Abstract
Deletions of chromosome 1p36 affect approximately 1 in 5,000 newborns and are associated with developmental delay, intellectual disability, and defects involving the brain, eye, ear, heart, and kidney. Arginine-glutamic acid dipeptide repeats (RERE) is located in the proximal 1p36 critical region. RERE is a widely-expressed nuclear receptor coregulator that positively regulates retinoic acid signaling. Animal models suggest that RERE deficiency might contribute to many of the structural and developmental birth defects and medical problems seen in individuals with 1p36 deletion syndrome, although human evidence supporting this role has been lacking. In this report, we describe ten individuals with intellectual disability, developmental delay, and/or autism spectrum disorder who carry rare and putatively damaging changes in RERE. In all cases in which both parental DNA samples were available, these changes were found to be de novo. Associated features that were recurrently seen in these individuals included hypotonia, seizures, behavioral problems, structural CNS anomalies, ophthalmologic anomalies, congenital heart defects, and genitourinary abnormalities. The spectrum of defects documented in these individuals is similar to that of a cohort of 31 individuals with isolated 1p36 deletions that include RERE and are recapitulated in RERE-deficient zebrafish and mice. Taken together, our findings suggest that mutations in RERE cause a genetic syndrome and that haploinsufficiency of RERE might be sufficient to cause many of the phenotypes associated with proximal 1p36 deletions.
Main Text
Deletions of chromosome 1p36 constitute the most common group of terminal deletions in humans—affecting approximately 1 in 5,000 newborns—and are recognized as an important cause of intellectual disability.1, 2, 3 Additional phenotypes that characterize the 1p36 deletion syndrome (MIM: 607872) include developmental delay, seizures, brain anomalies, vision problems, hearing loss, orofacial clefting, congenital heart defects, cardiomyopathy, renal anomalies, short stature, and a recognizable constellation of dysmorphic features—microbrachycephaly; large, late-closing anterior fontanel; posteriorly rotated, low-set, abnormally shaped ears; straight eyebrows; deep-set eyes; epicanthal folds; midface hypoplasia; a wide and depressed nasal bridge; a long philtrum; and a pointed chin.4, 5, 6 By analyzing the clinical presentations of individuals with terminal 1p36 deletions, Wu et al. determined that haploinsufficiency of genes located distal to marker D1S2870 (chr1:6,289,764–6,289,973; hg19) was sufficient to cause most of these phenotypes.7 This region has since been termed the distal or classical 1p36 critical region.8
Subsequently, Kang et al. delineated a non-overlapping proximal 1p36 critical region (chr1:8,395,179–11,362,893; hg19) which, when deleted, was associated with a similar constellation of phenotypes—intellectual disability, developmental delay, vision problems, hearing loss, orofacial clefting, congenital heart defects, cardiomyopathy, and short stature.9 However, these individuals had distinctive facial features—frontal and parietal bossing, low-set, posteriorly-rotated ears, epicanthal folds, anteverted nares, broad eyebrows, and hirsutism.
Animal models have implicated several genes located in the distal and proximal 1p36 critical regions as potential contributors to the neurodevelopmental and structural birth defects associated with the 1p36 deletion syndrome.8 However, human evidence supporting the role of these genes in the development of specific phenotypes is often lacking. Uncertainty regarding the clinical effects caused by haploinsufficiency of specific genes makes it difficult for physicians to provide prognostic information to families and to generate individualized care plans for their patients solely on the basis of the location and extent of their 1p36 deletions.
Arginine-glutamic acid dipeptide repeats (RERE [MIM: 605226]) is located in the proximal 1p36 critical region.8, 9 RERE encodes a widely-expressed nuclear receptor coregulator that positively regulates retinoic acid signaling.10, 11, 12 Data available from the Exome Aggregation Consortium (ExAC) Browser suggests that RERE has a high probability of loss-of-function intolerance (pLI = 1.0) given that 47.5 loss-of-function variants were expected based on gene size and GC content but only three were observed. Data from zebrafish and mouse models suggest that haploinsufficiency of RERE might contribute to the intellectual disability, developmental delay, structural brain anomalies, vision problems, hearing loss, congenital heart defects, cardiomyopathy, and renal anomalies seen in individuals with 1p36 deletions.13, 14, 15, 16, 17 However, the exact role that RERE deficiency plays in 1p36 deletion syndrome, and more generally in human disease, remains unclear.
Here, we describe ten individuals with neurodevelopmental phenotypes and congenital anomalies who carry rare, putatively deleterious, sequence changes in RERE. Clinical data were obtained through written informed consent. In all cases, the procedures followed were in accordance with the ethical standards of the respective institution’s committee on human research and were in keeping with national standards. RERE sequence changes were identified in nine of these individuals (subjects 1–9) by clinically-based exome sequencing performed in CLIA- or ISO15189-certified laboratories and confirmed by Sanger sequencing. Changes in subject 10 were identified by the Simons Foundation Autism Research Initiative (SFARI) and reported in SFARI Base—a central database of clinical and genetic information about families affected by autism and other neurodevelopmental disorders. In all cases where parental DNA samples were available, these changes were found to be de novo. The clinical phenotypes of these individuals and a description of their RERE sequence changes—including in silico prediction of effects on protein function via PolyPhen-2, SIFT, and MutationTaster and allele frequencies in control populations reported in the NHLBI Exome Variant Server and the ExAC Browser—are summarized below and in Table 1, Tables S1 and S2, Figure 1, and the Supplemental Note.
Table 1.
