Unfortunately, the original version of this article [1] contained an error. Figures 2, 4 and 5 were incorrect and the captions for Figs. 4 and 5 were incorrect. Below are the correct figures and captions:
Fig. 2.
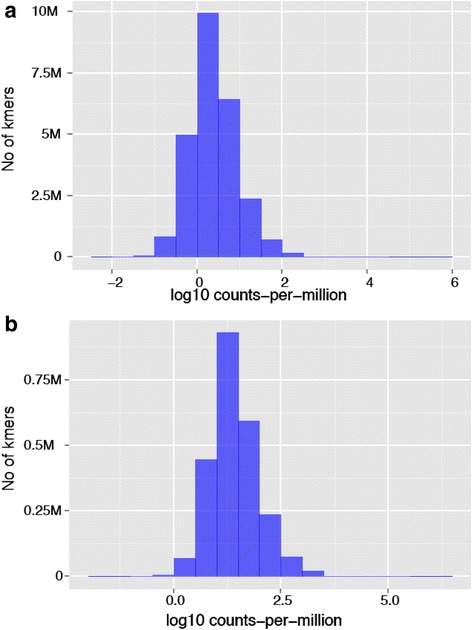
Histogram of k-mer relative abundances. Both 20- and 25-mer relative abundance densities appear log-laplacian. These data included 20- and 25-mers found in all tumor cells. a Histogram of 20-mer relative abundances in log10 scale. b Histogram of 25-mer relative abundances in log10 scale
Fig. 4.
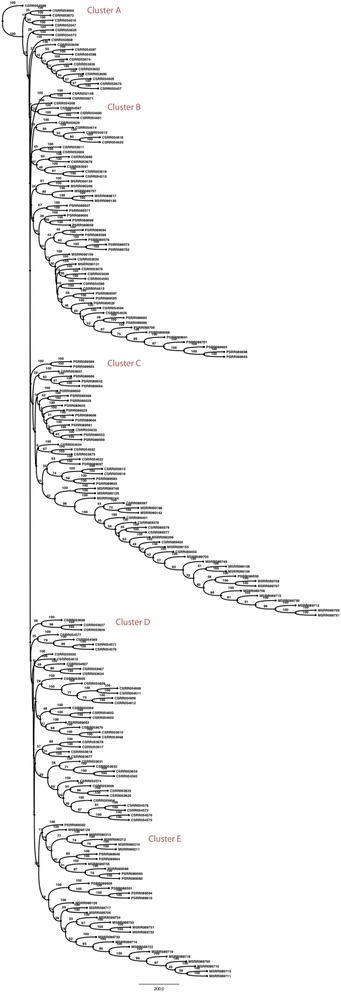
20-mer bootstrap consensus neighbor-joining tree built from T10 primary breast tumor cells (prefix C), T16 primary (prefix P) and metastatic data (prefix M). Distinct groupings of cells are labeled as clusters
Fig. 5.
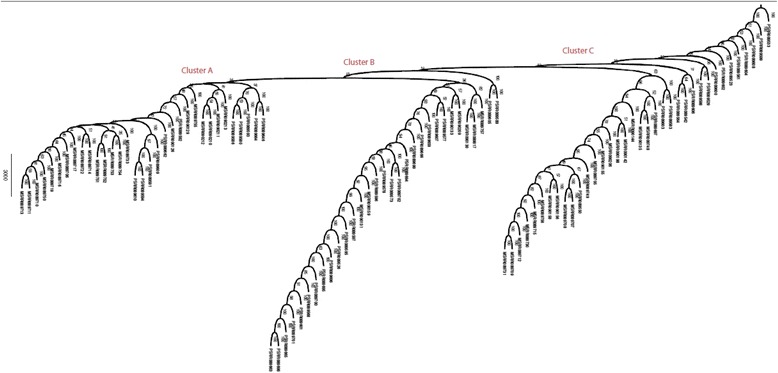
20-mer bootstrap consensus neighbor-joining tree built from T16 primary (prefix P) and metastatic data (prefix M). Distinct groupings of cells are labeled as clusters
Footnotes
The online version of the original article can be found under doi:10.1186/s12864-015-2292-8.
Reference
- 1.Schwartz R, Subramanian A. Reference-free inference of tumor phylogenies from single-cell sequencing data’. BMC Genomics. 2015;16(Suppl 11):S7. doi: 10.1186/1471-2164-16-S11-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]


