Abstract
Complement Factor H has recently come to the fore with variant forms implicated in a range of serious disease states. This review aims to bring together recent data concerning the structure and biological activity of this molecule to highlight the way in which a molecular understanding of function may open novel therapeutic possibilities. In particular we examine the evidence for and against the hypothesis that sequence variations in factor H may predispose to disease if they perturb its ability to recognise and respond appropriately to polyanionic carbohydrates on host surfaces that require protection from complement-mediated damage.
1 Introduction
The human complement factor H (CFH) protein is a key regulator of the alternative pathway of the complement system. The alternative pathway entails ubiquitous and continuous generation of relatively small quantities of C3b molecules. These bind indiscriminately and have the potential to self-amplify. Any surface exposed to complement, and not protected by regulatory molecules, will therefore quickly become coated in C3b and subject to multiple potentially destructive complement-mediated processes. CFH acts to specifically prevent C3b amplification on self-surfaces while permitting complement to proceed unchecked on foreign surfaces. It thereby selectively protects host tissues and it appears to be essential for the healthy functioning of at least some organs.
This approximately 155-kDa (Ripoche et al. 1988) plasma (~150–550 μg/ml) (Esparza-Gordillo et al. 2004) glycoprotein is encoded by the CFH gene within the regulators of complement activation (RCA) gene cluster on chromosome 1q32 (Rodriguez de Cordoba et al. 1985) where it is linked closely to genes CFHR1-CFHR5, which code for five smaller and less abundant CFH-related proteins (Zipfel et al. 1999). A seventh protein – FH-like or CFHT – is a splice-variant of CFH present at appreciable levels in the blood (Schwaeble et al. 1987). Some of the CFH-related proteins and CFH-like-1 also exhibit complement regulatory activity (Zipfel and Skerka 1994) although their physiological properties have not yet been clearly defined. Most CFH in the circulation has a hepatic origin but many cell-types are endogenous sources (Schlaf et al. 2001).
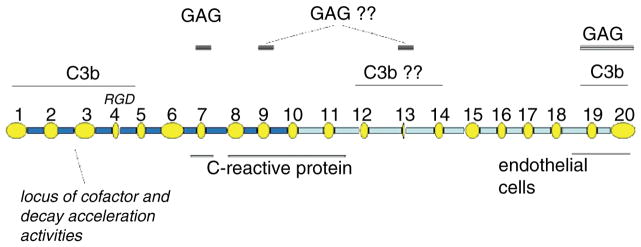
The CFH protein is composed from a total of 20 domains, each containing approximately 60 amino acid residues and termed “complement control protein modules” (CCPs) or short consensus repeats (Ripoche et al. 1986), that are joined by short linkers consisting of 3–8 residues (Fig. 1). Each module is encoded by a single exon out of the 23 in CFH, except CCP module 2 that is encoded by two exons (Male et al. 2000). Exon one encodes an 18 amino acid signal peptide that is cleaved during processing. Exon ten does not contribute to CFH but encodes the last of seven modules in the splice-variant, CFH-like-1, plus its unique C-terminal sequence Ser-Phe-Leu-Thr (Estaller et al. 1991).
Fig. 1.
Modular composition of CFH and putative binding sites. In this schematic, the CCPs (ovals) of CFH are drawn to emphasize their variation in size (51–63 residues) and the linkers (rectangles) are proportional to their lengths (3–8 residues). Functional/binding sites are indicated. The question marks are used where no direct proof is available
By utilising its multiple protein and carbohydrate binding sites (Schmidt et al. 2008), CFH binds to C3b and its complexes C3bBb and C3b2Bb – the C3- and C5-convertases of the alternative pathway. It engages with these binding partners both in fluid phase and when they are immobilised. Critically, CFH has the remarkable property of acting selectively on C3b and C3bBb on self vs. non-self surfaces (Meri and Pangburn 1990). It competes for binding of complement factor B (CFB) to C3b (Farries et al. 1990), acts as a cofactor for factor I-catalysed proteolytic cleavage of C3b (Pangburn et al. 1977), and it accelerates the irreversible dissociation of C3bBb and C3b2Bb into their separate components (Pangburn and Muller-Eberhard 1983). Thus CFH not only inhibits formation of the convertases but it also shortens the lifespan of any convertase complex that forms.
The discovery of mutations and single nucleotide polymorphisms (SNPs) in CFH that can be linked to human diseases has led to a surge of interest in this protein; an update is presented in the next section. We will then summarise the current state of knowledge of structure-function relationships of CFH, with an emphasis on recent structural studies of a key disease-linked, glycosamioglycan (GAG)-binding region in CCPs 6, 7 and 8. Finally, both established and emerging biochemical attributes/properties of CFH will be discussed in the context of the consequences for human health of its genetically encoded sequence variations.
2 Involvement of Factor H in Human Disease
Mutations and polymorphisms that lead to amino-acid residue substitutions (or a deletion in one case) within CFH have been linked to several human diseases, including atypical haemolytic uremic syndrome (aHUS), age-related macular degeneration (AMD), and membranoproliferative glomulonephritis type II (MPGNII also known as dense deposit disease) (de Cordoba and de Jorge 2008). More tentatively and controversially the AMD-linked Y402H SNP (see below) has been linked to an altered risk of three other late-onset conditions – myocardial infarction (Kardys et al. 2006; Mooijaart et al. 2007; Nicaud et al. 2007; Pai et al. 2007; Stark et al. 2007), CAD/CHD (Meng et al. 2007; Pulido et al. 2007; Topol et al. 2006) and Alzheimer’s Disease (Hamilton et al. 2007; Zetterberg et al. 2007).
Hemolytic uremic syndrome (HUS) is a leading cause of pediatric kidney failures and is generally a sequela of bacterial infections. It is characterized by thrombocytopenia, microangiopathic hemolytic anemia and acute renal failure. The rare atypical HUS (aHUS) is not linked to infection, but is sporadic or familial. Full recovery from typical HUS is the norm, but the long-term diagnosis for sufferers of aHUS – 5–10 % of cases of HUS – is unfavorable. Single and double amino-acid changes that occur predominantly in the C-terminal segment of CFH (CCPs 19 and 20) have been identified in 10–15% of patients with aHUS (Buddles et al. 2000; Heinen et al. 2006; Richards et al. 2001; Warwicker et al. 1998) (http://www.fh-hus.org/). In many cases the variant form of CFH is present in plasma at approximately normal levels (Kavanagh et al. 2007). Mutations in membrane cofactor protein and factor I have also been linked to aHUS. This topic has been the subject of a recent review (Kavanagh et al. 2007), hence our emphasis here is on AMD.
Age-related macular degeneration is characterized by a progressive loss of central vision attributable to degenerative and, in advanced cases, neovascular changes in the macula, a highly specialized region of the ocular retina comprising only 4% of total retinal surface area (Gehrs et al. 2006). This region contains the highest density of cone photoreceptor cells, is the only portion of the retina where 20/20 vision is attainable, and accounts for approximately 10% of the visual field. Thus, the pathological lesions that develop in this region have a major impact on visual function and productivity.
Age-related macular degeneration is characterized in its earliest clinical stages by the accumulation of drusen, hallmark extracellular deposits that accumulate between the retinal pigmented epithelium (RPE) and Bruch’s membrane, a unique basement membrane complex separating the retina and choroid. In recent years, a variety of complement activators, complement components, and complement regulatory proteins have been identified as molecular constitutents of drusen (Anderson et al. 2001; Crabb et al. 2002; Hageman et al. 1999; Johnson et al. 2000, 2001; Mullins et al. 2000). This compositional profile of drusen formed the basis for the general conclusion that drusen are a byproduct of chronic, local inflammatory events at the level of the RPE-Bruch’s membrane interface and that complement activation and immune responsiveness are important facets of AMD pathogenesis (Hageman et al. 2001; Penfold et al. 2001; Zarbin 2004). In this model, RPE atrophy and the subsequent deposition of RPE-derived debris in the sub-RPE space is construed as a local pro-inflammatory event, leading to activation and amplification of the complement alternative pathway which, in turn, induces substantial bystander damage to macular cells and tissues over time.
Strong support for this new paradigm of AMD pathobiology emerged from subsequent genetic discoveries reported in early 2005 by four independent research teams (Edwards et al. 2005; Hageman et al. 2005; Haines et al. 2005; Klein et al. 2005). These studies revealed a highly significant association between AMD and common variants in the CFH gene on human chromosome 1q31. Three of these studies focused on a single SNP in the coding region of CFH, Y402H, as the causal variant (Edwards et al. 2005; Haines et al. 2005; Klein et al. 2005). However, it was clear from the fourth study (Hageman et al. 2005) that multiple CFH variants (and their tagged haplotypes), rather than the single Y402H variant, confer either an elevated or reduced risk for one’s propensity to develop AMD. Three significant haplotypes; a major risk haplotype, tagged by the Y402H coding SNP (cSNP) and two protective haplotypes, tagged by intronic variants and/or cSNPs, were identified (Hageman et al. 2005). Iwata and colleagues subsequently identified an additional risk haplotype in the Japanese population (Okamoto et al. 2006). More recent studies of Caucasian populations have provided evidence for an additional risk haplotype, tagged by rs2274700 [A473A] or an intronic rs1410996 SNP (Li et al. 2006; Seddon et al. 2006) and a strongly protective haplotype tagged by a complete deletion of the two CFH-related genes, CFHR1 and CFHR3 (Hageman et al. 2006; Hughes et al. 2006). Interestingly, the same AMD-associated risk haplotype has also been shown to be associated with MPGNII, a rare renal disease characterized by uncontrolled activation of the alternative complement pathway and the development of ocular drusen and neovascularization that are indistinguishable from those that occur in AMD (Hageman et al. 2005). These data provided additional strong support for the notion that specific CFH variants are associated with drusen deposition and AMD.
