Abstract
Genome-wide association studies (GWAS) have provided a rich collection of ~58 CAD loci that suggest the existence of previously unsuspected new biology relevant to atherosclerosis. However, these studies only identify genomic loci associated with CAD and many questions remain even after a genomic locus is definitively implicated, including the nature of the causal variant(s) and the causal gene(s), as well as the directionality of effect. There are a number of tools that can be employed for investigation of the functional genomics of these loci, and progress has been made on a limited number of novel CAD loci. New biology regarding atherosclerosis and CAD will be learned through the functional genomics of these loci and the hope is that at least some of these new pathways relevant to CAD pathogenesis will yield new therapeutic targets for the prevention and treatment of CAD.
Keywords: atherosclerosis, genome-wide, functional, coronary artery disease, genomics
Introduction
Efforts to perform unbiased discovery using the tools of human genetics to uncover novel pathways underlying complex diseases and traits have been pursued extensively over the last decade. In particular, genome-wide association studies (GWAS), reviewed elsewhere in this compendium, have capitalized on the millions of common single nucleotide polymorphisms (SNPs) to identify those SNPs that are genome-wide significantly associated with a disease or trait. More than 20 thousand genetic loci have been identified to be associated with diseases or traits (http://www.ebi.ac.uk/gwas December 2015). In particular, myocardial infarction (MI), and more generally coronary artery disease (CAD), have been the focus of intense discovery using GWAS. In the most recent and largest meta-analysis of GWAS for CAD1, 10 new CAD loci were identified, bringing the total number of CAD loci to 58 (Table 1).
Table 1.
Overview of 58 previously reported loci with genome-wide significance for Coronary Artery Disease1
| reported gene |
first reported lead SNP |
chr | location* | 1000G p-value |
new lead SNP | updated location* |
updated 1000G p- value |
reported phenotypes** |
|---|---|---|---|---|---|---|---|---|
| PCSK9 | rs11206510 | 1 | intergenic | 2.34E-08 | MI,188 LDL-C2 | |||
| PPAP2B | rs17114036 | 1 | PPAP2B | 2.22E-13 | rs9970807 | PPAP2B | 5.00E-14 | MI163 |
| SORT1 | rs646776 | 1 | CELSR2 | 9.01E-19 | rs7528419 | CELSR2, PSRC | 1.97E-23 | MI,188 LDL-C,124 Phospholipase A2189 |
| IL6R | rs4845625 | 1 | IL6R | 3.93E-08 | rs6689306 | IL6R | 2.60E-09 | MI22 |
| MIA3 | rs17464857 | 1 | TAF1A | 4.18E-05 | rs67180937 | MIA3 | 1.01E-12 | MI22,190 |
| rs17465637 | 1 | MIA3 | 3.52E-12 | |||||
| AK097927 | rs16986953 | 2 | intergenic | 1.45E-08 | MI22 | |||
| APOB | rs515135 | 2 | intergenic | 3.09E-08 | chr2:21378433:D | intergenic | 2.89E-08 | MI,22 LDL-C,124 |
| ABCG5-ABCG8 | rs6544713 | 2 | ABCG8 | 8.88E-07 | chr2:44074126:D | ABCG8 | 2.60E-08 | MI,22 LDL-C124 |
| VAMP5-VAMP8-GGCX | rs1561198 | 2 | VAMP5, VAMP8 | 6.37E-10 | rs7568458 | GGCX, VAMP8 | 3.62E-10 | MI22 |
| ZEB2-ACO74093.1 | rs2252641 | 2 | TEX41 | 5.16E-04 | rs17678683 | LINC01412, ZEB2 | 3.00E-09 | MI22 |
| WDR12 | rs6725887 | 2 | WDR12 | 9.51E-18 | chr2:203828796:I | ALS2CR8 | 2.15E-18 | MI188 |
| MRAS | rs9818870 | 3 | MRAS | 2.21E-06 | chr3:138099161:I | MRAS | 2.89E-09 | MI191 |
| REST-NOA1 | rs17087335 | 4 | NOA1 | 4.6E-08 | MI1 | |||
| EDNRA | rs1878406 | 4 | intergenic | 1.24E-06 | rs4593108 | intergenic | 8.82E-10 | plaque,192 stroke10 |
| GUCY1A3 | rs7692387 | 4 | GUCY1A3 | 7.35E-09 | rs72689147 | GUCY1A3 | 6.07E-09 | MI,22 |
| SLC22A4-SLC22A5 | rs273909 | 5 | SLC22A4, LOC553103 | 1.24E-04 | MI22 | |||
| ADTRP-C6orf105*** | rs6903956 | 6 | ADTRP | 0.96 | MI193 | |||
| PHACTR1 | rs12526453 | 6 | PHACTR1 | 2.14E-20 | rs9349379 | PHACTR1 | 1.81E-42 | MI,188 calcification,112 artery dissection,194 migraine195 |
| ANKS1A | rs17609940 | 6 | ANKS1A | 0.03 | MI163 | |||
| KCNK5 | rs10947789 | 6 | KCNK5 | 1.63E-06 | rs56336142 | intergenic | 1.85E-08 | MI22 |
| TCF21 | rs12190287 | 6 | TCF21,TARID | 1.07E-03 | rs12202017 | TARID, LINC01312 | 1.98E-11 | MI163 |
| SLC22A3-LPAL2-LPA | rs2048327 | 6 | SLC22A3 | 2.46E-09 | rs55730499 | LPA | 5.39E-39 | MI196,163 |
| rs3798220 | 6 | LPA | 4.66E-09 | |||||
| PLG | rs4252120 | 6 | PLG | 3.32E-03 | rs4252185 | PLG | 1.64E-32 | MI22 |
| HDAC9 | rs2023938 | 7 | HDAC9 | 1.36E-04 | rs2107595 | intergenic | 8.05E-11 | stroke,197 MI22 |
| 7q22 | rs10953541 | 7 | BCAP29 | 1.02E-05 | MI164 | |||
| ZC3HC1 | rs11556924 | 7 | ZC3HC1 | 5.34E-11 | MI163 | |||
| NOS3 | rs3918226 | 7 | NOS3 | 1.7E-09 | MI1 | |||
| LPL | rs264 | 7 | LPL | 1.06E-05 | MI22 | |||
| TRIB1 | rs2954029 | 8 | RP11-136O12.2 | 2.61E-06 | Triglycerides,124 LDL-C,4 HDL-C,4 total cholesterol,4 MI22 | |||
| 9p21 | rs3217992 | 8 | CDKN2B, CDKN2B-AS1 | 1.03E-42 | rs2891168 | CDKN2B-AS1 | 2.29E-98 | MI22,195,188 |
| rs4977574 | 9 | CDKN2B-AS1 | 6.35E-98 | |||||
| ABO | rs579459 | 9 | ABO | 1.14E-10 | rs2519093 | ABO | 1.19E-11 | MI,163 venous thromboembolism198 |
| KIAA1462 | rs2505083 | 9 | KIAA1462 | 1.57E-10 | rs2487928 | KIAA1462 | 4.41E-11 | MI164 |
| CXCL12 | rs2047009 | 9 | intergenic | 2.75E-11 | rs1870634 | intergenic | 5.55E-15 | MI22,190 |
| rs501120 | 10 | RP11-20J15.2 | 1.39E-11 | |||||
| LIPA | rs11203042 | 10 | LIPA | 1.22E-04 | rs1412444 | LIPA | 5.15E-12 | MI,22,164 |
| rs1412444 | 10 | LIPA | 5.15E-12 | |||||
| CYP17A1-CNNM2-NT5C2 | rs12413409 | 10 | CNNM2 | 1.07E-07 | rs11191416 | PFN1P11 | 4.65E-09 | MI,163 intracranial aneurysm199 |
| SWAP70 | rs10840293 | 10 | SWAP70 | 1.3E-08 | MI1 | |||
| PDGFD | rs974819 | 10 | RP11-563P16.1 | 2.44E-10 | rs2128739 | RP11-563P16.1 | 7.05E-11 | MI164 |
| ZNF259-APOA5-APOA1 (ZPR1) | rs964184 | 11 | ZNF259 | 5.60E-05 | MI,163 Triglycerides,124 HDL-C,124 LDL-C4 | |||
| ATP2B1 | rs7136259 | 11 | ATP2B1 | 2.45E-05 | rs2681472 | ATP2B1 | 6.17E-11 | MI, blood pressure200 |
| SH2B3 | rs3184504 | 11 | SH2B3 | 1.03E-09 | MI,163 blood pressure200 | |||
| KSR2 | rs11830157 | 12 | KSR2 | 2.12E-09 | MI1 | |||
| FLT1 | rs9319428 | 12 | FLT1 | 7.13E-05 | MI22 | |||
| COL4A1/A2 | rs4773144 | 12 | COL4A1, COL4A2 | 3.87E-07 | rs11838776 | COL4A2 | 1.83E-10 | MI163,22 |
| rs9515203 | 13 | COL4A2 | 9.33E-10 | |||||
| HHIPL1 | rs2895811 | 13 | HHIPL1 | 1.86E-05 | rs10139550 | HHIPL1 | 1.38E-08 | MI163 |
| SMAD3 | rs56062135 | 13 | SMAD3 | 4.5E-09 | MI1 | |||
| ADAMTS7-MORF4L1 | rs7173743 | 14 | MORF4L1 | 5.55E-16 | rs4468572 | MORF4L1 | 4.44E-16 | MI22 |
| MFGE8-ABHD2 | rs8042271 | 15 | intergenic | 3.7E-08 | MI1 | |||
| FURIN-FES | rs17514846 | 15 | FURIN | 3.10E-07 | MI22 | |||
| SMG6-SRR | rs216172 | 15 | SMG6 | 5.07E-07 | MI163 | |||
| RASD1, SMCR3, PEMT | rs12936587 | 15 | intergenic | 8.24E-04 | MI163 | |||
| UBE2Z, GIP, ATP5G1, SNF8 | rs46522 | 15 | UBE2Z | 1.84E-05 | MI163 | |||
| BCAS3 | rs7212798 | 15 | BCAS3 | 1.9E-08 | MI1 | |||
| PMAIP1-MC4R | rs663129 | 17 | intergenic | 3.2E-08 | MI1 | |||
| LDLR | rs1122608 | 17 | SMARCA4 | 2.73E-11 | rs56289821 | intergenic | 4.44E-15 | MI188 |
| ZNF507-LOC400684 | rs12976411 | 17 | LOC400684, ZNF507 | 1.18E-14 | MI1 | |||
| APOE-APOC1 | rs2075650 | 17 | TOMM40, PVRL2 | 1.61E-06 | rs4420638 | APOC1 | 7.07E-11 | Total Cholesterol,201 LDL-C,202,203 Phospholipase A2,204 plaque,192 Triglycerides205 |
| rs445925 | 18 | APOC1, APOE | 4.23E-06 | |||||
| SLC5A3-MRPS6-KCNE2 | rs9982601 | 19 | intergenic | 1.33E-13 | rs28451064 | AP000320.7 | 1.33E-15 | MI188 |
| POM121L9P-ADORA2A | rs180803 | 19 | POM121L9P | 1.6E-10 | MI1 | |||
gene body +-5kb (hg19, UCSC genes, Ensembl genes, Refseq genes)
CAD lead SNPs with genome-wide significance in published GWAS studies for CAD relevant phenotypes
only significant in East Asians

It is interesting to examine this list of loci in light of known risk factors for CAD. For example, 10 of the loci are also genome-wide significantly associated with LDL-cholesterol (LDL-C)2–5, a known causal risk factor for CAD. The causal genes at these loci exert their effects through their expression in hepatocytes or enterocytes, consistent with their role in regulating LDL metabolism. Another locus, LPL, harbors the gene encoding the enzyme lipoprotein lipase (LPL), the most important regulator of triglyceride-rich lipoprotein (TRL) metabolism. In addition, the APOA1/C3/A4/A5 locus associated with LDL-C is also associated with triglycerides (TG) and harbors two genes, APOC3 and APOA5, in which coding variants have been shown to be associated with both TG levels and CAD.6–9 These and other observations have helped to confirm the causal role of TRLs in CAD. Another five loci are genome-wide significant for association with blood pressure, consistent with the causal role of elevated blood pressure in CAD. Also there is some overlap with GWAS studies for other vascular diseases such as stroke10. Interestingly, none of the 58 loci are associated with type 2 diabetes mellitus (T2DM), raising interesting questions regarding the genetic overlap between T2DM and CAD and whether T2DM per se is causally related to CAD. Importantly, the majority of CAD GWAS loci are not associated with known risk factors for CAD (Table 1) and thus have the potential to provide novel insights into the biology and pathophysiology of CAD.
Issues and challenges for functional genomics of CAD GWAS loci
Common variant GWAS studies only identify genomic loci associated with disease or trait. However, many questions remain even after a genomic locus is definitively implicated, including the nature of the causal variant(s) and the causal gene(s), as well as the directionality of effect. For the majority of the CAD GWAS loci, the answers to these questions are unknown, and after excluding the loci associated with lipids or blood pressure virtually none of the remaining loci have answers to these fundamental questions. Identification of the causal variant is challenging, because of linkage disequilibrium (LD) and the possibility that the variant(s) at a given locus with the lowest p-values for association with CAD may simply be proxies for the causal variant (see Figure 1). Furthermore, although the majority of variants with the lowest p-values fall in non-coding intergenic regions, they usually do not fall within a well-established cis-regulatory element such as a known promoter, and thus challenge predictions of their impact on regulatory elements, like disruption of transcription factor binding or function of a long non-coding RNA (lncRNA). Below we discuss approaches to elucidating the causal variant at a GWAS locus.
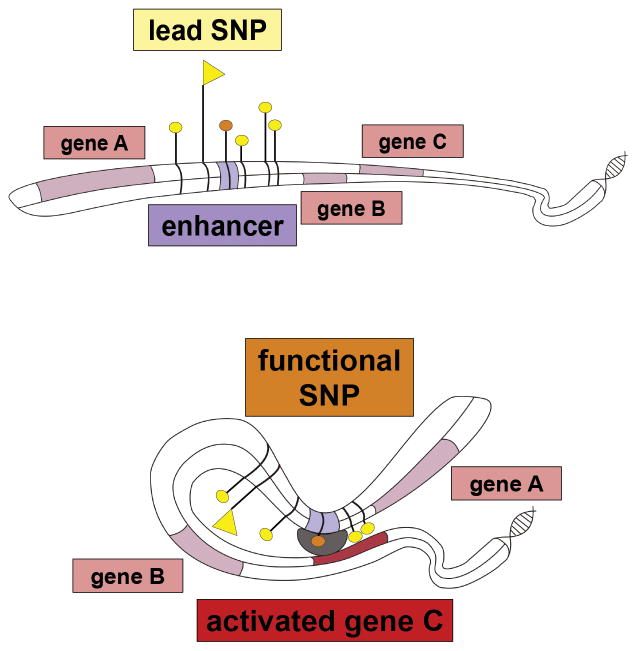
Figure 1. Mechanism by which non-coding risk SNP can affect phenotype.
Top: multiple SNPs associated with disease are located in the intergenic region proximal to genes A, B and C. One of the SNPs with genome-wide significance is situated within a cis- regulatory element (orange tag). The lowest P-value SNP (‘lead SNP’, flag tag) lies outside the regulatory element.
Bottom: Through bending of the DNA molecule the regulatory element gets into physical contact with the promoter of its target gene, in this case gene C which is not the gene in closest proximity, leading to regulation of its expression (‘activation’ or upregulation in case of an enhancer element). The SNP located within the regulatory element (‘functional SNP’, orange tag) can now affect transcription by for instance altering transcription factor (TF) binding affinity based on genotype via disruption of a TF binding motif
Arguably the most important biological question to be addressed at each CAD GWAS locus is what the causal gene(s) at the locus are. By convention, GWAS loci are tabulated by the coding gene closest to the ‘lead SNP’ with the lowest p-value. However, it is becoming clear that this approach does not always identify the causal gene11. Because of chromatin looping that places regulatory enhancer elements in proximity to the promoters of genes that may be quite distant on the physical map, a causal variant may influence expression of distant genes (Figure 1). Furthermore, the ‘causal gene’ at a GWAS locus need not necessarily be a protein-coding gene, but could be a lncRNA (for example ANRIL, a lncRNA at the 9p21 CAD locus), a microRNA, or some other transcribed or regulatory element. Finally, some loci may not have a single ‘causal gene’ but in fact may be characterized by the coordinate regulation of several genes, potentially in different contributing cell types, that have additive effects on disease phenotype. Below we discuss in some detail the methodological approaches to solving these critical biological conundrums.
The directionality of effect at the locus is a critically important issue, particularly with regard to the question of whether the biology represented by that locus can be approached from a therapeutic targeting standpoint. For example, if the minor allele at a locus is associated with protection from CAD, it is essential to know if the minor allele is associated with increased or decreased expression of the causal gene in the relevant cell type. Approaches such as expression quantitative trait loci (eQTL) and allele-specific expression (ASE) can be used to establish directionality of effect. However, the effects of many variants on differential gene expression are cell type specific and in most cases we don’t know with any confidence the relevant cell type for the genetic effect. Again, below we discuss the experimental approach to establishing directionality of effect given these challenges.
The tools of functional genomics
There is large variety of experimental tools available to investigate the mechanism by which GWAS loci exert their effect on biological phenotype. They are tailored to the scale of interrogation, sampling on the genome-wide level, targeting a gene, a genomic region or even a specific SNP of interest. Figure 2 gives an overview of the experimental techniques which will be discussed in detail below.
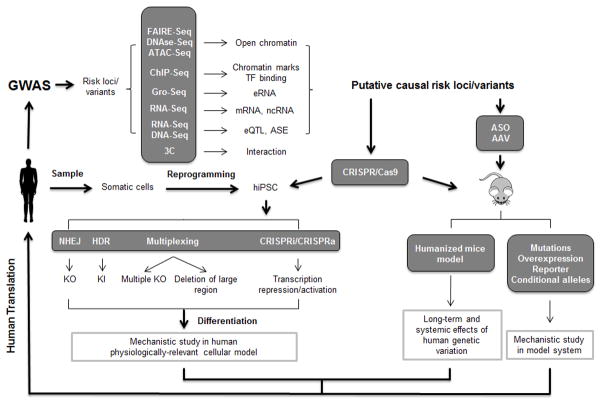
Figure 2. Experimental tools for GWAS functional follow-up studies.
