in biotechnological production of fuel ethanol from different feedstocks. Bioresour. Technol. 99, 5270–5295. 10.1016/j.biortech.2007.11.013 [DOI] [PubMed] [Google Scholar]
- Sauer B. (1987). Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. J. Mol. Cell Biol. 7, 2087–2096. 10.1128/MCB.7.6.2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage M. D., Drake H. L. (1986). Adaptation of the acetogen Clostridium thermoautotrophicum to minimal medium. J. Bacteriol. 165, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage M. D., Wu Z. R., Daniel S. L., Lundie L. L., Drake H. L. (1987). Carbon monoxide-dependent chemolithotrophic growth of Clostridium thermoautotrophicum. Appl. Environ. Microbiol. 53, 1902–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena J., Tanner R. S. (2011). Effect of trace metals on ethanol production from synthesis gas by the ethanologenic acetogen, Clostridium ragsdalei. J. Ind. Microbiol. Biotechnol. 38, 513–521. 10.1007/s10295-010-0794-6 [DOI] [PubMed] [Google Scholar]
- Saxena J., Tanner R. S. (2012). Optimization of a corn steep medium for production of ethanol from synthesis gas fermentation by Clostridium ragsdalei. World J. Microbiol. Biotechnol. 28, 1553–1561. 10.1007/s11274-011-0959-0 [DOI] [PubMed] [Google Scholar]
- Schiel-Bengelsdorf B., Dürre P. (2012). Pathway engineering and synthetic biology using acetogens. FEBS Lett. 586, 2191–2198. 10.1016/j.febslet.2012.04.043 [DOI] [PubMed] [Google Scholar]
- Schill S. R. (2013). Ineos declares commercial cellulosic ethanol online in Florida, in Ethanol Prod. Mag. Available online at: http://www.ethanolproducer.com/articles/10096/ineos-declares-commercial-cellulosic-ethanol-online-in-florida [Accessed March 30, 2016].
- Schink B. (1984). Clostridium magnum sp. nov, a non-autotrophic homoacetogenic bacterium. Arch. Microbiol. 137, 250–255. 10.1007/BF00414553 [DOI] [Google Scholar]
- Schuchmann K., Müller V. (2012). A bacterial electron-bifurcating hydrogenase. J. Biol. Chem. 287, 31165–31171. 10.1074/jbc.M112.395038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchmann K., Müller V. (2014). Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821. 10.1038/nrmicro3365 [DOI] [PubMed] [Google Scholar]
- Schwartz T. J., Shanks B. H., Dumesic J. A. (2016). Coupling chemical and biological catalysis: a flexible paradigm for producing biobased chemicals. Curr. Opin. Biotechnol. 38, 54–62. 10.1016/j.copbio.2015.12.017 [DOI] [PubMed] [Google Scholar]
- Scotcher M. C., Rudolph F. B., Bennett G. N. (2005). Expression of abrB310 and sinR, and effects of decreased abrB310 expression on the transition from acidogenesis to solventogenesis, in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 71, 1987–1995. 10.1128/AEM.71.4.1987-1995.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCS Global Services (2013). Beijing Shougang LanzaTech New Energy Science and Technology Company Earns Roundtable on Sustainable Biomaterials (RSB) Certification. Available online at: https://www.scsglobalservices.com/beijing-shougang-lanzatech-new-energy-science-technology-company-earns-roundtable-on-sustainable [Accessed January 27, 2016]. [Google Scholar]
- Seedorf H., Fricke W. F., Veith B., Bruggemann H., Liesegang H., Strittimatter A., et al. (2008). The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. U.S.A. 105, 2128–2133. 10.1073/pnas.0711093105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C. M., Vogel J. (2014). Differential RNA-seq: the approach behind and the biological insight gained. Curr. Opin. Microbiol. 19, 97–105. 10.1016/j.mib.2014.06.010 [DOI] [PubMed] [Google Scholar]
- Shima S., Ataka K. (2011). Isocyanides inhibit [Fe]-hydrogenase with very high affinity. FEBS Lett. 585, 353–356. 10.1016/j.febslet.2010.12.014 [DOI] [PubMed] [Google Scholar]
- Siedlecki M., De Jong W., Verkooijen A. H. M. (2011). Fluidized Bed gasification as a mature and reliable technology for the production of bio-syngas and applied in the production of liquid transportation fuels—a review. Energies 4, 389–434. 10.3390/en4030389 [DOI] [Google Scholar]
- Siemens (2013). Silyzer - A Proton-Exchange-Membrane (PEM) Technology Based on Electrolysis System. Available online at: http://www.industry.siemens.com/topics/global/en/pem-electrolyzer/silyzer/Pages/silyzer.aspx [Accessed March 30, 2016].
