Abstract
Heart failure is a morbid disorder characterized by progressive cardiomyocyte (CM) dysfunction and death. Interest in cell-based therapies is growing, but sustainability of injected CMs remains a challenge. To mitigate this, we developed an injectable biomimetic Reverse Thermal Gel (RTG) specifically engineered to support long-term CM survival. This RTG biopolymer provided a solution-based delivery vehicle of CMs, which transitioned to a gel-based matrix shortly after reaching body temperature. In this study we tested the suitability of this biopolymer to sustain CM viability. The RTG was biomolecule-functionalized with poly-l-lysine or laminin. Neonatal rat ventricular myocytes (NRVM) and adult rat ventricular myocytes (ARVM) were cultured in plain-RTG and biomolecule-functionalized-RTG both under 3-dimensional (3D) conditions. Traditional 2D biomolecule-coated dishes were used as controls. We found that the RTG-lysine stimulated NRVM to spread and form heart-like functional syncytia. Regarding cell contraction, in both RTG and RTG-lysine, beating cells were recorded after 21 days. Additionally, more than 50% (p value < 0.05; n = 5) viable ARVMs, characterized by a well-defined cardiac phenotype represented by sarcomeric cross-striations, were found in the RTG-laminin after 8 days. These results exhibit the tremendous potential of a minimally invasive CM transplantation through our designed RTG-cell therapy platform.
Introduction
Heart failure continues to be a leading cause of death, characterized by high mortality and morbidity, affecting over 5 million persons in the United States. Cardiomyopathies are a clinically and genetically heterogeneous group of heart muscle disorders characterized by life-threatening arrhythmias, sudden death, and progression toward heart failure.1,2 Most cardiomyopathies at the late stage of heart failure exhibit CM loss.3 In addition, the adult mammalian heart does not sufficiently regenerate CMs after injury.4 Therefore, at the end-stage of heart failure, heart transplantation remains the best option.5 However, replacing the failed heart with a healthy one raises several limitations such as shortage of organs available for transplantation, immune rejection, surgical complications, and so on.6,7 Due to these restrictions, innovative alternatives are urgently needed to repair the wounded heart and to permanently restore its function.8
Direct injection of single cells or small clusters of cells into the cardiac muscle has been used as a therapeutic approach to heal damaged heart tissue.9 However, cell therapy faces several limitations, such as low rate of cell survival and poor retention of transplanted cells in the injured tissue; in addition, the cell source, the route of administration, and cell post-transplantation arrhythmias are other major limitations that affect the treatment success.10−12
To ensure effective cell transplantation, tissue engineering approaches have been used to develop engineered scaffolds, which were able to provide unique microenvironments to heart-specific cell types.13−16 These scaffolds support cell differentiation and expansion, protect cells injected into the damage heart and guide the regeneration of the injured tissue. However, most of the scaffold approaches require invasive surgeries for implantation.17−19 For many patients with severe heart failure, surgical implantation is not an option due to various comorbidities that prohibit them from undergoing surgical procedures.20 In such patients, cell-scaffold implantation is a challenge and an unmet clinical need.
To overcome this limitation, in situ gelling injectable systems are advantageous in that their initial liquid state permits delivery through a minimally invasive injection immediately at the desired location.21,22 Once injected, the system will transform into a physical gel and relocate cells at the injury site, protecting them from the external harsh environment.10 Thermally induced gelling systems (thermogels), a class of in situ gelling injectable systems, are defined by their ability to transition from a solution to a gel solely through temperature stimuli.23 By utilizing a physical gelation platform, these systems can circumvent harmful free radicals, high polymerization temperatures, and potentially irritating solvents that potentially could increase the health risk.
For this project, we have developed injectable biomimetic reverse thermogel (RTG), poly(serinol hexamethylene urea)-co-poly(N-isopropylacylamide) or PSHU-PNIPAAm,24,25 specially designed to support long-term CMs survival. We first began by synthesizing a functionalizable biomimetic poly(urea) backbone (PSHU) capable of attaching a large quantity of functional groups (18 potential linkages per molecule). Poly(N-isopropylacrylamide) (PNIPAAm), a thermosensitive water-soluble homopolymer, was then conjugated to functionalize PSHU and form the PSHU-PNIPAAm copolymer or RTG. PSHU-PNIPAAm undergoes a reversible phase transition from a low viscosity solution state at room temperature to a physical gel upon reaching body temperature. This unique thermal gelling property allows for CM incorporation simply through mixing at room temperature and easy deployment through injection at the target area with minimal surgical intervention. Transition of the RTG polymer to a physical gel at body temperature localizes CMs to the target (injection) site. In addition, the remaining functional groups of PSHU can still be functionalized (e.g., extracellular matrix (ECM)-based peptides and proteins can be chemically incorporated) to improve cell survival, support cell adhesion, and promote proliferation. For this project, we designed different RTG systems by functionalizing the PSHU backbone with the chemical conjugation of laminin or poly-l-lysine, which are proteins commonly used as a culture substrate for CMs. We further assessed the long-term viability of neonatal rat ventricular myocytes (NRVM) and adult rat ventricular myocytes (ARVM) cultured in plain PSHU-PNIPAAm and biomolecule-functionalized PSHU-PNIPAAm (PSHU-PNIPAAm-laminin and PSHU-PNIPAAm-l-lysine).
Materials and Methods
Materials
N-Isopropylacrylamide (NIPAAm), anhydrous N,N-dimethylformamide (DMF), 4,4′-azobis(4-cyanovaleric acid) (CVA), urea, N-BOC-serinol, hexamethylene diisocyanate (HDI), diethyl ether, trifluoroacetic acid (TFA), dichloromethane (DCM), N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide hydrochloride (EDC-HCl), and l-lysine monohydrochloride were purchased from Sigma-Aldrich, (St. Louis, MO, U.S.A.). N-Hydroxysuccinimide (NHS) was purchased from Alfa Aesar (Ward Hill, MA, U.S.A.).
Equipment
Morphological characterization was carried out by scanning electron microscopy (SEM) using a Field Emission SEM JEOL JSM 7401F. Fourier Transform (FT) IR spectra were collected on a Nicolet 6700 (Thermo Fisher Scientific, Waltham, MA) using polyethylene-windowed cards. Rheological measurements were performed on a stress-controlled rheometer Rheostress Haake RS 150 using a cone-and-plate geometry (angle 1°, diameter 60 mm) and a solvent trap to improve thermal control and limit evaporation. Polymers were dissolved in PBS (pH 7.4) at 1 wt %, and temperature sweep tests composed of heating ramps (at 1 °C/min) were conducted at constant frequency (1 Hz) and stress (0.05 Pa) between 25 and 45 °C. Thermogravimetric analyses (TGA) were performed under a nitrogen flow (60 mL/min) using a TA Instruments Q500 on sample sizes from 0.7 to 1 mg, and the mass was recorded as a function of temperature. The samples were heated at 100 °C for 20 min and then to 800 °C at 10 °C/min. Data was interpreted using Thermal Advantage v1.1A software.
