Analysis of an Arabidopsis mutant with a defect in photosystem II reveals that the integral thylakoid membrane protein PAM71 is involved in manganese and calcium homeostasis in the chloroplast.
Abstract
In plants, algae, and cyanobacteria, photosystem II (PSII) catalyzes the light-driven oxidation of water. The oxygen-evolving complex of PSII is a Mn4CaO5 cluster embedded in a well-defined protein environment in the thylakoid membrane. However, transport of manganese and calcium into the thylakoid lumen remains poorly understood. Here, we show that Arabidopsis thaliana PHOTOSYNTHESIS AFFECTED MUTANT71 (PAM71) is an integral thylakoid membrane protein involved in Mn2+ and Ca2+ homeostasis in chloroplasts. This protein is required for normal operation of the oxygen-evolving complex (as evidenced by oxygen evolution rates) and for manganese incorporation. Manganese binding to PSII was severely reduced in pam71 thylakoids, particularly in PSII supercomplexes. In cation partitioning assays with intact chloroplasts, Mn2+ and Ca2+ ions were differently sequestered in pam71, with Ca2+ enriched in pam71 thylakoids relative to the wild type. The changes in Ca2+ homeostasis were accompanied by an increased contribution of the transmembrane electrical potential to the proton motive force across the thylakoid membrane. PSII activity in pam71 plants and the corresponding Chlamydomonas reinhardtii mutant cgld1 was restored by supplementation with Mn2+, but not Ca2+. Furthermore, PAM71 suppressed the Mn2+-sensitive phenotype of the yeast mutant Δpmr1. Therefore, PAM71 presumably functions in Mn2+ uptake into thylakoids to ensure optimal PSII performance.
INTRODUCTION
The multiprotein and pigment complex photosystem II (PSII), which is embedded in the thylakoid membranes of chloroplasts, provides the high redox potential needed to oxidize water. Nearly all carbon fixation on Earth depends on the electrons and protons derived from light-driven water oxidation reactions catalyzed by PSII, and as a by-product molecular oxygen powers the aerobic metabolism utilized by all complex organisms. PSII is the most intricately structured multiprotein assembly found in the thylakoid membrane, containing more than 20 subunits in plants (Shi et al., 2012).
The D1/D2 heterodimer that forms the core of PSII binds all the redox components essential for primary charge separation, namely, chlorophyll a, pheophytin, and plastoquinone, and the processes mediated by PSII are highly conserved from cyanobacteria to plants (Nelson and Junge, 2015). The electron donor chlorophyll a P680 in its excited state (P680*) donates an electron to pheophytin. Pheophytin reduces the primary plastoquinone electron acceptor, QA, which in turn reduces QB. Plastoquinone QB accepts two protons and two electrons and diffuses as plastoquinol out of its binding pocket. During this process, the protons are transported into the thylakoid lumen and the electrons flow into the cytochrome b6f complex (Cytb6f). At the donor side of PSII, P680+ receives an electron from the redox-active Tyr-161 of the D1 protein (D1-Y161) (Debus et al., 1988), which is subsequently reduced upon water oxidation at the oxygen-evolving complex (OEC). The catalytic center for water oxidation is the OEC, where O2 is produced and protons are released into the lumen. Detailed structural models of cyanobacterial PSII including the OEC have been reported at a resolution of 1.9 Å and recently at 1.95 Å in its native state (Umena et al., 2011; Suga et al., 2015). The OEC consists of an inorganic Mn4CaO5 cluster that is bound to six amino acids from the D1 protein and to one amino acid from CP43 (Ferreira et al., 2004; Umena et al., 2011; Kawakami et al., 2011). O2 is produced via a four-step mechanism, in which the OEC cycles through four light-generated intermediates known as S states (Kok et al., 1970).
In plants, the nonligating protein environment of the OEC requires an additional set of at least three lumen-exposed extrinsic proteins (PsbO, PsbP, and PsbQ), each of which is encoded by two separate genes in Arabidopsis thaliana. Numerous studies have shown that these PSII subunits play important roles in supporting oxygen evolution by maintaining optimal manganese, calcium, and chloride concentrations and protecting/stabilizing the manganese cluster (Suorsa and Aro, 2007). Using T-DNA insertional mutagenesis, it has been shown that PsbP1, but not PsbP2, is essential for photoautotrophic growth (Allahverdiyeva et al., 2013). PsbP purified from spinach (Spinacia oleracea) contains Mn2+ ions, suggesting its involvement in providing Mn2+ to PSII during the process of OEC assembly and/or during PSII repair (Bondarava et al., 2005, 2007). In contrast, complete loss of PsbQ in a psbq1 psbq2 double mutant has only minor effects on oxygen evolution and does not alter photoautotrophic growth under normal lighting conditions (Allahverdiyeva et al., 2013). The largest extrinsic subunit, PsbO, is known as the manganese-stabilizing protein but is also thought to bind Ca2+ ions (Murray and Barber, 2006; Guskov et al., 2009). There are two distinct isoforms in Arabidopsis, PsbO1 and PsbO2, with the PsbO1 isoform being by far the more abundant of the two (Murakami et al., 2002). The psbo1 mutant exhibits a distinctive phenotype, with pale-green leaves, reduced rosette size, and slower growth rate, which underlines the importance of the PsbO1 isoform for the structure and function of PSII (Murakami et al., 2002; Lundin et al., 2007; Bricker and Frankel, 2008; Allahverdiyeva et al., 2009).
Calcium and manganese are structural components of PSII and are essential for efficient oxygen evolution. Uptake of Ca2+ into the chloroplast (Kreimer et al., 1985) and its further transport into the thylakoid lumen (Ettinger et al., 1999) have been shown to be light dependent; however, the molecular mechanism of the transport processes remains elusive. Furthermore, Johnson et al. (1995) observed a Ca2+ flux into the chloroplast stroma upon shift from light to darkness, which peaked after ∼20 to 30 min. They assumed that Ca2+ was taken up during illumination, sequestered in the thylakoid lumen and/or thylakoid membrane, and released from these stores after the light was switched off. The question of how Mn2+ is delivered to PSII and assembled into the Mn4CaO5 cluster has been addressed in Synechocystis sp PCC 6803. Two distinct systems for cellular Mn2+ uptake have been described, the so-called MntABC transporter and a second pathway whose components have not yet been identified (Bartsevich and Pakrasi, 1995, 1996). Synechocystis can store Mn2+ in the periplasmic space (Keren et al., 2002), and efficient delivery to and preloading of PSII with Mn2+ are mediated by the periplasmic tetratricopeptide repeat protein PratA (Stengel et al., 2012). PratA is a cyanobacterial protein, and no obvious PratA homologs have been identified in eukaryotes (Klinkert et al., 2004). However, LOW PSII ACCUMULATION1 (LPA1) in Arabidopsis and PSII repair protein REP27 in Chlamydomonas reinhardtii, which both belong to the tetratricopeptide repeat family and directly interact with D1, share some features with PratA (Peng et al., 2006; Park et al., 2007). Interestingly, recombinant LPA1 has Mn2+ binding activity, suggesting that LPA1 might have a PratA-related function in plants (Stengel et al., 2012).
Ion fluxes across the thylakoid membrane have been studied in the past and were proposed to counterbalance the electron transport-coupled pumping of H+ into the lumen, thus altering the proton motive force (PMF) (Hind et al., 1974). The PMF, which is made up of the proton chemical potential (ΔpH) and the transmembrane electrical potential (Δψ), is the total energy available for ATP biosynthesis. Recently, K+ was reported to participate in the modulation of PMF via the two-pore K+ channel TPK3 and the K+/H+ exchanger KEA3, both of which are localized to the thylakoid membrane of Arabidopsis (Carraretto et al., 2013; Armbruster et al., 2014; Kunz et al., 2014). It was proposed that TPK3 drives K+ transport out of the lumen (down the electrical potential gradient) into the stroma, whereas KEA3 presumably functions as a K+/H+ antiporter, driving K+ uptake into the lumen. Since tpk3 knockdown plants exhibit a diminished capacity for building up ΔpH, as well as a higher Δψ in the light (relative to the wild type), Carraretto et al. (2013) concluded that TPK3 plays a role in the dissipation of Δψ and enhancement of ΔpH. In the same report, it was shown that the TPK3 channel activity is sensitive to Ca2+ ions.
In this work, we analyzed photosynthesis affected mutant71 (pam71) mutant lines of Arabidopsis. The PAM71 gene encodes a protein that is well conserved in the green lineage and shares homology with putative Ca2+/H+ exchangers from yeast (Saccharomyces cerevisiae) (GDT1) and human (Homo sapiens) (TMEM165) (Demaegd et al., 2013, 2014). We found that PAM71 belongs to a small gene family in Arabidopsis comprising five members and that PAM71 localizes to the thylakoid membrane. Knockout mutants for PAM71 were found to be impaired in photosynthesis, with a primary defect in PSII and enhanced PMF across thylakoids. Analysis of metal ion contents and PSII activity after growth under enrichment with different metal ions, together with the outcome of yeast complementation experiments, allows us to propose a critical role for PAM71 in calcium and manganese homeostasis in the chloroplast.
RESULTS
Mutations in PAM71 Affect PSII Function
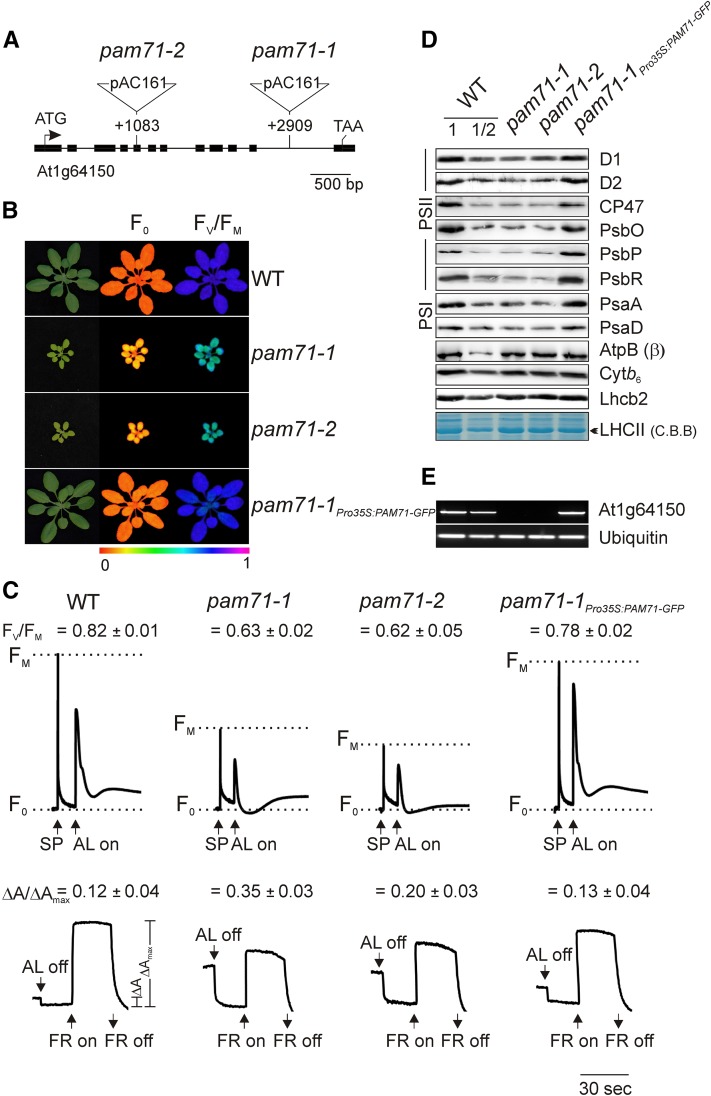
The Arabidopsis GABI-Kat collection (Rosso et al., 2003) was screened for lines with T-DNA insertions in genes coding for “uncharacterized membrane proteins with putative chloroplast localization” listed in the ARAMEMNON database (http://aramemnon.botanik.uni-koeln.de). Both of the lines recovered, GK-166A05 and GK-094C03, carried insertions in At1g64150. Because they showed defects in photosynthesis and reduced growth rates, the mutants were designated as photosynthesis affected mutant71-1 (pam71-1) and pam71-2, respectively, in the style of previously identified pam mutants (Varotto et al., 2000; Armbruster et al., 2010) (Figure 1A). The phenotypic defects in both lines could be complemented by introducing a cDNA copy of the PAM71 gene under the control of the 35S promoter (Supplemental Figure 1). In addition, expression of a PAM71-GFP fusion in the pam71-1 background rescued the mutant phenotypes (Figure 1B).
Figure 1.
Isolation, Characterization, and Complementation of the Arabidopsis Mutants pam71-1 and pam71-2.
(A) T-DNA tagging of the PAM71/At1g64150 locus. Exons are shown as black boxes and the introns as black lines. Start and stop codons are indicated. The pam71-1 and pam71-2 alleles were identified in the GABI-Kat collection (GK-166A05 and GK-094C03). Locations of T-DNA insertions are indicated with respect to the start codon. Note that the insertions are not drawn to scale.
(B) Four-week-old wild-type plants, mutants (pam71-1 and pam71-2), and the complemented line pam71-1Pro35S:PAM71-GFP were grown in a 12-h/12-h dark/light cycle at 21°C/18°C and at 90 µmol photons m−2 s−1, and the minimal chlorophyll a fluorescence (F0) (middle panel) and maximum quantum yield of PSII (FV/FM) (right panel) were recorded. The color scale at the bottom indicates the signal intensities.
(C) Chlorophyll a fluorescence induction curves (top panel) and P700 redox kinetics (bottom panel) were recorded from leaves of the different genotypes. FV/FM values are indicated for each genotype. For chlorophyll a fluorescence measurement, the minimal level of fluorescence of dark-adapted leaves is indicated by dashed lines. Saturating pulses (SP) and actinic light (AL; 100 µmol photons m−2 s−1) were applied as indicated. For P700 absorption measurements, leaves were first illuminated with actinic light (100 µmol photons m−2 s−1) for 4 min, and absorbance changes (at 830 nm) induced by exposure to far-red light (FR; 720 nm) were recorded. Mean values (±sd) are each based on five individual plants.
(D) Immunodetection of thylakoid proteins from the different genotypes using antibodies raised against individual subunits of PSII (D1, D2, CP47, PsbO, PsbP, and PsbR), LHCII (Lhcb2), PSI (PsaA and PsaD), the Cytb6f complex (Cytb6), and chloroplast ATP synthase [AtpB (β) subunit]. Samples equivalent to 15 µg protein were loaded in all lanes, except in those marked WT 1/2, where only half as much was loaded. As a loading control, LHCII was visualized by Coomassie blue (C.B.B.) staining.
(E) Expression of PAM71/At1g64150 in the different genotypes, as determined by RT-PCR (30 cycles). Primers used for amplification were complementary to sequences within the 3rd and 12th exons. The expression of Ubiquitin (At4g36800) was used as a control for RNA integrity.
The increase in minimal chlorophyll a fluorescence (F0) and concomitant decrease in the maximum quantum yield of PSII (FV/FM) observed in both mutant lines indicated either a defect in electron transfer within PSII and/or a partial disconnection of the LHCII antenna. Moreover, the fluorescence yield of chlorophyll a transiently dropped below F0 after the initial rise upon exposure of the pam71 lines to actinic light (Figure 1C), which is characteristic for mutants defective in PSII (Peng et al., 2006, Ma et al., 2007; Armbruster et al., 2010).
