PIFs directly activate HEC expression in the dark, and HECs oppositely regulate PIF activity by heterodimerization, forming a negative feedback loop to fine-tune photomorphogenesis.
Abstract
The phytochrome interacting factors (PIFs), a small group of basic helix-loop-helix transcription factors, repress photomorphogenesis both in the dark and light. Light signals perceived by the phytochrome family of photoreceptors induce rapid degradation of PIFs to promote photomorphogenesis. Here, we show that HECATE (HEC) proteins, another small group of HLH proteins, antagonistically regulate PIFs to promote photomorphogenesis. HEC1 and HEC2 heterodimerize with PIF family members. PIF1, HEC1, and HEC2 genes are spatially and temporally coexpressed, and HEC2 is localized in the nucleus. hec1, hec2, and hec3 single mutants and the hec1 hec2 double mutant showed hyposensitivity to light-induced seed germination and accumulation of chlorophyll and carotenoids, hallmark processes oppositely regulated by PIF1. HEC2 inhibits PIF1 target gene expression by directly heterodimerizing with PIF1 and preventing DNA binding and transcriptional activation activity of PIF1. Conversely, PIFs directly activate the expression of HEC1 and HEC2 in the dark, and light reduces the expression of these HECs possibly by degrading PIFs. HEC2 is partially degraded in the dark through the ubiquitin/26S-proteasome pathway and is stabilized by light. HEC2 overexpression also reduces the light-induced degradation of PIF1. Taken together, these data suggest that PIFs and HECs constitute a negative feedback loop to fine-tune photomorphogenesis in Arabidopsis thaliana.
INTRODUCTION
Phytochrome interacting factors (PIFs) belong to the basic helix-loop-helix (bHLH) superfamily of transcription factors (Toledo-Ortiz et al., 2003; Duek and Fankhauser, 2005; Leivar and Quail, 2011; Leivar and Monte, 2014). The autographic feature of the bHLH factors is the presence of a bipartite signature domain, the bHLH domain, which contains an N-terminal DNA binding basic region (b) and a C-terminal dimerization region (HLH). The DNA binding region is composed of ∼15 amino acids with a high percentage of basic residues. The HLH region consists of ∼60 amino acids containing two α-helices joined by a variable loop and mediates homodimerization and/or heterodimerization with other bHLH proteins (Littlewood and Evans, 1998). bHLH proteins can bind to cis-acting regulatory elements found in the promoter regions of target genes either as homodimers and/or heterodimers. One subclass of bHLH factors (group D) lacks the basic DNA binding region of the bHLH domain and is designated as HLH proteins (Benezra et al., 1990). Predictably, these proteins lack the ability to bind DNA but are still able to heterodimerize with other bHLH proteins through the HLH domains. Heterodimerization between HLH and other bHLH proteins prevents the DNA binding and transcriptional activation activities of the bound bHLH protein. Consequently, HLH proteins are considered dominant-negative regulators of bHLH proteins (Benezra et al., 1990) and are involved in a number of developmental processes in eukaryotic systems (Littlewood and Evans, 1998; Toledo-Ortiz et al., 2003).
PIFs consist of seven family members (PIF1 and PIF3-8) from group 15 of the Arabidopsis thaliana bHLH superfamily (Toledo-Ortiz et al., 2003; Duek and Fankhauser, 2005; Castillon et al., 2007). In addition to the bHLH domain, PIFs have either an active phytochrome B (phyB) binding (APB) domain and/or active phyA binding (APA) domain located at the N terminus (Castillon et al., 2007; Leivar and Monte, 2014). PIFs interact with the biologically active form of phytochromes (phys), the red/far-red (R/FR) light photoreceptors, using the APA and/or APB domains (Leivar and Quail, 2011; Leivar and Monte, 2014). Being members of the same clade of the bHLH family, PIFs display high degree of sequence similarity and can homo- and heterodimerize among themselves (Toledo-Ortiz et al., 2003; Bu et al., 2011a). Moreover, pif single mutants display distinct visible phenotypes (Castillon et al., 2007; Leivar and Quail, 2011; Jeong and Choi, 2013). pif3 and pif7 single mutants displayed short hypocotyl phenotypes under red and/or FR light conditions, while pif1 mutants showed strong effects on seed germination, chlorophyll, and carotenoid accumulation in response to light (Huq et al., 2004; Oh et al., 2004; Toledo-Ortíz et al., 2010). pif1 mutants germinate after FR light exposure due to misregulation of various hormone biosynthetic and signaling genes (Oh et al., 2009). pif1 and pif3 seedlings exhibit photooxidative damage (bleaching) and reduced greening when dark-grown seedlings are transferred to light primarily due to misregulation of chlorophyll and carotenoid biosynthetic genes in the dark (Moon et al., 2008; Stephenson et al., 2009; Toledo-Ortíz et al., 2010). A quadruple pif1 pif3 pif4 pif5 mutant displayed constitutive photomorphogenic phenotypes both morphologically and at the gene expression level in the dark (Leivar et al., 2008, 2009; Shin et al., 2009), suggesting that PIFs repress photomorphogenic growth in the dark. In addition to photomorphogenesis, PIFs regulate many other pathways, including circadian clock, flowering time in response to temperature, stomatal development, and senescence, suggesting that PIFs act as signaling hubs in regulating plant growth and development (Leivar and Monte, 2014; Sakuraba et al., 2014).
In contrast to PIFs, the phytochrome family of photoreceptors (phyA-phyE in Arabidopsis) perceives R/FR/blue light signals in surrounding environment and promotes photomorphogenic development of plants (Rockwell et al., 2006; Bae and Choi, 2008; Quail, 2010; Galvão and Fankhauser, 2015). The phys are synthesized as the Pr form in the cytosol. Upon perceiving light signals using the bilin chromophore attached to the N-terminal domains, phys make an allosteric change in their structure, which produces a biologically active Pfr form and translocates into the nucleus as either a homodimer or heterodimer (Fankhauser and Chen, 2008; Clack et al., 2009; Pfeiffer et al., 2012). One mode of phy function is to interact with PIFs through the APA and/or APB domains within the nucleus and inhibit PIF functions, thus promoting photomorphogenesis (Leivar and Quail, 2011). Recent data suggest that phys inhibit PIF functions by at least two mechanisms. First, phys directly interact with PIFs and induce rapid phosphorylation, polyubiquitylation, and 26S proteasome-mediated degradation of PIFs (Castillon et al., 2007; Henriques et al., 2009; Leivar and Quail, 2011; Xu et al., 2015). Second, phyB sequesters PIF3 in response to light by direct interaction (Park et al., 2012). Moreover, phys also inhibit the activity of CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) by directly interacting with SUPPRESSOR OF PHYA-105 (SPA1) and modulating the COP1-SPA complex in response to light, thereby increasing the level of positive regulators (e.g., HY5, HFR1, LAF1, and others) (Lu et al., 2015; Sheerin et al., 2015; Xu et al., 2015). This dual level of regulation under light largely reduces the repressive functions of PIFs and COP1 to promote photomorphogenesis.
The light-induced degradation of PIFs is mediated through the ubiquitin/26S proteasome pathway. Except PIF7 and PIF8, all other PIFs (PIF1 and PIF3-PIF6) are rapidly degraded in response to light signals with differential kinetics (Castillon et al., 2007; Leivar and Quail, 2011). Light signals induce rapid phosphorylation of PIFs (Leivar and Quail, 2011) and the light-induced phosphorylation is necessary for PIF3 degradation (Ni et al., 2013). A polyubiquitin-recognizing factor (HEMERA) is also necessary for degradation of PIF1 and PIF3 under deetiolated conditions possibly by coupling PIFs transcriptional activation activity with proteasomal degradation (Chen et al., 2010; Galvão et al., 2012; Qiu et al., 2015). PIF1 and PIF4 are phosphorylated by Arabidopsis Casein Kinase 2 and BRASSINOSTEROID INSENSITIVE2, respectively, and this phosphorylation affects the light-induced degradation of PIF1 and PIF4 (Bu et al., 2011b; Bernardo-García et al., 2014). Recently, both CUL3- and CUL4-based E3 ligases have been shown to mediate PIF degradation (Xu et al., 2015). Three LIGHT RESPONSE BRIC-A-BRACK/TRAMTRACK/BROAD (LRB1, 2, and 3) E3 ligases induce both PIF3 and phyB codegradation in response to light (Ni et al., 2014; Zhu and Huq, 2014). In addition, the CUL4-COP1-SPA complex induces rapid light-induced degradation of PIF1 (Zhu et al., 2015). However, PIFs are still degraded in these E3 ligase mutant backgrounds under prolonged light, suggesting additional E3 ligases are necessary for PIF degradation.
In addition to light-induced degradation and sequestration, PIF activity is also regulated by other factors (Leivar and Monte, 2014). For example, PIF4 interacts with BRASSINAZOLE-RESISTANT1, and this interaction promotes light and brassinosteroid-mediated plant growth and development (Oh et al., 2012). By contrast, DELLA proteins interact with PIFs and inhibit their DNA binding ability and subsequent target gene expression (de Lucas et al., 2008; Feng et al., 2008). LONG HYPOCOTYL IN FAR-RED1 (HFR1), PACLOBUTRAZOL-RESISTANT (PRE6/KIDARI), and PHY RAPIDLY REGULATED (PAR1 and PAR2) interact with PIF family members and inhibit their DNA binding ability and target gene expression (Fairchild et al., 2000; Hyun and Lee, 2006; Roig-Villanova et al., 2006, 2007; Hornitschek et al., 2009; Bu et al., 2011a; Bai et al., 2012; Hao et al., 2012; Oh et al., 2012; Shi et al., 2013). PIL1 also interacts with PIFs and HFR1 and inhibits PIF function to promote photomorphogenesis in an additive manner with HFR1 (Luo et al., 2014). ELF3 also interacts with PIF4 and regulates plant growth independent of the evening complex (Nieto et al., 2015). Moreover, PIF4 and PIF5 interact with the blue light photoreceptors CRY1 and CRY2 to regulate shade avoidance under low blue light or hypocotyl elongation under high temperature (Ma et al., 2016; Pedmale et al., 2016). In addition, the bZIP transcription factor HY5 interacts with PIFs and functions antagonistically to regulate photomorphogenesis (Chen et al., 2013; Toledo-Ortiz et al., 2014; Xu et al., 2014). Strikingly, PIF1 functions as a cofactor of the COP1 E3 Ubiquitin ligase and promotes substrate recruitment and auto- and transubiquitylation of HY5, thereby synergistically represses photomorphogenesis in the dark (Xu et al., 2014). In contrast, DET1 interacts with PIFs and stabilize them in the dark (Dong et al., 2014; Shi et al., 2015). Overall, PIFs interact with multiple unrelated proteins to modulate various signaling pathways.
Although the Arabidopsis genome has >160 bHLH transcription factors and >27 are predicted to be HLH factors (Bailey et al., 2003; Toledo-Ortiz et al., 2003), only four (HFR1, PRE6/KIDARI, and PAR1/2) have been shown to interact with PIFs and regulate PIF function (Hyun and Lee, 2006; Roig-Villanova et al., 2006; Hornitschek et al., 2009; Hao et al., 2012; Shi et al., 2013; Zhou et al., 2014). In an effort to identify additional factors interacting with PIF1, we performed a yeast two-hybrid screening (Bu et al., 2011a). We identified a new family of HLH proteins named HECATE (HEC) in addition to other PIF family members and HFR1 from this screen. In this study, we show that PIFs and HECs constitute a negative feedback loop to fine-tune photomorphogenesis.