The AMD-CFH discovery was followed by the identification of an association of AMD with three additional complement regulators – Complement Factor B and component 2 (CFB/C2) on chromosome 6p21 (Gold et al. 2006) and C3 on chromosome 19p13 (Maller et al. 2007; Yates et al. 2007) – as well as a significant association of three tightly linked genes (PLEKHA1, LOC387715 and PRSS11/HTRA1) on human chromosome 10q26 (Jakobsdottir et al. 2005; Rivera et al. 2005; Weeks et al. 2004). Genetic variation at all four loci defines a major proportion of AMD disease burden, making this disease one of the most well-defined complex traits.
All accumulated data at this time support the concept that dysregulation of the complement alternative pathway is an early step in AMD (Gehrs et al. 2006) regardless of ocular phenotype. This contrasts with genes within the 10q26 locus, which is associated mainly with the advanced stages of AMD. The specific variants, or their combinations, within the CFH, CFB/C2 and C3 genes, which result in functional consequences at the protein and functional levels, must be more precisely defined. Nonetheless, it seems clear from the available data that genetically predetermined variation in genes associated with the complement pathway, when combined with an as yet undefined triggering event, underlie a major proportion of AMD in the human population.
Thus, although aHUS and MPGNII affect glomeruli while AMD is an ocular disease, it is MPGNII and AMD that share pathological characteristics. Both MPGNII and AMD involve the deposition of complement-containing material (Mullins et al. 2000) onto specialised regions of extracellular matrix (the glomerular basement membrane in the case of the kidney or Bruch’s membrane in the macula) accessible to plasma via fenestrations within the endothelial lining of the microvasculature; and indeed MPGNII patients develop ocular drusen as mentioned earlier (Mullins et al. 2001). The phenotype of CFH−/− knockout mice includes both MPGNII (Pickering et al. 2002) and abnormalities in retinal ultrastructure (Coffey et al. 2007). On the other hand aHUS, which is characterised by endothelial cell activation, swelling and detachment from the basement membrane, does not arise in CFH−/− mice unless a gene encoding CFH- 1–15 (i.e. a CFHΔ16–20 mutant) is re-introduced (Pickering et al. 2007). As discussed below, such a mutant will retain fluid-phase C3b-binding activity but will be unable to recognise surfaces. This suggests that the fluid-phase convertase-regulating activity of CFH is required for development of aHUS while surface-specific convertase regulation by CFH inhibits the development of mouse model of aHUS. This is discussed further below in the light of recent structural and functional results.
3 CFH Binding Sites
Early work identified the N-terminal five or seven CCPs of CFH as the locus of its fluid-phase C3b-binding and cofactor activities (Alsenz et al. 1984, 1985; Misasi et al. 1989). Subsequent studies localised this functionality (Gordon et al. 1995; Kuhn et al. 1995), along with a proportion of the native decay accelerating activity (Kuhn and Zipfel 1996), to CCP modules 1–4. Additional C3b-binding sites were identified using module-deletion CFH mutants (Sharma and Pangburn 1996). Thus, while CFHΔ1–5 retained some binding affinity for cell-surface -bound C3b (csbC3b), deletions of CCPs 16–20 decimated its binding affinity for csbC3b. This observation implicated the C-terminal region of CFH as a second C3b-binding site, which was later pinpointed to CCPs 19 and 20 REF. A third C3b-binding site has been suggested on the basis that CFHΔ6–10 exhibited a decreased affinity for csbC3b, similar to that of CFHΔ1–5. Additional studies (Jokiranta et al. 2000) implied but did not confirm a third C3b binding site in CCPs 12–14. This information is summarised in Fig. 1.
Fundamental to the ability of CFH to regulate efficiently complement on surfaces is its affinity for polyanions (Meri et al. 1990). A photoaffinity-tagging analogue (Pangburn et al. 1991) implicated the very basic CCP 13 in heparin binding. But while deletion of CCP 13 (Sharma et al. 1996) resulted in only very slightly reduced ability to bind a heparin-agarose column, deletion of CCPs 6–10 showed significantly weaker heparin affinity (Sharma et al. 1996) implying that a stronger GAG/sialic acid-binding site exists in the CCPs 6–10 region; CCP 7 was subsequently demonstrated to play a prominent role (Blackmore et al. 1996). A CFHΔ7Δ13 deletion mutant still bound heparin but a CFHΔ7Δ20 construct did not (Blackmore et al. 1998) implicating CCP 20 as a further locus for polyanion interaction. Convincingly, non-heparin-binding CFH-1–5 was converted to heparin-binding CFH-1–5,20 by inclusion of CCP 20 in the construct (Blackmore et al. 1998). Moreover, highly purified, structurally characterised CFH-19,20 (Herbert et al. 2006) binds well to a heparin-agarose column. More recently, a set of short recombinant CFH constructs having in common the presence of CCP 9 were all found to bind heparin (Ormsby et al. 2006), but the additional presence of an N-terminal cloning artefact containing arginine residues might have contributed to this affinity. In summary (Fig. 1), while GAG-binding sites in module 7 (with contributions likely from CCPs 6 and 8) and module 20 (with possible contributions from CCP 19) are well-established, current evidence for involvement of either CCP 9 or CCP 13 is inconclusive. Finally, it should be mentioned that the N-linked glycans of CFH could theoretically have an elecrostatic influence on CFH-polyanion interactions although they are dispensable for complement regulation (Jouvin et al. 1984).
The importance of the C-terminal heparin-binding site for self vs. non-self discrimination was clearly demonstrated by Ferreira et al. (2006) who found that highly purified CFH-19,20 competitively inhibited the action of CFH on cell surfaces. This double-module construct was able to overcome the protective effects of full-length CFH and thereby promote aggressive complement-mediated lysis of sheep erythrocytes. This finding is reinforced by studies showing the ability of monoclonal CCP 20-specific antibodies (Oppermann et al. 2006) to block interactions of CFH with endothelial cells. Thus this C-terminal polyanion- and C3b-binding site is critical for the ability of CFH to recognise and protect host cells bearing sialic acids and GAGs.
4 The Structure of Factor H
The multiple, flexibly-linked modules of CFH render it a problematic target for high-resolution structural methods. Below we outline low-resolution structural studies of intact, full-length CFH molecule, which have provided us with valuable insights into its overall architecture. Our understanding of the atomic-resolution structure of CFH derives from studies of constructs consisting of only a few CCPs. Some of these studies will be described subsequently, with an emphasis on recent work on the AMD-related and GAG-binding CCP 7 and its neighbouring modules.
4.1 Low Resolution Structural Information on CFH
Determination of 3-D structure from extended, multi-domain, proteins is a significant challenge and the level of accuracy of such models is low compared to the atomic structures of fragments. However, major progress has been made by the group of Perkins using a combination of biophysical techniques (primarily neutron and X-ray solution-scattering combined with analytical ultracentrifugation, reviewed in (Perkins et al. 2002)) capable of giving low-resolution information about molecular shape combined with using homology models of the domains to aid interpretation of the shape information. Although these studies lead to sets of atomic coordinates for the proteins studied it is important to reflect on the fact that the data can only inform about overall shape and have no ability to validate the atomic detail of the homology models used, as discussed further below. When applied to study of full length CFH (Aslam and Perkins 2001) the data revealed that the maximum length of the CFH in solution was approximately 40 nm compared to a length of 73 nm theoretically possible for a fully extended structure. These data strongly suggest that, on the average, the 20 CCP modules within CFH are folded back upon themselves in solution. Such folding back has been supported by later studies (Okemefuna et al. 2008) using the same techniques, which suggest short truncated segments of CFH are likewise not entirely linear with regard to their module arrangements.
4.2 Atomic Structure of the CFH Carboxy-Terminus
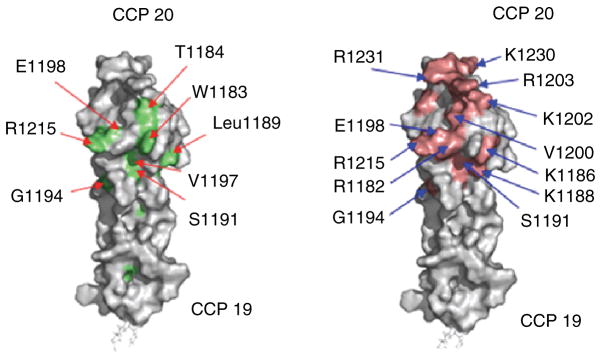
Both NMR (Herbert et al. 2006) and X-ray crystallography (Jokiranta et al. 2006) have been employed to study the 3-D structure of the C-terminal pair of CCPs, CFH-19,20 (Fig. 2a). Module 19 is similar in structure to other CCPs of known structure such as CFH modules 5 (Barlow et al. 1992), 15 (Barlow et al. 1993) and 16 (Barlow et al. Campbell 1991). In all these CCPs, the polypeptide chain contributes to five extended regions that are joined by turns or loops. The extended regions, which form short beta-strands and small sheets to an extent that varies between CCP structures (Soares et al. 2005), are approximately aligned forming an oblate structure with N and C termini at opposite poles. Each 40 Å-long module has its own small hydrophobic core containing highly conserved non-polar side-chains and is further stabilised by two disulfides that form between four invariant Cys residues. CCP 19 is tightly coupled, in a head-to-tail arrangement, to CCP 20 via a three-residue linking sequence – the shortest linker in CFH. This C-terminal module also has many features typical of CCP modules but it is shorter and broader than any other CCP of known structure. When CFH-19,20 is titrated with the GAG-like compound, dp4, perturbations are observed of CCP 20 NMR chemical shifts but not CCP 19 ones (Herbert et al. 2006), thus identifying module 20 as the dp4-binding site. Closer scrutiny of which residues within CCP 20 are most affected in terms of their chemical shifts upon addition of dp4 implicates the presence of a dp4-binding patch on one face of CCP 20 (Fig. 2b). There is a notable degree of overlap between this putative GAG-binding region, which is predominantly electropositive, and a set of residues considered important in aHUS (see above).