Redox kinetic studies of P700, the reaction center chlorophyll in photosystem I (PSI), revealed that the absorbance changes at 830 nm induced by exposure to far-red light in the two pam71 alleles resulted in an increased ΔA/ΔAmax value (Figure 1C), indicating that electron flow toward PSI was reduced. In this respect, both pam71 alleles behave like the psbo1 psbo2 mutant, which has a reduced amount of PsbO (Steinberger et al., 2015), in contrast to the PSI mutant psad1-1, which displayed a reduced ΔA/ΔAmax value (Supplemental Figure 2A). An increased ΔA/ΔAmax value is also a characteristic feature of other PSII mutants (Meurer et al., 1996; Peng et al., 2006).
The defect in PSII functionality could be accompanied by altered levels of thylakoid membrane protein(s) in pam71 alleles. Therefore, the steady state levels of subunits of PSII, LHCII, Cytb6f, ATP synthase, and PSI were examined by immunoblot analysis. The amounts of PSII subunits (D1, D2, CP47, PsbO, PsbP, and PsbR) were reduced to ∼50% of wild-type levels in pam71-1 and pam71-2. Only the amount of PSII subunit CP43 was found to behave differently presumably due to the occurrence of a larger fraction of CP43 in low molecular weight complexes in pam71-1 (Supplemental Figure 3). We verified that the partial loss of PSII subunits was not attributable to lower rates of transcription or translation of the respective plastid-encoded genes (Supplemental Figure 4). The levels of Lhcb2, Cytb6, and the β subunit of ATP synthase were unchanged (Figure 1D). Interestingly, the steady state levels of PSI subunits (PsaA and PsaD) were also reduced to ∼50% of wild-type levels in both pam71 alleles (Figure 1D). This most probably reflects a secondary adjustment of photosystem stoichiometry to accommodate the deficiency in PSII function (Dietzel et al., 2008).
Further analysis confirmed that both pam71-1 and pam71-2 are knockout alleles of At1g64150 (Figure 1E). In the complemented line pam71-1Pro35S:PAM71-GFP, expression of PAM71 and all other defects were corrected (Figures 1B to 1E).
Changes in Thylakoid Composition and Structure in pam71
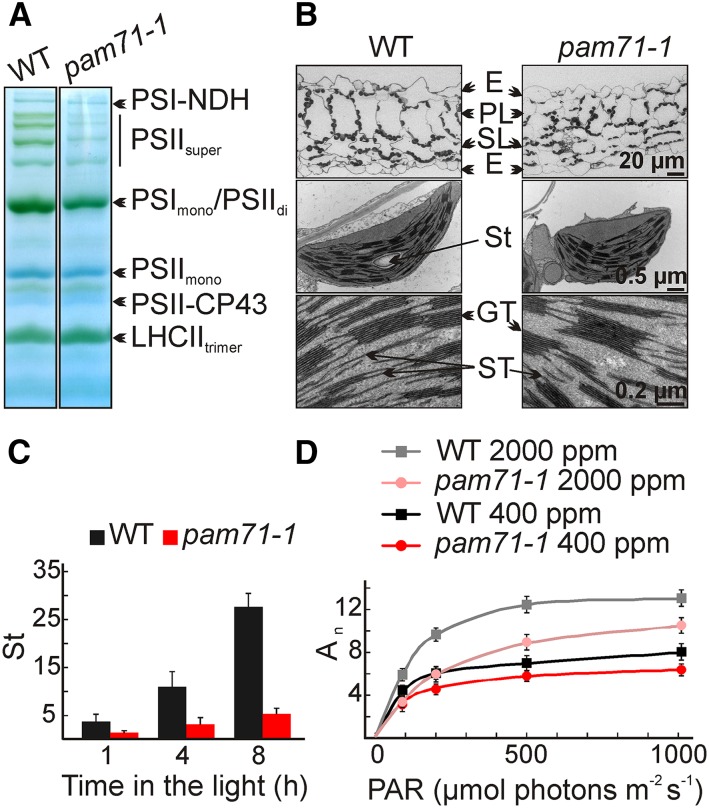
Next, we investigated the effect of deleted PAM71 function on the formation of thylakoid multiprotein complexes. Thylakoids were isolated from wild-type and pam71-1 leaves, solubilized, and fractionated by Blue-Native PAGE. Several prominent bands were assigned to different complexes based on earlier reports (Granvogl et al., 2006; Peng et al., 2006; Armbruster et al., 2010). The abundance of LHCII trimer complexes, as well as CP43-less PSII complexes and PSII monomer complexes, were similar in both genotypes, whereas the amounts of PSII supercomplexes and the PSII dimer/PSI monomer fraction were markedly reduced (Figure 2A) in pam71-1 in accordance with lower steady state levels of PSII and PSI subunits (Figure 1D). The thylakoid membrane complex formation in pam71-1 resembles that of psbo1 psbo2 (Supplemental Figure 2B). The abundance of the PSI-NDH complex was comparable in the wild type and pam71-1, whereas, for instance, the level of this complex was reduced in the PSI mutant psad1-1 (Supplemental Figure 2B). We also investigated whether these changes were reflected in chloroplast ultrastructure in pam71. However, the overall organization of pam71 chloroplasts and the disposition of thylakoid membranes in grana and stroma lamellae were found to be unaltered compared with the wild type (Figure 2B), which is comparable to psbo1psbo2 and psad1-1 (Supplemental Figure 2C). Only the numbers of starch granules were greatly reduced in pam71-1 (Figure 2B), especially at the end of the light phase (Figure 2C). The reduced amount of starch is most probably due to a reduction in CO2 assimilation, which was particularly pronounced under high CO2 conditions (Figure 2D).
Figure 2.
Physiological Effects of pam71-1 on Chloroplasts.
(A) Accumulation of thylakoid membrane complexes in the wild type and pam71-1. Thylakoid membranes (equivalent to 25 µg of chlorophyll) were solubilized with 1% (w/v) β-dodecyl maltoside, and protein complexes were fractionated by Blue-Native gel electrophoresis. The positions of protein complexes (trimeric LHCII = LHCIItrimer, CP43-free PSII complexes = PSII-CP43, PSII monomers = PSIImono, PSII dimers = PSIIdi, PSI monomers = PSImono, PSII supercomplexes = PSIIsuper and PSI-NDH) are indicated.
(B) Electron micrographs of wild-type and pam71-1 leaf sections (top panel), chloroplasts (middle panel), and views of wild-type and pam71-1 chloroplasts at higher magnification (bottom panel). Epidermis (E), palisade layer (PL), spongy layer (SL), starch (St), grana thylakoids (GT), and stroma thylakoids (ST) are indicated.
(C) Starch contents (St; µmol C6 g fresh weight−1) of wild-type and pam71-1 leaves harvested at three different time points during the day were determined enzymatically. Mean values (±sd) are each based on three individual plants.
(D) Net CO2 assimilation rate An (µmol CO2 m−2 s−1). The rate of net CO2 fixation was recorded in the range of 0 to 1000 µmol photons m−2 s−1 PAR (photosynthetically active radiation) under atmospheric (400 ppm) or high CO2 (2000 ppm) conditions. Mean values (±sd) are each based on five to six individual plants.
The OEC as the Primary Site of Diminished PSII Function
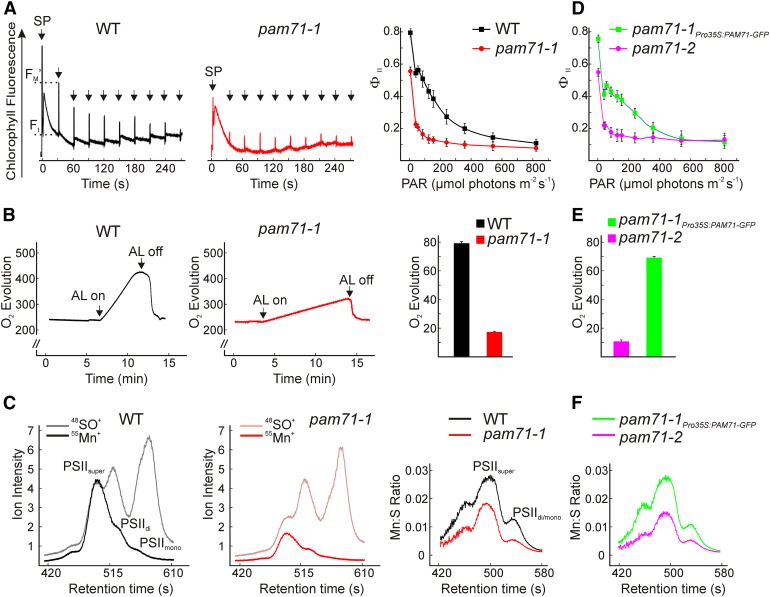
The effective quantum yield of PSII (ΦII) in pam71-1 was clearly reduced, particularly at low and medium actinic light intensities (Figure 3A), indicating reduced linear electron flow through the mutant thylakoid membrane. This eventually results in the production of less reducing power for CO2 assimilation, thus explaining the reduced growth observed in pam71. The ΦII changes may be the direct consequence of the reduced FV/FM value (Figures 1B and 1C). The changes in ΦII were accompanied by a higher nonphotochemical quenching (NPQ) in pam71-1 under the conditions tested (30-s illumination periods; Supplemental Table 1). In plants, the NPQ is induced under high light and functions in dissipation of excess excitation energy as heat (Holt et al., 2004). We therefore investigated the steady state NPQ upon high light exposure of pam71-1 leaves compared with the wild type. In both the wild type and pam71-1, the NPQ was clearly enhanced upon high light treatment compared with moderate light conditions, but in pam71-1, it remained significantly below wild-type levels (Table 1).
Figure 3.
Effective Quantum Yield of PSII, Oxygen Evolution, and Multielement Chromatogram of the Wild Type, pam71-1, pam71-2, and pam71-1Pro35S:PAM71-GFP.
(A) Chlorophyll a fluorescence induction curves were recorded in the range 0 to 856 µmol photons m−2 s−1 for the wild type (left panel) and pam71-1 (middle panel) and plotted as values from eight individual plants (±sd) (right panel) displayed as light response curve of the effective quantum yield of PSII ΦII [(FM′ − Ft)/FM′]. Saturating pulses (SP) were applied as indicated. The plants were dark-adapted for 30 min prior to measurement. ΦII was recorded following illumination for 30 s at the appropriate PAR. FM′ and Ft are indicated for the wild type at 37 µmol photons m−2 s−1 (left panel).
(B) Oxygen evolution (nmol O2 mL−1) of the wild type (left panel) and pam71-1 (middle panel) was measured over the indicated time in isolated thylakoids adjusted to 30 µg chlorophyll mL−1 and using DCBQ as an external electron acceptor. Reactions were started with the application of actinic light (AL) at 500 µmol photons m−2 s−1. Oxygen evolution rate (µmol O2 mg chlorophyll−1 h−1) was calculated from two independent thylakoid preparations including three replicates (right panel). The data are means ± sd (n = 6).
(C) Size exclusion profiles were recorded for the nonoxide ion 55Mn+ and the oxide ion 48SO+ in the wild type (left panel) and pam71-1 (middle panel) and given as ion intensities (counts 105 s−1). Individual 55Mn+ fractions were assigned to PSII dimers/monomers = PSIIdi/mono and PSII supercomplexes = PSIIsuper according to Schmidt et al. (2015). Quantification of the Mn:S stoichiometric ratio (as described in Methods) is shown for a representative sample (right panel) out of five samples derived from independent thylakoid preparations.
(D) to (F) Analysis of pam71-2 and pam71-1Pro35S:PAM71-GFP was performed as in (A) to (C), respectively.
Table 1. NPQ under Steady State Conditions in Wild-Type and pam71 Leaves.
| PAR | Wild Type | pam71-1 | pam71-2 | pam71-1Pro35S:PAM71-GFP |
|---|---|---|---|---|
| 72 | 0.16 ± 0.04 | 0.13 ± 0.03 | 0.13 ± 0.04 | 0.16 ± 0.06 |
| 532 | 1.72 ± 0.17 | 1.01 ± 0.28* | 1.01 ± 0.21* | 1.69 ± 0.19 |
NPQ was recorded from plants that were dark-adapted overnight by exposing them to 72 or 532 µmol photons m−2 s−1 PAR. The NPQ was recorded following illumination for 15 min at the appropriate PAR. The NPQ parameter was calculated according to the following equation: NPQ = (FM − FM′)/FM′ (where FM and FM′ represent the maximal fluorescence of dark-adapted or illuminated samples, respectively). At least five different plants were measured (n = 5 upper row; n = 7 lower row). Mean values ± sd are provided. Asterisks indicate statistical significance (t test, P < 0.01) of difference between wild-type and mutant plants.
To identify the site of PSII malfunction in pam71, we determined oxygen evolution rates according to Hakala et al. (2005). Thylakoids were isolated from pam71-1 and wild-type leaves and, as a control, from psbo1 psbo2 and psad-1 leaves. Oxygen evolution was measured under nonsaturating light conditions in the presence of the artificial electron acceptor 2,6-dichloro-1,4-benzoquinone (DCBQ) (Figure 3B, left and middle panels), and the oxygen evolution rate was calculated (Figure 3B, right panel). The rate of oxygen evolution was reduced to ∼20% of wild- type levels in pam71-1, similar to the decline in oxygen evolution observed in psbo1 psbo2 thylakoids (Supplemental Figure 2D). Taking into account the reduction of PSII subunit levels to ∼50%, we concluded that the OEC activity in pam71-1 is particular affected.
In addition, fast chlorophyll a fluorescence kinetic measurements were performed, which give several kinetic peaks in the wild type called the OJIP transients. The development of an additional peak, termed K, at ∼300 µs in pam71-1 and psbo1 psbo2 (Supplemental Figure 5) can be regarded as an indicator of OEC malfunction (Bukhov et al., 2004; Tóth et al., 2007). To investigate whether PAM71 could be involved in the assembly of the OEC protein environment, a coimmunoprecipitation experiment was performed, showing that PAM71-GFP does not interact with either PsbO or PsbP (Supplemental Figure 6).
Next, the intactness of the inorganic Mn4CaO5 cluster in pam71 was studied by fractionation of thylakoid protein complexes using size-exclusion chromatography (SEC) combined with inductively coupled plasma mass spectrometry (ICP-MS) to analyze the ion intensity of manganese and sulfur (Schmidt et al., 2015). This allowed us to analyze manganese binding in various PSII super- and subcomplexes in wild-type and pam71-1 thylakoids. Manganese binding to PSIIsuper, PSIIdimer, and PSIImonomer complexes was assigned according to Schmidt et al. (2015) (Figure 3C). A marked decrease in the ion intensity of manganese was observed in pam71-1 samples compared with the wild type (Figure 3C, left and middle panels). The decrease in manganese binding in the pam71-1 mutant was not only a response to an overall decrease in PSII levels (50% reduction in the amount of PSII subunits; Figure 1D). When analyzing the manganese to sulfur (Mn:S) ratio, as an indicator of the relative manganese incorporation per unit PSII protein (using sulfur as proxy for protein), the Mn cluster in pam71-1 was shown to be severely affected. Much less manganese was incorporated per unit sulfur in pam71-1 compared with the wild type (Figure 3C, right panel). It is noteworthy that the total manganese concentrations of leaves were not significantly different between the wild type and pam71-1 (84.5 ± 4.6 µg Mn g dry weight−1 in the wild type and 84.7 ± 5.5 µg Mn g dry weight−1 in pam71-1). To exclude any eventuality in pam71-1 that might have arisen independently of the mutated PAM71 gene, all experiments were repeated with pam71-2 and the complemented pam71-1Pro35S:PAM71-GFP line (Figures 3D to 3F, Table 1). The mutant line pam71-2 showed similar defects, and all defects were restored to nearly wild-type levels in pam71-1Pro35S:PAM71-GFP (Figures 3D to 3F, Table 1).
