Abstract
Genome-wide association studies (GWASs) have identified hundreds of susceptibility genes, including shared associations across clinically distinct autoimmune diseases. We performed an inverse χ2 meta-analysis across ten pediatric-age-of-onset autoimmune diseases (pAIDs) in a case-control study including more than 6,035 cases and 10,718 shared population-based controls. We identified 27 genome-wide significant loci associated with one or more pAIDs, mapping to in silico–replicated autoimmune-associated genes (including IL2RA) and new candidate loci with established immunoregulatory functions such as ADGRL2, TENM3, ANKRD30A, ADCY7 and CD40LG. The pAID-associated single-nucleotide polymorphisms (SNPs) were functionally enriched for deoxyribonuclease (DNase)-hypersensitivity sites, expression quantitative trait loci (eQTLs), microRNA (miRNA)-binding sites and coding variants. We also identified biologically correlated, pAID-associated candidate gene sets on the basis of immune cell expression profiling and found evidence of genetic sharing. Network and protein-interaction analyses demonstrated converging roles for the signaling pathways of type 1, 2 and 17 helper T cells (TH1, TH2 and TH17), JAK-STAT, interferon and interleukin in multiple autoimmune diseases.
Autoimmune diseases affect 7–10% of individuals living in Europe and North America1 and represent a significant cause of chronic morbidity and disability. High rates of familial clustering and comorbidity across autoimmune diseases suggest that genetic predisposition underlies disease susceptibility. GWASs and immune-focused fine-mapping studies of autoimmune thyroiditis (THY)2, psoriasis (PSOR)3, juvenile idiopathic arthritis (JIA)4, primary biliary cirrhosis (PBC)5, primary sclerosing cholangitis (PSC)6, rheumatoid arthritis (RA)7, celiac disease (CEL)8, inflammatory bowel disease (IBD, which includes Crohn’s disease (CD) and ulcerative colitis (UC)9), and multiple sclerosis (MS)10,11 have identified hundreds of autoimmune disease–associated SNPs across the genome12–14. SNP associations in certain pan-autoimmune loci, such as PTPN22 c.1858C>T (rs2476601), are evident in independent GWASs across multiple autoimmune diseases15–18, whereas others have been uncovered through large-scale meta-analyses (for example, CEL-RA and type 1 diabetes (T1D)-CD) or by searches for known loci from one disease in another (for example, systemic lupus erythematosus (SLE))19. These studies demonstrate that more than half of genome-wide significant (GWS) autoimmune disease associations are shared by at least two distinct autoimmune diseases20,21. However, the degree to which common, shared genetic variations may similarly affect the risk of different pAIDs and whether these effects are heterogeneous have not been systematically examined at the genotype level across multiple diseases simultaneously.
RESULTS
Shared genetic risk associations across ten pediatric autoimmune diseases
We performed whole-genome imputation on a combined cohort of more than 6,035 pediatric subjects across ten clinically distinct pAIDs (Supplementary Table 1) and 10,718 population-based control subjects without prior history of autoimmune or immune-mediated disorders. We performed whole-chromosome phasing and used the 1,000 Genomes Project Phase I Integrated cosmopolitan reference panel (1KGP-RP) for imputation as previously described (SHAPEIT and IMPUTE2)22,23. Only individuals of self-reported European ancestry and confirmed by principal-component analysis (Supplementary Figs. 1 and 2) were included (Online Methods). Rare (minor allele frequency (MAF) < 1%) and poorly imputed (INFO score < 0.8) SNPs were removed, leaving a total of 7,347,414 variants.
Whole-genome case-control association testing was done using case samples from each of the ten pAIDs and the shared controls, and additive logistic regression was applied with SNPTESTv2.5 (ref. 24). There was no evidence of genomic inflation. To identify shared pAID-association loci, we performed an inverse χ2 meta-analysis, accounting for sample-size variation and the use of a shared control across the ten pAIDs25. We identified 27 linkage disequilibrium (LD)-independent loci, consisting of associated SNPs with r2 > 0.05 within a 1-Mb window where at least one lead SNP reached a conventionally defined GWS threshold (P < 5 × 10−8; Fig. 1 and Supplementary Fig. 1b). An additional 19 loci reached a genome-wide marginally significant (GWM) threshold at or below PMETA < 1 × 10−6, of which 12 mapped to previously reported autoimmune loci and 7 mapped to putatively novel autoimmune loci (Fig. 1 and Supplementary Table 2a).
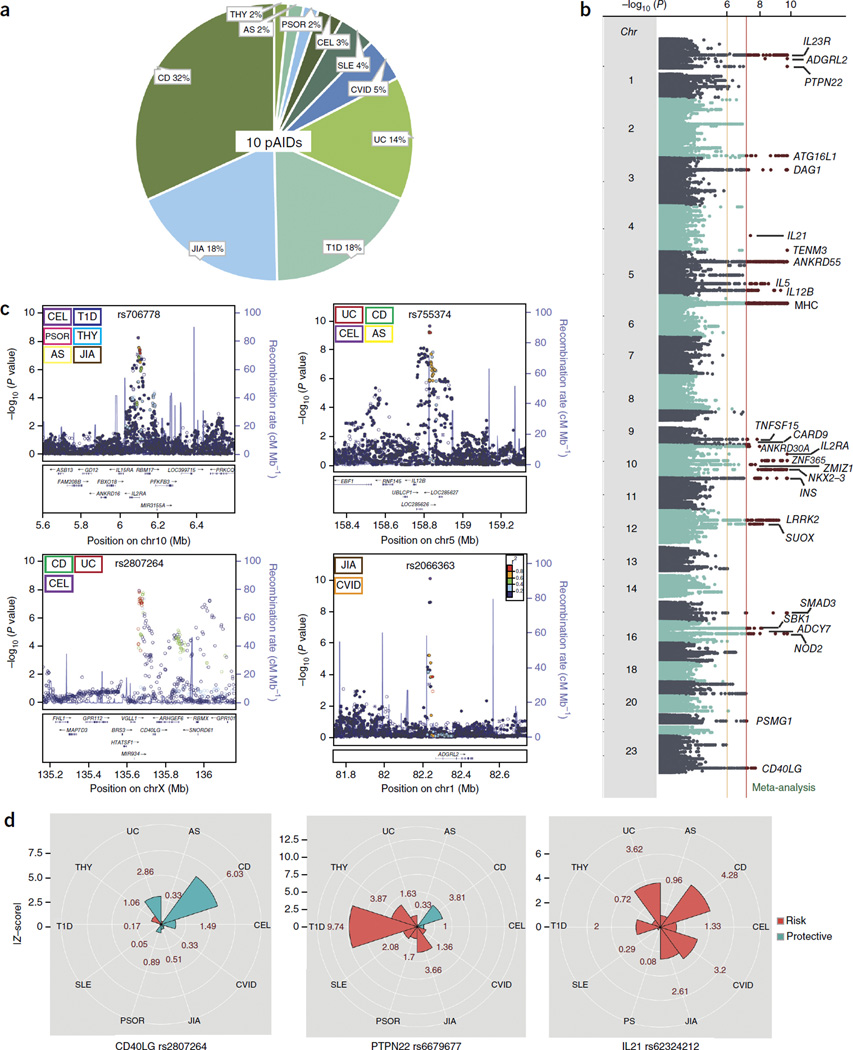
Figure 1.
The ten pAID case cohorts and top pAID-association loci identified. (a) Percentage and relative contribution of cases for the ten pediatric autoimmune diseases studied. (b) Top pAID-association signals identified by inverse χ2 meta-analysis. The top 27 loci (where at least one lead SNP reached genome-wide significance: PMETA < 5 × 10−8) are annotated with the candidate gene symbol. (c) Novel and established pAID-association loci. Top left: rs706778 (chr10p15.1) is a known DNase I peak and an intronic SNP in IL2RA and was associated with THY, AS, PSOR, CEL, T1D and JIA. Top right: rs755374 (chr5q33.3) is an intergenic SNP upstream of IL12B and was associated with AS, CEL, UC and CD. Bottom left: rs2807264 (chrXq26.3), mapping near CD40LG, was associated with CEL, UC and CD, and chr15q22.33 (rs72743477), also mapping to an intronic position in SMAD3, was associated with UC, CD and AS. Bottom right: SNPs are colored according to pairwise LD (r2) with respect to the most strongly associated lead SNP in the locus. Associated pAIDs are indicated at the upper left. pAID associations are color-coded according to the key in each plot. (d) Pleiotropic candidate genes have pleiotropic effect sizes and directions across pAIDs. Although a few pleiotropic SNPs had consistent effect directions across diseases (e.g., IL21), for many loci (e.g., PTPN22 and CLEC16A), the candidate SNP had variable effect directions across diseases. The radii of the wedges correspond to the absolute values of the Z-scores (beta/s.e.) for each pAID, and the color indicates whether the SNP is protective (green) or risk-associated (red) for each disease.
We identified five putatively novel GWS loci: CD40LG (PMETA < 8.38 × 10−11), ADGRL2 (PMETA < 8.38 × 10−11), TENM3 (PMETA < 8.38 × 10−11), ANKRD30A (PMETA < 8.38 × 10−11) and ADCY7 (PMETA < 5.99 × 10−9). For each lead association locus, we identified the corresponding combination of pAIDs contributing to the association signal by enumerating all 1,023 unique disease combinations (for example, one disease, T1D; two diseases, T1D and SLE; or four diseases, UC, CD, CEL and SLE) and performing association testing to identify the disease combination that yielded the maximum logistic regression Z-score (Online Methods)26. With the exception of ANKRD30A, the loci were jointly associated with at least two or more pAIDs; for example, CD40LG was shared by CEL, CD and UC (Fig. 1 and Table 1). Among the 27 GWS lead SNPs, 22 had been reported previously as GWS for at least one of the associated pAIDs (specifically, for the corresponding adult phenotypes) identified by our analysis (Supplementary Tables 1b and 2b)12,27. The most widely shared locus, chr4q27:rs62324212, mapping to an intronic SNP in IL21-AS1 and residing just upstream of IL21, was shared across all ten diseases, and three of these associations were novel (THY, ankylosing spondylitis (AS) and common variable immunodeficiency (CVID)). For more than 50% of previously known GWS loci in adult-onset or generalized autoimmune disease, we identified at least one previously unrecognized pAID association (Supplementary Table 2c,d).
Table 1.