RERE Sequence Changes Identified in Subjects 1–10
| Subject No. | Sequence Changes in REREa | PolyPhen-2 | SIFT | MutationTaster | Alleles in Exome Variant Server | Alleles in ExAC Browser |
|---|---|---|---|---|---|---|
| 1 | c.3466 G>A, p.Gly1156Arg | probably damaging | damaging | disease causing | none | 2/66,082 European; 2/120,478 totalb |
| 2 | c.4313_4318dupTCCACC, p.Leu1438_His1439dup | NA | NA | polymorphism | none | none |
| 3 | c.3785C>G, p.Pro1262Arg | probably damaging | damaging | disease causing | none | none |
| 4 | c.4293C>G, p.His1431Gln | probably damaging | damaging | disease causing | none | none |
| 5 | c.4293C>A, p.His1431Gln | probably damaging | damaging | disease causing | none | none |
| 6 | c.3122delC, p.Pro1041Lysfs∗40 | NA | NA | disease causing | none | none |
| 7 | c.1411G>A, p.Val471Ile | possibly damaging | tolerated | disease causing | 1, African American | none |
| 8 | c.1104delA, p.Leu369Cysfs∗16 | NA | NA | NAc | none | none |
| 9 | c.2249_2270dup p.Thr758Serfs∗36 | NA | NA | disease causing | none | none |
| 10d | c.2278 C>T, p.Gln760∗ | NA | NA | disease causing | none | none |
NA, not applicable.
Based on RERE transcript variant 1 [RefSeq: NM_012102.3].
Alleles found only in the heterozygous state.
Change affects the last nucleotide of an exon.
Subject 10 was previously reported by Krumm et al. as subject 11654.p1.18
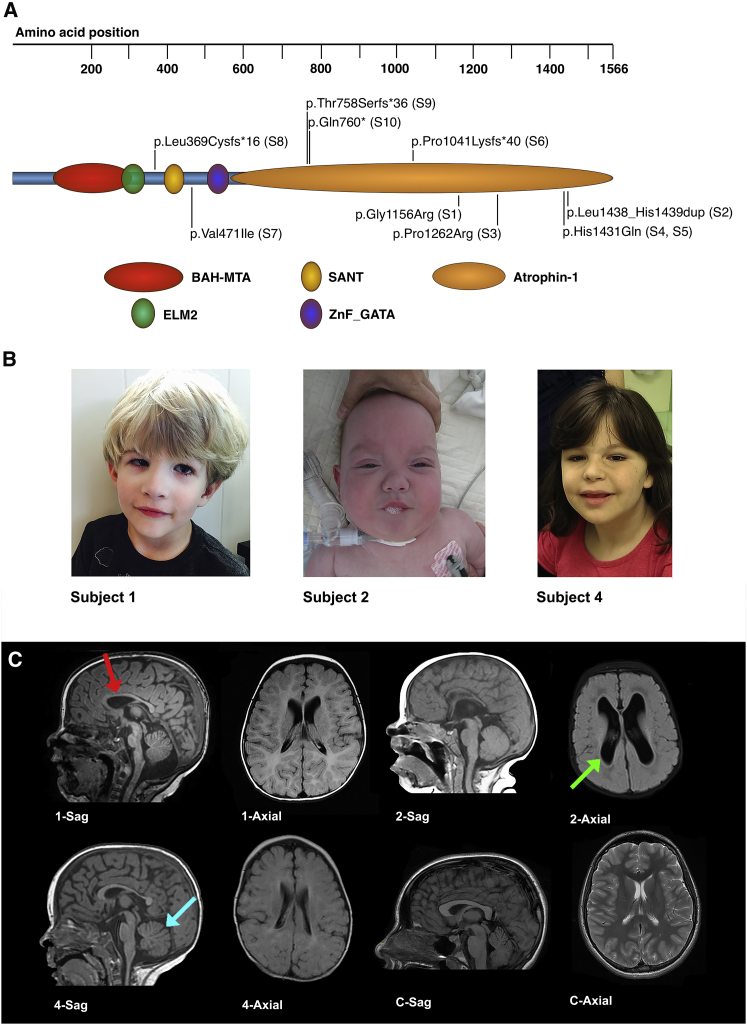
Figure 1.
Molecular Changes and Selected Clinical Findings for Individuals with Putatively Deleterious Changes in RERE
(A) The predicted locations of domains within RERE are presented along with the locations of the RERE changes seen in subjects 1–10.
(B) Craniofacial changes noted in subjects. Subject 1, at 3 years of age, presents with bilateral optic colobomas, unilateral micropthalmia, low-set ears, and micrognathia. Subject 2, at 11 months of age, presents with a unilateral iris coloboma and simple ears. Subject 4, at 9 years of age, presents with deep-set eyes, epicanthal folds, and abnormal ears.
(C) Sagital (Sag) and axial brain MRI scans of subjects 1, 2, and 4 demonstrating characteristic findings. Subject 1, at 7 months of age, had a thin corpus callosum (red arrow), a small anterior vermis, a small pons with a ventral cleft at the ponto-medullary junction, delayed myelination, and severely decreased white matter volume. Subject 2, at 4 months of age, had a thin corpus callosum, ventriculomegaly (green arrow), incompletely folded hippocampi, and severely diminished white matter volume. Subject 4, at 1 year and 2 months of age, had a thin corpus callosum, a diminished cerebellar vermis with a deep fissures (blue arrow), and significantly diminished white matter volume. For comparison, we have shown a mid-sagittal T1-weighted image showing a normal sized corpus callosum and intact cerebellar vermis and an axial T2-weighted image showing normal sized ventricles without increased extra-axial space.