PAM71 Is Evolutionarily Conserved and Located in the Thylakoid Membrane
PAM71 belongs to the UPF0016 family of membrane proteins, which is present in nearly all organisms (Demaegd et al., 2014) and is encoded by a gene belonging to a small gene family in Arabidopsis comprising five members (Supplemental Table 2). PAM71 and its closest homolog (which we named PAM71-HL) are both predicted to possess an N-terminal chloroplast transit peptide, whereas the other three homologs were predicted to be substrates of the secretory pathway. Interestingly, we identified exactly two homologous PAM71/PAM71-HL proteins in other plant species, whereas the number of “secretory pathway targeted” homologs was variable (Supplemental Table 2). Furthermore, the existence of two homologous PAM71/PAM71-HL proteins appears to be conserved in the green lineage (Supplemental Figure 7 and Supplemental Data Set 1).
The yeast and human homologs were shown to be functionally related and to be involved in calcium homeostasis (Demaegd et al., 2013). Thus, a sequence alignment of PAM71 with PAM71-HL, TMEM165 (human), and GDT1 (yeast) was created, which showed that both Arabidopsis proteins possess N-terminal extensions that are predicted to include the chloroplast transit peptides (Figure 4). The two highly conserved E-x-G-D-(KR)-(TS) motifs, each with two negatively charged acidic residues (Glu/E and Asp/D), were found in transmembrane domains TM1 and TM4. The central loops between TM3 and TM4 are also enriched in Asp/Glu and may be involved in binding divalent cations (Demaegd et al., 2014).
Figure 4.
Sequence Alignment of At-PAM71, At-PAM71-HL, and Its Homologs from Yeast and Human.
The protein sequence alignment was generated using Clustal Omega (version 1.2.1). Black and gray boxes indicate identical residues and conservative exchanges (scoring > 0.5), respectively. Six putative transmembrane spans were manually adjusted and numbered TM1 to TM6 using Hs-TMEM165 and Sc-GDT1 as references (Demaegd et al., 2013). An additional transmembrane domain predicted by TmHMM (version 2) is indicated by a dashed line. The two highly conserved internal E-x-G-D-(KR)-(TS) motifs are highlighted by red lines. The putative sites of cleavage of the N-terminal chloroplast transit peptide (cTP) of PAM71 and PAM71-HL, as predicted by TargetP (version1), are indicated by a green arrow and a light-green arrow, respectively. The central loop is indicated by a dotted line. NCBI accession codes are as follows: Homo sapiens NP_060945.2 (transmembrane protein 165, TMEM165); Saccharomyces cerevisiae AHY74663.1 (GDT1); Arabidopsis thaliana NP_564825.1 (At1g64150, PAM71); and Arabidopsis thaliana NP_193095.2 (At4g13590, PAM71-HL).
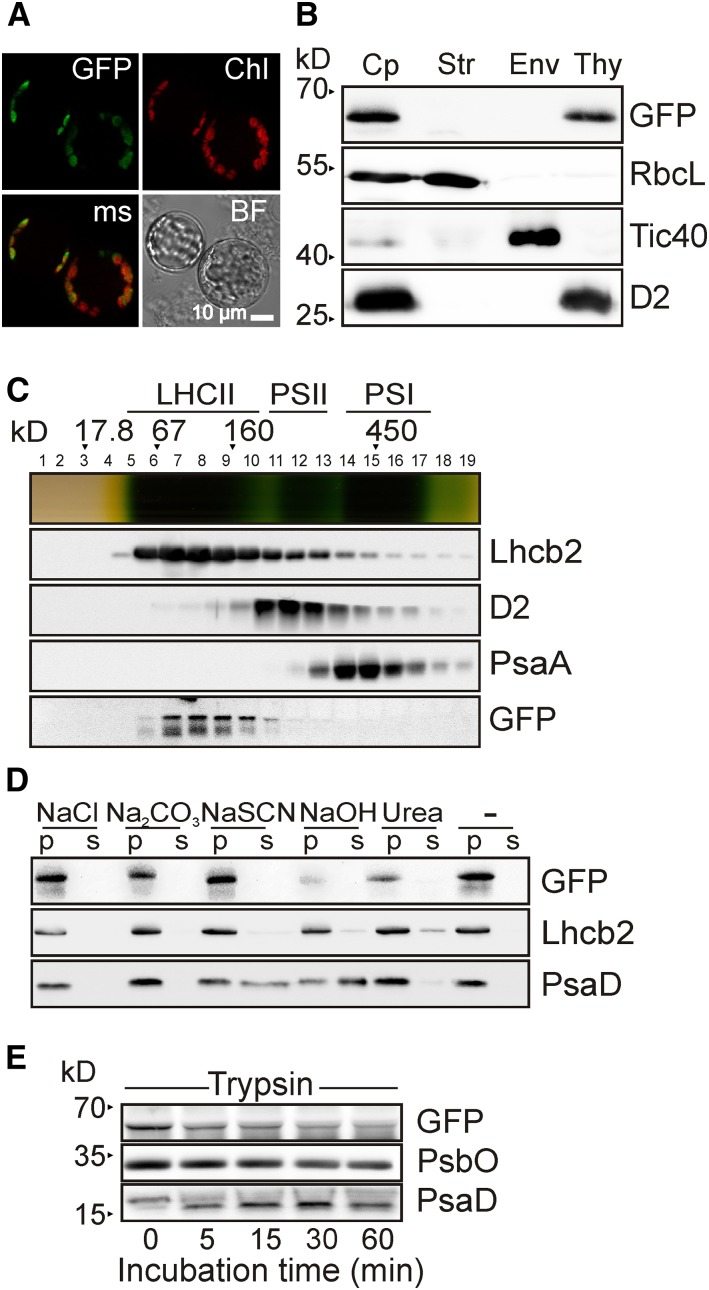
The introduction of a functional Pro35S:PAM71-GFP construct into the pam71 mutant background (Figure 1) enabled us to verify its subcellular localization. In contrast to PAM71-HL, which was found to be enriched in the chloroplast envelope fraction of Arabidopsis (Ferro et al., 2003, 2010), no experimental evidence for the subcellular localization of PAM71 was available. Therefore, protoplasts of pam71-1Pro35S:PAM71-GFP leaf cells were isolated and analyzed by fluorescence microscopy. We found that PAM71-GFP localized exclusively to chloroplasts (Figure 5A); more specifically, it was found only in the thylakoid membrane fraction (Figure 5B). When the pam71-1Pro35S:PAM71-GFP line was used for sucrose gradient fractionation experiments (Figure 5C), the PAM71-GFP signal was found in fractions between the 67- and 160-kD markers and did not colocalize with either the PSII or PSI complexes. Because the calculated molecular mass of PAM71-GFP is ∼60 kD, it appears to be present as a homodimer and/or in a small complex. Moreover, it behaved like an integral thylakoid membrane when subjected to various salt treatments (Figure 5D). We also assessed the accessibility of the C-terminal GFP tag to trypsin. Some GFP remained undigested even after 60 min, which is similar to the behavior of PsbO, which is exposed on the lumenal side of the thylakoid (Figure 5E). Based on these results, and in agreement with Demaegd et al. (2014), we propose a topological model including at least six TM domains, with an additional predicted TM domain at the N terminus and the C terminus exposed on the opposite side of the membrane.
Figure 5.
Thylakoid Localization and Topology of PAM71.
(A) Subcellular localization of PAM71-GFP. Protoplasts were isolated from pam71-1Pro35S:PAM71-GFP, and the fusion protein was localized by fluorescence microscopy. GFP fluorescence was excited at 470 ± 40 nm, and the emission was recorded at 525 ± 50 nm. Chlorophyll autofluorescence (Chl) was excited at 450 to 490 nm, and emission was recorded at >515 nm. The obtained signals were merged (ms); a bright-field (BF) photograph showed intactness of the protoplasts.
(B) Suborganellar localization of PAM71-GFP. Total chloroplasts (Cp) were isolated from pam71-1Pro35S:PAM71-GFP and fractionated into stroma (Str), envelope membranes (Env), and thylakoid membranes (Thy). Immunodetection was performed with anti-RbcL (a marker for the stroma fraction), anti-Tic40 (envelope membrane fraction), anti-D2 (thylakoid membrane fraction), and anti-GFP to detect PAM71-GFP.
(C) Thylakoid membrane fractionation of PAM71-GFP. Thylakoids (1 mg chlorophyll mL−1) of pam71-1Pro35S:PAM71-GFP were solubilized with 1% (w/v) β-dodecyl maltoside, and complexes were separated in a linear 0.1 to 1 M sucrose gradient. Nineteen fractions were collected, proteins from each fraction were precipitated, and immunoblot analysis was performed using anti-Lhcb2, anti-D2, anti-PsaA, and anti-GFP antibodies. The positions of molecular mass markers and of the major complexes (LHCII, PSII, and PSI) are indicated.
(D) PAM71-GFP is an integral membrane protein. Thylakoid membranes were isolated from pam71-1Pro35S:PAM71-GFP and exposed to chaotropic salts or alkaline pH. Membranes were resuspended at 0.5 mg chlorophyll mL−1 in 10 mM HEPES (pH 7.5) containing either 2 M NaCl, 0.1 M Na2CO3, 2 M NaSCN, 0.1 M NaOH, 6 M urea, or no additive. After incubation for 30 min on ice, supernatants containing soluble proteins (S) or pelleted proteins (P) were fractionated by SDS-PAGE, and immunoblot analysis was performed using anti-GFP, anti-Lhcb2, and anti-PsaD antibodies. Note that PAM71-GFP behaves like the integral membrane protein Lhcb2.
(E) Membrane accessibility of PAM71-GFP. Isolated thylakoid membranes from pam71-1Pro35S:PAM71-GFP were treated with trypsin for the indicated times. Immunoblot analysis was performed using anti-GFP, anti-PsbO, and anti-PsaD antibodies. Note that only ∼2 kD of PsaD is susceptible to proteolysis, in agreement with observations by Minai et al. (1996).
Partitioning of the PMF in pam71 Is Altered in Favor of the Δψ Component
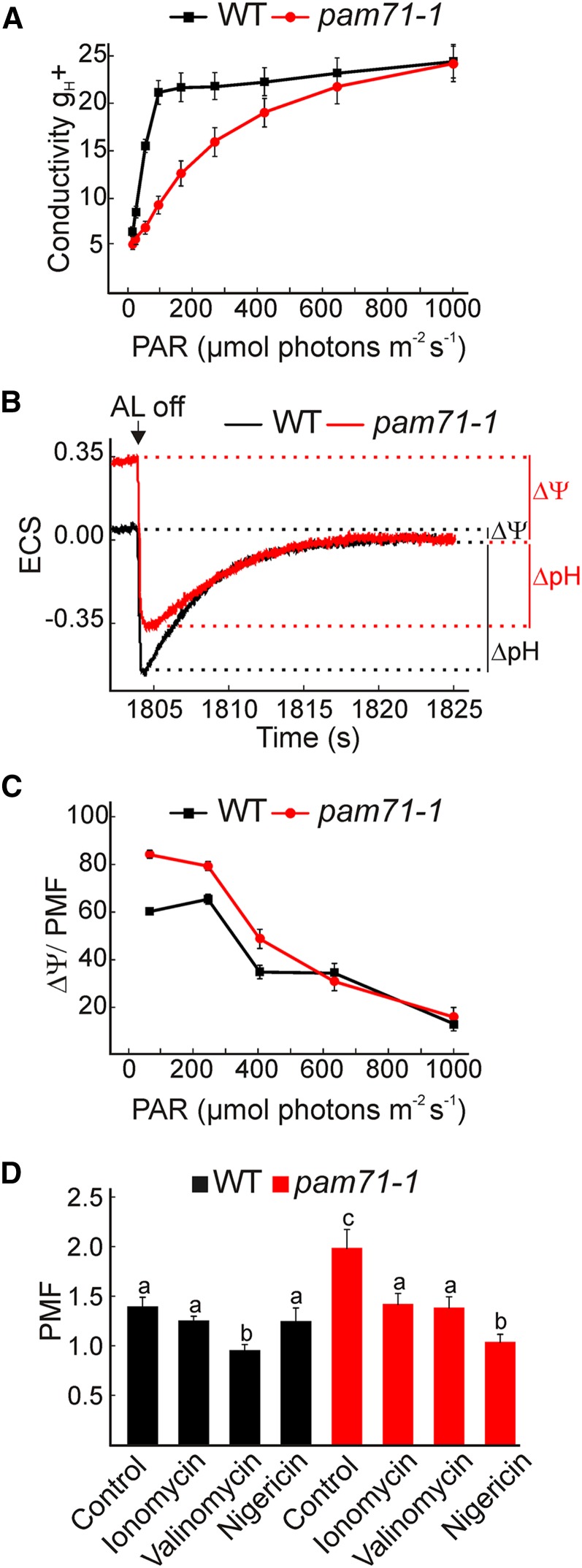
Based on its homology to yeast GDT1 and human TMEM165 proteins, an involvement of PAM71 in divalent cation transport across the thylakoid membrane, which could affect thylakoid ion conductivity (gH+) and PMF size and composition, seemed reasonable. Both parameters (gH+ and PMF) were determined from electrochromic shift (ECS) measurements, a method that determines pigment (mainly carotenoids and Chlb) absorbance changes induced by an electric field (Sacksteder and Kramer, 2000). The gH+ parameter indicates the rate at which cations (mainly H+) move from the thylakoid lumen to the stroma when briefly (100 ms) switching off the light. We found gH+ to be significantly reduced in pam71-1 under low and medium light intensities, reaching wild-type values only at high light intensities (Figure 6A). This finding most probably reflects a reduced H+ conductivity through the ATP synthase, which arises from a diminished OEC functionality (Figures 3B and 3C) and, hence, less H+ availability.
Figure 6.
Membrane Conductivity and PMF Formation of pam71-1 and the Wild Type.
(A) Thylakoid membrane conductivity gH+ (s−1) was determined from plants that were dark-adapted overnight. Leaves were illuminated for 2 min at increasing light intensities in the range of 13 to 1028 µmol photons m−2 s−1 before membrane conductivity was determined from the ECS signal upon switching off the light after each time point. The data are means ± se (n = 9).
(B) ECS (515 to 550 nm ΔI/I 103) was determined after actinic light (AL) was switched off. Before actinic light was switched off, leaves were illuminated for 30 min at 90 µmol photons m−2 s−1. Partitioning of PMF to Δψ and ΔpH is indicated with bars.
(C) PMF partitioning to Δψ (%) was determined after actinic light was switched off following 2 min illumination at the indicated PAR. The data are means ± se (n = 9).
(D) Magnitude of the PMF (ECSt) in the presence of ionophores. Detached leaves were incubated for 30 min in water (control), the ionophores ionomycin (30 µM) and valinomycin (30 µM), or in the presence of nigericin (50 µM) under a growth light, then kept in the dark for 30 min before measurement. To determine ECSt, the treated leaves were illuminated for 2 min at 90 µmol photons m−2 s−1. The data are means ± se (n = 5 to 6). Different letters indicate statistical significance according to ANOVA (P < 0.05).