Twenty-seven independent loci reaching genome-wide significance (PMETA < 5 × 10−8) after adjustment for the use of shared controls using an inverse χ2 meta-analysis across the pAIDs
| Chr | Pos (Mb) | SNP | Region | Gene | A1 | MAF | PMETA | Known P* | pAIDs |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 67.7 | rs11580078 | 1p31.3 | IL23R | G | 0.43 | 8.4 × 10−11 | 1.0 × 10−146 | CD# |
| 1 | 82.2 | rs2066363 | 1p31.1 | ADGRL2 | C | 0.34 | 8.4 × 10−11 | Novel | CVID, JIA |
| 1 | 114.3 | rs6679677 | 1p13.2 | PTPN22 | A | 0.09 | 8.4 × 10−11 | 1.1 × 10−88 | THY#, PSOR, T1D#, JIA# |
| 2 | 234.2 | rs36001488 | 2q37.1 | ATG16L1 | C | 0.48 | 8.4 × 10−11 | 1.0 × 10−12 | PSOR, CD# |
| 3 | 49.6 | rs4625 | 3p21.31 | DAG1 | G | 0.31 | 8.4 × 10−11 | 1.0 × 10−47 | PSOR#, CEL, UC#, CD# |
| 4 | 123.6 | rs62324212 | 4q27 | IL21 | A | 0.42 | 2.6 × 10−8 | 1.0 × 10−9 | THY, AS, CEL#, CVID, UC#, T1D#, JIA#, CD# |
| 4 | 183.7 | rs7660520 | 4q35.1 | TENM3 | A | 0.26 | 8.4 × 10−11 | Novel | THY, AS, CEL, SLE, CVID, JIA |
| 5 | 40.5 | rs7725052 | 5p13.1 | PTGER4 | C | 0.43 | 8.4 × 10−11 | 1.4 × 10−10 | CD# |
| 5 | 55.4 | rs7731626 | 5q11.2 | ANKRD55 | A | 0.39 | 1.4 × 10−10 | 2.7 × 10−11 | JIA#, CD# |
| 5 | 131.8 | rs11741255 | 5q31.1 | IL5 | A | 0.42 | 1.6 × 10−9 | 1.4 × 10−52 | PSOR#, CEL, CD# |
| 5 | 158.8 | rs755374 | 5q33.3 | IL12B | T | 0.32 | 2.3 × 10−10 | 1.4 × 10−42 | AS#, CEL, UC#, CD# |
| 9 | 117.6 | rs4246905 | 9q32 | TNFSF15 | T | 0.28 | 9.5 × 10−9 | 1.2 × 10−17 | UC#, CD# |
| 9 | 139.3 | rs11145763 | 9q34.3 | CARD9 | C | 0.40 | 3.3 × 10−8 | 1.0 × 10−6 | AS#, UC#, CD# |
| 10 | 6.1 | rs706778 | 10p15.1 | IL2RA | T | 0.41 | 6.3 × 10−9 | 1.7 × 10−12 | THY, AS, PSOR#, CEL, T1D#, JIA# |
| 10 | 37.6 | rs7100025 | 10p11.21 | ANKRD30A | G | 0.34 | 8.4 × 10−11 | Novel | JIA |
| 10 | 64.4 | rs10822050 | 10q21.2 | ZNF365 | C | 0.39 | 8.4 × 10−11 | 5.0 × 10−17 | SLE, CD# |
| 10 | 81.0 | rs1250563 | 10q22.3 | ZMIZ1 | C | 0.29 | 1.3 × 10−8 | 1.1 × 10−30 | PSOR#, CD# |
| 10 | 101.3 | rs1332099 | 10q24.2 | NKX2-3 | T | 0.46 | 9.1 × 10−11 | 1.0 × 10−54 | UC#, CD# |
| 11 | 2.2 | rs17885785 | 11p15.5 | INS | T | 0.20 | 8.4 × 10−11 | 4.4 × 10−48 | T1D# |
| 12 | 40.8 | rs17466626 | 12q12 | LRRK2 | G | 0.02 | 3.2 × 10−10 | 3.0 × 10−10 | AS, CD# |
| 12 | 56.4 | rs1689510 | 12q13.2 | SUOX | C | 0.31 | 4.0 × 10−9 | 1.1 × 10−10 | PSOR#, T1D# |
| 15 | 67.5 | rs72743477 | 15q22.33 | SMAD3 | G | 0.21 | 8.4 × 10−11 | 2.7 × 10−19 | AS, UC, CD# |
| 16 | 28.3 | rs12598357 | 16p11.2 | SBK1 | G | 0.39 | 4.4 × 10−9 | 1.0 × 10−8 | THY, AS#, PSOR, CEL, UC, CD# |
| 16 | 50.3 | rs77150043 | 16q12.1 | ADCY7 | T | 0.23 | 6.0 × 10−9 | Novel | PSOR, CD |
| 16 | 50.7 | rs117372389 | 16q12.1 | NOD2 | T | 0.02 | 8.4 × 10−11 | 2.9 × 10−69 | CD# |
| 21 | 40.5 | rs2836882 | 21q22.2 | PSMG1 | A | 0.27 | 4.8 × 10−8 | 2.8 × 10−14 | UC#, CD# |
| 23 | 135.7 | rs2807264 | Xq26.3 | CD40LG | C | 0.21 | 1.3 × 10−8 | Novel | CEL, UC, CD |
Chr, chromosome; Pos (Mb), position in hg19; Region, cytogenetic band; A1, alternative allele; MAF, minor allele frequency (controls);
Known P*, lowest P value from published association studies.
Pound symbols (#) denote previously reported disease-associated SNPs.
“Novel” denotes new loci (bolded) that reached genome-wide significance for the first time in the present study (to our knowledge).
A number of the pAIDs were significantly associated with disease-specific signals mapping to or near the locus encoding HLA-DRB1. However, even the two most significant LD-independent variants that mapped to this locus and were associated with T1D and JIA, respectively, were disease specific (Supplementary Fig. 3), which suggests that the variants associated with a given disease are distinct. Although some of these associations were shared by at least two diseases, in no instance was a single signal associated with any of the diseases shared across all other diseases, which further underscores the complexity of signal sharing across the major histocompatibility complex (MHC) (Supplementary Fig. 3b).
Disease-specific and cross-autoimmune replication support for pAID-associated loci
We performed in silico analysis to test whether the reported associations could be replicated in an independent data set. We observed nominally significant replication support for four of the five putatively novel GWS loci, including three instances of disease-specific replication (Supplementary Table 1d). Among the replicated loci, chrXq26.3 (rs2807264), mapping within 70 Kb upstream of CD40LG, was notable, as we observed disease-specific replication in both UC (P < 4.66 × 10−5) and CD (P < 5.81 × 10−4), as well as cross-autoimmune replication in AS (P < 9.54 × 10−3). Although rs2807264 was not identified in our analysis as associated with pediatric AS, it is well documented that adult-onset AS and pediatric AS may be biologically different diseases with independent genetic etiologies28,29. A third disease-specific replication (P < 5.99 × 10−6) was identified in CD for the chr16q12.1 (rs77150043) signal mapping to an intronic position in ADCY7. This third instance and the replication of the CD40LG locus in UC were both significant, even after a very conservative Bonferonni adjustment for 156 tests (P < 3.21 × 10−4). A nominally significant pan-autoimmune replication signal (P < 1.69 × 10−2) was also observed at chr1p31.1 (rs2066363) near LPHN2 in UC, and a replication signal (P < 3.65 × 10−3) was also observed at the chr4q35.1 locus (rs77150043) in PSOR (Supplementary Tables 1d and 2e).
Sharing of pAID-associated SNPs and bidirectional effects of some SNPs on disease-specific risk
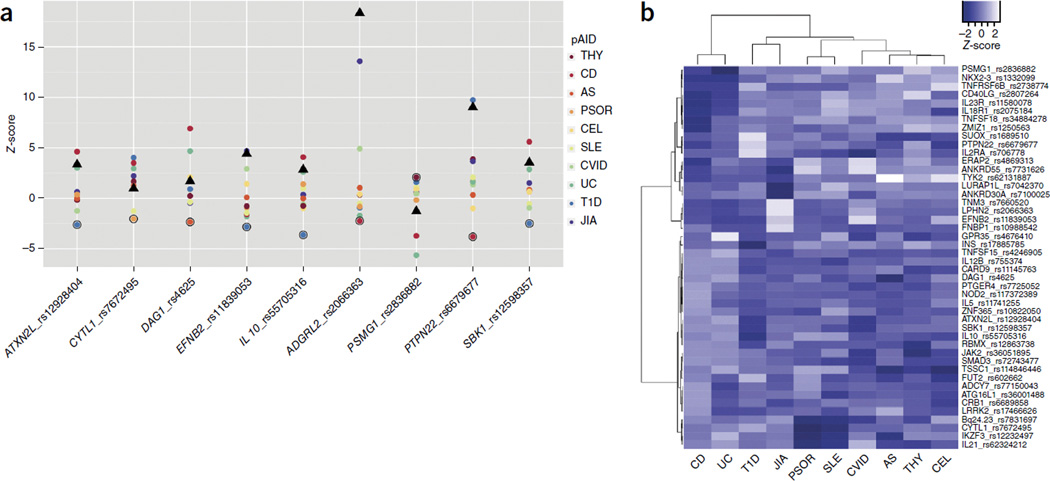
Of the 27 GWS loci, 81% (22) showed evidence of being shared among multiple pAIDs. These mapped to 77 different SNP-pAID combinations, 44 of which had been previously reported at or near genome-wide significance (P < 1 × 10−6), whereas 33 represented potentially novel disease-association signals (Table 1 and Supplementary Table 1). Although PTPN22 c.1858C>T (rs2476601) increases the risk for T1D, the variant is protective against CD17,30–32. We identified eight other instances (P < 0.05) where the risk allele shared by the model pAID combination was associated with protection against another pAID (Fig. 2 and Supplementary Fig. 4).
Figure 2.
Pleiotropic loci with heterogeneous effect directions across pAIDs. (a) Disease-specific Z-scores (beta/s.e.) for each SNP identified as having different effect directions across the ten pAIDs and as detailed in the figure. Circles (color-coded by disease as in key) denote diseases where the indicated SNP had an opposite effect compared with that of the group of pAIDs identified as sharing the lead association on the basis of results of the model search (black triangles). (b) Clustering of pAIDs across the lead loci on the basis of disease-specific effect sizes. Agglomerative hierarchical clustering across ten pAIDs on the basis of normalized directional Z-scores (beta/s.e.) resulting from logistic regression analysis in each disease for the 27 lead loci based on those disease combinations identified by the model search analysis as producing the strongest association-test statistics.
Biological support of associated loci from the public domain
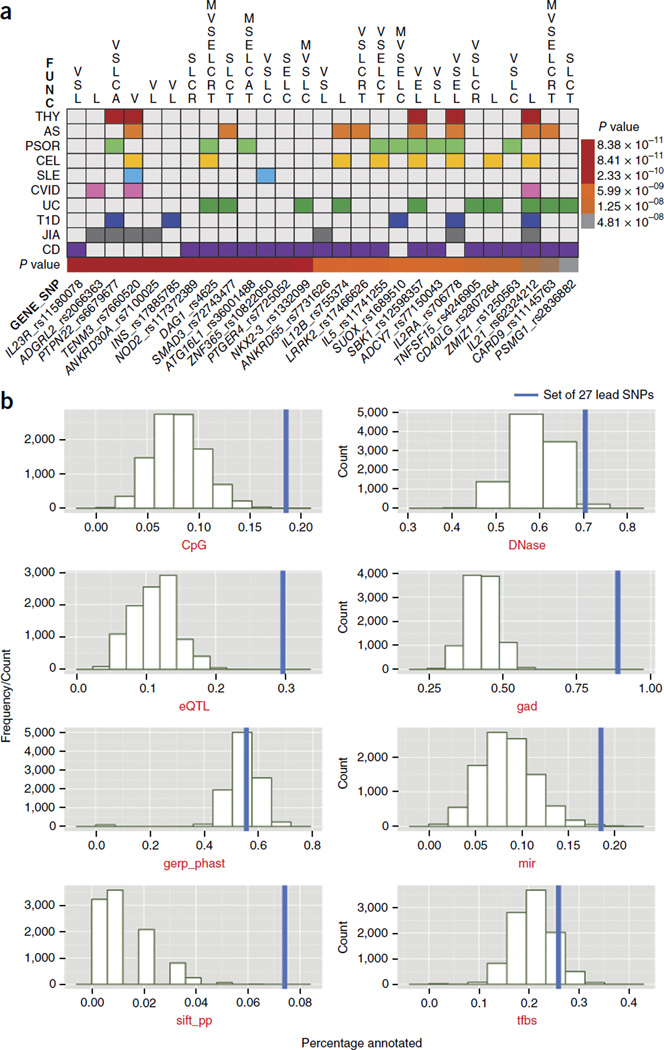
To integrate our results with experimental and predictive biological data, we curated four categories of SNP annotations: (1) functional: variants that are exonic, affect transcription, are miRNA targets or tag copy-number polymorphic regions; (2) regulatory: transcription factor (TF)-binding sites and DNase-hypersensitivity sites or eQTL SNPs; (3) conserved: variants with evolutionarily constrained positions or CpG islands; or (4) prior literature support: a gene or locus previously reported to be associated with autoimmune diseases or immune function. Indeed, 100% of the GWS lead SNPs or their nearby LD proxies (r2 > 0.8 on the basis of 1KGP-RP within 500 Kb up- or downstream) belonged to one or more of these categories (Fig. 3a). Nevertheless, the majority of the 27 GWS SNPs did not confer direct transcriptional consequences (51% were intronic variants and 28% were intergenic or up- or downstream gene variants), which suggests that many of these SNPs either tag the true causal variants or affect disease risk through regulatory and/or epigenetic mechanisms (Fig. 3b).