RESULTS
PIF1 Interacts with HECATE Proteins
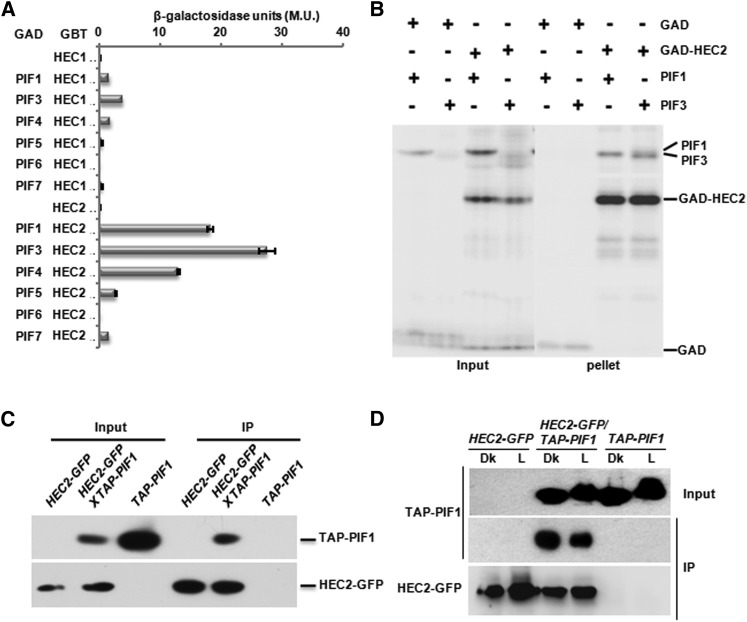
To identify and characterize potential regulators of PIF1, we performed a yeast two-hybrid screening using a truncated form of PIF1, as previously described (Bu et al., 2011a). We identified two members of a small family of bHLH proteins named HECATE (HEC1 and HEC2) from this screen. HEC proteins have previously been shown to regulate the female reproductive tract as well as shoot apical meristem development in Arabidopsis (Gremski et al., 2007; Crawford and Yanofsky, 2011; Schuster et al., 2014). The bHLH domains of HEC1 and HEC2 displayed high similarity to the bHLH domains of PIFs and HFR1, bHLH proteins previously shown to function in light signaling pathway (Supplemental Figure 1). To examine whether HEC1 and HEC2 interact with all the PIFs, we performed yeast two-hybrid interaction assays. Figure 1 shows that HEC1 weakly interacted with PIF1, PIF3, and PIF4, while HEC2 interacted PIF1, PIF3, PIF4, PIF5, and PIF7 as measured by a yeast two-hybrid liquid β-galactosidase assay (Figure 1A).
Figure 1.
HEC Proteins Interact with PIFs.
(A) Quantitative yeast two-hybrid assay shows the interactions among the HEC1/HEC2 and members of the PIF family. LacZ assays were performed in triplicate and the data represent mean ± se. β-Galactosidase units are Miller units (M.U.). GAD, GAL4 activation domain; GBD, GAL4 DNA binding domain.
(B) HEC2 heterodimerizes with PIF1 and PIF3 in vitro. The gel photograph shows the input and the pellet fractions. Full-length HEC2 ORF fused to GAD was used for this coimmunoprecipitation assay as described (Huq and Quail, 2002; Toledo-Ortiz et al., 2003; Shen et al., 2005). All proteins were synthesized as 35S-Met-labeled products in TnT reactions.
(C) PIF1 interacts with HEC2 in in vivo coimmunoprecipitation assays. The input and pellet fractions are indicated. Total protein was extracted from 4-d-old dark-grown seedlings. Coimmunoprecipitations were performed using the anti-GFP antibody, and the immunoprecipitated samples were probed with both anti-myc and anti-GFP antibodies.
(D) HEC2 preferentially interacts with the unphosphorylated form of PIF1 in in vivo coimmunoprecipitation assays. The input and pellet fractions are indicated. Total protein was extracted from 4-d-old dark-grown seedlings or dark-grown seedlings exposed R light. Coimmunoprecipitations were performed using the anti-GFP antibody, and the immunoprecipitated samples were probed with both anti-myc and anti-GFP antibodies.
To independently verify the physical interaction between HEC and PIF proteins, we cloned HEC2 into an in vitro expression vector (pET17b) as a fusion protein with the GAL4 activation domain (GAD). We coexpressed either GAD alone with PIF1 and PIF3 or GAD-HEC2 with PIF1 and PIF3 in an in vitro transcription/translation system as described (Huq and Quail, 2002; Toledo-Ortiz et al., 2003; Huq et al., 2004), and coimmunoprecipitated using antibody against GAD. Figure 1B shows that GAD-HEC2 efficiently coimmunoprecipitated both PIF1 and PIF3, which is consistent with the yeast two-hybrid assay results shown in Figure 1A.
To demonstrate that HEC and PIF proteins interact in vivo, we made transgenic plants expressing HEC2-GFP fusion protein expressed from a constitutively active 35S promoter. We crossed HEC2-GFP line with TAP-PIF1 expressed from the endogenous PIF1 promoter (Bu et al., 2011b) and produced homozygous lines expressing both fusion proteins. These transgenic lines were used to perform in vivo coimmunoprecipitation assays using α-GFP antibody. The results show that HEC2-GFP efficiently coimmunoprecipitated TAP-PIF1 from extracts of plants grown in the dark (Figure 1C). Because PIFs are phosphorylated in response to light, HEC2 might interact with phosphorylated or unphosphorylated PIF1, or with both forms. To dissect these possibilities, we performed coimmunoprecipitation assays using plants grown in the dark and dark-grown seedlings exposed to R light. Results show that HEC2 preferentially interacted with the unphosphorylated form of PIF1 (Figure 1D). Taken together, these data suggest that HEC1 and HEC2 interact with PIFs and might function in light signaling pathways by regulating the function of PIF1, PIF3, and possibly other PIFs.
HEC Proteins Positively Regulate Seed Germination
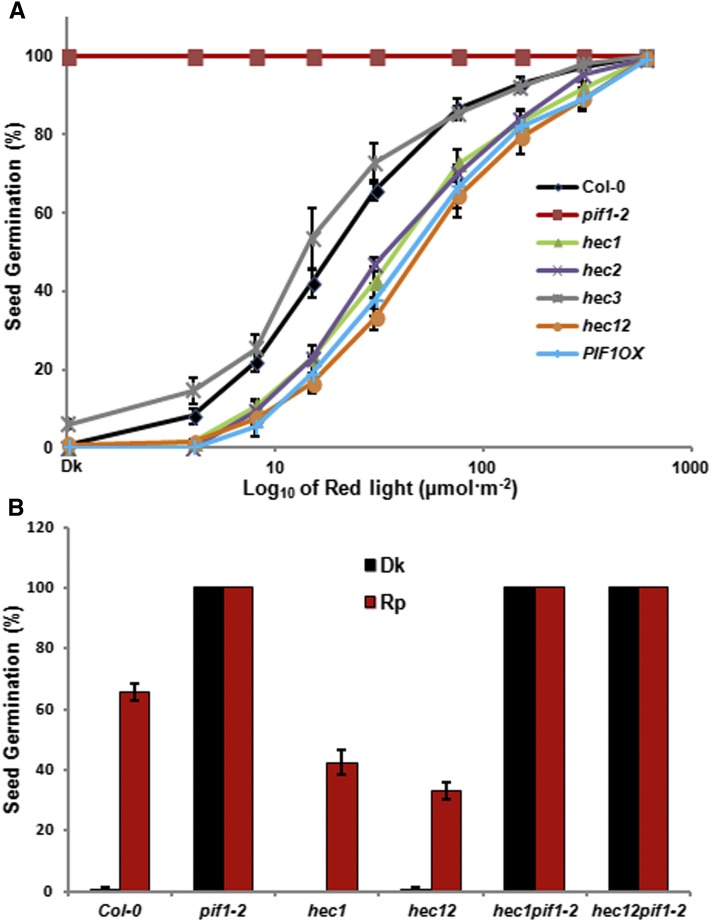
To investigate the biological functions of HECs in light signaling pathways, we obtained hec1, hec2, and hec3 single mutants and hec1 hec2 double mutants, which were previously described (Gremski et al., 2007; Crawford and Yanofsky, 2011). Because PIF1 has an exclusive role in regulating seed germination, we used hec single and double mutants to investigate the seed germination phenotypes in response to R light (Oh et al., 2004, 2006, 2009; Shen et al., 2005). While the germination of hec3 single mutants was similar to the wild-type control (Col-0), hec1, hec2, and hec1 hec2 double mutants displayed much reduced levels of seed germination compared with the wild type under increasing amount of R light similar to the TAP-PIF1 overexpression (PIF1OX) line (Figure 2A). These seeds eventually germinated under 600 µmol R light (Figure 2A), suggesting that they are not permanently dormant. To assess whether HEC1 and HEC2 promote seed germination through inhibition of PIF1 function, we created hec1 pif1 and hec1 hec2 pif1 mutant combinations and examined the seed germination phenotype. Results show that the reduced seed germination of hec1 and hec1 hec2 mutants in response to light is eliminated in the pif1 background. The double and triple mutant seeds germinated similar to the pif1 single mutant (Figure 2B), suggesting that pif1 is epistatic to hec in regulating seed germination.
Figure 2.
HEC Proteins Promote Seed Germination in Arabidopsis in a PIF1-Dependent Manner.
(A) hec1, hec2, and hec12 showed reduced seed germination in response to light similar to PIF1 overexpression lines and opposite to pif1 mutant. Seed germination assays were performed as described (Oh et al., 2004; Shen et al., 2007). Seeds of all the genotypes were surface sterilized within 1 h of imbibition, exposed to 5 min of FR light (34 μmol m−2 s−1) before being exposed to different amount of R light indicated. After light exposure, each plate was wrapped in aluminum foil and kept at 21°C for 5 d in the dark. All the plates were scored for radical emergence, and percentage of seeds germinated was plotted against the amount of R light exposed.
(B) Reduced seed germination of hec1 and hec12 is eliminated in the pif1 background. The seed germination assays were performed as described in (A). After FR pulse (FRp), the seeds were either kept in dark or exposed to R pulse (Rp) (30 μmol m−2 s−1) followed by dark incubation for 5 d.
HEC Proteins Inhibit Hypocotyl Elongation in the Dark
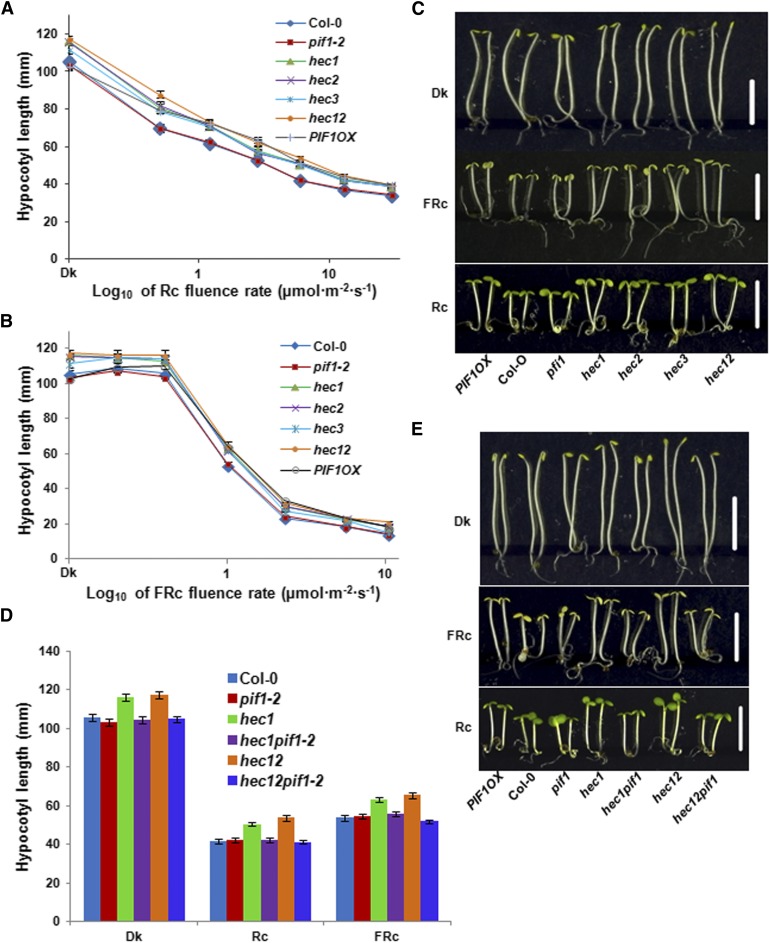
Since hec mutants showed hyposensitive seed germination phenotypes (Figure 2), we investigated the seedling deetiolation phenotypes of these lines in response to R and FR light conditions. Results showed that the hypocotyl lengths for the hec1, hec2, hec3, and hec1 hec2 were longer than that of the wild type under both R and FR light conditions similar to TAP-PIF1 overexpression line (Figures 3A to 3C). The cotyledon areas were largely similar to the wild type (Figure 3C). However, the hypocotyl lengths of hec1, hec2, hec3, and hec1 hec2 seedlings were already longer than the wild type in the dark (Figures 3A to 3C), suggesting that HECs inhibit hypocotyl elongation in the dark. We also measured hypocotyl lengths for seedlings grown in the dark over time. The difference in hypocotyl lengths between the wild type and the hec1 and hec1 hec2 mutants appeared after 2 d of growth (Supplemental Figure 3). To investigate if HECs positively regulate seedling deetiolation through inhibiting PIF1 function, we measured the hypocotyl lengths of hec1 pif1 and hec1 hec2 pif1 mutant combinations grown under dark, R, and FR light conditions. Similar to the data from seed germination assays, the elongated hypocotyl phenotype in hec1 and hec1 hec2 was eliminated in the hec1 pif1 and hec1 hec2 pif1 mutant combinations (Figures 3D and 3E). The hec1 pif1 and hec1 hec2 pif1 mutant seedlings grew similar to the wild-type seedlings compared with the hec1 and hec1 hec2 mutants. These data suggest that pif1 is epistatic to hec mutants in regulating seedling deetiolation.