Fig. 2.
Surface representations of the structure of FH-19,20. Highlighting: (a) The positions of mutations in CFH found in aHUS patients. (b) The residues that are involved in interacting with a model GAG compund, dp4
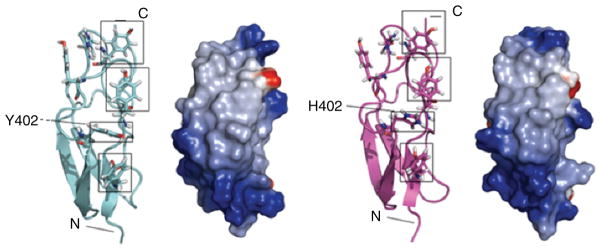
4.3 Atomic Structures for both Tyr and His Variants of CCP 7
The first detailed structural information on an AMD-associated CCP 7, emerged from NMR-derived structures of both the Tyr402 and His402 allotypic variants of the recombinantly expressed single module (Herbert et al. 2007) (Fig. 3) i.e. CFH-7. This study revealed that both variants of CFH-7 adopt the classical CCP-fold (Norman et al. 1991), and that the side-chain of residue 402 lies on the surface between two Tyr side-chains (Tyr390 and Tyr393). With the exception of the altered side-chain, the two variants are identical in structure; indeed even the orientations of the altered side-chain are the same. The authors further demonstrated that residue 402 lies on the edge of a patch of amino acid residues whose NMR chemical shifts are sensitive to addition of a fully sulphated heparin-derived tetrasaccharide, dp4, which is a model-GAG compound. Thus they concluded that the AMD-linked substitution could alter the interaction between CCP 7 and GAGs on the self-surfaces.
Fig. 3.
Comparison of structures of FH-7. Secondary structure and electrostatic surface representations (red = negative, blue = positive) are shown for the Y402 (left-hand panels) and H402 (right-hand panels) allotypic variants. Residue 402 is labeled and boxed and three tyrosine side-chains on the same face of the module are also boxed
4.4 Low-Resolution Models for CFH-6,7,8
Subsequent insights into the structure of this AMD-associated region of CFH came from low-resolution techniques: small-angle X-ray scattering (SAXS) measurements in solution and analytical ultracentrifugation. These methods were employed (Fernando et al. 2007) to investigate the structure of a recombinantly expressed construct (Clark et al. 2006) consisting of the seventh CCP flanked by its immediate neighbours i.e. CFH-6,7,8. As pointed out earlier, neither of these techniques is able to yield atomic-level information and both are dependent in their interpretation upon construction of homology-based computer-generated models for the domains or modules under investigation. Their potential advantage is that they can provide information about the dimensions of the molecule and hence inferences may be made regarding orientation of the domains with respect to each other and the overall molecular architecture. Both His402 and Tyr402 variants of CFH-6,7,8 were shown to adopt similar, bent conformations (Fernando et al. 2007). The authors noted that the His402 variant had a slightly higher tendency to self-associate (Fernando et al. 2007; Nan et al. 2008), although there is no evidence that this occurs in vivo.
By exploring numerous possible relative arrangements of the three homology-modelled CCPs for best fit to the data, restrained models of CFH-6,7,8 were created. These allowed the tentative mapping of positively charge and therefore putative GAG-interacting residues, and led to the claim that many of these are exposed on the convex surfaces of the models. Additionally, study of CFH-6,7,8 in complex with a heparin decamer by SAXS (Fernando et al. 2007) suggested that subtle structural rearrangements resulting in a more linear arrangement of the three CCPs might accompany the binding of heparin-like ligands. Presumably these could arise from the involvement of more than one module in contacting a single heparin molecule.
4.5 Atomic Structure for CFH-678
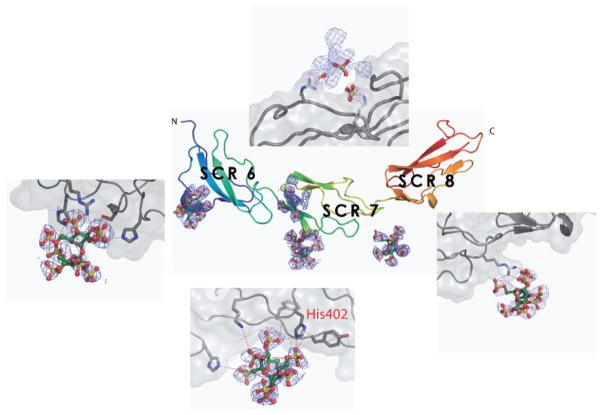
While low-resolution studies inform the overall shape of CFH-6,7,8 they are difficult or impossible to interpret reliably in terms of individual side-chains. To gain true insight into the implications of the Y402H substitution for this region, high-resolution data were required. In particular, there was a need for structural information on the interaction of CFH-6,7,8 with anionic carbohydrates. Such information has become available in the form of a near-atomic resolution crystal structure (Prosser et al. 2007b) of the His402 variant of CFH-6,7,8 in complex with sucrose octasulfate (SOS), a highly sulfated sugar analogue of GAGs (Fig. 4). Justification for the use in this study of SOS as a structurally amenable mimic of GAGs was provided by NMR work on CFH-7,8, which indicated similar binding modes for both the heparin-derived tetrasaccharide, dp4, and SOS.
Fig. 4.
Centre panel shows a cartoon representation of the structure of CFH-6,7,8402H (Prosser et al. 2007b) with the sulphated sugar ligands displayed as electron density and modeled atoms (ball and stick representation). The panels arranged around the centre show close up views of the GAG binding sites
As was predicted on the basis of low-resolution methods (Fernando et al. 2007) The CFH-6,7,8402H structure (Prosser et al. 2007b) adopts an extended but slightly curved conformation. The crystal structure reveals that the overall conformation is maintained by a tight interface between CCPs 6 and 7, but there are weaker interactions between CCPs 7 and 8 that may allow some flexibility between the domains. However, the scattering and homology based model was found to be wrong with respect to inter-modular angles, the structures of individual modules, and the details of how side-chains are arranged at the modular surfaces. These discrepancies likely reflect the difficulties of making detailed interpretations of inherently low-resolution data and there is not necessarily a conflict between the crystal structure and the solution scattering and sedimentation data of Fernando et al. (Fernando et al. 2007).
Within the crystal structure of CFH-6,7,8402H (Prosser et al. 2007b) the structure of CCP 7 is identical to the NMR-derived solution structure of CFH-7 (Herbert et al. 2007), which increases confidence in the reliability of both solution and crystal structures. Comparison of the NMR structure (solved in absence of a GAG-like ligand) and the crystal structure (solved in the presence of bound SOS) reveals that the His402 side-chain adopts the same conformation in the absence or presence of bound SOS. The Tyr402 side-chain in the NMR structure of CFH-7402Y also adopts this conformation. These observations justified the generation of a reliable model for the CFH-6,7,8402Y structure, which unfortunately could not be crystallized (Prosser et al. 2007a).
4.6 Sulfated Sugar Recognition at the Polymorphic Residue
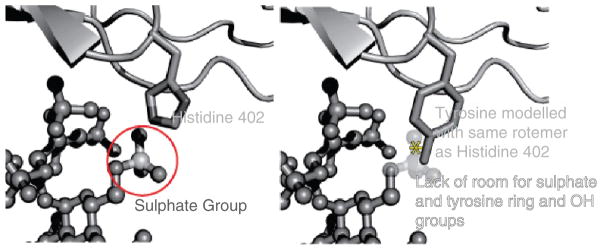
The earlier NMR chemical-shift perturbation studies had implied a direct role for the polymorphic residue in recognition of GAGs (Herbert et al. 2007). This was confirmed by the crystallographic structure of the complex that revealed a direct interaction between the AMD-associated His402 residue in CCP 7 and the sulfated-sugar ligand; one of the histidine ring nitrogens is hydrogen-bonded to a sulfate group of the ligand (Figs. 4 and 5). Together with His360 from CCP 6 His402 forms a histidine-clamp around the SOS sulfate groups, docking the ligand into the CCP 6–7 interface. This mode of SOS recognition by His402 is incompatible – both chemically and sterically – with the presence of a bulkier tyrosine at the same position. This is not to imply that the Tyr402 variant is incapable of interacting with other sulfated-sugar ligands that are subtly different at the molecular level; indeed experimental data suggests CFH-6,7,8402Y can bind to some GAGs more tightly than CFH-6,7,8402Y depending upon GAG-type and level of sulfation (Clark et al. 2006; Herbert et al. 2007; Skerka et al. 2007). Other protein-sugar contacts in this region consist mainly of hydrogen-bonds and Van der Waals contacts with the protein backbone; there is an additional contribution from the side chain of the more distant Lys405, which has been previously implicated in heparin binding by mutagenesis (Clark et al. 2006; Giannakis et al. 2003).
Fig. 5.