Polymer Synthesis
PSHU-PNIPAAm was synthesized as described previously.24 Briefly, PSHU was obtained by reacting N-BOC-serinol (1.147 g, 6 mmol), urea (0.36 g, 6 mmol), and HDI (2.018 g, 12 mmol) for 7 days at 90 °C under a nitrogen atmosphere. Anhydrous DMF (6 mL) was used as solvent. The mixture was precipitated into excess anhydrous diethyl ether three times. Unreacted urea was removed by washing with water, and the polymer was lyophilized at −45 °C for 48 h. PSHU was deprotected using 30 mL of a TFA/DCM (1:1, v/v) mixture. The deprotection reaction was performed for 45 min at room temperature. The resulting polymer was purified by three precipitations into diethyl ether. PNIPAAm-COOH was synthesized by reacting NIPAAm (5 g, 800 mmol) and CVA (0.06 g, 4 mmol) for 3 h at 68 °C under a nitrogen atmosphere. Anhydrous DMF (10 mL) was used as solvent. Then, the mixture was precipitated into hot water (60 °C). The polymer was then dissolved in milli-Q water and dialyzed (MWCO: 12000–14000 Da) for 3 days. The conjugation of PNIPAAm-COOH onto PSHU-NH2 was performed as follows: PNIPAAm-COOH (0.75 g, 1.21 mmol) was dissolved in 5 mL of anhydrous DMF with five molar excess of EDC-HCl and NHS at room temperature under a nitrogen atmosphere for 24 h. A 1 mL aliquot of PSHU-NH2 solution (0.125 g/mL) prepared in anhydrous DMF was added, and the reaction was performed for 48 h at room temperature under a nitrogen atmosphere. The mixture was precipitated into excess diethyl three times. The polymer was then dissolved in milli-Q water and dialyzed (MWCO: 12000–14000 Da) for 5 days at room temperature and then filtered through a 2 μm filter. The filtered solution was lyophilized at −45 °C for 48 h.
Synthesis of PSHU-PNIPAAm-Lysine
Poly-l-lysine was synthesized by dissolving l-lysine (0.034 g, 5 mmol) in 5 mL of PBS 1× with five molar excess of EDC-HCl and NHS in a 25 mL round-bottom flask. The mixture was stirred for 15 min at 4 °C. A 10 mL aliquot of PSHU-PNIPAAm solution (0.1 g/mL) prepared in PBS 1× was added dropwise, and the reaction was performed for 48 h at room temperature. The polymer was dialyzed (MWCO: 12000–14000 Da) for 5 days at room temperature and then filtered through a 2 μm filter. The filtered solution was lyophilized at −45 °C for 48 h.
Synthesis of PSHU-PNIPAAm-Laminin
Laminin (1 mg, 2.5 × 10–3 mmol) was dissolved in 5 mL of PBS 1× with five molar excess of EDC-HCl and NHS in a 25 mL round-bottom flask. The mixture was stirred for 15 min at 4 °C. A total of 10 mL of PSHU-PNIPAAm solution (0.1 g/mL) prepared in PBS 1× was added dropwise, and the reaction was performed for 48 h at room temperature. The polymer was dialyzed (MWCO: 12000–14000 Da) for 5 days at room temperature and then filtered through a 2 μm filter. The filtered solution was lyophilized at −45 °C for 48 h.
Adult Rat Ventricular Myocytes (ARVMs) Culture
ARVMs were isolated from adult Sprague–Dawley rats, as described previously.26 All animal studies were performed according to the guidelines of the University of Colorado Denver Animal Care and Use Committee. Briefly, ketamine and xylazine were administered according to the weight of the animal. Hearts were rapidly removed and retrograde perfused with perfusion buffer containing 120.4 mM NaCl, 14.7 mM KCl, 0.6 mM KH2PO4, 0.6 mM Na2HPO4, 1.2 mM MgSO4, 4.6 mM NaHCO3, 10 mM sodium-HEPES, 30 mM taurine, 10 mM 2,3-butanedione monoxime, and 5.5 mM glucose (pH 7.4) for 8 min at 37 °C. The perfusion solution with collagenase II (2 mg/mL, Worthington) was then infused for 22 min for enzymatic digestion. The heart was then cut into small pieces, and the cells were dispersed by shaking for an additional 10 min in fresh perfusion solution with collagenase II (2 mg/mL, Worthington). The slurry was filtered through a sterile 150 nm mesh, allowed to settle (20 min), resuspended in DMEM (GIBCO) serum-free, and layered over 60 g/mL bovine serum albumin (Sigma) to separate ventricular myocytes from nonmyocytes. ARVMs were collected and used for 2D (laminin-coated dishes) and 3D polymeric cell cultures and subjected to the different treatments and subsequent analyses. All experimental conditions were tested in triplicate on at least three independent cell cultures, and the ratio of ARVM viability was normalized to control groups (all the control groups were normalized to one). Data was presented as mean ± SD (n = 5) with a p value < 0.05 (Student’s test).
Neonatal Rat Ventricular Myocytes Culture (NRVMs)
NRVMs were prepared from six 1–3 day old pups. Briefly, ventricles were separated from the atria using scissors and then dissociated in calcium and bicarbonate-free Hanks with Hepes (CBFHH) buffer containing 500 mg/mL of collagenase type 2 (Worthington, Biochemical Corporation) and 1 mg/mL of pancreatine. Cardiomyocytes were enriched (>90% purity) over nonmyocytes by two sequential preplating steps on 100 mm dishes in DMEM, 4.5 g of glucose supplemented with 10% horse serum, 5% bovine calf serum, and 2 mg/mL vitamin B12 and cultured as previously described by Martinelli et al.4 Unattached cells (predominantly myocytes) were collected and cultured in 2D gelatin-coated dishes and into 3D polymeric matrices and then subjected to the different treatments and subsequent analyses. All experimental conditions were tested in triplicate on at least three independent cell cultures, and the ratio of NRVM was normalized to control groups (all the control groups were normalized to one). Data was presented as mean ± SD (n = 5) with a p value < 0.05 (Student’s test).
3D In Vitro Cell Culture
In vitro 3D culture experiments were performed with the freshly isolated ARVM and NRVM by mixing 3 × 103 or 3 × 104 cells, respectively, with 50 μL of polymeric solution and allowed to form a gel at 37 °C. Then 100 μL of warm cell culture medium was added on top.
Immunocytochemical Staining
Immunocytochemistry was performed after 8, 14, and 21 days of culture using the cardiac-specific marker α-sarcomeric actinin 1:100 (SIGMA) to assess the contractile apparatus of CMs, CD3 1:100 (abcam) as a marker of endothelial and vimentin 1:100 (abcam) being a cytoskeleton marker commonly used for fibroblast staining. Goat antimouse antibody conjugated to Alexa Fluor 488 (Invitrogen), goat antichicken Cy5 (abcam), and goat antirabbit antibody conjugated to TRITC (sigma) were used as secondary antibodies at 1:300. Connexin 43 1:100 (sigma) was assessed at 21 days to determine the gap junction area between CMs. Goat antirabbit antibody conjugated to Alexa Fluor 594 was used as secondary antibody 1:300 (Invitrogen). 3D and 2D (control) cell cultures were washed twice with PBS 1× and then fixed in PBS containing 4% PFA for 15 min at 37 °C. Cells were permeabilized with 1% Triton X-100 for 1.5 h, blocked in 2% BSA in PBS (blocking buffer) for 45 min, and incubated with primary antibodies overnight (all the steps were performed at 37 °C). Secondary antibodies were incubated for 45 min at 37 °C. Cell nuclei were stained with DAPI, and samples were mounted in Vectashield (Vector Laboratories). When indicated, cells were further processed using the Click-IT EdU 555 Imaging kit (Life Technologies) to reveal EdU incorporation, according to the manufacturer’s instructions, and stained with DAPI. Fluorescent images were taken from four regions of each sample (n = 3) using an inverted Leica DFC 450 C microscope and a Zeiss LSM780 spectral, FLIM, 2P, SHG confocal. Within each experiment, instrument settings were kept constant.