The PMF size and composition were determined after 30 and 2 min illumination under growth light conditions, and the ECS signal was obtained after switching off the light (20 s) (Figure 6B; Supplemental Figure 8A). The total PMF size was higher in pam71-1 relative to the wild type in both experiments and was more pronounced after a short illumination period (Supplemental Figure 8A). Both components (Δψ and ΔpH) of the PMF were altered in pam71-1, with Δψ being enhanced and ΔpH being decreased (Figure 6B). The total PMF was higher in pam71-1 across all tested light intensities (Supplemental Figure 8B), which at low and medium light intensities (90 to 500 µmol photons m−2 s−1) could be explained by the higher Δψ (Figure 6C). However, at light intensities of 600 to 1000 µmol photons m−2 s−1, Δψ reached similar values to the wild type (Figure 6C).
A higher Δψ contribution to the thylakoid PMF in pam71-1 suggests ion imbalance between the two sides of the thylakoid membrane. To further investigate the nature of ion imbalance across the thylakoid membrane, we used ionophore treatments. The total PMF in pam7-1 decreased to wild-type values in the presence of ionomycin, a divalent cation ionophore (Figure 6D). PMF size in pam71-1 was also reduced upon treatment with valinomycin, a monovalent cation ionophore with high affinity for K+, but remained significantly higher than in treated wild-type plants. In the presence of nigericin, a H+/K+ uncoupler, the PMF decreased further in pam71-1, suggesting that the ion imbalance across the membrane is also associated with an altered ΔpH, which may be a direct or indirect effect. Taken together, these results suggest that pam71-1 displayed an increased PMF and Δψ due to an altered ion distribution in the chloroplast, with cation accumulation inside the thylakoid lumen.
Altered Ca2+ and Mn2+ Partitioning in pam71 Chloroplasts
We used radiolabeled 45Ca2+ assays to investigate calcium partitioning in the chloroplast following illumination. Intact chloroplasts were used for two reasons: (1) Thylakoids isolated from wild-type Arabidopsis take up very little 45Ca2+ (Supplemental Figure 9). (2) More importantly, total chloroplasts (stroma plus thylakoids) and thylakoids alone could be obtained from the same source material, allowing the relative distribution of 45Ca2+ to be estimated. Light-dependent uptake of Ca2+, according to Kreimer et al. (1985), is shown in Figure 7A. 45Ca2+ was taken up into both wild-type and pam71-1 chloroplasts during illumination. The levels of radioactivity recovered in subsequently isolated thylakoids clearly differed between the wild type and pam71-1. The pam71-1 thylakoids accumulated ∼70%, while the wild type accumulated only ∼30%, of the total amount of 45Ca2+ enclosed in chloroplasts after 30 min incubation (Figures 7A and 7C). These results are in agreement with the observed PMF effects (Figure 6D) and suggest that Ca2+ accumulation in the lumen may contribute to those effects.
Figure 7.
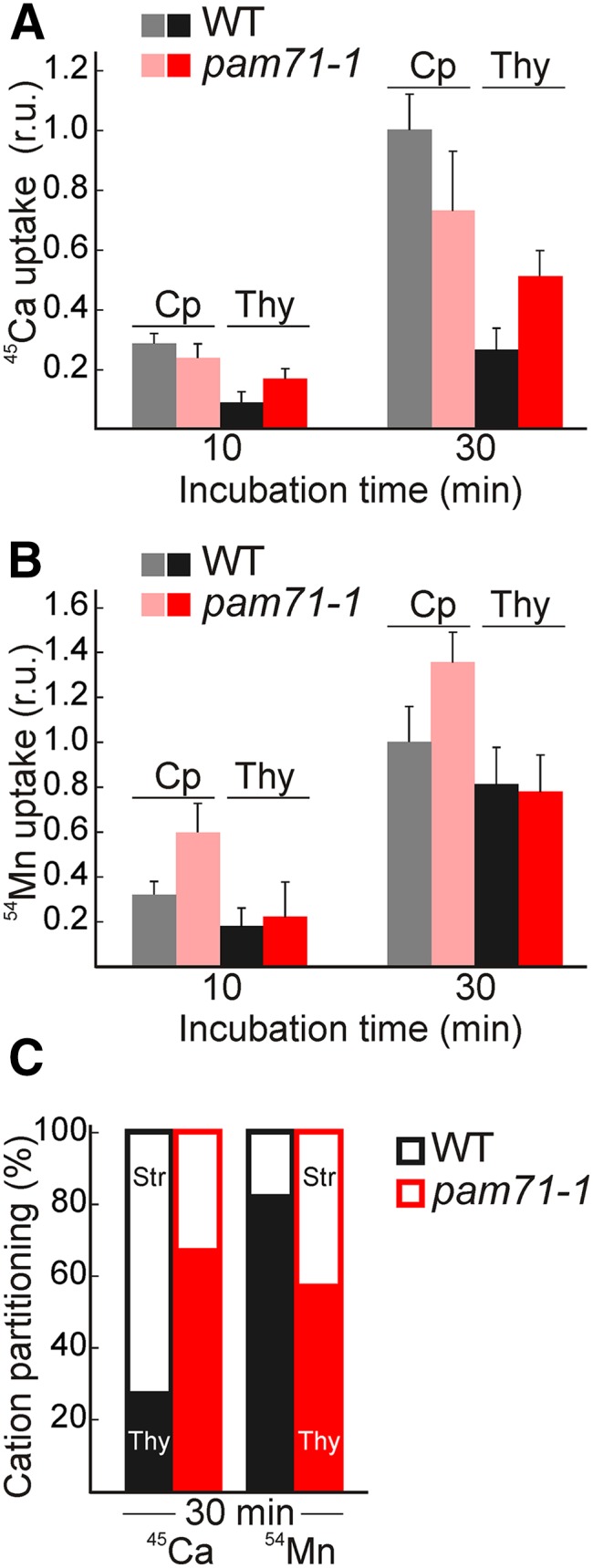
Cation Partitioning in pam71-1 and the Wild Type.
(A) 45Ca2+ uptake (r.u.) into intact chloroplasts and recovery of radioactivity from thylakoids. Reactions were initiated by adding 45Ca2+ (1.5 µM final concentration) to intact chloroplasts adjusted to 0.3 mg chlorophyll mL−1 and then the chloroplasts (Cp) were either kept intact for 10 min on ice in sorbitol (0.33 M) or osmotically lysed for 10 min on ice and vortexed every minute to release thylakoids (Thy), indicated by 10 min incubation. Light incubation was performed for 20 min at 90 µmol photons m−2 s−1 and then the chloroplasts (Cp) were either kept intact for 10 min on ice in sorbitol (0.33 M) or osmotically lysed for 10 min on ice and vortexed every minute to release thylakoids (Thy), indicated by 30 min incubation. Reactions were stopped by the addition of 50 mM EGTA.45Ca2+ uptake values are given as relative units. The mean cpm (counts per minute) value after 30 min of incubation for wild-type chloroplasts was set as a relative unit to one in individual experiments. Mean cpm values were 0.8 × 105, 1.2 × 105, and 1.5 × 105. Measurements were performed with three independent chloroplast isolations including three replicates each. The data are means ± sd (n = 9).
(B) 54Mn2+ uptake (r.u.) into chloroplasts and recovery of radioactivity from thylakoids. Reactions were initiated by adding 54Mn2+ (0.15 µM final concentration) to intact chloroplasts adjusted to 0.3 mg chlorophyll mL−1. Further processing was performed as in (A). Mean cpm values were 1.4 × 103 and 1.6 × 103. Measurements were performed with two independent chloroplast isolations including three replicates each. The data are means ± sd (n = 6).
(C) 45Ca2+ and 54Mn2+ partitioning between stroma (Str) and thylakoid (Thy) in wild-type and pam71-1 chloroplasts (%). Values are deduced from (A) and (B) for samples that were incubated for 30 min.
We also analyzed 54Mn2+ partitioning in wild-type and pam71-1 chloroplasts using the same experimental setup. Light-dependent uptake of Mn2+ was observed in both wild-type and mutant chloroplasts (Figure 7B). Under our experimental conditions, the mutant chloroplasts took up 1.4-times the wild-type amount during the illumination period (Figure 7B). The proportion of the total uptake of 54Mn2+ found in the thylakoids was also determined. In the wild type, more than 80% of the radiolabeled Mn2+ within the chloroplasts was found in the thylakoid fraction (Figures 7B and 7C). In contrast, only 58% of radiolabeled Mn2+ was recovered in the pam71-1 thylakoid fraction (Figures 7B and C). Similar observations were made with pam71-2 (Supplemental Figure 10). Based on these findings, we assume that the Ca2+ and Mn2+ ions are differently sequestered in pam71. While Ca2+ accumulates in the thylakoid lumen of pam71, Mn2+ accumulates in the stroma. Mn2+ uptake into thylakoids is not abolished in pam71. However, the fraction allocated to the thylakoids is less than in the wild type. This finding is in good accordance with the results obtained by SEC-ICP analysis, where a clear reduction in manganese bound to PSII complexes was observed (Figures 3C and 3F).
Supplementation with Mn2+ Restores Photosynthesis in pam71-1
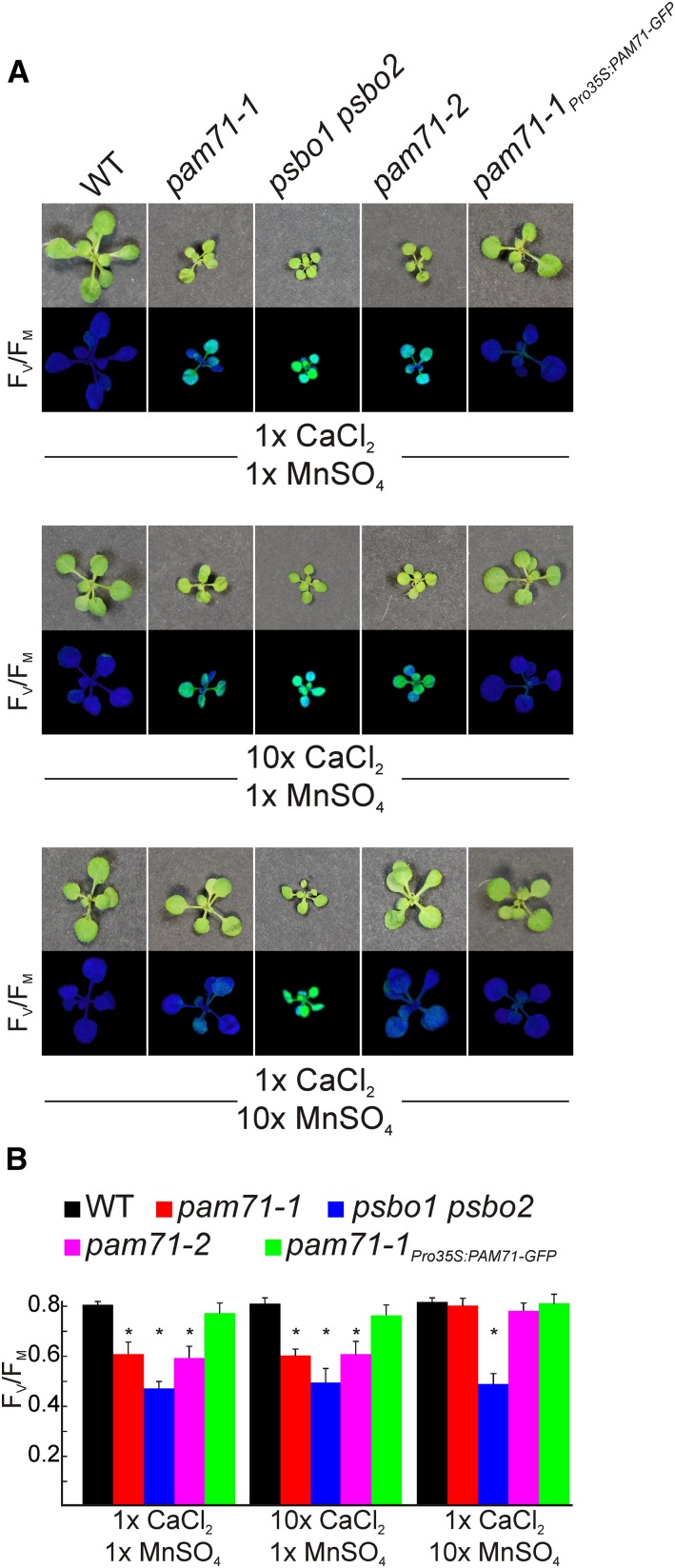
To further study the effect of pam71 on cation homeostasis, wild-type and mutant plants were cultured on Murashige and Skoog (MS) medium supplemented with either calcium or manganese. The addition of appropriate ions to the medium has been shown to restore defects caused by perturbations in metal ion homeostasis, e.g., for the iron transporter IRT1 (Varotto et al., 2002). As outlined above, the pam71 phenotype resembles that of the psbo1 psbo2 mutant; hence, the latter was included as a control in the feeding experiment. The expected drop in FV/FM, comparable to that seen in soil-grown plants, was observed in both pam71 alleles and psbo1 psbo2 grown on control medium, whereas pam71-1Pro35S:PAM71-GFP behaved similar to the wild type. The FV/FM phenotype in pam71 and psbo1 psbo2 was unaffected by supplementation with calcium (Figures 8A and 8B). Likewise, deprivation of calcium did not restore the FV/FM phenotype in pam71 alleles (Supplemental Figure 11). In contrast, when pam71 plants were grown on medium supplemented with manganese, the FV/FM value increased significantly to attain the value typical of wild-type plants (Figures 8A and 8B), while it remained unchanged in psbo1 psbo2 plants. Thus, the possibility that the addition of Mn2+ leads to a generalized improvement of PSII function in PSII-deficient mutants can be ruled out. This result indicates that the diminished PSII functionality in pam71 caused by manganese depletion in thylakoids (Figure 7) can be rescued by increasing the manganese availability in pam71 chloroplasts.
Figure 8.
Manganese Supplementation Restores Photosynthesis in pam71.
(A) Seeds of the wild type, pam71-1, psbo1 psbo2, pam71-2, and pam71-1Pro35S:PAM71-GFP were surface sterilized and grown for 2 weeks on MS medium containing either 1× CaCl2 (0.332 g L−1) and 1× MnSO4 (0.017 g L−1), 10× CaCl2 (3.32 g L−1) and 1× MnSO4, or 1× CaCl2 and 10× MnSO4 (0.17 g L−1). At the end of the growth period, the maximum quantum yield of PSII (FV/FM) was recorded.
(B) FV/FM values are given for the wild type, pam71-1, psbo1 psbo2, pam71-2, and pam71-1Pro35S:PAM71-GFP grown on the indicated MS medium. Bars represent mean values (±sd) of three experiments, each including five individual plants (n = 15). Asterisks indicate statistical significance (t test, P < 0.001) of differences between wild-type and mutant plants.