- Sikkema J., de Bont J. A., Poolman B. (1995). Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59, 201–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim J. H., Kamaruddin A. H. (2008). Optimization of acetic acid production from synthesis gas by chemolithotrophic bacterium–Clostridium aceticum using statistical approach. Bioresour. Technol. 99, 2724–2735. 10.1016/j.biortech.2007.07.004 [DOI] [PubMed] [Google Scholar]
- Simpson S. D., Collet C., Tran P. L., Al-Sinawi B., Forster R. L. S., Rowe M. J., et al. (2009a). Microbial Alcohol Production Process. US8119378. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Simpson S. D., Forster R. L., Tran P., Rowe M. J., Warner I. L. (2009b). Novel Bacteria and Methods of Use Thereof. US8222013. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Simpson S. D., Koepke M., Liew F. M. (2011). Recombinant Microorganisms and Methods of Use Thereof. US2011/0256600. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Simpson S. D., Koepke M., Smart K. F., Tran L. P., Sechrist P. (2014). System and Method for Controlling Metabolite Production in a Microbial Fermentation. US20140273115 A1. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Simpson S. D., Warner I. L., Fung J. M. Y., Köpke M. (2010). Optimised Fermentation Media. US8354269. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Soucaille P., Figge R., Croux C. (2008). Process for Chromosomal Integration and DNA Sequence Replacement In Clostridia. US8628967. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Spirito C. M., Richter H., Rabaey K., Stams A. J. M., Angenent L. T. (2014). Chain elongation in anaerobic reactor microbiomes to recover resources from waste. Curr. Opin. Biotechnol. 27, 115–122. 10.1016/j.copbio.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Sreekumar S., Baer Z. C., Gross E., Padmanaban S., Goulas K., Gunbas G., et al. (2014). Chemocatalytic upgrading of tailored fermentation products toward biodiesel. ChemSusChem 7, 2445–2448. 10.1002/cssc.201402244 [DOI] [PubMed] [Google Scholar]
- Sreekumar S., Baer Z. C., Pazhamalai A., Gunbas G., Grippo A., Blanch H. W., et al. (2015). Production of an acetone-butanol-ethanol mixture from Clostridium acetobutylicum and its conversion to high-value biofuels. Nat. Protoc. 10, 528–537. 10.1038/nprot.2015.029 [DOI] [PubMed] [Google Scholar]
- Steinbusch K. J. J., Hamelers H. V. M., Buisman C. J. N. (2008). Alcohol production through volatile fatty acids reduction with hydrogen as electron donor by mixed cultures. Water Res. 42, 4059–4066. 10.1016/j.watres.2008.05.032 [DOI] [PubMed] [Google Scholar]
- Steinbusch K. J. J., Hamelers H. V. M., Plugge C. M., Buisman C. J. N. (2011). Biological formation of caproate and caprylate from acetate: fuel and chemical production from low grade biomass. Energy Environ. Sci. 4, 216–224. 10.1039/c0ee00282h [DOI] [Google Scholar]
- Stephanopoulos G. (2011). Bioprocess And Microbe Engineering for Total Carbon Utilization in Biofuel Production. US20110177564. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Strätz M., Sauer U., Kuhn A., Dürre P. (1994). Plasmid transfer into the homoacetogen Acetobacterium woodii by electroporation and conjugation. Appl. Environ. Microbiol. 60, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub M., Demler M., Weuster-Botz D., Dürre P. (2014). Selective enhancement of autotrophic acetate production with genetically modified Acetobacterium woodii. J. Biotechnol. 178, 67–72. 10.1016/j.jbiotec.2014.03.005 [DOI] [PubMed] [Google Scholar]
