Spontaneous chloroplast mutants mostly occur by replication slippage and display a mutation spectrum very different from that in the eukaryotic nucleus and that of bacteria.
Abstract
Spontaneous plastome mutants have been used as a research tool since the beginning of genetics. However, technical restrictions have severely limited their contributions to research in physiology and molecular biology. Here, we used full plastome sequencing to systematically characterize a collection of 51 spontaneous chloroplast mutants in Oenothera (evening primrose). Most mutants carry only a single mutation. Unexpectedly, the vast majority of mutations do not represent single nucleotide polymorphisms but are insertions/deletions originating from DNA replication slippage events. Only very few mutations appear to be caused by imprecise double-strand break repair, nucleotide misincorporation during replication, or incorrect nucleotide excision repair following oxidative damage. U-turn inversions were not detected. Replication slippage is induced at repetitive sequences that can be very small and tend to have high A/T content. Interestingly, the mutations are not distributed randomly in the genome. The underrepresentation of mutations caused by faulty double-strand break repair might explain the high structural conservation of seed plant plastomes throughout evolution. In addition to providing a fully characterized mutant collection for future research on plastid genetics, gene expression, and photosynthesis, our work identified the spectrum of spontaneous mutations in plastids and reveals that this spectrum is very different from that in the nucleus.
INTRODUCTION
Mutations in the chloroplast genome (plastome) are typically recognized by pale or bleached leaf areas in variegated plants (Kirk and Tilney-Bassett, 1978; Greiner, 2012; Figure 1A). Such material has been used as a genetic tool since the beginning of genetics. For example, the use of plastome mutants by Correns and Baur formed the foundation of cytoplasmic genetics (Baur, 1909; Correns, 1909; Baur, 1910; Hagemann, 2010). Plastome mutants continue to play an important role in elucidating the rules of non-Mendelian inheritance (Chiu and Sears, 1985; Azhagiri and Maliga, 2007; Ruf et al., 2007) and in studying chloroplast gene expression and photosynthesis (Schaffner et al., 1995; Landau et al., 2009). Moreover, plastid mutants provide selectable markers for antibiotic or herbicide resistance that are widely used in molecular biology, biotechnology, and agriculture (Schmitz-Linneweber et al., 2005; Powles and Yu, 2010).
Figure 1.
Representative Phenotypes in the Oenothera Chloroplast Mutant Collection.
For details, see text and Table 1.
(A) I-ny, variegated individual with completed sorting-out giving rise to homoplasmic white (mutated) and green (nursery plastome) sectors.
(B) Developmental series of homoplasmic IV-epsilon leaves, showing age-dependent bleaching.
(C) Age-dependent greening and mottled pattern of leaves of mutant I-iota. From left to right: homoplasmic young nearly white leaf, homoplasmic medium-age mottled leaves, and older green leaf. Note that the oldest leaf displays variegation. As a result of a completed sorting-out, the leaf blade is homoplasmic for the nearly regreened I-iota plastome. The margin is homoplasmic for the wild-type chloroplast (indicated by the arrow).
(D) I-tau, growing photoautotrophically in soil as a homoplasmic plant. It displays a reversible bleaching (virescent) phenotype, alternately forming bleached and green leaves. The homoplasmic state of all mutants was confirmed by Illumina deep sequencing. For molecular causes of the mutations, see Table 1.
Bars = 4 cm in (A), (B), and (D). Leaf sizes in (C) from left to right: ∼5, 10, 12, and 20 cm.
Over several decades, a substantial number of chlorotic chloroplast mutants have been identified and characterized from multiple plant species (Kirk and Tilney-Bassett, 1978; Börner and Sears, 1986; Somerville, 1986; Hagemann, 1992; Greiner, 2012). Importantly, phenotypes of plastome mutants go beyond simple bleaching. Some plastome mutants display mild chlorotic effects, are developmentally impaired or sensitive to environmental factors, and show mottled phenotypes and/or bleach reversibly (Kirk and Tilney-Bassett, 1978; Stubbe and Herrmann, 1982; Chia et al., 1986; Archer and Bonnett, 1987; Colombo et al., 2008; Hirao et al., 2009; Landau et al., 2009; Greiner, 2012; Figures 1B to 1D, Table 1). A number of mutant lines have been described that display unexpected phenotypes, such as mutant plastids having a phenotypic effect on wild-type plastids in the same cell (Michaelis, 1957) or mutations that confer drought and temperature tolerance (Usatov et al., 2004; Mashkina et al., 2010). Collectively these works show that although chlorosis is a common feature of plastome mutants, plastid mutations can have diverse effects on the plant phenotype.
Table 1. Phenotypes, Major Defects, and Mutated Loci of the Mutant Lines in the Oenothera Plastome Mutant Collection.
| Mutanta | Defect in | Gene or Locus | Leaf Phenotypeb |
Description | Mutation and Predicted Molecular Consequence | ||||
|---|---|---|---|---|---|---|---|---|---|
| Young | Medium | Old | |||||||
| M1/M4 | I-alpha/I-delta | PSII | psbD | 4 | 3-4 | 4 | Very old leaves with narrow whitish zones, older leaves sometimes darker green | 5-bp duplication (CTTGG), +18 to +22 psbD, truncated PsbD, alternative start codon [?] | |
| M2 | I-beta | Cyt b6f | petD | 4 | 3 | 1 | Old leaves often with small green spots | 4-bp duplication (ATTT), +998 to +1001 petD, truncated PetD | |
| M2 | I-beta | – | SSC-IRA junction | 4 | 3 | 1 | Old leaves often with small green spots | 66-bp deletion (AATTT…TATGG), −258 to −323 ycf1 (tic214)c | |
| M3/M5 | I-gamma/I-epsilon | PSII | psbA | 3 | 2-3 | 1 | Older leaves with enhanced greening close to vascular tissue | 2-bp duplication (GT), +82 to +83 psbA, truncated PsbA | |
| M6 | I-zeta | PSI | psaA | 1-2 | 1 | 1 | – | 5-bp duplication (GTTTT), +1350 to +1354 psbA, truncated PsaA | |
| M7 | I-eta | PSI | psaA | 2 | 1-2 | 1 | – | 4-bp duplication (GATT), +1088 to +1090 psaA, truncated PsaA | |
| M8 | I-theta | PSI | psaB | 2 | 1-2 | 1 | – | 32-bp duplication (TTCCA…ATTAG), +2132 to +2163 psaB, truncated PsaB | |
| M9 | I-iota | ATP synthase | atpB | (1) | (1-4) | (4) | Mottled, young leaves white, old leaves nearly green | 1-bp insertion (A), +58 atpB, truncated AtpB [?] | |
| M10/M15 | I-kappa/I-omikron | PSI | psaA | 1-2 | 1-2 | 1-2 | – | 70-bp duplication (ACGAT…CCCGT), +1437 to +1506 psaA, truncated PsaA | |
| M11 | I-lambda | PSII | psbA | 3-4 | 1-2 | 1 | – | 4-bp deletion (GCTT), +859 to +862 psbA, truncated PsbA | |
| M12 | I-my | Cyt b6f | ccsA | 4 | 3-4 | 1 | Old leaves often with small green spots | 31-bp duplication (GAATT…TTAAA), +540 to +570 ccsA; truncated CcsA | |
| M13 | I-ny | PSI | psaB | 2 | 1-2 | 1 | – | 4-bp deletion (AGAT), +399 to +402 psaB, truncated PsaB | |
| M14 | I-xi | PSII | psbD | 4 | 3-4 | 1 | – | 5-bp deletion (CAGCT), +530 to +534 psbD, truncated PsbD | |
| M16 | I-pi | PSII | psbA | 3-4 | 2-3 | 1 | – | 5-bp deletion (TCTCT), +208 to +212 psbA, truncated PsbA | |
| M17 | I-rho | PSII | psbA | 3-4 | 2 | 1 | Greening declining from leaf base to tip | 5-bp deletion (ACAAC), +797 to +801 psbA, truncated PsbA | |
| M17 | I-rho | – | Upstream accD | 3-4 | 2 | 1 | Greening declining from leaf base to tip | 30-bp deletion (CCTTT…CTTTA), −30 to −1 accDc | |
| M18d | I-sigma | Rubisco | rbcL | 4 | 4 | 4 | – | 5-bp deletion (TTAAC), +808 to +812 rbcL, truncated RbcL | |
| M19 | I-tau | Translation | rps7 | (1-4) | (1-4) | (1-4) | Periodical bleaching/regreening (virescent), viable on soil at homoplasmic state, retarded in cell growth | 4-bp duplication (ATTT), +462 to +465 rps7, last 2 aa of Rps7 modified and 2 aa added | |
| M20 | I-ypsilon | ATP synthase | atpB | (1) | (1) | (1) | Mottled, but spots very rare | 34-bp duplication (GATAC…AATTA), +461 to +494 atpB, truncated AtpB | |
| M20 | I-ypsilon | – | ycf1 (tic214) | (1) | (1) | (1) | Mottled, but spots very rare | 21-bp deletion (AAAAG…ACGTC), +2404 to +2424 ycf1 (tic 214), 21-bp duplication (TCAAA…CGAAA), +2486 to +2406 ycf1 (tic214), 3 aa exchanges in the variable part of Ycf1 (Tic214)c | |
| M21 | I-phi | Cyt b6f | ccsA | 3-4 | 2 | 2 | – | 4-bp duplication (ATTT), +10 to +13 ccsA, truncated CcsA | |
| M22 | I-chi | PSI | psaA | 1-2 | 1 | 1 | – | 5-bp duplication (CCGCT), +1420 to +1424 psaA, truncated PsaA | |
| M23 | I-psi | PSI | ycf3 (intron) | 1-2 | 2 | 2 | – | 5-bp duplication (ATTGA), +316 to +320 ycf3 (intron), splicing deficiency | |
| M24 | I-omega | PSI | psaA | 1-2 | 1 | 1 | – | 4-bp duplication (TCTT), +2226 to +2229 psaA, changed/extended PsaA | |
| M25 | I-do | Cyt b6f | ccsA | 3 | 2 | 2 | – | 6-bp deletion (ACAATA), +298 to +303 ccsA, deletion of 2 aa in CcsA | |
| M26/M27 | I-re/I-mi | Cyt b6f | petA | 2-3 | 1-2 | 1-2 | – | 1-bp insertion (G), +667 petA, truncated PetA | |
| M28 | II-alpha | PSI | ycf4 | 3 | 2-3 | 1 | Old leaves of field grown plants with characteristic yellowish/whitish tip | 4-bp duplication (AAAA), +442 to +445 ycf4; truncated Ycf4 | |
| M29 | III-epsilone | PSI | ycf3 | 2 | 1 | 1 | – | 29-bp duplication (ACAAG…GCTTT), +996 to +1024 ycf3, truncated Ycf3 | |
| M30d | II-gamma | PSII | psbE | 4 | 2-3 | 1-2 | Old leaves often with small green spots | 5-bp duplication (ATTAT), +42 to +46 psbE, truncated PsbE | |
| M31 | II-delta | – | ndhE-psaC spacer | 4 | 4 | 4 | No change during development | 24-bp deletion (TATAT…AATCA), −60 to −83 psaC, transcript processing affected | |
| M32 | II-epsilon | PSI | psaA | 2 | 1 | 1 | – | 4-bp duplication (TGAG), +124 to +127 psaA, truncated PsaA | |
| M33 | II-zeta | PSII | psbB | 4 | 2-3 | 1 | Old leaves with enhanced greening close to vascular tissue | 4-bp duplication (CTTT), +117 to +120 psbB, truncated PsbB | |
| M34 | II-eta | Cyt b6f | petB | 4 | 3 | 1 | Old leaves often with small green spots | 5-bp duplication (ATTAC), +1346 to +1350 petB, truncated PetB, transcript processing/stability affected | |
| M35d | II-theta | Cyt b6f | petB (intron) | 4 | 3 | 1 | Old leaves often with small green spots | 2-bp deletion (AC), +430 to +431 petB (intron), splicing deficiency | |
| M36/M47 | II-iota/IV-delta | PSI | psaA | 2 | 2 | 2 | – | 5-bp duplication (TTAAC), +667 to +671 psaA, truncated PsaA | |
| M37 | II-kappa | PSI | ycf3 (intron) | 3-4 | 3-4 | 4 | Old leaves in parts darker than younger leaves, viable on soil at homoplasmic state | transversion G/T, +1438 ycf3 (intron), splicing deficiency | |
| M37 | II-kappa | – | ndhF-rpl32 spacer | 3-4 | 3-4 | 4 | Old leaves in parts darker than younger leaves, viable on soil at homoplasmic state | Single-bp insertion/deletion, results in a transversion (C/T), −511 rpl32c | |
| M38 | II-lambda | Cyt b6f | petA | 4 | 3-4 | 2-3 | – | 39-bp deletion (GGTTT...CCGAG), +337 to +375 petA, 13 aa deleted in PetA | |
| M39 | II-my | Cyt b6f | petA | 3-4 | 3-4 | 1-2 | – | 5-bp duplication (TAGCC), +352 to +356 petA, truncated PetA | |
| M39 | II-my | – | ndhG-ndhI spacer | 3-4 | 3-4 | 1-2 | – | Single-bp deletion (A), −310 ndhGc | |
| M40 | II-ny | PSI | ycf4 | Mutation of M28 (II-alpha) | |||||
| M40 | II-ny | PSII | psbB | 2 | 1-2 | 1 | – | 2-bp deletion (CT), +791 to +792 psbB, truncated PsbBf | |
| M41g | III-alpha | PSII | psbD | 4 | 3-4 | 1 | – | 6-bp duplication (TATATT), +540 to +545 psbD, 2 additional aa in PsbD | |
| M42 | III-beta | PSI | psaB | 1-3 | 1-3 | 1-3 | Bleaches under higher light conditions, sometimes inhomogeneous | 5-bp duplication (ATAAA), +1246 to +1250 psaB, truncated PsaB | |
| M43 | III-gamma | PSI | psaA | 1-2h | 1-2 | 1 | – | 8-bp duplication (AATTTAGT), +1540 to +1547 psaA, truncated PsaA | |