Figure 3.
Integrated annotation of pAID-association loci using existing predictive and experimental data sets. (a) Biological, functional and literature annotations for the 27 loci reaching genome-wide significance in meta-analysis. Loci (identified by the lead SNPs and candidate genes) are organized by column; the colors in the table denote the associated pAIDs, functional annotations are presented at the top of the table, and the color bar at the bottom represents the meta-analysis Pmeta values (according to key at right). For each locus, the lead SNP and proxy SNPs (r2 > 0.8) were included in the annotation protocol (Online Methods). (b) Distribution and enrichment of experimental and predicted annotations for the top 27 GWS SNPs. The annotation frequencies were used to calculate the relative enrichment of pAID SNPs (blue bars) as compared with that of 10,000 random 100-SNP sets drawn from the genome in each annotation category. CpG, CpG islands; DNase, DNase-hypersensitivity I sites; gad, known genetic association; gerp_phast, conserved positions; mir, miRNAs; sift_pp, functional mutations in SIFT; tfbs, TF–binding sites.
To determine whether the set of pAID-associated SNPs was enriched for specific annotation categories, we compared its annotation percentage with the percentages of 10,000 simulated sets of SNPs with MAF > 0.01 drawn from 1KGP-RP for each category. We found that pAID-associated SNPs were enriched for CpG islands (Pperm < 1.0 × 10−4), TF-binding sites (Pperm < 3.4 × 10−3) and miRNA-binding sites (Pperm < 1.0 × 10−4), among other findings of biological disease relevance (Supplementary Fig. 1d,e).
Candidate pAID genes share expression profiles across immune cell types and tissues
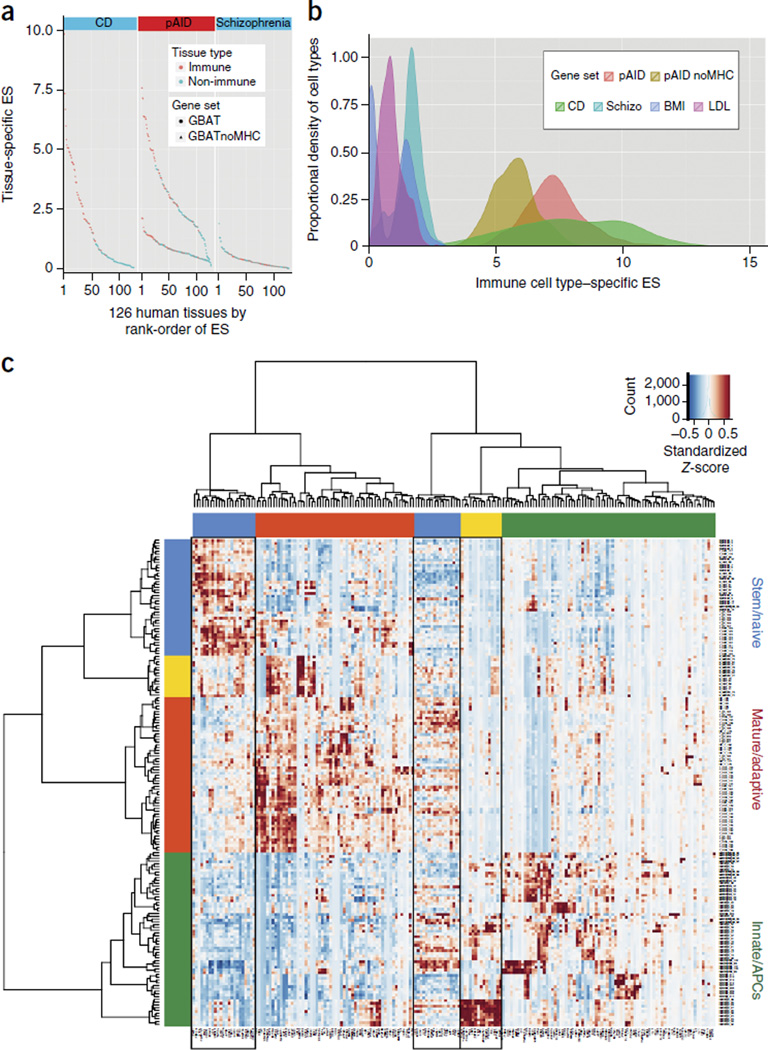
Recent studies show that gene-based association testing (GBAT) may boost the power of genetic discovery33–35. We performed GBAT (with VEGAS33) using genome-wide summary-level PMETA values. We identified 182 significant pAID-associated genes (simulation-based Psim < 2.80 × 10−6) on the basis of a Bonferonni adjustment for ~17,500 protein-coding genes in the genome (Supplementary Table 3a). To illustrate the biological relevance of this set of genes, we examined their transcript levels in a human gene expression microarray data set consisting of 12,000 genes and 126 tissue and/or cell types36. pAID-associated gene expression across immune tissues or cell types (ES-I, 4.05) was notably higher than that across non-immune types (ES-NI, 2.10) on the basis of a one-tailed Wilcoxon rank-sum test (P < 1.66 × 10−10). When all extended MHC genes were excluded, the average expression of pAID-associated genes remained significantly higher (P < 1.27 × 10−7) for immune (1.043) than for non-immune (0.648) tissues and cell types. The immune-specific enrichment of pAID-associated gene transcripts was comparable to that observed in adult cohorts12; comparatively, schizophrenia-associated genes showed no such enrichment (Fig. 4a and Supplementary Table 3b). We observed similar results when we used the Kolmogorov-Smirnov test (Supplementary Fig. 5).
Figure 4.
Tissue-specific gene set enrichment analysis (TGSEA) of pediatric and adult autoimmune data sets identifies autoimmune-associated gene expression patterns across immune cells and tissues. (a) Expression enrichment of autoimmune-associated genes across human tissues. Distribution of TGSEA enrichment score (ES) values across 126 tissues for pAID-associated genes (center) either with (circles, top curve) or without (triangles, bottom curve) the extended MHC. Results for the pAID gene set are compared with those obtained for known genes associated with CD (left) and schizophrenia (right). Tissue and cell types are classified as immune (red) or non-immune (blue) and are ranked left to right on the basis of the magnitude of the ES test statistic. (b) Enrichment of pAID-associated gene expression across diverse murine immune cell types. Distribution of pAID-associated gene ES values across murine immune cell types either including (red) or excluding the genes within the MHC (gold); results are compared with those for genes associated with CD, schizophrenia (Schizo, turquoise), LDL cholesterol (LDL, magenta) or body mass index (BMI, blue) abstracted from the National Human Genome Research Institute (NHGRI) GWAS Catalog. (c) Hierarchical clustering based on the expression of pleiotropic candidate genes associated with three or more autoimmune diseases across the murine immune cells. Boxes outlined in black denote gene clusters enriched for specific disease associations discussed in the text. An enlarged version of c is presented in Supplementary Figure 8.
We examined the expression of pAID genes across a whole-transcriptome data set comprising more than 200 murine immune cell types isolated by flow cytometry (ImmGen37; Online Methods and Supplementary Table 3c). Genes associated with pAIDs demonstrated differential expression across immune cell types (Supplementary Fig. 6) and showed higher expression than genes associated with non-immune traits, similar to results observed from human tissue data (Fig. 4b). As the expression levels of these ‘pleiotropic’ genes varied diversely across immune cell types, we performed agglomerative hierarchical clustering to identify sets of genes sharing similar profiles. Genes that belonged to the same cluster (and thus shared similar expression profiles) were found to be enriched for association with specific individual or multiple autoimmune diseases (Fig. 4c). For example, cluster 1 genes, such as ICAM1, CD40, JAK2, TYK2 and IL12B, with known roles in immune effector cell activation and proliferation, were enriched for association with PSC and UC and were associated with both diseases (P < 6.82 × 10−4, one-tailed Fisher’s exact test), and the expression of these genes was highest in a small subset of CD11b+ dendritic cells6. These findings are consistent with the clinical observation that as many as 80% of patients diagnosed with PSC have been diagnosed with UC, and that the risk of PSC is approximately 600-fold higher in patients with UC38,39. Cluster 2 genes included genes encoding a number of cytokines and cytokine-response factors, such as IL19, IL20, STAT5A and IL2RA, the products of which regulate effector T cell activation, differentiation and proliferation. All of these were more broadly expressed across mature natural killer (NK) cells, NK T cells and T cells, as well as neutrophils. This cluster of genes was enriched for association with MS (P < 9.8 × 10−4), with CEL (marginally) (P < 0.062) and with both diseases (P < 3.41 × 10−4). Genes encoding nucleic acid–binding proteins, such as ILF3, CENPO, MED1 and NCOA3, were enriched in cluster 3. Genes in this cluster were jointly associated with SLE and PSOR (P < 0.03), which is consistent with experimental and clinical data demonstrating that early defects in B cell40,41 and T cell42–44 clonal selection, respectively, may have important roles in the etiology of these diseases.
Quantification of genetic risk factors shared across pAIDs
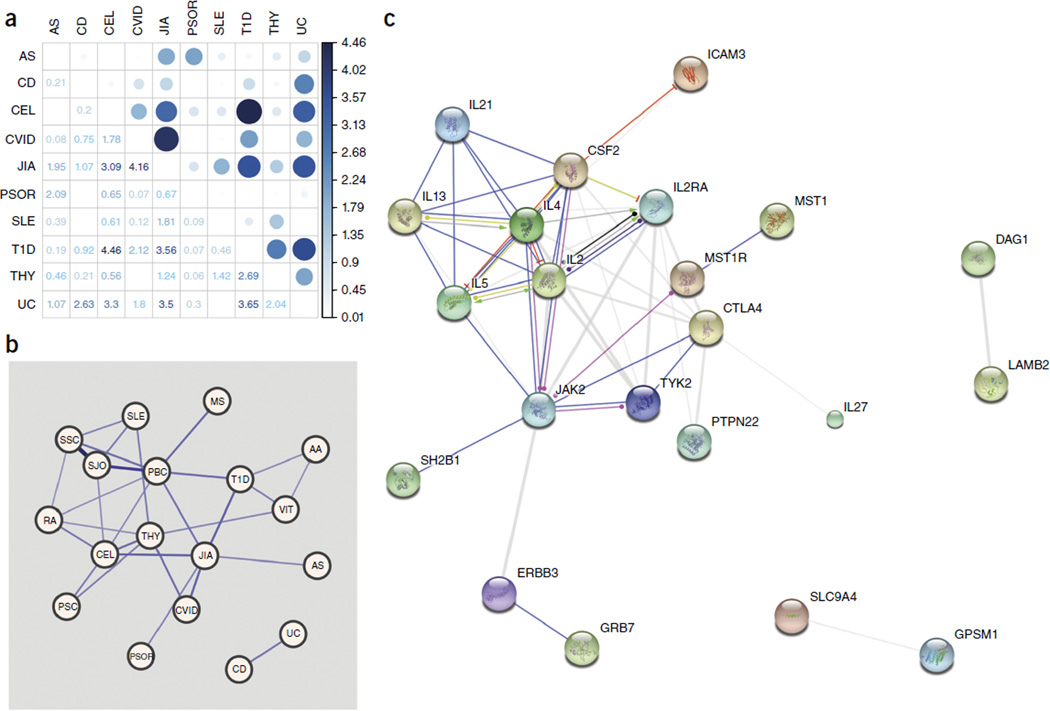
We developed a novel method to specifically examine genome-wide pairwise-association signal sharing (referred to as a GPS test) across the pAIDs (Online Methods). Only data from the genotyped pAID cohort were used for this analysis. After Bonferroni adjustment for 45 pairwise combinations, the GPS test identified evidence of sharing between a number of pAID pairs at marginal levels of significance, as reported previously, including T1D-CEL (Pgps < 3.44 × 10−5), T1D-THY (Pgps < 2.03 × 10−3) UC-CD (Pgps < 2.36 × 10−3) and AS-PS (Pgps < 8.15 × 10−3). We also identified a strong GPS score for JIA-CVID (Pgps < 6.88 × 10−5). The correlations between JIA-CVID (Pgps < 7.30 × 10−5) and UC-CD (Pgps < 7.32 × 10−4) were more significant after the exclusion of markers from within the MHC region (Supplementary Fig. 4b).