Figure 3.
HEC Proteins Inhibit Hypocotyl Elongation in the Dark in a PIF1-Dependent Manner.
(A) and (B) Fluence-rate response curves of mean hypocotyl lengths of wild-type Col-0, pif1-2, hec1, hec2, hec3, hec12, and PIF1 overexpression line grown for 4 d under either continuous R (Rc) (A), continuous FR (FRc) (B), or dark.
(C) Photographs of wild-type Col-0, pif1-2, hec1, hec2, hec3, hec12, and PIF1 overexpression line grown under dark (Dk), R (Rc; 7.8 μmol m−2 s−1) and FR light (FRc; 0.5 μmol m−2 s−1) conditions for 4 d. Bars = 5 mm.
(D) and (E) pif1 is epistatic to hec1 and hec12 in regulating seedling deetiolation.
(D) Bar graph shows the hypocotyl length of wild-type Col-0, pif1-2, hec1, hec1 hec2, hec1 pif1, and hec1 hec2 pif1 mutant combinations grown under dark (Dk), R (Rc; 6 μmol m−2 s−1), and FR light (FRc; 1 μmol m−2 s−1) conditions for 4 d. For each genetic background under each condition, at least 40 seedlings were measured using ImageJ. Error bars = se.
(E) Photographs of wild type Col-0, pif1-2, hec1, hec1 hec2, hec1 pif1 and hec1 hec2 pif1 mutant combinations grown under dark, R and FR light. The growth and light conditions are similar to (D). White bar = 5 mm.
HEC Proteins Positively Regulate Chlorophyll and Carotenoid Biosynthesis
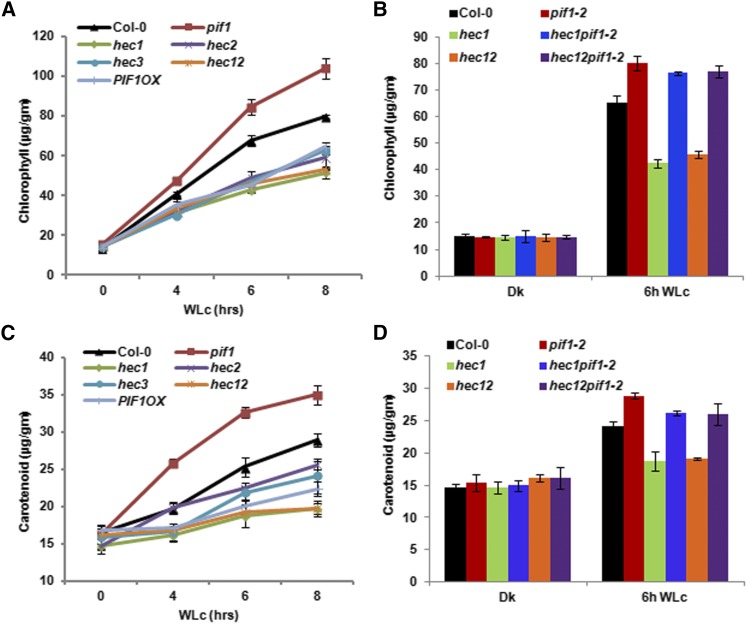
Chlorophyll and carotenoid biosynthesis is coordinately regulated in Arabidopsis in response to light, and PIFs play a critical role in directly regulating both of these pathways (Huq et al., 2004; Moon et al., 2008; Stephenson et al., 2009; Toledo-Ortíz et al., 2010). To assess the roles of HECs in regulating these pathways, we measured chlorophyll and carotenoid levels in hec1, hec2, hec3, and hec1 hec2 mutants along with controls. Seedlings were grown for 2.5 d in the dark and then exposed to white light over time before harvesting for pigment measurement. Results show that all three hec single and hec1 hec2 double mutants display reduced level of both chlorophyll and carotenoid in response to light similar to the PIF1OX line (Figures 4A and 4B). By contrast, pif1 mutants displayed much higher levels of chlorophyll and carotenoid levels compared with the wild-type seedlings, as previously shown (Huq et al., 2004; Moon et al., 2008; Stephenson et al., 2009; Toledo-Ortíz et al., 2010). We also examined chlorophyll and carotenoid content for the hec1 pif1 and hec1 hec2 pif1 along with controls to examine genetic relationship for this phenotype. Results show that the reduced pigment content of the hec1 and hec1 hec2 double mutants is largely eliminated in the hec1 pif1 and hec1 hec2 pif1 (Figures 4B and 4D), suggesting that pif1 is epistatic to hec1 and hec1 hec2 in these phenotypes. These data suggest that HEC and PIF proteins function antagonistically to regulate the pigment biosynthetic pathways. Overall, these data also suggest that HECs function as positive regulators of phy signaling pathways. This is in contrast to the PIF functions, where a majority of the PIFs function as negative regulators of phy signaling pathways.
Figure 4.
HEC Proteins Promote Chlorophyll and Carotenoid Biosynthesis in Arabidopsis.
(A) and (C) hec1, hec2, hec3, and hec12 were grown with Col-0, pif1-2, and PIF1OX for 2.5 d in the dark and then transferred to 75 μmol m−2 s−1 of white light for various times as indicated. Total chlorophyll (A) and carotenoid (C) contents were determined as described by Huq et al. (2004) or Toledo-Ortíz et al. (2010), respectively.
(B) and (D) Bar graph showing total chlorophyll (B) and carotenoid (D) contents measured from Col-0, pif1-2, hec1, hec1 pif1-2, hec12, and hec12 pif1-2 seedlings. The growth conditions and assay procedure are similar to (A) and (C). All assays were performed in triplicate, and the data represent mean ± se.
HEC Proteins Regulate the Indirect and Direct Target Genes of PIFs
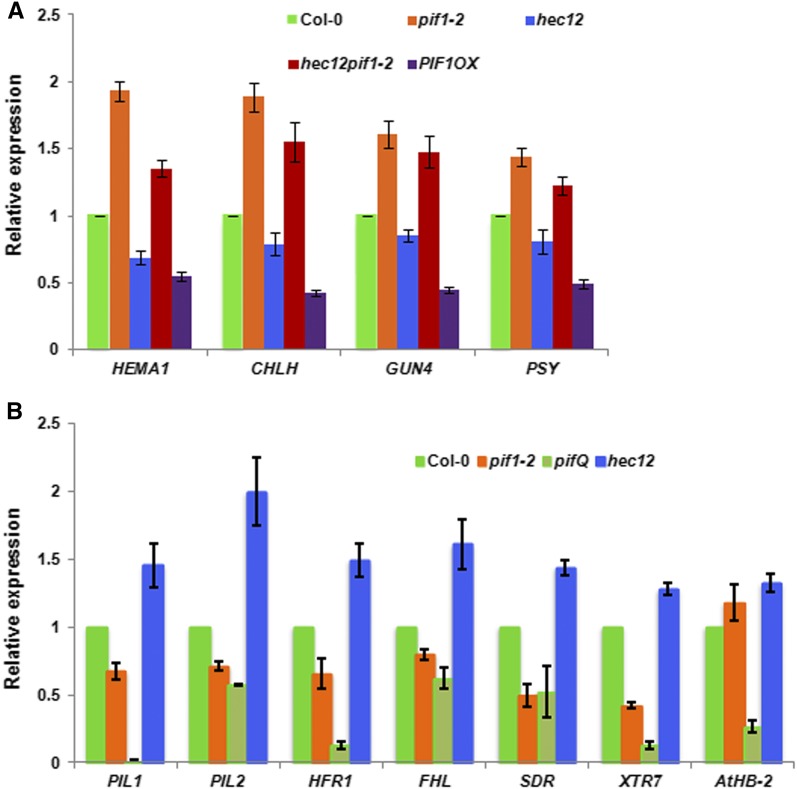
To investigate the molecular phenotypes of hec mutants, we selected a group of genes in the chlorophyll and carotenoid biosynthetic pathways that are also regulated by PIFs indirectly and/or directly. As previously shown, the expression of these genes is upregulated in the pif1 mutant compared with the wild type (Figure 5A). Conversely, the expression of these genes is downregulated in the hec1 hec2 double mutant similar to the PIF1OX lines. Moreover, the reduced expression of these genes in the hec1 hec2 double mutant is largely restored to above the wild-type level in the hec1 hec2 pif1 background, suggesting pif1 is epistatic to hec1 hec2 in gene expression phenotype as well. The incomplete rescue of gene expression might be due to the presence of other PIFs that regulate the expression of these genes.
Figure 5.
HEC Proteins Regulate the Direct and Indirect Target Genes of PIFs.
(A) HEC1 and HEC2 promote expression of biosynthetic genes in the chlorophyll and carotenoid pathways. RNA was extracted from 3-d-old dark-grown seedlings of wild-type Col-0, pif1-2, hec12, hec12 pif1-2, PIF1OX, and reverse transcribed to cDNA. RT-qPCR data showing relative expression of selected chlorophyll and carotenoid biosynthetic pathway genes in the wild type and various mutants (n = 3 biological repeats, each with three technical replicates, ±se).
(B) HEC1 and HEC2 oppositely control the expression of PIF1 direct target genes. RNA was extracted from 3-d-old dark-grown seedlings of wild-type Col-0, pif1-2, pifQ, and hec1 hec2 and reverse transcribed to cDNA. RT-qPCR data show relative expression of PIF1 direct target genes in the wild type and various mutants (n = 3 biological repeats, each with three technical replicates, ±se).
To examine whether HECs function antagonistically to regulate direct target genes of PIFs, we performed RT-qPCR assays for selected PIF target genes at the seedling stage. RNA was isolated from 3-d-old dark-grown wild-type Col-0, pif1, pifQ, and hec1 hec2 seedlings. RT-qPCR was performed for PIL1, PIL2, HFR1, FHL, SDR, XRT7, and AtHB-2, which are the direct targets of PIFs as previously shown (Zhang et al., 2013; Pfeiffer et al., 2014). Results show that the expression of all those PIF target genes is stimulated in the hec1 hec2 mutant compared with the wild type in the dark (Figure 5B). By contrast, the expression of the same genes is modestly reduced in pif1 and more strongly reduced in pifQ mutant compared with the wild type. Overall, these data suggest that HEC1/HEC2 and PIFs function antagonistically to regulate the expression of both indirect and direct target genes in the dark.