Cartoon to illustrate incompatibility of sulphated sugar recognition by the Histidine at position 402 with a tyrosine at the same position
It is noteworthy that the SOS at this site is present in two, slightly different conformations in the native protein crystals, related by a rotation of the sulfated fructose ring (a major conformation representing 65% of the population and a minor conformation representing 35%). This conformational difference between two forms of SOS is accompanied by a reorientation of the side-chain of Tyr390 to accommodate a repositioned sulfate group. In the ligand-free solution structure of CFH-7, only the minor conformation of the tyrosine side-chain is observed (Herbert et al. 2007), implying that movement of this side-chain may contribute to recognition of specific ligands. Such a role for Tyr390 in binding heparin fits well with NMR chemical shift perturbation studies on CFH-7 (Herbert et al. 2007) that also implicated this residue in GAG binding.
4.7 Additional Binding Sites for Sulfated-Sugar Within CFH-6,7,8
In addition to a SOS-binding site centred on the polymorphic residue 402, the co-crystal structure of CFH-6,7,8 showed that other parts of the surface of this region of CFH could be involved in specific GAG recognition. Furthermore, earlier work (Clark et al. 2006; Giannakis et al. 2003; Herbert et al. 2007), along with mutagenesis experiments designed specifically to test these additional sites (Prosser et al. 2007b), support a novel picture of GAG binding that incorporates extensive interactions over this region.
A secondary SOS-binding site is observed on the opposite face of CCP 7 in which the sugar sulfates contact the side-chains of Arg404 and Lys410, previously implicated in heparin binding by mutagenesis (Clark et al. 2006; Giannakis et al. 2003), and consistent with NMR chemical-shift perturbation studies on CFH-7 (Herbert et al. 2007). In addition to the involvement of H360 from CCP 6 in the major CCP 7-centered SOS-binding site, the structure also revealed a novel SOS-binding site contained entirely within CCP 6. This third site is chemically similar to that seen in CCP 7, with a central positively charged residue (Arg341) flanked by two histidine sidechains (His337 and His371) that directly coordinate two ligand sulfate groups. Finally, a fourth SOS-binding site in the crystal was observed in the linker between CCPs 7 and 8, and involves a salt bridge between sulfate and Arg444.
Previously, GAG-binding sites have been localised primarily to single modules of CFH – see above (Blackmore et al. 1996, 1998; Ormsby et al. 2006; Pangburn et al. 1991). Calculation of the electrostatic potential of the crystal structure reveals that the binding sites for SOS observed in the complex occupy a positively charged groove extending over all three modules of CFH-6,7,8402H with the AMD-related polymorphism lying within this channel. Many other residues (e.g. Arg387, Lys388, Lys405) previously implicated in polyanion recognition by mutagenesis (Clark et al. 2006; Giannakis et al. 2003) also line this groove. Taken together this suggests an extended mode of interaction between GAGs and this region of CFH with the polyanion lying along the positively charged groove to form a large interaction surface. This hypothesis was supported by an analytical ultracentrifugation analysis of CFH-6,7,8 in complex with heparin dp18 or dp24.
5 Structure: Insights into Disease Mechanism
As discussed above, three regions of the CFH molecule have been highlighted in respect of its role in disease. To summarise: single and double amino-acid changes occurring predominantly in the C-terminal segment of factor H (CCPs 19 and 20) have been identified in some aHUS patients. (Buddles et al. 2000; Heinen et al. 2006; Richards et al. 2001; Warwicker et al. 1998) (http://www.fh-hus.org/); an SNP at the CFH locus (rs1061170) corresponding to the presence of His at position 402 (in CCP 7 of CFH), is one component of a major risk haplotype for AMD (Edwards et al. 2005; Hageman et al. 2005; Haines et al. 2005; Klein et al. 2005) There is evidence for at least one aHUS-linked mutation (Kavanagh et al. 2007) and an AMD-protective SNP in the N-terminal portion of factor H (CCPs 1–2) (Hageman et al. 2005). The SNPs linked to AMD (Abrera-Abeleda et al. 2006) are also linked to DDD/MPGNII (Hageman et al. 2005) along with a deletion in CCP 4 (Licht et al. 2006).
The results of structural and functional work carried out on various regions of CFH (outlined in the previous sections) afford the opportunity to explore the molecular basis of some of these diseases. A “GAG recognition” hypothesis is emerging as one mechanism amongst several possibilities for linking molecular changes to disease (Schmidt et al. 2008). This is founded upon the putative capability of CFH to discriminate between subtly different GAG molecules through specific contacts with, for example, sugar rings or sulfate groups. For the host, the advantage is that a specific recognition capability of CFH deters subversion by a pathogen that adopts a molecular mimicry strategy (Lindahl et al. 2000). A potential disadvantage is that GAG-specific CFH would have a diminished capacity to bind to the diversity of GAGs that is necessitated by unrelated aspects of cell and tissue physiology or that may arise from loss of homeostasis in stressed or ageing individuals (Verdugo and Ray 1997). GAGs form an extremely diverse group of molecules that vary according to tissue, cell type and age (Turnbull et al. 2001). Thus according to the GAG recognition hypothesis, CFH has different affinities for different tissues and this will vary over the lifetime of an individual. It is difficult to prove or disprove the notion that CFH can discriminate between closely related GAGs due to the difficulty of preparing a panel of chemically pure model GAG compounds with which to test the idea. Nonetheless, the available structural and functional data can be interpreted in a way that is broadly supportive of this model as follows:
Different allotypic variations (at 402) of CFH (in the context of the CFH-6,7,8 fragment) have different affinities for certain types of GAG (Clark et al. 2006).
Disease-linked amino-acid substitutions in modules 7 and 20 coincide with discrete patches on the surface of these modules that have been identified as GAG-interaction sub-sites (Herbert et al. 2006, 2007). Therefore they are likely to modulate specific aspects of factor H-GAG interactions.
The two modules, CCP 7 and CCP 20, each lie within different segments of CFH that are unique in their proven affinities for heparin (Schmidt et al. 2008), which is a widely accepted although crude model for GAGs. It seems unlikely that the correspondence of the two principal GAG interaction sites with the two principal sites of disease-linked sequence variations is a mere coincidence.
The structure of CFH-6,7,8 (Prosser et al. 2007b) shows an interaction between His402 and a sulfate group in the ligand, SOS – another GAG model compound – that is incompatible with the presence of a Tyr at this position. This is direct evidence for the ability of CFH to “read” a pattern of sulfates and for such a pattern recognition functionality to be modulated by a disease-linked amino-acid switch.
The presence of several sub-sites able to contact SOS within CFH-6,7,8 (Prosser et al. 2007b) implies binding of a GAG molecule across multiple modules and a consequent alteration in the shape of this segment of the protein. Scattering studies support this idea to some extent (Fernando et al. 2007). This scenario creates the potential for CFH to respond in different ways to different GAGs – thus adding an extra layer of specificity or selectivity to this interaction.
The absence of proven binding functionality amongst the CCPs 8–18 (Schmidt et al. 2008) of CFH implies they could have an architectural rather than a binding role. The preponderance of long linkers and small modules in the CCPs 12–14 region is consistent with a role as a “hinge” promoting the bending back upon itself of CFH. This could bring the two GAG-interaction sites into proximity creating the potential for higher specificity via a multi-site interaction.
Scattering studies indicate that CFH does indeed have a bent back structure (Aslam et al. 2001). Other studies indicate that antibodies to the C-terminus can modulate complement regulatory activity at the N-terminus adding further support to functionally critical proximity of the two ends of CFH (Oppermann et al. 2006).
Thus an attractive model, supported by most data but requiring further testing, is that the likelihood of excessive complement activation and attendant inflammation is inversely correlated to the affinity of CFH for self-surfaces. A diminishment in affinity could arise either from an amino-acid substitution in CFH; or it could stem from inappropriate levels or patterns of sulfation amongst surface GAGs. A third factor to consider is the level of complement activation in the immediate vicinity generating short-lived C3b molecules that will stretch local complement-regulatory capacity including that contributed by CFH. Some combination of two or all of these factors may be required for pathogenic levels of complement activation to occur. The variation of GAGs between tissue-types might explain organ-to-organ variations in the consequences (or lack of consequences) of CFH sequence variations; and given that GAG composition also changes with age, this would additionally explain the late onset of some symptoms. Moreover, certain locations may be particularly susceptible due to higher levels of apoptotic or necrotic activity –the macular RPE-choroid interface is one such region since Bruch’s membrane separates the RPE from the epithelial cells of the choriocapallaris, thereby creating a potential subRPE entrapment site that accumulates debris (Sivaprasad et al. 2005).
Not all observations, however, fully support a GAG-recognition model. Several aHUS-linked mutations – within the context of the double-module fragment CFH-19,20 – do not appear to affect binding to heparin columns or defined-length, sulphated, heparin fragments (Jokiranta et al. 2006). This may simply reflect the inadequacy of heparin as an emulator of the physiological GAG ligand. On the other hand, several of these mutants appear to exhibit lower affinity for C3b (Jokiranta et al. 2006). Given that CFH probably binds to a composite binding surface consisting both of C3b and the surrounding GAGs, it is not unreasonable to propose that disturbance of either its protein binding or carbohydrate binding components could have similar physiological outcomes. Thus it is not necessary to abandon the idea of specific GAG recognition to accommodate these results. A more radical suggestion originating from the observation of a crystallographic tetramer for CFH-19,20 (Jokiranta et al. 2006) is that some mutations could interfere with a putative ologomerisation of CFH via the C-terminal region. There is little support so far in the literature for oligomerisation of CFH but it is an enticing idea (Aslam et al. 2001; Perkins et al. 1991). CFH recognition of CRP (Jarva et al. 1999) (Aronen et al. 1990) is also mapped to this region and, in the case of Y402H in module 7, there are multiple reports of defective C-reactive protein (CRP) recognition (Laine, Jarva et al. 2007; Sjoberg et al. 2007; Skerka et al. 2007; Yu et al. 2007) but these have been challenged on the basis that some were performed on CRP in a physiologically questionable non-pentameric state (Hakobyan 2007). On the other hand a “CRP hypothesis” to explain the mechanism of the AMD pathogenesis is seductive in that the putative CRP-CFH interaction has been reported to form part of a strategy for the recruitment of CFH to apoptotic cells so as to protect against alternative pathway activation and inflammation (Gershov et al. 2000). The presence of higher levels of CRP in the choroids of AMD patients (Johnson et al. 2006) further fuels this theory.