Finally, we examined evidence of sharing across the full range of autoimmune diseases using ImmunoBase27. We identified significant associations between UC-CD (P < 2.15 × 10−4) and JIA-CVID (P < 1.44 × 10−6), along with a number of novel pairwise relationships that included autoimmune diseases other than the ten in this study, such as that between Sjogren’s disease (SJO)–systemic sclerosis (SS) (P < 1.30 × 10−28) and PBC-SJO (P < 3.86 × 10−12). We plotted those relationships that were significant after Bonferroni adjustment for 153 pairwise tests using an undirected weighted network (Fig. 5 and Supplementary Table 4). Collectively, these results support genetic sharing between the various autoimmune diseases and allow for further refinement of the shared signals, potentially enabling the application of targeted therapeutic interventions at multiple levels, such as along the CD40L-CD40, JAK-STAT and TH1/TH2-TH17-interleukin signaling pathways.
Figure 5.
Genetic variants shared across the ten pAIDs reveal autoimmune disease networks. (a) Quantification of pAID genetic sharing by GPS test including SNPs within the extended MHC. Correlation plot of results of the pairwise pAID GPS test; the color intensity and the size of each circle are proportional to the strength of the correlation as the negative base ten logarithms of the GPS test P values (color-coded numbers in squares). (b) Quantification of autoimmune disease genetic sharing by locus-specific pairwise sharing. Undirected weighted network graph depicting results from the LPS test. Edge size represents the magnitude of the LPS test statistic; labeled nodes for each of the 17 autoimmune diseases are positioned on the basis of a force-directed layout. Edges represent significant pairs after Bonferroni adjustment (Padj < 0.05). (c) Protein-protein interaction network analysis of the top pAID-associated protein candidates in STRING; action view of protein interactions observed across the top 46 GWM (P < 1 × 10−6) signals, of which 44 could be mapped to corresponding proteins. Views were generated on the basis of results for known and predicted protein interactions produced by the STRING DB Homo sapiens database. The plots shown are results of the ‘action’ view, where the molecular actions (stimulatory, repressive or binding) are illustrated by arrows.
DISCUSSION
A major goal of this study was to identify shared genetic etiologies across pAIDs and illustrate how they jointly and disparately affect pAID susceptibility. Knowledge of shared genetic etiologies may help pinpoint common therapeutic mechanisms, especially since certain pAIDs (for example, THY, CEL and T1D) exhibit high rates of comorbidity and concordance in twins, and others (for example, CD and UC) cluster in families9,19,45,46.
Of the 27 GWS pAID-association loci identified, 81% were shared by at least two pAIDs (Table 1 and Supplementary Table 1). Moreover, 5 of the 27 loci were novel signals not previously reported at GWS levels in association with autoimmune diseases, including chr1p31.1 (rs2066363), mapping near ADGRL2, a gene that encodes a member of the latrophilin subfamily of G protein–coupled receptors that regulates exocytosis. Although this signal was associated with JIA and CVID, a microsatellite study of PBC in a Japanese cohort localized an association signal to a 100-Kb region enclosing ADGRL2 (ref. 47). Nominally significant replication support at this locus was identified in the adult UC cohort from the International IBD Genetics Consortium. Both JIA and CVID are among the six pAIDs (THY, AS, CEL, SLE, CVID and JIA) associated with the chr4q35.1 locus (rs7660520), which resides just downstream of TENM3. The observed association with a broad range of pAIDs may be related to eQTL signals in TENM3 SNPs that correlate with serum eosinophil counts48 and immunoglobulin G (IgG) glycosylation rates; the latter was referenced in a study showing a pleiotropic role for IgG glycosylation–associated SNPs in autoimmune-disease risk susceptibility49. The third novel association was identified near chr10p11.21 (rs7100025), mapping to TF gene ANKRD30A, which encodes an antigen recognized by CD8+ T cell clones50. The fourth signal was associated with the inflammatory diseases PSOR and CD near chr16q12.1 (rs77150043), an intronic SNP in ADCY7. ADCY7 encodes a member of the adenylate cyclase enzyme family; is strongly expressed in peripheral leukocytes, spleen, thymus and lung tissues51; and it is supported by data from studies in mice52. The fifth novel signal, rs34030418, mapping near CD40LG and associated with CEL, UC and CD, is the ligand of the prominent TNF superfamily receptor CD40 (refs. 53,54). The CD40 ligand is a particularly compelling candidate, as the locus encoding the CD40 receptor is an established GWAS locus in RA and MS, has been functionally studied in cell culture and animal models, and was the focus of a recent large-scale RA drug-screening effort55.
A set of GWS candidate SNPs were enriched for miRNA and TF-binding sites. We performed a gene-set enrichment analysis56 using GBAT and identified 39 significant (PBH < 0.05 (BH, Benjamini-Hochberg)) miRNAs, including as top candidates two well-known miRNA families, miR-22 and miR-135a (Supplementary Table 5a). miR-135a has been shown to target IRS2, a regulator of insulin signaling and glucose uptake, in model systems57. Our candidate genes were enriched for targets of dozens of TFs, with the most prominent being SP1 (PBH < 2.30 × 10−12), NFAT (PBH < 8.54 × 10−9) and NFKB (PBH < 1.03 × 10−8) (Supplementary Table 5b).
Using GBAT with DAVID58, GSEA36, IPA59 and Pathway Commons60, among others, we identified strong enrichment for proteins that act in cytokine signaling; antigen processing and presentation; T cell activation; JAK-STAT activation; and TH1-, TH2- and TH17-associated cytokine signaling (Supplementary Tables 6 and 7). Of these pathways, JAK2 signaling was particularly compelling (PBH < 6.93 × 10−5), consistent with the enrichment of known protein- protein interactions (PSTRING < 1 × 10−20) (Supplementary Fig. 7). We also uncovered evidence supporting shared genetic susceptibility for disease pairs that have not yet been well established (for example, JIA-CVID). The association between JIA and CVID is noteworthy, given that CVID actually represents a group of complex immunodeficiencies rather than a classic autoimmune disease. When we examined the overlap between CVID and each of the other pAIDs using both GPS (Padj < 3.10 × 10−3) and locus-specific pairwise sharing (LPS) (Padj < 1.47 × 10−8) network analysis tests, we consistently observed overrepresentation of interaction between CVID and JIA (Fig. 5 and Supplementary Fig. 4b). Our results show that more than 70% (19) of the 27 GWS loci we identified were shared by at least three autoimmune diseases (Table 1), including both previously reported (for example, IL2RA (six diseases) and IL12B (four diseases)) and novel (for example, TENM3 (six diseases) and CD40LG (three diseases)) signals. Moreover, using tissue-specific gene set enrichment analysis, we not only highlighted the expected enrichment of genes associated with CEL and SLE in γδ T cells, CD4+ T cells and NK T cells but also identified interesting joint enrichment of genes associated with PSC and UC in a set of CD11b+ dendritic cells (Fig. 4c).
Many of the shared risk variants in pAIDs affect genes encoding proteins that are established therapeutic targets (for example, CD40L and CD40 (refs. 54,55)), and a number of the genes identified here have diverse biological effects and are currently being explored for clinical uses. Consequently, drug-repurposing approaches may present feasible options in pAIDs, where these gene networks and pathways could be targeted in an expedited manner.
Methods
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Study population
Affected subjects and controls were identified either directly as described in prior studies61–70 or from de-identified samples and associated electronic medical records (EMRs) in the genomics biorepository at The Children’s Hospital of Philadelphia (CHOP). The predominant majority (>80%) of the included cases for IBD, T1D and CVID have been described in previous publications.
Details of each study population are outlined below. EMR searches were conducted with previously described algorithms based on phenotype mapping established using phenome-wide association study (PheWAS) ICD-9 code mapping tables61–63,70 in consultation with qualified physician specialists for each disease cohort. All DNA samples were assessed for quality control (QC) and genotyped on the Illumina HumanHap550 or HumanHap610 platform at the Center for Applied Genomics (CAG) at CHOP. Note that the patient counts below refer to the total recruited sample size from which we excluded non-qualified samples or genotypes that did not pass QC criteria required for inclusion in the genetic analysis (for example, because of relatedness or poor genotyping rate).
The IBD cohort comprised 2,796 individuals between the ages of 2 and 17, of European ancestry, and with biopsy-proven disease, including 1,931 with CD and 865 with UC and excluding all patients with unclassified IBD. Affected individuals were recruited from multiple centers from four geographically discrete countries and were diagnosed before their 19th birthday according to standard IBD diagnostic criteria, as previously reported63,65.
The T1D cohort consisted of 1,120 subjects from nuclear family trios (one affected child and two parents), including 267 independent Canadian T1D patients collected in pediatric diabetes clinics in Montreal, Toronto, Ottawa and Winnipeg and 203 T1D patients recruited at CHOP since September 2006. All patients were Caucasian by self-report and between 3 and 17 years of age, with a median age at onset of 7.9 years. All patients had been treated with insulin since diagnosis. Disease diagnosis was based on these clinical criteria, rather than on any laboratory tests.
The JIA cohort was recruited in the United States, Australia and Norway and comprised a total of 1,123 patients with onset of arthritis at less than 16 years of age. JIA diagnosis and JIA subtype were determined according to the International League of Associations for Rheumatology (ILAR) revised criteria71 and confirmed using the JIA Calculator72 (http://www.jra-research.org/JIAcalc/), an algorithm-based tool adapted from the ILAR criteria. Prior to standard QC procedures and exclusion of non-European ancestry, the JIA cohort comprised 464 subjects of self-reported European ancestry from Texas Scottish Rite Hospital for Children (Dallas, Texas, USA) and the Children’s Mercy Hospitals and Clinics (Kansas City, Missouri, USA); 196 subjects from CHOP; 221 subjects from the Murdoch Children’s Research Institute (Royal Children’s Hospital, Melbourne, Australia); and 504 subjects from Oslo University Hospital (Oslo, Norway).
The CVID study population consisted of 223 patients from Mount Sinai School of Medicine (MSSM; New York, New York, USA), 76 patients from University of Oxford, (London, England), 47 patients from CHOP, and 27 patients from University of South Florida (USF; Tampa, Florida, USA). The diagnosis in each case was validated against the ESID-PAGID diagnostic criteria, as previously described73. Although the diagnosis of CVID is most commonly made in young adults (ages 20–40), all of the CHOP and USF subjects had pediatric-age-of-onset disease, whereas the majority of the subjects from MSSM and Oxford had onset in young adulthood. We note that as the number of individuals with adult-onset CVID is so small (less than 5% of all cases presented) and all ten diseases studied here can present with pediatric age of onset, we elected to refer to the cohort material as pAID.