- Sun Y., Cheng J. (2002). Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour. Technol. 83, 1–11. 10.1016/S0960-8524(01)00212-7 [DOI] [PubMed] [Google Scholar]
- Swanson R. M., Platon A., Satrio J. A., Brown R. C. (2010). Techno-economic analysis of biomass-to-liquids production based on gasification. Fuel 89, S11–S19. 10.1016/j.fuel.2010.07.027 [DOI] [Google Scholar]
- Tan Y., Liu J., Chen X., Zheng H., Li F. (2013). RNA-seq-based comparative transcriptome analysis of the syngas-utilizing bacterium Clostridium ljungdahlii DSM 13528 grown autotrophically and heterotrophically. Mol. Biosyst. 9, 2775–2784. 10.1039/c3mb70232d [DOI] [PubMed] [Google Scholar]
- Tanner R. S., Miller L. M., Yang D. (1993). Clostridium ljungdahlii sp. nov., an acetogenic species in clostridial rRNA homology group I. Int. J. Syst. Bacteriol. 43, 232. 10.1099/00207713-43-2-232 [DOI] [PubMed] [Google Scholar]
- Temme K., Hill R., Segall-Shapiro T. H., Moser F., Voigt C. A. (2012a). Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 40, 8773–8781. 10.1093/nar/gks597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme K., Zhao D., Voigt C. A. (2012b). Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc. Natl. Acad. Sci. U.S.A. 109, 7085–7090. 10.1073/pnas.1120788109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R. K. (2007). Microbiology - a fifth pathway of carbon fixation. Science 318, 1732–1733. 10.1126/science.1152209 [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Jungermann K., Decker K. (1977). Energy conservation in chemotrophic anerobic bacteria. Bacteriol. Rev. 41, 100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason M. K., Bischler T., Eisenbart S. K., Förstner K. U., Zhang A., Herbig A., et al. (2015). Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 197, 18–28. 10.1128/JB.02096-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trawick J. D., Burk M. J., Burgard P. A. (2012). Microorganisms and Methods for Conversion of Syngas and Other Carbon Sources to Useful Products. US8129155. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Tremblay P.-L., Zhang T. (2015). Electrifying microbes for the production of chemicals. Front. Microbiol. 6, 1–10. 10.3389/fmicb.2015.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay P., Zhang T., Dar S. A., Leang C., Lovley D. R. (2012). The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin:NAD+ oxidoreductase essential for autotrophic growth. MBio 4:e00406–12. 10.1128/mBio.00406-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala S. B., Welker N. E., Papoutsakis E. T. (1999). Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65, 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala S. B., Welker N. E., Papoutsakis E. T. (2003). Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J. Bacteriol. 185, 1923–1934. 10.1128/JB.185.6.1923-1934.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T., Nevin K. P., Woodard T. L., Lovley D. R. (2014). Converting carbon dioxide to butyrate with an engineered strain of Clostridium ljungdahlii. MBio 5:e01636–14–e4. 10.1128/mbio.01636-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerman A. J., Heindel T. J. (2007). Carbon monoxide mass transfer for syngas fermentation in a stirred tank reactor with dual impeller configurations. Biotechnol. Prog. 23, 613–620. 10.1021/bp060311z [DOI] [PubMed] [Google Scholar]
- United Nations (2015). Adoption of the Paris Agreement. 21932, 32. Available online at: http://unfccc.int/resource/docs/2015/cop21/eng/l09r01.pdf [Accessed March 30, 2016].