| M43 | III-gamma | – | trnQ-UUG-accD spacer | 1-2h | 1-2 | 1 | – | 12-bp deletion (CTCGGCTGATGA), −689 to −678 accDc | |
| M44 | III-delta | PSI | psaB | 1 | 1 | 1 | – | Mutation of M42 (III-beta) | |
| M44 | III-delta | ATP synthase | atpB | 1 | 1 | 1 | – | 1-bp deletion (A), +57 atpB, truncated AtpBf | |
| M45 | IV-alpha | PSI | psaA | 3 | 1-2 | 1 | – | 5-bp duplication (GTACT), +2173 to +2177 psaA, truncated PsaA | |
| M464) | IV-beta | Rubisco | rbcL | 4 | 4 | 3-4 | – | Transversion (G/C), +337 rbcL, single aa exchange | |
| M48 | IV-epsilon | PSII | psbA | 3 | 2-3 | 1-2 | Greening declining from leaf base to tip | 8-bp deletion (AGTGGGAA), +389 to +396 psbA, truncated PsbA | |
| M49 | V-alpha | Rubisco (tentative)i | rbcL (tentative)i | 4 | 4 | 4 | Grayish green phenotype, older leaves in parts darker | 6-bp deletion (GATGAA), +750 to +755 rbcL, deletion of 2 aa in RbcL | |
| M50 | V-delta | PSI | ycf3 | 4 | 3 | 1 | – | 1364-bp deletion in ycf3 affecting 11 bp of the 3′-end of exon 1, the full intron 1, the full exon 2, and 403 bp of the 5′-end of intron 2 | |
| M50 | V-delta | – | accD | 4 | 3 | 1 | – | 153-bp deletion +208 to +360 accD, deletion of 51 aa in the variable part of AccDc | |
| M51 | I-fa | Cyt b6f | ccsA | 3 | 2-3 | 1-2 | – | 5-bp duplication (TTAAC), +702 to +705 ccsA, truncated CcsA | |
Mutants are designated according to their basic plastome genotype (I to V) with Greek letters indicating the sequence of mutant isolation (with alpha representing the first mutant isolated). In the case of plastome I, for which more mutants have been isolated than letters exist in the Greek alphabet, additional mutants are designated by the seven tone syllables. For details, see Kutzelnigg and Stubbe (1974) and Stubbe (1989).
1 = nearly white, 2 = yellowish cream-colored, 3 = light green, and 4 = nearly green.
Potentially phenotypically neutral second site mutation. For details, see text.
Mutations in the mutants M18 (I-sigma), M30 (II-gamma), M35 (II-theta), and M46 (IV-beta) were determined previously and could be verified in this study. For summary and references, see Greiner (2012).
This mutant was originally obtained under the name II-beta, but redesignated in this work. For details, see Supplemental Table 1.
Second-site mutation isolated based on a phenotypic change in the primary mutant. For details, see text.
With respect to its reference plastome (deserens de Vries; GenBank accession number KT881172.1), the mutant M41 (III-alpha) carries a 6-bp deletion (TAATCA) in the rbcL-atpB intergenic spacer, −113 to −118 of rbcL. However, this mutation must have arisen in the wild-type reference after the isolation of M41 (III-alpha). Hence, it is not treated as second site mutation in M41 (III-alpha). For details, see text.
The phenotype of the mutant M43 (III-gamma) was described as “lighter green (3)” in young tissue by W. Stubbe. However, in our hands, the mutant appeared to be nearly white to yellowish (1-2) at most developmental stages. It is possible that this phenotypic change is due to a second site mutation upstream of accD which was not present when W. Stubbe initially assessed the phenotypes. For details, see text.
The corresponding wild-type strain (argillicola Douthat 4b) of the mutant M50 (V-alpha) is no longer present in our genetic stocks. For mutant allele identification, the two available O. argillicola plastomes (EU262887.2 and KT881177.1) were used. See Stubbe and Herrmann (1982) for assignment of this mutant to a physiological defect in Rubisco.
Although chloroplast mutants can provide insight into chloroplast gene function and regulation, technical limitations have limited wider application in plant research. Disregarding mutants resistant to herbicides or antibiotics, only 12 plastome mutations have been identified in vascular plants (Greiner, 2012). Since such mutations cannot be identified by conventional linkage mapping, direct sequence analysis is required to pinpoint a mutated locus. Past approaches relied on physiological and biochemical analyses of the mutant, which allowed predictions about the mutated gene. This strategy was applied to most of the plastome mutations that have been identified by molecular approaches (Greiner, 2012) and is still successfully used in screens of plastid-encoded herbicide resistance (Thiel et al., 2010). However, the approach of combining physiological characterization with local sequence analysis suffers from limitations: First, the candidate gene needs to be predictable, which implies that its biochemical and physiological attributes are already well characterized. Consequently, identifying mutations in genes of unknown function is very difficult. Second, the presence of a second-site mutation cannot be ruled out. Full chloroplast genome sequencing by next-generation sequencing can overcome these technical constraints.
Another reason why chloroplast mutants are currently understudied is the challenge of maintaining plastome mutants in standard model species with uniparental plastid inheritance. Many plastome mutants are not viable on soil. They must therefore be kept in as variegated plants during propagation because wild-type plastomes are essential to nourish the mutant tissue (Figure 1A). Variegation occurs because of random chloroplast sorting during cell division, which gives rise to homoplastic mutants and wild-type tissue. Sorting-out is a statistical process and is very rapid (Michaelis, 1955; Kirk and Tilney-Bassett, 1978). It is typically completed early in leaf development (Schötz, 1954). Hence, in most plant species, variegation cannot be maintained through sexual reproduction because it is challenging to identify egg cells that are heteroplasmic and could give raise to variegated plants in the next generation (Kirk and Tilney-Bassett, 1978; Greiner, 2012). Therefore, chloroplast mutants are typically maintained by cutting variegated stems or in tissue culture, but these approaches are infeasible for many vascular plants. Using a model organism displaying biparental plastid inheritance, like Oenothera, overcomes the technical limitation.
The genus Oenothera (evening primrose, Onagraceae) is an excellent model for the study of chloroplast mutants, including their mutation spectrum, molecular basis, and phenotypic effects. Past research shows that spontaneous chloroplast mutants occur with a frequency of 0.3% in Oenothera (Kutzelnigg and Stubbe, 1974). Biparental transmission allows the directed generation of variegated plants by crossing, using individuals with mutated and wild-type plastomes as crossing partners. Thus, plastome mutants can be reliably propagated from seeds and kept as variegated plants in soil (Figure 1A; see Methods). This approach has allowed the generation of a large collection of spontaneous chloroplast mutants. Based on chlorotic phenotypes, a total of 51 spontaneous plastome mutants were isolated and maintained through efforts of Wilfried Stubbe during the past century (Kutzelnigg and Stubbe, 1974; Stubbe and Herrmann, 1982; Table 1; Supplemental Table 1). All mutations arose in the shoot apical meristem, since they entered the germline and are sexually transmitted. Hence, the collection represents the spectrum of heritable mutations that lead to visibly impaired chloroplast function. Although the collection has been characterized extensively at the phenotypic level, technical limitations have prevented molecular analysis (Greiner, 2012).
Here, we describe the mutation spectrum of disruptive spontaneous chloroplast mutations using the Oenothera plastome mutant collection. Based on full plastome sequencing, we suggest a molecular mechanism of their occurrence. Employing statistical methods, we test to what extent mutation events and recovery are biased toward the mutated target gene, the local sequence context, and the location of a mutation on the chloroplast genome.
RESULTS
Phenotypes of Chloroplast Mutants in Homoplasmic Leaf Tissue
The 51 mutants of the collection show various types of chlorophyll deficiency, which typically occurred in an age-dependent manner. Some of the phenotypes are sensitive to environmental factors (Figure 1, Table 1). Nonuniform leaf phenotypes in homoplasmic mutant tissue occur either constitutively or are restricted to a certain developmental stage in 16 mutants. Three of those display a pigmentation gradient from the leaf base to the tip (Figure 1B). In mature rosette leaves, 40 mutants fully bleach to a nearly white or yellowish cream-colored phenotype, whereas light-green tissue remains throughout development in 11 mutants. Two of those are viable in soil and need no nourishment from wild-type tissue (Table 1; see Methods). Particularly interesting phenotypes are seen in the mutants I-tau and I-iota. In the mutant I-iota, an age-dependent mosaic pattern of pigmentation is observed where young leaves are white and slightly mottled. The green spots increase in size and number during development, leading to nearly green mature rosette leaves (Figure 1C). I-tau displays a reversible bleaching (virescent) phenotype (Figure 1D). It can grow photoautotrophically and forms bleached and green leaves in an alternating order.
Wild-Type Plastomes
For identification of the underlying mutations, the full sequence of the corresponding wild-type reference plastome was obtained. The mutants of the collection are derived from 13 wild-type strains (Supplemental Tables 1 and 2). The complete plastome sequences of four of the wild-type lines had been determined previously (Greiner et al., 2008a). In this study, we resequenced all five previously determined Oenothera plastomes using Illumina next-generation DNA sequencing technology (EMBL/GenBank accessions: AJ271079.4, EU262887.2, EU262889.2, EU262890.2, and EU262891.2). In addition to correcting a few sequencing errors (mostly single nucleotides), we detected two larger insertions of 147 and 24 bp compared with the previously published chloroplast genome of Oenothera elata ssp hookeri strain johansen Standard. This likely reflects genetic variation in our seed stocks within unstable highly repetitive genomic regions (Figure 2; Greiner et al., 2008a; see below). Newly determined were the complete chloroplast genome sequence of eight reference strains and an additional line to address plastome stability (see below; GenBank accession numbers KT881169.2 to KT881177.1; for summary, see Supplemental Table 3). The wild-type reference strain of one mutant (V-alpha) is no longer present in our collections (see Table 1 for details). Overall, the Oenothera chloroplast genome is well conserved in structure and size (spanning 163,367 to 166,545 bp) and harbors an identical gene set of 113 unique genes, as described earlier (Greiner et al., 2008a, 2008b).
Figure 2.
Distribution of the Mutated Alleles of the Plastome Mutant Collection across the Oenothera Plastome.
The physical map of the plastome is shown, and the origins of replication (oriA and oriB), the GC content (gray inner circle), and the macro repeats (red inner circle) are indicated. Blue circles denote mutations conferring a mutant phenotype; orange circles denote second site mutations which may be phenotypically neutral. Green circles refer to phenotypically neutral mutations in the closely related O. glazioviana and O. elata strains. Note clustering of these mutations in the highly repetitive regions around accD and ycf1 (tic214). Numbers refer to the number of individual mutations identified for a gene or locus. Class I genes are indicated in color and Class II genes and Class III genes in gray or white. For details on the definition of the gene classes, see text. For macro repeat identification, see Greiner et al. (2008a), and for location of the origins of replication, see Chiu and Sears (1992).