The balance of the pediatric subjects’ (THY, AS, PSOR, CEL and SLE ; a full list of phenotype abbreviations is provided in Supplementary Table 8) samples were derived from our biorepository at CHOP, which includes more than 50,000 pediatric patients recruited and enrolled by CAG at CHOP (Supplementary Table 9a includes details of genotyped subjects within the CAG pediatric biobank). These individuals were confirmed for diagnosis of THY, SPA, PSOR, CEL and SLE in the age range of 1–17 years at the time of diagnosis and were required to fulfill the clinical criteria for these respective disorders, as confirmed by a specialist. Only patients that upon EMR search were confirmed to have at least two or more in-person visits, at least one of which was with the specified ICD-9 diagnosis code(s), were pursued for clinical confirmation (Supplementary Table 9b presents ICD-9 inclusion and exclusion codes). We used ICD-9 codes previously identified and used for PheWASs or EMR-based GWASs and agreed upon by board-certified physicians62,63.
Age- and gender-matched control subjects were identified from the CHOP-CAG biobank and selected by exclusion of any patient with any ICD-9 codes for disorders of autoimmunity or immunodeficiency61 (http://icd9.chrisendres.com/). Research ethics boards of CHOP and other collaborating centers approved this study, and written informed consent was obtained from all subjects (or their legal guardians). Genomic DNA extraction and sample QC before and after genotyping were performed using standard methods as described previously64. All samples were genotyped at CAG on HumanHap550 and 610 BeadChip arrays (Illumina, CA). To minimize confounding due to population stratification, we included only individuals of European ancestry (as determined by both self-reported ancestry and principal-component analysis (PCA)) for the present study. Details of the PCA are provided below.
Genotyping, imputation, association testing and QC
Disease-specific QC
We merged the genotyping results from each disease-specific cohort with data from the shared controls before extracting the genotyping results from SNPs common to both Infinium HumanHap550 and 610 BeadChip array platforms and performing genotyping QC. SNPs with a low genotyping rate (<95%) or low MAF (<0.01) or those significantly departing from the expected Hardy-Weinberg equilibrium (HWE; P < 1 × 10−6) were excluded. Samples with low overall genotyping call rates (<95%) or determined to be of outliers of European ancestry by PCA (>6.0 s.d. as identified by EIGENSTRAT74) were removed. In addition, one of each pair of related individuals as determined by identity-by-state analysis (PI_HAT > 0.1875) was excluded, with cases preferentially retained where possible.
Merged-cohort QC
To prepare for whole-genome imputation across the entire study cohort, we combined case samples across the 10 pAIDs with the shared control samples. We repeated the genotyping and sample QC with the same criteria as described above, leaving a final set of ~486,000 common SNPs passing individual-cohort and merged-cohort QC. We again performed identity-by-state analysis and removed related samples (in order to remove related subjects that may have been recruited for different disease studies). We also repeated the PCA and removed population outliers. The final cohort, after the application of all QC metrics mentioned above, included a total of 6,035 patients representing ten pAIDs and 10,718 population-matched controls.
Note that because of the merged QC, compared with the sum of all ten disease-specific GWASs, the final case and control counts in the merged cohort were smaller than the “sum of all cases and controls” (Supplementary Table 1a). In addition, to avoid the potential for confounding due to the presence of duplicated samples, we assigned individuals fitting the diagnostic criteria for two or more pAIDs to whichever disease cohort had the smaller (or smallest) sample size. No subject was included twice. A total of 160 subjects in the study cohort fulfilled criteria for two or more diseases but were counted only once in our reported total of 6,035 unique subjects.
Whole-genome phasing and imputation
We used SHAPEIT75 for whole-chromosome prephasing and IMPUTE2 (ref. 76) for imputation to the 1KGP-RP (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html, June 2014 haplotype release). For both, we used parameters suggested by the developers of the software and described elsewhere75–77. Imputation was done for each 5-Mb regional chunk across the genome, and data were subsequently merged for association testing. Prior to imputation, all SNPs were filtered using the criteria described above.
To verify the imputation accuracy, we validated randomly selected SNPs that reached a nominally significant P value after imputation. Because commercially designed genotyping probes were not readily available, we performed Sanger sequencing by designing primers to amplify and sequence the 200-bp region around the imputed SNP markers for two separate 96-well plates. We manually visualized and examined sequences and chromatograms using SeqTrace78. Results from this are presented in Supplementary Table 1e, showing >99% mean imputation accuracy.
In addition, a subset of the IBD and CVID subjects were subsequently genotyped on the Immunochip (Illumina) platform. We compared the genotype concordance of all pAID GWAS imputed SNPs that were directly genotyped on the Immunochip after performing sample and marker QC as described above. Results are shown in Supplementary Table 1f.
Disease-specific association testing
We performed whole-genome association testing using post-imputation genotype probabilities with the software SNPTEST (v2.5)24. We used logistic regression to estimate odds ratios and betas, 95% confidence intervals and P values for trend, using additive coding for genotypes (0, 1 or 2 minor alleles). For autosomal regions, we used a score test, whereas for regions on ChrX we used the ChrX-specific SNPTEST method Newml. QC was performed directly after association testing, excluding any SNPs with an INFO score of <0.80, HWE P < 1 × 10−6, and MAF < 0.01 (overall).
In all analyses, we adjusted for both gender and ancestry by conditioning on gender and the first ten principal components derived from EIGENSTRAT PCA79. The λGC values for all cohorts were within acceptable limits; the highest was observed for the cohort with the largest case sample size, namely, CD (λGC < 1.07), consistent with what was previously reported for this data set65. In fact, we have previously reported on all the non-CHOP cases included in the present analysis in individual studies using CHOP controls and shown that these individual case-control analyses were well controlled for genomic inflation61–70. A QQ plot is provided for each independent cohort in Supplementary Figure 2a.
Meta-analysis to identify shared pAID association loci
To identify association loci shared across pAIDs, we meta-analyzed the summary-level test statistics from each of the study cohorts after extracting those markers that passed post-association testing QC for all ten individual disease-specific analyses. To adjust for confounding due to the use of a shared or pooled control population, we applied a previously published method to perform an inverse weighted χ2 meta-analysis80.
We LD-clumped the results of the meta-analysis (PLINK) and identified 27 LD-independent associations (r2 < 0.05 within 500 kB up- or downstream of the lead or most strongly associated SNP) reaching a conventional genome-wide significance threshold of PMETA < 5 × 10−8. We observed that the calculated meta-analysis λGC was less than 1.09. As recently discussed by de Bakker and colleagues and shown in a number of large-scale GWAS publications, λGC is related to sample size81. As discussed by Yang et al., λGC depends on the relative contribution of variance due to population structure and true associations versus sampling variance: with no population structure or systematic error, inflation would still depend on heritability, genetic architecture and study sample size82. On the basis of de Bakker et al.’s recommendations, we also calculated a sample-size-adjusted λ1000 by interpolating the λGC that would have been expected if this study had included only 1,000 cases and 1,000 controls. We performed this only for the meta-analysis results, as the case and control counts for the meta-analysis were both significantly greater than 1,000 (Supplementary Table 1a).
Model search to identify pAIDs associated with the lead signals
The meta-analysis identified SNPs significantly associated with at least one pAID. To determine which pAIDs each SNP was most strongly associated with, we performed a model or ‘disease-combination’ search. For the lead SNP in each pAID-association locus, we searched for the pAID disease combination that, when the corresponding cases were merged in a mega-analysis, yielded the largest association test statistic.
To identify the disease phenotypes most likely contributing to each identified association signal, we applied the “h.types” method as implemented in the R statistical software package ASSET83 to perform an exhaustive disease-subtype model search. Note that ASSET provides both a method for genotype-level association testing (h.types used in this study) and a summary-level modified fixed-effect meta-analysis approach (“h.traits”) that allows for heterogeneity of SNP effects across different phenotypes. Both methods exhaustively enumerate each combination of phenotypes that are jointly considered, and therefore test a total of
where r is the total number of disease subtypes assigned to cases (for example, ranging from one to ten pAIDs) and n is the total number of disease subtypes (i.e., ten pAIDs). Note that this reduces to 2n − 1 (or 1,023 unique combinations here), as in this case we considered all possibilities of r across n of ten diseases. The ASSET algorithm iteratively tests each pAID case combination using logistic regression to determine whether there is an association between genotype counts and case status. For each SNP tested, the ‘optimal’ subtype model is the combination of pAIDs that, when tested against the shared controls in the logistic regression analysis produced the best test statistic after the DLM method had been used to correct for multiple testing across all subtype combinations.
Identification of lead associated variants showing opposite direction of effect
For each of the top 46 associating loci (PMETA< 1 × 10−6), we identified those loci for which the lead SNP had an effect direction (on the basis of logistic regression betas) opposite that reported for the disease combination identified by the subtype model search and whose corresponding association P value reached at least nominal significance (P < 0.05). We identified nine instances.
Candidate gene prioritization
To annotate the lead SNPs to candidate genes, we prioritized the mapping to candidate genes systematically in the following manner:
If the SNP or locus was previously reported in autoimmune diseases at genome-wide significance, we provided the candidate gene symbol, where available, as identified in the GWAS Catalog84 or ImmunoBase83.
If an SNP was annotated as coding or fell within the coding DNA sequence (i.e., intronic or in the UTRs), we reported that gene as identified by the variant effect predictor (VEP)85.
If the SNP was upstream, downstream, or intergenic, we prioritized the gene by using the best candidate gene identified with the network tool DAPPLE86.
If none of the above was feasible, we manually curated the most ‘likely’ gene on the basis of the observed LD block and evidence of prior association signals with autoimmune diseases or other immune-related phenotypes as presented in the dbSNP or GWAS catalog.
Functional or biological annotations and enrichment analysis using publicly accessible resources
We annotated the lead pAID-associated SNPs using publicly available functional and biological databases and resources. We considered the top imputed lead SNP for each locus and, in addition, any of its near-perfect proxies (defined as r2 > 0.8 within 500 kB up- or downstream) on the basis of the 1KGP-RP.
We included annotation, expression, interaction and network data from the following resources:
Genomic mapping and annotation: SNAP87, SNP-Nexus88, Ensemble89 and UCSC90.
Regulatory annotations: EnCODE (TF-binding sites and DNase-hypersensitivity sites)91, GTex92 (eQTLs), and a published lymphoblastoid cell line eQTL data set93.
Functional annotations: SIFT94, Polyphen95, miRNA target site polymorphisms96,97.
Conservational or evolutionary predictions: GERP98, PHAST++99, CpG islands100.
Literature search: GAD101, NHGRI GWAS catalog102, dbGAP103, or published Immunochip studies104 (http://www.immunobase.org) for literature support.
Gene expression and enrichment analysis: ImmGen102 (murine) and whole-transcriptome analysis across 126 tissues104 (human).
Protein-protein interaction (PPI) database: DAPPLE86, STRING105.
Pathway-based and gene set enrichment analysis: Gene Ontogeny106, Webgestalt107, Wikipathways108, IPA109, DAVID110, GSEA111, and Pathways Commons112.
Gene network analysis and visualization: DAPPLE86 and VEP85 to prioritize candidate causal genes and Grail113 for text-mining of PubMed database for coassociations.