Subject 1 is a 3-year-old male of European descent who carries a de novo c.3466G>A, p.Gly1156Arg missense change in RERE (GenBank: NM_012102.3). He was noted to have intrauterine growth retardation, was born prematurely at 35 weeks of gestation, and was small for gestation age with a birth weight of 1.64 kg (<3rd percentile). Multiple regions of infarction were found in the placenta. His clinical features include global developmental delay, mild spastic quadriparesis, dysarthric speech, swallowing difficulties, bilateral optic colobomas, optic nerve hypoplasia, unilateral microphthalmia, anisometropia, mild sensorineural hearing loss, micrognathia, a ventricular septal defect (VSD), a patent foramen ovale, hypospadias with penile chordee, and gastresophageal reflux disease (GERD). A brain MRI obtained at 7 months of age revealed a thin corpus callosum, small anterior vermis, small pons with a ventral cleft at the ponto-medullary junction, delayed myelination, and severely decreased white matter volume (Figure 1).
Subject 2 is a 15-month-old male of European descent who carries a de novo c.4313_4318dupTCCACC, p.Leu1438_His1439dup change in RERE. This change results in the addition of a leucine and histidine to an already histidine-rich compositional bias region of RERE (UniProt: Q9P2R6). This type of change is not amenable to evaluation by PolyPhen-2 or SIFT, and MutationTaster predicts this change to be a polymorphism. However, this duplication—like the amino acid change in subject 1—is found in a conserved region of the atrophin domain of RERE. He was also found to have a de novo c.4211G>A, p.Arg1404His (GenBank: NM_001252100.1) change in kinesin family member 21B (KIF21B [MIM: 608322]) and a de novo c.1433C>T, p.Pro478Leu (GenBank: NM_170744.4) change in unc-5 netrin receptor B (UNC5B [MIM: 607870]). Both of these changes were predicted to be probably damaging by PolyPhen-2, damaging by SIFT, and disease causing by MutationTaster. KIF21B encodes a member of the kinesin family of proteins that acts as a plus-end-directed microtubule motor, participating in transport of cellular cargo in neurons as well as in other cell types.19 Variants in KIF21B have been associated with multiple sclerosis and ankylosing spondylitis in genome-wide association studies.20, 21 The UNC5 family of proteins plays a critical role in a number of cellular processes, including axonal guidance, angiogenesis, and apoptosis. However, mutations in UNC5B have not been associated with a genetic disorder in humans.22
Subject 2 was born prematurely at 30 weeks and 6 days of gestation via C-section for symmetric intrauterine growth retardation and fetal distress. His birth weight was 0.94 kg (∼3rd percentile). His clinical features include developmental delay, post-natal growth retardation, short stature with a height of 64 cm at 15 months (<1st percentile), microcephaly with an occipital frontal circumference (OFC) of 42.5 cm at 15 months (<1st percentile), a unilateral iris coloboma, choanal atresia, a VSD, a patent ductus arteriosus, unilateral multicystic kidney, and cryptorchidism. A brain MRI obtained at 4 months revealed a thin corpus callosum, ventriculomegaly, incompletely folded hippocampi, and severely diminished white matter volume. This subject was initially presumed to have CHARGE syndrome. However, CHD7 sequencing and deletion and duplication analysis revealed no pathologic variants.
Subject 3 is a 2-year-old Hispanic male who carries a de novo c.3785C>G, p.Pro1262Arg missense change in RERE that was found to be probably damaging by PolyPhen-2, damaging by SIFT, and disease causing by MutationTaster. Pregnancy was complicated by polyhydramnios secondary to duodenal atresia. His other clinical features include post-natal growth retardation with a weight of 10.3 kb (1st percentile), a height of 82 cm (2nd percentile), and an OFC of 46.4 cm (4th percentile) at 2 years and 4 months of age, developmental delay, hypotonia, staring spells, dysmorphic features, unilateral Peter anomaly, bilateral iris abnormalities, a VSD, bilateral hydronephrosis with grade 2–4 vesicoureteral reflux, an annular pancreas, and syndactyly. A brain MRI obtained at 1 year and 6 months showed a thin corpus callosum, deep primary fissure of the cerebellar vermis, ventriculomegaly, and severely diminished white matter volume.
Subject 4 is a 9-year-old female of European descent who carries a de novo c.4293C>G, p.His1431Gln missense change in RERE that was found to be probably damaging by PolyPhen-2, damaging by SIFT, and disease causing by MutationTaster. She was also found to be mosaic for a de novo c.2464G>A, p.Gly822Ser (GenBank: NM_021007.2) change in sodium channel, voltage-gated, type II alpha subunit (SCN2A [MIM: 182390]). This variant is present in 14/95 reads for 14.7% mosaicism with a 95% confidence interval of 8.3%–23.5%. This SCN2A change is predicted to be probably damaging by PolyPhen-2, damaging by SIFT, and disease causing by MutationTaster. Mutations in SCN2A have been implicated in two autosomal-dominant disorders: epileptic encephalopathy, early infantile, 11 (MIM: 613721) and seizures, benign familial infantile, 3 (MIM: 607745). Her clinical features included hypotonia, moderate intellectual disability, severe behavioral issues and seizures, dysmorphic features, GERD, developmental hip dysplasia, and severe vesicoureteral reflux requiring surgery. A brain MRI obtained at 1 year and 2 months showed a thin corpus callosum, a diminished cerebellar vermis with deep fissures, and significantly diminished white matter volume—findings not reported in prior studies of SCN2A.23, 24
Subject 5 is a 12-year, 8-month-old Dutch male (reported previously by Bosch et al. as patient 22) with a de novo c.4293C>A change in RERE that results in the same amino acid change, p.His1431Gln, caused by the c.4293C>G change documented in subject 4.25 His clinical features include severe developmental delay, seizures, feeding problems, blepharophimosis, strabismus, bilateral optic atrophy, cortical visual impairment, dysmorphic features, GERD, and pyloric hypertrophy requiring surgical intervention. A brain MRI obtained at 3 years and 6 months of age showed an abnormal corpus callosum with blunting of the rostrum, mild to moderate ventriculomegaly, a small cerebellar vermis, and globally diminished white matter volume.