HEC2 Blocks the DNA Binding and Transcriptional Activation Activity of PIF1
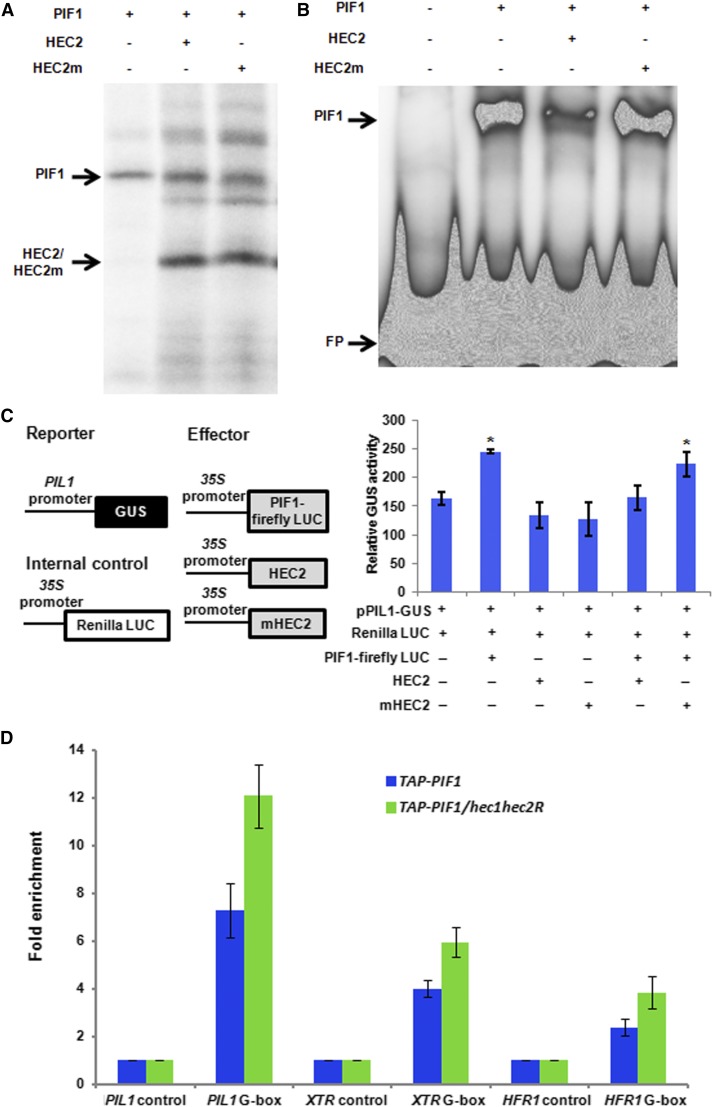
Previously, we have shown that PIF1 binds to a G-box motif present in PORC and PSY promoters using a gel-shift assay (Moon et al., 2008; Toledo-Ortíz et al., 2010). To determine if HEC2 can block the DNA binding ability of PIF1, we coexpressed PIF1 and HEC2 using a coupled transcription-translation system (Figure 6A) and performed a gel-shift assay as described (Huq and Quail, 2002; Huq et al., 2004; Moon et al., 2008). Results show that HEC2 prevents the binding of PIF1 to the PORC G-box fragment (Figure 6B). To examine the specificity of this inhibition, we mutated two residues within the HLH domain that has been shown to be necessary for dimerization (Hornitschek et al., 2009). Yeast two-hybrid assays showed that the mutant version of HEC2 has strongly reduced affinity for PIF1 and PIF3 compared with wild-type HEC2 (Supplemental Figure 3). We coexpressed the mutant form of HEC2 (HEC2m) and used it as a control in these binding assays. Results showed that the mutant form of HEC2 failed to reduce the DNA binding ability of PIF1 compared with the wild-type HEC2 (Figure 6B). These results suggest that HEC2 heterodimerizes with PIF1 and prevents PIF1 from binding to its target promoters.
Figure 6.
HEC2 Inhibits the DNA Binding and Transcriptional Activation Activity of PIF1.
(A) A gel photograph shows the amount of protein used for the EMSAs. PIF1 and the wild type or mutant form of HEC2 clones were coexpressed in TnT system with 35S-Met labeling.
(B) EMSA showing PIF1 binding to PORC G-box is inhibited by HEC2, but not by the mutant version of HEC2. The same amount of TnT mix shown in (A) without the 35S-Met labeling was used in EMSA. A total of 30,000 cpm of labeled probe was used in each lane. EMSA conditions are as described (Moon et al., 2008).
(C) HEC2 inhibits the transcriptional activation activity of PIF1. Left: Schematic illustration of reporter, effectors, and internal control constructs used in transient promoter activation assay. Right: 2.5-d-old pif1-2 seedlings were transiently transformed with the different combinations of constructs indicated below. Relative expression of GUS activity was measured. The data were normalized by protein concentration and Renilla luciferase activity. The GUS activities of the control group, which was transiently expressed pPIL1:GUS and 35S:Renilla Luciferase, are set to 1 (n = 3 trials, each with four technical replicates, ±se, P < 0.01).
(D) HEC2 inhibits the promoter occupancy of PIF1 in vivo. ChIP assays show TAP-PIF1 binding to the G-box motif of PIF1 target promoters. The ChIP assay was performed on 3-d-old dark-grown seedlings expressing the TAP-PIF1 fusion protein either in pif1-2 or pif1-2 hec1 hec2RNAi line. Anti-MYC antibodies were used to immunoprecipitate TAP-PIF1 and associated DNA fragment. DNA was amplified using primers specific to the G-box fragments or control regions in PIL1, XTR, and HFR1 promoters.
As a transcription factor, PIF1 binds to the promoter regions of target genes and activates their expression. To determine whether HEC1 and HEC2 inhibit the transcriptional activation activity of PIF1, we performed in vivo transient transcription assays as described previously (Moon et al., 2008; Hornitschek et al., 2009; Shi et al., 2013). A schematic demonstration of the plasmids used in the transient bombardment assay is shown in Figure 6C (left panel). pPIL1:GUS and 35S:PIF1-Firefly LUC were cobombarded with either the wild-type version of HEC2 or the mutant version of HEC2 driven by the constitutively active 35S promoter into 3-d-old dark-grown Arabidopsis seedlings. Renilla LUC driven by 35S promoter was used as a control. The results showed that PIF1 activates pPIL1 driving GUS gene expression (Figure 6C, right). The addition of the wild-type version of HEC2 in the cobombardment reduced the level of GUS activity, suggesting that HEC2 blocks PIF1 transcription activation from the PIL1 promoter. By contrast, the mutant version of HEC2 did not affect the pPIL1 driving GUS expression (Figure 6C, right). Moreover, the wild-type and mutant HEC2 showed GUS activity similar to the reporter construct only in this assay. These data suggest that HEC2 inhibits PIF1-mediated activation of pPIL1:GUS expression by direct heterodimerization.
We also tested if HEC1 and HEC2 block the promoter occupancy of PIF1 in vivo using chromatin immunoprecipitation (ChIP) assays. To eliminate HEC function as much as possible, we previously produced hec2 RNAi plants and crossed hec1 with one of the hec2 RNAi lines (hec2 RNAi 41-7). We used quantitative and regular RT-PCR to examine HEC2 mRNA levels in two independent hec2 RNAi lines (Supplemental Figure 4). Then, TAP-PIF1 (described previously) was crossed with hec1 hec2R (Bu et al., 2011b). We performed ChIP assays to examine PIF1 binding to the cis-acting regulatory elements, particularly G-box regions, found in the promoters of the PIF target genes. The results showed that TAP-PIF1 has increased binding to the G-box region of PIL1, XTR, and HFR1 promoters in the hec1 hec2R background compared with the wild-type background (Figure 6D). These data indicate that HEC1 and HEC2 block PIF1 binding to its downstream target gene promoters in vivo.
PIF1, HEC1, and HEC2 Are Coexpressed, and the Expression of HEC1 and HEC2 Is Reduced in Response to Light
Because PIF1 and HEC proteins heterodimerize, coexpression of PIF1 and HEC genes is expected for such heterodimers to be functionally relevant in vivo. We analyzed the spatial regulation of expression of PIF1 and HEC genes using the eFP browser (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Strikingly, PIF1 and HEC1 are coexpressed in the seeds imbibed for 24 h (Supplemental Figure 5). Because the HEC2 probe is absent from the microarray chips, data for HEC2 were not available. To compare tissue-specific or developmental expression patterns of PIF1 and HEC genes, we used a promoter:reporter fusion strategy. We cloned ∼2-kb promoter region upstream of the ATG start codon of PIF1, HEC1, and HEC2 genes into a pENTRY vector and then recombined with a Gateway-compatible destination vector containing the GUS gene as a transcriptional fusion (Karimi et al., 2005). These constructs have been transformed into the wild-type Arabidopsis, and single-insert homozygous transgenic plants have been selected. Histochemical GUS assays have been performed using X-gluc as a substrate, as described (Shen et al., 2007). Results showed that all three genes are coexpressed at the seedling stage in a tissue-specific manner (Supplemental Figure 6A). Moreover, these genes are coexpressed in seedlings grown in the dark or light (R, FR, and white light) conditions. The striking coexpression of PIF1, HEC1, and HEC2 in the imbibed seeds (Supplemental Figure 5) as well as seedlings grown under different conditions (Supplemental Figure 6A) suggests that these genes might function together in a tissue- and developmental stage-specific manner.
To examine the kinetics of light regulation of HEC1 and HEC2 expression, we performed quantitative RT-PCR under various light regimens for different time periods. Four-day-old dark-grown seedlings were exposed to either continuous R or FR light for 1, 6, and 24 h or kept in darkness. Total RNA was isolated from these samples for RT-PCR experiments. Results showed that both HEC1 and HEC2 mRNA levels were downregulated under R and FR light conditions. PIF1 transcript level was modestly affected under FR light conditions (Supplemental Figure 6B). Thus, HEC1 and HEC2 are light-repressed genes.
HEC2 Is Localized in the Nucleus
To investigate the subcellular localization of HEC1 and HEC2 proteins, we transformed wild-type Arabidopsis with 35S:HEC1-YFP and 35S:HEC2-GFP constructs. Homozygous transgenic lines for HEC1-YFP were lethal. However, single-insert homozygous HEC2-GFP lines were viable. We investigated the subcellular localization in stable transgenic background using fluorescence microscopy. The results showed that HEC2-GFP is localized in the nucleus (Supplemental Figure 6C). This is also consistent with the predicted subcellular localization of HEC2 using PRORT (http://psort.ims.u-tokyo.ac.jp/form.html; version 6.4).
PIFs Directly Activate HEC1 and HEC2 Expression in the Dark
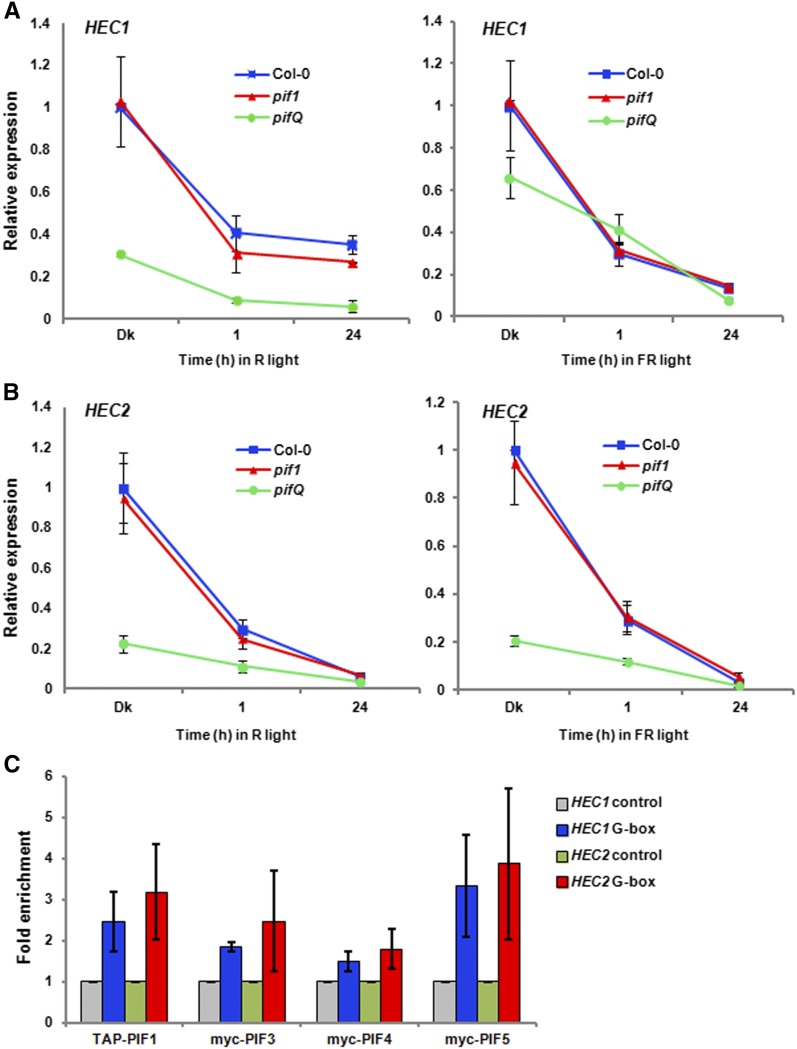
Since both HEC1 and HEC2 have the highest expression in the dark, and PIFs are more abundant in the dark, we examined if PIFs can regulate HEC1 and HEC2 gene expression. The expression of HEC1 and HEC2 is reduced in the pifQ mutant compared with the wild-type Col-0 in the dark, although the downregulation is not significant in the pif1 single mutant (Figures 7A and 7B). The promoter regions of both HEC1 and HEC2 have G-boxes, a putative binding site of PIFs. To examine direct binding, we performed ChIP assays using tagged version of PIFs. The results showed that all four PIFs (PIF1, PIF3, PIF4, and PIF5) bind to the G-box region of both HEC1 and HEC2 promoters in the dark (Figure 7C). Overall, the data suggest that PIFs activate the expression of HEC1 and HEC2 in the dark, and light-induced degradation of PIFs might result in reduced expression of HEC1 and HEC2 under R and FR light conditions.