Functional and biological annotations (categories 1–5) for the 27 lead SNPs are illustrated in Figure 3a; annotations are also provided for the 46 GWM loci in Supplementary Figure 5. The following annotation types were used:
Regulatory: EnCODE consensus TF-binding sites (T), DNase I hypersensitivity sites (S), or published eQTL signals (E)
Functional: known mutations in PolyPhen or SIFT (A), experimentally validated (miRBASE 18.0) and predicted (mirSNP) miRNA target sites (R), or SNPs that tag regions containing common copy-number variation regions reported by the database of genomic variants (DGV) (V)
Conserved: conserved nucleotide sequences based on GERP++/phastCon (C) or known CpG islands that correlate with epigenetic methylation patterns (M)
Literature-supported: published association with immune or inflammatory diseases or immune-related endophenotypes from candidate studies or GWASs catalogued in the Genetic Association Database, NHGRI GWAS catalog, dbGAP, or Immunochip studies (L)
In addition to determining whether the 27 GWS pAID-associated SNPs were enriched for a given annotation type, we performed Monte Carlo simulations to resample 10,000 times the SNPs (MAF > 0.01 in Europeans) from all SNPs in 1KGP-RP. As for the 27 lead SNPs, for each set of 100 randomly sampled SNPs, we expanded the list by first identifying all nearby SNPs in strong LD (i.e., LD proxies with r2 > 0.8 within 500 kB up- or downstream) within the 1KGP-RP data set filtered for only SNPs with MAF > 0.01 in the European population. We then annotated each original and any proxy SNPs as above for each major annotation category. We collapsed the information for all proxies identified for a given lead such that for any given category, if the lead SNP or any of its proxies were annotated, the lead SNP was marked as annotated. We then calculated the frequency of annotation for the 100 SNPs in each set. After sampling and annotating 100-SNP sets 10,000 times, we use the permutation- derived distribution of annotation percentages for each annotation type to calculate an enrichment P value such that
where N is the number of permutations, f is the percentage of SNPs in the pAID set that are annotated and F is the distribution of the percentage of SNPs annotated across 10,000 sets of 100 SNPs resampled from the 1KGP-RP using only markers with MAF > 0.01 in Europeans.
Hierarchical clustering based on effect size and direction of association
We performed agglomerative hierarchical clustering across the top 27 independent loci using the directional Z-score obtained from logistic regression analysis in each of the ten disease-specific GWASs, defined as
where beta is the effect size. The standardized and normalized Z-scores were used as inputs to the agglomerative hierarchical clustering. We used Ward’s minimal-variance method to identify relatively consistent gene and locus cluster sizes.
Gene-based association testing
Given our interest in genetic overlap across pAIDs, we sought to identify genes associated with pAIDs in a disease-agnostic manner that was insensitive to locus and phenotypic heterogeneity. We used VEGAS114, a set-based method, to perform GBAT.
As input, we used the nominal PMETA values from the pooled, inverse χ2 meta-analysis for the ten pAIDs across the genome as the input summary statistics for VEGAS, without considering which specific diseases were identified in the model search analysis. We assigned SNPs to gene regions and performed 107 simulations to estimate the gene-based P value as described in VEGAS’s documentation. We used two thresholds: Psim < 2.8 × 10−6 to identify significant candidate genes, on the basis of a Bonferroni adjustment for approximately 17,500 genes tested, and a false discovery rate (FDR) of <2%, which corresponds to a q value of <0.0205, which was used only for pathway and gene set enrichment analysis.
Tissue-specific gene set enrichment analysis
With few exceptions, most genes that are known to have a causative role in autoimmune disease have been shown to regulate molecular or subcellular processes in immune or immune-related tissues. If candidate pAID-associated genes are relevant to autoimmune-disease biology, then expression of these genes would be expected to be, on average, higher across immune or immune-related tissues (as compared with expression in non-immune-related tissues). Thus, we compared the expression of candidate pAID-associated genes identified by GBAT with that of non-candidate genes in a variety of tissues.
We curated the expression of the transcriptome in a broad spectrum of human tissues using a publicly available data set consisting of summary-level, normalized gene expression levels for more than 12,000 unique genes across 126 tissues and/or cell types, including a large number of immune tissues and cells104. We downloaded the processed data set “mean expression data matrix.”
Across the 126 unique tissues, we tested whether the median or cumulative distribution of expression of pAID-associated gene transcripts as identified by GBAT was higher than that of the remaining transcripts in the data set using a one-sided Wilcoxon rank test or a one-sided Kolmogorov-Smirnov (KS) test, respectively. We calculated a tissue-specific gene expression ES value, which is the −log10 (P value) obtained from comparing the relative enrichment in transcript expression of pAID-associated genes versus the transcripts of the remaining genes in the data set. The tests were done on a per-tissue basis to derive a set of KS and a set of Wilcoxon ES values. We performed this per tissue analysis (1) for the total set of pAID-associated genes from GBAT and (2) when genes across the extended MHC (chr6: 25–34 Mb) were excluded.
We performed the secondary immune–versus–non-immune comparative analysis by plotting the ES values obtained from either Wilcoxon or KS tests in descending rank order of the respective test statistics, as shown in Figure 4a and Supplementary Figure 6a for all 126 tissue types. In those figures each point represents a single tissue and is colored according to its classification as either immune (red) or non-immune (blue), as described previously86. To formally test whether the overall ES values were higher among immune tissues than among non-immune tissues, we performed both the Wilcoxon rank sum test and the KS test on the vector of per-tissue ES values, comparing those derived from immune and non-immune tissues. We found that the enrichment observed across immune tissues was specific and not general to any GWAS-identified signals. We repeated this analysis in two sets of candidate genes, one for CD and another for schizophrenia, by identifying all associated genes for the two phenotypes from the NHGRI GWAS Catalog.
Immune cell gene set enrichment analysis
Cells of the immune system are extremely diverse in function and gene expression. To more precisely assess the expression of pAID-associated genes, we examined the mRNA expression of pAID candidate genes across specific immune cell subtypes, as well as during different developmental time points.
ImmGen provides a publicly available, high-quality murine gene expression data set. The ImmGen data set consists of 226 murine immune cell types across different lineages at multiple developmental stages, sorted by FACS and assayed at least in triplicate. Standard QC and quantile-normalization methods were applied to the data set as described by ImmGen102. The total set of transcripts mapped to 14,624 homologs in the human transcriptome on the basis of genes annotated in the hg18/build36 of the human reference genome, which were used to query the gene expression data.
Some of the cell types were derived from genetically altered animals, and the results from analysis of those cell types would have been difficult to interpret, so we removed those cell lines from the analysis. The complete list of cell types used in the analysis and the category to which we assigned each cell type for the categorical analysis are presented in Supplementary Table 3c. A total of 176 unique cell lines remained for subsequent analyses using this data set.
As with the human data set, we calculated the ES values by comparing the expression of the pAID-associated candidate gene transcripts to that of the remaining transcripts assayed in the data set for each immune cell type examined. We plotted the distribution of relative gene expression ES values as a density plot across the range of ES values from all of the examined cell types available. We compared the results obtained using the full set of candidate pAID genes identified by GBAT or obtained when we excluded the genes within the extended MHC. To ensure that this was not simply a result of selection bias (as GWASs may be biased toward regions or genes across the genome that are better sampled or more densely genotyped), we compared the results to those obtained with the curated gene lists from the GWAS catalog (as above) for CD, schizophrenia, body mass index and LDL cholesterol.
To determine whether pAID-associated candidate genes are expressed at higher levels (relative to the rest of the genes in the transcriptome) in some immune cell types than in others, we defined immune cell types according to surface marker expression and tissue isolation details provided by ImmGen. Some categories were further divided into subcategories (for example, B and T cells) on the basis of developmental stage or lineage into a total of 16 non-overlapping cell-type categories. To compare the results across the cell-type categories, we plotted the distribution of ES value ranks for each cell type, binning the results according to the category each cell type belonged to (again, we performed the analysis either with or without the extended MHC region).
Expression profiling of pleiotropic autoimmune disease–associated genes across specific immune cell types
We profiled the expression of genes that had been identified in at least three autoimmune diseases in our subtype model search, previously published Immunochip fine-mapping studies, or a combination thereof (for example, identified as associated with JIA and UC in our analysis but previously identified as a candidate gene from an Immunochip analysis of alopecia areata). We identified 217 candidate pleiotropic genes, of which 191 could be mapped to unique gene transcripts within the ImmGen data sets.
We performed agglomerative hierarchical clustering with the matrix of gene expression levels from the 191 candidate gene transcripts using Ward’s minimal-variance method across all 176 immune cell types. The genes and cell types shown in dendrograms are based on the results of unsupervised hierarchical clustering analysis and represent four major groups of cells and six major groups of genes.
We examined whether genes that were clustered on the basis of similar immune cell–expression profiles were likely to be associated with the same disease(s). Specifically, given a set of genes associated with one or more autoimmune diseases grouped in cluster i (Ci), we asked whether there is an increased likelihood (i.e., more so than expected by chance as compared with genes not found within this cluster) that these genes are also associated with disease j (Dj), such that
| Ci (yes) | Ci (no) | |
|---|---|---|
| Dj (yes) | a | b |
| Dj (no) | c | d |
where the expected probability of the values observed under the null is given by the hypergeometric distribution. As some of the cell counts were small and we were interested only in identifying instances where a >> b, c or d, we used a one-sided Fisher’s exact test. We first tested each of the 18 autoimmune diseases across all identified clusters, declaring nominal and Bonferonni-adjusted significance at P < 0.05 and P < 5.6 × 10−4, respectively. For any clusters where at least two diseases reached nominal or marginal significance, we also tested whether there was an overrepresentation of genes associated with both diseases at P < 0.05.
PPI and network analysis
DAPPLE86: PPIs among the set of either 27 GWS or 46 GWM candidate regions were identified; the input seeds were defined as the 100-kB sequences up- and downstream of the most significantly associated SNP (based on hg19) in each candidate region. Other input parameters included 50-kB regulatory region length, a common interactor binding degree cutoff of 2, and the following specified known genes: IL23R, PTPN22, INS, NOD2, DAG1, SMAD3, ATG16L1, ZNF365, PTGER4, NKX2-3, ANKRD55 and IL12B. We performed 10,000 permutations to accurately calculate enrichment network statistics. Seed scores Pdapple were used to color the protein nodes in the network plot.
STRING105: We used the Homo sapiens PPI database to query one of three lists: (1) the GWS loci, (2) GWS and GWM loci or (3) the list of genes identified by GBAT shown to be enriched for key proteins in the JAK-STAT pathway. We assessed and reported the evidence of PPI enrichment on the basis of these queries as compared to the results expected for the rest of the genes in the human genome. We generated network plots for the directly connected protein candidates (Supplementary Fig. 7a–c represents the “evidence” plot option).
Pathway and gene set enrichment analysis
Webgestalt107: For pathway and gene set analysis, we used the web-based tool Webgestalt to examine evidence of shared TF binding, miRNA target–binding sites, and enrichment in specific Gene Ontology and Pathway Commons categories. The inputs for this analysis included all lead genes (FDR < 2%) from the GBAT (similar to that for the other pathway annotation databases below for consistency).
DAVID110: We used the bioinformatics web tool DAVID (v6.7, available at http://david.abcc.ncifcrf.gov) for functional-annotation analysis of the significant genes. Significant genes with FDR < 2% in VEGAS, the gene-based association analysis, were used as input for DAVID. DAVID performed overrepresentation analysis of functional-annotation terms on the basis of hypergeometric testing and adjusted for multiple testing. To compare the results of this analysis with results obtained via other methods, we used BioCarta, KEGG pathways and GO_BP_FAT as gene set definition files.
IPA109: We used IPA software (http://www.ingenuity.com/) for canonical pathway and network analysis. We inputted all the significant genes in the VEGAS output (FDR < 2%) for IPA analysis. In the IPA core analysis, we selected the Ingenuity Knowledge Base (Genes Only) as the reference set, including both direct and indirect relationships. We used the filter setting of relationships in human and experimentally observed only. Information regarding canonical pathways was obtained from IPA output.