Subject 6 is a 6-year-old adopted female of mixed European and Hispanic descent with a c.3122delC, p.Pro1041Lysfs∗40 frameshift change in RERE. Parental DNA is not available. Given that this change occurs in the 17th of 21 RERE coding exons, it might trigger nonsense-mediated mRNA decay. Pregnancy and birth history are limited, but there was reported alcohol use during the pregnancy. Her family history is positive for intellectual disability and mental illness in her biological mother and intellectual disability in both of her maternal grandparents and a maternal half-uncle. Her clinical features include developmental delay, hypotonia, behavior problems, dysmorphic features, lumbar lordosis, and macrocephaly with an OFC of 54.1 cm (>98th percentile).
Subject 7 is an 11-year-old Dutch male who carries a de novo c.1411G>A, p.Val471Ile missense change in RERE that was estimated to be possibly damaging by PolyPhen-2 and disease causing by MutationTaster. He was also found to have a compound heterozygous pair of mutations in fibrous sheath interacting protein 2 (FSIP2 [MIM: 615796]): c.2602del, p.Ser868Hfs∗12 and c.4583C>G, p.Thr1528Arg (GenBank: NM_173651.2). The c.2602del deletion is inherited from the father, and the c.4583C>G missense mutation is inherited from the mother and is a previously reported variant, rs192957612. This gene encodes for a protein localized to the sperm flagellum and is not associated with any known human disease.26 The subject was born prematurely at 34 weeks of gestation and weighed 2.24 kg (∼5th percentile). This twin pregnancy was complicated by maternal hypertension for which the mother used medication. The subject has an older sister who is healthy, but his twin sister was recently diagnosed with KBG syndrome caused by a de novo c.3382_3383delGA, p.Asp1128Glnfs∗41 (GenBank: NM_001256182.1) change in ankyrin repeat domain-containing protein 11 (ANKRD11 [MIM: 611192]), which was not found in this individual.27 His clinical features include developmental delay, hypotonia, ADHD, mild intellectual disability, growth retardation, dysmorphic features, and a cleft lip. A brain MRI obtained at 7 years, 9 months of age showed an abnormal corpus callosum with blunting of the rostrum, a small anterior commissure, and diminished white matter volume.
Subject 8 is a 10-year-old male of European descent who carries a c.1104delA, p.Leu369Cysfs∗16 change in RERE. This variant was not present in his mother, but a paternal DNA sample could not be obtained for testing. Because this change occurs in the last nucleotide of the 9th of 21 RERE coding exons, it might trigger nonsense-mediated mRNA decay. Pregnancy was complicated by anemia, a vaginal infection, and maternal smoking and alcohol consumption until eight weeks of gestation, when the pregnancy was recognized. He was born prematurely at 36 weeks of gestation. His clinical features included fine motor and speech delay, autism spectrum disorder, and dysmorphic features. A brain MRI ordered for developmental delay was normal.
Subject 9 is a 7-year-old Dutch male who carries a de novo c.2249_2270dup, p.Thr758Serfs∗36 frameshift change in RERE. This mutation was predicted to be disease causing by MutationTaster. Pertinent findings in his family history include a younger sister with autism, a paternal uncle who died of Duchenne muscular dystrophy, and a second paternal uncle who died of acute heart failure at 20 years of age. Clinical features include severe speech and language delay, intellectual disability, autism, dysmorphic features, and a VSD. He has macrocephaly and an OFC of 57.3 cm (>99th percentile) at 6 years, 10 months of age. However, this might be familial given that his mother’s OFC is 59.7 cm (+ 2.5 SD).
While investigating the genetic causes of autism spectrum disorder in a large cohort, Krumm et al. identified a 14-year-old non-Hispanic female (subject 11654.p1) with autism spectrum disorder who carried a de novo c.2278C>T, p.Gln760∗ change in RERE.18 This mutation was estimated to be disease causing by MutationTaster. Here, we refer to her as subject 10. Her clinical features include autism spectrum disorder, sleep apnea, and nocturnal enuresis.
All of the subjects presented here have neurodevelopmental disorders—intellectual disability, developmental delay, and/or autism (Table 2, Table S2). Recurrent neurological abnormalities reported in these subjects include seizures, hypotonia, behavioral problems, ADHD, and problems with feeding and swallowing. Brain anomalies were documented in 88% (7/8) of subjects who had an MRI and included abnormalities and/or thinning of the corpus callosum (6/8, or 75%), diminished white matter volume (6/8, or 75%), abnormal cerebellar vermis (4/8, or 50%), and ventriculomegaly (3/8, or 38%). Structural eye defects were seen in 40% (4/10) of subjects and included coloboma, optic atrophy and/or hypoplasia, microphthalmia, Peter anomaly, iris anomalies, and blepharophimosis. Congenital heart defects were seen in 40% (4/10) of subjects, and VSDs were the most common defect. Genitourinary defects were seen in 50% (5/10), and vesicoureteral reflux was documented in 30% (3/10) of subjects.
Table 2.