GWAS findings can be functionally annotated using genome-wide methods, which help prioritize loci with likely biological function. These putative risk loci can be further interrogate by genome editing using the CRISPR/Cas9 system in vitro and in vivo. Additionally, adeno-associated virus (AAV) and antisense oligos (ASO) can be employed to study candidate gene knockdown and overexpression in the mouse model. TF: transcription factor; eRNA: enhancer RNA; ncRNA: non-coding RNA; eQTL: expression quantitative trait loci; ASE: allele-specific expression; iPSC: induced pluripotent stem cells; NHEJ: non-homologous end joining; HDR: homology-directed repair; KO: knockout; KI: knockin.
Next-generation sequencing
Most polymorphisms associated with risk for disease lie in non protein-coding regions of the human genome. A major mechanism of how these SNPs affect disease phenotype is by disruption of a transcription factor binding site within a cis-regulatory element. The subsequent dysregulation of gene expression leads to a changed cellular response. The first step in establishing a causal chain of evidence from association SNP to disease phenotype lies in the definition of genomic regions with cis-regulatory potential in a disease-relevant cell type. Genome-wide methods such as ‘Formaldehyde-Assisted Isolation of Regulatory Elements’ (FAIRE-Seq)12, DNase Hypersensitivity sequencing13 and the novel ‘Assay for Transposase-Accessible Chromatin using Sequencing’ (ATAC-Seq)14 assess the general accessibility of chromatin. However, these patterns only portray a general ‘openness’ of the genome and include cis-regulatory regions as well as promoters and protein-coding regions. Also, these techniques cannot distinguish between poised, primed and actively functional regulatory regions – they only map the general regulatory potential.
In contrast, several different histone modifications have been shown to sample distinct fractions of the human genome, such as promoters (Histone 3 Lysine 4 trimethylation - H3K4me3), transcribed genes (Histone 3 Lysine 36 trimethylation - H3K36me3), primed (Histone 3 Lysine 4 monomethylation - H3K4me1) and active (Histone 3 Lysine 27 acetylation - H3K27ac) enhancers.15 Chromatin immunoprecipitation with massively parallel sequencing (ChIP-Seq) is the method of choice to identify these chromatin marks. This technique can also be used to identify the binding events of specific transcription factors of interest.
Active enhancers are identified with the highest confidence when datasets are intersected, for example by combining H3K27ac abundance with Polymerase II binding patterns from ChIP-seq as well as Gro-Seq. The latter technique maps actively transcribed RNAs from enhancers (eRNAs)16 through the isolation of newly synthesized RNA from nuclei incubated with bromouridine. Large-scale consortium data for histone marks and transcription factor binding is now publicly available from the ENCODE project (https://genome.ucsc.edu/ENCODE/dataMatrix/encodeChipMatrixHuman.html). The experimental data stems from in vitro cultured cells, many of them immortalized human cell lines. Of particular interest for the annotation of CAD GWAS loci is data generated from HepG2 cells (a hepatocellular carcinoma line that serves as a proxy for hepatocytes), endothelial HUVECs, CD14+ monocytes, and aortic smooth muscle cells. Similarly, the Roadmap Epigenomics Project (www.roadmapepigenomics.org/data/tables/adult) compiled a large portfolio of histone marks from human tissues, including data from liver, aorta and primary CD14+ monocytes.
In addition to revealing transcription factor occupancy and histone modification at sites of interest, ChIP-Seq data can also be interrogated for allelic imbalance on an individual basis, if paired with corresponding genotype information.17,18 In this case sequencing reads from ChIP-Seq experiments are assigned to either the maternal or paternal allele. A significant difference in coverage on either allele is indicative for sequence dependent differential transcription factor binding or histone abundance in this region and provides direct evidence for the functional role of a polymorphic site.
A second step in elucidating the mechanism by which non-coding polymorphisms affect disease risk lies in the analysis of transcriptome data from disease-relevant cells and tissues. Whole-transcriptome RNA-Seq data provides useful information on the transcriptional repertoire of a cell of interest. No meaningful association of regulatory variants with disease risk genes is possible without knowledge of the cell’s transcriptome. Publicly available expression data from relevant cells and tissues is provided by the ENCODE consortium and to a larger extent the GTEx consortium (www.gtexportal.org), which generated transcriptome data from primary human post mortem tissue.19
Technical advances such as single-cell based RNA-Seq20 and single-cell ChIP-Seq21 allow for the interrogation of the transcriptional machinery in single cells. Future applications may allow simultaneous derivation of both datasets from the same cell, which would greatly reduce the information lost due to sampling heterogeneous pools of cells at different cell cycle and developmental stages. This could be particularly useful for the characterization of the diverse cell populations within the atherosclerotic neointima.
An important consideration in functional transcriptomic analysis is that since both transcriptional regulation and gene expression are tissue-specific processes, particular attention should be paid to the suitability of cell type from which the data is generated. Early datasets have depended heavily on samples generated in cell culture for practical reasons, namely the accessibility of long-established cell lines, and the ability to generate homogeneous material, reproducibly, and in large scale. However, all cultured cells, whether they be immortalized or cancer cell lines, induced pluripotent stem cell-derived cells, or cultured primary cells, display to some extent rather an immature, precursor-like phenotype in comparison to the corresponding fully differentiated primary cell in vivo. Additionally, cancer cell lines in particular, often carry genomic rearrangements leading to artifacts that are not representative of their cell type of origin. On the other hand, primary ex vivo tissues, whilst displaying the most ‘authentic’ transcriptional profiles, are often comprised of multiple cell types which complicates analysis and can mask subtle effects within data noise, or due to numerically underrepresented cell types. The best approach when dealing with tissues of mixed cell types such as coronary arteries may be to focus on effects also observed in in vitro cultured pure cell populations.
Furthermore primary tissues need to be extensively phenotyped as to their disease status to enable identification of changes within the transcriptome between the healthy and diseased state. Additionally, the exact tissue origin can play an important role. For example aortic, coronary or femoral artery transcriptomes, although often very similar, can show distinct differences in their expression profiles which may be based on their differing tissue environments or developmental origins. Differences in gene expression are well established along the different section of the aorta itself and follow embryological and hemodynamic patterns.
For meaningful correlation of ChIP-Seq and RNA-Seq data to identify SNP-to-gene interactions, data should be generated from the same cell type, if possible even from the same individual culture. In case of primary tissue, combined data sets from the same individual are particularly informative. The association of cis-regulatory regions with one or multiple nearby regulated targeted gene or genes is however problematic. Frequently employed distance-based methods do not adequately reflect the true biology. More meaningful approaches to directly link SNP genotype with gene expression levels include expression quantitative trait loci (eQTL) and allele-specific expression (ASE) studies.
eQTLs and ASE
Quantitative trait loci are polymorphic sites within a genome which show significant association with a quantitative trait such as plasma lipid levels22, carotid intima-media thickness23 or gene expression levels (eQTL) 24, 25. eQTL studies combine genotype with gene expression level information - the latter usually assessed using expression microarrays or RNA-seq - and can detect both local cis-effects as well as distal trans-effects of regulatory elements on gene expression. eQTL data has been generated from multiple human cells and tissues26–28. Of particular relevance is data derived from liver, which has proven especially successful for loci involved in dyslipidemia, as well as data from peripheral blood monocytes29 and in vitro cultured human aortic endothelial cells30.
eQTL studies require sampling from several hundreds of individuals to identify the majority of loci with statistical significance as inter-individual noise arises from differences in genetic background, host factors such as age or gender as well as environmental factors like diet and lifestyle. The large sample sizes needed for eQTL analysis are particularly problematic when interrogating tissues that are rare or difficult to sample. Additional challenges such as insufficient platform coverage and batch variation stem from the use of microarrays to determine gene expression levels. These issues can be circumvented in large part by the use of RNA-Seq as a data source, which greatly increases statistical power, and is more compatible with meta-analysis.
eQTL studies have been successfully used to identify functional SNPs and directionality from GWAS studies for a variety of diseases, due to changes in the expression of their downstream putative risk genes31. Since CAD is a complex disease involving multiple tissues, recent studies have aimed at integrating expression data from several tissues for a more comprehensive annotation of CAD GWAS loci32,33. However, the portfolio of tissues used included several human tissues not relevant to CAD, which may have introduced bias in the SNP-to-gene association process. Fortunately, the GTEx consortium is generating publicly available RNA-Seq data from currently under represented CAD-relevant tissues, including coronary artery and aorta.
Allele-specific expression has emerged as an alternative to eQTL analysis for linking genetic variation in cis-regulatory regions to gene expression34–37. It is based on the identification of allelic imbalance, showing differences in gene expression levels between the two alleles in a single heterozygous individual. The within person allelic analyses greatly reduces impact of inter-individual variation from environmental and genetic trans effects thus enhancing statistical power. ASE requires allele-specific transcriptomic data such as stranded RNA-Seq data, but in contrast to eQTL analysis, a relatively small number of samples that carry the same heterozygous site of interest are needed. The information value of each sample is limited by the number of heterozygous sites of its genome. Additionally, the data sets that are generated need to be of high read coverage to ensure presence of multiple reads at interrogated sites of interest. With regards to CAD, to date ASE has for been applied to assess general principles of the regulation of gene expression in mouse liver38, 39.
Additionally, Chromosome Conformation Capture (3C) can be used as a complementary technique to associate cis-regulatory regions with their target genes. This method captures the physical interaction between two genomic regions such as enhancers with promoters, and has recently been applied to study human liver and aorta40. Several different variations of the technique are in use, with Hi-C and 5C as genome-wide methods. A major obstacle is however their low resolution of tens to hundreds of kilobases. Targeted approaches such as Capture-C41 and Capture Hi-C42 that can interrogate hundreds of select loci simultaneously are displaying higher resolutions of down to 1 kilobase (kb) and single cell approaches are starting to emerge43. The improved availability of tissues relevant to CAD and increasingly sensitive methods at hand to link association SNPs to downstream risk genes, suggest that a comprehensive re-annotation of all 58 known CAD association loci in disease-relevant cells and tissues using the latest methods may be worthwhile.
Non-coding RNAs
A substantial proportion of trait-associated SNPs identified by GWAS lie outside of protein coding regions and map to the non-coding intervals44. However, the mechanistic relationship of trait-associated SNPs with the non-coding functional genome is poorly understood. Since protein-coding genes account for only a very small proportion of the transcribed human genome, non-coding (nc) RNA are now emerging as alternative functional genomic elements underlying GWAS hits. Along with microRNAs, long non-coding RNAs (lncRNAs), ncRNAs defined as transcripts larger than 200 nucleotides (nt) in length, are emerging as important regulators involved in cancer as well as in neurological, cardiovascular, developmental and other human diseases45, highlighting the need to investigate the possible contributions of variations in ncRNAs to human diseases.
Systematic analyses are emerging that evaluate the potential association of regulatory ncRNAs, with complex traits. miRNAs comprise a class of short (20–24 nt) regulatory RNAs that modulate mRNA translation and turnover. A recent study leveraged GWAS meta-analysis in more than 188,000 individuals to identify 69 miRNAs located in genomic regions associated with abnormal blood lipid levels46. The work identified four miRNAs (miR-128-1, miR-148a, miR-130b and miR-301b) that are associated with LDL-C uptake and cholesterol efflux by possibly controlling the expression of the LDL receptor (LDLR) and the ATP-binding cassette transporter A1, respectively. miR-QTL studies using liver tissue from 424 morbidly obese individuals revealed an association of miR-128-1 and miR-148a expression with SNPs linked to abnormal human blood lipid levels, suggesting the relevance of these miRNAs identified by GWAS studies to human cardiometabolic disorders46.
A subset of lncRNAs, intergenic lncRNAs (lincRNAs), represents a rapidly evolving catalog of lncRNA species that does not overlap with exons of protein-coding genes.47 A number of studies examined the implication of lincRNAs in complex diseases based on GWAS studies (LincPoly48, LincSNP49). These studies represent initial efforts to integrate disease-associated SNPs and human lincRNAs, but both datasets do not include complete GWAS SNP data, and focus on only a few thousand lincRNAs. A recent study identified 495,729 and 777,095 SNPs in more than 30,000 lncRNA transcripts in human and mouse, respectively. A large number of SNPs were predicted to impact the lncRNA secondary structure and modulate lncRNA–miRNA interactions. By mapping these SNPs to GWAS results, 142 human lncRNA SNPs are GWAS tagSNPs and 197,827 lncRNA SNPs are within these GWAS LD regions.50 Kumar et al. examined the association of SNPs with expression of lincRNAs in human blood, and identified 112 cis-regulated lincRNAs. A considerable number of the observed lincRNA cis-eQTLs had disease- or trait-associations51 suggesting that intergenic GWAS-associated SNPs may act by modulating expression of specific lincRNAs.
Although functional roles of most lincRNAs remain elusive, mechanistic insights into distinct nuclear and cytoplasmic actions47, 52, 53 for a small number of well-studied lincRNAs strongly suggest that some lincRNAs play major regulatory roles in a variety of cellular processes, such as X chromosome inactivation54, embryogenesis55, cell pluripotency56, cell development and differentiation57. Depending on their subcellular localization, lincRNAs can mediate gene expression through distinct mechanisms. In the nucleus, they are involved in co-transcriptional regulation, recruitment of proteins complexes to specific loci for cis or trans regulation of gene expression or scaffolding of nuclear complexes47, 52. In the cytoplasm, lincRNAs can function as competitive endogenous RNAs that bind miRNAs and inhibit their activity, pair with mRNAs to trigger post-transcriptional regulation or interact with target proteins to modulate their function47, 52. LincRNAs are increasingly implicated in human diseases, including cancer,58 neurological diseases59 and cardiovascular disorders60–66, and also modulate physiology and pathophysiology in cells relevant to cardiometabolic disease. For example, cardiac lncRNAs like Braveheart, CHRF and Mhrt regulate cardiomyocyte differentiation and cardiac hypertrophy60, 63, 64. LincRNAs in smooth muscle and endothelial cells (MALAT, linc-p21) regulate proliferation65, 66. A few lincRNAs have been implicated in macrophage functions: lincRNA-Cox2 in mouse represses the basal expression of interferon-stimulated genes (ISGs) by partnering with the heterogeneous nuclear ribonucleoproteins (hnRNPs) hnRNPA/B and hnRNPA2/B167 and a human monocytic THP-1 lincRNA called TNF and hnRNPL-related immunoregulatory lincRNA (THRIL) regulates expression of tumor necrosis factor (TNF) through its interactions with hmRNPL68. Conserved adipose lincRNAs, such as Firre and Blnc169–71, and species-specific lincRNAs, such as lnc-BATE1 and ADINR72, 73, regulate adipogenesis of white and brown adipocytes in mouse and human.
Several studies have tried to identify and validate the causal GWAS variants that regulate lncRNA expression and function. A well-known example on chromosome 9p21 that encompasses an antisense lncRNA, ANRIL (antisense ncRNA of the INK4 locus) has been significantly associated with susceptibility to coronary disease as well as abdominal aortic and intracranial aneurysms74. Some associated SNPs in this region have been shown to alter the transcription and processing of ANRIL transcripts75.
To further uncover the effects of GWAS-associated genetic variants on ncRNAs function, future studies are needed to: 1) Define a comprehensive genome-wide set of human lncRNAs across all disease-relevant tissues. Because lncRNAs have lower and more tissue-specific expression patterns than mRNAs, this requires deeper RNA sequencing of multiple human cells and tissue than is currently available in GENCODE76 or the human bodymap77; 2) Investigate ncRNA enrichment and association in targeted yet genome-wide approaches within catalogued GWAS and particularly whole genome sequencing projects as they emerge, for example NHLBI’s Trans-Omics for Precision Medicine (TOPMed) Program (https://www.nhlbiwgs.org); 3) establish bioinformatics pipelines to prioritize trait-associated ncRNAs (systematically using synteny and conservation, tissue expression, eQTL and ASE, and ChIP-Seq promoter, enhancer and transcription factors marks at trait-associated lncRNAs); and 4) Establish high-throughput pipelines for human-relevant functional follow-up of trait-associated ncRNAs in disease-relevant cell types and in in vivo animal models.
Taken together, ncRNAs, such as miRNAs and lncRNAs may be important for interpreting GWAS data and may in many cases act as the causal genomic element in contributing to human cardiometabolic diseases. The therapeutic tractability of potent and specific antisense technologies targeting single or multiple ncRNAs implicated in human cardiometabolic diseases may thus have important clinical ramifications for the treatment of these diseases.
Genome editing
The putative functional variants identified by fine mapping overlapping with regulatory marks, eQTL and ASE analysis, and other bioinformatic approaches require further experimental validation to establish causality. Genome engineering and human induced pluripotent stem cells (hiPSCs), when combined, represent powerful tools to accelerate GWAS-driven functional validation of causal variants at trait-associated loci.
Genome engineering represents strategies and techniques developed in recent years for the targeted modification of the genetic information. The type II clustered regularly interspaced short palindromic repeats (CRISPR) and the CRISPR-associated protein 9 (Cas9), an RNA-guided nuclease, are based on a bacterial system that has been modified for genome engineering in mammalian cells78, 79. Due to its ease of adaptability and improved efficiency, CRISPR/Cas9 has rapidly become one of the most popular approaches for genome engineering80. Consisting of Cas9 and a short guide RNA (gRNA), it generates site-specific DNA breaks, which are repaired by either non-homologous end-joining (NHEJ), creating insertions or deletions at the site of the break, or homology-directed repair (HDR) by precise change of a genomic sequence using an exogenously introduced donor template81. In addition to the disruption of genomic sequence through nucleases, the CRISPR-associated catalytically inactive Cas9 protein, termed dCas9, can be fused to repressor82 or activator domains82–84, named CRISPRi82 and CRISPRa82–84, respectively. Such modified CRISPR/dCas9-fusion proteins, together with guide RNA, can then be introduced to control the expression or activity of candidate gene or regulatory elements that harbor GWAS signals85.