GSEA115,116: We conducted gene set enrichment analysis with the software GSEA (http://www.broadinstitute.org/gsea) using as input the pre-ranked gene list generated on the basis of the −log(P value) from VEGAS using all genes. We selected the following settings for our analysis: number of permutations, 5,000; enrichment statistic, weighted; maximum size of gene set, 500; minimum size of gene set, 15; and with normalization.
Interdisease genetic sharing analysis
To examine the degree of overlap in genetic risk susceptibility between any two autoimmune diseases, we developed and/or implemented the following statistical measures to quantify interdisease genetic sharing:
LPS test, optimized to evaluate whether two pAIDs share more loci in common than would be expected to occur by chance; the score ‘penalizes’ disease pairs if many of the loci are disease specific. The test is helpful if only data on whether diseases share specific candidate genes or association loci in common are known.
GPS test, optimized to assess the correlation between the set of association test statistics observed genome-wide across any two pAIDs. This test is valuable because it is independent of the gene sets chosen and thus does not require the use of any arbitrary method to define a significance ‘threshold’ of input data.
LPS analysis
To quantify the similarity between any two diseases D1 and D2 on the basis of the degree to which D1 and D2 share independent genetic risk associations (i.e., loci, SNPs or candidate genes), we considered the following model.
We began with a list of candidate genes, association loci or LD-independent SNPs nr identified as having reached a predefined GWAS significance threshold (e.g., GWS or GWM) across one or more SNPs from nr for a set of diseases with expected or hypothesized sharing (i.e., all autoimmune diseases in this study and those reported on by the Immunochip studies catalogued by ImmunoBase83).
For any two diseases D1 and D2, a given candidate gene or SNP xi could be uniquely classified in one of four ways: associated with D1 and D2 (n11), associated only with D1 (n12) or D2 (n21), or associated with neither D1 nor D2 (n22). For any given list of TOP associations (i.e., nr), the distribution across the four possible categories can be tabulated as follows:
| Locus xi | D2 (yes) | D2 (no) |
|---|---|---|
| D1 (yes) | n11 | n12 |
| D1 (no) | n21 | n22 |
where n11 + n12 + n21 + n22 = nr and D1 (yes) or (no) means the SNP xi is or is not associated with that marker, respectively.
The probability Px that an SNP xi from the list nr is associated with either D1 or D2 can be expressed as
for any two pAIDs D1 and D2.
Thus, the frequency at which xi should truly be associated with two distinct disease subtypes is given by nr(P1P2), and the observed number of overlapping associations is represented by n11. Therefore, under the null hypothesis H0, for a given pair of diseases D1 and D2, the variance of the difference between the numbers of expected and observed associations of all those tested (nT) shared by both D1 and D2 should follow a normal distribution.
We used the one-sided Z-test to examine whether the degree of overlap was significantly greater than expected, assuming a normal distribution under the null hypothesis that D1 and D2 do not share more associations than they would by chance. We used a Bonferroni adjustment to correct for 45 pairwise disease-combination tests.
GPS analysis
The GPS test determines whether two pAIDs are genetically related. For the ith SNP, let Xi = 1 if the SNP is truly associated with one disease, and let Xi = 0 otherwise. Similarly, define Yi as the indicator of whether the SNP is associated with the other disease in the pair. We can therefore consider the diseases to be genetically related if there are more SNPs with (Xi,Yi) = (1,1) than would be expected to occur by chance. This amounts to testing the independence of Xi and Yi.
However, we do not directly observe Xi and Yi and instead observe P values Ui and Vi, which come from the two GWAS studies for the two diseases. When Xi = 1, the P value Ui will tend to be small, and otherwise Ui will be uniformly distributed; the same is true of Yi and Vi. If Ui and Vi are independent, then Xi and Yi must be as well. We can therefore test for genetic relatedness by testing whether the P values are dependent.
Most existing methods may not take advantage of the availability of the full genome data set for testing genetic sharing using Ui and Vi. To address this limitation, we developed a novel, threshold-free method to detect genetic relatedness. Our test statistic is defined by
where n is the total number of SNPs, Fuv(u,v) is the empirical bivariate distribution function of (Ui, Vi), and Fu(u) and Fv(v) are the empirical univariate distribution functions of Ui and Vi, respectively. Intuitively, the numerator of D is motivated by the fact that if Ui and Vi are truly independent, their bivariate distribution is equal to the product of their univariate distributions. The denominator of D makes the test capable of detecting even very weak correlations. Under the null hypothesis of no genetic sharing, it can be shown that D is approximately distributed like the inverse square root of a standard exponential random variable. This gives us an analytic expression for calculating P values. Note that no significance threshold is required.
The asymptotic null distribution of D is derived under the assumption that the genetic markers examined across the genome are statistically independent. We therefore pruned the SNPs for each pair of diseases before applying our test. We conducted inverse χ2 meta-analyses separately for each pair of diseases and pruned the resulting P values using a threshold of r2 < 0.5 within a 500-kB up- and downstream region. This left about 800,000 SNPs for each disease pair analyzed. The use of more stringent r2 thresholds (for example, r2 < 0.3 or 0.2) gave comparable results.
Undirected weighted cyclic network visualization of results from the locus-specific sharing test
In graphic representations, pairwise relationships between autoimmune diseases (nodes) are represented by edges, whose weights are determined by the magnitude of the LPS test statistic (R statistical software package q-graph). Specifically, the width and density of the edges are the standardized transformations of the test statistic, and the colors denote whether the direction of the test statistic is positive (blue, meaning more sharing than expected) or negative (red, meaning less sharing than expected). Although graphs are constructed from all 45 pairwise interactions, for simplicity and improved visualization, we showed only those edges that represented a pairwise interaction that reached a Bonferroni-adjusted or nominal (Supplementary Fig. 4c) significance threshold (P < 0.05). The nodes are positioned on the basis of a force-directed layout based on the Fruchterman-Reingold algorithm.
In silico replication of novel pAID-association loci using previously published autoimmune disease cohort data sets
Replication set I: The following data sets were used in the first replication set: CASP117, CIDR Celiac Disease118, NIDDK Crohn’s Disease119, Wellcome Trust Case Control Consortium (WT) Crohn’s Disease and Type 1 Diabetes120, WT Ulcerative Colitis121 and WT Ankylosing Spondylitis122. These data sets were obtained via dbGaP or the Wellcome Trust Case Control Consortium. In order to maximize the power, we sought replication for each of the 12 significant SNPs in all of the seven available data sets. Full results are summarized in Supplementary Table 2e.
Each data set was subjected to strict QC filtering as follows: we removed individuals that were inferred to be related on the basis of genetic data, individuals with >10% missing data, individuals with a reported sex that did not match the observed heterozygosity rates on chromosome X, and individuals not of European ancestry. We further removed variants with >10% missingness, variants not in HWE, variants with missingness significantly correlated to phenotype, and variants with MAF < 0.005. Variants to be replicated that were not observed in the original data set were imputed using IMPUTE2 (ref. 123) and the 1KGP-RP haplotype data124. Markers across the X chromosome, which were previously considered by most of these studies, were reanalyzed using the XWAS toolset125,126.
Replication-association analysis was carried out by logistic regression implemented in PLINK127. The first ten principal components calculated using EIGENSOFT128 were added as covariates for all data sets except CASP, where no population stratification was observed.
Replication set II: The second replication set consisted of the following data sets: Rheumatoid Arthritis meta-analysis129, IBDG Ulcerative Colitis meta-analysis130, IBDG Crohn’s Disease meta-analysis131, Systemic Lupus Erythematosus GWAS132, and SLEGEN133. Individuals from these data sets were of European ancestry. Summary statistics from the original studies were publicly available and were used for the replication analysis. Details regarding QC procedures and association analysis can be obtained from the original studies129–133.
LD-based replication for replication sets I and II: We further assessed replication in SNPs that were in LD with the significant SNPs in the discovery set. For each associated SNP, a list of SNPs in LD (r2 > 0.5) within 500 kb of the original SNP was obtained from SNAP87 using the 1KGP-RP.
Supplementary Material
Acknowledgments
We thank the subjects and their families for their participation in genotyping studies and the Biobank Repository at the Center for Applied Genomics at the Children’s Hospital of Philadelphia. We acknowledge M.V. Holmes, H. Matsunami, L. Steel and E. Carrigan for their technical assistance and review of the manuscript. We are also thankful for the contributions of the Italian IBD Group, including S. Cucchiara (Roma), P. Lionetti (Firenze), G. Barabino (Genova), G.L. de Angelis (Parma), G. Guariso (Padova), C. Catassi (Ancona), G. Lombardi (Pescara), A.M. Staiano (Napoli), D. De Venuto (Bari), C. Romano (Messina), R. D’incà (Padova), M. Vecchi (Milano), A. Andriulli and F. Bossa (S. Giovanni Rotondo). The data sets used for the replication analyses were obtained through dbGaP accession numbers phs000344, phs000127, phs000274, phs000171, phs000224, phs000130, phs000019, phs000091, phs000206, phs000168, phs000138, phs000125 and phs000092. We thank the NIH data repository, the investigators who contributed the phenotype data and DNA samples from their original studies, and the primary funding organizations that supported these contributing investigators. This study made use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113. Y.R.L. is supported by the Paul and Daisy Soros Fellowship for New Americans and the NIH F30 Individual NRSA Training Grant. This study was supported by Institutional Development Funds from The Children’s Hospital of Philadelphia and by DP3DK085708, RC1AR058606, U01HG006830, the Crohn’s & Colitis Foundation of America, the Juvenile Diabetes Research Foundation, NIH grant CA127334 (to H.L. and S.D.Z.), the UK National Institutes of Healthcare Research (to H.C.) and a grant from the Lupus Research Institute (to E.T.L.P.). This work was supported in part by the NIH (grant R01-HG006849 to A.K.). F.G. is a Howard Hughes Medical Institute International Student Research fellow.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Y.R.L. and H.H. were leading contributors in the design, analysis and writing of this study. D.J.A. contributed to data collection and literature review. B.F., Ø.F., L.A.D., S.D.T., M.L.B., S.L.G., A.L., E.P., E.R., C.S., A.S., E.M., M.S.S., B.A.L., M.P., R.K.R., D.C.W., H.C., C.C.-R., J.S.O., E.M.B., K.E.S., S.K., A.M.G., J. Snyder, T.H.F., C.P., R.N.B., J.E.M. and J.A.E. contributed samples and phenotypes. F.D.M., K.A.T., H.Q., R.M.C., C.E.K., F.W. and J. Satsangi provided assistance with samples, genotyping and data processing. S.D.Z., J.P.B., J.L. and H.L. contributed to, advised on and supervised statistical analysis. E.T.L.P., J.A.E. and B.J.K. assisted in composing and revising the manuscript. A.K., C.A.W., C.H., C.J.C., C.K., D.C., D.L., D.S.M., F.G., J.J.C., J.T.G., M.B., M.C.D., M.D.R., P.M.A.S., S.F.A.G., S.M.M., V.A., Y.G. and Z.W. read, edited and approved of the manuscript, along with all other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Cooper GS, Bynum ML, Somers EC. Recent insights in the epidemiology of autoimmune diseases: improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun. 2009;33:197–207. doi: 10.1016/j.jaut.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper JD, et al. Seven newly identified loci for autoimmune thyroid disease. Hum. Mol. Genet. 2012;21:5202–5208. doi: 10.1093/hmg/dds357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinks A, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat. Genet. 2013;45:664–669. doi: 10.1038/ng.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu JZ, et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 2012;44:1137–1141. doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu JZ, et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat. Genet. 2013;45:670–675. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eyre S, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhernakova A, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International Multiple Sclerosis Genetics Consortium et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beecham AH, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Human Genome Research Institute. Published Genome-Wide Associations through 08/01/2014. NHGRI GWAS Catalog. 2014 https://www.genome.gov/26525384.