Overlapping Features Seen in Our Subjects, Individuals with Isolated 1p36 Deletions that Include RERE, and Zebrafish and Mice Models
| Phenotype | Subjects 1–10 Documented with Phenotype (% and No.) | Individuals with Isolated 1p36 Deletions Involving RERE Documented with Phenotype (% and No.) | Related Phenotype Documented in Rerea-Deficient Zebrafish?13, 17 | Related Phenotype Documented in RERE-Deficient Mice?14, 15 |
|---|---|---|---|---|
| Neurologic problems | 100% (10/10) | 97% (30/31) | no | no |
| CNS anomalies | 88% (7/8) | 29% (9/31) | no | yes |
| Ophthalmologic abnormalities | 40% (4/10) | 42% (13/31) | yes | yes |
| Hearing loss | 10% (1/10) | 26% (8/31) | yes | yes |
| Orofacial clefting | 10% (1/10) | 16% (5/31) | no | no |
| Congenital heart defects | 40% (4/10) | 71% (22/31) | no | yes |
| Genitourinary anomalies | 50% (5/10) | 13% (4/31) | no | yes |
In addition to these examples of extra-neuronal organ involvement, a variety of dysmorphic features were also observed in our subjects (Table S2), including some features previously associated with deletions of the proximal 1p36 critical region, such as frontal bossing, low-set, posteriorly rotated ears, epicanthic folds, anteverted nares, and broad eyebrows (Figure 1).9 Our cohort also demonstrates a variety of growth patterns—some subjects have short stature (3/9, or 33%) or microcephaly (2/9, or 22%), whereas others have tall stature (2/9, or 22%) or macrocephaly (2/9, or 22%). This suggests that physicians should not rely heavily on the identification of a characteristic constellation of physical features but rather on a grouping of medical conditions, such as brain, eye, and cardiac developmental defects, when considering the possible diagnosis of an RERE-related disorder.
To determine the subset of 1p36 deletion phenotypes that could possibly be explained by RERE haploinsufficiency, we compared the spectrum of structural birth defects seen in our subjects to a cohort of 31 individuals with isolated 1p36 deletions involving RERE (Table 2, Table S1, Table S3, and Figure S1). These individuals were either previously described in the literature or provided informed consent and were enrolled in an institutional-review-board-approved research study.9, 16, 28, 29, 30, 31, 32, 33 In all cases, the procedures followed were in accordance with the ethical standards of the institution’s committee on human research and were in keeping with national standards. Within this cohort, 24 individuals carried larger terminal or interstitial deletions that included all or part of both the distal and proximal 1p36 deletion critical regions, and seven individuals carried interstitial 1p36 deletions that did not include the distal critical region (Figure S1). We found that the majority of recurrent neurologic, ophthalmologic, cardiac, and gastrointestinal phenotypes seen our subjects were also seen in individuals in the 1p36 deletion cohort. Overlap was also seen for sensorineural hearing loss, cleft lip, duodenal atresia, cryptorchidism, hip dysplasia, and scoliosis, all of which were identified in only one of our subjects. This overlap suggests that haploinsufficiency of RERE might also be sufficient to cause these additional phenotypes.
The phenotypic overlap between Rerea-deficient zebrafish and RERE-deficient mice and our subjects provides further evidence that the deleterious sequence changes observed in RERE are the cause of many of the phenotypes observed in our subjects. This overlap also suggests that these animals represent useful models in which to study RERE’s role in the development of many organ systems (Table 2). Specifically, we note that zebrafish carrying homozygous mutations in rerea—the zebrafish homolog of RERE—have microphthalmia, inconsistent startle response, and decreased microphonic potentials.13, 17 This provides further evidence that changes in RERE are likely to be responsible for the eye anomalies and sensorineural hearing loss seen in our subjects.
Mice that are compound heterozygous for an Rere-null allele (om) and a hypomorphic allele (eyes3) have an even greater phenotypic overlap with our subjects. We have previously shown that these mice have microphthalmia, optic nerve hypoplasia, sensorineural hearing loss, congenital heart defects, and post-natal growth deficiency.14 They also have abnormal brain development including a reduced brain size and weight, decreased numbers of NeuN-positive hippocampal neurons, cerebellar foliation defects, and delayed Purkinje cell maturation and migration in the cerebellum.14, 15
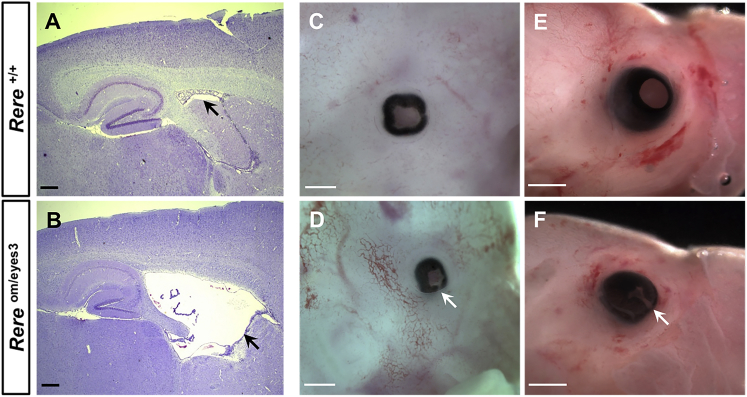
Ventriculomegaly is seen in 38% (3/8) of our subjects. To determine whether Rereom/eyes3 mice also have ventriculomegaly, we compared cresyl-violet-stained sagittal brain sections from adult Rereom/eyes3 mice and their wild-type littermates. Obtaining large numbers of Rereom/eyes3 mice for study is difficult because only a small fraction live into adulthood.14 However, in the three adult Rereom/eyes3 brains examined, all had enlarged lateral ventricles in comparison to those of their littermate controls (Figures 2A and 2B). Given that 20% (2/10) of our subjects have colobomas, we also looked for evidence of incomplete closure of the optic fissure—a precursor to the development of coloboma—in Rereom/eyes3 embryos. Although the optic fissures of wild-type mice close between embryonic days 11 (E11) and E13, we found incomplete closure of the optic fissure in 80% (8/10) of Rereom/eyes3 eyes examined at E13.5 and 100% (6/6) of the Rereom/eyes3 eyes examined at E17.5 (Figures 2C–2F).34 In contrast, the optic fissures of all wild-type control littermates were closed at these time points.