Live/Dead Assay
Gels were washed twice with warm PBS and then a live/dead assay (life technologies) was used according to manufacturer instruction. Fluorescent images were taken from 10 randomly selected areas (n = 3) using an inverted EVOS FL microscope to determine cell viability. Within each experiment, instrument settings were kept constant.
Statistical Analysis
Two-tailed Student’s t test was used to determine differences in the percentage of cell viability between samples. Differences between groups were considered significant when p < 0.05.
Results and Discussion
PSHU-PNIPAAm Synthesis and Characterization
With the aim to create a biomimetic polymer that not only supports CM survival, but also presents the advantages of an injectable RTG, we developed a novel functionalizable RTG formulation using a PSHU-PNIPAAm copolymer.24 Due to its thermal gelling properties allowing minimally invasive applications, PNIPAAm has gained significant interest for injectable materials in drug and cell delivery systems. Li et al., proposed the use of poly(ethylene glycol) (PEG)-PNIPAAm-poly(e-caprolactone) (PCL) hydrogels with LCST around 31 °C as potential systems for long-term drug delivery.27 In addition, PCL-PNIPAAm amphiphilic graft copolymers were also synthesized for the same drug delivery application.28
Although several investigators reported promising results, the key to successfully use RTG polymers for biological applications is to functionalize the platform with proteins and peptides to promote biocompatibility.24 This continues to be a limiting factor for most existing RTGs. For this study, we used the PSHU as a main copolymer backbone, in which every repeating unit has a BOC-protected amine functionality. While the primary amine in PSHU is initially protected by the BOC group, it is easily deprotected to a primary amine in the presence of TFA/DCM. These primary amines can then be used for further modifications. This particular characteristic allows our RTG system to be very versatile and adaptable for various applications. Moreover, PSHU presents amide ester bonding in its structure that provides biomimetic characteristics and high biocompatibility. We have previously reported the use of PSHU-PNIPAAm for different biomedical applications: Yun et al. reported the use of this polymer for 3D neural tissue engineering;29 Famili et al. combined micelles and PSHU-PNIPAAm for controlled ocular drug delivery system;25 In addition, Pena et al. modified PSHU-PNIPAAm to obtain a heparin-mimicking RTG for positively charged therapeutic protein delivery.24
With these experiences, for this study, we functionalized the PSHU-PNIPAAm with laminin and poly-l-lysine via EDC/NHS chemistry to support long-term NRVM and ARVM CM viability. These proteins were selected because they are extensively used to promote CM attachment and survival in culture.30−36 As laminin is a component of the extracellular matrix (ECM), it is commonly used as a culture substrate to promote strong ARVM adherence, through the binding of β1-integrin surface receptors.30 Similar to laminin, poly-l-lysine, is a cationic amino acid which is also frequently used in vitro for NRVM culture.37 Furthermore, laminin was chosen as the biomolecule to functionalize PSHU-PNIPAAm for all the ARVM studies and poly-l-lysine for all the NRVM studies.
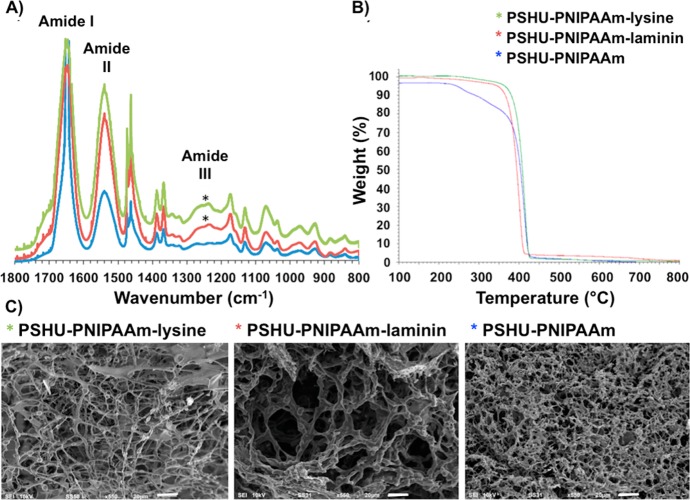
The presence of laminin and poly-l-lysine in PSHU-PNIPAAm was confirmed by FTIR (Figure 1A). Both PSHU-PNIPAAm-laminin and PSHU-PNIPAAm-lysine showed amide III (C–H stretching) vibrations at 1230–1280 cm–1, which are characteristic of proteins, while no signals of amide III were observed in PSHU-PNIPAAm. The amide III band is usually weak in the FTIR spectroscopy but can be found in the region from 1235 to 1350 cm–1.38,39 Nagai et al. reported amide III signal was confirmed at 1235 cm–1 in deer tendon collagen samples. Jeevithan et al. analyzed type II collagen and gelatin from shark cartilage samples, in which amide III signals were observed in the rage of 1237 to 1240 cm–1.38 Furthermore, Rahman et al. reported the presence of amide III signals at 1262 cm–1 in the chitin-containing samples.39
Figure 1.
Characterization of PSHU-PNIPAAm systems. (A) FTIR of PSHU-PNIPAAm-lysine (green), PSHU-PNIPAAm-laminin (red), and PSHU-PNIPAAm (blue). (*) Amide III (C–H stretching) vibrations, which are characteristic of proteins, were observed at 1230–1280 cm–1 in both PSHU-PNIPAAm-lysine and PSHU-PNIPAAm-laminin, while no amide III vibrations were observed in plain PSHU-PNIPAAm. (B) TGA of PSHU-PNIPAAm-lysine (green), PSHU-PNIPAAm-laminin (red), and PSHU-PNIPAAm (blue). The chemical incorporation of proteins makes the polymeric system more thermally stable. (C) From left to right, SEM of PSHU-PNIPAAm-lysine 1% (w/w) gel, PSHU-PNIPAAm-laminin 1% (w/w) gel, and PSHU-PNIPAAm 1% (w/w) gel. The chemical incorporation of proteins enlarged the pore size of the polymeric gels, facilitating nutrients and waste transportation. Scale bar: 20 μm.
Therefore, the presence of amide III corroborates a successful chemical incorporation of proteins in PSHU-PNIPAAm. Moreover, in all the PSHU-PNIPAAm systems, amide I (C=O stretching vibration) and amide II (N–H in-plane bending vibration coupled with C–N stretching vibration) signals were observed at 1645 and 1500 cm–1, respectively. Similar signals can also be found in natural polymers such as collagen or gelatin,38 making PSHU-PNIPAAm a highly biocompatible material.
To corroborate the chemical linkage of laminin and lysine in PSHU-PNIPAAm, a TGA analysis was performed (Figure 1B). A single weight loss event, without component separation, was observed in all the samples suggesting that proteins are chemically attached to PSHU-PNIPAAm. On the other hand, all the polymers presented different decomposition kinetics. After the isothermal treatment (100 °C during 20 min), a 4% weight loss was observed in PSHU-PNIPAAm, which can be attributed to a moisture weight loss. PSHU-PNIPAAm starts decomposition around 200 °C and finally losing a total mass of 96% at 430 °C, while PSHU-PNIPAAm-lysine and PSHU-PNIPAAm-laminin present a more stable thermal material degradation: 430 and 410 °C, respectively. PSHU-PNIPAAm-lysine has a very similar mass loss as PSHU-PNIPAAm, around 96%. However, PSHU-PNIPAAm-laminin has a mass loss of 94%. In general, the chemical incorporation of proteins makes PSHU-PNIPAAm system thermally more stable.