Figure 7.
PIFs Directly Regulate HEC1 and HEC2 Gene Expression in the Dark.
(A) The expression of HEC1 and HEC2 is downregulated in pifQ mutant compared with wild-type Col-0 in the dark. Four-day-old dark-grown wild-type Col-0, pif1-2, and pifQ seedlings were either kept in darkness or exposed to R (7.8 μmol m−2 s−1) or FR light (1 μmol m−2 s−1) for the indicated times. RNA was extracted and reverse transcribed to cDNA. RT-qPCR was performed using HEC1 and HEC2 gene-specific primers (n = 3 biological repeats, each with three technical replicates, ±se).
(B) In vivo ChIP assay shows PIF1, PIF3, PIF4, and PIF5 binding to the G-box motif of HEC1 and HEC2 promoters. The ChIP assay was performed on 3-d-old dark-grown seedlings expressing the TAP-PIF1, myc-PIF3, myc-PIF4, or myc-PIF5 fusion proteins. The proteins and associated DNA fragments were immunoprecipitated using anti-MYC antibody. qPCR was performed to amplify either the G-box region or control region on HEC1/2 promoter. For individual genes in all the ChIPs, the control region qPCR data was set to 1 (n = 3 biological repeats, each with three technical replicates, ±se).
HEC2 Is Partially Degraded in the Dark and Stabilized under Light
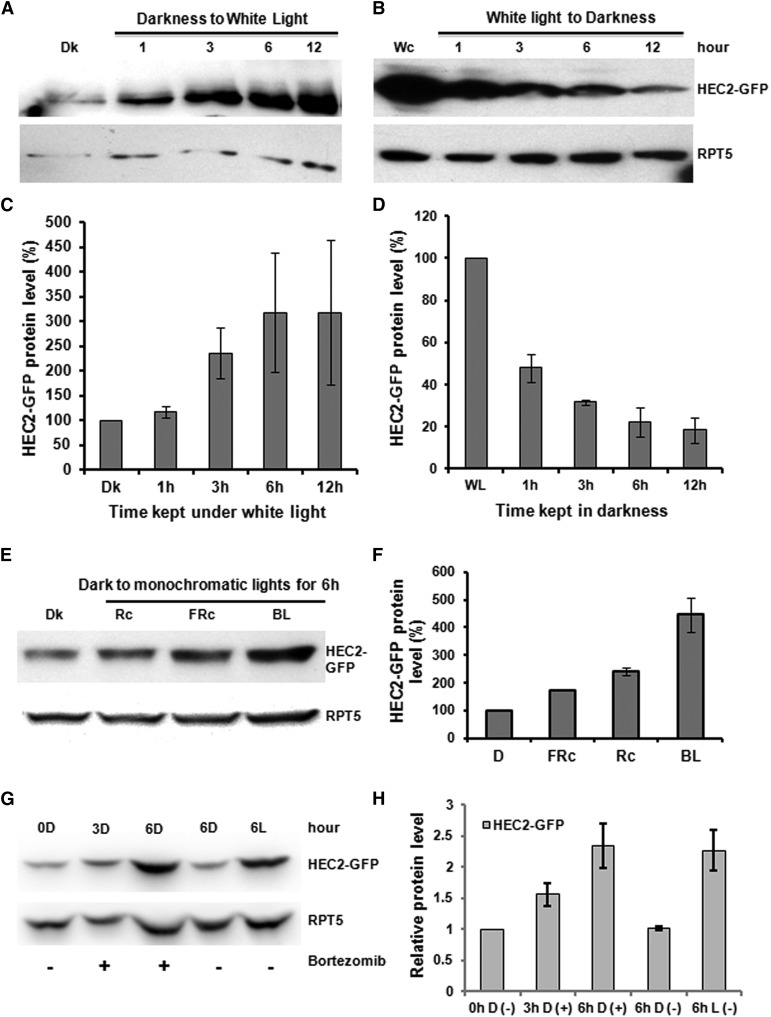
Posttranslational regulation of oppositely acting transcription factors has been shown to be central in light signaling pathways (Huq, 2006). For example, HY5, LAF1, and HFR1 (positive regulators) are degraded in the dark to repress photomorphogenesis, while PIFs (negative regulators) are degraded in light to promote photomorphogenesis (Huq, 2006; Henriques et al., 2009). To investigate the effect of light on HEC protein levels, we used antibody against GFP to examine the HEC2-GFP protein levels in the dark and light. We grew seedlings for 4 d in the dark or continuous white light. Then, the dark-grown seedlings were exposed to white light over time, and the light-grown seedlings were exposed to dark over time. Samples were collected at different times as indicated for immunoblot using anti-GFP antibody. Results show that HEC2-GFP is stabilized in response to prolonged white light conditions (Figures 8A and 8C). Conversely, HEC2-GFP is degraded in the dark over time (Figures 8B and 8D). We also examined the effect of monochromatic lights on HEC2 level by growing seedlings under continuous R, FR, and blue light conditions. Results show that HEC2-GFP is stabilized under all three monochromatic lights with strongest effect under blue light conditions (Figures 8E and 8F). To examine if the degradation of HEC2-GFP in darkness is mediated by the 26S-proteasomal pathway, we treated dark-grown seedlings with and without Bortezomib, a 26S proteasome inhibitor, for 3 and 6 h before sample collection. Seedlings grown under white light for 6 h were used as a control. Results show that the HEC2-GFP protein accumulated in the dark with the treatment of Bortezomib to a similar level as the white light-treated sample (Figures 8G and 8H). These data suggest that HEC2 is degraded through the 26S-proteasome pathway in the dark and is stabilized in response to light exposure.
Figure 8.
HEC2 Is Partially Degraded in the Dark, and Light Stabilizes HEC2-GFP Posttranslationally.
(A) HEC2-GFP protein is accumulated upon white light exposure. Four-day-old dark-grown seedlings of HEC2-GFP transgenic seedlings were either kept in darkness or exposed to 75 μmol m−2 s−1 white light for the time indicated before samples harvested for protein extraction. Around 30 μg total proteins per sample were separated on 8% polyacrylamide gel and transferred onto PVDF membrane for immunoblot analysis. Anti-GFP and anti-RPT5 antibodies were used for detecting HEC2-GFP level or RPT5 level as a control.
(B) HEC2-GFP is degraded in darkness. Seven-day-old white light-grown seedlings of HEC2-GFP transgenic lines were kept under the same white light condition or in the dark for the time indicated before protein extraction. The immunoblot process was done as described in (A).
(C) and (D) Quantification of HEC2-GFP level in the conditions indicated in (A) and (B). Three biological repeats were performed. The band intensities were measured with ImageJ. The HEC2-GFP protein level in each sample has been normalized using the RPT5 level.
(E) HEC2-GFP protein accumulated upon exposure to all three monochromatic lights. Four-day-old dark-grown seedlings of HEC2-GFP transgenic lines were kept either in darkness or exposed to R light (Rc; 20 μmol m−2 s−1), FR light (FRc; 10 μmol m−2 s−1), or blue light (BL; 20 μmol m−2 s−1) for 6 h. The immunoblot process was done similar to that in (A).
(F) Quantification of HEC2-GFP level in the conditions indicated in (E). Three biological repeats were performed. The band intensities were measured with ImageJ. The HEC2-GFP protein level in each sample has been normalized using the RPT5 level.
(G) HEC2-GFP is stabilized in darkness by the 26S proteasome inhibitor Bortezomib. Four-day-old dark-grown seedlings of HEC2-GFP transgenic lines were treated with 40 μmol Bortezomib for either 3 or 6 h in the dark. Dark samples without treatment before and after 6 h were used as control to indicate the HEC2-GPF protein level in darkness. Four-day-old dark-grown seedlings plus 6 h white light treatment (75 μmol m−2 s−1) were used to present HEC2-GPF under light condition. The immunoblot was done as described in (A).
(H) Quantification of HEC2-GFP level in the conditions indicated in (G). Three biological repeats were performed. The band intensities were measured with ImageJ. The HEC2-GFP protein level in each sample has been normalized using the RPT5 level.
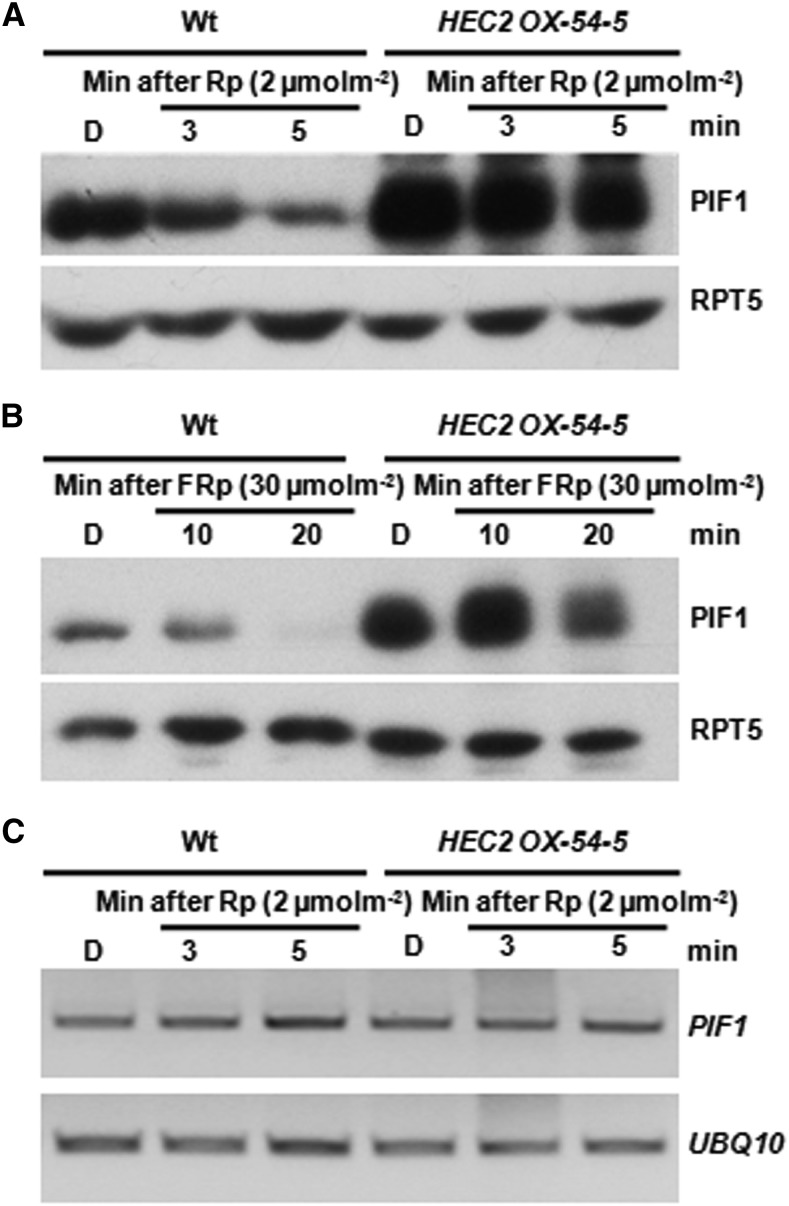
HEC2 Reduces the Light-Induced Degradation of PIF1
Because HEC proteins interact with PIF1, we examined whether this heterodimerization prevents the light-induced degradation of native PIF1. We examined light-induced degradation of native PIF1 in the wild type and hec1 hec2 double mutant over time. However, the degradation kinetics of native PIF1 under light is similar in both the wild type and hec1 hec2 double mutant. Reasoning that this might be due to extremely rapid degradation of native PIF1 (Shen et al., 2008; Zhu et al., 2015), we used the HEC2 overexpression plants where both quantitative and regular RT-PCR assays showed increased level of HEC2 mRNA in two independent HEC2 overexpression lines (Supplemental Figure 4). Results show that the light-induced degradation of PIF1 is greatly reduced in HEC2 overexpression line under both dark and light conditions (Figures 9A and 9B). This stabilization is at the posttranslational level, as the mRNA for PIF1 is not altered in the HEC2 overexpression lines under the identical conditions (Figure 9C). PIF1 is rapidly degraded in the wild-type background under the same conditions, suggesting that HEC2 stabilizes PIF1 at the protein level under both dark and light.