Figure 2.
RERE-Deficient Rereom/eyes3 Mice and Embryos Have Ventriculomegaly, Microphthalmia, and Coloboma
(A and B) Representative cresyl-violet-stained sagittal brain sections from a wild-type mouse (A) and its Rereom/eyes3 littermate (B) show enlargement of the lateral ventricles (black arrows). Ventriculomegaly was seen in 3/3 Rereom/eyes3 adult mice harvested between 5 and 10 months of age. Scale bar represents 200 μm.
(C–F) Representative eyes from wild-type embryos harvested at E13.5 (C) and E17.5 (E) and their Rereom/eyes3 littermates (D and F) demonstrate microphthalmia and incomplete closure of the optic fissure (white arrows in D and F). Failure of closure was seen in 8/10 (80%) of Rereom/eyes3 eyes examined at E13.5 and in 6/6 (100%) of Rereom/eyes3 eyes examined at E17.5. Scale bar represents 0.5 mm (C and D) or 1 mm (E and F).
Phenotype-genotype correlations are often not possible to discern with a cohort of ten individuals, given that genetic and non-genetic influences outside of the gene can also affect the development of the final phenotype. However, we have observed a partial association between missense mutations in the atrophin 1 domain and clinical severity. Thus, subjects 1–3 and subject 5, all of whom have missense mutations in this domain, have anatomic abnormalities of both the eye and brain. Subjects 1–3 all have VSDs. Subjects 3–5 all have vesicoureteral reflux, and subjects 1 and 2 present with other genitourinary malformations (Table S1). However, it is also possible that the phenotypic differences seen between subjects are due to the effects of other genetic, epigenetic, environmental, and/or stochastic factors. This hypothesis is supported by subjects 4 and 5 whose missense mutations in RERE result in the same amino acid change. Although they share many phenotypes in common—intellectual disability, developmental delay, seizures, abnormal and/or thin corpora callosa, abnormal cerebellar vermis, vesicoureteral reflux, deep-set eyes, and abnormal ears—they have other features that are discordant. This hypothesis is also consistent with the phenotypic variation seen in RERE-deficient mice on different genetic backgrounds.14
RERE has previously been shown to positively regulate retinoic acid signaling during somite development.11 Perturbations of retinoic acid signaling could also be the underlying cause of RERE deficiency’s effects on eye and heart development. We note that autosomal-recessive mutations in stimulated by retinoic acid 6 (STRA6 [MIM: 610745])—which encodes a transmembrane protein that transports retinol into cells—have been shown to cause microphthalmia, isolated with colobomas 8 (MIM: 601186), and microphthalmia, syndromic 9 (MIM: 601186), also known as Matthew-Wood syndrome. This syndrome is characterized by congenital heart defects and eye anomalies—microphthalmia and coloboma—that are similar to those seen in subjects 1 and 2.35, 36, 37 Similarly, recessive loss-of-function mutations in aldehyde dehydrogenase 1 family, members A3 (ALDH1A3 [MIM: 600463]), which encodes an enzyme that converts retinaldehyde to retinoic acid, cause microphthalmia, isolated 8 (MIM: 615113).38
In conclusion, we have shown that mutations in RERE cause an autosomal-dominant genetic syndrome characterized by neurodevelopmental defects, hypotonia, seizures, behavioral problems, structural CNS anomalies—abnormalities and/or thinning of the corpus callosum, diminished white matter volume, abnormal cerebellar vermis, and ventriculomegaly—ophthalmologic anomalies, congenital heart defects, and genitourinary abnormalities. Many of these phenotypes are seen in individuals with proximal 1p36 deletions that include RERE. This suggests that haploinsufficiency of RERE might be sufficient to cause many of the phenotypes associated with proximal 1p36 deletions. These phenotypes are also recapitulated in RERE-deficient zebrafish and mice, which might serve as effective models for elucidating the molecular mechanisms by which RERE acts in various organ systems.
Acknowledgments
The authors thank the family members for participating in this research. This work was supported by the National Institute of Neurological Disorders and Stroke (grant R01 NS058721 to E.H.S.), the Netherlands Organization for Health Research and Development (grant 912-12-109 to B.B.A.d.V), and ODAS Stichting (to D.G.M.B. and B.B.A.d.V). E.H.S is a member of the clinical advisory board of InVitae and consults for Personalis. M.T.C, R.S., K.G.M, and J.J. are all employees of GeneDx, which provides exome sequencing on a clinical basis. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from clinical laboratory testing conducted at Baylor Miraca Genetics Laboratories, which provides exome sequencing on a clinical basis.
Published: April 14, 2016
Footnotes
Supplemental Data include a Supplemental Note, one figure, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.002.
Contributor Information
Daryl A. Scott, Email: dscott@bcm.edu.
Elliott H. Sherr, Email: elliott.sherr@ucsf.edu.
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SFARI-base, http://sfari.org/resources/sfari-base.