Identification of Mutated Alleles and Second-Site Mutations in the Plastome Mutant Collection
From the 51 spontaneous mutants, 46 unique chloroplast genomes were recovered (Table 1), and five mutant pairs share an identical sequence. It is conceivable that contamination during propagation of the collection is responsible for these cases of seemingly identical chloroplast genomes (the collection was maintained for decades without the benefit of molecular markers for tracking). However, since the distribution of mutated alleles was found to be biased within the Oenothera plastome (Figure 3; see below), we cannot exclude the possibility that at least some of the identical mutations might have arisen independently. This view is supported by the fact that in lines with different wild-type plastomes, the same locus sometimes experienced independent mutations. However, those resulted in distinct mutated alleles (mutant pairs I-iota/III-delta and I-xi/III-alpha; Supplemental Figures 1 to 3). All mutants sharing an identical mutation also share the same wild-type background. Also, in previous analyses, their phenotypic descriptions were essentially identical (Table 1; see Methods for details). In this context, it is noteworthy that the oldest viable seed lots that we could investigate for potential propagation errors date back to 1985.
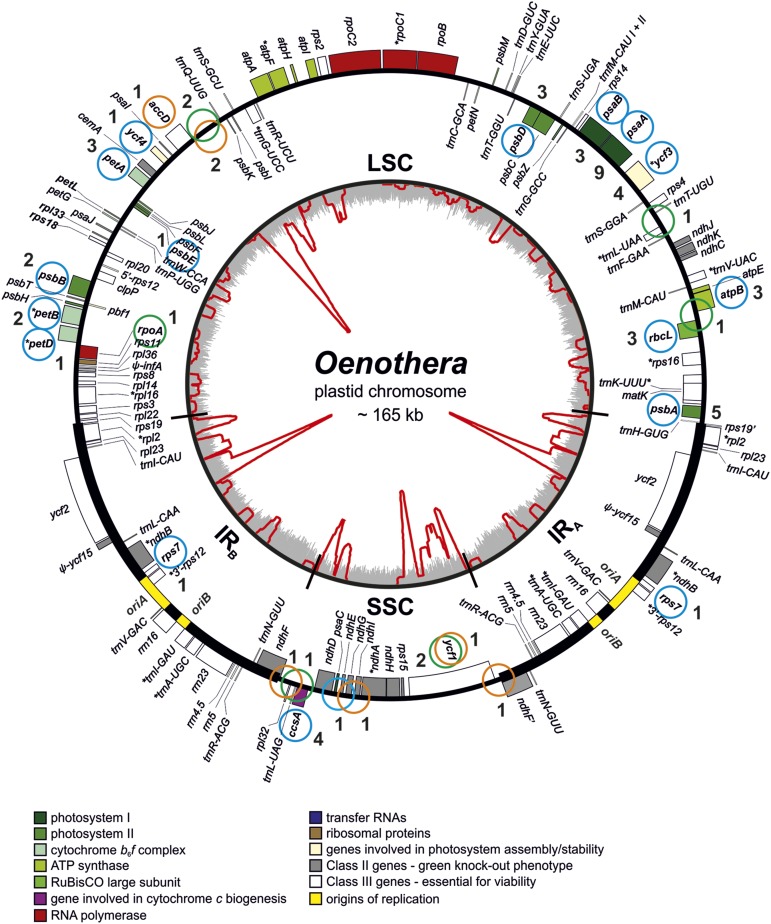
Figure 3.
Distribution of Mutations in the Mutational Hot Spots psaA, psbA, and ccsA.
Note that solely duplications could be identified in psaA. Also note the uneven distribution of mutations within the genes. For molecular causes of the mutations and associated repetitive elements, see Table 1, Supplemental Data Set 1, and Supplemental Figures 2 to 4.
Within the 46 unique mutated chloroplast genomes of the collection, 37 genomes carry a mutation at a single site (Table 1). The identification of a single mutation in V-alpha is tentative, since the original wild-type reference is no longer present in our strain collection (see above; Table 1). Two mutated sites are present in each of the remaining nine mutants. Two of these mutants represent known second-site mutations in that they were isolated based on a phenotypic change arising in a pr-existing chloroplast mutant. Thus, in total 53 (37 + 9 + 7) unique mutated sites could be detected in the entire collection (Table 1; Supplemental Data Set 1). The second-site mutations of the remaining seven mutants are located in regions of intrinsic instability associated with larger tandem repeats or oligo(N) stretches in intergenic regions (Figure 2). Their phenotypic relevance is discussed below.
Types of Mutations in the Collection
The 53 mutant sites in the collection essentially fall into five classes: (1) single-base pair transversions (two mutations), (2) single-base pair, double-base pair, and micro insertions/deletions (indels, <10 bp) at oligo(N) stretches (six mutations), (3) micro indels at micro repeats (<10 bp; 32 mutations, 21 duplications, and 11 deletions), (4) macro indels (mostly between 12 and 70 bp, one exceptional deletion of 153 bp) at macro repeats (>10 bp; 12 mutations, five duplications, six deletions, and one mutation event most likely caused by an insertion and a deletion), and (5) one large deletion (1364 bp), which could not be associated with a repetitive element (see Supplemental Data Set 1 and Supplemental Figures 1 to 6 for detailed description of the individual mutations). Within the micro indels (<10 bp), the major class of mutations detected in this study, a bias toward duplications is not present (χ2: P = 0.3113). However, a clear bias to indels conferred by tandem/direct repeats is present (χ2: P = 0.0003; 32 mutations, 30 associated with tandem/direct repeats, two with palindrome/inverted repeats; Supplemental Data Set 1). In summary, nearly all chloroplast mutations are associated with repetitive elements. These repeats can be very small, in some cases with two repetitive elements of 3 bp (Supplemental Figures 1 to 6 and Supplemental Data Set 1).
Molecular Cause of Mutations and Phenotypic Relevance of the Second-Site Mutations
The detected chloroplast mutations have a bias toward coding sequence. Most mutations are located in protein-coding regions (44 of 53) of which 37 induce frameshifts and six represent in-frame indels. One mutation is conferred by a transversion, leading to a single amino acid change (Table 1). At least five of the mutations affect chloroplast mRNA metabolism. Three of these occur in introns where they result in splicing defects. The mutations in I-psi and II-kappa are located in intron 1 and intron 2 of ycf3, respectively, and affect splicing of these introns (compared with Landau et al., 2009). The gene is involved in photosystem I assembly (Ruf et al., 1997). A similar situation is found for the mutant II-theta where the mutation is located in the intron of petB. The pet genes encode subunits of the cytochrome b6f complex. Here, unspliced transcripts overaccumulate. The mutant II-delta occurs in the intergenic region of the ndhE-psaC spacer (ndh genes encode subunits of the chloroplast NADH dehydrogenase complex; psa genes encode subunits of photosystem I). The mutation in the ndhE-psaC spacer is the sole mutation in this line and it is not located in a known chloroplast promoter (see Greiner et al., 2008a for searched motifs). The mutant has an mRNA processing defect in the ndhH-ndhA-ndhI-ndhG-ndhE-psaC-ndhD operon (Figure 4). Interestingly, defects in mRNA metabolism were not only found in the four mutants where one might have expected them (i.e., mutants with mutations in introns or intergenic regions). The mutant II-eta, harboring a mutation in exon 2 of petB, was found to be affected in processing and/or stability of the psbB (petB) operon transcripts. The primary transcripts from this operon undergo a series of processing and maturation steps, resulting in a highly complex pattern of partially processed transcripts and mature mRNAs (Westhoff and Herrmann, 1988). As judged from transcript sizes and the previous characterization of RNA species (Westhoff and Herrmann, 1988), a major spliced RNA species comprising psbH, petB, and petD is hardly detectable in II-eta (psbH encodes a subunit of photosystem II), whereas a transcript likely containing the spliced petB/petD dicistron overaccumulates (Figure 4).
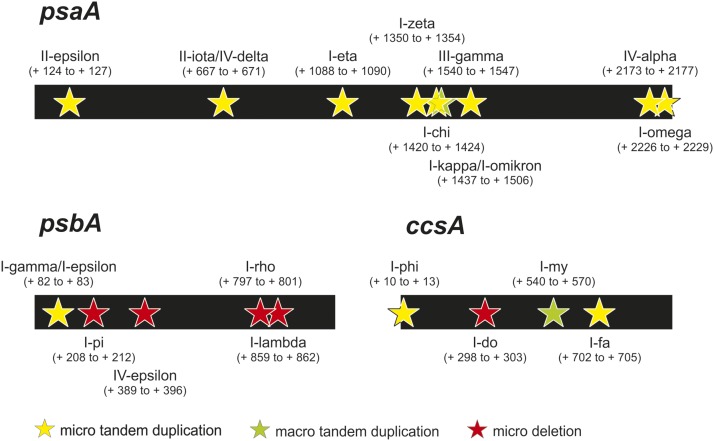
Figure 4.
Defects in mRNA Maturation Due to Mutations in Introns, a Coding Region, or an Intergenic Region in the Chloroplast Mutants I-psi, II-kappa, II-eta, II-theta, and II-delta.
(A) Schematic overview of the ycf3, psaC, and petB regions of the plastome. Positions of the mutations are marked with stars, and hybridization probes are indicated as green bars.
(B) RNA gel blot analyses of mRNA accumulation and RNA processing patterns. Upper row, left and middle panel: Splicing defects in ycf3 as found in the mutants I-psi and II-kappa, respectively. In I-psi, a 5-bp duplication in intron 1 leads to loss of splicing of this intron, correlating with a severe bleaching phenotype (Table 1). In II-kappa, a transversion in intron 2 results in a reduced splicing activity. Note that some mature transcript (0.7 kb) is still produced, consistent with the homoplasmic mutant being viable upon cultivation in soil (Table 1). Upper row, right panel: Processing defect in the psaC operon of the mutant II-delta. The mutant harbors a 24-bp deletion in the psaC-ndhE intergenic spacer. Whereas a precursor transcript of ∼8.0 kb overaccumulates in the mutant, accumulation of processed mRNA species is strongly reduced. Some mature monocistronic mRNA (0.6 kb) is produced in the mutant, consistent with its mild phenotype (Table 1). Lower row: Splicing and processing defects in transcripts from the psbB operon (containing the petB gene) in the mutants II-eta and II-theta. A 2-bp deletion is present in the petB intron in II-theta. In II-eta, a 5-bp duplication is present in the second exon of petB. Left panel: Hybridization to a petB exon probe; right panel: intron probe. In II-theta, a petB splicing defect is clearly visible. Transcripts overaccumulating in the mutant (i.e., the 2.2- and 1.5-kb RNA species) likely represent the unspliced petB/petD dicistron and the unspliced petB, respectively. The putative mature transcript (0.8 kb) does not accumulate in the mutant, in agreement with its severe phenotype (Table 1). Interestingly, in II-eta, the mutation in exon 2 leads to loss of the spliced psbH/petB/petD tricistron (1.7 kb in the wild type; Westhoff and Herrmann, 1988) and instead to accumulation of a 1.4-kb transcript likely containing the spliced petB/petD dicistron. The mature transcript (0.8 kb) seems to accumulate, which, however, carries a frameshift. This leads to a largely identical phenotype as II-theta (Table 1). For details on the mutations, see Table 1. 1 and 2 represent biological replicates of all plant lines analyzed here. To confirm equal loading, the ethidium bromide-stained agarose gel prior to blotting is also shown.
The remaining five mutations of the collection are in noncoding regions and are second-site mutations. Two are located within oligo(N) stretches in intergenic regions; three are upstream of ycf1 (tic214) or accD in regions of intrinsic instability due to the presence of large complex tandem repeats (Figure 2; see below). The accD gene encodes a subunit of the plastidic acetyl-CoA carboxylase; ycf1 (tic214) is currently considered to encode a subunit of the chloroplast protein import machinery (Kikuchi et al., 2013).
Among the mutations in genic regions, three are associated with ATP synthase, 12 with photosystem I (nine of them within psaA, exclusively representing duplications; Figure 3), 11 with photosystem II, and six within the cytochrome b6f complex. Furthermore, four mutations were found in ycf3 and one in ycf4 (as ycf3 involved in photosystem I assembly; Ruf et al., 1997; Krech et al., 2012), three within rbcL (encoding the large subunit of Rubisco), and four within ccsA (a gene involved in cytochrome c biogenesis; Xie and Merchant, 1996). These functional gene classes affect photosynthesis and are considered to be nonessential for cell viability (Scharff and Bock, 2014). An additional mutation occurred in rps7, which is likely an essential gene in dicots encoding a protein for the small (30S) subunit of the plastid ribosome (Scharff and Bock, 2014; Tiller and Bock, 2014). Viability of this mutation is likely due to the particular mutated allele, which modifies the very C-terminal sequence of the protein. Additional second-site mutations include in-frame indels in the intrinsically unstable regions of accD and ycf1 (tic214). Those are again associated with large complex tandem repeats (Figure 2).