6 Options for Therapy
Activation of complement is part of a normal homeostatic mechanism that includes opsonization and lysis of microorganisms, removal of foreign particles and dead cells, recruitment and activation of inflammatory cells, regulation of antibody production, and the elimination of immune complexes (Kinoshita 1991; Markiewski and Lambris 2007). Under normal conditions, local complement activation is beneficial by promoting the rapid clearance of cell debris and facilitating the removal of toxic protein aggregates. Uncontrolled or aberrant activation and/or regulation of this system, however, can lead to bystander damage of host cells and tissues, thereby contributing significantly to the pathogenesis of a number of diseases in addition to those discussed above including rheumatoid arthritis, IgA nephropathy, asthma, systemic lupus erythematosus, and ischemia reperfusion injury (Markiewski et al. 2007; Thurman 2007; Thurman and Holers 2006). Much of the tissue damage in these diseases results from complement-mediated lysis of bystander host cells by the membrane attack complex or downstream effects resulting from excess production of the anaphylatoxin C3a by C3 convertases.
Based upon a surge of complement-related research over the last two decades, it has become apparent that modulation of the complement system is a promising strategy for drug discovery (Liszewski and Atkinson 1998; Ricklin and Lambris 2007; Ryan 1995; Sahu and Lambris 2000). Additional evidence supporting the development of complement modulating strategies comes from studies showing that complement inhibitors can reduce inflammation and tissue destruction and ameliorate disease progression in a variety of animal models of human disease (Bao et al. 2003; Holers 2003; Holers et al. 2002; Thurman et al. 2006). Importantly, the first complement pathway-directed therapeutic, a humanized monoclonal antibody directed against complement component C5 (Eculizumab; Soliris) was approved in March 2007 for use in the treatment of paroxysmal nocturnal hemoglobinuria (PNH), a debilitating disease characterized by chronic complement-mediated intravascular hemolysis (Rother et al. 2007). This breakthrough validates the complement system as a realistic therapeutic target and has provided a strong rationale for the investigation of other indications for this and other drugs in complement-related diseases.
Many promising complement pathway-directed therapeutics have been developed; these include various recombinant human complement inhibitors, monoclonal antibodies, synthetic peptides, and peptidomimetics designed to block activation of specific complement components, neutralize complement activation fragments, or antagonize complement receptors (Ricklin et al. 2007). Additional therapeutic compounds are currently in various investigational and developmental stages (Ricklin et al. 2007). None of the agents developed to date have been designed to specifically target CFH in diseases such as AMD, MPGNII, and aHUS. The more comprehensive understanding of the functional attributes of the risk and protective forms of CFH that has emerged from studies conducted over the past 2 years will no doubt hasten the rapid conception and development of CFH-directed drug candidates. That being said, important forethought will have to be given regarding length of administration (chronic vs. acute), mode of delivery (systemic vs. local) and method of delivery. These parameters will certainly affect decisions related to the nature (i.e. small molecule, large protein) of candidate drugs.
In contrast to a strategy of inhibition, one interesting operational paradigm being developed is based upon the concept that replacement or augmentation of the complement modulating activity of dysfunctional “risk” CFH protein with functional “protective” protein might be an effective strategy for preventing or delaying the pathology associated with CFH-mediated disease. Indeed, replacement of defective CFH with normal CFH protein through plasma exchange has been shown to be effective in preventing further disease episodes and normalizing kidney function in MPGN II and atypical hemolytic uremic syndrome (aHUS) patients harboring loss of function CFH mutations (Gerber et al. 2003; Licht et al. 2005; Pickering and Cook 2008; Stratton and Warwicker 2002). Similar results have been obtained in Yorkshire pigs with an inherited deficiency of CFH that develop lethal glomerulonephritis (Hegasy et al. 2002; Jansen et al. 1998).
Collectively, the robust efforts being expended to understand the structures, functions and roles of CFH and other complement-associated molecules will likely expedite the transition from early stage discovery and identification of therapeutic targets, to the translation of this information into clinically effective diagnostics and pharmaceutical treatment modalities for a number of diseases associated with aberrant regulation of the complement system.
Contributor Information
Paul N. Barlow, Email: Paul.Barlow@ed.ac.uk.
Gregory S. Hageman, Email: gregory-hageman@uiowa.edu.
Susan M. Lea, Email: susan.lea@path.ox.ac.uk.
References
- Abrera-Abeleda MA, Nishimura C, Smith JL, Sethi S, McRae JL, Murphy BF, Silvestri G, Skerka C, Jozsi M, Zipfel PF, et al. Variations in the complement regulatory genes factor H (CFH) and factor H related 5 (CFHR5) are associated with membranoproliferative glomerulonephritis type II (dense deposit disease) Journal of Medical Genetics. 2006;43:582–589. doi: 10.1136/jmg.2005.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsenz J, Lambris JD, Schulz TF, Dierich MP. Localization of the complement-component-C3b-binding site and the cofactor activity for factor I in the 38kDa tryptic fragment of factor H. The Biochemical Journal. 1984;224:389–398. doi: 10.1042/bj2240389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsenz J, Schulz TF, Lambris JD, Sim RB, Dierich MP. Structural and functional analysis of the complement component factor H with the use of different enzymes and monoclonal antibodies to factor H. The Biochemical Journal. 1985;232:841–850. doi: 10.1042/bj2320841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DH, Ozaki S, Nealon M, Neitz J, Mullins RF, Hageman GS, Johnson LV. Local cellular sources of apolipoprotein E in the human retina and retinal pigmented epithelium: implications for the process of drusen formation. American Journal of Ophthalmology. 2001;131:767–781. doi: 10.1016/s0002-9394(00)00961-2. [DOI] [PubMed] [Google Scholar]
- Aronen M, Leijala M, Meri S. Value of C-reactive protein in reflecting the magnitude of complement activation in children undergoing open heart surgery. Intensive Care Medicine. 1990;16:128–132. [PubMed] [Google Scholar]
- Aslam M, Perkins SJ. Folded-back solution structure of monomeric factor H of human complement by synchrotron X-ray and neutron scattering, analytical ultracentrifugation and constrained molecular modelling. Journal of Molecular Biology. 2001;309:1117–1138. doi: 10.1006/jmbi.2001.4720. [DOI] [PubMed] [Google Scholar]
- Bao L, Haas M, Kraus DM, Hack BK, Rakstang JK, Holers VM, Quigg RJ. Administration of a soluble recombinant complement C3 inhibitor protects against renal disease in MRL/lpr mice. Journal of American Society of Nephrology. 2003;14:670–679. doi: 10.1097/01.asn.0000051597.27127.a1. [DOI] [PubMed] [Google Scholar]
- Barlow PN, Baron M, Norman DG, Day AJ, Willis AC, Sim RB, Campbell ID. Secondary structure of a complement control protein module by two-dimensional 1H NMR. Biochemistry. 1991;30:997–1004. doi: 10.1021/bi00218a016. [DOI] [PubMed] [Google Scholar]
- Barlow PN, Norman DG, Steinkasserer A, Horne TJ, Pearce J, Driscoll PC, Sim RB, Campbell ID. Solution structure of the fifth repeat of factor H: a second example of the complement control protein module. Biochemistry. 1992;31:3626–3634. doi: 10.1021/bi00129a011. [DOI] [PubMed] [Google Scholar]
- Barlow PN, Steinkasserer A, Norman DG, Kieffer B, Wiles AP, Sim RB, Campbell ID. Solution structure of a pair of complement modules by nuclear magnetic resonance. Journal of Molecular Biology. 1993;232:268–284. doi: 10.1006/jmbi.1993.1381. [DOI] [PubMed] [Google Scholar]
- Blackmore TK, Sadlon TA, Ward HM, Lublin DM, Gordon DL. Identification of a heparin binding domain in the seventh short consensus repeat of complement factor H. Journal of Immunology. 1996;157:5422–5427. [PubMed] [Google Scholar]
- Blackmore TK, Hellwage J, Sadlon TA, Higgs N, Zipfel PF, Ward HM, Gordon DL. Identification of the second heparin-binding domain in human complement factor H. Journal of Immunology. 1998;160:3342–3348. [PubMed] [Google Scholar]
- Buddles MR, Donne RL, Richards A, Goodship J, Goodship TH. Complement factor H gene mutation associated with autosomal recessive atypical hemolytic uremic syndrome. American Journal of Human Genetics. 2000;66:1721–1722. doi: 10.1086/302877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Higman VA, Mulloy B, Perkins SJ, Lea SM, Sim RB, Day AJ. His-384 allotypic variant of factor H associated with age-related macular degeneration has different heparin binding properties from the non-disease-associated form. The Journal of Biological Chemistry. 2006;281:24713–24720. doi: 10.1074/jbc.M605083200. [DOI] [PubMed] [Google Scholar]