Supplementation with Mn2+ Also Restores Photosynthesis in the Chlamydomonas Mutant cgld1
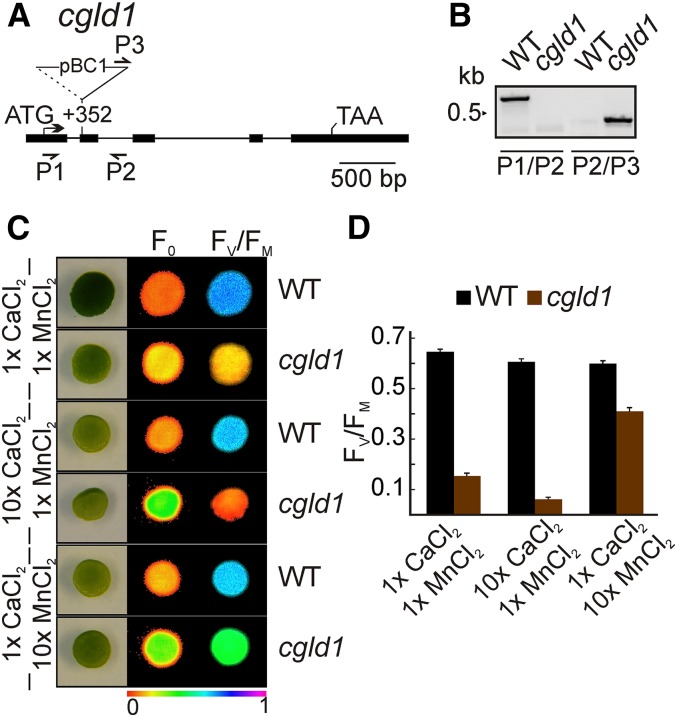
We speculated whether the function of PAM71 is conserved in green algae. Based on phylogenetic analysis (Supplemental Figure 7), a PAM71 homolog exists in Chlamydomonas that shares 45% identity/59% similarity with PAM71 over its entire length. We made use of the publicly available insertional mutant collection described recently (Dent et al., 2015) and obtained the mutant strain cgld1 from the Chlamydomonas Resource Center (http://chlamycollection.org/). This strain contains an insertion in exon 2 of the CGLD1 gene, at position +352 bp relative to the start codon (Figure 9A) and genotyping confirmed the presence of the mutation (Figure 9B). The mutant cgld1 exhibits a higher NPQ (Dent et al., 2015), a light-green phenotype when grown on standard medium, and a higher F0 and a reduced FV/FM value compared with the wild type (Figure 9C), indicating reduced PSII function, similar to the Arabidopsis mutant pam71. We therefore grew cgld1 on medium supplemented with additional calcium or manganese. The FV/FM value was partially restored only on medium supplemented with 10× manganese (Figures 9C and 9D). Although the FV/FM value in cgld1 did not reach wild type levels under these conditions, it was significantly increased compared with standard growth conditions (t test, P < 0.001). Thus, taken together, our results indicate that PSII function in cgld1 can be restored by excess Mn2+, as in the case of the Arabidopsis pam71 mutant; thus, the function of the corresponding gene products is presumably conserved in the green lineage.
Figure 9.
Manganese Supplementation Partially Restores Photosynthesis in the cgld1 Mutant of Chlamydomonas.
(A) Tagging of the CGLD1 locus (mRNA model XM_001701863). Exons are shown as black boxes and introns as black lines. Start and stop codons are indicated. The cgld1 allele was identified by Dent et al. (2015) (CAL029_02_05). The location of the insertion is indicated with respect to the start codon. The insertion is not drawn to scale. Binding sites of primers used for genotyping are indicated by arrows.
(B) The position of the insertion of the paromomycin resistance cassette was verified by PCR using wild-type (P1/P2) and mutant-specific (P2/P3) primer pairs.
(C) Test of PSII function in the presence of increased CaCl2 and MnCl2 concentrations. Cells were grown photoheterotrophically after spotting (105 cells per spot) onto TAP plates containing 0.34 mM (1×) or 3.4 mM (10×) CaCl2 and 25 µM (1×) or 250 µM (10×) MnCl2 in the indicated combinations, and the minimal chlorophyll a fluorescence (F0) (middle panel) and maximum quantum yield of PSII (FV/FM) (right panel) were recorded. The colored scale at the bottom indicates the signal intensities.
(D) FV/FM values are indicated for the wild type and cgld1 grown on the indicated TAP medium in (C). Bars represent values from five biological replicates (±sd).
PAM71 Confers Manganese Tolerance to Δpmr1 Yeast Cells
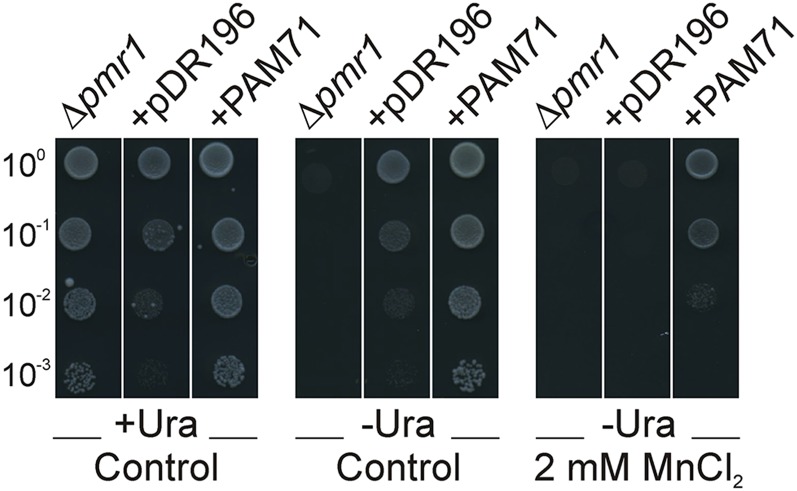
Our results so far suggested an involvement of PAM71 in manganese transport from the chloroplast stroma into the thylakoid lumen of Arabidopsis. However, it could not be ruled out that PAM71 only regulates manganese transport systems and that the phenotypic changes in pam71 resulted from a deactivation of this as yet unidentified manganese transport system. Therefore, PAM71 was cloned into the yeast expression vector pDR196 and then introduced into the Δpmr1 yeast strain. The P2-type Ca-ATPase, PMR1 (Plasma Membrane ATPase related), pumps Ca2+ and Mn2+ into the Golgi, and when PMR1 is defective, yeast cells are more sensitive to high concentrations of Mn2+ (Dürr et al., 1998). As expected, we observed Δpmr1 growth on control medium, but not on medium supplemented with manganese (Figure 10). When Δpmr1 cells were transformed with the empty pDR196 vector, uracil auxotrophy was recovered, but sensitivity to Mn2+ remained. However, when PAM71 was expressed in Δpmr1 cells, uracil auxotrophy was recovered and sensitivity to high Mn2+ was suppressed (Figure 10). These results suggest that PAM71 is directly involved in manganese transport. Therefore, we propose that the phenotypic changes are a direct consequence of defective manganese uptake into the thylakoid lumen in pam71 mutant alleles.
Figure 10.
Suppression of the Mn2+-Sensitive Phenotype of the Δpmr1 Yeast Mutant via Expression of PAM71.
Cells of the yeast mutant in the P-type Ca2+/Mn2+-ATPase (Δpmr1) containing the empty vector pDR196 (+pDR196) or vector with PAM71 (+PAM71) were verified by PCR and dropped on synthetic medium (control) and on synthetic medium supplemented with 2 mM MnCl2. For selection, uracil was added (+) or omitted (−). Undiluted cells (10°) corresponded to an OD600 = 3.0. Photographs were taken after 4 d at 30°C.
DISCUSSION
In this article, we described the phenotypic changes in Arabidopsis plants that lack PAM71. PAM71 is a thylakoid membrane protein and a member of the UPF0016 family of membrane proteins, with homologs in prokaryotes and eukaryotes (Figure 4; Supplemental Figure 7, Supplemental Table 2, and Supplemental Data Set 1). The pam71 mutant plants showed a clear defect in PSII. Numerous auxiliary proteins involved in PSII assembly have been described in recent years (Mulo et al., 2008; Chi et al., 2012), but PAM71 is apparently not one of these classical factors. Instead, our data implicate PAM71 in manganese uptake at the thylakoid membrane, which is needed for correct OEC function, and they indicate that PAM71 is beneficial for dissipation of Δψ.
Role of PAM71 in Calcium Homeostasis
Ca2+ ions, which are important for multiple processes in chloroplasts, are redistributed between the thylakoid lumen and the stroma upon transfer from light to dark and vice versa (Hochmal et al., 2015). PAM71 is well conserved through evolution and shares homology with proteins from yeast (GDT1) and human (TMEM165) (Figure 4), which might form a new group of Ca2+/H+ exchangers involved in calcium and pH homeostasis (Demaegd et al., 2013, 2014). Hence, a possible explanation for the PSII phenotype in pam71 could be reduced calcium accumulation in the thylakoid lumen, owing to a lack of ΔpH-coupled Ca2+ uptake. Indeed, a Ca2+/H+ exchanger operating in the thylakoid membrane of pea (Pisum sativum) has been found (Ettinger et al., 1999) (Supplemental Figure 9). However, PMF measurements in this study revealed that, in comparison to the wild type, the pam71 mutant displayed altered divalent cation distribution between the stroma and the thylakoid lumen (Figure 6D). In fact, by employing light-stimulated uptake assays, Ca2+ was found to be enriched in the thylakoid lumen of pam71 (Figure 7) and, hence, presumably contributes to the misbalanced cation distribution. We therefore conclude that the PSII phenotype of pam71 plants is not caused by diminished calcium uptake from the chloroplast stroma into the thylakoid lumen. A higher Ca2+ content in the lumen of pam71 could affect the structure of the OEC, as PsbO detaches from the PSII complex when excess Ca2+ is present (Ono and Inoue, 1983). However, PSII functionality in pam71 could not be rescued by deprivation of calcium from the growth medium (Supplemental Figure 11). Accordingly, higher calcium levels failed to have strong effects on PSII functionality in wild-type plants. It cannot be ruled out that a minor adverse effect on the OEC structure in pam71 is caused by higher calcium contents; however, the strong pam71 phenotype is most likely due to other factors.
In some respects, pam71 resembles the tpk3 mutant. In both mutants, diminished capacity to generate a normal PMF is coupled with a higher Δψ/ΔpH ratio (Figure 6; Carraretto et al., 2013). In the case of tpk3, it was concluded that this arises from defective K+ homeostasis due to reduced efflux of K+ from the lumen. Despite the similarity in PMF partitioning, the FV/FM value, which reflects the functional competence of PSII, is less severely affected in tpk3 mutant lines (Carraretto et al., 2013) than in pam71. In the latter, the maximum quantum yield of PSII is clearly reduced (Figure 1), which allowed us to conclude that the impairment of PSII function was not a consequence of altered PMF partitioning. Nevertheless, the question remains how a higher Δψ/ΔpH ratio in pam71 can be explained. Manganese is needed by the plant cell only as a micronutrient and thus the free stromal Mn2+ concentration might be at least one magnitude below that of Ca2+. Any altered partitioning of Mn2+ might thus be negligible in terms of Δψ. Instead, calcium is needed by the plant cell as a macronutrient. The predominant portion of the chloroplastic Ca2+ is bound to the negatively charged thylakoid membrane or to calcium binding proteins (Kreimer et al., 1987), and free stromal Ca2+ was determined to be in the range of 2 to 6 µM in spinach leaves (Kreimer et al., 1988). Thus, it is conceivable that an altered partitioning of calcium (and perhaps other monovalent and divalent cations) in pam71 could contribute to a higher Δψ at growth light intensities (Figure 6; Supplemental Figure 8).
Role of PAM71 in Manganese Homeostasis
In almost every PSII mutant with a reduced maximum quantum yield of PSII, growth is retarded (Peng et al., 2006; Ma et al., 2007; Armbruster et al., 2010; Schneider et al., 2014), and pam71 is no exception (Figure 1). Starch accumulation and carbon fixation are also depressed in pam71, presumably as a direct consequence of impaired PSII activity (Figures 2C and 2D). The primary site of the defect in PSII was narrowed down to the OEC (Figures 3B and 3E). The OEC is an inorganic Mn4CaO5 cluster, and several lines of evidence suggest a depletion of Mn2+ ions in thylakoids of pam71. Manganese deficiency is associated with the reduction in PSII supercomplexes in barley (Hordeum vulgare) (Schmidt et al., 2015), and this feature is readily observed in pam71 (Figures 2A). Second, Mn2+ ions were differentially distributed, in favor of the stroma, in pam71 chloroplasts during illumination (Figure 7C). Third, growth of pam71 on manganese-supplemented medium resulted in recovery of the maximum quantum yield of PSII (Figure 8). This is in line with our analysis of the Chlamydomonas mutant cgld1 (Figure 9). As in plants, manganese deficiency in Chlamydomonas results in the loss of PSII function (Allen et al., 2007), and in cgld1, this loss could partially be rescued by the addition of manganese (Figures 9C and 9D). Fourth, the decreased manganese incorporation per unit PSII in pam71, despite having a similar total leaf manganese concentration when compared with the wild type, clearly demonstrates that the allocation of manganese within the plant is significantly altered. In particular, in PSII supercomplexes, the decreased manganese incorporation per unit PSII was pronounced in pam71 (Figures 3C and 3F). Fifth, expression of the PAM71 protein in Δpmr1 yeast cells suppresses their Mn2+ sensitivity (Figure 10).
Together, these findings indicate that a deficiency of Mn2+ ions in thylakoids is responsible for the PSII phenotype of pam71. The primary effect of diminished OEC function is reduced water splitting, which in turns results in reduced lumen acidification, as reflected in a reduced ΔpH (Figure 6B) and reduced H+ conductivity through ATP synthase in pam71 (Figure 6A). These results are in good agreement with the reduced NPQ observed in pam71 compared with the wild type under steady state illumination conditions (Table 1), as the major component of NPQ, energy-dependent quenching, is strictly dependent on an acidic lumenal pH (Ruban et al., 2012; Zaks et al., 2013). Accordingly, the increase in NPQ in pam71 leaves exposed to short illumination periods (Supplemental Table 1) is similar to observations made with a psbo1 mutant (Allahverdiyeva et al., 2009). In pam71, a reduced linear electron flow is evident from the reduced effective quantum yield of PSII (Figures 3A and 3D). We propose that during the dark-light transition, the reduced linear electron flow in pam71 causes a decelerated activation of the Calvin cycle (Michelet et al., 2013) and/or ATP synthase (Nalin and McCarty, 1984; Buchert et al., 2015), both regulated by the ferredoxin/thioredoxin system (Schürmann and Buchanan, 2008). In this scenario, the resulting transient tailback (pileup) of protons in the lumen might trigger a transient NPQ increase.