The phenotypic relevance of the second site mutations is not entirely clear. Mutations within oligo(N) stretches in intergenic regions are likely to be phenotypically neutral. By contrast, the contribution to the phenotype is uncertain for the mutations in the intrinsically unstable repetitive regions: The location of these mutations is upstream or in the coding regions of accD and ycf1 (tic214), respectively, which are both essential in dicots (Drescher et al., 2000; Kode et al., 2005; Supplemental Data Set 2). However, the phenotype of all mutants carrying such mutations can be fully explained by the second mutated allele present in these lines (Table 1; Supplemental Data Set 2). This is likely also the case with mutated sites in repetitive regions that represent indels within the reading frames of accD and ycf1 (tic214; Figure 2). These indels are in frame and the amino acid sequences in these regions are poorly conserved among Oenothera species (Greiner et al., 2008a). In addition, very similar polymorphisms, including in-frame indels within the coding region of ycf1 (tic214), were also detected upon comparison of very closely related wild-type strains, as described below.
Statistical Distribution of Mutant Alleles within the Chloroplast Genome
An important question addressed by our study is whether mutations are randomly or nonrandomly distributed within the plastome and whether these patterns reflect real mutational biases or biases related to our selection process. We tested for biological and methodological biases by assessing the influence of the expected phenotype, gene length, and the location of the gene in the plastome on the probability of isolating a mutation. We assigned the genes of the Oenothera plastome to one of three classes based on phenotypes observed in systematic knockout analyses of chloroplast genes in Nicotiana tabacum (Supplemental Data Set 2). Class I genes included those in which spontaneous mutations are easily detected. This class combines genes that are nonessential for heterotrophic growth and whose knockouts display either white or pale phenotypes (subgroups: “White” and “Pale”). Examples for such genes are psaA and rbcL. Class II genes do not display a visible phenotype (e.g., ndhB and all other subunits of the plastid NDH complex) and unsurprisingly the collection does not contain any mutants in that class, which excludes it from the following analyses. Class III genes are essential for cellular viability (e.g., rps7 or ycf1 [tic214]). Only a limited number of specific (nonlethal) mutations could be isolated from this class (see above). Class I genes are exclusively located in the two single-copy regions of the plastome (large single-copy [LSC] or small single-copy [SSC] region). However, class III genes are also located in the inverted repeat (IR; Figure 2). The IR is a typical structure of chloroplast genomes and contains exclusively Class II and Class III genes. Due to recombination and gene conversion between the two large inverted repeats (Palmer, 1983), mutations in the IR are constantly purged. Hence, Class III genes need to be subdivided based on their physical location on the plastome (LSC and SSC versus IR). It also should be noted that regions of intrinsic instability are found only in Class III genes (Figure 2).
The first assumption we tested was if mutations in nongenic regions (52,922 bp in the genome; six mutations in the collection) were more common than mutations within genes (112,977 bp in the genome; 47 mutations in the collection). Unsurprisingly, this was the case (χ2: P = 0.0022). Second, we confirmed our expectation that mutations in Class I genes were more common than mutations in Class III genes (Class I, 44 mutations within 37,080 bp of genic sequence; Class III, three mutations within 56,920 bp; χ2, P < 0.0001). We found no difference in the abundance of mutations between the two subgroups of Class III genes (IR, one mutation within 33,672 bp; LSC and SSC, two mutations within 23,248 bp; χ2, P = 0.7470), but this nonsignificance may reflect the small sample size.
Next, we asked if gene length affected the recovery of mutations in Class I genes, represented by 44 mutations within 37,080 bp of genic sequence. χ2 test yielded P = 0.0002 for the likelihood of accumulating nine mutations in psaA (2253 bp), P = 0.0254 for four mutations in ccsA (960 bp), but P = 0.5287 for three mutations in rbcL (1428 bp). This strongly suggests a gene-specific bias for mutation recovery beyond the differences observed between gene classes (Figure 3). The lack of relationship with gene length is further supported by a large cluster of Class I genes in the LSC (13,969 bp) that lack any mutation. The cluster contains ATP synthase genes (atpA, F, H, and I) and genes for the chloroplast bacterial-type RNA polymerase (rpoB, C1, and C2; Figure 2; Supplemental Data Set 2). The joint random probability that among 44 independent mutations not a single one is located in this cluster is P = 9.2 × 10−10.
Finally, we assessed whether bias in mutant recovery can be explained by the strength of the chlorotic effect of a mutation, since strong (white) phenotypes might be easier to detect by eye during the selection procedure. Of the 51 mutant lines, 40 displayed an essentially white phenotype and 11 a pale phenotype (Supplemental Table 4). Surprisingly, compared with the combined lengths of the coding sequences of “White” and “Pale” genes (35,119 and 1961 bp, respectively), white phenotypes are significantly underrepresented, whereas pale phenotypes are overrepresented (χ2: P < 0.0001). This difference is most likely explained by many mutations not representing full knockouts (see Discussion).
Addressing Plastome Stability
The finding that chloroplast mutations are biased toward replication slippage and spatially clustered in the genome raises the question: To what extent is the natural mutation spectrum of the chloroplast represented by our collection? To address this point, we sequenced and compared four wild-type Oenothera plastomes from O. glazioviana (=O. lamarckiana; Cleland, 1972; Harte, 1994; GenBank accessions: EU262890.2, KT881171.1, KT881172.1, and KT881174.1). This is a young species that arose in Europe in the mid 19th century by hybridization (Cleland, 1972; Dietrich et al., 1997). We investigated the Swedish strain of O. lamarckiana, a line that has been cultivated since 1906, as well as three independent descendants of O. lamarckiana de Vries (strains blandina, deserens, and vetaurea). Those were isolated in 1908, 1913, and 1921 from the original line of de Vries, collected in the Netherlands in 1886 (see Supplemental Table 2 for details on the O. lamarckiana lines). Sequence comparison of the plastome of O. lamarckiana Sweden with those of the three descendants of O. lamarckiana de Vries revealed one transition in the intergenic spacer between trnL-UAA and trnT-UGU, one 48-bp indel in the region of larger complex repeats upstream of accD, and a single base pair insertion at a tandem oligo(C+T) stretch between trnL-UAG and ccsA. The three O. lamarckiana de Vries plastomes differ by two 21-bp indels in the repetitive region of the coding frame of ycf1 (tic214). These arose independently at different sites in blandina and deserens. Only the vetaurea strain fully resembles the ycf1 (tic214) sequence of O. lamarckiana Sweden. Lastly, the deserens strain has a 6-bp deletion in the intergenic spacer between rbcL and atpB that is associated with a micro repeat. The mutation is not present in the corresponding plastome mutant III-alpha. Hence, it must have occurred in the deserens plastome after isolation of the mutant in 1954 (compared with Table 1).
Further evidence that certain regions in the chloroplast genome of Oenothera can evolve very rapidly was provided by comparison of plastome sequences of O. elata ssp hookeri strain johansen Standard (assembled in 2007; Greiner et al., 2008a) with that obtained in this work (EMBL accessions: AJ271079.3 versus AJ271079.4). A 147-bp insertion was detected in the region of large complex repeats upstream of accD, and a 24-bp insertion was identified at the 3′ end of rpoA, associated with a larger tandem repeat. Inspection of the original Sanger sequencing chromatograms revealed that our 2007 sequence lacked these features. The current version of our johansen Standard plastome is based on multiple sequencing methods (Illumina, 454, PacBio, and Sanger) and ∼50 individuals. It is further supported by the independent chloroplast mutants I-zeta, I-xi, and I-fa, which had been isolated in 1962, 1968, and 2011, respectively. It is therefore clear that the particular material sequenced in 2007 must have been polymorphic for the two plastomes.
In summary, all but one mutation detected in the closely related Oenothera plastome under study have arisen by replication slippage. They are found at very similar locations and/or sequence contexts as the detect mutations of the collection and are often located in regions of intrinsic instability associated with oligo(N) stretches or large repetitive elements (Figure 2).
DISCUSSION
Spontaneous Mutations Are Largely Limited to Replication Slippage-Based Events
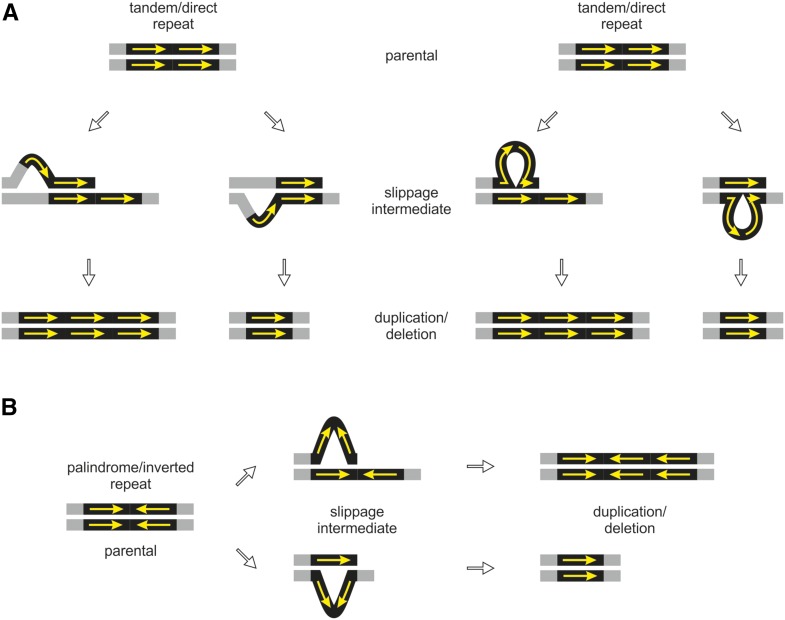
Strikingly, nearly all detected mutations could be associated with repetitive elements (Supplemental Data Set 1). Therefore, they must have occurred by a replication slippage-based mechanism, conferred by misalignment or formation of hairpin structures (Levinson and Gutman, 1987; Wolfson et al., 1991; Leach, 1994; Lovett, 2004; Figure 5). Consistent with this reasoning is the observation that, with only one exception, all duplications are perfect tandem duplications, which strongly implicates replication slippage as a template-based mechanism responsible for the occurrence of spontaneous chloroplast mutations. Although tandem, direct, inverted, or palindrome repeats can induce indels in the chloroplast genome (compared with Lovett, 2004; Darmon and Leach, 2014), tandem/direct repeats dominated as mutation-inducing sequence motives. Interestingly, these repeats can be very small, spanning only two repetitive elements, sometimes no larger than 3 bp (compared with Kondrashov and Rogozin, 2004; Tanay and Siggia, 2008; Supplemental Figures 1 to 6 and Supplemental Data Set 1; see Results).
Figure 5.
Polymerase Slippage during DNA Replication at Repetitive Elements Leading to Duplication or Deletions.
Arrows indicate repeats and pairing homology.
(A) Slippage conferred by misalignment at tandem or direct repeats. If slippage occurs at the nascent (upper) strand, replication leads to duplication. Insertions are a result of slippage at the leading (lower) strand.
(B) Slippage conferred by a hairpin structure at palindrome or inverted repeats. Duplications are a result of a hairpin at the nascent (upper) strand. A hairpin at the leading (lower) strand confers a deletion.
We only rarely detected mutations suggestive of DNA damage by reactive oxygen species (resulting in transversions; Imlay, 2008) or UV light (likely causing transitions; Pfeifer et al., 2005). This also holds true for the types of mutations that are caused by double-strand breaks (DSBs) followed by nonhomologous end-joining (NHEJ), microhomology-mediated end-joining (MMEJ), or microhomology-mediated break-induced replication (MMBIR) pathways (McVey and Lee, 2008; Hastings et al., 2009). NHEJ typically results in small indels of 1 to 4 bp and require 0 to 5 bp microhomologies at the joining ends. Deletions of variable size are common for MMEJ (between 3.1 and 4.6 kb, as determined for the chloroplast genome) and microhomologies are utilized for end joining (6 to 12 bp for perfect and 10 to 16 bp for imperfect homology in the chloroplast; Kwon et al., 2010). However, such signatures are not found in the indels identified in this study (Supplemental Data Set 1). Furthermore, no plastome rearrangements suggesting MMBIR repair (Cappadocia et al., 2010) or U-turn-like inversions (Zampini et al., 2015) were found. One possible exception is the large 1364-bp deletion in the mutant V-delta, which is not associated with a repetitive element (Supplemental Figure 6). It might have arisen from a NHEJ-like pathway (compared with Kwon et al., 2010).