- Coffey PJ, Gias C, McDermott CJ, Lundh P, Pickering MC, Sethi C, Bird A, Fitzke FW, Maass A, Chen LL, et al. Complement factor H deficiency in aged mice causes retinal abnormalities and visual dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16651–16656. doi: 10.1073/pnas.0705079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, et al. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cordoba SR, de Jorge EG. Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H. Clinical and Experimental Immunology. 2008;151:1–13. doi: 10.1111/j.1365-2249.2007.03552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO, Ritter R, III, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Esparza-Gordillo J, Soria JM, Buil A, Almasy L, Blangero J, Fontcuberta J, Rodriguez de Cordoba S. Genetic and environmental factors influencing the human factor H plasma levels. Immunogenetics. 2004;56:77–82. doi: 10.1007/s00251-004-0660-7. [DOI] [PubMed] [Google Scholar]
- Estaller C, Koistinen V, Schwaeble W, Dierich MP, Weiss EH. Cloning of the 1.4-kb mRNA species of human complement factor H reveals a novel member of the short consensus repeat family related to the carboxy terminal of the classical 150-kDa molecule. Journal of Immunology. 1991;146:3190–3196. [PubMed] [Google Scholar]
- Farries TC, Seya T, Harrison RA, Atkinson JP. Competition for binding sites on C3b by CR1, CR2, MCP, factor B and Factor H. Complement and Inflammation. 1990;7:30–41. doi: 10.1159/000463124. [DOI] [PubMed] [Google Scholar]
- Fernando AN, Furtado PB, Clark SJ, Gilbert HE, Day AJ, Sim RB, Perkins SJ. Associative and structural properties of the region of complement factor H encompassing the Tyr402His disease-related polymorphism and its interactions with heparin. Journal of Molecular Biology. 2007;368:564–581. doi: 10.1016/j.jmb.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Ferreira VP, Herbert AP, Hocking HG, Barlow PN, Pangburn MK. Critical role of the C-terminal domains of factor H in regulating complement activation at cell surfaces. Journal of Immunology. 2006;177:6308–6316. doi: 10.4049/jimmunol.177.9.6308. [DOI] [PubMed] [Google Scholar]
- Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration – emerging pathogenetic and therapeutic concepts. Annals of Medicine. 2006;38:450–471. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A, Kirchhoff-Moradpour AH, Obieglo S, Brandis M, Kirschfink M, Zipfel PF, Goodship JA, Zimmerhackl LB. Successful (?) therapy of hemolytic-uremic syndrome with factor H abnormality. Pediatric Nephrology (Berlin, Germany) 2003;18:952–955. doi: 10.1007/s00467-003-1192-3. [DOI] [PubMed] [Google Scholar]
- Gershov D, Kim S, Brot N, Elkon KB. C-Reactive protein binds to apoptotic cells, protects the cells from assembly of the terminal complement components, and sustains an antiinflammatory innate immune response: implications for systemic autoimmunity. The Journal of Experimental Medicine. 2000;192:1353–1364. doi: 10.1084/jem.192.9.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis E, Jokiranta TS, Male DA, Ranganathan S, Ormsby RJ, Fischetti VA, Mold C, Gordon DL. A common site within factor H SCR 7 responsible for binding heparin, C-reactive protein and streptococcal M protein. European Journal of Immunology. 2003;33:962–969. doi: 10.1002/eji.200323541. [DOI] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nature Genetics. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin DM. Identification of complement regulatory domains in human factor H. Journal of Immunology. 1995;155:348–356. [PubMed] [Google Scholar]
- Hageman GS, Mullins RF, Russell SR, Johnson LV, Anderson DH. Vitronectin is a constituent of ocular drusen and the vitronectin gene is expressed in human retinal pigmented epithelial cells. FASEB J. 1999;13:477–484. doi: 10.1096/fasebj.13.3.477. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Progress in Retinal and Eye Research. 2001;20:705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Hancox LS, Taiber AJ, Gehrs KM, Anderson DH, Johnson LV, Radeke MJ, Kavanagh D, Richards A, Atkinson J, et al. Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications. Annals of Medicine. 2006;38:592–604. [PMC free article] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science (New York, NY) 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Hakobyan S, Harris CL, van den Berg C, Pepys MB, Morgan BP. Binding of factor H to C-reactive protein occurs only when the latter has undergone non-physiologic denaturation. Molecular Immunology. 2007;44:3983–3984. [Google Scholar]
- Hamilton G, Proitsi P, Williams J, O’Donovan M, Owen M, Powell J, Lovestone S. Complement factor H Y402H polymorphism is not associated with late-onset Alzheimer’s disease. Neuromolecular Medicine. 2007;9:331–334. doi: 10.1007/s12017-007-8013-y. [DOI] [PubMed] [Google Scholar]
- Hegasy GA, Manuelian T, Hogasen K, Jansen JH, Zipfel PF. The molecular basis for hereditary porcine membranoproliferative glomerulonephritis type II: point mutations in the factor H coding sequence block protein secretion. The American Journal of Pathology. 2002;161:2027–2034. doi: 10.1016/S0002-9440(10)64481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen S, Sanchez-Corral P, Jackson MS, Strain L, Goodship JA, Kemp EJ, Skerka C, Jokiranta TS, Meyers K, Wagner E, et al. De novo gene conversion in the RCA gene cluster (1q32) causes mutations in complement factor H associated with atypical hemolytic uremic syndrome. Human Mutation. 2006;27:292–293. doi: 10.1002/humu.9408. [DOI] [PubMed] [Google Scholar]
- Herbert AP, Uhrin D, Lyon M, Pangburn MK, Barlow PN. Disease-associated sequence variations congregate in a polyanion recognition patch on human factor H revealed in three-dimensional structure. The Journal of Biological Chemistry. 2006;281:16512–16520. doi: 10.1074/jbc.M513611200. [DOI] [PubMed] [Google Scholar]
- Herbert AP, Deakin JA, Schmidt CQ, Blaum BS, Egan C, Ferreira VP, Pangburn MK, Lyon M, Uhrin D, Barlow PN. Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism. The Journal of Biological Chemistry. 2007;282:18960–18968. doi: 10.1074/jbc.M609636200. [DOI] [PubMed] [Google Scholar]
- Holers VM. The complement system as a therapeutic target in autoimmunity. Clinical Immunology (Orlando, Fla) 2003;107:140–151. doi: 10.1016/s1521-6616(03)00034-2. [DOI] [PubMed] [Google Scholar]
- Holers VM, Girardi G, Mo L, Guthridge JM, Molina H, Pierangeli SS, Espinola R, Xiaowei LE, Mao D, Vialpando CG, et al. Complement C3 activation is required for antiphospholipid antibody-induced fetal loss. The Journal of Experimental Medicine. 2002;195:211–220. doi: 10.1084/jem.200116116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nature Genetics. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. American Journal of Human Genetics. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JH, Hogasen K, Harboe M, Hovig T. In situ complement activation in porcine membranoproliferative glomerulonephritis type II. Kidney International. 1998;53:331–349. doi: 10.1046/j.1523-1755.1998.00765.x. [DOI] [PubMed] [Google Scholar]
- Jarva H, Jokiranta TS, Hellwage J, Zipfel PF, Meri S. Regulation of complement activation by C-reactive protein: targeting the complement inhibitory activity of factor H by an interaction with short consensus repeat domains 7 and 8–11. Journal of Immunology. 1999;163:3957–3962. [PubMed] [Google Scholar]
- Johnson LV, Ozaki S, Staples MK, Erickson PA, Anderson DH. A potential role for immune complex pathogenesis in drusen formation. Experimental Eye Research. 2000;70:441–449. doi: 10.1006/exer.1999.0798. [DOI] [PubMed] [Google Scholar]
- Johnson LV, Leitner WP, Staples MK, Anderson DH. Complement activation and inflammatory processes in Drusen formation and age related macular degeneration. Experimental Eye Research. 2001;73:887–896. doi: 10.1006/exer.2001.1094. [DOI] [PubMed] [Google Scholar]
- Johnson PT, Betts KE, Radeke MJ, Hageman GS, Anderson DH, Johnson LV. Individuals homozygous for the age-related macular degeneration risk-conferring variant of complement factor H have elevated levels of CRP in the choroid. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17456–17461. doi: 10.1073/pnas.0606234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokiranta TS, Hellwage J, Koistinen V, Zipfel PF, Meri S. Each of the three binding sites on complement factor H interacts with a distinct site on C3b. The Journal of Biological Chemistry. 2000;275:27657–27662. doi: 10.1074/jbc.M002903200. [DOI] [PubMed] [Google Scholar]
- Jokiranta TS, Jaakola VP, Lehtinen MJ, Parepalo M, Meri S, Goldman A. Structure of complement factor H carboxyl-terminus reveals molecular basis of atypical haemolytic uremic syndrome. The EMBO Journal. 2006;25:1784–1794. doi: 10.1038/sj.emboj.7601052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvin MH, Kazatchkine MD, Cahour A, Bernard N. Lysine residues, but not carbohydrates, are required for the regulatory function of H on the amplification C3 convertase of complement. Journal of Immunology. 1984;133:3250–3254. [PubMed] [Google Scholar]
- Kardys I, Klaver CC, Despriet DD, Bergen AA, Uitterlinden AG, Hofman A, Oostra BA, Van Duijn CM, de Jong PT, Witteman JC. A common polymorphism in the complement factor H gene is associated with increased risk of myocardial infarction: the Rotterdam Study. Journal of the American College of Cardiology. 2006;47:1568–1575. doi: 10.1016/j.jacc.2005.11.076. [DOI] [PubMed] [Google Scholar]