Although the CRISPR/Cas9 system is highly efficient in human cell lines, gene editing in primary human cells is very challenging. Human iPSCs have the potential to be differentiated to all adult cell types, including rare or inaccessible human cell populations, for reliable disease modeling. By generating clonal lines carrying desired genetic modification introduced by CRISPR/Cas9 and then differentiating to somatic cells relevant to atherosclerosis (for example hepatocytes, macrophages, smooth muscle cells, endothelial cells, cardiomyocytes), genome editing in hiPSCs provides a unique platform for functional validation of GWAS CAD loci across multiple disease-relevant cell types. Applications include: 1) Double strand break repaired by NHEJ pathway may introduce frame-shift mutation or early stop codon, which, if in a critical coding exon, likely causes nonsense-mediated decay of the mRNA and effectively eliminates gene function for loss-of function (LOF) studies; 2) By HDR-mediated precise nucleotide alteration using a donor template, it is feasible to generate hiPSC lines in which the disease-associated SNP is the sole experimental variable, thereby investigating the causal role of genetic variants - for instance the targeted alteration of specific transcription factor binding site motifs in otherwise intact loci could reveal the functional contribution of transcription factor binding to the function of a regulatory element; 3) The unique multiplexing capabilities of the CRISPR/Cas9 system facilitate the deletion of a large stretch of genomic DNA, enabling the functional interrogation of non-coding regulatory elements and non-coding transcripts. In addition, because most disease-associated SNPs confer only modest risk, the relevance of multiple monoallelic and biallelic combination can be addressed by multiplexing of CRISPR/Cas9 gene editing.
One example of such an interrogation investigates the role of an intronic variant (rs9349379) in the CAD/MI risk locus PHACTR1 prioritized by genetic fine-mapping and eQTL in human coronary arteries. The study using endothelial cell extracts first showed that alleles at rs9349379 are differentially bound by the transcription factors myocyte enhancer factor-2 (MEF2). The deletion of this MEF2-binding site using CRISPR/Cas9 in hiPSC and subsequent differentiation to endothelial cells then revealed that heterozygous endothelial cells carrying the deletion express 35 percent less PHACTR1 transcript86.
Despite being powerful, precise editing of human genomes in pluripotent stem cells by HDR of targeted nuclease-induced cleavage has been hindered by the low efficiency of HDR over NHEJ, making the screening of clones containing desired genotypes time-consuming and labor-intensive. However, great strides have been made to improve the efficiency of HDR. A Cas9D10A mutant functioning as a nickase yields similar HDR but lower NHEJ mutation rates81. The establishment of the iCRISPR platform through targeting of inducible Cas9 expression cassettes into the AAVS1 locus in human embryonic stem (ES) and iPS cell lines has increased markedly the efficiency of genetic modifications for both knockout or knockin of genetic variants87. The overall gene editing efficiency can be further enhanced by transfection of ribonuclear protein (RNP) complex that is comprised of the recombinant Cas9 protein and synthetic gRNAs88, either in vitro transcribed or chemically synthesized and modified89, the latter further improving efficiency. Several studies have also found that inhibition of DNA ligase IV90 and DNA-PKcs (DNA-dependent protein kinase, catalytic subunit)91, key players in the NHEJ pathway, promote HDR while reducing the frequency of NHEJ90–92.
Future studies need to: 1) Further improve the efficiency and reduce the cost of hiPSC differentiation; 2) Optimize differentiation protocols to produce mature cells phenotypically, functionally and transcriptomically highly similar to primary somatic cells; 3) Improve the efficiency of HDR-mediated precise nucleotide alteration over NHEJ; 4) Apply CRISPRi and CRISPRa in hiPSC for dynamic and precise control of expression of individual transcripts in hiPSC and differentiated cells; 5) Adapt conditional knockout for the assessment of gene function in different lineages of differentiation; 6) Establish more advanced techniques to facilitate rapid screening of rare iPS clones carrying the desired genotypes93, and thoroughly evaluate potential off-target effects.
In summary, facile high efficiency genome-editing coupled with hiPSC differentiation can pave the way for functional interrogation of GWAS variants and loci of complex non-Mendelian diseases such as CAD, and can help delineate human genotype-phenotype relationship in human cellular disease models, and potentially, in genetically-modified mice carrying mutations, reporter or conditional alleles for in vivo modeling using CRISPR/Cas9-mediated genome engineering94, 95.
Somatic gene targeting (siRNA, ASO, and AAV)
As mentioned above, transgenic mouse models that are genetically predisposed to develop atherosclerosis due to partial or complete loss of ApoE or Ldlr function, or that have been modified to have plasma lipid profiles which more closely reflect human biology are invaluable in the functional study of candidate GWAS genes. Mouse lines with ‘humanized’ lipid profiles include the Apobec knockout and the human apolipoprotein B(100) transgenic mouse models, which have been combined with haploinsufficient Ldlr deficiency in the LAhB-H mouse strain96. Genetic ablation remains the gold standard for the characterization of gene function: combining conditional aproaches and tissue-specific Cre drivers allow precise interrogation of the potential role of a gene to the phenotype of interest. On the other hand, even with the advent of facile CRISPR/Cas9 genome editing, the development of genetic models is laborious, expensive, and time-consuming.
Alternatives to genetic approaches include the use of small interfering RNA (siRNA) or antisense oligonucleotide (ASO) inhibitors. In addition, adeno-associated virus (AAV) platforms can be used for either overexpression, or for permanent loss of function by expression of short hairpin RNAs (shRNA). Combining these approaches with the existing genetic atherosclerosis models is a potent way to accelerate GWAS functional analysis, but compared to more rigorous genetic approaches involves some compromises and limitations in interpretability. These approaches are therefore not a substitute for subsequent genetic validation of promising putative causal genes, but rather a way to quickly prioritize candidate genes for further study. ASO and siRNA have in common that they are systemically delivered, modified nucleic acids that target the gene of interest through complementary base-pairing between their primary sequences and those of their target transcripts. Despite these similarities, the two forms differ in their mechanism of action. In the case of siRNA, the technology takes advantage of the RNA-direct RNA endonuclease activity of Argonaute2 (Ago2), the miRNA binding component of the RNA-interference Silencing Complex (RISC). The cytoplasmic RNase III endonuclease, Dicer1, cleaves double-stranded or short-hairpin RNA, and concomitantly loads one strand of the circa 22 basepair cleavage product into a binding cleft in Ago2. The solvent-exposed bases of the Ago-loaded RNA serve to target RISC to complementary sequences97. Endogenous microRNAs in mammals have imperfect complementarity with their targets: down regulation occurs by a combination of mRNA destabilization due to the recruitment of decapping and de-adenylation factors, and to the inhibition of translational initiation 98. In contrast, artificial shRNA and siRNA systems take advantage of an evolutionary remnant activity of Ago2, which cleaves the paired target strand where perfect complementarity exists between itself and the loaded RNA 99. The interfering RNA serves only as a targeting component and is not cleaved itself. The two types of RNA interference most significantly differ in their entry points to the endogenous system: while exogenous siRNAs can be transfected directly in cell culture, the shRNA are supplied as transgenes (usually along with a reporter gene, such as eGFP). As such, the shRNA genes must be transcribed in the nucleus, and the hairpin RNAs exported to the cytoplasm, cleaved by Dicer and loaded into RISC. The advantage of the shRNA approach is that candidate inhibitory RNAs can be validated in cell culture, and then readily adapted to viral or transgenic applications in vivo. However, the nuclear export protein, Exportin-5, which shuttles shRNA to the cytoplasm has been shown to be limiting in the biogenesis of shRNA, leading to initial limitations of this approach100. A number of subsequent innovations have alleviated these concerns, which were partly due to saturation of the miRNA biogenesis pathway, but also to off-target effects of the passenger strands of the shRNAs101–103.
In contrast to shRNA, siRNA bypasses the requirement for nuclear transcription and export: upon entry into the cytoplasm; the siRNA is rapidly loaded into RISC and interference begins. Since naked RNA has an extremely low half-life in plasma, and since endocytosed RNA is targeted to the lysosome and degraded, a variety of strategies have been developed to evade these obstacles to in vivo use104. In addition to its usefulness in basic research, RNA interference has already shown therapeutic potential in CVD, for example by targeting PCSK9105.
ASOs are also short nucleic acids, but do not depend on the RISC complex for their action. In the context of functional analysis of GWAS hits, ASOs may be deployed in three ways: 1) targeted to a transcript of a protein-coding gene to interfere with the initiation of translation; 2) targeted to intron-containing genes to block splicing, and 3) targeted against microRNAs to block their inhibitory effects on target genes. In each of these cases, pairing of the ASO with its target physically precludes the interaction of the target RNA with another molecule, elongation initiation factors, splicing factors, or target mRNAs respectively. In addition, the specialized class of ASOs termed ‘gapmers’ are designed with a central stretch of unmodified DNA nucleotides, which when base-paired to a complementary RNA target yields a heteroduplex that is recognized as a substrate by the ubiquitous intracellular ribonuclease, RNase H1. The resulting cleavage of the RNA strand of the heteroduplex by RNase H1 is analogous to that of siRNA, albeit by a completely different mechanism, and induces rapid turnover of the cleaved RNA. It should be noted that mipomersen, a therapeutic ASO targeting apoB, which is approved for the treatment of homozygous familial hypercholesterolemia, is based on a gapmer strategy106.
The advantage of RNA-targeting strategies as an approach, irrespective of the precise mechanism, is that they are relatively straightforward in design and validation. There are however key limitations of this approach. Systemic delivery of nucleic acids has been shown to induce an inflammatory response - for any given inhibitor there is a possibility of off-target effects of the artificial RNA, and the biodistribution of the oligonucleotides can be influenced slightly, but not tightly controlled. The inflammatory side effects have been largely mitigated by successive innovations in the chemistry of the synthetic nucleic acids used, and concerns over off-target effects can be addressed by the separate use of independent ASOs targeting a given gene. The limitations concerning the delivery and distribution are harder to address: the fact that most oligonucleotides end up by default in the liver (predominantly in hepatocytes) is less of a concern in the functional analysis of CVD GWAS candidate risk genes that are liver-expressed, than it might be in other biological contexts. In addition, both siRNA and ASO approaches only permit gene knockdown but not upregulation.
Adeno-associated virus (AAV) vectors provide a tractable system to perform the reciprocal experiment: overexpression of candidate causal genes for functional analysis in vivo107. As noted above, they can also be adapted for corresponding loss of function experiments using shRNA expression cassettes. As the name suggests, AAV was identified as a coinfecting parvovirus with adenovirus. To date AAV has no identified role in any human disease, and does not replicate in the absence of adenovirus. These characteristics have made it an attractive platform for candidate gene therapy development, which has been a boon for parallel uses in basic science. AAV induces minimal immune response compared to other viral vectors commonly used for somatic gene expression (specifically adenovirus and lentivirus). An additional advantage is the availability of multiple serotypes with varying tropisms, which addresses a significant shortfall in the application of system siRNA/ASOs: combining serotyped-limited tropism with tissue-restricted promoters allows a significantly nuanced expression of genes or shRNAs that target them. Furthermore, significant efforts have been made to improve and refine the naturally isolated serotypes by repeated rounds of in vivo selection and expansion, suggesting that even more tissue-selective versions will be available in the near future108. A significant validation of this platform is that AAV-driven expression of PCSK9 when coupled with a dietary stress (high fat diet) induces atherosclerosis in mice109.
Hepatocyte-expressed genes are particularly amenable to study using AAV: AAV8 transduction rates of hepatocytes are high and the relatively quiescent nature of the adult liver allows expression to be maintained for many months. One significant limitation is however that the packaging capacity of AAV is not very large: approximately 4.8 kb for AAV and about 2.4 kb for its self-complementarity derivative. While this precludes the use of AAV for some genes, the platform is nonetheless very powerful and widely used. To date, clinical AAV gene therapy has been approved only in Europe, and only for the treatment of familial lipoprotein lipase deficiency110, but homozygous familial hypercholesterolemia is a CVD-relevant condition that is an excellent candidate for an AAV-based therapeutic111.
Examples of functional genomics at selected CAD GWAS loci
Recent progress has been made in understanding the biology underlying some of the genes which have been implicated in risk for CAD. Being expressed in distinct cell types involved in atherosclerosis (Figure 3), these genes exert their effects in a cell type specific manner, which determines their specific contributions to disease. We provide here five selected examples of CAD GWAS loci for which the tools described above have been variably used to identify the causal gene at the locus and probe the underlying biology linking the gene to CAD.
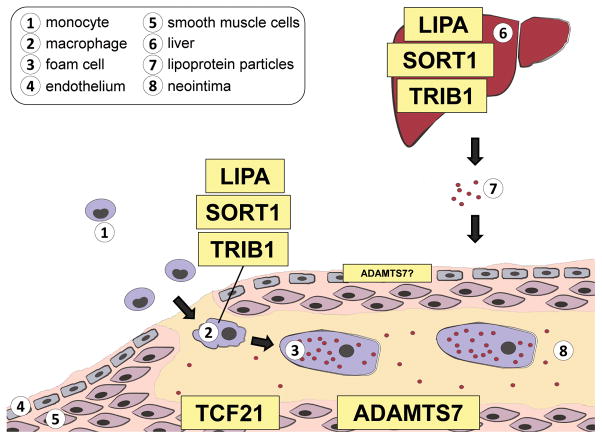
Figure 3. CHD GWAS risk genes are active in selective cell types involved in atherosclerosis.
Coronary Heart Disease follow-up studies have demonstrated roles for LIPA, SORT1 and TRIB1 as plasma lipid regulators in the liver, as well as in macrophages biology. Within the vessel wall, TCF21 is upregulated in de-differentiated smooth muscle cells which migrate to the forming fibrous cap. Adamts7 is also a regulator of smooth muscle migration but also a role in endothelial cells has been suggested
SORT1 (sortilin)
A compelling and now widely replicated novel locus associated with plasma lipid traits is the chromosome 1p13 locus, which had the lowest p-value of association in the Global Lipids Genetics Consortium (GLGC) study4. Notably, this locus had been independently and genome-wide significantly associated with MI/CAD, suggesting that it is of high importance to human cardiovascular health112–115. The locus harbors a high density of genes that might plausibly contribute to the phenotype, which necessitated thorough functional analysis. Fine-scale mapping of the locus refined the signal to a 6.1 kb genomic region containing six SNPs in high LD. Cloning of this region into a luciferase reporter construct, and the separate replacement of each SNP with the corresponding minor allele variant identified rs12740374 as the causal SNP. The mechanism by which it exerts its effect is due to the creation of a novel C/EBPa binding site, which was functionally validated by gel shift assays115. Nonetheless, the causal gene remained ambiguous: SORT1 and PSRC1 both had strong eQTLs in liver and a priori, neither gene could be eliminated as causal. To address this, the genes were separately overexpressed using the hepatocyte-tropic AAV8 system. SORT1 overexpression, but not that of PSRC1, substantially decreased plasma LDL-C in a mouse model with a humanized lipid profile (LahB, as described above), identifying SORT1 as the causal gene115.
Sortilin is a type I transmembrane multi-ligand receptor that is synthesized in the ER as a propeptide and is further processed to an active, mature form in the Golgi. It localizes to both the Golgi and plasma membranes, and facilitates trafficking of a variety of proteins bidirectionally between the Golgi lumen and the extracellular environment. Sortilin can also facilitate protein degradation by shuttling proteins from the Golgi to the lysosome through the endolysosome. Preliminary characterization of the role of sortilin in regulating very low-density lipoprotein (VLDL) secretion was performed through a series of Sort1 overexpression studies in hepatocytes and hepatocyte-like cell lines, and in a variety of mouse models. Sort1 expression was shown both to decrease VLDL secretion rates and increase plasma LDL turnover, thereby reducing plasma cholesterol additively116. Surface plasmon resonance demonstrated a high affinity pH-dependent interaction between sortilin and apoB-containing lipoproteins; and mutants defective in their ability to traffic to the endolysosomal system were used to show that sortilin serves as a bona fide cell surface LDL receptor. Wild-type sortilin binds LDL at the cell surface in an LDL receptor-independent manner and delivers the LDL to the endolysosomal system for degradation115, 116.
Based on the concordance of the human GWAS and mouse overexpression data it was hypothesized that genetic knockout or knockdown of Sort1 would have the opposite effect, increasing plasma cholesterol and VLDL secretion. However, the reported effects of the genetic loss of sortilin on VLDL secretion have been contradictory and perplexing: loss of sortilin has been shown in different studies to result in either increased and decreased VLDL secretion116–118. These discrepancies likely reflect the differences in models, methods of ablating sortilin function, lengths of time under diet-induced lipid overload, and technical approaches to measuring outcomes. Further complicating the story, Sort1−/− mice have recently been shown to be more insulin sensitive than wild-type mice on a high fat diet119, and loss of sortilin in 3T3-L1 adipocytes and C2C12 myotubes decreases insulin-stimulated glucose uptake due to decreased transport of Glut4 to the plasma membrane120, 121. While these data suggest that sortilin influences insulin signaling, the sortilin protein is itself regulated by insulin signaling122. Increased insulin sensitivity in extrahepatic tissues in Sort1−/− mice could be responsible for a decrease in FFA flux to the liver, a major contributor to hepatic lipid accumulation and a driver of VLDL secretion during insulin resistance. The role of sortilin in the tissues other than the liver under these conditions is unknown and confounds the interpretation of the effect of sortilin knockout and knockdown in the liver on VLDL secretion. In addition, there may be other aspects of extrahepatic sortilin biology that could influence disease risk: in a mouse model with a humanized plasma lipid profile, whole body knockout Sort1−/− had no effect on plasma lipids, but nonetheless was associated with decreased atherosclerosis. This effect was found to be attributable to the ability of sortilin to serve as a receptor of LDL: macrophages lacking sortilin have reduced LDL uptake, which led to decreased foam cell formation123.
The detailed molecular mechanisms by which sortilin influences the complex processes of hepatic and plasma lipid metabolism, VLDL secretion, and MI/CVD risk have not yet been elucidated, but under conditions consistent with western lifestyle it clearly impacts ApoB100 secretion, LDL clearance and foam cell formation. While the roles of sortilin in diverse CVD-relevant cell and tissue types have confounded the analysis of its biological function, this underscores the importance of the gene, and likely explains its robustness as a GWAS signal.