Our data strongly suggest that mutations caused by processes other than replication slippage are rare in the germline and largely absent from our collection. This observation might reflect protection of the plastid DNA (ptDNA) from reactive oxygen species and UV light mutagenesis in the shoot apical meristem (Greiner et al., 2015). Harder to explain is the nearly complete lack of signatures resulting from the repairing of DSB in the chloroplast genome. Appropriate pathways exist for DSB repair (Maréchal et al., 2009; Cappadocia et al., 2010; Kwon et al., 2010), but they are likely of minor biological relevance. Similar arguments apply to U-turn-like rearrangements. NHEJ-like repair mechanisms, if present in chloroplasts at all, occur at very low frequencies (Kohl and Bock, 2009; Cappadocia et al., 2010; Kwon et al., 2010). However, neither MMEJ nor MMBIR repair nor U-turn-like rearrangements appear to have produced mutations in our material. This is in line with the observation that extensive x-ray radiation experiments failed to cause recoverable chloroplast mutations (Greiner, 2012). Therefore, it seems reasonable to propose an integrity-sensing mechanism for ptDNA in the germline. This could explain the strong structural conservation of higher plant chloroplast genomes during evolution, as reflected in the rarity of large sequence inversions or rearrangements (Ruhlman and Jansen, 2014).
Mutation Spectrum of the Chloroplast Mutant Collection
The mutation spectrum in the Oenothera plastome is in sharp contrast to that detected in the nuclear genome of Arabidopsis thaliana (Ossowski et al., 2010; Yang et al., 2015) or Escherichia coli (Lee et al., 2012). Mutations in nuclear genomes are biased toward single base pair G:C to A:T transitions (with a lower frequency of transversions) that are explained by deamination of methylated cytosine and UV light-induced DNA damage. If single base pair substitutions were common in plastid genomes, we would expect more than just the two single-nucleotide mutations present in our collection. The paucity of single-nucleotide mutations in the plastome compared with the nuclear genome may reflect that substitution rates in chloroplast DNA are substantially lower than in the nuclear genome (Wolfe et al., 1987; Drouin et al., 2008). Our results clearly show that chloroplast ptDNA replication machinery is more prone to mutations from replication slippage than misincorporation of single nucleotides. Small indels in E. coli also arise in repetitive sequences but they represent only 10% of spontaneous mutations (Lee et al., 2012). Hence, the chloroplast DNA replication and/or repair machinery differs in its error spectrum from that of bacteria. Although the chloroplast originated from an endosymbiotic event, the difference mutation spectrum likely reflects the fact that the DNA replication system is a mosaic of proteobacterial, cyanobacterial, and eukaryotic components (Moriyama and Sato, 2014). The fact that indels in chloroplast genomes are generated with much higher frequency than single base substitutions should be taken into account when performing phylogenetic analyses based on chloroplast sequences. The two classes of mutations cannot be weighted equally as phylogenetically informative characters.
Reasons for Biased Mutation Recovery
Some regions in the chloroplast genome of Oenothera are more prone to mutation than others. Although oligo(N) stretches in coding regions are rare, they are preferred targets of mutation (Supplemental Figure 1 and Supplemental Data Set 1). Other hot spots reside in large tandem repeats (Figure 2). Strikingly, compared with the second-site mutations found in the plastome mutant collection, very similar mutations could be detected between the closely related wild-type plastomes of O. glazioviana. We therefore conclude that specific repetitive regions in the Oenothera chloroplast genomes are intrinsically unstable and prone to mutation. This finding is in line with earlier studies on Oenothera plastome organization, since these reparative regions are major sources of phylogenetic divergence in evening primrose (compared with Gordon et al., 1982; Greiner et al., 2008a, 2008b). Many mutations in these regions might be phenotypically neutral and can be fixed over experimental time frames, at least in cultured evening primroses.
Our work also shows a strong mutation bias toward certain nonessential genes. Most mutations appear to be mediated by micro repeats, but several lines of evidence point to additional factors contributing to the unequal distribution of mutated alleles. First, many of the micro repeats are very small (Supplemental Data Set 1), and they are abundant throughout the chloroplast DNA. From a statistical perspective, they might occur at random. For example, the nested micro tandem repeat TATAT independently induced an insertion and a deletion at exactly the same site in the mutants I-xi and III-alpha (Supplemental Figures 2 and 3). However, this is the only TATAT site in the genome where mutations were found, although the same motif is present altogether 114 times in the sequences of Class I genes. Within the atp-rpo cluster of Class I genes, this motif occurs 45 times and again with no mutations. In psaA, nine mutations could be recovered independently of this element (Figure 3; Supplemental Data Set 1 and Supplemental Figures 2 and 4). It is present there three times. Hence, although this element is distributed randomly in the genome (χ2: P = 0.4942 for the full set of Class I genes versus the atp-rpo subset cluster), its potential to induce a recoverable mutation seems to be confined to particular positions in the plastome. It seems conceivable that the surrounding sequence context governs whether TATAT motifs are susceptible to DNA polymerase slippage (compared with Figure 3).
We were not able to deduce a consensus sequence for the mutation-inducing micro repeats. The 32 identified sites with mutations consist of many different types of repeats (perfect or imperfect, [nested] tandem, direct, or inverted/palindromic repeats). Also, in a few cases, a combination of repeat types is present. The individual repeat elements are 3 to 9 bp long, and the spacer regions, if present, are between 1 and 9 bp (Supplemental Data Set 1). A much larger data set would be needed to deduce any consensus that may exist. Nevertheless, we noticed that mutation-inducing micro repeats tend to be more A/T rich (mutation-inducing sites in total: 364 bp, with 252 bp A+T; Class I genes: 37,080 bp, with 22,031 bp A+T; χ2: P = 0.0694; compared with Tanay and Siggia, 2008). However, this tendency cannot explain the lack of mutations in the atp-rpo cluster, when compared with Class I genes (13,969 bp, with 8424 bp A+T; χ2: P = 0.3631).
Beside the sequence context, the relative positions of genes to each other or within the chloroplast genome might generate mutational hotspots. For example, DNA polymerase processivity could vary with distance relative to the origins of replication. Furthermore, collision of the replication with the transcription complexes or other DNA binding proteins might differ depending on gene position or the usage of the nuclear- or plastid-encoded RNA polymerase (Börner et al., 2014). Whereas the interference of the DNA replication complex with other DNA binding proteins is a matter of speculation, the relative position of a gene to an origin of replication does not seem to have an influence on a mutational hot spot. For example, the hot spot in ccsA is rather close to the origin of replication, where the one in psaA is not. On the other hand, the atp-rpo gene cluster, fully lacking mutations, is most distantly located from the replication origins (Figure 2).
Differences in gene expression and mutation rate have been reported to be correlated in E. coli (Martincorena et al., 2012; but also see Lee et al., 2012). However, this observation is unlikely to explain the patterns of mutation in our material. Since all chloroplast mutations are present in the germ line, they must have arisen in the meristem of the plants. Although gene expression data from proplastids in meristematic tissues are largely lacking, it is extremely unlikely that the expression rates of photosynthesis genes differ substantially in meristems. Therefore, we favor the alternative explanation that mutations in the atp-rpo gene cluster are more difficult to recover than genes with elevated mutation rates such as psaA or ccsA (see above). Possible reasons include biased gene conversion (Khakhlova and Bock, 2006) or recombinational hot spots that increase the efficiency of recombination repair by homologous recombination with an unmutated genome copy. Such hot spots of recombination were reported for the chloroplast DNA of the green alga Chlamydomonas reinhardtii (Newman et al., 1992). It seems conceivable that molecular mechanisms like biased gene conversion or biased recombination repair eliminate de novo mutations in some plastome regions more efficiently than in others.
Mutation Rates in the Chloroplast Genome
Comparison of the closely related O. glazioviana plastomes allows a rough estimation of the mutation rates for chloroplast genomes. We used the reasonable assumptions that O. glazioviana is monophyletic. The species arose by hybridization in the middle of the 19th century (Cleland, 1972; Dietrich et al., 1997). Hence, we can estimate the maximum number of generations separating the Swedish strain from the lines of Hugo de Vries to be ∼150 generations with annual reproduction. Since three to five polymorphisms distinguish these lines (see Results), we can assume one fixed mutation per 30 to 50 generations, equivalent to 2.0 to 3.3 × 10−2 mutations per generation and genome. Per site (∼165,000, with each base pair representing a site), the mutation rate would be 1.2 to 2.0 × 10−7 per site and generation. Interestingly, compared with the nuclear genome of Arabidopsis (7 × 10−9; Ossowski et al., 2010), and the genome of E. coli (2.2 × 10−10; Lee et al., 2012), this rate is rather high. Nevertheless, taking into account the small genome size of the plastid, the actual number of mutations that effectively accumulate is still small.
These approximations should be treated with caution for three reasons. First, the number of mutations is low; thus, the precision of our estimate is compromised. Second, a previous study assessing intraspecific plastome variation in tomato (Solanum lycopersicum) over the time span of approximately one century found no polymorphisms (Kahlau et al., 2006). Third, substitution rates calculated based on phylogenetic data suggest a lower mutation rate in chloroplast genomes than in plant nuclear genomes (Wolfe et al., 1987; Drouin et al., 2008). However, it is also known that organellar mutation rates can vary significantly in different lineages of seed plant evolution, even within a genus (Cho et al., 2004; Guisinger et al., 2008; Sloan et al., 2009). Last, it should be noted that three of the mutations in the O. glazioviana lines are associated with large repetitive elements repeats. These regions are a major source of phylogenetic divergence (Greiner et al., 2008a) and evolve relatively quickly. Therefore, an accurate mutation rate for plastid DNA in Oenothera remains to be determined by genome sequencing at a larger scale.
Chloroplast Mutants as a Tool to Understand Chloroplast Gene Functions and Gene Regulation
The finding that most chloroplast mutants in our collection show only one mutated site makes them a valuable resource to understand chloroplast gene function and regulation. Although not all genes in the genome are covered, chloroplast mutants for all photosynthetic complexes and also for the chloroplast translation machinery are represented. For the psaA, psbA, and ccsA genes, allelic series are available, with nine, five, and four mutants, respectively (Table 1, Figure 3). Although rare, the collection also contains mutations in essential genes, such as the mutant I-tau, which carries a defect in rps7. Since this mutant displays a reversible bleaching (virescent) phenotype (Figure 1D), its characterization should provide new insights into the developmental regulation of chloroplast translation. Also of particular interest are the five mutants identified so far that display defects in chloroplast RNA metabolism (Figure 4). Such mutants are not readily obtainable by targeted approaches. A particularly interesting finding in this respect was that mutations in the protein-coding region can influence transcript processing, as discovered in the mutant II-eta (Figure 4). The molecular basis of these effects needs to be further investigated.
It is also important to note that many of the mutants (I-alpha, I-delta, I-iota, I-tau, I-omega, I-do, II-delta, II-theta, II-kappa, II-lambda, III-alpha, III-beta, IV-beta, and V-alpha) do not harbor full gene knockouts, as judged by their phenotype and genotype (Table 1). This renders them a rich platform for functional analyses. Most null mutants of photosynthesis genes are essentially white (Schaffner et al., 1995; Ruf et al., 1997; Hager et al., 1999; Supplemental Data Set 2 and Supplemental Table 4), lacking any photosynthetic activity and, thus, of limited value for functional studies. The mutations in our Oenothera collection that affect chloroplast mRNA metabolism are located in photosynthesis genes. At least two of them (II-kappa and II-delta) display mild phenotypes. This is due to reduced abundance of the mature transcript (Figure 4), leading to a less severe phenotype than a frameshift mutation in the coding region (Table 1). An additional set of five mutants that are affected in the coding region of photosynthesis genes (I-alpha, I-delta, I-iota, II-lambda, and III-beta) are pale green rather than white. With one exception (II-lambda), the mutations cause a frameshift. In some of these cases, the pale phenotype of the mutants is likely caused by the position of the frame-shifting indel within the gene. For example, the mutants I-alpha and I-delta carry a 5-bp duplication just downstream of the translation initiation codon of psbD (Table 1). We hypothesize that these mutations are compensated for by an alternative in-frame start codon present downstream of the mutation. By contrast, in the mutant III-beta, a 5-bp duplication exists in the middle of psaB (encoding an essential reaction center protein of photosystem I; Table 1), which makes its pale-green phenotype more difficult to understand (compared with Table 1). A possible clue may be provided by one of the most striking mutant phenotypes observed in the collection: the mottled phenotype of the mutant I-iota, which largely disappears with age, giving rise to a nearly green phenotype (Figure 1C). The underlying mutation is a single A insertion located in a oligo(A) stretch very early in atpB (Table 1; Supplemental Data Set 1). The recovery of the mutant over time strongly points to the presence of a frame-correcting mechanism in chloroplasts, such as ribosomal frameshifting (Kohl and Bock, 2009).