The morphological characterization of PSHU-PNIPAAm-laminin, PSHU-PNIPAAm-lysine, and PSHU-PNIPAAm was achieved by SEM (Figure 1C). Cross-sectional images of the gels revealed a highly porous configuration with an interconnected porous structure in all the PSHU-PNIPAAm systems. In addition, the chemical incorporation of laminin and poly-l-lysine greatly increased the pore size within the gel matrices, which is beneficial to facilitate the transport of nutrients and waste materials required for cell growth, interaction, and differentiation. A notable increase in the pore size was observed in PSHU-PNIPAAm-laminin that may be due to the repulsion between the hydrophilic groups of the large protein chain and the hydrophobic groups of the polymer. Thus, the conjugation of proteins can modify the morphology of PSHU-PNIPAAm scaffolds. These findings can be used to control cell protection during injection, modulate cell migration to a desired location, and aid in cell survival.
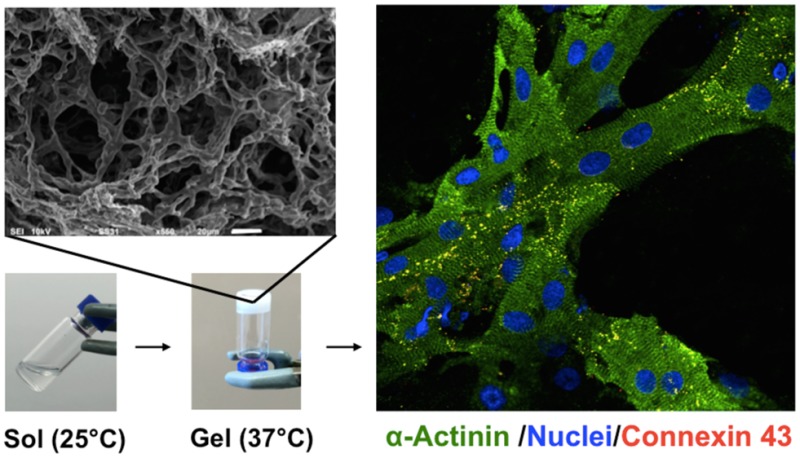
The sol–gel phase transition of PSHU-PNIPAAm-laminin, PSHU-PNIPAAm-lysine, and PSHU-PNIPAAm was determined rheologically by measuring the elastic modulus upon temperature sweep (Figure 2C).
Figure 2.
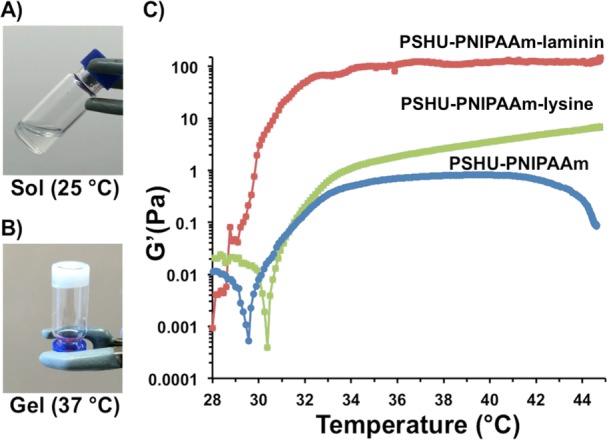
Temperature-dependent phase transition and elastic modulus. (A) Transparent aqueous solution of RTG at room temperature (B) turns to physical gel (white) at 37 °C. (C) Temperature-dependent elastic modulus of 1% (w/w) gels of PSHU-PNIPAAm-laminin (red), PSHU-PNIPAAm-lysine (green), and PSHU-PNIPAAm (blue). The chemical incorporation of laminin greatly increases the elastic modules of the polymeric system.
A sharp increase in the elastic modulus was observed between 28 and 33 °C corresponding to the sol (Figure 2A) to gel (Figure 2B) phase transition, in which the polymeric solution turns to a physical gel as the temperature increases. Further heating over 41 °C led to a decrease in elastic modulus of PSHU-PNIPAAm, corresponding to phase separation. On the other hand, no decrease in the elastic modulus after 41 °C was observed with both PSHU-PNIPAAm-laminin and PSHU-PNIPAAm-lysine. Thus, the chemical incorporation of proteins seems to stabilize PSHU-PNIPAAm rheologically as well as thermally. The fact that these polymers present sol-to-gel transition temperatures close to body temperature make them ideal for biomedical application. The chemical incorporation of both laminin and lysine increase the elastic modulus of plain PSHU-PNIPAAm. It has been previously reported that the elastic modulus of materials increase with the chemical incorporation of proteins.40,41 These findings indicate that the mechanical properties of PSHU-PNIPAAm can be easily tuned by chemically incorporating different proteins that can be extremely beneficial to support cell differentiation, as reported by Wen et al.42 and Jacot et al.43
Effect of 3D PSHU-PNIPAAm and 3D PSHU-PNIPAAm-Lysine Scaffolds on NRVM Culture
The effect of the 3D PSHU-PNIPAAm systems on the viability of NRVMs was first assessed using live/dead staining. Cells were analyzed in complete medium at different time points during 3 weeks. Traditional 2D gelatin coating culture dishes, prepared as previously described, were used as controls.4,44,45Figure 3 shows the live/dead results.
Figure 3.
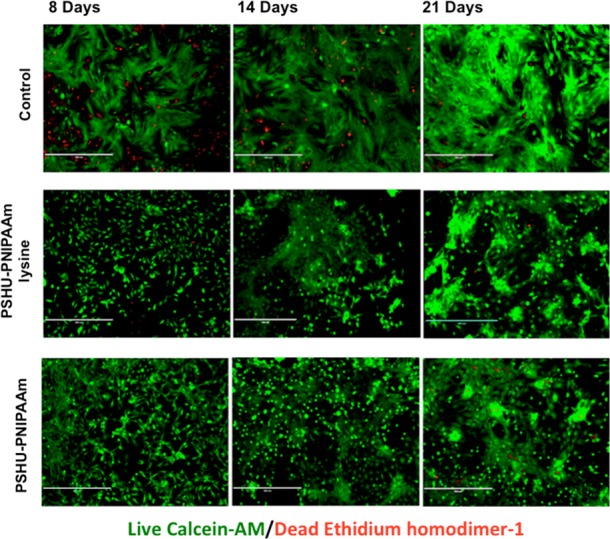
Live/dead (green/red) staining of NRVM after 8, 14, and 21 days of culture in different conditions: Top-row panels, 2D tissue culture plate coated with gelatin (control). Middle-row panels, NRVM cultured in 3D PSHU-PNIPAAm-lysine. Bottom-row panels: NRMV cultured in 3D PSHU-PNIPAAm. Compared to control groups, NRVM cultured in both 3D matrices showed a well-developed cell-interconnected network that may allow better impulse propagation between cells. Scale bar: 400 μm.
Compared with the 2D controls, cells growing in both 3D polymeric develop a better cell-interconnected contractile network, where a prolonged and more rapid coordinated contraction was observed in PSHU–PNIPAAm-lysine system (see videos in Supporting Information). On the other hand, no contraction was detected in the 2D gelatin controls after 21 days of culture. In addition, a smaller amount of dead cells (red fluorescent nuclei) were observed in both 3D polymeric systems compared with the 2D controls. Therefore, both 3D matrices induce a more rapid and synchronized contraction with improved survival and functionality when compared with standard 2D gelatin-control.