Molecular Function(s) of the PAM71 Protein
The PAM71 protein contains two highly conserved E-x-G-D-(KR)-(TS) motifs with two negatively charged acidic residues (Glu/E and Asp/D) and an acidic loop (Figures 4), which provide a suitable microenvironment for binding of divalent cations (Demaegd et al., 2014). These features, together with its proposed topology (Figure 5), are reminiscent of the cation/Ca2+ exchanger superfamily (Emery et al., 2012; Demaegd et al., 2014), although there is no notable primary sequence homology. PAM71 belongs to the UPF0016 family of membrane proteins. Its yeast and human homologs, Sc-GDT1 and Hs-TMEM165, were postulated to function as Ca2+/H+ exchangers since they transport calcium from the cytosol into the lumen of an acidic organelle, the Golgi apparatus (Demaegd et al., 2013). In analogy to this, PAM71 could act as a Mn2+/H+ exchanger, which transports Mn2+ from the chloroplast stroma into the acidic thylakoid lumen. As outlined above, the physiological impairments in pam71 resulted from depletion of manganese in the thylakoid lumen, and expression of the PAM71 protein in Δpmr1 yeast cells suppressed their Mn2+ sensitivity (Figure 10). A number of putative Mn2+ transporters in plants have been described and can be grouped based on the direction of manganese transport either into or out of the cytosol (Socha and Guerinot, 2014). Manganese transport from the cytosol into the vacuole mediated by CATION EXCHANGER2 (CAX2), CAX4, and CAX5 (Hirschi et al., 2000; Cheng et al., 2002; Edmond et al., 2009) or into the prevacuolar and Golgi-like compartment mediated by the Mn2+ transporter MTP11 (Delhaize et al., 2007; Peiter et al., 2007) presumably also involves a Mn2+/H+ antiport mechanism in Arabidopsis. Most plant Mn2+ transporters characterized to date have a broader specificity for other divalent cations (Socha and Guerinot, 2014), for instance, CAX2 (Hirschi et al., 2000; Schaaf et al., 2002; Shigaki et al., 2003). Therefore, it cannot be excluded that PAM71 also has a broader specificity, and further experiments are needed to firmly establish the substrate affinity of PAM71 toward manganese and other divalent cations, e.g., by employing electrophysiological measurements in Xenopus laevis oocytes.
It is interesting to note that pam71, despite having a dysfunctional OEC, can still grow photoautotrophically (Figure 1B) and complete its life cycle. Therefore, we anticipate that other low-affinity systems involved in manganese and calcium transport at the thylakoid membrane exist. In this context, it is conceivable that other such low-affinity systems are able to compensate for the defect in pam71 by pumping higher amounts of cations into the lumen. This could provide an explanation for the higher Δψ and the enrichment of calcium in the lumen of pam71 (Figures 6 and 7A) and would further explain the observation that the addition of increasing manganese concentrations rescued the pam71 PSII phenotype (Figure 8). It is tempting to speculate that during evolution of the green lineage, PAM71 might have evolved in order to meet the high demand of manganese in PSII and at the same time to balance PMF generation. This idea is supported by the observation that PAM71 homologs are strictly conserved in Viridiplantae. PAM71 homologs are encoded in the genomes of embryophytes and green algae (Supplemental Figure 7), and its function is presumably conserved in green algae (Figure 9). The challenge for future work is to identify additional thylakoid proteins facilitating the transport of divalent cations from the chloroplast stroma into the lumen; the pam71 mutant offers a promising platform for such studies.
METHODS
Plant Material and Vector Construction
The Arabidopsis thaliana pam71 mutants (GK-166A05 and GK-094C03, accession Columbia-0) were identified in the GABI-Kat collection (Rosso et al., 2003). Genotyping of both lines was performed using PCR with appropriate primer combinations (Supplemental Table 3) and PCR products were sequenced. The psad1-1 (Ihnatowicz et al., 2004) and psbo1-3 psbo2-2 (here named psbo1 psbo2; Steinberger et al., 2015) mutant lines were used in control experiments. Wild-type plants were accession Columbia-0.
Cloning of pPro35S:PAM71 and pPro35S:PAM71-GFP started with PCR primers (Supplemental Table 3) bearing a 5′ CACC overhang and a 3′ primer with or without a stop codon, together with Arabidopsis first-strand cDNA. The amplified full-length PAM71 cDNAs were ligated into pENTR-TOPO (Life Technologies). The Entry clones were subsequently recombined into pB7FWG2 (Karimi et al., 2002), yielding the plasmid pPro35S:PAM71-GFP or into pGWB2 (Nakagawa et al., 2007), yielding pPro35S:PAM71.
Both plasmids were introduced into Arabidopsis by floral dip transformation (Clough and Bent, 1998). The Pro35S:PAM71 construct was stably introduced into pam71-1 and pam71-2 strains, and individual transgenic plants (pam71-1Pro35S:PAM71 and pam71-2Pro35S:PAM71) were selected on the basis of their resistance to kanamycin (50 mg L−1). The Pro35S:PAM71-GFP construct was stably introduced into pam71-1, and several individual transgenic plants (pam71-1Pro35S:PAM71-GFP) were selected on the basis of resistance to BASTA by spraying with 0.5 mL L−1 BASTA (which contains 200 g L−1 ammonium glufosinate). Successful integration of the transgene was verified by PCR. Expression of PAM71 in different genotypes was analyzed by RT-PCR using an appropriate primer combination (Supplemental Table 3), first-strand cDNA, and 30 PCR cycles. The absence or presence of a PCR product was evaluated using DNA gel electrophoresis.
Plant Growth Conditions
Unless stated otherwise, Arabidopsis wild-type and mutant lines were grown for 4 to 5 weeks in a growth chamber (equipped with 17-W cool white fluorescent lamps; CLF Plant Climatics) in a 12-h/12-h day-night cycle at 90 µmol photons m−2 s−1. Plants were cultivated on soil substrate A210 (Stender AG) without further fertilization and watered with tap water. For growth on medium with different metal ion contents, surface-sterilized wild-type and mutant seeds were grown on 1× MS medium supplemented with 1% (w/v) sucrose and CaCl2/MnSO4 (as indicated in the main text) at 90 µmol photons m−2 s−1 white light and in a 16-h/18-h day-night cycle. Arabidopsis plants used for microscopy and kanamycin selection were surface sterilized and grown on standard MS medium. Arabidopsis plants used for transformation and BASTA selection were grown in a temperature-controlled greenhouse in a 16-h/8-h light/dark cycle.
Pea plants (Pisum sativum var Arvica) were grown for 2 weeks on vermiculite (3 to 8 mm) in a growth chamber with a 14-h/10-h day-night cycle at 20°C/15°C and 100 µmol photons m−2 s−1.
Chlamydomonas reinhardtii Material and Cultivation
Wild-type Chlamydomonas (strain CC-4051) and the cgld1 mutant (strain CAL029_02_05) (Dent et al., 2015) were obtained from the Chlamydomonas Resource Center (http://chlamycollection.org/) and grown under continuous light (20 to 40 µmol photons m−2 s−1) in Tris-acetate-phosphate (TAP) medium, which contains 0.34 mM Ca2+ and 25 µM Mn2+ cations (Harris, 1989). Strain cgdl1 was grown on TAP plates (1.5% [w/v] agar) supplemented with 10 µg mL−1 paromomycin for genotyping. DNA was extracted as described by Newman et al. (1990), genotyping was performed using PCR with appropriate primer combinations (Supplemental Table 3), and PCR products were sequenced. For analysis of chlorophyll a fluorescence, strains were grown to a chlorophyll concentration of 10 to 15 µg mL−1. Cell number was determined in a Thoma cell-counting chamber, and ∼105 cells were spotted onto TAP medium enriched with CaCl2 (3.4 mM, 10×) or MnCl2 (250 µM, 10×). Colonies were exposed to low light (1 to 5 µmol photons m−2 s−1) for 48 h and their chlorophyll a fluorescence was determined.
Heterologous Expression of PAM71 in Yeast
The coding sequence of PAM71 was PCR amplified from Arabidopsis cDNA using the primers PAM71-F and PAM71-R (Supplemental Table 3). The PCR product was subcloned into pJET1.2 (Thermo Fisher Scientific) and verified by sequencing. After restriction digestion with SalI and SpeI, the fragment was cloned into the vector pDR196 (Rentsch et al., 1995; Loqué et al., 2007). The yeast mutant in the P-type Ca2+/Mn2+ ATPase, Δpmr1 (YGL167C) (Dürr et al., 1998) was transformed with the pDR196 vector containing PAM71 as insert and pDR196 vector without insert, respectively, according to Gietz and Schiestl (1991). After verification by PCR, yeast cells were streaked twice to preclude revertants and finally tested for manganese sensitivity. Cultures were precultivated overnight in synthetic medium (SC) at 30°C, 160 rpm. For selection, the medium was supplemented with G418 (Sigma-Aldrich) and uracil was omitted. Cells were harvested by centrifugation (2500g, 10 min, room temperature) and washed twice with 1× TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA). Then, the cells were resuspended in 1× TE buffer and cell density was adjusted to OD600 = 3.0. The different yeast strains were dropped undiluted, and as the dilution series 1:10, 1:100, and 1:1000, onto control SC plates, with uracil, without uracil, and without uracil supplemented with 2 mM MnCl2. Growth was photographically documented after 4 d incubation at 30°C.
Chlorophyll a Fluorescence and P700 Absorption Measurements
The photosynthetic performance of PSII of Arabidopsis was assessed by chlorophyll a fluorescence measurements using the Imaging-PAM or the Dual-PAM (Walz) as described for plant leaves (Schneider et al., 2014; Steinberger et al., 2015). To measure the performance of PSII in Chlamydomonas, colonies were dark-adapted for 5 min and exposed to a blue measuring beam to determine the minimal fluorescence (F0). A saturating 0.8-s light flash (2800 µmol photons m−2 s−1) was then applied, maximum fluorescence (FM) was determined with the Imaging-PAM, and the maximum quantum yield of PSII (FV/FM) was calculated (Maxwell and Johnson, 2000).
Fast chlorophyll a fluorescence [O-(K)-J-I-P] kinetics were recorded using a Handy PEA (Hansatech) on 10-min dark-adapted leaves (Strasser et al., 2004). The functionality of the PSII oxygen-evolving complex was studied based on the formation of the K peak as described previously (Tóth et al., 2007).
P700 absorption measurements were performed using the Dual-PAM, and ΔA/ΔAmax was determined according to Meurer et al. (1996).
Carbon Fixation and Starch Content
Responses of net CO2 assimilation rate (An) to light intensity were measured at either atmospheric (400 ppm) or saturating CO2 concentrations (2000 ppm) using a LI-6400XT portable photosynthesis system (Li-COR). Leaves from at least 2-h light-adapted plants were illuminated in the gas exchange chamber with stepwise increase in light intensity (six points ranging from 0 to 1000 µmol photons m−2 s−1 with 2 to 4 min of adaptation at each intensity). CO2 fixation data were normalized to leaf area.
Starch was isolated from leaves and assayed according to the method described by Lin et al. (1988).
Cation Partitioning
For isolation of intact chloroplasts, leaf samples (12 g fresh weight) were homogenized in 0.4 M sorbitol, 20 mM Tricine-NaOH (pH 8.4), 10 mM EDTA, 0.1% (w/v) BSA, 5 mM NaHCO3, 1 mM MgCl2, and 1 mM MnCl2 using a mixer (Waring Laboratory). The extract was filtered through two layers of Miracloth and concentrated by centrifugation for 5 min at 1500g. The pellet was resuspended in 80 mM sorbitol, 4 mM Tricine-NaOH (pH 8.4), 0.5 mM EDTA, and 1 mM MgCl2 and then layered onto a discontinuous 40% (w/v)/ 80% (w/v) Percoll gradient. After centrifugation for 15 min at 7000g, intact chloroplasts were isolated from the interface of the two layers and washed in the same buffer to remove residual Percoll.
Intact chloroplasts were resuspended at a concentration of 0.3 mg chlorophyll mL−1 in 0.33 M sorbitol, 50 mM Tricine/NaOH (pH 8.0), and 5 mM MgCl2. Import reactions were initiated by adding 45Ca2+ (specific activity > 10 Ci g−1; 1:20 dilution) or 54Mn2+ (specific activity > 20 Ci g−1; 1:20 dilution) to the reaction (Perkin-Elmer). Samples were divided and incubated for 10 min on ice in either 0.33 M sorbitol, 50 mM Tricine/NaOH (pH 8.0), and 5 mM MgCl2 (to keep chloroplasts intact) or in 10 mM HEPES/NaOH (pH 7.6) and 5 mM MgCl2 with vortexing (to release thylakoids). For light treatment, samples were exposed to 90 µmol photons m−2 s−1 provided by an FL-440 fiber illuminator (Walz) prior to splitting and incubation on ice. Samples were centrifuged for 1 min at 4°C, and the reactions were terminated by resuspending the pellets in 50 mM EGTA, 0.33 M sorbitol, and 50 mM Tricine/NaOH (pH 8.0). EGTA was used as a chelating agent. After centrifugation for 1 min at 4°C, the pellets were resuspended in 0.1% (w/v) SDS and transferred to scintillation cocktail before counting. Before converting counts per minute into relative units, the background radiation was subtracted. Background radiation was determined by adding 45Ca2+ (specific activity > 10 Ci g−1) or 54Mn2+ (specific activity > 20 Ci g−1) to chloroplasts as above and resuspending the chloroplasts in 50 mM EGTA, 0.33 M sorbitol, and 50 mM Tricine/NaOH (pH 8.0) after centrifugation. After a second centrifugation for 1 min at 4°C, pellets were resuspended in 0.1% (w/v) SDS and transferred to scintillation cocktail for counting.
Protein Analysis
Thylakoid membranes were isolated according to Schmidt et al. (2015). Protein concentration was determined using the Pierce BCA Protein Assay (Thermo Fisher Scientific). Protein samples were resuspended in 45 mM Tris-HCl (pH 6.8), 50 mM DTT, 0.1% (w/v) SDS, 10% (v/v) glycerol, and 0.01% (w/v) bromophenol blue, incubated at 65°C for 10 min, and fractionated on Tris-glycine SDS gels (12% acrylamide).
Sucrose gradient fractionation, Blue-Native PAGE analysis, and determination of protein topology were performed as described previously (Armbruster et al., 2010; Schneider et al., 2014).
For preparation of chloroplast subfractions, intact chloroplasts (see above) were broken by osmotic lysis in 10 mM HEPES-KOH (pH 7.6) and 5 mM MgCl2, incubated on ice for 10 min and vortexed periodically. The mixture was layered onto a discontinuous sucrose gradient consisting of 1.2, 1.0, and 0.46 M sucrose and centrifuged at 58,000g for 2 h. The stroma was obtained from the top fraction, the envelopes were collected from the middle fraction, and the thylakoids were collected as a pellet. The envelope fraction was further concentrated by centrifugation at 135,200g for 1 h at 4°C.
Coimmunoprecipitation assays were performed as described by Schneider et al. (2014).
Individual proteins of interest were detected using antibodies raised against D1 (Agrisera 05 084; lot 1412; 1:10,000 diluted), PsbO (Agrisera 06 142-33; lot 0912; 1:4000 diluted), PsbP (Agrisera 06 142-23; lot 0702; 1:4000 diluted), PsbR (Agrisera 05 059; 1:5000 diluted), Lhcb2 (Agrisera 01 003; lot 1301; 1:5000 diluted), PsaA (Agrisera 06 172; lot 1211; 1:1000 diluted), Cytb6 (Agrisera 03 034; lot 0612; 1:10,000 diluted), AtpB (β) (Agrisera 05 085; lot 0901; 1:4000 diluted), RbcL (Agrisera 03 037-200; lot 1307; 1:10,000 diluted), PsaD (Agrisera 04 046; lot 0510; 1:4000 diluted), GFP (Life Technologies A6455; lot 1692915; 1:5000 diluted), D2 (1:5000 diluted), CP43 (1:5000 diluted), CP47 (1:5000 diluted), and Tic40 (1:5000 diluted), in combination with α-rabbit IgG HRP (Sigma-Aldrich A9169; 1:25,000 diluted) using the Pierce enhanced chemiluminescence system (Thermo Fisher Scientific).