- Kavanagh D, Richards A, Fremeaux-Bacchi V, Noris M, Goodship T, Remuzzi G, Atkinson JP. Screening for complement system abnormalities in patients with atypical hemolytic uremic syndrome. Clinical Journal of American Society of Nephrology. 2007;2:591–596. doi: 10.2215/CJN.03270906. [DOI] [PubMed] [Google Scholar]
- Kinoshita T. Biology of complement: the overture. Immunology today. 1991;12:291–295. doi: 10.1016/0167-5699(91)90001-A. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn S, Zipfel PF. Mapping of the domains required for decay acceleration activity of the human factor H-like protein 1 and factor H. European Journal of Immunology. 1996;26:2383–2387. doi: 10.1002/eji.1830261017. [DOI] [PubMed] [Google Scholar]
- Kuhn S, Skerka C, Zipfel PF. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H1. Journal of Immunology. 1995;155:5663–5670. [PubMed] [Google Scholar]
- Laine M, Jarva H, Seitsonen S, Haapasalo K, Lehtinen MJ, Lindeman N, Anderson DH, Johnson PT, Jarvela I, Jokiranta TS, et al. Y402H polymorphism of complement factor H affects binding affinity to C-reactive protein. Journal of Immunology. 2007;178:3831–3836. doi: 10.4049/jimmunol.178.6.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, Li Y, Liang L, Zareparsi S, Swaroop A, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nature Genetics. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht C, Weyersberg A, Heinen S, Stapenhorst L, Devenge J, Beck B, Waldherr R, Kirschfink M, Zipfel PF, Hoppe B. Successful plasma therapy for atypical hemolytic uremic syndrome caused by factor H deficiency owing to a novel mutation in the complement cofactor protein domain 15. American Journal of Kidney Disease. 2005;45:415–421. doi: 10.1053/j.ajkd.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Licht C, Heinen S, Jozsi M, Loschmann I, Saunders RE, Perkins SJ, Waldherr R, Skerka C, Kirschfink M, Hoppe B, et al. Deletion of Lys224 in regulatory domain 4 of Factor H reveals a novel pathomechanism for dense deposit disease (MPGN II) Kidney International. 2006;70:42–50. doi: 10.1038/sj.ki.5000269. [DOI] [PubMed] [Google Scholar]
- Lindahl G, Sjobring U, Johnsson E. Human complement regulators: a major target for pathogenic microorganisms. Current Opinion in Immunology. 2000;12:44–51. doi: 10.1016/s0952-7915(99)00049-7. [DOI] [PubMed] [Google Scholar]
- Liszewski MK, Atkinson JP. Novel complement inhibitors. Expert Opinion on Investigational Drugs. 1998;7:323–331. doi: 10.1517/13543784.7.3.323. [DOI] [PubMed] [Google Scholar]
- Male DA, Ormsby RJ, Ranganathan S, Giannakis E, Gordon DL. Complement factor H: sequence analysis of 221 kb of human genomic DNA containing the entire fH, fHR-1 and fHR-3 genes. Molecular Immunology. 2000;37:41–52. doi: 10.1016/s0161-5890(00)00024-9. [DOI] [PubMed] [Google Scholar]
- Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nature Genetics. 2007;39:1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. The American Journal of Pathology. 2007;171:715–727. doi: 10.2353/ajpath.2007.070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Hughes A, Patterson CC, Belton C, Kamaruddin MS, Horan PG, Kee F, McKeown PP. Genetic variants of complement factor H gene are not associated with premature coronary heart disease: a family-based study in the Irish population. BMC Medical Genetics. 2007;8:62. doi: 10.1186/1471-2350-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meri S, Pangburn MK. Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:3982–3986. doi: 10.1073/pnas.87.10.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi R, Huemer HP, Schwaeble W, Solder E, Larcher C, Dierich MP. Human complement factor H: an additional gene product of 43 kDa isolated from human plasma shows cofactor activity for the cleavage of the third component of complement. European Journal of Immunology. 1989;19:1765–1768. doi: 10.1002/eji.1830190936. [DOI] [PubMed] [Google Scholar]
- Mooijaart SP, Koeijvoets KM, Sijbrands EJ, Daha MR, Westendorp RG. Complement Factor H polymorphism Y402H associates with inflammation, visual acuity, and cardiovascular mortality in the elderly population at large. Experimental Gerontology. 2007;42:1116–1122. doi: 10.1016/j.exger.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14:835–846. [PubMed] [Google Scholar]
- Mullins RF, Aptsiauri N, Hageman GS. Structure and composition of drusen associated with glomerulonephritis: implications for the role of complement activation in drusen biogenesis. Eye (London, England) 2001;15:390–395. doi: 10.1038/eye.2001.142. [DOI] [PubMed] [Google Scholar]
- Nan R, Gor J, Perkins SJ. Implications of the progressive self-association of wild-type human factor H for complement regulation and disease. Journal of Molecular Biology. 2008;375:891–900. doi: 10.1016/j.jmb.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Nicaud V, Francomme C, Ruidavets JB, Luc G, Arveiler D, Kee F, Evans A, Morrison C, Blankenberg S, Cambien F, et al. Lack of association between complement factor H polymorphisms and coronary artery disease or myocardial infarction. Journal of Molecular Medicine. 2007;85:771–775. doi: 10.1007/s00109-007-0185-2. [DOI] [PubMed] [Google Scholar]
- Norman DG, Barlow PN, Baron M, Day AJ, Sim RB, Campbell ID. Three-dimensional structure of a complement control protein module in solution. Journal of Molecular Biology. 1991;219:717–725. doi: 10.1016/0022-2836(91)90666-t. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Umeda S, Obazawa M, Minami M, Noda T, Mizota A, Honda M, Tanaka M, Koyama R, Takagi I, et al. Complement factor H polymorphisms in Japanese population with age-related macular degeneration. Molecular Vision. 2006;12:156–158. [PubMed] [Google Scholar]
- Okemefuna AI, Gilbert HE, Griggs KM, Ormsby RJ, Gordon DL, Perkins SJ. The regulatory SCR-1/5 and cell surface-binding SCR-16/20 fragments of factor H reveal partially folded-back solution structures and different self-associative properties. Journal of Molecular Biology. 2008;375:80–101. doi: 10.1016/j.jmb.2007.09.026. [DOI] [PubMed] [Google Scholar]
- Oppermann M, Manuelian T, Jozsi M, Brandt E, Jokiranta TS, Heinen S, Meri S, Skerka C, Gotze O, Zipfel PF. The C-terminus of complement regulator Factor H mediates target recognition: evidence for a compact conformation of the native protein. Clinical and Experimental Immunology. 2006;144:342–352. doi: 10.1111/j.1365-2249.2006.03071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormsby RJ, Jokiranta TS, Duthy TG, Griggs KM, Sadlon TA, Giannakis E, Gordon DL. Localization of the third heparin-binding site in the human complement regulator factor H1. Molecular Immunology. 2006;43:1624–1632. doi: 10.1016/j.molimm.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Pai JK, Manson JE, Rexrode KM, Albert CM, Hunter DJ, Rimm EB. Complement factor H (Y402H) polymorphism and risk of coronary heart disease in US men and women. European Heart Journal. 2007;28:1297–1303. doi: 10.1093/eurheartj/ehm090. [DOI] [PubMed] [Google Scholar]
- Pangburn MK, Muller-Eberhard HJ. Kinetic and thermodynamic analysis of the control of C3b by the complement regulatory proteins factors H and I. Biochemistry. 1983;22:178–185. doi: 10.1021/bi00270a026. [DOI] [PubMed] [Google Scholar]
- Pangburn MK, Schreiber RD, Muller-Eberhard HJ. Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein beta1H for cleavage of C3b and C4b in solution. The Journal of Experimental Medicine. 1977;146:257–270. doi: 10.1084/jem.146.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangburn MK, Atkinson MA, Meri S. Localization of the heparin-binding site on complement factor H. The Journal of Biological Chemistry. 1991;266:16847–16853. [PubMed] [Google Scholar]
- Penfold PL, Madigan MC, Gillies MC, Provis JM. Immunological and aetiological aspects of macular degeneration. Progress in Retinal and Eye Research. 2001;20:385–414. doi: 10.1016/s1350-9462(00)00025-2. [DOI] [PubMed] [Google Scholar]
- Perkins SJ, Nealis AS, Sim RB. Oligomeric domain structure of human complement factor H by X-ray and neutron solution scattering. Biochemistry. 1991;30:2847–2857. doi: 10.1021/bi00225a017. [DOI] [PubMed] [Google Scholar]
- Perkins SJ, Gilbert HE, Aslam M, Hannan J, Holers VM, Goodship TH. Solution structures of complement components by X-ray and neutron scattering and analytical ultracentrifugation. Biochemical Society Transactions. 2002;30:996–1001. doi: 10.1042/bst0300996. [DOI] [PubMed] [Google Scholar]
- Pickering MC, Cook HT. Translational mini-review series on complement factor H: renal diseases associated with complement factor H: novel insights from humans and animals. Clinical and Experimental Immunology. 2008;151:210–230. doi: 10.1111/j.1365-2249.2007.03574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MC, Cook HT, Warren J, Bygrave AE, Moss J, Walport MJ, Botto M. Uncontrolled C3 activation causes membranoproliferative glomerulonephritis in mice deficient in complement factor H. Nature Genetics. 2002;31:424–428. doi: 10.1038/ng912. [DOI] [PubMed] [Google Scholar]