TRIB1 (Tribbles-1)
GWAS have consistently associated variants at the 8q24 locus containing the gene TRIB1 with multiple human metabolic phenotypes. The TRIB1 locus was first implicated in plasma lipid metabolism by two papers published simultaneously that showed non-coding variation in the TRIB1 gene locus was associated with circulating TG levels in humans2, 124. The landmark GLGC meta-analysis of more than 100,000 individuals further illustrated the importance of TRIB1 in lipid metabolism by associating the locus with not only TGs but also total cholesterol (TC), HDL cholesterol (HDL-C), LDL cholesterol (LDL-C), and CAD4, making TRIB1 the only novel locus from these studies to be associated with all four lipid traits and CAD. The plasma lipid associations were replicated in a GLGC follow-up meta-analysis of about 200,000 individuals125, while the CAD association was confirmed in two recent GWAS from the CARDIoGRAMplusC4D consortium, in which researchers investigated the association of genome-wide sequence variation with atherosclerosis regardless of plasma lipid phenotype1, 22. The significantly associated SNPs in all instances fall around 20 kb upstream of the TRIB1 gene, suggesting a role in the regulation of TRIB1 gene expression. One recent study showed that significantly associated SNPs in the TRIB1 locus alter the expression of a long non-coding RNA named TRIBAL, although the role of TRIBAL in any disease pathology is currently unclear126.
The TRIB1 gene encodes a protein known as Tribbles-1, which was originally identified in a drosophila mutagenesis screen which revealed that the protein Trbl (the drosophila homolog of TRIB1) participates in oogenesis via promoting the proteasomal degradation of String, Twine and Slbo, the latter of which is the Drosophila homolog of the human transcription factor CCAAT/enhancer binding protein alpha (C/EBPa)127–130. Subsequent work in the myeloblast 32D cell line showed that human Tribbles-1 can induce the proteasomal degradation of C/EBPa and C/EBPb by promoting their ubiquitination by the E3 ligase COP1 through direct binding to both targets and the ligase131, 132. This function is critical for Tribbles-mediated leukemogenesis133, 134, and also coordinates Tribbles regulation of macrophage polarization and differentiation135. Prior to the GWAS described above, TRIB1 had not been implicated in cardiometabolic disease pathology.
The human genetic findings have spurred a great deal of research into TRIB1 aimed at elucidating the mechanism through which it may participate in CAD pathogenesis. Studies using AAV-mediated overexpression of mouse Trib1 (AAV_mTrib1) to investigate this association found that increasing levels of hepatic Trib1 decreased plasma TC, HDL-C, LDL-C, and TG levels in a dose-dependent manner96. AAV-treated mice showed a decrease in hepatic lipogenic gene expression, and ex vivo studies of primary hepatocytes from those mice showed reduced cellular TG production and secretion. Furthermore, LAhB mice treated with AAV_mTrib1 had decreased plasma ApoB levels, and HepG2 cells overexpressing TRIB1 had decreased ApoB secretion. These data suggest that TRIB1 can modulate VLDL secretion from the liver, presumably by affecting the level of Triglycerides available for efficient VLDL assembly. More recent work from our group in a liver-specific KO of Trib1 (Trib1_LSKO) established that C/EBPa is the mechanistic link between TRIB1 and hepatic lipogenesis136. Trib1_LSKO mice have increased hepatic TG content, lipogenic gene expression, and de novo lipogenesis. They also display increased hepatic C/EBPa protein, and this increase is both necessary and sufficient to drive the lipogenic phenotype. The Trib1_LSKO mice also have increased plasma lipids, however this appears to be a C/EBPa-independent affect and suggests that TRIB1 regulates plasma lipid metabolism via other mechanisms independent of lipogenesis. TRIB1 has also recently been shown via in vitro overexpression assays to interact with the transcription factor ChREBP137 as well as SAP18, a component of the Sin3A-HDAC co-repressor complex138. The role of these interactions in vivo and the extent to which they all participate in plasma lipid regulation remains to be determined.
The association of the TRIB1 gene locus with CAD is likely driven in large part by its putative regulation of VLDL secretion. However, it remains possible that other mechanistic links between the gene and CAD contribute to this genetic association. The aforementioned role of TRIB1 in macrophage polarization135 is one potential link, as the M1/M2 status of macrophages in the lesion can contribute to plaque progression139.
Human genetics have implicated the TRIB1 locus in a host of other human phenotypes, including levels of circulating adiponectin140 and liver enzymes141 - an association functionally confirmed by the Trib1_LSKO mouse - as well as the onset of metabolic syndrome in humans142. Each of these human traits could by themselves contribute to CAD, either directly or indirectly. Thus it is possible that pleiotropic effects of TRIB1 contribute to CAD, and careful work in animal models of metabolic disease with TRIB1 tissue-specific deletion will be required to determine the specific contribution to disease burden by each specific function of TRIB1.
LIPA
A number of GWAS studies have identified LIPA as a novel locus for CAD18, 171, 172. Meta-analyses revealed that LIPA CAD risk alleles rs1412444T and rs2246833T (clustered in introns 2 and 3 in high linkage disequilibrium, r2=0.985) were associated with higher LIPA expression in monocytes143 but not in liver26, nor did they alter plasma lipids125. Fine mapping of the LIPA region by the CARDIoGRAM+C4D consortium22, 144 failed to reveal additional variants with stronger signals than the original GWAS SNPs, and rs2246833 had the strongest CAD association (P=4.9x10−12). Both aforementioned SNPs show strong H3K27Ac enrichment, and are in and near DNase I hypersensitivity site and TF binding sites (ENCODE),145, 146 suggesting possible regulatory roles.
LIPA encodes an enzyme called lysosomal acid lipase (LAL) which catalyzes the hydrolysis of cholesteryl ester (CE) and triglycerides in intracellular lysosomes after their internalization via receptor-mediated endocytosis of lipoprotein particles. Human LAL is encoded by the LIPA gene on chromosome 10q23.2–23.3147, and is a 46 kDa glycoprotein. After undergoing co-translational glycosylation in the endoplasmic reticulum and attachment of mannose-6-phosphate residues in the Golgi apparatus, LAL is targeted to pre-lysosomal compartments148, 149.
Prior to the GWAS discovery for CAD, loss-of-function (LOF) mutations in LIPA were identified as causes of rare lysosomal disorders. Wolman disease is an infantile-onset disorder with massive infiltration of CE/TG filled macrophages in multiple organs due to complete LIPA LOF. Cholesteryl ester storage disease (CESD) is a later-onset disorder with incomplete LIPA LOF mutations resulting in hepatomegaly, hyperlipidemia and premature atherosclerosis150, 151. The most common mutation seen in CESD patients is a splice junction mutation at exon 8 of LIPA, which leads to about 3 to 5 percent of normally spliced LAL protein and similar low levels of LAL activity152. Data of CESD fibroblasts suggest that LIPA deficiency leads to lysosomal CE accumulation that limits lysosomal FC release153, 154 and cytosolic cholesterol esterification21, 22, and impaired ATP binding cassette transporter A1 (ABCA1)-mediated cholesterol efflux155, 156. In a phase 3 trial of enzyme-replacement therapy in children and adults with lysosomal acid lipase deficiency, recombinant human LAL Sebelipase Alfa resulted in a reduction in multiple disease-related hepatic and lipid abnormalities157; with the long term effects of Sebelipase Alfa on cardiovascular events undetermined.
Although recombinant human LAL (rhLAL) enzyme replacement therapy is likely to exert protective effects against premature atherosclerosis in CESD patients, it is unclear what the effects of rhLAL treatment would be in CAD patients without LAL deficiency. Indeed, our understanding of the role of LIPA in the progression of atherosclerosis is far from complete. eQTL studies have suggested that the GWAS risk alleles for CAD are associated with increased LIPA mRNA in monocytes.143 How the higher LIPA mRNA in monocytes relates to increased risk of CAD is unclear: whether the GWAS CAD variants associated with higher LIPA mRNA is correlated with higher LAL protein levels or enzymatic activity, and if it is a true gain of function (GOF) mutation has yet to be determined. The GWAS CAD alleles are in LD with a missense coding variant (rs1051338) in the signal peptide that may alter post-translational trafficking and secretion, so it is possible that the CAD signal marks a loss of normal LAL processing and function despite higher mRNA. Thus, whether increased or decreased monocyte-macrophage activity of LAL beyond the normal cellular response, in the general population and in CAD patients is atherogenic or protective remains a completely open and controversial question.
A comprehensive understanding of the impact of LIPA on CAD pathogenesis relies on in vivo modeling. Lipa knockout mice (Lipa−/−) display shortened life span, tissue CE accumulation and hepatosplenomegaly.158 Furthermore, Lipa−/− accelerates atherosclerosis in the ApoE−/− hyperlipidemic mouse model.159 These findings recapitulate the pathological phenotypes of human LIPA LOF in CESD.160 Although systemic rhLAL administration reduces hyperlipidemia and atherosclerosis in Ldlr−/− mice,159 the atheroprotection was most likely attributable to the reduction in plasma lipids.159 Surprisingly, transgenic (Tg) mice with whole body Lipa overexpression apparently have elevated plasma VLDL-C and hepatocellular lipids on western diet.161 In context of the lack of association between GWAS CAD risk alleles and plasma lipid levels or liver LIPA expression,125 the whole body knockout or Tg mice do not serve as an appropriate model of the CAD-associated locus identified in GWAS studies. Indeed, because the CAD risk alleles are specifically associated with higher monocyte LIPA mRNA, it is now imperative to define the monocyte/macrophage-specific role of LIPA CAD risk alleles in vitro in macrophage function using isogenic hiPSC lines carrying risk or non-risk alleles with subsequently differentiation to macrophages - or in vivo in the progression of atherosclerosis using monocyte/macrophage-specific GOF of LIPA in mice models of atherosclerosis.
ADAMTS7
The association of the ADAMTS7 locus with CAD risk has been identified and replicated through GWAS.1, 162–164 This locus for coronary atherosclerosis was discovered in the PennCath cohort using angiographic CAD as the primary outcome,162 and subsequent studies have shown that ADAMTS7 also relates to MI.1, 163, 164 Its association is most robust for angiographic CAD, a marker of coronary atherosclerotic burden, suggesting that ADAMTS7 is likely to relate to clinical events through the development and progression of atherosclerosis. Recent findings in mouse vascular injury and atherosclerosis models165, 166 are consistent with such an action in the humans. Genetic variation at the ADAMTS7 locus has no relationship with traditional risk factors or mechanistic biomarkers;163, 164 hence the directional impact of ADAMTS7 expression on CAD risk and the underlying biological mechanisms have been unclear. Functional studies suggest that ADAMTS7, a metalloproteinase expressed in vascular smooth muscle cells (VSMC) and endothelial cells (ECs), is the probable causal proatherogenic gene at this locus.165–168 Briefly, the top CAD-risk SNPs at this locus are eQTLs for higher ADAMTS7 expression while allelic variation at a non-synonymous variant (rs3825807, Ser214Pro) in ADAMTS7 associates with reduced CAD risk and may impair ADAMTS7 function in VSMC (7). Our recent work demonstrated that deletion of Adamts7 is atheroprotective in both Ldlr−/− and ApoE−/− mouse models.166 Thus, blockade of ADAMTS7 expression or inhibition of its function presents novel therapeutic opportunities for prevention and treatment of CAD.
Understanding the relationship between CAD risk alleles at the ADAMTS7 chr15q21.1 region and expression levels of ADAMTS7 in human disease-relevant cells has not been straightforward. Interestingly, in available eQTL datasets with large sample sizes169 the lead SNPs from the PennCath (rs1994016), CARDIoGRAM (rs3825807), and C4D (rs4380028) GWAS studies demonstrate a significant association with ADAMTS7 expression and match the directionality and causality of in vivo data using mouse model, with the CAD risk alleles being associated with higher ADAMTS7 expression. However, currently there are no large eQTL or RNA-Seq based ASE data that provide adequate power to determine eQTL directionality in the most pertinent human vascular cells and tissues. Individual laboratories and the GTEx consortia170 are now generating datasets from a large enough sample pool of human vasculature to address this question. In the ENCODE Project,171 the NIH Roadmap Epigenomics Mapping data and in our own ChIP-Seq experiments, the top CAD SNPs fall in ADAMTS7 5′ and 3′ regions that overlap regulatory elements in VSMC and aortic tissues and are, for instance, close to binding sites for TCF21, a VSMC transcription factor that regulates coronary development and is itself a GWAS locus for CHD.163, 172, 173 In unpublished data, several of these regions have been found to have enhancer activity in rat A7r5 VSMC suggesting that this chr15q21.1 CHD SNPs may act on CHD by regulating human coronary arterial smooth muscle cells (HCASMC) ADAMTS7 expression.
ADAMTS7 (or the A disintegrin and metalloproteinase with thrombospondin motifs-7) is a member of the ADAMTS family of secreted zinc metalloproteases with characteristic protein domain composition including at least one thrombospondin type I repeat (TSPI).174–177 The family of ADAMTS proteases degrades extra-cellular matrix and several ADAMTS family members have been implicated in human diseases including thrombotic thrombocytopenic purpura (TTP),178 Weill-Marchesani syndrome179 and atherosclerosis.180 Unlike other metalloproteinases, ADAMTS family members demonstrate narrow substrate specificity due to their C-terminal exosites (13–16).174–177 Previous research on ADAMTS7 has mainly centered on its role in bone and cartilage growth because cartilage oligomeric matrix protein (COMP) has been identified as a substrate.181 ADAMTS7 can regulate endochondral bone formation through interactions with COMP. COMP is also expressed in VSMC and vasculature and additional studies with viral-mediated overexpression and knockdown in vivo and in vitro suggests that ADAMTS7 might modulate VSMC phenotype switching and migration via interactions with COMP.180
Most domains in human and mouse Adamts7 are highly conserved rendering the mouse as a useful model for actions in human disease. The first evidence that Adamts7 deficiency (Adamts7−/−) attenuates atherosclerosis in vivo, in both the ApoE−/− and Ldlr−/− mouse models, and Adamts7−/− confers a specific loss of VSMC migration in response to inflammatory signals was recently published.166 It has been shown that Adamts7−/− also reduces vascular response to mechanical injury.165, 166 Adamts7 gene expression was induced transiently in the mouse vasculature in response to stress, both in the wire injury model and in the atherosclerosis experiments, that TNFa induces Adamts7 expression in primary VSMCs, and that VSMC of Adamts7−/− mice show reduced TNFa-induced migration.166 Immunostaining in human diseased coronary arteries reveals colocalization of ADAMTS7 with cells positive for VSMC markers, and immunofluorescence in human aortic smooth muscle cells shows subcellular localization with leading edges of migrating VSMCs. These data suggests that Adamts7 modulates VSMC phenotype and migration during inflammatory stress and mechanical injury and that Adamts7 deficiency markedly reduces atherosclerotic lesions in hyperlipidemic mice.
Human eQTL interrogations reveal that common alleles that relate to lower CAD risk are also associated with reduced ADAMTS7 expression. This is consistent with rodent studies and supports a pro-atherogenic role of ADAMTS7 in humans. Because ADAMTS7 has narrow substrate specificity, it has promise as a potentially safe drug target. Thus, inhibition of ADAMTS7 is a potential novel therapeutic strategy for CAD in humans. Howevver, several important questions still need to be addressed in order to accelerate clinical and therapeutic translation related to this locus.
TCF21 (Pod1, capsulin, epicardin)
The TCF21 gene codes for a basic helix-loop-helix transcription factor known to bind cis-regulatory elements as heterodimers with TCF3 or TCF12. It is expressed in the mesenchyme of developing organs including the lung, kidney, gut and heart, and constitutive Tcf21 knockout mice die at birth due to missing alveoli in their lungs182. In addition, Tcf21 has been shown to play a role in vascular development: Tcf21 is highly expressed in the proepicardial organ which contains progenitor cells of coronary artery smooth muscle and endothelial cells and cardiac fibroblasts. It is necessary for epithelial-to-mesenchymal transition of epicardial fibroblasts and their subsequent migration into the cardiac interstitium183, 184.
The TCF21 gene locus has been linked to CAD risk by GWAS which first reported rs12190287 as the association lead SNP163. This polymorphism lies in the 3′UTR of one of the two TCF21 transcript variants. It has been shown to disrupt an AP-1 binding site inside an enhancer in vascular smooth muscle cells172. Interestingly, this variant also alters a mir-224 binding site inside a TCF21 transcript variant, suggesting additional miRNA-dependent regulation of TCF21 on the post-transcriptional level185. More recently, the 1000 Genomes based CARDIoGRAMplusC4D GWAS meta-analysis reported rs12202017, which lies 3.7kB upstream of the TCF21 gene within the TARID lncRNA locus, as the lowest p-value association SNP1. This locus also harbors a separately reported CAD association signal in Han Chinese, with rs12524865 as the strongest association SNP in this study186. The potential role of TARID in the vasculature has so far not been interrogated and presents an attractive target for future study. The contribution of TCF21 to disease appears to be coronary artery specific: a meta-analysis for shared susceptibility reported that the TCF21 association signal is confined to CAD and is not implicated in risk for ischemic stroke10.
The role of the Tcf21 protein in CAD has recently been investigated in vivo using a lacZ reporter and a lineage tracing model in mouse. These studies show that Tcf21 expressing cells migrate into the forming atherosclerotic lesion and contribute to the fibrous cap187. Further studies are needed to investigate how lack of TCF21 contributes to lesion size and composition. To identify TCF21 transcriptional target genes, ChIP-Seq studies have been carried out in HCASMCs173. Enrichment analysis showed that TCF21 binding sites are enriched for CAD GWAS association SNPs. This suggests a role of TCF21 as regulator of genomic loci such as ADAMTS7 conferring risk for atherosclerosis in coronary artery SMCs.