In summary, the mild chlorotic and developmental stage-dependent phenotypes of several mutants in the collection provide interesting new avenues for the study of plastid gene function and expression.
METHODS
Plant Material
Throughout this work, the terms Oenothera or evening primrose refer to subsection Oenothera (genus Oenothera section Oenothera; Dietrich et al., 1997). Plant material used here is derived from the Oenothera germplasm collection harbored at the Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm, Germany (Greiner and Köhl, 2014). Part of this collection is represented by the 51 spontaneous plastome mutants that were collected systematically during the past century in Germany (Kutzelnigg and Stubbe, 1974; Stubbe and Herrmann, 1982; Stubbe, 1989; Supplemental Table 1). The plastome mutants were mostly compiled by Wilfried Stubbe and coworkers, maintained by Reinhold G. Herrmann since the end of the 1980s and later taken over by the authors. The mutants are designated according to their basic plastome genotype (I to V) and Greek letters, in order of the respective isolation date (i.e., I-alpha, I-beta, II-alpha, II-beta, etc.). In the case of basic plastome I, of which more mutants were isolated than Greek letters exist, further mutants are designated by the seven tone syllables. To maintain the material, a substitution of the mutant chloroplast by introgression into the suitable johansen Standard strain of O. elata ssp hookeri was performed by W. Stubbe. Except for the lines containing plastome V, which are still maintained in the background of O. argillicola, the collection has been propagated in the johansen Standard background since then (Greiner, 2012). Due to the introgression of most of the lines into the johansen Standard strain, chloroplast substitution lines with this nuclear background and the wild-type plastomes of the investigated mutant chloroplasts were used for RNA gel blot analyses as wild-type controls. For a summary of the genetic stocks, see Supplemental Tables 1 and 2. For further details see Kutzelnigg and Stubbe (1974), Stubbe (1989), Rauwolf et al. (2008), and Greiner (2012).
Plant Cultivation, Maintenance, Tissue Harvest, and Phenotyping of Chloroplast Mutants
Chloroplast mutants, the wild type, and introgression strains were cultivated as described previously (Greiner and Köhl, 2014). In Oenothera, photosynthetically incompetent chloroplast mutants can be kept as variegated plants, containing green wild-type as well as mutated chloroplasts (Figure 1A). These plants are viable in soil because the green tissue feeds the bleached tissue. The combination of two chloroplast types in one plant is possible by taking advantage of biparental chloroplast transmission, which is the rule in evening primroses. In such experiments, a maintainer strain containing a green nursery plastome is crossed to the chloroplast mutant. This results in variegated offspring because sorting-out of the two chloroplast types leads to tissues homoplasmic for either wild-type or mutant chloroplasts (Figures 1A and 1B; also see Introduction). Homoplasmic mutant tissue generated this way was used for sequence and molecular analysis, as well as phenotyping in this work. For details on the genetics, see Kutzelnigg and Stubbe (1974), Kirk and Tilney-Bassett (1978), Stubbe and Herrmann (1982), and Greiner (2012).
Phenotyping of mutants was performed in an unheated polyethylene greenhouse. Plants were analyzed after the appearance of the first two true leaves (young), during the early rosette stage (medium), and during the mature rosette stage (old). Observed phenotypes largely resembled those reported in the laboratory documentation of W. Stubbe that was based on field-grown material. However, minor discrepancies were observed. Different degrees of bleaching were assigned to a numerical code (1 = nearly white, 2 = yellowish cream-colored, 3 = light green, and 4 = nearly green). Numbers in parentheses in Table 1 indicate mottled or heterogeneous chlorotic phenotypes.
Chloroplast Isolation
An improved chloroplast isolation protocol for Oenothera was developed for this study. It overcomes severe technical limitations caused by the high amounts of mucilage present in Oenothera leaf tissue and does not require ultracentrifugation steps. As judged from sequencing analysis, ptDNA can be enriched to up to 90% by employing the following procedure at 4°C: 50 g of leaf material was homogenized in a blender adding 1 liter of BoutHomX buffer (0.4 M sucrose, 50 mM Tris, 25 mM boric acid, 10 mM EGTA, 10 mM KH2PO4, 1% [w/v] BSA, 0.1% [w/v] PVP-40, pH 7.6 with KOH, and 5 mM freshly added β-mercaptoethanol). Aliquots of the homogenate were filtered through a double layer of cheese cloth (Hartmann), followed by filtering through a single layer of Miracloth (Merck). The filtrate was then centrifuged for 15 min at 5000g. The chloroplast pellet was resuspended in 150 mL ChloroWash (2 mM EDTA, 1 mM MgCl2, 1 mM MnCl2, 50 mM HEPES, 330 mM sorbitol, pH 7.6 with KOH, and 5 mM freshly added sodium ascorbate) using a 30-cm3 Potter homogenizer (mill chamber tolerance: 0.15 to 0.25 mm; VWR). The suspension was again filtered in aliquots through a double layer of Miracloth and adjusted with ChloroWash to a volume of ∼700 mL followed by another filtering step. Next, chloroplasts were pelleted at 2400g, resuspended in 9 mL of ChloroWash, and transferred to three 85%/45% Percoll step gradients (PBF-Percoll stock solution: 3% [w/v] polyethylene glycol 6000, 1% [w/v] BSA, 1% [w/v] Ficoll 400, dissolved in Percoll; 45%/85% Percoll: 45%/85% PBF-Percoll stock solution, 330 mM sorbitol, 50 mM HEPES, 2 mM EDTA, 1 mM MgCl2, pH 7.6 with KOH; Percoll step gradient: 8 mL 85% Percoll, 16 mL 45% Percoll in a 30-mL Corex tube). The gradients were centrifuged at 10,000g for 10 min. Intact chloroplasts were recovered from the gradient’s interphase and washed in 200 mL ChloroWash. After repeating the washing steps in smaller volumes, the isolated chloroplasts were resuspended in a suitable buffer for further use.
DNA Isolation
Total DNA for Illumina sequencing was isolated from rosette leaves using a phenol-based DNA isolation protocol developed for Oenothera (Ågren et al., 2015). In contrast to the previously described protocol, we used 100 μL of a 10 mg/mL RNase A solution (50 units/mg; Roche Diagnostics). For PCR analyses, we used total DNA obtained with the DNeasy Plant Mini Kit (Qiagen; Rauwolf et al., 2008). For certain Oenothera strains, especially when older material was used, DNA was additionally purified by an anion-exchange column (Nucleobond AX 20; Macherey-Nagel). This procedure was found to be very effective in removing remaining phenolic components, mucilage, and/or polysaccharides that potentially inhibit enzymatic reactions from Oenothera DNA isolations in general.
Total high molecular weight DNA for PacBio sequencing was obtained from etiolated seedlings. To this end, seeds were sown on a wet sterile filter paper in Petri dishes and supplemented with 0.05% (v/v) of Plant Preservative Mixture (Plant Cell Technology Store) to prevent fungal contamination. Subsequently, seeds were incubated at 27°C in the light. Immediately after germination (when root tips became visible), Petri dishes were wrapped with aluminum foil, and after 3 d, material was harvested and frozen in liquid nitrogen. One hundred milligrams of seedlings were ground in liquid nitrogen, subsequently supplemented with 1550 μL CTAB buffer (130 mM Tris/HCl, pH 8.0, 1.8 M NaCl, 1.3% [w/v] PVP-40, and 2.6% [w/v] CTAB) and 250 μL 1-thioglycerol. Throughout the procedure, samples were mixed by gentle inversion. The aqueous DNA solution was pipetted with a cut tip. After adding 100 μL of a 10 mg/mL RNase A solution (see above), samples were incubated for 30 min at 60°C. Afterwards, 100 μL of proteinase K (10 mg/mL; Sigma-Aldrich) was added and samples were incubated at the same conditions as described above. To remove cell debris, samples were centrifuged at 20,000g for 5 min at room temperature. The supernatant was treated with 2 mL phenol/chloroform/isoamyl alcohol (25:25:1, pH 7.5) and 2 mL chloroform/isoamyl alcohol (24:1). DNA was precipitated with one-tenth volume of 5 M NH4-acetate and 1 volume of isopropanol. Further purification was achieved using Qiagen Genomic-tip 20/G columns and (optionally) with a High Salt Phenol Chloroform Cleanup recommended by Pacific Biosciences. Deviating from the Pacific Biosciences protocol, we omitted EDTA from our buffers. As judged from pulsed-field gel electrophoresis, our procedure yields DNA fragments between 20 and 200 kb, with ODs of ∼1.8 (A260/A280) and ∼2.1 (A260/A230). Prior to library preparation, the DNA was subjected to an additional AMPure PB purification step (Pacific Biosciences).
To obtain ptDNA for 454 sequencing, isolated chloroplasts were resuspended in 400 μL of TENTS buffer (100 mM Tris/HCl, pH 8.0, 50 mM EDTA, 0.5 M NaCl, 0.2% [v/v] Triton X-100, and 1% [w/v] SDS) and incubated at 60°C for 15 min. Next, the lysate was supplemented with 100 μL RNase A (10 mg/mL; see above) and incubated for 1.5 h at 37°C. Subsequently, 100 μL of 10 mg/mL proteinase K (see above) solution was added and samples were placed overnight at room temperature. After extraction with phenol/chloroform/isoamyl alcohol (25:24:1, pH 7.5) and with chloroform/isoamylalcohol (24:1), the DNA was precipitated at −20°C overnight as described above.
RNA Isolation
Total RNA for RNA gel blot analyses was isolated from Oenothera leaves using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). Columns and buffers used were included by the manufacturer. To cope with mucilage and high amounts of phenolic components in the tissue, minor modifications were introduced to the provided protocol: Up to 25 mg of frozen tissue was ground in liquid nitrogen. Material was lysed in 750 μL of freshly prepared lysis solution (50 μL β-mercaptoethanol for 1 mL of supplied lysis solution). The lysate was incubated at room temperature for 5 min and filtrated on a filtration column. Centrifugation time was extended up to 5 min. Binding solution (750 μL) was mixed with the clarified lysate, mixed gently through pipetting, and centrifuged on a binding column. An additional elution step was performed to increase of RNA yields.
Detection of RNA via RNA Gel Blot
Total RNA (1.5 to 2.0 µg) was separated on 1% agarose gels (w/v) under denaturing conditions. RNA was transferred to a nylon membrane (Hybond-XL; GE Healthcare) by capillary transfer using 10× SSC buffer (1× SSC: 0.015 M sodium citrate and 0.15 M NaCl). The RNA was covalently linked to the membrane using a UV cross-linker (Vilber Lourmat). Radiolabeled probes were used for strand-specific detection of transcripts in RNA gel blot hybridization. For this purpose, transcription of gene-specific PCR products was performed with the MAXIscript T7 kit (Ambion) using radiolabeled [32P]dUTP (Hartman Analytic). Gene-specific PCR products for ycf3, petB, and psaC were obtained employing the following primers in a PCR with total Oenothera DNA as a template: ycf3ex2_north_fr, 5′-GGATGTCGGCTCAGTCAG-3′; ycf3ex2_north_rv, 5′-TAATACGACTCACTATAGGGGTAAGAACGGGTTTCGTTCTAGTG-3′; petBex2_north_fr, 5′-GTCTCGAGATTCAGGCGATTGC-3′; petBex2_north_rv, 5′-TAATACGACTCACTATAGGGAATACAGCTAGAACCACGCCTGTG-3′; petBint_north_fr, 5′-GTCGGATATTCATTCTCGTATCTGGAGC-3′; petBint_north_rv, 5′-TAATACGACTCACTATAGGGAGTTGCTCGTTCCAGGCCTC-3′; psaC_north_fr, 5′-TAATACGACTCACTATAGGGACCCATACTTCGAGTTGTTTCATGC-3′; and psaC_north_rv, 5′-ATGTCGCATTCAGTAAAGATTTATGATACCTG-3′. RNA gel blots were performed according to a standard protocol. Nylon membrane with RNA was incubated in Church buffer (0.5 M NaH2PO4, 7% SDS [w/v], and 1 mM EDTA, pH 7.2) for 1 h. After this, time radioactive probes were transferred to hybridization tube and incubated overnight in 65°C. The next day, three washing steps were performed: once in 1× SSC supplemented with 0.2% SDS (w/v) for 20 min and twice in 0.5× SSC supplemented with 0.2% SDS (w/v) for 20 min each. Visualization of radioactive signals was performed at different time points, depending on the strengths of the mRNA signal.