UniProtKB, http://www.uniprot.org/help/uniprotkb
Supplemental Data
References
- 1.Shaffer L.G., Lupski J.R. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 2.Heilstedt H.A., Ballif B.C., Howard L.A., Kashork C.D., Shaffer L.G. Population data suggest that deletions of 1p36 are a relatively common chromosome abnormality. Clin. Genet. 2003;64:310–316. doi: 10.1034/j.1399-0004.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- 3.Giraudeau F., Taine L., Biancalana V., Delobel B., Journel H., Missirian C., Lacombe D., Bonneau D., Parent P., Aubert D. Use of a set of highly polymorphic minisatellite probes for the identification of cryptic 1p36.3 deletions in a large collection of patients with idiopathic mental retardation. J. Med. Genet. 2001;38:121–125. doi: 10.1136/jmg.38.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slavotinek A., Shaffer L.G., Shapira S.K. Monosomy 1p36. J. Med. Genet. 1999;36:657–663. [PMC free article] [PubMed] [Google Scholar]
- 5.Battaglia A., Hoyme H.E., Dallapiccola B., Zackai E., Hudgins L., McDonald-McGinn D., Bahi-Buisson N., Romano C., Williams C.A., Brailey L.L. Further delineation of deletion 1p36 syndrome in 60 patients: a recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics. 2008;121:404–410. doi: 10.1542/peds.2007-0929. [DOI] [PubMed] [Google Scholar]
- 6.Heilstedt H.A., Ballif B.C., Howard L.A., Lewis R.A., Stal S., Kashork C.D., Bacino C.A., Shapira S.K., Shaffer L.G. Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am. J. Hum. Genet. 2003;72:1200–1212. doi: 10.1086/375179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y.Q., Heilstedt H.A., Bedell J.A., May K.M., Starkey D.E., McPherson J.D., Shapira S.K., Shaffer L.G. Molecular refinement of the 1p36 deletion syndrome reveals size diversity and a preponderance of maternally derived deletions. Hum. Mol. Genet. 1999;8:313–321. doi: 10.1093/hmg/8.2.313. [DOI] [PubMed] [Google Scholar]
- 8.Jordan V.K., Zaveri H.P., Scott D.A. 1p36 deletion syndrome: an update. Appl. Clin. Genet. 2015;8:189–200. doi: 10.2147/TACG.S65698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang S.H., Scheffer A., Ou Z., Li J., Scaglia F., Belmont J., Lalani S.R., Roeder E., Enciso V., Braddock S. Identification of proximal 1p36 deletions using array-CGH: a possible new syndrome. Clin. Genet. 2007;72:329–338. doi: 10.1111/j.1399-0004.2007.00876.x. [DOI] [PubMed] [Google Scholar]
- 10.Zoltewicz J.S., Stewart N.J., Leung R., Peterson A.S. Atrophin 2 recruits histone deacetylase and is required for the function of multiple signaling centers during mouse embryogenesis. Development. 2004;131:3–14. doi: 10.1242/dev.00908. [DOI] [PubMed] [Google Scholar]
- 11.Vilhais-Neto G.C., Maruhashi M., Smith K.T., Vasseur-Cognet M., Peterson A.S., Workman J.L., Pourquié O. Rere controls retinoic acid signalling and somite bilateral symmetry. Nature. 2010;463:953–957. doi: 10.1038/nature08763. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S., Duester G. Retinoic acid controls body axis extension by directly repressing Fgf8 transcription. Development. 2014;141:2972–2977. doi: 10.1242/dev.112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plaster N., Sonntag C., Schilling T.F., Hammerschmidt M. REREa/Atrophin-2 interacts with histone deacetylase and Fgf8 signaling to regulate multiple processes of zebrafish development. Dev. Dyn. 2007;236:1891–1904. doi: 10.1002/dvdy.21196. [DOI] [PubMed] [Google Scholar]
- 14.Kim B.J., Zaveri H.P., Shchelochkov O.A., Yu Z., Hernández-García A., Seymour M.L., Oghalai J.S., Pereira F.A., Stockton D.W., Justice M.J. An allelic series of mice reveals a role for RERE in the development of multiple organs affected in chromosome 1p36 deletions. PLoS ONE. 2013;8:e57460. doi: 10.1371/journal.pone.0057460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim B.J., Scott D.A. Mouse model reveals the role of RERE in cerebellar foliation and the migration and maturation of Purkinje cells. PLoS ONE. 2014;9:e87518. doi: 10.1371/journal.pone.0087518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaveri H.P., Beck T.F., Hernández-García A., Shelly K.E., Montgomery T., van Haeringen A., Anderlid B.M., Patel C., Goel H., Houge G. Identification of critical regions and candidate genes for cardiovascular malformations and cardiomyopathy associated with deletions of chromosome 1p36. PLoS ONE. 2014;9:e85600. doi: 10.1371/journal.pone.0085600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilling T.F., Piotrowski T., Grandel H., Brand M., Heisenberg C.P., Jiang Y.J., Beuchle D., Hammerschmidt M., Kane D.A., Mullins M.C. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development. 1996;123:329–344. doi: 10.1242/dev.123.1.329. [DOI] [PubMed] [Google Scholar]
- 18.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marszalek J.R., Weiner J.A., Farlow S.J., Chun J., Goldstein L.S. Novel dendritic kinesin sorting identified by different process targeting of two related kinesins: KIF21A and KIF21B. J. Cell Biol. 1999;145:469–479. doi: 10.1083/jcb.145.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Zhang H., Li J., Zhao H., Xin Q., Shan S., Dang J., Bian X., Liu Q. Association of common variants in KIF21B and ankylosing spondylitis in a Chinese Han population: a replication study. Immunogenetics. 2013;65:835–839. doi: 10.1007/s00251-013-0733-6. [DOI] [PubMed] [Google Scholar]