Summary
GWAS have provided a rich collection of CAD loci that suggest the existence of exciting new biology relevant to atherosclerosis that we never suspected but that require extensive functional follow-up studies. However, causality of a specific gene cannot be inferred solely based on proximity to a region of statistical association with disease. A thorough and meticulous annotation of the region with data generated in relevant cell types is paramount. Specific targeting of the SNP or region of association needs to be combined with the interrogation of changes in transcription of all genes in the haplotype block of interest. Far too often are candidate genes chosen for functional studies without solid evidence for their definitive association with the GWAS signal, and poorly characterized or long-distance genes are rarely followed up upon. Combinatorial effects of more than one gene within a locus, possibly across multiple tissues, could be one mechanism by which common, non protein-coding variation contributes to CAD. We may eventually be surprised by the complexity with which the implicated genomic regions modulate the CAD phenotype. In any case, it remains early days for the functional genomics of CAD GWAS loci. The likelihood is very high that fundamentally new biology regarding atherosclerosis and CAD will be learned through the interrogation of CAD GWAS loci. Furthermore, the hope remains that at least some of these new pathways relevant to CAD pathogenesis will yield new therapeutic targets for the prevention and treatment of CAD.
Supplementary Material
Acknowledgments
Sources of Funding: This work was supported by a Transatlantic Network of Excellence Award from Foundation Leducq.
Non-standard Abbreviations and Acronyms
- 3C
Chromosome Conformation Capture
- AAV
adeno-associated virus
- ASE
allele-specific expression
- ASO
antisense oligonucleotide
- ATAC-Seq
Assay for Transposase-Accessible Chromatin using Sequencing
- Cas9
CRISPR-associated protein 9
- ChIP-Seq
Chromatin immunoprecipitation with massively parallel sequencing
- CRISPR
type II clustered regularly interspaced short palindromic repeats
- CRISPRa
activation of gene transcription using the CRISPR/Cas9 technology
- CRISPRi
inhibition of gene transcription using the CRISPR/Cas9 technology
- eGFP
enhanced Green Fluorescent Protein
- eRNA
enhancer RNA
- eQTL
expression quantitative trait loci
- FAIRE
Formaldehyde-Assisted Isolation of Regulatory Elements
- GOF
gain of function
- gRNA
guide RNA
- GWAS
genome-wide association studies
- H3K4me1
Histone 3 Lysine 4 monomethylation
- H3K4me3
Histone 3 Lysine 4 trimethylation
- H3K27ac
Histone 3 Lysine 27 acetylation
- H3K36me3
Histone 3 Lysine 36 trimethylation
- HDR
homology-directed repair
- hiPSC
human induced pluripotent stem cell
- hnRNP
heterogeneous nuclear ribonucleoprotein
- iCRISPR
inducible genome editing using the CRISPR/Cas9 technology
- LD
linkage disequilibrium
- lincRNA
intergenic lncRNA
- lncRNA
long non-coding RNA
- LOF
loss-of function
- LSKO
liver-specific gene knockout
- NHEJ
non-homologous end-joining
- shRNA
short hairpin RNA
- SNP
single nucleotide polymorphism
Footnotes
Disclosures: None
References
- 1.Consortium CAD. A comprehensive 1000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nature genetics. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nature genetics. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kathiresan S, Musunuru K, Orho-Melander M. Defining the spectrum of alleles that contribute to blood lipid concentrations in humans. Current opinion in lipidology. 2008;19:122–127. doi: 10.1097/MOL.0b013e3282f70296. [DOI] [PubMed] [Google Scholar]
- 4.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, Bennett F, Bowden DW, Chakravarti A, Dreisbach A, Farlow DN, Folsom AR, Fornage M, Forrester T, Fox E, Haiman CA, Hartiala J, Harris TB, Hazen SL, Heckbert SR, Henderson BE, Hirschhorn JN, Keating BJ, Kritchevsky SB, Larkin E, Li M, Rudock ME, McKenzie CA, Meigs JB, Meng YA, Mosley TH, Newman AB, Newton-Cheh CH, Paltoo DN, Papanicolaou GJ, Patterson N, Post WS, Psaty BM, Qasim AN, Qu L, Rader DJ, Redline S, Reilly MP, Reiner AP, Rich SS, Rotter JI, Liu Y, Shrader P, Siscovick DS, Tang WH, Taylor HA, Tracy RP, Vasan RS, Waters KM, Wilks R, Wilson JG, Fabsitz RR, Gabriel SB, Kathiresan S, Boerwinkle E. Genome-wide association study of coronary heart disease and its risk factors in 8,090 african americans: The nhlbi care project. PLoS genetics. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Loss-of-function mutations in apoc3 and risk of ischemic vascular disease. The New England journal of medicine. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 7.Tg, Hdl Working Group of the Exome Sequencing Project NHL. Blood I, Crosby J, Peloso GM, Auer PL, Crosslin DR, Stitziel NO, Lange LA, Lu Y, Tang ZZ, Zhang H, Hindy G, Masca N, Stirrups K, Kanoni S, Do R, Jun G, Hu Y, Kang HM, Xue C, Goel A, Farrall M, Duga S, Merlini PA, Asselta R, Girelli D, Olivieri O, Martinelli N, Yin W, Reilly D, Speliotes E, Fox CS, Hveem K, Holmen OL, Nikpay M, Farlow DN, Assimes TL, Franceschini N, Robinson J, North KE, Martin LW, DePristo M, Gupta N, Escher SA, Jansson JH, Van Zuydam N, Palmer CN, Wareham N, Koch W, Meitinger T, Peters A, Lieb W, Erbel R, Konig IR, Kruppa J, Degenhardt F, Gottesman O, Bottinger EP, O’Donnell CJ, Psaty BM, Ballantyne CM, Abecasis G, Ordovas JM, Melander O, Watkins H, Orho-Melander M, Ardissino D, Loos RJ, McPherson R, Willer CJ, Erdmann J, Hall AS, Samani NJ, Deloukas P, Schunkert H, Wilson JG, Kooperberg C, Rich SS, Tracy RP, Lin DY, Altshuler D, Gabriel S, Nickerson DA, Jarvik GP, Cupples LA, Reiner AP, Boerwinkle E, Kathiresan S. Loss-of-function mutations in apoc3, triglycerides, and coronary disease. The New England journal of medicine. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Do R, Stitziel NO, Won HH, Jorgensen AB, Duga S, Angelica Merlini P, Kiezun A, Farrall M, Goel A, Zuk O, Guella I, Asselta R, Lange LA, Peloso GM, Auer PL, Project NES, Girelli D, Martinelli N, Farlow DN, DePristo MA, Roberts R, Stewart AF, Saleheen D, Danesh J, Epstein SE, Sivapalaratnam S, Hovingh GK, Kastelein JJ, Samani NJ, Schunkert H, Erdmann J, Shah SH, Kraus WE, Davies R, Nikpay M, Johansen CT, Wang J, Hegele RA, Hechter E, Marz W, Kleber ME, Huang J, Johnson AD, Li M, Burke GL, Gross M, Liu Y, Assimes TL, Heiss G, Lange EM, Folsom AR, Taylor HA, Olivieri O, Hamsten A, Clarke R, Reilly DF, Yin W, Rivas MA, Donnelly P, Rossouw JE, Psaty BM, Herrington DM, Wilson JG, Rich SS, Bamshad MJ, Tracy RP, Cupples LA, Rader DJ, Reilly MP, Spertus JA, Cresci S, Hartiala J, Tang WH, Hazen SL, Allayee H, Reiner AP, Carlson CS, Kooperberg C, Jackson RD, Boerwinkle E, Lander ES, Schwartz SM, Siscovick DS, McPherson R, Tybjaerg-Hansen A, Abecasis GR, Watkins H, Nickerson DA, Ardissino D, Sunyaev SR, O’Donnell CJ, Altshuler D, Gabriel S, Kathiresan S. Exome sequencing identifies rare ldlr and apoa5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khetarpal SA, Rader DJ. Triglyceride-rich lipoproteins and coronary artery disease risk: New insights from human genetics. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:e3–9. doi: 10.1161/ATVBAHA.114.305172. [DOI] [PubMed] [Google Scholar]
- 10.Dichgans M, Malik R, Konig IR, Rosand J, Clarke R, Gretarsdottir S, Thorleifsson G, Mitchell BD, Assimes TL, Levi C, O’Donnell CJ, Fornage M, Thorsteinsdottir U, Psaty BM, Hengstenberg C, Seshadri S, Erdmann J, Bis JC, Peters A, Boncoraglio GB, Marz W, Meschia JF, Kathiresan S, Ikram MA, McPherson R, Stefansson K, Sudlow C, Reilly MP, Thompson JR, Sharma P, Hopewell JC, Chambers JC, Watkins H, Rothwell PM, Roberts R, Markus HS, Samani NJ, Farrall M, Schunkert H Consortium M, Consortium CA, Consortium CD, International Stroke Genetics C. Shared genetic susceptibility to ischemic stroke and coronary artery disease: A genome-wide analysis of common variants. Stroke; a journal of cerebral circulation. 2014;45:24–36. doi: 10.1161/STROKEAHA.113.002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung HK, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui CC, Gomez-Skarmeta JL, Nobrega MA. Obesity-associated variants within fto form long-range functional connections with irx3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. Faire (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome research. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: From properties to genome-wide predictions. Nature reviews. Genetics. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 16.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer rnas and regulated transcriptional programs. Trends in biochemical sciences. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDaniell R, Lee BK, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, Keefe D, Collins FS, Willard HF, Lieb JD, Furey TS, Crawford GE, Iyer VR, Birney E. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, Lewellen N, Myrthil M, Gilad Y, Pritchard JK. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium GT. Human genomics. The genotype-tissue expression (gtex) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. Mrna-seq whole-transcriptome analysis of a single cell. Nature methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 21.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Single-cell chip-seq reveals cell subpopulations defined by chromatin state. Nature biotechnology. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium CAD; Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, Konig IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikainen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Muller-Nurasyid M, Mu TC, Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schafer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrieres J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kahonen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lee JY, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Tregouet DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvanen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimaki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O’Donnell C, Reilly MP, Marz W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ Consortium D, Consortium C, Wellcome Trust Case Control C. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature genetics. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boardman-Pretty F, Smith AJ, Cooper J, Palmen J, Folkersen L, Hamsten A, Catapano AL, Melander O, Price JF, Kumari M, Deanfield JE, Kivimaki M, Gertow K, Baragetti A, Norata GD, Humphries SE. Functional analysis of a carotid intima-media thickness locus implicates bcar1 and suggests a causal variant. Circulation. Cardiovascular genetics. 2015;8:696–706. doi: 10.1161/CIRCGENETICS.115.001062. [DOI] [PubMed] [Google Scholar]
- 24.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 25.Rockman MV, Kruglyak L. Genetics of global gene expression. Nature reviews. Genetics. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 26.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS biology. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Innocenti F, Cooper GM, Stanaway IB, Gamazon ER, Smith JD, Mirkov S, Ramirez J, Liu W, Lin YS, Moloney C, Aldred SF, Trinklein ND, Schuetz E, Nickerson DA, Thummel KE, Rieder MJ, Rettie AE, Ratain MJ, Cox NJ, Brown CD. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS genetics. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenawalt DM, Dobrin R, Chudin E, Hatoum IJ, Suver C, Beaulaurier J, Zhang B, Castro V, Zhu J, Sieberts SK, Wang S, Molony C, Heymsfield SB, Kemp DM, Reitman ML, Lum PY, Schadt EE, Kaplan LM. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome research. 2011;21:1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, Eleftheriadis M, Sinning CR, Schnabel RB, Lubos E, Mennerich D, Rust W, Perret C, Proust C, Nicaud V, Loscalzo J, Hubner N, Tregouet D, Munzel T, Ziegler A, Tiret L, Blankenberg S, Cambien F. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PloS one. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanoski CE, Che N, Yin F, Mai N, Pouldar D, Civelek M, Pan C, Lee S, Vakili L, Yang WP, Kayne P, Mungrue IN, Araujo JA, Berliner JA, Lusis AJ. Network for activation of human endothelial cells by oxidized phospholipids: A critical role of heme oxygenase 1. Circulation research. 2011;109:e27–41. doi: 10.1161/CIRCRESAHA.111.241869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nature reviews. Genetics. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 32.Braenne I, Civelek M, Vilne B, Di Narzo A, Johnson AD, Zhao Y, Reiz B, Codoni V, Webb TR, Foroughi Asl H, Hamby SE, Zeng L, Tregouet DA, Hao K, Topol EJ, Schadt EE, Yang X, Samani NJ, Bjorkegren JL, Erdmann J, Schunkert H, Lusis AJ Leducq Consortium CADGd. Prediction of causal candidate genes in coronary artery disease loci. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:2207–2217. doi: 10.1161/ATVBAHA.115.306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asl HF, Talukdar HA, Kindt ASD, Jain RK, Ermel R, Ruusalepp A, Nguyen KDH, Dobrin R, Reilly DF, Schunkert H, Samani NJ, Braenne I, Erdmann J, Melander O, Qi JL, Ivert T, Skogsberg J, Schadt EE, Michoel T, Bjorkegren JLM Consortium C. Expression quantitative trait loci acting across multiple tissues are enriched in inherited risk for coronary artery disease. Circ-Cardiovasc Gene. 2015;8:305. doi: 10.1161/CIRCGENETICS.114.000640. [DOI] [PubMed] [Google Scholar]
- 34.Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, Le J, Koka V, Lam KC, Gagne V, Dias J, Hoberman R, Montpetit A, Joly MM, Harvey EJ, Sinnett D, Beaulieu P, Hamon R, Graziani A, Dewar K, Harmsen E, Majewski J, Goring HH, Naumova AK, Blanchette M, Gunderson KL, Pastinen T. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nature genetics. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 35.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK. Understanding mechanisms underlying human gene expression variation with rna sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlof J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HP, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Geuvadis C, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Hasler R, Syvanen AC, van Ommen GJ, Brazma A, Meitinger T, Rosenstiel P, Guigo R, Gut IG, Estivill X, Dermitzakis ET. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goncalves A, Leigh-Brown S, Thybert D, Stefflova K, Turro E, Flicek P, Brazma A, Odom DT, Marioni JC. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome research. 2012;22:2376–2384. doi: 10.1101/gr.142281.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagarrigue S, Martin L, Hormozdiari F, Roux PF, Pan C, van Nas A, Demeure O, Cantor R, Ghazalpour A, Eskin E, Lusis AJ. Analysis of allele-specific expression in mouse liver by rna-seq: A comparison with cis-eqtl identified using genetic linkage. Genetics. 2013;195:1157–1166. doi: 10.1534/genetics.113.153882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, Yen CA, Lin S, Lin Y, Qiu Y, Xie W, Yue F, Hariharan M, Ray P, Kuan S, Edsall L, Yang H, Chi NC, Zhang MQ, Ecker JR, Ren B. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature genetics. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 42.Jager R, Migliorini G, Henrion M, Kandaswamy R, Speedy HE, Heindl A, Whiffin N, Carnicer MJ, Broome L, Dryden N, Nagano T, Schoenfelder S, Enge M, Yuan Y, Taipale J, Fraser P, Fletcher O, Houlston RS. Capture hi-c identifies the chromatin interactome of colorectal cancer risk loci. Nature communications. 2015;6:6178. doi: 10.1038/ncomms7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell hi-c reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science (New York, NY ) 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esteller M. Non-coding rnas in human disease. Nature reviews. Genetics. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 46.Wagschal A, Najafi-Shoushtari SH, Wang L, Goedeke L, Sinha S, deLemos AS, Black JC, Ramirez CM, Li Y, Tewhey R, Hatoum I, Shah N, Lu Y, Kristo F, Psychogios N, Vrbanac V, Lu YC, Hla T, de Cabo R, Tsang JS, Schadt E, Sabeti PC, Kathiresan S, Cohen DE, Whetstine J, Chung RT, Fernandez-Hernando C, Kaplan LM, Bernards A, Gerszten RE, Naar AM. Genome-wide identification of micrornas regulating cholesterol and triglyceride homeostasis. Nat Med. 2015;21:1290–1297. doi: 10.1038/nm.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulitsky I, Bartel DP. Lincrnas: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ning S, Wang P, Ye J, Li X, Li R, Zhao Z, Huo X, Wang L, Li F, Li X. A global map for dissecting phenotypic variants in human lincrnas. European journal of human genetics : EJHG. 2013;21:1128–1133. doi: 10.1038/ejhg.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ning S, Zhao Z, Ye J, Wang P, Zhi H, Li R, Wang T, Li X. Lincsnp: A database of linking disease-associated snps to human large intergenic non-coding rnas. BMC bioinformatics. 2014;15:152. doi: 10.1186/1471-2105-15-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gong J, Liu W, Zhang J, Miao X, Guo AY. Lncrnasnp: A database of snps in lncrnas and their potential functions in human and mouse. Nucleic acids research. 2015;43:D181–186. doi: 10.1093/nar/gku1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, Almeida R, Zhernakova A, Reinmaa E, Vosa U, Hofker MH, Fehrmann RS, Fu J, Withoff S, Metspalu A, Franke L, Wijmenga C. Human disease-associated genetic variation impacts large intergenic non-coding rna expression. PLoS genetics. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris KV, Mattick JS. The rise of regulatory rna. Nature reviews. Genetics. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding rna. Journal of molecular biology. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Augui S, Nora EP, Heard E. Regulation of x-chromosome inactivation by the x-inactivation centre. Nature reviews. Genetics. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 55.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, Schier AF. Systematic identification of long noncoding rnas expressed during zebrafish embryogenesis. Genome research. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. Lincrnas act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincrnas in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding rna: An emerging paradigm of cancer research. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 59.Ziats MN, Rennert OM. Aberrant expression of long noncoding rnas in autistic brain. Journal of molecular neuroscience : MN. 2013;49:589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien HC, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HS, Quertermous T, Chang CP. A long noncoding rna protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H, Schuler G, Thiery J, Teupser D. Anril expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 62.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Saito S, Nakamura Y, Tanaka T. Identification of a novel non-coding rna, miat, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 63.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding rna required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. The long noncoding rna chrf regulates cardiac hypertrophy by targeting mir-489. Circ Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 65.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding rna malat1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 66.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C. Lincrna-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding rna mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding rna thril regulates tnfalpha expression through its interaction with hnrnpl. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, Yuan B, Kellis M, Lodish HF, Rinn JL. Long noncoding rnas regulate adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding rna firre. Nature structural & molecular biology. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding rna transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Molecular cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao T, Liu L, Li H, Sun Y, Luo H, Li T, Wang S, Dalton S, Zhao RC, Chen R. Long noncoding rna adinr regulates adipogenesis by transcriptionally activating c/ebpalpha. Stem cell reports. 2015 doi: 10.1016/j.stemcr.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alvarez-Dominguez JR, Bai Z, Xu D, Yuan B, Lo KA, Yoon MJ, Lim YC, Knoll M, Slavov N, Chen S, Chen P, Lodish HF, Sun L. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding rna regulators of brown adipocyte development. Cell metabolism. 2015;21:764–776. doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pasmant E, Sabbagh A, Vidaud M, Bieche I. Anril, a long, noncoding rna, is an unexpected major hotspot in gwas. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 75.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an ink4/arf-associated non-coding rna correlates with atherosclerosis risk. PLoS genetics. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The gencode v7 catalog of human long noncoding rnas: Analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding rnas reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using crispr/cas systems. Science (New York, NY) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. Rna-guided human genome engineering via cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaj T, Gersbach CA, Barbas CF., 3rd Zfn, talen, and crispr/cas-based methods for genome engineering. Trends in biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-rna-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, NY ) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. Crispr-mediated modular rna-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. Crispr rna-guided activation of endogenous human genes. Nature methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F. Rna-guided gene activation by crispr-cas9-based transcription factors. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mali P, Esvelt KM. Cas9 as a versatile tool for engineering biology. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beaudoin M, Gupta RM, Won HH, Lo KS, Do R, Henderson CA, Lavoie-St-Amour C, Langlois S, Rivas D, Lehoux S, Kathiresan S, Tardif JC, Musunuru K, Lettre G. Myocardial infarction-associated snp at 6p24 interferes with mef2 binding and associates with phactr1 expression levels in human coronary arteries. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:1472–1479. doi: 10.1161/ATVBAHA.115.305534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An icrispr platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell stem cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient rna-guided genome editing in human cells via delivery of purified cas9 ribonucleoproteins. Genome research. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH. Chemically modified guide rnas enhance crispr-cas genome editing in human primary cells. Nature biotechnology. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with crispr-cas9 by inhibition of nonhomologous end joining. Nature biotechnology. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robert F, Barbeau M, Ethier S, Dostie J, Pelletier J. Pharmacological inhibition of DNA-pk stimulates cas9-mediated genome editing. Genome medicine. 2015;7:93. doi: 10.1186/s13073-015-0215-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chu VT, Weber T, Wefers B. Increasing the efficiency of homology-directed repair for crispr-cas9-induced precise gene editing in mammalian cells. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 93.Miyaoka Y, Chan AH, Judge LM, Yoo J, Huang M, Nguyen TD, Lizarraga PP, So PL, Conklin BR. Isolation of single-base genome-edited human ips cells without antibiotic selection. Nature methods. 2014;11:291–293. doi: 10.1038/nmeth.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by crispr/cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by crispr/cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burkhardt R, Toh SA, Lagor WR, Birkeland A, Levin M, Li X, Robblee M, Fedorov VD, Yamamoto M, Satoh T, Akira S, Kathiresan S, Breslow JL, Rader DJ. Trib1 is a lipid- and myocardial infarction-associated gene that regulates hepatic lipogenesis and vldl production in mice. The Journal of clinical investigation. 2010;120:4410–4414. doi: 10.1172/JCI44213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microrna targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huntzinger E, Izaurralde E. Gene silencing by micrornas: Contributions of translational repression and mrna decay. Nature reviews. Genetics. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 99.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of drosophila argonautes and its involvement in risc formation. Genes & development. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microrna/short hairpin rna pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 101.Mockenhaupt S, Grosse S, Rupp D, Bartenschlager R, Grimm D. Alleviation of off-target effects from vector-encoded shrnas via codelivered rna decoys. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E4007–4016. doi: 10.1073/pnas.1510476112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grimm D, Wang L, Lee JS, Schurmann N, Gu S, Borner K, Storm TA, Kay MA. Argonaute proteins are key determinants of rnai efficacy, toxicity, and persistence in the adult mouse liver. The Journal of clinical investigation. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Giering JC, Grimm D, Storm TA, Kay MA. Expression of shrna from a tissue-specific pol ii promoter is an effective and safe rnai therapeutic. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 104.Wang J, Lu Z, Wientjes MG, Au JL. Delivery of sirna therapeutics: Barriers and carriers. The AAPS journal. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fitzgerald K, Frank-Kamenetsky M, Shulga-Morskaya S, Liebow A, Bettencourt BR, Sutherland JE, Hutabarat RM, Clausen VA, Karsten V, Cehelsky J, Nochur SV, Kotelianski V, Horton J, Mant T, Chiesa J, Ritter J, Munisamy M, Vaishnaw AK, Gollob JA, Simon A. Effect of an rna interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (pcsk9) and the concentration of serum ldl cholesterol in healthy volunteers: A randomised, single-blind, placebo-controlled, phase 1 trial. Lancet. 2014;383:60–68. doi: 10.1016/S0140-6736(13)61914-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Geary RS, Baker BF, Crooke ST. Clinical and preclinical pharmacokinetics and pharmacodynamics of mipomersen (kynamro((r))): A second-generation antisense oligonucleotide inhibitor of apolipoprotein b. Clinical pharmacokinetics. 2015;54:133–146. doi: 10.1007/s40262-014-0224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lagor WR, Johnston JC, Lock M, Vandenberghe LH, Rader DJ. Adeno-associated viruses as liver-directed gene delivery vehicles: Focus on lipoprotein metabolism. Methods in molecular biology. 2013;1027:273–307. doi: 10.1007/978-1-60327-369-5_13. [DOI] [PubMed] [Google Scholar]
- 108.Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, Kay MA. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. Journal of virology. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Roche-Molina M, Sanz-Rosa D, Cruz FM, Garcia-Prieto J, Lopez S, Abia R, Muriana FJ, Fuster V, Ibanez B, Bernal JA. Induction of sustained hypercholesterolemia by single adeno-associated virus-mediated gene transfer of mutant hpcsk9. Arteriosclerosis, thrombosis, and vascular biology. 2015;35:50–59. doi: 10.1161/ATVBAHA.114.303617. [DOI] [PubMed] [Google Scholar]
- 110.Ferreira V, Petry H, Salmon F. Immune responses to aav-vectors, the glybera example from bench to bedside. Frontiers in immunology. 2014;5:82. doi: 10.3389/fimmu.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Somanathan S, Jacobs F, Wang Q, Hanlon AL, Wilson JM, Rader DJ. Aav vectors expressing ldlr gain-of-function variants demonstrate increased efficacy in mouse models of familial hypercholesterolemia. Circulation research. 2014;115:591–599. doi: 10.1161/CIRCRESAHA.115.304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.O’Donnell CJ, Kavousi M, Smith AV, Kardia SL, Feitosa MF, Hwang SJ, Sun YV, Province MA, Aspelund T, Dehghan A, Hoffmann U, Bielak LF, Zhang Q, Eiriksdottir G, van Duijn CM, Fox CS, de Andrade M, Kraja AT, Sigurdsson S, Elias-Smale SE, Murabito JM, Launer LJ, van der Lugt A, Kathiresan S, Krestin GP, Herrington DM, Howard TD, Liu Y, Post W, Mitchell BD, O’Connell JR, Shen H, Shuldiner AR, Altshuler D, Elosua R, Salomaa V, Schwartz SM, Siscovick DS, Voight BF, Bis JC, Glazer NL, Psaty BM, Boerwinkle E, Heiss G, Blankenberg S, Zeller T, Wild PS, Schnabel RB, Schillert A, Ziegler A, Munzel TF, White CC, Rotter JI, Nalls M, Oudkerk M, Johnson AD, Newman AB, Uitterlinden AG, Massaro JM, Cunningham J, Harris TB, Hofman A, Peyser PA, Borecki IB, Cupples LA, Gudnason V, Witteman JC Consortium CA. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011;124:2855–2864. doi: 10.1161/CIRCULATIONAHA.110.974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saade S, Cazier JB, Ghassibe-Sabbagh M, Youhanna S, Badro DA, Kamatani Y, Hager J, Yeretzian JS, El-Khazen G, Haber M, Salloum AK, Douaihy B, Othman R, Shasha N, Kabbani S, Bayeh HE, Chammas E, Farrall M, Gauguier D, Platt DE, Zalloua PA Consortium F. Large scale association analysis identifies three susceptibility loci for coronary artery disease. PloS one. 2011;6:e29427. doi: 10.1371/journal.pone.0029427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang AZ, Li L, Zhang B, Shen GQ, Wang QK. Association of snp rs17465637 on chromosome 1q41 and rs599839 on 1p13.3 with myocardial infarction in an american caucasian population. Annals of human genetics. 2011;75:475–482. doi: 10.1111/j.1469-1809.2011.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ. From noncoding variant to phenotype via sort1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Strong A, Ding Q, Edmondson AC, Millar JS, Sachs KV, Li X, Kumaravel A, Wang MY, Ai D, Guo L, Alexander ET, Nguyen D, Lund-Katz S, Phillips MC, Morales CR, Tall AR, Kathiresan S, Fisher EA, Musunuru K, Rader DJ. Hepatic sortilin regulates both apolipoprotein b secretion and ldl catabolism. The Journal of clinical investigation. 2012;122:2807–2816. doi: 10.1172/JCI63563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ai D, Baez JM, Jiang H, Conlon DM, Hernandez-Ono A, Frank-Kamenetsky M, Milstein S, Fitzgerald K, Murphy AJ, Woo CW, Strong A, Ginsberg HN, Tabas I, Rader DJ, Tall AR. Activation of er stress and mtorc1 suppresses hepatic sortilin-1 levels in obese mice. The Journal of clinical investigation. 2012;122:1677–1687. doi: 10.1172/JCI61248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kjolby M, Andersen OM, Breiderhoff T, Fjorback AW, Pedersen KM, Madsen P, Jansen P, Heeren J, Willnow TE, Nykjaer A. Sort1, encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab. 2010;12:213–223. doi: 10.1016/j.cmet.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 119.Rabinowich L, Fishman S, Hubel E, Thurm T, Park WJ, Pewzner-Jung Y, Saroha A, Erez N, Halpern Z, Futerman AH, Zvibel I. Sortilin deficiency improves the metabolic phenotype and reduces hepatic steatosis of mice subjected to diet-induced obesity. Journal of hepatology. 2015;62:175–181. doi: 10.1016/j.jhep.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 120.Shi J, Kandror KV. Sortilin is essential and sufficient for the formation of glut4 storage vesicles in 3t3-l1 adipocytes. Developmental cell. 2005;9:99–108. doi: 10.1016/j.devcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 121.Tsuchiya Y, Hatakeyama H, Emoto N, Wagatsuma F, Matsushita S, Kanzaki M. Palmitate-induced down-regulation of sortilin and impaired glut4 trafficking in c2c12 myotubes. The Journal of biological chemistry. 2010;285:34371–34381. doi: 10.1074/jbc.M110.128520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li J, Matye DJ, Li T. Insulin resistance induces posttranslational hepatic sortilin 1 degradation in mice. The Journal of biological chemistry. 2015;290:11526–11536. doi: 10.1074/jbc.M115.641225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Patel KM, Strong A, Tohyama J, Jin X, Morales CR, Billheimer J, Millar J, Kruth H, Rader DJ. Macrophage sortilin promotes ldl uptake, foam cell formation, and atherosclerosis. Circulation research. 2015;116:789–796. doi: 10.1161/CIRCRESAHA.116.305811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nature genetics. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Global Lipids Genetics C. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Doring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR. Discovery and refinement of loci associated with lipid levels. Nature genetics. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Douvris A, Soubeyrand S, Naing T, Martinuk A, Nikpay M, Williams A, Buick J, Yauk C, McPherson R. Functional analysis of the trib1 associated locus linked to plasma triglycerides and coronary artery disease. Journal of the American Heart Association. 2014;3:e000884. doi: 10.1161/JAHA.114.000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rorth P, Szabo K, Texido G. The level of c/ebp protein is critical for cell migration during drosophila oogenesis and is tightly controlled by regulated degradation. Molecular cell. 2000;6:23–30. doi: 10.1016/s1097-2765(05)00008-0. [DOI] [PubMed] [Google Scholar]
- 128.Seher TC, Leptin M. Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during drosophila gastrulation. Current biology : CB. 2000;10:623–629. doi: 10.1016/s0960-9822(00)00502-9. [DOI] [PubMed] [Google Scholar]
- 129.Grosshans J, Wieschaus E. A genetic link between morphogenesis and cell division during formation of the ventral furrow in drosophila. Cell. 2000;101:523–531. doi: 10.1016/s0092-8674(00)80862-4. [DOI] [PubMed] [Google Scholar]
- 130.Mata J, Curado S, Ephrussi A, Rorth P. Tribbles coordinates mitosis and morphogenesis in drosophila by regulating string/cdc25 proteolysis. Cell. 2000;101:511–522. doi: 10.1016/s0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 131.Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, Rodriguez CG, Maillard I, Tobias JW, Valk P, Carroll M, Aster JC, Delwel R, Pear WS. Tribbles homolog 2 inactivates c/ebpalpha and causes acute myelogenous leukemia. Cancer cell. 2006;10:401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dedhia PH, Keeshan K, Uljon S, Xu L, Vega ME, Shestova O, Zaks-Zilberman M, Romany C, Blacklow SC, Pear WS. Differential ability of tribbles family members to promote degradation of c/ebpalpha and induce acute myelogenous leukemia. Blood. 2010;116:1321–1328. doi: 10.1182/blood-2009-07-229450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Keeshan K, Bailis W, Dedhia PH, Vega ME, Shestova O, Xu L, Toscano K, Uljon SN, Blacklow SC, Pear WS. Transformation by tribbles homolog 2 (trib2) requires both the trib2 kinase domain and cop1 binding. Blood. 2010;116:4948–4957. doi: 10.1182/blood-2009-10-247361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yoshida A, Kato JY, Nakamae I, Yoneda-Kato N. Cop1 targets c/ebpalpha for degradation and induces acute myeloid leukemia via trib1. Blood. 2013;122:1750–1760. doi: 10.1182/blood-2012-12-476101. [DOI] [PubMed] [Google Scholar]