Figure 9.
HEC2 Reduces the Light-Induced Degradation of PIF1.
(A) and (B) Immunoblots showing PIF1 level in the HEC2 OX line and wild-type Col-0. Four-day-old dark-grown seedlings were either kept in darkness or exposed to R (A) or FR (B) pulse and incubated in the dark for the time indicated. Total protein was extracted, separated in 8% SAD-PAGE gel, and transferred to PVDF membranes. Native PIF1 antibody was used to detect PIF1 protein level and anti-RPT5 antibody was used as control. Amount of light pulse is shown on the figure.
(C) The expression of PIF1 was not altered upon light treatment. RNAs were extracted from samples under the same treatment as in (A) and reverse transcribed. RT-PCR was performed to detect PIF1 expression level. UBQ10 was used as control.
DISCUSSION
PIFs have been shown to function as cellular hubs for various signaling pathways, including their central roles in phy signaling (Castillon et al., 2007; Leivar and Quail, 2011; Leivar and Monte, 2014). Therefore, other proteins that regulate PIF function are of great importance in understanding how PIFs modulate these diverse pathways. In this study, we identify factors that regulate PIFs, with a focus on PIF1. The genetic, photobiological, and biochemical data presented here provide strong evidence that HEC1, HEC2, and possibly HEC3 function positively in phy signaling pathways, a behavior opposite to the roles of PIFs in this pathway. First, hec1, hec2, and hec1 hec2 double mutants showed reduced seed germination in response to R light, and pif1 is epistatic to hec1 and hec1 hec2 in this process (Figure 2). Second, hec1, hec2, hec3 single and hec1 hec2 double mutants have reduced levels of chlorophyll and carotenoid compared with wild-type seedlings (Figure 4). hec1, hec2, and hec3 single and hec1 hec2 double mutants displayed longer hypocotyls compared with the wild type under dark, R, and FR light (Figure 3). Although the hypocotyl length phenotype is not light dependent, the above hyposensitive phenotypes of the hallmark biological processes strongly suggest that the HEC proteins are positively acting components in phy signaling pathways.
Previously, positively acting components in phy signaling pathways have been described (Huq and Quail, 2005). Among those, HFR1, PRE6/KIDARI, PAR1, and PAR2 encode HLH proteins that function positively in light signaling pathways (Fairchild et al., 2000; Fankhauser and Chory, 2000; Duek and Fankhauser, 2003; Hyun and Lee, 2006; Roig-Villanova et al., 2006; Zhou et al., 2014). All four proteins play major roles in shade avoidance response (Roig‐Villanova et al., 2007; Lorrain et al., 2008). In addition, HFR1 has recently been shown to promote seed germination under R light by inhibiting PIF1 (Shi et al., 2013). Although HEC1/2 function positively in the same pathways, the expression pattern of these genes suggests a division of labor among HLH proteins. HEC1 and HEC2 are highly expressed in the dark and the HEC2 protein is present in the dark (Figures 7 and 8; Supplemental Figure 6). In contrast, HFR1, PAR1, and PAR2 are induced in response to light (Fairchild et al., 2000; Roig-Villanova et al., 2006). HFR1 is also degraded in the dark and stabilized in response to light (Duek et al., 2004; Yang et al., 2005). Therefore, HECs are expected to function more in the dark and dark-to-light transition, while HFR1, PAR1, and PAR2 are expected to function under prolonged light conditions. The phenotypes of these mutants are consistent with this prediction. For example, hec1, hec2, and hec1 hec2 mutants displayed hyposensitivity to light-induced seed germination and accumulation of chlorophyll and carotenoid levels but did not display light-dependent hypocotyl phenotype, a behavior observed under prolonged light conditions (Figures 2 and 4). The hec1, hec2, and hec1 hec2 mutants also displayed phenotypes in the PIF target gene expression and hypocotyl lengths in the dark (Figures 3 and 5). Thus, HECs represent new class of positively acting factors functioning early in the light signaling pathways.
HEC proteins function antagonistically to PIF1 and possibly other PIFs in light signaling pathways. HEC2 interacts with PIF1 in yeast two-hybrid assays, in vitro, and in vivo coimmunoprecipitation assays (Figure 1). The hallmark biological processes that are regulated by PIF1 (e.g., inhibition of seed germination and inhibition of chlorophyll and carotenoid biosynthesis) are oppositely regulated by HEC1/HEC2 proteins (Figures 2 to 4). PIF1 and HEC1/HEC2 genes are expressed in the same tissues at similar developmental stages, and PIF1 and HEC2 are localized in the same subcellular compartment, the nucleus. The DNA binding activity of PIF1 is inhibited by wild-type HEC2 in vitro, but not by a mutant HEC2 that does not interact with PIF1 (Figures 6A and 6B; Supplemental Figure 3). The transcriptional activation activity of PIF1 is inhibited by wild-type HEC2, but not by a mutant HEC2, in vivo in a transient assay in Arabidopsis seedlings (Figure 6C). The promoter occupancy of PIF1 is also increased in vivo in the hec1 hec2R lines compared with the wild-type control (Figure 6D). In addition, the expression of PIF direct target genes are oppositely regulated in hec1 hec2 double mutants compared with pif1 and pifQ seedlings (Figure 5). Taken together, these data strongly suggest that HEC1 and HEC2 directly bind to PIF1 and other PIFs and prevent PIF functions. The mechanisms by which HEC1 and HEC2 oppose PIFs is similar to a well-established relationship between bHLH-HLH proteins found in many eukaryotic systems including Arabidopsis (Benezra et al., 1990; Littlewood and Evans, 1998; Hornitschek et al., 2009; Hao et al., 2012; Shi et al., 2013; Zhou et al., 2014). For example, HFR1, PAR1, and PAR2 bind to PIF1, PIF4, and PIF5 and prevent their DNA binding and transcriptional regulation of their target genes (Hornitschek et al., 2009; Hao et al., 2012; Shi et al., 2013; Zhou et al., 2014). Therefore, we identified HEC proteins as components of the light signaling pathways that function through PIF1 and potentially other PIFs in Arabidopsis.
Previously, HLH proteins have been shown to regulate the activity of bHLH proteins by forming a dominant-negative heterodimer complex. However, this heterodimerization has not been shown to regulate the abundance of the partner bHLH proteins. Our data showing that HEC2 overexpression strongly reduces the light-induced degradation of PIF1 in vivo (Figure 9) suggest that HLH proteins not only inhibit the DNA binding activity, but also might regulate the stability of their interacting bHLH partners. Although the mechanism of this stabilization is still unknown, our data along with previous reports shed light on this process. Previously, PIF3 has been shown to interact with the Pfr form of phyB in a 1:1 stoichiometry, suggesting that PIF3 dimer interacts with the phyB dimer (Zhu et al., 2000). In addition, direct interaction with the Pfr forms of phyA and/or phyB is necessary for the light-induced phosphorylation and subsequent degradation of PIFs (Castillon et al., 2007; Henriques et al., 2009; Leivar and Quail, 2011; Xu et al., 2015). The fact that HEC2 preferentially interacts with the unphosphorylated form of PIF1 even under light (Figure 1D) suggests that HEC2 might inhibit the phosphorylation of PIF1 by forming a HEC2-PIF1 heterodimer and thereby reducing PIF1 degradation. Alternatively, the HEC2-PIF1 heterodimer may not be recognized by the E3 ligase for polyubiquitination and degradation. Further studies are necessary to understand the mechanism of HEC2-mediated stabilization of PIF1.
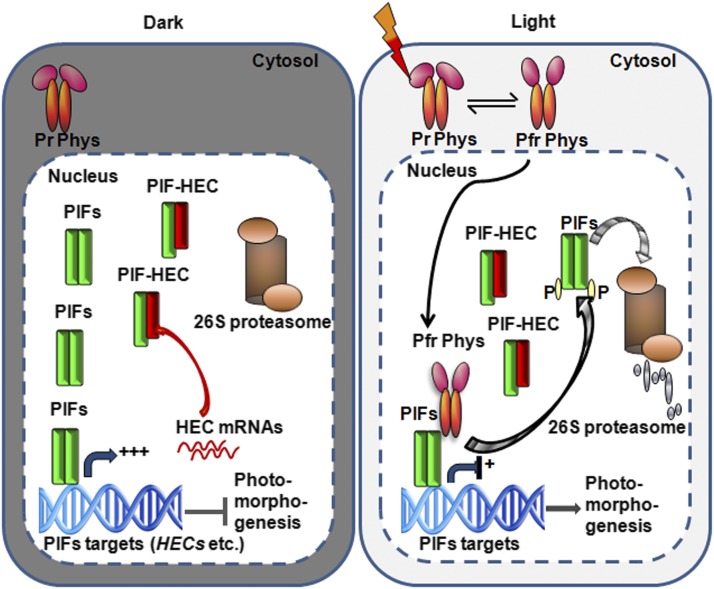
PIFs and HECs appear to form a negative feedback loop to fine-tune photomorphogenesis. On the one hand, all four PIFs directly bind to the G-box region present in the HEC1/HEC2 promoters and transcriptionally activate HEC1/HEC2 in the dark, and light signal reduces the expression of HEC1/HEC2 possibly due to the light-induced degradation of PIFs (Figures 7 and 10). On the other hand, PIF-induced accumulation of HECs prevents PIF function to reduce the expression of HEC and other PIF target genes in the dark (Figures 5 to 7 and 10). Therefore, PIFs essentially autoregulate their activities through HECs, and the PIF:HEC ratio determines the expression of PIF target genes. This stoichiometry appears to be regulated by light quality and quantity. Although HEC2 and possibly other HECs are degraded in the dark through the 26S proteasome pathway, HEC genes are highly expressed in the dark and light stabilizes HEC2 ∼3-4-fold posttranscriptionally (Figure 8). In contrast, PIFs are degraded in response to light through the 26S proteasome pathways (Leivar and Quail, 2011; Xu et al., 2015). Therefore, this dynamic PIF:HEC ratio fine-tunes photomorphogenesis both in the dark and in response to a constantly changing light environment (Figure 10).
Figure 10.
Simplified Model of the Negative Feedback Loop between PIFs and HECs Shows Fine-Tuning of Photomorphogenesis.