- 13.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res. Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakonarson H, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 16.Hinks A, et al. Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: further support that PTPN22 is an autoimmunity gene. Arthritis Rheum. 2005;52:1694–1699. doi: 10.1002/art.21049. [DOI] [PubMed] [Google Scholar]
- 17.Smyth DJ, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos PS, et al. A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. PLoS Genet. 2011;7:e1002406. doi: 10.1371/journal.pgen.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotsapas C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotsapas C, Hafler DA. Immune-mediated disease genetics: the shared basis of pathogenesis. Trends Immunol. 2013;34:22–26. doi: 10.1016/j.it.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaneau O, Coulonges C, Zagury J-F. Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinformatics. 2008;9:540. doi: 10.1186/1471-2105-9-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchini J. SNPTEST (v2.5) 2007 https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html. [Google Scholar]
- 25.Zaykin DV, Kozbur DO. P-value based analysis for shared controls design in genome-wide association studies. Genet. Epidemiol. 2010;34:725–738. doi: 10.1002/gepi.20536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharjee S, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am. J. Hum. Genet. 2012;90:821–835. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Institute for Systems Biology and Juvenile Diabetes Research Foundation–Wellcome Trust Diabetes and Inflammation Laboratory. ImmunoBase. 2013 http://www.immunobase.org. [Google Scholar]
- 28.Gensler LS, et al. Clinical, radiographic and functional differences between juvenile-onset and adult-onset ankylosing spondylitis: results from the PSOAS cohort. Ann. Rheum. Dis. 2008;67:233–237. doi: 10.1136/ard.2007.072512. [DOI] [PubMed] [Google Scholar]
- 29.Lin Y-C, Liang T-H, Chen W-S, Lin H-Y. Differences between juvenile-onset ankylosing spondylitis and adult-onset ankylosing spondylitis. J. Chin. Med. Assoc. 2009;72:573–580. doi: 10.1016/S1726-4901(09)70432-0. [DOI] [PubMed] [Google Scholar]
- 30.Anaya J-M, Gómez L, Castiblanco J. Is there a common genetic basis for autoimmune diseases? Clin. Dev. Immunol. 2006;13:185–195. doi: 10.1080/17402520600876762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Jager PL, et al. Evaluating the role of the 620W allele of protein tyrosine phosphatase PTPN22 in Crohn’s disease and multiple sclerosis. Eur. J. Hum. Genet. 2006;14:317–321. doi: 10.1038/sj.ejhg.5201548. [DOI] [PubMed] [Google Scholar]
- 32.Zhernakova A, et al. Differential association of the PTPN22 coding variant with autoimmune diseases in a Dutch population. Genes Immun. 2005;6:459–461. doi: 10.1038/sj.gene.6364220. [DOI] [PubMed] [Google Scholar]
- 33.Liu JZ, et al. A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M-X, Gui H-S, Kwan JSH, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am. J. Hum. Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H, Chanda P, Alonso A, Bader JS, Arking DE. Gene-based tests of association. PLoS Genet. 2011;7:e1002177. doi: 10.1371/journal.pgen.1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benita Y, et al. Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood. 2010;115:5376–5384. doi: 10.1182/blood-2010-01-263855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heng TSP, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 38.Olsson R, et al. Prevalence of primary sclerosing cholangitis in patients with ulcerative colitis. Gastroenterology. 1991;100:1319–1323. [PubMed] [Google Scholar]
- 39.Feld JJ, Heathcote EJ. Epidemiology of autoimmune liver disease. J. Gastroenterol. Hepatol. 2003;18:1118–1128. doi: 10.1046/j.1440-1746.2003.03165.x. [DOI] [PubMed] [Google Scholar]
- 40.Yurasov S, et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J. Exp. Med. 2005;201:703–711. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cappione A, et al. Germinal center exclusion of autoreactive B cells is defective in human systemic lupus erythematosus. J. Clin. Invest. 2005;115:3205–3216. doi: 10.1172/JCI24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evenou J-P, et al. The potent protein kinase C-selective inhibitor AEB071 (sotrastaurin) represents a new class of immunosuppressive agents affecting early T-cell activation. J. Pharmacol. Exp. Ther. 2009;330:792–801. doi: 10.1124/jpet.109.153205. [DOI] [PubMed] [Google Scholar]
- 43.Jegasothy BV. Tacrolimus (FK 506)—a new therapeutic agent for severe recalcitrant psoriasis. Arch. Dermatol. 1992;128:781–785. [PMC free article] [PubMed] [Google Scholar]
- 44.Nograles KE, Krueger JG. Anti-cytokine therapies for psoriasis. Exp. Cell Res. 2011;317:1293–1300. doi: 10.1016/j.yexcr.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 45.Ergür AT, et al. Celiac disease and autoimmune thyroid disease in children with type 1 diabetes mellitus: clinical and HLA-genotyping results. J. Clin. Res. Pediatr. Endocrinol. 2010;2:151–154. doi: 10.4274/jcrpe.v2i4.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eyre S, et al. Overlapping genetic susceptibility variants between three autoimmune disorders: rheumatoid arthritis, type 1 diabetes and coeliac disease. Arthritis Res. Ther. 2010;12:R175. doi: 10.1186/ar3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joshita S, et al. A2BP1as a novel susceptible gene for primary biliary cirrhosis in Japanese patients. Hum. Immunol. 2010;71:520–524. doi: 10.1016/j.humimm.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Pruitt K, Brown G, Tatusova T, Maglott D. The Reference Sequence (RefSeq) database. 2012 doi: 10.1093/nar/gki025. http://www.ncbi.nlm.nih.gov/books/NBK21091/ [DOI] [PMC free article] [PubMed]
- 49.Lauc G, et al. Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet. 2013;9:e1003225. doi: 10.1371/journal.pgen.1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jäger D, et al. Humoral and cellular immune responses against the breast cancer antigen NY-BR-1: definition of two HLA-A2 restricted peptide epitopes. Cancer Immun. 2005;5:11. [PubMed] [Google Scholar]
- 51.Ludwig M-G, Seuwen K. Characterization of the human adenylyl cyclase gene family: cDNA, gene structure, and tissue distribution of the nine isoforms. J. Recept. Signal Transduct. Res. 2002;22:79–110. doi: 10.1081/rrs-120014589. [DOI] [PubMed] [Google Scholar]
- 52.Jiang LI, Sternweis PC, Wang JE. Zymosan activates protein kinase A via adenylyl cyclase VII to modulate innate immune responses during inflammation. Mol. Immunol. 2013;54:14–22. doi: 10.1016/j.molimm.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson DM, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390:175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 54.Miyashita T, et al. Bidirectional regulation of human B cell responses by CD40–CD40 ligand interactions. J. Immunol. 1997;158:4620–4633. [PubMed] [Google Scholar]
- 55.Li G, et al. Human genetics in rheumatoid arthritis guides a high-throughput drug screen of the CD40 signaling pathway. PLoS Genet. 2013;9:e1003487. doi: 10.1371/journal.pgen.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agarwal P, Srivastava R, Srivastava AK, Ali S, Datta M. miR-135a targets IRS2 and regulates insulin signaling and glucose uptake in the diabetic gastrocnemius skeletal muscle. Biochim. Biophys. Acta. 2013;1832:1294–1303. doi: 10.1016/j.bbadis.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 58.Huang DW, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ingenuity Systems. Ingenuity Pathway Analysis. 2015 http://www.ingenuity.com/products/ipa. [Google Scholar]
- 60.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 61.Denny JC, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ritchie MD, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am. J. Hum. Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao KP, et al. Associations of autoantibodies, autoimmune risk alleles, and clinical diagnoses from the electronic medical records in rheumatoid arthritis cases and non-rheumatoid arthritis controls. Arthritis Rheum. 2013;65:571–581. doi: 10.1002/art.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hakonarson H, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 65.Imielinski M, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kugathasan S, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orange JS, et al. Genome-wide association identifies diverse causes of common variable immunodeficiency. J. Allergy Clin. Immunol. 2011;127:1360.e6–1367.e6. doi: 10.1016/j.jaci.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Behrens EM, et al. Association of the TRAF1-C5 locus on chromosome 9 with juvenile idiopathic arthritis. Arthritis Rheum. 2008;58:2206–2207. doi: 10.1002/art.23603. [DOI] [PubMed] [Google Scholar]
- 69.Grant SF, et al. Association of the BANK 1 R61H variant with systemic lupus erythematosus in Americans of European and African ancestry. Appl. Clin. Genet. 2009;2:1–5. doi: 10.2147/tacg.s4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao KP, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res. (Hoboken) 2010;62:1120–1127. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petty RE, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J. Rheumatol. 2004;31:390–392. [PubMed] [Google Scholar]
- 72.Behrens EM, et al. Evaluation of the presentation of systemic onset juvenile rheumatoid arthritis: data from the Pennsylvania Systemic Onset Juvenile Arthritis Registry (PASOJAR) J. Rheumatol. 2008;35:343–348. [PubMed] [Google Scholar]
- 73.Conley ME, Notarangelo LD, Etzioni A. Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies) Clin. Immunol. 1999;93:190–197. doi: 10.1006/clim.1999.4799. [DOI] [PubMed] [Google Scholar]
- 74.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 75.Delaneau O, Coulonges C, Zagury J-F. Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinformatics. 2008;9:540. doi: 10.1186/1471-2105-9-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stucky BJ. SeqTrace: a graphical tool for rapidly processing DNA sequencing chromatograms. J. Biomol. Tech. 2012;23:90–93. doi: 10.7171/jbt.12-2303-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 80.Zaykin DV, Kozbur DO. P-value based analysis for shared controls design in genome-wide association studies. Genet. Epidemiol. 2010;34:725–738. doi: 10.1002/gepi.20536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Institute for Systems Biology and Juvenile Diabetes Research Foundation–Wellcome Trust Diabetes and Inflammation Laboratory. ImmunoBase. 2013 http://www.immunobase.org. [Google Scholar]
- 84.NHGRI. Published GWAS through 08/01/2014. NHGRI GWA Catalog. 2014 http://www.genome.gov/multimedia/illustrations/GWAS_2011_3.pdf.
- 85.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chelala C, Khan A, Lemoine NR. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25:655–661. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.National Institutes of Health Genotype-Tissue Expression (GTEx) 2015 http://commonfund.nih.gov/GTEx/index.
- 93.Liang L, et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 2013;23:716–726. doi: 10.1101/gr.142521.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 95.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;Chapter 7(Unit 7.20) doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu C, et al. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics. 2012;13:661. doi: 10.1186/1471-2164-13-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP. PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nguyen D-Q, et al. Reduced purifying selection prevails over positive selection in human copy number variant evolution. Genome Res. 2008;18:1711–1723. doi: 10.1101/gr.077289.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 101.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat. Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 102.Heng TSP, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 103.Mailman MD, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Benita Y, et al. Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood. 2010;115:5376–5384. doi: 10.1182/blood-2010-01-263855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kelder T, et al. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40:D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ingenuity Systems Ingenuity Pathway Analysis. 2015 http://www.ingenuity.com/products/ipa. [Google Scholar]
- 110.Huang DW, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu JZ, et al. A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 117.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ahn R, et al. Association analysis of the extended MHC region in celiac disease implicates multiple independent susceptibility loci. PLoS ONE. 2012;7:e36926. doi: 10.1371/journal.pone.0036926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wellcome Trust Case Control Consortium et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barrett JC, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Evans DM, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 124.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gao F, et al. XWAS: a software toolset for genetic data analysis and association studies of the X chromosome. bioRxiv. doi: 10.1093/jhered/esv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chang D, et al. Accounting for eXentricities: analysis of the X chromosome in GWAS reveals X-linked genes implicated in autoimmune diseases. PLoS One. 2014;9:e113684. doi: 10.1371/journal.pone.0113684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 2008;358:900. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 133.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.