- Pickering MC, de Jorge EG, Martinez-Barricarte R, Recalde S, Garcia-Layana A, Rose KL, Moss J, Walport MJ, Cook HT, de Cordoba SR, et al. Spontaneous hemolytic uremic syndrome triggered by complement factor H lacking surface recognition domains. The Journal of Experimental Medicine. 2007;204:1249–1256. doi: 10.1084/jem.20070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser BE, Johnson S, Roversi P, Clark SJ, Tarelli E, Sim RB, Day AJ, Lea SM. Expression, purification, cocrystallization and preliminary crystallographic analysis of sucrose octasulfate/human complement regulator factor H SCRs 6–8. Acta Crystallographica. 2007a;63:480–483. doi: 10.1107/S1744309107020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser BE, Johnson S, Roversi P, Herbert AP, Blaum BS, Tyrrell J, Jowitt TA, Clark SJ, Tarelli E, Uhrin D, et al. Structural basis for complement factor H linked age-related macular degeneration. The Journal of Experimental Medicine. 2007b;204:2277–2283. doi: 10.1084/jem.20071069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido JS, McConnell JP, Lennon RJ, Bryant SC, Peterson LM, Berger PB, Somers V, Highsmith WE. Relationship between age-related macular degeneration-associated variants of complement factor H and LOC387715 with coronary artery disease. Mayo Clinic Proceedings. 2007;82:301–307. doi: 10.4065/82.3.301. [DOI] [PubMed] [Google Scholar]
- Richards A, Buddles MR, Donne RL, Kaplan BS, Kirk E, Venning MC, Tielemans CL, Goodship JA, Goodship TH. Factor H mutations in hemolytic uremic syndrome cluster in exons 18–20, a domain important for host cell recognition. American Journal of Human Genetics. 2001;68:485–490. doi: 10.1086/318203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Lambris JD. Complement-targeted therapeutics. Nature Biotechnology. 2007;25:1265–1275. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoche J, Day AJ, Willis AC, Belt KT, Campbell RD, Sim RB. Partial characterization of human complement factor H by protein and cDNA sequencing: homology with other complement and non-complement proteins. Bioscience Reports. 1986;6:65–72. doi: 10.1007/BF01145180. [DOI] [PubMed] [Google Scholar]
- Ripoche J, Erdei A, Gilbert D, Al Salihi A, Sim RB, Fontaine M. Two populations of complement factor H differ in their ability to bind to cell surfaces. The Biochemical Journal. 1988;253:475–480. doi: 10.1042/bj2530475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, Meitinger T, Weber BH. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Human Molecular Genetics. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- Rodriguez de Cordoba S, Lublin DM, Rubinstein P, Atkinson JP. Human genes for three complement components that regulate the activation of C3 are tightly linked. The Journal of Experimental Medicine. 1985;161:1189–1195. doi: 10.1084/jem.161.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother RP, Rollins SA, Mojcik CF, Brodsky RA, Bell L. Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nature Biotechnology. 2007;25:1256–1264. doi: 10.1038/nbt1344. [DOI] [PubMed] [Google Scholar]
- Ryan US. Complement inhibitory therapeutics and xenotransplantation. Nature Medicine. 1995;1:967–968. doi: 10.1038/nm0995-967. [DOI] [PubMed] [Google Scholar]
- Sahu A, Lambris JD. Complement inhibitors: a resurgent concept in anti-inflammatory therapeutics. Immunopharmacology. 2000;49:133–148. doi: 10.1016/s0162-3109(00)80299-4. [DOI] [PubMed] [Google Scholar]
- Schlaf G, Demberg T, Beisel N, Schieferdecker HL, Gotze O. Expression and regulation of complement factors H and I in rat and human cells: some critical notes. Molecular Immunology. 2001;38:231–239. doi: 10.1016/s0161-5890(01)00045-1. [DOI] [PubMed] [Google Scholar]
- Schmidt CQ, Herbert AP, Hocking HG, Uhrin D, Barlow PN. Translational mini-review series on complement factor H: structural and functional correlations for factor H. Clinical and Experimental Immunology. 2008;151:14–24. doi: 10.1111/j.1365-2249.2007.03553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaeble W, Zwirner J, Schulz TF, Linke RP, Dierich MP, Weiss EH. Human complement factor H: expression of an additional truncated gene product of 43 kDa in human liver. European Journal of Immunology. 1987;17:1485–1489. doi: 10.1002/eji.1830171015. [DOI] [PubMed] [Google Scholar]
- Seddon JM, George S, Rosner B, Klein ML. CFH gene variant, Y402H, and smoking, body mass index, environmental associations with advanced age-related macular degeneration. Human Heredity. 2006;61:157–165. doi: 10.1159/000094141. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Pangburn MK. Identification of three physically and functionally distinct binding sites for C3b in human complement factor H by deletion mutagenesis. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10996–11001. doi: 10.1073/pnas.93.20.10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprasad S, Chong NV, Bailey TA. Serum elastin-derived peptides in age-related macular degeneration. Investigative Ophthalmology and Visual Science. 2005;46:3046–3051. doi: 10.1167/iovs.04-1277. [DOI] [PubMed] [Google Scholar]
- Sjoberg AP, Trouw LA, Clark SJ, Sjolander J, Heinegard D, Sim RB, Day AJ, Blom AM. The factor H variant associated with age-related macular degeneration (His-384) and the non-disease-associated form bind differentially to C-reactive protein, fibromodulin, DNA, and necrotic cells. The Journal of Biological Chemistry. 2007;282:10894–10900. doi: 10.1074/jbc.M610256200. [DOI] [PubMed] [Google Scholar]
- Skerka C, Lauer N, Weinberger AA, Keilhauer CN, Suhnel J, Smith R, Schlotzer-Schrehardt U, Fritsche L, Heinen S, Hartmann A, et al. Defective complement control of factor H (Y402H) and FHL-1 in age-related macular degeneration. Molecular Immunology. 2007;44:3398–3406. doi: 10.1016/j.molimm.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Soares DC, Gerloff DL, Syme NR, Coulson AF, Parkinson J, Barlow PN. Large-scale modelling as a route to multiple surface comparisons of the CCP module family. Protein Engineering Design and Selection. 2005;18:379–388. doi: 10.1093/protein/gzi039. [DOI] [PubMed] [Google Scholar]
- Stark K, Neureuther K, Sedlacek K, Hengstenberg W, Fischer M, Baessler A, Wiedmann S, Jeron A, Holmer S, Erdmann J, et al. The common Y402H variant in complement factor H gene is not associated with susceptibility to myocardial infarction and its related risk factors. Clinical Science (Lond) 2007;113:213–218. doi: 10.1042/CS20070028. [DOI] [PubMed] [Google Scholar]
- Stratton JD, Warwicker P. Successful treatment of factor H-related haemolytic uraemic syndrome. Nephrology Dialysis Transplantation. 2002;17:684–685. doi: 10.1093/ndt/17.4.684. [DOI] [PubMed] [Google Scholar]
- Thurman JM. Triggers of inflammation after renal ischemia/reperfusion. Clinical Immunology (Orlando, Fla) 2007;123:7–13. doi: 10.1016/j.clim.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman JM, Holers VM. The central role of the alternative complement pathway in human disease. Journal of Immunology. 2006;176:1305–1310. doi: 10.4049/jimmunol.176.3.1305. [DOI] [PubMed] [Google Scholar]
- Topol EJ, Smith J, Plow EF, Wang QK. Genetic susceptibility to myocardial infarction and coronary artery disease. Human Molecular Genetics. 2006;15(Spec No 2):R117–R123. doi: 10.1093/hmg/ddl183. [DOI] [PubMed] [Google Scholar]
- Turnbull J, Powell A, Guimond S. Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends in Cell Bbiology. 2001;11:75–82. doi: 10.1016/s0962-8924(00)01897-3. [DOI] [PubMed] [Google Scholar]
- Verdugo ME, Ray J. Age-related increase in activity of specific lysosomal enzymes in the human retinal pigment epithelium. Experimental Eye Research. 1997;65:231–240. doi: 10.1006/exer.1997.0325. [DOI] [PubMed] [Google Scholar]
- Warwicker P, Goodship TH, Donne RL, Pirson Y, Nicholls A, Ward RM, Turnpenny P, Goodship JA. Genetic studies into inherited and sporadic hemolytic uremic syndrome. Kidney International. 1998;53:836–844. doi: 10.1111/j.1523-1755.1998.00824.x. [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai HJ, Mah TS, Schmidt S, Postel EA, Agarwal A, Haines JL, Pericak-Vance MA, Rosenfeld PJ, et al. Age-related maculopathy: a genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. American Journal of Human Genetics. 2004;75:174–189. doi: 10.1086/422476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, et al. Complement C3 variant and the risk of age-related macular degeneration. The New England Journal of Medicine. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- Yu J, Wiita P, Kawaguchi R, Honda J, Jorgensen A, Zhang K, Fischetti VA, Sun H. Biochemical analysis of a common human polymorphism associated with age-related macular degeneration. Biochemistry. 2007;46:8451–8461. doi: 10.1021/bi700459a. [DOI] [PubMed] [Google Scholar]
- Zarbin MA. Current concepts in the pathogenesis of age-related macular degeneration. Archives of Ophthalmology. 2004;122:598–614. doi: 10.1001/archopht.122.4.598. [DOI] [PubMed] [Google Scholar]
- Zetterberg M, Tasa G, Palmer MS, Juronen E, Teesalu P, Blennow K, Zetterberg H. Apolipoprotein E polymorphisms in patients with primary open-angle glaucoma. American Journal of Ophthalmology. 2007;143:1059–1060. doi: 10.1016/j.ajo.2007.01.031. [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Skerka C. Complement factor H and related proteins: an expanding family of complement-regulatory proteins? Immunology Today. 1994;15:121–126. doi: 10.1016/0167-5699(94)90155-4. [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Jokiranta TS, Hellwage J, Koistinen V, Meri S. The factor H protein family. Immunopharmacology. 1999;42:53–60. doi: 10.1016/s0162-3109(99)00015-6. [DOI] [PubMed] [Google Scholar]