- 135.Satoh T, Kidoya H, Naito H, Yamamoto M, Takemura N, Nakagawa K, Yoshioka Y, Morii E, Takakura N, Takeuchi O, Akira S. Critical role of trib1 in differentiation of tissue-resident m2-like macrophages. Nature. 2013;495:524–528. doi: 10.1038/nature11930. [DOI] [PubMed] [Google Scholar]
- 136.Bauer RC, Sasaki M, Cohen DM, Cui J, Smith MA, Yenilmez BO, Steger DJ, Rader DJ. Tribbles-1 regulates hepatic lipogenesis through posttranscriptional regulation of c/ebpalpha. The Journal of clinical investigation. 2015;125:3809–3818. doi: 10.1172/JCI77095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ishizuka Y, Nakayama K, Ogawa A, Makishima S, Boonvisut S, Hirao A, Iwasaki Y, Yada T, Yanagisawa Y, Miyashita H, Takahashi M, Iwamoto S Jichi Medical University Promotion Team of Large-Scale Human Genome Bank for All over J. Trib1 downregulates hepatic lipogenesis and glycogenesis via multiple molecular interactions. Journal of molecular endocrinology. 2014;52:145–158. doi: 10.1530/JME-13-0243. [DOI] [PubMed] [Google Scholar]
- 138.Makishima S, Boonvisut S, Ishizuka Y, Watanabe K, Nakayama K, Iwamoto S. Sin3a-associated protein, 18 kda, a novel binding partner of trib1, regulates mttp expression. Journal of lipid research. 2015;56:1145–1152. doi: 10.1194/jlr.M057802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hanna RN, Shaked I, Hubbeling HG, Punt JA, Wu R, Herrley E, Zaugg C, Pei H, Geissmann F, Ley K, Hedrick CC. Nr4a1 (nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circulation research. 2012;110:416–427. doi: 10.1161/CIRCRESAHA.111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dastani Z, Hivert MF, Timpson N, Perry JR, Yuan X, Scott RA, Henneman P, Heid IM, Kizer JR, Lyytikainen LP, Fuchsberger C, Tanaka T, Morris AP, Small K, Isaacs A, Beekman M, Coassin S, Lohman K, Qi L, Kanoni S, Pankow JS, Uh HW, Wu Y, Bidulescu A, Rasmussen-Torvik LJ, Greenwood CM, Ladouceur M, Grimsby J, Manning AK, Liu CT, Kooner J, Mooser VE, Vollenweider P, Kapur KA, Chambers J, Wareham NJ, Langenberg C, Frants R, Willems-Vandijk K, Oostra BA, Willems SM, Lamina C, Winkler TW, Psaty BM, Tracy RP, Brody J, Chen I, Viikari J, Kahonen M, Pramstaller PP, Evans DM, St Pourcain B, Sattar N, Wood AR, Bandinelli S, Carlson OD, Egan JM, Bohringer S, van Heemst D, Kedenko L, Kristiansson K, Nuotio ML, Loo BM, Harris T, Garcia M, Kanaya A, Haun M, Klopp N, Wichmann HE, Deloukas P, Katsareli E, Couper DJ, Duncan BB, Kloppenburg M, Adair LS, Borja JB, Mu TC, Wilson JG, Musani S, Guo X, Johnson T, Semple R, Teslovich TM, Allison MA, Redline S, Buxbaum SG, Mohlke KL, Meulenbelt I, Ballantyne CM, Dedoussis GV, Hu FB, Liu Y, Paulweber B, Spector TD, Slagboom PE, Ferrucci L, Jula A, Perola M, Raitakari O, Florez JC, Salomaa V, Eriksson JG, Frayling TM, Hicks AA, Lehtimaki T, Smith GD, Siscovick DS, Kronenberg F, van Duijn C, Loos RJ, Waterworth DM, Meigs JB, Dupuis J, Richards JB, Voight BF, Scott LJ, Steinthorsdottir V, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Hofmann OM, Segre AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bostrom KB, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Crawford G, Doney AS, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PR, Jorgensen T, Kao WH, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Petersen AK, Platou C, Proenca C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparso T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, van Vliet-Ostaptchouk JV, Walters GB, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Morris AD, Palmer CN, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Pedersen O, Barroso I, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn CM, Stefansson K, Altshuler D, Boehnke M, McCarthy MI, Soranzo N, Wheeler E, Glazer NL, Bouatia-Naji N, Magi R, Randall J, Elliott P, Rybin D, Dehghan A, Hottenga JJ, Song K, Goel A, Lajunen T, Doney A, Cavalcanti-Proenca C, Kumari M, Timpson NJ, Zabena C, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Roccasecca RM, Pattou F, Sethupathy P, Ariyurek Y, Barter P, Beilby JP, Ben-Shlomo Y, Bergmann S, Bochud M, Bonnefond A, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Crisponi L, Day IN, de Geus EJ, Delplanque J, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Grundy S, Gwilliam R, Hallmans G, Hammond N, Han X, Hartikainen AL, Hayward C, Heath SC, Hercberg S, Hillman DR, Hingorani AD, Hui J, Hung J, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Mahley R, Mangino M, Martinez-Larrad MT, McAteer JB, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Mukherjee S, Naitza S, Neville MJ, Orru M, Pakyz R, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tonjes A, Uitterlinden AG, van Dijk KW, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Ward KL, Watkins H, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Global BPC, Borecki IB, Meneton P, Magnusson PK, Nathan DM, Williams GH, Silander K, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Serrano-Rios M, Lind L, Palmer LJ, Hu FBs, Franks PW, Ebrahim S, Marmot M, Kao WH, Pramstaller PP, Wright AF, Stumvoll M, Hamsten A, Procardis C, Buchanan TA, Valle TT, Rotter JI, Penninx BW, Boomsma DI, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Peltonen L, Mooser V, Sladek R, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Chasman DI, Johansen CT, Fouchier SW, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Feitosa MF, Orho-Melander M, Melander O, Li X, Li M, Cho YS, Go MJ, Kim YJ, Lee JY, Park T, Kim K, Sim X, Ong RT, Croteau-Chonka DC, Lange LA, Smith JD, Ziegler A, Zhang W, Zee RY, Whitfield JB, Thompson JR, Surakka I, Spector TD, Smit JH, Sinisalo J, Scott J, Saharinen J, Sabatti C, Rose LM, Roberts R, Rieder M, Parker AN, Pare G, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, McArdle W, Masson D, Martin NG, Marroni F, Lucas G, Luben R, Lokki ML, Lettre G, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Konig IR, Khaw KT, Kaplan LM, Johansson A, Janssens AC, Igl W, Hovingh GK, Hengstenberg C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Groop LC, Gonzalez E, Freimer NB, Erdmann J, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Faire U, Crawford G, Chen YD, Caulfield MJ, Boekholdt SM, Assimes TL, Quertermous T, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Taylor HA, Jr, Gabriel SB, Holm H, Gudnason V, Krauss RM, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Strachan DP, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, Kathiresan S investigators M, Investigators G, Consortium G, Consortium D, Consortium G, Consortium D, Consortium M. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: A multi-ethnic meta-analysis of 45,891 individuals. PLoS genetics. 2012;8:e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, Eiriksdottir G, Garcia ME, Launer LJ, Nalls MA, Clark JM, Mitchell BD, Shuldiner AR, Butler JL, Tomas M, Hoffmann U, Hwang SJ, Massaro JM, O’Donnell CJ, Sahani DV, Salomaa V, Schadt EE, Schwartz SM, Siscovick DS, Nash CRN, Voight BF, Carr JJ, Feitosa MF, Harris TB, Fox CS, Smith AV, Kao WH, Hirschhorn JN, Borecki IB Consortium G, Consortium G, Investigators M. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS genetics. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kraja AT, Chasman DI, North KE, Reiner AP, Yanek LR, Kilpelainen TO, Smith JA, Dehghan A, Dupuis J, Johnson AD, Feitosa MF, Tekola-Ayele F, Chu AY, Nolte IM, Dastani Z, Morris A, Pendergrass SA, Sun YV, Ritchie MD, Vaez A, Lin H, Ligthart S, Marullo L, Rohde R, Shao Y, Ziegler MA, Im HK, Schnabel RB, Jorgensen T, Jorgensen ME, Hansen T, Pedersen O, Stolk RP, Snieder H, Hofman A, Uitterlinden AG, Franco OH, Ikram MA, Richards JB, Rotimi C, Wilson JG, Lange L, Ganesh SK, Nalls M, Rasmussen-Torvik LJ, Pankow JS, Coresh J, Tang W, Linda Kao WH, Boerwinkle E, Morrison AC, Ridker PM, Becker DM, Rotter JI, Kardia SL, Loos RJ, Larson MG, Hsu YH, Province MA, Tracy R, Voight BF, Vaidya D, O’Donnell CJ, Benjamin EJ, Alizadeh BZ, Prokopenko I, Meigs JB, Borecki IB Cross Consortia Pleiotropy G, Cohorts for Heart a, Aging Research in Genetic E, Genetic Investigation of Anthropometric Traits C, Global Lipids Genetics C, Meta-Analyses of G, Insulin-related traits C, Global BC, Consortium AD, Women’s Genome Health S, Howard University Family S. Pleiotropic genes for metabolic syndrome and inflammation. Molecular genetics and metabolism. 2014;112:317–338. doi: 10.1016/j.ymgme.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wild PS, Zeller T, Schillert A, Szymczak S, Sinning CR, Deiseroth A, Schnabel RB, Lubos E, Keller T, Eleftheriadis MS, Bickel C, Rupprecht HJ, Wilde S, Rossmann H, Diemert P, Cupples LA, Perret C, Erdmann J, Stark K, Kleber ME, Epstein SE, Voight BF, Kuulasmaa K, Li M, Schafer AS, Klopp N, Braund PS, Sager HB, Demissie S, Proust C, Konig IR, Wichmann HE, Reinhard W, Hoffmann MM, Virtamo J, Burnett MS, Siscovick D, Wiklund PG, Qu L, El Mokthari NE, Thompson JR, Peters A, Smith AV, Yon E, Baumert J, Hengstenberg C, Marz W, Amouyel P, Devaney J, Schwartz SM, Saarela O, Mehta NN, Rubin D, Silander K, Hall AS, Ferrieres J, Harris TB, Melander O, Kee F, Hakonarson H, Schrezenmeir J, Gudnason V, Elosua R, Arveiler D, Evans A, Rader DJ, Illig T, Schreiber S, Bis JC, Altshuler D, Kavousi M, Witteman JC, Uitterlinden AG, Hofman A, Folsom AR, Barbalic M, Boerwinkle E, Kathiresan S, Reilly MP, O’Donnell CJ, Samani NJ, Schunkert H, Cambien F, Lackner KJ, Tiret L, Salomaa V, Munzel T, Ziegler A, Blankenberg S. A genome-wide association study identifies lipa as a susceptibility gene for coronary artery disease. Circulation. Cardiovascular genetics. 2011;4:403–412. doi: 10.1161/CIRCGENETICS.110.958728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, Burtt NP, Fuchsberger C, Li Y, Erdmann J, Frayling TM, Heid IM, Jackson AU, Johnson T, Kilpelainen TO, Lindgren CM, Morris AP, Prokopenko I, Randall JC, Saxena R, Soranzo N, Speliotes EK, Teslovich TM, Wheeler E, Maguire J, Parkin M, Potter S, Rayner NW, Robertson N, Stirrups K, Winckler W, Sanna S, Mulas A, Nagaraja R, Cucca F, Barroso I, Deloukas P, Loos RJ, Kathiresan S, Munroe PB, Newton-Cheh C, Pfeufer A, Samani NJ, Schunkert H, Hirschhorn JN, Altshuler D, McCarthy MI, Abecasis GR, Boehnke M. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS genetics. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Consortium EP; Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Program NCS, Broad I, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ Baylor College of Medicine Human Genome Sequencing C, Washington University Genome Sequencing C, Children’s Hospital Oakland Research I. Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Anderson RA, Rao N, Byrum RS, Rothschild CB, Bowden DW, Hayworth R, Pettenati M. In situ localization of the genetic locus encoding the lysosomal acid lipase/cholesteryl esterase (lipa) deficient in wolman disease to chromosome 10q23.2-q23.3. Genomics. 1993;15:245–247. doi: 10.1006/geno.1993.1052. [DOI] [PubMed] [Google Scholar]
- 148.Sando GN, Henke VL. Recognition and receptor-mediated endocytosis of the lysosomal acid lipase secreted by cultured human fibroblasts. Journal of lipid research. 1982;23:114–123. [PubMed] [Google Scholar]
- 149.Sheriff S, Du H, Grabowski GA. Characterization of lysosomal acid lipase by site-directed mutagenesis and heterologous expression. The Journal of biological chemistry. 1995;270:27766–27772. doi: 10.1074/jbc.270.46.27766. [DOI] [PubMed] [Google Scholar]
- 150.Bernstein DL, Hulkova H, Bialer MG, Desnick RJ. Cholesteryl ester storage disease: Review of the findings in 135 reported patients with an underdiagnosed disease. Journal of hepatology. 2013;58:1230–1243. doi: 10.1016/j.jhep.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 151.Du H, Schiavi S, Levine M, Mishra J, Heur M, Grabowski GA. Enzyme therapy for lysosomal acid lipase deficiency in the mouse. Human molecular genetics. 2001;10:1639–1648. doi: 10.1093/hmg/10.16.1639. [DOI] [PubMed] [Google Scholar]
- 152.Scott SA, Liu B, Nazarenko I, Martis S, Kozlitina J, Yang Y, Ramirez C, Kasai Y, Hyatt T, Peter I, Desnick RJ. Frequency of the cholesteryl ester storage disease common lipa e8sjm mutation (c.894g>a) in various racial and ethnic groups. Hepatology (Baltimore, Md ) 2013;58:958–965. doi: 10.1002/hep.26327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Brown MS, Sobhani MK, Brunschede GY, Goldstein JL. Restoration of a regulatory response to low density lipoprotein in acid lipase-deficient human fibroblasts. The Journal of biological chemistry. 1976;251:3277–3286. [PubMed] [Google Scholar]
- 154.Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein. Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. The Journal of biological chemistry. 1975;250:8487–8495. [PubMed] [Google Scholar]
- 155.Bowden KL, Bilbey NJ, Bilawchuk LM, Boadu E, Sidhu R, Ory DS, Du H, Chan T, Francis GA. Lysosomal acid lipase deficiency impairs regulation of abca1 gene and formation of high density lipoproteins in cholesteryl ester storage disease. The Journal of biological chemistry. 2011;286:30624–30635. doi: 10.1074/jbc.M111.274381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Choi HY, Karten B, Chan T, Vance JE, Greer WL, Heidenreich RA, Garver WS, Francis GA. Impaired abca1-dependent lipid efflux and hypoalphalipoproteinemia in human niemann-pick type c disease. The Journal of biological chemistry. 2003;278:32569–32577. doi: 10.1074/jbc.M304553200. [DOI] [PubMed] [Google Scholar]
- 157.Burton BK, Balwani M, Feillet F, Baric I, Burrow TA, Camarena Grande C, Coker M, Consuelo-Sanchez A, Deegan P, Di Rocco M, Enns GM, Erbe R, Ezgu F, Ficicioglu C, Furuya KN, Kane J, Laukaitis C, Mengel E, Neilan EG, Nightingale S, Peters H, Scarpa M, Schwab KO, Smolka V, Valayannopoulos V, Wood M, Goodman Z, Yang Y, Eckert S, Rojas-Caro S, Quinn AG. A phase 3 trial of sebelipase alfa in lysosomal acid lipase deficiency. The New England journal of medicine. 2015;373:1010–1020. doi: 10.1056/NEJMoa1501365. [DOI] [PubMed] [Google Scholar]
- 158.Du H, Heur M, Duanmu M, Grabowski GA, Hui DY, Witte DP, Mishra J. Lysosomal acid lipase-deficient mice: Depletion of white and brown fat, severe hepatosplenomegaly, and shortened life span. Journal of lipid research. 2001;42:489–500. [PubMed] [Google Scholar]
- 159.Du H, Schiavi S, Wan N, Levine M, Witte DP, Grabowski GA. Reduction of atherosclerotic plaques by lysosomal acid lipase supplementation. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:147–154. doi: 10.1161/01.ATV.0000107030.22053.1e. [DOI] [PubMed] [Google Scholar]
- 160.Valle D. Metabolic and molecular bases of inherited disease. Lysosomal Acid Lipase Deficiencies: The Wolman Disease/Cholesteryl Ester Storage Disease Spectrum. 2000;142 [Google Scholar]
- 161.Zschenker O, Illies T, Ameis D. Overexpression of lysosomal acid lipase and other proteins in atherosclerosis. Journal of biochemistry. 2006;140:23–38. doi: 10.1093/jb/mvj137. [DOI] [PubMed] [Google Scholar]
- 162.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, Burnett MS, Devaney JM, Knouff CW, Thompson JR, Horne BD, Stewart AF, Assimes TL, Wild PS, Allayee H, Nitschke PL, Patel RS, Martinelli N, Girelli D, Quyyumi AA, Anderson JL, Erdmann J, Hall AS, Schunkert H, Quertermous T, Blankenberg S, Hazen SL, Roberts R, Kathiresan S, Samani NJ, Epstein SE, Rader DJ Myocardial Infarction Genetics C, Wellcome Trust Case Control C. Identification of adamts7 as a novel locus for coronary atherosclerosis and association of abo with myocardial infarction in the presence of coronary atherosclerosis: Two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O’Donnell CJ, McPherson R, Erdmann J, Samani NJ Consortium CA, Cardiogenics. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature genetics. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Coronary Artery Disease Genetics C. A genome-wide association study in europeans and south asians identifies five new loci for coronary artery disease. Nature genetics. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 165.Wang L, Zheng J, Bai X, Liu B, Liu CJ, Xu Q, Zhu Y, Wang N, Kong W, Wang X. Adamts-7 mediates vascular smooth muscle cell migration and neointima formation in balloon-injured rat arteries. Circulation research. 2009;104:688–698. doi: 10.1161/CIRCRESAHA.108.188425. [DOI] [PubMed] [Google Scholar]
- 166.Bauer RC, Tohyama J, Cui J, Cheng L, Yang J, Zhang X, Ou K, Paschos GK, Zheng XL, Parmacek MS, Rader DJ, Reilly MP. Knockout of adamts7, a novel coronary artery disease locus in humans, reduces atherosclerosis in mice. Circulation. 2015;131:1202–1213. doi: 10.1161/CIRCULATIONAHA.114.012669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pu X, Xiao Q, Kiechl S, Chan K, Ng FL, Gor S, Poston RN, Fang C, Patel A, Senver EC, Shaw-Hawkins S, Willeit J, Liu C, Zhu J, Tucker AT, Xu Q, Caulfield MJ, Ye S. Adamts7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. American journal of human genetics. 2013;92:366–374. doi: 10.1016/j.ajhg.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kessler T, Zhang L, Liu Z, Yin X, Huang Y, Wang Y, Fu Y, Mayr M, Ge Q, Xu Q, Zhu Y, Wang X, Schmidt K, de Wit C, Erdmann J, Schunkert H, Aherrahrou Z, Kong W. Adamts-7 inhibits re-endothelialization of injured arteries and promotes vascular remodeling through cleavage of thrombospondin-1. Circulation. 2015;131:1191–1201. doi: 10.1161/CIRCULATIONAHA.114.014072. [DOI] [PubMed] [Google Scholar]