DNA Sequencing
Sanger sequencing was performed with PCR products using standard methods (at Eurofins MWG Operon). Primers were obtained from the same source.
Illumina paired-end sequencing (375-bp insert size) was performed as described previously (Ågren et al., 2015). To improve sequence quality at very long oligo(N) stretches and to avoid a bias in the Illumina libraries against certain (TA)n rich sequences in some regions of the Oenothera chloroplast genome, “PCR free” Illumina libraries were employed at a later stage of the project. In most cases, ∼10 to 30% of the sequenced DNA was found to be ptDNA. The observed variance results from the developmental stage of the leaf tissue used and/or the particular genotype of the line (compared with Rauwolf et al., 2010). From these data, we could deduce that roughly five million total DNA 100-bp Illumina paired-end reads are sufficient to sequence a chloroplast genome of Oenothera at 500× to 1500× coverage. In this context, it is relevant to mention that the mutant tissue (obtained from random sorting-out of mutant and nursery plastomes) used in this work for Illumina sequencing (see above) was confirmed to be fully homoplasmic for the mutant chloroplast genome.
Prior to 454 sequencing, 2 to 5 μg of isolated ptDNA was preamplified to 20 µg using the GenomiPhi HY DNA amplification kit (GE Healthcare). Single-end read libraries were sequenced on a Roche/GS FLX Titanium platform to an approximate coverage of 20× per chloroplast genome. The 454 sequencing was performed at Eurofins MWG Operon. To correct sequence uncertainties that are frequent in 454 data sets and a potential bias in the template DNA introduced by the rolling cycle amplification with the GenomiPhi Kit, all chloroplast genomes obtained from 454 data were resequenced by Illumina.
PacBio sequences were obtained from a 7-kb cutoff insert library using P4-C2 chemistry. Size selection, library preparation, and sequencing were performed at the Max Planck-Genome-Centre in Cologne, Germany. PacBio sequencing is part of the Oenothera genome project and will be communicated elsewhere.
Plastome Assembly
In total, the complete sequences of 65 chloroplast genomes were determined in this study, many of them through multiple methods. For a summary of the different sequencing and assembly methods used, see Supplemental Tables 3 and 5. De novo assemblies of the chloroplast genomes and reference-guided mutation mapping were performed using the standard parameters of SeqMan NGen v.12.1.0 or earlier (DNASTAR) and/or CLC Genomic Workbench v.7.5.1 or earlier (CLC bio). First, we determined whether Illumina libraries obtained from total DNA contain plastome sequences translocated to the nuclear genome (NUPTs; Kleine et al., 2009). Translocated sequences, potentially containing polymorphisms compared with the true cytoplasmic ptDNA, might interfere with the assembly/mutation identification. Therefore, we independently de novo assembled 454 reads from ptDNA of isolated chloroplasts and Illumina reads from total DNA. For this, we used three wild-type lines and 11 chloroplast mutants. Both approaches (Illumina and 454 of purified ptDNA from the same materials) gave comparable results, and NUPTs could not be detected with certainty in the Illumina sequences. Hence, the remaining chloroplast genomes were solely obtained from Illumina reads of total DNA. However, both assemblers and sequencing techniques did not yield satisfactory results in the de novo assemblies of the notoriously repetitive regions of the Oenothera plastome, such as upstream and within accD and ycf1 (tic214), within ycf2, and in the rrn16-trnI-GAU spacer. In these regions, individual repetitive elements can span more than the read length and/or the insert size of a paired-end library. In the wild-type reference plastomes, those gaps were closed by Sanger sequencing and even consistent assemblies were routinely verified with the same technique. In the johansen Standard line, these regions were additionally verified by single PacBio reads, fully spanning the repetitive regions. All repetitive regions discussed in this work are supported by Sanger reads. Chloroplast mutations, if not identified from a de novo assembly, were identified by a reference-guided assembly using the chloroplast genome of the reference strain. Misassemblies, indicating larger indels, were identified by paired-end mapping. Smaller mutations could be directly identified in the reference-guided assembly. The notoriously repetitive regions as well as some of the mutant alleles, especially those harboring larger indels, were verified at random by Sanger sequencing.
Sequence Annotation
Wild-type chloroplast genomes were annotated using GeSeq v.0.2.1, and GenBank submission files were generated by GenBank 2 Sequin (https://chlorobox.mpimp-golm.mpg.de). These software tools were developed for this project to annotate and process organelle sequences (P. Lehwark and S. Greiner, unpublished data; M. Tillich, P. Lehwark, E.S. Ulbricht-Jones, A. Fischer, T. Pellizzer, R. Bock, and S. Greiner, unpublished data). The existing annotation of the reference Oenothera chloroplast genome from O. elata ssp hookeri strain johansen Standard (AJ271079) was used as a reference. Also, in the course of this work, the annotation of our previously sequenced Oenothera plastomes (Greiner et al., 2008a) was revised, and the exon/intron borders of protein-coding genes are now verified by cDNA sequences. Furthermore, some names of genes and gene products were updated (see AJ271079.4, EU262887.2, EU262889.2, EU262890.2, and EU262891.2). For Illumina transcriptome sequencing, total RNA extractions, sequencing, and assemblies, see Johnson et al. (2012) and Hollister et al. (2015).
Repeat Analysis
Longer repetitive elements were identified manually or using the EMBOSS suite (Rice et al., 2000). Larger imperfect palindromes were identified with the standard parameters of mfold (Zuker, 2003). For macro repeat analyses incorporated into Figure 2, refer to Greiner et al. (2008a). Since we found that repetitive elements of no larger than 3 bp are able to induce replication slippage in the chloroplast, a sequence inspection by eye was necessary to identify these elements. For this reason, the sequence context 20 bp upstream and downstream of a micro indel (<10 bp) was inspected for (perfect/imperfect/nested) micro (<10 bp) tandem, direct, palindrome, and inverted repeats or combinations thereof.
Global Sequence Analyses and Visualization of the Oenothera Plastome
For global sequence analyses, including statistical analysis, GC content determination, repeat analyses, and search for sequence motif abundance, the reference Oenothera chloroplast genome of the johansen Standard strain (O. elata ssp hookeri) was used (AJ271079.4). Coding regions of the Oenothera plastome are largely identical between Oenothera species (Greiner et al., 2008a). Chloroplast genome maps were drawn by OrganellarGenomeDRAW (Lohse et al., 2013) and edited in CorelDarw X5 (Corel).
Statistical Analyses
χ2 tests were performed using GraphPad (http://www.graphpad.com) with Yates' correction. Two-tailed P values are given.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ271079.4, EU262887.2, EU262889.2 to EU262891.2, and KT881169.2 to KT881177.1.
Supplemental Data
Supplemental Figure 1. Sequence context of the detected mutations at oligo(N) stretches.
Supplemental Figure 2. Sequence context of the detected micro tandem duplications.
Supplemental Figure 3. Sequence context of the detected micro deletions.
Supplemental Figure 4. Sequence context of the detected macro tandem duplications.
Supplemental Figure 5. Sequence context of the detected macro tandem duplication and deletion.
Supplemental Figure 6. Sequence context of the detected macro deletions.
Supplemental Table 1. Origin of the mutant lines in the Oenothera plastome mutant collection.
Supplemental Table 2. Oenothera wild-type, nuclear genome mutants, and chloroplast substitution lines used this work.
Supplemental Table 3. Summary on the wild-type Oenothera chloroplast genomes determined de novo or resequenced in this work.
Supplemental Table 4. Major phenotypic classes of the Oenothera plastome mutants.
Supplemental Table 5. Sequencing and assembly methods of the full chloroplast genomes determined in this study.
Supplemental Data Set 1. Summary of the sequence context of the identified mutations in the chloroplast mutant collection.
Supplemental Data Set 2. Macroscopic phenotypes of deletion mutants in the chloroplast genome.
Supplementary Material
Acknowledgments
We thank Wilfried Stubbe for compiling and previously maintaining the Oenothera plastome mutant collection and Reinhold G. Herrmann, Herbert Hopf, and Elisabeth Gerick for maintaining the material after the retirement of Wilfried Stubbe. We also thank Dirk Walther for advice in statistical analysis and Jörg Meurer and Serena Schwenkert for sharing information on the macroscopic phenotypes of psbI and psbK knockout plants. We thank Barbara B. Sears for providing her mutant material, which served as control samples. She obtained her material earlier from Wilfried Stubbe’s collection. Barbara B. Sears and two anonymous reviewers are acknowledged for fruitful discussion and helpful comments on the manuscript. Transcriptome sequencing was made possible through Gane Ka-Shu Wong, the 1kp initiative (1kp.org) and BGI-Shenzhen. The work was supported by grants from the Deutsche Forschungsgemeinschaft (GR 4193/1-1) to S.G. and R.B. (BO 1482/17-1) and by the Max Planck Society. M.T.J.J. is supported by an NSERC Discovery Grant and an Early Researcher Award from the Ontario Government. S.I.W. is funded by a Natural Sciences and Engineering Research Council of Canada grant.
AUTHOR CONTRIBUTIONS
A.M., J.S., L.Y.-R., E.S.U.-J., A.Z., T.P., J.S., and S.G. performed experiments. All authors analyzed and discussed the data. S.G. designed the study and wrote the manuscript. R.B. participated in writing.
Glossary
- LSC
large single-copy
- SSC
small single-copy
- IR
inverted repeat
- NHEJ
nonhomologous end-joining
- DSB
double-strand break
- MMEJ
microhomology-mediated end-joining
- MMBIR
microhomology-mediated break-induced replication
- ptDNA
plastid DNA
Footnotes
Articles can be viewed without a subscription.