Oxygen Evolution Rate
The oxygen evolution measurement was performed as reported by Hakala et al. (2005). Briefly, Arabidopsis leaves (10 g fresh weight) were harvested and ground in 50 mL of buffer containing 40 mM HEPES (pH 7.4), 0.3 M sorbitol, 10 mM MgCl2, 1 mM EDTA, 1 M betaine, and 1% (w/v) BSA. The suspension was filtered through two layers of Miracloth and centrifuged at 1100g for 5 min. The pellet was resuspended in 50 mL of osmotic shock buffer (10 mM HEPES, pH 7.4, 5 mM sorbitol, and 10 mM MgCl2) and centrifuged at 2000g for 5 min. The thylakoid pellet was resuspended in storage buffer (10 mM HEPES, pH 7.4, 0.5 M sorbitol, 10 mM MgCl2, and 5 mM NaCl) to 30 µg chlorophyll mL−1. Oxygen evolution was measured with an oxygen electrode (Oxygraph; Hansatech) using 0.5 mM DCBQ as an external electron acceptor. Thylakoids were illuminated with 500 µmol photons m−2 s−1 provided by an FL-440 fiber illuminator.
Total Manganese Concentration in Leaves
The manganese concentration in leaves was determined using ICP-MS (Agilent 8800 ICP-QQQ-MS). Harvested leaves were freeze-dried and homogenized. Subsequently, a representative sample (∼40 mg) was digested using a pressurized microwave digestion system (UltraWAVE; Milestone) (Hansen et al., 2013). The accuracy and precision of the measurements were estimated by the analysis of certified reference material (apple leaf, NIST 1515, National Institute of Standards and Technology).
SEC-ICP-MS Measurements
Thylakoids membranes were isolated and solubilized as described by Schmidt et al. (2015). In short, thylakoids were centrifuged at 7000g at 4°C for 2 min and resuspended in ice-cold buffer A (25 mM Bis-Tris/HCl, pH 7.0, 12.5% [w/v] glycerol, 2 M betaine, and 0.25 mg mL−1 Pefabloc) to a protein concentration of 2 mg mL−1. An equal volume of detergent solution prepared in buffer A was added to a final concentration of 1% (w/v) α-dodecyl maltoside (Anatrace Products). Thylakoids were solubilized in darkness for 10 min on ice and insoluble material was removed by centrifugation at 18,000g at 4°C for 15 min.
Solubilized thylakoid proteins were passed through a 0.45-µm nylon membrane filter (Q-max RR syringe filters; Frisenette), and 50 µg of total protein was applied to a size-exclusion column (BioBasic SEC 1000; Thermo Scientific) using an inert HPLC system (Ultimate 3000; Dionex, Thermo Scientific). The column temperature was kept at 6°C during analysis. Protein elution was performed with 25 mM Bis-Tris (Sigma-Aldrich; BioXtra; pH 7.0, adjusted with trifluoroacetic acid) and 0.03% (w/v) α-dodecyl maltoside as the mobile phase. The outlet of the column was coupled to a triple quadrupole (QQQ) ICP-MS (Agilent 8800 ICP-QQQ-MS) for online detection of manganese binding in the size-fractionated PSII complexes. The ICP-QQQ-MS was operated in MS/MS scan mode with oxygen as the reaction gas, enabling simultaneous analysis of manganese and sulfur as the parent ion 55Mn+ and oxide product ion 48SO+, respectively. The integration time was 0.1 s per element. Quantification of the Mn:S stoichiometric ratio in photosynthetic complexes was performed by external calibration according to Schmidt et al. (2015). The plasma conditions and ion lenses were tuned on a daily basis for robust plasma conditions and maximum sensitivity.
Microscopy
Leaves of agar-grown Arabidopsis lines used for transmission electron microscopy were processed as described before (Schneider et al., 2014). For fluorescence microscopy, leaves of soil-grown Arabidopsis lines were cut into small pieces and incubated in 20 mM KCl, 10 mM MES (pH 5.7), 10 mM CaCl2, 0.5 M mannitol, 0.1% (w/v) BSA, and 0.1 g mL−1 macerozyme in the dark for 3 h. The isolated protoplasts were collected by centrifugation at 50g for 5 min, washed, and analyzed under an Axio-Imager fluorescence microscope (Carl Zeiss).
ECS Measurements
ECS measurements were performed using a Walz Dual PAM-100 equipped with a P515/535 module (Walz) (Schreiber and Klughammer, 2008). PMF size (ECSt) and partitioning into ΔpH and ΔΨ were determined using dark-interval relaxation kinetics as described (Cruz et al., 2001). The PMF data were normalized to a PSII single turnover determined by recording the ECS induced by a 5-µs flash at 200,000 µmol photons m−2 s−1. For ionophore treatments, detached leaves were incubated with ionomycin (30 µM), valinomycin (30 µM), or nigericin (50 µM) for 30 min in the light (90 µmol photons m−2 s−1), followed by a 30-min incubation in the dark before the magnitude of the PMF was determined.
Thylakoid membrane conductivity to cations (gH+) was calculated as gH+ = 1/τ, where τ is the timer constant for decay determined by fitting a single exponential decay function to the ECS decay signal obtained after the light was switched off (Cruz et al., 2005).
Transcription and Translation Analysis
Total RNA was isolated from Arabidopsis leaves using TRI reagent (Molecular Research Center), and RNA gel blot analysis was performed under stringent conditions using standard protocols. PCR amplification products were generated using specific primer combinations (Supplemental Table 3) and total DNA of Arabidopsis. Probes were labeled with [32P]dCTP and hybridization was performed at 65°C overnight.
Polysome isolation was performed by grinding 200 mg frozen leaf tissue in 0.2 M Tris-HCl (pH 9.0), 0.4 M KCl, 25 mM MgCl2, 25 mM EGTA, 0.2 M sucrose, and 1% (v/v) Triton X-100. Microsomal membranes were solubilized with 1% (v/v) Triton X-100 and 0.5% (w/v) sodium deoxycholate for 5 min on ice. The solubilized material was layered onto 15% (w/v) to 55% (w/v) sucrose gradients and centrifuged at 250,000g for 65 min at 4°C. The gradient was divided into 11 fractions from which RNA was extracted. All samples were subjected to RNA gel blot analysis.
For in vivo labeling, four to five leaves were incubated in 1 mCi of [35S]Met for 20 min in the presence of 20 µg mL−1 cycloheximide under constant illumination (60 µmol photons m−2 s−1). Subsequently, thylakoid proteins were prepared and fractionated on denaturing Tris-glycine SDS gradient gels (8 to 12% acrylamide) and signals were detected using a Typhoon imager (GE Healthcare).
Phylogenetic and Sequence Analysis
Alignments were generated using Clustal Omega (Sievers and Higgins, 2014). The phylogenetic tree was constructed and bootstrap values calculated using the program MEGA6 as described (Hall, 2013; Tamura et al., 2013). The evolutionary history was inferred using the maximum likelihood method based on the General Reverse Transcriptase + Freq. model (Dimmic et al., 2002). Bootstrap values were calculated based on 1000 iterations, and the tree with the highest log likelihood was chosen. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value. Sequence identities and similarities were calculated using NCBI blastp (Tatusova and Madden, 1999). Putative chloroplast transit peptide sequences were predicted by TargetP (Nielsen et al., 1997; Emanuelsson et al., 2000), and transmembrane helix prediction was performed with TmHMM (Krogh et al., 2001) and adjusted manually afterwards.
Accession Numbers
Sequence data were obtained from the National Center for Biotechnology Information under the following accession numbers: Homo sapiens NP_060945.2 (transmembrane protein 165, TMEM165); Saccharomyces cerevisiae AHY74663.1 (GDT1); Arabidopsis thaliana-1 NP_564825.1 (At1g64150, PAM71); Glycine max-1 NP_001241025.1; Vitis vinifera-1 XP_002272191.1; Oryza sativa-1 EEC70197.1; Brachypodium distachyon-1 XP_003565685.1; Zea mays-1 XP_008655787.1; Physcomitrella patens-1 XP_001756332.1; Chlamydomonas reinhardtii-1 XP_001701915.1/XM_001701863 (CGLD1); Volvox carteri-1 XP_002953583.1; Galdieria sulphuraria EME31602.1; Arabidopsis thaliana-2 NP_193095.2 (At4g13590, PAM71-HL); Oryza sativa-2 NP_001068053.1; Glycine max-2 XP_003535962.1; Vitis vinifera-2 XP_002285620.1; Zea mays-2 ACG35639.1; Brachypodium distachyon-2 XP_003570533.1; Physcomitrella patens-2 XP_001770923.1; Chlamydomonas reinhardtii-2 XP_001697928.1; Volvox carteri-2 XP_002949806.1; Synechocystis sp WP_010873575.1; Thermosynechococcus elongatus NP_681169.1; Anabaena sp WP_016950136.1; Mus musculus AAI41082.1; Drosophila melanogaster ACR20070.1; Caenorhabditis elegans NP_497567.1; Neurospora crassa CAE76363.1; Aspergillus nidulans XP_662418.1; Geobacter sulfurreducens NP_951914.1; Mycobacterium vaccae EJZ12059.1; Thermomonospora curvata WP_012855029.1; Arabidopsis thaliana NP_568535.1 (At5g36290), Arabidopsis thaliana NP_177032.1 (At1g68650); and Arabidopsis thaliana NP_173923.1 (At1g25520).
Supplemental Data
Supplemental Figure 1. Complementation of pam71-1 and pam71-2.
Supplemental Figure 2. Characterization of psbo1 psbo2 and psad1-1 with respect to chlorophyll a fluorescence parameters, thylakoid protein complex accumulation and ultrastructure, and oxygen evolution.
Supplemental Figure 3. Analysis of the CP43 subunit in pam71-1.
Supplemental Figure 4. Transcription and translation of chloroplast-encoded proteins.
Supplemental Figure 5. Fast chlorophyll a fluorescence kinetics.
Supplemental Figure 6. Coimmunoprecipitation analysis using pam71-1Pro35S:PAM71-GFP.
Supplemental Figure 7. Phylogenetic analysis.
Supplemental Figure 8. Proton motive force size and composition.
Supplemental Figure 9. Light-dependent accumulation of 45Ca2+ across thylakoid membranes of pea and Arabidopsis.
Supplemental Figure 10. Cation partitioning in pam71-2 and pam71-1Pro35S:PAM71-GFP.
Supplemental Figure 11. Calcium deprivation does not restore photosynthesis in pam71.
Supplemental Table 1. Nonphotochemical quenching under transient illumination in wild-type and pam71-1 plants.
Supplemental Table 2. Homologous PAM71 genes/proteins found in plants.
Supplemental Table 3. Primers used in this study.
Supplemental Data Set 1. Text file of the alignment used for the phylogenetic analysis in Supplemental Figure 7.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gabi Burkhard and Reinhild Rothe for their excellent technical assistance. We also thank Sarah Kühn for participating as an undergraduate student in this project, D. Marquardt for generating Pro35S:PAM71 plants, J. Soll for the Tic40 antibody and pea plants, R. Barbato for providing D2, CP43, and CP47 antibodies, N. Manavski for protoplast photographs, and Paul Hardy for critical reading of the manuscript. This research was supported by the German Science Foundation (Deutsche Forschungsgemeinschaft; LE 1265/20-1 to D.L., EXC 1028 to A.P.M.W. and U.-I.F., and EI 945/3-1 to M.E.), the Danish Council for Independent Research-Technology and Production Sciences (Grant DFF-5054-00042 to S.B.S.), and the Swedish Research Council (to C.S.).
AUTHOR CONTRIBUTIONS
A.S., I.S., A.M., A.H., T.R., M.L., C.G., N.H., S.K., S.B.S., M.E., and S.G., performed the research. A.S., A.M., M.E., S.B.S., S.H., and A.H. analyzed the data. A.S., U.-I.F., C.S., M.E., A.P.M.W., and D.L. designed the experiments. A.S., C.S., and D.L. wrote the article.