- Utturkar S. M., Klingeman D. M., Bruno-Barcena J. M., Chinn M. S., Grunden A. M., Köpke M., et al. (2015). Sequence data for Clostridium autoethanogenum using three generations of sequencing technologies. Sci. Data 2, 150014. 10.1038/sdata.2015.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakkilainen E., Kuparinen K., Heinimö J. (2013). Large Industrial Users of Energy Biomass. Available online at: http://www.bioenergytrade.org/downloads/t40-large-industrial-biomass-users.pdf
- van der Drift A., van Doorn J., Vermeulen J. W. (2001). Ten residual biomass fuels for circulating fluidized-bed gasification. Biomass Bioenergy 20, 45–56. 10.1016/S0961-9534(00)00045-3 [DOI] [Google Scholar]
- Vasudevan D., Richter H., Angenent L. T. (2014). Upgrading dilute ethanol from syngas fermentation to n-caproate with reactor microbiomes. Bioresour. Technol. 151, 378–382. 10.1016/j.biortech.2013.09.105 [DOI] [PubMed] [Google Scholar]
- Vega J. L., Holmberg V. L., Clausen E. C., Gaddy J. L. (1988). Fermentation parameters of Peptostreptococcus productus on gaseous substrates (CO, H2/CO2). Arch. Microbiol. 151, 65–70. 10.1007/BF00444671 [DOI] [Google Scholar]
- Vega J. L., Klasson K. T., Kimmel D. E., Clausen E. C., Gaddy J. L. (1990). Sulfur gas tolerance and toxicity of co-utilizing and methanogenic bacteria. Appl. Biochem. Biotechnol. 24–25, 329–340. 10.1007/BF02920257 [DOI] [Google Scholar]
- Walker D. F., Nagaraju S., Koepke M., Mueller A. P. (2015). Recombinant Microorganisms and Methods of Use Thereof. US20150072395. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Walker D. J. F., Köpke M. (2015). Method of Producing a Recombinant Microorganism. US 9315830. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Wang S., Huang H., Kahnt J., Mueller A. P., Köpke M., Thauer R. K. (2013a). NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J. Bacteriol. 195, 4373–4386. 10.1128/JB.00678-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Huang H., Kahnt J., Thauer R. K. (2013b). A reversible electron-bifurcating ferredoxin- and NAD-dependent [FeFe]-hydrogenase (HydABC) in Moorella thermoacetica. J. Bacteriol. 195, 1267–1275. 10.1128/JB.02158-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Huang H., Moll J., Thauer R. K. (2010). NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri. J. Bacteriol. 192, 5115–5123. 10.1128/JB.00612-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang Z. T., Seo S. O., Choi K., Lu T., Jin Y. S., et al. (2015). Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J. Biotechnol. 200, 1–5. 10.1016/j.jbiotec.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Weghoff M. C., Bertsch J., Müller V. (2014). A novel mode of lactate metabolism in strictly anaerobic bacteria. Environ. Microbiol. 17, 670–677. 10.1111/1462-2920.12493 [DOI] [PubMed] [Google Scholar]
- Whitham J. M., Tirado-Acevedo O., Chinn M. S., Pawlak J. J., Grunden A. M. (2015). Metabolic response of Clostridium ljungdahlii to oxygen exposure. Appl. Environ. Microbiol. 81, 8379–8391. 10.1128/AEM.02491-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H. G. (1991). Life with CO or CO2 and H2 as a source of carbon and energy. Fed. Am. Soc. Exp. Biol. J. 5, 156–163. [DOI] [PubMed] [Google Scholar]
- Woolley R. C., Pennock A., Ashton R. J., Davies A., Young M. (1989). Transfer of Tn1545 and Tn916 to Clostridium acetobutylicum. Plasmid 22, 169–174. 10.1016/0147-619X(89)90027-9 [DOI] [PubMed] [Google Scholar]