- 169.Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, Bell JT, Yang TP, Meduri E, Barrett A, Nisbett J, Sekowska M, Wilk A, Shin SY, Glass D, Travers M, Min JL, Ring S, Ho K, Thorleifsson G, Kong A, Thorsteindottir U, Ainali C, Dimas AS, Hassanali N, Ingle C, Knowles D, Krestyaninova M, Lowe CE, Di Meglio P, Montgomery SB, Parts L, Potter S, Surdulescu G, Tsaprouni L, Tsoka S, Bataille V, Durbin R, Nestle FO, O’Rahilly S, Soranzo N, Lindgren CM, Zondervan KT, Ahmadi KR, Schadt EE, Stefansson K, Smith GD, McCarthy MI, Deloukas P, Dermitzakis ET, Spector TD Multiple Tissue Human Expression Resource C. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nature genetics. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Consortium GT. The genotype-tissue expression (gtex) project. Nature genetics. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Consortium EP. A user’s guide to the encyclopedia of DNA elements (encode) PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Miller CL, Anderson DR, Kundu RK, Raiesdana A, Nurnberg ST, Diaz R, Cheng K, Leeper NJ, Chen CH, Chang IS, Schadt EE, Hsiung CA, Assimes TL, Quertermous T. Disease-related growth factor and embryonic signaling pathways modulate an enhancer of tcf21 expression at the 6q23.2 coronary heart disease locus. PLoS genetics. 2013;9:e1003652. doi: 10.1371/journal.pgen.1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sazonova O, Zhao Y, Nurnberg S, Miller C, Pjanic M, Castano VG, Kim JB, Salfati EL, Kundaje AB, Bejerano G, Assimes T, Yang X, Quertermous T. Characterization of tcf21 downstream target regions identifies a transcriptional network linking multiple independent coronary artery disease loci. PLoS genetics. 2015;11:e1005202. doi: 10.1371/journal.pgen.1005202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hurskainen TL, Hirohata S, Seldin MF, Apte SS. Adam-ts5, adam-ts6, and adam-ts7, novel members of a new family of zinc metalloproteases. General features and genomic distribution of the adam-ts family. The Journal of biological chemistry. 1999;274:25555–25563. doi: 10.1074/jbc.274.36.25555. [DOI] [PubMed] [Google Scholar]
- 175.Somerville RP, Longpre JM, Apel ED, Lewis RM, Wang LW, Sanes JR, Leduc R, Apte SS. Adamts7b, the full-length product of the adamts7 gene, is a chondroitin sulfate proteoglycan containing a mucin domain. The Journal of biological chemistry. 2004;279:35159–35175. doi: 10.1074/jbc.M402380200. [DOI] [PubMed] [Google Scholar]
- 176.Porter S, Clark IM, Kevorkian L, Edwards DR. The adamts metalloproteinases. The Biochemical journal. 2005;386:15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gao W, Zhu J, Westfield LA, Tuley EA, Anderson PJ, Sadler JE. Rearranging exosites in noncatalytic domains can redirect the substrate specificity of adamts proteases. The Journal of biological chemistry. 2012;287:26944–26952. doi: 10.1074/jbc.M112.380535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Moake JL. Thrombotic microangiopathies. The New England journal of medicine. 2002;347:589–600. doi: 10.1056/NEJMra020528. [DOI] [PubMed] [Google Scholar]
- 179.Dagoneau N, Benoist-Lasselin C, Huber C, Faivre L, Megarbane A, Alswaid A, Dollfus H, Alembik Y, Munnich A, Legeai-Mallet L, Cormier-Daire V. Adamts10 mutations in autosomal recessive weill-marchesani syndrome. American journal of human genetics. 2004;75:801–806. doi: 10.1086/425231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Salter RC, Ashlin TG, Kwan AP, Ramji DP. Adamts proteases: Key roles in atherosclerosis? Journal of molecular medicine. 2010;88:1203–1211. doi: 10.1007/s00109-010-0654-x. [DOI] [PubMed] [Google Scholar]
- 181.Liu CJ, Kong W, Ilalov K, Yu S, Xu K, Prazak L, Fajardo M, Sehgal B, Di Cesare PE. Adamts-7: A metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:988–990. doi: 10.1096/fj.05-3877fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126:5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- 183.Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, Olson EN, Tallquist MD. The bhlh transcription factor tcf21 is required for lineage-specific emt of cardiac fibroblast progenitors. Development. 2012;139:2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Braitsch CM, Combs MD, Quaggin SE, Yutzey KE. Pod1/tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Developmental biology. 2012;368:345–357. doi: 10.1016/j.ydbio.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Miller CL, Haas U, Diaz R, Leeper NJ, Kundu RK, Patlolla B, Assimes TL, Kaiser FJ, Perisic L, Hedin U, Maegdefessel L, Schunkert H, Erdmann J, Quertermous T, Sczakiel G. Coronary heart disease-associated variation in tcf21 disrupts a mir-224 binding site and mirna-mediated regulation. PLoS genetics. 2014;10:e1004263. doi: 10.1371/journal.pgen.1004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, Cheng J, Zhang L, Gu CC, Huang J, Wu T, Ma Y, Li J, Cao J, Chen J, Ge D, Fan Z, Li Y, Zhao L, Li H, Zhou X, Chen L, Liu D, Chen J, Duan X, Hao Y, Wang L, Lu F, Liu Z, Yao C, Shen C, Pu X, Yu L, Fang X, Xu L, Mu J, Wu X, Zheng R, Wu N, Zhao Q, Li Y, Liu X, Wang M, Yu D, Hu D, Ji X, Guo D, Sun D, Wang Q, Yang Y, Liu F, Mao Q, Liang X, Ji J, Chen P, Mo X, Li D, Chai G, Tang Y, Li X, Du Z, Liu X, Dou C, Yang Z, Meng Q, Wang D, Wang R, Yang J, Schunkert H, Samani NJ, Kathiresan S, Reilly MP, Erdmann J, Coronary ADG-WR, Meta-Analysis C, Peng X, Wu X, Liu D, Yang Y, Chen R, Qiang B, Gu D. Genome-wide association study in han chinese identifies four new susceptibility loci for coronary artery disease. Nature genetics. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nurnberg ST, Cheng KR, Raiesdana A, Kundu R, Miller CL, Kim JB, Arora K, Carcamo-Oribe I, Xiong YQ, Tellakula N, Nanda V, Murthy N, Boisvert WA, Hedin U, Perisic L, Aldi S, Maegdefessel L, Pjanic M, Owens GK, Tallquist MD, Quertermous T. Coronary artery disease associated transcription factor tcf21 regulates smooth muscle precursor cells that contribute to the fibrous cap. PLoS genetics. 2015;11 doi: 10.1371/journal.pgen.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Myocardial Infarction Genetics C; Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O’Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Ardissino D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Mannucci PM, Schwartz SM, Siscovick DS, Yee J, Friedlander Y, Elosua R, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Kathiresan S, Meigs JB, Williams G, Nathan DM, MacRae CA, O’Donnell CJ, Salomaa V, Havulinna AS, Peltonen L, Melander O, Berglund G, Voight BF, Kathiresan S, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Musunuru K, Daly MJ, Purcell S, Voight BF, Purcell S, Nemesh J, Korn JM, McCarroll SA, Schwartz SM, Yee J, Kathiresan S, Lucas G, Subirana I, Elosua R, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Samani NJ, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall A, Schunkert H, Erdmann J, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Schunkert H, Samani NJ, Erdmann J, Ouwehand W, Hengstenberg C, Deloukas P, Scholz M, Cambien F, Reilly MP, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser V, Epstein SE, Rader DJ, Scheffold T, Berger K, Stoll M, Huge A, Girelli D, Martinelli N, Olivieri O, Corrocher R, Morgan T, Spertus JA, McKeown P, Patterson CC, Schunkert H, Erdmann E, Linsel-Nitschke P, Lieb W, Ziegler A, Konig IR, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Engert JC, Do R, Xie C, Anand S, Kathiresan S, Ardissino D, Mannucci PM, Siscovick D, O’Donnell CJ, Samani NJ, Melander O, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Altshuler D Wellcome Trust Case Control C. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nature genetics. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Grallert H, Dupuis J, Bis JC, Dehghan A, Barbalic M, Baumert J, Lu C, Smith NL, Uitterlinden AG, Roberts R, Khuseyinova N, Schnabel RB, Rice KM, Rivadeneira F, Hoogeveen RC, Fontes JD, Meisinger C, Keaney JF, Jr, Lemaitre R, Aulchenko YS, Vasan RS, Ellis S, Hazen SL, van Duijn CM, Nelson JJ, Marz W, Schunkert H, McPherson RM, Stirnadel-Farrant HA, Psaty BM, Gieger C, Siscovick D, Hofman A, Illig T, Cushman M, Yamamoto JF, Rotter JI, Larson MG, Stewart AF, Boerwinkle E, Witteman JC, Tracy RP, Koenig W, Benjamin EJ, Ballantyne CM. Eight genetic loci associated with variation in lipoprotein-associated phospholipase a2 mass and activity and coronary heart disease: Meta-analysis of genome-wide association studies from five community-based studies. European heart journal. 2012;33:238–251. doi: 10.1093/eurheartj/ehr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H Wtccc, the Cardiogenics C. Genomewide association analysis of coronary artery disease. The New England journal of medicine. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Tregouet DA, Cambien F, Bruse P, Aherrahrou Z, Wagner AK, Stark K, Schwartz SM, Salomaa V, Elosua R, Melander O, Voight BF, O’Donnell CJ, Peltonen L, Siscovick DS, Altshuler D, Merlini PA, Peyvandi F, Bernardinelli L, Ardissino D, Schillert A, Blankenberg S, Zeller T, Wild P, Schwarz DF, Tiret L, Perret C, Schreiber S, El Mokhtari NE, Schafer A, Marz W, Renner W, Bugert P, Kluter H, Schrezenmeir J, Rubin D, Ball SG, Balmforth AJ, Wichmann HE, Meitinger T, Fischer M, Meisinger C, Baumert J, Peters A, Ouwehand WH, Cardiogenics C, Deloukas P, Thompson JR, Ziegler A, Samani NJ, Schunkert H Italian Atherosclerosis T, Vascular Biology Working G, Myocardial Infarction Genetics C, Wellcome Trust Case Control C. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nature genetics. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bis JC, Kavousi M, Franceschini N, Isaacs A, Abecasis GR, Schminke U, Post WS, Smith AV, Cupples LA, Markus HS, Schmidt R, Huffman JE, Lehtimaki T, Baumert J, Munzel T, Heckbert SR, Dehghan A, North K, Oostra B, Bevan S, Stoegerer EM, Hayward C, Raitakari O, Meisinger C, Schillert A, Sanna S, Volzke H, Cheng YC, Thorsson B, Fox CS, Rice K, Rivadeneira F, Nambi V, Halperin E, Petrovic KE, Peltonen L, Wichmann HE, Schnabel RB, Dorr M, Parsa A, Aspelund T, Demissie S, Kathiresan S, Reilly MP, Taylor K, Uitterlinden A, Couper DJ, Sitzer M, Kahonen M, Illig T, Wild PS, Orru M, Ludemann J, Shuldiner AR, Eiriksdottir G, White CC, Rotter JI, Hofman A, Seissler J, Zeller T, Usala G, Ernst F, Launer LJ, D’Agostino RB, Sr, O’Leary DH, Ballantyne C, Thiery J, Ziegler A, Lakatta EG, Chilukoti RK, Harris TB, Wolf PA, Psaty BM, Polak JF, Li X, Rathmann W, Uda M, Boerwinkle E, Klopp N, Schmidt H, Wilson JF, Viikari J, Koenig W, Blankenberg S, Newman AB, Witteman J, Heiss G, Duijn C, Scuteri A, Homuth G, Mitchell BD, Gudnason V, O’Donnell CJ Consortium CA. Meta-analysis of genome-wide association studies from the charge consortium identifies common variants associated with carotid intima media thickness and plaque. Nature genetics. 2011;43:940–947. doi: 10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, Cheng X, Li C, Yang R, Wang CC, Wu G, Lu QL, Bai Y, Huang YF, Yin D, Yang Q, Wang XJ, Dai DP, Zhang RF, Wan J, Ren JH, Li SS, Zhao YY, Fu FF, Huang Y, Li QX, Shi SW, Lin N, Pan ZW, Li Y, Yu B, Wu YX, Ke YH, Lei J, Wang N, Luo CY, Ji LY, Gao LJ, Li L, Liu H, Huang EW, Cui J, Jia N, Ren X, Li H, Ke T, Zhang XQ, Liu JY, Liu MG, Xia H, Yang B, Shi LS, Xia YL, Tu X, Wang QK. Genome-wide association identifies a susceptibility locus for coronary artery disease in the chinese han population. Nature genetics. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 194.Debette S, Kamatani Y, Metso TM, Kloss M, Chauhan G, Engelter ST, Pezzini A, Thijs V, Markus HS, Dichgans M, Wolf C, Dittrich R, Touze E, Southerland AM, Samson Y, Abboud S, Bejot Y, Caso V, Bersano A, Gschwendtner A, Sessa M, Cole J, Lamy C, Medeiros E, Beretta S, Bonati LH, Grau AJ, Michel P, Majersik JJ, Sharma P, Kalashnikova L, Nazarova M, Dobrynina L, Bartels E, Guillon B, van den Herik EG, Fernandez-Cadenas I, Jood K, Nalls MA, De Leeuw FE, Jern C, Cheng YC, Werner I, Metso AJ, Lichy C, Lyrer PA, Brandt T, Boncoraglio GB, Wichmann HE, Gieger C, Johnson AD, Bottcher T, Castellano M, Arveiler D, Ikram MA, Breteler MM, Padovani A, Meschia JF, Kuhlenbaumer G, Rolfs A, Worrall BB, Ringelstein EB, Zelenika D, Tatlisumak T, Lathrop M, Leys D, Amouyel P, Dallongeville J group C, group C, International Stroke Genetics C. Common variation in phactr1 is associated with susceptibility to cervical artery dissection. Nature genetics. 2015;47:78–83. doi: 10.1038/ng.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Freilinger T, Anttila V, de Vries B, Malik R, Kallela M, Terwindt GM, Pozo-Rosich P, Winsvold B, Nyholt DR, van Oosterhout WP, Artto V, Todt U, Hamalainen E, Fernandez-Morales J, Louter MA, Kaunisto MA, Schoenen J, Raitakari O, Lehtimaki T, Vila-Pueyo M, Gobel H, Wichmann E, Sintas C, Uitterlinden AG, Hofman A, Rivadeneira F, Heinze A, Tronvik E, van Duijn CM, Kaprio J, Cormand B, Wessman M, Frants RR, Meitinger T, Muller-Myhsok B, Zwart JA, Farkkila M, Macaya A, Ferrari MD, Kubisch C, Palotie A, Dichgans M, van den Maagdenberg AM International Headache Genetics C. Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nature genetics. 2012;44:777–782. doi: 10.1038/ng.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tregouet DA, Konig IR, Erdmann J, Munteanu A, Braund PS, Hall AS, Grosshennig A, Linsel-Nitschke P, Perret C, DeSuremain M, Meitinger T, Wright BJ, Preuss M, Balmforth AJ, Ball SG, Meisinger C, Germain C, Evans A, Arveiler D, Luc G, Ruidavets JB, Morrison C, van der Harst P, Schreiber S, Neureuther K, Schafer A, Bugert P, El Mokhtari NE, Schrezenmeir J, Stark K, Rubin D, Wichmann HE, Hengstenberg C, Ouwehand W, Cardiogenics C, Ziegler A, Tiret L, Thompson JR, Cambien F, Schunkert H, Samani NJ Wellcome Trust Case Control C. Genome-wide haplotype association study identifies the slc22a3-lpal2-lpa gene cluster as a risk locus for coronary artery disease. Nature genetics. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 197.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, Fornage M, Ikram MA, Malik R, Bevan S, Thorsteinsdottir U, Nalls MA, Longstreth W, Wiggins KL, Yadav S, Parati EA, Destefano AL, Worrall BB, Kittner SJ, Khan MS, Reiner AP, Helgadottir A, Achterberg S, Fernandez-Cadenas I, Abboud S, Schmidt R, Walters M, Chen WM, Ringelstein EB, O’Donnell M, Ho WK, Pera J, Lemmens R, Norrving B, Higgins P, Benn M, Sale M, Kuhlenbaumer G, Doney AS, Vicente AM, Delavaran H, Algra A, Davies G, Oliveira SA, Palmer CN, Deary I, Schmidt H, Pandolfo M, Montaner J, Carty C, de Bakker PI, Kostulas K, Ferro JM, van Zuydam NR, Valdimarsson E, Nordestgaard BG, Lindgren A, Thijs V, Slowik A, Saleheen D, Pare G, Berger K, Thorleifsson G, Hofman A, Mosley TH, Mitchell BD, Furie K, Clarke R, Levi C, Seshadri S, Gschwendtner A, Boncoraglio GB, Sharma P, Bis JC, Gretarsdottir S, Psaty BM, Rothwell PM, Rosand J, Meschia JF, Stefansson K, Dichgans M, Markus HS International Stroke Genetics C, Australian Stroke Genetics Collaborative WTCCC. Genetic risk factors for ischaemic stroke and its subtypes (the metastroke collaboration): A meta-analysis of genome-wide association studies. The Lancet. Neurology. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Heit JA, Armasu SM, Asmann YW, Cunningham JM, Matsumoto ME, Petterson TM, De Andrade M. A genome-wide association study of venous thromboembolism identifies risk variants in chromosomes 1q24.2 and 9q. Journal of thrombosis and haemostasis : JTH. 2012;10:1521–1531. doi: 10.1111/j.1538-7836.2012.04810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yasuno K, Bilguvar K, Bijlenga P, Low SK, Krischek B, Auburger G, Simon M, Krex D, Arlier Z, Nayak N, Ruigrok YM, Niemela M, Tajima A, von und zu Fraunberg M, Doczi T, Wirjatijasa F, Hata A, Blasco J, Oszvald A, Kasuya H, Zilani G, Schoch B, Singh P, Stuer C, Risselada R, Beck J, Sola T, Ricciardi F, Aromaa A, Illig T, Schreiber S, van Duijn CM, van den Berg LH, Perret C, Proust C, Roder C, Ozturk AK, Gaal E, Berg D, Geisen C, Friedrich CM, Summers P, Frangi AF, State MW, Wichmann HE, Breteler MM, Wijmenga C, Mane S, Peltonen L, Elio V, Sturkenboom MC, Lawford P, Byrne J, Macho J, Sandalcioglu EI, Meyer B, Raabe A, Steinmetz H, Rufenacht D, Jaaskelainen JE, Hernesniemi J, Rinkel GJ, Zembutsu H, Inoue I, Palotie A, Cambien F, Nakamura Y, Lifton RP, Gunel M. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nature genetics. 2010;42:420–425. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nature genetics. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin MR, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga JJ, de Geus EJ, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJ, Uitterlinden AG, Witteman JC, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Doring A, Wichmann HE, Smit JH, McCarthy MI, van Duijn CM, Peltonen L Consortium E. Loci influencing lipid levels and coronary heart disease risk in 16 european population cohorts. Nature genetics. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Middelberg RP, Ferreira MA, Henders AK, Heath AC, Madden PA, Montgomery GW, Martin NG, Whitfield JB. Genetic variants in lpl, oasl and tomm40/apoe-c1-c2-c4 genes are associated with multiple cardiovascular-related traits. BMC medical genetics. 2011;12:123. doi: 10.1186/1471-2350-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Smith EN, Chen W, Kahonen M, Kettunen J, Lehtimaki T, Peltonen L, Raitakari OT, Salem RM, Schork NJ, Shaw M, Srinivasan SR, Topol EJ, Viikari JS, Berenson GS, Murray SS. Longitudinal genome-wide association of cardiovascular disease risk factors in the bogalusa heart study. PLoS genetics. 2010;6:e1001094. doi: 10.1371/journal.pgen.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chu AY, Guilianini F, Grallert H, Dupuis J, Ballantyne CM, Barratt BJ, Nyberg F, Chasman DI, Ridker PM. Genome-wide association study evaluating lipoprotein-associated phospholipase a2 mass and activity at baseline and after rosuvastatin therapy. Circulation. Cardiovascular genetics. 2012;5:676–685. doi: 10.1161/CIRCGENETICS.112.963314. [DOI] [PubMed] [Google Scholar]
- 205.Diabetes Genetics Initiative of Broad Institute of H; Mit LU, Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S Novartis Institutes of BioMedical R. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.