To confirm the cardiac phenotype a specific cardiac maker, the contractile protein α-sarcomeric actinin, was examined together with vimentin, a cytoskeleton marker commonly used for fibroblast staining, and CD31, a marker for endothelial cells, in order to distinguish α-sarcomeric actinin positive cardiomyocytes from vimentin positive fibroblast and CD31 positive endothelial cells. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain all cell nuclei. 2D gelatin coating culture dishes prepared as previously described were used as controls.4,44,45 The number of α-actinin-positive cells, vimentin-positive cells, and CD31 positive cells was analyzed in complete medium and then quantified at different time points during 3 weeks, as shown in Figure 4. Although it was tested CD31 as a marker for endothelial cells, no positive CD31 cells were observed in the cultures. On the other hand, a clear cardiac sarcomere represented by cross-striations can be observed in both NRVMs grown in 3D PSHU-PNIPAAm and PSHU-PNIPAAm-lysine systems. Even though fibroblasts were observed in both polymeric substrates, most of the cells show a α-actinin-positive cardiac phenotype. The presence of cardiac fibroblast is essential to mimic native cardiac tissues. Therefore, the development of a 3D matrix, which can sustain both CMs and cardiac fibroblasts holds tremendous potential as an emerging artificial cardiac tissue.
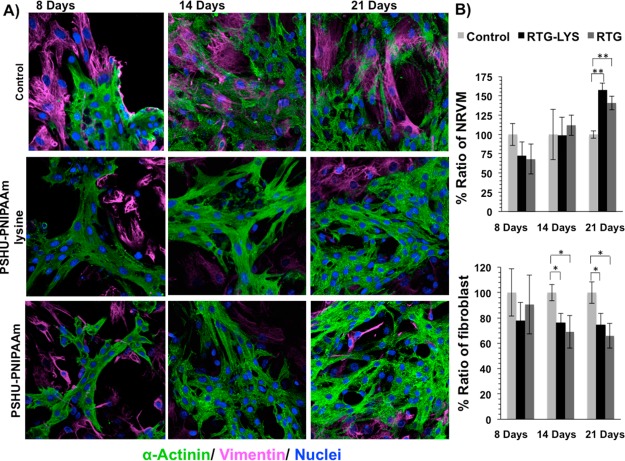
Figure 4.
Fluorescence staining of NRVM and fibroblast growing in different substrates. sarcomeric α-actinin (green), vimentin (pink), and DAPI (blue). (A) Top-row panels: NRVM cultured on 2D tissue culture plate coated with gelatin (control) after 8, 14, and 21 days. Middle-row panels: NRVM cultured in 3D PSHU-PNIPAAm-lysine after 8, 14, and 21 days. Bottom-row panels: NRMV cultured in 3D PSHU-PNIPAAm after 8, 14, and 21 days. Compared to controls, the 3D matrices stimulate NRVM to spread and form heart-like functional syncytia. (B) Quantification histogram of the ratio of NRVM and fibroblast growing in gelatin-control and RTG biopolymers. Values were normalized to controls. Significant differences on the % ratio of NRVM can be observed after 21 days between gelatin control group and RTG biopolymers. ** indicates p value < 0.01 (Student’s test) (PNIPAAM-lysine p value: 0.0005; n = 5; PSHU–PNIPAAM p value: 0.002; n = 5). Significant differences on the % ratio of fibroblast was also observed after 14 and 21 days between gelatin control group and RTG biopolymers. * indicates p value < 0.05 (Student’s test; 14 days: PSHU-PNIPAAM-lysine p value: 0.013; n = 5; PSHU-PNIPAAM p value: 0.02; n = 5. 21 days: PSHU-PNIPAAM-lysine p value: 0.023; n = 5; PSHU–PNIPAAM p value: 0.01; n = 5). Data are presented as mean ± SD (n = 5).
A cardiac phenotype was also observed in the 2D gelatin-controls after 21 days. However, as shown in Figure 4B, the amount of NRVM significantly decreased compared with the RTG biopolymers (PSHU-PNIPAAM-lysine p value: 0.0005; n = 5; PSHU-PNIPAAM p value: 0.002; n = 5); in contrast, the amount of fibroblast significantly increased in the 2D gelatin-controls compared with the polymeric systems (14 days: PSHU-PNIPAAM-lysine p value: 0.013; n = 5; PSHU-PNIPAAM p value: 0.02; n = 5. 21 days: PSHU-PNIPAAM-lysine p value: 0.023; n = 5; PSHU-PNIPAAM p value: 0.01; n = 5). No significant differences, in terms of NRVM or fibroblast ratio, were observed between PSHU-PNIPAAm-lysine and plain PSHU-PNIPAAm.
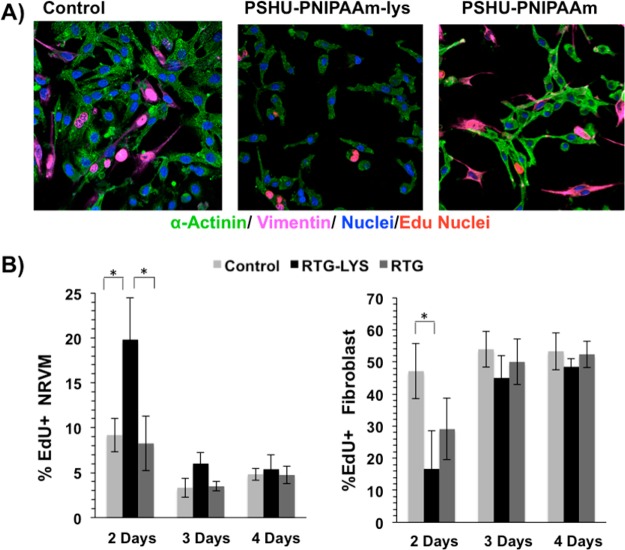
The effect of the 3D PSHU-PNIPAAm systems on the proliferation capacity of NRVMs and fibroblast was also assessed using 5-ethynyl-2′-deoxyuridine (EdU), a nucleoside analog of thymidine that is incorporated into DNA during active DNA synthesis. Cells were analyzed in complete medium at three different time points, specifically on postplating days 2, 3, and 4. 2D gelatin coating culture dishes were used as controls. The cells were subsequently fixed and immunostained for α-sarcomeric actinin, vimentin and DAPI. Figure 5 shows the proliferative results. At all time points, a percentage of cells showed evidence of EdU incorporation (see Figure 5A, top panel for representative images, and Figure 5B, bottom panel for quantification). On day 2, ∼19% of nuclei in NRVM scored positive for EdU incorporation on the PSHU-PNIPAAm-lysine scaffold, whereas only ∼9 and 8% of nuclei were found positive for EdU on the gelatin controls and the PSHU-PNIPAAm system, respectively (Figure 5B). The percentage of EdU positivity progressively decreased to ∼6%, ∼3.3%, and ∼3.5% at day 3 and to ∼5.3%, ∼4.8%, and ∼4.7% at day 4 on PSHU-PNIPAAm-lys, PSHU-PNIPAAm, and gelatin controls, respectively (Figure 5B, bottom panel). Only at day 2 it was observed a significant difference of dividing NRVMs on the PSHU-PNIPAAm-lysine system than on the gelatin controls (P = 0.02) and on the PSHU-PNIPAAm scaffold (P = 0.02). On the other hand, a significant increment of dividing fibroblast was observed at day 2 on the gelatin controls compared with PSHU-PNIPAAm-lysisne (P = 0.02). No significant differences of dividing fibroblast where observed between PSHU-PNIAAm and the gelatin controls.
Figure 5.