Glossary
- OEC
oxygen-evolving complex
- PMF
proton motive force
- NPQ
nonphotochemical quenching
- DCBQ
2,6-dichloro-1,4-benzoquinone
- ICP-MS
inductively coupled plasma mass spectrometry
- ECS
electrochromic shift
- MS
Murashige and Skoog
- TAP
Tris-acetate-phosphate
References
- Allahverdiyeva Y., Mamedov F., Holmström M., Nurmi M., Lundin B., Styring S., Spetea C., Aro E.M. (2009). Comparison of the electron transport properties of the psbo1 and psbo2 mutants of Arabidopsis thaliana. Biochim. Biophys. Acta 1787: 1230–1237. [DOI] [PubMed] [Google Scholar]
- Allahverdiyeva Y., et al. (2013). Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 75: 671–684. [DOI] [PubMed] [Google Scholar]
- Allen M.D., Kropat J., Tottey S., Del Campo J.A., Merchant S.S. (2007). Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143: 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U., Carrillo L.R., Venema K., Pavlovic L., Schmidtmann E., Kornfeld A., Jahns P., Berry J.A., Kramer D.M., Jonikas M.C. (2014). Ion antiport accelerates photosynthetic acclimation in fluctuating light environments. Nat. Commun. 5: 5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U., Zühlke J., Rengstl B., Kreller R., Makarenko E., Rühle T., Schünemann D., Jahns P., Weisshaar B., Nickelsen J., Leister D. (2010). The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsevich V.V., Pakrasi H.B. (1995). Molecular identification of an ABC transporter complex for manganese: analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 14: 1845–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsevich V.V., Pakrasi H.B. (1996). Manganese transport in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 271: 26057–26061. [DOI] [PubMed] [Google Scholar]
- Bondarava N., Beyer P., Krieger-Liszkay A. (2005). Function of the 23 kDa extrinsic protein of Photosystem II as a manganese binding protein and its role in photoactivation. Biochim. Biophys. Acta 1708: 63–70. [DOI] [PubMed] [Google Scholar]
- Bondarava N., Un S., Krieger-Liszkay A. (2007). Manganese binding to the 23 kDa extrinsic protein of Photosystem II. Biochim. Biophys. Acta 1767: 583–588. [DOI] [PubMed] [Google Scholar]
- Bricker T.M., Frankel L.K. (2008). The psbo1 mutant of Arabidopsis cannot efficiently use calcium in support of oxygen evolution by photosystem II. J. Biol. Chem. 283: 29022–29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchert F., Konno H., Hisabori T. (2015). Redox regulation of CF1-ATPase involves interplay between the γ-subunit neck region and the turn region of the βDELSEED-loop. Biochim. Biophys. Acta 1847: 441–450. [DOI] [PubMed] [Google Scholar]
- Bukhov N.G., Egorova E.A., Govindachary S., Carpentier R. (2004). Changes in polyphasic chlorophyll a fluorescence induction curve upon inhibition of donor or acceptor side of photosystem II in isolated thylakoids. Biochim. Biophys. Acta 1657: 121–130. [DOI] [PubMed] [Google Scholar]
- Carraretto L., Formentin E., Teardo E., Checchetto V., Tomizioli M., Morosinotto T., Giacometti G.M., Finazzi G., Szabó I. (2013). A thylakoid-located two-pore K+ channel controls photosynthetic light utilization in plants. Science 342: 114–118. [DOI] [PubMed] [Google Scholar]
- Cheng N.H., Pittman J.K., Shigaki T., Hirschi K.D. (2002). Characterization of CAX4, an Arabidopsis H(+)/cation antiporter. Plant Physiol. 128: 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Ma J., Zhang L. (2012). Regulatory factors for the assembly of thylakoid membrane protein complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 3420–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cruz J.A., Avenson T.J., Kanazawa A., Takizawa K., Edwards G.E., Kramer D.M. (2005). Plasticity in light reactions of photosynthesis for energy production and photoprotection. J. Exp. Bot. 56: 395–406. [DOI] [PubMed] [Google Scholar]
- Cruz J.A., Sacksteder C.A., Kanazawa A., Kramer D.M. (2001). Contribution of electric field (Delta psi) to steady-state transthylakoid proton motive force (pmf) in vitro and in vivo. control of pmf parsing into Delta psi and Delta pH by ionic strength. Biochemistry 40: 1226–1237. [DOI] [PubMed] [Google Scholar]
- Debus R.J., Barry B.A., Sithole I., Babcock G.T., McIntosh L. (1988). Directed mutagenesis indicates that the donor to P+680 in photosystem II is tyrosine-161 of the D1 polypeptide. Biochemistry 27: 9071–9074. [DOI] [PubMed] [Google Scholar]
- Delhaize E., Gruber B.D., Pittman J.K., White R.G., Leung H., Miao Y., Jiang L., Ryan P.R., Richardson A.E. (2007). A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 51: 198–210. [DOI] [PubMed] [Google Scholar]
- Demaegd D., Colinet A.S., Deschamps A., Morsomme P. (2014). Molecular evolution of a novel family of putative calcium transporters. PLoS One 9: e100851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaegd D., Foulquier F., Colinet A.S., Gremillon L., Legrand D., Mariot P., Peiter E., Van Schaftingen E., Matthijs G., Morsomme P. (2013). Newly characterized Golgi-localized family of proteins is involved in calcium and pH homeostasis in yeast and human cells. Proc. Natl. Acad. Sci. USA 110: 6859–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R.M., Sharifi M.N., Malnoë A., Haglund C., Calderon R.H., Wakao S., Niyogi K.K. (2015). Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J. 82: 337–351. [DOI] [PubMed] [Google Scholar]
- Dietzel L., Bräutigam K., Pfannschmidt T. (2008). Photosynthetic acclimation: state transitions and adjustment of photosystem stoichiometry--functional relationships between short-term and long-term light quality acclimation in plants. FEBS J. 275: 1080–1088. [DOI] [PubMed] [Google Scholar]
- Dimmic M.W., Rest J.S., Mindell D.P., Goldstein R.A. (2002). rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J. Mol. Evol. 55: 65–73. [DOI] [PubMed] [Google Scholar]
- Dürr G., Strayle J., Plemper R., Elbs S., Klee S.K., Catty P., Wolf D.H., Rudolph H.K. (1998). The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol. Biol. Cell 9: 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmond C., Shigaki T., Ewert S., Nelson M.D., Connorton J.M., Chalova V., Noordally Z., Pittman J.K. (2009). Comparative analysis of CAX2-like cation transporters indicates functional and regulatory diversity. Biochem. J. 418: 145–154. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emery L., Whelan S., Hirschi K.D., Pittman J.K. (2012). Protein phylogenetic analysis of Ca(2+)/cation antiporters and insights into their evolution in plants. Front. Plant Sci. 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger W.F., Clear A.M., Fanning K.J., Peck M.L. (1999). Identification of a Ca2+/H+ antiport in the plant chloroplast thylakoid membrane. Plant Physiol. 119: 1379–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838. [DOI] [PubMed] [Google Scholar]
- Ferro M., et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9: 1063–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., Salvi D., Brugière S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., Rolland N. (2003). Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2: 325–345. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. (1991). Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7: 253–263. [DOI] [PubMed] [Google Scholar]
- Granvogl B., Reisinger V., Eichacker L.A. (2006). Mapping the proteome of thylakoid membranes by de novo sequencing of intermembrane peptide domains. Proteomics 6: 3681–3695. [DOI] [PubMed] [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16: 334–342. [DOI] [PubMed] [Google Scholar]
- Hakala M., Tuominen I., Keränen M., Tyystjärvi T., Tyystjärvi E. (2005). Evidence for the role of the oxygen-evolving manganese complex in photoinhibition of Photosystem II. Biochim. Biophys. Acta 1706: 68–80. [DOI] [PubMed] [Google Scholar]
- Hall B.G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30: 1229–1235. [DOI] [PubMed] [Google Scholar]
- Hansen T.H., de Bang T.C., Laursen K.H., Pedas P., Husted S., Schjoerring J.K. (2013). Multielement plant tissue analysis using ICP spectrometry. Methods Mol. Biol. 953: 121–141. [DOI] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press; ). [DOI] [PubMed] [Google Scholar]
- Hind G., Nakatani H.Y., Izawa S. (1974). Light-dependent redistribution of ions in suspensions of chloroplast thylakoid membranes. Proc. Natl. Acad. Sci. USA 71: 1484–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi K.D., Korenkov V.D., Wilganowski N.L., Wagner G.J. (2000). Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 124: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochmal A.K., Schulze S., Trompelt K., Hippler M. (2015). Calcium-dependent regulation of photosynthesis. Biochim. Biophys. Acta 1847: 993–1003. [DOI] [PubMed] [Google Scholar]
- Holt N.E., Fleming G.R., Niyogi K.K. (2004). Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry 43: 8281–8289. [DOI] [PubMed] [Google Scholar]
- Ihnatowicz A., Pesaresi P., Varotto C., Richly E., Schneider A., Jahns P., Salamini F., Leister D. (2004). Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J. 37: 839–852. [DOI] [PubMed] [Google Scholar]
- Johnson C.H., Knight M.R., Kondo T., Masson P., Sedbrook J., Haley A., Trewavas A. (1995). Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science 269: 1863–1865. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Umena Y., Kamiya N., Shen J.R. (2011). Structure of the catalytic, inorganic core of oxygen-evolving photosystem II at 1.9 Å resolution. J. Photochem. Photobiol. B 104: 9–18. [DOI] [PubMed] [Google Scholar]
- Keren N., Kidd M.J., Penner-Hahn J.E., Pakrasi H.B. (2002). A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry 41: 15085–15092. [DOI] [PubMed] [Google Scholar]
- Klinkert B., Ossenbühl F., Sikorski M., Berry S., Eichacker L., Nickelsen J. (2004). PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 279: 44639–44644. [DOI] [PubMed] [Google Scholar]
- Kok B., Forbush B., McGloin M. (1970). Cooperation of charges in photosynthetic O2 evolution-I. A linear four step mechanism. Photochem. Photobiol. 11: 457–475. [DOI] [PubMed] [Google Scholar]
- Kreimer G., Melkonian M., Holtum J.A., Latzko E. (1985). Characterization of calcium fluxes across the envelope of intact spinach chloroplasts. Planta 166: 515–523. [DOI] [PubMed] [Google Scholar]
- Kreimer G., Melkonian M., Holtum J.A., Latzko E. (1988). Stromal free calcium concentration and light-mediated activation of chloroplast fructose-1,6-bisphosphatase. Plant Physiol. 86: 423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer G., Surek B., Woodrow I.E., Latzko E. (1987). Calcium binding by spinach stromal proteins. Planta 171: 259–265. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Kunz H.H., Gierth M., Herdean A., Satoh-Cruz M., Kramer D.M., Spetea C., Schroeder J.I. (2014). Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 7480–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.P., Caspar T., Somerville C., Preiss J. (1988). Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADPglucose pyrophosphorylase activity. Plant Physiol. 86: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D., Lalonde S., Looger L.L., von Wirén N., Frommer W.B. (2007). A cytosolic trans-activation domain essential for ammonium uptake. Nature 446: 195–198. [DOI] [PubMed] [Google Scholar]
- Lundin B., Hansson M., Schoefs B., Vener A.V., Spetea C. (2007). The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein. Plant J. 49: 528–539. [DOI] [PubMed] [Google Scholar]
- Ma J., Peng L., Guo J., Lu Q., Lu C., Zhang L. (2007). LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence--a practical guide. J. Exp. Bot. 51: 659–668. [DOI] [PubMed] [Google Scholar]
- Meurer J., Meierhoff K., Westhoff P. (1996). Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396. [DOI] [PubMed] [Google Scholar]
- Michelet L., Zaffagnini M., Morisse S., Sparla F., Pérez-Pérez M.E., Francia F., Danon A., Marchand C.H., Fermani S., Trost P., Lemaire S.D. (2013). Redox regulation of the Calvin-Benson cycle: something old, something new. Front. Plant Sci. 4: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L., Cohen Y., Chitnis P.R., Nechushtai R. (1996). The precursor of PsaD assembles into the photosystem I complex in two steps. Proc. Natl. Acad. Sci. USA 93: 6338–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P., Sirpiö S., Suorsa M., Aro E.M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98: 489–501. [DOI] [PubMed] [Google Scholar]
- Murakami R., Ifuku K., Takabayashi A., Shikanai T., Endo T., Sato F. (2002). Characterization of an Arabidopsis thaliana mutant with impaired psbO, one of two genes encoding extrinsic 33-kDa proteins in photosystem II. FEBS Lett. 523: 138–142. [DOI] [PubMed] [Google Scholar]
- Murray J.W., Barber J. (2006). Identification of a calcium-binding site in the PsbO protein of photosystem II. Biochemistry 45: 4128–4130. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nalin C.M., McCarty R.E. (1984). Role of a disulfide bond in the gamma subunit in activation of the ATPase of chloroplast coupling factor 1. J. Biol. Chem. 259: 7275–7280. [PubMed] [Google Scholar]
- Nelson N., Junge W. (2015). Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 84: 659–683. [DOI] [PubMed] [Google Scholar]
- Newman S.M., Boynton J.E., Gillham N.W., Randolph-Anderson B.L., Johnson A.M., Harris E.H. (1990). Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events. Genetics 126: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Ono T.A., Inoue Y. (1983). Mn-preserving extraction of 33-Kda, 24-Kda and 16-Kda proteins from O2-evolving PSII particles by divalent salt washing. FEBS Lett. 164: 255–260. [Google Scholar]
- Park S., Khamai P., Garcia-Cerdan J.G., Melis A. (2007). REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol. 143: 1547–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E., Montanini B., Gobert A., Pedas P., Husted S., Maathuis F.J., Blaudez D., Chalot M., Sanders D. (2007). A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Natl. Acad. Sci. USA 104: 8532–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D., Laloi M., Rouhara I., Schmelzer E., Delrot S., Frommer W.B. (1995). NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett. 370: 264–268. [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259. [DOI] [PubMed] [Google Scholar]
- Ruban A.V., Johnson M.P., Duffy C.D. (2012). The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta 1817: 167–181. [DOI] [PubMed] [Google Scholar]
- Sacksteder C.A., Kramer D.M. (2000). Dark-interval relaxation kinetics (DIRK) of absorbance changes as a quantitative probe of steady-state electron transfer. Photosynth. Res. 66: 145–158. [DOI] [PubMed] [Google Scholar]
- Schaaf G., Catoni E., Fitz M., Schwacke R., Schneider A., von Wiren N., Frommer W.B. (2002). A putative role for the vacuolar calcium/manganese proton antiporter AtCAX2 in heavy metal detoxification. Plant Biol. (Stuttg.) 4: 612–618. [Google Scholar]
- Schmidt S.B., Persson D.P., Powikrowska M., Frydenvang J., Schjoerring J.K., Jensen P.E., Husted S. (2015). Metal binding in photosystem II super- and subcomplexes from barley thylakoids. Plant Physiol. 168: 1490–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Steinberger I., Strissel H., Kunz H.H., Manavski N., Meurer J., Burkhard G., Jarzombski S., Schünemann D., Geimer S., Flügge U.I., Leister D. (2014). The Arabidopsis Tellurite resistance C protein together with ALB3 is involved in photosystem II protein synthesis. Plant J. 78: 344–356. [DOI] [PubMed] [Google Scholar]
- Schreiber U., Klughammer C. (2008). New accessory for the DUAL-PAM-100: The P515/535 module and examples of its application. PAM Application Notes 1: 1–10. [Google Scholar]
- Schürmann P., Buchanan B.B. (2008). The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 10: 1235–1274. [DOI] [PubMed] [Google Scholar]
- Shi L.X., Hall M., Funk C., Schröder W.P. (2012). Photosystem II, a growing complex: updates on newly discovered components and low molecular mass proteins. Biochim. Biophys. Acta 1817: 13–25. [DOI] [PubMed] [Google Scholar]
- Shigaki T., Pittman J.K., Hirschi K.D. (2003). Manganese specificity determinants in the Arabidopsis metal/H+ antiporter CAX2. J. Biol. Chem. 278: 6610–6617. [DOI] [PubMed] [Google Scholar]
- Sievers F., Higgins D.G (2014). Clustal omega. Curr. Protoc. Bioinformatics 48: 3.13.11–13.13.16. [DOI] [PubMed] [Google Scholar]
- Socha A.L., Guerinot M.L. (2014). Mn-euvering manganese: the role of transporter gene family members in manganese uptake and mobilization in plants. Front. Plant Sci. 5: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger I., Egidi F., Schneider A. (2015). Chlorophyll fluorescence measurements in Arabidopsis wild-type and photosystem II mutant leaves. Bio Protoc. 5: e1532. [Google Scholar]
- Stengel A., Gügel I.L., Hilger D., Rengstl B., Jung H., Nickelsen J. (2012). Initial steps of photosystem II de novo assembly and preloading with manganese take place in biogenesis centers in Synechocystis. Plant Cell 24: 660–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R.J., Tsimilli-Michael M., Srivastava A. (2004). Analysis of the chlorophyll a fluorescence transient. In Chlorophyll a Fluorescence: A Signature of Photosynthesis, G.C. Papageorgiou and Govindjee, eds (Dordrecht, The Netherlands: Springer-Verlag: ), pp. 321–362. [Google Scholar]
- Suga M., Akita F., Hirata K., Ueno G., Murakami H., Nakajima Y., Shimizu T., Yamashita K., Yamamoto M., Ago H., Shen J.R. (2015). Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 517: 99–103. [DOI] [PubMed] [Google Scholar]
- Suorsa M., Aro E.M. (2007). Expression, assembly and auxiliary functions of photosystem II oxygen-evolving proteins in higher plants. Photosynth. Res. 93: 89–100. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T.A., Madden T.L. (1999). BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247–250. [DOI] [PubMed] [Google Scholar]
- Tóth S.Z., Schansker G., Garab G., Strasser R.J. (2007). Photosynthetic electron transport activity in heat-treated barley leaves: the role of internal alternative electron donors to photosystem II. Biochim. Biophys. Acta 1767: 295–305. [DOI] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J.R., Kamiya N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60. [DOI] [PubMed] [Google Scholar]
- Varotto C., Maiwald D., Pesaresi P., Jahns P., Salamini F., Leister D. (2002). The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J. 31: 589–599. [DOI] [PubMed] [Google Scholar]
- Varotto C., Pesaresi P., Meurer J., Oelmüller R., Steiner-Lange S., Salamini F., Leister D. (2000). Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 22: 115–124. [DOI] [PubMed] [Google Scholar]
- Zaks J., Amarnath K., Sylak-Glassman E.J., Fleming G.R. (2013). Models and measurements of energy-dependent quenching. Photosynth. Res. 116: 389–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.