References
- Ågren J.A., Greiner S., Johnson M.T.J., Wright S.I. (2015). No evidence that sex and transposable elements drive genome size variation in evening primroses. Evolution 69: 1053–1062. [DOI] [PubMed] [Google Scholar]
- Archer E.K., Bonnett H.T. (1987). Characterization of a virescent chloroplast mutant of tobacco. Plant Physiol. 83: 920–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhagiri A.K., Maliga P. (2007). Exceptional paternal inheritance of plastids in Arabidopsis suggests that low-frequency leakage of plastids via pollen may be universal in plants. Plant J. 52: 817–823. [DOI] [PubMed] [Google Scholar]
- Baur E. (1909). Das Wesen und die Erblichkeitsverhältnisse der “Varietates albomarginatae hort.” von Pelargonium zonale. Z. Indukt. Abstamm. Vererbungsl. 1: 330–351. [Google Scholar]
- Baur E. (1910). Untersuchungen über die Vererbung von Chromatophorenmerkmalen bei Melandrium, Antirrhinum und Aquilegia. Z. Indukt. Abstamm. Vererbungsl. 4: 81–102. [Google Scholar]
- Börner T., Sears B. (1986). Plastome mutants. Plant Mol. Biol. Report. 4: 69–92. [Google Scholar]
- Börner T., Zhelyazkova P., Legen J., Schmitz-Linneweber C. (2014). Chloroplast gene expression: RNA synthesis and processing. In Plastid Biology, Theg S.M., Wollman F.-A., eds (New York: Springer: Science+Business Media; ), pp. 3–47. [Google Scholar]
- Cappadocia L., Maréchal A., Parent J.-S., Lepage E., Sygusch J., Brisson N. (2010). Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22: 1849–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia C.P., Duesing J.H., Arntzen C.J. (1986). Developmental loss of photosystem II activity and structure in a chloroplast-encoded tobacco mutant, lutescens-1. Plant Physiol. 82: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W.-L., Sears B.B. (1985). Recombination between chloroplast DNAs does not occur in sexual crosses of Oenothera. Mol. Gen. Genet. 198: 525–528. [DOI] [PubMed] [Google Scholar]
- Chiu W.-L., Sears B.B. (1992). Electron microscopic localization of replication origins in Oenothera chloroplast DNA. Mol. Gen. Genet. 232: 33–39. [DOI] [PubMed] [Google Scholar]
- Cho Y., Mower J.P., Qiu Y.-L., Palmer J.D. (2004). Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc. Natl. Acad. Sci. USA 101: 17741–17746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland R.E. (1972). Oenothera - Cytogenetics and Evolution. (London, New York: Academic Press; ). [Google Scholar]
- Colombo N., Emanuel C., Lainez V., Maldonado S., Prina A.R., Börner T. (2008). The barley plastome mutant CL2 affects expression of nuclear and chloroplast housekeeping genes in a cell-age dependent manner. Mol. Genet. Genomics 279: 403–414. [DOI] [PubMed] [Google Scholar]
- Correns C. (1909). Vererbungsversuche mit blass (gelb) grünen und buntblättrigen Sippen bei Mirabilis Jalapa, Urtica pilulifera und Lunaria annua. Z. Indukt. Abstamm. Vererbungsl. 1: 291–329. [Google Scholar]
- Darmon E., Leach D.R.F. (2014). Bacterial genome instability. Microbiol. Mol. Biol. Rev. 78: 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich W., Wagner W.L., Raven P.H. (1997). Systematics of Oenothera section Oenothera subsection Oenothera (Onagraceae). Syst. Bot. Monogr. 50: 1–234. [Google Scholar]
- Drescher A., Ruf S., Calsa T. Jr., Carrer H., Bock R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22: 97–104. [DOI] [PubMed] [Google Scholar]
- Drouin G., Daoud H., Xia J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49: 827–831. [DOI] [PubMed] [Google Scholar]
- Gordon K.H.J., Crouse E.J., Bohnert H.J., Herrmann R.G. (1982). Physical mapping of differences in chloroplast DNA of the five wild-type plastomes in Oenothera subsection Euoenothera. Theor. Appl. Genet. 61: 373–384. [DOI] [PubMed] [Google Scholar]
- Greiner S. (2012). Plastome mutants of higher plants. In Genomics of Chloroplasts and Mitochondria, Bock R., Knoop V., eds (Dordrecht, Heidelberg, New York, London: Springer Netherlands; ), pp. 237–266. [Google Scholar]
- Greiner S., Köhl K. (2014). Growing evening primroses (Oenothera). Front. Plant Sci. 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S., Sobanski J., Bock R. (2015). Why are most organelle genomes transmitted maternally? BioEssays 37: 80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S., Wang X., Rauwolf U., Silber M.V., Mayer K., Meurer J., Haberer G., Herrmann R.G. (2008a). The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. sequence evaluation and plastome evolution. Nucleic Acids Res. 36: 2366–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S., Wang X., Herrmann R.G., Rauwolf U., Mayer K., Haberer G., Meurer J. (2008b). The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Mol. Biol. Evol. 25: 2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger M.M., Kuehl J.V., Boore J.L., Jansen R.K. (2008). Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. USA 105: 18424–18429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann R. (1992). Plastid genetics in higher plants. In Cell Organelles: Plant Gene Research, Knowledge B., Application R.G.H., eds (Berlin, Heidelberg, New York: Springer; ), pp. 65–96. [Google Scholar]
- Hagemann R. (2010). The foundation of extranuclear inheritance: plastid and mitochondrial genetics. Mol. Genet. Genomics 283: 199–209. [DOI] [PubMed] [Google Scholar]
- Hager M., Biehler K., Illerhaus J., Ruf S., Bock R. (1999). Targeted inactivation of the smallest plastid genome-encoded open reading frame reveals a novel and essential subunit of the cytochrome b6f complex. EMBO J. 18: 5834–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harte C. (1994). Oenothera - Contributions of a Plant to Biology. (Berlin, Heidelberg, New York: Springer; ). [Google Scholar]
- Hastings P.J., Ira G., Lupski J.R. (2009). A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao T., Watanabe A., Kurita M., Kondo T., Takata K. (2009). A frameshift mutation of the chloroplast matK coding region is associated with chlorophyll deficiency in the Cryptomeria japonica virescent mutant Wogon-Sugi. Curr. Genet. 55: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister J.D., Greiner S., Wang W., Wang J., Zhang Y., Wong G.K.-S., Wright S.I., Johnson M.T.J. (2015). Recurrent loss of sex is associated with accumulation of deleterious mutations in Oenothera. Mol. Biol. Evol. 32: 896–905. [DOI] [PubMed] [Google Scholar]
- Imlay J.A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77: 755–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.T.J., et al. (2012). Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS One 7: e50226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S., Aspinall S., Gray J.C., Bock R. (2006). Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J. Mol. Evol. 63: 194–207. [DOI] [PubMed] [Google Scholar]
- Khakhlova O., Bock R. (2006). Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 46: 85–94. [DOI] [PubMed] [Google Scholar]
- Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M., Takase M., Ide T., Nakai M. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574. [DOI] [PubMed] [Google Scholar]
- Kirk J.T.O., Tilney-Bassett R.A.E. (1978). The Plastids. Their Chemistry, Structure, Growth and Inheritance. (Amsterdam, New York, Oxford: Elsevier; ). [Google Scholar]
- Kleine T., Maier U.G., Leister D. (2009). DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 60: 115–138. [DOI] [PubMed] [Google Scholar]
- Kode V., Mudd E.A., Iamtham S., Day A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44: 237–244. [DOI] [PubMed] [Google Scholar]
- Kohl S., Bock R. (2009). Transposition of a bacterial insertion sequence in chloroplasts. Plant J. 58: 423–436. [DOI] [PubMed] [Google Scholar]
- Kondrashov A.S., Rogozin I.B. (2004). Context of deletions and insertions in human coding sequences. Hum. Mutat. 23: 177–185. [DOI] [PubMed] [Google Scholar]
- Krech K., Ruf S., Masduki F.F., Thiele W., Bednarczyk D., Albus C.A., Tiller N., Hasse C., Schöttler M.A., Bock R. (2012). The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 159: 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzelnigg H., Stubbe W. (1974). Investigation on plastome mutants in Oenothera: 1. General considerations. Subcell. Biochem. 3: 73–89. [Google Scholar]
- Kwon T., Huq E., Herrin D.L. (2010). Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 13954–13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau A.M., Lokstein H., Scheller H.V., Lainez V., Maldonado S., Prina A.R. (2009). A cytoplasmically inherited barley mutant is defective in photosystem I assembly due to a temperature-sensitive defect in ycf3 splicing. Plant Physiol. 151: 1802–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D.R.F. (1994). Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays 16: 893–900. [DOI] [PubMed] [Google Scholar]
- Lee H., Popodi E., Tang H., Foster P.L. (2012). Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 109: E2774–E2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G.A. (1987). Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4: 203–221. [DOI] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Kahlau S., Bock R. (2013). OrganellarGenomeDRAW - a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41: W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S.T. (2004). Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52: 1243–1253. [DOI] [PubMed] [Google Scholar]
- Maréchal A., Parent J.-S., Véronneau-Lafortune F., Joyeux A., Lang B.F., Brisson N. (2009). Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I., Seshasayee A.S.N., Luscombe N.M. (2012). Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature 485: 95–98. [DOI] [PubMed] [Google Scholar]
- Mashkina E.V., Usatov A.V., Skorina M.V. (2010). [Comparative analysis of thermotolerance of sunflower chlorophyll mutants]. Genetika 46: 203–210. [PubMed] [Google Scholar]
- McVey M., Lee S.E. (2008). MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24: 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis P. (1955). Über Gesetzmäßigkeiten der Plasmon-Umkombination und über eine Methode zur Trennung einer Plastiden-, Chondriosomen-, resp. Sphaerosomen-, (Mikrosomen)- und einer Zytoplasmavererbung. Cytologia (Tokyo) 20: 315–338. [Google Scholar]
- Michaelis P. (1957). Genetische, entwicklungsgeschichtliche und cytologische Untersuchungen zur Plasmavererbung: II. Mitteilung. Über eine Plastidenmutation mit intrazellularer Wechselwirkung der Plastiden, zugleich ein Beitrag zur Methodik der Plasmonanalyse und zur Entwicklungsgeschichte von Epilobium. Planta 50: 60–106. [Google Scholar]
- Moriyama T., Sato N. (2014). Enzymes involved in organellar DNA replication in photosynthetic eukaryotes. Front. Plant Sci. 5: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman S.M., Harris E.H., Johnson A.M., Boynton J.E., Gillham N.W. (1992). Nonrandom distribution of chloroplast recombination events in Chlamydomonas reinhardtii: evidence for a hotspot and an adjacent cold region. Genetics 132: 413–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J.I., Warthmann N., Clark R.M., Shaw R.G., Weigel D., Lynch M. (2010). The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J.D. (1983). Chloroplast DNA exists in two orientations. Nature 301: 92–93. [Google Scholar]
- Pfeifer G.P., You Y.-H., Besaratinia A. (2005). Mutations induced by ultraviolet light. Mutat. Res. 571: 19–31. [DOI] [PubMed] [Google Scholar]
- Powles S.B., Yu Q. (2010). Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61: 317–347. [DOI] [PubMed] [Google Scholar]
- Rauwolf U., Golczyk H., Greiner S., Herrmann R.G. (2010). Variable amounts of DNA related to the size of chloroplasts III. Biochemical determinations of DNA amounts per organelle. Mol. Genet. Genomics 283: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauwolf U., Golczyk H., Meurer J., Herrmann R.G., Greiner S. (2008). Molecular marker systems for Oenothera genetics. Genetics 180: 1289–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet. 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Ruf S., Kössel H., Bock R. (1997). Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139: 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Karcher D., Bock R. (2007). Determining the transgene containment level provided by chloroplast transformation. Proc. Natl. Acad. Sci. USA 104: 6998–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T.A., Jansen R.K. (2014). The plastid genomes of flowering plants. In Chloroplast Biotechnology: Methods and Protocols, Methods in Molecular Biology, Maliga P., ed (New York: Springer: Science+Business Media; ), pp. 3–38. [DOI] [PubMed] [Google Scholar]
- Schaffner C., Laasch H., Hagemann R. (1995). Detection of point mutations in chloroplast genes of Antirrhinum majus L. I. Identification of a point mutation in the psaB gene of a photosystem I plastome mutant. Mol. Gen. Genet. 249: 533–544. [DOI] [PubMed] [Google Scholar]
- Scharff L.B., Bock R. (2014). Synthetic biology in plastids. Plant J. 78: 783–798. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Kushnir S., Babiychuk E., Poltnigg P., Herrmann R.G., Maier R.M. (2005). Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase alpha-subunit mRNA. Plant Cell 17: 1815–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schötz F. (1954). Über Plastidenkonkurrenz bei Oenothera. Planta 43: 182–240. [Google Scholar]
- Sloan D.B., Oxelman B., Rautenberg A., Taylor D.R. (2009). Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol. Biol. 9: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C.R. (1986). Analysis of photosynthesis with mutants of higher plants and algae. Annu. Rev. Plant Physiol. 37: 467–507. [Google Scholar]
- Stubbe W. (1989). Oenothera - An ideal system for studying the interaction of genome and plastome. Plant Mol. Biol. Report. 7: 245–257. [Google Scholar]
- Stubbe W., Herrmann R.G. (1982). Selection and maintenance of plastome mutants and interspecific genome/plastome hybrids from Oenothera. In Methods in Chloroplast Molecular Biology, Edelman M., Hallick R.B., Chua N.-H., eds (Amsterdam, New York, Oxford: Elsevier; ), pp. 149–165. [Google Scholar]
- Tanay A., Siggia E.D. (2008). Sequence context affects the rate of short insertions and deletions in flies and primates. Genome Biol. 9: R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel H., Kluth C., Varrelmann M. (2010). A new molecular method for the rapid detection of a metamitron-resistant target site in Chenopodium album. Pest Manag. Sci. 66: 1011–1017. [DOI] [PubMed] [Google Scholar]
- Tiller N., Bock R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant 7: 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usatov A.V., Razoriteleva E.K., Mashkina E.V., Ulitcheva I.I. (2004). Spontaneous and nitrosomethylurea-induced reversions in plastome chlorophyll mutants of sunflower Helianthus annuus L. Russ. J. Genet. 40: 186–192. [PubMed] [Google Scholar]
- Westhoff P., Herrmann R.G. (1988). Complex RNA maturation in chloroplasts. The psbB operon from spinach. Eur. J. Biochem. 171: 551–564. [DOI] [PubMed] [Google Scholar]
- Wolfe K.H., Li W.H., Sharp P.M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 84: 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson R., Higgins K.G., Sears B.B. (1991). Evidence for replication slippage in the evolution of Oenothera chloroplast DNA. Mol. Biol. Evol. 8: 709–720. [DOI] [PubMed] [Google Scholar]
- Xie Z., Merchant S. (1996). The plastid-encoded ccsA gene is required for heme attachment to chloroplast c-type cytochromes. J. Biol. Chem. 271: 4632–4639. [DOI] [PubMed] [Google Scholar]
- Yang S., Wang L., Huang J., Zhang X., Yuan Y., Chen J.-Q., Hurst L.D., Tian D. (2015). Parent-progeny sequencing indicates higher mutation rates in heterozygotes. Nature 523: 463–467. [DOI] [PubMed] [Google Scholar]
- Zampini É., Lepage É., Tremblay-Belzile S., Truche S., Brisson N. (2015). Organelle DNA rearrangement mapping reveals U-turn-like inversions as a major source of genomic instability in Arabidopsis and humans. Genome Res. 25: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.