Proliferation assay of NRVM and fibroblast. Fluorescence staining: sarcomeric α-actinin (green), vimentin (magenta), DAPI (blue), and EdU (pink nuclei). (A) From right to left: NRVM and fibroblast cultured on 2D tissue culture plate coated with gelatin (control), 3D PSHU-PNIPAAm-lysine and 3D PSHU-PNIPAAm. (B) Significant differences of dividing NRVMs on the PSHU-PNIPAAm-lysine system were observed at day 2 when compared with the gelatin controls (P = 0.02) and with the PSHU-PNIPAAm scaffold (P = 0.02). Significant differences of dividing fibroblast were observed at day 2 between the gelatin controls and the RTG biopolymers. * indicates p value < 0.05 (Student’s test). Data are presented as mean ± SD (n = 5).
As mentioned before, regarding cell contraction (see videos in Supporting Information), PSHU-PNIPAAm-lysine demonstrated a clear prolonged, more rapid and coordinated contraction compared to gelatin-controls and plain PSHU-PNIPAAm. This may be due to a better intercellular communication of CMs growing in PSHU-PNIPAAm-lysine. Since intercellular communication is one of the most important organizational features of the heart and gap junction channels form the basis of direct intercellular communication, we examined the level and localization of connexin (Cx)-43 as a marker of functionality and differentiation for CMs grown on gelatin-controls and PSHU-PNIPAAm-lysine scaffolds. We decided to test only the PSHU-PNIPAAm-lysine as it presented a clear difference in terms of cell contraction as compare to control. Thus, we investigated the organization of the cellular communication, specifically the gap-junctions localization, through Cx-43 immunostaining 21 days after culturing NRMV in 3D-PSHU-PNIPAAm-lysine and gelatin controls.
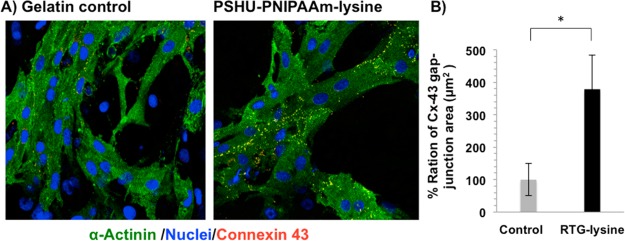
To distinguish between cardiomyocytes and non-CM cells, α-sarcomeric actinin was analyzed together with DAPI. The area of Cx-43 in α-actinin-positive cells was then quantified in both PSHU-PNIPAAm-lysine biopolymer and gelatin controls. Figure 6A shows representative images of NRVM stained for Cx-43 (yellow dots) and α-actinin-positive cells (green) after 21 days of culture in gelatin-control (left panel) and PSHU-PNIPAAm-lysine (right panel). Quantitative analysis of Cx-43-positive area (Figure 6B) indicated the amount of Cx-43 and relative gap-junction were significantly higher on the PSHU-PNIPAAm-lysine compared to control (p value: 0.014; n = 5); thus, demonstrating that CMs growing in 3D PSHU-PNIPAAm-lysine present a better impulse propagation compared to controls. It has been previously reported that cardiac fibroblasts, provide mechanotransductive cues and paracrine factors that influence CM assembly and maturation.46 Thavandiran et al. reported that the combination of a 3D matrix-based microenvironment and the mixture of CMs and fibroblast improve the performance of in vitro engineered cardiac tissues.47 Since a mix-culture of a large amount of CMs and fibroblast grew perfectly together in the 3D PSHU-PNIPAAm-lysine, it is possible that the intercellular interactions were favored, leading to more mature functional syncytia.
Figure 6.
Gap junctions are increased in cardiomyocytes grown in PSHU-PNIPAAm-lysine (RTG-LYS). (A) Fluorescence connexin 43 (yellow dots), sarcomeric α-actinin (green), and DAPI (blue) staining of NRVM. (B) Quantification histogram of Cx-43 gap junction ratio after 21 days of culture in gelatin-control group and PSHU-PNIPAAm-lysine. Values were normalized to controls. P = 0.014, significance PSHU-PNIPAAm-lysine vs gelatin-control. * indicates p value < 0.05 (Student’s test). Data are presented as mean ± SD (n = 5).
Effect of 3D PSHU-PNIPAAm and 3D PSHU-PNIPAAm-Laminin Scaffolds on ARVM Culture
Live/dead staining was also used to evaluate ARVM survival in 3D PSHU-PNIPAAm biopolymers. Cells were cultured in serum-free medium to avoid nonmyocytes cell proliferation and analyzed at different time points. Traditional 2D laminin-coated culture dishes were used as controls.
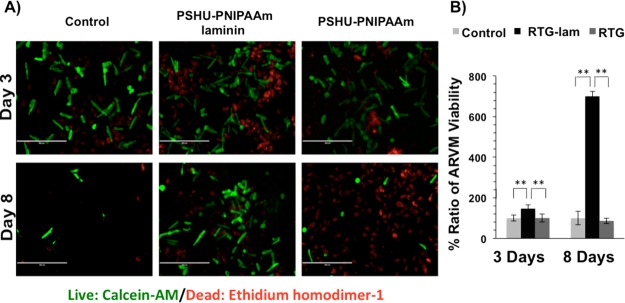
Figure 7 shows the live/dead results. A significant improvement of ARVM viability was observed in the 3D PSHU-PNIPAAm-laminin matrix compared with the laminin-controls and the PSHU-PNIPAAm biopolymer after 3 days of culture (PSHU-PNIPAAm-laminin compared to (1) control p value: 0.0006, n = 5; (2) PSHU-PNIPAAm p value: 0.0005, n = 5). In contrast, no significant differences were observed in the 3D PSHU-PNIPAAm matrix compared to control groups.
Figure 7.
Live/dead (green/red) staining of ARVM after 3 and 8 days of culture in different conditions: (A) from left to right 2D tissue culture plate coated with laminin (control), 3D PSHU-PNIPAAm-laminin, and 3D PSHU-PNIPAAm. Top-row panels ARVM after 3 days of culture. Bottom-row panels ARVM after 8 days of culture. (B) Compared to control groups, a notable improvement of ARVM viability was observed in the PSHU-PNIPAAm-aminin after 3 and 8 days of culture. ** indicates p value < 0.01 (Student’s test). Three days of culture: PSHU-PNIPAAm-laminin compared to (1) control p value: 0.0006 and (2) PSHU-PNIPAAm p value: 0.0005. Eight days of culture: PSHU-PNIPAAm-laminin compared to (1) control p value: 0.0004 and (2) plain PSHU-PNIPAAm p value: 0.0003. Data are presented as mean ± SD (n = 5).
A large number of dead cells can be seen in both 3D cultures; however, this is likely due to the cell preparation procedure. Since ARVM are large cells26 it is not possible to eliminate dead cells and debris produced during the cell preparation. Only after culturing the cells, unattached cells can be removed in the 2D cell culture. However, it is not an option for the 3D cultures that entrap both alive and dead cells as well as debris. The encapsulation of dead cells and debris creates a harsh environment for the viable cells. However, due to the high porosity of the scaffolds, nutrients and cell waste can diffuse through the polymeric matrix. Compared with PSHU-PNIPAAm, PSHU-PNIPAAm-laminin has a larger pore size that facilitates better nutrients and waste transportation, leading to enhanced cell viability.
After 8 days of culture, a significant number of the ARVMs in the PSHU-PNIPAAm-laminin group were rod shaped and viable, while the PSHU-PNIPAAm and 2D laminin-controls groups showed more than 90% round-shaped and nonviable ARVMs (PSHU-PNIPAAm-laminin compared to (1) control p value: 0.0004, n = 5 and (2) PSHU–PNIPAAm p value: 0.0003, n = 5).