In the dark (left panel), phytochromes are present in the cytosol as Pr form, while PIFs are constitutively localized to the nucleus. PIFs activate their target genes, including HEC1 and HEC2 expression, in the dark. HECs in turn repress the DNA binding and transcriptional activation activity of PIFs reducing their own expression as well as other PIF target genes. Phytochromes perceive R light signal, undergo allosteric changes in conformation into the Pfr form, and migrate into the nucleus. Within the nucleus, phys interact with PIFs and induce rapid degradation of PIFs while stabilizing HEC2. PIFs repress photomorphogenesis, while HECs promote photomorphogenesis. However, the balance between PIF:HEC stoichiometry determines the level of photomorphogenesis in a dynamic environmental light condition.
In Arabidopsis, there are >162 bHLH proteins of which >27 are predicted to be non-DNA binding HLH proteins (Bailey et al., 2003; Toledo-Ortiz et al., 2003). Plants have more bHLH and HLH proteins compared with animals, and these proteins function in many signaling pathways in plants (Leivar and Monte, 2014). It is possible that these antagonistically acting pairs of proteins have coevolved in multiple signaling pathways in plants to fine-tune these pathways. Furthers studies are necessary to assess whether the bHLH and HLH proteins have coevolved in plants.
METHODS
Plant Materials, Growth Conditions, Light Treatments, and Phenotypic Analyses
Seeds of Arabidopsis thaliana hec1 and hec1 hec2 hec3+/− mutants were previously described (Crawford and Yanofsky, 2011) and kindly shared by M.F. Yanofsky from UC San Diego. We isolated hec1 hec2 double mutants from the above population. Homozygous hec3 T-DNA insertional mutant (SALK_005294C) was obtained from the Arabidopsis Biological Resource Center (Alonso et al., 2003). Seeds were sterilized with 10% bleach + 0.3% SDS for 10 min, washed five times with water, and then plated on Murashige and Skoog growth medium containing 0.9% agar without sucrose (GM-Suc). After 4 d of stratification at 4°C in the dark, seeds were exposed to 1 h white light at room temperature to induce germination and kept in darkness for 23 h. After this time, the plates were transferred to growth chambers under R, FR, or blue light conditions for additional 3 d. Light fluence rates were measured using a spectroradiometer (Model EPP2000; StellarNet) as described (Shen et al., 2005). Plants were grown in Metro-Mix 200 soil (Sun Gro Horticulture) under continuous light at 24°C ± 0.5°C.
For quantitation of hypocotyl lengths, digital photographs of seedlings were taken and at least 30 seedlings were measured using the publicly available software ImageJ (http://rsbweb.nih.gov/ij/). The seed germination assays and chlorophyll and carotenoid measurements were performed as described (Oh et al., 2004; Shen et al., 2005; Toledo-Ortíz et al., 2010). Experiments were repeated at least three times.
Quantitative β-Galactosidase Assay
HEC1 and HEC2 open reading frames (ORFs) were amplified using PCR and then cloned into pGBT9 vector (Clontech Laboratories) using the restriction sites included in the PCR primers. Prey constructs encoding full-length PIF1, PIF3, PIF4, PIF5, PIF6, and PIF7 were as described (Huq et al., 2004; Bu et al., 2011a). The specific amino acid mutations in HEC2 were introduced using a site-directed mutagenesis kit (Stratagene). Procedures for the yeast two-hybrid quantitative interaction assays were performed according to the manufacturer’s instructions (Matchmaker Two-Hybrid System; Clontech Laboratories).
In Vitro and in Vivo Coimmunoprecipitation Assays
The HEC2 ORF was cloned into pET17b as a fusion protein with the GAL4 activation domain (GAD) using the same pair of primers for cloning the yeast two-hybrid assay constructs. PIF1 and PIF3 constructs are as described (Toledo-Ortiz et al., 2003; Huq et al., 2004). HEC2, PIF1, and PIF3 were cotranslated using the TnT system (Promega), and the in vitro coimmunoprecipitation assays were performed as previously described (Huq and Quail, 2002; Toledo-Ortiz et al., 2003). For in vivo coimmunoprecipitation assay, the HEC2-GFP-expressing transgenic line was crossed into TAP-PIF1 expressed from the endogenous PIF1 promoter (Bu et al., 2011b), and homozygous transgenic plants were selected using antibiotic selection. The in vivo coimmunoprecipitation assay was performed as previously described (Shen et al., 2008). Briefly, total proteins were extracted from ∼0.4 g dark-grown seedlings with 1 mL native extraction buffer (100 mM NaH2PO4, pH 7.8, 100 mM NaCl, 0.1% Nonidet P-40, 1× Protease inhibitor cocktail [Sigma-Aldrich; catalog no. P9599], 1 mM phenylmethylsulfonyl fluoride, 20 µM MG132, 25 mM β-glycerophosphate, 10 mM NaF, and 2 mM Na orthovanadate) and cleared by centrifugation at 16,000g for 15 min at 4°C. Anti-GFP antibody (Invitrogen; catalog no. A11120) was incubated with Dynabead protein A (Invitrogen; catalog no. 10002D] (1 µg antibody and 20 μL beads per sample) for 30 min at 4°C. The beads were washed twice with the extraction buffer to remove the unbound antibody. The bound antibody beads were added to a total of 1 mg protein extracts and rotated for another 3 h at 4°C in the dark. The beads were collected using a magnet, washed three times with wash buffer, dissolved in 1× SDS-loading buffer, and heated at 65°C for 5 min. The immunoprecipitated samples were separated on an 8% SDS-PAGE gel, blotted onto PVDF membrane, and probed with anti-myc antibody (Calbiochem/EMD; catalog no. OP10) to detect TAP-PIF1 and anti-GPF antibody (Santa Cruz; catalog no. sc-9996) to detect HEC2-GFP.
Construction of Vectors and Generation of Transgenic Plants
DNA sequence from nucleotide 59 to 359 of HEC2 did not show any significant similarity (≥20 bp) to any other Arabidopsis sequence; therefore, this region has been used to construct RNAi vectors for HEC2. The above region was amplified by PCR and cloned into pENTRY vector (Invitrogen), sequence verified, and recombined into pB7GWIWG2 (II) vector (Karimi et al., 2005) to produce binary plasmid for HEC2 RNAi. To construct overexpression and GFP fusion vectors, full-length HEC2 ORF was cloned into pENTRY vector and recombined with pB7WG2 (for overexpression) and pB7FWG2 (for GFP fusion) (Karimi et al., 2005). A stop codon was included in the overexpression vector, but not in the GFP fusion vector, to allow C-terminal fusion protein expression. These constructs were then transformed into the wild type using the Agrobacterium tumefaciens-mediated transformation protocol as described (Clough and Bent, 1998). Single-locus transgenic plants were selected based on antibiotic resistance, and several homozygous lines were produced for analyses for each construct. TAP-PIF1 transgenic plants have been described (Bu et al., 2011b). myc-PIF3 and myc-PIF5 were previously described (Kim et al., 2003; Sakuraba et al., 2014). For myc-PIF4, the ORF of PIF4 was PCR amplified and cloned into pENTR vector (Invitrogen). After sequence verification, PIF4 ORF was recombined into pGWB17 vector (Nakagawa et al., 2007). The construct was transformed into pif4-2 mutant, and single insertion transgenic lines were selected using antibiotic selection. One homozygous line expressing myc-PIF4 protein was used in the ChIP assays.
RNA Isolation, RT-PCR, and Quantitative RT-PCR Assays
Total RNA was isolated from materials indicated in the figure legends using the Sigma-Aldrich plant RNA isolation kit as described (Oh et al., 2009). One microgram of total RNA was reverse transcribed using SuperScript III (Invitrogen) as per the manufacturer’s protocol. For the RT-PCR, gene-specific primers listed in Supplemental Table 1 were used to detect mRNA levels. UBQ10 (At4g05320) was used as a housekeeping gene control for all RT-PCR assays. The RT-qPCR assays used the Power SYBR Green RT-PCR Reagents Kit (Applied Biosystems). Primer sequences used for RT-qPCR and RT-PCR assays are listed (Supplemental Table 1). PP2A (At1g13320) was used as a control for normalization of the expression data. The cycle threshold (Ct) values were used for calculation of the levels of expression of different genes relative to PP2A as follows: 2∆CT where ∆CT = CT(PP2A) − CT(specific gene).
Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift assays (EMSAs) were conducted according to Moon et al. (2008). For the experiment, PIF1 and HEC2 recombinant proteins were produced using the TnT kit (Promega) with or without the 35S-labeled Met. The group with the 35S-labeled Met was directly loaded into 8% SDS-PAGE gel to indicate the amount of protein being expressed. The group without the 35S-labeled Met was incubated with a PORC promoter fragment containing the G-box motif labeled with 32P-dCTP as described (Moon et al., 2008). A total of 30,000 cpm was used per lane. The samples were separated on 5% native PAGE gel. The gel was fixed, dried, and exposed to a phosphor imager for visualization.
ChIP Assays
ChIP assays were performed as described (Moon et al., 2008). Briefly, 3-d-old dark-grown seedlings were vacuum infiltrated with 1% formaldehyde for 15 min at 21°C, and cross-linking was quenched by vacuum infiltration with 0.125 M glycine for 5 min. Samples were washed with large amounts of water, dried on filter papers, and ground into powder in liquid nitrogen. Three times volume of nuclei isolation buffer (0.25 M sucrose, 15 mM PIPES, pH 6.8, 5 mM MgCl2, 60 mM KCl, 15 mM NaCl, 0.9% Triton X-100, 1 mM PMSF, and 1× Protease inhibitor cocktail [Sigma-Aldrich; catalog no. P9599]) was added to the powder (around 1 mL) and incubated on ice for 15 min. Samples were centrifuged at 16,000g for 10 min at 4°C. The pellets were resuspended with 1.5 mL lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, 0.1% SDS, 1 mM PMSF, and 1× Protease inhibitor cocktail) prior to sonication. Sonicated samples were clarified by centrifugation at 16,000g at 4°C for 5 min. Monoclonal antibody against MYC tag (EMD Millipore) was used for immunoprecipitation at 4°C for overnight. Forty microliters of Dynabead protein A was added into each sample and rotated for another hour at 4°C. Immunoprecipitated samples were sequentially washed by low-salt wash buffer (150 mM NaCl, 0.2% SDS, 0.5% Triton X-100, 2 mM EDTA, and 20 mM Tris-Cl, pH 8.0), high-salt wash buffer (500 mM NaCl, 0.2% SDS, 0.5% Triton X-100, 2 mM EDTA, and 20 mM Tris-Cl, pH 8.0), LiCl wash buffer (0.25 M LiCl, 0.5% Nonidet P-40, 0.5% deoxycholate sodium salt, 1 mM EDTA, and 10 mM Tris-Cl, pH 8.0), and TE buffer (10 mM Tris-Cl, pH 8.0, and 1 mM EDTA). One milliliter of buffer per sample was used for all washes, and each wash requires 5 min rotation at 4°C. Immune complexes were eluted in 150 μL elution buffer (1% SDS and 0.1 M NaHCO3) twice. Each time samples were gently rotated in elution buffer at room temperature for 30 min. Total 300 μL eluted sample was incubated with 10 μL 5 M NaCl at 65°C overnight for de-cross-linking. DNA was extracted the next day using the QIAEX II gel extraction kit (Qiagen; catalog no. 20051). RT-qPCR was performed to measure the amount of DNA immunoprecipitated at the different promoter regions of binding target genes.
Transient Transfection of Promoter-GUS Fusion Constructs
pPIL1-GUS and 35S:PIF1-firefly LUC constructs are as described (Moon et al., 2008; Hornitschek et al., 2009). To construct 35S:HEC2, the coding region of HEC2 gene was cloned into the pENTR vector (Invitrogen) previously to make HEC2 overexpression lines. For the mutant version of HEC2, the specific amino acid mutations in HEC2 were introduced using a site-directed mutagenesis kit (Stratagene). After sequence verification, both the wild type and mutant HEC2 were recombined into p2GW7.0 destination vector (Karimi et al., 2005). The particle bombardment experiment was performed as previously developed (Moon et al., 2008). Briefly, different combinations of DNA were incubated with gold particles at 4°C for half an hour. The DNA coated particles were bombarded into 3-d-old dark-grown pif1-2 seedlings and treated with saturated FR light. Total proteins were extracted after 18 h dark incubation. Renilla luciferase, protein concentration, and GUS activity were measured as described (Huq et al., 2004; Shen et al., 2005). The GUS activities were normalized by Renilla luciferase and protein concentration. The relative GUS activities were determined by normalizing with the GUS activity of the control group: pPIL1:GUS and 35S: Renilla LUC. The experiment was repeated at least three times with four technical repeats for each experiment.