- Wu P., Wang G., Wang G., Børresen B. T., Liu H., Zhang J. (2016). Butanol production under microaerobic conditions with a symbiotic system of Clostridium acetobutylicum and Bacillus cereus. Microb. Cell Fact. 15, 8. 10.1186/s12934-016-0412-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Li Y., Shi Z., Hemme C. L., Li Y., Zhu Y., et al. (2015). Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl. Environ. Microbiol. 81, 4423–4431. 10.1128/aem.00873-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Wan C., Street J., Yan D. W., Han J., Yu F. (2013). Catalytic removal of oxygen from biomass-derived syngas. Bioresour. Technol. 147, 117–123. 10.1016/j.biortech.2013.08.036 [DOI] [PubMed] [Google Scholar]
- Ye C. Z., Ariya P. A. (2015). Co-adsorption of gaseous benzene, toluene, ethylbenzene, m-xylene (BTEX) and SO2 on recyclable Fe3O4 nanoparticles at 0-101% relative humidities. J. Environ. Sci. (China) 31, 164–174. 10.1016/j.jes.2014.10.019 [DOI] [PubMed] [Google Scholar]
- Zahn J. (2015). Scale-up of renewable chemical manufacturing processes, in Recent Advances in Fermentation Technology (RAFT) (Clearwater Beach, FL: ). Available online at: https://sim.confex.com/sim/raft11/webprogram/Paper31231.html. [Google Scholar]
- Zahn J. A., Saxena J. (2012). Ethanologenic Clostridium Species, Clostridium coskatii. US2012/015, US8143037. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Zeikus J. G., Lynd L. H., Thompson T. E., Krzycki J. A., Weimer P. J., Hegge P. W. (1980). Isolation and characterization of a new, methylotrophic, acidogenic anaerobe, the marburg strain. Curr. Microbiol. 3, 381–386. 10.1007/BF02601907 [DOI] [Google Scholar]
- Zhang H., Pereira B., Li Z., Stephanopoulos G. (2015a). Engineering Escherichia coli coculture systems for the production of biochemical products. Proc. Natl. Acad. Sci. U.S.A. 112, 8266–8271. 10.1073/pnas.1506781112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Yu Y. (2013). Dehydration of ethanol to ethylene. Ind. Eng. Chem. Res. 52, 9505–9514. 10.1021/ie401157c [DOI] [Google Scholar]
- Zhang T. (2015). More efficient together. Science 350, 738–739. 10.1126/science.aad6452 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Grosse-Honebrink A., Minton N. P. (2015b). A universal mariner transposon system for forward genetic studies in the genus Clostridium. PLoS ONE 10:e0122411. 10.1371/journal.pone.0122411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu S., Chai C., Yang S., Jiang W., Minton N. P., et al. (2016). Development of an inducible transposon system for efficient random mutagenesis in Clostridium acetobutylicum. FEMS Microbiol. Lett. [Epub ahead of print]. 10.1093/femsle/fnw065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. L., Zhu M. Q., Wen J. L., Sun R. C. (2016). Gasification of bio-oil: effects of equivalence ratio and gasifying agents on product distribution and gasification efficiency. Bioresour. Technol. 211, 164–172. 10.1016/j.biortech.2016.03.088 [DOI] [PubMed] [Google Scholar]
- Zhou K., Qiao K., Edgar S., Stephanopoulos G. (2015). Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 33, 377–383. 10.1038/nbt.3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. T., Johnson E. A. (1993). Genetic transformation of Clostridium botulinum hall A by electroporation. Biotechnol. Lett. 15, 121–126. 10.1007/BF00133010 [DOI] [Google Scholar]