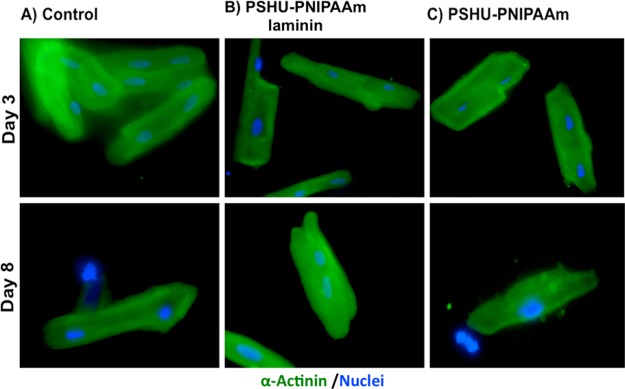
The cardiac phenotype of the ARVMs cultured on the 2D laminin-controls and the 3D matrices was also evaluated using the α-sarcomeric actinin cardiac maker with DAPI for cell nuclei. Cells were cultured in serum free medium to avoid nonmyocytes cell proliferation.
Figure 8 shows the α-sarcomeric actinin and DAPI immunofluorescence staining of ARVM after 3 and 8 days of culture. After 3 days of culture, all the samples showed a well-defined cardiac phenotype represented by a sarcomere striation. Similar results were observed in the remaining cells after 8 days of culture. Compared with control groups, PSHU-PNIPAAm-laminin showed promising capability to support ARVM survival while also preserving their cardiac phenotype.
Figure 8.
Fluorescence sarcomeric α-actinin (green) and DAPI (blue) staining of ARVM in different conditions: (A) 2D tissue culture plate coated with laminin (control), (B) 3D PSHU-PNIPAAm-laminin, and (C) 3D PSHU-PNIPAAm. Top-row panels ARVM after 3 days of culture. Bottom-row panels ARVM after 8 days of culture. Compared to control groups we found that the 3D PSHU-PNIPAAm-laminin matrix allows a long-term ARVM survival with a well-defined cardiac phenotype represented by a sarcomere striation.
As previously reported, ARVMs in culture prove to have a low viability over time.30 Previous investigations demonstrated that ARVM loss their rod-shaped phenotype and start showing intracellular signaling and morphology similar to neonatal myocytes after 4 days in culture. In addition, there is a significant cell loss after 5 days of culture.48 Therefore, the fact that a large number of ARVM preserves their cardiac phenotype after 8 days of culture in the 3D PSHU-PNIPAAm-laminin matrix makes this approach very promising not only for cardiac tissue engineering but also for further investigations in an in vitro analysis.
Conclusions
PSHU-PNIPAAm was successfully functionalized with the chemical incorporation of laminin or lysine to support adult and neonatal CM survival. Our results demonstrate excellent NRVM viability in both 3D PSHU-PNIPAAm and 3D PSHU-PNIPAAm-lysine for up to 21 days. Moreover, PSHU-PNIPAAm-lysine exhibited a significantly greater intercellular communication capacity, which may facilitate CM maturation. On the other hand, a large number of viable ARVMs were found in the PSHU-PNIPAAm-laminin matrix after 8 days of culture. As ARVM proved to be difficult to sustain viability in culture, this may suggest that our novel system has resounding and ubiquitous effect in treating cardiomyopathies. The ability to functionalize PSHU-PNIPAAm makes this system very versatile, providing tremendous potential for engineered materials specifically tuned for cardiac regeneration.
Acknowledgments
This study was supported by a generous Grant of the John Patrick Albright Foundation (L.M. and M.T.), NIH/NHLBI R01 HL116905 (L.M.), PDS HL116906 (B.P.), and 1R01HL109209-01A1 (M.T.). This work was supported in part by a Trans-Atlantic Network of Excellence Grant from the Leducq Foundation (14 CVD 03). The authors would like to thank Dr. Carmen Sucharov for providing NRVM cells for the experiments here presented and for her guidance. As well, Dr. Pena would like to specially thank Karen Dockstader, Penny Nelson, and Jennifer Mahaffey for their valuable support in the cells preparation.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biomac.5b01734.
The authors declare no competing financial interest.
Supplementary Material
References
- Cahill T. J.; Ashrafian H.; Watkins H. Circ. Res. 2013, 113 (6), 660–675. 10.1161/CIRCRESAHA.113.300282. [DOI] [PubMed] [Google Scholar]
- Mestroni L.; Rocco C.; Gregori D.; Sinagra G.; Di Lenarda A.; Miocic S.; Vatta M.; Pinamonti B.; Muntoni F.; Caforio A. L. P.; McKenna W. J.; Falaschi A.; Giacca M.; Camerini F. J. Am. Coll. Cardiol. 1999, 34 (1), 181–190. 10.1016/S0735-1097(99)00172-2. [DOI] [PubMed] [Google Scholar]
- Li X.; Zhou J.; Liu Z.; Chen J.; Lu S.; Sun H.; Li J.; Lin Q.; Yang B.; Duan C.; Xing M.; Wang C. Biomaterials 2014, 35 (22), 5679–5688. 10.1016/j.biomaterials.2014.03.067. [DOI] [PubMed] [Google Scholar]
- Martinelli V.; Cellot G.; Toma F. M.; Long C. S.; Caldwell J. H.; Zentilin L.; Giacca M.; Turco A.; Prato M.; Ballerini L.; Mestroni L. Nano Lett. 2012, 12 (4), 1831–1838. 10.1021/nl204064s. [DOI] [PubMed] [Google Scholar]
- Li S. L.; Loganathan S.; Korkmaz S.; Radovitss T.; Hegedus P.; Zhou Y.; Karck M.; Szabo G. J. Surg. Res. 2015, 195 (1), 315–324. 10.1016/j.jss.2014.12.038. [DOI] [PubMed] [Google Scholar]
- Dunn D. A.; Hodge A. J.; Lipke E. A. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2014, 6 (1), 15–39. 10.1002/wnan.1241. [DOI] [PubMed] [Google Scholar]
- Garbade J.; Barten M. J.; Bittner H. B.; Mohr F. W. Clin. Cardiol. 2013, 36 (7), 378–382. 10.1002/clc.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventrelli L.; Ricotti L.; Menciassi A.; Mazzolai B.; Mattoli V. J. J. Nanomater. 2013, 2013, 1. 10.1155/2013/108485. [DOI] [Google Scholar]
- Camci-Unal G.; Annabi N.; Dokmeci M. R.; Liao R.; Khademhosseini A. NPG Asia Mater. 2014, 6, 12. 10.1038/am.2014.19. [DOI] [Google Scholar]
- Li Z. Q.; Guan J. J. Polymers 2011, 3 (2), 740–761. 10.3390/polym3020740. [DOI] [Google Scholar]
- Dixit P.; Katare R. Stem Cell Res. Ther. 2015, 6, 26. 10.1186/s13287-015-0010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menasche P. Circulation 2009, 119 (20), 2735–2740. 10.1161/CIRCULATIONAHA.108.812693. [DOI] [PubMed] [Google Scholar]