- 21.International Multiple Sclerosis Genetics Consortium (IMSGC) Comprehensive follow-up of the first genome-wide association study of multiple sclerosis identifies KIF21B and TMEM39A as susceptibility loci. Hum. Mol. Genet. 2010;19:953–962. doi: 10.1093/hmg/ddp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R., Wei Z., Jin H., Wu H., Yu C., Wen W., Chan L.N., Wen Z., Zhang M. Autoinhibition of UNC5b revealed by the cytoplasmic domain structure of the receptor. Mol. Cell. 2009;33:692–703. doi: 10.1016/j.molcel.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Touma M., Joshi M., Connolly M.C., Grant P.E., Hansen A.R., Khwaja O., Berry G.T., Kinney H.C., Poduri A., Agrawal P.B. Whole genome sequencing identifies SCN2A mutation in monozygotic twins with Ohtahara syndrome and unique neuropathologic findings. Epilepsia. 2013;54:e81–e85. doi: 10.1111/epi.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukasawa T., Kubota T., Negoro T., Saitoh M., Mizuguchi M., Ihara Y., Ishii A., Hirose S. A case of recurrent encephalopathy with SCN2A missense mutation. Brain Dev. 2015;37:631–634. doi: 10.1016/j.braindev.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Bosch D.G., Boonstra F.N., de Leeuw N., Pfundt R., Nillesen W.M., de Ligt J., Gilissen C., Jhangiani S., Lupski J.R., Cremers F.P., de Vries B.B. Novel genetic causes for cerebral visual impairment. Eur. J. Hum. Genet. 2015 doi: 10.1038/ejhg.2015.186. Published online September 9, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown P.R., Miki K., Harper D.B., Eddy E.M. A-kinase anchoring protein 4 binding proteins in the fibrous sheath of the sperm flagellum. Biol. Reprod. 2003;68:2241–2248. doi: 10.1095/biolreprod.102.013466. [DOI] [PubMed] [Google Scholar]
- 27.Monroe G.R., Frederix G.W., Savelberg S.M., de Vries T.I., Duran K.J., van der Smagt J.J., Terhal P.A., van Hasselt P.M., Kroes H.Y., Verhoeven-Duif N.M. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet. Med. 2016 doi: 10.1038/gim.2015.200. Published online February 4, 2016. [DOI] [PubMed] [Google Scholar]
- 28.Nicoulaz A., Rubi F., Lieder L., Wolf R., Goeggel-Simonetti B., Steinlin M., Wiest R., Bonel H.M., Schaller A., Gallati S., Conrad B. Contiguous ∼16cMb 1p36 deletion: Dominant features of classical distal 1p36 monosomy with haplo-lethality. Am. J. Med. Genet. A. 2011;155A:1964–1968. doi: 10.1002/ajmg.a.33210. [DOI] [PubMed] [Google Scholar]
- 29.Shimada S., Shimojima K., Okamoto N., Sangu N., Hirasawa K., Matsuo M., Ikeuchi M., Shimakawa S., Shimizu K., Mizuno S. Microarray analysis of 50 patients reveals the critical chromosomal regions responsible for 1p36 deletion syndrome-related complications. Brain Dev. 2015;37:515–526. doi: 10.1016/j.braindev.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Paciorkowski A.R., Thio L.L., Rosenfeld J.A., Gajecka M., Gurnett C.A., Kulkarni S., Chung W.K., Marsh E.D., Gentile M., Reggin J.D. Copy number variants and infantile spasms: evidence for abnormalities in ventral forebrain development and pathways of synaptic function. Eur. J. Hum. Genet. 2011;19:1238–1245. doi: 10.1038/ejhg.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campeau P.M., Ah Mew N., Cartier L., Mackay K.L., Shaffer L.G., Der Kaloustian V.M., Thomas M.A. Prenatal diagnosis of monosomy 1p36: a focus on brain abnormalities and a review of the literature. Am. J. Med. Genet. A. 2008;146A:3062–3069. doi: 10.1002/ajmg.a.32563. [DOI] [PubMed] [Google Scholar]
- 32.Bursztejn A.C., Bronner M., Peudenier S., Grégoire M.J., Jonveaux P., Nemos C. Molecular characterization of a monosomy 1p36 presenting as an Aicardi syndrome phenocopy. Am. J. Med. Genet. A. 2009;149A:2493–2500. doi: 10.1002/ajmg.a.33051. [DOI] [PubMed] [Google Scholar]
- 33.Arndt A.K., Schafer S., Drenckhahn J.D., Sabeh M.K., Plovie E.R., Caliebe A., Klopocki E., Musso G., Werdich A.A., Kalwa H. Fine mapping of the 1p36 deletion syndrome identifies mutation of PRDM16 as a cause of cardiomyopathy. Am. J. Hum. Genet. 2013;93:67–77. doi: 10.1016/j.ajhg.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hero I. Optic fissure closure in the normal cinnamon mouse. An ultrastructural study. Invest. Ophthalmol. Vis. Sci. 1990;31:197–216. [PubMed] [Google Scholar]
- 35.Pasutto F., Sticht H., Hammersen G., Gillessen-Kaesbach G., Fitzpatrick D.R., Nürnberg G., Brasch F., Schirmer-Zimmermann H., Tolmie J.L., Chitayat D. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am. J. Hum. Genet. 2007;80:550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golzio C., Martinovic-Bouriel J., Thomas S., Mougou-Zrelli S., Grattagliano-Bessieres B., Bonniere M., Delahaye S., Munnich A., Encha-Razavi F., Lyonnet S. Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am. J. Hum. Genet. 2007;80:1179–1187. doi: 10.1086/518177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casey J., Kawaguchi R., Morrissey M., Sun H., McGettigan P., Nielsen J.E., Conroy J., Regan R., Kenny E., Cormican P. First implication of STRA6 mutations in isolated anophthalmia, microphthalmia, and coloboma: a new dimension to the STRA6 phenotype. Hum. Mutat. 2011;32:1417–1426. doi: 10.1002/humu.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fares-Taie L., Gerber S., Chassaing N., Clayton-Smith J., Hanein S., Silva E., Serey M., Serre V., Gérard X., Baumann C. ALDH1A3 mutations cause recessive anophthalmia and microphthalmia. Am. J. Hum. Genet. 2013;92:265–270. doi: 10.1016/j.ajhg.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.