Histochemical GUS Analysis and Subcellular Localization of HEC2
Histochemical GUS analysis was performed on intact seedlings. Transgenic plant samples were incubated with X-gluc buffer for 2 h at 37°C and then rinsed and cleared of chlorophyll by 75% (v/v) ethanol. The stained tissues were photographed under a Leica S6D stereomicroscope with a Leica DFC 320 color camera (Leica Instruments). For subcellular localization of HEC2, 4-d-old dark-grown 35S:HEC2-GFP homozygous lines were carefully transferred to glass slides under dim light. The GFP signal was examined under a Zeiss Axiovert 200 M microscope (Carl Zeiss). After the fluorescent signal and the bright-field signal were captured, 50 μL of 0.005 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) in DAPI solution (1× PBS, 50% glycerol, and 0.001% Triton X-100) was added on top of the seedling with 5-min incubation. The DAPI-stained nuclear signal was captured under UV light.
Protein Extraction and Immunoblotting
Protein samples from seedlings were extracted with denaturing buffer (100 mM MOPS, pH 7.6, 5% SDS, 10% glycerol, 4 mM EDTA, 40 mM β-mercaptoethanol, 1 mM PMSF, and 1× Protease inhibitor cocktail). Buffer was preheated and added with a 1:3 (w/v) ratio. Thirty microliters of total protein supernatants were separated on 8% SDS-PAGE gel, blotted onto PVDF membrane, and probed with anti-GFP (Santa Cruz Biotechnology), anti-PIF1 (Shen et al., 2008), and anti-RPT5 (Enzo Life Sciences) antibodies. The publicly available software ImageJ was used for the quantification of the immunoblots.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: PIF1 (At2g20180), PIF3 (At1g09530), PIF4 (At2g43010), PIF5 (At3g59060), PIF6/PIL2 (At3g62090), PIF7 (At5g61270), HEC1 (At5g67060), HEC2 (At3g50330), HEC3 (AT5g09750), PHYA (At1g09570), PHYB (At2g18790), PORC (At1g03630), PSY (AT5g17230), PIL1 (At2g46970), HFR1 (At1g02340), FHL (At5g02200), SDR (At1g01800), XTR7 (At4g14130), AtHB-2 (At4g16780), UBQ10 (At4g05320), and PP2A (At1g13320).
Supplemental Data
Supplemental Figure 1. Sequence alignment of the bHLH domain of various PIFs, HFR1, and HECATE proteins.
Supplemental Figure 2. hec1 and hec12 double mutants are longer than the wild type in the dark.
Supplemental Figure 3. Yeast two-hybrid interactions assays between the wild type and mutant HEC2 with PIF1 and PIF3.
Supplemental Figure 4. HEC2 mRNA level in wild-type, HEC2 RNAi, and overexpression lines.
Supplemental Figure 5. Coexpression analyses of PIF1 and HEC1 in Arabidopsis.
Supplemental Figure 6. Coexpression analyses of PIF1 and HEC1/HEC2 and subcellular localization of HEC2-GFP.
Supplemental Table 1. Primer sequences used in experiments described in text.
Supplemental Data Set 1. Text file of the alignment used for the phylogenetic analysis in Supplemental Figure 1.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Huq laboratory for critical reading of the manuscript, Justina Hill, Jimmy Wang, James C. O’Brien, and Judy Liang for technical assistance, Martin F. Yanofsky for sharing hec mutants, and G. Choi for sharing myc-PIF3 and myc-PIF5 seeds. This work was supported by grants from the National Science Foundation (MCB-1543813), the National Institutes of Health (1R01 GM-114297), the Human Frontier Science Program (HFSP# RGP0025/2013), and a research grant from UT Austin to E.H.
AUTHOR CONTRIBUTIONS
L.Z, R.X., H.S., and E.H. designed experiments. L.Z., R.X., H.S., Q.B., and J.D. carried out experiments. L.Z., R.X., H.S., Q.B., and E.H. analyzed data. L.Z., R.X., and E.H. wrote the manuscript. L.Z., R.X., H.S., and Q.B. commented on the manuscript.
Glossary
- R
red
- FR
far-red
- ChIP
chromatin immunoprecipitation
- ORF
open reading frame
- EMSA
electrophoretic mobility shift assay
- DAPI
4′,6-diamidino-2-phenylindole
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59: 281–311. [DOI] [PubMed] [Google Scholar]
- Bai M.-Y., Fan M., Oh E., Wang Z.-Y. (2012). A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P.C., Martin C., Toledo-Ortiz G., Quail P.H., Huq E., Heim M.A., Jakoby M., Werber M., Weisshaar B. (2003). Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R., Davis R.L., Lockshon D., Turner D.L., Weintraub H. (1990). The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61: 49–59. [DOI] [PubMed] [Google Scholar]
- Bernardo-García S., de Lucas M., Martínez C., Espinosa-Ruiz A., Davière J.-M., Prat S. (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28: 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Castillon A., Chen F., Zhu L., Huq E. (2011a). Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol. Biol. 77: 501–511. [DOI] [PubMed] [Google Scholar]
- Bu Q., Zhu L., Yu L., Dennis M., Lu X., Person M., Tobin E., Browning K., Huq E. (2011b). Phosphorylation by CK2 enhances the rapid light-induced degradation of PIF1. J. Biol. Chem. 286: 12066–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12: 514–521. [DOI] [PubMed] [Google Scholar]
- Chen D., Xu G., Tang W., Jing Y., Ji Q., Fei Z., Lin R. (2013). Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Galvão R.M., Li M., Burger B., Bugea J., Bolado J., Chory J. (2010). Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T., Shokry A., Moffet M., Liu P., Faul M., Sharrock R.A. (2009). Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crawford B.C.W., Yanofsky M.F. (2011). HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development 138: 2999–3009. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Dong J., Tang D., Gao Z., Yu R., Li K., He H., Terzaghi W., Deng X.W., Chen H. (2014). Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26: 3630–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2003). HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 34: 827–836. [DOI] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2005). bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 10: 51–54. [DOI] [PubMed] [Google Scholar]
- Duek P.D., Elmer M.V., van Oosten V.R., Fankhauser C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14: 2296–2301. [DOI] [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A., Quail P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14: 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. (2000). RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol. 124: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chen M. (2008). Transposing phytochrome into the nucleus. Trends Plant Sci. 13: 596–601. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão R.M., Li M., Kothadia S.M., Haskel J.D., Decker P.V., Van Buskirk E.K., Chen M. (2012). Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev. 26: 1851–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão V.C., Fankhauser C. (2015). Sensing the light environment in plants: photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 34: 46–53. [DOI] [PubMed] [Google Scholar]
- Gremski K., Ditta G., Yanofsky M.F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601. [DOI] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z.-Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5: 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R., Jang I.-C., Chua N.-H. (2009). Regulated proteolysis in light-related signaling pathways. Curr. Opin. Plant Biol. 12: 49–56. [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E. (2006). Degradation of negative regulators: a common theme in hormone and light signaling networks? Trends Plant Sci. 11: 4–7. [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2005). Phytochrome signaling. In Handbook of Photosensory Receptors, Briggs W.R., Spudich J.L., eds (Weinheim, Germany: Wiley-VCH; ), pp. 151–170. [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Hyun Y., Lee I. (2006). KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61: 283–296. [DOI] [PubMed] [Google Scholar]
- Jeong J., Choi G. (2013). Phytochrome-interacting factors have both shared and distinct biological roles. Mol. Cells 35: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105. [DOI] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood T., Evans G.I. (1998). Helix-Loop-Helix Transcription Factors. (New York: Oxford University Press; ). [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Lu X.-D., Zhou C.-M., Xu P.-B., Luo Q., Lian H.-L., Yang H.-Q. (2015). Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8: 467–478. [DOI] [PubMed] [Google Scholar]
- Luo Q., Lian H.-L., He S.-B., Li L., Jia K.-P., Yang H.-Q. (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26: 2441–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Li X., Guo Y., Chu J., Fang S., Yan C., Noel J.P., Liu H. (2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 113: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Zhu L., Shen H., Huq E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 9433–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Ni W., Xu S.-L., Chalkley R.J., Pham T.N.D., Guan S., Maltby D.A., Burlingame A.L., Wang Z.-Y., Quail P.H. (2013). Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.-L., Tepperman J.M., Stanley D.J., Maltby D.A., Gross J.D., Burlingame A.L., Wang Z.-Y., Quail P.H. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C., López-Salmerón V., Davière J.-M., Prat S. (2015). ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr. Biol. 25: 187–193. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.-Y., Wang Z.-Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.-I., Choi G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47: 124–139. [DOI] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J.C., Choi G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale U.V., Huang S.S., Zander M., Cole B.J., Hetzel J., Ljung K., Reis P.A., Sridevi P., Nito K., Nery J.R., Ecker J.R., Chory J. (2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J.M., Zhang Y., Quail P.H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7: 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Nagel M.-K., Popp C., Wüst F., Bindics J., Viczián A., Hiltbrunner A., Nagy F., Kunkel T., Schäfer E. (2012). Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl. Acad. Sci. USA 109: 5892–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Li M., Pasoreck E.K., Long L., Shi Y., Galvão R.M., Chou C.L., Wang H., Sun A.Y., Zhang Y.C., Jiang A., Chen M. (2015). HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. Plant Cell 27: 1409–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. (2010). Phytochromes. Curr. Biol. 20: R504–R507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Su Y.-S., Lagarias J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57: 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou J., Sorin C., Devlin P.F., Martínez-García J.F. (2006). Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 141: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou-Torrent J., Galstyan A., Carretero-Paulet L., Portolés S., Rodríguez-Concepción M., Martínez-García J.F. (2007). Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 26: 4756–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Jeong J., Kang M.-Y., Kim J., Paek N.-C., Choi G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5: 4636. [DOI] [PubMed] [Google Scholar]
- Schuster C., Gaillochet C., Medzihradszky A., Busch W., Daum G., Krebs M., Kehle A., Lohmann J.U. (2014). A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 28: 438–449. [DOI] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.-D., Huq E., Hiltbrunner A. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize seedling photomorphogenesis in Arabidopsis. Plant J. 44: 1023–1035. [DOI] [PubMed] [Google Scholar]
- Shen H., Luong P., Huq E. (2007). The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol. 145: 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhong S., Mo X., Liu N., Nezames C.D., Deng X.W. (2013). HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wang X., Mo X., Tang C., Zhong S., Deng X.W. (2015). Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc. Natl. Acad. Sci. USA 112: 3817–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P.G., Fankhauser C., Terry M.J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 106: 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortíz G., Huq E., Rodríguez-Concepción M. (2010). Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 107: 11626–11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Johansson H., Lee K.P., Bou-Torrent J., Stewart K., Steel G., Rodríguez-Concepción M., Halliday K.J. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Huq E. (2015). Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 20: 641–650. [DOI] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Bu Q., Huang X., Deng X.W., Huq E. (2014). PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell 26: 1992–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Song M., Yang Q., Su L., Hou P., Guo L., Zheng X., Xi Y., Meng F., Xiao Y., Yang L., Yang J. (2014). Both PAR1 and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol. 164: 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Huq E. (2014). Suicidal co-degradation of the phytochrome interacting factor 3 and phytochrome B in response to light. Mol. Plant 7: 1709–1711. [DOI] [PubMed] [Google Scholar]
- Zhu L., Bu Q., Xu X., Paik I., Huang X., Hoecker U., Deng X.W., Huq E. (2015). CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 6: 7245. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Tepperman J.M., Fairchild C.D., Quail P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 97: 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.