- Kharaziha M.; Shin S. R.; Nikkhah M.; Topkaya S. N.; Masoumi N.; Annabi N.; Dokmeci M. R.; Khademhosseini A. Biomaterials 2014, 35 (26), 7346–54. 10.1016/j.biomaterials.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Guo X.; Guan J. Biomaterials 2012, 33 (25), 5914–5923. 10.1016/j.biomaterials.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Nguyen D. T.; Barham W.; Zheng L.; Shillinglaw B.; Tzou W. S.; Neltner B.; Mestroni L.; Bosi S.; Ballerini L.; Prato M.; Sauer W. H. J. Cardiovasc. Electrophysiol. 2014, 25 (12), 1385–1390. 10.1111/jce.12509. [DOI] [PubMed] [Google Scholar]
- Nichol J. W.; Koshy S. T.; Bae H.; Hwang C. M.; Yamanlar S.; Khademhosseini A. Biomaterials 2010, 31 (21), 5536–5544. 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.-K.; Cho C.-S. Int. J. Stem Cells 2010, 3 (2), 96–102. 10.15283/ijsc.2010.3.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton A. J.; Alsberg E.; Albertelli M.; Mooney D. J. Transplantation 2004, 77 (12), 1798–1803. 10.1097/01.TP.0000131152.71117.0E. [DOI] [PubMed] [Google Scholar]
- Wang L.; Cao L.; Shansky J.; Wang Z.; Mooney D.; Vandenburgh H. Mol. Ther. 2014, 22 (8), 1441–1449. 10.1038/mt.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.; Webb J.; Verheye S.; Moss R.; Boone R.; Leipsic J.; Ree R.; Banai S. J. Am. Coll. Cardiol. 2014, 64 (17), 1814–1819. 10.1016/j.jacc.2014.06.1208. [DOI] [PubMed] [Google Scholar]
- Hatefi A.; Amsden B. J. J. Controlled Release 2002, 80 (1–3), 9–28. 10.1016/S0168-3659(02)00008-1. [DOI] [PubMed] [Google Scholar]
- Packhaeuser C. B.; Schnieders J.; Oster C. G.; Kissel T. Eur. J. Pharm. Biopharm. 2004, 58 (2), 445–455. 10.1016/j.ejpb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Park M. H.; Joo M. K.; Choi B. G.; Jeong B. Acc. Chem. Res. 2012, 45 (3), 424–433. 10.1021/ar200162j. [DOI] [PubMed] [Google Scholar]
- Pena B.; Shandas R.; Park D. J. Biomed. Mater. Res., Part A 2015, 103 (6), 2102–8. 10.1002/jbm.a.35345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famili A.; Kahook M. Y.; Park D. Macromol. Biosci. 2014, 14 (12), 1719–1729. 10.1002/mabi.201400250. [DOI] [PubMed] [Google Scholar]
- Jeong M. Y.; Walker J. S.; Brown R. D.; Moore R. L.; Vinson C. S.; Colucci W. S.; Long C. S. Am. J. Physiol.: Heart Circ. Physiol. 2010, 298 (6), H1719–H1726. 10.1152/ajpheart.00937.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.-X.; Lu M.-G.; Wu K.; Zhang Y.-F.; Miao L.; Li Y.-W.; Guo H.-L.; Zheng J. Polym. Eng. Sci. 2015, 55 (1), 223–230. 10.1002/pen.23877. [DOI] [Google Scholar]
- Li M.; Pan P.; Shan G.; Bao Y. Polym. Int. 2015, 64 (3), 389–396. 10.1002/pi.4800. [DOI] [Google Scholar]
- Yun D.; Laughter M. R.; Park D. Austin J. Biomed. Eng. 2014, 1, 1019. [Google Scholar]
- Louch W. E.; Sheehan K. A.; Wolska B. M. J. Mol. Cell. Cardiol. 2011, 51 (3), 288–298. 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M. L.; Graham B. T.; Pabon L. M.; Han S. J.; Murry C. E.; Sniadecki N. J. J. Biomech. Eng. 2014, 136 (5), 051005. 10.1115/1.4027145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird S. D.; Doevendans P. A.; van Rooijen M. A.; de la Riviere A. B.; Hassink R. J.; Passier R.; Mummery C. L. Cardiovasc. Res. 2003, 58 (2), 423–434. 10.1016/S0008-6363(03)00253-0. [DOI] [PubMed] [Google Scholar]
- Sander V.; Sune G.; Jopling C.; Morera C.; Izpisua Belmonte J. C. Nat. Protoc. 2013, 8 (4), 800–809. 10.1038/nprot.2013.041. [DOI] [PubMed] [Google Scholar]
- Reinecke H.; Zhang M.; Bartosek T.; Murry C. E. Circulation 1999, 100 (2), 193–202. 10.1161/01.CIR.100.2.193. [DOI] [PubMed] [Google Scholar]
- Banyasz T.; Lozinskiy I.; Payne C. E.; Edelmann S.; Norton B.; Chen B.; Chen-Izu Y.; Izu L. T.; Balke C. W. Exp. Physiol. 2008, 93 (3), 370–382. 10.1113/expphysiol.2007.040659. [DOI] [PubMed] [Google Scholar]
- McDevitt T. C.; Angello J. C.; Whitney M. L.; Reinecke H.; Hauschka S. D.; Murry C. E.; Stayton P. S. J. Biomed. Mater. Res. 2002, 60 (3), 472–479. 10.1002/jbm.1292. [DOI] [PubMed] [Google Scholar]
- Zaika O.; Zhang J.; Shapiro M. S. J. Physiol. (Oxford, U. K.) 2011, 589 (10), 2559–2568. 10.1113/jphysiol.2010.204768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeevithan E.; Bao B.; Bu Y.; Zhou Y.; Zhao Q.; Wu W. Mar. Drugs 2014, 12 (7), 3852–3873. 10.3390/md12073852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. A.; Halfar J. Sci. Rep. 2014, 4, 6162. 10.1038/srep06162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I.; Georges P. C.; Janmey P. A. Soft Matter 2007, 3 (3), 299–306. 10.1039/B610522J. [DOI] [PubMed] [Google Scholar]
- Georges P. C.; Miller W. J.; Meaney D. F.; Sawyer E. S.; Janmey P. A. Biophys. J. 2006, 90 (8), 3012–3018. 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J. H.; Vincent L. G.; Fuhrmann A.; Choi Y. S.; Hribar K. C.; Taylor-Weiner H.; Chen S. C.; Engler A. Nat. Mater. 2014, 13 (10), 979–987. 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot J. G.; Martin J. C.; Hunt D. L. J. Biomech. 2010, 43 (1), 93–98. 10.1016/j.jbiomech.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli V.; Cellot G.; Toma F. M.; Long C. S.; Caldwell J. H.; Zentilin L.; Giacca M.; Turco A.; Prato M.; Ballerini L.; Mestroni L. ACS Nano 2013, 7 (7), 5746–5756. 10.1021/nn4002193. [DOI] [PubMed] [Google Scholar]
- Martinelli V.; Cellot G.; Fabbro A.; Bosi S.; Mestroni L.; Ballerini L. Front. Physiol. 2013, 4, n/a. 10.3389/fphys.2013.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panakova D.; Werdich A. A.; MacRae C. A. Nature 2010, 466 (7308), 874–U109. 10.1038/nature09249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thavandiran N.; Dubois N.; Mikryukov A.; Masse S.; Beca B.; Simmons C. A.; Deshpande V. S.; McGarry J. P.; Chen C. S.; Nanthakumar K.; Keller G. M.; Radisic M.; Zandstra P. W. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (24), E3151–E3151. 10.1073/pnas.1509023112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter B. J.; Smith J. R.; Buja L. M.; Bick R. J. Cell Calcium 2001, 30 (6), 373–382. 10.1054/ceca.2001.0249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.